- 1Department of Clinical Laboratory, Xiangya Hospital, Central South University, Changsha, China
- 2State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
Pseudomonas aeruginosa is noted for its intrinsic antibiotic resistance and capacity of acquiring additional resistance genes. In this study, the genomes of nine clinical P. aeruginosa isolates were fully sequenced. An extensive genetic comparison was applied to 18 P. aeruginosa accessory genetic elements (AGEs; 13 of them were sequenced in this study and located within P. aeruginosa chromosomes) that were divided into four groups: five related integrative and conjugative elements (ICEs), four related integrative and mobilizable elements (IMEs), five related unit transposons, and two related IMEs and their two derivatives. At least 45 resistance genes, involved in resistance to 10 different categories of antibiotics and heavy metals, were identified from these 18 AGEs. A total of 10 β-lactamase genes were identified from 10 AGEs sequenced herein, and nine of them were captured within class 1 integrons, which were further integrated into ICEs and IMEs with intercellular mobility, and also unit transposons with intracellular mobility. Through this study, we identified for the first time 20 novel MGEs, including four ICEs Tn6584, Tn6585, Tn6586, and Tn6587; three IMEs Tn6853, Tn6854, and Tn6878; five unit transposons Tn6846, Tn6847, Tn6848, Tn6849, and Tn6883; and eight integrons In1795, In1778, In1820, In1784, In1775, In1774, In1789, and In1799. This was also the first report of two resistance gene variants blaCARB-53 and catB3s, and a novel ST3405 isolate of P. aeruginosa. The data presented here denoted that complex transposition and homologous recombination promoted the assembly and integration of AGEs with mosaic structures into P. aeruginosa chromosomes.
Introduction
Pseudomonas aeruginosa is a major opportunistic pathogen and a leading cause of morbidity and mortality in cystic fibrosis patients and immunocompromised individuals (Alvarez-Ortega et al., 2011). P. aeruginosa displays resistance to multiple classes of antibiotics (De Oliveira et al., 2020), being in the critical priority list of antibiotic resistant bacteria created by the World Health Organization (Tacconelli et al., 2018). One of the main mechanisms driving to the antibiotic resistance in P. aeruginosa is the production of β-lactamases, which are capable of hydrolyzing chemical compounds containing a β-lactam ring (Bush, 2018) and are the most common determinant for resistance to bacterial β-lactam antibiotics (Bush and Bradford, 2020). β-lactamases can be divided into four Ambler classes A to D, where classes A/C/D β-lactamases utilize a serine moiety while class B β-lactamases (also known as metallo-β-lactamases) need a zinc ion at its active site (Ambler, 1980). Besides the intrinsic narrow-spectrum β-lactamase (NSBL) gene blaOXA-50 (Girlich et al., 2004), P. aeruginosa has evolved to acquire various extended-spectrum β-lactamase (ESBL) and even carbapenemase genes through horizontal gene transfer mediated by different mobile genetic elements (MGEs) (Botelho et al., 2019). MGEs play critical roles in the accumulation and spread of antibiotic resistance genes in P. aeruginosa (Botelho et al., 2019).
Among these genetic platforms, the integrative and conjugative elements (ICEs) are transposons which encode self-integration and -conjugation modules, typically including attL (attachment site at the left end), int (integrase), xis (excisionase), rlx (relaxase), oriT (origin of conjugative replication), cpl (coupling protein), a P (TivB)- or F (TivF)-type T4SS gene set (mating pair formation), and attR (attachment site at the right end) (Botelho and Schulenburg, 2020). Conversely, the integrative and mobilizable elements (IMEs) (that typically have attL, int, rlx, oriT, and attR) do not encode the T4SS machinery, and their mobility needs a helper conjugative element (Guédon et al., 2017). The Tn3-family unit transposons carry a core transposition module typically composed of tnpA (transposase), tnpR (resolvase), and res (resolution site), with a pair of terminal inverted repeats IRL/IRR (Nicolas et al., 2015).
Various ICEs, IMEs, and unit transposons have been identified and found to be important vehicles for antibiotic resistance genes in P. aeruginosa (Stokes et al., 2007; Mathee et al., 2008; Roy Chowdhury et al., 2016; Botelho et al., 2018a; Botelho et al., 2018b; van der Zee et al., 2018; Chew et al., 2019). Deep understanding of the MGEs at a genomic level will provide the theoretical basis for inhibiting the emergence of multidrug-resistant P. aeruginosa and overcoming the challenge of limited antimicrobial chemotherapy measures. However, few studies have dedicated to detailedly and accurately dissect the genetic structures of these MGEs from P. aeruginosa.
Our previous studies (Zhan et al., 2018; Zeng et al., 2019) have dissected the genetic characteristics of three carbapenemase-encoding novel transposons (blaIMP-1-containing ISPa17-based transposition unit Tn6394, blaVIM-4-containing ICE Tn6413, and blaIMP-1-containing unit transposon Tn6411) located within P. aeruginosa chromosomes. This follow-up study presented the complete sequences of 10 β-lactamase-encoding accessory genetic elements (AGEs; encoding four NSBLs CARB-2/-53, TEM-1B, and OXA-101; four ESBLs GES-1, PER-1, VEB-3 and OXA-10; and two carbapenemases GES-6/-15) along with three additional ones that did not encode any β-lactamase genes (but harbored dfrA12 and aphA6 and strAB) from the chromosomes of the sequenced P. aeruginosa strains. A detailed genetic dissection was applied to these 13 AGEs together with additional five reference/prototype ones from GenBank. Data presented here provided a deeper understanding of diversification of chromosomal MGEs (especially those carrying β-lactamase genes) in P. aeruginosa.
Materials and Methods
Bacterial Strains
Nine P. aeruginosa isolates (Table S1) were recovered either from sputum, airway secretions, urine, or blood of nine patients with nosocomial infections in five different Chinese public hospitals from 2011 to 2019. Bacterial species identification was performed using genome sequence-based average nucleotide identity analysis (http://www.ezbiocloud.net/tools/ani) (Richter and Rossello-Mora, 2009).
Sequencing and Sequence Assembly
Bacterial genomic DNA was isolated using the UltraClean Microbial Kit (Qiagen, NW, Germany) and sequenced from a sheared DNA library with average size of 15 kb (ranged from 10 to 20 kb) on a PacBio RSII sequencer (Pacific Biosciences, CA, USA), as well as a paired-end library with an average insert size of 350 bp (ranged from 150 to 600 bp) on a HiSeq sequencer (Illumina, CA, USA). The paired-end short Illumina reads were used to correct the long PacBio reads utilizing proovread (Hackl et al., 2014), and then the corrected PacBio reads were assembled de novo utilizing SMARTdenovo (https://github.com/ruanjue/smartdenovo).
