- 1Department of Ecosystem and Public Health, Faculty of Veterinary Medicine, University of Calgary, Calgary, AB, Canada
- 2Animal Policy and Epidemiology Section, Animal Health Branch, Animal Health and Assurance Division, Alberta Agriculture and Forestry, Edmonton, AB, Canada
- 3Western College of Veterinary Medicine, University of Saskatchewan, Saskatoon, SK, Canada
- 4Center for Foodborne, Environmental and Zoonotic Infectious Diseases, Public Health Agency of Canada, Guelph, ON, Canada
Horizontal gene transfer is an important mechanism which facilitates bacterial populations in overcoming antimicrobial treatment. In this study, a total of 120 Escherichia coli and 62 Salmonella enterica subsp. enterica isolates were isolated from broiler chicken farms in Alberta. Fourteen serovars were identified among Salmonella isolates. Thirty one percent of E. coli isolates (37/120) were multiclass drug resistant (resistant to ≥ 3 drug classes), while only about 16% of Salmonella isolates (10/62) were multiclass drug resistant. Among those, eight E. coli isolates had an AmpC-type phenotype, and one Salmonella isolate had an extended-spectrum beta-lactamase (ESBL)-type beta-lactamase phenotype. We identified both AmpC-type (blaCMY-2) and ESBL-type (blaTEM) genes in both E. coli and Salmonella isolates. Plasmids from eight of nine E. coli and Salmonella isolates were transferred to recipient strain E. coli J53 through conjugation. Transferable plasmids in the eight E. coli and Salmonella isolates were also transferred into a lab-made sodium azide-resistant Salmonella recipient through conjugation. The class 1 integrase gene, int1, was detected on plasmids from two E. coli isolates. Further investigation of class 1 integron cassette regions revealed the presence of an aadA gene encoding streptomycin 3’’-adenylyltransferase, an aadA1a/aadA2 gene encoding aminoglycoside 3’’-O-adenyltransferase, and a putative adenylyltransferase gene. This study provides some insight into potential horizontal gene transfer events of antimicrobial resistance genes between E. coli and Salmonella in broiler chicken production.
Introduction
For decades, antimicrobial resistance (AMR) has been a global issue of grave concern. Understanding potential mechanisms and driving forces for dissemination of genes encoding antimicrobial resistance between bacteria will help reduce the prevalence of resistant bacteria and thereby reduce risks to human and animal health. Acquisition of new resistance genes occurs frequently and naturally among bacterial communities from humans, animals and environments as outlined in the model known as ‘the epidemiology of AMR’ (Prescott, 2006). However, the mechanism of dissemination of resistance genes is not yet fully understood.
Escherichia coli and Salmonella spp. are common bacterial causes of foodborne disease in humans as well as gastrointestinal disease in animals (Folster et al., 2011; Ghodousi et al., 2015). E. coli is a genetically diverse species which has both commensal and pathogenic strains (Leimbach et al., 2013). Salmonella enterica are enteric pathogens, and are closely related to commensal E. coli, sharing ~85% of their genomes in common at the nucleotide level (Mcclelland and Wilson, 1998; McClelland et al., 2000). Multi-drug resistant (MDR) E. coli and Salmonella could lead to the antibiotic choices of last resort for treating multidrug-resistant Gram-negative infections. Therefore, MDR E. coli and Salmonella, especially the ones that exhibit AmpC/extended-spectrum beta-lactamase (ESBL) phenotypes, have become a major cause of clinical concern (Mohanty et al., 2010). However, these isolates are not always detected in routine susceptibility tests (Mohanty et al., 2010).
AmpC-type CMY beta-lactamase genes (blaCMY) have been found on both the chromosome and plasmids of many gram negative bacteria such as Klebsiella spp., Escherichia coli, and Salmonella spp. CMY-2 is reported to be the most common plasmid-carried AmpC-type CMY in both E. coli and Salmonella isolates from various global regions including Asia, North America and Europe (Guo et al., 2014). Extended spectrum beta-lactamases (ESBLs) are beta-lactamases belonging mainly to Ambler class A, which includes TEM-, SHV-, CTX-M, GES, VEB enzyme families. ESBLs also include one enzyme family, the OXA- family, belonging to class D (Cantón et al., 2012). Isolates carrying plasmid-encoded AmpC can be easily misidentified as ESBLs due to their overlapping activity against beta-lactam antimicrobials. The inability to distinguish them could have significant treatment consequences (Hanson, 2003).
Mobile genetic elements, such as plasmids or DNA transposons, are the main mechanisms facilitating horizontal genetic transfer (HGT). Plasmid-mediated blaCMY-2 has been found to be the most predominant among other acquired ampC genes (Mata et al., 2012). The plasmids carrying blaCTX-M or blaCMY beta-lactamase genes have been associated with transferable replicon types IncA/C or IncI1 (Hopkins et al., 2006; Guo et al., 2014).
Antimicrobial use in the poultry industry improves animal health, welfare and production by preventing and treating animal disease resulting in lowered mortality, but may lead to the selection of AMR organisms (Diarra and Malouin, 2014). In Canada, the preventive use of ceftiofur in broiler chicken was voluntarily eliminated by the poultry industry in May 2014; the broiler chicken antimicrobial use (AMU)-AMR surveillance component of the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) detected a significant drop in ceftiofur use at the hatcheries between 2013 and 2014 (Agunos et al., 2017). The three most frequently reported antimicrobials by producers participating in the CIPARS farm surveillance between 2013 and 2015 were bacitracin, virginiamycin and avilamycin (Agunos et al., 2017). Avilamycin was approved around May 2013 and was first reported by producers in 2014.
The isolates used in this study were a subset of those described in the 2015 CIPARS annual report (Public Health Agency of Canada, 2015). The objectives of the study were threefold: 1. To further investigate AMR phenotypes in E. coli and Salmonella isolates from broiler chickens, 2. To identify genes responsible for ESBL/AmpC phenotypes in multi-drug resistant E. coli and Salmonella isolates and 3. To conduct conjugation assays to initially assess the potential for horizontal gene transfer between E. coli and Salmonella.
Materials and Methods
Sampling, Bacterial Isolation, and Isolates Used in This Study
Fecal samples were taken from a single production unit on each of 30 registered premises/establishments (farms) participating in the CIPARS broiler chicken farm AMU/AMR surveillance in Alberta in 2015. Participating sentinel veterinarians were responsible for enrolling farms and collecting samples. Farms were chosen based on the veterinary practice profile and using specific inclusion and exclusion criteria. Samples were collected at pre-harvest, approximately one week prior to slaughter (broilers >30 days of age). Fecal samples consisted of 10 fecal droppings from each of the four quadrants of the chosen barn/floor, pooled to represent the chosen production unit.
For the recovery of organisms, four pooled fresh fecal samples (equivalent to at least 10 individual droppings) were collected from four quadrants of the barn. Laboratory techniques are as follows (in brief): 1) Salmonella: fecal samples were weighed and Buffered Peptone Water (BPW) was added (1:10) and incubated at 35°C for 24 hours. A loopful was inoculated into a Modified Semisolid Rappaport Vassiliadis (MSRV) plate and incubated at 42°C for 24 to 72 hours. 2) E. coli: A drop of BPW aliquot prepared from above was inoculated on MacConkey agar and incubated at 35°C for 24 hours. It is important to note that the isolates recovered here were part of the CIPARS farm surveillance component and thus no selective media were used. The intent was to harmonize with the isolation/characterization procedures outlined in the CIPARS Report (Public Health Agency of Canada, 2015).
A total number of 120 E. coli and 62 Salmonella were isolated, banked, and shipped to the University of Calgary frozen on dry ice, by the Agri Food Laboratories Section of Alberta Agriculture and Forestry (Public Health Agency of Canada, 2015; FoodNet Canada, 2017). E. coli J53 (KACC 16628), a recipient isolate for the conjugation experiment, was received from the Korean Agricultural Culture Collection (KACC), Agricultural Microbiology Division, National Academy of Agricultural Science. E. coli HB101carrying plasmid pRK600, used as a helper strain, was received from the Dong lab, University of Calgary.
Susceptibility Tests
Minimal Inhibitory Concentrations (MICs) of various antimicrobial agents were determined using Sensititre™ (TREK Diagnostic Systems, Inc.) Gram negative plates (CMV3AGNF) designed by the National Antimicrobial Resistance Monitoring System (Public Health Agency of Canada, 2015). The same panel of antimicrobial agents was used for both E. coli and Salmonella isolates (Tables 2–4). Antimicrobial resistance assays were conducted by the National Microbiology Laboratory (NML) St. Hyacinthine, and NML Guelph (Public Health Agency of Canada, 2015).
