- Department of Parasitology, Biomedical Primate Research Center, Rijswijk, Netherlands
Recent studies of liver stage malaria parasite-host interactions have provided exciting new insights on the cross-talk between parasite and its mammalian (predominantly rodent) host. We review the latest state of the art and and zoom in on new technologies that will provide the tools necessary to investigate host-parasite interactions of relapsing parasites. Interactions between hypnozoites and hepatocytes are particularly interesting because the parasite can remain in a quiescent state for prolonged periods of time and triggers for reactivation have not been irrefutably identified. If we learn more about the cross-talk between hypnozoite and host we may be able to identify factors that encourage waking up these dormant parasite reservoirs and help to achieve the total eradication of malaria.
Introduction
Malaria, caused by Plasmodium parasites, remains a very serious infectious disease, killing over 400,000 people per year (WHO, 2019). With a complex life-cycle in mosquito and vertebrate hosts, the parasite has to interact with its hosts to be able to survive and multiply. In the vertebrate host, the parasite has life-cycle stages initially in the liver and subsequently in the blood. Five malaria species can infect humans, Plasmodium 8falciparum, vivax, ovale, malariae (Hay et al., 2004), and the recently added zoonotic parasite P. knowlesi (Yegneswaran et al., 2009). Most of the fatal malaria infections are caused by P. falciparum, but also infection with P. vivax, the second most important human malaria parasite, can result in death (Price et al., 2007). P vivax, as well as a few other primate malarias form dormant stages called hypnozoites in the liver, that after months or even years can re-activate to yield new malaria episodes, without new infections through mosquito bites. This hidden reservoir of parasites complicates future malaria eradication. Liver stage biology, and especially hypnozoite biology, remains obscure as the liver stages are relatively inaccessible. Here we review recent progress in studies on liver stage parasite-host interactions in general and zoom in on new technologies that will allow detailed biological studies on dormant liver stages and their interaction with the host.
Parasite-Host Interactions Inside the Liver; the Rodent Models
The rodent malarias Plasmodium berghei, Plasmodium yoelii, Plasmodium chabaudi, and Plasmodium vinckei have played a pivotal role in understanding malaria biology [reviewed in (De Niz and Heussler, 2018)]. In the absence of prolonged blood stage cultures, these systems are almost exclusively in vivo based. Different mouse strains each have their own characteristics and the creation of transgenic mice has enabled studies that pinpoint specific parasite-host interactions (Liehl et al., 2014). The combination with highly efficient transfection systems for these parasite species (Mikolajczak et al., 2008; Matz and Kooij, 2015) renders the rodent malarias invaluable for studying parasite-host interactions in vivo.
Particularly liver stage research has greatly benefitted from the rodent models. Once sporozoites have switched from traversal to invasion (Coppi et al., 2007), the rodent malaria parasites show unprecedented multiplication inside hepatocytes. Within about 2 days, liver stage development is completed and thousands of merozoites are formed. Characterization of the liver stage parasites has been technically challenging due to their inaccessible location for experimentation. With robust in vitro parasite liver stage cultures established in many labs, more information has become available as to how the parasite manages to perform this daunting task.
The first comprehensive transcriptomic analysis for liver stages, which was combined with a proteomic survey, was described for P. yoelii (Tarun et al., 2008). The development of genetically engineered parasites allowed FACS isolation of liver stage infected hepatocytes. This revealed that liver stage schizonts express a wide range of metabolic pathways, including the liver stage-specific FASII pathway. Further in-depth RNAseq analyses performed at different timepoints during P. berghei liver stage development identified genes predominantly expressed in liver stages and showed that liver stage development was accompanied by differential expression of hundreds of parasite genes which may be regulated by a variety of posttranscriptional and posttranslational mechanisms (Caldelari et al., 2019; Shears et al., 2019).
While malaria liver stage development is a clinically silent phase of the life cycle, it has become clear that the hepatocyte does respond to the presence of the parasite in various ways. Transcriptional profiling of hepatoma cells early after infection with P. berghei showed an initial stress response of the cell to the presence of the parasite, which was followed by altered host cell metabolic responses to meet the requirements of parasite multiplication while maintaining parasite survival (Albuquerque et al., 2009). The parasite appears to prolong survival of the host cell by protecting it against extrinsic apoptosis (van de Sand et al., 2005; Kaushansky et al., 2013a), for example by suppression of host cell p53 (Kaushansky et al., 2013b) and by upregulating the “cellular inhibitor of apoptosis” protein (cIAP) (Ebert et al., 2020). Furthermore, the parasite has developed mechanisms to protect itself against elimination by autophagy of the host cell (Prado et al., 2015; Agop-Nersesian et al., 2017; Real et al., 2018). While autophagy can have detrimental effects, the liver stage parasite also appears to benefit from a non-canonical form of autophagy, termed Plasmodium Associated Autophagic-Response (PAAR) which was shown to support liver stage development (Agop-Nersesian et al., 2017; Coppens, 2017; Wacker et al., 2017; Evans et al., 2018).
That the malaria parasite is sensed by its host has also become evident by the specific type I interferon response that is triggered by liver stage malaria parasites following rodent liver infection (Liehl et al., 2014). In a rodent malaria model, it was shown that this response could inhibit malaria reinfections (Liehl et al., 2015). Similarly, host responses elicited during blood stage infection may impair liver stage infection. This appears to be mediated by the iron-regulatory hormone hepcidin, which restricts iron availability in the liver and thereby inhibits liver stage growth of the parasite (Portugal et al., 2011). This points to the importance of metal homeostasis during liver stage development, which is also highlighted by detrimental effects on liver stage parasites caused by gene knockouts of parasite metal transporters (Sahu et al., 2014; Kenthirapalan et al., 2016).
The parasite has been shown to recruit various host cell proteins in order to sustain its development, including GLUT1 (Meireles et al., 2017), aquaporin-3 (Posfai et al., 2018; Posfai et al., 2020) and protein traffic modulators such as COPB2 and GGA1 (Raphemot et al., 2019). Many of the host factors involved are recruited to the host-parasite interface in the liver cell, the parasitophorous vacuole membrane (PVM). Most interactions between host cell proteins and parasite proteins located at the PVM have remained elusive. To date, only a few connections between parasite antigens and hepatocyte proteins have been described. “Up-regulated in sporozoites protein” UIS3 has been shown to interact with liver fatty acid binding protein 1 (LFABP1) (Mueller et al., 2005; Mikolajczak et al., 2007), suggesting fatty acid scavenging from the host cell. Furthermore, P. berghei Exported protein 1 (EXP-1) was shown to interact with host Apolipoprotein H (ApoH) (Sa et al., 2017). These interactions have only been described for rodent malaria and possibly different parasite-host combinations require different interactions. This is highlighted by the finding that P. falciparum EXP-1 did not appear to interact with human ApoH (Sa et al., 2017).
Clearly, rodent models have been and will continue to play an important role in dissecting various aspects of malaria liver stage biology. However, it will be important to determine how the findings in these models translate to the human parasites, as has already been done for only a limited set of proteins, such as aquaporin-3 (Posfai et al., 2020) and Mucin-13 (LaMonte et al., 2019). While the rodent malaria studies allow investigations into the biology of developing liver stages, these parasite species do not develop into hypnozoites and thus do not enable studies on hypnozoite biology and hypnozoite-hepatocyte interaction.
P. vivax Parasite-Host Interactions Inside the Liver
Studying P. vivax parasite-host interactions in the liver is hampered by the fact that P. vivax develops only in primates. P. vivax liver stages were studied in humans, as described by Shortt and Garnham in 1948 (Shortt et al., 1948), in which liver biopsies were taken from a volunteer at day 6/7 post infection by the bites of ~1700 mosquitoes. They demonstrated the existence of a liver tissue stage in the P. vivax malaria life-cycle, similar to their observations in monkeys infected with P. cynomolgi sporozoites that they published shortly prior to the human experiment (Shortt and Garnham, 1948a). They observed large liver schizonts responsible for primary disease, but unfortunately did not find the hypnozoites, which were described only in 1980 by Krotoski (in liver biopsies from P. cynomolgi-infected monkeys) (Krotoski et al., 1980). A lot of knowledge has been gained from early experimental human infections regarding relapse patterns and the clinical profile of P. vivax, but these types of experiments are nowadays restricted. Although sporozoite-derived controlled human infections with P. vivax are allowed (under strict supervision) and can be highly significant to test new drugs or vaccines, volunteers are usually cured at low blood stage parasitemia (Herrera et al., 2011; Arevalo-Herrera et al., 2016). Relapses are not studied in this model and studying the liver stage parasites by taking liver biopsies from these volunteers is not performed. The best non-human model for P. vivax infections used to be the chimpanzee, and after finding the first hypnozoites in P. cynomolgi-infected rhesus monkeys, P. vivax hypnozoites were discovered in liver biopsies taken from P. vivax-infected chimps (Krotoski et al., 1982a). Nowadays animal experiments on apes are banned, so this model is no longer available (Hutson, 2010).
