
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell. Infect. Microbiol. , 13 April 2021
Sec. Bacteria and Host
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.638058
 Jonathon Keck1
Jonathon Keck1 James P. Chambers1
James P. Chambers1 Jieh-Juen Yu1
Jieh-Juen Yu1 Xingguo Cheng2†
Xingguo Cheng2† Lane K. Christenson3
Lane K. Christenson3 M. N. Guentzel1
M. N. Guentzel1 Rishein Gupta1
Rishein Gupta1 Bernard P. Arulanandam1*
Bernard P. Arulanandam1*Previously, our laboratory established the role of small, noncoding RNA species, i.e., microRNA (miRNA) including miR-135a in anti-chlamydial immunity in infected hosts. We report here chlamydial infection results in decreased miR-135a expression in mouse genital tissue and a fibroblast cell line. Several chemokine and chemokine receptor genes (including CXCL10, CCR5) associated with chlamydial pathogenesis were identified in silico to contain putative miR-135a binding sequence(s) in the 3’ untranslated region. The role of miR-135a in the host immune response was investigated using exogenous miR-135a mimic to restore the immune phenotype associated with decreased miR-135a following Chlamydia muridarum (Cm) infection. We observed miR-135a regulation of Cm-primed bone marrow derived dendritic cells (BMDC) via activation of Cm-immune CD4+ T cells for clonal expansion and CCR5 expression. Using a transwell cell migration assay, we explore the role of miR-135a in regulation of genital tract CXCL10 expression and recruitment of CXCR3+ CD4+ T cells via the CXCL10/CXCR3 axis. Collectively, data reported here support miR-135a affecting multiple cellular processes in response to chlamydial infection.
Chlamydia trachomatis (Ct) is the leading cause of bacterial sexually transmitted infection (STI), and preventable blindness worldwide. In the U.S. alone, over 1.8 million cases were reported in 2018 (CDC, 2019). An accurate number of infected individuals is not known due to the high level of asymptomatic infections in men and women, i.e., ~50 and 75%, respectively (Darville and Hiltke, 2010). If left untreated in symptomatic and asymptomatic women, Ct induces pathology leading to a whole host of reproductive sequelae including cervicitis, pelvic inflammatory disease (PID), ectopic pregnancy, and infertility (Darville and Hiltke, 2010). The lack of organized healthcare programs in several developing or under-developed countries (Lanjouw et al., 2015), absence of a licensed vaccine (De la Maza et al., 2017), and compounding risk factors (Singh and Marrazzo, 2013; Rey-Ladino et al., 2014; Falasinnu et al., 2015) have collectively resulted in the rising incidence of Ct globally (Yu et al., 2016).
Interest in the effect(s) of non-coding RNA on control of immune signaling has increased. MicroRNA (miRNA/miRs) has been shown to play a significant role in innate and adaptive immune signaling in response to invading bacteria (Keck et al., 2017). The influence of miRs in optimizing the immune response is analogous to other intracellular regulatory events, e.g., phosphorylation and histone modification. Typically, miRs modulate gene function post transcriptionally by binding to target mRNAs leading to increased mRNA degradation with concomitant reduction of translation (Bartel, 2004).
The role of specific miRs in genital Ct infection has been established by our group as well as others (Arkatkar et al., 2015; Gupta et al., 2015; Gupta et al., 2016; Yeruva et al., 2017). Several reports suggest miR-135a may play an important role in proliferation and migration of immune and non-immune cells within the inflammatory milieu (Navarro et al., 2009; Wan et al., 2016; Zeng et al., 2016). In this study, we sought to examine involvement of miR-135a in chlamydial infection using the murine Chlamydia muridarum (Cm) infection model. Identification of host response immune regulator molecules may lead to new and effective treatment strategies for this leading global cause of bacterial STIs.
Cm seed stocks were propagated in HeLa 229 cells. At 24 hours post infection, HeLa cells were mechanically disrupted, and following centrifugation, bacterial pellets were purified on a Renografin (E.R.Squibb and Sons, Inc., Princeton, NJ, USA) gradient as previously described (Gupta et al., 2016). The same Cm seed stock was used throughout this study.
All experiments utilizing animals were performed in accordance with the Animal Welfare Act, the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, and approved guidelines and protocols set forth by the University of Texas at San Antonio Institutional Animal Care and Use Committee (IACUC, approved experimental protocol document MU012). Animals were euthanized by CO2 inhalation followed by cervical dislocation when the experimental endpoints were reached. Female (4-6 week old) wild type (WT) C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME, USA) were inoculated with 5 × 104 inclusion forming units (IFUs) of live Cm elementary bodies (EBs) suspended in 40 μl sucrose-phosphate-glutamic acid (SPG) buffer by intravaginal route 6 days post depo-progesterone administration as previously described (Gupta et al., 2015). Mice challenged with SPG alone were used as mock control.
At the indicated time following infection, mice were euthanized, and genital tract tissue (cervix, uterine horns, and oviducts) was removed aseptically for either immediate cell isolation or storage of tissues at -80°C for downstream RNA isolation and analysis.
Genital tract tissue was minced, transferred to 5 mL culture media containing collagenase (500 U/mL), and vigorously stirred for 1 hour to dissociate tissue as previously described (Jiang and Kelly, 2012). The resulting heterogenous tissue cell suspension (comprised primarily of mucosal epithelial cells, tissue endothelial cells, and immune cells such as macrophages, dendritic, and CD4+ T cells) was passed through a 100 µm cell strainer, and cells were harvested by centrifugation (200 x g for 5 minutes). Bone marrow derived dendritic cells (BMDC) and splenic CD4+T cells were prepared as previously described (Gupta et al., 2016).
Genital tract tissue was snap-frozen, pulverized using a mortar and pestle, and the resulting genital tract tissue frozen powder served as source of total RNA (Gupta et al., 2015).
Cells were suspended in Fc Block (BD Biosciences, San Jose, CA, USA) and PBS containing 0.5% (v/v) FBS for 15 minutes. Following incubation, cells were surface stained with mouse specific antibodies (BD Biosciences); anti-mouse: CD3 (BV605), CD4 (perCP Cy5.5), and CXCR3 (PE) for 30 minutes. Cell suspensions were washed twice, resuspended in PBS containing 0.5% (v/v) FBS and FACS solution, and phenotype determination assessed using an LSR II instrument, and FACSDiva software (BD Biosciences).
