
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol. , 10 March 2021
Sec. Bacteria and Host
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.624094
This article is part of the Research Topic Bacterial Effectors as Drivers of Human Disease: Models, Methods, Mechanisms View all 16 articles
Type I and III interferons (IFNs) are archetypally antiviral cytokines that are induced in response to recognition of foreign material by pattern recognition receptors (PRRs). Though their roles in anti-viral immunity are well established, recent evidence suggests that they are also crucial mediators of inflammatory processes during bacterial infections. Type I and III IFNs restrict bacterial infection in vitro and in some in vivo contexts. IFNs mainly function through the induction of hundreds of IFN-stimulated genes (ISGs). These include PRRs and regulators of antimicrobial signaling pathways. Other ISGs directly restrict bacterial invasion or multiplication within host cells. As they regulate a diverse range of anti-bacterial host responses, IFNs are an attractive virulence target for bacterial pathogens. This review will discuss the current understanding of the bacterial effectors that manipulate the different stages of the host IFN response: IFN induction, downstream signaling pathways, and target ISGs.
The first interferons (IFNs) were discovered in 1957 by Alick Isaacs and Jean Lindenmann when they noticed that tissues inoculated with inactivated virus produced a soluble substance that “interfered” with subsequent viral infection (Isaacs and Lindenmann, 1957). Over 20 years before Charles Janeway’s (Janeway, 1989) predictions on pattern recognition, Isaacs and Lindenmann had recognized the fundamental properties of IFN: a cytokine produced in response to detection of pathogen associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs), that can protect tissues from subsequent microbial infections.
There are three families of IFN: type I, II, and III, which bind to the IFNα receptor (IFNAR), IFNγ receptor (IFNGR) and IFNλ receptor (IFNLR), respectively. Type I IFNs include IFNα, IFNβ, and the lesser-studied IFNϵ, κ, τ, δ, ζ, and ω and are expressed by almost all cells (Pestka et al., 2004; Hertzog and Williams, 2013). IFNγ is the sole type II IFN family member and is not induced in response to pattern recognition, but rather by immune cells in response to other cytokines (Pien et al., 2000; Salazar-Mather et al., 2000; Schindler et al., 2001). Type III IFNs were discovered most recently (Sheppard et al., 2002; Kotenko et al., 2003) and include four members in humans: IFNλ1-4. IFNAR and IFNGR are expressed on all nucleated cells, however the expression of IFNLR is restricted to epithelial cells (ECs) and some immune cells, including neutrophils (Blazek et al., 2015; Broggi et al., 2017). As such, type I and II IFNs have systemic functions (De Weerd and Nguyen, 2012; Chen et al., 2017; Lazear et al., 2019) while type III IFNs are crucial for host defense at barrier sites including the gut and lung (Durbin et al., 2013; Broggi et al., 2017; Lazear et al., 2019; Broggi et al., 2020). IFNs signal in an autocrine and paracrine manner through their respective receptors to activate janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling cascades, resulting in the expression of hundreds of genes, known collectively as interferon-stimulated genes (ISGs). Although the roles of IFNs and ISGs have been extensively studied in the context of viral infection, there is a dearth of knowledge of their role during bacterial infections. Type I and III IFNs are strongly induced upon recognition of bacterial ligands, and play diverse and context-dependent roles during infection (Kagan et al., 2008; Pandey et al., 2009).
To promote their survival within the host, bacteria have evolved virulence factors such as secretion systems; apparatuses that translocate effector proteins across host membranes (Green and Mecsas, 2016). These effectors enable the pathogen to evade and perturb the host response or even use it to their advantage. This review will examine the interplay between bacterial effectors and the IFN response. As modulation of IFNγ has been previously discussed (Kak et al., 2018) we have focussed our discussion on the lesser understood type I and III IFNs. We will first summarize the current understanding of the complex functions of type I and III IFNs in the defense against pathogenic bacteria.
Type I and III IFNs have anti-bacterial properties in most in vitro tissue culture models. For example, treatment of polarized ECs by type I and/or III IFNs protects epithelial barriers from damage caused by enteropathogenic Escherichia coli (EPEC) (Long et al., 2014), Salmonella enterica serovar Typhimurium, and Shigella flexneri (Odendall et al., 2017). In addition, type I IFN restricts the replication of several intracellular bacteria, including S. Typhimurium and S. flexneri (Bukholm et al., 1984; Niesel et al., 1986; Duménil et al., 1998; Helbig et al., 2019), Chlamydia trachomatis (Snyder et al., 2017), Mycobacterium tuberculosis (Ranjbar et al., 2015), Legionella pneumophila (Lippmann et al., 2011), Listeria monocytogenes (Zwaferink et al., 2008; Radoshevich et al., 2015), Francisella novicida (Henry et al., 2007; Henry et al., 2010), and Rickettsia parkeri (Burke et al., 2020). The inhibition of bacterial replication is often the result of the action of ISGs. For example, viperin is a highly evolutionarily conserved ISG that restricts S. flexneri infection in HeLa cells (Helbig et al., 2019).
The roles of type I and III IFNs are more complex to delineate in vivo and can vary depending on the pathogen and biological context. Type I IFNs can restrict infections by pathogens such as Bacillus anthracis (Walberg et al., 2008), L. pneumophila (Lippmann et al., 2011; Naujoks et al., 2016), Helicobacter pylori (Watanabe et al., 2010), Streptococcus pyogenes (Castiglia et al., 2016), group B Streptococcus, E. coli (Mancuso et al., 2007), and Streptococcus pneumoniae (LeMessurier et al., 2013). In particular, IFNs prevented bacterial migration across endothelial and epithelial barriers (LeMessurier et al., 2013). Whether this protection extends to type III IFNs is unclear, but type III IFNs were demonstrated to be protective in murine models of colitis (Rauch et al., 2015; Broggi et al., 2017). In contrast, in other infection models, type I IFNs can be detrimental to hosts infected with bacterial pathogens. When compared to wild-type (WT) mice, Ifnar−/− animals were more resistant to systemic infection with L. monocytogenes (Auerbuch et al., 2004; Brzoza-Lewis et al., 2012), and S. Typhimurium (Robinson et al., 2012; Perkins et al., 2015; Wilson et al., 2019; Zhang et al., 2020). Similarly, IFNβ treatment exacerbated infection with M. tuberculosis (Manca et al., 2001; Manca et al., 2005) and B. anthracis (Gold et al., 2007). The detrimental nature of type I IFN responses during M. tuberculosis infection is reflected in human disease. The blood transcriptome of patients with active disease has demonstrated a correlation between the abundance of Type I IFN-inducible transcripts and disease pathogenesis (Berry et al., 2010; Maertzdorf et al., 2011; Ottenhoff et al., 2012; Cliff et al., 2013). In addition, individuals with an inherited defect in ISG15 have an increased susceptibility to mycobacterial, but not viral, disease (Bogunovic et al., 2012).
Finally, type I and III IFNs induced by viral infection were shown to exacerbate subsequent respiratory superinfections with S. pneumoniae or Staphylococcus aureus. Indeed, Ifnar−/− and Ifnlr−/− mice showed improved bacterial control in virus-bacteria superinfection models (Shahangian et al., 2009; Broggi et al., 2020; Major et al., 2020). Similarly, administration of recombinant type I or III IFN resulted in increased bacterial burdens following viral infection or activation of antiviral pathways with viral ligands. Recent work has shown that uncontrolled type III IFN responses, such as those observed during SARS-CoV-2 infection, led to damage of epithelial barriers and increased susceptibility to bacterial superinfection, a known complication of COVID19 and Influenza (Broggi et al., 2020; Langford et al., 2020; Major et al., 2020).
In order to control their host, pathogenic bacteria secrete effectors that manipulate different stages of the IFN response, from its production to signaling and even ISG functions.