The disc diffusion method was used to compare antimicrobial resistance profiles of isolate donors and E. coli recipients (Tendencia, 2004). Antibiotic discs were purchased from either BD BBL™ or Oxoid companies. The diameters of the zones of inhibition were recorded and interpreted according to Clinical and Laboratory Standards Institute (CLSI) guidelines (Clinical and Laboratory Standards Institute, 2013). For the purposes of this study, isolates displaying intermediate resistance were categorized as sensitive.
Phenotype and Genotype Confirmation of ESBL/AmpC Genes
Two different ESBL/AmpC detection disc sets have been used to confirm ESBL/AmpC phenotypes. The first set is a combination of 4 individual discs of Cefotaxime/Cefotaxime + Clavulanic acid/Ceftazidime/Ceftazidime + Clavulanic acid, purchased from either BD BBL™ or Oxoid company. The second set is a combination of 4 individual discs of Cefpodoxime/Cefpodoxime + ESBL inhibitor/Cefpodoxime + AmpC inhibitor/Cefpodoxime + ESBL inhibitor + AmpC inhibitor, purchased from Mast Group company (D68C set).
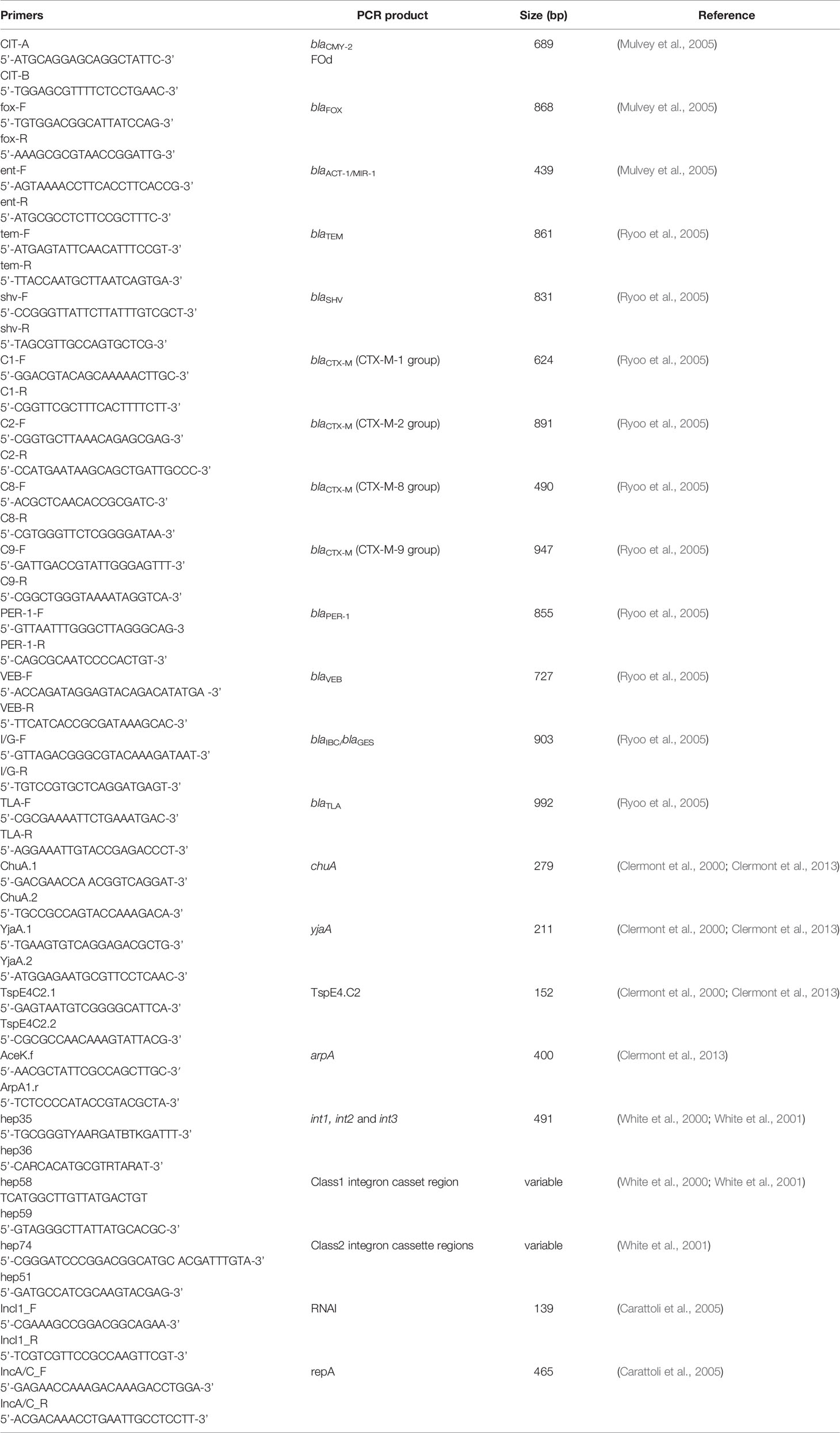
In addition, AmpC and ESBL beta-lactamase genes were detected using PCR assays. A total of three AmpC (blaCMY-2, blaFOX, blaACT-1/MIR-1) and ten ESBL (blaTEM, blaSHV blaCTX-M-1, blaCTX-M-2, blaCTX-M-8, blaCTX-M-9, blaPER-1, blaVEB, blaIBC/blaGES, blaTLA) beta-lactamase genes were screened in AmpC/ESBL positive isolates. Primers used in the PCR assays are listed in Table 1. PCR products at desired sizes were purified and sent for Sanger sequencing (http://www.ucalgary.ca/dnalab/sequencing) to confirm their sequences
Multi-drug resistant (MDR) E. coli and Salmonella isolates that were resistant to at least three drug classes and were confirmed to exhibit ESBL/AmpC phenotypes were selected for additional experiments. Seven drug classes were used in this study: beta-lactams (AMC = Amoxycillin + Clavulanic acid, AMP = Ampicillin, FOX = Cefoxitin, CRO = Ceftriaxone, TIO = Ceftiofur), aminoglycosides (GEN = Gentamicin, STR = Streptomycin), quinolones (NAL = Nalidixic acid, CIP = Ciprofloxacin), sulfonamides (SSS = Sulfisoxazole, SXT = Trimethoprim sulfamethoxazole), macrolides (AZM = Azithromycin), phenicols (CHL = Chloramphenicol), tetracyclines (TET =Tetracycline).
Plasmid Characterization/Replicon Typing
Plasmid miniprep was performed using an alkaline lysis method (Birnboim and Doly, 1984). The plasmid size was evaluated using agarose gel electrophoresis.
Replicon typing was performed using PCR assay as described previously, with primers listed in Table 1 (Carattoli et al., 2005).
E. coli Phylogroups/Salmonella Serovars
MDR E. coli isolates exhibiting ESBL/AmpC phenotypes were assigned into one of four main phylogenetic groups by using a simplified two-step triplex polymerase reaction (Clermont et al., 2000). The results were confirmed using a quadruplex PCR assay which enabled us to classify isolates into a broader range of E. coli phylogroups as well as distinguish them from the cryptic clades II to V (Clermont et al., 2013).
An assay to classify Salmonella serovars was performed by the PHAC serotyping laboratory as described previously (Public Health Agency of Canada, 2015).
Detection of Integrons/Integrases
To further study other mobile genetic elements, int1, int2 and int3 genes encoding integrase of class 1, 2 and 3 integrons, respectively, were investigated by PCR assays using primers listed in Table 1 as described previously (White et al., 2001). Primers for amplifying the class 1 and class 2 integron cassette regions were used to detect the presence of resistance gene cassettes (Table 1) (White et al., 2001). PCR products were purified and sent for Sanger sequencing (http://www.ucalgary.ca/dnalab/sequencing) to confirm their sequences.
Conjugation Experiment
Conjugation experiments were conducted using MDR isolates of interest as donors and E. coli J53 as the recipient with or without the presence of helper strain HB101/pRK600. E. coli J53 (F- met pro Azir Amps), an E. coli K-12 derivative strain, is resistant to sodium azide but sensitive to beta-lactams (Yi et al., 2012). This strain is also sensitive to other drugs (e.g. tetracycline, chloramphenicol, gentamicin, streptomycin) (Lei et al., 2019). Recipient and donor strains were inoculated into LB broth and cultured overnight at 37°C. The next day, cells were harvested, washed with saline, and mixed together in a ratio of 1:1, and spotted on to LB plates. They were also spotted individually on LB plates as controls. After overnight incubation at 37°C, mating spots were washed and resuspended in saline; and different dilutions were plated on LB media containing sodium azide (0.2 gL-1) and ampicillin (100 µgml-1) to select transconjugants. Control spots were transferred to the same selective media to make sure that no growth was observed. Conjugation frequency was calculated by taking the ratio of the number of colonies counted on selective plates (LB supplemented with sodium azide (0.2 gL-1) + ampicillin (100 µgml-1)) for transconjugants over the number of colonies on selective plates (LB supplemented with sodium azide (0.2 gL-1)) for recipients. If there were no transconjugants obtained, a helper strain (HB101/pRK600) was added into the mating mix in the proportion of 1:1:0.5 (donor: recipient: helper strain) and spotted on LB plates as described. If there was no growth on plates selected for recipients, trypsin was added to the media to reverse the effect of colicin produced by the donors, and recover the recipients (Nomura and Nakamura, 1962; Dankert et al., 1980).