Total RNA was obtained from genital tissue powder, cell lines, and murine primary cells using either a miRNeasy RNA extraction Kit (Qiagen, Hilden, DE, USA) or Aurum Total RNA Fatty and Fibrous Tissue Kit (Bio-Rad Laboratories, Hercules, CA, USA) according to manufacturer’s instructions. Extracted RNA was assessed using a Nanodrop Spectrophotometer (ThermoScientific, Asheville, NC). RNA samples (1 µg with A260/280 and A260/230 values of ≅ 2.0 and 1.8 or higher, respectively) were converted to cDNA.
RNA was converted to cDNA using a miScript-II-RT (Qiagen) or iScript Advanced cDNA Synthesis Kit for RT-qPCR according to manufacturer’s recommendations. Amplification was carried out using the Bio-Rad PrimePCR™ SYBR Green Assay (BioRad Laboratories, Hercules, CA, USA), with proprietary forward and reverse primers for mouse CXCL10 (qMmuCED0001068), CXCL12 (qMmuCID0019961), and CCR5 (qMmuCED0051515). MiR-135a expression was determined using the Qiagen hsa-miR-135a-5p miRCURY LNA miRNA (SYBR Green) PCR Assay. Amplification was carried out using a BioRad CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories). Quantitation-normalization of gene expression was achieved using BioRad CFX Maestro software (Bio-Rad Laboratories). MiRNA expression was normalized using Snord68 small nucleolar RNA (Qiagen) which has been shown to exhibit stable expression across different tissues and cell types; whereas, Gapdh and HSP90 housekeeping genes were used for determination of expression of all other gene targets. Differences in miR and gene expression with respect to respective control groups (mock infected, mock treated, and media alone) were determined using the 2(−Average ΔΔCq) method of Livak and Schmittgen (2001).
Transfection of cells with MiR-135a mimic (5’-UAUGGCUUUUUAUUCCUAUGUGA-3’, Qiagen) was conducted using 20 µM Attractene transfection reagent per manufacturer’s (Qiagen) recommendations. Using this established protocol, transfection efficiencies >80% (data not shown) have been previously reported by this laboratory (Gupta et al., 2015). Transfection was carried out for 18-24 hours prior to downstream applications.
BMDC CD11c+ and splenic CD4+ T cells were enriched using cell specific isolation kits per manufacturer’s instructions (Stemcell Technologies, Vancouver, WA, USA). Purified CD11c+ cells were plated at 2 x 105 cells per well, treated with media alone or mmu-miR-135a-mimic for 24 hours, and subsequently infected with Cm (MOI = 1) for 24 hours. Enriched CD4+ T cells from SPG (mock) or Cm infected mice (12 days post infection) were stained with BD Horizon™ Violet Proliferation Dye 450 (VPD, BD Biosciences, San Jose, CA, USA) for 30 minutes followed by co-culturing with BMDCs for 72 hours. Following cell division, the VPD450 dye is distributed uniformly amongst daughter cells with each daughter cell retaining approximately one-half of the parental cell VPD450 fluorescence intensity. The number of VPD450 positive CD4+ T cells and fluorescence intensity were determined by flow cytometry and % T cell proliferation calculated.
Cell migration was assessed as previously described (Gomez-Lopez et al., 2011) using a modified Boyden Chamber. Genital tract tissue from Day 6 mock infected or Cm infected C57BL/6 mice was disassociated (cf., tissue isolation), and cells were seeded at 5 x 105 per well. Cells were transfected with mmu-miR-135a mimic for 24 hours or left untreated. Splenic CD4+ T cells were isolated from Day 6 mock and Cm infected C57BL/6 mice using an EasySep Mouse CD4+ T Cell Isolation Kit per manufacturer’s instructions (Stemcell Technologies, Vancouver, WA, USA). The upper well of the Boyden Chamber containing a 3 μm screen (Corning, NY, USA) was seeded with 5 x 105 cells. An upper insert containing isolated splenic CD4+ T cells or no cells (as control) was introduced into respective wells containing genital tract cells, and incubated for 14 hours. Lower well contents were collected, and subjected to flow cytometry. The number of CD4+ CXCR3+ T cells that migrated from the upper chamber was determined by subtraction of the number of CD4+ CXCR3+ T cells obtained from the respective ‘no cell’ control. The contents of the lower well were used for determination of 1) gene expression by qRT-PCR and 2) cytokine (IFN-γ) production by ELISA (BD OptEIA Mouse IFN-γ ELISA Set).
Bioinformatic analysis for putative related miR binding sites in genes encoding proteins observed to be modulated by C. muridarum infection was accomplished using a miR target predictive algorithm (www.microRNA.org, Memorial Sloan-Kettering Cancer Center, NY). GraphPad Prism 5 (La Jolla, CA) was used to perform all tests of significance. The in silico predicted Mir-135a regulated gene sequences are listed in Table I.
Using an 88 immuno-pathologic miR array, we previously identified 11 miRs (including miR-135a) that were modulated in the genital tract post chlamydial challenge (Gupta et al., 2015). In order to ascertain the role of miR-135a in Chlamydia pathogenesis, we first confirmed that its expression is regulated by Cm infection. qRT-PCR quantification of miR-135a expression in genital tract tissue revealed a 75% decrease at day 6 post intravaginal Cm challenge (Figure 1). Furthermore, in vitro chlamydial infection (24 hours) of genital tract single-cell suspensions and McCoy cells (a murine fibroblast cell line commonly employed to propagate chlamydial strains) resulted in a 76 and 78% reduction of miR-135a expression, respectively (Figure 1). These results suggest Cm infection down-regulates miR-135a. Subsequent in silico analysis of potential miR-135a target genes resulted in identification of several chemokine and chemokine receptor genes including CCR5, CXCL10 and CXCL12 containing putative miR-135a binding sequences in their respective 3’ untranslated region (Table 1). Host response down-regulation of miR-135a following Cm infection may prevent degradation of these gene transcripts thus allowing disease resolution. In order to assess restoration of decreased miR-135a following Cm infection, an in vitro approach using primary cells isolated from mock and Cm infected mice with and without administered miR-135a mimic was employed.