Other primate models for P. vivax, like Saimiri or Aotus monkeys have been primarily used for schizonticidal drugs and vaccine efficacy studies [reviewed in (Joyner et al., 2015)] but to be able to detect (low level) blood stage parasites caused by relapses animals need to be splenectomized, and pattern and frequency of relapses appear to be difficult to predict in these animal models (Joyner et al., 2015).
An important step for in vivo P. vivax research was achieved by the development of the FRG huHep chimeric mouse. This model is suitable to study the pre-erythrocytic stages of P. vivax, including hypnozoites, and can be used to test the activity of potential radical cure drugs that would kill hypnozoites (Mikolajczak et al., 2015). Blood stage parasites can be observed when injecting the FRG huHep mice with human reticulocytes around the time that the merosomes will be released from the mature liver schizonts, but this becomes very complicated if one wants to study relapses. Also, these mice are severely immunocompromised and will not completely reflect the human response to malaria infection or drug treatment. Thus, this model may not be suitable to study parasite-host interactions of relapsing malaria.
Studying parasite-host interactions in vitro may sound a bit counterintuitive, but some processes can be fairly easily studied in an in vitro setting, like the hepatocyte’s response to infection, activity of anti-relapse compounds and monitoring reactivation of the dormant stages. Primary human hepatocytes or hepatoma-derived cell lines (Hollingdale et al., 1984; Mazier et al., 1984; Sattabongkot et al., 2006; Chattopadhyay et al., 2010) are used as monoculture to study P. vivax liver stages in vitro. Recently, Roth et al. (Roth et al., 2018) have described a system using cryopreserved human primary hepatocytes and patient-derived sporozoites with high infection rates. This system should be helpful in the identification of new hypnozoite targeting compounds, as well as studying hypnozoites and reactivation as well as hepatocyte responses to infection. However, all of these models are dependent on patient material for the infection of mosquitoes. This means that there can be large variation between the different experiments, both in infection rate as well as in hypnozoite ratios (Roth et al., 2018). The variation caused by the different lots of hepatocytes is reduced when using cryopreserved cells, but variation caused by different patient-derived parasite isolates can’t be tackled. This can be circumvented by using the P. cynomolgi-monkey model for relapsing malaria (described below). Additional advantages for this model are that in vitro and in vivo experiments can be performed with the same well-characterized parasite and the availability of a robust transfection procedure for this parasite. P. cynomolgi can be genetically modified using episomes, centromere-containing constructs (for stable retention of the episome) and by single crossover integration into the genome (Kocken et al., 1999; Akinyi et al., 2012; Voorberg-van der Wel et al., 2013; Voorberg-van der Wel et al., 2020). Transfection of P.vivax is also possible, but is more difficult due to the restrictions (as described above) when working with small monkeys like Aotus and Saimiri (Pfahler et al., 2006; Moraes Barros et al., 2015), and so far the papers describing P. vivax transfection only show proof of concept.
P. cynomolgi, the Monkey Sister Parasite of P. vivax
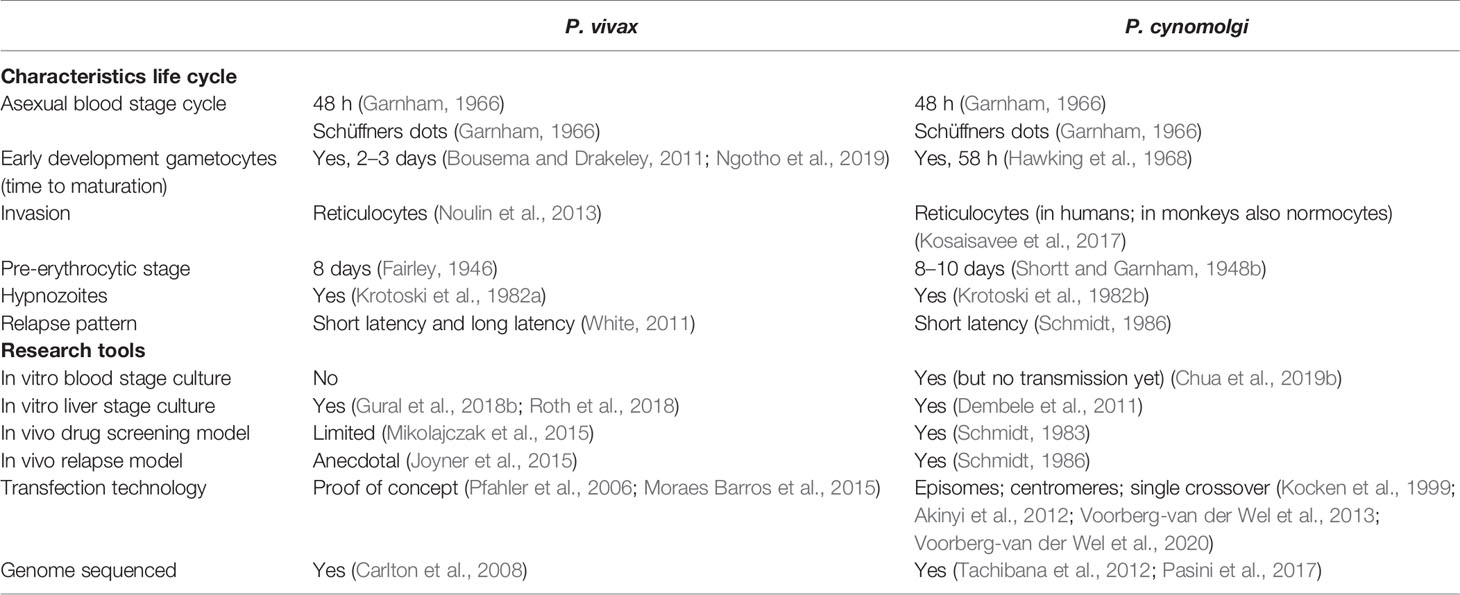
The monkey malaria parasite P. cynomolgi is considered to be an important model for the relapsing human malaria P. vivax, as it is phylogenetically closely related (Tachibana et al., 2012) and shares many biological characteristics (Table 1). Not only liver stage parasites, but also hypnozoites were first identified in the liver of rhesus monkeys that had been infected with high numbers of P. cynomolgi sporozoites (Shortt and Garnham, 1948a; Krotoski, 1985).
Monkeys infected with P. cynomolgi sporozoites have shown similar pathology as P. vivax-infected humans, including anemia and thrombocytopenia (Joyner et al., 2016; Joyner et al., 2017). In addition, it was shown that P. cynomolgi relapses can be clinically silent. This is likely to be due to the rapid development of memory B cell responses that help to clear asexual blood stage parasites but not gametocytes (Joyner et al., 2019).
Moreover, P. cynomolgi showed drug activity profiles that were highly similar to P. vivax (Schmidt et al., 1982b). This led to large scale drug screening studies with P. cynomolgi sporozoite-induced infections in rhesus monkeys as central step in efforts [which initially also used patients undergoing P. vivax malaria therapy as well as prison inmate volunteers (Coatney, 1985)] to find new hypnozoite-killing drugs (Davidson et al., 1981; Schmidt et al., 1982b; Schmidt, 1983; Dutta et al., 1989; Deye et al., 2012).
The in vivo data have been crucial for the discovery of the liver stages and for assessing the effects of drugs targeting these stages. However, experimentation and throughput are limited for ethical and economic reasons and, apart from the 8-aminoquinolines, other compounds killing hypnozoites have not been identified. Therefore, higher throughput approaches in order to find new, more potent and less toxic drugs that cure relapsing infections are needed (Wells et al., 2010; Campo et al., 2015). Knowledge of liver stage biology may reveal new targets for drug development, which may be more efficient than random screening approaches.
