
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 17 March 2021
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.532989
This article is part of the Research Topic Current Perspectives of Antimicrobial Resistance in Campylobacteraceae and Helicobacteraceae View all 6 articles
Aliarcobacter cibarius and Aliarcobacter thereius are two rarely detected Aliarcobacter species. In the study, we analyzed the antimicrobial susceptibility and provide detailed insights into the genotype and phylogeny of both species using whole-genome sequencing. Thermophilic Campylobacter species are the most common bacterial foodborne pathogens causing gastroenteritis in humans worldwide. The genus Aliarcobacter is part of the Campylobacteraceae family and includes the species Aliarcobacter butzleri, Aliarcobacter cryaerophilus, Aliarcobacter skirrowii, and the rarely described Aliarcobacter cibarius, Aliarcobacter faecis, Aliarcobacter lanthieri, Aliarcobacter thereius, and Acrobarter trophiarum. Aliarcobacter are emergent enteropathogens and potential zoonotic agents. Here, we generated, analyzed, and characterized whole-genome sequences of Aliarcobacter cibarius and Aliarcobacter thereius. They were isolated from water poultry farms in Germany, cultured and identified by MALDI-TOF MS. With PCR the identity was verified. Antibiotic susceptibility testing was carried out with erythromycin, ciprofloxacin, doxycycline, tetracycline, gentamicin, streptomycin, ampicillin, and cefotaxime using the gradient strip method (E-test). Whole-genome sequences were generated including those of reference strains. Complete genomes for six selected strains are reported. These provide detailed insights into the genotype. With these, we predicted in silico known AMR genes, virulence-associated genes, and plasmid replicons. Phenotypic analysis of resistance showed differences between the presence of resistance genes and the prediction of phenotypic resistance profiles. In Aliarcobacter butzleri, the nucleotide sequence of the gyrA gene (DQ464331) can show a signature mutation resulting in an amino acid change T85>I. Acrobarter cibarius and Acrobarter thereius showed the same gene as assessed by similarity annotation of the mutations 254C>G. Most of the isolates were found to be sensitive to ciprofloxacin. The ciprofloxacin-resistant Aliarcobacter thereius isolate was associated with the amino acid change T85>I. But this was not predicted with antibiotic resistance databases, before. Ultimately, a phylogenetic analysis was done to facilitate in future outbreak analysis.
Thermophilic Campylobacter (C.) are the most common bacterial cause of human gastroenteritis in the world (WHO, 2018). The genus Aliarcobacter (A.) within the family Campylobacteriaceae was created to accommodate Gram-negative, curved-shaped bacteria belonging to atypical Campylobacter spp. classified by their ability to grow at lower temperatures (15–30°C) and without microaerophilic conditions (Vandamme et al., 1991). The genus Aliarcobacter (formerly Arcobacter) consists of A. butzleri, A. cibarius, A. cryaerophilus, A. faecis, A. lanthieri, A. skirrowii, A. thereius and A. trophiarum (Pérez-Cataluña et al., 2017; Pérez-Cataluña et al., 2018a; Pérez-Cataluña et al., 2018b; Pérez-Cataluña et al., 2018c). Aliarcobacter spp. are present in the digestive tract of healthy animals (Uenver et al., 2013), but have also been associated with enteritis and reproductive disorders (De Smet et al., 2012). Aliarcobacter in contaminated food (e.g. poultry products) and water can be transmitted to humans and can cause diarrhea and in rare cases bacteremia (Collado and Figueras, 2011; Hanel et al., 2016).
The species A. butzleri, A. cryaerophilus, and A. skirrowii are considered emergent enteropathogens and potential zoonotic agents (Prouzet-Mauleon et al., 2006; Levican et al., 2013; Van den Abeele et al., 2014; Van den Abeele et al., 2016; Hänel et al., 2018). A. cibarius and A. thereius are present in farm animals and have been isolated from food of animal origin (Chinivasagam et al., 2007; Pentimalli et al., 2009; Adam et al., 2014a; Yesilmen et al., 2014) but have not yet been isolated from human specimens. The pathogenicity and virulence mechanisms of Aliarcobacter spp. are still poorly understood. Adhesion, invasion, and cytotoxicity capacity have been studied only in A. skirrowii, A. butzleri, A. cryaerophilus, and A. cibarius (Collado and Figueras, 2011; Pérez-Cataluña et al., 2018b), and isolates have been rarely sequenced.
Isolation and taxonomic identification of A. cibarius and A. thereius are often difficult. Only a few representative genomes have been described so far (Adam et al., 2014a; Adam et al., 2014b; Rovetto et al., 2017). There are few accessible data providing the antibiotic susceptibility of A. cibarius and A. thereius (Abdelbaqi et al., 2007; Fanelli et al., 2019).
The aim of this study was to determine the antimicrobial susceptibility of A. cibarius and A. thereius isolated from domestic water poultry to antibiotics commonly used in treatments of humans. Additionally, the genomic features of A. cibarius and A. thereius isolates were analyzed to improve diagnostic and antibiotic treatment options. The study presents a full genome analysis of A. cibarius and A. thereius isolates and their reference strains, available from a public culture collection. The genomes of reference strains were analyzed with a focus on virulence-associated genes and antibiotic resistance genes. The strains were analyzed for the gyrA gene known to be causal for ciprofloxacin resistance in A. butzleri. Data were compared to the A. butzleri strain D2686 DNA gyrase subunit A gene (DQ464331) (Abdelbaqi et al., 2007). Subsequently, phylogenetic strain variation in A. cibarius and A. thereius was assessed to analyze the previously reported high heterogeneity (De Smet et al., 2012; Pérez-Cataluña et al., 2018b).
The A. cibarius and A. thereius isolates were handled as before reported for A. skirrowii (Hänel et al., 2018). Aliarcobacter isolates were cultivated from fecal samples collected in two water poultry farms in Thuringia, Germany. A two-step procedure was done in Arcobacter broth (Oxoid GmbH, Wesel, Germany) supplemented with antibiotics cefoperazone, amphotericin, and teicoplanin (CAT; Oxoid GmbH) under microaerophilic conditions (5% O2, 10% CO2 and 85% N2) for 48 h at 30°C. Subsequently, the broth was streaked on plates (Mueller-Hinton agar/CAT/5 % defibrinated bovine blood) and incubated under microaerophilic conditions for another 24–48 h at 30°C. Suspicious colonies were recultivated and identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) as described before (El-Ashker et al., 2015; Busch et al., 2018). IVD Bacterial Test Standard, Biotyper 3.1 software and the database DB 4613 (all Bruker Daltonik GmbH, Bremen, Germany) containing spectra of all Aliarcobacter species were used. Confirmation of the species identification was performed with multiplex PCR as described before (Hänel et al., 2018).