Salmonella isolate 112.2 was screened for spontaneously mutated colonies resistant to sodium azide (AziR) by plating on LB supplemented with sodium azide (0.2 gL-1). Then this AziR Salmonella was used as a recipient in conjugation with MDR isolates of interest as donors. Conjugation protocol was performed as described above.
Data Visualization Tools
Data visualization in this study was performed using following programs: Microsoft Excel 2013, R programming (R version 3.4.1).
Results
Sampling, Isolation and Identification of Bacterial Strains
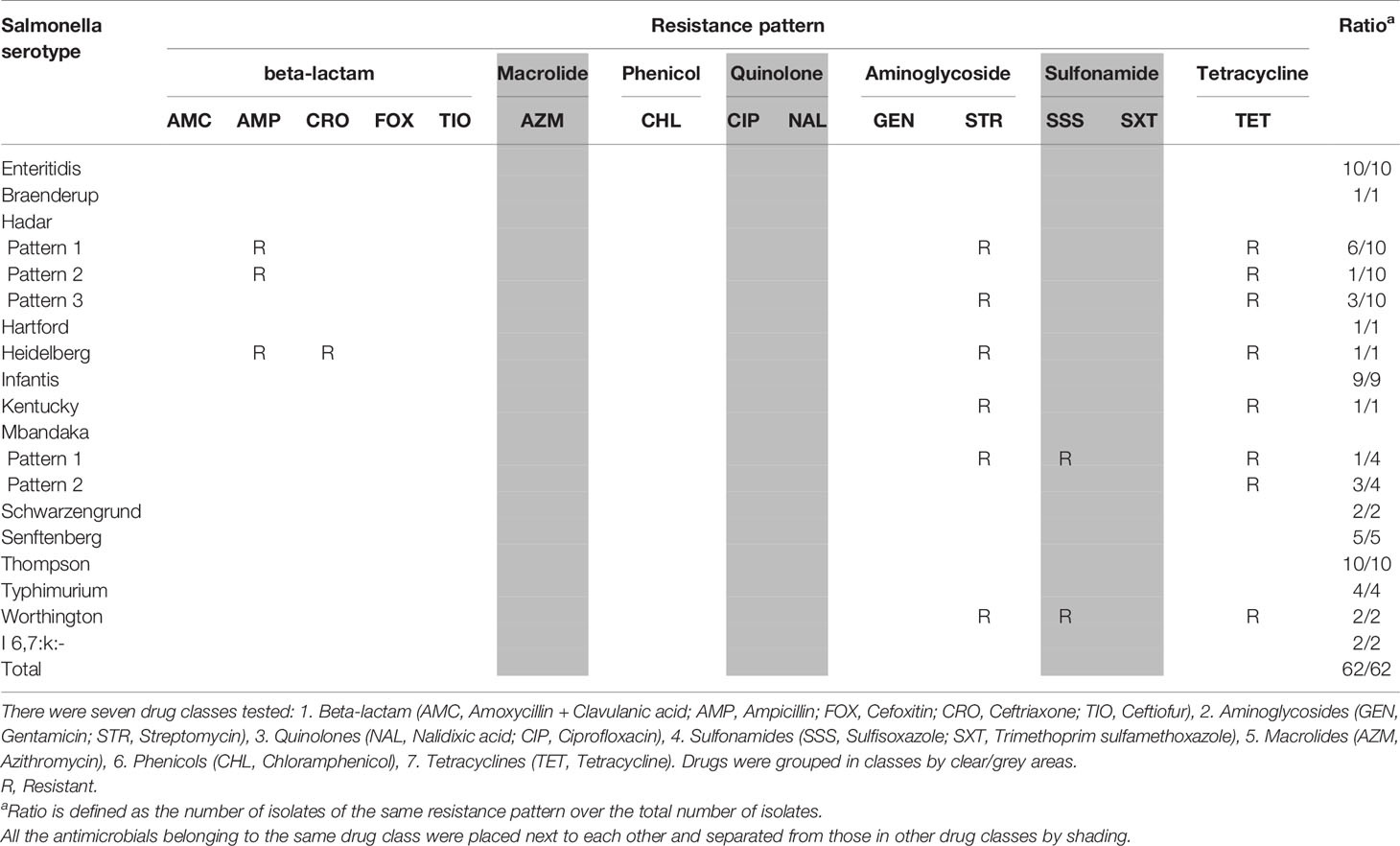
Four E. coli isolates were obtained from each farm, resulting in 120 E. coli isolates from 30 farms with a recovery rate of 100% (30/30). There were 23 of 30 farms Salmonella positive, with between one and four isolates identified per farm. Hence the recovery rate for Salmonella was about 77% (23/30). There were 14 different serovars identified among 62 Salmonella isolates (Table 2). The three most prevalent Salmonella serovars in our study were Enteritidis, Hadar, and Thompson.
Antimicrobial Susceptibility Testing
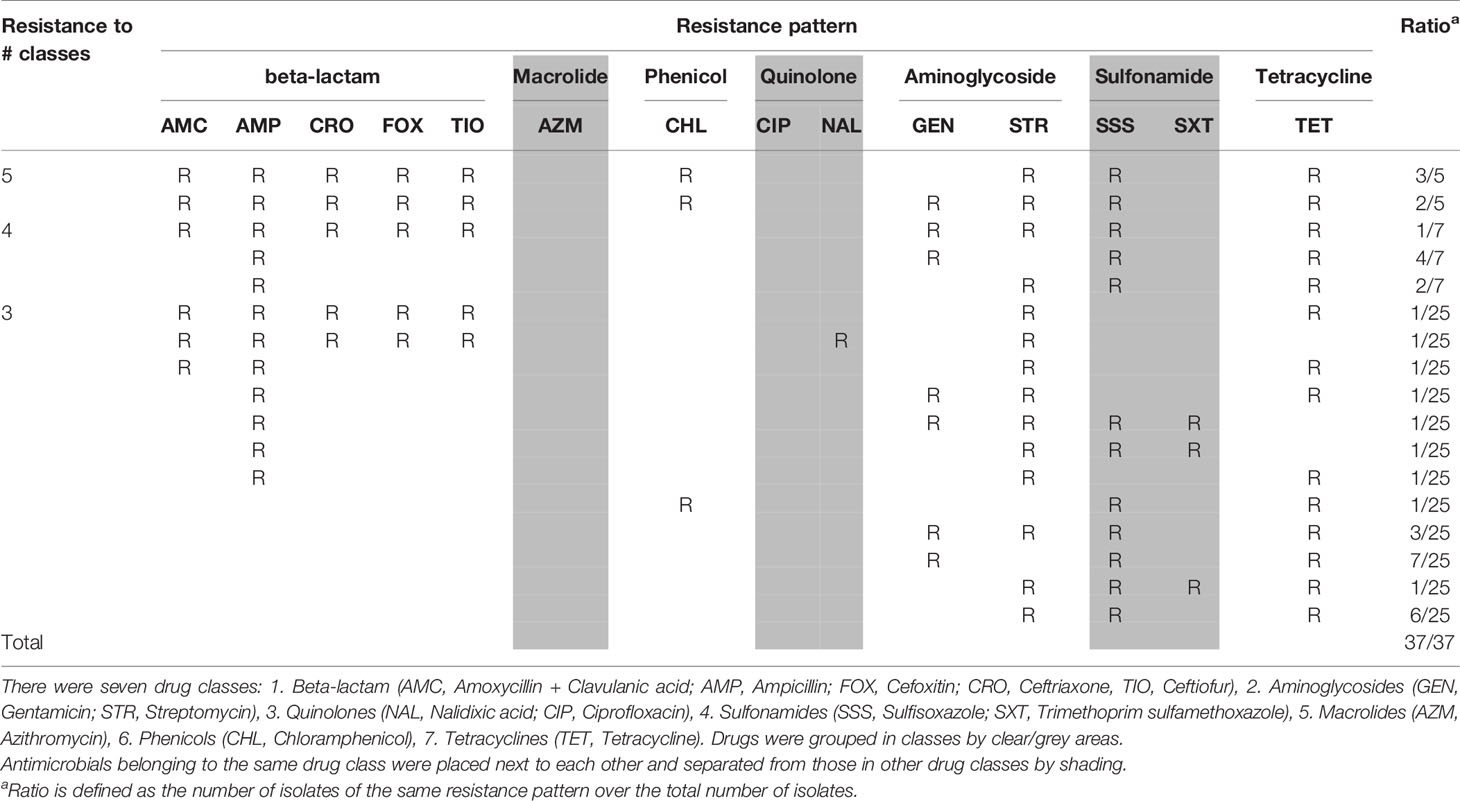
Antimicrobial resistance in E. coli and Salmonella isolates is described as follows. Isolates that were resistant to three or more antimicrobial classes were considered MDR. Thirty-one percentage of E. coli (37/120) were MDR and 16% of Salmonella (10/62) were MDR. About 4% of E. coli (5/120) were resistant to five antimicrobial classes, while none of Salmonella were resistant to five classes.