The advent of in vitro culture techniques for malaria liver stage parasites, including P. cynomolgi, (Millet et al., 1988) has greatly increased opportunities for the development of drug screening platforms and to begin to study parasite-host interactions. For P. cynomolgi, a low-throughput 96-well based assay system which enabled testing of compounds that are active against hypnozoites was developed, in which hypnozoites could be distinguished from developing forms (schizonts) by their size and differential sensitivity against selected drugs (Dembele et al., 2011).
Using this assay, a PI4 kinase inhibitor (McNamara et al., 2013) was identified showing high activity against early hypnozoites (Zeeman et al., 2014). This translated to in vivo prophylactic, but not radical cure activity (Zeeman et al., 2016), illustrating that young hypnozoites may be different from maturing hypnozoites. This is in line with earlier in vivo work which showed that when P. cynomolgi infected rhesus monkeys were treated at different timepoints with only 1 or 2 dosages of primaquine, it appeared that some phases (mainly early stages) of liver stage development were more vulnerable for the activity of the drug than others (Schmidt et al., 1982a).
The culture system was further improved through the addition of a Matrigel cover, which makes it possible to culture the P. cynomolgi exoerythrocytic forms for prolonged periods of time, revealing possible events of hypnozoite reactivation (Dembele et al., 2014). Recently, a 3D spheroid-culture system was reported that allows long-term cultivation of P. cynomolgi liver stages including full maturation of liver schizonts and invasion of red blood cells. While mimicking the in vivo microenvironment of the liver the 3D-structure of the spheroids renders it difficult to image and quantitate parasite load, presenting an obstacle for the use of this technology for high-throughput screening (Chua et al., 2019a). However, such a 3D-platform may be suitable for studying parasite-host interactions, with optimal in vitro hepatocyte quality, mimicking the in vivo situation.
In an attempt to characterize hypnozoites at the transcript level, P. cynomolgi day 7 hypnozoites and schizonts were collected by Laser Capture Microdissection (LCM) (Cubi et al., 2017). Two hypnozoite samples were obtained, containing a total of 45 and 59 hypnozoites, respectively (Cubi et al., 2017). Given the low levels of hypnozoite RNA in these small-sized samples, low read counts were obtained. Some ApiAP2 transcription factors were identified that were upregulated in hypnozoites. Further functional studies are needed to confirm the roles of these proteins.
P. cynomolgi has the advantage that it can be genetically manipulated (Kocken et al., 1999; Akinyi et al., 2012). By including a centromere (Iwanaga et al., 2010) in the construct, reporter lines have been developed which enable live visualization and purification of hypnozoites and liver stage schizonts (Voorberg-van der Wel et al., 2013). This has allowed a comprehensive transcriptomics analysis of day 6/7 and day 9 hypnozoites and schizonts (Voorberg-van der Wel et al., 2017; Bertschi et al., 2018). This revealed that developing schizonts are metabolically highly active, while hypnozoites continue to shut down transcription, except for pathways involved in the maintenance of genome stability, glycolysis and the pentose phosphate pathway. A marker for hypnozoites was not identified but Liver Stage Protein-2 was found to be schizont-specific and to be expressed very early on during schizogeny (Gupta et al., 2019). Rhesus host responses to P. cynomolgi infection and development in cultured hepatocytes have not yet been reported.
Using a P. cynomolgi reporter line that constitutively expresses GFP and shows mCherry expression when schizogony occurs, reactivation of hypnozoites in vitro was observed (Voorberg-van der Wel et al, 2020). This provides strong proof for the hypnozoite theory of relapse and allows screening of compounds that induce activation. If such compounds can be identified, “wake-and-kill” strategies can be envisaged in which hypnozoite activation is evoked, followed by killing of developing forms by currently available drugs.
The trigger for hypnozoite activation has remained enigmatic (Box 1) and the parasite-host interactions involved are elusive. Hypnozoite activation may be epigenetically controlled (Dembele et al., 2014). Furthermore, it has been suggested that activation may be triggered by mosquito bites (Hulden and Hulden, 2011), infectious disease (Shanks and White, 2013; Commons et al., 2019), or blood transfusion (Shanks and Waller, 2019). The P. cynomolgi fluorescent reporter line now offers the opportunity to investigate if/which molecules may stimulate hypnozoite activation. It must be realized, however, that in the context of the current in vitro platform it may be difficult to mimic the complex bodily reactions possibly involved in this. Moereover, reactivation events in culture are rare, making it challenging to isolate reactivating hypnozoites to study parasite and host transcriptomics.
Another question mark is how the hypnozoite survives for such a long time in a hepatocyte. Under normal conditions, the life-span of hepatocytes is estimated to be 6–12 months (Seeger and Mason, 2000). If the late recurrences [800–1,000 days after infection; (Schmidt, 1986)] of P. cynomolgi sporozoite induced infections in rhesus monkeys derive from activated hypnozoites, then how is this possible? Does the hypnozoite extend the longevity of the hepatocyte, or does it end up in a new hepatocyte after cell division?
Box 1. Outstanding questions:
Hypnozoite-host interaction
*How do the findings in the rodent liver stage models with respect to parasite-host interactions relate to the primate malarias?
*Which parasite-host interactions occur in the liver stage development of primate malarias?
*What are the differences between in vitro and in vivo parasites in terms of parasite-host interactions?
*How does the hypnozoite hide from the host immune system?
Hypnozoite-dormancy
*Do hypnozoites preferentially develop in a certain type of hepatocyte?
*How can a hypnozoite remain in the liver for prolonged times (longer than the generally estimated lifespan of hepatocytes)
*When does hypnozoite commitment occur?
*Which parasite/host molecules are involved in maintaining hypnozoite dormancy?
Hypnozoite-reactivation
*What is the mechanism behind hypnozoite activation?
*Is there a trigger involved or is it stochastic, via a biological clock, or a combination of this?
Newly Emerging Technologies
Liver stage parasites reside inside hepatocytes, located inside the liver. Given this multilayered, inaccessible location it has proven difficult to study this stage of the parasite life cycle. Furthermore, the existence of two forms of the parasite in some primate species, hypnozoites and schizonts, adds another layer of complexity to this. Much knowledge of parasite-host interactions of liver stage parasites has already been gained in the rodent malarias, although most likely this information represents only the tip of the iceberg. It will be important to determine whether this information can be translated to the primate malaria species. On top of this, virtually nothing is known about the interactions that take place between the hypnozoite stage of development and its host cell (Figure 1). Tools to study this are vital and have only recently begun to emerge, benefitting from technologies that have already been developed for the rodent malarias.
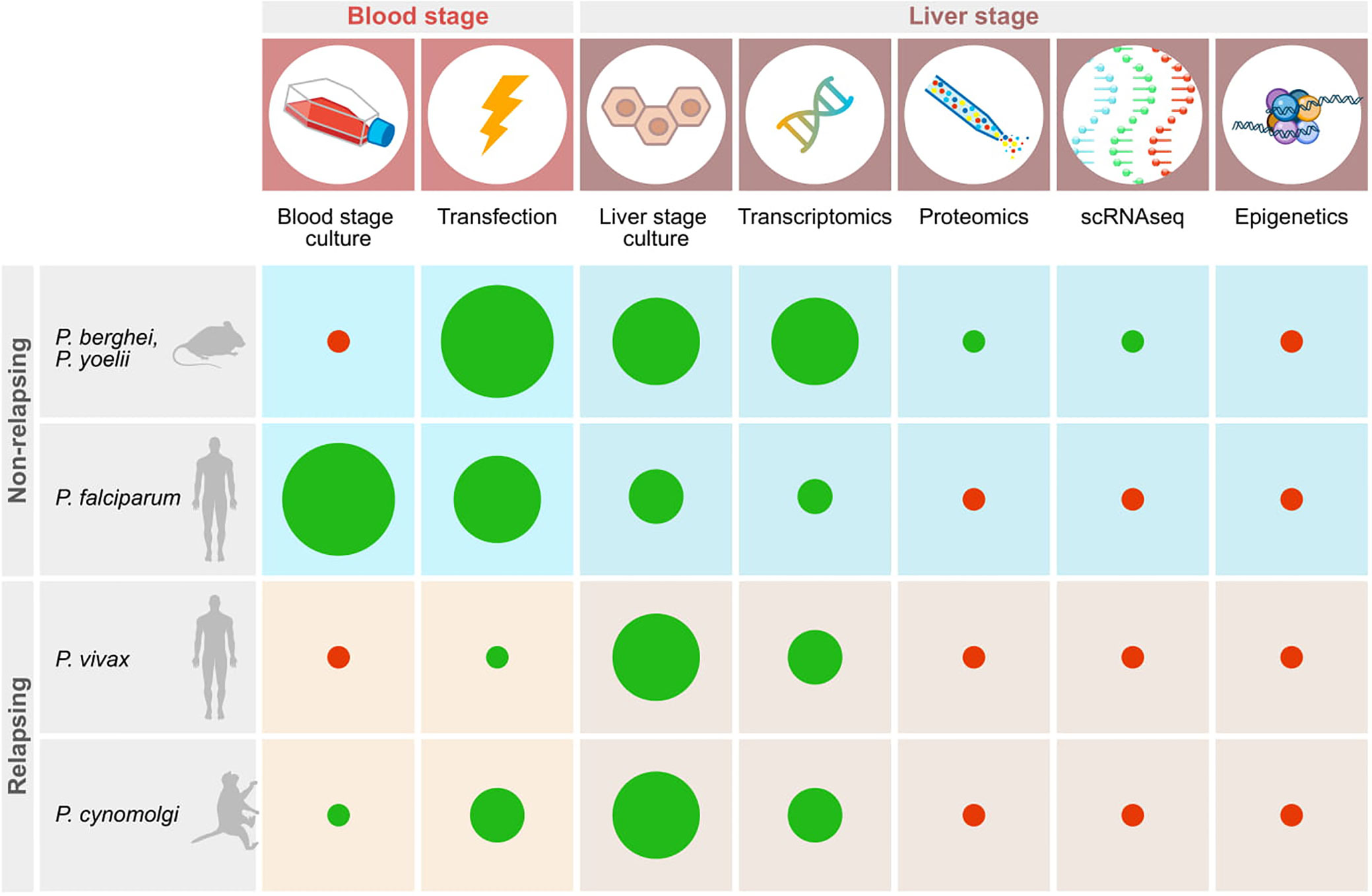
Figure 1 Schematic representation of important technologies available for studying relapsing and non-relapsing malaria parasites. Red dots indicate the absence of tools, green dots show that techniques have been established. The size of the diameter of the dots schematically indicates how widely the technology has been adopted based on published reports.