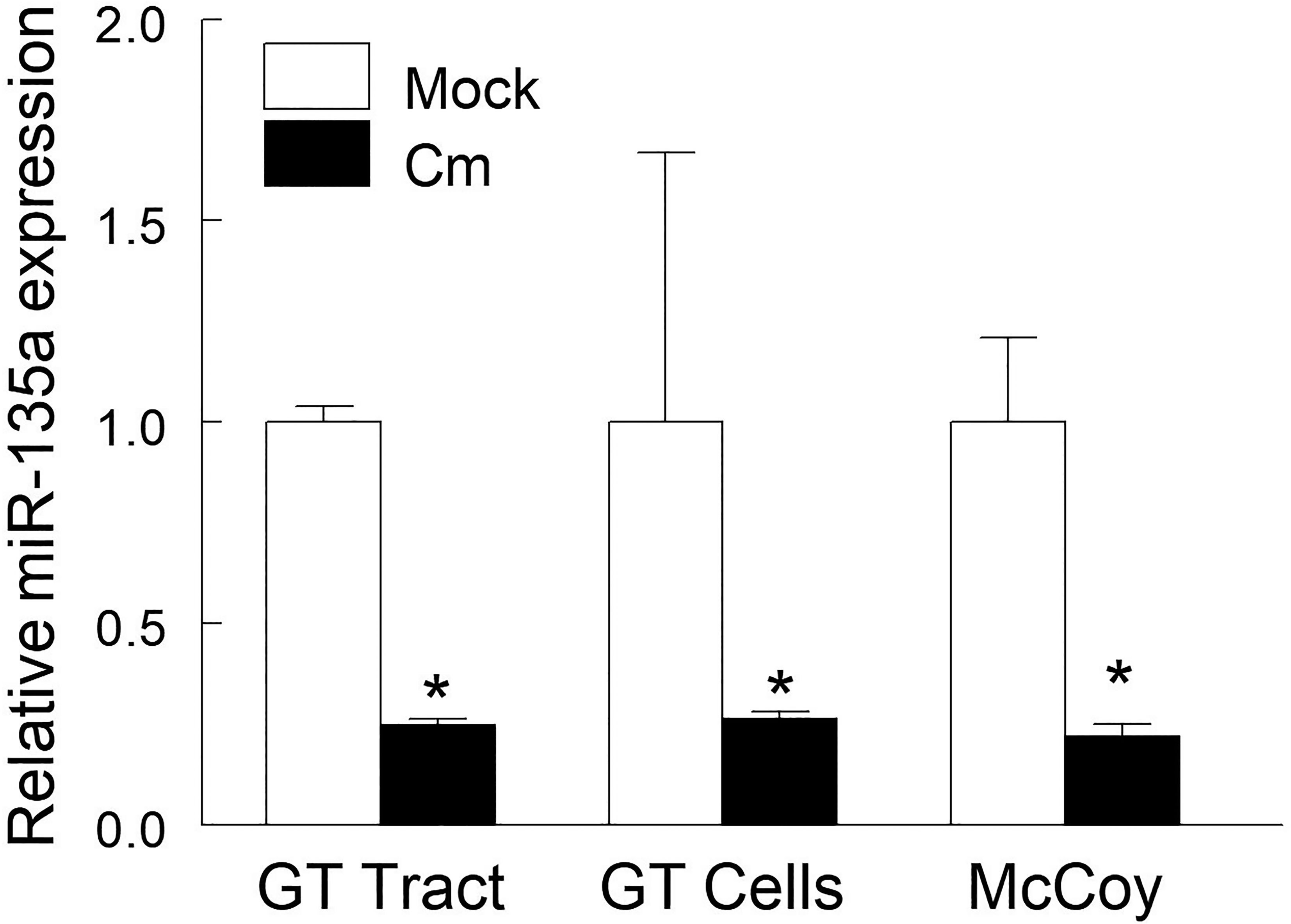
Figure 1 Chlamydia muridarum infection down-regulates miR-135a. C57BL/6 mice (n = 3) were intravaginally challenged with Cm (5 × 104 IFUs in SPG buffer) or SPG buffer alone (mock). Genital tract tissue was collected at day 6 post-challenge and miR-135a expression was measured. Cell suspensions of naïve mouse genital tract and fibroblast cell line (McCoy) cells were exposed to Cm (2 x 105 IFUs, MOI = 1) for 24 hours in a 24-well microplate. Relative expression of miR135a in Cm infected tissue/cells compared to the respective uninfected tissue/cells was determined by qRT-PCR and calculated using the 2−ΔΔCT method. Data shown are representative of 3 experiments. *p < 0.05 Student t test.
Dendritic and Th1 cells are important for immune defense against genital chlamydial infection. It has been reported that CCR5 expressing CD4+ T cells are critical for host protection against chlamydial infection (Belay et al., 2002; Barr et al., 2005; Olive et al., 2011). Therefore, we assessed in BMDC the contribution of miR-135a in T cell proliferation and CCR5 gene expression by co-culturing Cm-exposed BMDC with CD4+ T cells from Cm infected mice. Specifically, BMDC prepared from naïve mice and transfected ormock-transfected with miR-135a mimic were exposed to Cm for 24 hours followed by co-culturing with CD4+ T cells enriched from Cm infected or uninfected (mock) spleen. As shown in Figure 2A, Cm-exposed naïve BMDCs with or without miR135a mimic transfection minimally activate naive CD4+ T cells for proliferation (less than 12%). In contrast, 67.07% ± 4.13 of Cm-primed CD4+ T cell population underwent proliferation following activation by Cm-primed BMDCs. This antigen specific BMDC-mediated T cell proliferation was abrogated when BMDCs were transfected with miR135a mimic (10.60% ± 3.77, p<0.05). Furthermore, CCR5 gene expression in naïve CD4+ T cells was not altered by incubation with Cm-primed BMDCs with or without transfected miR-135a mimic (Figure 2B). In contrast, Cm-exposed BMDC activated Cm-primed T cells indicated a 1.44 ± 0.01fold-increase in CCR5 gene expression in the absence of miR-135a mimic but a 56% decrease in the presence of miR-135a mimic compared to the corresponding naïve (mock) T cell control (Figure 2B).