Sequence data used in this study had the following accession numbers: A. anaerophilus PDKO01, A. butzleri RM4018, CP000361.1, A. halophilus DSM 18005, GenBank: NXIF00000000., A. aquimarinus CECT 8442, NCBI Reference Sequence NZ_NXIJ00000000.1, A. bivalviorum LMG 26154 NZ_CP031217.1, A. cibarius LMG 21996, NCBI Reference Sequence: NZ_JABW00000000.1, A. cloacae CECT 7834, NZ_NXII00000000.1, A. cryaerophilus L406, NZ_LRUV00000000.1.
Antimicrobial susceptibility to eight antibiotics (erythromycin, ciprofloxacin, doxycycline, tetracycline, gentamicin, streptomycin, ampicillin, and cefotaxime) was determined using the gradient strip diffusion method (E-test™, bioMérieux, Nürtingen, Germany) following the instructions (Table 1). The bacterial suspensions for the E-test were adjusted to an optical density of 0.1 at 600 nm (approximately 3 to 5 × 108 cfu/ml) in phosphate-buffered saline (PBS). 750 µl was spread on a Mueller-Hinton agar plate and a single strip was put on each plate. After 48 h of incubation at 30°C under microaerophilic conditions, the minimum inhibitory concentration (MIC) was determined. The type strain of A. skirrowii DSM 7302 was used as control. For erythromycin, ciprofloxacin, doxycycline, and tetracycline, interpretative criteria were based upon EUCAST breakpoints for Campylobacter (2019). For gentamicin, ampicillin, and cefotaxime (EUCAST, 2019) Enterobacterales breakpoints were applied (2019). For streptomycin, the cut-off value for C. jejuni was used as suggested by the EFSA–working group (EFSA, 2008). The phenotypes were classified as sensitive (S), and resistant (R) (Table 1).
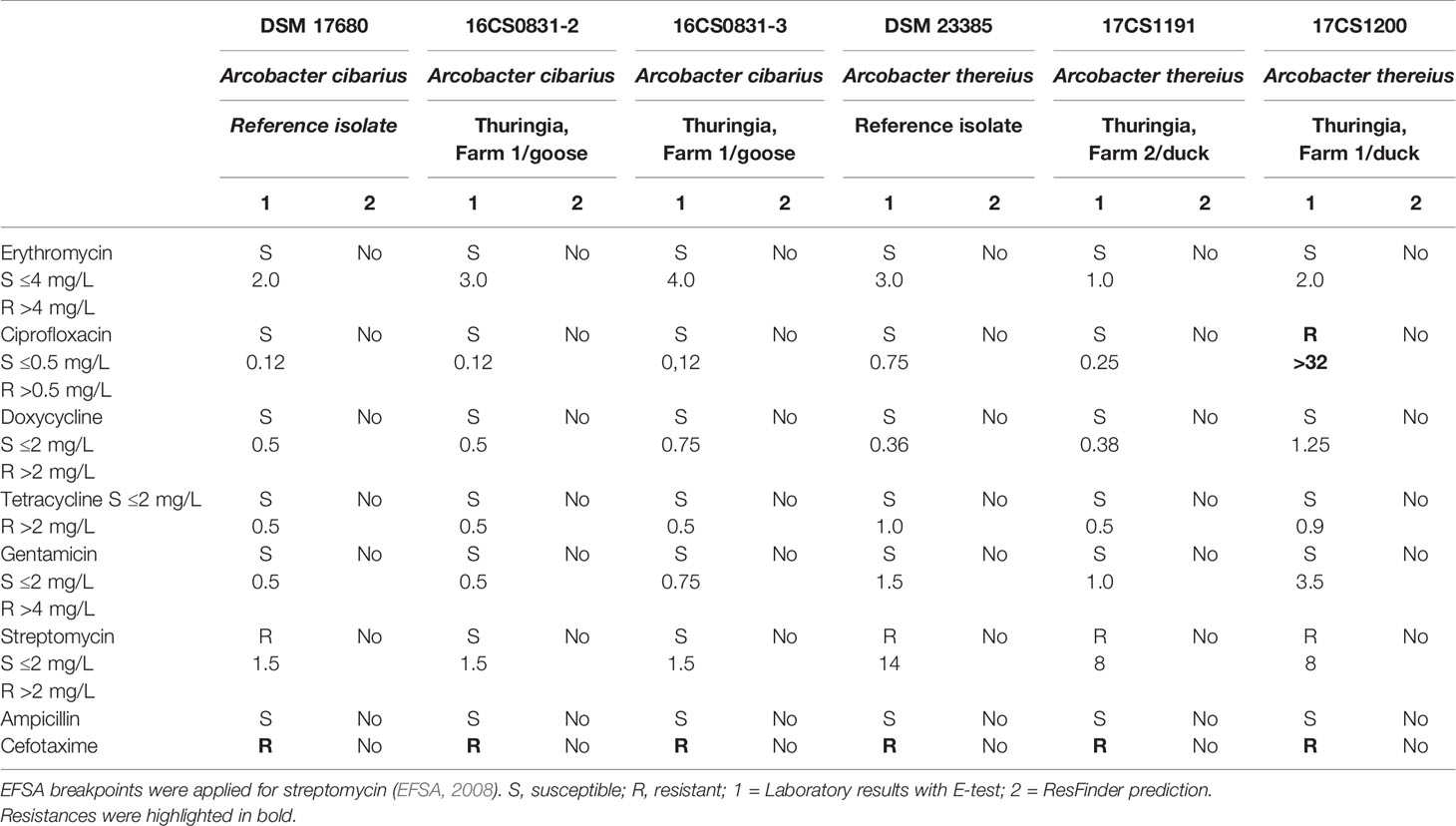
Table 1 Antibiotic susceptibility of Arcobacter spp. EUCAST breakpoints for Campylobacter jejuni was used for erythromycin, ciprofloxacin, doxycycline, tetracycline, and gentamicin.
DNA for whole-genome sequencing (WGS) was prepared from colonies harvested from plates. DNA was purified (High Pure PCR Template Preparation Kit; Roche Diagnostics, Mannheim, Germany) and sequencing libraries were generated using the Nextera XT DNA Library Prep Kit (Illumina, Inc., San Diego, CA, USA). From an Illumina MiSeq run paired-end reads were generated to mean sequencing depth between 50 and 100 reads. The assignment of the taxonomic labels to all reads was performed with MetaPhlAn (Segata et al., 2012) and Kraken version 0.10.6 (Wood and Salzberg, 2014). Further, read processing included quality trimming and assembly with SPAdes v3.12.0 (–careful) (Bankevich et al., 2012), and filtering by removing contigs with a coverage <5 and a length <500. Annotation was performed with Prokka using the recommended standard settings (Seemann, 2014). The report of the presence of antibiotic resistance genes in silico was compared with results from in vivo susceptibility testing.