Host recognition of bacterial PAMPs occurs via PRR ligation, leading to the assembly of signaling complexes and the activation of intracellular adaptor proteins [reviewed in (Odendall and Kagan, 2017)]. Cytosolic RNA sensors, retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), signal via the adaptor mitochondria-antiviral signaling protein (MAVS) (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005; Dixit et al., 2010), while the DNA sensor cyclic GMP–AMP synthase (cGAS) signals via stimulator of interferon genes (STING), localized on the endoplasmic reticulum (ER) (Gao et al., 2012; Wu et al., 2017). Toll-like receptors (TLRs) present on the cell surface and endosomes, signal via the adaptors MyD88 and/or TRIF (Kawasaki and Kawai, 2014). Although only endosomal TLRs efficiently induce type I IFNs, all TLRs were shown to strongly induce type III IFNs in response to bacterial ligands (Odendall et al., 2017). These signaling pathways culminate in the activation and nuclear translocation of transcription factors, including interferon regulatory factors (IRFs) that control IFN expression (Takeuchi and Akira, 2010; Odendall and Kagan, 2017; Odendall and Kagan, 2019). IFN expression also requires the NFκB and MAP-kinase activated transcription factor AP-1 (Odendall and Kagan, 2017).
As IFNs can have both beneficial and detrimental effects on bacterial pathogens, some species have evolved effectors that promote or inhibit their production (summarized in Figure 1 and Table 1). For example, L. monocytogenes secretes Zea, a ribonucleoprotein, that binds RIG-I and potentiates type I IFN responses in ECs (Pagliuso et al., 2019). In contrast, many bacterial effectors block the induction of IFN. The effects of some of these effectors on IFN induction may be considered as indirect as they either interfere with PAMP detection or block general innate sensing pathways. L. monocytogenes secretes an effector, PgdA, that deacetylates peptidoglycan, conferring a resistance to host lysozyme. This prevents the release of PAMPs and dampens the induction of cytokines and type I IFN (Boneca et al., 2007). Similarly, the L. pneumophila effector SdhA inhibits RLR activation in mouse Bone marrow-derived macrophage (BMDM) by maintaining the integrity of the Legionella-containing vacuole (Monroe et al., 2009). The cGAS/STING pathway has since been shown to mediate IFN expression following Legionella infection (Ruiz-Moreno et al., 2018). Although this has not been formally tested, it is likely that SdhA also protects from detection by cGAS. Yersinia pseudotuberculosis YopJ targets MAP-kinase signaling in dendritic cells and macrophages, disrupting MyD88- and TRIF-dependent signaling downstream of TLR4, which prevents type I IFN and pro-inflammatory cytokine expression (Paquette et al., 2012; Rosadini et al., 2015). EPEC is able to counteract the protective effects of type I IFN on epithelial barriers; its effector NleD inhibits RNase L, an endoribonuclease that enhances RLR-mediated production of IFNβ (Long et al., 2014).
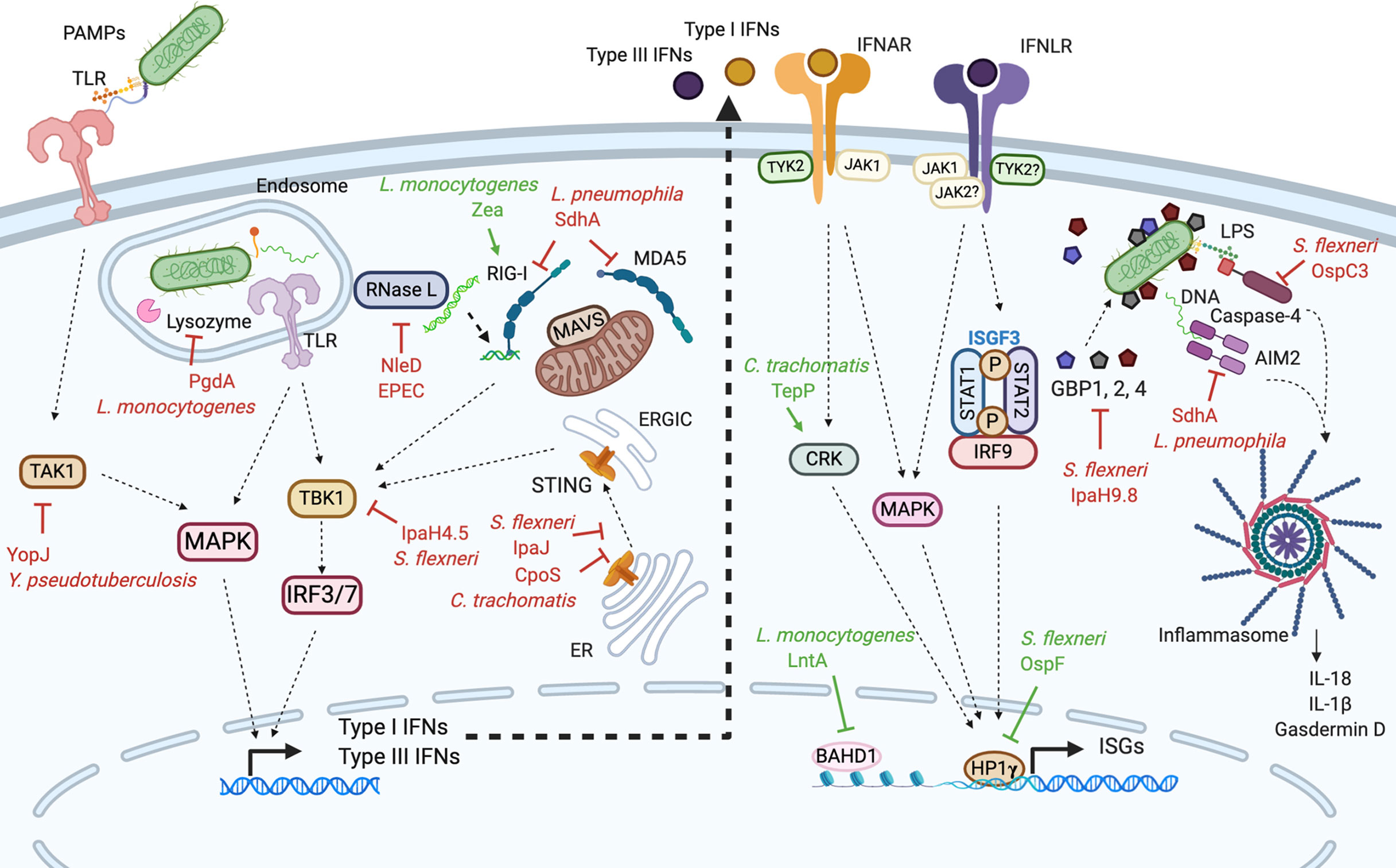
Figure 1 Modulation of types I and III interferon responses by bacterial effectors. In many different cell types, the recognition of bacterial PAMPs by PRRs leads to the production of type I and III IFNs (left side). Type I and III IFNs bind their respective receptors initiating signaling pathways which trigger the expression of ISGs (right side). Bacteria have evolved effectors that either inhibit (depicted in red) or stimulate (depicted in green) type I and III IFN responses. Yersinia pseudotuberculosis YopJ effector interferes with the MAP kinase activator TAK1. When internalized in endosomes, Listeria monocytogenes secretes PgdA which modifies its peptidoglycan, hiding it from the host lysozyme. Listeria monocytogenes also secretes Zea which binds RIG-I and potentiates the production of IFNs. The Legionella pneumophila protein SdhA blocks RIG-I and MDA5 cytosolic PRRs preventing them to interact with MAVS. The Shigella flexneri protease IpaJ interferes with STING translocation to the ERGIC while IpaH4.5 inhibits TBK1 kinase. Similarly, Chlamydia trachomatis CpoS effector affects STING migration. EPEC, through the secretion of NleD inhibits RNase L which counteracts the protective effects of type I IFN on epithelial barriers. Additionally, some bacteria perturb type I and III IFN signaling. L. monocytogenes LntA and S. flexneri OspF effectors enter the cell nucleus and potentiate ISGs transcription. LntA sequesters BAHD1, a negative regulator of ISGs transcription therefore stimulating ISGs expression. OspF interacts with HP1γ preventing its phosphorylation, which promotes ISGs expression. C. trachomatis TepP effector inhibits CRK, a component and regulators of type I and III IFN signaling, stimulating ISGs transcription. Finally, S. flexneri IpaH9.8 and OspC3 as well as L. pneumophila SdhA effectors inhibit inflammasome components; GBP1, caspase-4, and AIM2 respectively. PRRs, pattern recognition receptors; PAMPs, pathogen-associated molecular patterns; TLR, Toll like receptor; STING, stimulator of interferon genes; ER, endoplasmic reticulum; ERGIC, ER-Golgi intermediate compartments; IRF, interferon regulatory factor; RIG-I, retinoic acid-inducible gene-I; MDA5, melanoma differentiation-associated protein 5; MAVS, mitochondria-antiviral signaling protein; TAK1, TGFbeta activated kinase 1; TYK2, tyrosine kinase 2; JAK1, Janus kinase 1; STAT, signal transducer and activator of transcription; ISGF3, interferon-stimulated gene factor 3; MAPK, mitogen-activated protein kinase; GBP, guanylate-binding protein; AIM2, absent in melanoma-2; LPS, lipopolysaccharide; ISG, interferon stimulated gene; IFNAR, IFNα receptor; IFNLR,IFNλ receptor. Figure 1 was created with Biorender.com.