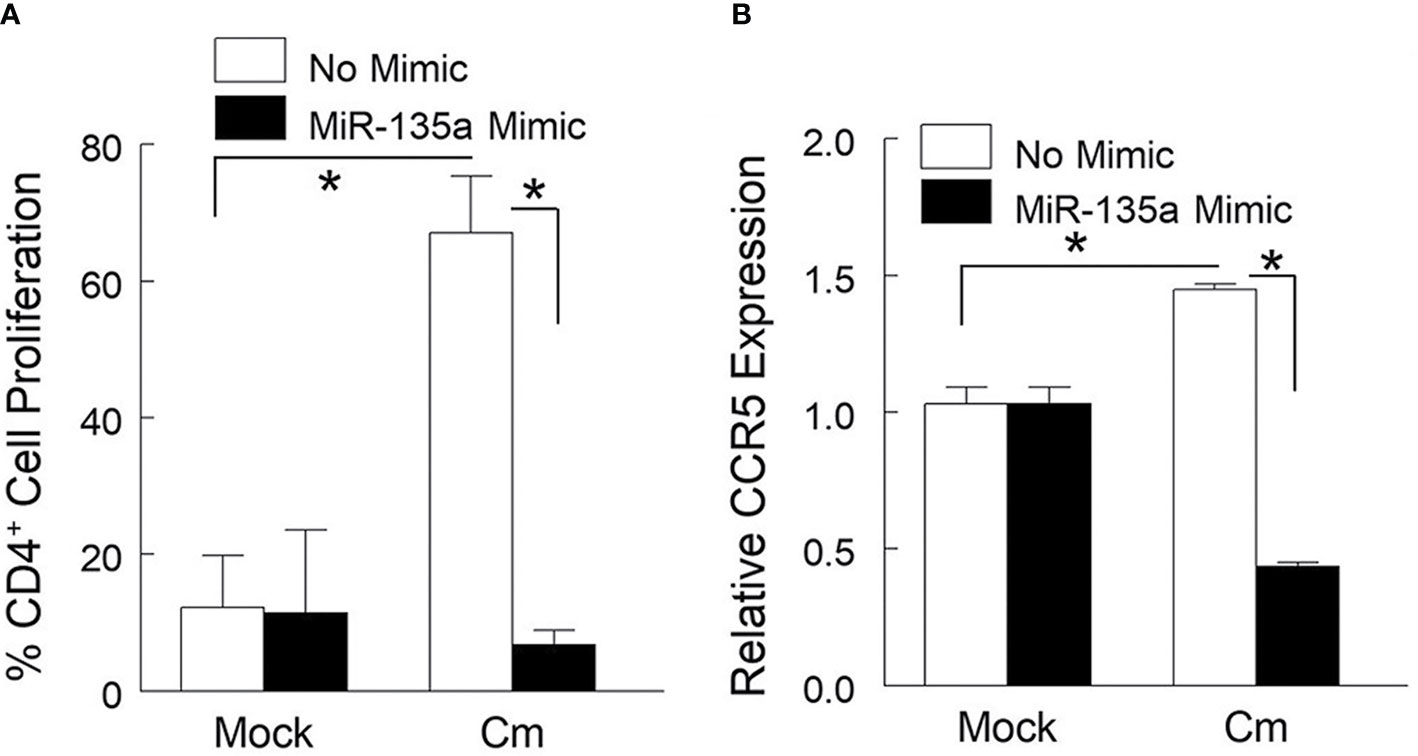
Figure 2 miR-135a affects dendritic cell mediated Cm-primed CD4+ T cell proliferation and CCR5 gene expression. Bone marrow derived dendritic cells (BMDC) were stimulated with media alone, or media containing miR-135a mimic for 24 hours followed by exposure to Cm (MOI = 1). Cm primed BMDCs were co-cultured with splenic CD4+ T cells from day 6-post intravaginally challenged mice (5 x 104 IFU) or mock infected. (A) Effect of miR-135a mimic on CD4+ T cell proliferation. (B) Effect of miR-135a mimic on CCR5 gene expression. Data shown are representative of 3 experiments. *p < 0.05 Student t test.
In primary Cm infection, the initial immune response in the mouse genital tract is dominated by myeloid cell infiltrates, including neutrophils, followed by recruitment of T cells (CD4+ T cells in particular) during the resolution stage (Morrison and Morrison, 2000). Considering the presence of a predictive miR-135a binding target sequence in the 3’ UTR of chemokine CXCL10 mRNA (Table 1), we hypothesized that miR-135a could disrupt CD4+ T cell migration through the CXCL10/CXCR3 axis. To test this hypothesis, we measured migration of CXCR3+ CD4+ T cells to CXCL10 secreting genital cells using a combined Boyden chamber-Flow Cytometry Assay. Specifically, mouse genital tract cells isolated from day 6 mock and Cm infected animals were seeded in the lower well of Boyden chamber, and either transfected with miR-135a mimic or left untreated. Splenic CD4+ T cells isolated from Day 6 Cm infected mice were placed in the upper well of the Boyden chamber. As shown in Figure 3A using a gating strategy (Supplementary Figure 1), migration of splenic CD3+CD4+ T cells expressing the CXCR3 marker increased approximately 61% after incubation with Cm-infected genital cells (6873 ± 839) compared to incubation with uninfected cells (4269 ± 854). Transfection of Cm-infected genital tract cells with miR-135a mimic significantly reduced CXCR3+ CD4+ T cell migration approximately 56%, i.e., from 6873 ± 839 to 3047 ± 493 cells. Furthermore, we analyzed the total cell population present in lower wells following T cell migration (14 hours) for chemotaxis gene expression and effector cytokine secretion. As shown in Figure 3B, Cm-infected genital tract cells interacted with Cm-primed CD4+ T cells resulting in increased CXCL10 gene expression (7.49 ± 0.11) compared to the uninfected cell control (normalized as 1). The observed increase in CXCL10 gene expression was reduced in Cm-infected genital tract cells transfected with miR-135a mimic (from 7.49 ± 0.11 to 5.18 ± 0.04-fold) which correlated with reduction of CXCR3+ CD4+ T cell migration (Figure 3A). Other Cm infection induced cell migration/homing immune molecules were also affected by miR135-a mimic transfection including reduced CXCL12 (6.33 ± 0.84 to 2.65 ± 0.42), and CCR5 (29.13 ± 0.89 to 23.43 ± 0.11) (Figure 3B). Effector Th1 T cells producing INF-γ have been shown to be critical in resolving genital tract Cm infection (Perry et al., 1997; Morrison and Caldwell, 2002; Li et al., 2008). Interaction of genital tract cells collected from Cm infected mice with CD4+ T cells induced IFN-γ production secretion (268 ± 61 pg/ml, p < 0.05), and transfection of these Cm-infected genital tract cells with miR135a mimic significantly reduced IFN-γ production (192 ± 35 pg/ml) which may partially be due to the reduced CD4+ T cell migration to the lower chamber (Figure 3C).
