
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Cell. Infect. Microbiol., 07 January 2021
Sec. Microbiome in Health and Disease
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.601573
This article is part of the Research TopicViruses: Drivers or Passengers of the Human Gut Microbiome?View all 8 articles
In recent years, our understanding of the importance of microorganisms on and within our bodies has been revolutionized by the ability to characterize entire microbial communities. No more so is this true than in cases of disease. Community studies have revealed strong associations between microbial populations and disease states where such concomitance was previously absent from aetiology: including in cancers. The study of viruses, in particular, has benefited from the development of new community profiling techniques and we are now realising that their prominence within our physiology is nearly as broad as the diversity of the organisms themselves. Here, we examine the relationship between viruses and colorectal cancer (CRC), the leading cause of gastrointestinal cancer-related death worldwide. In CRC, viruses have been suggested to be involved in oncogenesis both directly, through infection of our cells, and indirectly, through modulating the composition of bacterial communities. Interestingly though, these characteristics have also led to their examination from another perspective—as options for treatment. Advances in our understanding of molecular and viral biology have caused many to look at viruses as potential modular biotherapeutics, where deleterious characteristics can be tamed and desirable characteristics exploited. In this article, we will explore both of these perspectives, covering how viral infections and involvement in microbiome dynamics may contribute to CRC, and examine ways in which viruses themselves could be harnessed to treat the very condition their contemporaries may have had a hand in creating.
Our bodies are host to a fascinatingly complex community of microorganisms, termed the microbiome. Each of us contains a unique mix of viruses; bacteria; archaea; and eukaryotes, such as fungi, protists, and nematodes. From birth to death, the constituents found within our microbiomes, and how they are configured, are interwoven within nearly every facet of our existence. This is particularly true of organisms within the gastrointestinal tract, where microorganism density is greatest (Lynch and Pedersen, 2016; Sender et al., 2016). Dietary processes, such as digestion and nutrient absorption, all rely upon microorganisms to function normally (Conlon and Bird, 2015; Basolo et al., 2020). However, so do non-dietary processes, including cell proliferation and angiogenesis (Reinhardt et al., 2012; Ijssennagger et al., 2015). Critical traits from our metabolic profiles, immune responses, to even our moods are all shaped by microbes in the gut (Turnbaugh et al., 2006; Steenbergen et al., 2015; Spencer et al., 2019). Yet, their paramountcy in health also gives way to a reciprocal role in disease.
Microbial-associated diseases can arise from several origins. For example, as infections, the presence or abundance of a pathogenic organism can directly lead to the production of a disease state [e.g. Vibrio cholerae in cholera; Baker-Austin et al. (2018)]. Alternatively, more complex population-level changes can occur that lead to disease. Here, alterations in the taxonomic composition of the microbiome away from that found during the “healthy” state lead to disease. This can involve the loss of important commensal organisms, depleting the functional pool of the microbiome, reducing its ability to support normal physiological processes (Levy et al., 2017; Durack and Lynch, 2019). Non-communicable disorders, such as obesity, hypertension, and rheumatoid arthritis, have been linked to changes in microbial populations (Mar Rodríguez et al., 2015; Zhang et al., 2015; Li et al., 2017).
Much of the focus on gastrointestinal microbiome-associated disease has been towards its bacterial component, yet, viruses are also important drivers of disease. Viruses of eukaryotes, those that infect our cells (or other eukaryotic organisms, e.g. fungi), can considerably impact health; most notably as infectious agents. Norovirus, for instance, is the leading cause of foodborne gastroenteritis worldwide (Kirk et al., 2015). There is also growing evidence implicating viruses in conditions involving bacterial community changes. Viruses of prokaryotes, termed bacteriophages, are the most abundant viruses within the human gastrointestinal tract, and regulate bacterial populations across ecological niches (Wigington et al., 2016; Argov et al., 2017; Shkoporov and Hill, 2019). Conditions such as Crohn’s disease and ulcerative colitis, are believed to have a marked bacteriophage component in their aetiology (Norman et al., 2015).
One set of conditions that have long held an association with viruses are cancers. Viral involvement in cancers can be seen across the body, where they contribute to a significant proportion of incidence. In 2018, around 2.2 million cancers worldwide (~13% of all cancers) were caused by carcinogenic infections: the majority attributed to viruses (de Martel et al., 2020). The remainders of infection-associated cases are primarily linked to bacterial activity (e.g. Helicobacter pylori in gastric cancer). Given what we are beginning to understand about the importance of bacteriophages in bacterial-associated diseases, such as their involvement in driving immune dampening and selecting for pathogenic bacterial phenotypes (Hosseinidoust et al., 2013; Sweere et al., 2019), it is not implausible that some of these cases, too, may have a viral relation. In colorectal cancer (CRC), both direct and indirect involvement of viruses in disease has been suggested. Numerous studies have indicated the presence of oncogenic viruses within tumors, including viruses that have been linked to other cancers (Chen et al., 2015), while there is also evidence linking bacteriophages to CRC through their interactions with other members of the gastrointestinal microbiome (Emlet et al., 2020).
