- 1Department of Microbiology and Immunology, Tulane University School of Medicine, New Orleans, LA, United States
- 2Center for Microbial Pathogenesis, Abigail Wexner Research Institute at Nationwide Children’s Hospital, Columbus, OH, United States
- 3Infectious Diseases Institute, The Ohio State University, Columbus, OH, United States
- 4Department of Pediatrics, College of Medicine, The Ohio State University, Columbus, OH, United States
Within the species of Salmonella enterica, there is significant diversity represented among the numerous subspecies and serovars. Collectively, these account for microbes with variable host ranges, from common plant and animal colonizers to extremely pathogenic and human-specific serovars. Despite these differences, many Salmonella species find commonality in the ability to form biofilms and the ability to cause acute, latent, or chronic disease. The exact outcome of infection depends on many factors such as the growth state of Salmonella, the environmental conditions encountered at the time of infection, as well as the infected host and immune response elicited. Here, we review the numerous biofilm lifestyles of Salmonella (on biotic and abiotic surfaces) and how the production of extracellular polymeric substances not only enhances long-term persistence outside the host but also is an essential function in chronic human infections. Furthermore, careful consideration is made for the events during initial infection that allow for gut transcytosis which, in conjunction with host immune functions, often determine the progression of disease. Both typhoidal and non-typhoidal salmonellae can cause chronic and/or secondary infections, thus the adaptive immune responses to both types of bacteria are discussed with particular attention to the differences between Salmonella Typhi, Salmonella Typhimurium, and invasive non-typhoidal Salmonella that can result in differential immune responses. Finally, while strides have been made in our understanding of immunity to Salmonella in the lymphoid organs, fewer definitive studies exist for intestinal and hepatobiliary immunity. By examining our current knowledge and what remains to be determined, we provide insight into new directions in the field of Salmonella immunity, particularly as it relates to chronic infection.
Introduction
Most bacteria can grow as either individual planktonic cells or as communities within a biofilm (Flemming and Wuertz, 2019). Biofilms are surface-attached aggregates of cells surrounded by a self-produced extracellular polymeric substances (EPSs) (Costerton et al., 1999; Branda et al., 2005; Hall-Stoodley and Stoodley, 2009; Flemming, 2016). The development of these structures often occurs in response to perceived negative environmental stimuli such as shifts in pH, temperature, oxygen, and nutrient availability (O’Toole and Kolter, 1998; O’Toole et al., 2000a; O’Toole et al., 2000b). Individual cells adhere to a surface, biotic or abiotic, producing EPSs such as proteins, exopolysaccharides, and nucleic acids. In this niche, they grow and mature, at which point some cells detach and return to planktonic growth to repeat the process (Khatoon et al., 2018). While the bacteria reside within a biofilm they are protected from a variety of challenges such as UV exposure (Espeland and Wetzel, 2001), host defense mechanisms, and antibiotics (Mah and O’Toole, 2001; Stewart and Costerton, 2001). As a result, biofilms play an essential role in the environment, industry, and medicine.
In the natural environment, biofilms are important for microbial survival. Biofilms provide a stable environment for the bacteria by fostering symbiotic relationships with other organisms, such as settlement of marine invertebrate larvae (Nelson et al., 2020), degrading harmful chemicals in the soil (Ahmad et al., 2017), and promoting plant growth (Liu et al., 2013). However, biofilms are less desirable in industries such as food, water, energy, and medicine, where they can cause harm by developing on food and food processing equipment (Flemming, 2016; Galie et al., 2018), contaminating water pipelines (Wingender and Flemming, 2011), corroding underwater metal surfaces (de Carvalho, 2018), and persisting on medical devices (Percival et al., 2015).
Here we focus on biofilms in the field of medicine. Biofilm communities can enhance recalcitrance to host defense mechanisms. As a result, biofilms are estimated to be involved in approximately 80% of chronic infections, which increase hospitalization rates, healthcare costs, and morbidity and mortality (Davies, 2003). Examples of biofilm-associated diseases include several respiratory diseases, chronic otitis media, periodontitis, and chronic wound infections (Høiby et al., 2010; Wessman et al., 2014; Bao et al., 2015; Wu et al., 2015). In cystic fibrosis (CF), persistent lung infections are often caused by Pseudomonas aeruginosa biofilms or aggregates. Due to the ability of the biofilm to resist phagocytosis via polysaccharide synthesis locus (Psl) (Mishra et al., 2012) and other EPS components, chronic inflammation causes lung tissue damage (Høiby et al., 2010). Additionally, CF patients co-infected with Staphylococcus aureus have been shown to have a greater decline in lung function than when infected with P. aeruginosa alone (Limoli et al., 2016; Maliniak et al., 2016; Limoli and Hoffman, 2019). Such interspecies interactions alter the efficacy of antibiotics and lead to worse patient outcomes.
Biofilms also develop on abiotic medical devices such as prosthetics, stents, catheters, implants, and dentures (Percival et al., 2015). Biofilms on indwelling medical devices are estimated to cause 50% of persistent nosocomial infections (Paredes et al., 2014). The most commonly identified biofilms are formed by Staphylococcus epidermidis, S. aureus, and P. aeruginosa (Hall-Stoodley et al., 2004; Khatoon et al., 2018). Staphylococcus epidermidis is the model bacterial species for dental implant-related biofilms, which are different than the biofilms that form on teeth (Daubert and BF, 2019). The shape of the dental implant makes complete elimination almost impossible (Müsken et al., 2018). The bacteria that survive treatment result in persistent biofilm infections, implant failure, and promote the development of antibiotic resistance.
Multidrug resistant bacteria have become a serious medical concern, accounting for over 2.8 million infections and 35,000 deaths annually in the United States (CDC, 2019). Bacteria residing within a biofilm are also recalcitrant to antibiotics, surviving levels 10–1,000 times greater than planktonic cells (Høiiby et al., 2010; Mah, 2012). The close association of bacteria in a biofilm can also promote antibiotic gene transfer (Christensen et al., 1998; Hausner and Wuertz, 1999; Li et al., 2018). Biofilm recalcitrance to antibiotics is due to complex factors that are incompletely understood and often involve multiple species, which limits the efficacy of antibiotic clearance (Jiang et al., 2020). While improvements have been made in preventing biofilm formation, such as the development of medical devices made or coated with antibacterial materials (Vasilev et al., 2018), surgical removal of the infected tissue or medical device is often required for complete resolution (Høiiby et al., 2010).
Infections due to biofilm-associated bacteria are a significant clinical problem. Therefore, further studies are needed to identify the key aspects of biofilm formation and persistence in order to develop novel preventative and therapeutic strategies to enhance the efficacy of treatment.
Biofilms and Chronic Infections
Whether comprised of a single species or polymicrobial, biofilms demonstrate the remarkable ability of bacteria to coordinate functions and develop complex behaviors. While the discovery of biofilms coincides with the discovery of microorganisms by Antoni van Leeuwenhoek in 1683, this unique modality of growth was originally overlooked in a medical context, gaining prominence only in the past 50 years. However, the study of infectious biofilms has overwhelmingly demonstrated their important role in chronic infections, with, as mentioned previously, estimates by the National Institutes of Health that 80% of all chronic infections are related to biofilms (Davies, 2003).
Whether opportunistic or primary pathogens, many bacteria are capable of causing acute or chronic infections, depending on circumstance. While we do not wish to downplay the medical importance of acute infections, we will mention them only briefly in this review as our focus is chronic infections. Acute infections coincide with rapid growth and dissemination in the host leading to overt clinical signs days after onset of disease (Furukawa et al., 2006; Thakur et al., 2019). Acute infections generally lead to one of three consequences: successful clearance (either naturally by the immune system or with medical intervention), death of the host, or development into a latent infection. In this third outcome, the acute phase of infection is followed by a dormant phase with long-term infection that can span the life of the host and is characterized by repeated spells of reactivation (Thakur et al., 2019). This is a type of chronic infection. Reactivation is associated with production of infectious agents that can be transmitted to others although this activity may or may not renew symptoms in the host (Thakur et al., 2019).
While latent infections only develop after an initial acute phase, many pathogens do not cause acute disease and in these cases may directly result in a chronic infection. These chronic infections can arise opportunistically from a compromised immune system, an altered microbiota, a breach in skin or mucosal immune barriers, or from contamination at surgical sites and on implanted medical devices (Furukawa et al., 2006; Thakur et al., 2019). Both bacteria and host responses to their environment can result in genetic or phenotypic alterations that foster chronic infection. Several recent reviews have catalogued various chronic infections by body site (Vestby et al., 2020), organism and immune response (Thakur et al., 2019), or survival strategy (facultative intracellular, small colony variants, persister populations, environmentally induced antibiotic indifference, and biofilm formation) (Grant and Hung, 2013).
A common theme of chronic infections is their ability to last at least a few months, often years, and even up to the entire lifetime of the host. While a long-term infection in the absence of acute disease is the most rudimentary definition, other characteristics are fundamental to chronic infections. Similar to latent infections, chronic infections are generally asymptomatic and usually do not pose immediate risk to host health (Grant and Hung, 2013; Thakur et al., 2019). However, the possibility of future reactivation of latent/chronic infections into clinically significant disease, disease dissemination, or the onset of malignancy all pose long-term risks (Grant and Hung, 2013). There is often a direct link between chronic infections and biofilm formation (Parsek and Singh, 2003; Hall-Stoodley et al., 2004; Furukawa et al., 2006) as long-term survival requires pathogen stealth and/or a protected niche for the pathogen. Both properties are enhanced by biofilms and while the niche could be provided by granulomas or intracellular compartments, survival in these locations is associated with slowed growth and reduced replicative capacity. However, unlike latent infections, chronic infections often have a high level of bacterial replication and concomitant burden of the pathogen (albeit less than acute infection) (Furukawa et al., 2006; Thakur et al., 2019). This activity is possible by forming a biofilm, which alters the bacterial growth physiology to allow for significant tolerance to elevated levels and prolonged administration of antibiotics as well as tolerance to other host responses such as the complement system, antimicrobial peptides, antibodies, and phagocytic activity by neutrophils and macrophages as further discussed below (Furukawa et al., 2006; Grant and Hung, 2013; Pletzer et al., 2017; Thakur et al., 2019). Furthermore, biofilm formation permits shedding of planktonic bacteria and/or biologically active molecules into the host (Zegans et al., 2002; Furukawa et al., 2006) which has pleiotropic effects on the host immune state and is important for maintaining a favorable niche by altering the activation of innate immune receptors, inhibiting apoptosis, inducing an inappropriate immune response, or causing immunosuppression (Thakur et al., 2019). Chronic infections often maintain their niche by simultaneous activation of both the innate and adaptive immune responses (Vestby et al., 2020), but interestingly, neither of these responses eliminate the pathogen but instead result in collateral damage to host cells and tissue (Moser et al., 2017; Vestby et al., 2020). Accumulation of macrophages and neutrophils at the site of infection is characteristic of chronic infection (Laskin et al., 2011; Pletzer et al., 2017). Prolonged reactive oxygen species and reactive nitrogen species production by these cells represent a significant source of inflammation (Grant and Hung, 2013; Mittal et al., 2014; Pletzer et al., 2017). This activity ultimately benefits the pathogen as chronic inflammation from an aberrant immune response makes host nutrients available to the biofilm (Pletzer et al., 2017; Thakur et al., 2019), obstructs would healing, and facilitates cellular invasion for dissemination, access to other protected sites, or acute infection (Vestby et al., 2020). Chronic inflammation is also linked to many malignancies associated with chronic infection, including gastric adenocarcinoma and gastric lymphoma from Helicobacter pylori infections (Wotherspoon et al., 1991; Nomura et al., 1994) or gallbladder carcinoma from Salmonella Typhi infections (Caygill et al., 1994; Grant and Hung, 2013; Vestby et al., 2020).
To highlight an example, P. aeruginosa, mentioned earlier, is well-suited for opportunistic chronic infections. While ubiquitous in soil and water and a common colonizer of animals and humans, P. aeruginosa rarely infects healthy individuals but can infect multiple body sites in the immunocompromised (Wu and Li, 2015). While P. aeruginosa may have the most notoriety for infecting CF patients, it is also consequential in ventilator-associate pneumonia (Bergmans and Bonten, 1999; Mulcahy et al., 2014), on orthopedic implants and joint replacement surgical sties (Mulcahy et al., 2014), catheters (Cole et al., 2014), and in severe soft tissue wounds (Lochab et al., 2020). Characteristic of chronic biofilm infections, P. aeruginosa infections are difficult to detect with clinical microbiology techniques (Costerton et al., 2003; Mulcahy et al., 2014) and are extremely recalcitrant to antibiotics and the host environment (Mulcahy et al., 2014; Lochab et al., 2020). In 2015, hospitalized cancer, CF, or burn wound patients with P. aeruginosa infection experienced a 50% mortality rate (Gómez and Prince, 2007; Wu and Li, 2015) and the CDC reports that P. aeruginosa is the fourth-most isolated pathogen in hospitals, accounting for 10% of all nosocomial infections (Wu and Li, 2015). There is ample evidence to conclude P. aeruginosa relies on a biofilm forming strategy for survival in the host and that P. aeruginosa biofilms control the host response to establish a favorable environment. In most instances of P. aeruginosa disease, the sustained collateral damage from frustrated neutrophil phagocytosis at the site of biofilm infection is the primary cause of major disease sequellae (Jensen et al., 2010; Mulcahy et al., 2014). Remarkably, studies of CF infants have demonstrated P. aeruginosa biofilms remodel host immunity from a balanced Th1/Th2 response to a Th2 response with reduced levels of IFN-γ (Moser et al., 2000; Mulcahy et al., 2014).