Additionally, parSNP and kSNP3.0 (Treangen et al., 2014; Gardner et al., 2015) were used to assign microbial phylogeny and results were visualized with Dendroscope (Huson et al., 2007) (Figure 1). Based on ARIBA (Hunt et al., 2017) several databases were used to identify single nucleotide polymorphisms (SNPs) directly from short reads. For detection of resistance genes the ResFinder (Zankari et al., 2012) and for plasmid finding the PlasmidFinder (Carattoli et al., 2014) were used. Multilocus Sequence Typing (MLST) was done using the MLST database (Carattoli et al., 2014) and for detection of virulence factors VFDB_full was used (Chen et al., 2005) (Table 2). Virulence-associated genes ciaB (HF935951), cj1349 (HF935963), and cadF (HF935942) were mapped with Geneious (Kearse et al., 2012) on all assemblies and to the known A. skirrowii sequence LRUX01000036.1. All the reads of all isolates were mapped to a set of six putative virulence-associated genes (ciaB, pldA, tlyA, mviN, cadF, and cj1349) (Karadas et al., 2013; Douidah et al., 2014; Sekhar et al., 2017). Further analysis of plasmids was done with Bandage (Wick et al., 2015). Reads were mapped against selected antibiotic genes known to be responsible to resistance to ampicillin and cefotaxime. The following genes were selected from databases with the following locus tags: A7H1H_0053-16806891, A7H1H_1295-16807124, A7H1H_1494-16808301, ABED_1213-12128208, ABED_1379-12128538, Abu_0578-5624058, Abu_1299-5623980, Abu_1478-5624289, blaOXA-24305112, CHAB381_1693-5408367, cj0299-904623, cmeD-12249850, cmeE-12249851, cmeF-12249852, ftsI-16808465, pbpC-5624113, pbpC-16807438, ABU_RS00920-24304134.
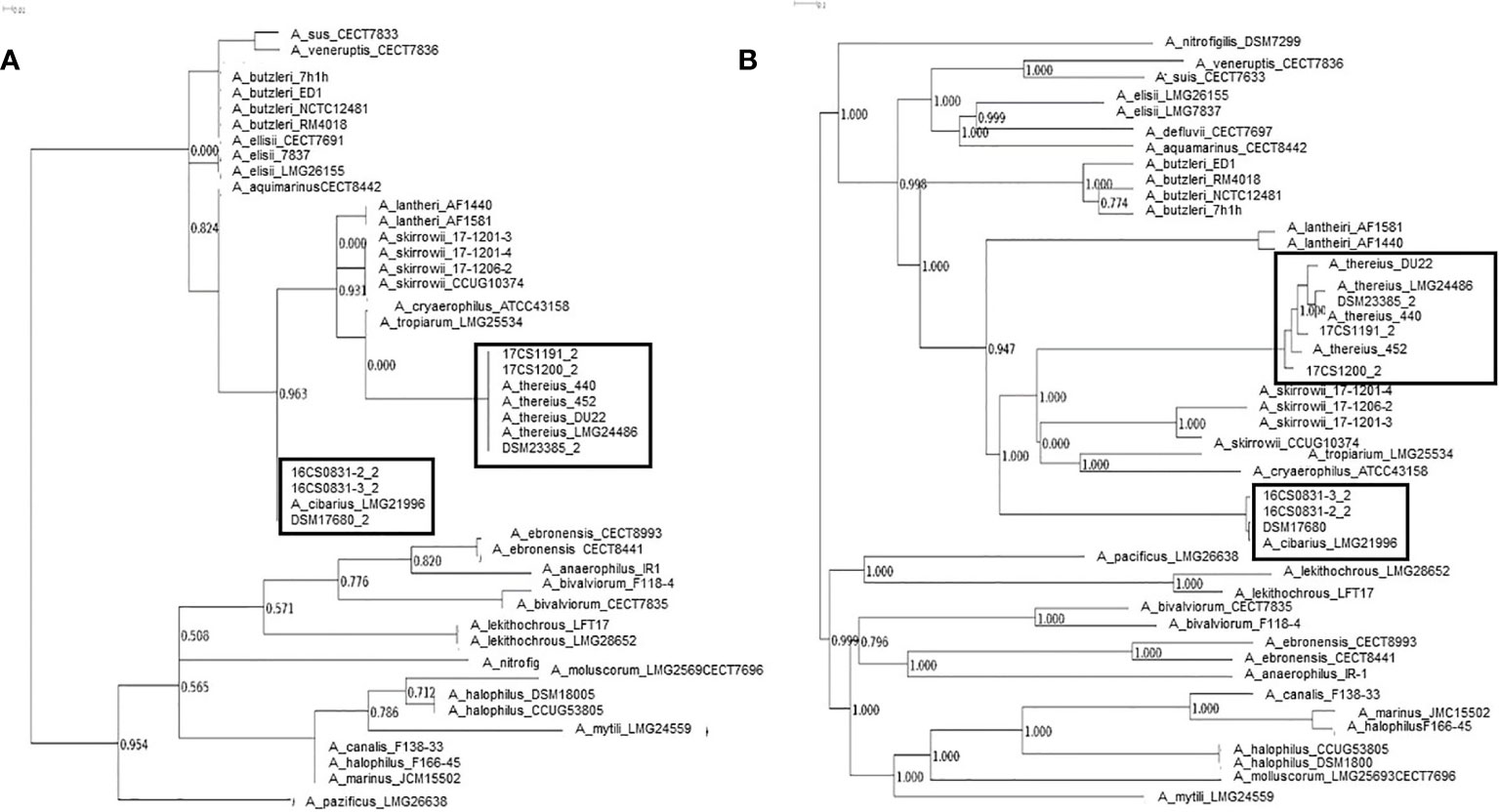
Figure 1 Phylogeny based on all identified SNPs and phylogenetic analysis without genome alignment or the requirement for reference genomes all SNPs and reference independent performed with kSNP3.0. (A) Core genome analysis and (B) parsimony tree based on whole-genome SNPs.
A. cibarius and A. thereius isolates were cultivated and identified by MALDI-TOF MS due to scores >2,3 (spectra are available upon request). Identification was confirmed by PCR. Taxonomic analysis of the WGS data with MetaPhlAn and Kraken resulted for A. cibarius and A. thereius in “unclassified Arcobacter”. Also, analyses based on the genomic features by sequence identity with RAST, Rapid Annotation using Subsystem Technology, (Aziz et al., 2008) showed only a relationship to A. butzleri RM4018 for all isolates. A correct taxonomic differentiation of the species with the reference strains was possible with SNP detection using parSNP and kSNP 3.0.
The phenotypic antibiotic susceptibility resulting by E-test for the Aliarcobacter isolates is shown in Table 1. All A. cibarius and A. thereius isolates were susceptible to erythromycin, doxycycline, tetracycline, streptomycin, ampicillin, and gentamicin and showed resistance to cefotaxime. The isolate A. thereius 17CS1200 was resistant to ciprofloxacin. No resistance determinants were predicted with the ResFinder (see Table 1).
