Abstract
Urinary tract infections (UTIs) are frequent in humans, affecting the upper and lower urinary tract. Present diagnosis relies on the positive culture of uropathogenic bacteria from urine and clinical markers of inflammation of the urinary tract. The bladder is constantly challenged by adverse environmental stimuli which influence urinary tract physiology, contributing to a dysbiotic environment. Simultaneously, pathogens are primed by environmental stressors such as antibiotics, favoring recurrent UTIs (rUTIs), resulting in chronic illness. Due to different confounders for UTI onset, a greater understanding of the fundamental environmental mechanisms and microbial ecology of the human urinary tract is required. Such advancements could promote the tandem translation of bench and computational studies for precision treatments and clinical management of UTIs. Therefore, there is an urgent need to understand the ecological interactions of the human urogenital microbial communities which precede rUTIs. This review aims to outline the mechanistic aspects of rUTI ecology underlying dysbiosis between both the human microbiome and host physiology which predisposes humans to rUTIs. By assessing the applications of next generation and systems level methods, we also recommend novel approaches to elucidate the systemic consequences of rUTIs which requires an integrated approach for successful treatment. To this end, we will provide an outlook towards the so-called ‘uncomplicated environment of UTIs’, a holistic and systems view that applies ecological principles to define patient-specific UTIs. This perspective illustrates the need to withdraw from traditional reductionist perspectives in infection biology and instead, a move towards a systems-view revolving around patient-specific pathophysiology during UTIs.
Introduction
Recent advances in DNA and RNA sequencing contradicts prior assumptions that human urine is sterile; instead it harbors a unique microbiome with ecological interactions in health and disease (Wolfe et al., 2012; Hilt et al., 2014; Alteri and Mobley, 2015; Whiteside et al., 2015; de Vos et al., 2017; Aragón et al., 2018). As the physiological role of the bladder is to store nutrients and waste products in the form of urine, healthcare practitioners must consider the unique metabolism of bladder-associated microbial communities within this niche (Subashchandrabose et al., 2014; Alteri and Mobley et al., 2015; Conover et al., 2016; Horsley et al., 2018; Martín-Rodríguez et al., 2020). More so, with the emergence of pathogens that are resistant to first-line antibiotics, there is a pressing need to reinterpret uncomplicated UTIs, as an ever-changing and complicated pathology in humans (Mediavilla et al., 2016; Zilberberg et al., 2017). With the development of novel systems biology approaches and next generation methods, new viewpoints of UTIs and human-associated infections are being uncovered.
Most recently, the American Urological Association estimates that 150 million UTIs occur worldwide annually and cost up to $6 billion USD in healthcare costs (Flores-Mireles et al., 2015; AUA, 2016). This has led to the misapplication of antibiotics, likely resulting in long-term effects upon the interconnected gastrointestinal tract, vagina, and general urinary system (Kostakioti et al., 2012; Bartoletti et al., 2016; Nielsen et al., 2016; Gottschick et al., 2017; Thomas-White et al., 2018). Uropathogenic Escherichia coli (UPEC) primarily causes UTIs and is isolated from approximately 80% of patients (Flores-Mireles et al., 2015). Other pathogens such as Enterococcus faecalis, Klebsiella pneumoniae, or Proteus mirabilis can also be isolated from UTI patients (Abat et al., 2015; Thänert et al., 2019). UTIs begin at the urethra, colonize the bladder, and ascend to the kidneys through a multitude of mechanisms such as evading host protective factors or inhibiting host immunoglobulin A transport (Rice et al., 2005; Ashkar et al., 2008). The primary microbial strain causing the rUTI may originate from new colonizers deriving from various environmental reservoirs such as: a sexual partner, contaminated foods, or fecal/gut contamination of the urinary tract (Scholes et al., 2000; Nordstrom et al., 2013; Foxman, 2014; Gilbert et al., 2017; Thänert et al., 2019). While strains which initiate infection may cause a rUTI relapse, it is suggested that the initial infection caused by one UPEC strain primes the bladder for a new strain (or slightly similar) within several hours, which is diagnosed by two separate cultures over a period of six months (Andersen et al., 2012; Luo et al., 2012; Schreiber et al., 2017; Anger et al., 2019). Specifically, uncomplicated UTIs are acute infections of urinary tracts without anatomical or physiological defects that would make a patient more susceptible to initial urinary tract infections (Hooton, 2012). Women are significantly at a higher risk for contracting a UTI as compared to men. Previous reports suggest that one-third of all women under the age of 26 will experience a UTI and 50% of those women will experience a subsequent UTI episode (Foxman, 2002; Brumbaugh et al., 2013).: it is estimated that (This figure has potentially increased due to the emergence of multidrug resistance (MDR) UTIs (; Hooton et al., 2004; Nordstrom et al., 2013; Zilberberg et al., 2017).
As knowledge in modeling the fine-scale and dynamic changes in biological organisms increases, the context of personalized treatment strategies must also update with observations in molecular systems biology and microbiome sciences (Thiele et al., 2013; Thiele et al., 2020). Ideally, this includes withdrawing from reductionist approaches to diagnose UTIs and moving towards models that input multiple human data sources to create patient-specific models to characterize infection in a holistic view (Figure 1). Overall, our understanding of both host-microbiome interactions and rUTI pathophysiology elicits an update to the present concept of infection biology by redefining infectious diseases through a modern lens.
Figure 1

A healthy host and associated commensal microbiota enforces an environment that prevents colonization by invasive pathogens. However dysregulation of host physiology and native microbial communities is influenced by environmental factors. Through adverse community stressors such as DNA or protein impairment, poor diet, depleted microbial diversity, excessive metals, an inactive lifestyle, antibiotic usage, and adverse environment. The two-pronged attack of dysregulation of both microbial communities and host homeostasis leads to dysbiosis of beneficial bacteria, thus facilitating disease outcomes of individuals and formation of new microbial communities reflective of the pathology.
Microbiota of the Female Urogenital System
A culturable urinary tract microbial community exists within healthy individuals (Hilt et al., 2014). Specifically, the urobiome has been hypothesized to shift both microbial abundance and predicted metabolic pathways associated with various patient-specific urological morbidities or infections (Shoskes et al., 2016; Aragón et al., 2018). While less reports of the male urinary microbiome exist, both sexes share a similar core microbiome with genera from Lactobacillus, Streptococcus, and Corynebacterium; the latter is more common in men and is regularly associated with the skin microbiome (Fouts et al., 2012; Nelson et al., 2012; Lewis et al., 2013). Specifically, the urine of healthy females is characterized by the presence of Corynebacterium, Lactobacillus, Staphylococcus, and Streptococcus that tends to fluctuate abundance during periods of health and disease (Fouts et al., 2012; Aragón et al., 2018). Additionally when assessed by microbiome sequencing, it has been found that female urinary tract samples mainly consist of organisms from the phyla Actinobacteria (Actinomyces & Arthrobacter) and Bacteroidetes (Bacteroides), which are typically absent from their male counter parts (Lewis et al., 2013). More so, E. coli is readily cultured from 91% of healthy women and only 25% of men, highlighting a stark difference in E. coli that is cultured as a residential bacteria from the female urobiome (Ipe et al., 2013). Generally, these differences between males and females leading to unique microbiomes could simply be due to both anatomical and hormone differences between sexes (Whiteside et al., 2015).
When assessing cohorts by age, it becomes clear aging affects normal physiology and disease types (Irizar et al., 2018). Specifically a core female urine microbiota which parallels aging and age-related morbidities was identified and is manifested as asymptomatic bacteriuria (Lewis et al., 2013). In the case of patients with so-called asymptomatic bacteriuria, E. coli typically acts as symbionts (Godaly et al., 2016). Therefore, unnecessary and excessive application of antibiotics to “treat” asymptomatic bacteriuria in differing age groups leads to long-term consequences by depleting the resilient urinary system’s microbiota, thus driving an increased prevalence of multidrug resistance (MDR) pathogens within the urinary tract across patients (Cai et al., 2015; Ipe et al., 2016; Zilberberg et al., 2017). This suggests that the definition of UTI diagnosis: ‘the detection of a pathogen from the not be sterile during all periods of health. Table 1 summarizes the known microbial diversity of urine of a symptomatic patient’, must be modified in acknowledgement that human urine may is both the male and female urogenital system during periods of health, disease, and the asymptomatic colonization of microbiota.
Table 1
| Male | Female | Common to Male & Female | Location | Citation |
|---|---|---|---|---|
| Actinobaculum, | Actinobaculum, | Actinobaculum Aerococcus Atopobium Azospira Butyricicoccus Campylobacter Catonella Corynebacterium Dialister Finegoldia Fusobacterium Gardnerella Lactobacillus Mobiluncus Murdochiella Peptococcus Peptoniphilus Peptostreptococcus Porphyromonas Prevotella Proteiniphilum Saccharofermentans Sneathia Soehngenia Staphylococcus | Asymptomatic Urine | Lewis et al. (2013) |
| Aminobacterium, | Actinomyces, | |||
| Anaerococcus, | Aerococcus, | |||
| Anaerophaga, | Anaerococcus, | |||
| Anaerosphaera, | Anaerosphaera, | |||
| Aerococcus, | Anaerovorax, | |||
| Anaerotruncus, | Arcanobacterium, | |||
| Atopobium, | Arthrobacter, | |||
| Atopostipes, | Atopobium, | |||
| Azospira, | Azospira, | |||
| Butyricicoccus, | Brevibacterium, | |||
| Campylobacter, | Brooklawnia, | |||
| Catonella, | Butyricicoccus, | |||
| Corynebacterium, | Campylobacter, | |||
| Dialister, | Catonella, | |||
| Eubacterium, | Caulobacter, | |||
| Filifactor, | Coriobacterium, | |||
| Finegoldia, | Corynebacterium, | |||
| Fusobacterium, | Dialister, | |||
| Gardnerella, | Enterobacter, | |||
| Gemella, | Enterobacter, | |||
| Gordonibacter, | Enterococcus, | |||
| Kocuria, | Facklamia, | |||
| Lactobacillus, | Fastidiosipila, | |||
| Lactonifactor, | Finegoldia, | |||
| Marixanthomonas, | Flavonifractor, | |||
| Megasphaera, | Friedmanniella, | |||
| Microvirgula, | Fusobacterium, | |||
| Mobiluncus, | Gallicola, | |||
| Murdochiella, | Gardnerella, | |||
| Mycoplasma, | Gulosibacter, | |||
| Peptococcus, | Helcococcus, | |||
| Peptoniphilus, | Howardella, | |||
| Peptostreptococcus, | Incertae Sedis, | |||
| Porphyromonas, | Jonquetella, | |||
| Prevotella, | Lachnospiracea, | |||
| Pseudomonas, | Lactobacillus, | |||
| Rikenella, | Methylovirgula, | |||
| Proteiniphilum, | Microvirgula, | |||
| Pseudoramibacter, | Mobiluncus, | |||
| Saccharofermentans, | Modestobacter, | |||
| Sediminitomix, | Murdochiella, | |||
| Sneathia, | Negativicoccus, | |||
| Soehngenia, | Neisseria, | |||
| Staphylococcus, | Oligella, | |||
| Paraprevotella, | ||||
| Parvimonas, | ||||
| Pelomonas, | ||||
| Peptococcus, | ||||
| Peptoniphilus, | ||||
| Peptostreptococcus, | ||||
| Porphyromonas, | ||||
| Prevotella, | ||||
| Propionimicrobium , | ||||
| Proteiniphilum, | ||||
| Rhodococcus, | ||||
| Rhodopila, | ||||
| Saccharofermentans, | ||||
| Sneathia, | ||||
| Soehngenia, | ||||
| Sporanaerobacter, | ||||
| Staphylococcus , | ||||
| Stenotrophomonas, | ||||
| Streptococcus, | ||||
| Streptophyta, | ||||
| Sutterella, | ||||
| Tepidimonas, | ||||
| Tessaracoccus, | ||||
| Thermoleophilum , | ||||
| Varibaculum, | ||||
| Atopobium Corynebacterium Staphylococcus Streptococcus Veillonella | Corynebacterium, Escherichia coli Lactobacillus Prevotella Staphylococcus aureus Streptococcus (beta-hemolytic) | Corynebacterium, Staphylococcus Streptococcus | Healthy Urine | Fouts et al., 2012 |
| Atopobium vaginae Lactobacillus species (Lactobacillus iners, L. crispatus, L. gasseri, or L. jensenii) Leptotrichia spp. Megasphaera | N/A | Healthy Vagina | Lamont et al., 2011 | |
| N/A | Lactobacillus spp. Pseudomonas spp. Pseudomonadaceae | N/A | Non-Pregnant Urine | Yoo et al., 2016 |
| N /A | Bacillus spp. Lactobacillus spp. Ureaplasma spp. Veillonellaceae | N /A | Pregnant Urine | Yoo et al., 2016 |
| N /A | Atopobium spp. Fusobacterium spp. Lactobacillus spp. Megasphaera spp. Megasphaera spp. Prevotella spp. Sneathia spp. Ureaplasma spp Ureaplasma urealyticum Veillonellaceae | N /A | Preterm Delivery Urine | Yoo et al., 2016 |
| Enterococcus faecalis Escherichia coli Klebsiella pneumoniae | Gardnerella Klebsiella oxytoca | N /A | Neuropathic Urine - Void spontaneously | Fouts et al., 2012 |
| Escherichia coli Klebsiella pneumoniae Proteus | Escherichia coli | Escherichia coli | Neuropathic Urine - Intermittent Catheter | Fouts et al., 2012 |
| Enterococcus faecalis Escherichia coli (ESBL) Klebsiella pneumoniae Providencia stuartii Pseudomonas aeruginosa | Citrobacter koseri (diversus) Enterococcus faecalis Escherichia coli Gram Negative Rods, | Enterococcus faecalis | Neuropathic Urine - Foley Catheter | Fouts et al., 2012 |
| Actinobaculum, Actinomyces, Aerococcus, Anaerococcus, Arthrobacter, Bifidobacterium spp., Campylobacter, Clostridium Corynebacterium, Enterococcus, Escherchia, Lactobacillus, Gardnerella, Prevotella, Proteus, Psuedomonas, Serratia, Staphylococcus, Streptococcus | N /A | Overactive Bladder | Hilt et al., 2014; Curtiss et al., 2017 | |
| N /A | Atopobium vaginae, Chlamydia trachomatis, Gardnerella vaginalis, Mobiluncus species, Mycoplasma hominis, Trichomonas vaginalis, | N /A | Bacterial Vaginosis | Lamont et al., 2011 |
| Chlamydia trachomatis, Neisseria gonorrhoeae, Treponema pallidum | Atopobium vaginae Bifidobacterium, C. trachomatis, Dialister, Gardnerella vaginalis, Leptotrichia, M. genitalium, Megasphaera elsdenii, Mobiluncus spp. Mycoplasma hominis, N. gonorrhoeae, Prevotella spp. | Chlamydia trachomatis, N. gonorrhoeae, | Male Penis or Female Vagina w/ HIV Infections | Saxena et al., 2012 |
| Anaerococcus, Bifidobacterium, Coriobacteriaceae, Dialister, Lactobacillus, Megasphaera, Prevotella, Rhodococcus, Shuttleworthia, Streptococcus, | N /A | Interstital Cystitis / Bladder Pain (Urine) | Meriwether et al., 2019 | |
| Bifidobacterium, Coriobacteriaceae, Dialister, Finegoldia, Lactobacillus, Megasphaera, Mobiluncus, Peptoniphilus, Prevotella, Shuttleworthia, | N /A | Interstital Cystitis / Bladder Pain (Vagina) | Meriwether et al., 2019 | |
| Candida Albicans, Enterobacter spp., Enterococcus, E. coli, Klebesilla spp. Proteus mirabilis, Pseudomonas aeurignosa | Blautia, Klebsiella, Prevotella, | Klebsiella spp | Type 2 diabetes Patients w/ Asymptomatic Bacteriuria (Urine) Bladder Cancer | Balamuruganvelu et al., 2017; Liu et al., 2017 |
| Acinetobacter baylyi, Anaerococcus, Atopostipes, Candidatus Limnoluna, Carnobacteriaceae, Rickettsiales, Rothia, Rubrobacteria, Rubrobacterales, Sphingobacteriaceae | N /A | Bladder Cancer | Wu et al., 2018 | |
| Alloscardovia omnicolens, Anaerococcus lactolyticus, Anaerococcus murdochii, Auritidibacter ignavus Corynebacterium coyleae, Corynebacterium genitalium Corynebacterium minutissimum, Enterobacteriaceae, Gammaproteobacteria, Gardnerella vaginalis Haemophilus haemolyticus Propionimicrob um lymphophilum, Streptococcus, Ureaplasma parvum Ureaplasma urealyticum, | N /A | Prostate Cancer | Shrestha et al., 2018 |
Comparison of known microbial diversity of human male and female urogenital systems during the onset of physiological disequilibrium.
N/A, Not Applicable.