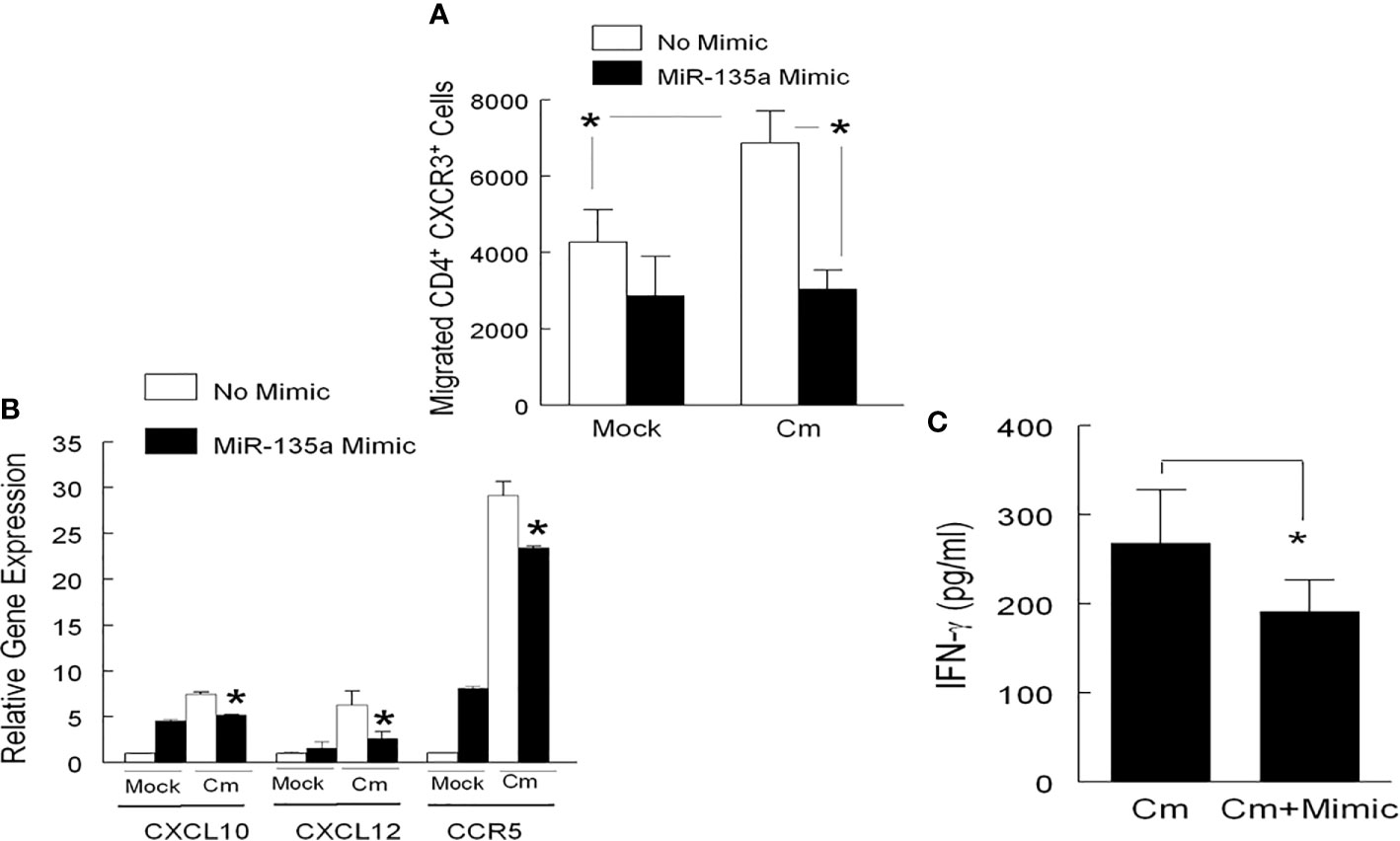
Figure 3 miR-135a affects CD4+ T cell migration. T cell migration was determined using a modified 24-well Boyden Chamber. Genital tract tissue cell suspensions from mock or Cm infected (day 6) mice were added to the lower chamber (5 x 105 per well), and transfected with miR-135a mimic or media alone for 24 hours prior to determination of migration. Enriched CD4+ T cells (from day 6 Cm infected splenocytes) were added to the upper chamber (5 x 105 cell per well). Fourteen hours post addition of cells to the upper chamber, the lower chamber contents were used for (A) enumeration of migrated CXCR3-expressing CD4+ T cells by Flow cytometry, (B) comparison of relative CXCL10, CXCL12, and CCR5 gene expression in Cm infected cells transfected with and without miR-135a mimic to the uninfected/non-transfected control group (expression level normalized to 1), and (C) determination of IFN-γ secretion. Data shown are representative of 3 experiments. *p < 0.05 Student t test.
The miR-135a gene (mir135a-2) is located on chromosome 10 (Chr10:92072086-92072185 bp, - strand), and is known for its role as an oncogenic miRNA which controls tumor aggressiveness in several cancers including colon, melanoma, breast, and prostate (Wu et al., 2012; Zhou et al., 2012; Yamada et al., 2013; Kroiss et al., 2015; Zeng et al., 2016; Fukagawa et al., 2017). The regulatory role of miR-135a in cancer progression by either promoting or suppressing tumor cell growth/migration has been well documented (Wu et al., 2012; Yamada et al., 2013; Zeng et al., 2016; Xie et al., 2019; Zhang et al., 2019); however, its role in infectious diseases is largely unknown.
The proliferative responses observed in inflamed microenvironments has been reported to be controlled by FOXO1 in a miR-135a dependent fashion (Mao et al., 2015; Ren et al., 2015; Zeng et al., 2016). Importantly, because of the putative regulation of transcription factor STAT-6 (Pandey et al., 2016), or proliferation factors such as ROCK1, HOXA10, and BCL-2, the cellular therapeutic potential of miR-135a may hold promise (Yang and Yin, 2017). To this end, the in vivo regulation of GATA3, a cellular Th2 lineage marker for miR-135a has been used in correcting the Th1/Th2 imbalance in acute allergic rhinitis (Luo et al., 2014), and is thus proposed as a therapeutic agent for intervention of mast cell and allergen induced inflammation (Deng et al., 2015).
Pursuing our previous findings (Arkatkar et al., 2015; Gupta et al., 2015; Keck et al., 2017), we report here an unrecognized but suggested role for miR-135a, i.e., regulation of immune gene expression following chlamydial infection. Intravaginal (i. vag) infection of mice by Cm significantly reduced overall miR-135a expression in genital tract tissue. This Cm-mediated reduction of miR-135a was demonstrated in vitro using genital tract tissue cell preparations and a murine fibroblast cell line. Although absent bacterial burden data needed to conclusively show increasing disease severity in the presence of a miR-135a mimic countering the suggested Cm-induced down-regulation of miR-135a, we do demonstrate the immune-regulatory role of miR-135a as evidenced by alteration of effector T cell activation and infection site homing. Specifically, we observed that miR-135a can regulate 1) CCR5 expression and cell proliferation of Cm-primed CD4+ T cells when activated by Cm-exposed BMDC, and 2) migration of Cm-primed CD4+ T cells to Cm-infected genital tract cells via CXCL10/CXCR3 axis mediated immune activation. These miR-135a mediated effects on immune cell proliferation and/or migration are consistent with previously implicated chemokine and chemokine receptor function in Ct/Cm infection (Belay et al., 2002; Olive et al., 2011). The ability of miR-135a to disrupt CD4+ T cell proliferation in concert with expression of CCR5 (Figures 2 and 3) is suggestive of its potentially important role in immune cell trafficking following infection. Additionally, in the presence of miR-135a mimic, a decrease in the migratory index of splenic CD4+ CXCR3+ T cells were observed (Figure 3 and Supplementary Figure 1). Expression of CXCR3 in CD4+ T cells has been shown to alter migration patterns in Chlamydia infected hosts (Olive et al., 2011). This observation is congruent with that of others defining the important role for CXCL10 in migrating T cells (Bonecchi et al., 1998; Dufour et al., 2002).