The mean coverage using WGS obtained for the isolates was >100 (see Table 3). All sequences were assembled and annotated. Genome assemblies had 11 to 78 contigs. Sequence length prediction was 1,897,659–2,233,860 bases. All assemblies had a mean coverage of 102–136 resulting in high-quality genome drafts. The genome sizes were estimated between 1.9–2.2 million bases with approximately 1,980 coding sequences. The programs as used in Busch et al. (Busch et al., 2020a; Busch et al., 2020b) were applied with standard settings. No reads mapped to reference genes in the database, therefore, no local assemblies were run and no SNPs were detected. However, the known virulence-associated genes cj1349 (HF935963), ciaB (HF935951), cadF (HF935942) and hecA (HF935064) were found and could be mapped to all assemblies as in the published A. skirrowii sequence LRUX01000036.1 (Hänel et al., 2018) (Table 3). However, all genes mapped with a low sequence identity (49.9 to 87.0%). For the ampicillin and cefotaxime genes, the following genes could be mapped with more than 95% identity: A7H1H_0053-16806891, A7H1H_1295-16807124, ftsI-16808465, pbpC-5624113, pbpC-16807438, and AA347_RS09755-39300940. No mapping was detected with assemblies with the exclusion of AA347_RS01370-39299273 deriving from A. thereius LMG24486. From the sequencing data, MLST results were extracted. Sequence types were assigned (Supplementary Table 3). The results were not verified by traditional sequencing methods for MLST. As alternative method phylogenetic analysis was done (Figure 1). The first approach was based on all identified SNPs of the core genome (Treangen et al., 2014). The second approach was done without reference genome alignment kSNP3.0 (Gardner et al., 2015) with subsequent parsimony tree based on all SNPs detected in the genomes. Both approaches confirmed the classification of the isolates obtained by MALDI-TOF MS and PCR and proofed as suitable tools. Outbreak analysis, diagnostics and characterization of unknown isolates can be done with these methods.

Table 3 Reads of all Aliarcobacter spp. mapped to known virulence-associated genes of Arcobacter butzleri RM4018 (Karadas et al., 2013).
A. butzleri, A. cryaerophilus, and A. skirrowii have been associated with gastrointestinal diseases. For A. cibarius and A. thereius, pathogenicity is unknown (Collado and Figueras, 2011; Isidro et al., 2020). It can be assumed that Aliarcobacter infections in human are so far underestimated. Thus, we assessed genomic and phenotypic characteristics of the rare species A. cibarius and A. thereius. Here, we present cultivation protocols for A. cibarius and A. thereius. We evaluated MALDI-TOF MS spectra and made them available. Genome sequencing data were published at the NCBI so that the software tools based on the RefSeq (such as BLAST and Kraken) will allow quick identification in the future.
Data on antimicrobial susceptibility of A. cibarius and A. thereius are scarce. A. cibarius and A. thereius are reported mostly susceptible to many antimicrobials (Houf et al., 2001; Yesilmen et al., 2014) in concordance with the hereby reported isolates. Antibiotic testing is not standardized, yet. Microdilution assays are usually favored, but gradient strips are more robust (Van den Abeele et al., 2014; Hänel et al., 2018). A. butzleri or A. cryaerophilus isolates from Europe showed susceptibility to gentamicin, tetracycline, erythromycin, ciprofloxacin, and doxycycline and some of them resistance to ampicillin (Van den Abeele et al., 2016). Phenotypic analysis of resistance showed differences between the prediction of resistance genes and the phenotypic resistance profile. The here investigated A. cibarius and A. thereius isolates were mostly sensitive to ciprofloxacin but the isolate 17CS1200 was resistant. This could not be predicted with antibiotic resistance databases, ResFinder (Zankari et al., 2012), yet. Resistance to ciprofloxacin is reported for almost 25 % of human Campylobacter isolates (CDC, 2015). In A. butzleri, the nucleotide sequence of the gyrA gene (DQ464331) has the signature mutation resulting in an amino acid change T85>I (Abdelbaqi et al., 2007). The isolates of A. cibarius and A. thereius that were sensitive to ciprofloxacin showed a nucleotide change 254C>G that resulted in T85>S changing on protein basis. The ciprofloxacin-resistant isolate 17CS1200 had an amino acid change T85>I. This is the described signature mutation as reported in A. butzleri. It can be assumed that this, as a universally functional mechanism, is likely to affect DNA supercoiling, and the expression of several virulence factors and proteins as described in different species is the same (Ozeki et al., 1997; Aubry et al., 2006; Abdelbaqi et al., 2007; Hashimi et al., 2008; Lau et al., 2011; Malik et al., 2012; Yamachika et al., 2012; Sutera et al., 2017). Ciprofloxacin resistance usually occurs due to specific point mutations within the DNA gyrase A (gyrA) and/or topoisomerase IV parC and parE genes, often in combination (Casin et al., 2003; Johnning et al., 2015; Onseedaeng and Ratthawongjirakul, 2016). Annotation of mutation in the gyrase A gene is considered not to be sufficient to produce resistance to ciprofloxacin (Hooper and Jacoby, 2015). For example mutations in gyrA and parE subunits of the respective enzymes can reduce drug-binding to the enzyme–DNA complex. But also plasmid-encoded resistance due to Qnr proteins can shield from quinolone action. On the other hand, single mutations in gyrA are known to generate resistance to nalidixic acid, a first-generation quinolone. However, additional mutations in other type II topoisomerase genes are often necessary to generate resistance to later generations of fluoroquinolones, such as ciprofloxacin (Johnning et al., 2015). A mutation as in the parE gene as in Salmonella could not be detected.
The antibiotic susceptibility results we report here are in agreement with antibiotic resistance or susceptibility prevalence reported for other Aliarcobacter isolates from seafood and water sources (Levican et al., 2013; Levican et al., 2014; Levican et al., 2015; Rathlavath et al., 2017a; Rathlavath et al., 2017b; Silha et al., 2017). Aliarcobacter resistance is reported in food sources as meat, milk, shellfish, slaughterhouses, dairy plants, and humans, although rarely done for rare Aliarcobacter comparing phenotype and genotype (Atabay et al., 2008; Ferreira et al., 2013; Ferreira et al., 2014a; Ferreira et al., 2014b; Ferreira et al., 2016; Shirzad Aski et al., 2016; Ferreira et al., 2017; Ferreira et al., 2018; Isidro et al., 2020).
In Campylobacter infections macrolides are used as preferred therapeutic agents. Tetracycline was proposed for severe cases only (Yan et al., 2000; Kayman et al., 2012; Arguello et al., 2015). Here investigated isolates were susceptible to both classes of antibiotics and would guarantee effectiveness in treatment of infections. Monitoring and reporting of antimicrobial resistance data even in rare species, as well as whole-genome sequencing data analysis of Aliarcobacter from domestic animals are important to monitor the evolution of antimicrobial resistance. This can be used to optimize diagnostics and therapy.
Gene transfer or plasmid transfer events can induce antibiotic resistance (Sundin and Bender, 1996). In the Enterobacteriaceae family, antibiotic resistance is often mediated by plasmids (Nikaido, 2009). Indication is here that the antimicrobial resistance determinants are located on the chromosome. Plasmids play an important role in the genetic evolution and adaptation. In A. butzleri 9.9% of isolates carried plasmids (Harrass et al., 1998; Toh et al., 2011). PlasmidFinder could not predict plasmids nor plasmids could be detected by the graphical assembly graphs, allowing visual inspection (Wick et al., 2015). In agarose gels, no plasmids could be detected in A. cibarius and A. thereius comparable with A. skirrowii (Becker et al., 2016; Hänel et al., 2018).