UTIs and Vaginal Microbial Communities
The dynamic exchange of microbiota, between the vagina and urinary tract illustrates a complex environment which is dependent on microbial composition. Probiotic strains of Lactobacillus (L. crispatus, L. gasseri, L. iners, and L. jensenii) that are the dominant bacteria of the vagina are demonstrated to repel or suppress non-native pathogens in environments through the production of secondary metabolites and control of environmental pH (Mirmonsef et al., 2014; Brubaker and Wolfe, 2017; Tachedijian et al., 2017; O’ Hanlon et al., 2019). Specifically, recent metagenomic sequencing of both the female urinary tract an vagina have highlighted highly similar microbiota between both systems such as E. coli, Lactobacillus spp. and Streptococcus anginosus suggesting a degree of cross-talk between both environments (Thomas-White et al., 2018). These findings suggest a dynamic continuous interplay between beneficial commensals and invasive pathogens (Vodstrcil et al., 2017). For example, loss or depletion of one species of Lactobacillus spp. leads to other organisms invading the previously occupied niche (van de Wijgert et al., 2014). More so, loss of commensal microbial communities consisting of probiotic strains likely leads to dysbiosis and an increased risk for UTIs, rUTIs, or infections from other bacteria and viruses (Sewankambo et al., 1997; Martin et al., 1999; Lamont et al., 2011; Stapleton et al., 2011; Gilbert et al., 2017). While microbial dysbiosis is a known mechanism underlying bacterial vaginosis, it remains unknown how dysbiosis of microbial communities impacts the urinary tract (Zozaya et al., 2016; Liu et al., 2017).
While microbiota of the host urogenital system are ultimately connected, microbiota can also be shared between the urogenital systems of two hosts.Interestingly, microbiome studies of patients with bacterial vaginosis highlighted a shared microbiome between the penile/urethral microbiota and vaginal microbiota (Zozaya et al., 2016). Specifically, penile-vaginal sex is the primary driver for the sexual exchange and increase of G. vaginalis in individuals with or without bacterial vaginosis (Vodstrcil et al., 2017). Furthermore, the increased presence of G. vaginalis in tandem with biofilm-producing communities drives an intense competition for resources, leading to a decreased presence of Lactobacillus spp. and an increased risk for acquiring urogenital infections (Machado et al., 2013). This loss of Lactobacillus spp. shifts the host vagina towards a more alkaline environment to include more diverse microbial communities such as Anaerococcus, Atopobium, Bacteroides spp. Gardnerella, Mobiluncus spp, Mycoplasma, Peptoniphilus, Peptostreptococcus spp. Prevotella, and Streptococcus that is found in patients with bacterial vaginosis (Lamont et al., 2011; Ravel et al., 2011; Mirmonsef et al., 2014; Onderdonk et al., 2016).
Host Gut and Urogenital Microbiome Crosstalk Underlying rUTIs
Gut microbiota play a unique role in the hypothetical fecal-perianal-urethra transmission route:fecal-associated microbiota can contaminate patient urine during a UTI. (Yamamoto et al., 1997; Jantunen et al., 2001; Paalanne et al., 2018). Microbiome sequencing of human feces revealed that high abundance of either Escherichia or Enterococcus is a risk factor for bacteriuria and symptomatic UTIs (Magruder et al., 2019). While previous evidence suggested that women with E. coli UTIs have a different strains in the urine and gut; Magruder and colleagues observed that E. coli strains are closely related, further supporting the hypothesis of a gut microbiota-UTI axis (Bahadori et al., 2019; Magruder et al., 2019). Interestingly, a study investigating genomic diversity of UTI patients between urine and feces strains observed two major patterns: the first case being that rUTIs are caused by the same strains and the opposite, there is a rapid and complete overtake of the bladder environment by another strain (Chen et al., 2013). These studies highlight the remaining lack of clarity in developing theories to describe the origins of gut-associated UTIs. Furthermore, all present studies lack longitudinal data that would be useful in describing potential reservoirs that link gut microbiota changes before and after the onset of uncomplicated UTIs (Chen et al., 2013). With these observations in the interconnected urogenital ecosystem, we can begin to decipher ecological interactions and control mechanisms between hosts and microbes, which drive changes in microbiota composition. For example, Thänert et al. (2019) deciphered the core genomic relatedness among E. coli, suggesting that E.coli is well-adapted to transit between various environments within a host, such as between the human gut and bladder. For example, in a study investigating pairwise interactions from patient fecal-associated E. coli and UTI E. coli, the authors identified unique mutations for virulence and nutrient-uptake in addition to proteins for biofilm formation, these factors are necessary for transition between the gut and urinary tract (Nielsen et al., 2016). Figure 2 describes the normal female urogenital microbiome during health and how gut-associated pathogens transit to both the bladder and vagina during dysbiosis.
Figure 2
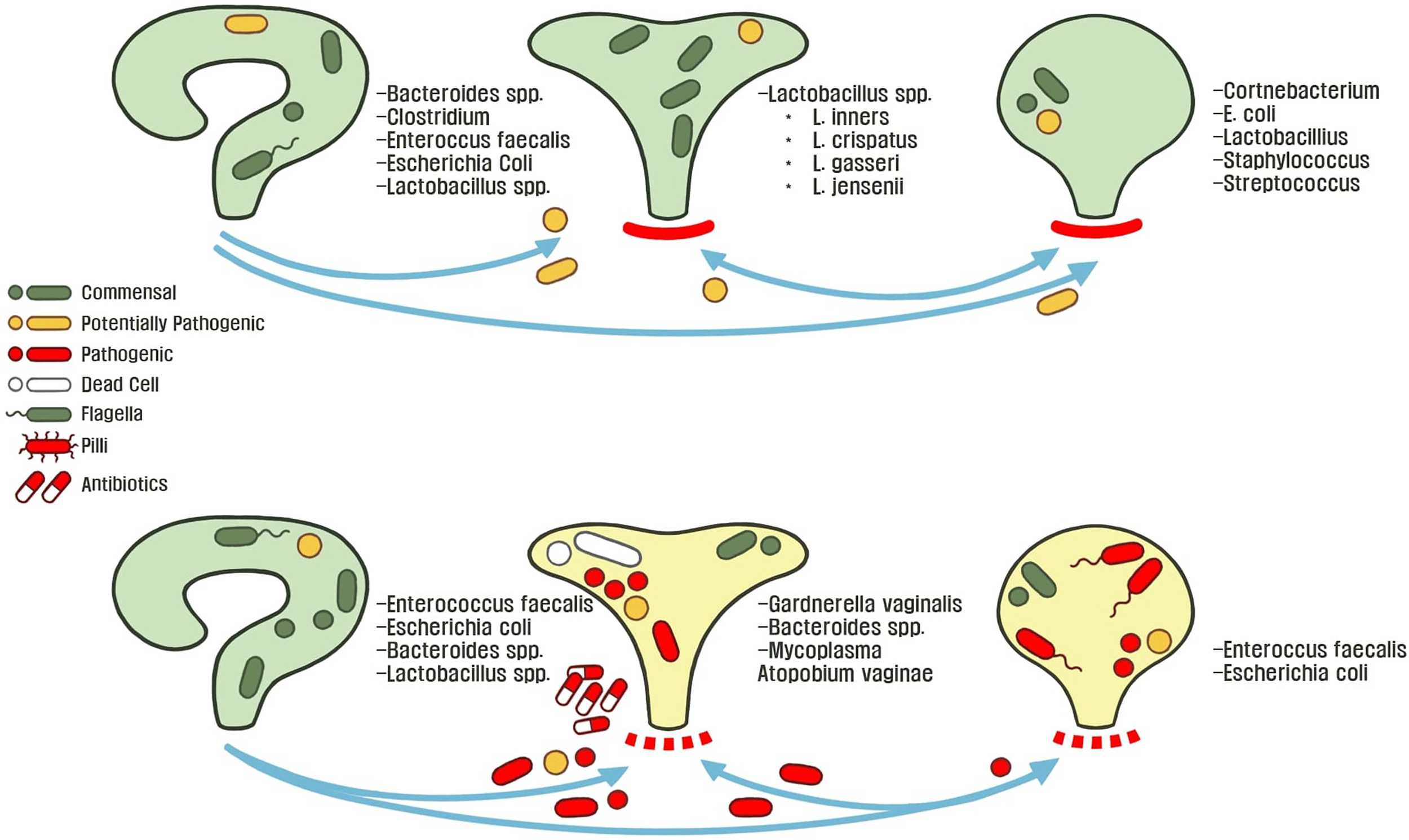
The healthy or asymptomatic gut, vagina, and bladder microbiome is stable across individuals conditions. These microbiota protect their host-associated niche from foreign pathogens by controlling abiotic factors and outcompeting potentially invasive microbiota. Microbial dysbiosis and an adverse environment disrupts host homeostasis through various mechanisms such as: poor hygiene, metabolic changes (menopause, metabolic diseases, etc.), exposure to environmental metals or antibiotics, and consumption of food-borne pathogens. This allows for microbiota to transit between the perianal-urogenital pathway and in turn shapes other microbial communities towards dysbiosis and predisposes individuals to UTI pathophysiology.
A different aspect of the microbial reservoir route was revealed when Gottschick et al. (2017) hypothesized that the bladder and urethra have distinct microbiotas, in comparison to the vagina. It was found that abundant bacteria of the vaginal fluid from bacterial vaginosis patients were also increased in the urine. This suggests that biofilms in the urine persist for extended times as a reservoir for recurrent bacterial vaginosis infections (Gottschick et al., 2017). More so, this study highlights the need to clarify the underlying crosstalk and mechanisms of microbiota transition between the urethral and vagina. For example, it is understood that uropathogenic strains rapidly form biofilms to rapidly grow in human urine, as compared to other gut-associated E. coli strains (Anderson et al., 2003; Justice et al., 2004; Forsyth et al., 2018). This evidence points to a major question: is there a microbial reservoir for the initial contamination and recurrence of UTIs? If so, what mechanisms is UPEC employing to invade the urinary tract when transiting from the gastrointestinal tract?
Antibiotic Resistance Mechanisms
The emergence of so-called “superbugs” is prevalent in global hospitals (Liu et al., 2016; Malhotra-Kumar et al., 2016; Versporten et al., 2018). The Antimicrobial Resistance Epidemiological Survey on Cystitis (ARESC) conducted a multi-national survey on various nations, in which 3018 pathogens were isolated from 4264 female patients (Schito et al., 2009). In total, 2315 (76.7%) were E. coli with an acquired complete resistance to ampicillin (Schito et al., 2009). These microbiota become MDR through the acquisition of genotypes through horizontal gene transfer or mutations to resist treatments (Davies and Davies, 2010; Mathers et al., 2015; Salverda et al., 2017; van der Zee et al., 2018). Increased prevalence of MDR globally is likely attributed to wide-spread and indiscriminate application of broad-spectrum antibiotics, which do not eliminate MDR pathogens (Cai et al., 2015; Goneau et al., 2015). There is a great need for the application of antibiotic stewardship surrounding UTI treatment (Rossolini et al., 2014; Bartoletti et al., 2016).
Subinhibitory Antibiotic Treatments
In face of the emergence of horizontally-transferred MDR genes globally, UTIs are one of the most common sources for human-associated MDR pathogens due to inappropriate antibiotic usage which do not eliminate pathogens (Cullen et al., 2012; Vellinga et al., 2012; Nordstrom et al., 2013; Bartoletti et al., 2016; Salverda et al., 2017; Zilberberg et al., 2017; van der Zee et al., 2018). Through antibiotic treatment, microbiota regulate a multitude of stress response mechanisms such as mutation/modification of the genome to alter translation, produce enzymes to degrade antibiotics, and modifications in membrane permeability to counteract antibiotic stress (Dantas et al., 2008; Gniadkowski, 2008; Davies and Davies, 2010). These observations demonstrate a wide variety of mechanisms that pathogens employ to tolerate stress. Chronic usage of both low-dose and full-course antibiotics disturbs commensal microbiota, dysbiosis drives virulence and biofilm-associated resistance mechanisms by foreign pathogens, leading to recurrent infections (Figure 3).To map bacterial responses and adaptation, Erikson and colleagues (2017) created a database assessing transcriptome-level expression of genes coding for E. coli resistance and tolerance to various stressors in the environment. The authors identified that 12% of all transcript changes across all E. coli experiments were due to antibiotic treatment. The multiple antibiotic resistance transcriptional regulator (marA) was found to be overexpressed in the database and acts as a key-driver for MDR phenotypes in E. coli (Ruiz and Levy, 2010; Erikson et al., 2017). The increased expression of marA in uropathogenic bacteria is associated with expression of flagella proteins, biofilm production, and enhanced planktonic aggregation; all are essential for pathogenic colonization of urothelial tissue (Hadjifrangiskou and Hultgren, 2012). Furthermore, marA is a global regulator during antibiotic stress and activates transcription of the multidrug efflux system, which is observed in UTI patients (Schuster et al., 2017; Atac et al., 2018; Chowdhury et al., 2019; Praski Alzrigat et al., 2021).
Figure 3
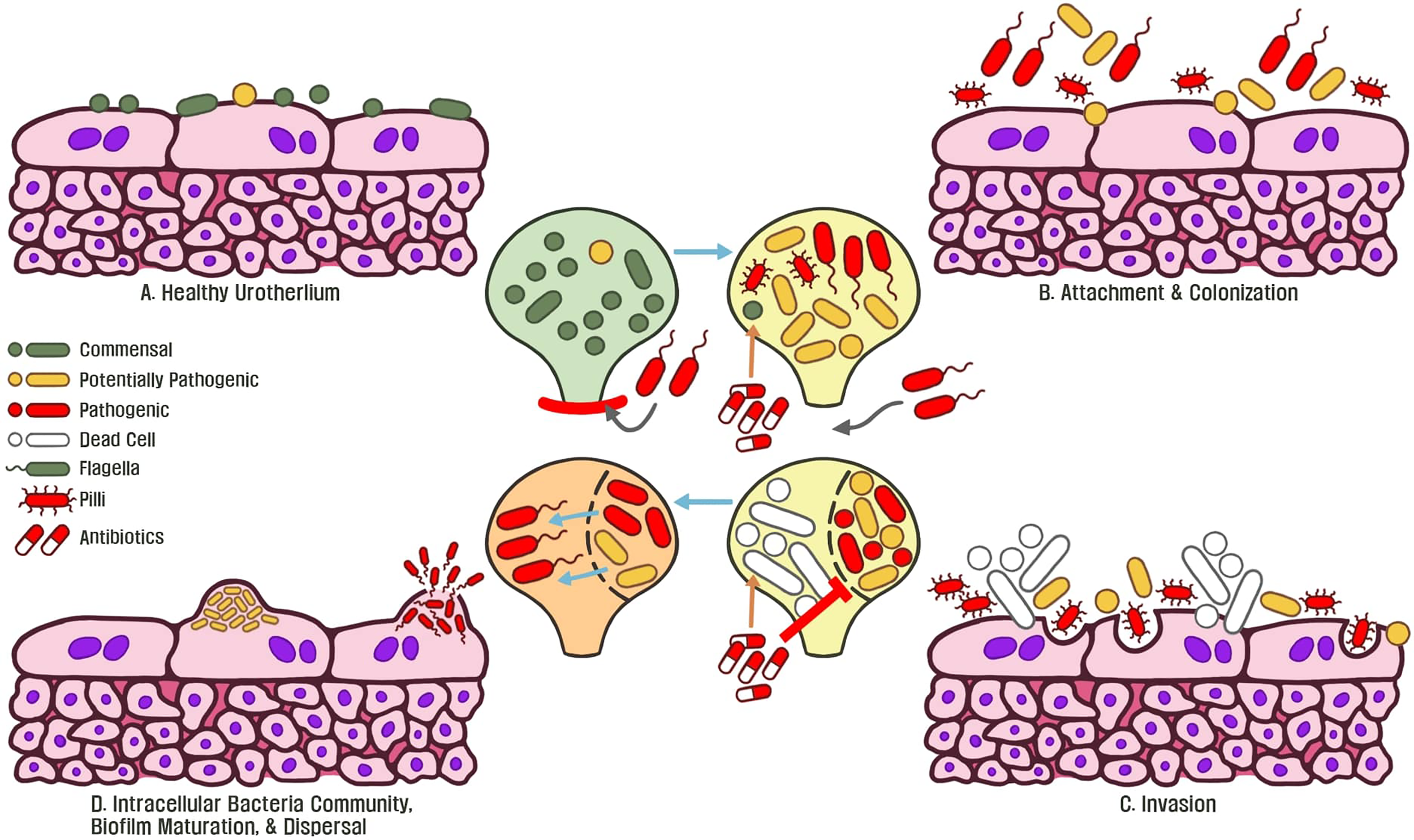
(A) Commensal microbiota are associated with a healthy normal human urothelium and protect the host from invasive pathogens. (B) An initial course of antibiotics enforces bacteria to express flagella, type 1 pili, and adhesin proteins to attach and colonize the urothelium. (C) Continued antibiotic exposure eliminates commensal bacteria and surviving microbiota exposed to this stressor become resistant. Invasive bacteria begin to express virulence genes to colonize and replicate within tissue. (D) Following replication, intracellular bacteria communities are formed and antibiotic resistant biofilms become matured. Continued exposure to stressors such as antibiotics allows for the emergence of pathogenic, MDR biofilms, which underlines recurrent chronic UTIs of the host.