Sequence Annotation and Comparison
Open reading frames (ORFs) and pseudogenes were predicted using RAST 2.0 (Brettin et al., 2015) combined with BLASTP/BLASTN searches (Boratyn et al., 2013) against the UniProtKB/Swiss-Prot database (Boutet et al., 2016) and the RefSeq database (O’Leary et al., 2016). Annotation of resistance genes, MGEs, and other features was carried out using the online databases including CARD (Jia et al., 2017), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and Tn Number Registry (Roberts et al., 2008). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 1.0 (https://inkscape.org/en/). Heatmaps were plotted with MeV 4.9.0 (Saeed et al., 2003).
Multi-Locus Sequence Typing
The sequence types (STs) of P. aeruginosa isolates were identified according to the online P. aeruginosa MLST scheme (https://pubmlst.org/paeruginosa/).
Conjugal Transfer
Conjugal transfer experiments were carried out with rifampin-resistant P. aeruginosa PAO1 being used as a recipient, and the indicated wild-type P. aeruginosa isolate as a donor. Three milliliters of overnight cultures of each of donor and recipient bacteria was mixed together, harvested, and resuspended in 80 μL of Brain Heart Infusion (BHI) broth (BD Biosciences). The mixture was spotted on a 1 cm2 hydrophilic nylon membrane filter with a 0.45 µm pore size (Millipore) that was placed on BHI agar (BD Biosciences) plate and then incubated for mating at 37°C for 12 to 18 h. Bacteria were washed from filter membrane and spotted on Muller–Hinton (MH) agar (BD Biosciences) plates, for selecting a blaGES- or blaCARB-carrying PAO1 transconjugant. 1500 mg/mL rifampin (for PAO1), together with 80 mg/mL ceftazidime (for blaGES) or 200 mg/L carbenicillin (for blaCARB) was used for transconjugant selection.
Cloning Experiments
The blaCARB-53 coding region together with its 365-bp upstream (promoter) region and 305-bp downstream (terminator) region from strain 201330 was cloned into the cloning vector pUC57-Kan (pUC57K). Similarly, the catB3s coding region together with its 309-bp upstream (promoter) region and 260-bp downstream (terminator) region from strain YT12746 was cloned into pUC57K. The resulting recombinant plasmid pUC57K-CARB or pUC57K-catB was transformed through electroporation into Escherichia coli TOP10, generating the E. coli electroporant TOP10/pUC57K-CARB or TOP10/pUC57K-catB, respectively. 200 μg/mL ampicillin (for blaCARB-53) or 25 μg/mL chloramphenicol (for catB3s) was used for electroporant selection.
Bacterial Antimicrobial Susceptibility Test
Bacterial antimicrobial susceptibility was tested by VITEK 2, E-test, or the classic broth microdilution method and interpreted as per the 2020 Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2020).
Nucleotide Sequence Accession Numbers
The complete chromosome sequences of the SE5352, SE5331, 31130, 201330, T12726, YTSEY8, SE5429, SE5458, and SE5357 isolates were submitted to GenBank under accession numbers CP054843, CP046402, CP060392, CP054623, CP045552, CP054581, CP054845, CP046406, and CP054844 respectively.
Results and Discussion
Identification of Four STs From the Nine Clinical P. aeruginosa Isolates
Four different STs, namely ST235, ST244, ST292, and ST3405, were identified from the nine P. aeruginosa isolates analyzed in this work (Table S1). The five isolates SE5352, SE5331, 31130, SE5429, and SE5458, and the SE5357 isolate belonged to ST235 and ST244, respectively, which were recognized as the high-risk clones with ST235 being the most widely disseminated (Oliver et al., 2015). The YT12746 isolate belonged to ST292, a P. aeruginosa clone type identified in Asian countries only such as Korea (Lee et al., 2011), China (Fan et al., 2016), and Thailand (Khuntayaporn et al., 2019). The 201330 isolate belonged to a novel ST variant ST3405, with an allelic profile 17-5-5-4-137-4-3 corresponding to the seven housekeeping genes acsA, aroE, guaA, mutL, nuoD, ppsA, and trpE.
Identification of 13 Chromosomal AGEs From the Nine Clinical P. aeruginosa Isolates
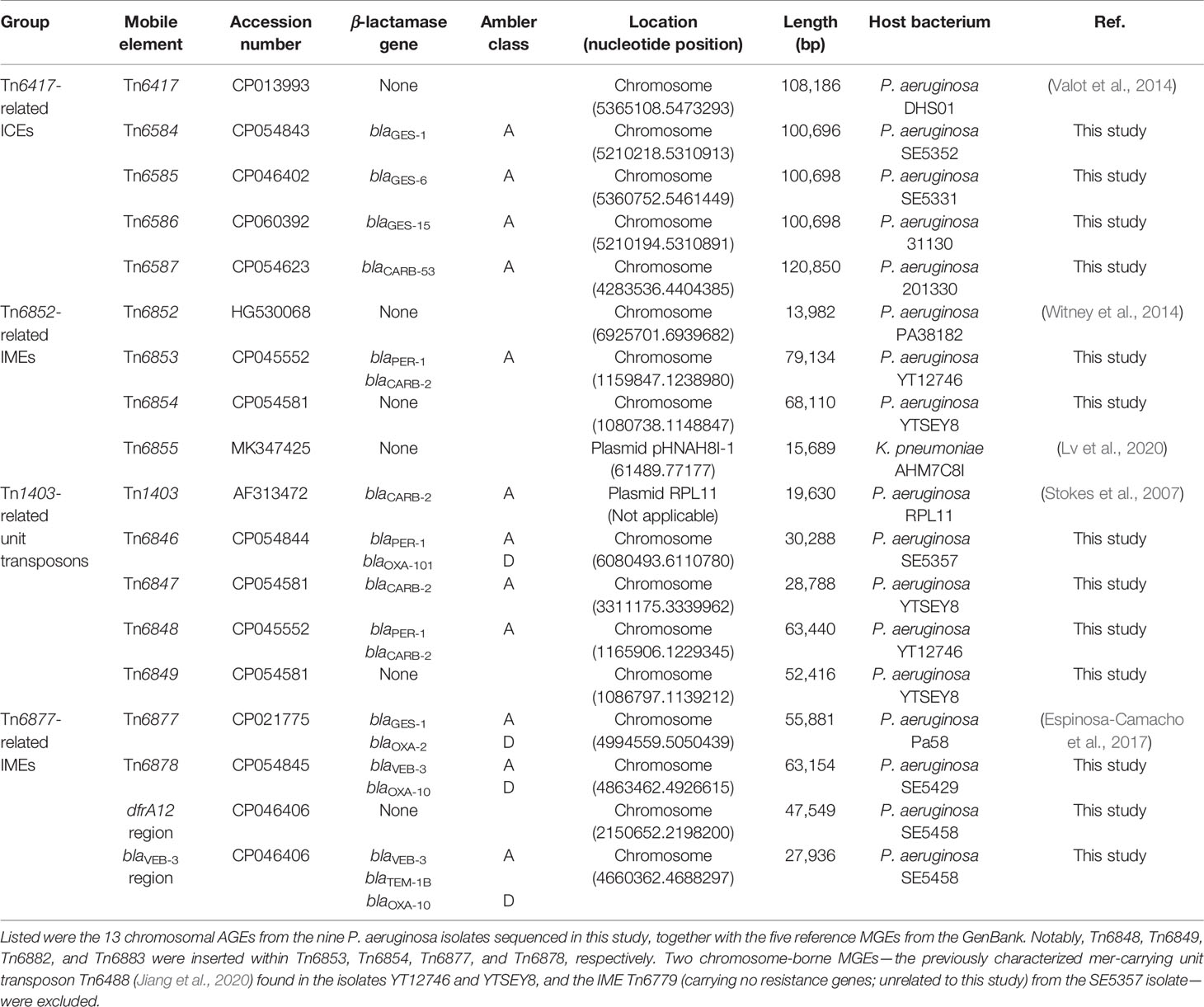