MLST characterizes isolates using allelic DNA sequences of several housekeeping genes and is a traditional technique. Here, no MLST genes could be predicted for both rarely found Aliarcobacter species. As a consequence, MLST might be replaced by phylogeny based on all identified SNPs of the core or whole genomes.
Virulence factors were not predicted. But genes like ciaB, pldA, tlyA, mviN, cadF, and cj1349 are known to be involved in host adherence and invasion. They could be mapped with low sequence identity (Levican et al., 2013). The protein CiaB has been shown to be responsible for host cell invasion in Campylobacter. The protein TlyA is a hemolysin with a role in cellular adherence in C. jejuni. The protein CadF and cj1349 are fibronectin-binding proteins, that promote bacteria to cell contact and PldA is a phospholipase associated with lysis of erythrocytes (Miller et al., 2007; Sekhar et al., 2017; Isidro et al., 2020). The low sequence identity was also an indication that A. cibarius and A. thereius might show more variability within the nucleotide sequences. This will benefit to be included the databases.
As a suitable tool, we evaluated whole-genome sequencing with phylogenetic analysis. A parsimony tree analysis based on SNPs in the genomes proofed to be specifically useful. With these outbreak analysis, diagnostics and characterization can be approached even for rare species and classify new, not yet unclassified species correctly. A phylogenetic tree revealed the relatedness of these isolates and will facilitate in future outbreak analysis. With the provided genomes a core genome analysis and whole-genome SNP phylogeny analysis provided exact classification of identified strains and reference strains. This phylogenetic analysis has the advantage that it is reference-free and alignment-independent. This allows for the usage of the method for the previously reported high heterogeneity of isolates and a correct placement within the phylogenetic tree, even when the strains are previously unknown (De Smet et al., 2012; Pérez-Cataluña et al., 2018b; Pérez-Cataluña et al., 2019).
The application of open-source software allows an economic and transparent assessment of the sequencing data. The diagnostic capabilities can be improved and contributes to epidemiological, pathogenic and functional analysis of these rarely recognized bacteria. The mechanisms underlying the pathogenicity of Aliarcobacter may thus be further elucidated. The optimized database will serve bioinformatics tools to do comprehensive bioinformatics analysis. It remains speculative that the eradication of Campylobacter spp. from the microbiome of farmed animals could foster the replacement by Aliarcobacter subspecies. In such case, any basic assessment of the pathogenicity of Aliarcobacter spp. will be advantageous.
In conclusion, A. cibarius and A. thereius are rarely detected but potentially harmful bacteria. Due to the fastidious growth of A. cibarius and A. thereius the antibiotic susceptibility testing is preferably done using the gradient strip method. Prediction of antibiotic susceptibility, plasmid sequences, MLST or virulence factors based on WGS data should be carefully assessed. The present study adds the aspect of antimicrobial phenotyping of the A. cibarius and A. thereius species into Aliarcobacter comparative genomics field. In fact, we here provide extended data on core and full genome phylogenetics and some gene-by-gene assignment of these bacteria showing that further functional and expressional studies are needed to understand the genotypic and phenotypic correlations of genomes and genes. Our data supports the notion that some aspects of the virulome and resistome diversity, environmental/host adaptation and pathogenicity need to be adapted for rare species. This is highlighting the need to implement more robust species-oriented diagnostics towards an enhanced global surveillance and control of this emerging pathogen.
The datasets generated for this study can be found in the Data availability: Arcobacter cibarius and Arcobacter thereius sequences were deposited at BioProject under the accession number PRJNA542783, and as BioSample under the accession number SAMN11638312 (VBUC00000000, Arcobacter cibarius 16CS0831-2), SAMN11638336 (VBUD00000000, Arcobacter cibarius 16CS0831-3), SAMN11638387 (VBUE00000000, Arcobacter cibarius DSM17680), SAMN11638388 (VBUF00000000, Arcobacter thereius 17CS1191_2), SAMN11638390 (VBUG00000000, Arcobacter thereius 17CS1200_2), SAMN11638433 (VBUH00000000, Arcobacter thereius DSM23385). The version described in this paper is the first version.
IH, HH, BGS, AB and HT have jointly conceived the study. IH provided strains, strain information, and metadata and antibiotic testing to the samples. EM provided further antibiotic testing to the samples and bioinformatics analysis. AB performed bioinformatics analysis of genomes, assembly, and phylogenetic relationships. All authors contributed to the article and approved the submitted version.
For this work, IH and EM were supported by an in-house project of Friedrich-Loeffler-Institut. AB was supported by a grant of the German Federal Ministry of Education and Research within the framework of the project Ess-B.A.R. (FKZ 13N13983) and by the Deutsche Forschungsgemeinschaft (German Research Foundation, DFG) under Germany’s Excellence Strategy–EXC 2051–Project-ID 390713860.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Sandra Thierbach, Anja Hackbarth, Peggy Methner, and Byrgit Hofmann for skillful technical assistance and Sebastian Weis for proofreading.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.532989/full#supplementary-material
Abdelbaqi K., Menard A., Prouzet-Mauleon V., Bringaud F., Lehours P., Megraud F. (2007). Nucleotide sequence of the gyrA gene of Arcobacter species and characterization of human ciprofloxacin-resistant clinical isolates. FEMS Immunol. Med. Microbiol. 49, 337–345. doi: 10.1111/j.1574-695X.2006.00208.x
Adam Z., Whiteduck-Leveillee K., Cloutier M., Chen W., Lewis C. T., Levesque C. A., et al. (2014a). Draft Genome Sequence of Arcobacter cibarius Strain LMG21996T, Isolated from Broiler Carcasses. Genome Announc. 2 (1), e00034–14. doi: 10.1128/genomeA.00034-14
Adam Z., Whiteduck-Leveillee K., Cloutier M., Tambong J. T., Chen W., Lewis C. T., et al. (2014b). Draft genome sequences of three arcobacter strains of pig and dairy cattle manure origin. Genome Announc. 2 (1), e00377–14. doi: 10.1128/genomeA.00377-14
Arguello E., Otto C. C., Mead P., Babady N. E. (2015). Bacteremia caused by Arcobacter butzleri in an immunocompromised host. J. Clin. Microbiol. 53, 1448–1451. doi: 10.1128/JCM.03450-14
Atabay H. I., Unver A., Sahin M., Otlu S., Elmali M., Yaman H. (2008). Isolation of various Arcobacter species from domestic geese (Anser anser). Vet. Microbiol. 128, 400–405. doi: 10.1016/j.vetmic.2007.10.010
Aubry A., Veziris N., Cambau E., Truffot-Pernot C., Jarlier V., Fisher L. M. (2006). Novel gyrase mutations in quinolone-resistant and -hypersusceptible clinical isolates of Mycobacterium tuberculosis: functional analysis of mutant enzymes. Antimicrob. Agents Chemother. 50, 104–112. doi: 10.1128/AAC.50.1.104-112.2006
Aziz R. K., Bartels D., Best A. A., Dejongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75. doi: 10.1186/1471-2164-9-75
Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Becker L., Steglich M., Fuchs S., Werner G., Nubel U. (2016). Comparison of six commercial kits to extract bacterial chromosome and plasmid DNA for MiSeq sequencing. Sci. Rep. 6, 28063. doi: 10.1038/srep28063
Busch A., Thomas P., Zuchantke E., Brendebach H., Neubert K., Gruetzke J., et al. (2018). Revisiting Francisella tularensis subsp. holarctica, Causative Agent of Tularemia in Germany With Bioinformatics: New Insights in Genome Structure, DNA Methylation and Comparative Phylogenetic Analysis. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00344
Busch A., Homeier-Bachmann T., Abdel-Glil M. Y., Hackbart A., Hotzel H., Tomaso H. (2020a). Using affinity propagation clustering for identifying bacterial clades and subclades with whole-genome sequences of Francisella tularensis. PloS Negl. Trop. Dis. 14 (9), e0008018. doi: 10.1371/journal.pntd.0008018
Busch A., Hotzel H., Methner U. (2020b). Complete genome and plasmid sequences of a multidrug-resistant Salmonella enterica subsp. enterica serovar Panama isolate from a German cattle farm. J. Genomics 8, 71–75. doi: 10.7150/jgen.48656
Carattoli A., Zankari E., Garcia-Fernandez A., Voldby Larsen M., Lund O., Villa L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Casin I., Breuil J., Darchis J. P., Guelpa C., Collatz E. (2003). Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica typhimurium isolates in humans. Emerg. Infect. Dis. 9, 1455–1457. doi: 10.3201/eid0911.030317
CDC (2015). “National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2013. Atlanta, GA: US Department of Health and Human Services, CDC, 2015,” in National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2013 (Atlanta, GA: US Department of Health and Human Services, CDC, 2015).
