- 1School of Public Health, University of South China, Hengyang, China
- 2Shenzhen Center for Disease Control and Prevention, Shenzhen, China
- 3School of Public Health (Shenzhen), Sun Yat-sen University, Guangzhou, China
- 4Shenzhen Key Laboratory of Unknown Pathogen Identification, BGI-Shenzhen, Shenzhen, China
- 5School of Life Sciences, Xiamen University, Xiamen, China
The serotyping of Vibrio parahaemolyticus, which is crucial to the surveillance and detection of outbreaks of vibriosis infection, has been widely used in many countries. In this study, we developed a molecular assay, named multiplex ligation reaction based on probe melting curve analysis (MLMA), for simultaneous identification of V. parahaemolyticus 57 K-serogroups. Based on the previous genomes of 418 strains including 39 K-serogroups and the 18 K-serogroups sequences from public databases, we obtained 57 K-serogroups specific gene sequences for designing primers and probes. The developed MLMA assay for identifying the V. parahaemolyticus 57 K-serogroups showed high reproducibility, with the intra- and inter-assay standard deviations and coefficients of variation of no more than 1°C and 1%, respectively. The limit of detection for all gene targets ranged from 0.1 to 1.0 ng/µl. We validated the MLMA assay with a double-blind test identifying 595 V. parahaemolyticus isolates using conventional serotyping methods for comparison. The results showed the kappa value between the MLMA assay and the traditional serological method was 0.936 and that there was a 96.97% consistency rate with conventional serotyping methods for all detected isolates. Additionally, five rare K-serogroups were identified using the MLMA assay, as well as 18 strains that could not be identified using the traditional serotyping method. Thus, the MLMA assay provides a rapid, robust, and promising tool for the molecular serotyping of V. parahaemolyticus K-serogroups and has the potential application to the detection of outbreaks and surveillance of V. parahaemolyticus infection.
Introduction
Vibrio parahaemolyticus is a Gram-negative, motile bacterium commonly found in marine and estuarine environments worldwide (Letchumanan et al., 2014; Wang et al., 2015). Investigation of V. parahaemolyticus infection has revealed that raw or undercooked seafood contaminated by V. parahaemolyticus strains is commonly responsible for acute gastroenteritis, and that in rare cases such as wound infection, ear infection or septicemia (Letchumanan et al., 2014). Between 2011 and 2016, 790 outbreaks of foodborne diseases were reported in more than 703 hospitals of China, causing 13,013 individuals to become ill. Of these cases, V. parahaemolyticus was the most common pathogen being responsible for foodborne disease outbreaks that caused most cases reported (42.3%). Therefore, V. parahaemolyticus is recognized as the leading cause of foodborne disease in China (Liu et al., 2018). Recent studies have found that pandemic V. parahaemolyticus strains are responsible for approximately 50%–80% of all V. parahaemolyticus cases in many countries, including India, Peru, Mexico and Chile (Han et al., 2019). In addition, a newly emerging shrimp disease, acute hepatopancreatic necrosis disease (AHPND), is known to be caused by strains of V. parahaemolyticus that contain a unique virulence plasmid. It has caused significant economic loss in global shrimp industry (Lai et al., 2015). Recent genomic analysis of 233 V. parahaemolyticus strains isolated from diseased shrimp, humans and environmental samples suggested that the spread of AHPND is mainly via horizontal transfer of the AHPND-associated plasmid, which highlighted a significant transmission route of V. parahaemolyticus between aquaculture and human communities (Fu et al., 2020). Thus, V. parahaemolyticus has become a large public health issue on a global scale, especially in coastal regions (Han et al., 2016).
V. parahaemolyticus is a multiserotype bacterium found in foodborne diseases or external environments that includes at least 13 O-serogroups and 71 K-serotypes (Iguchi et al., 1995; Bhuiyan et al., 2002). In 1996, the epidemiology of V. parahaemolyticus displayed a radical change with a new serotype, O3:K6, that appeared abruptly (Nair et al., 2007; Baker-Austin et al., 2018). The pandemic O3:K6 strains have specific genetic markers such as positivity for the thermostable direct hemolysin (tdh) gene, negativity for the TDH-related hemolysin (trh) gene and positivity for a toxRS/new gene (Matsumoto et al., 2000; Han et al., 2016). Since then, the serotype O3:K6 and its serovariants have spread rapidly throughout the world (Han et al., 2016). Additionally, a new emerging serotype with enhanced acid resistance, O4:KUT-recAin, has become the second most common serotype following O3:K6. In China, O4:KUT-recAin has become widespread during a short period of time (Chen et al., 2020). Accordingly, serotyping has been widely applied to provide a vital information for epidemiological investigation.
Serotyping is an essential method for identification of pathogens, foodborne disease outbreak investigations and source tracing that has been widely used to obtain an accurate biological phenotype of many pathogens. The serotyping of V. parahaemolyticus is generally accomplished using traditional methods based on agglutination with certain antisera. However, serological methods are limited in that they are time-consuming, expensive and labor-intensive. As understanding of antigens and their gene clusters has improved, some molecular approaches have been developed as alternatives to the traditional method. For example, polymerase chain reaction (PCR) assays are often used to identify and distinguish specific serotype. In 2012, an O-serogroup-specific PCR based assay was first used for the identification and detection of V. parahaemolyticus pathogens in clinical and environmental samples (Chen et al., 2012). Later, the use of the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) enabled detection of a specific serotype, O4:K8, but failed to identify other serotypes (Li et al., 2018). Recently, a microsphere-based suspension array was established for the detection and identification of 55 V. parahaemolyticus K-serogroups based on specific genes of the capsular polysaccharide (CPS) gene cluster (Pang et al., 2019). However, these methods are limited in that they require labor-intensive post-PCR manipulations, expensive instruments, or involve the preparation of microsphere beads, which are complicated and time-consuming.