Based on the determined complete genome sequences of the nine P. aeruginosa isolates, a total of 13 chromosomal AGEs were identified for further genetic characterization: blaGES-1/-6/-15-carrying Tn6584, Tn6585, and Tn6586 from SE5352, SE5331, and 31130, respectively; blaCARB-2/-53-carrying Tn6847 and Tn6587 from YTSEY8 and 201330, respectively; blaVEB-3-carrying Tn6878 and ‘blaVEB-3 region’ from SE5429 and SE5458, respectively; blaPER-1-carrying Tn6846 and Tn6853 (containing Tn6848) from SE5357 and YT12746, respectively; dfrA12- and aphA6-carrying ‘dfrA12 region’ from SE5458; and strAB-carrying Tn6854 (containing Tn6849) from YTSEY8 (Table 1).
Collection of 18 AGEs for Sequence Comparison
A detailed sequence comparison was then applied to a collection of four groups of 18 AGEs including the above 13 chromosomal AGEs together with five additional reference/prototype ones Tn6417, Tn6852, Tn6855, Tn6877, and Tn1403 from the GenBank: five related ICEs Tn6417, Tn6584, Tn6585, Tn6586, and Tn6587; four related IMEs Tn6852, Tn6853, Tn6854, and Tn6855; five related unit transposons Tn1403, Tn6846, Tn6847, Tn6848, and Tn6849; and two related IMEs Tn6877 and Tn6878 and their two derivatives dfrA12 region and blaVEB-3 region (Table 1). At least 45 resistance genes, involved in resistance to 10 different categories of antibiotics and heavy metals, were identified in 17 of these 18 AGEs (Figure 1 and Table S2).

Figure 1 Heatmap of prevalence of resistance genes. The original data are shown in Table S2.
Comparison of Five Related ICEs Tn6417, Tn6584, Tn6585, Tn6586, and Tn6587
Tn6417 (108.2 kb in length) was used as reference ICE (Zeng et al., 2019) and initially described in P. aeruginosa DHS01 (Valot et al., 2014). The backbones of Tn6417, Tn6584, Tn6585, Tn6586, and Tn6587 varied in size from about 71.5 kb to nearly 103.3 kb, but all contained attL/R, int, cpl, rlx, and a F-type T4SS gene set (Figure 2). In addition, their backbones had at least four major modular differences: i) presence of unique orf672, orf306, piL1–to–orf381 region, orf3336–to–ftsk region, and orf231–to–orf2514 region in only Tn6587; ii) presence of unique xerC–to–orf1068 region in only Tn6417; iii) absence of orf1371 and orf582 from only Tn6587; and iv) orf384–to–orf765 region and orf1419–to–rlx region from Tn6587 displayed <90% nucleotide identity to the counterparts of Tn6417, Tn6584, Tn6585, and Tn6586 (Figure 2). Tn6417, Tn6584, Tn6585, and Tn6586 were integrated into the chromosomal tRNAGly gene, while Tn6587 into the chromosomal gene tolC (outer membrane protein). The attL/R sequences of these five ICEs somewhat showed differences but shared a core motif ‘TTCGCCCGCTCCA’.

Figure 2 Comparison of five related ICEs Tn6417, Tn6584, Tn6585, Tn6586, and Tn6587. Genes are denoted by arrows. Genes, MGEs, and other features are colored based on their functional classification. Shading in light blue denotes regions of homology (nucleotide identity ≥90%), light orange (nucleotide identity <90%). The accession number of Tn6417 (Valot et al., 2014) used as reference is EU696790.
Each of these five ICEs carried a single accessory module: Tn6807, Tn6808, Tn6809, Tn6532, and In1784 in Tn6584, Tn6585, Tn6586, Tn6417, and Tn6587, respectively (Figure 2). The former four were integrated at a site upstream of the ICE backbone gene orf582 and identified as derivatives of Tn6346 (Ng et al., 2009), while the last one into the ICE backbone gene ftsk (cell division protein).
Tn6346, a prototype Tn3-family unit transposon originally identified in Achromobacter spp. AO22 (Ng et al., 2009), was a hybrid of the core transposition module tnpAR–res from Tn5051 and the mer region from Tn501 (Figure 3). Tn6532, Tn6807, Tn6808, and Tn6809 differed from Tn6346 because of the interruption of tnpA due to the insertion of IS1071 at the same position, and that of urf2 due to the insertion of four different concise class 1 integrons In159, In1795, In1778, and In1820, respectively, at the same site; the four transposons Tn6532, Tn6807, Tn6808, and Tn6809 were bracketed by the same 5-bp direct repeats (DRs; target site duplication signals for transposition).