Other effectors affect the localization of adaptor proteins that are crucial to PRR signaling cascades. For example, STING translocates from the ER to the ER-Golgi intermediate compartments (ERGIC) to mediate cGAS signaling (Ishikawa et al., 2009; Burdette and Vance, 2013). IpaJ is a S. flexneri protease that targets the small guanosine triphosphatases (GTPases) required for this migration event. This blocks STING translocation, abolishing type I IFN and ISG expression (Dobbs et al., 2015). Similarly CpoS from C. trachomatis affects STING migration and blocks type I and III IFN and ISG induction (Sixt et al., 2017).
The kinase TBK1 lies downstream of many PRRs and phosphorylates IRF3 and IRF7, leading to the expression of type I and III IFNs (Clark et al., 2011; Tanaka and Chen, 2012; Schneider et al., 2014; Liu et al., 2015; Bakshi et al., 2017). It can also activate IRF1, which may specifically drive type III IFN induction (Odendall et al., 2014). The S. flexneri effector IpaH4.5 is an E3 ubiquitin ligase that targets TBK1 for proteasomal degradation, leading to the suppression of IFN production (Zheng et al., 2016). Interestingly, mice infected with ΔipaH4.5 mutants had lower bacterial burdens than those infected with WT S. flexneri.
Finally, some bacteria modulate IFN production via yet-unknown mechanisms. Salmonella represses TRIF-dependent type I IFN induction in macrophages (Owen et al., 2016). Likewise, Chlamydia pneumoniae infection of ECs does not lead to IFNβ production and very efficiently blocks IRF3 phosphorylation. Although the effector responsible was not identified, Chlamydia was shown to induce the degradation of TRAF3, a crucial TBK1 activator (Wolf and Fields, 2013). Conversely, some pathogens such as Legionella (Ruiz-Moreno et al., 2018) or Burkholderia pseudomallei (Ku et al., 2020), were shown to induce IFNβ by activating the cGAS-STING pathway in a secretion system-dependent manner.
Although the pathways that drive type I and III IFN expression overlap significantly, the majority of this research has focused on type I IFNs. It will be interesting to investigate whether bacterial effectors can specifically target the production of one family or the other.
JAK/STAT signaling activated by type I or III IFN receptor binding leads to the formation of a STAT1/STAT2/IRF9 complex called the interferon-stimulated gene factor 3 (ISGF3). This complex translocates to the nucleus and binds interferon stimulated response elements (ISREs) to transcribe ISGs. Instead of targeting IFN production, some bacterial effectors target signaling cascades downstream of IFN receptors to alter the expression of ISGs. For example, B. anthracis inhibits IFNβ-mediated STAT1 phosphorylation and the formation of ISGF3, but the bacterial effectors remain to be identified (Gold et al., 2004).
Bacteria can also affect transcription of ISGs by altering epigenetic regulators of transcription. Heterochromatin protein 1 (HP1) family members are epigenetic regulators that bind methylated histone H3 to influence the expression of a wide variety of genes (Saint-André et al., 2011; Ameyar-Zazoua et al., 2012; Smallwood et al., 2012). In a guinea pig model of Shigella enterocolitis, S. flexneri inhibited HP1γ phosphorylation via OspF. Next-generation sequencing of mouse embryonic fibroblast-derived cell lines revealed that HP1γ represses the expression of some IFN-regulated genes, namely Ifi44, Ifit3, and Oas1b (Harouz et al., 2014), confirming previous observations (Lavigne et al., 2009). This suggests that S. flexneri effectors are capable of modifying epigenetic regulators of subsets of ISGs to alter the outcome of IFN signaling. Likewise, the C. trachomatis effector TepP, was shown to bind the adaptor protein CRK and affect expression of ISGs in human cell lines (Chen et al., 2014).
Some bacterial effectors enter the nucleus to alter chromatin remodeling in their mammalian hosts (Bhavsar et al., 2007; Lebreton et al., 2011). Infection of cells with an L. monocytogenes strain constitutively expressing the effector LntA resulted in enrichment of expression of a number of genes, 83% of which were ISGs. Interestingly, when lntA was deleted, there was a reduction in bacterial burden in the spleens and livers of mice. LntA was shown to interact with BAHD1; a heterochromatin protein that acts as a transcriptional repressor (Bierne et al., 2009). Chromatin immunoprecipitation analysis showed that association of BAHD1 with key ISGs was reduced in cells infected with L. monocytogenes constitutively expressing LntA. Therefore, Listeria induces the expression of ISGs by inhibiting the negative regulator BAHD1 (Lebreton et al., 2011).
All three families of IFNs induce the expression of ISGs. As discussed above, the set of 300–600 genes transcribed by type I and III IFNs contain ISRE sequences. In contrast, IFNγ induces ISGs containing a gamma interferon activation site (GAS) element. ISGs can contain both ISRE and GAS sequences and therefore be induced by all three families. As they are induced in concert, the individual roles of many ISGs are not well understood. Although more is known about the ISGs targeted by viral virulence factors, there are few examples of bacterial factors that can alter their integrity or function.
Among ISGs are several components of inflammasomes. These are large multi-protein complexes that are assembled upon recognition of PAMPs or changes in host homeostasis. Inflammasome activation leads to pyroptosis, a highly inflammatory form of cell death. Inflammasomes are key to host defense against intracellular bacteria. As such, some bacteria have evolved strategies to evade inflammasome activation. For example, absent in melanoma-2 (AIM2) is an IFN-inducible inflammasome receptor that detects the presence of bacterial DNA in the cytosol (Man et al., 2015; Meunier et al., 2015). The L. pneumophila effector SdhA, in addition to suppressing type I IFN responses (Monroe et al., 2009; Creasey and Isberg, 2012), negatively regulates AIM2 inflammasome activation in human macrophages (Ge et al., 2012).