Polymicrobial UTI Biofilm Formation and Persistence
Biofilms are microbial communities that are formed through a complex matrix of extracellular polymeric substances, such as proteins, polysaccharides, and DNA, attached to a surface (Stewart et al., 2013). Biofilm formation is initiated when planktonic cells attach to both biotic and abiotic surfaces in an environment. Through the secretion and intake of autoinducers (small molecules or peptides), bacteria chemically interact and interpret changes in cellular density among the biofilm, in a process described as ‘quorum sensing’ (Azeredo et al., 2017). Lack of space within an environment triggers self-imposed mechanical stress for the entire biofilm community, driving biofilm formation and uptake of nutrients at the periphery of the colony (Chu et al., 2018). Cells sense both surfaces and intermingling microbiota, thus biofilm communities in tandem are able to dynamically orchestrate gene expression to produce adhesins for forming multicellular communities (Papenfort and Bassler, 2016).
Most cases of lower UTIs with UPEC infections are diagnosed as monomicrobial (Pruetpongpun et al., 2017). Specifically, UPEC strains harbor numerous virulence factors to enable survival and persistence of polymicrobial communities to persist for months against harsh host environments, thus acting as a reservoir for recurring infections in various human niches (Meier et al., 2008; Luo et al., 2012; Bjarnsholt, 2013). UPEC strains generally have a higher number of fimbrial gene clusters such as adhesins and motility complexes, such as type 1 fimbriae or pili, and flagella which greatly enhance the ability of UPEC to form biofilms (Pratt and Kolter 1998; Spurbeck et al., 2011; Luo et al., 2012; Sharma et al., 2016). In the case of the human urinary tract, the formation of polymicrobial biofilms occurs as a reaction to stressful conditions such as sub-inhibitory antibiotic exposure, to shelter bacteria from harm (Penesyan et al., 2019). Thus, biofilms are heavily regulated by environmental signaling and pathways, especially as a defensive response to antibiotics (Hoffman et al., 2005; Karatan and Watnick, 2009). For example, rapA enhances biofilm gene regulation to increase antibiotic resistance, both ymgB and yafQ initiates E. coli leads to biofilm formation under antibiotic stress (Sharma et al., 2016).
Intracellular communities of biofilms are inhabited by multiple species that are dependent on polymicrobial interactions (chemical, physical, spatial organization) (Kostakioti et al., 2013; Marchal et al., 2017) between mixed or segregated communities. Whether multispecies interactions in polymicrobial UTI biofilms generally are synergistic or competitive remains unclear. It is difficult to paint a clear overall picture of the polymicrobial ecological interactions in UTIs, especially during antagonistic interactions when exposed to antimicrobial agents (de Vos et al., 2017). However, it is understood that organisms involved in a mutualistic symbiosis will evolve towards the benefit of each other, leading to mixing of cellular populations (Marchal et al., 2017). In contrast, competitive microorganisms tend to dissociate, thus leading to layered or otherwise divided populations (Momeni et al., 2013). The mechanisms for various species interact with one another is likely within a system to support one another through cross-feeding, co-exist in neutralism, or rigorously compete in a multispecies system respectively (Bauer et al., 2017; Ferreiro et al., 2018; Giri et al., 2019). Determination of the dominating bacterium in a polymicrobial biofilm can so far only be done in a pair-wise manner, as there is insufficient information about the entire picture of polymicrobial UTIs (de Vos et al., 2017). However, analysis of multispecies communities may determine the nature of underlying host-microbiota interactions.These challenging observations of the microbial world, involving complex species interactions, are presently being interpreted through in silico modeling of microbial communities and will be discussed later (Wootton and Emmerson, 2005; Bauer et al., 2017; García-Jiménez et al., 2018).
Animal-Associated Pathogens as a Potential Reservoir for rUTIs
Over time, recent evidence has uncovered that the infection route of both UTIs and rUTIs goes far beyond the fecal-perianal-urethra transmission route. UPEC specifically is an intermingling pathogen which switches between humans, animals, and the environment. This paradigm described as One Health requires the standardization of new studies to better identify sources of transmission by integrating an ecological and epidemiological framework that can be translated into clinical settings (Ewers et al., 2012; Singer, 2015; Davis et al., 2017). For example, zoonotic pathogens that may or may not be MDR are reintroduced as a probable environmental reservoir for rUTIs (Nordstrom et al., 2013). Preliminary studies suggesting a potential link between animals, E. coli, in which women who consume chicken or pork were at a 3.2 and 3.7 greater risk respectively, to acquire MDR UTIs, suggesting that dietary habits are an important risk factor for E. coli associated UTIs (Shooter et al., 1970; Bettelheim et al., 1974; Linton et al., 1977; Manges et al., 2007; Vincent et al., 2010; Nordstrom et al., 2013). Furthermore, there is evidence of a sharing of E. coli and K. pneumoniae between humans and companion animals (Stenske et al., 2009; Harada et al., 2012; Naziri et al., 2016; Wang et al., 2018; Marques et al., 2019). Zoonotic gut-associated E. coli presents a risk to humans by transferring from farm or companion animals, through the environment, into humans is a significant concern with regards to MDR pathogens (Cantas and Suer, 2014; Pomba et al., 2017; Bourne et al., 2019).
Recently, the first MDR E. coli classified as a superbug on US soil acquired the mobilized colistin resistance (MCR-1) gene from a commercial hog farm and manifested itself as a clinical UTI in a human patient (McGann et al., 2016). The MCR-1 gene is readily transferred through horizontal gene transfer and translates to a resistance against colistin, a last-resort antibiotic (van der Zee et al., 2018; Wang et al., 2018; He et al., 2019). MCR-1 was observed when profiling one of the most common MDR UTI strains: ST131 (Hasman et al., 2015). The authors assessed 1,923 meat and 1,188 human clinical isolates and observed that 1.3% and 15.3% strains respectively were identified as ST131. Specifically, nearly all meat isolates of ST131 were from the H22 lineage, which corresponds with fimH22 which is involved in the expression of adhesion and host invasion genes (Hasman et al., 2015; Liu et al., 2018).Additionally, a recent study of MCR-1 was found to be wide-spread in companion animals (Wang et al., 2018). Shockingly, evidence is mounting towards the potential that microbial strains isolated from the feces of animals may be directly linked to human UTIs by identifying shared strains between both species and feces or urine (Johnson et al., 2008; Osugui et al., 2014; Marques et al., 2019).
These studies illustrate a rapidly evolving story of MDR pathogens in which the ever-increasing emergence of UTIs that are associated with the interactions of contaminated animals, presenting a significant threat to public health in both communities and clinical settings. However, the correlation between farm or companion animals should be interpreted with caution. Identifying similarities between human/animal isolates does not include the transmission of microbiota and the routes that are employed to cause disease in humans and therefore requires an integrated One Health approach to better understand the issue (Ewers et al., 2012; Singer, 2015; Davis et al., 2017).
Host-Microbial Interactions Leading to UTI
Urine is the primary source of information for identifying UTIs in patients. Analytical chemistry techniques use urine to detect biomarkers, such as excreted metabolites from pathogens (Smart et al., 2019). Specifically, urine is a waste product from various metabolic end-points of secondary metabolism whose concentration is determined by an individual’s diet, lifestyle, and a variety of other environmental factors (Bouatra et al., 2013; Playdon et al., 2016; Tang, 2017). With regards to sex, the urine of females generally contains greater quantities of citrate, but not as much calcium or oxalate as males (Ipe et al., 2016). These metabolic differences likely favors a cohort of specific microbes which thrive in these niches. A large-scale multi-experimental mass spectrometry study of human urine samples was conducted to create the database of “The Human Urine Metabolome” to identify 2651 different metabolites and unique ionic species in healthy human urine (Bouatra et al., 2013). Overall, this study preliminary assesses the diversity of the urine metabolome in healthy humans and can be useful to predict metabolites which predisposes individuals to UTIs. However, there is a lack of data detailing the metabolic state of urine in various pathophysiologies and lack of knowledge regarding biomarkers that may predispose patients to various infection outcomes.
Host Metabolome & Bacteria of the Urogenital System
Urine is a hostile environment for most bacterial species due to a pH range of 5.5 to 7, with an average of 6.2 (Rose et al., 2015). This biofluid is a patient-specific excretion containing a metabolomic profile, strongly dependent on health or disease (Beger et al., 2016). Advances in quantitative mass spectrometry have enabled the identification of specific metabolites as biomarkers of infections or inflammation. For example, a study identified unique biomarkers of UPEC-specific UTIs by identifying an increased ratio of acetic acid to creatinine and trimethylamine concentration in urine (Lam et al., 2014). While traditional methods such as a urine dipstick urinalysis have a variable sensitivity of 68 to 88% across different patient groups, employment of mass spectrometry was found to have a 92% true positive predictive value to diagnose urinary tract infections (Devillé et al., 2004); Lam et al., 2014). (Table 2 illustrates directional changes in microbial abundances of the urinary tract associated with disturbances in host-metabolism. Microbiota adapt and uptake various energy sources from the host diet, likely leading to modifications of urine composition and metabolic profile (David et al., 2014; Playdon et al., 2016; Thiele et al., 2020). Generally, high quantities of glucose in the urine tends to favor bacterial growth and eventually a UTI (Wilke et al., 2015). For example E. faecalis is able to grow in urine containing greater concentrations of glucose and has enhanced recurrent biofilm formation potential, primarily in diabetic patients (Pillai et al., 2004). Such environments exacerbate the development of pathogenesis in patients who are affected by metabolic disorders.
Table 2
| State of dysbiosis | Metabolic Change | Microbiota Change | Citation |
|---|---|---|---|
| Follicular fluid during IVF | ↑ Estradiol & progesterone | ↑ Lactobacillus gassen, L. crispatus, L. jensenii | Pelzer et al., 2012 |
| Kidney Transplant | ↑ Folate metabolism | ↑ E. coli & E. faecalis | Rani et al., 2017 |
| N/A | ↑ Levels of free glycogen | ↑ Lactobacillus spp. | Mirmonsef et al., 2014 |
| Post-Menopausal (Symptomatic) | ↓ Estrogen & low glycogen | ↓ Lactobacillus spp. | Muhleisen and Herbst-Kralovetz, 2016 |
| Premenopausal | ↑ Estrogen & glycogen | ↑ Lactobacillus spp. | Muhleisen and Herbst-Kralovetz, 2016 |
| Renal Tubular Acidosis | Hypercalciuria | ↑ E. coli | Jamshidian et al., 2018 |
| Type Two Diabetes Mellitus | ↑ Fasting Glucose & hyperlipidemia | ↑ Prevotella, Lactobacillus, and Shuttleworthia | Liu et al., 2016 |
| Urinary Tract Infections | ↑ Acetic Acid & trimethylamine | ↑ E. coli | Lam et al., 2014 |
| Urinary Tract Infections | ↑ Ethanolamine | ↑ E. coli | Sintsova et al., 2018 |
| Urinary Tract Infections | ↑ Acetate & Creatinine | ↑ E. coli | Grochocki et al., 2017 |
| Urosepsis | ↑ Procalcitonin/Albumin ratio | ↑ Increased E. coli | Luo et al., 2018 |
Microbial changes linked to host metabolic dysregulation.
N/A, Not Applicable.
Hormonal changes that impact specific host metabolic functions play a major role in the onset of female urogenital infections. Menopause influences metabolite availability and may increase the risk of UTIs (Mody and Juthani-Mehta, 2014). Menopausal women typically have lower levels of available estrogen, thereby lowering the level of glycogen within vaginal fluids (Arnold et al., 2016). This change in the metabolic composition of vaginal fluids, leads to substantial differences in substrates that microbiota employ to attach to the vaginal epithelium (Chan et al., 1984). Therefore menopausal individuals with low estradiol levels tend to have an increased risk for rUTIs (Mody and Juthani-Mehta, 2014). In contrast, increased exposure to estrogen modulates urothelium growth and differentiation to supplement epithelial defense mechanisms against UPEC invasion and UTI recurrence (Lüthje et al., 2013; Mody and Juthani-Mehra, 2014). Clinical observations found that the application of low-doses of estriol (0.03 mg) with probiotic Lactobacillus acidophilus can restore vaginal microflora (Donders et al., 2010). This therapy was demonstrated to restore normal vaginal physiology and mitigated bacteria linked to bacterial vaginosis (O’Hanlon et al., 2011; Mueck et al., 2018). In a recent review of non-antibiotic treatments for the treatment of UTIs in postmenopausal women, the authors suggested that the application of topical estrogen not only normalizes vaginal microbial composition, but also represent a non-antimicrobial therapy for the prevention and treatment of UTIs (Caretto et al., 2017).
Impact of Dietary Transition Metals on UTI Pathophysiology
Metals and their ions play major roles in human health and disease through electron exchange or redox reactions, thereby promoting cellular stress through reactive oxygen species. Chronic dietary exposure to metals in the food-chain has the potential to negatively impact human health, leading to a variety of nephron- and urinary-tract associated diseases (Zhao et al., 2017; Xu et al., 2018; Yen et al., 2018). Excessive exposure to dietary metals eventually reach the kidneys and bladder causing pathologies; heavy metals are also taken up by various microbiota to incite host infection (Subashchandrabose et al., 2014; Habibi et al., 2017; Hyre et al., 2017; Zhang et al. 2019). Therefore, interpretation of rUTIs must be updated to include the possibility that metal exposure from inhalation, diet, and cosmetics are potential risk factors for UTIs onset and recurrence. On the other hand, mitigation of urinary levels of heavy metals may prove to be an alternative treatment to rUTIs by depriving pathogens use of essential metals or nutrients in comparison to antibiotic usage (Subashchandrabose and Mobley, 2015; Bauckman et al., 2019).
Free iron (Fe) is necessary for most biological processes such as cellular respiration, DNA replication, and oxygen transport via hemoglobin. Therefore, there is an intense competition between the host and pathogens for Fe, Fe deprivation from uropathogens may be a primary mechanism to mitigate virulence (Subashchandrabose and Mobley, 2015; Bauckman et al., 2019). For E. coli pathogenesis, invasion and disruption of the host urothelium to increase the availability of free Fe2+/Fe3+ is essential to initiate and maintain virulence (Gao et al., 2012). This occurs through the promotion of the UPEC toxin 𝛂-hemolysin, which ruptures and degrades host membranes to promote hemoglobin release, thereby promoting bacterial growth and virulence (Dhakal and Mulvey, 2012). UPEC opportunistically adapts to Fe abundant conditions by increasing the expression of siderophores, to scavenge environmental metals(ireA, irp-2, iucC) (Zhao et al., 2009; Shields-Cutler et al., 2015 Additionally, the induction of evolved virulence genes (chuA, fepA, fyuA, iroN, iucA, iutA, and sitA) are required for Fe uptake and transport systems to incite UPEC virulence mechanisms (Subashchandrabose and Mobley, 2015; Khasheii et al., 2016; Habibi et al., 2017; Tang et al., 2018). Siderophore biosynthesis and Fe acquisition from the host-environment allows UPEC to promote asymptomatic growth by overexpressing genes for growth, fitness, and colonization (Watts et al., 2012). Uptake and use of metals by uropathogens enforces an environmental niche for invasive microbiota to resist natural and artificial stressors, thus leading to persistent host infections (Hancock et al., 2008; Subashchandrabose and Mobley, 2015; Ipe et al., 2016). During UPEC-associated UTIs, Cu levels in the urine are elevated and play an underappreciated role in UTI pathophysiology (Subashchandrabose et al., 2014). Bacterial Cu-associated virulence is not as common as Fe-associated virulence in UTIs, though it is still observed in numerous organisms. The micronutrient Cu is necessary for a variety of aerobic organisms such as bacteria, fungi, plants, and animals by supporting metabolic processes through the maintenance of proteins and metalloenzymes; conversely, the interaction of Cu ions and free oxygen radicals can damage proteins (Festa and Thiele, 2011). Hyre et al. (2017) demonstrated that the gain or loss of electrons of Cu through oxidation-reduction reactions constitutes an active host response mechanism to infections with UPEC, Klebsiella pneumoniae, and Proteus mirabilis. Ceruloplasmin, a transport protein for Cu, serves as a molecular source for Cu in the urine (Hyre et al., 2017). This molecular mechanism can be observed in clinical patients who are affected by Menkes disease, a lethal hereditary disorder of Cu metabolism leading to Cu deficiency (Tümer and Moller, 2010). Consequently, those with this Cu deficiency are especially prone to developing rUTIs. (Kim et al., 2019). In contrast, recent studies demonstrate that host-mediated mobilization of Cu into the urinary tract during UTIs occurs, this illustrates a potential novel approach to reduce bacteria which rely on Cu-virulence within the urinary tract (Hyre et al., 2017). Table 3 describes uropathogenic mechanisms of siderophores and other transport mechanisms to acquire free metals from hosts, to elicit behaviors to colonize the urothelium.