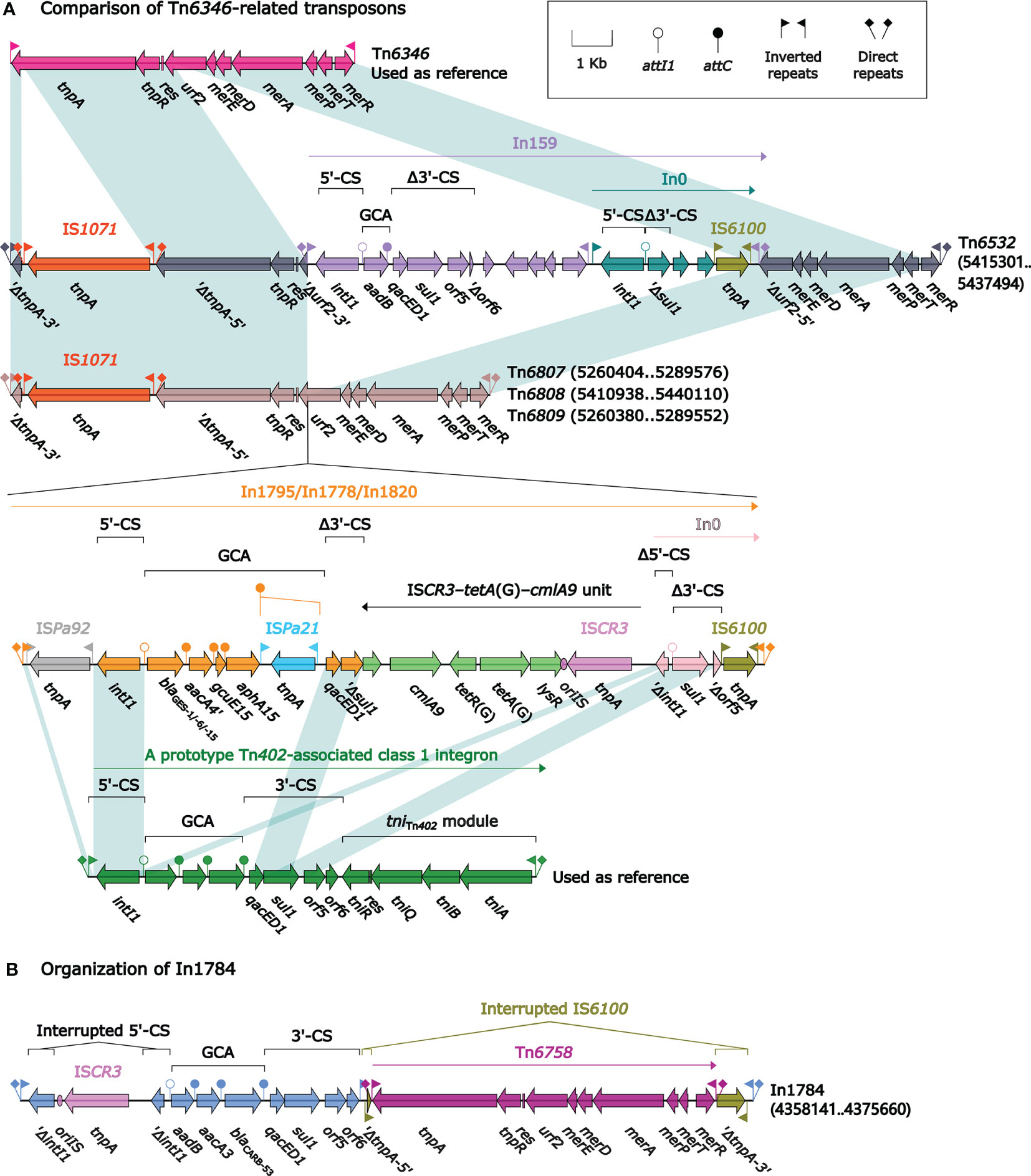
Figure 3 Comparison of Tn6532, Tn6807, Tn6808 and Tn6809, and organization of In1784. Shown are five Tn6346-related transposons (A) and one integron In1784 (B). Genes are denoted by arrows. Genes, MGEs, and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within the chromosomes of strains DHS01, SE5352, SE5331, 31130, and 201330, respectively. The accession number of Tn6346 (Ng et al., 2009) used as reference is EU696790.
In159 harbored the single-gene gene cassette array (GCA) aadB. In1795, In1778, and In1820 had the GCA blaGES–aacA4′gcuE15–aphA15 but with differences in blaGES subtypes: blaGES-1, blaGES-6, and blaGES-15, respectively. There were additional resistance loci besides GCAs integrated into these four integrons: an integron In0 into In159 and an ISCR3–tetA(G)–cmlA9 unit connected with a different In0 into In1795/In1778/In1820. All these four integrons were bracketed by the same 5-bp DRs (Figure 3).
In1784 from Tn6587 was a concise class 1 integron with the GCA aadB–aacA3–blaCARB-53. Its 5′-conserved segment (5′-CS: intI1–attI1) was interrupted by the insertion of ISCR3, leading to a 106-bp deletion of intI1. In addition, IS6100 downstream of 3′-CS (qacED1–sul1–orf5–orf6) was interrupted by the insertion of Tn6758 (Hu et al., 2015), which was a prototype Tn3-family unit transposon initially found in Achromobacter xylosoxidans X02736 and contained a core transposition module tnpAR–res plus a mer locus.
Comparison of Four Related IMEs Tn6852, Tn6853, Tn6854, and Tn6855
Tn6852 (14.0 kb in length) was used as the prototype IME and initially found in P. aeruginosa PA38182 (Witney et al., 2014). Tn6852 carried the backbone markers attL/R, int, and the chrA region (Figure 4). Conversely, Tn6853, Tn6854, and Tn6855 (Lv et al., 2020) carried a multidrug efflux locus nfxB–mexCD–oprJ instead of the chrA region. Both the chrA region and the nfxB–mexCD–oprJ region were considered as the IME backbone components, since none of the associated MGEs were identified for them. Tn6852 thereby had no accessory modules, but two Tn3-family unit transposons Tn6848 and Tn6849 as accessory modules were integrated at the same site within the backbone gene mexC of Tn6853 and Tn6854, respectively. Tn6852, Tn6853, and Tn6854 were integrated into the chromosomal gene umuC (DNA polymerase V subunit) in P. aeruginosa, while Tn6855 into Tn5393c of plasmid pHNAH8I-1 in K. pneumoniae. Although plasmid-borne, Tn6855 was included in this study because it was a prototype MGE carrying an intact nfxB–mexCD–oprJ locus that was often found in Pseudomonas species.

Figure 4 Organization of four related IMEs Tn6852, Tn6853, Tn6854, and Tn6855. Genes are denoted by arrows. Genes, MGEs, and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within the chromosomes of strains PA38182, YT12746 and YTSEY8, and the plasmid pHNAH8I-1 of strain AHM7C8I, respectively. The accession number of Tn6852 (Witney et al., 2014) used as reference is HG530068.