Another inflammasome ‘receptor’ induced by IFNs is murine caspase 11, which detects intracellular LPS and induces the non-canonical inflammasome (Ding and Shao, 2017). The human orthologs of caspase 11 are caspases 4 and 5, but although caspase 4 was shown to be transcriptionally induced by type I IFN, whether this occurs at the protein level is still poorly understood (Knodler et al., 2014; Casson et al., 2015; Schmid-Burgk et al., 2015). However, it is clear that the caspase 4 inflammasome is “primed” by IFN via the action of guanylate-binding proteins (GBPs) (Pilla et al., 2014; Feng and Man, 2020; Santos et al., 2020). GBPs are among the best-characterized antibacterial ISGs. This family of GTPases has an ever-growing list of functions, many of which contribute to defenses against intracellular pathogens. There are seven GBP genes in humans, whose expression is most robustly induced by IFNγ, but can also be induced by treatment with type I IFNs (Cheng et al., 1985; Kim et al., 2011; Tretina et al., 2019). Some GBPs can also be produced in response to treatment with type III IFNs (Alase et al., 2015; Tretina et al., 2019). GBPs are recruited to intracellular gram-negative bacteria (S. Typhimurium, S. flexneri, Burkholderia thailandensis, Brucella abortus, F. novicida, C. trachomatis, and L. pneumophila) and liberate LPS into the cytosol (Kim et al., 2011; Kim et al., 2012; Shenoy et al., 2012; Haldar et al., 2014; Meunier et al., 2014; Man et al., 2015; Meunier and Broz, 2015; Pilla et al., 2014; Shi et al., 2014; Man et al., 2016; Feeley et al., 2017; Finethy et al., 2017; Lindenberg et al., 2017; Cerqueira et al., 2018; Fisch et al., 2019; Bass and Shin, 2020; Santos et al., 2020). Recent work on S. flexneri and S. Typhimurium has shown that GBP1 directly binds bacterial LPS through electrostatic interactions and assembles a signaling platform containing other GBPs. This platform recruits and activates human caspase 4, leading to assembly of the non-canonical inflammasome (Kutsch et al., 2020; Santos et al., 2020; Wandel et al., 2020). In addition, GBPs inhibit intracellular motility and cell-to-cell spread of B. thailandensis and S. flexneri (Ostler et al., 2014; Piro et al., 2017; Wandel et al., 2017).
S. flexneri has evolved at least two effector proteins to bypass these important host defense mechanisms; firstly, OspC3, which binds and inhibits caspase 4. ospC3 mutants have a growth defect in IFNγ-treated cells and induce rapid cell death. These mutants were also attenuated in WT mice, but not in mice lacking GBP1, GBP2, or caspase 11 (Kobayashi et al., 2013; Wandel et al., 2020). Whether they are more sensitive to type I or III IFNs is unclear. Secondly, the E3 ubiquitin ligase IpaH9.8 targets hGBP1, hGBP2, and hGBP4 for proteasomal degradation (Li et al., 2017; Wandel et al., 2017). Shigella strains lacking ipaH9.8 displayed an increased recruitment of GBP1, 2, 3, and 4, as well as caspase 4, compared to WT (Wandel et al., 2017; Wandel et al., 2020). In addition, cell-to-cell spread of ipaH9.8 mutant bacteria was lowered (Wandel et al., 2017). These data highlight the importance of GBPs to entrap intracellular bacteria and act as platforms for the activation of innate signaling. It is therefore highly probable that other intracellular bacteria have evolved mechanisms to counteract GBPs.
IFNs are crucial mediators of inflammation, playing complex, yet key roles in both systemic and localized bacterial infections. In order to survive and cause disease, bacteria secrete effectors that interfere with type I and III IFN production and signaling, as well as ISG expression and function. As there is still much to learn about the role of IFN and ISGs, perhaps understanding the mechanisms that bacterial pathogens have evolved to evade or enhance these responses will lead to insights into their function in host defense. These discussions certainly mandate further studies into the interactions between IFNs and bacteria.
NA and RD contributed equally. All authors contributed to the article and approved the submitted version.
NA is supported by a studentship from the King’s College London/Francis Crick Institute partnership, which receives its core funding from the Cancer Research UK (FC001206), the UK Medical Research Council (FC001206), and the Wellcome Trust (FC001206). RD is supported by a studentship from the UK Medical Research Council (MR/N013700/1). CO is supported by a Sir Henry Dale Fellowship from the Royal Society and the Wellcome Trust (206200/Z/17/Z).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank our colleagues Joseph Wanford, Andrew Voak and Abderrahman Hachani for ideas and critical reading of this manuscript.
Alase A. A., El-Sherbiny Y. M., Vital E. M., Tobin D. J., Turner N. A., Wittmann M. (2015). IFNλ stimulates MxA production in human dermal fibroblasts via a MAPK-dependent STAT1-independent mechanism. J. Invest. Dermatol. 135, 2935–2943. doi: 10.1038/jid.2015.317
Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R., et al. (2012). Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 19, 998–1005. doi: 10.1038/nsmb.2373
Auerbuch V., Brockstedt D. G., Meyer-Morse N., O’Riordan M., Portnoy D. A. (2004). Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J. Exp. Med. 200, 527–533. doi: 10.1084/jem.20040976
Bakshi S., Taylor J., Strickson S., McCartney T., Cohen P. (2017). Identification of TBK1 complexes required for the phosphorylation of IRF3 and the production of interferon β. Biochem. J. 474, 1163–1174. doi: 10.1042/BCJ20160992
Bass A. R., Shin S. (2020). Human GBP1 promotes pathogen vacuole rupture and inflammasome activation during. doi: 10.1101/2020.05.27.120477
Berry M. P. R., Graham C. M., McNab F. W., Xu Z., Bloch S. A. A., Oni T., et al. (2010). An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977. doi: 10.1038/nature09247
Bhavsar A. P., Guttman J. A., Finlay B. B. (2007). Manipulation of host-cell pathways by bacterial pathogens. Nature 449, 827–834. doi: 10.1038/nature06247
Bierne H., Tham T. N., Batsche E., Dumay A., Leguillou M., Kernéis-Golsteyn S., et al. (2009). Human BAHD1 promotes heterochromatic gene silencing. Proc. Natl. Acad. Sci. U. S. A. 106, 13826–13831. doi: 10.1073/pnas.0901259106
Blazek K., Eames H. L., Weiss M., Byrne A. J., Perocheau D., Pease J. E., et al. (2015). IFN-λ resolves inflammation via suppression of neutrophil infiltration and IL-1β production. J. Exp. Med. 212, 845–853. doi: 10.1084/jem.20140995
Bogunovic D., Byun M., Durfee L. A., Abhyankar A., Sanal O., Mansouri D., et al. (2012). Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science 337, 1684–1688. doi: 10.1126/science.1224026