Multiplex ligation reaction based on probe melting curve analysis (MLMA) is a high-throughput, high-accuracy, low-cost method that has been successfully used for the simultaneous identification of 10 bacterial pathogens and applied to the simultaneous identification of 30 common Salmonella serotypes (Jiang et al., 2017; Zuo et al., 2019). In MLMA, a fluorescence signal and melting temperature (Tm) are combined and used as a virtual 2D label that enables homogenous detection of one order of magnitude more targets (Liao et al., 2013). Previously, we presented a molecular assay based on the principles of MLMA to simultaneously identify the 11 clinically most common V. parahaemolyticus serotypes (Li et al., 2019). In this study, to further increase the number of target K-serogroups in multiplexity, we established a three-tube system to serotype V. parahaemolyticus 57 K-serogroups that could be applied as an alternative for the surveillance and control of vibriosis infections.
Materials and Methods
Bacterial Strains
There were 3,826 strains stored that were isolated from the stool specimens of infectious diarrheal patients and food samples over 15 years (2003–2018) in Shenzhen Center for Disease Control and Prevention, 3,590 strains were K-serogroups typeable and 236 strains were K-serogroups untypeable (KUT). Among 3,826 strains, 418 strains were previously sequenced (Li et al., 2019; Bian et al., 2021). Among 418 strains, 338 strains were selected to develop the MLMA assay (Table 1), which included 328 strains representing 39 K-serogroups and 10 strains of untyped K-serogroups for detection of cross-reactions. In addition, 18 rare K-serogroups were incorporated into the assay using Escherichia coli TOP10 strains (n=18, Table S1) containing serogroup-specific genes cloned into a pUC57 vector (Sangon Biotech Co. Ltd., Shanghai, China). Among 3,826 strains, 595 strains which included 10% of 3,590 K-serogroups typeable strains (n=359, Table S2) and all 236 untypeable strains (Table S3) were selected to validate the MLMA assay.

Table 1 Reference isolates (n = 338) representing V. parahaemolyticus K-serogroups over a 12-year period (2006–2018) for the development of multiplex ligation reaction based on probe melting curve analysis (MLMA) assay.
Bacterial Culture, DNA Extraction, and Conventional Serotyping
All V. parahaemolyticus strains were revived and cultured in Vibrio chromogenic agar (Guangdong Huankai Microbial Science and Technology, Guangzhou, China) at 37°C for 12 h to isolate single colonies. Individual colonies were then selected and cultured in 3 ml of alkaline peptone water (Guangdong Huankai Microbial Science and Technology), after which they were incubated at 37°C for 16 h while shaking at 200 rpm. Escherichia coli TOP10 strains were cultured in normal LB medium under the same conditions. Genomic DNA templates were extracted using the boiled lysates method, in which 1 ml of each culture was boiled at 100°C for 8 min. After boiling, the suspension was centrifuged for 10 min at 12,000 rpm. The supernatant was then used as the DNA template. Plasmid DNA templates were extracted using a SanPrep Plasmid MiniPrep Kit (Sangon Biotech Co. Ltd.). K-serogroups of all V. parahaemolyticus strains were identified using commercial antisera based on agglutination tests (Denka Seiken, Tokyo, Japan) according to the manufacturer’s protocol and the Chinese National Food Safety Standards: Food Microbiological Examination Vibrio parahaemolyticus Testing, GB 4789.7-2013.
Primer and Probe Design
In this study, the primers were designed as previously described (Li et al., 2019). Primers designed for development of the MLMA assay included two parts (Table 2). (1) The whole genome sequences of 418 V. parahaemolyticus strains were sequenced in the previous study (Li et al., 2019; Bian et al., 2021) and 39 pairs of primers of different K-serogroups were designed based on specific serogroup antigen genes sequences of the 418 genomes which the bioproject numbers could be obtained from the GenBank database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA677930). Among 39 pairs of primers, the eight pairs of primers were previously reported (Li et al., 2019). (2) Additionally, 18 pairs of primers targeting 18 rare K-serogroup gene sequences were designed based on the wzy or wzx gene as previously described (Pang et al., 2019) using sequence information obtained from the GenBank database (https://www.ncbi.nlm.nih.gov/). The left hybridization-ligation oligonucleotide probe (L) was purified using polyacrylamide gel electrophoresis, while the right hybridization-ligation oligonucleotide probe (R) was labeled at the 5′ end with the phosphate group (5′P) and purified using high-performance liquid chromatography. Universal primer and fluorophore-labeled probes with the reporter fluorophores carboxy-X-rhodamine (ROX), carboxyfluorescein (FAM), or indodicarbocyanine-5 (Cy5) at the 5′ end and with Black Hole Quencher at the 3′ end were obtained as described in our previous study (Jiang et al., 2017) (Table S4). All primers and probes were synthesized by Sangon Biotech Co. Ltd.