Chen L., Yang J., Yu J., Yao Z., Sun L., Shen Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
Chinivasagam H. N., Corney B. G., Wright L. L., Diallo I. S., Blackall P. J. (2007). Detection of Arcobacter spp. in piggery effluent and effluent-irrigated soils in southeast Queensland. J. Appl. Microbiol. 103, 418–426. doi: 10.1111/j.1365-2672.2007.03275.x
Collado L., Figueras M. J. (2011). Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microbiol. Rev. 24, 174–192. doi: 10.1128/CMR.00034-10
De Smet S., De Zutter L., Houf K. (2012). Spatial distribution of the emerging foodborne pathogen Arcobacter in the gastrointestinal tract of pigs. Foodborne Pathog. Dis. 9, 1097–1103. doi: 10.1089/fpd.2012.1184
Douidah L., De Zutter L., Van Nieuwerburgh F., Deforce D., Ingmer H., Vandenberg O., et al. (2014). Presence and analysis of plasmids in human and animal associated arcobacter species. PloS One 9, e85487. doi: 10.1371/journal.pone.0085487
EFSA (2008). Harmonised monitoring of antimicrobial resistance in Salmonella and Campylobacter isolates from food animals in the European Union. Clin. Microbiol. Infect. 14, 522–533. doi: 10.1111/j.1469-0691.2008.02000.x
El-Ashker M., Gwida M., Tomaso H., Monecke S., Ehricht R., El-Gohary F., et al. (2015). Staphylococci in cattle and buffaloes with mastitis in Dakahlia Governorate, Egypt. J. Dairy Sci. 98, 7450–7459. doi: 10.3168/jds.2015-9432
EUCAST (2019). The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Available at: http://www.eucast.org, Version 9.0.
Fanelli F., Di Pinto A., Mottola A., Mule G., Chieffi D., Baruzzi F., et al. (2019). Genomic Characterization of Arcobacter butzleri Isolated From Shellfish: Novel Insight Into Antibiotic Resistance and Virulence Determinants. Front. Microbiol. 10, 670. doi: 10.3389/fmicb.2019.00670
Ferreira S., Fraqueza M. J., Queiroz J. A., Domingues F. C., Oleastro M. (2013). Genetic diversity, antibiotic resistance and biofilm-forming ability of Arcobacter butzleri isolated from poultry and environment from a Portuguese slaughterhouse. Int. J. Food Microbiol. 162, 82–88. doi: 10.1016/j.ijfoodmicro.2013.01.003
Ferreira S., Julio C., Queiroz J. A., Domingues F. C., Oleastro M. (2014a). Molecular diagnosis of Arcobacter and Campylobacter in diarrhoeal samples among Portuguese patients. Diagn. Microbiol. Infect. Dis. 78, 220–225. doi: 10.1016/j.diagmicrobio.2013.11.021
Ferreira S., Queiroz J. A., Oleastro M., Domingues F. C. (2014b). Genotypic and phenotypic features of Arcobacter butzleri pathogenicity. Microb. Pathog. 76, 19–25. doi: 10.1016/j.micpath.2014.09.004
Ferreira S., Queiroz J. A., Oleastro M., Domingues F. C. (2016). Insights in the pathogenesis and resistance of Arcobacter: A review. Crit. Rev. Microbiol. 42, 364–383. doi: 10.3109/1040841X.2014.954523
Ferreira S., Oleastro M., Domingues F. C. (2017). Occurrence, genetic diversity and antibiotic resistance of Arcobacter sp. in a dairy plant. J. Appl. Microbiol. 123, 1019–1026. doi: 10.1111/jam.13538
Ferreira S., Correia D. R., Oleastro M., Domingues F. C. (2018). Arcobacter butzleri Ciprofloxacin Resistance: Point Mutations in DNA Gyrase A and Role on Fitness Cost. Microb. Drug Resist. 24, 915–922. doi: 10.1089/mdr.2017.0295
Gardner S. N., Slezak T., Hall B. G. (2015). kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31, 2877–2878. doi: 10.1093/bioinformatics/btv271
Hanel I., Tomaso H., Neubauer H. (2016). [Arcobacter - an underestimated zoonotic pathogen?]. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz 59, 789–794. doi: 10.1007/s00103-016-2350-7
Hänel I., Hotzel H., Tomaso H., Busch A. (2018). Antimicrobial Susceptibility and Genomic Structure of Arcobacter skirrowii Isolates. Front. Microbiol. 9, 3067. doi: 10.3389/fmicb.2018.03067
Harrass B., Schwarz S., Wenzel S. (1998). Identification and characterization of Arcobacter isolates from broilers by biochemical tests, antimicrobial resistance patterns and plasmid analysis. Zentralbl. Veterinarmed. B 45, 87–94. doi: 10.1111/j.1439-0450.1998.tb00770.x
Hashimi S. M., Huang G., Maxwell A., Birch R. G. (2008). DNA gyrase from the albicidin producer Xanthomonas albilineans has multiple-antibiotic-resistance and unusual enzymatic properties. Antimicrob. Agents Chemother. 52, 1382–1390. doi: 10.1128/AAC.01551-07
Hooper D. C., Jacoby G. A. (2015). Mechanisms of drug resistance: quinolone resistance. Ann. N. Y. Acad. Sci. 1354, 12–31. doi: 10.1111/nyas.12830
Houf K., Devriese L. A., De Zutter L., Van Hoof J., Vandamme P. (2001). Susceptibility of Arcobacter butzleri, Arcobacter cryaerophilus, and Arcobacter skirrowii to antimicrobial agents used in selective media. J. Clin. Microbiol. 39, 1654–1656. doi: 10.1128/JCM.39.4.1654-1656.2001
Hunt M., Mather A. E., Sanchez-Buso L., Page A. J., Parkhill J., Keane J. A., et al. (2017). ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genom. 3, e000131. doi: 10.1099/mgen.0.000131
Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., Rupp R. (2007). Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8, 460. doi: 10.1186/1471-2105-8-460
Isidro J., Ferreira S., Pinto M., Domingues F., Oleastro M., Gomes J. P., et al. (2020). Virulence and antibiotic resistance plasticity of Arcobacter butzleri: Insights on the genomic diversity of an emerging human pathogen. Infect. Genet. Evol. 80, 104213. doi: 10.1016/j.meegid.2020.104213