Comparison of Five Related Unit Transposons Tn1403, Tn6846, Tn6847, Tn6848, and Tn6849
The Tn3-family prototype unit transposon Tn1403 was originally found in P. aeruginosa plasmid RPL11 (Levesque and Jacoby, 1988) and displayed a backbone structure IRL–tnpAR–res–sup–uspA–dksA–yjiK–IRR, with integration of two accessory modules In28 and Tn5393c into res and uspA, respectively (Stokes et al., 2007). Four Tn1403 derivatives Tn6846, Tn6847, Tn6848, and Tn6849 were identified in this study: the former two were chromosome-borne, Tn6846 being integrated at a site upstream of the chromosomal gene orf540 (thioesterase) while Tn6847 being inserted into the chromosomal gene orf1128 (glycerophosphodiester phosphodiesterase); the last two were located within the above-mentioned IMEs Tn6853 and Tn6854, respectively (Figure 5). The primary modular difference in Tn6846, Tn6847, Tn6848, and Tn6849 relative to Tn1403 was the integration of different class 1 integrons In1079Tn6846, In1789Tn6847, In1775Tn6848 (containing In1774Tn6848), and In1774Tn6849, instead of In28, into res. Another major modular difference was the occurrence of a 5.1-kb inversion in only Tn6846, and this inversion resulted from the insertion of IS6100 into tnpRTn5393c and led to the disruption of Tn5393c (strAB remained intact).

Figure 5 Organization of five related unit transposons Tn1403, Tn6846, Tn6847, Tn6848, and Tn6849. Shown are five Tn1403-related regions (A) and two Tn6489b-related transposons (B). Genes are denoted by arrows. Genes, MGEs, and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within the chromosomes of strains SE5357, YTSEY8, YT12746, and YTSEY8, respectively. The accession numbers of Tn1403 (Stokes et al., 2007) and Tn6489b (Jiang et al., 2020) used as reference are AF313472 and MF344569, respectively.
In1079Tn6846 was a complex class 1 integron containing two resistance loci: GCA aacA4-12–blaOXA-101–aacA5 (VR1: variable region 1), and ISCR1–blaPER-1 unit (VR2). The two concise class 1 integrons In1789Tn6847 and In1775Tn6848 harbored the distinct GCAs aacA4′-17–blaCARB-2–aadA2 and aacA4–blaCARB-2–aadA2b, respectively and exhibited two additional major modular differences: i) a disrupted ISCR3–tetA(G)–cmlA9 unit and another truncated version were inserted at the same site downstream of 3′-CS1, respectively; and ii) Tn6830–In1774Tn6848 were integrated into the 3′-terminal region of In1775Tn6848 but not In1789Tn6847. The two complex class 1 integrons In1774Tn6848 and In1774Tn6849 harbored the same GCA aadB–aacA64–catB3s–aacA3e–catB3 (VR1), but contained two different VR2 regions: ISCR1–blaPER-1 unit and ΔISCR1, respectively. Besides, a 31.9-kb fragment composed of Tn6830, a truncated ISCR3–tetA(G)–cmlA9 unit, an empty integron In0 and IS6100 was inserted into the 3′-terminal region of In1774Tn6849. Tn6830 belonged to the Tn4651 (Tsuda et al., 1989) subgroup of Tn3-family unit transposons, and it highly resembled Tn6489b (Jiang et al., 2020): Tn6830 and Tn6489b shared a core transposition structure IRL–tnpAC..tnpS–res–tnpT–IRR, but Tn6346 was integrated into Tn6489b but not into Tn6830.
Comparison of Two Related IMEs Tn6877 and Tn6878 and Their Derivatives dfrA12 Region and blaVEB-3 Region
Tn6877 (55.9 kb in length) was used as the prototype IME and initially found in P. aeruginosa Pa58 (Espinosa-Camacho et al., 2017). Tn6877, Tn6878, and a presumed primordial Tn6877-related element were all integrated at the same site between the two chromosomal genes orf2892 (α subunit of ribonucleotide reductase) and orf1248 (ß subunit), and they had essentially identical backbones, which was 21.6 kb in length and contained attL/R and two different int genes (Figure 6). Three Tn21-related accessory modules, namely Tn6882, Tn6883, and a presumed primordial Tn21-related element were inserted at the same site within the above three AGEs, respectively. In addition, Tn6877 acquired the second accessory module ISPa1635.

Figure 6 Organization of two related IMEs Tn6877 and Tn6878, and their derivatives dfrA12 region and blaVEB-3 region. Genes are denoted by arrows. Genes, MGEs, and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). The accession number of Tn6877 (Espinosa-Camacho et al., 2017) used as reference is CP021775.
A huge DNA region composed of the dfrA12 region plus a 2,462.2-kb chromosomal region underwent an inversion, which was likely mediated by two copies of IS26: one located within the primordial Tn6877-related element and another inserted into the chromosomal gene orf1113 (chain length determinant protein) (Figure 6). This inversion split the primordial Tn6877-related element into two separate parts: the dfrA12 region and the blaVEB-3 region; correspondingly, the primordial Tn21-related element in it was disrupted into the 5′-fragment and 3′-fragment.
Tn21 was a Tn3-family prototype unit transposon initially found in Shigella flexneri plasmid R100 and contained a core transposition module tnpAR–res plus a mer locus (Partridge et al., 2001). Tn6882, Tn6883, and the primordial Tn21-related element differed from Tn21 by acquisition of In1404 + In173, In1799, and In576 + In27, respectively, instead of In2-3 (a class 2 integron with a single-gene GCA dfrA1); additionally, the tnpR gene of the primordial Tn21-related element was interrupted by the integration of a 4.4-kb truncated version of IS26–rmtB–qepA–IS26 unit (Figure 7).

Figure 7 Comparison of Tn6882, Tn6883, and related regions. Shown are five Tn21-related regions (A) and VR3In1799/VR2In27 (B). Genes are denoted by arrows. Genes, MGEs, and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity ≥95%). Numbers in brackets indicate nucleotide positions within the chromosomes of strains Pa58, SE5429, and SE5458, respectively. The accession numbers of Tn21 (Partridge et al., 2001) and IS26–rmtB–qepA–IS26 unit (Yamane et al., 2007) used as reference are AF071413 and AB263754, respectively.
The concise class 1 integrons In1404 and In173 contained two different GCAs aadB4–aacA7–gcu177a–gcu177b–blaOXA-2–aadA1b and blaGES-1, respectively, as their sole resistance locus (Figure 7). The complex class 1 integrons In1799, In57, and In27 each carried multiple resistance loci. In1799 carried VR1 (GCA ΔaadB–ΔcmlA1a–blaOXA-10–aadA1a), VR2 (ISCR1–blaVEB-3 unit), and VR3 (a 17.5-kb region containing ISAba14–aphA6–ISAba14 unit). In27 carried GCA dfrA12–gcuF–aadA2 (VR1) and the 17.5-kb VR3In1799 region (VR2). In576 harbored GCA aadB4–cmlA1a–blaOXA-10–aadA1 (VR1) and ISCR1–blaVEB-3 unit (VR2).