Table 3
| Metal | Pathogen | Gene Name (s) | Function | Citation |
|---|---|---|---|---|
| Cu II | UPEC | Ybt | Pathogenic siderophore | Chaturvedi et al., 2012 |
| Cu 1+/2+ | UPEC | cusC | Cu resistance & virulence mechanisms | Subashchandrabose and Mobley, 2015 |
| Fe | UPEC | Iha | Fe siderophore uptake and virulence of bladder and kidney | Léveillé et al., 2006 |
| Fe | UPEC | FyuA | Fe-Ybt siderophore uptake & increased virulence through biofilm in bladder | Hancock et al., 2008; Brumbaugh et al., 2013 |
| Fe (Haem) | UPEC | ChuA | Heme receptor for iron transport and kidney invasion during UTI | Hagan and Mobley, 2009 |
| Fe | UPEC | fepA, iroN*, iutA*, fyuA, chuA, hma, sitA | Iron uptake and metal transport* | Subashchandrabose and Mobley, 2015 |
| Ni2+ & | UPEC | nikA | Nickel acquisition & urofitness | Subashchandrabose et al., 2014 |
| Ni(II) | UPEC | Ybt | Metal acquisition, unknown purpose | Robinson et al., 2018 |
| Zn2+ | UPEC | ZnuACB & ZupT | Uropathogenic fitness & metal transport | Sabri et al., 2009 |
| Ni | Uropathogenic S. aureus | NixA, NikA | Urinary tract colonization and increased fitness | Hiron et al., 2010 |
| Ni & Co | Uropathogenic S. aureus | Cnt (Opp1) | Colonization of bladder & kidneys during UTI | Remmy et al., 2013 |
Uropathogenic uptake of various metals to incite virulence in host urothelium.
*Both the IutA and IroN have been observed to lead to bladder colonization.
Exploring the Urinary Tract Environment With Systems Biological Approaches
The following represents recent advances in next-generation models and methods to examine UTI pathomechanisms. These methods reach endpoints that cannot be typically reached through traditional in vivo or in vitro models. To understand the underlying environmental mechanisms which control host invasion, these methods are useful to develop novel treatment strategies for rUTIs. To achieve precision treatment options for UTIs, the application of next-generation sequencing, genome-scale metabolic modeling, and the development of novel microfluidic or stem cells technologies to mimic the bladder is needed based on patient specifics, rather than generalizations. These next-generation methodologies foreshadow a future for treating UTIs from an ecological and systemic perspective, rather than through reductionist approaches. These examples bolster the argument for a necessary adjustment towards clinical diagnosis of infections and progression of host-specific diseases.
Transcriptomics and the Tolerome
E. coli are flexible microbes that thrive in various niches and environments by tuning gene expression. These modifications allow E. coli to rapidly adapt to stressful environments for successive colonization during environmental stressors, like antibiotic exposure, to incite host pathogenicity (Schwartz et al., 2016; Erickson et al., 2017; Zhang et al., 2017). While details of UPEC function in human urine exist, there is a lack of consistent information about UPEC acclimation to a rapidly changing environment Hagan et al., 2010; Sintsova et al., 2019). Assessing UPEC gene expression from patients becomes convoluted due to patient-specific host factors which may induce various bacterial genes, bacterial behavioral responses, and gene expression in differing host environments (Subashchandrabose et al., 2014; Schreiber et al., 2017). Furthermore, a recent study which employed UPEC strains from humans, compared pathogenesis mechanisms from humans in mice and found a similar gene expression pattern associated with metabolic machinery (Frick-Cheng et al., 2020).The complexity to assess human UPEC infections requires updating and clarifying definitions of bacterial virulence in heterogeneous human patients for optimized precision health outcomes (Mobley 2015). This aspect of biology underlies the deepening complexity of systems-oriented interaction needed to understand patient-specific disease outcomes.
Presently, the “Tolerome” describes transcriptome-level information detailing E. coli tolerance and resistance response to over 89 different stressful conditions (Erickson et al., 2017). Understanding the Tolerome and environmental stress response of uropathogens at a global transcriptomic level can enable a realistic understanding of the functions underpinning microbial colonization in urine. The undertaking of this study translates to 56263 events of up- or down-regulation in 5049 different genes (Erickson et al., 2017). Comparative studies of this nature can further elucidate unique bacterial signatures of microbial communities necessary for maintaining physiological equilibrium in human hosts. For example, global pathogenic transcriptional responses are potential biomarkers of infection. For example, a recent study by Sintsova and colleagues (2019) identified a novel and universal transcriptional response which controls various transcriptional regulators, thus driving rapid growth of UPEC during infection. While these studies assess E. coli response to various known or stable environments, investigations of rapidly changing environments is not well understood. E. coli response to environments is not well-characterized(i.e during dynamic disease processes and pathophysiologies), there is a need to quantify microbial communities phenotypic reactions to host-specific perturbations and interpret how these changes impact human health or disease in real-time.
Shotgun metatranscriptomics profiles the transcriptome of all present strains of a microbial community in an environment and their functions (Shakya et al., 2019). While metagenomic sequencing provides information on the assembled genome and potential genes of organisms which comprise microbial communities, it cannot provide real-time information on the functions or metabolic diversity of these communities (Mick and Sorek, 2014). More so, while metagenomics can identify active or inactive microbial members of a community, does not provide much information towards the function of a microbial community like metatranscriptomics (Brown et al., 2017; Shakya et al., 2019). While metagenomics can provdie information on all functional possibilities of the microbiome, metatranscriptomics provides a snapshot of the present infection state of the patients. Particularly, application of metatranscriptomics deciphers both function and diversity of microbial communities in responsive and non-responsive individuals. Further advances in next-generation sequencing has led to the development of scRNA sequencing of microbial communities as a novel approach to elucidate functional interactions among microbial species, mixed microbial communities, and within the host-associated microbiome (Lloyd et al., 2018; Imdahl et al., 2020; Kuchina et al., 2021). While scRNA sequencing for microbial communities is developing, metatranscriptomics is becoming readily available to identify present organisms in microbial communities from human samples in tandem with microbe-specific gene expression changes host responses (Shakya et al., 2019). Metatranscriptomics was applied to study the vaginal microbiota during bacterial vaginosis, following antibiotic treatment (Deng et al., 2018). Subjects were divided into responsive and non-responsive groups to identify potential mechanisms underlying response to the drug. The authors observed that G. vaginalis upregulated CRISPR-Cas genes as a stress response to mitigate DNA strand breakage from metronidazole by inducing DNA repair mechanisms. Female patients colonized by G. vaginalis with this unique change in gene expression were shown to be non-responsive to antibiotic treatment. Overall the observations from this study cast doubt on previous assumptions regarding microbial communities and pathogenesis progression. Metatranscriptomics for assessing the functional roles of diverse microbial strains in various environmental conditions of humans is growing in the literature (Shakya et al., 2019). But to date, metatranscriptomics has not been applied to urine or patient samples with UTIs in vitro or in vivo. Application of metranscriptomics for the bladder microbiome would clarify perspectives of not only the microbiota occupying the bladder, but also their community interactions in respect to UTI pathophysiology.
In Silico Modeling of Human-Like Bladder Environments
Organisms adapt their metabolism to maintain homeostasis in ideal or adverse environments by selecting either rapid growth or resistance phenotypes (Ewald et al., 2017). Specifically, the metabolic flux of prokaryotes dynamically adjusts during growth alterations, thus optimal adaptation of protein production depends on pathway expression that constrains or allows growth (Bartl et al., 2013). Fulfillment of metabolic requirements to maintain physiological balance is a strategy employed by pathogens in rapidly changing environments to thrive or survive (Wessely et al., 2011). Depending on required response time, E. coli employs transcriptional or post-transcriptional mechanisms to regulate metabolic pathways for stress adaptation (Wessely et al., 2011). Identification and understanding of stress-response regulatory networks illustrates the role of environmental sensing and non-genetic changes in the emergence of disease phenotypes or MDR in pathogens. During this phase, the rapid acclimation of bacteria can be exploited to innovate treatment options for infectious human diseases (Conover et al., 2016).
By assessing the remarkable flexibility and diversity of differentially expressed genes between different stressful conditions, we can decipher UPEC adaptation towards colonization and persistence within differing human hosts (López-Maury et al., 2008). Transcriptional responses to various environmental cues are understood, less is known about UPEC response to heterogenous humans with regards to UTIs (Erickson et al., 2017). Furthermore, transcriptional changes that are host-pathogen specific may not be readily replicated in biological experiments, due to complexities when attempting to control specific environments. Thus, the application of genome-scale metabolic models for uropathogens which predicts an organisms’ transcriptional and metabolic changes when primed by environmental stresses is a promising approach to unravel the complexities pathophysiology and predict treatment strategies to prevent or intervene against rUTIs (Józefczuk et al., 2010). The mapped genomes of various wild-type and mutant E. coli strains have been defined and applied to predict or engineer various metabolic capabilities through reconstruction of constraint-based genome-scale models. Modeling metabolic systems has been constantly updated since the original metabolic network reconstruction of E. coli in 2000 (Edwards and Palsoon, 2000; Orth et al., 2011; King et al., 2015; O’Brien et al., 2015). Genome-scale metabolic modeling is useful for creating genome-wide transcriptional networks to illustrate transcriptional response to various environmental stresses. By integrating both transcriptomics and metabolomics to identify conserved responses for various environmental stressors, such as oxidative stress, acid, cold, heat, and shifting glucose to lactose in media which are necessary for the maintenance of homeostasis (Józefczuk et al., 2010; Seo et al., 2015; Du et al., 2019). Application of genome-scale metabolic models that incorporate environmental stressors provides inspiration to interpret uropathogenic transcriptomes which are associated with chronic infections.
In silico community metabolic models of microbiota provide valuable insights into the ecological interactions within a microbial consortia. In brief, metabolic models of microbial species are constructed based on the organism’s annotated genome sequence. Enzymes, for which predicted genes are assigned to reactions, which the enzymes can catalyze. By connecting the individual reactions to a metabolic network, in silico representations of the organism’s catalytic capabilities allow predictions of metabolic phenotypes, for instance environmental context-dependent growth rates, nutrient utilization, and metabolic by-products release (O’Brien et al., 2015). Models of community metabolism are constructed by connecting species-specific models and allowing the exchange of metabolites between cells as well as the competition for shared resources. To our knowledge, there are no applications of community metabolic models of the urinary tract microbiome. However, in silico modeling of the human gastrointestinal microbiota revealed specific metabolic processes, including fermentation and metabolic cross-feeding interactions that are altered in disease when compared to healthy controls (Bauer and Thiele, 2018; Graspeuntner et al., 2018; Aden et al., 2019; Pryor et al., 2019). The rationale behind such models is that microorganisms not only adjust their metabolism in response to abiotic stresses and chemical composition of the environment, but also depend on the presence or absence of other microbial cells in their vicinity (Klitgord and Segrè, 2011).
In systems medicine, these models are applicable to generate hypotheses of factors that promote ecosystem colonization with a potential pathogen. Thereby providing potential treatment options, as well as prevention strategies against resistant pathogens. Thus with the growing appreciation of the role of the urinary tract microbiome in rUTIs, in silico models of community metabolism can expand clinical understanding of rUTI pathophysiology and foster the development of novel therapeutic strategies to target the urobiome specifically.
In Vitro Modeling of Human-Like Bladder Environments
For designing an in vitro model reflective of human rUTIs that yields meaningful data, mimicking human conditions realistically is necessary. Identification of a relevant urine medium, characterization of the physical bladder environment, and understanding the patient-specific urobiome function is required. Standard artificial urine medium that was first detailed in 1961 for UTI modeling is based on the 17 most prevalent urine substances (Altman, 1961; Brooks and Keevil, 1997). However, Boutra and colleagues identified more than 2600 unique metabolite species in healthy urine by aggregating urine from multiple donors to compensate for missing metabolites (Boutra et al., 2013). Consequently, this approach to urine modeling further complicates reproducibility due to urine metabolite composition and variability. To our knowledge, there is a deficiency of studies detailing uropathogen adaptation to various environmental conditions of healthy urine or during pathophysiological changes over time.
Due to the advancing need to model and replicate human disease outside of human systems and away from animals in a translational way towards precision medicine, a plethora of methods have been developed to interpret complex human diseases in vitro. Generally, preliminary animal models have provided the investigative foundation to understand human systems, but consistently fail to replicate and provide specific insights into the complexity of the human system, let alone the intricacy of the human microbiome over a lifetime. The nature of precision medicine requires that advances move away from the testing of in vivo models which do not reflect human health. There is a rapid drive to develop in vitro models which accurately mimic human systems.For instance, a flow chamber culture system was developed to model human bladder infection in real-time to observe the phenotypic changes. These dynamic UPEC changes demonstrated a switching between filamentous or rod-shape forms to facilitate secondary infections, which could not be observed in standard in vivo or static in vitro models (Andersen et al., 2010; Andersen et al., 2012). (). In a follow-up study of the same cultured flow chamber system, E. coli was allowed to flow through a microfluidic system, in comparison to static growth (Stærk et al., 2015). A key difference was flowing E. coli expressed the adhesive type-1 fimbriae, as an essential regulator for bladder cell adhesion,colonization, biofilm formation, and eventually persistence in urothelium (Stærk et al., 2015). Recently, a novel microfluidic biosensor for monitoring and measuring electrical fluctuations caused by mechanical movements of single bacteria was developed (Kara et al., 2018).This model can uniquely employ patient-derived urine to study UPEC movement and susceptibility to various host-derived factors in real-time (Kara et al., 2018). These works illustrate the benefits of engineered microfluidic-based systems for UPEC time-course studies, in comparison to static models which do not accurately reflect infection in the human host.
Stem cells used to create human-like systems to model disease pathophysiology, towards drug discovery, and toxicity testing by developing cell lines relevant to humans for precision medicine (Singh et al., 2015). Advances in UTI modeling is becoming closer to reality as researchers design in vitro approaches to employ induced pluripotent stem cells from donors to differentiate stem cells into various cell types for creating organ-like structures. One patient-specific method surgically collects human urothelium from donors to create organoid cultures, relevant to a patient-specific bladder and urinary tract (Varley and Southgate, 2011). More recently, Horsley et al. (2018) presented a new method for organoid development which relies on urine for characterizing various infection types. This novel organoid model employs a human-based urothelium model as a platform to study the relationship between hosts and pathogens in a variety of environments (Horsley et al., 2018). Overall, these advances describe the rapid movement towards redefining human UTIs by transplanting patients into a laboratory setting, and vice-versa. In principle, this paves a path towards studying the interactions of host-specific cells with microbiota in application to personalized medicine.
Conclusion
This review challenges previous interpretations of UTI treatment and diagnosis to discuss UTIs as a host-centric disease, requiring a holistic approach. There is an overarching need to understand the uncomplicated UTIs as a dynamic system, complicated by host-associated environments and dysbiotic microbial communities. Integrating the ecological interactions between the host and microbial factors is necessary to progress rUTI diagnosis and therapy in the age of antimicrobial resistance. This integration includes observations of the female bladder microbiome in health and disease, host metabolic dysregulation, and dietary contaminants such as antibiotics or transition metals. This information can be processed through the combination of transcriptomics, in silico metabolic modeling or systems biology methods, and culturing induced pluripotent stem cells with microfluidic systems which are representative of the host. Overall, the presentation of the ‘uncomplicated environment of UTIs’ perspective can become an essential framework to further assess the effects of UTIs and their treatments on entire patients, rather than only the urinary tract. Thus advancing systems biology applications and precision medicine practices towards understanding UPEC pathology and other urogenital colonizing pathogens is ultimately necessary to effectively treat patients. The broad aspect of systems medicine in tandem with omics-based analysis and in vitro modeling can pave the way towards a mechanistic understanding of evidence-based diagnosis for rUTIs. To date, there are no systems medicine works on UTIs. Systems medicine is evolving more rapidly as available data becomes available and is translated into patient-specific models of biological systems. In particular, UTIs are not only one of the most common types of human infections but it is also poorly understood as a complicated system. That is why presently, scientists, biomedical engineers, physicians, and bioinformaticians have the best chance together, for combinatorial efforts towards employing precision treatments for rUTIs while maximizing benefits to patients.
Funding
We acknowledge financial support from the Land Schleswig-Holstein within the funding program Open Access Publication Fund”.
Statements
Author contributions
JJ-S drafted the manuscript. TK, HR, SG, ML, MC, SW, JR, JM-J, and CK provided extensive input and materials into the support of this work. JJ-S and TK drafted and refined the figures. All authors contributed to the article and approved the submitted version.
Acknowledgments
CK and JR acknowledge support by the German Research Foundation (Excellence Cluster “Precision Medicine in Chronic Inflammation” [EXC 2167] and Research Unit miTarget [FOR5042]). The authors thank Dr. Erik Beeler, Dr. Meghan F. Davis, and the Research Group Medical Systems Biology for insightful comments during the preparation of this manuscript. JJ-S also thanks Melissa Josephs-Spaulding and Thrity Spaulding for editing this manuscript and providing valuable feedback. The authors also thank Améline Lagasse for substantially redesigning and improving the quality of all figures in this manuscript
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AbatC.RaoultD.RolainJ.-M. (2015). Low Level of Resistance in Enterococci Isolated in Four Hospitals, Marseille, France. Microb. Drug Resist.22, 218–222. doi: 10.1089/mdr.2015.0121
2
AdenK.RehmanA.WaschinaS.PanW.-H.WalkerA.LucioM.et al. (2019). Metabolic Functions of Gut Microbes Associate With Efficacy of Tumor Necrosis Factor Antagonists in Patients With Inflammatory Bowel Diseases. Gastroenterology157 (5), 1279–1292. doi: 10.1053/j.gastro.2019.07.025
3
AlteriC. J.Mobley,. H. L. T. (2015). Metabolism and Fitness of Urinary Tract Pathogens. Microbiol. Spectr.3, 1–20. doi: 10.1128/microbiolspec.mbp-0016-2015
4
AltmanP. L. (1961). Physical Properties and Chemical Composition of Urine: Mammals. Part 1: Man. Blood other bodily fuids Fed. Am. Societies Exp. Biol Washington DC, 363–369.