Johnning A., Kristiansson E., Fick J., Weijdegard B., Larsson D. G. (2015). Resistance Mutations in gyrA and parC are Common in Escherichia Communities of both Fluoroquinolone-Polluted and Uncontaminated Aquatic Environments. Front. Microbiol. 6, 1355. doi: 10.3389/fmicb.2015.01355
Karadas G., Sharbati S., Hanel I., Messelhausser U., Glocker E., Alter T., et al. (2013). Presence of virulence genes, adhesion and invasion of Arcobacter butzleri. J. Appl. Microbiol. 115, 583–590. doi: 10.1111/jam.12245
Kayman T., Abay S., Hizlisoy H., Atabay H. I., Diker K. S., Aydin F. (2012). Emerging pathogen Arcobacter spp. in acute gastroenteritis: molecular identification, antibiotic susceptibilities and genotyping of the isolated arcobacters. J. Med. Microbiol. 61, 1439–1444. doi: 10.1099/jmm.0.044594-0
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Lau R. W., Ho P. L., Kao R. Y., Yew W. W., Lau T. C., Cheng V. C., et al. (2011). Molecular characterization of fluoroquinolone resistance in Mycobacterium tuberculosis: functional analysis of gyrA mutation at position 74. Antimicrob. Agents Chemother. 55, 608–614. doi: 10.1128/AAC.00920-10
Levican A., Alkeskas A., Gunter C., Forsythe S. J., Figueras M. J. (2013). Adherence to and invasion of human intestinal cells by Arcobacter species and their virulence genotypes. Appl. Environ. Microbiol. 79, 4951–4957. doi: 10.1128/AEM.01073-13
Levican A., Collado L., Yustes C., Aguilar C., Figueras M. J. (2014). Higher water temperature and incubation under aerobic and microaerobic conditions increase the recovery and diversity of Arcobacter spp. from shellfish. Appl. Environ. Microbiol. 80, 385–391. doi: 10.1128/AEM.03014-13
Levican A., Rubio-Arcos S., Martinez-Murcia A., Collado L., Figueras M. J. (2015). Arcobacter ebronensis sp. nov. and Arcobacter aquimarinus sp. nov., two new species isolated from marine environment. Syst. Appl. Microbiol. 38, 30–35. doi: 10.1016/j.syapm.2014.10.011
Malik S., Willby M., Sikes D., Tsodikov O. V., Posey J. E. (2012). New insights into fluoroquinolone resistance in Mycobacterium tuberculosis: functional genetic analysis of gyrA and gyrB mutations. PloS One 7, e39754. doi: 10.1371/journal.pone.0039754
Miller W. G., Parker C. T., Rubenfield M., Mendz G. L., Wosten M. M., Ussery D. W., et al. (2007). The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri. PloS One 2, e1358. doi: 10.1371/journal.pone.0001358
Nikaido H. (2009). Multidrug resistance in bacteria. Annu. Rev. Biochem. 78, 119–146. doi: 10.1146/annurev.biochem.78.082907.145923
Onseedaeng S., Ratthawongjirakul P. (2016). Rapid Detection of Genomic Mutations in gyrA and parC Genes of Escherichia coli by Multiplex Allele Specific Polymerase Chain Reaction. J. Clin. Lab. Anal. 30, 947–955. doi: 10.1002/jcla.21961
Ozeki S., Deguchi T., Yasuda M., Nakano M., Kawamura T., Nishino Y., et al. (1997). Development of a rapid assay for detecting gyrA mutations in Escherichia coli and determination of incidence of gyrA mutations in clinical strains isolated from patients with complicated urinary tract infections. J. Clin. Microbiol. 35, 2315–2319. doi: 10.1128/JCM.35.9.2315-2319.1997
Pentimalli D., Pegels N., Garcia T., Martin R., Gonzalez I. (2009). Specific PCR detection of Arcobacter butzleri, Arcobacter cryaerophilus, Arcobacter skirrowii, and Arcobacter cibarius in chicken meat. J. Food Prot. 72, 1491–1495. doi: 10.4315/0362-028X-72.7.1491
Pérez-Cataluña A., Tapiol J., Benavent C., Sarvisé C., Gómez F., Martínez B., et al. (2017). Antimicrobial susceptibility, virulence potential and sequence types associated with Arcobacter strains recovered from human faeces. J. Med. Microbiol. 66, 1736–1743. doi: 10.1099/jmm.0.000638
Pérez-Cataluña A., Collado L., Salgado O., Lefiñanco V., Figueras M. J. (2018a). A Polyphasic and Taxogenomic Evaluation Uncovers Arcobacter cryaerophilus as a Species Complex That Embraces Four Genomovars. Front. Microbiol. 9, 805. doi: 10.3389/fmicb.2018.00805
Pérez-Cataluña A., Salas-Massó N., Diéguez A. L., Balboa S., Lema A., Romalde J. L., et al. (2018ba). Revisiting the Taxonomy of the Genus Arcobacter: Getting Order From the Chaos. Front. Microbiol. 9, 2077. doi: 10.3389/fmicb.2018.02077
Pérez-Cataluña A., Salas-Masso N., Dieguez A. L., Balboa S., Lema A., Romalde J. L., et al. (2018b). Corrigendum: revisiting the taxonomy of the genus Arcobacter: getting order from the chaos. Front. Microbiol. 9, 3123. doi: 10.3389/fmicb.2018.02077
Pérez-Cataluña A., Salas-Massó N., Figueras M. J. (2018c). Arcobacter canalis sp. nov., isolated from a water canal contaminated with urban sewage. Int. J. Syst. Evol. Microbiol. 68, 1258–1264. doi: 10.1099/ijsem.0.002662
Pérez-Cataluña A., Salas-Masso N., Dieguez A. L., Balboa S., Lema A., Romalde J. L., et al. (2019). Corrigendum (2): revisiting the taxonomy of the genus Arcobacter: getting order from the chaos. Front. Microbiol. 10, 2253. doi: 10.3389/fmicb.2019.02253
Prouzet-Mauleon V., Labadi L., Bouges N., Menard A., Megraud F. (2006). Arcobacter butzleri: underestimated enteropathogen. Emerg. Infect. Dis. 12, 307–309. doi: 10.3201/eid1202.050570
Rathlavath S., Kohli V., Singh A. S., Lekshmi M., Tripathi G., Kumar S., et al. (2017a). Virulence genotypes and antimicrobial susceptibility patterns of Arcobacter butzleri isolated from seafood and its environment. Int. J. Food Microbiol. 263, 32–37. doi: 10.1016/j.ijfoodmicro.2017.10.005