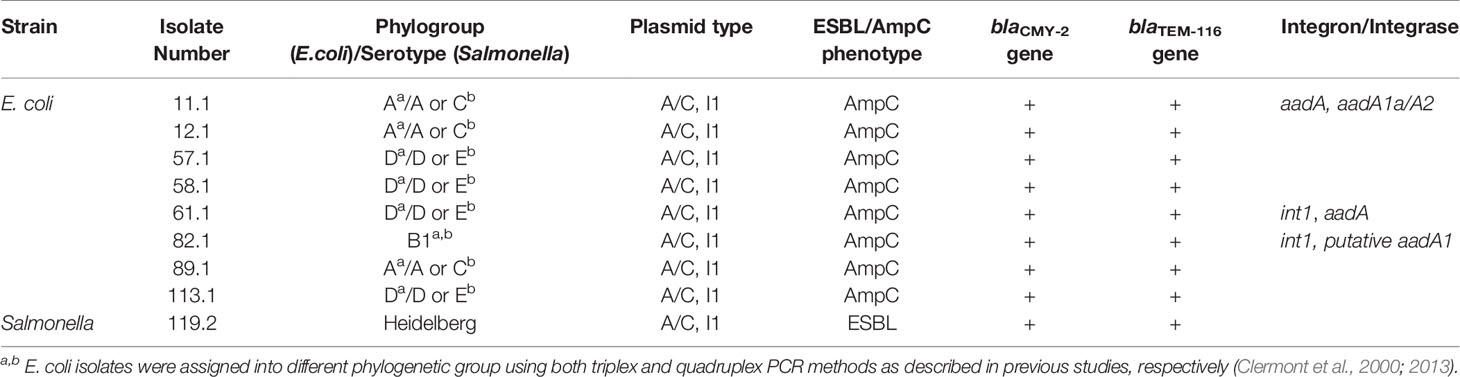
The majority of Salmonella isolates were resistant to streptomycin and tetracycline (Table 2). There were 8 Salmonella serotypes that were sensitive to all tested antimicrobials (Enteritidis, Typhimurium, Braenderup, Hartford, Infantis, Schwarzengrund, Senftenberg, Thompson). In addition to streptomycin and tetracycline, the majority of MDR E. coli showed resistance to sulfisoxazole (Table 3). Among E. coli that were phylo-typed, those belonging to groups D or E had diverse AMR patterns (Table 4). Overall, E. coli isolates showed more diversity in resistance phenotype between farms than did Salmonella (Figure 1).
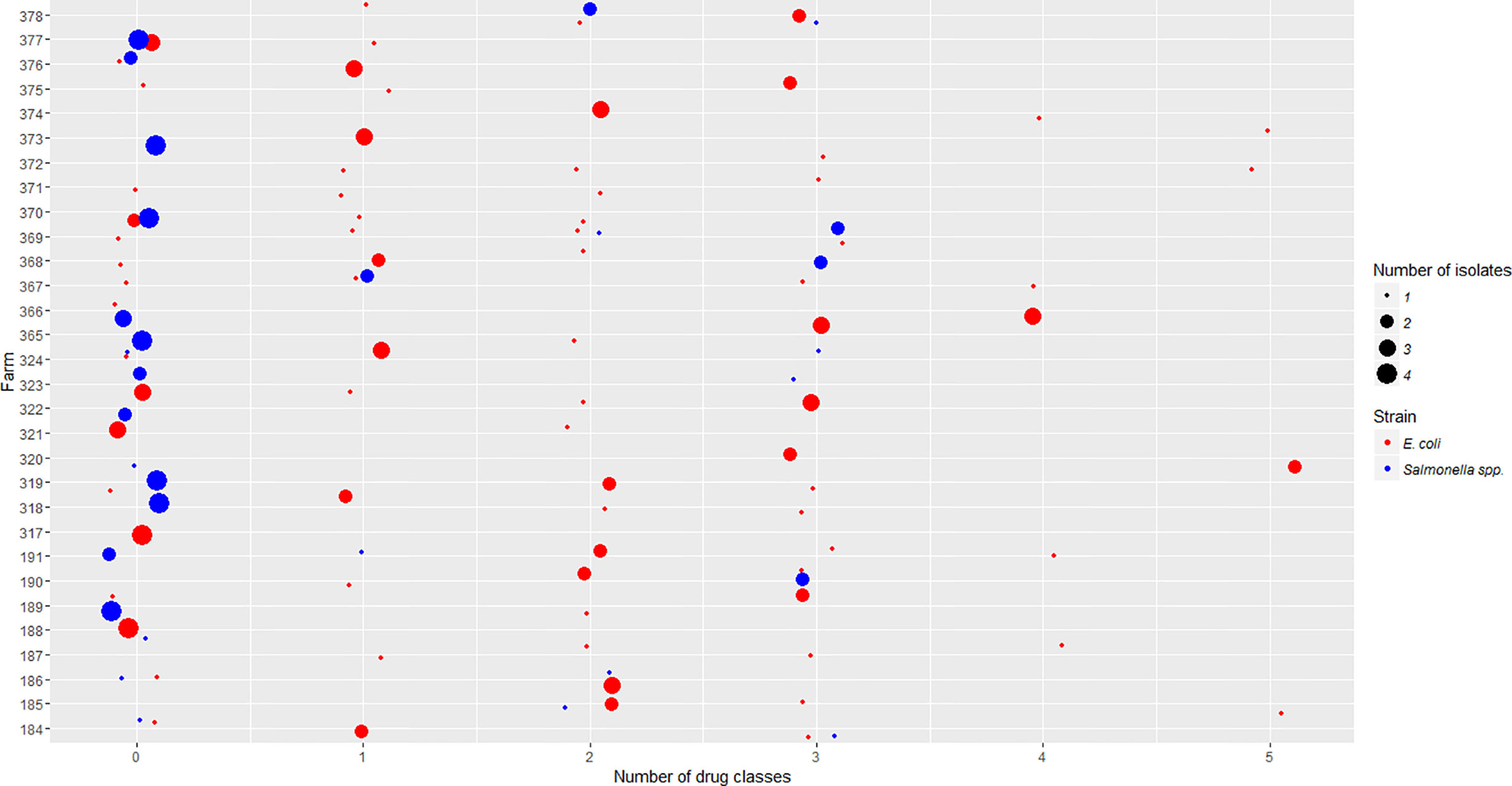
Figure 1 Salmonella isolates resistant to differing number of drug classes were compared to E. coli isolates across participating farms. The x-axis is the number of drug classes to which isolates showed resistance. The y-axis is the code of individual farms participating in this project. The size of each dot represents the number of isolates obtained in each farm. The color represents whether the isolates are E. coli or Salmonella strains.
Among MDR isolates, 19 E. coli and 10 Salmonella isolates with resistance to the beta-lactam class of antimicrobials and at least two other drug classes, were selected for further study. These isolates came from 19 farms.
ESBL/AmpC Phenotypes and Genotypes
Eight out of 19 MDR E. coli isolates were resistant to both penicillin and cephalosporin beta-lactam sub-classes and were confirmed as AmpC phenotype (Table 5). A unique Salmonella was resistant to both penicillin and cephalosporin sub-classes and was potentially an ESBL phenotype; in the presence of an ESBL inhibitor (clavulanic acid) the isolate showed sensitivity to penicillin (specifically amoxicillin). The ESBL phenotype was subsequently confirmed by disc diffusion method.