5
AndersenT. E.KhandigeS.MadelungM.BrewerJ.KolmosH. J.Møller-JensenJ. (2012). Escherichia Coli Uropathogenesis In Vitro: Invasion, Cellular Escape, and Secondary Infection Analyzed in a Human Bladder Cell Infection Model. Infect Immun.80 (5), 1858–1867. doi: 10.1128/IAI.06075-11
6
AndersenT. E.KingshottP.PalarasahY.BenterM.AleiM.KolmosH. Jørn (2010). A Flow Chamber Assay for Quantitative Evaluation of Bacterial Surface Colonization Used to Investigate the Influence of Temperature and Surface Hydrophilicity on the Biofilm Forming Capacity of Uropathogenic Escherichia Coli. J. microbiol Methods81 (2), 135–140. doi: 10.1016/j.mimet.2010.02.009
7
AndersonG. G.PalermoJ. J.SchillingJ. D.RothR.HeuserJ.Hultgren,. S. J. (2003). Intracellular Bacterial Biofilm-Like Pods in Urinary Tract Infections. Science301, 105–107. doi: 10.1126/science.1084550
8
AngerJ.LeeU.Lenore AckermanA.ChouR.ChughtaiB.Quentin ClemensJ.et al. (2019). Recurrent Uncomplicated Urinary Tract Infections in Women: Aua/Cua/Sufu Guideline. J. Urol.202, 10–1097. doi: 10.1097/JU.0000000000000296
9
AragónI. M.Herrera-ImbrodaB.Queipo-OrtuñoMaríaI.CastilloE.Sequeira-García Del MoralJ.Gómez-MillánJ.et al. (2018). The Urinary Tract Microbiome in Health and Disease. Eur. Urol. Focus4 (1), 128–138. doi: 10.1016/j.euf.2016.11.001
10
ArnoldJ. J.HehnL. E.KleinD. A. (2016). Common Questions About Recurrent Urinary Tract Infections in Women. Am. Fam. Physician93, 560–569.
11
AshkarA. A.MossmanK. L.CoombesB. K.GylesC. L.Mackenzie,. R. (2008). Fimh Adhesin of Type 1 Fimbriae is a Potent Inducer of Innate Antimicrobial Responses Which Requires TLR4 and Type 1 Interferon Signalling. PloS Pathog.4, e1000233. doi: 10.1371/journal.ppat.1000233
12
AtacN.Kurt-AzapO.DolapciI.YesilkayaA.ErgonulO.GonenM.et al. (2018). The Role of AcrAB–TolC Efflux Pumps on Quinolone Resistance of E. Coli ST131. Curr. Microbiol.75, 1661–1666. doi: 10.1007/s00284-018-1577-y
13
American Urological Association, Adult UTI, (2016). Available at: https://www.auanet.org/education/adult-uti.cfm.
14
AzeredoJ.AzevedoN. F.BriandetR.CercaN.CoenyeT.CostaA. R.et al. (2017). Critical Review on Biofilm Methods. Crit. Rev. Microbiol.43 (3), 313–351. doi: 10.1080/1040841X.2016.1208146
15
BalamuruganveluS.GeethavaniB.ReddyS. V.SrikumarR.SundaramV. K. G. (2017). Incidence Microbiological Profile and Drug Resistance Pattern of Uropathogens Causing Asymptomatic Bacteriuria Among Below Poverty Line Diabetic Male Patients. Afr. J. Clin. Exp. Microbiol.18 (3), 145–153. doi: 10.4314/ajcem.v18i3.3
16
BartlM.KötzingM.SchusterS.LiPuKaletaC. (2013). Dynamic Optimization Identifies Optimal Programmes for Pathway Regulation in Prokaryotes. Nat. Commun.4, 2243. doi: 10.1038/ncomms3243
17
BartolettiR.CaiT.WagenlehnerF. M.NaberK.JohansenT. E. B. (2016). Treatment of Urinary Tract Infections and Antibiotic Stewardship. Eur. Urol. Suppl.15 (4), 81–87. doi: 10.1016/j.eursup.2016.04.003
18
BauckmanK. A.MatsudaR.HigginsC. B.DeBoschB. J.WangC.MysorekarI. U. (2019). Dietary Restriction of Iron Availability Attenuates UPEC Pathogenesis in a Mouse Model of Urinary Tract Infection. Am. J. Physiol. Renal Physiol.316, F814–F822. doi: 10.1152/ajprenal.00133.2018
19
BauerE.ThieleI. (2018). From Metagenomic Data to Personalized in Silico Microbiotas: Predicting Dietary Supplements for Crohn’s Disease. NPJ Syst. Biol. Appl.4 (1), 27. doi: 10.1038/s41540-018-0063-2
20
BauerE.ZimmermannJ.BaldiniF.ThieleI.KaletaC. (2017). BacArena: Individual-Based Metabolic Modeling of Heterogeneous Microbes in Complex Communities. PloS Comput. Biol.13 (5), e1005544. doi: 10.1371/journal.pcbi.1005544
21
BegerR. D.DunnW.SchmidtM. A.GrossS. S.KirwanJ. A.CascanteM.et al. (2016). Metabolomics Enables Precision Medicine:”A White Paper, Community Perspective. Metabolomics12 (9), 149. doi: 10.1007/s11306-016-1094-6
22
BettelheimK. A.BushrodF. M.ChandlerM. E.Mary CookeE.O’FarrellS.ShooterR. A. (1974). Escherichia Coli Serotype Distribution in Man and Animals. J. hygiene73 (3), 467. doi: 10.1017/s0022172400024086
23
Bjarnsholt,.T. (2013). The Role of Bacterial Biofilms in Chronic Infections. Apmis121, 1–58. doi: 10.1111/apm.12099
24
BouatraS.AziatF.MandalR.GuoAn C.WilsonM. R.KnoxC.et al. (2013). The Human Urine Metabolome. PloS One8 (9), e73076. doi: 10.1371/journal.pone.0073076
25
BourneJ. A.ChongW. L.Gordon,. D. M. (2019). Genetic Structure, Antimicrobial Resistance and Frequency of Human Associated Escherichia Coli Sequence Types Among Faecal Isolates From Healthy Dogs and Cats Living in Canberra, Australia. PloS One14, 1–13. doi: 10.1371/journal.pone.0212867
26
BrooksT.KeevilC. W. (1997). A Simple Artificial Urine for the Growth of Urinary Pathogens. Lett. Appl. Microbiol.24 (3), 203–206. doi: 10.1046/j.1472-765X.1997.00378.x
27
BrownC. T.OlmM. R.ThomasB. C.BanfieldJ. F.et al. (2016) Analysis Measurement of bacterial replication rates in microbial communitiesNat. Biotechnol.34, 1256–1263. doi: 10.1038/nbt.3704
28
BrubakerL.WolfeA. J. (2017). The Female Urinary Microbiota/Microbiome: Clinical and Research Implications. Rambam Maimonides Med. J.8 (2), 1–11. doi: 10.5041/RMMJ.10292
29
BrumbaughA. R.SmithS. N.Mobley,. H. L. (2013). Immunization With the Yersiniabactin Receptor, FyuA, Protects Against Pyelonephritis in a Murine Model of Urinary Tract Infection. Infect. Immun.81, 3309–3316. doi: 10.1128/IAI.00470-13
30
CaiT.NesiG.MazzoliS.MeacciF.LanzafameP.CaciagliP.et al. (2015). Asymptomatic Bacteriuria Treatment is Associated With a Higher Prevalence of Antibiotic Resistant Strains in Women With Urinary Tract Infections. Clin. Infect. Dis.61 (11), 1655–1661. doi: 10.1093/cid/civ696
31
CantasL.SuerK. (2014). The Important Bacterial Zoonoses in “One Health” Concept. Front. Public Health2, 144. doi: 10.3389/fpubh.2014.00144
32
CarettoM.GianniniA.RussoE.SimonciniT. (2017). Preventing Urinary Tract Infections After Menopause Without Antibiotics. Maturitas99, 43–46. doi: 10.1016/j.maturitas.2017.02.004
33
ChanR. C.BruceA. W.ReidG. (1984). Adherence of Cervical, Vaginal and Distal Urethral Normal Microbial Flora to Human Uroepithelial Cells and the Inhibition of Adherence of Gram-Negative Uropathogens by Competitive Exclusion. J. Urol.131, 596–601. doi: 10.1016/s0022-5347(17)50512-1
34
ChaturvediK. S.HungC. S.CrowleyJ. R.StapletonA. E.HendersonJ. P. (2012). The Siderophore Yersiniabactin Binds Copper to Protect Pathogens During Infection. Nat. Chem. Biol.8 (8), 731. doi: 10.1038/nchembio.1020
35
ChenS. L.WuM.HendersonJ. P.HootonT. M.HibbingM. E.HultgrenS. J.et al. (2013). Genomic Diversity and Fitness of E. Coli Strains Recovered From the Intestinal and Urinary Tracts of Women With Recurrent Urinary Tract Infection. Sci. Transl. Med.5, 184ra60. doi: 10.1126/scitranslmed.3005497
36
ChowdhuryN.SuhaniS.PurkaysthaA.BegumM. K.RaihanT.AlamM. J.et al. (2019). Identification of AcrAB-TolC Efflux Pump Genes and Detection of Mutation in Efflux Repressor AcrR From Omeprazole Responsive Multidrug-Resistant Escherichia Coli Isolates Causing Urinary Tract Infections. Microbiol. insights12:1178636119889629. doi: 10.1177/1178636119889629
37
ChuE. K.KilicO.ChoH.GroismanA.LevchenkoA. (2018). Self-Induced Mechanical Stress can Trigger Biofilm Formation in Uropathogenic Escherichia Coli. Nat. Commun.9 (1), 4087. doi: 10.1038/s41467-018-06552-z
38
ConoverM. S.HadjifrangiskouM.PalermoJ. J.HibbingM. E.DodsonK. W.HultgrenS. J. (2016). Metabolic Requirements of Escherichia Coli in Intracellular Bacterial Communities During Urinary Tract Infection Pathogenesis. MBio7 (2), e00104–e00116. doi: 10.1128/mBio.00104-16
39
CullenI. M.ManeckshaR. P.McCullaghE.AhmadS.O’KellyF.FlynnR. J.et al. (2012). The Changing Pattern of Antimicrobial Resistance Within 42,033 Escherichia Coli Isolates From Nosocomial, Community and Urology Patient-Specific Urinary Tract Infections, Dubli-2009. BJU Int.109, 1198–1206. doi: 10.1111/j.1464-410X.2011.10528
40
CurtissN.BalachandranA.KrskaL.Peppiatt–WildmanC.WildmanS.DuckettJ.et al. (2017). A Case Controlled Study Examining the Bladder Microbiome in Women With Overactive Bladder (OAB) and Healthy Controls. Eur. J. Obstetrics Gynecol Reprod. Biol.214, 31–355. doi: 10.1016/j.ejogrb.2017.04.040
41
DantasG.SommerM. O. A.OluwasegunR. D.Church,. G. M. (2008). Bacteria Subsisting on Antibiotics. Sci. (80-. )320, 100 LP – 103. doi: 10.1126/science.1155157
42
DavidL. A.MauriceC. F.CarmodyR. N.GootenbergD. B.ButtonJ. E.WolfeB. E.et al. (2014). Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature505, 559–563. doi: 10.1038/nature12820
43
DaviesJ.Davies,. D. (2010). Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev.74, 417 LP – 433. doi: 10.1128/MMBR.00016-10
44
DavisM. F.RankinS. C.SchurerJ. M.ColeS.ContiL.RabinowitzP.et al. (2017). Checklist for One Health Epidemiological Reporting of Evidence (COHERE). One Health4, 14–215. doi: 10.1016/j.onehlt.2017.07.001
45
DengZ.-L.GottschickC.BhujuS.MasurC.AbelsC.Wagner-DoeblerI. (2018). Metatranscriptome Analysis of the Vaginal Microbiota Reveals Potential Mechanisms for Protection Against Metronidazole in Bacterial Vaginosis. mSphere3 (3), e00262–e00218. doi: 10.1128/mSphereDirect.00262-18
46
DevilléW. L. J. M.YzermansJ. C.DuijnN. P.V.Dick BezemerP.van der WindtDaniëlleA. W. M.BouterL. M. (2004). The Urine Dipstick Test Useful to Rule Out Infections. A Meta-Analysis of the Accuracy. BMC Urol.4 (1), 4. doi: 10.1186/1471-2490-4-4
47
de VosM. G. J.ZagorskiM.McNallyA.BollenbachT. (2017). Interaction Networks, Ecological Stability, and Collective Antibiotic Tolerance in Polymicrobial Infections. Proc. Natl. Acad. Sci.114 (40), 10666–10671. doi: 10.1073/pnas.1713372114
48
DhakalB. K.MulveyM. A. (2012). The UPEC Pore-Forming Toxin α-Hemolysin Triggers Proteolysis of Host Proteins to Disrupt Cell Adhesion, Inflammatory, and Survival Pathways. Cell Host Microbe11 (1), 58–69. doi: 10.1016/j.chom.2011.12.003
49
DondersG. G. G.Van BulckB.Van de WalleP.KaiserR. R.PohligG.GonserS.et al. (2010). Effect of Lyophilized Lactobacilli and 0.03 Mg Estriol (Gynoflor®) on Vaginitis and Vaginosis With Disrupted Vaginal Microflora: A Multicenter, Randomized, Single-Blind, Active-Controlled Pilot Study. Gynecol Obstet Invest70, 264–272. doi: 10.1159/000314016
50
DuB.YangL.LloydC. J.FangX.Palsson,. B. O. (2019). Genome-Scale Model of Metabolism and Gene Expression Provides a Multi-Scale Description of Acid Stress Responses in Escherichia Coli. PloS Comput. Biol.15, e1007525. doi: 10.1371/journal.pcbi.1007525
51
EdwardsJ. S.PalssonB. O. (2000). The Escherichia Coli MG1655 in Silico Metabolic Genotype: its Definition, Characteristics, and Capabilities. Proc. Natl. Acad. Sci.97 (10), 5528–5533. doi: 10.1073/pnas.97.10.5528
52
EricksonK. E.WinklerJ. D.NguyenD. T.GillR. T.ChatterjeeA. (2017). The Tolerome: A Database of Transcriptome-Level Contributions to Diverse Escherichia Coli Resistance and Tolerance Phenotypes. ACS synthetic Biol.6 (12), 2302–2315. doi: 10.1021/acssynbio.7b00235
53
EwaldJ.BartlM.DandekarT.KaletaC. (2017). Optimality Principles Reveal a Complex Interplay of Intermediate Toxicity and Kinetic Efficiency in the Regulation of Prokaryotic Metabolism. PloS Comput. Biol.13 (2), e1005371. doi: 10.1371/journal.pcbi.1005371
54
EwersC.BetheA.SemmlerT.GuentherS.Wieler,. L. H. (2012). Extended-Spectrum β-Lactamase-Producing and AmpC-producing Escherichia Coli From Livestock and Companion Animals, and Their Putative Impact on Public Health: A Global Perspective. Clin. Microbiol. Infect. Off. Publ Eur. Soc Clin. Microbiol. Infect. Dis.18, 646–655. doi: 10.1111/j.1469-0691.2012.03850.x
55
FerreiroA.CrookN.GasparriniA. J.DantasG. (2018). Multiscale Evolutionary Dynamics of Host-Associated Microbiomes. Cell172, 1216–1227. doi: 10.1016/j.cell.2018.02.015
56
FestaR. A.ThieleD. J. (2011). Copper: An Essential Metal in Biology. Curr. Biol.21 (21), R877–R883. doi: 10.1016/j.cub.2011.09.040
57
Flores-MirelesA. L.WalkerJ. N.CaparonM.HultgrenS. J. (2015). Urinary Tract Infections: Epidemiology, Mechanisms of Infection and Treatment Options. Nat. Rev. Microbiol.13 (5), 269. doi: 10.1038/nrmicro3432
58
ForsythV. S.ArmbrusterC. E.SmithS. N.PiraniA.Cody SpringmanA.WaltersM. S.et al. (2018). Rapid Growth of Uropathogenic Escherichia Coli During Human Urinary Tract Infection. MBio9 (2), e00186–e00118. doi: 10.1128/mBio.00186-18
59
FoutsD. E.PieperR.SzpakowskiS.PohlH.KnoblachS.SuhM.-J.et al. (2012). Integrated Next-Generation Sequencing of 16S rDNA and Metaproteomics Differentiate the Healthy Urine Microbiome From Asymptomatic Bacteriuria in Neuropathic Bladder Associated With Spinal Cord Injury. J. Trans. Med.10 (1), 174. doi: 10.1186/1479-5876-10-174
60
FoxmanB. (2002). Epidemiology of Urinary Tract Infections: Incidence, Morbidity, and Economic Costs. Am. J. Med.113, 5–13. doi: 10.1016/S0002-9343(02)01054-9
61
FoxmanB. (2014). Urinary Tract Infection Syndromes: Occurrence, Recurrence, Bacteriology, Risk Factors, and Disease Burden. Infect. Dis. Clin. North Am.28, 1–13. doi: 10.1016/j.idc.2013.09.003
62
Frick-ChengA. E.SintsovaA.SmithS. N.KrauthammerM.EatonK. A.Mobley,. H. L. T. (2020). The Gene Expression Profile of Uropathogenic Escherichia Coli in Women With Uncomplicated Urinary Tract Infections Is Recapitulated in the Mouse Model. MBio11, e01412–e01420. doi: 10.1128/mBio.01412-20
63
GaoQ.WangX.XuH.XuY.LingJ.ZhangD.et al. (2012). Roles of Iron Acquisition Systems in Virulence of Extraintestinal Pathogenic Escherichia Coli: Salmochelin and Aerobactin Contribute More to Virulence Than Heme in a Chicken Infection Model. BMC Microbiol.12, 143. doi: 10.1186/1471-2180-12-143
64
García-JiménezB.GarcíaJoséL.NogalesJ. (2018). FLYCOP: Metabolic Modeling-Based Analysis and Engineering Microbial Communities. Bioinformatics34 (17), i954–i963. doi: 10.1093/bioinformatics/bty561
65
GilbertN. M.O’BrienV. P.LewisA. L. (2017). Transient Microbiota Exposures Activate Dormant Escherichia Coli Infection in the Bladder and Drive Severe Outcomes of Recurrent Disease. PloS Pathog.13 (3), e1006238. doi: 10.1371/journal.ppat.1006238
66
GiriS.WaschinaS.KaletaC.Kost,. C. (2019). Defining Division of Labor in Microbial Communities. J. Mol. Biol.431, 4712–4731. doi: 10.1016/j.jmb.2019.06.023
67
GniadkowskiM. (2008). Evolution of Extended-Spectrum β-Lactamases by Mutation. Clin. Microbiol. Infect.14, 11–32. doi: 10.1111/j.1469-0691.2007.01854.x
68
GodalyG.AmbiteI.PuthiaM.NadeemA.HoJ.NagyK.et al. (2016). Urinary Tract Infection Molecular Mechanisms and Clinical Translation. Pathogens5 (1), 24. doi: 10.3390/pathogens5010024