Salmonella Typhi is an especially interesting pathogen because of its ability to cause disease in all three mechanisms discussed. As the primary etiologic agent of typhoid fever (Stanaway et al., 2019) (a systemic disease that can be deadly) the organism must be considered an acute pathogen. However, the only known reservoir of S. Typhi is human carriers who have long-term infections primarily by biofilms on cholesterol gallstones in the gallbladder. Interestingly, about 5% of acute infection survivors will develop latent infections but as many as 25% of chronic carriers have gallstone biofilms without ever expressing symptoms of acute disease (Parry et al., 2002; Gonzalez-Escobedo et al., 2010; Gunn et al., 2014). These different disease strategies (acute, latent, and chronic) highlight the complex nature of host-pathogen interactions during chronic infection.
Salmonella Chronic Infections
In addition to S. Typhi, many other serovars of Salmonella enterica possess the ability to colonize and potentially cause disease in humans and/or other animal hosts. Biofilm formation in these serovars is highly conserved—especially in the serovars that are able to colonize multiple hosts—suggesting that the ability to form a biofilm serves as an evolutionarily advantage during the cycle of infection and transmission (Römling et al., 2003; MacKenzie et al., 2017; MacKenzie et al., 2019). As with other biofilm-forming bacteria, biofilm-associated Salmonella are encased within a matrix of EPSs consisting of proteins, carbohydrates, and extracellular DNA (eDNA) (Maruzani et al., 2019). The exact composition of the Salmonella biofilm EPSs may vary depending on both serovar and environmental conditions, but primarily consists of a network of proteinaceous curli fimbriae fibers, cellulose, and eDNA (Zogaj et al., 2001; Römling, 2005; Wang et al., 2014). Additional EPS components may include cellular appendages such as flagella or various adhesive fimbriae (Römling and Rohde, 1999; Boddicker et al., 2002; Prouty et al., 2002; Prouty and Gunn, 2003; Ledeboer et al., 2006; Dwyer et al., 2011); cell surface proteins such as the large biofilm-associated surface protein BapA (Latasa et al., 2005); and other exopolysaccharides such as colanic acid or the O-antigen capsule (Ledeboer and Jones, 2005; Crawford et al., 2008). A self-secreted “common good,” these EPSs collectively allow cells to securely adhere to both surfaces and other cells within the biofilm, help retain moisture in dry environments, and possibly slow the diffusion of harmful molecules such as antimicrobial peptides and antibiotics (Dieltjens et al., 2020).
The transition of planktonic Salmonella into a biofilm state may be influenced by a variety of environmental triggers, including temperature fluctuations, changes in nutrient availability, and exposure to harsh or harmful substances (De Oliveira et al., 2014; Paytubi et al., 2017; González et al., 2019b). Accordingly, the regulatory network controlling the expression of biofilm genes is highly complex. Numerous global regulators, two-component response systems, and other regulatory proteins and small RNAs contribute to regulate expression of key biofilm genes, primarily through expression of csgD (Simm et al., 2014). An orphan response regulator encoded within the curli biosynthesis operon, CsgD is often regarded as the master biofilm regulator in Salmonella (Gerstel and Römling, 2003). CsgD induces the production of the two major EPS components, curli fimbriae and cellulose, by directly binding the csgB promoter within the csgBAC curli biosynthesis operon and by inducing expression of the diguanylate cyclase AdrA, which subsequently regulates cellulose biosynthesis through the modulation of the secondary messenger (3’-5’)-cyclic-diguanosine monophosphate (c-di-GMP) (Römling et al., 2000; Zakikhany et al., 2010). Evidence suggests CsgD may also regulate the expression of other EPS genes, including those responsible for the production of BapA and the O-antigen capsule (Latasa et al., 2005; Gibson et al., 2006).
Outside of the host, biofilm formation allows for Salmonella to attach to a variety of biotic and abiotic surfaces, thereby enabling the bacteria to persist in a viable but relatively dormant state until ultimately ingested by a host. Because Salmonella are enteric pathogens, non-typhoidal Salmonella biofilms are of particular concern in the agricultural and food processing and packaging industries where they contaminate both fresh and processed food products (Yaron and Römling, 2014; Pande et al., 2016; Galie et al., 2018; Lamas et al., 2018; Merino et al., 2019). Contamination of food products with Salmonella typically originates from colonized or infected livestock including chickens, pigs, and cattle (Lamas et al., 2018). This contamination may be direct, e.g., if the intestinal contents of infected animal carcasses are released during processing, but is more often indirect, occurring when uncompromised products come into contact with previously contaminated processing surfaces or machinery (Reij and Den Aantrekker, 2004; Giaouris et al., 2012). Salmonella has been shown to adhere as a biofilm to multiple abiotic materials used in industrial settings, including stainless steel, polystyrene, and glass (Joseph et al., 2000; Giaouris and Nychas, 2006; Chia et al., 2009; Kim and Wei, 2009). While sanitation practices are implemented to disinfect potential sources of cross-contamination, Salmonella within a biofilm are much more recalcitrant to disinfectants and other antimicrobials compared to their planktonic counterparts, rendering such practices ineffective (Joseph et al., 2000; Sheffield et al., 2009; Corcoran et al., 2014; Galie et al., 2018). Additionally, after a product has been contaminated, Salmonella can also form a biofilm directly on the surface of the food itself. Raw meat and poultry, fresh fruits and vegetables, and even low-moisture processed foods such as cereal products have been shown to support Salmonella biofilm growth, supporting the theory that biofilm formation is a critical method of persistence outside of the host (Tamblyn et al., 1997; Podolak et al., 2010; Yaron and Römling, 2014; Lamas et al., 2018).
Salmonella Biofilms Within the Infected Host
While the association of the Salmonella biofilm phenotype with persistence outside of the host has long been established, additional research was required to fully interrogate the role of Salmonella biofilms in chronic infection within the host. While many researchers originally doubted biofilm formation was possible in vivo due to the lack of significant csgD expression observed in the majority of isolates tested at temperatures ≥37°C (Römling et al., 1998; Römling et al., 2003; White et al., 2008) there is now ample evidence to show Salmonella produces EPSs in vivo and that chronic infections are mediated by biofilms. Regarding the effect of temperature on csgD expression, further investigations revealed the true complexity of the regulatory network governing csgD expression; some postulated that other environmental signals encountered in vivo might be able to override the effects of temperature on csgD expression (Simm et al., 2014). While the intricacies of such a regulatory hierarchy are still under investigation, numerous research groups have also published evidence of Salmonella biofilm formation in various in vivo systems. Perhaps most well-documented is the contribution of biofilms to chronic gallbladder carriage of the human-specific S. Typhi (Parry et al., 2002; Bhan et al., 2005; Gonzalez-Escobedo et al., 2011; Gunn et al., 2014). As mentioned previously, while infection with S. Typhi most commonly results in acute disease (typhoid fever), chronic infections may also develop (Parry et al., 2002; Bhan et al., 2005; Gunn et al., 2014). The majority of these chronic infections are localized to the gallbladder and progression to chronic disease is associated with the presence of gallstones or other gallbladder abnormalities as this host environment provides an appropriate surface for S. Typhi to attach and establish robust biofilms (Schioler et al., 1983; Lai et al., 1992). Initial attachment of S. Typhi to gallstone cholesterol surfaces is mediated by FliC on flagellar appendages (Crawford et al., 2010a). However, S. Typhi can also anchor itself to the gallbladder epithelium, suggesting additional biofilm-forming capabilities in vivo (Crawford et al., 2010b; Gonzalez-Escobedo and Gunn, 2013; Marshall et al., 2014). This mechanism of chronic infection has also been recapitulated in mice by intraperitoneal infection of NRAMP+/+ mice with S. Typhimurium (Crawford et al., 2010b). Importantly, the chronic mouse model of infection involves treatment of the mice for 8 weeks prior to infection with a lithogenic diet to induce gallstone formation, thus providing an optimal environment for Salmonella biofilm formation. In this context, biofilms provide Salmonella with increased recalcitrance to the harsh gallbladder environment (which includes detergent-like bile salts) and other potential threats such as the host immune system or antibiotic therapy, allowing for long-term survival within the host and prolonged transmissibility via intermittent fecal shedding (Jolivet-Gougeon and Bonnaure-Mallet, 2014; González et al., 2018; Hahn and Gunn, 2020; Tsai et al., 2020). Chronic gallstone biofilm carriers can shed S. Typhi for at least a year or longer (Gonzalez-Escobedo et al., 2011). This population represents the only known reservoir of S. Typhi, with subsequent transmission to others either directly or through contamination of food and water, which is a significant public health challenge for addressing endemic typhoid fever.
The extent to which non-typhoidal Salmonella (NTS) serovars utilize biofilm formation during the natural course of infection is less apparent. In vitro, numerous NTS isolates exhibit biofilm-like adherence to both human and chicken epithelial cells; however, the effects of biofilm-mediated adherence on virulence were variable (Solano et al., 2001; Ledeboer and Jones, 2005; Ledeboer et al., 2006; Lamprokostopoulou et al., 2010). Whereas S. Typhimurium biofilm formation on HT-29 cells was associated with a relative decrease in virulence characterized by reduced invasion and production of pro-inflammatory IL-8, the level of S. Enteritidis biofilm formation on HEp-2 and Caco-2 cells was positively correlated with virulence as measured by the extent of epithelial barrier disruption (Solano et al., 2001; Lamprokostopoulou et al., 2010). While the transition to the biofilm phenotype is likely to have pleiotropic effects on the Salmonella virulence program, evidence from in vivo studies suggests that, overall, biofilm formation is utilized to simultaneously reduce virulence and increase persistence. Within the cecum of newborn chickens, biofilm formation is a key asset for both S. Typhimurium and S. Pullorum, allowing the bacteria to successfully overcome colonization resistance and establish persistent asymptomatic infection (Sheffield et al., 2009; El Hag et al., 2017). Salmonella Typhimurium biofilms have also been implicated in the formation of extracellular aggregates in vivo. In the C. elegans gut, CsgD-dependent formation of aggregates was associated with relative increases in both bacterial persistence and host survival, a phenotypic shift that likely contributes to the high transmissibility of similar aggregative populations within the cecum and colon of mice (Lam and Monack, 2014; Desai and Kenney, 2019; Desai et al., 2019). While persistent and/or recurrent NTS infections of the human intestine have been documented, reports of cases are limited and do not provide any concrete evidence that would implicate the involvement of biofilms (Musher and Rubenstein, 1973; Kazemi et al., 1974; Buchwald and Blaser, 1984; Marzel et al., 2016; Gal-Mor, 2019). However, antibodies to curli have been detected in the serum of patients recovering from NTS infection and recent work in mice orally infected with S. Typhimurium demonstrated the production of curli amyloids in the gastrointestinal tract, suggesting that expression of biofilm genes does occur on some level during human infection (Tursi and Tukel, 2018; Miller et al., 2020).
In summary, the ability to form a biofilm is a conserved trait found in numerous serovars of S. enterica. Biofilm formation allows Salmonella to persistently colonize sites both inside and outside of the animal host, ultimately enhancing both bacterial survival and transmission (Figure 1).
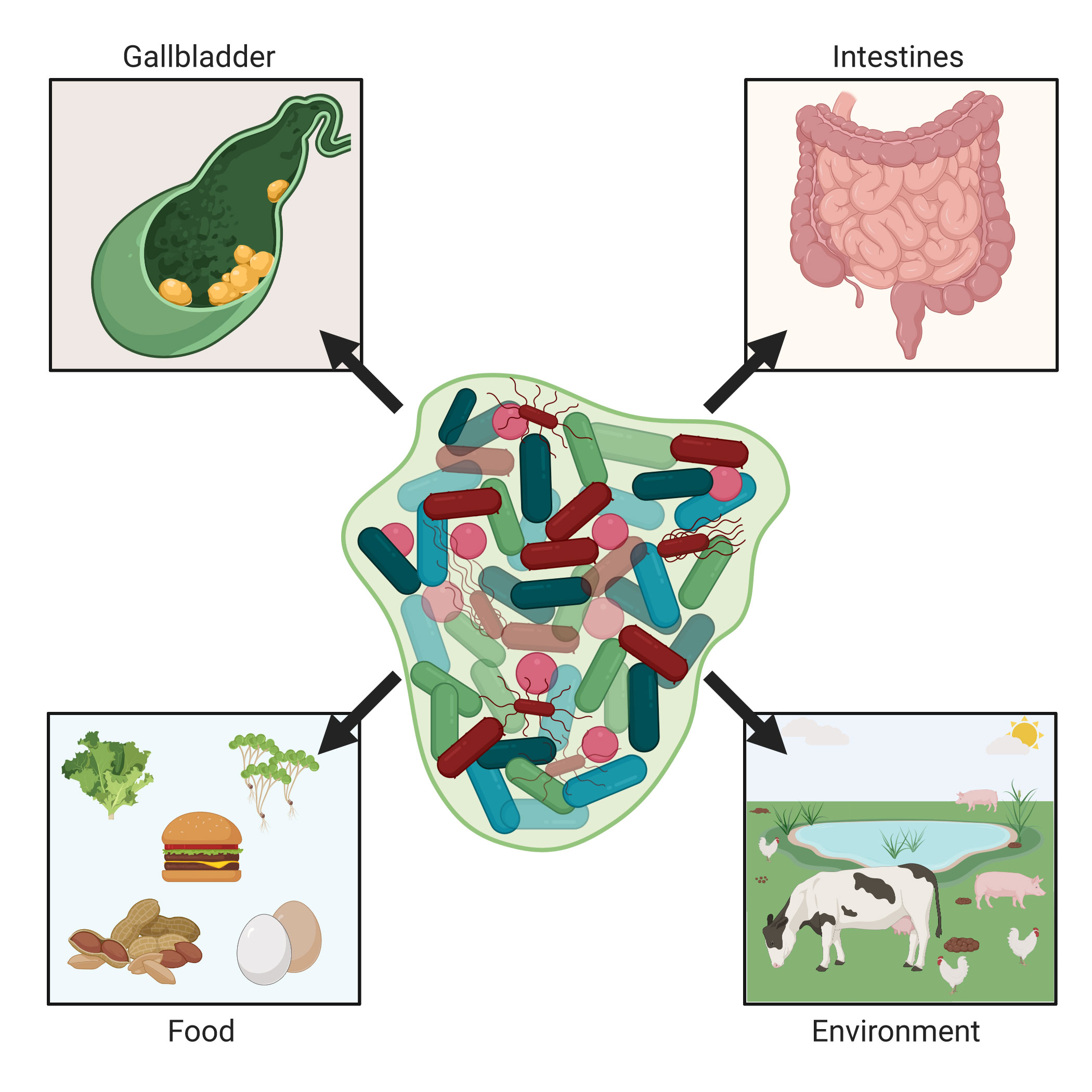
Figure 1 Salmonella biofilms in the environment and humans. Non-typhoidal and typhoidal salmonellae are naturally acquired by humans through environmental or food sources. Salmonella Typhimurium colonizes and can form biofilms on various types of produce, while S. Enteritidis is often found within and on eggs. When within the intestines of various non-human animal species, salmonellae can cause either diarrheal disease or be a non-pathogenic inhabitant (e.g., in the chicken). This can lead to contaminated meat upon animal processing or shedding resulting in contaminated animal feces. Once ingested by the host, Salmonella spp. are capable of forming biofilms in the intestines and for S. Typhi, after systemically gaining access to the liver, can pass into the gallbladder and form biofilms on cholesterol gallstones. These biofilms within humans facilitate chronic Salmonella infection as well as continual shedding of Salmonella from the host. Images created with BioRender.com.