The development of liver stage cultures has greatly facilitated liver stage research. Some systems use hepatoma-derived cell lines (Hollingdale et al., 1984; Mazier et al., 1984; Sattabongkot et al., 2006; Chattopadhyay et al., 2010). While this provides a constant source of host cells, these cells differ in a number of aspects from primary hepatocytes (Tripathi et al., 2020), including a lower metabolic activity (Castell et al., 2006) and a high dependence on glucose uptake (Meireles et al., 2017). Therefore, care should be taken to validate results that mimic the natural situation. The importance of metabolic activity of hepatocytes was investigated for P. falciparum (Yang et al., 2020 BioRXiv, non peer-reviewed paper). This study indicates that P. falciparum liver stage development is strongly influenced by the differential metabolic activity of human hepatocytes derived from different zones of the liver.
The drawback of cultures using primary hepatocytes is that after about 12 days of culture the hepatocyte quality starts to deteriorate (Voorberg-van der Wel et al., 2020), which precludes analyses of hypnozoite activation. Approaches to overcome this issue include the addition of a Matrigel cover (Dembele et al., 2014), co-cultivation of human primary hepatocytes with fibroblasts (March et al., 2013; Gural et al., 2018a) or through the use of specific 384-well plates coated with collagen (Roth et al., 2018). Nucleic-acid mediated gene silencing has been successful in this type of systems, having the potential of exploring functional parasite-host interactions (Mancio-Silva L et al., 2019).
The advent of three-dimensional (3D) cell culture methods has opened up ways to develop cultures that mimic the in vivo physiological conditions to a greater extent. Proof-of-concept of this type of technology has already been shown, involving the use of hepatic spheroids using various hepatoma cell lines for culturing P. berghei (Arez et al., 2019) and using primary hepatocytes for P. cynomolgi (Chua et al., 2019a). Full development was shown for both parasite species, and cultures could be maintained for prolonged periods of time [up to 60 days in case of the simian spheroids (Chua et al., 2019a)]. While further improvements in terms of infection rate and by adding more cell types to create organoid like features is warranted, this type of systems provide new opportunities to study hypnozoite activation in vitro under conditions that are resembling the vivo situation.
More continuous, stable sources with truly hepatocyte features may be derived using newly emerging stem cell technologies. Proof of concept liver stage infections have already been shown using human induced Pluripotent Stem Cell (iPSC) derived hepatocyte-like cells (Ng et al., 2015) and chemically differentiated mouse embryonic stem cell (ESC)-based cells (Tripathi et al., 2020). These systems are attractive, since they not only provide a virtually unlimited source of hepatocytes, but the stem cells are also amenable to genetic manipulation thus allowing validation of genes important for parasite-host interactions in liver cells. In this way it was shown that the host adipose triglyceride lipase gene was dispensable for P. berghei liver stage development (Tripathi et al., 2020).
Little is known about host molecules involved in hypnozoite/liver stage development. Given that the first hypnozoite transcriptomes have become available (Cubi et al., 2017; Voorberg-van der Wel et al., 2017; Bertschi et al., 2018), a Dual-RNAseq approach can be envisaged whereby not only the transcriptome of the parasite, but also that of the host cell can be determined (LaMonte et al., 2019).
Information about transcriptional profiles of individual parasites can be obtained by a new technique called single cell RNA sequencing (scRNA-seq). Although technically challenging, researchers have accomplished (Poran et al., 2017) and optimized (Reid et al., 2018) a method for single cell RNA sequencing of malaria parasites. Using this new technique, individual parasites of all stages of the P. berghei life cycle were sorted and a transcriptional profile was generated, including difficult samples such as rings, which have low levels of RNA, and ookinetes, which are hard to sort (Howick et al., 2019). When application of scRNA-seq and other newly emerging “omics” approaches [e.g. lipidomics, metabolomics, proteomics, epigenomics (Cowell and Winzeler, 2018)] to relapsing malaria species (and more specifically to dormant liver stages) becomes feasible, these studies will likely shed more light onto genes involved in hypnozoite dormancy/activation.
The capacity to genetically modify parasite genes is key to study genes that may be involved in parasite host-interactions essential for hypnozoites. At the moment, such studies can only realistically be envisaged using the primate malaria P. cynomolgi. Lines that express reporter genes in P. cynomolgi liver stages have already been engineered (Voorberg-van der Wel et al., 2020), opening up studies that investigate phenotypic consequences of overexpression of gene candidates that may be involved in hypnozoite development. However, the P. cynomolgi transfection system is still in its infancy and only limited studies have been reported. Further development is warranted, because full exploitation of the capacity to genetically modify a relapsing parasite species will be vital for studying parasite-host interactions of hypnozoites with their host cell. Transfection systems have already been further optimized in other malaria species and it is expected that tools successful in these parasites, such as Crispr/Cas9 gene modification reviewed in (Lee et al., 2019), conditional (over)expression using DiCre (Jones et al., 2016) will be applicable to P. cynomolgi as well. Development of transfection tools may greatly benefit from the recently developed blood stage culture for this parasite (Chua et al., 2019b), extending the range of conditions that can be tested avoiding the use of donor and recipient monkeys.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
Some of the work presented here was supported by funding from the Medicines for Malaria Venture, and the Bill and Melinda Gates Foundation, grant number OPP1141292.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
P., plasmodium; FACS, fluorescence-activated cell sorting; FASII, fatty acid synthesis (in apicoplast); RNAseq, Ribonucleic acid sequencing; PVM, parasitophorous vacuolar membrane; UIS3, Up-regulated in sporozoites; huHep, human hepatocytes; 3D, three dimensional.