Plasmids extracted from nine isolates which either showed the AmpC or ESBL phenotype were used as templates in PCR reactions to detect a variety of AmpC and ESBL genes. The blaCMY-2 gene, an AmpC-type gene, and the blaTEMgene, an ESBL-type gene, were identified on plasmids from all nine E. coli and Salmonella isolates. The sequence of the blaTEM gene identified in this study shared 100% identity with the sequence of the blaTEM-116 gene found in E. coli strain MRC3 (accession no. KJ923009.1)
Plasmid Characterization
Nine isolates were found to carry IncI1 and IncA/C-type replicon plasmids (Table 5). The plasmids varied in size from approximately 7kb to larger than 20kb. The plasmid in one Salmonella isolate has a size larger than 20 kb with I1 and A/C-type replicon. Two E. coli isolates, 58.1 and 61.1, carried small plasmids (<10kb), while the rest carried larger ones (≥20 kb). All plasmids in E. coli isolates had IncI1 and IncA/C-type replicons.
E. coli Phylogroups/Salmonella Serovars of ESBL/AmpC-Positive Isolates
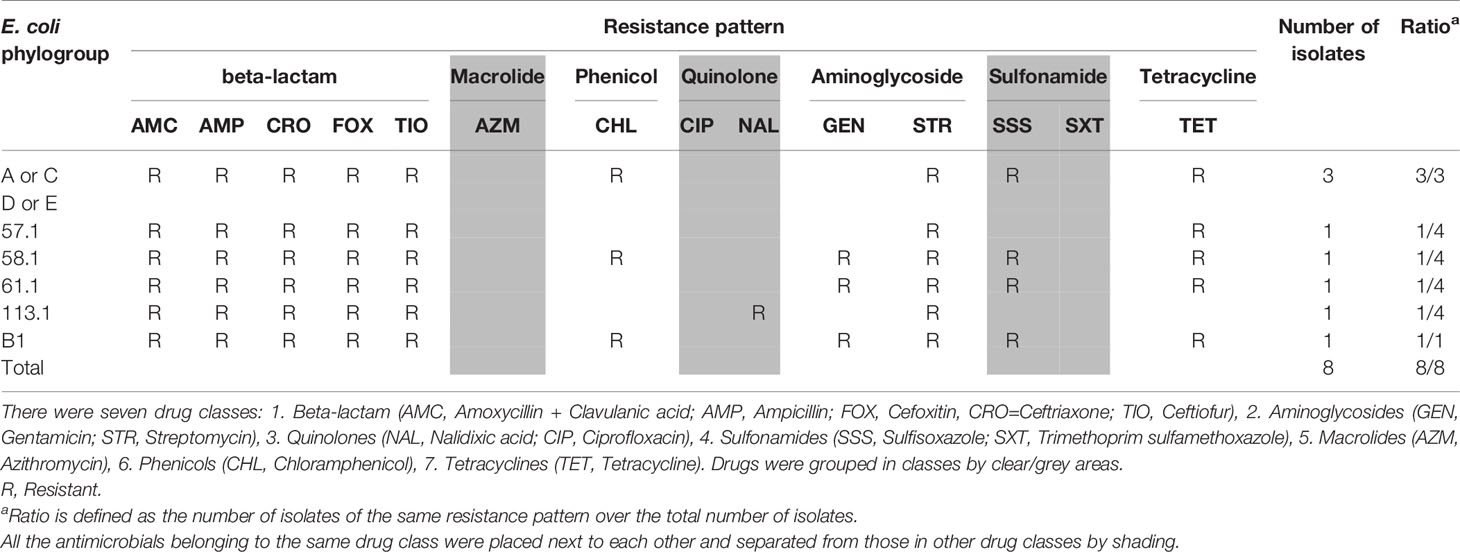
Both PCR methods confirmed that none of the E. coli isolates belonged to the group B2 (a group with high potential for pathogenicity) (Tables 4 and 5). Phylogroups of E. coli identified in this study were A, B1 and D (triplex PCR) or A, B1, C, D and E (quadruplex PCR). One MDR Salmonella isolate exhibiting the ESBL phenotype was identified as Heidelberg serovar.
Detection of Integrons/Integrases
Using DNA from plasmid extraction as a template in PCR reactions, the class 1 integrase gene int1 was detected in two E. coli isolates that were from 2 different farms (Table 5). The class 1 integron cassette region was also detected in three isolates by PCR (Table 5). The PCR products were sequenced, and the results were confirmed by blasting the sequence against the NCBI database. The sequence matched the aminoglycoside resistance genes, aadA encoding streptomycin 3’’-adenylyltransferase, aadA1a/aadA2 encoding aminoglycoside 3’’-O-adenyltransferase. The class 1 integron cassettes were also amplified from plasmid extraction from E. coli isolate 82.1. The results were confirmed by blasting the sequence against the NCBI database. The sequences matched the sequence of a putative adenylyltransferase found on a plasmid isolated from the Salmonella Heidelberg strain N418 (Accession no. CP009409).
Transfer of Plasmids Carrying Resistance Genes by Conjugation
Plasmids were mobilized from all, but one E. coli isolate 89.1, to an E. coli recipient J53 (Table 6). No growth was observed on selective plates of either the transconjugants or recipients when attempting to conjugate E. coli isolate 61.1. However, the conjugation experiment of this isolate was successful when the media were supplemented with trypsin to reverse the effect of colicin produced by the donors. Additionally, the E. coli isolate 113.1 required the presence of the helper E. coli strain HB101 carrying helper plasmid pRK600 to enable movement of the plasmid to recipients. All transconjugants were resistant to ampicillin which was used as a selective marker in conjugation experiments. Interestingly, their resistance phenotypes to other drugs, which were presented as “transferable AMR” in Table 6, were distinctly different, except for recipients that were conjugated with either donor 12.1 or 113.1. Although transconjugants obtained from conjugation assays with either donor 12.1 or 113.1 were resistant to the same drugs including amoxicillin, cefoxitin, ceftriaxone and ceftiofur, donor 12.1 produced E. coli transconjugants more efficiently (higher conjugation frequency) and did not require the presence of the helper strain (Table 6).
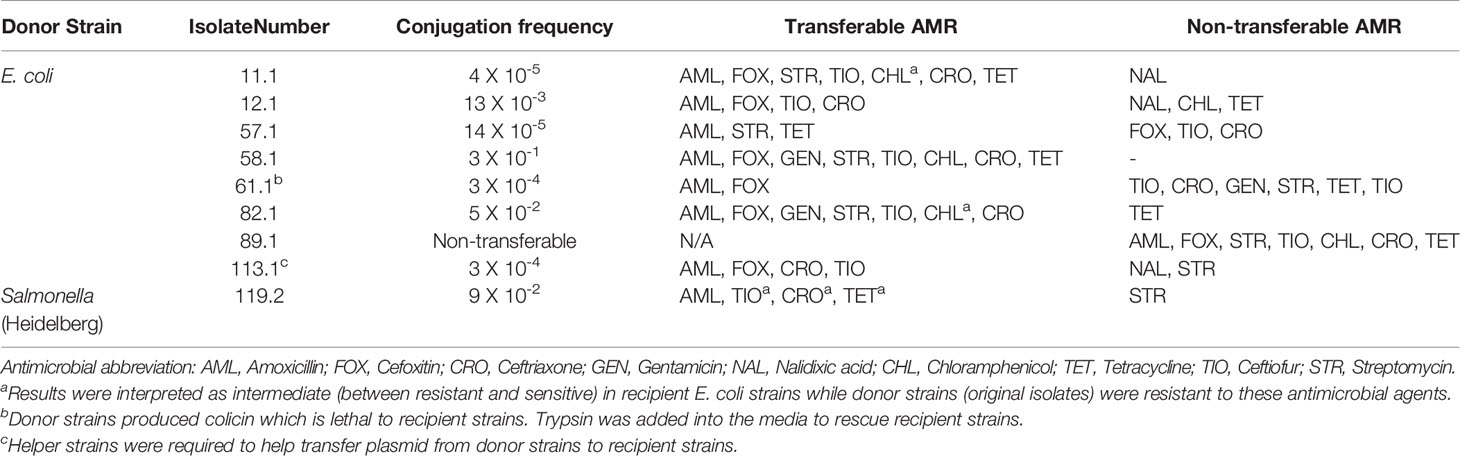
Table 6 Conjugation frequency and AMR profile of transconjugants compared to donors (tested isolates) using E. coli J53 as a recipient.
When using a lab-engineered sodium azide-resistant Salmonella as a recipient, we observed that plasmids from eight E. coli isolates with the exception of the isolate 89.1 mentioned above, were able to move to Salmonella with variable conjugation frequency (Table 7).