Salmonella Typhi Early Infection and Invasion of the Gastrointestinal Tract
After ingestion, the first site of infection is typically the small intestine where S. Typhi uses its SPI-1 type III secretion system (T3SS-1) to enable penetration of the epithelial layer of the small intestine, typically via microfold (M) cells of the Peyer’s patches (Kohbata et al., 1986). Salmonella Typhi can be found within the Peyer’s patches 3 to 6 h post infection (Carter and Collins, 1974). This invasion of M cells leads to their destruction, further disrupting the intestinal barrier and promoting increased entry of Salmonella (Jones et al., 1994). Salmonella is also capable of disrupting the tight junctions found between epithelial cells which further increases permeability (Boyle et al., 2006). Following entry at Peyer’s patches, the primary innate immune response is initiated in resident dendritic cells (DCs) and macrophages. These cells recognize Salmonella via the pattern recognition receptors called toll-like receptors (TLRs) and NOD-like receptors (NLRs) that detect pathogen associated molecular patterns (PAMPs), such as lipopolysaccharide (LPS). These PAMPs trigger an immune response through a multitude of TLRs expressed on the surface of immune cells, notably TLR4, TLR5, and TLR9 (Zeng et al., 2003; Lahiri et al., 2010; Rathinam et al., 2019). LPS, particularly the lipid A portion, is recognized by TLR4 on DCs or macrophages. Salmonella can adapt in vivo to modify its LPS, which promotes immune evasion via decreased colonization and pro-inflammatory cytokine production as well as by eliciting a change in the type I interferon response (Kong et al., 2012; Avraham et al., 2015).
Salmonella Typhi expresses a polysaccharide capsule with incredible antigenic capability (Parry et al., 2002). The Vi antigen is an important virulence factor of typhoid-inducing Salmonella and is encoded on the Salmonella pathogenicity island (SPI) SPI-7 (Seth-Smith, 2008). This capsular antigen resists complement deposition and reduces phagocytic death by decreasing PAMP expression (Sharma and Qadri, 2004; Atif et al., 2014; Wangdi et al., 2014). Additionally, Vi antigen and other biofilm exopolysaccharides are advantageous in that they can mask LPS, allowing the pathogen to evade TLR4 recognition by both DCs and macrophages (Wilson et al., 2008). An additional virulence factor important for S. Typhi pathogenesis is typhoid toxin, which has been shown to cause DNA damage that can suppress the intestinal inflammatory response, which could potentially increase the frequency of chronic asymptomatic carriers. An additional sequela of chronic colonization of the gallbladder is the development of gallbladder cancer, which could also potentially be related to the typhoid toxin (Del Bel Belluz et al., 2016).
Once recognized by innate immune cells, Salmonella is readily phagocytosed by both DCs and macrophages, in which they can reside within a specialized intracellular structure called a Salmonella containing vacuole (SCV). DCs infected with Salmonella traffic to the gut-draining mesenteric lymph nodes (mLNs) where they present antigen to naïve T cells. Activation of T cells in either the mLNs or Peyer’s patches requires CD11c+CCR6+ DCs (Salazar-Gonzalez et al., 2006). The mLNs are an important site of infection control, as it is known that mice lacking mLNs demonstrate increased bacterial burdens in systemic tissues following relapse of the primary infection due to increased colonization of the liver and spleen (Voedisch et al., 2009). Once phagocytosed, NLRs recognize bacterial components in the cytosol and trigger an inflammasome response (Ferrand and Ferrero, 2013). The response of the innate immune system leads to downstream effects in the adaptive immune response, which is essential for long term infection control of persistent Salmonella infection.
Adaptive Immune Responses to Intestinal Salmonella Typhi Infection
The cell-mediated adaptive immune response, as opposed to the humoral response, is important for cytokine production by T cells that have a significant role in infection control. Both CD4+ and CD8+ T cells are important during Salmonella infection of humans due to their production of IFN-γ (Bao et al., 2000; McSorley et al., 2000). Results from controlled human infection studies have shown that regulatory CD4+ T cells (Tregs) are important in suppressing effector T cells (Clay et al., 2020). The activation of Tregs is likely important for balancing the pro-inflammatory response with the suppressive immune response, both of which are potentially damaging to the host if not carefully regulated (Johanns et al., 2010; McArthur et al., 2015). Salmonella Typhi genes, such as tviA, which encodes a protein involved in Vi antigen synthesis, can interfere with expansion of flagellin-specific CD4+ T cells at mucosal sites by reducing the amount of the dominant Salmonella antigen FliC; this change in gene expression helps S. Typhi to be able to disseminate more rapidly compared to S. Typhimurium (Atif et al., 2014). Several other studies highlight that Salmonella is capable of ubiquitinating surface major histocompatibility complex II (MHC-II), thereby inducing its degradation; the resulting loss of surface MHCII expression allows the bacterial invaders to evade the CD4+ T cell response (Cheminay et al., 2004; Mitchell et al., 2004; Halici et al., 2008; Lapaque et al., 2009; Jackson et al., 2013). A study in 2016 found that CD8+ T cells were critical for infection control of S. Typhi in humans; the T effector memory and T effector memory CD45RA+ cells were the subsets most associated with protection from typhoid-like disease, as those who were protected from typhoidal disease showed higher S. Typhi-specific responses at baseline (Fresnay et al., 2016).
The responses seen in both CD4+ and CD8+ T cells depend largely on initial stimulation. CD4+ T cell responses are initiated against soluble antigens, like flagellin, whereas CD8+ T cell responses are derived from S. Typhi-infected antigen presenting cells, predominantly DCs (McSorley et al., 2002; Jones-Carson et al., 2007; Bobat et al., 2011). In the presence of S. Typhi, DCs mature through suicide-cross presentation and are able to induce expansion of Salmonella-specific CD8+ T cells (Salerno-Goncalves and Sztein, 2009). The relationship between B cells and control of S. Typhi infection in humans is unclear. However, it is known that B cells are required to generate antibodies and to prime T cells in the context of Salmonella infections. Antibodies, specifically IgM, play a role in typhoidal disease protection and require help from IFN-γ producing CD4+ T cells to maintain a memory response (Perez-Shibayama et al., 2014). Furthermore, anti-Salmonella serum IgG and secreted IgA (sIgA) are found in patients infected with S. Typhi and B cells appear to be important during secondary infections as they may serve as antigen presenting cells to prime the CD4+ T cell response (Lopez-Medina et al., 2014).
Adaptive Immune Responses to Intestinal Salmonella Typhimurium Infection
While S. Typhi and S. Typhimurium share 90% of their genetic sequence, this difference is enough to alter host immune responses (Tang et al., 2013). Salmonella Typhimurium is one of the most common serovars of Salmonella to cause gastroenteritis infections in humans and is also able to establish typhoid-like infection in mice, providing an insightful infection model for the study of systemic and/or chronic Salmonella pathogenesis and immune evasion mechanisms (Santos et al., 2001). Salmonella Typhimurium notably lacks the Vi capsule and typhoid toxin expressed by S. Typhi, but does have alternative effector proteins enhance immune evasion. Like S. Typhi, S. Typhimurium is an oral-fecal pathogen and encounters the small intestine first, where it crosses the Peyer’s patches via M cells. Resident macrophages and DCs play a role in systemic spread of S. Typhimurium through their ability to phagocytose bacteria. Survival and replication of S. Typhimurium in SCVs relies on the SPI-2 encoded type III secretion system-2 (T3SS-2), which delivers more than 20 effectors through the SCV membrane into the cytosol of the host cell. Once in the cytosol, effectors can regulate the host environment so as to help the bacteria escape lysosomal degradation (Drecktrah et al., 2006; Figueira and Holden, 2012; Johnson et al., 2018). In mice, these evasion and adaptation mechanisms allow S. Typhimurium to survive and disseminate systemically and into the hepatobiliary system. Like S. Typhi, S. Typhimurium causes apoptotic disruption of the epithelial layer of the small intestine in humans, which is activated through the caspase-8 mediated inflammasome (Hefele et al., 2018). The production of IFN-γ by Natural Killer (NK) cells, especially during the initial stages of infection, helps control the systemic bacterial burden (Kupz et al., 2013; Ingram et al., 2017). Following invasion of the intestines, S. Typhimurium prevents DC antigen presentation by avoiding lysosomal degradation and causing DC dysfunction (Tobar et al., 2006; Bueno et al., 2008). Additionally, DCs require TLR stimulation and expression of the chemokine receptor CXCR1 to induce bacterial sampling (Rescigno et al., 2001; Niess et al., 2005). To control and clear S. Typhimurium infection in mice, Th1 and Th17 CD4+ T cells are important while CD8+ T cells may not be necessary in the early stages of the primary response (Lee et al., 2012a). Notably, early depletion of CD8+ T cells does not significantly affect splenic bacterial burden (Johanns et al., 2010). Several Salmonella proteins including flagellin and SseJ are responsible for inducing the Th1 and Th17 CD4+ T cell responses observed during infection, and B cell activation has been found to be required to activate Th1 and Th17 responses (Barr et al., 2010; Lee et al., 2012b). Following S. Typhimurium infection in mice, IgM, IgG, and IgA antibodies are produced, and passive transfer of these antibodies confers protection against infection, demonstrating that B cells help protect against secondary infection (Nanton et al., 2012). sIgA predominates in the gastrointestinal tract and aids in obstructing bacterial penetration of the intestinal epithelium while IgG opsonizes bacteria for increased uptake by macrophages (Uppington et al., 2006; Nanton et al., 2012). sIgA is primarily secreted from B cells found in the intestinal lamina propria and prevents bacteria from adhering to the mucosal surface (Mantis and Forbes, 2010).
Typically, S. Typhimurium and related NTS strains only cause gastroenteritis in humans. However, there is increasing prevalence of invasive non-typhoidal Salmonella (iNTS), particularly in sub-Saharan Africa. One distinct Salmonella strain in this region (ST313) appears genetically adapted to cause increased systemic disease in humans. This adaptation results in hyper-dissemination of Salmonella via CD11b+CCR7+ migratory DCs (migDCs) through the lymphatics (Carden et al., 2017). ST313 has evolved other mechanisms to promote invasive disease such as overexpression of ribB to avoid detection by Mucosal-associated Invariant T cells by reducing their activation (Preciado-Llanes et al., 2020). SseI, a known immunogen that induces a potent and sustained CD4+ T cell response, inhibits the migration of infected DCs (Kurtz et al., 2014; Carden et al., 2017). Notably, migDC-mediated dissemination is increasingly being observed with other Salmonella strains such as SL1344, demonstrating that iNTS is becoming more widespread (Carden et al., 2017).
Following the initial infection and transcytosis of M cells, S. Typhimurium uses its T3SSs to enter the lamina propria of the small intestine as soon as 8 h post infection (Muller et al., 2012). Approximately 20–40% of bacteria in the lamina propria are found within CD11c+CX3CR1high macrophages (Muller et al., 2012). Other studies have shown that CD64+ macrophages or F4/80+CD11b+ macrophages are important in the small intestine lamina propria and contribute to T cell activation in intestinal tissue (Carden et al., 2017; Yu et al., 2018). The lamina propria harbors a multitude of immune cell types including B cells, T cells, DCs, NK cells, and macrophages. Breach of this tissue by Salmonella triggers a local immune response that contains the infection in this site and prevents further dissemination. However, further breach of this tissue or subversion of immune function can lead to systemic dissemination of iNTS to organs rich in macrophages such as the liver and spleen. Because the liver is a target organ, systemic colonization generally involves the hepatobiliary system, where bacteria can further avoid the immune response and enter a persistent state.
Salmonella Typhi Microbial Pathogenesis and Immune Evasion Within the Hepatobiliary System
Kupffer cells, the liver-resident macrophages, phagocytose S. Typhi, and establish SCVs, which help evade immune surveillance (Dougan et al., 2011). From the liver, S. Typhi can go on to colonize the biliary tract via the ducts or vasculature that connect the liver to the gallbladder. As mentioned previously, studies have shown a strong correlation between the presence of gallstones and the chance of progressing to a chronic carrier state (Schioler et al., 1983).