References
Agop-Nersesian C., De Niz M., Niklaus L., Prado M., Eickel N., Heussler V. T. (2017). Shedding of host autophagic proteins from the parasitophorous vacuolar membrane of Plasmodium berghei. Sci. Rep. 7 (1), 2191. doi: 10.1038/s41598-017-02156-7
Akinyi S., Hanssen E., Meyer E. V., Jiang J., Korir C. C., Singh B., et al. (2012). A 95 kDa protein of Plasmodium vivax and P. cynomolgi visualized by three-dimensional tomography in the caveola-vesicle complexes (Schuffner’s dots) of infected erythrocytes is a member of the PHIST family. Mol. Microbiol. 84 (5), 816–831. doi: 10.1111/j.1365-2958.2012.08060.x
Albuquerque S. S., Carret C., Grosso A. R., Tarun A. S., Peng X., Kappe S. H., et al. (2009). Host cell transcriptional profiling during malaria liver stage infection reveals a coordinated and sequential set of biological events. BMC Genomics 10, 270. doi: 10.1186/1471-2164-10-270
Arevalo-Herrera M., Vasquez-Jimenez J. M., Lopez-Perez M., Vallejo A. F., Amado-Garavito A. B., Cespedes N., et al. (2016). Protective Efficacy of Plasmodium vivax Radiation-Attenuated Sporozoites in Colombian Volunteers: A Randomized Controlled Trial. PloS Negl. Trop. Dis. 10 (10), e0005070. doi: 10.1371/journal.pntd.0005070
Arez F., Rebelo S. P., Fontinha D., Simao D., Martins T. R., Machado M., et al. (2019). Flexible 3D Cell-Based Platforms for the Discovery and Profiling of Novel Drugs Targeting Plasmodium Hepatic Infection. ACS Infect. Dis. 5 (11), 1831–1842. doi: 10.1021/acsinfecdis.9b00144
Bertschi N. L., Voorberg-van der Wel A., Zeeman A. M., Schuierer S., Nigsch F., Carbone W., et al. (2018). Transcriptomic analysis reveals reduced transcriptional activity in the malaria parasite Plasmodium cynomolgi during progression into dormancy. Elife 7. doi: 10.7554/eLife.41081
Bousema T., Drakeley C. (2011). Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin. Microbiol. Rev. 24 (2), 377–410. doi: 10.1128/CMR.00051-10
Caldelari R., Dogga S., Schmid M. W., Franke-Fayard B., Janse C. J., Soldati-Favre D., et al. (2019). Transcriptome analysis of Plasmodium berghei during exo-erythrocytic development. Malar. J. 18 (1), 330. doi: 10.1186/s12936-019-2968-7
Campo B., Vandal O., Wesche D. L., Burrows J. N. (2015). Killing the hypnozoite–drug discovery approaches to prevent relapse in Plasmodium vivax. Pathog. Glob. Health 109 (3), 107–122. doi: 10.1179/2047773215Y.0000000013
Carlton J. M., Adams J. H., Silva J. C., Bidwell S. L., Lorenzi H., Caler E., et al. (2008). Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature 455 (7214), 757–763. doi: 10.1038/nature07327
Castell J. V., Jover R., Martinez-Jimenez C. P., Gomez-Lechon M. J. (2006). Hepatocyte cell lines: their use, scope and limitations in drug metabolism studies. Expert Opin. Drug Metab. Toxicol. 2 (2), 183–212. doi: 10.1517/17425255.2.2.183
Chattopadhyay R., Velmurugan S., Chakiath C., Andrews Donkor L., Milhous W., Barnwell J. W., et al. (2010). Establishment of an in vitro assay for assessing the effects of drugs on the liver stages of Plasmodium vivax malaria. PloS One 5 (12), e14275. doi: 10.1371/journal.pone.0014275
Chua A. C. Y., Ananthanarayanan A., Ong J. J. Y., Wong J. Y., Yip A., Singh N. H., et al. (2019a). Hepatic spheroids used as an in vitro model to study malaria relapse. Biomaterials 216, 119221. doi: 10.1016/j.biomaterials.2019.05.032
Chua A. C. Y., Ong J. J. Y., Malleret B., Suwanarusk R., Kosaisavee V., Zeeman A. M., et al. (2019b). Robust continuous in vitro culture of the Plasmodium cynomolgi erythrocytic stages. Nat. Commun. 10 (1), 3635. doi: 10.1038/s41467-019-11332-4
Coatney G. R. (1985). Reminiscences: my forty-year romance with malaria. Trans. Nebraska Acad. Sci. XIII, 5–11.
Commons R. J., Simpson J. A., Thriemer K., Hossain M. S., Douglas N. M., Humphreys G. S., et al. (2019). Risk of Plasmodium vivax parasitaemia after Plasmodium falciparum infection: a systematic review and meta-analysis. Lancet Infect. Dis. 19 (1), 91–101. doi: 10.1016/S1473-3099(18)30596-6
Coppens I. (2017). How Toxoplasma and malaria parasites defy first, then exploit host autophagic and endocytic pathways for growth. Curr. Opin. Microbiol. 40, 32–39. doi: 10.1016/j.mib.2017.10.009
Coppi A., Tewari R., Bishop J. R., Bennett B. L., Lawrence R., Esko J. D., et al. (2007). Heparan sulfate proteoglycans provide a signal to Plasmodium sporozoites to stop migrating and productively invade host cells. Cell Host. Microbe 2 (5), 316–327. doi: 10.1016/j.chom.2007.10.002
Cowell A., Winzeler E. (2018). Exploration of the Plasmodium falciparum Resistome and Druggable Genome Reveals New Mechanisms of Drug Resistance and Antimalarial Targets. Microbiol. Insights 11, 1178636118808529. doi: 10.1177/1178636118808529
Cubi R., Vembar S. S., Biton A., Franetich J. F., Bordessoulles M., Sossau D., et al. (2017). Laser capture microdissection enables transcriptomic analysis of dividing and quiescent liver stages of Plasmodium relapsing species. Cell Microbiol. 19 (8). doi: 10.1111/cmi.12735
Davidson D. E. Jr., Ager A. L., Brown J. L., Chapple F. E., Whitmire R. E., Rossan R. N. (1981). New tissue schizontocidal antimalarial drugs. Bull. World Health Organ 59 (3), 463–479.
De Niz M., Heussler V. T. (2018). Rodent malaria models: insights into human disease and parasite biology. Curr. Opin. Microbiol. 46, 93–101. doi: 10.1016/j.mib.2018.09.003
Dembele L., Gego A., Zeeman A. M., Franetich J. F., Silvie O., Rametti A., et al. (2011). Towards an in vitro model of Plasmodium hypnozoites suitable for drug discovery. PloS One 6 (3), e18162. doi: 10.1371/journal.pone.0018162
Dembele L., Franetich J. F., Lorthiois A., Gego A., Zeeman A. M., Kocken C. H., et al. (2014). Persistence and activation of malaria hypnozoites in long-term primary hepatocyte cultures. Nat. Med. doi: 10.1038/nm.3461
Deye G. A., Gettayacamin M., Hansukjariya P., Im-erbsin R., Sattabongkot J., Rothstein Y., et al. (2012). Use of a rhesus Plasmodium cynomolgi model to screen for anti-hypnozoite activity of pharmaceutical substances. Am. J. Trop. Med. Hyg. 86 (6), 931–935. doi: 10.4269/ajtmh.2012.11-0552
Dutta G. P., Puri S. K., Bhaduri A. P., Seth M. (1989). Radical curative activity of a new 8-aminoquinoline derivative (CDRI 80/53) against Plasmodium cynomolgi B in monkeys. Am. J. Trop. Med. Hyg. 41 (6), 635–637. doi: 10.4269/ajtmh.1989.41.635
Ebert G., Lopaticki S., O’Neill M. T., Steel R. W. J., Doerflinger M., Rajasekaran P., et al. (2020). Targeting the Extrinsic Pathway of Hepatocyte Apoptosis Promotes Clearance of Plasmodium Liver Infection. Cell Rep. 30 (13), 4343–4354.e4344. doi: 10.1016/j.celrep.2020.03.032
Evans R. J., Sundaramurthy V., Frickel E. M. (2018). The Interplay of Host Autophagy and Eukaryotic Pathogens. Front. Cell Dev. Biol. 6, 118. doi: 10.3389/fcell.2018.00118
Fairley N. H. (1946). Researches on paludrine (M.4888) in malaria; an experimental investigation undertaken by the L.H.Q. Medical Research Unit (A.I.F.) Cairns, Australia. Trans. R. Soc. Trop. Med. Hyg. 40 (2), 105–162. doi: 10.1016/0035-9203(46)90052-1
Gupta D. K., Dembele L., Voorberg-van der Wel A., Roma G., Yip A., Chuenchob V., et al. (2019). The Plasmodium liver-specific protein 2 (LISP2) is an early marker of liver stage development. Elife 8. doi: 10.7554/eLife.43362
Gural N., Mancio-Silva L., Miller A. B., Galstian A., Butty V. L., Levine S. S., et al. (2018a). In Vitro Culture, Drug Sensitivity, and Transcriptome of Plasmodium Vivax Hypnozoites. Cell Host. Microbe 23 (3), 395–406. doi: 10.1016/j.chom.2018.01.002
Gural N., Mancio-Silva L., Miller A. B., Galstian A., Butty V. L., Levine S. S., et al. (2018b). In Vitro Culture, Drug Sensitivity, and Transcriptome of Plasmodium Vivax Hypnozoites. Cell Host. Microbe 23 (3), 395–406 e394. doi: 10.1016/j.chom.2018.01.002
Hawking F., Worms M. J., Gammage K. (1968). 24- and 48-hour cycles of malaria parasites in the blood; their purpose, production and control. Trans. R. Soc. Trop. Med. Hyg. 62 (6), 731–765. doi: 10.1016/0035-9203(68)90001-1
Hay S. I., Guerra C. A., Tatem A. J., Noor A. M., Snow R. W. (2004). The global distribution and population at risk of malaria: past, present, and future. Lancet Infect. Dis. 4 (6), 327–336. doi: 10.1016/S1473-3099(04)01043-6S1473309904010436
Herrera S., Solarte Y., Jordan-Villegas A., Echavarria J. F., Rocha L., Palacios R., et al. (2011). Consistent safety and infectivity in sporozoite challenge model of Plasmodium vivax in malaria-naive human volunteers. Am. J. Trop. Med. Hyg. 84 (2 Suppl), 4–11. doi: 10.4269/ajtmh.2011.09-0498
Hollingdale M. R., Nardin E. H., Tharavanij S., Schwartz A. L., Nussenzweig R. S. (1984). Inhibition of entry of Plasmodium falciparum and P. vivax sporozoites into cultured cells; an in vitro assay of protective antibodies. J. Immunol. 132 (2), 909–913.