Boneca I. G., Dussurget O., Cabanes D., Nahori M.-A., Sousa S., Lecuit M., et al. (2007). A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proc. Natl. Acad. Sci. U. S. A. 104, 997–1002. doi: 10.1073/pnas.0609672104
Broggi A., Tan Y., Granucci F., Zanoni I. (2017). IFN-λ suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat. Immunol. 18, 1084–1093. doi: 10.1038/ni.3821
Broggi A., Ghosh S., Sposito B., Spreafico R., Balzarini F., Lo Cascio A., et al. (2020). Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science 369, 706–712. doi: 10.1126/science.abc3545
Brzoza-Lewis K. L., Jason Hoth J., Hiltbold E. M. (2012). Type I interferon signaling regulates the composition of inflammatory infiltrates upon infection with Listeria monocytogenes. Cell. Immunol. 273, 41–51. doi: 10.1016/j.cellimm.2011.11.008
Bukholm G., Berdal B. P., Haug C., Degre M. (1984). Mouse fibroblast interferon modifies Salmonella typhimurium infection in infant mice. Infect. Immun. 45, 62–66. doi: 10.1128/iai.45.1.62-66.1984
Burdette D. L., Vance R. E. (2013). STING and the innate immune response to nucleic acids in the cytosol. Nat. Immunol. 14, 19–26. doi: 10.1038/ni.2491
Burke T. P., Engström P., Chavez R. A., Fonbuena J. A., Vance R. E., Welch M. D. (2020). Inflammasome-mediated antagonism of type I interferon enhances Rickettsia pathogenesis. Nat. Microbiol. 5, 688–696. doi: 10.1038/s41564-020-0673-5
Casson C. N., Yu J., Reyes V. M., Taschuk F. O., Yadav A., Copenhaver A. M., et al. (2015). Human caspase-4 mediates noncanonical inflammasome activation against gram-negative bacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 112, 6688–6693. doi: 10.1073/pnas.1421699112
Castiglia V., Piersigilli A., Ebner F., Janos M., Goldmann O., Damböck U., et al. (2016). Type i Interferon Signaling Prevents IL-1β-Driven Lethal Systemic Hyperinflammation during Invasive Bacterial Infection of Soft Tissue. Cell Host Microbe 19, 375–387. doi: 10.1016/j.chom.2016.02.003
Cerqueira D. M., Gomes M. T. R., Silva A. L. N., Rungue M., Assis N. R. G., Guimarães E. S., et al. (2018). Guanylate-binding protein 5 licenses caspase-11 for Gasdermin-D mediated host resistance to Brucella abortus infection. PloS Pathog. 14, e1007519. doi: 10.1371/journal.ppat.1007519
Chen Y.-S., Bastidas R. J., Saka H. A., Carpenter V. K., Richards K. L., Plano G. V., et al. (2014). The Chlamydia trachomatis Type III Secretion Chaperone Slc1 Engages Multiple Early Effectors, Including TepP, a Tyrosine-phosphorylated Protein Required for the Recruitment of CrkI-II to Nascent Inclusions and Innate Immune Signaling. PloS Pathog. 10, e1003954. doi: 10.1371/journal.ppat.1003954
Chen C., Chen L., Chen C., Chen Q., Zhao Q., Dong Y. (2017). The Distribution Frequency of Interferon-Gamma Receptor 1 Gene Polymorphisms in Interferon- γ Release Assay-Positive Patients. Dis. Markers 2017, 5. doi: 10.1155/2017/4031671
Cheng Y. S. E., Becker-Manley M. F., Chow T. P., Horan D. C. (1985). Affinity purification of an interferon-induced human guanylate-binding protein and its characterization. J. Biol. Chem. 260, 15834–15839. doi: 10.1016/S0021-9258(17)36334-2
Clark K., Takeuchi O., Akira S., Cohen P. (2011). The TRAF-associated protein TANK facilitates cross-talk within the IκB kinase family during Toll-like receptor signaling. Proc. Natl. Acad. Sci. U. S. A. 108, 17093–17098. doi: 10.1073/pnas.1114194108
Cliff J. M., Lee J. S., Constantinou N., Cho J. E., Clark T. G., Ronacher K., et al. (2013). Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J. Infect. Dis. 207, 18–29. doi: 10.1093/infdis/jis499
Creasey E. A., Isberg R. R. (2012). The protein SdhA maintains the integrity of the Legionella-containing vacuole. Proc. Natl. Acad. Sci. U. S. A. 109, 3481–3486. doi: 10.1073/pnas.1121286109
De Weerd N. A., Nguyen T. (2012). The interferons and their receptors-distribution and regulation. Immunol. Cell Biol. 90, 483–491. doi: 10.1038/icb.2012.9
Ding J., Shao F. (2017). SnapShot: The Noncanonical Inflammasome. Cell 168, 544–544.e1. doi: 10.1016/j.cell.2017.01.008
Dixit E., Boulant S., Zhang Y., Lee A. S. Y., Odendall C., Shum B., et al. (2010). Peroxisomes Are Signaling Platforms for Antiviral Innate Immunity. Cell 141, 668–681. doi: 10.1016/j.cell.2010.04.018
Dobbs N., Burnaevskiy N., Chen D., Gonugunta V. K., Alto N. M., Yan N. (2015). STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 176, 139–148. doi: 10.1016/j.chom.2015.07.001
Duménil G., Olivo J. C., Pellegrini S., Fellous M., Sansonetti P. J., Van Tran Nhieu G. (1998). Interferon α inhibits a Src-mediated pathway necessary for Shigella- induced cytoskeletal rearrangements in epithelial cells. J. Cell Biol. 143, 1003–1012. doi: 10.1083/jcb.143.4.1003
Durbin R. K., Kotenko S. V., Durbin J. E. (2013). Interferon induction and function at the mucosal surface. Immunol. Rev. 255, 25–39. doi: 10.1111/imr.12101
Feeley E. M., Pilla-Moffett D. M., Zwack E. E., Piro A. S., Finethy R., Kolb J. P., et al. (2017). Galectin-3 directs antimicrobial guanylate binding proteins to vacuoles furnished with bacterial secretion systems. Proc. Natl. Acad. Sci. U. S. A. 114, E1698–E1706. doi: 10.1073/pnas.1615771114
Feng S., Man S. M. (2020). Captain GBP1: inflammasomes assemble, pyroptotic endgame. Nat. Immunol. 21, 823–834. doi: 10.1038/s41590-020-0727-0
Finethy R., Luoma S., Orench-Rivera N., Feeley E. M., Haldar A. K., Yamamoto M., et al. (2017). Inflammasome activation by bacterial outer membrane vesicles requires guanylate binding proteins. MBio 8, 1–11. doi: 10.1128/mBio.01188-17
Fisch D., Bando H., Clough B., Hornung V., Yamamoto M., Shenoy A. R., et al. (2019). Human GBP 1 is a microbe-specific gatekeeper of macrophage apoptosis and pyroptosis. EMBO J. 38, 1–19. doi: 10.15252/embj.2018100926
Gao D., Wu J., Wu Y., Du F., Aroh C., Yan N., et al. (2012). Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 339, 786–791. doi: 10.1126/science.1232458
Ge J., Gong Y. N., Xu Y., Shao F. (2012). Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. Proc. Natl. Acad. Sci. U. S. A. 109, 6193–6198. doi: 10.1073/pnas.1117490109
Gold J. A., Hoshino Y., Hoshino S., Jones M. B., Nolan A., Weiden M. D. (2004). Exogenous Gamma and Alpha/Beta Interferon Rescues Human Macrophages from Cell Death Induced by Bacillus anthracis. Infect. Immun. 72, 1291–1297. doi: 10.1128/IAI.72.3.1291-1297.2004
Gold J. A., Hoshino Y., Jones M. B., Hoshino S., Nolan A., Weiden M. D. (2007). Exogenous interferon-α and interferon-γ increase lethality of murine inhalational anthrax. PloS One 72, 1291–1297. doi: 10.1371/journal.pone.0000736
Green E. R., Mecsas J. (2016). Bacterial Secretion Systems: An Overview. Virulence Mech. Bact. Pathog. 4, 213–239. doi: 10.1128/9781555819286.ch8