Although recent studies have provided additional insight into asymptomatic carriage, the mechanisms behind this process in humans are still poorly understood. Information gained from S. Typhimurium infection in murine models is vital; however, relevance to human typhoidal infection cannot be automatically assumed. For example, one recent study found that exposure to bile causes an upregulation in S. Typhi genes associated with motility, lipid A modulation, and the virulence genes srfA and srfB. These same genes were significantly downregulated in response to bile in S. Typhimurium, highlighting differential responses to physiological changes and the potential mechanism for why these two serovars establish different types of infection (Johnson et al., 2018).
An intriguing finding that requires more investigation is that older females are at higher risk for S. Typhi carriage and the subsequent development of gallbladder cancer (Rakic et al., 2014). In a transcriptomic analysis of a chronic typhoid murine model using all female mice, some of the most downregulated genes at 7 and 21 days post infection were hormonal metabolism genes, a finding at odds with earlier studies showing an increase in steroid metabolism genes in the liver after Salmonella infection (Antunes et al., 2011; González et al., 2019a). Mice that were fed a lithogenic diet to induce gallstone development also had higher levels of inflammation and Salmonella-specific CD4+ T cells but were less capable of controlling bacterial infection, possibly because of an overreactive Treg response, which is known to be elevated in females in many inflammatory diseases (González et al., 2019a). A previous group has shown that estradiol can promote production of IFN-γ—an important Th1 cytokine for Salmonella clearance—by the immunoregulatory T cell subtype known as invariant natural killer cells (Gourdy et al., 2005). Women are also more susceptible to developing gallstones, and previous studies have shown that treatment with estradiols or progesterone can alter mouse susceptibility to intraperitoneal S. Typhimurium challenge (Kita et al., 1989; Stinton and Shaffer, 2012). The human sexual dimorphism seen in chronic S. Typhi carriage needs to be explored in depth to better develop informed therapeutics that would decrease the prevalence of asymptomatic spreaders and, ideally, reduce downstream sequalae such as potentially fatal gallbladder malignancies.
Salmonella Typhimurium Microbial Pathogenesis and Immune Evasion of the Hepatobiliary System
As with S. Typhi in humans, once disseminated to the liver iNTS in humans or S. Typhimurium in mice can take up residence in Kupffer cells, which play a role in maintaining immune homeostasis by triggering Tregs and attenuating activation of anti-bacterial inflammatory Th1 CD4+ T cells (Rathman et al., 1996; Klugewitz et al., 2002; Breous et al., 2009). Although Kupffer cells are known to prevent bacteria from entering the hepatobiliary system during non-disease states, the immunosuppressive environment of the liver may provide a permissive niche for S. Typhimurium. A recent study showed that during persistent infection in resistant mice, S. Typhimurium was able to adapt through distinct mutations that were liver- and spleen-specific (Sondberg and Jelsbak, 2016). Further supporting the idea of adaptation, Johnson et al. found that S. Typhimurium significantly downregulated many motility genes when grown in the presence of bile, showing that S. Typhimurium may establish chronic infection in the liver following localization and adaptation to the otherwise inhospitable environment of continuously flushing bacteriostatic bile salts (Johnson et al., 2018).
From the liver, S. Typhimurium can also travel to the gallbladder. During acute infection, S. Typhimurium can be found in the gallbladder lumen or tissue within 48 h. Active gallbladder epithelium invasion is followed by proliferative replication in SCVs. An inflammatory response initiated by neutrophils causes tissue damage and sloughing of host epithelial cells, which helps released S. Typhimurium invade additional cells (Menendez et al., 2009). As with S. Typhi, colonization of the gallbladder by S. Typhimurium can result in asymptomatic chronic carriage leading to the unwitting spread of bacteria by seemingly convalescent patients through the fecal-oral route (Sirinavin et al., 2004). Chronic carriage of iNTS is particularly a problem in sub-Saharan Africa (Kingsley et al., 2009).
As discussed above, a robust CD4+ Th1 response is essential during the systemic infection phase with S. Typhimurium. However, it has also been shown that other adaptive immunity processes are important. Th17 cells produce IL-17 which is needed to recruit neutrophils that aid in controlling infection (Raffatellu et al., 2008). We recently showed that in a biofilm-mediated chronic murine typhoid model, the immune response shifts to a less effective Th2 response by 21 days post infection that was characterized by increased yet ineffective antibody production and increased expression of GATA3, a Th2 transcriptional regulator (González et al., 2019a). Furthermore, the Salmonella-specific T cell compartment differs greatly between the lymphoid tissues, such as the mLNs, and the liver (Kurtz et al., 2020). In this case, liver T cells appear to be far more immunosuppressive and permissive of disease compared to lymphoid T cells. They also produced far more of the immunoregulatory cytokine IL-10 compared to lymphoid CD4+ T cells, which were more likely to produce IFN-γ. In addition, lack of thymic output prior to persistent S. Typhimurium infection results in a loss of control over infection, despite the fact that Salmonella specific CD4+ T cells are elevated (Goggins et al., 2020). It will be important to explore whether the thymus contributes more T cells to the lymphoid compartment or the hepatobiliary tissue. The compounded virulence and immune evasion techniques of S. Typhimurium and S. Typhi, along with inappropriate or manipulated immune responses of the host, make them formidable pathogens in both humans and mice.
Discussion/Summary
The immune response to Salmonella infection is complex and multifaceted and relies on coordination between the early innate response and the subsequent adaptive response. While adaptive immunity to Salmonella has been extensively studied in the lymphoid organs (e.g., mLNs and spleen) the response in the intestine and hepatobiliary system, particularly the gallbladder, is less clear (Figure 2). It is notable that there are differences in infection and immune responses between the two major Salmonella serovars that cause human disease. As summarized in this review, these differences are attributable to differential subversion of the immune response by the bacteria itself, influenced by its planktonic or biofilm lifestyle, as well as where and how the immune system first encounters the bacteria. While progress has been made in our understanding of how Salmonella interacts with the host gut and hepatobiliary system, important questions remain regarding these types of interactions:
1. Does biofilm formation affect the development, function, and subversion of the immune response in the gut and gallbladder? Our group has already shown that gallstones and chronic Salmonella carriage in the gallbladder recruits both T and B cells to the gallbladder itself (González et al., 2019a). Over time, these immune cells develop a Th2 phenotype in the gallbladder. It remains to be explored whether this Th2 shift is permissive of infection in the gallbladder and thus contributes to persistence. Future studies could examine this question by depleting T or B cells and assessing the respective contributions to gallbladder immunity over time. Surprisingly little is known regarding intestinal immunity during the chronic phases of infection and so these studies could also be extended to the intestine.
2. How does the presence of gallstones and biofilms regulate persistence and bacterial shedding early and late in infection? It will also be important to determine whether gallbladder immunity creates an environment that promotes increased bacterial shedding in the feces. It is possible that the immunological shift toward a Th2 phenotype not only permits bacterial persistence, but somehow elicits a greater shedding phenotype, potentially via production of anti-inflammatory cytokines such as IL-10 or IL-4. We have already shown that Salmonella specific T cells in the liver produce IL-10 and are permissive of infection and so extending this to the gallbladder and gut would be an important extension of this work (Kurtz et al., 2020).
3. What immunological mechanisms allow the gut and hepatobiliary system to become permissive to chronic infection? While the Th2 shift is fairly generalized, it is not known how Salmonella specific T cells specifically shift over time. As previously discussed, some of these cells may adopt a Treg phenotype, either in the liver, the gallbladder, or the intestinal tissue itself. While this remains to be explored, it would offer a logical explanation for why these tissues appear to be more tolerant of infection compared to other tissues such as the spleen.
4. Does the immune response in the hepatobiliary system and gut differ between chronic typhoidal infection compared to iNTS or acute infection? This review touches upon some of the major differences between typhoidal and non-typhoidal Salmonella infection; however, it is not well known whether, or how, the immune responses differ between these two types of infections, especially in the hepatobiliary tract and the intestine. Does chronic typhoidal infection drive a more regulatory immune response and acute infection elicits a more robust antibacterial response? These are important questions that must be addressed in order to answer the final outstanding question below.
5. How can we eliminate chronic carriage? Understanding the immune response in the major tissues that harbor bacteria, especially the gut or hepatobiliary system, would greatly aid in the fight to eliminate chronic carriage. The ultimate goal of this understanding would be combatting bacterial carriage, thus eliminating human to human spread of infection. It is possible that using biofilm dispersal agents would allow the immune system adequate access to bacteria ensuring their elimination. Alternatively, a vaccination strategy that preferentially targets the gut, as we have previously shown to be possible using the appropriate adjuvant, might be sufficient to eliminate bacteria (Frederick et al., 2018).
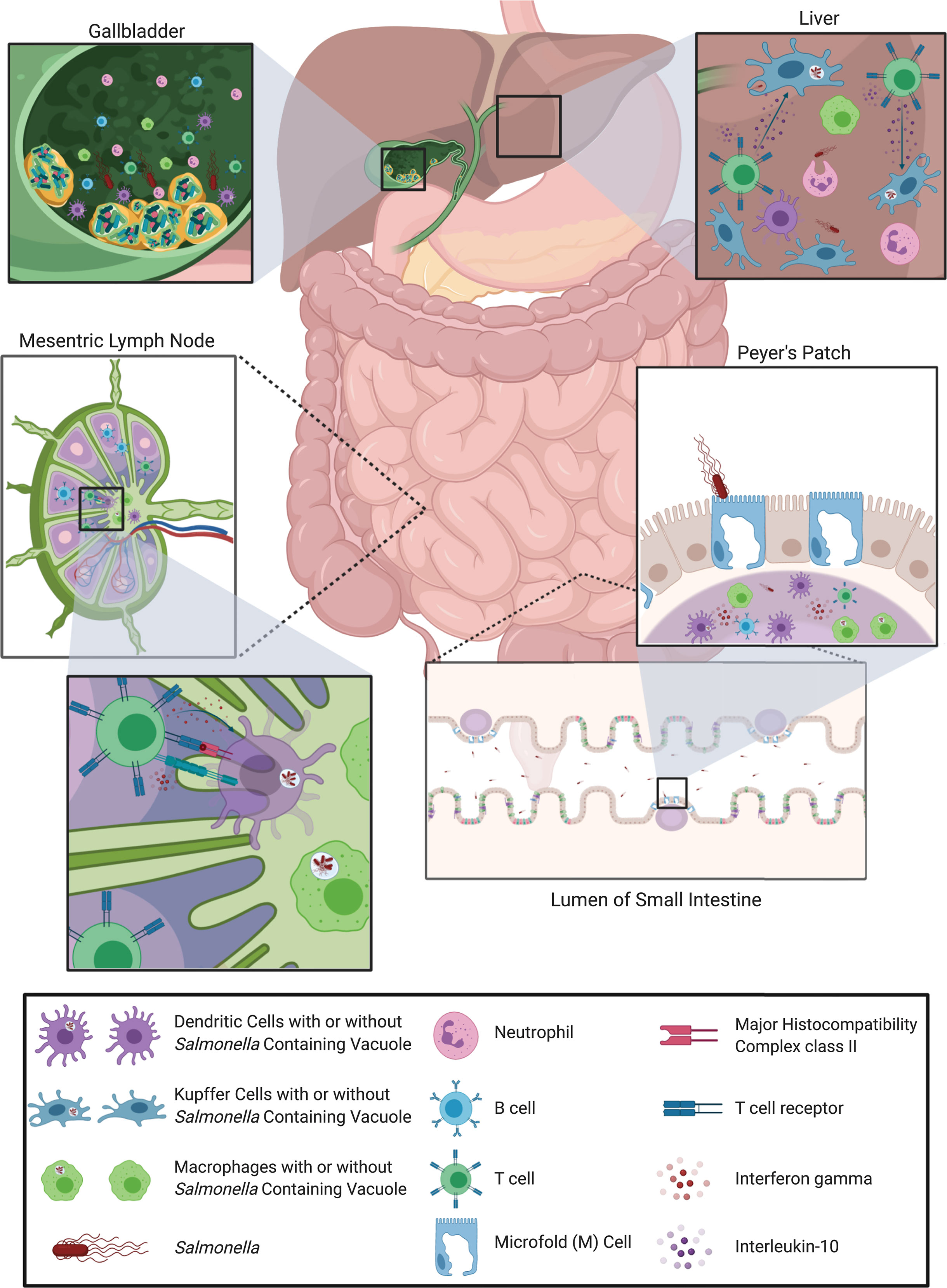
Figure 2 Overview of host immune response to Salmonella infection. Salmonella, infecting via the oral-fecal route, first passes through the stomach and into the lumen of the small intestine. There, Salmonella uses its SPI-1 type III secretion system to preferentially transcytose across M cells to gain access to the Peyer’s Patches. Immune cells in the Peyer’s Patches, such as macrophages and dendritic cells, sense Salmonella via toll-like receptors and begin to trigger an immune response. Macrophages and dendritic cells then migrate to the gut-draining mesenteric lymph nodes and the spleen (not shown) harboring Salmonella antigens as well as whole bacteria in Salmonella-containing vacuoles. In the lymph nodes and spleen, dendritic cells and macrophages present antigen on MHC class II to activate Salmonella-specific helper T cells which can then subsequently activate Salmonella-specific B cells. These adaptive immune cells then traffic back into the intestine to fight infection by releasing the antimicrobial cytokine interferon gamma or neutralizing antibodies. Any Salmonella that escape the immune response of the Peyer’s patches and mesenteric lymph nodes or spleen, can travel to, and invade, the liver and subsequently the gallbladder via the bile duct. In the liver, Kupffer cells phagocytose Salmonella and activate Salmonella-specific T cells to produce the anti-inflammatory cytokine interleukin-10, likely preventing bacterial clearance. The immune environment in the gallbladder is less clear, primarily because of the presence of Salmonella biofilms on gallstones, which can hinder/alter the immune response. The liver and gallbladder are both sites where the host can experience chronic infection with Salmonella. Images created with BioRender.com.