CRC encompasses both cancers of the colon and rectum. Cancer of the colon is the most common of the two, though incidence rates can differ based on factors such as gender, race, and age (Siegel et al., 2020a). The majority of cases are sporadic (~60–65%), but can be influenced by hereditary factors; either linked to a prior familial occurrence or an inherited cancer syndrome (Keum and Giovannucci, 2019). While CRC is the third most common form of cancer globally, it is responsible for the second-highest level of cancer-related death worldwide (Bray et al., 2018). Despite seeing reductions in the total numbers of CRC cases in many countries in the last decade, some countries are seeing rates of decline slowing, including the US, in large part due to increases in the incidences of early-onset CRC (cases in individuals < 50 years old) (Siegel et al., 2019; Siegel et al., 2020b). Therefore, not only is it imperative that all aspects of its aetiology are considered, but also all potential options for treatment. Interestingly, viruses have been suggested as one such possible avenue, with the aim of exploiting the onco- and bacteriolytic properties of some viruses (Choi et al., 2016; Paule et al., 2018). In this article, we will explore the potential involvement of viruses in CRC aetiology, and examine how they could be applied in its mitigation.
An array of viruses have been demonstrated to contribute to cancer development across the body. For example, human papillomavirus (HPV) infections are associated with cervical and oral cancers (Crosbie et al., 2013; Conway et al., 2018), hepatitis B and C viruses (HBV and HCV respectively) are linked with liver cancers (Lin et al., 2015; Xie, 2017), human herpes viruses are connected to the development of Kaposi sarcomas (Mariggiò et al., 2017), and Eppstein-Barr virus (EBV) is linked to nasopharyngeal cancers, Hodgkin’s lymphomas, and Burkitt’s lymphomas (Farrell, 2019). While the associations between these viruses and their respective cancers are well established, in CRC the involvement of viral infection in disease is less defined. Evidence does exist though — probably the most deeply examined connections involve members of the Polyomaviridae family, particularly the JC polyomavirus (JCPyV), although other viruses have also been linked to CRC, including HBV, HPV, and EBV (Su et al., 2020; Fernandes et al., 2020).
Polyomaviruses are regularly faced by humans, highlighted by the frequency with which antibodies targeting them are found within the population (Kean et al., 2009). Infection is normally asymptomatic, likely occurring following the consumption of virus-contaminated water or food (Bofill-Mas et al., 2001). Suggestions of their involvement in CRC began after JCPyV DNA was observed in colonic tumor samples. Although also found in the surrounding normal epithelia, JCPyV-loads were around 10-fold higher in cancerous tissues (Laghi et al., 1999). Subsequent studies corroborated these observations. Theodoropoulos et al. (2005) for example, found that 49/80 colon adenocarcinomas samples they examined were positive for JCPyV DNA, with mean viral load around 60-fold higher in carcinomas than in the adjacent non-cancerous epithelia. Other members of the Polyomaviridae, including BK polyomavirus and simian virus 40, have also been identified in CRC tumor tissue (Casini et al., 2005; Giuliani et al., 2008; Jarzyński et al., 2017).
The oncogenic potential of polyomaviruses is believed to stem mainly from their large T-antigen (T-Ag; Figure 1A). In the polyomavirus life cycle, T-Ag serves multiple purposes, including interacting with host proteins to alter cell cycle conditions and to replicate the viral genome (Eash et al., 2006). T-Ag can also interact with other cellular proteins though, including those involved in pathways related to CRC development. One example is β-catenin, a transcriptional activator and componentof the Wnt signalling pathway that, when in the nucleus, activates transcription factors associated with the proliferation of colonic epithelial cells and apoptosis (Zhang and Shay, 2017; Agrawal et al., 2019). When mutations occur that increase levels of β-catenin in the cell and the nucleus [e.g inactivation of the negative regulator adenomatous polyposis coli (APC)], increased transcription of key tumor progression genes results, such as c-myc and cyclin D1, leading to abhorrent cell growth and ultimately tumorigenesis (Powell et al., 1992; Morin et al., 1997; He et al., 1998; Tetsu and McCormick, 1999; Henderson, 2000). T-Ag can interact directly with β-catenin and facilitate its translocation into the nuclei, significantly increasing transcription of carcinogenic β-catenin targets (Enam et al., 2002; Ripple et al., 2014). In vitro, Ricciardiello et al. (2003) noted that exposure of colonic epithelial cell lines to JCPyV produced cells with aberrant chromosomes with signs of chromosomal breakages and aneuploidy, correlating with nuclear accumulation of β-catenin and T-Ag—chromosomal changes that are often observed in CRC (Bakhoum and Cantley, 2018). T-Ag has also been linked to metastasis, with one study finding T-Ag expression in >70% of metastatic primary tumors and their matching liver metastasis, and that T-Ag expression in vitro could increase cell migration (Link et al., 2009).