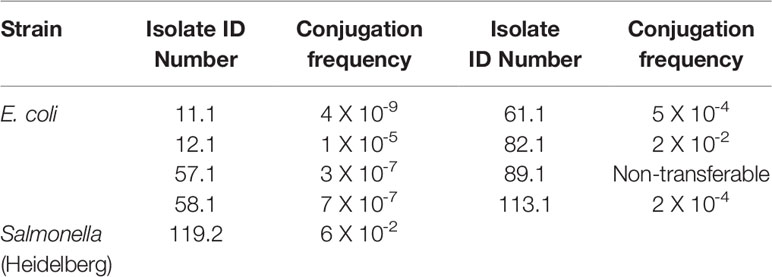
Table 7 Conjugation frequency between donors (tested isolates) and a recipient strain (sodium azide-resistant Salmonella).
Farm Characteristics for Nine Isolates From Which Plasmids Were Mobilized
Seven ‘conventional’ (i.e., antimicrobials were used to some extent in all flocks) farms under the veterinary care of one practice were represented by the nine ESBL/AmpC phenotyped isolates in this study. All of the farms, excluding the farm providing isolate 89.1, received their chicks from the same hatchery. All birds were Ross 308 strain. The most frequently used antimicrobials were bacitracin and salinomycin administered via feed (n = 5), followed by the combination of penicillin and streptomycin administered via water (n = 3). In addition, the feed were also reportedly medicated with the following antimicrobials avilamycin (n = 1), tylosin (n = 2), and coccidiostats such as decoquinate (n = 1), monensin (n = 1), the ionophore-chemical coccidiostat combination, narasin and nicarbazin (n = 2).
The number of chicks sampled per flock ranged from 14,790 to 55,000 within a single production unit. Age on the day of sampling ranged from 30 to 35 days old with an average weight ranging from 1.7 kg to 2.2 kg. The recorded floor space in the barns ranged from 8000 ft2 to 30550 ft2 and stocking density ranged from 0.54 to 0.67 ft2 per bird. Reported percentage of mortality ranged from 2.47% to 7.19% of the birds placed within the barn.
Hydrogen peroxide was used on three of the farms for cleaning of water lines between flocks. Five of the farms also used chlorine for treatment of their water lines during the production cycle. Footbaths (n = 3), dedicated farm clothes (n = 4) and gloves (n =2) were methods of farm biosecurity utilized. Manure was stored onsite in the vicinity of the barn on three farms. The most frequently reported method of cleaning the barns after each production cycle was washing only (n = 6) and chlorine products were used for disinfection on four farms.
Discussion
The three most prevalent Salmonella serovars in our study were Enteritidis, Hadar, and Thompson. Serovars Typhimurium and Heidelberg were also identified. According to the National Enteric Surveillance Program (NESP) 2013 Annual Report, the three most commonly reported serovars from human cases in Canada, which has remained unchanged since 2008, were Enteritidis, Heidelberg and Typhimurium (Government of Canada, 2015). Serovar Enteritidis is known as one of the most common Salmonella serovars found in broiler chickens, and the second most prevalent cause of Salmonella infection in humans after the serovar Typhimurium (Suzuki, 1994; Porwollik et al., 2005; Trampel et al., 2014). Serovars Hadar, Heidelberg, Mbandaka and Worthington were MDR. Of the 14 serovars in our study, the Salmonella serovar Heidelberg was the only MDR isolate with an ESBL phenotype. This is a of concern because in 2013-2014, a national outbreak of MDR Salmonella Heidelberg infections in the United States resulted in 200 hospitalized cases of 528 total cases (38%) (Gieraltowski et al., 2016). This outbreak was linked to chicken products from a single poultry company. In our study, six of the seven farms had chicks sourced from the same hatchery. There is potential for widespread dissemination of virulent bacteria over a wide geographical region if such strains are present among eggs or chicks at the hatchery level. However, there was no evidence of this in our study.
Urinary tract infections are one of the most common bacterial infections reported in primary care and the emergency department in Canada (Sanyal et al., 2019). Phylogroup B2 has been considered to be the most extra-intestinal virulent group (Clermont et al., 2000), and shown to have a strong association with the uropathogenic subpathotype (Hutton et al., 2018). To determine whether the MDR E. coli isolates that exhibited AmpC or ESBL phenotypes belonged to phylogroup B2, both triplex and quadruplex PCR assays were performed. Both assays confirmed that these isolates did not belong to group B2, therefore less likely caused urinary tract infections.
Both blaCMY-2 and blaTEM genes may be present on plasmids isolated from AmpC/ESBL positive MDR E. coli and Salmonella isolates. The blaCMY-2 and blaTEM genes had identical sequences in both E. coli and Salmonella isolates in this study. A previous study showed the evidence for the transfer of blaCMY-2- carrying plasmids between E. coli and Salmonella isolates (Winokur et al., 2001). Interestingly, E. coli isolates had the AmpC beta-lactamase phenotype while Salmonella had the ESBL beta-lactamase phenotype. Even though both species carried both blaCMY-2 and blaTEM genes, the phenotype differences suggest differential expression of these genes in these E. coli and Salmonella isolates.
The blaCMY-2 gene is the most common AmpC-type gene found in both E. coli and Salmonella from various sources: food, animals, and hospitals in multiple countries (Mulvey et al., 2005; Hiki et al., 2013; Cejas et al., 2014; Guo et al., 2014; Ghodousi et al., 2015). This gene has been hypothesized to have originated on the chromosome of E. coli and it could be induced with beta-lactams in some Enterobacteriaceae such as Enterobacter cloacae, Citrobacter freundii, Serratia marcescens, and Pseudomonas aeruginosa (Sanders, 1987; Philippon et al., 2002). Unlike these bacteria, E. coli and Salmonella lack systems to produce inducible AmpC enzymes. Mutations in the ampC promoter have increased the resistance to oxyimino-cephalosporins in E. coli (Caroff et al., 1999).
Plasmids are considered to be facilitators for disseminating beta-lactamase genes between various species such as P. mirabilis, Achromobacter, Salmonella and E. coli (Bobrowski et al., 1976; Levesque et al., 1982; Knothe et al., 1983; Bauernfeind et al., 1989). Molecular characterization of MDR plasmids is essential, yet complicated, because these plasmids are very diverse and promiscuous. The relatedness of plasmids can be analyzed using a PCR-based replicon typing method or whole genome sequence analysis (Carattoli et al., 2005). In previous studies, blaCMY-2-carrying plasmids found in either E. coli or S. enterica were most likely to belong to replicons I1 and A/C (Carattoli, 2009). Our results are also in accordance with these findings. blaTEM genes have been reported to be located on plasmids of various replicon types such as A/C, I1, K, ColE, H12, etc. (Carattoli, 2009). In our study, blaTEM gene was found on plasmids of replicons A/C or I1 in one Salmonella isolate and eight E. coli isolates. Previously, it was shown that plasmids encoding ESBL/AmpC genes in E. coli were highly promiscuous, resulting in the possibility of HGT between E. coli and related Enterobacteriaceae strains (Ewers et al., 2012).
In addition to plasmids, other mobile genetic elements (e.g. integrons containing gene cassettes and integrase) are also facilitating the spread of AMR genes (White et al., 2001). It was shown in multiple independent reports that there was an occurrence of integrons especially class 1 integrons and AMR genes (Leverstein-van Hall et al., 2002; Marashi et al., 2012; Di Cesare et al., 2016). More specifically, a significant association was found between integrons and resistance to certain antimicrobials including gentamicin, kanamycin, streptomycin, tobramycin, sulfafurazole, trimethoprim, ampicillin, chloramphenicol, and tetracycline (White et al., 2001). In our study, the integrase gene int1 was detected in two out of nine AmpC/ESBL-producing isolates. Using specific primers to amplify class 1 integron cassette regions revealed the presence of the aminoglycoside resistance genes aadA encoding streptomycin 3’’-adenylyltransferase, aadA1a/aadA2 encoding aminoglycoside 3’’-O-adenyltransferase, and a putative adenylyltransferase gene. E. coli isolate 11.1 carried a gene cassette in its variable region but did not carry the int1 gene of the classical class 1 integron. This might be due to the disruption caused by IS26 which was reported in a previous study (Dawes et al., 2010).
Conjugation in vitro showed that most of AmpC/ESBL positive MDR isolates carried transferable plasmids that can disseminate AMR phenotypes. The conjugation experiment on isolate 61.1 required the addition of trypsin into the media. Previous studies have shown that treatment of cells with trypsin reversed the inhibition activity caused by colicin (Nomura and Nakamura, 1962; Dankert et al., 1980). The isolate 61.1 likely harbored a colicin-producing plasmid which prevented conjugal transfer of the R-plasmid; hence, this would likely prevent conjugal transfer in the natural microbial community as well. It is interesting to note that in this study all ESBL/AmpC-producing MDR isolates, except for E. coli isolate 89.1, were from farms receiving their chicks from the same hatchery; and Salmonella serovar Heidelberg was one of them. In addition, we were able to transfer plasmids between strains: from E. coli to E. coli, from E. coli to Salmonella, from Salmonella to E. coli, and from Salmonella to Salmonella. Plasmids were transferable through conjugation from E. coli to E. coli or Salmonella to E. coli at higher frequencies compared to plasmids from E. coli to Salmonella or Salmonella to Salmonella.