69
GoneauL. W.HannanT. J.MacPheeR. A.SchwartzD. J.MacklaimJ. M.BloorG. B.et al. (2015). Subinhibitory Antibiotic Therapy Alters Recurrent Urinary Tract Infection Pathogenesis through Modulation of Bacterial Virulence and Host Immunity. MBio6, 1–13. doi: 10.1128/mBio.00356-15
70
GottschickC.DengZ.-L.VitalM.MasurC.AbelsC.PieperD. H.et al. (2017). The Urinary Microbiota of Men and Women and its Changes in Women During Bacterial Vaginosis and Antibiotic Treatment. Microbiome5 (1), 99. doi: 10.1186/s40168-017-0305-3
71
GraspeuntnerS.WaschinaS.KünzelS.TwisselmannN.RauschT. K.Cloppenborg-SchmidtK.et al. (2018). Gut Dysbiosis With Bacilli Dominance and Accumulation of Fermentation Products Precedes Late-Onset Sepsis in Preterm Infants. Clin. Infect. Dis.69, 268–277. doi: 10.1093/cid/ciy882
72
GrochockiW.MarkuszewskiMichałJ.QuirinoJ. P. (2017). Simultaneous Determination of Creatinine and Acetate by Capillary Electrophoresis With Contactless Conductivity Detector as a Feasible Approach for Urinary Tract Infection Diagnosis. J. Pharm. Biomed. Anal.137, 178–181. doi: 10.1016/j.jpba.2017.01.032
73
HabibiM.KaramM. R. A.BouzariS. (2017). Evaluation of Prevalence, Immunogenicity and Efficacy of FyuA Iron Receptor in Uropathogenic Escherichia Coli Isolates as a Vaccine Target Against Urinary Tract Infection. Microbial pathogenesis110, 477–483. doi: 10.1016/j.micpath.2017.07.037
74
HadjifrangiskouM.HultgrenS. J. (2012). What Does it Take to Stick Around? Molecular Insights Into Biofilm Formation by Uropathogenic Escherichia Coli. Virulence3, 231–233. doi: 10.4161/viru.19763
75
HaganE. C.LloydA. L.RaskoD. A.FaerberG. J.MobleyH. L. T. (2010). Escherichia Coli Global Gene Expression in Urine From Women With Urinary Tract Infection. PloS Pathog.6, 1–18. doi: 10.1371/journal.ppat.1001187
76
HaganE. C.MobleyH. L. T. (2009). Haem Acquisition is Facilitated by a Novel Receptor Hma and Required by Uropathogenic Escherichia Coli for Kidney Infection. Mol. Microbiol.71 (1), 79–91. doi: 10.1111/j.1365-2958.2008.06509.x
77
HancockV.FerrieresL.KlemmP. (2008). The Ferric Yersiniabactin Uptake Receptor FyuA is Required for Efficient Biofilm Formation by Urinary Tract Infectious Escherichia Coli in Human Urine. Microbiology154 (1), 167–175. doi: 10.1099/mic.0.2007/011981-0
78
HaradaK.OkadaE.ShimizuT.KataokaY.SawadaT.TakahashiT. (2012). Antimicrobial Resistance, Virulence Profiles, and Phylogenetic Groups of Fecal Escherichia Coli Isolates: A Comparative Analysis Between Dogs and Their Owners in Japan. Comp. Immunol. Microbiol. Infect. Dis.35, 139–144. doi: 10.1016/j.cimid.2011.12.005
79
HasmanH.HammerumA. M.HansenF.HendriksenR. S.OlesenB.AgersøY.et al. (2015). Detection of Mcr-1 Encoding Plasmid-Mediated Colistin-Resistant Escherichia Coli Isolates From Human Bloodstream Infection and Imported Chicken Meat, Denmark 2015. Euro Surveill20 (49), 1–5. doi: 10.2807/1560-7917.ES.2015.20.49.30085
80
HeY.-Z.LiX.-P.MiaoY.-Y.LinJ.SunR.-Y.WangX.-P.et al. (2019). The ISApl12 Dimer Circular Intermediate Participates in Mcr-1 Transposition. Front. Microbiol.10, 15. doi: 10.3389/fmicb.2019.00015
81
HiltE. E.McKinleyK.PearceM. M.RosenfeldA. B.ZillioxM. J.MuellerE. R.et al. (2014). Urine is Not Sterile: Use of Enhanced Urine Culture Techniques to Detect Resident Bacterial Flora in the Adult Female Bladder. J. Clin. Microbiol.52 (3), 871–876. doi: 10.1128/JCM.02876-13
82
HironA.PosteraroB.CarrièreM.RemyL.DelporteCécileLa SordaM.et al. (2010). A Nickel ABC-transporter of Staphylococcus Aureus is Involved in Urinary Tract Infection. Mol. Microbiol.77 (5), 1246–1260. doi: 10.1111/j.1365-2958.2010.07287.x
83
HoffmanL. R.D’ArgenioD. A.MacCossM. J.ZhangZ.JonesR. A.MillerS. I. (2005). Aminoglycoside Antibiotics Induce Bacterial Biofilm Formation. Nature436 (7054), 1171. doi: 10.1038/nature03912
84
HootonT. M. (2012). Uncomplicated Urinary Tract Infection. N. Engl. J. Med.366, 1028–1037. doi: 10.1056/NEJMcp1104429
85
HootonT. M.BesserR.FoxmanB.FritscheT. R.NicolleL. E. (2004). Acute Uncomplicated Cystitis in an Era of Increasing Antibiotic Resistance: A Proposed Approach to Empirical Therapy. Clin. Infect. Dis.39 (1), 75–80. doi: 10.1086/422145
86
HorsleyH.DharmasenaD.Malone-LeeJ.RohnJ. L. (2018). A Urine-Dependent Human Urothelial Organoid Offers a Potential Alternative to Rodent Models of Infection. Sci. Rep.8 (1), 1238. doi: 10.1038/s41598-018-19690-7
87
HyreA. N.KavanaghK.KockN. D.DonatiG. L.SubashchandraboseS. (2017). Copper is a Host Effector Mobilized to Urine During Urinary Tract Infection to Impair Bacterial Colonization. Infect Immun.85 (3), e01041–e01016. doi: 10.1128/IAI.01041-16
88
ImdahlF.VafadarnejadE.HombergerC.SalibaA.-E.VogelJ. (2020). Single-Cell RNA-sequencing Reports Growth-Condition-Specific Global Transcriptomes of Individual Bacteria. Nat. Microbiol.5, 1202–1206. doi: 10.1038/s41564-020-0774-1
89
IpeD. S.HortonE.UlettG. C. (2016). The Basics of Bacteriuria: Strategies of Microbes for Persistence in Urine. Front. Cell. infect Microbiol.6, 14. doi: 10.3389/fcimb.2016.00014
90
IpeD. S.SundacL.BenjaminW. H.JrMooreK. H.UlettG. C. (2013). Asymptomatic Bacteriuria: Prevalence Rates of Causal Microorganisms, Etiology of Infection in Different Patient Populations, and Recent Advances in Molecular Detection. FEMS Microbiol. Lett.346 (1), 1–10. doi: 10.1111/1574-6968.12204
91
IrizarP. A.SchäubleS.EsserD.GrothM.FrahmC.PriebeS.et al. (2018). Transcriptomic Alterations During Ageing Reflect the Shift From Cancer to Degenerative Diseases in the Elderly. Nat. Commun.9 (1), 1–11. doi: 10.1038/s41467-019-10559-5
92
JamshidianM.CoombsR. J.RatnamS.MalhotraD. (2018). Medullary Sponge Kidney With Distal Renal Tubular Acidosis: A Case Report and Review of the Literature. SAJ Case Rep. 5: 204 Abstract Case Rep. Open Access5 (2), 1–5.
93
JantunenM. E.SaxenH.LukinmaaS.Ala-HouhalaM.SiitonenA. (2001). Genomic Identity of Pyelonephritogenic Escherichia Coli Isolated From Blood, Urine and Faeces of Children With Urosepsis. J. Med. Microbiol.50 (7), 650–652. doi: 10.1099/0022-1317-50-7-650
94
JozefczukS.KlieS.CatchpoleG.SzymanskiJ.Cuadros-InostrozaA.SteinhauserD.et al. (2010). Metabolomic and Transcriptomic Stress Response of Escherichia Coli. Mol. Syst. Biol.6 (1), 364. doi: 10.1038/msb.2010.18
95
JusticeS. S.HungC.TheriotJ. A.FletcherD. A.AndersonG. G.FooterM. J.et al. (2004). Differentiation and Developmental Pathways of Uropathogenic Escherichia Coli in Urinary Tract Pathogenesis. Proc. Natl. Acad. Sci. U. S. A.101, 1333–1338. doi: 10.1073/pnas.0308125100
96
KaraV.DuanC.GuptaK.KurosawaS.Stearns-KurosawaD. J.EkinciK. L. (2018). Microfluidic Detection of Movements of Escherichia Coli for Rapid Antibiotic Susceptibility Testing. Lab. Chip18 (5), 743–753. doi: 10.1039/C7LC01019B
97
KaratanE.WatnickP. (2009). Signals, Regulatory Networks, and Materials That Build and Break Bacterial Biofilms. Microbiol. Mol. Biol. Rev.73 (2), 310–347. doi: 10.1128/MMBR.00041-08
98
KhasheiiB.AnvariS.JamalliA. (2016). Frequency Evaluation of Genes Encoding Siderophores and the Effects of Different Concentrations of Fe Ions on Growth Rate of Uropathogenic Escherichia Coli. Iranian J. Microbiol.8 (6), 359.
99
KimMi Y.KimJi H.ChoM. H.ChoiY. H.KimS. H.ImY. J.et al. (2019). Urological Problems in Patients With Menkes Disease. J. Korean Med. Sci.34 (1), 1–8. doi: 10.3346/jkms.2019.34.e4
100
KingZ. A.LloydC. J.FeistA. M.BernhardO. (2015). Palsson. “Next-Generation Genome-Scale Models for Metabolic Engineering. Curr. Opin. Biotechnol.35, 23–29. doi: 10.1016/j.copbio.2014.12.016
101
KlitgordN.SegrèD. (2011). Ecosystems Biology of Microbial Metabolism. Curr. Opin. Biotechnol.22 (4), 541–546. doi: 10.1016/j.copbio.2011.04.018
102
KostakiotiM.HadjifrangiskouM.HultgrenS. J. (2013). Bacterial Biofilms: Development, Dispersal, and Therapeutic Strategies in the Dawn of the Postantibiotic Era. Cold Spring Harb. Perspect. Med.3:a010306. doi: 10.1101/cshperspect.a010306
103
KostakiotiM.HultgrenS. J.HadjifrangiskouM. (2012). Molecular Blueprint of Uropathogenic Escherichia Coli Virulence Provides Clues Toward the Development of Anti-Virulence Therapeutics. Virulence3 (7), 592–593. doi: 10.4161/viru.22364
104
KuchinaA.BrettnerL. M.PaleologuL.RocoC. M.RosenbergA. B.CarignanoA.et al. (2021). Microbial Single-Cell RNA Sequencing by Split-Pool Barcoding. Sci. (80-. ).371, eaba5257. doi: 10.1126/science.aba5257
105
LamC.-W.LawC.-Y.ToK. K.-W.CheungS. K.-K.LeeK.-c.SzeK.-H.et al. (2014). NMR-Based Metabolomic Urinalysis: A Rapid Screening Test for Urinary Tract Infection. Clinica chimica Acta436, 217–223. doi: 10.1016/j.cca.2014.05.014
106
LamontR. F.SobelJ. D.AkinsR. A.HassanS. S.ChaiworapongsaT.KusanovicJ. P.et al. (2011). The Vaginal Microbiome: New Information About Genital Tract Flora Using Molecular Based Techniques. BJOG: Int. J. Obstetrics Gynaecol118 (5), 533–549. doi: 10.1111/j.1471-0528.2010.02840.x
107
LéveilléS.CazaMélissaJohnsonJ. R.ClabotsC.SabriM.DozoisC. M. (2006). Iha From an Escherichia Coli Urinary Tract Infection Outbreak Clonal Group A Strain is Expressed In Vivo in the Mouse Urinary Tract and Functions as a Catecholate Siderophore Receptor. Infect Immun.74 (6), 3427–3436. doi: 10.1128/IAI.00107-06
108
LewisD. A.BrownR.WilliamsJ.WhiteP.JacobsonS. K.MarchesiJ.et al. (2013). The Human Urinary Microbiome; Bacterial DNA in Voided Urine of Asymptomatic Adults. Front. Cell. infect Microbiol.3, 41. doi: 10.3389/fcimb.2013.00041
109
LintonA. H.HoweK.BennettP. M.RichmondM. H.WhitesideE. J. (1977). The Colonization of the Human Gut by Antibiotic Resistant Escherichia Coli From Chickens. J. Appl. Bacteriol43 (3), 465–469. doi: 10.1111/j.1365-2672.1977.tb00773.x
110
LiuF.LingZ.XiaoY.LvL.YangQ.WangB.et al. (2017). Dysbiosis of Urinary Microbiota is Positively Correlated With Type 2 Diabetes Mellitus. Oncotarget8 (3), 3798. doi: 10.18632/oncotarget.14028
111
LiuC. M.SteggerM.AzizM.JohnsonT. J.WaitsK.NordstromL.et al. (2018). Escherichia Coli ST131-H22 as a Foodborne Uropathogen. MBio9 (4), e00470–e00418. doi: 10.1128/mBio.00470-18
112
LiuY.-Y.WangY.WalshT. R.YiL.-X.ZhangR.SpencerJ.et al. (2016). Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis.16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
113
LloydK. G.SteenA. D.LadauJ.YinJ.CrosbyL. (2018). Phylogenetically Novel Uncultured Microbial Cells Dominate Earth Microbiomes. MSystems3 (5), e00055–e00018. doi: 10.1128/mSystems.00055-18
114
López-MauryL.MargueratS.BählerJürg (2008). Tuning Gene Expression to Changing Environments: From Rapid Responses to Evolutionary Adaptation. Nat. Rev. Genet.9 (8), 583. doi: 10.1038/nrg2398
115
LuoY.MaY.ZhaoQ.WangL.GuoL.YeL.et al. (2012). Similarity and Divergence of Phylogenies, Antimicrobial Susceptibilities, and Virulence Factor Profiles of Escherichia Coli Isolates Causing Recurrent Urinary Tract Infections That Persist or Result From Reinfection. J. Clin. Microbiol.50 (12), 1–6. doi: 10.1128/JCM.02086-12
116
LuoX.YangX.LiJ.ZouGeLinY.QingG.et al. (2018). The Procalcitonin/Albumin Ratio as an Early Diagnostic Predictor in Discriminating Urosepsis From Patients With Febrile Urinary Tract Infection. Medicine97 (28). doi: 10.1097/MD.0000000000011078
117
LüthjeP.BraunerH.RamosN. L.ÖvregaardA.GläserR.HirschbergA. L.et al. (2013). Estrogen Supports Urothelial Defense Mechanisms. Sci. Transl. Med.5, 190ra80 LP–190ra80. doi: 10.1126/scitranslmed.3005574
118
MachadoAntónioJeffersonK.CercaN. (2013). nteractions Between Lactobacillus Crispatus and Bacterial Vaginosis (BV)-Associated Bacterial Species in Initial Attachment and Biofilm Formation. Int. J. Mol. Sci.14 (6), 12004–12012. doi: 10.3390/ijms140612004
119
MagruderM.SholiA. N.GongC.ZhangL.EduseiE.HuangJ.et al. (2019). Gut Uropathogen Abundance is a Risk Factor for Development of Bacteriuria and Urinary Tract Infection. Nat. Commun.10, 1–9. doi: 10.1038/s41467-019-13467-w
120
Malhotra-KumarS.XavierB. B.DasA. J.LammensC.ButayeP.GoossensH. (2016). Colistin Resistance Gene Mcr-1 Harboured on a Multidrug Resistant Plasmid. Lancet Infect. Dis.16, 283–284. doi: 10.1016/S1473-3099(16)00012-8
121
MangesA. R.SmithS. P.LauB. J.NuvalC. J.EisenbergJ. N. S.DietrichP. S.et al. (2007). Retail Meat Consumption and the Acquisition of Antimicrobial Resistant Escherichia Coli Causing Urinary Tract Infections: A Case–Control Study. Foodborne Pathog. Dis.4 (4), 419–431. doi: 10.1089/fpd.2007.0026
122
MarchalM.GoldschmidtF.Derksen-MüllerS. N.PankeS.AckermannM.JohnsonD. R. (2017). A Passive Mutualistic Interaction Promotes the Evolution of Spatial Structure Within Microbial Populations. BMC Evol. Biol.17, 106. doi: 10.1186/s12862-017-0950-y
123
MarquesC.BelasA.AboimC.Cavaco-SilvaP.TrigueiroG.GamaL. T.et al. (2019). Evidence of Sharing of Klebsiella Pneumoniae Strains Between. J. Clin. Microbiol.57, 1–12. doi: 10.1128/JCM.01537-18
124
MartinH. L.RichardsonB. A.NyangeP. M.LavreysL.HillierS. L.ChohanB.et al. (1999). Vaginal Lactobacilli, Microbial Flora, and Risk of Human Immunodeficiency Virus Type 1 and Sexually Transmitted Disease Acquisition. J. Infect. Dis.180, 1863–1868. doi: 10.1086/315127
125
Martín-RodríguezA. J.RhenM.MelicanK.Richter-DahlforsA. (2020). Nitrate Metabolism Modulates Biosynthesis of Biofilm Components in Uropathogenic Escherichia coli and Acts as a Fitness Factor During Experimental Urinary Tract Infection. Front. Microbiol.11, 26. doi: 10.3389/fcimb.2020.00026
126
MathersA. J.PeiranoG.PitoutJ. D. D. (2015). The Role of Epidemic Resistance Plasmids and International High-Risk Clones in the Spread of Multidrug-Resistant Enterobacteriaceae. Clin. Microbiol. Rev.28, 565 LP – 591. doi: 10.1128/CMR.00116-14
127
McGannP.SnesrudE.MaybankR.CoreyB.OngA. C.CliffordR.et al. (2016). Escherichia Coli Harboring Mcr-1 and blaCTX-M on a Novel IncF Plasmid: First Report of Mcr-1 in the United States. Antimicrobial Agents Chemother60 (7), 4420–4421. doi: 10.1128/AAC.01103-16
128
MediavillaJ. R.PatrawallaA.ChenL.ChavdaK. D.MathemaB.VinnardC.et al. (2016). Colistin- and Carbapenem-Resistant Escherichia Coli Harboring Mcr-1 and Blandm-5, Causing a Complicated Urinary Tract Infection in a Patient From the United States. MBio7, e01191–e01116. doi: 10.1128/mBio.01191-16
129
MeierT. R.MauteC. J.CadillacJ. M.LeeJi Y.RighterD. J.HuguninK.et al. (2008). Quantification, Distribution, and Possible Source of Bacterial Biofilm in Mouse Automated Watering Systems. J. Am. Assoc. Lab. Anim. Sci.47 (2), 63–70.