Howick V. M., Russell A. J. C., Andrews T., Heaton H., Reid A. J., Natarajan K., et al. (2019). The Malaria Cell Atlas: Single parasite transcriptomes across the complete Plasmodium life cycle. Science 365 (6455). doi: 10.1126/science.aaw2619
Hulden L., Hulden L. (2011). Activation of the hypnozoite: a part of Plasmodium vivax life cycle and survival. Malar. J. 10, 90. doi: 10.1186/1475-2875-10-90
Hutson S. (2010). Following Europe’s lead, Congress moves to ban ape research. Nat. Med. 16 (10):1057. doi: 10.1038/nm1010-1057a
Iwanaga S., Khan S. M., Kaneko I., Christodoulou Z., Newbold C., Yuda M., et al. (2010). Functional identification of the Plasmodium centromere and generation of a Plasmodium artificial chromosome. Cell Host. Microbe 7 (3), 245–255. doi: 10.1016/j.chom.2010.02.010
Jones M. L., Das S., Belda H., Collins C. R., Blackman M. J., Treeck M. (2016). A versatile strategy for rapid conditional genome engineering using loxP sites in a small synthetic intron in Plasmodium falciparum. Sci. Rep. 6:21800. doi: 10.1038/srep21800
Joyner C., Barnwell J. W., Galinski M. R. (2015). No more monkeying around: primate malaria model systems are key to understanding Plasmodium vivax liver-stage biology, hypnozoites, and relapses. Front. Microbiol. 6, 145. doi: 10.3389/fmicb.2015.00145
Joyner C., Moreno A., Meyer E. V., Cabrera-Mora M., Ma H. C., Kissinger J. C., et al. (2016). Plasmodium cynomolgi infections in rhesus macaques display clinical and parasitological features pertinent to modelling vivax malaria pathology and relapse infections. Malar. J. 15 (1), 451. doi: 10.1186/s12936-016-1480-6
Joyner C. J., Consortium T. M., Wood J. S., Moreno A., Garcia A., Galinski M. (2017). Case Report: Severe and Complicated Cynomolgi Malaria in a Rhesus Macaque Resulted in Similar Histopathological Changes as Those Seen in Human Malaria. Am. J. Trop. Med. Hyg. 97 (2), 548–555. doi: 10.4269/ajtmh.16-0742
Joyner C. J., Brito C. F. A., Saney C. L., Joice Cordy R., Smith M. L., Lapp S. A., et al. (2019). Humoral immunity prevents clinical malaria during Plasmodium relapses without eliminating gametocytes. PloS Pathog. 15 (9), e1007974. doi: 10.1371/journal.ppat.1007974
Kaushansky A., Metzger P. G., Douglass A. N., Mikolajczak S. A., Lakshmanan V., Kain H. S., et al. (2013a). Malaria parasite liver stages render host hepatocytes susceptible to mitochondria-initiated apoptosis. Cell Death Dis. 4, e762. doi: 10.1038/cddis.2013.286
Kaushansky A., Ye A. S., Austin L. S., Mikolajczak S. A., Vaughan A. M., Camargo N., et al. (2013b). Suppression of host p53 is critical for Plasmodium liver-stage infection. Cell Rep. 3 (3), 630–637. doi: 10.1016/j.celrep.2013.02.010
Kenthirapalan S., Waters A. P., Matuschewski K., Kooij T. W. (2016). Functional profiles of orphan membrane transporters in the life cycle of the malaria parasite. Nat. Commun. 7:10519. doi: 10.1038/ncomms10519
Kocken C. H., van der Wel A., Thomas A. W. (1999). Plasmodium cynomolgi: transfection of blood-stage parasites using heterologous DNA constructs. Exp. Parasitol. 93 (1), 58–60. doi: 10.1006/expr.1999.4430
Kosaisavee V., Suwanarusk R., Chua A. C. Y., Kyle D. E., Malleret B., Zhang R., et al. (2017). Strict tropism for CD71(+)/CD234(+) human reticulocytes limits the zoonotic potential of Plasmodium cynomolgi. Blood 130 (11), 1357–1363. doi: 10.1182/blood-2017-02-764787
Krotoski W. A., Krotoski D. M., Garnham P. C. C., Bray R. S., Killick-Kendrick R., Draper C. C., et al. (1980). Relapses in primate malaria: discovery of two populations of exoerythrocytic stages. Br. Med. J. 280 (6208), 153–154. doi: 10.1136/bmj.280.6208.153-a
Krotoski W. A., Collins W. E., Bray R. S., Garnham P. C., Cogswell F. B., Gwadz R. W., et al. (1982a). Demonstration of hypnozoites in sporozoite-transmitted Plasmodium vivax infection. Am. J. Trop. Med. Hyg. 31 (6), 1291–1293. doi: 10.4269/ajtmh.1982.31.1291
Krotoski W. A., Garnham P. C., Bray R. S., Krotoski D. M., Killick-Kendrick R., Draper C. C., et al. (1982b). Observations on early and late post-sporozoite tissue stages in primate malaria. I. Discovery of a new latent form of Plasmodium cynomolgi (the hypnozoite), and failure to detect hepatic forms within the first 24 hours after infection. Am. J. Trop. Med. Hyg. 31 (1), 24–35. doi: 10.4269/ajtmh.1982.31.24
Krotoski W. A. (1985). Discovery of the hypnozoite and a new theory of malarial relapse. Trans. R. Soc. Trop. Med. Hyg. 79 (1), 1–11. doi: 10.1016/0035-9203(85)90221-4
LaMonte G. M., Orjuela-Sanchez P., Calla J., Wang L. T., Li S., Swann J., et al. (2019). Dual RNA-seq identifies human mucosal immunity protein Mucin-13 as a hallmark of Plasmodium exoerythrocytic infection. Nat. Commun. 10 (1), 488. doi: 10.1038/s41467-019-08349-0
Lee M. C. S., Lindner S. E., Lopez-Rubio J. J., Llinas M. (2019). Cutting back malaria: CRISPR/Cas9 genome editing of Plasmodium. Brief Funct. Genomics 18 (5), 281–289. doi: 10.1093/bfgp/elz012
Liehl P., Zuzarte-Luis V., Chan J., Zillinger T., Baptista F., Carapau D., et al. (2014). Host-cell sensors for Plasmodium activate innate immunity against liver-stage infection. Nat. Med. 20 (1), 47–53. doi: 10.1038/nm.3424
Liehl P., Meireles P., Albuquerque I. S., Pinkevych M., Baptista F., Mota M. M., et al. (2015). Innate immunity induced by Plasmodium liver infection inhibits malaria reinfections. Infect. Immun. 83 (3), 1172–1180. doi: 10.1128/IAI.02796-14
Mancio-Silva L F. H., Miller A. B., Milstein S., Liebow A., Haslett P., Sepp-Lorenzino L., et al. (2019). Improving Drug Discovery by Nucleic Acid Delivery in Engineered Human Microlivers. Cell Metab. 29 (3), 727–735. doi: 10.1016/j.cmet.2019.02.003
March S., Ng S., Velmurugan S., Galstian A., Shan J., Logan D. J., et al. (2013). A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax. Cell Host. Microbe 14 (1), 104–115. doi: 10.1016/j.chom.2013.06.005
Matz J. M., Kooij T. W. (2015). Towards genome-wide experimental genetics in the in vivo malaria model parasite Plasmodium berghei. Pathog. Glob. Health 109 (2), 46–60. doi: 10.1179/2047773215Y.0000000006
Mazier D., Landau I., Druilhe P., Miltgen F., Guguen-Guillouzo C., Baccam D., et al. (1984). Cultivation of the liver forms of Plasmodium vivax in human hepatocytes. Nature 307 (5949), 367–369. doi: 10.1038/307367a0
McNamara C. W., Lee M. C., Lim C. S., Lim S. H., Roland J., Nagle A., et al. (2013). Targeting Plasmodium PI(4)K to eliminate malaria. Nature 504 (7479), 248–253. doi: 10.1038/nature12782
Meireles P., Sales-Dias J., Andrade C. M., Mello-Vieira J., Mancio-Silva L., Simas J. P., et al. (2017). GLUT1-mediated glucose uptake plays a crucial role during Plasmodium hepatic infection. Cell Microbiol. 19 (2). doi: 10.1111/cmi.12646
Mikolajczak S. A., Jacobs-Lorena V., MacKellar D. C., Camargo N., Kappe S. H. (2007). L-FABP is a critical host factor for successful malaria liver stage development. Int. J. Parasitol. 37 (5), 483–489. doi: 10.1016/j.ijpara.2007.01.002