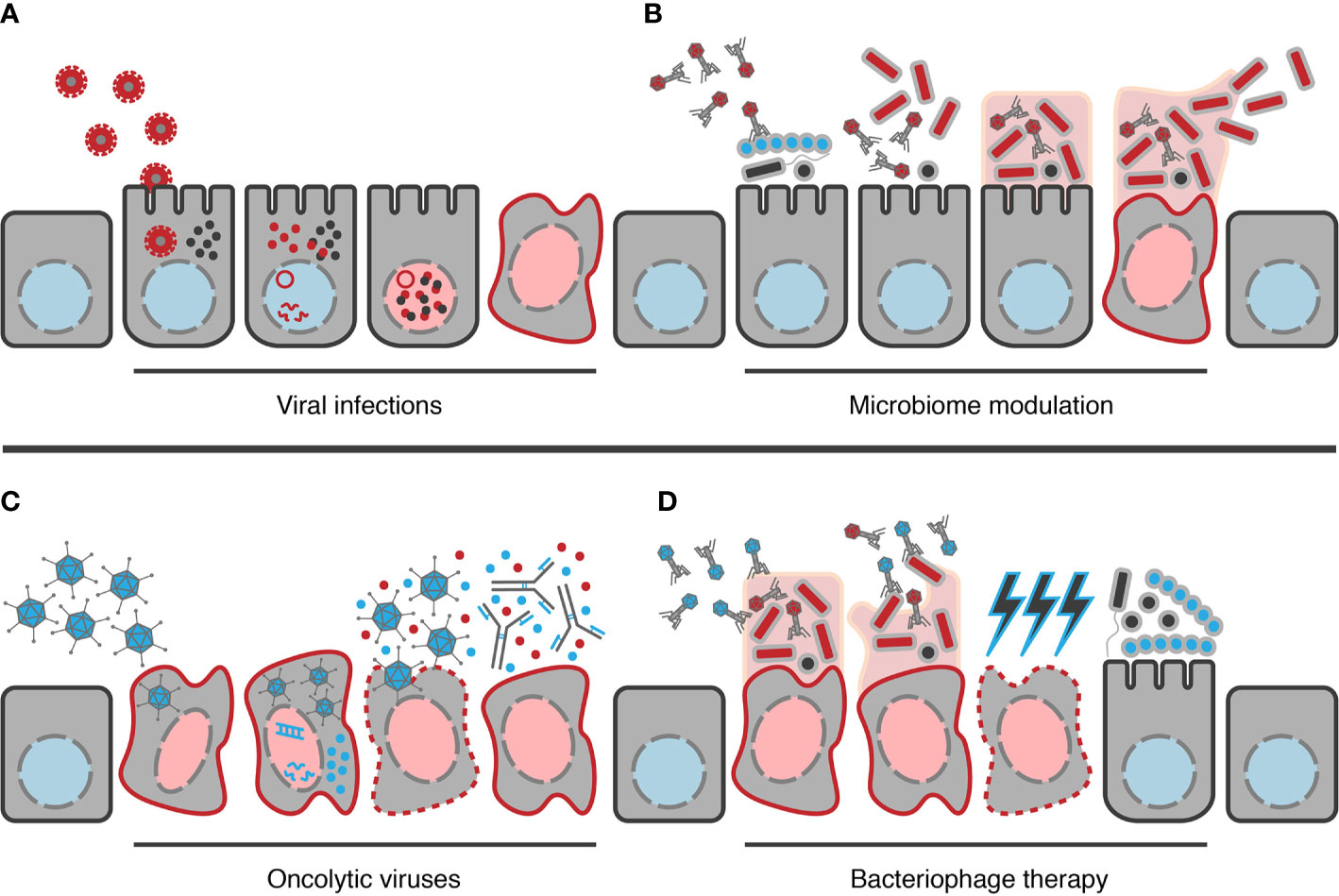
Figure 1 Examples of potential ways viruses can contribute to CRC development and how they could be applied in its treatment. (A) Example mechanism by which viral infections could contribute to disease, in this case polyomaviruses (red viruses). Following infection of the cell, viral DNA reaches the nucleus, leading to generation of viral transcripts and the production of viral proteins, e.g. T-Ag (red dots). These can interact with host effectors, e.g. β-catenin (black dots), resulting in changes in host gene expression from the healthy state (blue nucleus) to an oncogenic state (red nucleus). (B) Model example of how bacteriophages may contribute to cancer. Bacteriophages alter the composition of the gut bacterial community towards a configuration that allows colonization of oncogenic organisms (red bacteria). Following establishment, the oncogenic organisms can then form biofilms and begin to contribute towards transformation of the cell to a cancerous state. Once a cancerous state has developed, bacteria may enter a synergistic relationship with the cells, receiving peptides for their growth while continuing to contribute to maintenance of the cancerous state. In mature biofilms, bacteriophages will also contribute to dispersion of cells to establish similar cycles elsewhere. (C) Example of how oncolytic viruses (blue viruses) may be used in CRC treatment. Here, modified viruses are introduced proximal to the cancerous location, allowing them to locate and infect the target cells. When viral nucleic acid then reaches the nucleus, viral transcripts begin to be generated leading to production of progeny virions. If DNA sequences encoding therapeutic products have been inserted into the genome, e.g. GM-CSF (blue dots) here, these will be produced alongside viral proteins. Once assembled, progeny virions can then be released from the cell via lysis of the cancer cell, also causing the release of the therapeutic protein and antigens from the cancer cell itself (red dots); the latter of which can then stimulate the hosts immune response to target the cancer cell, possibly enhanced by the therapeutic product if it is an immunostimulatory product such as GM-CSF. (D) Model example of the use of bacteriophages therapeutically (blue bacteriophages) in CRC treatment. Bacteriophages are able to degrade biofilms through their depolymerases, providing access to the oncogenic organisms residing within. The bacteriophage can then infect and lyse these bacteria, selectively removing only the oncogenic organisms, reducing the oncogenic pressure on cells. As oncogenic bacteria can also confer chemoresistance with colonization, their removal may increase the sensitivity of the cancerous cell to chemotherapeutic treatments (blue lightning bolts). Selective removal of oncogenic organisms also clears the niche, allowing recolonization by other members of the microbiome or beneficial organisms introduced therapeutically.