130
MeriwetherK. V.LeiZ.SinghR.GaskinsJ.HobsonD. T. G.JalaV. (2019). The Vaginal and Urinary Microbiomes in Premenopausal Women With Interstitial Cystitis/Bladder Pain Syndrome as Compared to Unaffected Controls: A Pilot Cross-Sectional Study. Front. Cell. Infect Microbiol.9, 925. doi: 10.3389/fcimb.2019.00092
131
MickE.SorekR. (2014). High-Resolution Metagenomics. Nat. Biotechnol.32, 750–751. doi: 10.1038/nbt.2962
132
MirmonsefP.HottonA. L.GilbertD.BurgadD.LandayA.WeberK. M.et al. (2014). Free Glycogen in Vaginal Fluids is Associated With Lactobacillus Colonization and Low Vaginal Ph. PloS One9 (7), e102467. doi: 10.1371/journal.pone.0102467
133
MobleyH. (2016). Measuring Escherichia Coli Gene Expression During Human Urinary Tract Infections. Pathogens5 (1), 7. doi: 10.3390/pathogens5010007
134
ModyL.Juthani-MehtaM. (2014). Urinary Tract Infections in Older Women: A Clinical Review. Jama311 (8), 844–854. doi: 10.1001/jama.2014.303
135
MomeniB.BrileyaK. A.FieldsM. W.ShouW. (2013). Strong Inter-Population Cooperation Leads to Partner Intermixing in Microbial Communities. elife2, e00230. doi: 10.7554/eLife.00230
136
MueckA. O.RuanX.PrasauskasV.GrobP.OrtmannO. (2018). Treatment of Vaginal Atrophy With Estriol and Lactobacilli Combination: A Clinical Review. Climacteric21 (2), 140–147. doi: 10.1080/13697137.2017.1421923
137
MuhleisenA. L.Herbst-KralovetzM. M. (2016). Menopause and the Vaginal Microbiome. Maturitas91, 42–50. doi: 10.1016/j.maturitas.2016.05.015
138
NaziriZ.DerakhshandehA.FirouziR.MotamedifarM.Shojaee TabriziA. (2016). DNA Fingerprinting Approaches to Trace Escherichia Coli Sharing Between Dogs and Owners. J. Appl. Microbiol.120, 460–468. doi: 10.1111/jam.13003
139
NelsonD. E.DongQ.van der PolB.TohE.FanB.KatzB. P.et al. (2012). Bacterial Communities of the Coronal Sulcus and Distal Urethra of Adolescent Males. PloS One7, e36298. doi: 10.1371/journal.pone.0036298
140
NielsenK. L.SteggerM.GodfreyP. A.FeldgardenM.AndersenP. S.Frimodt-MøllerN. (2016). Adaptation of Escherichia Coli Traversing From the Faecal Environment to the Urinary Tract. Int. J. Med. Microbiol.306 (8), 595–603. doi: 10.1016/j.ijmm.2016.10.005
141
NordstromL.LiuC. M.PriceL. B. (2013). Foodborne Urinary Tract Infections: A New Paradigm for Antimicrobial-Resistant Foodborne Illness. Front. Microbiol.4, 29. doi: 10.3389/fmicb.2013.00029
142
O’BrienE. J.MonkJ. M.PalssonB. O. (2015). Using Genome-Scale Models to Predict Biological Capabilities. Cell161 (5), 971–987. doi: 10.1016/j.cell.2015.05.019
143
O’HanlonD. E.ComeR. A.MoenchT. R. (2019). Vaginal Ph Measured In Vivo: Lactobacilli Determine Ph and Lactic Acid Concentration. BMC Microbiol.19 (1), 13. doi: 10.1186/s12866-019-1388-8
144
O’HanlonD. E.MoenchT. R.ConeR. A. (2011). In Vaginal Fluid, Bacteria Associated With Bacterial Vaginosis can be Suppressed With Lactic Acid But Not Hydrogen Peroxide. BMC Infect. Dis.11 (1), 200. doi: 10.1186/1471-2334-11-200
145
OnderdonkA. B.DelaneyM. L.FichorovaR. N. (2016). The Human Microbiome During Bacterial Vaginosis. Clin. Microbiol. Rev.29 (2), 223–238. doi: 10.1128/CMR.00075-15
146
OrthJ. D.ConradT. M.NaJ.LermanJ. A.NamH.FeistA. M.et al. (2011). A Comprehensive Genome-Scale Reconstruction of Escherichia Coli Metabolism—2011. Mol. Syst. Biol.7 (1), 535. doi: 10.1038/msb.2011.65
147
OsuguiL.Pestana de CastroA. F.IovineR.IrinoK.CarvalhoV. M. (2014). Virulence Genotypes, Antibiotic Resistance and the Phylogenetic Background of Extraintestinal Pathogenic Escherichia Coli Isolated From Urinary Tract Infections of Dogs and Cats in Brazil. Vet. Microbiol.171, 242–247. doi: 10.1016/j.vetmic.2014.03.027
148
PaalanneN.HussoA.SaloJ.PieviläinenO.TejesviM. V.KoivusaariP.et al. (2018). Intestinal Microbiome as a Risk Factor for Urinary Tract Infections in Children. Eur. J. Clin. Microbiol. Infect. Dis.37 (10), 1881–1891. doi: 10.1007/s10096-018-3322-7
149
PapenfortK.BasslerB. L. (2016). Quorum Sensing Signal–Response Systems in Gram-negative Bacteria. Nat. Rev. Microbiol.14 (9), 576. doi: 10.1038/nrmicro.2016.89
150
PelzerE. S.AllanJ. A.TheodoropoulosC.RossT.BeagleyK. W.KnoxC. L. (2012). Hormone-Dependent Bacterial Growth, Persistence and Biofilm Formation–a Pilot Study Investigating Human Follicular Fluid Collected During IVF Cycles. PloS One7 (12), e49965. doi: 10.1371/journal.pone.0049965
151
PenesyanA.NagyS. S.KjellebergS.GillingsM. R.PaulsenI. T. (2019). Rapid Microevolution of Biofilm Cells in Response to Antibiotics. NPJ biofilms microbiomes5 (1), 1–14. doi: 10.1038/s41522-019-0108-3
152
PillaiS. K.SakoulasG.EliopoulosG. M.MoelleringR. C.JrMurrayB. E.InouyeR. T. (2004). Effects of Glucose on Fsr-Mediated Biofilm Formation in Enterococcus Faecalis. J. Infect. Dis.190 (5), 967–970. doi: 10.1086/423139
153
PlaydonM. C.SampsonJ. N.CrossA. J.SinhaR.GuertinK. A.MoyK. A.et al. (2016). Comparing Metabolite Profiles of Habitual Diet in Serum and Urine. Am. J. Clin. Nutr.104, 776–789. doi: 10.3945/ajcn.116.135301
154
PombaC.RantalaM.GrekoC.BaptisteK. E.CatryB.van DuijkerenE.et al. (2017). Public Health Risk of Antimicrobial Resistance Transfer From Companion Animals. J. Antimicrob. Chemother.72, 957–968. doi: 10.1093/jac/dkw481
155
PrattL. A.KolterR. (1998). Genetic Analysis of Escherichia Coli Biofilm Formation: Roles of Flagella, Motility, Chemotaxis and Type I Pili. Mol. Microbiol.30 (2), 285–293. doi: 10.1046/j.1365-2958.1998.01061.x
156
PruetpongpunN.KhawcharoenpornT.DamronglerdP.SuwantaratN.ApisarnthanarakA.RutjanawechS. (2017). Inappropriate Empirical Treatment of Uncomplicated Cystitis in Thai Women: Lessons Learned. Clin. Infect. Dis.64 (suppl_2), S115–S118. doi: 10.1093/cid/cix088
157
PryorR.NorvaisasP.MarinosG.BestL.ThingholmL. B.QuintaneiroL. M.et al. (2019). Host-Microbe-Drug-Nutrient Screen Identifies Bacterial Effectors of Metformin Therapy. Cell178 (6), 1299–1312. doi: 10.1016/j.cell.2019.08.003
158
RaniA.RanjanR.McGeeH. S.AndropolisK. E.PanchalD. V.HajjiriZ.et al. (2017). Urinary Microbiome of Kidney Transplant Patients Reveals Dysbiosis With Potential for Antibiotic Resistance. Trans. Res.181, 59–70. doi: 10.1016/j.trsl.2016.08.008
159
RavelJ.GajerP.AbdoZ.SchneiderG. M.KoenigS. S. K.McCulleS. L.et al. (2011). Vaginal Microbiome of Reproductive-Age Women. Proc. Natl. Acad. Sci. U. S. A.108 (Suppl 1), 4680–4687. doi: 10.1073/pnas.1002611107
160
RemyL.CarrièreM.Derré-BobillotAurélieMartiniCéciliaSanguinettiM.Borezée-DurantE. (2013). The Staphylococcus Aureus Opp1 ABC Transporter Imports Nickel and Cobalt in Zinc-Depleted Conditions and Contributes to Virulence. Mol. Microbiol.87 (4), 730–743. doi: 10.1111/mmi.12126
161
RiceJ. C.PengT.SpenceJ. S.WangH.-Q.GoldblumR. M.CorthésyB.et al. (2005). Pyelonephritic Escherichia Coli Expressing P Fimbriae Decrease Immune Response of the Mouse Kidney. J. Am. Soc Nephrol16, 3583 LP – 3591. doi: 10.1681/ASN.2005030243
162
RobinsonA. E.LoweJ. E.KohE.-I.HendersonJ. P. (2018). Uropathogenic Enterobacteria Use the Yersiniabactin Metallophore System to Acquire Nickel. J. Biol. Chem.293 (39), 14953–14961. doi: 10.1074/jbc.RA118.004483
163
RoseC.ParkerA.JeffersonB.CartmellE. (2015). The Characterization of Feces and Urine: A Review of the Literature to Inform Advanced Treatment Technology. Crit. Rev. Environ. Sci. Technol.45 (17), 1827–1879. doi: 10.1080/10643389.2014.1000761
164
RossoliniG. M.ArenaF.PecileP.PolliniS. (2014). Update on the Antibiotic Resistance Crisis. Curr. Opin. Pharmacol.18, 56–60. doi: 10.1016/j.coph.2014.09.006
165
RuizC.LevyS. B. (2010). Many Chromosomal Genes Modulate Mara-Mediated Multidrug Resistance in Escherichia Coli. Antimicrob. Agents Chemother.54, 2125–2134. doi: 10.1128/AAC.01420-09
166
SabriM.HouleSébastienDozoisC. M. (2009). Roles of the Extraintestinal Pathogenic Escherichia Coli ZnuACB and ZupT Zinc Transporters During Urinary Tract Infection. Infect Immun.77 (3), 1155–1164. doi: 10.1128/IAI.01082-08
167
SalverdaM. L. M.KoomenJ.KoopmanschapB.ZwartM. P.de VisserJ. A. G. M. (2017). Adaptive Benefits From Small Mutation Supplies in an Antibiotic Resistance Enzyme. Proc. Natl. Acad. Sci.114, 12773– 12778. doi: 10.1073/pnas.1712999114
168
SaxenaD.LiY.YangL.PeiZ.PolesM.AbramsW. R.et al. (2012). Human Microbiome and HIV/AIDS. Curr. HIV/AIDS Rep.9 (1), 44–51. doi: 10.1007/s11904-011-0103-7
169
SchitoG. C.NaberK. G.BottoH.PalouJ.MazzeiT.GualcoL.et al. (2009). The ARESC Study: An International Survey on the Antimicrobial Resistance of Pathogens Involved in Uncomplicated Urinary Tract Infections. Int. J. antimicrobial Agents34 (5), 407–413. doi: 10.1016/j.ijantimicag.2009.04.012
170
ScholesD.HootonT. M.RobertsP. L.StapletonA. E.GuptaK.StammW. E. (2000). Risk Factors for Recurrent Urinary Tract Infection in Young Women. J. Infect. Dis.182 (4), 1177–1182. doi: 10.1086/315827
171
SchreiberH. L.ConoverM. S.ChouW.-C.HibbingM. E.MansonA. L.DodsonK. W.et al. (2017). Bacterial Virulence Phenotypes of Escherichia Coli and Host Susceptibility Determine Risk for Urinary Tract Infections. Sci. Trans. Med.9 (382), eaaf1283. doi: 10.1126/scitranslmed.aaf1283
172
SchusterS.VavraM.SchweiggerT. M.RossenJ. W. A.MatsumuraY.KernW. V. (2017). Contribution of AcrAB-TolC to Multidrug Resistance in an Escherichia Coli Sequence Type 131 Isolate. Int. J. Antimicrob. Agents50, 477–481. doi: 10.1016/j.ijantimicag.2017.03.023
173
SchwartzM. H.WaldbauerJ. R.ZhangL.PanT. (2016). Global tRNA Misacylation Induced by Anaerobiosis and Antibiotic Exposure Broadly Increases Stress Resistance in Escherichia Coli. Nucleic Acids Res.44 (21), 10292–10303. doi: 10.1093/nar/gkw856
174
SeoS. W.KimD.SzubinR.PalssonB. O. (2015). Genome-Wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks Under Oxidative Stress in Escherichia Coli K-12 Mg1655. Cell Rep.12 (8), 1289–1299. doi: 10.1016/j.celrep.2015.07.043
175
SewankamboN.GrayR. H.WawerM. J.PaxtonL.McNaimD.Wabwire-MangenF.et al. (1997). HIV-1 Infection Associated With Abnormal Vaginal Flora Morphology and Bacterial Vaginosis. Lancet (London England)350, 546–550. doi: 10.1016/s0140-6736(97)01063-5
176
ShakyaM.LoC. C.ChainP. S. G. (2019). Advances and Challenges in Metatranscriptomic Analysis. Front. Genet.10:904. doi: 10.3389/fgene.2019.00904
177
SharmaG.SharmaS.SharmaP.ChandolaD.DangS.GuptaS.et al. (2016). Escherichia Coli Biofilm: Development and Therapeutic Strategies. J. Appl. Microbiol.121, 309–319. doi: 10.1111/jam.13078
178
Shields-CutlerR. R.CrowleyJ. R.HungC. S.StapletonA. E.AldrichC. C.MarschallJ.et al. (2015). Human Urinary Composition Controls Antibacterial Activity of Siderocalin*, ♦. J. Biol. Chem.290, 15949–15960. doi: 10.1074/jbc.M115.645812
179
ShooterR. A.RousseauS. A.Mary CookeE.BreadenA. (1970). Animal Sources of Common Serotypes of Escherichia Coli in the Food of Hospital Patients Possible Significance in Urinary-Tract Infections. Lancet296 (7666), 226–228. doi: 10.1016/s0140-6736(70)92583-3
180
ShoskesD. A.AltemusJ.PolackwichA. S.TuckyB.WangH.EngC. (2016). The Urinary Microbiome Differs Significantly Between Patients With Chronic Prostatitis/Chronic Pelvic Pain Syndrome and Controls as Well as Between Patients With Different Clinical Phenotypes. Urology92, 26–32. doi: 10.1016/j.urology.2016.02.043
181
ShresthaE.WhiteJ. R.YuS.-H.KulacI.ErtuncO.De MarzoA. M.et al. (2018). Profiling the Urinary Microbiome in Men With Positive Versus Negative Biopsies for Prostate Cancer. J. Urol.199 (1), 161–171. doi: 10.1016/j.juro.2017.08.001
182
SingerR. S. (2015). Urinary Tract Infections Attributed to Diverse ExPEC Strains in Food Animals : Evidence and Data Gaps. Front. Microbiol6, 1–9. doi: 10.3389/fmicb.2015.00028