Further studies will better elucidate immunological connections between the gut and hepatobiliary system, particularly during chronic infection, and how this might contribute to Salmonella pathogenesis, and how to eliminate it from the human population.
Author Contributions
JH, MH, SD’S, EV, and JS all wrote sections of the manuscript and edited the final version. JH created the figures. MH referenced the manuscript. JG and JM provided funding for the work and edited the text and figures. All authors contributed to the article and approved the submitted version.
Funding
The work presented here was funded by grants AI116917, AI099525, AI153752 and AI156328 from the National Institutes of Health to JG and National Institutes of Health grant U01 AI124289 and a grant from the WM Keck Foundation to JM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahmad I., Khan M., Altaf M., Qais F., A. F. A., Rumbaugh K. (2017). “Biofilms: an overview of their significance in plant and soil health,” in Biofilms in Plant and Soil Health (Chichester, UK: John Wiley & Sons, Ltd), 1–25. doi: 10.1002/9781119246329.ch1
Antunes L. C., Arena E. T., Menendez A., Han J., Ferreira R. B., Buckner M. M., et al. (2011). Impact of salmonella infection on host hormone metabolism revealed by metabolomics. Infect. Immun. 79 (4), 1759–1769. doi: 10.1128/IAI.01373-10
Atif S. M., Winter S. E., Winter M. G., McSorley S. J., Baumler A. J. (2014). Salmonella enterica serovar Typhi impairs CD4 T cell responses by reducing antigen availability. Infect. Immun. 82 (6), 2247–2254. doi: 10.1128/IAI.00020-14
Avraham R., Haseley N., Brown D., Penaranda C., Jijon H. B., Trombetta J. J., et al. (2015). Pathogen Cell-to-Cell Variability Drives Heterogeneity in Host Immune Responses. Cell 162 (6), 1309–1321. doi: 10.1016/j.cell.2015.08.027
Bao S., Beagley K. W., France M. P., Shen J., Husband A. J. (2000). Interferon-gamma plays a critical role in intestinal immunity against Salmonella typhimurium infection. Immunology 99 (3), 464–472. doi: 10.1046/j.1365-2567.2000.00955.x
Bao K., Papadimitropoulos A., Akgul B., Belibasakis G., Bostanci N. (2015). Establishment of an oral infection model resembling the periodontal pocket in a perfusion bioreactor system. Virulence 6), 265–273. doi: 10.4161/21505594.2014.978721
Barr T. A., Brown S., Mastroeni P., Gray D. (2010). TLR and B cell receptor signals to B cells differentially program primary and memory Th1 responses to Salmonella enterica. J. Immunol. 185 (5), 2783–2789. doi: 10.4049/jimmunol.1001431
Bergmans D., Bonten M. (1999). “Colonization and infection with Pseudomonas aeruginosa in Intensive care: Endogenous or exogenous origin?,” in Yearbook of Intensive Care and Emergency Medicine 1999. Ed. Vincent J. L. (Berlin, Heidelberg: Springer), 131–140. doi: 10.1007/978-3-662-13453-5_13
Bhan M. K., Bahl R., Bhatnagar S. (2005). Typhoid and paratyphoid fever. Lancet 366 (9487), 749–762. doi: 10.1016/s0140-6736(05)67181-4
Bobat S., Flores-Langarica A., Hitchcock J., Marshall J. L., Kingsley R. A., Goodall M., et al. (2011). Soluble flagellin, FliC, induces an Ag-specific Th2 response, yet promotes T-bet-regulated Th1 clearance of Salmonella typhimurium infection. Eur. J. Immunol. 41 (6), 1606–1618. doi: 10.1002/eji.201041089
Boddicker J. D., Ledeboer N. A., Jagnow J., Jones B. D., Clegg S. (2002). Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica Serovar Typhimurium is dependent on allelic variation in the fimH gene of the fim gene cluster. Mol. Microbiol. 45 (5), 1255–1265. doi: 10.1046/j.1365-2958.2002.03121.x
Boyle E. C., Brown N. F., Finlay B. B. (2006). Salmonella enterica serovar Typhimurium effectors SopB, SopE, SopE2 and SipA disrupt tight junction structure and function. Cell. Microbiol. 8 (12), 1946–1957. doi: 10.1111/j.1462-5822.2006.00762.xPMID-16869830
Branda S., Vik S., Friedman L., Kolter R. (2005). Biofilms: The Matrix Revisited. Trends Microbiol. 13), 20–26. doi: 10.1016/j.tim.2004.11.006
Breous E., Somanathan S., Vandenberghe L. H., Wilson J. M. (2009). Hepatic regulatory T cells and Kupffer cells are crucial mediators of systemic T cell tolerance to antigens targeting murine liver. Hepatology 50 (2), 612–621. doi: 10.1002/hep.23043
Buchwald D. S., Blaser M. J. (1984). A review of human Salmonellosis: II. Duration of infection with nontyphi Salmonella. Rev. Infect. Dis. 6 (3), 345–356. doi: 10.1093/clinids/6.3.345
Bueno S. M., Gonzalez P. A., Carreno L. J., Tobar J. A., Mora G. C., Pereda C. J., et al. (2008). The capacity of Salmonella to survive inside dendritic cells and prevent antigen presentation to T cells is host specific. Immunology 124 (4), 522–533. doi: 10.1111/j.1365-2567.2008.02805.x
Carden S. E., Walker G. T., Honeycutt J., Lugo K., Pham T., Jacobson A., et al. (2017). Pseudogenization of the Secreted Effector Gene sseI Confers Rapid Systemic Dissemination of S. Typhimurium ST313 within Migratory Dendritic Cells. Cell Host Microbe 21 (2), 182–194. doi: 10.1016/j.chom.2017.01.009
Carter P. B., Collins F. M. (1974). The route of enteric infection in normal mice. J. Exp. Med. 139 (5), 1189–1203. doi: 10.1084/jem.139.5.1189
Caygill C. P., Hill M. J., Braddick M., Sharp J. C. (1994). Cancer mortality in chronic typhoid and paratyphoid carriers. Lancet 343 (8889), 83–84. doi: 10.1016/S0140-6736(94)90816-8
CDC (2019). Antibiotic Resistance Threats in the United States, 2019 (Atlanta, GA: U.S. Department of Health and Human Services, CDC).
Cheminay C., Chakravortty D., Hensel M. (2004). Role of neutrophils in murine salmonellosis. Infect. Immun. 72 (1), 468–477. doi: 10.1128/iai.72.1.468-477.2004
Chia T. W. R., Goulter R. M., McMeekin T., Dykes G. A., Fegan N. (2009). Attachment of different Salmonella serovars to materials commonly used in a poultry processing plant. Food Microbiol. 26 (8), 853–859. doi: 10.1016/j.fm.20
Christensen B. B., Sternberg C., Andersen J. B., Eberl L., Møller S., Givskov M., et al. (1998). Establishment of new genetic traits in a microbial biofilm community. Appl. Environ. Microbiol. 64 (6), 2247–2255. doi: 10.1128/AEM.64.6.2247-2255.1998
Clay S. L., Bravo-Blas A., Wall D. M., MacLeod M. K. L., Milling S. W. F. (2020). Regulatory T cells control the dynamic and site-specific polarization of total CD4 T cells following Salmonella infection. Mucosal. Immunol. 13, 946–957. doi: 10.1038/s41385-020-0299-1
Cole S. J., Records A. R., Orr M. W., Linden S. B., Lee V. T. (2014). Catheter-associated urinary tract infection by Pseudomonas aeruginosa is mediated by exopolysaccharide-independent biofilms. Infect. Immun. 82 (5), 2048–2058. doi: 10.1128/IAI.01652-14
Corcoran M., Morris D., De Lappe N., O’Connor J., Lalor P., Dockery P., et al. (2014). Commonly used disinfectants fail to eradicate Salmonella enterica biofilms from food contact surface materials. Appl. Environ. Microbiol. 80 (4), 1507–1514. doi: 10.1128/AEM.03109-13
Costerton J., Stewart P., Greenberg E. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284), 1318–1322. doi: 10.1126/science.284.5418.1318
Costerton W., Veeh R., Shirtliff M., Pasmore M., Post C., Ehrlich G. (2003). The application of biofilm science to the study and control of chronic bacterial infections. J. Clin. Invest. 112 (10), 1466–1477. doi: 10.1172/JCI200320365
Crawford R. W., Gibson D. L., Kay W. W., Gunn J. S. (2008). Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect. Immun. 76 (11), 5341–5349. doi: 10.1128/IAI.00786-08
Crawford R. W., Reeve K. E., Gunn J. S. (2010a). Flagellated but Not Hyperfimbriated Salmonella enterica Serovar Typhimurium Attaches to and Forms Biofilms on Cholesterol-Coated Surfaces. J. Bacteriol. 192 (12), 2981–2990. doi: 10.1128/jb.01620-09
Crawford R. W., Rosales-Reyes R., Ramírez-Aguilar M., Chapa-Azuela O., Alpuche-Aranda C., Gunn J. S. (2010b). Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc. Natl. Acad. Sci. U.S.A. 107 (9), 4353–4358. doi: 10.1073/pnas.1000862107
Daubert D., BF W. (2019). Biofilm as a risk factor in implant treatment. Periodontology 81), 29–40. doi: 10.1111/prd.12280
Davies D. (2003). Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discovery 2 (2), 114–122. doi: 10.1038/nrd1008
de Carvalho C. (2018). Marine biofilms: a successful microbial strategy with economic implications. Front. Mar. Sci. 5), 126. doi: 10.3389/fmars.2018.00126
De Oliveira D. C., Fernandes Junior A., Kaneno R., Silva M. G., Araujo Junior J. P., Silva N. C., et al. (2014). Ability of Salmonella spp. to produce biofilm is dependent on temperature and surface material. Foodborne Pathog. Dis. 11 (6), 478–483. doi: 10.1089/fpd.2013.1710
Del Bel Belluz L., Guidi R., Pateras I. S., Levi L., Mihaljevic B., Rouf S. F., et al. (2016). The Typhoid Toxin Promotes Host Survival and the Establishment of a Persistent Asymptomatic Infection. PloS Pathog. 12 (4), e1005528. doi: 10.1371/journal.ppat.1005528
Desai S. K., Kenney L. J. (2019). Switching Lifestyles Is an in vivo Adaptive Strategy of Bacterial Pathogens. Front. Cell Infect. Microbiol. 9:421:421. doi: 10.3389/fcimb.2019.00421
Desai S. K., Padmanabhan A., Harshe S., Zaidel-Bar R., Kenney L. J. (2019). Salmonella biofilms program innate immunity for persistence in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 116 (25), 12462–12467. doi: 10.1073/pnas.1822018116
Dieltjens L., Appermans K., Lissens M., Lories B., Kim W., Van der Eycken E. V., et al. (2020). Inhibiting bacterial cooperation is an evolutionarily robust anti-biofilm strategy. Nat. Commun. 11 (1), 107. doi: 10.1038/s41467-019-13660-x
Dougan G., John V., Palmer S., Mastroeni P. (2011). Immunity to salmonellosis. Immunol. Rev. 240 (1), 196–210. doi: 10.1111/j.1600-065X.2010.00999.x
Drecktrah D., Knodler L. A., Ireland R., Steele-Mortimer O. (2006). The mechanism of Salmonella entry determines the vacuolar environment and intracellular gene expression. Traffic 7 (1), 39–51. doi: 10.1111/j.1600-0854.2005.00360.x
Dwyer B. E., Newton K. L., Kisiela D., Sokurenko E. V., Clegg S. (2011). Single nucleotide polypmorphisms of fimH associated with adherence and biofilm formation by serovars of Salmonella enterica. Microbiol. (Reading) 157, 3162–3171. doi: 10.1099/mic.0.051425-0
El Hag M., Feng Z., Su Y., Wang X., Yassin A., Chen S., et al. (2017). Contribution of the csgA and bcsA genes to Salmonella enterica serovar Pullorum biofilm formation and virulence. Avian Pathol. 46 (5), 541–547. doi: 10.1080/03079457.2017.1324198
Espeland E. M., Wetzel R. G. (2001). Complexation, stabilization, and UV photolysis of extracellular and surface-bound glucosidase and alkaline phosphatase: implications for biofilm microbiota. Microb. Ecol. 42), 572–585. doi: 10.1007/s00248-001-1023-7
Ferrand J., Ferrero R. L. (2013). Recognition of Extracellular Bacteria by NLRs and Its Role in the Development of Adaptive Immunity. Front. Immunol. 4:344:344. doi: 10.3389/fimmu.2013.00344
Figueira R., Holden D. W. (2012). Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiol. (Reading) 158 (Pt 5), 1147–1161. doi: 10.1099/mic.0.058115-0
Flemming H. C., Wuertz S. (2019). Bacteria and archaea on Earth and their abundance in biofilm. Nat. Rev. Microbiol. 17), 247–260. doi: 10.1038/s41579-019-0158-9