We observed miR-135a regulatory binding sites for genes previously implicated in anti-chlamydial immunity (Table 1) and regulation of IFN-γ (Figure 3C). Based on signaling axes in which these genes are involved and additional, ongoing preliminary investigations including in silico and wet laboratory analyses (data not shown), we hypothesize that miR-135a modulates multiple signaling pathways using the following mechanisms: (1) Type-1 and Type-2 IFN signaling through molecules such as Jak1 an IFNαR2, IL-33, CXCL10, and/or (2) alteration of canonical (MyD88) and non-canonical (Tram) signaling pathways, thereby collectively contributing to regulation of anti-chlamydial immunity via interferon signaling. Thus, additional gain and loss of function studies are warranted to investigate these pathways.
Using our previously established dendritic effector cell: co-culture model (Gupta et al., 2016), and a modified Boyden Chamber assay (Gomez-Lopez et al., 2011), we determined the effects of miR-135a on cell proliferation (cognate interaction between dendritic cells and antigen specific CD4+ T cells), and cell migration in antigen specific CD4+ T cells, respectively (Figures 2 and 3). Focusing on the initial stage of i.vag. infection (day 6 post infection) (Gupta et al., 2015; Gupta et al., 2016), we interrogated the effect of miR-135a on CD4+ T cell function (Figures 2 and 3) which has been implicated in early stages of genital chlamydial infection (Maxion et al., 2004; Roan et al., 2006; Jayarapu et al., 2009; Farris et al., 2010; Lee et al., 2010; Yu et al., 2010; Olive et al., 2011; Jiang et al., 2017; Naglak et al., 2017; Lijek et al., 2018).
While our findings suggest changes at the mRNA level consistent with miR regulation of gene expression, we cannot rule out possible miR-135a independent events, e.g., the presence of mRNA variants, post-transcriptional silencing, and/or compensatory effects by other immune cell populations. Furthermore, we assume that the differences in immune gene function following Cm infection is at minimum antigen-based, and not Cm antigen specific. The lack of a specific cell type in vivo model, i.e., conditional knockout, fate-mapped or inducible mouse model (Mak et al., 2001; Poltorak and Schraml, 2015; Shi et al., 2018) necessitated use of differing cell lines/cell types which may exhibit differential immune presentation. Nevertheless, this is the first report of immune regulatory mechanisms in which the host response to chlamydial infection occurs via miR-135a. Although data reported here are primarily in vitro derived, they can serve as a starting point for in vivo study into the role of miR-135a in chlamydial pathogenesis either by intraperitoneal injection of liposome encapsulated miR-135a mimic or use of miR-135a gene knockout mice when such animals become available.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
The animal study was reviewed and approved by The University of Texas at San Antonio Institutional Animal Care and Use Committee (IACUC).
JPC, MNG, RG, and BPA conceived and designed the experiments. JK carried out the experiments. XC provided reagents for experiments. JK, J-JY, MNG, JPC, RG, and BPA analyzed the data. JK, J-JY, JPC, LC, RG, and BPA wrote and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Institutes of Health (NIH) grant IR03AI11771401A1, and the Army Research Office (Department of Defense contract No. W911NF-11-1-0136).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We gratefully acknowledge the technical assistance of Dona Haj Bashir, Laura Henley, Aravind Kancharla, Tanvi Arkatkar, and Dr. Irma Yadira Izaguirre Hernández.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.638058/full#supplementary-material
Arkatkar, T., Gupta, R., Li, W., Yu, J. J., Wali, S., Neal Guentzel, M., et al. (2015). Murine MicroRNA-214 regulates intracellular adhesion molecule (ICAM1) gene expression in genital Chlamydia muridarum infection. Immunology 145, 534–542. doi: 10.1111/imm.12470
Barr, E. L., Ouburg, S., Igietseme, J. U., Morre, S. A., Okwandu, E., Eko, F. O., et al. (2005). Host inflammatory response and development of complications of Chlamydia trachomatis genital infection in CCR5-deficient mice and subfertile women with the CCR5delta32 gene deletion. J. Microbiol. Immunol. Infect. 38, 244–254.
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Belay, T., Eko, F. O., Ananaba, G. A., Bowers, S., Moore, T., Lyn, D., et al. (2002). Chemokine and chemokine receptor dynamics during genital chlamydial infection. Infect. Immun. 70, 844–850. doi: 10.1128/IAI.70.2.844-850.2002
Bonecchi, R., Bianchi, G., Bordignon, P. P., D’ambrosio, D., Lang, R., Borsatti, A., et al. (1998). Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 187, 129–134. doi: 10.1084/jem.187.1.129
CDC (2019). Sexually Transmitted Disease Surveillance 2018 (Altanta, GA: US Department of Health and Human Services).