However, while some studies have found T-Ag expression in as many as 35–60% of CRC carcinomas examined (Enam et al., 2002; Nosho et al., 2009) and others have observed a significant association between its presence in CRC tissues and genetic changes commonly observed in CRC (Goel et al., 2006) the involvement of polyomaviruses in CRC remains controversial. For example, studies have been unable to detect its expression in any CRC samples, even when 26.1% of samples tested positive for T-Ag DNA (Hori et al., 2005). While, in other studies, little to no polyomavirus DNA was detected in CRC samples at all (Newcomb et al., 2004; Campello et al., 2010; Gock et al., 2020). Yet, there is the possibility that these observations could be justified by a temporal change in viral expression in what has been referred to as a “hit and run” style dynamic. In this dynamic, it is proposed an initial infection leads to the initiation of events that result in cellular transformation towards a cancerous phenotype (e.g. chromosomal breakages), but, after this event, viruses then face selection pressures that either alter gene expression levels or begin to select against viral presence altogether (Niller et al., 2011). Therefore in the case of polyomaviruses, viral infection and expression of T-Ag may trigger initial dysplasia but is no longer required or actively selected against in later disease. This idea is supported by observations showing equivalent JCPyV DNA loads in carcinomas as in adenomas, benign tumors of the epithelium that normally serve as the precursor for carcinomas, and where T-Ag expression can also be found (Theodoropoulos et al., 2005; Jung et al., 2008).
Regardless of whether viral infection is associated with CRC, viruses may still contribute to CRC in other ways, such as through their ability to alter microbial population structures. The gut microbiome is believed to play an important role in CRC aetiology. For example, studies have observed that the CRC intestinal microbiome is often characterized by reductions in butyrate-producing organisms, such as Faecalibacterium spp. and Roseburia spp., but is enriched for the likes of enterotoxigenic Bacteroides fragilis and Escherichia coli, or other pathobionts including Fusobacterium spp., Selenomonas spp., and Porphyromonas spp. (Chen et al., 2012; Wang et al., 2012; Wu et al., 2013; Hibberd et al., 2017). Members of the enriched sets of organisms are believed to promote cancer development in CRC. For example, enterotoxigenic B. fragilis and strains of E. coli harboring the polyketide synthase (pks) genomic island have been shown to produce tumorigenesis in murine models of CRC (Wu et al., 2009; Arthur et al., 2012). Evidence is particularly strong for the involvement of Fusobacterium spp. in CRC, in particular Fusobacterium nucleatum. F. nucleatum is commonly found as part of the oral microbiota of humans, as well as in other mucosal locations, where it serves important roles in community biofilm organization (Brennan and Garrett, 2019). In CRC, studies have found Fusobacterium spp. to be enriched within tumor sites relative to the surrounding non-tumorous tissue and that its abundance can correlate with patient survival, suggesting a link between this organism and the disease state (Kostic et al., 2012; Castellarin et al., 2012; Mima et al., 2016). Furthermore, when studied in isolation, colonization of ApcMin/+ mice with F. nucleatum leads to the development of significantly more colonic tumors than that seen in untreated mice (Kostic et al., 2013). However, recently it has been suggested that the ability of such organisms to promote cancer may involve viral assistance.
In a study of the bacterial and viral populations in CRC, Hannigan et al. (2018) noted that there were strong associations between both the presence of specific bacterial “operational taxonomic units” (OTUs) and viral “operational viral units” (OVUs) with cancerous state; the OTU with the highest overall association to the cancerous state belonging to Fusobacterium spp. and the OVUs most strongly associated with disease belonging to bacteriophages—particularly the order Caudovirales. Nakatsu et al. (2018) also noted enrichment of bacteriophages in CRC faecal samples, and that, of 22 viral genera they found could maximally discriminate CRC samples from healthy controls, the majority were bacteriophages from the order Caudovirales. Hannigan and colleagues further observed that their OVU and OTU abundances were unrelated, leading them to examine if bacteriophages were “community hubs”, instead interacting with multiple organisms within the CRC microbiome. This was the case, with those bacteriophages interacting with the most organisms correlating positively with disease, suggesting that the bacteriophages acting as community hubs were important drivers of CRC. The authors then proposed a model of bacteriophage involvement in CRC wherein, bacteriophages with broad host-ranges can infect a wide range of bacteria within the gut microbiome, leading to initial dysbiosis (Figure 1B). This allows niche expansion of other untargeted organisms e.g. F. nucleatum, eventually resulting in the formation of a multi-species biofilm. Bacteriophages then assist within the biofilm through mechanisms such as contributing to nutrient/substrate availability and biofilm dispersal (Rossmann et al., 2015; Abedon, 2020). This provides a sufficient environment for bacteria to transform colonic epithelia, disrupt tight junctions, and infiltrate cells to produce a pro-cancer inflammatory response. Although not included in their model, Hannigan et al. (2018) do not exclude the possibility that direct interactions between bacteriophages and human cells may also contribute to the pro-cancer inflammatory responses seen in CRC. For example, members of the Caudovirales have been observed to directly interact with human cells (Lehti et al., 2017), cross epithelial barriers (Nguyen et al., 2017), and produce pro-inflammatory responses (Van Belleghem et al., 2017). Other bacteriophage taxa associated with CRC have also been shown to interact with the human host. Nakatsu and others observed Inovirus to be the genus of bacteriophage able to provide the greatest level of differentiation between CRC samples and the healthy controls in their study (Nakatsu et al., 2018). Members of this genus are known to play prominent roles in biofilm formation in some bacteria (Secor et al., 2015), decorate themselves with potentially immunogenic bacterial proteins (Piekarowicz et al., 2020), and even directly alter immune responses (Sweere et al., 2019).