183
SinghV. K.KalsanM.KumarN.SainiA.ChandraR. (2015). Induced Pluripotent Stem Cells: Applications in Regenerative Medicine, Disease Modeling, and Drug Discovery. Front. Cell Dev. Biol.3, 2. doi: 10.3389/fcell.2015.00002
184
SintsovaFrick-ChengA. E.SmithS.PiraniA. (2019). Subashchandrabose Sargurunathan, and Harry Mobley. “Genetically Diverse Uropathogenic Escherichia Coli Adopt a Common Transcriptional Program in Patients With Utis. eLife8, 1–26. doi: 10.7554/eLife.49748
185
SintsovaA.SmithS.SubashchandraboseS.MobleyH. L. (2018). Role of Ethanolamine Utilization Genes in Host Colonization During Urinary Tract Infection. Infect Immun.86 (3), e00542–e00517. doi: 10.1128/IAI.00542-17
186
SmartA.de Lacy CostelloB.WhiteP.AvisonM.BattyC.TurnerC.et al. (2019). Sniffing Out Resistance – Rapid Identification of Urinary Tract Infection-Causing Bacteria and Their Antibiotic Susceptibility Using Volatile Metabolite Profiles. J. Pharm. Biomed. Anal.167, 59–65. doi: 10.1016/j.jpba.2019.01.044
187
SpurbeckR. R.StapletonA. E.JohnsonJ. R.WalkS. T.HootonT. M.MobleyH. L. T. (2011). Fimbrial Profiles Predict Virulence of Uropathogenic Escherichia Coli Strains: Contribution of Ygi and Yad Fimbriae. Infect Immun.79 (12), 4753–4763. doi: 10.1128/IAI.05621-11
188
StærkK.KhandigeS.KolmosH. JørnMøller-JensenJ.AndersenT. E. (2015). Uropathogenic Escherichia Coli Express Type 1 Fimbriae Only in Surface Adherent Populations Under Physiological Growth Conditions. J. Infect. Dis.213 (3), 386–394. doi: 10.1093/infdis/jiv422
189
StapletonA. E.Au-YeungM.HootonT. M.FredricksD. N.RobertsP. L.CzajaC. A.et al. (2011). Randomized, Placebo-Controlled Phase 2 Trial of a Lactobacillus Crispatus Probiotic Given Intravaginally for Prevention of Recurrent Urinary Tract Infection. Clin. Infect. Dis.52, 1212–1217. doi: 10.1093/cid/cir183
190
StenskeK. A.BemisD. A.GillespieB. E.D’SouzaD. H.OliverS. P.DraughonF. A.et al. (2009). Comparison of Clonal Relatedness and Antimicrobial Susceptibility of Fecal Escherichia Coli From Healthy Dogs and Their Owners. Am. J. Vet. Res.70, 1108–1116. doi: 10.2460/ajvr.70.9.1108
191
StewartT. J.TraberJ.KrollA.BehraR.SiggL. (2013). Characterization of Extracellular Polymeric Substances (EPS) From Periphyton Using Liquid Chromatography-Organic Carbon Detection–Organic Nitrogen Detection (LC-OCD-OND). Environ. Sci. Pollution Res.20 (5), 3214–3223. doi: 10.1007/s11356-012-1228-y
192
SubashchandraboseS.HazenT. H.BrumbaughA. R.HimpslS. D.SmithS. N.ErnstR. D.et al. (2014). Host-Specific Induction of Escherichia Coli Fitness Genes During Human Urinary Tract Infection. Proc. Natl. Acad. Sci.111 (51), 18327–18332. doi: 10.1073/pnas.1415959112
193
SubashchandraboseS.MobleyH. L. T. (2015). Back to the Metal Age: Battle for Metals at the Host–Pathogen Interface During Urinary Tract Infection. Metallomics7 (6), 935–942. doi: 10.1039/C4MT00329B
194
TachedjianG.AldunateM.BradshawC. S.ConeR. A. (2017). The Role of Lactic Acid Production by Probiotic Lactobacillus Species in Vaginal Health. Res. Microbiol.168 (9-10), 782–792. doi: 10.1016/j.resmic.2017.04.001
195
TangJ. (2017). Microbiome in the Urinary System—a Review. AIMS Microbiol. (2017) 32:143–154. doi: 10.3934/microbiol.2017.2.143
196
TangM.ChenZ.WuDiChenL. (2018). Ferritinophagy/Ferroptosis: Iron-related Newcomers in Human Diseases. J. Cell. Physiol.233 (12), 9179–9190. doi: 10.1002/jcp.26954
197
ThänertR.ReskeK. A.HinkT.WallaceM. A.WangB.SchwartzD. J.et al. (2019). Comparative Genomics of Antibiotic-Resistant Uropathogens Implicates Three Routes for Recurrence of Urinary Tract Infections. mBio10 (4), e01977–e01919. doi: 10.1128/mBio.01977-19
198
ThieleI.HeinkenA.FlemingR. M. T. (2013). A Systems Biology Approach to Studying the Role of Microbes in Human Health. Curr. Opin. Biotechnol.24, 4–12. doi: 10.1016/j.copbio.2012.10.001
199
ThieleI.SahooS.HeinkenA.HertelJ.HeirendtL.AurichM. K.et al. (2020). Personalized Whole-Body Models Integrate Metabolism, Physiology, and the Gut Microbiome. Mol. Syst. Biol.16, e8982. doi: 10.15252/msb.20198982
200
Thomas-WhiteK.ForsterS. C.KumarN.Van KuikenM.PutontiC.StaresM. D.et al. (2018). Culturing of Female Bladder Bacteria Reveals an Interconnected Urogenital Microbiota. Nat. Commun.9, 1557. doi: 10.1038/s41467-018-03968-5
201
TümerZ.MøllerL. B. (2010). Menkes Disease. Eur. J. Hum. Genet.18 (5), 511. doi: 10.1038/ejhg.2009.187
202
van der ZeeA.KraakW. B.BurggraafA.GoessensW. H. F.PirovanoW.OssewaardeJ. M.et al. (2018). Spread of Carbapenem Resistance by Transposition and Conjugation Among Pseudomonas Aeruginosa. Front. Microbiol.9, 2057. doi: 10.3389/fmicb.2018.02057
203
van de WijgertJ. H. H. M.BorgdorffH.VerhelstR.CrucittiT.FrancisS.VerstraelenH.et al. (2014). The Vaginal Microbiota: What Have We Learned After a Decade of Molecular Characterization? PloS One9 (8), e105998. doi: 10.1371/journal.pone.0105998
204
VarleyC. L.SouthgateJ. (2011). “Organotypic and 3D Reconstructed Cultures of the Human Bladder and Urinary Tract,” in 3d Cell CultureTotowa, New Jersey (Humana Press), 197–211.
205
VellingaA.TanseyS.HanahoeB.BennettK.MurphyA. W.CormicanM. (2012). Trimethoprim and Ciprofloxacin Resistance and Prescribing in Urinary Tract Infection Associated With Escherichia Coli: A Multilevel Model. J. Antimicrob. Chemother.67, 2523–2530. doi: 10.1093/jac/dks222
206
VersportenA.ZarbP.CaniauxI.GrosMarie-FrançoiseDrapierN.MillerM.et al. (2018). Antimicrobial Consumption and Resistance in Adult Hospital Inpatients in 53 Countries: Results of an Internet-Based Global Point Prevalence Survey. Lancet Global Health6 (6), e619–e629. doi: 10.1016/S2214-109X(18)30186-4
207
VincentC.BoerlinP.DaignaultD.DozoisC. M.DutilL.GalanakisC.et al. (2010). Food Reservoir for Escherichia Coli Causing Urinary Tract Infections. Emerging Infect. Dis.16 (1), 88. doi: 10.3201/eid1601.091118
208
VodstrcilL. A.TwinJ.GarlandS. M.FairleyC. K.HockingJ. S.LawM. G.et al. (2017). The Influence of Sexual Activity on the Vaginal Microbiota and Gardnerella Vaginalis Clade Diversity in Young Women. PloS One12 (2), e0171856. doi: 10.1371/journal.pone.0171856
209
WangJ.HuangX.-Y.XiaY.-B.GuoZ.-W.MaZ.-B.YiM.-Y.et al. (2018). Clonal Spread of Escherichia Coli ST93 Carrying Mcr-1-Harboring IncN1-IncHI2/ST3 Plasmid Among Companion Animals, China. Front. Microbiol.9, 2989. doi: 10.3389/fmicb.2018.02989
210
WattsR. E.TotsikaM.ChallinorV. L.MabbettA. N.UlettG. C.De VossJ. J.et al. (2012). Contribution of Siderophore Systems to Growth and Urinary Tract Colonization of Asymptomatic Bacteriuria Escherichia Coli. Infect Immun.80 (1), 333–344. doi: 10.1128/IAI.05594-11
211
WesselyF.BartlM.GuthkeR.LiPuSchusterS.KaletaC. (2011). Optimal Regulatory Strategies for Metabolic Pathways in Escherichia Coli Depending on Protein Costs. Mol. Syst. Biol.7 (1), 515. doi: 10.1038/msb.2011.46
212
WhitesideS. A.RazviH.DaveS.ReidG.BurtonJ. P. (2015). The Microbiome of the Urinary Tract—a Role Beyond Infection. Nat. Rev. Urol.12 (2), 81. doi: 10.1038/nrurol.2014.361
213
WilkeT.BoettgerB.BergB.GrothA.MuellerS.BottemanM.et al. (2015). Epidemiology of Urinary Tract Infections in Type 2 Diabetes Mellitus Patients: An Analysis Based on a Large Sample of 456,586 German T2DM Patients. J. Diabetes Complications29, 1015–1023. doi: 10.1016/j.jdiacomp.2015.08.021
214
WolfeA. J.TohE.ShibataN.RongR.KentonK.FitzGeraldM.et al. (2012). Evidence of Uncultivated Bacteria in the Adult Female Bladder. J. Clin. Microbiol.50 (4), 1376–1383. doi: 10.1128/JCM.05852-11
215
Wootton,.J.T.EmmersonM. (2005). Measurement of Interaction Strength in Nature. Annu. Rev. Ecol. Evol. Syst.36, 419–444. doi: 10.1146/annurev.ecolsys.36.091704.175535
216
WuP.ZhangG.ZhaoJ.ChenJ.ChenY.HuangW.et al. (2018). Profiling the Urinary Microbiota in Male Patients With Bladder Cancer in China. Front. Cell. Infect Microbiol.8, 1–10. doi: 10.3389/fcimb.2018.00167
217
XuX.NieS.DingH.HouF. F. (2018). Environmental Pollution and Kidney Diseases. Nat. Rev. Nephrol.14 (5), 313. doi: 10.1038/nrneph.2018.11
218
YamamotoS.TsukamotoT.TeraiA.KurazonoH.TakedaY.YoshidaO. (1997). Genetic Evidence Supporting the Fecal-Perineal-Urethral Hypothesis in Cystitis Caused by Escherichia Coli. J. Urol.157 (3), 1127–1129. doi: 10.1097/00005392-199703000-00119
219
YenY.ChengB.ChanC.LinC.ChenH. (2018). Heavy Metal Components in Blood and Urinary Stones of Urolithiasis Patients. Biol. Trace element Res.185 (2), 266–274. doi: 10.1007/s12011-018-1253-x
220
YooJ. Y.RhoM.YouY.-A.KwonE. J.KimM.-H.KymS.et al. (2016). 16s rRNA Gene-Based Metagenomic Analysis Reveals Differences in Bacteria-Derived Extracellular Vesicles in the Urine of Pregnant and non-Pregnant Women. Exp. Mol. Med.48 (2), e208. doi: 10.1038/emm.2015.110
221
ZhangY.-F.HanK.ChandlerC. E.TjadenB.ErnstR. K.LoryS. (2017). Probing the sRNA Regulatory Landscape of P. Aeruginosa: Post-Transcriptional Control of Determinants of Pathogenicity and Antibiotic Susceptibility. Mol. Microbiol.106 (6), 919–937. doi: 10.1111/mmi.13857
222
ZhangT.RuanJ.ZhangB.LuS.GaoC.HuangL.et al. (2019). Heavy Metals in Human Urine, Foods and Drinking Water From an E-Waste Dismantling Area: Identification of Exposure Sources and Metal-Induced Health Risk. Ecotoxicol Environ. Saf169, 707–713. doi: 10.1016/j.ecoenv.2018.10.039
223
ZhaoL.GaoS.HuanH.XuX.ZhuX.YangW.et al. (2009). Comparison of Virulence Factors and Expression of Specific Genes Between Uropathogenic Escherichia Coli and Avian Pathogenic E. Coli in a Murine Urinary Tract Infection Model and a Chicken Challenge Model. Microbiology155 (5), 1634–1644. doi: 10.1099/mic.0.024869-0
224
ZhaoDiLiuR.-Y.XiangP.JuhaszA. L.HuangL.LuoJ.et al. (2017). Applying Cadmium Relative Bioavailability to Assess Dietary Intake From Rice to Predict Cadmium Urinary Excretion in Nonsmokers. Environ. Sci. Technol.51 (12), 6756–6764. doi: 10.1021/acs.est.7b00940
225
ZilberbergM. D.NathansonB. H.SulhamK.FanW.ShorrA. F. (2017). Carbapenem Resistance, Inappropriate Empiric Treatment and Outcomes Among Patients Hospitalized With Enterobacteriaceae Urinary Tract Infection, Pneumonia and Sepsis. BMC Infect. Dis.17, 279. doi: 10.1186/s12879-017-2383-z
226
ZozayaM.FerrisM. J.SirenJ. D.LillisR.MyersL.Jacques NsuamiM.et al. (2016). Bacterial Communities in Penile Skin, Male Urethra, and Vaginas of Heterosexual Couples With and Without Bacterial Vaginosis. Microbiome4 (1), 16. doi: 10.1186/s40168-016-0161-6
Summary
Keywords
recurrent urinary tract infections (rUTIs), human microbiome, UPEC, dysbiosis, microbial ecology
Citation
Josephs-Spaulding J, Krogh TJ, Rettig HC, Lyng M, Chkonia M, Waschina S, Graspeuntner S, Rupp J, Møller-Jensen J and Kaleta C (2021) Recurrent Urinary Tract Infections: Unraveling the Complicated Environment of Uncomplicated rUTIs. Front. Cell. Infect. Microbiol. 11:562525. doi: 10.3389/fcimb.2021.562525
Received
18 May 2020
Accepted
18 May 2021
Published
22 July 2021
Volume
11 - 2021
Edited by
Heather D. Bean, Arizona State University, United States
Reviewed by
Megan Behringer, Vanderbilt University, United States; Yanfei Chen, Zhejiang University, China
Updates
Copyright
© 2021 Josephs-Spaulding, Krogh, Rettig, Lyng, Chkonia, Waschina, Graspeuntner, Rupp, Møller-Jensen and Kaleta.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jonathan Josephs-Spaulding, j.josephs-spaulding@iem.uni-kiel.de
This article was submitted to Microbiome in Health and Disease, a section of the journal Frontiers in Cellular and Infection Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.