In conclusion, this study investigated antimicrobial resistance phenotypes of Escherichia coli and Salmonella isolates from 30 broiler farms, which were obtained through the Canadian Integrated Program for Antimicrobial Resistance Surveillance. The study subsequently identified MDR isolates of E. coli and Salmonella enterica with ESBL/AmpC phenotypes and examined the sequences of the ESBL/AmpC genes in these isolates. In addition, plasmids from these MDR isolates were isolated and shown to carry the identical replicon type. We also performed conjugation assays between E. coli and Salmonella isolates to initially assess the potential for HGT. Overall, results suggested there are MDR bacteria in broiler chicken environments with characteristics that could potentially allow them to flourish in the broiler environment, and the possibility of natural HGT by conjugation between E. coli and Salmonella may readily occur in the broiler chicken house environment.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
SC, KL, SG, and CM conceptualized the research idea and obtained research funding from AAF (PI: SC). SG and AA developed the CIPARS AMU-AMR farm surveillance framework, farm surveillance tools (questionnaire) and protocols, and validated the recovery and AMR datasets. Bacterial isolation and initial antimicrobial susceptibility testing were performed by RC. TT and KL were responsible for experimental design. TT conducted research and laboratory analysis. NC conducted statistical analysis. TT and NC designed and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by Alberta Agriculture and Forestry (AAF) [grant number 2015R025R] with significant in a kind support from PHAC and the AAF, Agri-Food Laboratories.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the poultry veterinarians and producers who voluntarily participated in the CIPARS farm surveillance program and enabled data and sample collection. We are grateful to the Chicken Farmers of Canada and the Alberta Chicken Producers for their valuable input to the framework development and technical discussions. We also greatly appreciate that Agricultural Microbiology Division, National Academy of Agricultural Science in South Korea provided the E. coli strain J53 from the Korean Agricultural Culture Collection. We sincerely thank Dr. Richard Moore formerly of the Dong lab for his technical support.
References
Agunos A., Léger D. F., Carson C. A., Gow S. P., Bosman A., Irwin R. J., et al. (2017). Antimicrobial use surveillance in broiler chicken flocks in Canad-2015. PloS One 12, 2013–2015. doi: 10.1371/journal.pone.0179384
Bauernfeind A., Schweighart S., Chong Y. (1989). Extended broad spectrum β-lactamase in Klebsiella pneumoniae including resistance to cephamycins. Infection 17, 316–321. doi: 10.1007/BF01650718
Birnboim H. C., Doly J. (1984). A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 12, 1513–1523. doi: 10.1093/nar/gkn907
Bobrowski M. M., Matthew M., Barth P. T., Datta N., Grinter N. J., Jacob A. E., et al. (1976). Plasmid-Determined Beta-Lactamase Indistinguishable from the Chromosomal Beta-Lactamase of Escherichia coli. J. Bacteriol. 125, 149–157. doi: 10.1128/JB.125.1.149-157.1976
Cantón R., González-Alba J. M., Galán J. C. (2012). CTX-M enzymes: Origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63, 219–228. doi: 10.1016/j.mimet.2005.03.018
Carattoli A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
Caroff N., Espaze E., Berard I., Richet H., Reynaud A. (1999). Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum β-lactamase production. FEMS Microbiol. Lett. 173, 459–465. doi: 10.1111/j.1574-6968.1999.tb13539.x
Cejas D., Vignoli R., Quinteros M., Marino R., Callejo R. (2014). First detection of CMY-2 plasmid mediated β -lactamase in Salmonella Heidelberg in South America. Rev. Argent. Microbiol. 46, 30–33. doi: 10.1016/S0325-7541(14)70044-6
Clermont O., Bonacorsi S., Bingen E. (2000). Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66, 4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000
Clermont O., Christenson J. K., Denamur E., Gordon D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Clinical and Laboratory Standards Institute (2013). Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. M100-S23 ed (Wayne, PA).
Dankert J., Hammond S. M., Cramer W. A. (1980). Reversal by Trypsin of the Inhibition of Active Transport by Colicin E1. J. Bacteriol. 143, 594–602. doi: 10.1128/JB.143.2.594-602.1980
Dawes F. E., Kuzevski A., Bettelheim K. A., Hornitzky M. A., Djordjevic S. P., Walker M. J. (2010). Distribution of Class 1 Integrons with IS26-Mediated Deletions in Their 3 9 -Conserved Segments in Escherichia coli of Human and Animal Origin. PloS One 5, 1–6. doi: 10.1371/journal.pone.0012754
Di Cesare A., Eckert E. M., D’Urso S., Bertoni R., Gillan D. C., Wattiez R., et al. (2016). Co-occurrence of integrase 1, antibiotic and heavy metal resistance genes in municipal wastewater treatment plants. Water Res. 94, 208–214. doi: 10.1016/j.watres.2016.02.049
Diarra M. S., Malouin F. (2014). Antibiotics in Canadian poultry productions and anticipated alternatives. Front. Microbiol. 5, 282. doi: 10.3389/fmicb.2014.00282
Ewers C., Bethe A., Semmler T., Guenther S., Wieler L. H. (2012). Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 18, 646–655. doi: 10.1111/j.1469-0691.2012.03850.x
Folster J. P., Pecic G., McCullough A., Rickert R., Whichard J. M. (2011). Characterization of blaCMY -Encoding Plasmids Among Salmonella Isolated in the United States in 2007. Foodborne Pathog. Dis. 8, 1289–1294. doi: 10.1089/fpd.2011.0944
FoodNet Canada (2017). FoodNet Canada 2015 Short Report. (Guelph, Ontario, Canada: Public Health Agency of Canada). Available at: http://www.phac-aspc.gc.ca/foodnetcanada/report-rapport-2014-eng.php.
Ghodousi A., Bonura C., Di Noto A. M., Mammina C. (2015). Extended-Spectrum ß -Lactamase, AmpC-Producing, and Fluoroquinolone-Resistant Escherichia coli in Retail Broiler Chicken Meat, Italy. Foodborne Pathog. Dis. 12, 619–625. doi: 10.1089/fpd.2015.1936
Gieraltowski L., Higa J., Peralta V., Green A., Schwensohn C., Rosen H., et al. (2016). National outbreak of multidrug resistant Salmonella Heidelberg infections linked to a single poultry company. PloS One 11, 1–13. doi: 10.1371/journal.pone.0162369
Government of Canada (2015). National Enteric Surveillance Program (NESP) 2013 Annual Report Executive Summary (Ontario, Canada: Public Health Agency of Canada. Guelph). Available at: https://www.nml-lnm.gc.ca/NESP-PNSME/surveillance-2013-eng.html.
Guo Y. F., Zhang W. H., Ren S. Q., Yang L., Lu D. H., Zeng Z. L., et al. (2014). IncA/C plasmid-mediated spread of CMY-2 in multidrug-resistant Escherichia coli from food animals in China. PloS One 9, 1–7. doi: 10.1371/journal.pone.0096738
Hanson N. D. (2003). AmpC ß-lactamases: what do we need to know for the future? J. Antimicrob. Chemother. 52, 2–4. doi: 10.1093/jac/dkg284
Hiki M., Usui M., Kojima A., Ozawa M., Ishii Y., Asai T. (2013). Diversity of Plasmid Replicons Encoding the blaCMY-2 Gene in Broad-Spectrum Cephalosporin-Resistant Escherichia coli from Livestock Animals in Japan. Foodborne Pathog. Dis. 10, 243–249. doi: 10.1089/fpd.2012.1306
Hopkins K. L., Liebana E., Villa L., Batchelor M., Threlfall E. J., Carattoli A. (2006). Replicon typing of plasmids carrying CTX-M or CMY β-lactamases circulating among Salmonella and Escherichia coli isolates. Antimicrob. Agents Chemother. 50, 3203–3206. doi: 10.1128/AAC.00149-06
Hutton T. A., Innes G. K., Harel J., Garneau P., Cucchiara A., Schifferli D. M., et al. (2018). Phylogroup and virulence gene association with clinical characteristics of Escherichia coli urinary tract infections from dogs and cats. J. Vet. Diagn. Invest. 30, 64–70. doi: 10.1177/1040638717729395
Knothe H., Shah P., Krcmery V., Antal M., Mitsuhashi S. (1983). Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11, 315–317. doi: 10.1007/BF01641355
Lei T., Zhang J., Jiang F., He M., Zeng H., Chen M. (2019). First detection of the plasmid-mediated colistin resistance gene mcr-1 in virulent Vibrio parahaemolyticus. Int. J. Food Microbiol. 308:108290. doi: 10.1016/j.ijfoodmicro.2019.108290
Leimbach A., Hacker J., Dobrindt U. (2013). “E. coli as an All-Rounder: The Thin Line Between Commensalism and Pathogenicity,” in Between Pathogenicity and Commensalism. Eds. Dobrindt U., Hacker J. H., Svanborg C. (Berlin, Heidelberg: Springer), 3–32. Current Topics in Microbiology and Immunology, vol 358.