Flemming H.-C. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14), 563. doi: 10.1038/nrmicro.2016.94
Frederick D. R., Goggins J. A., Sabbagh L. M., Freytag L. C., Clements J. D., McLachlan J. B. (2018). Adjuvant selection regulates gut migration and phenotypic diversity of antigen-specific CD4+ T cells following parenteral immunization. Mucosal. Immunol. 11 (2), 549–561. doi: 10.1038/mi.2017.70
Fresnay S., McArthur M. A., Magder L., Darton T. C., Jones C., Waddington C. S., et al. (2016). Salmonella Typhi-specific multifunctional CD8+ T cells play a dominant role in protection from typhoid fever in humans. J. Transl. Med. 14, 62. doi: 10.1186/s12967-016-0819-7
Furukawa S., Kuchma S., O’toole G. (2006). Keeping their options open: acute versus persistent infections. J. Bacteriol. 188 (4), 1211–1217. doi: 10.1128/JB.188.4.1211-1217.2006
Galie S., Garcia-Gutierrez C., Miguelez E. M., Villar C. J., Lombo F. (2018). Biofilms in the Food Industry: Health Aspects and Control Methods. Front. Microbiol. 9:898:898. doi: 10.3389/fmicb.2018.00898
Gal-Mor O. (2019). Persistent infection and long-term carriage of typhoidal and nontyphoidal Salmonellae. Clin. Microbiol. Rev. 32 (1), e00088–e00018. doi: 10.1128/CMR
Gerstel U., Römling U. (2003). The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res. Microbiol. 154 (10), 659–667. doi: 10.1016/j.resmic.2003.08.005
Giaouris E. D., Nychas G. J. (2006). The adherence of Salmonella Enteritidis PT4 to stainless steel: the importance of the air-liquid interface and nutrient availability. Food Microbiol. 23 (8), 747–752. doi: 10.1016/j.fm.2006.02.006
Giaouris E., Chorianopoulos N., Skandamis P., Nychas G.-J. (2012). “Attachment and biofilm formation by Salmonella in food processing environments,” in Salmonella: A dangerous foodborne pathogen. Ed. Mahmoud B. S. M. (Rijeka, Croatia), 157–180. doi: 10.5772/28107
Gibson D. L., White A. P., Snyder S. D., Martin S., Heiss C., Azadi P., et al. (2006). Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 188 (22), 7722–7730. doi: 10.1128/JB.00809-06
Goggins J. A., Kurtz J. R., McLachlan J. B. (2020). Control of Persistent Salmonella Infection Relies on Constant Thymic Output Despite Increased Peripheral Antigen-Specific T Cell Immunity. Pathogens 9 (8), 605. doi: 10.3390/pathogens9080605
Gómez M. I., Prince A. (2007). Opportunistic infections in lung disease: Pseudomonas infections in cystic fibrosis. Curr. Opin. Pharmacol. 7 (3), 244–251. doi: 10.1016/j.coph.2006.12.005
González J. F., Alberts H., Lee J., Doolittle L., Gunn J. S. (2018). Biofilm Formation Protects Salmonella from the Antibiotic Ciprofloxacin In Vitro and In Vivo in the Mouse Model of chronic Carriage. Sci. Rep. 8 (1), 222. doi: 10.1038/s41598-017-18516-2
González J. F., Kurtz J., Bauer D. L., Hitt R., Fitch J., Wetzel A., et al. (2019a). Establishment of Chronic Typhoid Infection in a Mouse Carriage Model Involves a Type 2 Immune Shift and T and B Cell Recruitment to the Gallbladder. mBio 10 (5), e02262–e02219. doi: 10.1128/mBio.02262-19
González J. F., Tucker L., Fitch J., Wetzel A., White P., Gunn J. S. (2019b). Human Bile-Mediated Regulation of Salmonella Curli Fimbriae. J. Bacteriol. 201 (18), e00055–e00019. doi: 10.1128/JB.00055-19
Gonzalez-Escobedo G., Gunn J. S. (2013). Gallbladder epithelium as a niche for chronic Salmonella carriage. Infect. Immun. 81 (8), 2920–2930. doi: 10.1128/IAI.00258-13
Gonzalez-Escobedo G., Marshall J. M., Gunn J. S. (2010). Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat. Rev. Microbiol. 9 (1), nrmicro2490. doi: 10.1038/nrmicro2490
Gonzalez-Escobedo G., Marshall J. M., Gunn J. S. (2011). Chronic and acute infection of the gall bladder by Salmonella Typhi: understanding the carrier state. Nat. Rev. Microbiol. 9 (1), 9–14. doi: 10.1038/nrmicro2490
Gourdy P., Araujo L. M., Zhu R., Garmy-Susini B., Diem S., Laurell H., et al. (2005). Relevance of sexual dimorphism to regulatory T cells: estradiol promotes IFN-gamma production by invariant natural killer T cells. Blood 105 (6), 2415–2420. doi: 10.1182/blood-2004-07-2819
Grant S. S., Hung D. T. (2013). Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence 4 (4), 273–283. doi: 10.4161/viru.23987
Gunn J. S., Marshall J. M., Baker S., Dongol S., Charles R. C., Ryan E. T. (2014). Salmonella chronic carriage: epidemiology, diagnosis, and gallbladder persistence. Trends Microbiol. 22 (11), 648–655. doi: 10.1016/j.tim.2014.06.007
Hahn M. M., Gunn J. S. (2020). Salmonella Extracellular Polymeric Substances Modulate Innate Phagocyte Activity and Enhance Tolerance of Biofilm-Associated Bacteria to Oxidative Stress. Microorganisms 8 (2), 19. doi: 10.3390/microorganisms8020253
Halici S., Zenk S. F., Jantsch J., Hensel M. (2008). Functional analysis of the Salmonella pathogenicity island 2-mediated inhibition of antigen presentation in dendritic cells. Infect. Immun. 76 (11), 4924–4933. doi: 10.1128/IAI.00531-08
Hall-Stoodley L., Stoodley P. (2009). Evolving concepts in biofilm infections. Cell Microbiol. 11), 1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x
Hall-Stoodley L., Costerton J. W., Stoodley P. (2004). Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2 (2), 95–108. doi: 10.1038/nrmicro821
Hausner M., Wuertz S. (1999). High rates of conjugation in bacterial biofilms as determined by quantitative in situ analysis. Appl. Environ. Microbiol. 65 (8), 3710–3713. doi: 10.1128/AEM.65.8.3710-3713.1999
Hefele M., Stolzer I., Ruder B., He G. W., Mahapatro M., Wirtz S., et al. (2018). Intestinal epithelial Caspase-8 signaling is essential to prevent necroptosis during Salmonella Typhimurium induced enteritis. Mucosal. Immunol. 11 (4), 1191–1202. doi: 10.1038/s41385-018-0011-x
Høiby N., Ciofu O., Bjarnsholt T. (2010). Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 5 (11), 1663–1674. doi: 10.2217/fmb.10.125
Høiiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. (2010). Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35), 322–332. doi: 10.1016/j.ijantimicag.2009.12.011
Ingram J. P., Brodsky I. E., Balachandran S. (2017). Interferon-gamma in Salmonella pathogenesis: New tricks for an old dog. Cytokine 98, 27–32. doi: 10.1016/j.cyto.2016.10.009
Jackson N. P., Kang Y. H., Lapaque N., Janssen H., Trowsdale J., Kelly A. P. (2013). Salmonella polarises peptide-MHC-II presentation towards an unconventional Type B CD4+ T-cell response. Eur. J. Immunol. 43 (4), 897–906. doi: 10.1002/eji.201242983
Jensen P.Ø., Givskov M., Bjarnsholt T., Moser C. (2010). The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Microbiol. 59 (3), 292–305. doi: 10.1111/j.1574-695X.2010.00706.x
Jiang Y., Geng M., Bai L. (2020). Targeting Biofilms Therapy: Current Research Strategies and Development Hurdles. Microorganisms (8), 1222. doi: 10.3390/microorganisms8081222
Johanns T. M., Ertelt J. M., Rowe J. H., Way S. S. (2010). Regulatory T cell suppressive potency dictates the balance between bacterial proliferation and clearance during persistent Salmonella infection. PloS Pathog. 6 (8), e1001043. doi: 10.1371/journal.ppat.1001043
Johnson R., Mylona E., Frankel G. (2018). Typhoidal Salmonella: Distinctive virulence factors and pathogenesis. Cell Microbiol. 20 (9), e12939. doi: 10.1111/cmi.12939
Jolivet-Gougeon A., Bonnaure-Mallet M. (2014). Biofilms as a mechanism of bacterial resistance. Drug Discovery Today Technol. 11, 49–56. doi: 10.1016/j.ddtec.2014.02.003
Jones B. D., Ghori N., Falkow S. (1994). Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J. Exp. Med. 180 (1), 15–23. doi: 10.1084/jem.180.1.15PMID-8006579
Jones-Carson J., McCollister B. D., Clambey E. T., Vazquez-Torres A. (2007). Systemic CD8 T-cell memory response to a Salmonella pathogenicity island 2 effector is restricted to Salmonella enterica encountered in the gastrointestinal mucosa. Infect. Immun. 75 (6), 2708–2716. doi: 10.1128/IAI.01905-06
Joseph B., Otta S. K., Karunasagar I., Karunasagar I. (2000). Biofilm formation by Salmonella spp. on food contact surfaces and their sensitivity to sanitizers. Int. J. Food Microbiol. 64 (3), 367–372. doi: 10.1016/S0168-1605(00)00466-9
Kazemi M., Gumpert G., Marks M. I. (1974). Clincal spectrum and carrier state of nontyphoidal Salmonella infections in infants and children. Can. Med. Assoc. J. 110, 1253–1257.
Khatoon Z., McTiernan C., Suuronen E., Mah T., Alarcon E. (2018). Bacterial biofilm formation on implantable devices and approaches to its treatment and prevention. Heliyon 4 (12), e01067. doi: 10.1016/j.heliyon.2018.e01067
Kim S.-H., Wei C.-I. (2009). Molecular characterization of biofilm formation and attachment of Salmonella enterica serovar Typhimurium DT104 on food contact surfaces. J. Food Prot. 72 (9), 1841–1847. doi: 10.4315/0362-028X-72.9.1841
Kingsley R. A., Msefula C. L., Thomson N. R., Kariuki S., Holt K. E., Gordon M. A., et al. (2009). Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 19 (12), 2279–2287. doi: 10.1101/gr.091017.109
Kita E., Yagyu Y., Nishikawa F., Hamuro A., Oku D., Emoto M., et al. (1989). Alterations of host resistance to mouse typhoid infection by sex hormones. J. Leukoc. Biol. 46 (6), 538–546. doi: 10.1002/jlb.46.6.538
Klugewitz K., Blumenthal-Barby F., Schrage A., Knolle P. A., Hamann A., Crispe I. N. (2002). Immunomodulatory effects of the liver: deletion of activated CD4+ effector cells and suppression of IFN-gamma-producing cells after intravenous protein immunization. J. Immunol. 169 (5), 2407–2413. doi: 10.4049/jimmunol.169.5.2407
Kohbata S., Yokoyama H., Yabuuchi E. (1986). Cytopathogenic Effect of Salmonella typhi GIFU 10007 on M cells of Murine Ileal Peyer’s Patches in Ligated Ileal Loops: An Ultrastructural Study. Microbiol. Immunol. 30 (12), 1225–1237. doi: 10.1111/j.1348-0421.1986.tb03055.xPMID-3553868
Kong Q., Six D. A., Liu Q., Gu L., Wang S., Alamuri P., et al. (2012). Phosphate groups of lipid A are essential for Salmonella enterica serovar Typhimurium virulence and affect innate and adaptive immunity. Infect. Immun. 80 (9), 3215–3224. doi: 10.1128/IAI.00123-12
Kupz A., Scott T. A., Belz G. T., Andrews D. M., Greyer M., Lew A. M., et al. (2013). Contribution of Thy1+ NK cells to protective IFN-gamma production during Salmonella typhimurium infections. Proc. Natl. Acad. Sci. U.S.A. 110 (6), 2252–2257. doi: 10.1073/pnas.1222047110
Kurtz J. R., Petersen H. E., Frederick D. R., Morici L. A., McLachlan J. B. (2014). Vaccination with a single CD4 T cell peptide epitope from a Salmonella type III-secreted effector protein provides protection against lethal infection. Infect. Immun. 82 (6), 2424–2433. doi: 10.1128/IAI.00052-14
Kurtz J. R., Nieves W., Bauer D. L., Israel K. E., Adcox H. E., Gunn J. S., et al. (2020). Salmonella Persistence and Host Immunity Are Dictated by the Anatomical Microenvironment. Infect. Immun. 88 (8). doi: 10.1128/IAI.00026-20
Lahiri A., Lahiri A., Das P., Vani J., Shaila M. S., Chakravortty D. (2010). TLR 9 activation in dendritic cells enhances salmonella killing and antigen presentation via involvement of the reactive oxygen species. PloS One 5 (10), e13772. doi: 10.1371/journal.pone.0013772
Lai C. W., Chan R. C. Y., Cheng A. F. B., Sung J. Y., Leung J. W. C. (1992). Common bile duct stones: a cause of chronic salmonellosis. Am. J. Gastroenterol. 87 (9), 1198-1199.