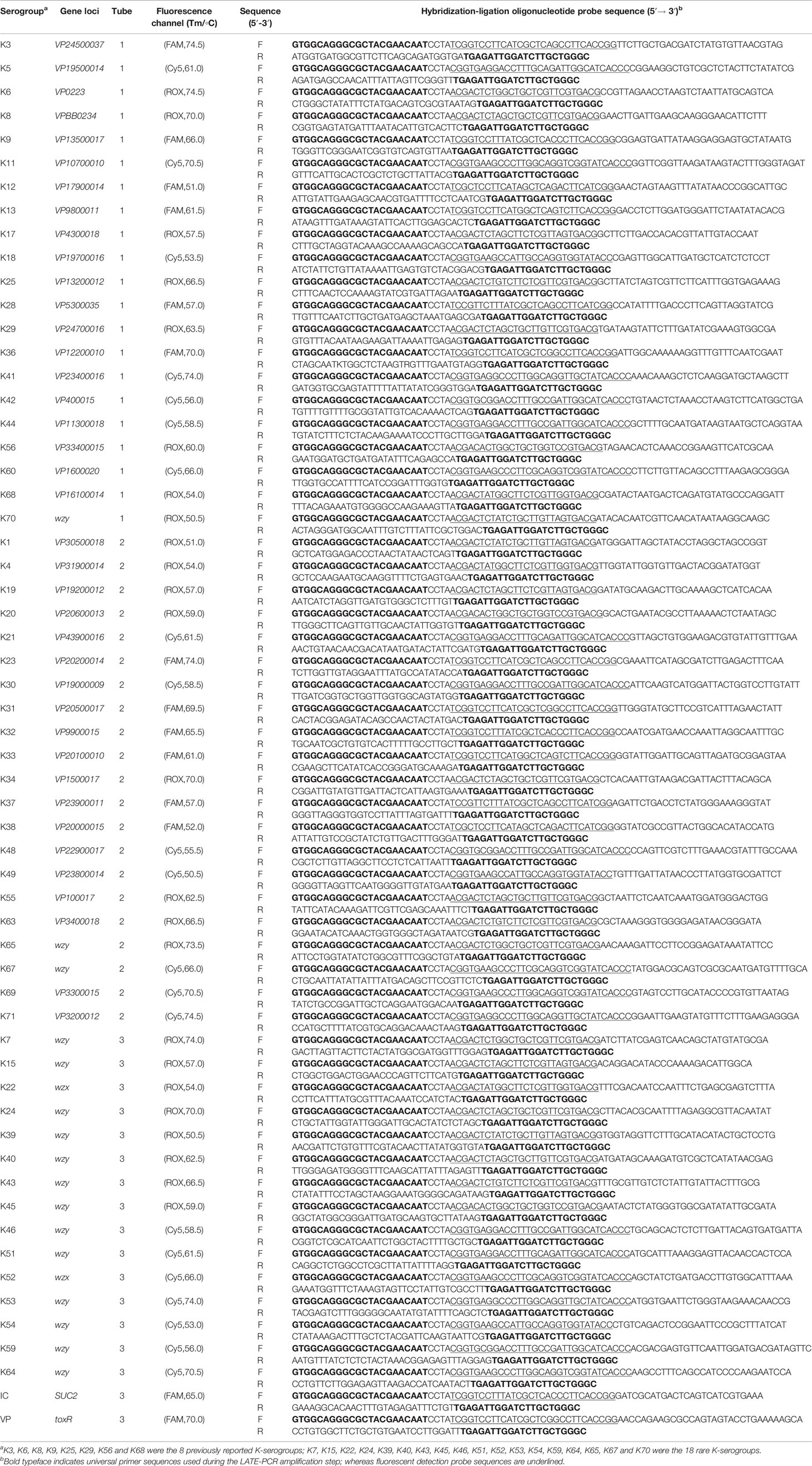
Table 2 Multiplex ligation reaction based on probe melting curve analysis (MLMA) assay for the identification of V. parahaemolyticus K- serogroups in a three-tube system.
Design of the MLMA Assay
The two key steps involved in the MLMA assay are (1) a hybridization-ligation process and (2) a PCR amplification and melt curve analysis (Li et al., 2019). In step 1, the reaction is conducted in a 10 µl reaction mixture containing 1.5 µl of ligation probe mix (0.63–10.0 nM of each hybridization oligonucleotide probe) (Table 2), 1 U Taq DNA ligase and 1 µl DNA ligase buffer (New England Biolabs, Beijing, China), 1.5 µl sterilized water, and 5 µl DNA template. The reaction was performed in a T3 Thermocycler (Biometra, Germany) under the following parameters: denaturation of the initial reaction, which only contained ligation probe mix and DNA template, at 95°C for 5 min, followed by a reduction to 75°C to pause the reaction, after which 3.5 µl containing the remainder of the reaction mixture were added and the samples were incubated at 60°C for 60 min, followed by 95°C for 5 min. In step 2, the reaction was conducted in a 50 µl reaction mixture containing 1×PCR buffer, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphate, 1 U Taq polymerase, 0.015 µM limiting primer, 0.3 µM excess primer, 0.12 µM fluorogenic probes, and 5 µl of ligation product from step 1. The reaction was performed in a BioRad CFX96 real-time PCR system (BioRad Inc., Hercules, CA, USA) under the following conditions: host start at 95°C for 3 min, then 38 cycles of 95°C for 10 s, 57°C for 20 s, and 72°C for 20 s, followed by 95°C for 1 min, 40°C for 2 min, and then an increase to 85°C in 1°C steps with 5 s between each step. All fluorescent signal intensity was captured by ROX, FAM, and Cy5 channels. The melting temperature (Tm) values of melt curve analysis were obtained automatically using CFX Manager 3.0 software.