Summary of Newly Identified or Designated MGEs
There were 20 newly identified MGEs, including four ICEs Tn6584, Tn6585, Tn6586, and Tn6587; three IMEs Tn6853, Tn6854, and Tn6878; five unit transposons Tn6846, Tn6847, Tn6848, Tn6849, and Tn6883; and eight integrons In1795, In1778, In1820, In1784, In1775, In1774, In1789, and In1799. Additional five MGEs (two IMEs Tn6852 and Tn6877, one unit transposon Tn6758, and two ISs ISPa97 and ISPa99) were newly designated (designated for the first time in this study but with previously determined sequences).
Characterization of Two Novel Variants of Antibiotic Resistance Genes
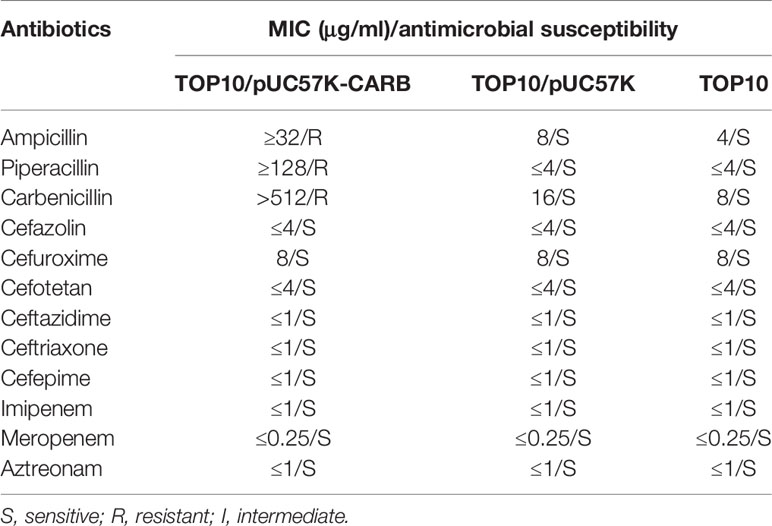
This was the first report of a novel blaCARB-2 variant blaCARB-53 and a novel catB3 variant catB3s. The deduced CARB-53 protein differs from CARB-2 by a single amino acid substitution Glu26Lys. catB3s differed from the catB3 reference gene by one amino acid substitution Val210Ile.The blaCARB-53 or catB3s gene fragments were cloned into the cloning vector pUC57K and then transformed into E. coli TOP10 to obtain the electroporant TOP10/pUC57K-CARB or TOP10/pUC57K-catB.
As expected, TOP10/pUC57K-CARB was highly resistant to ampicillin, piperacillin, and carbenicillin but remained susceptible to cephalosporins and carbapenems (Table 2). TOP10/pUC57K-catB showed a very high level of resistance to chloramphenicol with a minimum inhibitory concentration (MIC) value ≥256, while the two negative control strains TOP10/pUC57K and TOP10 were susceptible to chloramphenicol with a MIC value of 8.
Since the objective of this work was not to characterize these genes in terms of conferred phenotype, the cloning of the original genes was not done in parallel. Nevertheless, blaCARB-53 or catB3s had the drug resistance profile similar to its original variant blaCARB-2 (Korfhagen and Loper, 1975) or catB3 (Bunny et al., 1995), respectively.
ICE Transferability and Antimicrobial Susceptibility
A total of four ICEs Tn6584, Tn6585, Tn6586, and Tn6587 were identified from the nine P. aeruginosa isolates, and all these four ICEs had the essential conjugal transfer genes. After repeated conjugation experiment attempts, only Tn6587 was transferred from its rifampin-susceptible wild-type isolate into the rifampin-resistant P. aeruginosa PAO1, generating the transconjugant PAO1/Tn6587. PAO1/Tn6587 was highly resistant to carbenicillin with a MIC value >512 owing to the presence of blaCARB-53, whereas PAO1 was susceptible to chloramphenicol with a MIC value of 32.
Concluding Remarks
This work presented the complete sequences of 13 P. aeruginosa chromosomal AGEs, which could be divided into four groups: four Tn6417-related ICEs Tn6584, Tn6585, Tn6586 and Tn6587; two Tn6852-related IMEs Tn6853 and Tn6854; four Tn1403-related unit transposons Tn6846, Tn6847, Tn6848 and Tn6849; and one Tn6877-related IME Tn6878 and its two derivatives dfrA12 region and blaVEB-3 region.
Ten of these 13 chromosomal AGEs carried 10 β-lactamase genes in total: four NSBL genes blaCARB-2, blaCARB-53, blaOXA-101, and blaTEM-1B in Tn6847/Tn6853 (containing Tn6848), Tn6587, Tn6846, and blaVEB-3 region, respectively; four ESBL genes blaGES-1, blaPER-1, and blaVEB-3/blaOXA-10 in Tn6584, Tn6846/Tn6853 (containing Tn6848), and Tn6878/blaVEB-3 region, respectively; and two carbapenemase genes blaGES-6 and blaGES-15 in Tn6585 and Tn6586, respectively. Notably, Tn6853 (containing Tn6848), Tn6878, blaVEB-3 region, and Tn6846 each harbored two different β-lactamase genes.
Of the 10 β-lactamase genes in the 10 chromosomal AGEs sequenced in this study, nine (except for blaTEM-1B in a truncated IS26–rmtB–qepA–IS26 unit) were associated with class 1 integrons. The first seven β-lactamase genes blaCARB-2/-53, blaOXA-10/-101, and blaGES-1/-6/-15 were located in the GCAs of In1789/In1775/In1784, In1799/In1079, and In1795/In1778/In1820, respectively; conversely, the remaining two blaPER-1 and blaVEB-3 were located in the VRs of complex integrons In1079 and In576/In1799, respectively. These integrons, which captured various β-lactamase genes, were further integrated not only relevant into ICEs and IMEs with intercellular mobility but also into relevant unit transposons with intracellular mobility.
Besides the above 10 β-lactamase genes, there were additional 35 resistance genes identified in various subregions (including integrons, unit transposons, and putative resistance units) of these 13 chromosomal AGEs; thereby, most of these chromosomal AGEs had mosaic modular structures and encoded multiple drug resistance (Table S2).