There remain many other unknowns relating to the virome and disease in CRC that require further study to decipher. To give one important example, Nakatsu and colleagues found that the viral genus with the greatest discriminatory capacity between healthy and CRC samples was not a bacteriophage, but rather Orthobunyavirus, a viral taxon with no previously described role in human gastrointestinal disease (Nakatsu et al., 2018). Thus future studies must be conducted to resolve the relations between gut viral populations and CRC aetiology. These studies face considerable challenges though. Virome studies, for instance, must contend with considerable inter-individual variability (Shkoporov et al., 2019). Given that virome variability can be influenced by factors such as geography, diet, and ethnicity (Minot et al., 2011; Zuo et al., 2020), these studies should be longitudinal and recruit large numbers of individuals from diverse origins, cultures, and ethnic backgrounds. Recruitment of these large cohorts then will be in and of itself a considerable hurdle to overcome. Viromes also contain a vast amount of so-called “dark matter”—sequences with no known origin. This can account for as many as 90% of the reads within some virome data sets (Aggarwala et al., 2017). As such, viral isolation studies are also needed to assign a viral origin to the unknown sequences. This may require the development of new viral isolation methodologies for those viruses unable to be isolated through standard procedures. Methodological changes in bacterial isolation protocols have allowed “unculturable” bacteria previously only seen in metagenomic samples to be isolated and characterized (Browne et al., 2016). Overcoming these hurdles is no small feat, however, the potential clinical benefits are significant. Given that the existing studies of the CRC virome suggest the virome can discriminate between healthy individuals and those with CRC, and even predict both disease state and prognosis (Hannigan et al., 2018; Nakatsu et al., 2018), and that these associations are likely a reflection of underlying pathophysiological events, further insights could lead to important advances in both CRC diagnostics and treatment.
Viruses have a long history relating to the treatment of cancer, stemming from a series of reports where patients displayed periods of spontaneous remission after viral exposure. In an 1897 report, a patient previously diagnosed with myelogenous leukaemia was observed to undergo temporary disease regression following a believed influenza infection (Dock, 1904). Numerous accounts in the years that followed also reported improvements in patients after either viral infections or vaccination (e.g. De Pace (1912); Bierman et al. (1953); Bluming and Ziegler (1971); Hansen and Libnoch (1978)). This led viruses to be examined as potential therapeutic options against cancers. In an early example, after observing two cases where patients with Hodgkin’s lymphoma underwent a period of remission after viral hepatitis, Hoster et al. (1949) examined the effects of introducing sera and tissue containing the hepatitis virus to 21 Hodgkin’s lymphoma patients. Their tests bore success, albeit temporary, observing signs of improvement for at least one month in 7 of 13 patients in whom viral hepatitis developed. Today, viruses are still being examined for their therapeutic uses in cancers, including those akin to these early studies, namely oncolytic viruses (OVs).
The therapeutic benefits of OVs against cancers are two-fold. Firstly, OVs are able to preferentially infect cancerous cells, culminating in their lysis. Secondly, lysis of the infected cell results in the release of tumor antigens and viral particles into the surrounding environment, stimulating the host’s immune system and generating a complementary host-derived anti-cancer response (Melcher et al., 2011; Chiocca and Rabkin, 2014). Although early anti-cancer studies utilized wild-type viruses, oncolytic viral therapies have since been refined utilising our expanded knowledge of the underlying disease and advancements in molecular biology. This has allowed us to modify viruses to improve their safety and efficacy. Modifications can be in the form of mutations that broaden viral tropisms, remove unwanted characteristics, enhance immunogenicity, or any of a series of other therapeutically beneficial genetic alteration (Choi et al., 2016; Zheng M. et al., 2019).
One well-known example of a therapeutically modified virus is the talimogene laherparepvec (T-VEC) system (Figure 1C). T-VEC is a herpes simplex type-1 virus (HSV-1) that has been approved by both the US Food and Drug Administration (FDA) and the European Medicines Agency (EMA) for use in the treatment of metastatic melanomas but is also currently being trialed for use in CRC (Mullard, 2015; Raman et al., 2019). Three noteworthy modifications have been introduced in this system (Liu et al., 2003). The first involves the deletion of ICP34.5, which is associated with neurovirulence and antiviral evasion, to improve safety (Chou et al., 1990; Tallóczy et al., 2002; Orvedahl et al., 2007). While the safety benefits of reducing neurovirulence are self-explanatory, the removal of antiviral evasion improves safety by increasing selectivity of T-VEC towards cancer cells. ICP34.5 allows viral antagonism of protein kinase R (PKR), an important antiviral defence in healthy cells that is often dysfunctional in cancer cells (Kohlhapp and Kaufman, 2016; Gal-Ben-Ari et al., 2019). Removal of ICP34.5, therefore, attenuates antagonism in healthy cells, while in cancer cells viral infection can proceed despite the deletion. The second modification of T-VEC is the deletion of ICP47, a gene which allows HSV to reduce the hosts immune detection of infected cells by inhibiting antigen presentation (Hill et al., 1995). Its removal allows antigen presentation to occur, improving efficacy. Deletion of ICP47 also leads to the increased expression of US11 by shifting its expression to the control of the ICP47 promoter, which has been shown to improve viral growth in cancer cells with no impact to safety (Taneja et al., 2001; Liu et al., 2003). Lastly, two copies of the human granulocyte-macrophage colony-stimulating factor gene, GM-CSF, have been added to the T-VEC system, which recruits and activate antigen-presenting cells to the local area and induces anti-tumor T-cell responses (Liu et al., 2003; Kaufman et al., 2014).