Analytical Performance of the MLMA Assay
To determine the limit of detection (LOD) of the assay, a series of 10-fold dilutions in triplicates of purified DNA from 10.0 ng/µl to 0.01 ng/µl were analyzed. To evaluate the intra-assay and inter-assay reproducibility, two sets of 10-fold dilutions in triplicate from 10.0 ng/µl to 1.0 ng/µl were analyzed and the standard deviations and coefficient of variation values were calculated. Each concentration of the assay was analyzed in triplicate.
Evaluation of the MLMA Assay Using a Double-Blind Test
The 595 strains were selected to evaluate the specificity and sensitivity of the MLMA assay. The K-serogroups of the 595 isolates were distinguished based on the MLMA assay method compared with the agglutination tests (Denka Seiken) using commercial antisera according to the manufacturer’s instructions with a double-blind test. The genomic DNA was extracted from the 595 isolates using the boiled lysates protocol. Procedures were performed according to the protocols described above. Evaluation of consistency between the MLMA assay and traditional serological methods was performed using Kappa analysis.
Statistical Analysis
The SPSS 23.0 statistical software package was used for statistical analysis. The positive detection rate of the MLMA assay and traditional serological method was compared with the paired χ2 test, and a P<0.01 was considered to indicate statistical significance.
Ethics
V. parahaemolyticus isolates from the Shenzhen Center for Disease Control and Prevention were de-identified and anonymized to protect patient privacy and confidentiality. Therefore, ethical clearance was not required.
Results
Identification of 57 V. parahaemolyticus K-Serogroups
A three-tube system was set up with three fluorescence channels (ROX, FAM, and Cy5) in each tube. In the first tube, the ROX channel detected the K-serogroups K70, K68, K17, K56, K29, K25, K8, and K6, the Cy5 channel detected the K-serogroups K18, K42, K44, K5, K60, K11, and K41 and the FAM channel detected the K-serogroups K12, K28, K13, K9, K36, and K3. In the second tube, the ROX channel detected the K-serogroups K1, K4, K19, K20, K55, K63, K34, and K65, the Cy5 channel detected K49, K48, K30, K21, K67, K69, and K71 and the FAM channel detected the K-serogroups K38, K37, K33, K32, K31, and K23. In the third tube, the ROX channel detected the K-serogroups K39, K22, K15, K45, K40, K43, K24, and K7, the Cy5 channel detected the K-serogroups K54, K59, K46, K51, K52, K64, and K53 and the FAM channel detected the SUC2 gene as an internal control (IC) and the toxR gene as a confirmation of V. parahaemolyticus. The combined use of fluorescence channels and designed Tm values for targeting specific gene loci of K-serogroups in each tube are listed in Table 3.
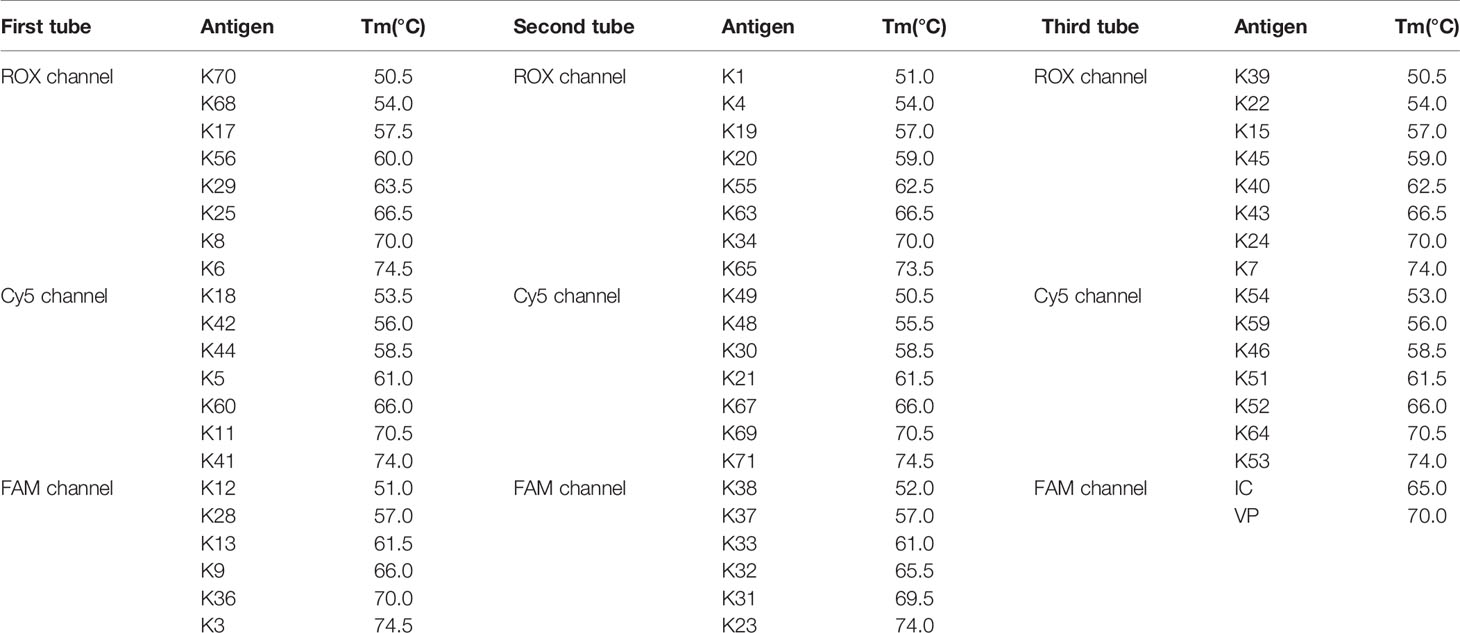
Table 3 Identification matrix of gene targets in a three-tube multiplex ligation reaction based on probe melting curve analysis (MLMA) system.