Data presented here denoted that complex events of transposition and homologous recombination promoted the assembly and further integration of these chromosomal AGEs, carrying a large amount of resistance genes, into P. aeruginosa chromosomes.
Data Availability Statement
The datasets generated for this study can be found in the complete nucleotide sequences of SE5352, SE5331, 31130, 201330, YT12726, YTSEY8, SE5429, SE5458 and SE5357 isolates, which were submitted to GenBank under accession numbers CP054843, CP046402, CP060392, CP054623, CP045552, CP054581, CP054845, CP046406 and CP054844, respectively.
Ethics Statement
This study uses the clinical bacterial isolates obtained from the Chinese public hospitals as listed in Table S1. The local legislation did not require the study to be reviewed or approved by an ethics committee because the bacterial isolates involved in this study were part of the routine hospital laboratory procedures. The research involving biohazards and all related procedures were approved by the Biosafety Committee of the Beijing Institute of Microbiology and Epidemiology.
Author Contributions
DZ and MZ conceived the study and designed experimental procedures. TY, YJ, and FC performed the experiments. TY, LH, and XL analyzed the data. HY, JL, and ZY contributed to reagents and materials. TY and HY wrote the original draft. DZ and MZ reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China under grant number 2018YFC1200100 and the Fundamental Research Funds for the Central Universities of Central South University under grant number 502211911 (2019zzts796).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.638087/full#supplementary-material
References
Alvarez-Ortega C., Wiegand I., Olivares J., Hancock R. E. W., Martínez J. L. (2011). The intrinsic resistome of Pseudomonas aeruginosa to β-lactams. Virulence 2, 144–146. doi: 10.4161/viru.2.2.15014
Ambler R. P. (1980). The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 289, 321–331. doi: 10.1098/rstb.1980.0049
Boratyn G. M., Camacho C., Cooper P. S., Coulouris G., Fong A., Ma N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Botelho J., Schulenburg H. (2020). The role of integrative and conjugative elements in antibiotic resistance evolution. Trends Microbiol. 29, 8–18. doi: 10.1016/j.tim.2020.05.011
Botelho J., Grosso F., Peixe L. (2018a). Unravelling the genome of a Pseudomonas aeruginosa isolate belonging to the high-risk clone ST235 reveals an integrative conjugative element housing a blaGES-6 carbapenemase. J. Antimicrob. Chemother. 73, 77–83. doi: 10.1093/jac/dkx337
Botelho J., Grosso F., Quinteira S., Brilhante M., Ramos H., Peixe L. (2018b). Two decades of blaVIM-2-producing Pseudomonas aeruginosa dissemination: an interplay between mobile genetic elements and successful clones. J. Antimicrob. Chemother. 73, 873–882. doi: 10.1093/jac/dkx517
Botelho J., Grosso F., Peixe L. (2019). Antibiotic resistance in Pseudomonas aeruginosa - mechanisms, epidemiology and evolution. Drug Resist. Update 44, 100640. doi: 10.1016/j.drup.2019.07.002
Boutet E., Lieberherr D., Tognolli M., Schneider M., Bansal P., Bridge A. J., et al. (2016). UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Brettin T., Davis J. J., Disz T., Edwards R. A., Gerdes S., Olsen G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. doi: 10.1038/srep08365
Bunny K. L., Hall R. M., Stokes H. W. (1995). New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob. Agents Chemother. 39, 686–693. doi: 10.1128/AAC.39.3.686
Bush K., Bradford P. A. (2020). Epidemiology of beta-lactamase-producing pathogens. Clin. Microbiol. Rev. 33, e00047–19. doi: 10.1128/CMR.00047-19
Bush K. (2018). Past and present perspectives on beta-lactamases. Antimicrob. Agents Chemother. 62, e01076–18. doi: 10.1128/AAC.01076-18
Chew K. L., Octavia S., Ng O. T., Marimuthu K., Venkatachalam I., Cheng B., et al. (2019). Challenge of drug resistance in Pseudomonas aeruginosa: clonal spread of NDM-1-positive ST-308 within a tertiary hospital. J. Antimicrob. Chemother. 74, 2220–2224. doi: 10.1093/jac/dkz169
CLSI (2020). Performance standards for antimicrobial susceptibility testing (Wayne, PA: Clinical and Laboratory Standards Institute).
De Oliveira D. M. P., Forde B. M., Kidd T. J., Harris P. N. A., Schembri M. A., Beatson S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33, e00181–19. doi: 10.1128/CMR.00181-19
Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Espinosa-Camacho L. F., Delgado G., Soberón-Chávez G., Alcaraz L. D., Castañon J., Morales-Espinosa R. (2017). Complete genome sequences of four extensively drug-resistant strains, isolated from adults with ventilator-associated pneumonia at a tertiary referral hospital in Mexico city. Genome Announc. 5, e00925–17. doi: 10.1128/genomeA.00925-17
Fan X., Wu Y., Xiao M., Xu Z.-P., Kudinha T., Bazaj A., et al. (2016). Diverse genetic background of multidrug-resistant Pseudomonas aeruginosa from mainland China, and emergence of an extensively drug-resistant ST292 clone in Kunming. Sci. Rep. 6, 26522. doi: 10.1038/srep26522
Girlich D., Naas T., Nordmann P. (2004). Biochemical characterization of the naturally occurring oxacillinase OXA-50 of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48, 2043–2048. doi: 10.1128/AAC.48.6.2043-2048.2004
Guédon G., Libante V., Coluzzi C., Payot S., Leblond-Bourget N. (2017). The obscure world of integrative and mobilizable elements, highly widespread elements that pirate bacterial conjugative systems. Genes 8, 337. doi: 10.3390/genes8110337
Hackl T., Hedrich R., Schultz J., Forster F. (2014). proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30, 3004–3011. doi: 10.1093/bioinformatics/btu392
Hu Y., Zhu Y., Ma Y., Liu F., Lu N., Yang X., et al. (2015). Genomic insights into intrinsic and acquired drug resistance mechanisms in Achromobacter xylosoxidans. Antimicrob. Agents Chemother. 59, 1152–1161. doi: 10.1128/AAC.04260-14
Jia B., Raphenya A. R., Alcock B., Waglechner N., Guo P., Tsang K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jiang X., Yin Z., Yuan M., Cheng Q., Hu L., Xu Y., et al. (2020). Plasmids of novel incompatibility group IncpRBL16 from Pseudomonas species. J. Antimicrob. Chemother. 75, 2093–2100. doi: 10.1093/jac/dkaa143