Darville, T., Hiltke, T. J. (2010). Pathogenesis of genital tract disease due to Chlamydia trachomatis. J. Infect. Dis. 201 Suppl 2, S114–S125. doi: 10.1086/652397
De la Maza, L. M., Zhong, G., Brunham, R. C. (2017). Update on Chlamydia trachomatis Vaccinology. Clin. Vaccine Immunol. 24 (85), e00409–17. doi: 10.1128/CVI.00543-16
Deng, Y. Q., Yang, Y. Q., Wang, S. B., Li, F., Liu, M. Z., Hua, Q. Q., et al. (2015). Intranasal Administration of Lentiviral miR-135a Regulates Mast Cell and Allergen-Induced Inflammation by Targeting GATA-3. PloS One 10, e0139322. doi: 10.1371/journal.pone.0139322
Dufour, J. H., Dziejman, M., Liu, M. T., Leung, J. H., Lane, T. E., Luster, A. D. (2002). IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J. Immunol. 168, 3195–3204. doi: 10.4049/jimmunol.168.7.3195
Falasinnu, T., Gilbert, M., Hottes, T. S., Gustafson, P., Ogilvie, G., Shoveller, J. (2015). Predictors identifying those at increased risk for STDs: a theory-guided review of empirical literature and clinical guidelines. Int. J. STD AIDS 26, 839–851. doi: 10.1177/0956462414555930
Farris, C. M., Morrison, S. G., Morrison, R. P. (2010). CD4+ T cells and antibody are required for optimal major outer membrane protein vaccine-induced immunity to Chlamydia muridarum genital infection. Infect. Immun. 78, 4374–4383. doi: 10.1128/IAI.00622-10
Fukagawa, S., Miyata, K., Yotsumoto, F., Kiyoshima, C., Nam, S. O., Anan, H., et al. (2017). MicroRNA-135a-3p as a promising biomarker and nucleic acid therapeutic agent for ovarian cancer. Cancer Sci. 108, 886–896. doi: 10.1111/cas.13210
Gomez-Lopez, N., Vadillo-Ortega, F., Estrada-Gutierrez, G. (2011). Combined boyden-flow cytometry assay improves quantification and provides phenotypification of leukocyte chemotaxis. PloS One 6, e28771. doi: 10.1371/journal.pone.0028771
Gupta, R., Arkatkar, T., Yu, J. J., Wali, S., Haskins, W. E., Chambers, J. P., et al. (2015). Chlamydia muridarum infection associated host MicroRNAs in the murine genital tract and contribution to generation of host immune response. Am. J. Reprod. Immunol. 73, 126–140. doi: 10.1111/aji.12281
Gupta, R., Arkatkar, T., Keck, J, Koundinya, G. K., Castillo, K., Hobel, S., et al. (2016). Antigen specific immune response in Chlamydia muridarum genital infection is dependent on murine microRNAs-155 and -182. Oncotarget 7, 64726–64742. doi: 10.18632/oncotarget.11461
Jayarapu, K., Kerr, M. S., Katschke, A., Johnson, R. M. (2009). Chlamydia muridarum-specific CD4 T-cell clones recognize infected reproductive tract epithelial cells in an interferon-dependent fashion. Infect. Immun. 77, 4469–4479. doi: 10.1128/IAI.00491-09
Jiang, J., Maxion, H., Champion, C. I., Liu, G., Kelly, K. A. (2017). Expression of CXCR3 on Adaptive and Innate Immune Cells Contributes Oviduct Pathology throughout Chlamydia muridarum Infection. J. Mucosal Immunol. Res. 1.
Jiang, J., Kelly, K. A. (2012). Isolation of lymphocytes from mouse genital tract mucosa. J. Vis. Exp. 67, e4391 10.3791/4391. doi: 10.3791/4391
Keck, J., Gupta, R., Christenson, L. K., Arulanandam, B. P. (2017). MicroRNA mediated regulation of immunity against gram-negative bacteria. Int. Rev. Immunol. 36, 287–299. doi: 10.1080/08830185.2017.1347649
Kroiss, A., Vincent, S., Decaussin-Petrucci, M., Meugnier, E., Viallet, J., Ruffion, A., et al. (2015). Androgen-regulated microRNA-135a decreases prostate cancer cell migration and invasion through downregulating ROCK1 and ROCK2. Oncogene 34, 2846–2855. doi: 10.1038/onc.2014.222
Lanjouw, E., Ouburg, S., De Vries, H. J., Stary, A., Radcliffe, K., Unemo, M. (2015). Background review for the ‘2015 European guideline on the management of Chlamydia trachomatis infections’. Int. J. STD AIDS. 24, 0956462415618838. doi: 10.1177/095646241561883
Lee, H. Y., Schripsema, J. H., Sigar, I. M., Murray, C. M., Lacy, S. R., Ramsey, K. H. (2010). A link between neutrophils and chronic disease manifestations of Chlamydia muridarum urogenital infection of mice. FEMS Immunol. Med. Microbiol. 59, 108–116. doi: 10.1111/j.1574-695X.2010.00668.x
Li, W., Murthy, A. K., Guentzel, M. N., Seshu, J., Forsthuber, T. G., Zhong, G., et al. (2008). Antigen-specific CD4+ T cells produce sufficient IFN-gamma to mediate robust protective immunity against genital Chlamydia muridarum infection. J. Immunol. 180, 3375–3382. doi: 10.4049/jimmunol.180.5.3375
Lijek, R. S., Helble, J. D., Olive, A. J., Seiger, K. W., Starnbach, M. N. (2018). Pathology after Chlamydia trachomatis infection is driven by nonprotective immune cells that are distinct from protective populations. Proc. Natl. Acad. Sci. U.S.A. 115, 2216–2221. doi: 10.1073/pnas.1711356115
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luo, Y., Deng, Y., Tao, Z., Chen, S., Xiao, B., Ren, J., et al. (2014). Regulatory effect of microRNA-135a on the Th1/Th2 imbalance in a murine model of allergic rhinitis. Exp. Ther. Med. 8, 1105–1110. doi: 10.3892/etm.2014.1855
Mak, T. W., Penninger, J. M., Ohashi, P. S. (2001). Knockout mice: a paradigm shift in modern immunology. Nat. Rev. Immunol. 1, 11–19. doi: 10.1038/3509551
Mao, X. P., Zhang, L. S., Huang, B., Zhou, S. Y., Liao, J., Chen, L. W., et al. (2015). Mir-135a enhances cellular proliferation through post-transcriptionally regulating PHLPP2 and FOXO1 in human bladder cancer. J. Transl. Med. 13, 86. doi: 10.1186/s12967-015-0438-8
Maxion, H. K., Liu, W., Chang, M. H., Kelly, K. A. (2004). The infecting dose of Chlamydia muridarum modulates the innate immune response and ascending infection. Infect. Immun. 72, 6330–6340. doi: 10.1128/IAI.72.11.6330-6340.2004
Morrison, R. P., Caldwell, H. D. (2002). Immunity to murine chlamydial genital infection. Infect. Immun. 70, 2741–2751. doi: 10.1128/IAI.70.6.2741-2751.2002
Morrison, S. G., Morrison, R. P. (2000). In situ analysis of the evolution of the primary immune response in murine Chlamydia trachomatis genital tract infection. Infect. Immun. 68, 2870–2879. doi: 10.1128/IAI.68.5.2870-2879.2000
Naglak, E. K., Morrison, S. G., Morrison, R. P. (2017). Neutrophils Are Central to Antibody-Mediated Protection against Genital Chlamydia. Infect. Immun. 85. doi: 10.1128/IAI.00409-17
Navarro, A., Diaz, T., Martinez, A., Gaya, A., Pons, A., Gel, B., et al. (2009). Regulation of JAK2 by miR-135a: prognostic impact in classic Hodgkin lymphoma. Blood 114, 2945–2951. doi: 10.1182/blood-2009-02-204842
Olive, A. J., Gondek, D. C., Starnbach, M. N. (2011). CXCR3 and CCR5 are both required for T cell-mediated protection against C. trachomatis infection in the murine genital mucosa. Mucosal Immunol. 4, 208–216. doi: 10.1038/mi.2010.58
Pandey, R. K., Sundar, S., Prajapati, V. K. (2016). Differential Expression of miRNA Regulates T Cell Differentiation and Plasticity During Visceral Leishmaniasis Infection. Front. Microbiol. 7, 206. doi: 10.3389/fmicb.2016.00206
Perry, L. L., Feilzer, K., Caldwell, H. D. (1997). Immunity to Chlamydia trachomatis is mediated by T helper 1 cells through IFN-gamma-dependent and -independent pathways. J. Immunol. 158, 3344–3352.