Lam L. H., Monack D. M. (2014). Intraspecies competition for niches in the distal gut dictate transmission during persistent Salmonella infection. PloS Pathog. 10 (12), e1004527. doi: 10.1371/journal.ppat.1004527
Lamas A., Regal P., Vazquez B., Miranda J. M., Cepeda A., Franco C. M. (2018). Salmonella and Campylobacter biofilm formation: a comparative assessment from farm to fork. J. Sci. Food Agric. 98 (11), 4014–4032. doi: 10.1002/jsfa.8945
Lamprokostopoulou A., Monteiro C., Rhen M., Römling U. (2010). Cyclic di-GMP signalling controls virulence properties of Salmonella enterica serovar Typhimurium at the mucosal lining. Environ. Microbiol. 12 (1), 40–53. doi: 10.1111/j.1462-2920.2009.02032.x
Lapaque N., Hutchinson J. L., Jones D. C., Meresse S., Holden D. W., Trowsdale J., et al. (2009). Salmonella regulates polyubiquitination and surface expression of MHC class II antigens. Proc. Natl. Acad. Sci. U.S.A. 106 (33), 14052–14057. doi: 10.1073/pnas.0906735106
Laskin D. L., Sunil V. R., Gardner C. R., Laskin J. D. (2011). Macrophages and tissue injury: agents of defense or destruction? Annu. Rev. Pharmacol. Toxicol. 51, 267–288. doi: 10.1146/annurev.pharmtox.010909.105812
Latasa C., Roux A., Toledo-Arana A., Ghigo J. M., Gamazo C., Penades J. R., et al. (2005). BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58 (5), 1322–1339. doi: 10.1111/j.1365-2958.2005.04907.x
Ledeboer N. A., Jones B. D. (2005). Exopolysaccharide sugars contribute to biofilm formation by Salmonella enterica serovar typhimurium on HEp-2 cells and chicken intestinal epithelium. J. Bacteriol. 187 (9), 3214–3226. doi: 10.1128/JB.187.9.3214-3226.2005
Ledeboer N. A., Frye J. G., McClelland M., Jones B. D. (2006). Salmonella enterica serovar Typhimurium requires the Lpf, Pef, and Tafi fimbriae for biofilm formation on HEp-2 tissue culture cells and chicken intestinal epithelium. Infect. Immun. 74 (6), 3156–3169. doi: 10.1128/IAI.01428-05
Lee S. J., Dunmire S., McSorley S. J. (2012a). MHC class-I-restricted CD8 T cells play a protective role during primary Salmonella infection. Immunol. Lett. 148 (2), 138–143. doi: 10.1016/j.imlet.2012.10.009
Lee S. J., McLachlan J. B., Kurtz J. R., Fan D., Winter S. E., Baumler A. J., et al. (2012b). Temporal expression of bacterial proteins instructs host CD4 T cell expansion and Th17 development. PloS Pathog. 8 (1), e1002499. doi: 10.1371/journal.ppat.1002499
Li B., Qiu Y., Zhang J., Huang X., Shi H., Yin H. (2018). Real-time study of rapid spread of antibiotic resistance plasmid in biofilm using microfluidics. Environ. Sci. Technol. 52 (19), 11132–11141. doi: 10.1021/acs.est.8b03281
Limoli D., Hoffman L. (2019). Help, hinder, hide and harm: what can we learn from the interactions between Pseudomonas aeruginosa and Staphylococcus aureus during respiratory infections? Thorax 74), 684–692. doi: 10.1136/thoraxjnl-2018-212616
Limoli D., Yang J., Khansaheb M., Helfman B., Peng L., Stecenko A., et al. (2016). Staphylococcus aureus and Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 35), 947–953. doi: 10.1007/s10096-016-2621-0
Liu F., Xing S., Ma H., Du Z., Ma B. (2013). Plant growth-promoting rhizobacteria affect the growth and nutrient uptake of fraxinus americana, container seedlings. Appl. Microbiol. Biotechnol. 97), 4617–4625. doi: 10.1007/s00253-012-4255-1
Lochab V., Jones T. H., Dusane D. H., Peters C. W., Stoodley P., Wozniak D. J., et al. (2020). Ultrastructure imaging of Pseudomonas aeruginosa lawn biofilms and eradication of the tobramycin-resistant variants under in vitro electroceutical treatment. Sci. Rep. 10 (1), 1–12. doi: 10.1038/s41598-020-66823-y
Lopez-Medina M., Perez-Lopez A., Alpuche-Aranda C., Ortiz-Navarrete V. (2014). Salmonella modulates B cell biology to evade CD8(+) T cell-mediated immune responses. Front. Immunol. 5:586:586. doi: 10.3389/fimmu.2014.00586
MacKenzie K. D., Palmer M. B., Köster W. L., White A. P. (2017). Examining the Link between Biofilm Formation and the Ability of Pathogenic Salmonella Strains to Colonize Multiple Host Species. Front. Vet. Sci. 4:138:138. doi: 10.3389/fvets.2017.00138
MacKenzie K. D., Wang Y., Musicha P., Hansen E. G., Palmer M. B., Herman D. J., et al. (2019). Parallel evolution leading to impaired biofilm formation in invasive Salmonella strains. PloS Genet. 15 (6), e1008233. doi: 10.1371/journal.pgen.1008233
Mah T., O’Toole G. (2001). Machanisms of biofilms resistance to antimicrobial agents. Trends Microbiol. 9), 34–39. doi: 10.1016/S0966-842X(00)01913-2
Mah T. F. (2012). Biofilm-specific antibiotic resistance. Future Microbiol. 7), 1061–1072. doi: 10.2217/fmb.12.76
Maliniak M., Stecenko A., McCarty N. (2016). A longitudinal analysis of chronic MRSA and Pseudomonas aeruginosa co-infection in cystic fibrosis: a single-center study. J. Cyst. Fibros. 15), 350–356. doi: 10.1016/j.jcf.2015.10.014
Mantis N. J., Forbes S. J. (2010). Secretory IgA: arresting microbial pathogens at epithelial borders. Immunol. Invest. 39 (4-5), 383–406. doi: 10.3109/08820131003622635
Marshall J. M., Flechtner A. D., La Perle K. M., Gunn J. S. (2014). Visualization of extracellular matrix components within sectioned Salmonella biofilms on the surface of human gallstones. PloS One 9 (2), e89243. doi: 10.1371/journal.pone.0089243
Maruzani R., Sutton G., Nocerino P., Marvasi M. (2019). Exopolymeric substances (EPS) from Salmonella enterica: polymers, proteins and their interactions with plants and abiotic surfaces. J. Microbiol. 57 (1), 1–8. doi: 10.1007/s12275-019-8353-y
Marzel A., Desai P. T., Goren A., Schorr Y. I., Nissan I., Porwollik S., et al. (2016). Persistent Infections by Nontyphoidal Salmonella in Humans: Epidemiology and Genetics. Clin. Infect. Dis. 62 (7), 879–886. doi: 10.1093/cid/civ1221
McArthur M. A., Fresnay S., Magder L. S., Darton T. C., Jones C., Waddington C. S., et al. (2015). Activation of Salmonella Typhi-specific regulatory T cells in typhoid disease in a wild-type S. Typhi challenge model. PloS Pathog. 11 (5), e1004914. doi: 10.1371/journal.ppat.1004914
McSorley S. J., Cookson B. T., Jenkins M. K. (2000). Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J. Immunol. 164 (2), 986–993. doi: 10.4049/jimmunol.164.2.986
McSorley S. J., Asch S., Costalonga M., Reinhardt R. L., Jenkins M. K. (2002). Tracking salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity 16 (3), 365–377. doi: 10.1016/s1074-7613(02)00289-3
Menendez A., Arena E. T., Guttman J. A., Thorson L., Vallance B. A., Vogl W., et al. (2009). Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. J. Infect. Dis. 200 (11), 1703–1713. doi: 10.1086/646608
Merino L., Procura F., Trejo F. M., Bueno D. J., Golowczyc M. A. (2019). Biofilm formation by Salmonella sp. in the poultry industry: Detection, control and eradication strategies. Food Res. Int. 119, 530–540. doi: 10.1016/j.foodres.2017.11.024
Miller A. L., Pasternak J. A., Medeiros N. J., Nicastro L. K., Tursi S. A., Hansen E. G., et al. (2020). In vivo synthesis of bacterial amyloid curli contributes to joint inflammation during S. Typhimurium infection. PloS Pathog. 16 (7), e1008591. doi: 10.1371/journal.ppat.1008591
Mishra M., Byrd M. S., Sergeant S., Azad A. K., Parsek M. R., McPhail L., et al. (2012). Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell. Microbiol. 14), 95–106. doi: 10.1111/j.1462-5822.2011.01704.x
Mitchell E. K., Mastroeni P., Kelly A. P., Trowsdale J. (2004). Inhibition of cell surface MHC class II expression by Salmonella. Eur. J. Immunol. 34 (9), 2559–2567. doi: 10.1002/eji.200425314
Mittal M., Siddiqui M. R., Tran K., Reddy S. P., Malik A. B. (2014). Reactive oxygen species in inflammation and tissue injury. Antioxid. Redox Signaling 20 (7), 1126–1167. doi: 10.1089/ars.2012.5149
Moser C., Kjaergaard S., Pressler T., Kharazmi A., Koch C., Høiby N. (2000). The immune response to chronic Pseudomonas aeruginosa lung infection in cystic fibrosis patients is predominantly of the Th2 type. Apmis 108 (5), 329–335. doi: 10.1034/j.1600-0463.2000.d01-64.x
Moser C., Pedersen H. T., Lerche C. J., Kolpen M., Line L., Thomsen K., et al. (2017). Biofilms and host response–helpful or harmful. Apmis 125 (4), 320–338. doi: 10.1111/apm.12674
Mulcahy L. R., Isabella V. M., Lewis K. (2014). Pseudomonas aeruginosa biofilms in disease. Microb. Ecol. 68 (1), 1–12. doi: 10.1007/s00248-013-0297-x
Muller A. J., Kaiser P., Dittmar K. E., Weber T. C., Haueter S., Endt K., et al. (2012). Salmonella gut invasion involves TTSS-2-dependent epithelial traversal, basolateral exit, and uptake by epithelium-sampling lamina propria phagocytes. Cell Host Microbe 11 (1), 19–32. doi: 10.1016/j.chom.2011.11.013
Musher D. M., Rubenstein A. D. (1973). Permanent carriers of nontyphosa Salmonellae. Arch. Intern. Med. 132, 869–872. doi: 10.1001/archinte.1973.03650120071013
Müsken M., Pawar V., Schwebs T., Bähre H., Felgner S., Weiss S., et al. (2018). Breaking the vicious cycle of antibiotic killing and regrowth of biofilm-residing Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 62), e01635–e01618. doi: 10.1128/AAC.01635-18
Nanton M. R., Way S. S., Shlomchik M. J., McSorley S. J. (2012). Cutting edge: B cells are essential for protective immunity against Salmonella independent of antibody secretion. J. Immunol. 189 (12), 5503–5507. doi: 10.4049/jimmunol.1201413
Nelson K. S., Baltar F., Lamare M. D., Morales S. E. (2020). Ocean acidifcation afects microbial community and invertebrate settlement on bioflms. Sci. Rep. (10), 3274. doi: 10.1038/s41598-020-60023-4
Niess J. H., Brand S., Gu X., Landsman L., Jung S., McCormick B. A., et al. (2005). CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307 (5707), 254–258. doi: 10.1126/science.1102901
Nomura A., Stemmermann G. N., Chyou P.-H., Perez-Perez G. I., Blaser M. J. (1994). Helicobacter pylori infection and the risk for duodenal and gastric ulceration. Ann. Internal Med. 120 (12), 977–981. doi: 10.7326/0003-4819-120-12-199406150-00001
O’Toole G., Kolter R. (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol. Microbiol. 28), 449–461. doi: 10.1046/j.1365-2958.1998.00797.x
O’Toole G., Gibbs K., Hager P., Phibbs P., Kolter R. (2000a). The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182), 425–431. doi: 10.1128/JB.182.2.425-431.2000
O’Toole G., Kaplan H., Kolter R. (2000b). Biofilm formation as microbial development. Ann. Rev. Microbiol. 54), 49–79. doi: 10.1146/annurev.micro.54.1.49
Pande V. V., McWhorter A. R., Chousalkar K. K. (2016). Salmonella enterica isolates from layer farm environments are able to form biofilm on eggshell surfaces. Biofouling 32 (7), 699–710. doi: 10.1080/08927014.2016.1191068
Paredes J., Alonso-Arce M., Schmidt C. (2014). Smart central venous port for early detection of bacterial biofilm related infections. BioMed. Microdevices 16), 365–374. doi: 10.1007/s10544-014-9839-3
Parry C. M., Hien T. T., Dougan G., White N. J., Farrar J. J. (2002). Typhoid fever. N. Engl. J. Med. 347 (22), 1770–1782. doi: 10.1056/NEJMra020201
Parsek M. R., Singh P. K. (2003). Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57 (1), 677–701. doi: 10.1146/annurev.micro.57.030502.090720
Paytubi S., Cansado C., Madrid C., Balsalobre C. (2017). Nutrient Composition Promotes Switching between Pellicle and Bottom Biofilm in Salmonella. Front. Microbiol. 8:2160. doi: 10.3389/fmicb.2017.02160
Percival S., Suleman L., Vuotto C., Donelli G. (2015). Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J. Med. Microbiol. 64), 323–334. doi: 10.1099/jmm.0.000032
Perez-Shibayama C., Gil-Cruz C., Pastelin-Palacios R., Cervantes-Barragan L., Hisaki E., Chai Q., et al. (2014). IFN-gamma-producing CD4+ T cells promote generation of protective germinal center-derived IgM+ B cell memory against Salmonella Typhi. J. Immunol. 192 (11), 5192–5200. doi: 10.4049/jimmunol.1302526
Pletzer D., Mansour S. C., Wuerth K., Rahanjam N., Hancock R. E. (2017). New mouse model for chronic infections by Gram-negative bacteria enabling the study of anti-infective efficacy and host-microbe interactions. MBio 8 (1), e00140-17. doi: 10.1128/mBio.00140-17
Podolak R., Enache E., Stone W., Black D. G., Elliott P. H. (2010). Sources and risk factors for contamination, survival, persistence, and heat resistance of Salmonella in low-moisture foods. J. Food Prot. 73 (10), 1919–1936. doi: 10.4315/0362-028X-73.10.1919