Mikolajczak S. A., Aly A. S., Dumpit R. F., Vaughan A. M., Kappe S. H. (2008). An efficient strategy for gene targeting and phenotypic assessment in the Plasmodium yoelii rodent malaria model. Mol. Biochem. Parasitol. 158 (2), 213–216. doi: 10.1016/j.molbiopara.2007.12.006
Mikolajczak S. A., Vaughan A. M., Kangwanrangsan N., Roobsoong W., Fishbaugher M., Yimamnuaychok N., et al. (2015). Plasmodium vivax liver stage development and hypnozoite persistence in human liver-chimeric mice. Cell Host. Microbe 17 (4), 526–535. doi: 10.1016/j.chom.2015.02.011
Millet P., Fisk T. L., Collins W. E., Broderson J. R., Nguyen-Dinh P. (1988). Cultivation of exoerythrocytic stages of Plasmodium cynomolgi, P. knowlesi, P. coatneyi, and P. inui in Macaca mulatta hepatocytes. Am. J. Trop. Med. Hyg. 39 (6), 529–534. doi: 10.4269/ajtmh.1988.39.529
Moraes Barros R. R., Straimer J., Sa J. M., Salzman R. E., Melendez-Muniz V. A., Mu J., et al. (2015). Editing the Plasmodium vivax genome, using zinc-finger nucleases. J. Infect. Dis. 211 (1), 125–129. doi: 10.1093/infdis/jiu423
Mueller A. K., Labaied M., Kappe S. H., Matuschewski K. (2005). Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature 433 (7022), 164–167. doi: 10.1038/nature03188
Ng S., Schwartz R. E., March S., Galstian A., Gural N., Shan J., et al. (2015). Human iPSC-derived hepatocyte-like cells support Plasmodium liver-stage infection in vitro. Stem Cell Rep. 4 (3), 348–359. doi: 10.1016/j.stemcr.2015.01.002
Ngotho P., Soares A. B., Hentzschel F., Achcar F., Bertuccini L., Marti M. (2019). Revisiting gametocyte biology in malaria parasites. FEMS Microbiol. Rev. 43 (4), 401–414. doi: 10.1093/femsre/fuz010
Noulin F., Borlon C., Van Den Abbeele J., D’Alessandro U., Erhart A. (2013). 1912-2012: a century of research on Plasmodium vivax in vitro culture. Trends Parasitol. 29 (6), 286–294. doi: 10.1016/j.pt.2013.03.012
Pasini E. M., Bohme U., Rutledge G. G., Voorberg-Van der Wel A., Sanders M., Berriman M., et al. (2017). An improved Plasmodium cynomolgi genome assembly reveals an unexpected methyltransferase gene expansion. Wellcome Open Res. 2, 42. doi: 10.12688/wellcomeopenres.11864.1
Pfahler J. M., Galinski M. R., Barnwell J. W., Lanzer M. (2006). Transient transfection of Plasmodium vivax blood stage parasites. Mol. Biochem. Parasitol. 149 (1), 99–101. doi: 10.1016/j.molbiopara.2006.03.018
Poran A., Notzel C., Aly O., Mencia-Trinchant N., Harris C. T., Guzman M. L., et al. (2017). Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature 551 (7678), 95–99. doi: 10.1038/nature24280
Portugal S., Carret C., Recker M., Armitage A. E., Goncalves L. A., Epiphanio S., et al. (2011). Host-mediated regulation of superinfection in malaria. Nat. Med. 17 (6), 732–737. doi: 10.1038/nm.2368
Posfai D., Sylvester K., Reddy A., Ganley J. G., Wirth J., Cullen Q. E., et al. (2018). Plasmodium parasite exploits host aquaporin-3 during liver stage malaria infection. PloS Pathog. 14 (5), e1007057. doi: 10.1371/journal.ppat.1007057
Posfai D., Maher S. P., Roesch C., Vantaux A., Sylvester K., Peneau J., et al. (2020). Plasmodium vivax Liver and Blood Stages Recruit the Druggable Host Membrane Channel Aquaporin-3. Cell Chem. Biol. 27 (6), 719–727 e715. doi: 10.1016/j.chembiol.2020.03.009
Prado M., Eickel N., De Niz M., Heitmann A., Agop-Nersesian C., Wacker R., et al. (2015). Long-term live imaging reveals cytosolic immune responses of host hepatocytes against Plasmodium infection and parasite escape mechanisms. Autophagy 11 (9), 1561–1579. doi: 10.1080/15548627.2015.1067361
Price R. N., Tjitra E., Guerra C. A., Yeung S., White N. J., Anstey N. M. (2007). Vivax malaria: neglected and not benign. Am. J. Trop. Med. Hyg. 77 (6 Suppl), 79–87. doi: 10.4269/ajtmh.2007.77.79
Raphemot R., Toro-Moreno M., Lu K. Y., Posfai D., Derbyshire E. R. (2019). Discovery of Druggable Host Factors Critical to Plasmodium Liver-Stage Infection. Cell Chem. Biol. 26 (9), 1253–1262.e1255. doi: 10.1016/j.chembiol.2019.05.011
Real E., Rodrigues L., Cabal G. G., Enguita F. J., Mancio-Silva L., Mello-Vieira J., et al. (2018). Plasmodium UIS3 sequesters host LC3 to avoid elimination by autophagy in hepatocytes. Nat. Microbiol. 3 (1), 17–25. doi: 10.1038/s41564-017-0054-x
Reid A. J., Talman A. M., Bennett H. M., Gomes A. R., Sanders M. J., Illingworth C. J. R., et al. (2018). Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. Elife 7. doi: 10.7554/eLife.33105
Roth A., Maher S. P., Conway A. J., Ubalee R., Chaumeau V., Andolina C., et al. (2018). A comprehensive model for assessment of liver stage therapies targeting Plasmodium vivax and Plasmodium falciparum. Nat. Commun. 9 (1), 1837. doi: 10.1038/s41467-018-04221-9
Sa E. C. C., Nyboer B., Heiss K., Sanches-Vaz M., Fontinha D., Wiedtke E., et al. (2017). Plasmodium berghei EXP-1 interacts with host Apolipoprotein H during Plasmodium liver-stage development. Proc. Natl. Acad. Sci. U. S. A. 114 (7), E1138–E1147. doi: 10.1073/pnas.1606419114
Sahu T., Boisson B., Lacroix C., Bischoff E., Richier Q., Formaglio P., et al. (2014). ZIPCO, a putative metal ion transporter, is crucial for Plasmodium liver-stage development. EMBO Mol. Med. 6 (11), 1387–1397. doi: 10.15252/emmm.201403868
Sattabongkot J., Yimamnuaychoke N., Leelaudomlipi S., Rasameesoraj M., Jenwithisuk R., Coleman R. E., et al. (2006). Establishment of a human hepatocyte line that supports in vitro development of the exo-erythrocytic stages of the malaria parasites Plasmodium falciparum and P. vivax. Am. J. Trop. Med. Hyg. 74 (5), 708–715. doi: 10.4269/ajtmh.2006.74.708
Schmidt L. H., Fradkin R., Genther C. S., Hughes H. B. (1982a). Delineation of the Potentials of Primaquine as a Radical Curative and Prophylactic Drug. Am. J. Trop. Med. Hyg. 31(3 (3 (Pt 2), 666–680. doi: 10.4269/ajtmh.1982.31.666
Schmidt L. H., Fradkin R., Genther C. S., Rossan R. N., Squires W. (1982b). Responses of Sporozoite-Induced and Trophozoite-Induced Infections to Standard Antimalarial Drugs. Am. J. Trop. Med. Hyg. 31(3 (3 (Pt 2), 646–665. doi: 10.4269/ajtmh.1982.31.646
Schmidt L. H. (1983). Appraisals of compounds of diverse chemical classes for capacities to cure infections with sporozoites of Plasmodium cynomolgi. Am. J. Trop. Med. Hyg. 32 (2), 231–257. doi: 10.4269/ajtmh.1983.32.231
Schmidt L. H. (1986). Compatibility of relapse patterns of Plasmodium cynomolgi infections in rhesus monkeys with continuous cyclical development and hypnozoite concepts of relapse. Am. J. Trop. Med. Hyg. 35 (6), 1077–1099. doi: 10.4269/ajtmh.1986.35.1077
Seeger C., Mason W. S. (2000). Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64 (1), 51–68. doi: 10.1128/mmbr.64.1.51-68.2000
Shanks G., Waller M. (2019). Blood transfusion as a possible initiator of Plasmodium vivax relapse through increased heme metabolism leading to hepatocyte apoptosis. Poster Presentation ICPVR Paris.