Khuntayaporn P., Yamprayoonswat W., Yasawong M., Chomnawang M. T. (2019). Dissemination of carbapenem-resistance among multidrug resistant carrying metallo-beta-lactamase genes, including the novel gene in Thailand. Infect. Chemother. 51, 107–118. doi: 10.3947/ic.2019.51.2.107
Korfhagen T. R., Loper J. C. (1975). RPL11, an R factor of Pseudomonas aeruginosa determining carbenicillin and gentamicin resistance. Antimicrob. Agents Chemother. 7, 69–73. doi: 10.1128/AAC.7.1.69
Lee J.-Y., Song J.-H., Ko K. S. (2011). Identification of nonclonal Pseudomonas aeruginosa isolates with reduced colistin susceptibility in Korea. Microb. Drug Resist. (Larchmont N.Y.) 17, 299–304. doi: 10.1089/mdr.2010.0145
Levesque R. C., Jacoby G. A. (1988). Molecular structure and interrelationships of multiresistance beta-lactamase transposons. Plasmid 19, 21–29. doi: 10.1016/0147-619X(88)90059-5
Lv L., Wan M., Wang C., Gao X., Yang Q., Partridge S. R., et al. (2020). Emergence of a plasmid-encoded resistance-nodulation-division efflux pump conferring resistance to multiple drugs, including tigecycline, in Klebsiella pneumoniae. mBio 11, e02930–19. doi: 10.1128/mBio.02930-19
Mathee K., Narasimhan G., Valdes C., Qiu X., Matewish J. M., Koehrsen M., et al. (2008). Dynamics of Pseudomonas aeruginosa genome evolution. Proc. Natl. Acad. Sci. U. S. A. 105, 3100–3105. doi: 10.1073/pnas.0711982105
Moura A., Soares M., Pereira C., Leitão N., Henriques I., Correia A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics (Oxford England) 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Ng S. P., Davis B., Palombo E. A., Bhave M. (2009). A Tn5051-like mer-containing transposon identified in a heavy metal tolerant strain Achromobacter sp. AO22. BMC Res. Notes 2, 38. doi: 10.1186/1756-0500-2-38
Nicolas E., Lambin M., Dandoy D., Galloy C., Nguyen N., Oger C. A., et al. (2015). The Tn3-family of replicative transposons. Microbiol. Spectr. 3. doi: 10.1128/microbiolspec.MDNA3-0060-2014
Oliver A., Mulet X., López-Causapé C., Juan C. (2015). The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist. Updat. 21-22, 41–59. doi: 10.1016/j.drup.2015.08.002
O’Leary N. A., Wright M. W., Brister J. R., Ciufo S., Haddad D., Mcveigh R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Partridge S. R., Brown H. J., Stokes H. W., Hall R. M. (2001). Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45, 1263–1270. doi: 10.1128/AAC.45.4.1263-1270.2001
Richter M., Rossello-Mora R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Roberts A. P., Chandler M., Courvalin P., Guedon G., Mullany P., Pembroke T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Roy Chowdhury P., Scott M., Worden P., Huntington P., Hudson B., Karagiannis T., et al. (2016). Genomic islands 1 and 2 play key roles in the evolution of extensively drug-resistant ST235 isolates of Pseudomonas aeruginosa. Open Biol. 6, 150175. doi: 10.1098/rsob.150175
Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378. doi: 10.2144/03342mt01
Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Stokes H. W., Elbourne L. D. H., Hall R. M. (2007). Tn1403, a multiple-antibiotic resistance transposon made up of three distinct transposons. Antimicrob. Agents Chemother. 51, 1827–1829. doi: 10.1128/AAC.01279-06
Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tsuda M., Minegishi K., Iino T. (1989). Toluene transposons Tn4651 and Tn4653 are class II transposons. J. Bacteriol. 171, 1386–1393. doi: 10.1128/JB.171.3.1386-1393.1989
Valot B., Rohmer L., Jacobs M. A., Miller S. I., Bertrand X., Hocquet D. (2014). Comparative genomic analysis of two multidrug-resistant clinical isolates of ST395 epidemic strain of Pseudomonas aeruginosa obtained 12 years apart. Genome Announc. 2, e00515–14. doi: 10.1128/genomeA.00515-14
van der Zee A., Kraak W. B., Burggraaf A., Goessens W. H. F., Pirovano W., Ossewaarde J. M., et al. (2018). Spread of carbapenem resistance by transposition and conjugation among Pseudomonas aeruginosa. Front. Microbiol. 9, 2057. doi: 10.3389/fmicb.2018.02057
Witney A. A., Gould K. A., Pope C. F., Bolt F., Stoker N. G., Cubbon M. D., et al. (2014). Genome sequencing and characterization of an extensively drug-resistant sequence type 111 serotype O12 hospital outbreak strain of Pseudomonas aeruginosa. Clin. Microbiol. Infect. 20, O609–O618. doi: 10.1111/1469-0691.12528
Yamane K., Wachino J., Suzuki S., Kimura K., Shibata N., Kato H., et al. (2007). New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob. Agents Chemother. 51, 3354–3360. doi: 10.1128/AAC.00339-07
Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zeng L., Zhan Z., Hu L., Jiang X., Zhang Y., Feng J., et al. (2019). Genetic characterization of a blaVIM-24-carrying IncP-7β plasmid p1160-VIM and a blaVIM-4-harboring integrative and conjugative element Tn6413 from clinical Pseudomonas aeruginosa. Front. Microbiol. 10, 213. doi: 10.3389/fmicb.2019.00213
Keywords: Pseudomonas aeruginosa, β-lactam resistance, mobile genetic elements, blaCARB-53, catB3s, ST3405
Citation: Yu T, Yang H, Li J, Chen F, Hu L, Jing Y, Luo X, Yin Z, Zou M and Zhou D (2021) Novel Chromosome-Borne Accessory Genetic Elements Carrying Multiple Antibiotic Resistance Genes in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 11:638087. doi: 10.3389/fcimb.2021.638087
Received: 05 December 2020; Accepted: 09 February 2021;
Published: 18 March 2021.
Edited by:
Elio Rossi, University of Milan, ItalyReviewed by:
Carlos Juan, Instituto de Investigación Sanitaria de Palma (IdISPa), SpainMariagrazia Perilli, University of L’Aquila, Italy
Copyright © 2021 Yu, Yang, Li, Chen, Hu, Jing, Luo, Yin, Zou and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongsheng Zhou, ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t; Mingxiang Zou, em91bWluZ3hpYW5nQGNzdS5lZHUuY24=
†These authors have contributed equally to this work
 Ting Yu1†
Ting Yu1† Huiying Yang
Huiying Yang Zhe Yin
Zhe Yin Mingxiang Zou
Mingxiang Zou Dongsheng Zhou
Dongsheng Zhou