Preciado-Llanes L., Aulicino A., Canals R., Moynihan P. J., Zhu X., Jambo N., et al. (2020). Evasion of MAIT cell recognition by the African Salmonella Typhimurium ST313 pathovar that causes invasive disease. Proc. Natl. Acad. Sci. U.S.A. 117 (34), 20717–20728. doi: 10.1073/pnas.2007472117
Prouty A. M., Gunn J. S. (2003). Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect. Immun. 71 (12), 7154–7158. doi: 10.1128/iai.71.12.7154-7158.2003
Prouty A. M., Schwesinger W. H., Gunn J. S. (2002). Biofilm formation and interaction with the surfaces of gallstones by Salmonella spp. Infect. Immun. 70 (5), 2640–2649. doi: 10.1128/iai.70.5.2640-2649.2002
Raffatellu M., Santos R. L., Verhoeven D. E., George M. D., Wilson R. P., Winter S. E., et al. (2008). Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat. Med. 14 (4), 421–428. doi: 10.1038/nm1743
Rakic M., Patrlj L., Kopljar M., Klicek R., Kolovrat M., Loncar B., et al. (2014). Gallbladder cancer. Hepatobiliary Surg. Nutr. 3 (5), 221–226. doi: 10.3978/j.issn.2304-3881.2014.09.03
Rathinam V. A. K., Zhao Y., Shao F. (2019). Innate immunity to intracellular LPS. Nat. Immunol. 20 (5), 527–533. doi: 10.1038/s41590-019-0368-3
Rathman M., Sjaastad M. D., Falkow S. (1996). Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect. Immun. 64 (7), 2765–2773. doi: 10.1128/IAI.64.7.2765-2773.1996
Reij M. W., Den Aantrekker E. D. (2004). Recontamination as a source of pathogens in processed foods. Int. J. Food Microbiol. 91 (1), 1–11. doi: 10.1016/s0168-1605(03)00295-2
Rescigno M., Urbano M., Valzasina B., Francolini M., Rotta G., Bonasio R., et al. (2001). Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2 (4), 361–367. doi: 10.1038/86373
Römling U., Rohde M. (1999). Flagella modulate the multicellular behavior of Salmonella typhimurium on the community level. FEMS Microbiol. Lett. 180 (1), 91–102. doi: 10.1016/S0378-1097(99)00469-3
Römling U., Bian Z., Hammar M., Sierralta W. D., Normark S. (1998). Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to opero structure and regulation. J. Bacteriol. 180 (3), 722–731. doi: 10.1128/JB.180.3.722-731.1998
Römling U., Rohde M., Olsén A., Normark S., Reinköster J. (2000). AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36 (1), 10–23. doi: 10.1046/j.1365-2958.2000.01822.x
Römling U., Bokranz W., Rabsch W., Zogaj X., Nimtz M., Tschape H. (2003). Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int. J. Med. Microbiol. 293 (4), 273–285. doi: 10.1078/1438-4221-00268
Römling U. (2005). Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol. Life Sci. 62 (11), 1234–1246. doi: 10.1007/s00018-005-4557-x
Salazar-Gonzalez R. M., Niess J. H., Zammit D. J., Ravindran R., Srinivasan A., Maxwell J. R., et al. (2006). CCR6-mediated dendritic cell activation of pathogen-specific T cells in Peyer’s patches. Immunity 24 (5), 623–632. doi: 10.1016/j.immuni.2006.02.015
Salerno-Goncalves R., Sztein M. B. (2009). Priming of Salmonella enterica serovar typhi-specific CD8(+) T cells by suicide dendritic cell cross-presentation in humans. PloS One 4 (6), e5879. doi: 10.1371/journal.pone.0005879
Santos R. L., Zhang S., Tsolis R. M., Kingsley R. A., Adams L. G., Baumler A. J. (2001). Animal models of Salmonella infections: enteritis versus typhoid fever. Microb. Infect. 3 (14-15), 1335–1344. doi: 10.1016/s1286-4579(01)01495-2
Schioler H., Christiansen E. D., Hoybye G., Rasmussen S. N., Greibe J. (1983). Biliary calculi in chronic Salmonella carriers and healthy controls: a controlled study. Scand. J. Infect. Dis. 15 (1), 17–19. doi: 10.3109/inf.1983.15.issue-1.04
Seth-Smith H. M. (2008). SPI-7: Salmonella’s Vi-encoding Pathogenicity Island. J. Infect. Dev. Ctries 2 (4), 267–271. doi: 10.3855/jidc.220
Sharma A., Qadri A. (2004). Vi polysaccharide of Salmonella typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc. Natl. Acad. Sci. U.S.A. 101 (50), 17492–17497. doi: 10.1073/pnas.0407536101
Sheffield C. L., Crippen T. L., Andrews K., Bongaerts R. J., Nisbet D. J. (2009). Planktonic and biofilm communities from 7-day-old chicken cecal microflora cultures: characterization and resistance to Salmonella colonization. J. Food Prot. 72 (9), 1812–1820. doi: 10.4315/0362-028X-72.9.1812
Simm R., Ahmad I., Rhen M., Le Guyon S., Römling U. (2014). Regulation of biofilm formation in Salmonella enterica serovar Typhimurium. Future Microbiol. 9 (11), 1261–1282. doi: 10.2217/fmb.14.88
Sirinavin S., Pokawattana L., Bangtrakulnondh A. (2004). Duration of nontyphoidal Salmonella carriage in asymptomatic adults. Clin. Infect. Dis. 38 (11), 1644–1645. doi: 10.1086/421027
Solano C., Sesma B., Alvarez M., Urdaneta E., Garcia-Ros D., Calvo A., et al. (2001). Virulent strains of Salmonella enteritidis disrupt the epithelial barrier of Caco-2 and HEp-2 cells. Arch. Microbiol. 175 (1), 46–51. doi: 10.1007/s002030000236
Sondberg E., Jelsbak L. (2016). Salmonella Typhimurium undergoes distinct genetic adaption during chronic infections of mice. BMC Microbiol. 16, 30. doi: 10.1186/s12866-016-0646-2
Stanaway J. D., Reiner R. C., Blacker B. F., Goldberg E. M., Khalil I. A., Troeger C. E., et al. (2019). The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 19 (4), 369–381. doi: 10.1016/S1473-3099(18)30685-6
Stewart P. S., Costerton J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358), 135–138. doi: 10.1016/S0140-6736(01)05321-1
Stinton L. M., Shaffer E. A. (2012). Epidemiology of gallbladder disease: cholelithiasis and cancer. Gut Liver 6 (2), 172–187. doi: 10.5009/gnl.2012.6.2.172
Tamblyn K. C., Conner D. E., Bilgili S. F. (1997). Utilization of the skin attachment model to determine the antibacterial efficacy of potential carcass treatments. Poult. Sci. 76 (9), 1318–1323. doi: 10.1093/ps/76.9.1318
Tang L., Wang C. X., Zhu S. L., Li Y., Deng X., Johnston R. N., et al. (2013). Genetic boundaries to delineate the typhoid agent and other Salmonella serotypes into distinct natural lineages. Genomics 102 (4), 331–337. doi: 10.1016/j.ygeno.2013.07.014
Thakur A., Mikkelsen H., Jungersen G. (2019). Intracellular pathogens: host immunity and microbial persistence strategies. J. Immunol. Res. 2019. doi: 10.1155/2019/1356540
Tobar J. A., Carreno L. J., Bueno S. M., Gonzalez P. A., Mora J. E., Quezada S. A., et al. (2006). Virulent Salmonella enterica serovar typhimurium evades adaptive immunity by preventing dendritic cells from activating T cells. Infect. Immun. 74 (11), 6438–6448. doi: 10.1128/IAI.00063-06
Tsai M. H., Liang Y. H., Chen C. L., Chiu C. H. (2020). Characterization of Salmonella resistance to bile during biofilm formation. J. Microbiol. Immunol. Infect. 53 (4), 518–524. doi: 10.1016/j.jmii.2019.06.003
Tursi S. A., Tukel C. (2018). Curli-containing enteric biofilms inside and out: matrix composition, immune recognition, and disease implications. Microbiol. Mol. Biol. R. 82 (4), e00028–e00018. doi: 10.1128/MMBR
Uppington H., Menager N., Boross P., Wood J., Sheppard M., Verbeek S., et al. (2006). Effect of immune serum and role of individual Fcgamma receptors on the intracellular distribution and survival of Salmonella enterica serovar Typhimurium in murine macrophages. Immunology 119 (2), 147–158. doi: 10.1111/j.1365-2567.2006.02416.x
Vasilev K., Cavallaro A., Zilm P. (2018). Special Issue: antibacterial materials and coatings. Molecules 23), 585. doi: 10.3390/molecules23030585
Vestby L. K., Grønseth T., Simm R., Nesse L. L. (2020). Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 9 (2), 59. doi: 10.3390/antibiotics9020059
Voedisch S., Koenecke C., David S., Herbrand H., Forster R., Rhen M., et al. (2009). Mesenteric lymph nodes confine dendritic cell-mediated dissemination of Salmonella enterica serovar Typhimurium and limit systemic disease in mice. Infect. Immun. 77 (8), 3170–3180. doi: 10.1128/IAI.00272-09
Wang H., Huang Y., Wu S., Li Y., Ye Y., Zheng Y., et al. (2014). Extracellular DNA inhibits Salmonella enterica Serovar Typhimurium and S. enterica Serovar Typhi biofilm development on abiotic surfaces. Curr. Microbiol. 68 (2), 262–268. doi: 10.1007/s00284-013-0468-5
Wangdi T., Lee C.-Y., Spees A. M., Yu C., Kingsbury D. D., Winter S. E., et al. (2014). The Vi capsular polysaccharide enables Salmonella enterica serovar Typhi to evade microbe-guided neutrophil chemotaxis. PloS Pathog. 10 (8), e1004306. doi: 10.1371/journal.ppat.1004306
Wessman M., Bjarnsholt T., Eickhardt-Sorensen S., Johansen H. K., Homøe P. (2014). Mucosal biofilm detection in chronic otitis media: a study of middle ear biopsies from Greenlandic patients. Eur. Arch. Otorhinolaryngol. 275 (5), 1079–1085. doi: 10.1007/s00405-00014-02886-00409
White A. P., Gibson D. L., Grassl G. A., Kay W. W., Finlay B. B., Vallance B. A., et al. (2008). Aggregation via the red, dry, and rough morphotype is not a virulence adaptation in Salmonella enterica serovar Typhimurium. Infect. Immun. 76 (3), 1048–1058. doi: 10.1128/IAI.01383-07
Wilson R. P., Raffatellu M., Chessa D., Winter S. E., Tukel C., Baumler A. J. (2008). The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell Microbiol. 10 (4), 876–890. doi: 10.1111/j.1462-5822.2007.01090.x
Wingender J., Flemming H.-C. (2011). Biofilms in drinking water and their role as reservoir for pathogens. Int. J. Hyg. Environ. Health 214), 417–423. doi: 10.1016/j.ijheh.2011.05.009
Wotherspoon A., Ortiz-Hidalgo C., Falzon M., Isaacson P. (1991). Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet 338 (8776), 1175–1176. doi: 10.1016/0140-6736(91)92035-Z
Wu M., Li X. (2015). “Klebsiella pneumoniae and Pseudomonas aeruginosa,” in Molecular medical microbiology. Ed. Tang Y. M., Sussman M., Liu D., Poxton I., Schwartzman J. (Waltham, Massachusetts: Elsevier), 1547–1564. doi: 10.1016/B978-0-12-397169-2.00087-1
Wu H., Moser C., Wang H., Hoiby N., Song Z. (2015). Strategies for combating bacterial biofilm infections. Int. J. Oral. Sci. 7), 1–7. doi: 10.1038/ijos.2014.65
Yaron S., Römling U. (2014). Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb. Biotechnol. 7 (6), 496–516. doi: 10.1111/1751-7915.12186
Yu X., Zhang H., Yu L., Liu M., Zuo Z., Han Q., et al. (2018). Intestinal lamina propria CD4+ T cells promote bactericidal activity of macrophages via galectin-9 and Tim-3 interaction during salmonella enterica serovar typhimurium infection. Infect. Immun. 86 (8), e00769-17. doi: 10.1128/IAI.00769-17
Zakikhany K., Harrington C. R., Nimtz M., Hinton J. C., Romling U. (2010). Unphosphorylated CsgD controls biofilm formation in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 77 (3), 771–786. doi: 10.1111/j.1365-2958.2010.07247.x
Zegans M. E., Becker H. I., Budzik J., O’Toole G. (2002). The role of bacterial biofilms in ocular infections. DNA Cell Biol. 21 (5-6), 415–420. doi: 10.1089/10445490260099700
Zeng H., Carlson A. Q., Guo Y., Yu Y., Collier-Hyams L. S., Madara J. L., et al. (2003). Flagellin is the major proinflammatory determinant of enteropathogenic Salmonella. J. Immunol. 171 (7), 3668–3674. doi: 10.4049/jimmunol.171.7.3668
Keywords: biofilm, chronic infection, immunity, hepatobiliary, Salmonella
Citation: Harrell JE, Hahn MM, D’Souza SJ, Vasicek EM, Sandala JL, Gunn JS and McLachlan JB (2021) Salmonella Biofilm Formation, Chronic Infection, and Immunity Within the Intestine and Hepatobiliary Tract. Front. Cell. Infect. Microbiol. 10:624622. doi: 10.3389/fcimb.2020.624622
Received: 31 October 2020; Accepted: 17 December 2020;
Published: 02 February 2021.
Edited by:
W. Edward Swords, University of Alabama at Birmingham, United StatesReviewed by:
Sargurunathan Subashchandrabose, Texas A&M University, United StatesAaron White, University of Saskatchewan, Canada
Copyright © 2021 Harrell, Hahn, D’Souza, Vasicek, Sandala, Gunn and McLachlan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James B. McLachlan, am1jbGFjaGxAdHVsYW5lLmVkdQ==; John S. Gunn, Sm9obi5HdW5uQG5hdGlvbndpZGVjaGlsZHJlbnMub3Jn
†These authors share first authorship
 Jaikin E. Harrell1†
Jaikin E. Harrell1† Shaina J. D’Souza
Shaina J. D’Souza Jenna L. Sandala
Jenna L. Sandala John S. Gunn
John S. Gunn James B. McLachlan
James B. McLachlan