Poltorak, M. P., Schraml, B. U. (2015). Fate mapping of dendritic cells. Front. Immunol. 6, 199. doi: 10.3389/fimmu.2015.00199
Ren, J. W., Li, Z. J., Tu, C. (2015). MiR-135 post-transcriptionally regulates FOXO1 expression and promotes cell proliferation in human malignant melanoma cells. Int. J. Clin. Exp. Pathol. 8, 6356–6366.
Rey-Ladino, J., Ross, A. G., Cripps, A. W. (2014). Immunity, immunopathology, and human vaccine development against sexually transmitted Chlamydia trachomatis. Hum. Vaccin Immunother. 10, 2664–2673. doi: 10.4161/hv.29683
Roan, N. R., Gierahn, T. M., Higgins, D. E., Starnbach, M. N. (2006). Monitoring the T cell response to genital tract infection. Proc. Natl. Acad. Sci. U.S.A. 103, 12069–12074. doi: 10.1073/pnas.0603866103
Shi, J., Hua, L., Harmer, D., Li, P., Ren, G. (2018). Cre Driver Mice Targeting Macrophages. Methods Mol. Biol. 1784, 263–275. doi: 10.1007/978-1-4939-7837-3_24
Singh, D., Marrazzo, J. M. (2013). Screening and management of genital chlamydial infections. Infect. Dis. Clin. North Am. 27, 739–753. doi: 10.1016/j.idc.2013.08.006
Wan, X., Pu, H., Huang, W., Yang, S., Zhang, Y., Kong, Z., et al. (2016). Androgen-induced miR-135a acts as a tumor suppressor through downregulating RBAK and MMP11, and mediates resistance to androgen deprivation therapy. Oncotarget 7, 51284–51300. doi: 10.18632/oncotarget.9992
Wu, H., Huang, M., Cao, P., Wang, T., Shu, Y., Liu, P. (2012). MiR-135a targets JAK2 and inhibits gastric cancer cell proliferation. Cancer Biol. Ther. 13, 281–288. doi: 10.4161/cbt.18943
Xie, Y., Li, F., Li, Z., Shi, Z. (2019). miR-135a suppresses migration of gastric cancer cells by targeting TRAF5-mediated NF-kappaB activation. Onco. Targets Ther. 12, 975–984. doi: 10.2147/OTT.S189976
Yamada, Y., Hidaka, H., Seki, N., Yoshino, H., Yamasaki, T., Itesako, T., et al. (2013). Tumor-suppressive microRNA-135a inhibits cancer cell proliferation by targeting the c-MYC oncogene in renal cell carcinoma. Cancer Sci. 104, 304–312. doi: 10.1111/cas.12072
Yang, G., Yin, B. (2017). Therapeutic effects of long-circulating miR-135a-containing cationic immunoliposomes against gallbladder carcinoma. Sci. Rep. 7, 5982. doi: 10.1038/s41598-017-06234-8
Yeruva, L., Pouncey, D. L., Eledge, M. R., Bhattacharya, S., Luo, C., Weatherford, E. W., et al. (2017). MicroRNAs Modulate Pathogenesis Resulting from Chlamydial Infection in Mice. Infect. Immun. 85. doi: 10.1128/IAI.00768-16
Yu, H., Jiang, X., Shen, C., Karunakaran, K. P., Jiang, J., Rosin, N. L., et al. (2010). Chlamydia muridarum T-cell antigens formulated with the adjuvant DDA/TDB induce immunity against infection that correlates with a high frequency of gamma interferon (IFN-gamma)/tumor necrosis factor alpha and IFN-gamma/interleukin-17 double-positive CD4+ T cells. Infect. Immun. 78, 2272–2282. doi: 10.1128/IAI.01374-09
Yu, H., Karunakaran, K. P., Jiang, X., Brunham, R. C. (2016). Subunit vaccines for the prevention of mucosal infection with Chlamydia trachomatis. Expert Rev. Vaccines 15, 977–988. doi: 10.1586/14760584.2016.1161510
Zeng, Y. B., Liang, X. H., Zhang, G. X., Jiang, N., Zhang, T., Huang, J. Y., et al. (2016). miRNA-135a promotes hepatocellular carcinoma cell migration and invasion by targeting forkhead box O1. Cancer Cell Int. 16, 63. doi: 10.1186/s12935-016-0328-z
Zhang, Y., Jiang, W. L., Yang, J. Y., Huang, J., Kang, G., Hu, H. B., et al. (2019). Downregulation of lysyl oxidase-like 4 LOXL4 by miR-135a-5p promotes lung cancer progression in vitro and in vivo. J. Cell Physiol. 234, 18679–18687. doi: 10.1002/jcp.28508
Keywords: Chlamydia, microRNA, miR135a, cell migration, dendritic cells
Citation: Keck J, Chambers JP, Yu J-J, Cheng X, Christenson LK, Guentzel MN, Gupta R and Arulanandam BP (2021) Modulation of Immune Response to Chlamydia muridarum by Host miR-135a. Front. Cell. Infect. Microbiol. 11:638058. doi: 10.3389/fcimb.2021.638058
Received: 04 December 2020; Accepted: 15 March 2021;
Published: 13 April 2021.
Edited by:
Vera Kozjak-Pavlovic, Julius Maximilian University of Würzburg, GermanyReviewed by:
Erika Ildiko Lutter, Oklahoma State University, United StatesCopyright © 2021 Keck, Chambers, Yu, Cheng, Christenson, Guentzel, Gupta and Arulanandam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bernard P. Arulanandam, QmVybmFyZC5hcnVsYW5hbmRhbUB1dHNhLmVkdQ==
†Present address: Xingguo Cheng, Dynamic Entropy Technology LLC, Devine, TX, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.