Shanks G. D., White N. J. (2013). The activation of vivax malaria hypnozoites by infectious diseases. Lancet Infect. Dis. 13 (10), 900–906. doi: 10.1016/S1473-3099(13)70095-1
Shears M. J., Sekhar Nirujogi R., Swearingen K. E., Renuse S., Mishra S., Jaipal Reddy P., et al. (2019). Proteomic Analysis of Plasmodium Merosomes: The Link between Liver and Blood Stages in Malaria. J. Proteome Res. 18 (9), 3404–3418. doi: 10.1021/acs.jproteome.9b00324
Shortt H. E., Garnham P. C., Covell G., Shute P. G., et al. (1948). The pre-erythrocytic stage of human malaria, Plasmodium vivax. Br. Med. J. 1 (4550):547. doi: 10.1136/bmj.1.4550.547
Shortt H. E., Garnham P. C. (1948a). Demonstration of a persisting exo-erythrocytic cycle in Plasmodium cynomolgi and its bearing on the production of relapses. Br. Med. J. 1 (4564), 1225–1228. doi: 10.1136/bmj.1.4564.1225
Shortt H. E., Garnham P. C. (1948b). The pre-erythrocytic development of Plasmodium cynomolgi and Plasmodium vivax. Trans. R. Soc. Trop. Med. Hyg. 41 (6), 785–795. doi: 10.1016/s0035-9203(48)80006-4
Tachibana S., Sullivan S. A., Kawai S., Nakamura S., Kim H. R., Goto N., et al. (2012). Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat. Genet. 44 (9), 1051–1055. doi: 10.1038/ng.2375
Tarun A. S., Peng X., Dumpit R. F., Ogata Y., Silva-Rivera H., Camargo N., et al. (2008). A combined transcriptome and proteome survey of malaria parasite liver stages. Proc. Natl. Acad. Sci. U. S. A. 105 (1), 305–310. doi: 10.1073/pnas.0710780104
Tripathi J., Segeritz C. P., Griffiths G., Bushell W., Vallier L., Skarnes W. C., et al. (2020). A Novel Chemically Differentiated Mouse Embryonic Stem Cell-Based Model to Study Liver Stages of Plasmodium berghei. Stem Cell Rep. 14 (6), 1123–1134. doi: 10.1016/j.stemcr.2020.04.010
van de Sand C., Horstmann S., Schmidt A., Sturm A., Bolte S., Krueger A., et al. (2005). The liver stage of Plasmodium berghei inhibits host cell apoptosis. Mol. Microbiol. 58 (3), 731–742. doi: 10.1111/j.1365-2958.2005.04888.x
Voorberg-van der Wel A., Zeeman A. M., van Amsterdam S. M., van den Berg A., Klooster E. J., Iwanaga S., et al. (2013). Transgenic fluorescent Plasmodium cynomolgi liver stages enable live imaging and purification of Malaria hypnozoite-forms. PloS One 8 (1), e54888. doi: 10.1371/journal.pone.0054888
Voorberg-van der Wel A., Roma G., Gupta D. K., Schuierer S., Nigsch F., Carbone W., et al. (2017). A comparative transcriptomic analysis of replicating and dormant liver stages of the relapsing malaria parasite Plasmodium cynomolgi. Elife 6. doi: 10.7554/eLife.29605
Voorberg-van der Wel A. M., Zeeman A. M., Nieuwenhuis I. G., van der Werff N. M., Klooster E. J., Klop O., et al. (2020). A dual fluorescent Plasmodium cynomolgi reporter line reveals in vitro malaria hypnozoite reactivation. Commun. Biol. Press. doi: 10.1038/s42003-019-0737-3.
Wacker R., Eickel N., Schmuckli-Maurer J., Annoura T., Niklaus L., Khan S. M., et al. (2017). LC3-association with the parasitophorous vacuole membrane of Plasmodium berghei liver stages follows a noncanonical autophagy pathway. Cell Microbiol. 19 (10). doi: 10.1111/cmi.12754
Wells T. N., Burrows J. N., Baird J. K. (2010). Targeting the hypnozoite reservoir of Plasmodium vivax: the hidden obstacle to malaria elimination. Trends Parasitol. 26 (3), 145–151. doi: 10.1016/j.pt.2009.12.005
White N. J. (2011). Determinants of relapse periodicity in Plasmodium vivax malaria. Malar. J. 10, 297. doi: 10.1186/1475-2875-10-297
WHO (2019). World Malaria Report 2019. Available at: https://www.who.int/publications/i/item/world-malaria-report-2019.
Yegneswaran B., Alcid D., Mohan J. (2009). Plasmodium knowlesi: an important yet overlooked human malaria parasite. Mayo. Clin. Proc. 84 (7), 664; author reply 664. doi: 10.4065/84.7.664
Zeeman A. M., van Amsterdam S. M., McNamara C. W., Voorberg-van der Wel A., Klooster E. J., van den Berg A., et al. (2014). KAI407, a Potent Non-8-Aminoquinoline Compound That Kills Plasmodium cynomolgi Early Dormant Liver Stage Parasites In Vitro. Antimicrob. Agents Chemother. 58 (3), 1586–1595. doi: 10.1128/AAC.01927-13
Zeeman A. M., Lakshminarayana S. B., van der Werff N., Klooster E. J., Voorberg-van der Wel A., Kondreddi R. R., et al. (2016). PI4 Kinase Is a Prophylactic but Not Radical Curative Target in Plasmodium vivax-Type Malaria Parasites. Antimicrob. Agents Chemother. 60 (5), 2858–2863. doi: 10.1128/AAC.03080-15
Keywords: malaria, parasite-host interaction, plasmodium, cynomolgi, hypnozoite, relapse
Citation: Voorberg-van der Wel A, Kocken CHM and Zeeman AM (2021) Modeling Relapsing Malaria: Emerging Technologies to Study Parasite-Host Interactions in the Liver. Front. Cell. Infect. Microbiol. 10:606033. doi: 10.3389/fcimb.2020.606033
Received: 14 September 2020; Accepted: 04 December 2020;
Published: 29 January 2021.
Edited by:
Jing-wen Lin, Sichuan University, ChinaReviewed by:
Glenn McConkey, University of Leeds, United KingdomFred David Mast, Seattle Children’s Research Institute, United States
Copyright © 2021 Voorberg-van der Wel, Kocken and Zeeman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anne-Marie Zeeman, emVlbWFuQGJwcmMubmw=
 Annemarie Voorberg-van der Wel
Annemarie Voorberg-van der Wel Clemens H. M. Kocken
Clemens H. M. Kocken Anne-Marie Zeeman
Anne-Marie Zeeman