Performance Characteristics of the MLMA Assay
With the use of 59 pairs of ligation oligonucleotides in a three-tube system, the MLMA assay was able to detect all target genes of 57 K-serogroups and one IC and the toxR gene, which yielded expected Tm values that represented distinguished K-serogroups (Figure 1). The specificity study showed no cross-reactivity among the 57 K-serogroups. The limit of detection (LOD) of the assay for all target genes of K-serogroups ranged from 0.1 to 1.0 ng/µl at the DNA level. The intra-assay and inter-assay reproducibility study demonstrated that the measurement of Tm values has highly reproducible traits. Specifically, the largest SD value of the mean Tm was no more than 1°C and the CV value ranged from 0 to 1% (Table 4).

Figure 1 Probe melting curve analysis for the identification of V. parahaemolyticus. 57 K-serogroups in the multiplex ligation reaction based on probe melting curve analysis (MLMA) assay. Color-coded melting curves represent the different antigen genes in each fluorophore channel (ROX, FAM, and Cy5) in a three-tube system. IC, SUC2 gene was used as a positive internal control (IC); NTC, negative control.

Table 4 Reproducibility of designated melting temperatures (Tm) and Limit of Identification values for each serogroup target loci in the assay.
Evaluation of the MLMA Assay
Of the 595 isolates, 377 were identified and classified to 29 K-serogroups, and the remaining 218 isolates were untypeable by using the MLMA assay. Among 377 isolates, the 18 could be serotyped using the MLMA assay, but could not be serotyped using the conventional serological method. Sanger sequencing of the 18 isolates demonstrated 100% concordance with the MLMA assay (Table S5). Evaluation using the McNemar test revealed that the difference was statistically significant (P=0.00), the positive detection rate of MLMA (63.36%) was higher than that of conventional serotyping (60.34%), and the MLMA assay sensitivity and specificity were 100% and 92.37%, respectively. Moreover, the MLMA assay showed 96.97% consistency with the conventional serological method for all isolates. The kappa value between the MLMA assay and the traditional serological method was 0.936 (Table 5). The results of identification using the MLMA assay and the traditional serological method are listed in Table S6. In addition, five rare K-serogroups, K24, K52, K54, K67, and K70, were identified using the established MLMA assay (Table S7).
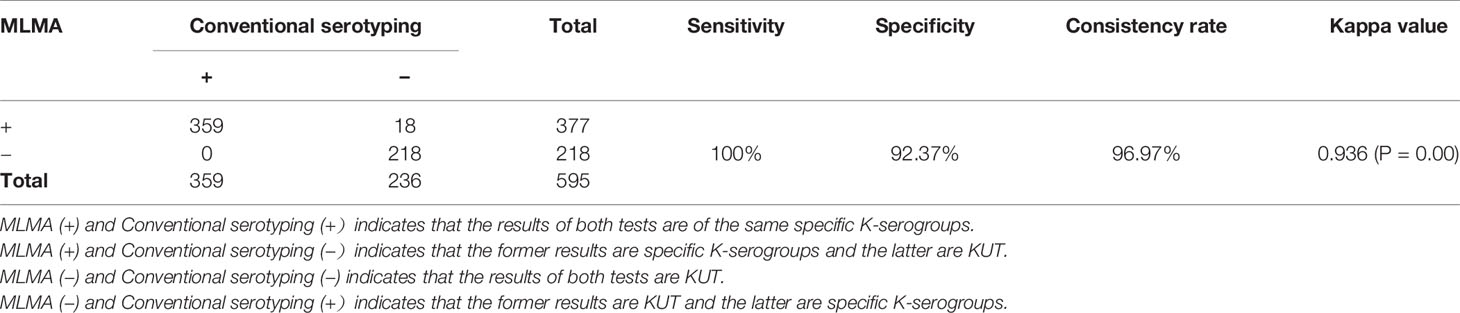
Table 5 The results of serotype identification by multiplex ligation reaction based on probe melting curve analysis (MLMA) assay and conventional serotyping tests among V. parahaemolyticus isolates (n = 595) from the double-blind study.