Leverstein-van Hall M. A., Paauw A., Box A. T. A., Blok H. E. M., Verhoef J., Fluit A. C. (2002). Presence of integron-associated resistance in the community is widespread and contributes to multidrug resistance in the hospital. J. Clin. Microbiol. 40, 3038–3040. doi: 10.1128/JCM.40.8.3038-3040.2002
Levesque R., Roy P. H., Letarte R., Pechère J. C. (1982). A plasmid-mediated cephalosporinase from Achromobacter species. J. Infect. Dis. 145, 753–761. doi: 10.1093/infdis/145.2.753
Marashi S. M. A., Esfahani B. N., Tavakoli A., Moghim S., Pourshafie M. R., Salehi M. (2012). Simultaneous detection of integrase and antibiotic resistance genes within SXT constin in Vibrio cholerae O1 El tor strains isolated from Iran using multiplex-PCR. Iranian J. Basic Med. Sci. 15, 885–889. doi: 10.22038/ijbms.2012.4874
Mata C., Miró E., Alvarado A., Garcillán-Barcia M. P., Toleman M., Walsh T. R., et al. (2012). Plasmid typing and genetic context of AmpC β-lactamases in enterobacteriaceae lacking inducible chromosomal ampC genes: Findings from a Spanish hospital 1999-2007. J. Antimicrob. Chemother. 67, 115–122. doi: 10.1093/jac/dkr412
Mcclelland M., Wilson R. K. (1998). Comparison of Sample Sequences of the Salmonella typhi Genome to the Sequence of the Complete Escherichia coli K-12 Genome. Infect. Immun. 66, 4305–4312. doi: 10.1128/IAI.66.9.4305-4312.1998
McClelland M., Florea L., Sanderson K., Clifton S. W., Parkhill J., Churcher C., et al. (2000). Comparison of the Escherichia coli K-12 genome with sampled genomes of a Klebsiella pneumoniae and three Salmonella enterica serovars, Typhimurium, Typhi and Paratyphi. Nucleic Acids Res. 28, 4974–4986. doi: 10.1093/nar/28.24.4974
Mohanty S., Gaind R., Ranjan R., Deb M. (2010). Use of the cefepime-clavulanate ESBL Etest for detection of extended-spectrum beta-lactamases in AmpC co-producing bacteria. J. Infect. Develop. Countries 4, 024–029. doi: 10.3855/jidc.493
Mulvey M. R., Bryce E., Boyd D. A., Ofner-agostini M., Land A. M., Simor A. E., et al. (2005). Molecular Characterization of Cefoxitin-Resistant Escherichia coli from Canadian Hospitals. Antimicrob. Agents Chemother. 49, 358–365. doi: 10.1128/AAC.49.1.358
Nomura M., Nakamura M. (1962). Reversibility of inhibition of nucleic acids and protein synthesis by colicin K. Biochem. Biophys. Res. Commun. 7, 306–309. doi: 10.1016/0006-291x(62)90196-1
Philippon A., Arlet G., Jacoby G. A. (2002). Plasmid-Determined AmpC-Type β-Lactamases. Antimicrob. Agents Chemother. 46, 1–11. doi: 10.1128/AAC.46.1.1
Porwollik S., Santiviago C. A., Cheng P., Florea L., Jackson S., Mcclelland M. (2005). Differences in Gene Content between Salmonella enterica Serovar Enteritidis Isolates and Comparison to Closely Related Serovars Gallinarum and Dublin. J. Bacteriol. 187, 6545–6555. doi: 10.1128/JB.187.18.6545
Prescott J. F. (2006). “History of Antimicrobial Usage in Agriculture: an Overview,” in Antimicrobial Resistance in Bacteria of Animal Origin. Ed. Aarestrup F. (Washington, DC: ASM Press), 19–28.
Public Health Agency of Canada (2015). Canadian Integrated Program for Antimicrobial (CIPARS) Annual Report 2015. Available at: http://publications.gc.ca/collections/collection_2017/aspc-phac/HP2-4-2015-eng.pdf.
Ryoo N. H., Kim E. C., Hong S. G., Park Y. J., Lee K., Bae I. K., et al. (2005). Dissemination of SHV-12 and CTX-M-type extended-spectrum β-lactamases among clinical isolates of Escherichia coli and Klebsiella pneumoniae and emergence of GES-3 in Korea. J. Antimicrob. Chemother. 56, 698–702. doi: 10.1093/jac/dki324
Sanders C. (1987). Chromosomal Cephalosporinases Responsible for Multiple Resistance To Newer β-Lactam Antibiotics. Annu. Rev. Microbiol. 41, 573–593. doi: 10.1146/annurev.mi.41.100187.003041
Sanyal C., Husereau D. R., Beahm N. P., Smyth D., Tsuyuki R. T. (2019). Cost-effectiveness and budget impact of the management of uncomplicated urinary tract infection by community pharmacists. BMC Health Serv. Res. 19, 1–13. doi: 10.1186/s12913-019-4303-y
Suzuki S. (1994). Pathogenicity of Salmonella enteritidis in poultry. Int. J. Food Microbiol. 21, 89–105. doi: 10.1016/0168-1605(94)90203-8
Tendencia E. A. (2004). “Chapter 2. Disk diffusion method,” in Laboratory manual of standardized methods for antimicrobial sensitivity tests for bacteria isolated from aquatic animals and environment (Aquaculture Department, Southeast Asian Fisheries Development Center), 13–29. Available at: http://repository.seafdec.org.ph/handle/10862/1635.
Trampel D. W., Holder T. G., Gast R. K. (2014). Integrated farm management to prevent Salmonella Enteritidis contamination of eggs. J. Appl. Poultry Res. J. Appl. Poultry Res. 23, 353–365. doi: 10.3382/japr.2014-00944
White P. A., McIver C. J., Deng Y. M., Rawlinson W. D. (2000). Characterisation of two new gene cassettes, aadA5 and dfrA17. FEMS Microbiol. Lett. 182, 265–269. doi: 10.1016/S0378-1097(99)00600-X
White P. A., Iver C. J. M. C., Rawlinson W. D. (2001). Integrons and Gene Cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45, 2658–2661. doi: 10.1128/AAC.45.9.2658
Winokur P. L., Vonstein D. L., Hoffman L. J., Uhlenhopp E. K., Doern G. V. (2001). Evidence for Transfer of CMY-2 AmpC β-Lactamase Plasmids between Escherichia coli and Salmonella Isolates from Food Animals and Humans. Antimicrob. Agents Chemother. 45, 2716–2722. doi: 10.1128/AAC.45.10.2716
Keywords: Escherichia coli, Salmonella, blaCMY-2, blaTEM, antimicrobial resistance genes
Citation: Tran T, Checkley S, Caffrey N, Mainali C, Gow S, Agunos A and Liljebjelke K (2021) Genetic Characterization of AmpC and Extended-Spectrum Beta-Lactamase Phenotypes in Escherichia coli and Salmonella From Alberta Broiler Chickens. Front. Cell. Infect. Microbiol. 11:622195. doi: 10.3389/fcimb.2021.622195
Received: 27 October 2020; Accepted: 18 February 2021;
Published: 12 March 2021.
Edited by:
Fangkun Wang, Shandong Agricultural University, ChinaReviewed by:
Mohamed Salah Abbassi, Tunis El Manar University, TunisiaXiaoping Liao, South China Agricultural University, China
Toshiyuki Murase, Tottori University, Japan
Copyright © 2021 Tran, Checkley, Caffrey, Mainali, Gow, Agunos and Liljebjelke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karen Liljebjelke, a2xpbGplYmpAdWNhbGdhcnkuY2E=; Tam Tran, dHRyYW4zMzJAdXdvLmNh
 Tam Tran
Tam Tran Sylvia Checkley
Sylvia Checkley Niamh Caffrey
Niamh Caffrey Chunu Mainali2
Chunu Mainali2 Sheryl Gow
Sheryl Gow Agnes Agunos
Agnes Agunos Karen Liljebjelke
Karen Liljebjelke