Other viruses are also being examined in clinical trial for use in CRC, with various different modifications. Examples include ONYX-015, an adenovirus that has been altered to remove the viral E1b gene (reducing its ability to infect non-cancerous cells) (Heise et al., 1997; Reid et al., 2001); Pexa-Vec (pexastimogene devacirepvec; JX-594), a vaccinia virus also modified to encode GM-CSF, but with the additional modifications to inactivate the viral thymidine kinase gene (reducing its ability to infect non-cancerous cells) and to add encoding of β-galactosidase (as a reporter) (Kim et al., 2006; Park et al., 2015); and even an adenovirus, Enadenotucirev (EnAd; ColoAd1), where modifications have not been directly introduced but rather are the result of recombination between different adenovirus serotypes following the passage of mixed viral pools through cancer cell lines (in this case HT-29 cells) in a process termed “Directed Evolution” (Kuhn et al., 2008; O’Cathail et al., 2020). Unmodified viruses, such as the reovirus Pelareorep (Reolysin), are also being examined in clinical trials for use in CRC treatment (Goel et al., 2020).
While the application of OVs could be useful in the treatment of cancer itself, there remains the issue of pro-cancer antagonism from members of the surrounding bacterial population, such as that seen for F. nucleatum (Kostic et al., 2013; Wu et al., 2019). Therefore, removal of the pro-CRC organisms would also be of therapeutic benefit. Bullman et al. (2017) examined the impact of antibiotic treatment on CRC tumor growth in a murine model using human CRC xenografts that were heavily associated with Fusobacterium spp. and found that antibiotic treatment reduced the rate of tumor cell proliferation and overall tumor growth. However, while antibiotic therapies could be a simple means of removing oncogenic organisms, the breadth of their action is not restricted to pro-cancer organisms (Lange et al., 2016). Other members of the colonic bacterial population can actually be anti-carcinogenic, and their collateral removal could have deleterious ramifications. For example, some bacteria can produce metabolites with anti-cancer properties following the fermentation of dietary fibres. One example is butyrate, a product of bacterial fermentation that is believed to protect the gut against development of CRC (Fung et al., 2012). In one study, colonization of BALB/c mice with Butyrivibrio fibrisolvens, a bacteria that can produce butyrate following fermentation of fibre, was shown to protect mice with high fibre diets more from CRC development than those mice without the bacteria (Donohoe et al., 2014). For this reason, a more selective approach to antimicrobial treatment in CRC would be desirable and could be achieved through the therapeutic use of bacteriophages (Figure 1D).
Bacteriophage-based therapeutics already exist and have existed for around 100 years (Sulakvelidze et al., 2001). Bacteriophage therapy (also known as phage therapy) is the most widely studied therapeutic application involving bacteriophages and describes the application of whole bacteriophage particles to treat bacterial infections (Kakasis and Panitsa, 2019). Although not common in Western medicine, bacteriophage therapy has been in use in Eastern Europe for decades, particularly in Georgia, where the Eliava Institute of Bacteriophages, Microbiology, and Virology has been examining their therapeutic potential since 1923 (Kutateladze, 2015). In the last two decades, the reinvigorated interest in bacteriophage therapy has primarily been motivated by the emergence of antibiotic-resistant microorganisms, for which it has been deployed successfully against in a number of high-profile examples [e.g. Schooley et al. (2017); Dedrick et al. (2019)]. Recently though, interest has grown for its application in microbiome manipulation, largely due to the specificity of bacteriophages (Lee et al., 2018). Bacteriophages can have small host-ranges, often only infecting a minute subset of strains within a particular species, negating the issue of indiscriminate action seen with broad-spectrum antibiotics (Flores et al., 2011; Ganeshan and Hosseinidoust, 2019). Bacteriophages also often encode depolymerases that allow them to degrade biofilms and gain access to the residing organisms (Alves et al., 2016; Knecht et al., 2020). Fusobacterium spp. are often found in biofilms in CRC, with this form of community structure believed to be important in the promotion of tumorigenesis (Dejea et al., 2014; Flynn et al., 2016; Bullman et al., 2017). Much like their oncolytic relatives, bacteriophages can also be engineered to carry additional therapeutic benefits (Kilcher and Loessner, 2019), which has been examined in CRC.
Zheng D. W. et al. (2019) examined the impact of a “bacteriophage-guided biotic–abiotic hybrid nanosystem” as a treatment option in mice bearing CT26 colorectal carcinomas. In this system, bacteriophages targeting Fusobacterium spp. are utilized along with two abiotic components; irinotecan, a first-line treatment for colorectal cancer, and dextran, a fermentable complex glucan (Olano-Martin et al., 2000; Fuchs et al., 2006). The bacteriophages are azide-modified, while irinotecan is encapsulated within azodibenzocyclooctyne-modified dextran nanoparticles. These modifications mean the irinotecan-dextran nanoparticles can link covalently to the bacteriophages, allowing the nanoparticles to concentrate within areas where Fusobacterium spp. are present, i.e. within the tumor sites. Irinotecan is used for its chemotherapeutic properties, while dextran is employed as a prebiotic due to the ability of fermentable carbohydrates to promote shifts in microbial colonization and metabolite production (Harris et al., 2017; Warren et al., 2018). Here, the use of dextran aimed to increase the abundance of butyrate and butyrate-producing bacteria within the gastrointestinal tract because of their anti-tumorigenic properties (Fung et al., 2012). It was observed that their bacteriophage-guided nanosystem was able to significantly decreases Fusobacterium spp. levels, increase butyrate production, and importantly, suppress tumor growth.