Discussion
V. parahaemolyticus is the leading causal agent of human acute gastroenteritis and has become a global public health issue (Letchumanan et al., 2014; Han et al., 2019). V. parahaemolyticus serotyping is considered to be the initial step for timely detection and source tracking of vibriosis outbreaks and can provide historical and comparable data for the surveillance of V. parahaemolyticus infections on a global scale (Han et al., 2016; Baker-Austin et al., 2018). Thus, the development of rapid and robust serotyping methods can facilitate national and international surveillance of V. parahaemolyticus infections.
In this study, we developed a molecular assay for the simultaneous identification of 57 V. parahaemolyticus K-serogroups based on the principles of MLMA. The assay can detect all gene targets with expected Tm values and no cross-reaction was observed. The detection limit of the assay ranged from 0.1 to 1.0 ng/µl. Moreover, the intra-assay and inter-assay reproducibility analyses showed that the assay had highly reproducible traits, with the largest SD value for the mean Tm being no more than 1°C and the CVs being less than 1%. Overall, the MLMA assay demonstrated a 96.97% consistency rate with the conventional serological method of all isolates and a kappa value of 0.936, which indicated the consistency of the two approaches is excellent.
With the growing number of genomic sequences and serogroup-specific gene sequences available in public databases, various molecular serotyping methods have been developed as alternatives for serotyping, such as common and real-time PCR, multiplex PCR-based microarrays, Luminex-based serotyping assays and sequencing-based approaches (Liu et al., 2015; Cabral-Castro et al., 2016; Afroj et al., 2017; Pang et al., 2019). Although crucial progress regarding antigen synthesis gene clusters and serogroup-specific genes for Salmonella and Escherichia coli, there has been little progress on the capsular polysaccharide gene clusters or genetic targets of V. parahaemolyticus K-antigens to date. Thus, the development of a molecular method for V. parahaemolyticus K-serogroups is highly dependent on the investigation of the genetic characteristics of V. parahaemolyticus K-antigen. Early studies of K-antigen genetic determinants in V. parahaemolyticus produced controversial results (Guvener and Mccarter, 2003 Okura et al., 2008). Later, in 2010, the detailed genetic determinants of K-antigen synthesis were confirmed to be between gmhD and rjg using an O3:K6 isolate based on the construction of gene deletions (Chen et al., 2010). We used the whole genome sequence of 418 V. parahaemolyticus strains to design the target genes primers and probes and then analyzed the genetic structure and evolutionary relationship of their 39 K-serogroups to identify the serogroup-specific genes of the capsular polysaccharide gene clusters (Bian et al., 2021). Additionally, another capsular polysaccharide gene clusters of 18 rare K-serogroups that we lack these data were analyzed (Pang et al., 2019). Thus, 57 K-serogroups specific sequences were selected for the development of V. parahaemolyticus serotyping assay.
Currently, only three assays have the ability to detect more than two V. parahaemolyticus K-serogroups, the first is a microsphere-based suspension array, the second is the sequence-based serotyping of V. parahaemolyticus 55 K-serogroups, and the third is the MLMA assay we presented for the simultaneous identification of nine K-serogroups (Li et al., 2019; Pang et al., 2019). However, the microsphere-based suspension array and sequence-based serotyping method require expensive equipment with related software and rely on bioinformatics analysis. In this study, we expanded the high throughput targets of our previously developed MLMA assay from nine K-serogroups to 57 K-serogroups and identified five rare K-serogroups using the method. To date, we have been able to detect 12 O-serogroups (Li et al., 2019) and 57 K-serogroups in four tubes using the established MLMA assay.
Upon the evaluation of the assay using 595 isolates over 15 years (2003–2018) from the Shenzhen Center for Disease Control and Prevention, the MLMA assay accurately detected 377 typeable isolates that belonged to 29 K-serogroups and 218 untypeable isolates. In addition, there were inconsistent results for 18 isolates generated by the MLMA assay and the serological method. The results of Sanger sequencing of the PCR amplicons of these isolates were consistent with the results of the MLMA assay, which indicates the MLMA assay has higher sensitivity and accuracy than the serological method. Among 377 typeable isolates, five rare K-serogroups were detected (2008–2017) by the MLMA assay, which suggested that V. parahaemolyticus with extremely rare K-serogroups emerged in Shenzhen as early as 2008, and that routine monitoring should be conducted. The current assay showed similar superior performances of the assay as previously reported. However, the entire MLMA assay could be completed within 3.5–4.0 h, which saves 12–18 h when compared with traditional serotyping (Li et al., 2019). Additionally, a simple boiled lysates protocol is used for the preparation of DNA templates, which also increases the flexibility of the MLMA assay. As demonstrated by the analytical studies, the MLMA assay could accurately and reproducibly detect 57 V. parahaemolyticus K-serogroups. Hence, the targets of the assay covered approximately 81% V. parahaemolyticus K-serogroups that have been identified worldwide, demonstrating that multiplex assays facilitate timely and effective detection.
However, this assay was unable to differentiate two closely related K-serogroup pairs, K56 and K57, K47, and K48, which could be attributed to their capsular polysaccharide gene clusters showing almost identical homology to the corresponding regions (Pang et al., 2019). Currently, genetic markers for the accurate identification of these two pairs are unavailable and some strains were still untypeable. Therefore, further research is necessary to enable differentiation of these serogroups at the molecular level. Furthermore, with the development of whole-genome sequencing, the increasing demand for the sequence-based serotyping method gives us an important consideration to combine the MLMA assay with the genotyping for more effective foodborne disease outbreak investigation, source tracing, and the surveillance of V. parahaemolyticus infection.