Rathlavath S., Kumar S., Nayak B. B. (2017b). Comparative isolation and genetic diversity of Arcobacter sp. from fish and the coastal environment. Lett. Appl. Microbiol. 65, 42–49. doi: 10.1111/lam.12743
Rovetto F., Carlier A., Van Den Abeele A. M., Illeghems K., Van Nieuwerburgh F., Cocolin L., et al. (2017). Characterization of the emerging zoonotic pathogen Arcobacter thereius by whole genome sequencing and comparative genomics. PloS One 12, e0180493. doi: 10.1371/journal.pone.0180493
Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C. (2012). Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814. doi: 10.1038/nmeth.2066
Sekhar M. S., Tumati S. R., Chinnam B. K., Kothapalli V. S., Sharif N. M. (2017). Virulence gene profiles of Arcobacter species isolated from animals, foods of animal origin, and humans in Andhra Pradesh, India. Vet. World 10, 716–720. doi: 10.14202/vetworld.2017.716-720
Shirzad Aski H., Tabatabaei M., Khoshbakht R., Raeisi M. (2016). Occurrence and antimicrobial resistance of emergent Arcobacter spp. isolated from cattle and sheep in Iran. Comp. Immunol. Microbiol. Infect. Dis. 44, 37–40. doi: 10.1016/j.cimid.2015.12.002
Silha D., Pejchalova M., Silhova L. (2017). Susceptibility to 18 drugs and multidrug resistance of Arcobacter isolates from different sources within the Czech Republic. J. Glob. Antimicrob. Resist. 9, 74–77. doi: 10.1016/j.jgar.2017.01.006
Sundin G. W., Bender C. L. (1996). Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol. Ecol. 5, 133–143. doi: 10.1111/j.1365-294X.1996.tb00299.x
Sutera V., Hoarau G., Renesto P., Caspar Y., Maurin M. (2017). In vitro and in vivo evaluation of fluoroquinolone resistance associated with DNA gyrase mutations in Francisella tularensis, including in tularaemia patients with treatment failure. Int. J. Antimicrob. Agents 50, 377–383. doi: 10.1016/j.ijantimicag.2017.03.022
Toh H., Sharma V. K., Oshima K., Kondo S., Hattori M., Ward F. B., et al. (2011). Complete genome sequences of Arcobacter butzleri ED-1 and Arcobacter sp. strain L, both isolated from a microbial fuel cell. J. Bacteriol. 193, 6411–6412. doi: 10.1128/JB.06084-11
Treangen T. J., Ondov B. D., Koren S., Phillippy A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524. doi: 10.1186/s13059-014-0524-x
Uenver A., Atabay H., Sahin M., Celebi Ö. (2013). Antimicrobial susceptibilities of various Arcobacter species. Turk. J. Med. Sci. 43 (2013), 548–552. doi: 10.3906/sag-1207-115
Van den Abeele A. M., Vogelaers D., Van Hende J., Houf K. (2014). Prevalence of Arcobacter species among humans, Belgium 2008-2013. Emerg. Infect. Dis. 20, 1731–1734. doi: 10.3201/eid2010.140433
Van den Abeele A. M., Vogelaers D., Vanlaere E., Houf K. (2016). Antimicrobial susceptibility testing of Arcobacter butzleri and Arcobacter cryaerophilus strains isolated from Belgian patients. J. Antimicrob. Chemother. 71, 1241–1244. doi: 10.1093/jac/dkv483
Vandamme P., Falsen E., Rossau R., Hoste B., Segers P., Tytgat R., et al. (1991). Revision of Campylobacter, Helicobacter, and Wolinella taxonomy: emendation of generic descriptions and proposal of Arcobacter gen. nov. Int. J. Syst. Bacteriol. 41, 88–103. doi: 10.1099/00207713-41-1-88
WHO (2018). Campylobacter Fact sheet. Available at: www.who.int/mediacentre/factsheets/fs255/en/.
Wick R. R., Schultz M. B., Zobel J., Holt K. E. (2015). Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31, 3350–3352. doi: 10.1093/bioinformatics/btv383
Wood D. E., Salzberg S. L. (2014). Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46. doi: 10.1186/gb-2014-15-3-r46
Yamachika S., Onodera Y., Hiramatsu K., Takase H. (2012). Plasmid integration method: a new tool for analysis of the essentiality and function of genes in S. aureus. J. Microbiol. Methods 90, 250–255. doi: 10.1016/j.mimet.2012.05.017
Yan J. J., Ko W. C., Huang A. H., Chen H. M., Jin Y. T., Wu J. J. (2000). Arcobacter butzleri bacteremia in a patient with liver cirrhosis. J. Formos. Med. Assoc. 99 (2), 166–169.
Yesilmen S., Vural A., Erkan M. E., Yildirim I. H. (2014). Prevalence and antimicrobial susceptibility of Arcobacter species in cow milk, water buffalo milk and fresh village cheese. Int. J. Food Microbiol. 188, 11–14. doi: 10.1016/j.ijfoodmicro.2014.07.006
Keywords: Aliarcobacter, genomic analysis, antibiotic susceptibility, ciprofloxacin resistance, sequencing
Citation: Hänel I, Müller E, Santamarina BG, Tomaso H, Hotzel H and Busch A (2021) Antimicrobial Susceptibility and Genomic Analysis of Aliarcobacter cibarius and Aliarcobacter thereius, Two Rarely Detected Aliarcobacter Species. Front. Cell. Infect. Microbiol. 11:532989. doi: 10.3389/fcimb.2021.532989
Received: 06 February 2020; Accepted: 19 January 2021;
Published: 17 March 2021.
Edited by:
Mónica Oleastro, Instituto Nacional de Saúde Doutor Ricardo Jorge (INSA), PortugalReviewed by:
Charles William Stratton, Vanderbilt University Medical Center, United StatesCopyright © 2021 Hänel, Müller, Santamarina, Tomaso, Hotzel and Busch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anne Busch, YW5uZS5idXNjaEBtZWQudW5pLWplbmEuZGU=; orcid.org/0000-0001-9560-0057
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.