Although viral therapies do show considerable promise they do have their drawbacks (Loc-Carrillo and Abedon, 2011; Zheng M. et al., 2019). In the case of bacteriophage therapy, although some studies have shown that the introduction of bacteriophages can have only minimal or transient effects on non-target bacterial populations (Reyes et al., 2013; Nale et al., 2018), other studies have shown that the impact could be broader (Hsu et al., 2019; Lange et al., 2019). There is also the issue of bacterial resistance development, and although currently this is mainly overcome by the use of bacteriophage “cocktails” (mixes containing multiple distinct bacteriophages; Nale et al. (2016)), resistance can still occur, meaning co-use with antibiotics can be required to completely eliminate the target organism (Oechslin et al., 2016). The ability to modify viruses though could allow us to overcome many of these issues (Twumasi-Boateng et al., 2018; Kilcher and Loessner, 2019). For example, in this latter case, alternative antimicrobials [e.g. silver nanoparticles; Dong et al. (2020)] could be attached to the bacteriophage particles to provide an additional means of locally-contained antibacterial action against resistant cells. As with the involvement of viral disease in CRC aetiology though, it should be echoed that much remains to be learned about host and microbiome dynamics within CRC, in particular at the individual level (Panagi et al., 2019). Further investigations are warranted to ensure laboratory promise can be translated to clinical success as although trials of viral therapies to treat other conditions and other cancers have brought success (Wright et al., 2009; Andtbacka et al., 2015), success has not been ubiquitous (Sarker et al., 2016; Guo, 2020).
Despite these points, viral therapeutics are an avenue worth exploring with huge potential through molecular engineering. One exciting proposition would be an “anti-cancer viral cocktail” combining engineered OVs and bacteriophages. This would allow the benefits of both oncolytic targeting of cancerous cells and promotion of anti-cancer immune responses from the OVs, while also providing the benefit of the antimicrobial action of bacteriophages. The viruses themselves could be modified to provide further benefits, be it increased immunogenicity through the addition of immunostimulatory products such as GM-CSF to the OVs, or addition of pre-biotics and chemotherapeutics to bacteriophages allowing targeted delivery to the site of the tumor, promoting the growth of beneficial organisms and impeding tumor growth. Whatever shape the desired modifications take, when it comes to viral therapies, the possibilities are endless.
All authors were involved in the conception of the review topic and focus. CT and AV contributed to the initial draft of the manuscript. CT generated the illustrations. All authors contributed to the article and approved the submitted version.
This work was supported by startup research funds provided by the University of California Merced School of Natural Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge and thank both reviewers for their feedback and suggested additions to this manuscript.
Abedon S. T. (2020). “Phage-Phage, Phage-Bacteria, and Phage-Environment Communication,” in Biocommunication of Phages (Cham: Springer International Publishing), 23–70. doi: 10.1007/978-3-030-45885-0_2
Aggarwala V., Liang G., Bushman F. D. (2017). Viral communities of the human gut: metagenomic analysis of composition and dynamics. Mobile DNA 8, 12. doi: 10.1186/s13100-017-0095-y
Agrawal S., Bhattacharya A., Manhas J., Sen S. (2019). “Molecular Diagnostics in Colorectal Cancer”. in Molecular Diagnostics in Cancer Patients. Eds. K. Shukla, P. Sharma, and S. Misra. 143–155. doi: 10.1007/978-981-13-5877-7_9
Alves D. R., Perez-Esteban P., Kot W., Bean J. E., Arnot T., Hansen L. H., et al. (2016). A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions. Microbial. Biotechnol. 9, 61–74. doi: 10.1111/1751-7915.12316
Andtbacka R. H., Kaufman H. L., Collichio F., Amatruda T., Senzer N., Chesney J., et al. (2015). Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma. J. Clin. Oncol. 33, 2780–2788. doi: 10.1200/JCO.2014.58.3377
Argov T., Azulay G., Pasechnek A., Stadnyuk O., Ran-Sapir S., Borovok I., et al. (2017). Temperate bacteriophages as regulators of host behavior. Curr. Opin. Microbiol. 38, 81–87. doi: 10.1016/j.mib.2017.05.002
Arthur J. C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J. M., Fan T. J., et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123. doi: 10.1126/science.1224820
Baker-Austin C., Oliver J. D., Alam M., Ali A., Waldor M. K., Qadri F., et al. (2018). Vibrio spp. infections. Nat. Rev. Dis. Primers 4, 8. doi: 10.1038/s41572-018-0005-8
Bakhoum S. F., Cantley L. C. (2018). The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 174, 1347–1360. doi: 10.1016/j.cell.2018.08.027
Basolo A., Hohenadel M., Ang Q. Y., Piaggi P., Heinitz S., Walter M., et al. (2020). Effects of underfeeding and oral vancomycin on gut microbiome and nutrient absorption in humans. Nat. Med. 26, 589–598. doi: 10.1038/s41591-020-0801-z