In a word, a novel molecular assay was developed for simultaneous detection of 57 K-serogroups of V. parahaemolyticus and five rare K-serogroups were found. This assay can provide a rapid, accurate, and highly sensitive identification of V. parahaemolyticus K-serogroups and is valuable for screening large samples during vibriosis outbreaks and surveillance.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material (Table S8 and Table S9).
Author Contributions
LLu and ML conceived and designed the experiments. LLu and ML performed the experiments and contributed to analysis. LLu, ML, and QH wrote the paper. YLi, MJ, and XS guided the selection of experimental strains. YQ and RC checked the information of strains. YJ, LZ, and LW guided the experimental operation. YL and QL provided experimental technical support. SB and LLi analyzed the gene sequence. LLu and ML contributed equally to this article. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the China National Science and Technology Major Projects Foundation (No. 2017ZX10303406), National Natural Science Foundation of China (No. 81773436), Sanming Project of Medicine in Shenzhen (No. SZSM201811071), Shenzhen Key Medical Discipline Construction Fund (SZXK064), and the Shenzhen Health and Family Planning Commission (SZGW2017004).
Conflict of Interest
SB and LLi was employed by BGI-Shenzhen.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.594808/full#supplementary-material
References
Afroj S., Aldahami K., Reddy G., Guard J., Adesiyun A., Samuel T., et al. (2017). Simultaneous detection of multiple Salmonella serovars from milk and chicken meat by real-time PCR using unique genomic target regions. J. Food Prot. 80, 1944–1957. doi: 10.4315/0362-028X.JFP-17-133
Baker-Austin C., Oliver J. D., Alam M., Ali A., Waldor M. K., Qadri F., et al. (2018). Vibrio spp. infections. Nat. Rev. Dis. Primers 4, 8. doi: 10.1038/s41572-018-0005-8
Bhuiyan N. A., Ansaruzzaman M., Kamruzzaman M., Alam K., Chowdhury N. R., Nishibuchi M., et al. (2002). Prevalence of the Pandemic Genotype of Vibrio parahaemolyticus in Dhaka, Bangladesh, and Significance of Its Distribution across Different Serotypes. J. Clin. Microbiol. 40, 284–286. doi: 10.1128/JCM.40.1.284-286.2002
Bian S., Wenhong Z., Li Q., Li Y., Wong N.-K., Jiang M., et al. (2021). Genetic structure, function and evolution of capsule biosynthesis loci in Vibrio parahaemolyticus. Front. Microbiol. 11, 3218. doi: 10.1101/2020.02.25.964247
Cabral-Castro M. J., Peralta R. H. S., Cavalcanti M. G., Puccioni-Sohler M., Carvalho V. L., Da Costa Vasconcelos P. F., et al. (2016). A Luminex-based single DNA fragment amplification assay as a practical tool for detecting and serotyping dengue virus. J. Virol. Methods 236, 18–24. doi: 10.1016/j.jviromet.2016.07.003
Chen Y., Dai J., Morris J. G. Jr., Johnson J. A. (2010). Genetic analysis of the capsule polysaccharide (K antigen) and exopolysaccharide genes in pandemic Vibrio parahaemolyticus O3:K6. BMC Microbiol. 10, 274. doi: 10.1186/1471-2180-10-274
Chen M., Guo D., Wong H. C., Zhang X., Liu F., Chen H., et al. (2012). Development of O-serogroup specific PCR assay for detection and identification of Vibrio parahaemolyticus. Int. J. Food Microbiol. 159, 122–129. doi: 10.1016/j.ijfoodmicro.2012.08.012
Chen X., Li Y., Yao W., Wu T., Zhu Q., Zhang Y., et al. (2020). A new emerging serotype of Vibrio parahaemolyticus in China is rapidly becoming the main epidemic strain. Clin. Microbiol. Infect. 26, 644.e1–644.e7. doi: 10.2139/ssrn.3294765
Fu S., Wei D., Yang Q., Xie G., Pang B., Wang Y., et al. (2020). Horizontal Plasmid Transfer Promotes the Dissemination of Asian Acute Hepatopancreatic Necrosis Disease and Provides a Novel Mechanism for Genetic Exchange and Environmental Adaptation. mSystems 5, e00799–e00719. doi: 10.1128/mSystems.00799-19
Guvener Z. T., Mccarter L. L. (2003). Multiple Regulators Control Capsular Polysaccharide Production in Vibrio parahaemolyticus. J. Bacteriol. 185, 5431–5441. doi: 10.1128/JB.185.18.5431-5441.2003
Han C., Tang H., Ren C., Zhu X., Han D. (2016). Sero-Prevalence and Genetic Diversity of Pandemic V. parahaemolyticus Strains Occurring at a Global Scale. Front. Microbiol. 7, 567. doi: 10.3389/fmicb.2016.00567
Han D., Yu F., Chen X., Zhang R., Li J. (2019). Challenges in Vibrio parahaemolyticus infections caused by the pandemic clone. Future Microbiol. 14, 437–450. doi: 10.2217/fmb-2018-0308