Haldar A. K., Piro A. S., Pilla D. M., Yamamoto M., Coers J. (2014). The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia- and Toxoplasma- containing vacuoles and host resistance. PloS One 9, e86684. doi: 10.1371/journal.pone.0086684
Harouz H., Rachez C., Meijer B. M., Marteyn B., Donnadieu F., Cammas F., et al. (2014). Shigella flexneri targets the HP 1γ subcode through the phosphothreonine lyase OspF. EMBO J. 33, 2606–2622. doi: 10.15252/embj.201489244
Helbig K. J., Teh M. Y., Crosse K. M., Monson E. A., Smith M., Tran E. N., et al. (2019). The interferon stimulated gene viperin, restricts Shigella. flexneri in vitro. Sci. Rep. 9, 1–12. doi: 10.1038/s41598-019-52130-8
Henry T., Brotcke A., Weiss D. S., Thompson L. J., Monack D. M. (2007). Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 204, 987–994. doi: 10.1084/jem.20062665
Henry T., Kirimanjeswara G. S., Ruby T., Jones J. W., Perret M., Ho L., et al. (2010). Type I Interferon signaling constrains IL-17A/F secretion by γδ T cells during bacterial infections. J. Immunol. 184, 3755–3767. doi: 10.4049/jimmunol.0902065
Hertzog P. J., Williams B. R. G. (2013). Fine tuning type I interferon responses. Cytokine Growth Factor Rev. 24, 217–225. doi: 10.1016/j.cytogfr.2013.04.002
Isaacs A., Lindenmann J. (1957). Virus interference. I. The interferon. Proc. R. Soc London Ser. B - Biol. Sci. 147, 258–267. doi: 10.1098/rspb.1957.0048
Ishikawa H., Ma Z., Barber G. N. (2009). STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461, 788–792. doi: 10.1038/nature08476
Janeway C. A. (1989). Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13. doi: 10.1101/sqb.1989.054.01.003
Kagan J. C., Su T., Horng T., Chow A., Akira S., Medzhitov R. (2008). TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat. Immunol. 9, 361–368. doi: 10.1038/ni1569
Kak G., Raza M., Tiwari B. K. (2018). Interferon-gamma (IFN-γ): Exploring its implications in infectious diseases. Biomol. Concepts 9, 64–79. doi: 10.1515/bmc-2018-0007
Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., et al. (2005). IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988. doi: 10.1038/ni1243
Kawasaki T., Kawai T. (2014). Toll-like receptor signaling pathways. Front. Immunol. 5, 461. doi: 10.3389/fimmu.2014.00461
Kim B. H., Shenoy A. R., Kumar P., Das R., Tiwari S., MacMicking J. D. (2011). A family of IFN-γ-inducible 65-kD GTPases protects against bacterial infection. doi: 10.1126/science.1201711
Kim B. H., Shenoy A. R., Kumar P., Bradfield C. J., MacMicking J. D. (2012). IFN-inducible GTPases in host cell defense. Cell Host Microbe 12, 432–444. doi: 10.1016/j.chom.2012.09.007
Knodler L. A., Crowley S. M., Sham H. P., Yang H., Wrande M., Ma C., et al. (2014). Non-canonical inflammasome activation of caspase-4/ caspase-11 mediates epithelial defenses against enteric bacterial pathogens. Cell Host Microbe 16, 249–256. doi: 10.1016/j.chom.2014.07.002
Kobayashi T., Ogawa M., Sanada T., Mimuro H., Kim M., Ashida H., et al. (2013). The Shigella OspC3 effector inhibits caspase-4, antagonizes inflammatory cell death, and promotes epithelial infection. Cell Host Microbe 13, 570–583. doi: 10.1016/j.chom.2013.04.012
Kotenko S. V., Gallagher G., Baurin V. V., Lewis-Antes A., Shen M., Shah N. K., et al. (2003). IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4, 69–77. doi: 10.1038/ni875
Ku J. W. K., Chen Y., Lim B. J. W., Gasser S., Crasta K. C., Gan Y. H. (2020). Bacterial-induced cell fusion is a danger signal triggering cGAS-STING pathway via micronuclei formation. Proc. Natl. Acad. Sci. U. S. A. 117, 15923–15934. doi: 10.1073/pnas.2006908117
Kutsch M., Coers J., Sistemich L., Lesser C. F., Goldberg M. B., Herrmann C. (2020). Direct binding of polymeric GBP1 to LPS disrupts bacterial cell envelope functions. Embo, 39, 1–22. doi: 10.15252/embj.2020104926
Langford B. J., So M., Raybardhan S., Leung V., Westwood D., MacFadden D. R., et al. (2020). Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin. Microbiol. Infect. 26, 1622–1629. doi: 10.1016/j.cmi.2020.07.016
Lavigne M., Eskeland R., Azebi S., Saint-André V., Jang S. M., Batsché E., et al. (2009). Interaction of HP1 and Brg1/Brm with the globular domain of histone H3 is required for HP1-mediated repression. PloS Genet. 5, e1000769. doi: 10.1371/journal.pgen.1000769
Lazear H. M., Schoggins J. W., Diamond M. S. (2019). Shared and Distinct Functions of Type I and Type III Interferons. Immunity 50, 907–923. doi: 10.1016/j.immuni.2019.03.025
Lebreton A., Lakisic G., Job V., Fritsch L., Tham T. N., Camejo A., et al. (2011). A bacterial protein targets the BAHD1 chromatin complex to stimulate type III interferon response. Science 331, 1319–1321. doi: 10.1126/science.1200120
LeMessurier K. S., Häcker H., Chi L., Tuomanen E., Redecke V. (2013). Type I Interferon Protects against Pneumococcal Invasive Disease by Inhibiting Bacterial Transmigration across the Lung. PloS Pathog. 9, e1003727. doi: 10.1371/journal.ppat.1003727
Li P., Jiang W., Yu Q., Liu W., Zhou P., Li J., et al. (2017). Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 551, 378–383. doi: 10.1038/nature24467
Lindenberg V., Mölleken K., Kravets E., Stallmann S., Hegemann J. H., Degrandi D., et al. (2017). Broad recruitment of mGBP family members to Chlamydia trachomatis inclusions. PloS One 12, 1–14. doi: 10.1371/journal.pone.0185273
Lippmann J., Müller H. C., Naujoks J., Tabeling C., Shin S., Witzenrath M., et al. (2011). Dissection of a type I interferon pathway in controlling bacterial intracellular infection in mice. Cell. Microbiol. 13, 1668–1682. doi: 10.1111/j.1462-5822.2011.01646.x
Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., et al. (2015). Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630. doi: 10.1126/science.aaa2630
Long T. M., Nisa S., Donnenberg M. S., Hassel B. A. (2014). Enteropathogenic Escherichia coli inhibits type I interferon- and RNase L-mediated host defense to disrupt intestinal epithelial cell Barrier function. Infect. Immun. 82, 2802–2814. doi: 10.1128/IAI.00105-14
Maertzdorf J., Repsilber D., Parida S. K., Stanley K., Roberts T., Black G., et al. (2011). Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 12, 15–22. doi: 10.1038/gene.2010.51
Major J., Crotta S., Llorian M., McCabe T. M., Gad H. H., Hartmann R., et al. (2020). Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science 369, 712–717. doi: 10.1126/science.abc2061
Man S. M., Karki R., Malireddi R. K. S., Neale G., Vogel P., Yamamoto M., et al. (2015). The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16, 467–475. doi: 10.1038/ni.3118
Man S. M., Karki R., Sasai M., Place D. E., Kesavardhana S., Temirov J., et al. (2016). IRGB10 Liberates Bacterial Ligands for Sensing by the AIM2 and Caspase-11-NLRP3 Inflammasomes. Cell 167, 382–396.e17. doi: 10.1016/j.cell.2016.09.012
Manca C., Tsenova L., Bergtold A., Freeman S., Tovey M., Musser J. M., et al. (2001). Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-α/β. Proc. Natl. Acad. Sci. U. S. A. 98, 5752–5757. doi: 10.1073/pnas.091096998
Manca C., Tsenova L., Freeman S., Barczak A. K., Tovey M., Murray P. J., et al. (2005). Hypervirulent M. tuberculosis W/Beijing strains upregulate Type I IFNs and increase expression of negative regulators of the jak-stat pathway. J. Interf. Cytokine Res. 25, 694–701. doi: 10.1089/jir.2005.25.694
Mancuso G., Midiri A., Biondo C., Beninati C., Zummo S., Galbo R., et al. (2007). Type I IFN Signaling Is Crucial for Host Resistance against Different Species of Pathogenic Bacteria. J. Immunol. 178, 3126–3133. doi: 10.4049/jimmunol.178.5.3126
Meunier E., Broz P. (2015). Interferon-inducible GTPases in cell autonomous and innate immunity. Cell. Microbiol. 18, 168–180. doi: 10.1111/cmi.12546
Meunier E., Dick M. S., Dreier R. F., Schürmann N., Broz D. K., Warming S., et al. (2014). Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature 509, 366–370. doi: 10.1038/nature13157
Meunier E., Wallet P., Dreier R. F., Costanzo S., Anton L., Rühl S., et al. (2015). Guanylate-binding proteins promote AIM2 inflammasome activation during Francisella novicida infection by inducing cytosolic bacteriolysis and DNA release. Nat. Immunol. 16, 476–484. doi: 10.1038/ni.3119
Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., et al. (2005). Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172. doi: 10.1038/nature04193
Monroe K. M., McWhirter S. M., Vance R. E. (2009). Identification of host cytosolic sensors and bacterial factors regulating the type I interferon response to Legionella pneumophila. PloS Pathog. 5, e1000665. doi: 10.1371/journal.ppat.1000665
Naujoks J., Tabeling C., Dill B. D., Hoffmann C., Brown A. S., Kunze M., et al. (2016). IFNs Modify the Proteome of Legionella-Containing Vacuoles and Restrict Infection Via IRG1-Derived Itaconic Acid. PloS Pathog. 12, e1005408. doi: 10.1371/journal.ppat.1005408
Niesel D. W., Hess C. B., Cho Y. J., Klimpel K. D., Klimpel G. R. (1986). Natural and recombinant interferons inhibit epithelial cell invasion by Shigella spp. Infect. Immun. 52, 828–833. doi: 10.1128/IAI.52.3.828-833.1986
Odendall C., Kagan J. C. (2017). Activation and pathogenic manipulation of the sensors of the innate immune system. Microbes Infect. 176, 139–148. doi: 10.1016/j.micinf.2017.01.003
Odendall C., Kagan J. C. (2019). Host-encoded sensors of bacteria—our windows into the microbial world. Microbiol. Spectr. 176, 139–148. doi: 10.1128/microbiolspec.BAI-0011-2019
Odendall C., Dixit E., Stavru F., Bierne H., Franz K. M., Durbin A. F., et al. (2014). Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 15, 717–726. doi: 10.1038/ni.2915
Odendall C., Voak A. A., Kagan J. C. (2017). Type III interferons are commonly induced by bacteria-sensing TLRs, and reinforce epithelial barriers during infection. J. Immunol. 176, 139–148. doi: 10.4049/jimmunol.1700250
Ostler N., Britzen-Laurent N., Liebl A., Naschberger E., Lochnit G., Ostler M., et al. (2014). Gamma Interferon-Induced Guanylate Binding Protein 1 Is a Novel Actin Cytoskeleton Remodeling Factor. Mol. Cell. Biol. 34, 196–209. doi: 10.1128/mcb.00664-13
Ottenhoff T. H. M., Dass R. H., Yang N., Zhang M. M., Wong H. E. E., Sahiratmadja E., et al. (2012). Genome-Wide Expression Profiling Identifies Type 1 Interferon Response Pathways in Active Tuberculosis. PloS One 7, e45839. doi: 10.1371/journal.pone.0045839
Owen K. A., Anderson C. J., Casanova J. E. (2016). Salmonella suppresses the TRIF-dependent type I interferon response in macrophages. MBio 7, 1–15. doi: 10.1128/mBio.02051-15
Pagliuso A., Tham T. N., Allemand E., Robertin S., Dupuy B., Bertrand Q., et al. (2019). An RNA-Binding Protein Secreted by a Bacterial Pathogen Modulates RIG-I Signaling. Cell Host Microbe 26, 823–835.e11. doi: 10.1016/j.chom.2019.10.004
Pandey A. K., Yang Y., Jiang Z., Fortune S. M., Coulombe F., Behr M. A., et al. (2009). Nod2, Rip2 and Irf5 play a critical role in the type I interferon response to Mycobacterium tuberculosis. PloS Pathog. 5, e1000500. doi: 10.1371/journal.ppat.1000500
Paquette N., Conlon J., Sweet C., Rus F., Wilson L., Pereira A., et al. (2012). Serine/threonine acetylation of TGFβ-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc. Natl. Acad. Sci. U. S. A. 109, 12710–12715. doi: 10.1073/pnas.1008203109
Perkins D. J., Rajaiah R., Tennant S. M., Ramachandran G., Higginson E. E., Dyson T. N., et al. (2015). Salmonella Typhimurium Co-opts the Host Type I Interferon System to Restrict Macrophage Innate Immune Transcriptional Responses Selectively. J. Immunol. 176, 139–148. doi: 10.4049/jimmunol.1500105
Pestka S., Krause C. D., Walter M. R. (2004). Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202, 8–32. doi: 10.1111/j.0105-2896.2004.00204.x
Pien G. C., Satoskar A. R., Takeda K., Akira S., Biron C. A. (2000). Cutting Edge: Selective IL-18 Requirements for Induction of Compartmental IFN-γ Responses During Viral Infection. J. Immunol. 165, 4787–4791. doi: 10.4049/jimmunol.165.9.4787
Pilla D. M., Hagar J. A., Haldar A. K., Mason A. K., Degrandi D., Pfeffer K., et al. (2014). Guanylate binding proteins promote caspase-11-dependent pyroptosis in response to cytoplasmic LPS. Proc. Natl. Acad. Sci. 111, 6046–6051. doi: 10.1073/pnas.1321700111
Piro A. S., Hernandez D., Luoma S., Feeley E. M., Finethy R., Yirga A., et al. (2017). Detection of cytosolic shigella flexneri via a C-terminal triple-arginine motif of GBP1 inhibits actin-based motility. MBio 8, e01979–17. doi: 10.1128/mBio.01979-17
Radoshevich L., Impens F., Ribet D., Quereda J. J., Tham T. N., Nahori M. A., et al. (2015). ISG15 counteracts Listeria monocytogenes infection. Elife 4, 1–23. doi: 10.7554/eLife.06848
Ranjbar S., Haridas V., Jasenosky L. D., Falvo J. V., Goldfeld A. E. (2015). A Role for IFITM Proteins in Restriction of Mycobacterium tuberculosis Infection. Cell Rep. 13, 874–883. doi: 10.1016/j.celrep.2015.09.048
Rauch I., Rosebrock F., Hainzl E., Heider S., Majoros A., Wienerroither S., et al. (2015). Noncanonical Effects of IRF9 in Intestinal Inflammation: More than Type I and Type III Interferons. Mol. Cell. Biol. 35, 2332–2343. doi: 10.1128/mcb.01498-14
Robinson N., McComb S., Mulligan R., Dudani R., Krishnan L., Sad S. (2012). Type i interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat. Immunol. 13, 954–962. doi: 10.1038/ni.2397
Rosadini C. V., Zanoni I., Odendall C., Green E. R., Paczosa M. K., Philip N. H., et al. (2015). A Single Bacterial Immune Evasion Strategy Dismantles Both MyD88 and TRIF Signaling Pathways Downstream of TLR4. Cell Host Microbe 18, 682–693. doi: 10.1016/j.chom.2015.11.006