Iguchi T., Kondo S., Hisatsune K. (1995). Vibrio parahaemolyticus O serotypes from 01 to 013 all produce R-type lipopolysaccharide:SDS-PAGE and compositional sugar analysis. FEMS Microbiol. Lett. 130, 287–292. doi: 10.1111/j.1574-6968.1995.tb07733.x
Jiang Y., He L., Wu P., Shi X., Jiang M., Li Y., et al. (2017). Simultaneous Identification of Ten Bacterial Pathogens Using the Multiplex Ligation Reaction Based on the Probe Melting Curve Analysis. Sci. Rep. 7, 5902. doi: 10.1038/s41598-017-06348-z
Lai H. C., Ng T. H., Ando M., Lee C. T., Chen I. T., Chuang J. C., et al. (2015). Pathogenesis of acute hepatopancreatic necrosis disease (AHPND) in shrimp. Fish Shellfish Immunol. 47, 1006–1014. doi: 10.1016/j.fsi.2015.11.008
Letchumanan V., Chan K. G., Lee L. H. (2014). Vibrio parahaemolyticus: a review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 5, 705. doi: 10.3389/fmicb.2014.00705
Li P., Xin W., Xia S., Luo Y., Chen Z., Jin D., et al. (2018). MALDI-TOF mass spectrometry-based serotyping of V. parahaemolyticus isolated from the Zhejiang province of China. BMC Microbiol. 18, 185. doi: 10.1186/s12866-018-1328-z
Li M., Jiang Y., Shi X., Li Y., Jiang M., Lin Y., et al. (2019). Simultaneous Identification of Clinically Common Vibrio parahaemolyticus Serotypes Using Probe Melting Curve Analysis. Front. Cell. Infect. Microbiol. 14, 385. doi: 10.3389/fcimb.2019.00385
Liao Y., Wang X., Sha C., Xia Z., Huang Q., Li Q. (2013). Combination of fluorescence color and melting temperature as a two-dimensional label for homogeneous multiplex PCR detection. Nucleic Acids Res. 41, e76. doi: 10.1093/nar/gkt004
Liu Y., Yan X., Debroy C., Fratamico P. M., Needleman D. S., Li R. W., et al. (2015). Escherichia coli O-antigen gene clusters of serogroups O62, O68, O131, O140, O142, and O163: DNA sequences and similarity between O62 and O68, and PCR-based serogrouping. Biosensors (Basel) 5, 51–68. doi: 10.3390/bios5010051
Liu J., Bai L., Li W., Han H., Fu P., Ma X., et al. (2018). Trends of foodborne diseases in China: lessons from laboratory-based surveillance since 2011. Front. Med. 12, 48–57. doi: 10.1007/s11684-017-0608-6
Matsumoto C., Okuda J., Ishibashi M., Bashi (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38, 578–585. doi: 10.1128/JCM.38.2.578-585.2000
Nair G. B., Ramamurthy T., Bhattacharya S. K., Dutta B., Takeda Y., Sack D. A. (2007). Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 20, 39–48. doi: 10.1128/CMR.00025-06
Okura M., Osawa R., Tokunaga A., Morita M., Arakawa E., Watanabe H. (2008). Genetic analyses of the putative O and K antigen gene clusters of pandemic Vibrio parahaemolyticus. Microbiol. Immunol. 52, 251–264. doi: 10.1111/j.1348-0421.2008.00027.x
Pang Y., Guo X., Tian X., Liu F., Wang L., Wu J., et al. (2019). Developing a novel molecular serotyping system based on capsular polysaccharide synthesis gene clusters of Vibrio parahaemolyticus. Int. J. Food Microbiol. 309, 108332. doi: 10.1016/j.ijfoodmicro.2019.108332
Wang R., Zhong Y., Gu X., Yuan J., Saeed A. F., Wang S. (2015). The pathogenesis, detection, and prevention of Vibrio parahaemolyticus. Front. Microbiol. 6, 144. doi: 10.3389/fmicb.2015.00144
Keywords: Vibrio parahaemolyticus, molecular identification, K-serogroups, vibriosis surveillance, multiplex ligation reaction based on probe melting curve analysis
Citation: Lu L, Li M, Li Y, Jiang M, Jiang Y, Shi X, Zuo L, Wang L, Bian S, Qiu Y, Cai R, Liao Y, Li Q, Li L and Hu Q (2021) A Novel Molecular Method for Simultaneous Identification of Vibrio parahaemolyticus 57 K-Serogroups Using Probe Melting Curve Analysis. Front. Cell. Infect. Microbiol. 11:594808. doi: 10.3389/fcimb.2021.594808
Received: 14 August 2020; Accepted: 18 January 2021;
Published: 26 February 2021.
Edited by:
Bin Liu, Nankai University, ChinaReviewed by:
Peng Li, Chinese Center for Disease Control and Prevention, ChinaSongzhe Fu, Dalian Ocean University, China
Copyright © 2021 Lu, Li, Li, Jiang, Jiang, Shi, Zuo, Wang, Bian, Qiu, Cai, Liao, Li, Li and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinghua Hu, aHVxaW5naHVhMDNAMTYzLmNvbQ==
 Linying Lu
Linying Lu Minxu Li
Minxu Li Yinghui Li2
Yinghui Li2 Min Jiang
Min Jiang Qingge Li
Qingge Li Liqiang Li
Liqiang Li Qinghua Hu
Qinghua Hu