Ruiz-Moreno J. S., Hamann L., Shah J. A., Verbon A., Mockenhaupt F. P., Puzianowska-Kuznicka M., et al. (2018). The common HAQ STING variant impairs cGAS-dependent antibacterial responses and is associated with susceptibility to Legionnaires’ disease in humans. PloS Pathog. 14, 1–22. doi: 10.1371/journal.ppat.1006829
Saint-André V., Batsché E., Rachez C., Muchardt C. (2011). Histone H3 lysine 9 trimethylation and HP1γ favor inclusion of alternative exons. Nat. Struct. Mol. Biol. 18, 337–344. doi: 10.1038/nsmb.1995
Salazar-Mather T. P., Hamilton T. A., Biron C. A. (2000). A chemokine-to-cytokine-to-chemokine cascade critical in antiviral defense. J. Clin. Invest. 105, 985–993. doi: 10.1172/JCI9232
Santos J. C., Boucher D., Schneider L. K., Demarco B., Dilucca M., Shkarina K., et al. (2020). Human GBP1 binds LPS to initiate assembly of a caspase-4 activating platform on cytosolic bacteria. Nat. Commun. 11, 3276. doi: 10.1038/s41467-020-16889-z
Schindler H., Lutz M. B., Röllinghoff M., Bogdan C. (2001). The Production of IFN-γ by IL-12/IL-18-Activated Macrophages Requires STAT4 Signaling and Is Inhibited by IL-4. J. Immunol. 166, 3075–3082. doi: 10.4049/jimmunol.166.5.3075
Schmid-Burgk J. L., Gaidt M. M., Schmidt T., Ebert T. S., Bartok E., Hornung V. (2015). Caspase-4 mediates non-canonical activation of the NLRP3 inflammasome in human myeloid cells. Eur. J. Immunol. 45, 2911–2917. doi: 10.1002/eji.201545523
Schneider W. M., Chevillotte M. D., Rice C. M. (2014). Interferon-Stimulated Genes: A Complex Web of Host Defenses. Annu. Rev. Immunol. 32, 513–545. doi: 10.1146/annurev-immunol-032713-120231
Seth R. B., Sun L., Ea C.-K., Chen Z. J. (2005). Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 122, 669–682. doi: 10.1016/j.cell.2005.08.012
Shahangian A., Chow E. K., Tian X., Kang J. R., Ghaffari A., Liu S. Y., et al. (2009). I IFNs mediate development of postinfluenza bacterial pneumonia in mice. J. Clin. Invest. 119, 1910–1920. doi: 10.1172/JCI35412.1910
Shenoy A. R., Wellington D. A., Kumar P., Kassa H., Booth C. J., Cresswell P., et al. (2012). GBP5 Promotes NLRP3 inflammasome assembly and immunity in mammals. Science 336, 481–485. doi: 10.1126/science.1217141
Sheppard P., Kindsvogel W., Xu W., Henderson K., Schlutsmeyer S., Whitmore T. E., et al. (2002). IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4, 63–68. doi: 10.1038/ni873
Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., et al. (2014). Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192. doi: 10.1038/nature13683
Sixt B. S., Bastidas R. J., Finethy R., Baxter R. M., Carpenter V. K., Kroemer G., et al. (2017). The Chlamydia trachomatis Inclusion Membrane Protein CpoS Counteracts STING-Mediated Cellular Surveillance and Suicide Programs. Cell Host Microbe 21, 113–121. doi: 10.1016/j.chom.2016.12.002
Smallwood A., Hon G. C., Jin F., Henry R. E., Espinosa J. M., Ren B. (2012). CBX3 regulates efficient RNA processing genome-wide. Genome Res. 22, 1426–1436. doi: 10.1101/gr.124818.111
Snyder D. T., Hedges J. F., Jutila M. A. (2017). Getting “inside” Type I IFNs: Type I IFNs in Intracellular Bacterial Infections. J. Immunol. Res. 2017, 17. doi: 10.1155/2017/9361802
Takeuchi O., Akira S. (2010). Pattern Recognition Receptors and Inflammation. Cell 140, 805–820. doi: 10.1016/j.cell.2010.01.022
Tanaka Y., Chen Z. J. (2012). STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5, ra20. doi: 10.1126/scisignal.2002521
Tretina K., Park E. S., Maminska A., MacMicking J. D. (2019). Interferon-induced guanylate-binding proteins: Guardians of host defense in health and disease. J. Exp. Med. 216, 482–500. doi: 10.1084/jem.20182031
Walberg K., Baron S., Poast J., Schwartz B., Izotova L., Pestka S., et al. (2008). Interferon protects mice against inhalation anthrax. J. Interf. Cytokine Res. 28, 597–601. doi: 10.1089/jir.2007.0143
Wandel M. P., Pathe C., Werner E. I., Ellison C. J., Boyle K. B., von der Malsburg A., et al. (2017). GBPs Inhibit Motility of Shigella flexneri but Are Targeted for Degradation by the Bacterial Ubiquitin Ligase IpaH9.8. Cell Host Microbe 22, 507–518.e5. doi: 10.1016/j.chom.2017.09.007
Wandel M. P., Kim B., Park E., Boyle K. B., Nayak K., Lagrange B., et al. (2020). Guanylate-binding proteins convert cytosolic bacteria into caspase-4 signaling platforms. Nat. Immunol. 21, 880–891. doi: 10.1038/s41590-020-0697-2
Watanabe T., Asano N., Fichtner-feigl S., Gorelick P. L., Tsuji Y., Matsumoto Y., et al. (2010). NOD1 contributes to mouse host defense against Helicobacter pylori via induction of type I IFN and activation of the ISGF3 signaling pathway. J. Clin. Invest. 120, 1645–1662. doi: 10.1172/JCI39481DS1
Wilson R. P., Tursi S. A., Rapsinski G. J., Medeiros N. J., Le L. S., Kotredes K. P., et al. (2019). STAT2 dependent type i interferon response promotes dysbiosis and luminal expansion of the enteric pathogen Salmonella Typhimurium. PloS Pathog. 15, 1–22. doi: 10.1371/journal.ppat.1007745
Wolf K., Fields K. A. (2013). Chlamydia pneumoniae Impairs the Innate Immune Response in Infected Epithelial Cells by Targeting TRAF3. J. Immunol. 190, 1695–1701. doi: 10.4049/jimmunol.1202443
Wu J., Sun L., Chen X., Du F., Shi H., Chen C., et al. (2017). Cyclic GMP-AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu. Rev. Biochem. 86, 541–566. doi: 10.1146/annurev-biochem-061516-044813
Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H.-B. (2005). VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740. doi: 10.1016/j.molcel.2005.08.014
Zhang H., Zoued A., Liu X., Sit B., Waldor M. K. (2020). Functional remodeling of lysosomes by type I interferon modifies host defense. bioRxiv Cell Biol. doi: 10.1101/2020.02.25.965061
Zheng Z., Wei C., Guan K., Yuan Y., Zhang Y., Ma S., et al. (2016). Bacterial E3 Ubiquitin Ligase IpaH4.5 of Shigella flexneri Targets TBK1 To Dampen the Host Antibacterial Response. J. Immunol. 196, 1199–1208. doi: 10.4049/jimmunol.1501045
Keywords: bacterial effectors, interferons, interferon-stimulated genes, janus kinase signal transducer and activator of transcription signaling, immunity, microbial pathogenesis, host-pathogen interactions
Citation: Alphonse N, Dickenson RE and Odendall C (2021) Interferons: Tug of War Between Bacteria and Their Host. Front. Cell. Infect. Microbiol. 11:624094. doi: 10.3389/fcimb.2021.624094
Received: 30 October 2020; Accepted: 05 January 2021;
Published: 10 March 2021.
Edited by:
Gunnar Neels Schroeder, Queen’s University Belfast, United KingdomReviewed by:
Bastian Opitz, Charité – Universitätsmedizin Berlin, GermanyCopyright © 2021 Alphonse, Dickenson and Odendall. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charlotte Odendall, Q2hhcmxvdHRlLm9kZW5kYWxsQGtjbC5hYy51aw==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.