Bierman H. R., Crile D. M., Dod K. S., Kelly K. H., Petrakis N. II, White L. P., et al. (1953). Remissions in leukemia of childhood following acute infectious disease. Staphylococcus and streptococcus, varicella, and feline panleukopenias. Cancer 6, 591–605. doi: 10.1002/1097-0142(195305)6:3<591::AID-CNCR2820060317>3.0.CO;2-M
Bluming A., Ziegler J. (1971). Regression of Burkitt’s lymphoma in association with measles infection. Lancet 298, 105–106. doi: 10.1016/S0140-6736(71)92086-1
Bofill-Mas S., Formiga-Cruz M., Clemente-Casares P., Calafell F., Girones R. (2001). Potential Transmission of Human Polyomaviruses through the Gastrointestinal Tract after Exposure to Virions or Viral DNA. J. Virol. 75, 10290–10299. doi: 10.1128/JVI.75.21.10290-10299.2001
Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. A. Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Brennan C. A., Garrett W. S. (2019). Fusobacterium nucleatum — symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 17, 156–166. doi: 10.1038/s41579-018-0129-6
Browne H. P., Forster S. C., Anonye B. O., Kumar N., Neville B. A., Stares M. D., et al. (2016). Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546. doi: 10.1038/nature17645
Bullman S., Pedamallu C. S., Sicinska E., Clancy T. E., Zhang X., Cai D., et al. (2017). Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448. doi: 10.1126/science.aal5240
Campello C., Comar M., Zanotta N., Minicozzi A., Rodella L., Poli A. (2010). Detection of SV40 in colon cancer: A molecular case-control study from Northeast Italy. J. Med. Virol. 82, 1197–1200. doi: 10.1002/jmv.21798
Casini B., Borgese L., Del Nonno F., Galati G., Izzo L., Caputo M., et al. (2005). Presence and incidence of DNA sequences of human polyomaviruses BKV and JCV in colorectal tumor tissues. Anticancer Res. 25, 1079–1085.
Castellarin M., Warren R. L., Freeman J. D., Dreolini L., Krzywinski M., Strauss J., et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306. doi: 10.1101/gr.126516.111
Chen W., Liu F., Ling Z., Tong X., Xiang C. (2012). Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients with Colorectal Cancer. PLoS One 7, e39743. doi: 10.1371/journal.pone.0039743
Chen H., Chen X. Z., Waterboer T., Castro F. A., Brenner H. (2015). Viral infections and colorectal cancer: A systematic review of epidemiological studies. Int. J. Cancer 137, 12–24. doi: 10.1002/ijc.29180
Chiocca E. A., Rabkin S. D. (2014). Oncolytic Viruses and Their Application to Cancer Immunotherapy. Cancer Immunol. Res. 2, 295–300. doi: 10.1158/2326-6066.CIR-14-0015
Choi A. H., O’Leary M. P., Fong Y., Chen N. G. (2016). From benchtop to bedside: A review of oncolytic virotherapy. Biomedicines 4, 1–20. doi: 10.3390/biomedicines4030018
Chou J., Kern E., Whitley R., Roizman B. (1990). Mapping of herpes simplex virus-1 neurovirulence to gamma 134.5, a gene nonessential for growth in culture. Science 250, 1262–1266. doi: 10.1126/science.2173860
Conlon M. A., Bird A. R. (2015). The impact of diet and lifestyle on gut microbiota and human health. Nutrients 7, 17–44. doi: 10.3390/nu7010017
Conway D. II, Purkayastha M., Chestnutt I. G. (2018). The changing epidemiology of oral cancer: Definitions, trends, and risk factors. Br. Dental J. 225, 867. doi: 10.1038/sj.bdj.2018.922
Crosbie E. J., Einstein M. H., Franceschi S., Kitchener H. C. (2013). Human papillomavirus and cervical cancer. Lancet 382, 889–899. doi: 10.1016/S0140-6736(13)60022-7
de Martel C., Georges D., Bray F., Ferlay J., Clifford G. M. (2020). Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Global Health 8, e180–e190. doi: 10.1016/S2214-109X(19)30488-7
De Pace N. (1912). Sulla scomparsa di un enorme cancro vegetante del collo dell’utero senza cura chirurgica. Ginecologia 9, 82–88.
Dedrick R. M., Guerrero-Bustamante C. A., Garlena R. A., Russell D. A., Ford K., Harris K., et al. (2019). Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 25, 730–733. doi: 10.1038/s41591-019-0437-z
Dejea C. M., Wick E. C., Hechenbleikner E. M., White J. R., Mark Welch J. L., Rossettid B. J., et al. (2014). Microbiota organization is a distinct feature of proximal colorectal cancers. Proceedings of the. Natl. Acad. Sci. U S A 111, 18321–18326. doi: 10.1073/pnas.1406199111
Dock G. (1904). The influence of complicating diseases upon leukaemia. Am. J. Med. Sci. 127, 563–592. doi: 10.1097/00000441-190412740-00001
Dong X., Pan P., Zheng D.-W., Bao P., Zeng X., Zhang X.-Z. (2020). Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci. Adv. 6, eaba1590. doi: 10.1126/sciadv.aba1590
Donohoe D. R., Holley D., Collins L. B., Montgomery S. A., Whitmore A. C., Hillhouse A., et al. (2014). A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov 4, 1387–1397. doi: 10.1158/2159-8290.CD-14-0501
Durack J., Lynch S. V. (2019). The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 216, 20–40. doi: 10.1084/jem.20180448
Eash S., Manley K., Gasparovic M., Querbes W., Atwood W. J. (2006). The human polyomaviruses. Cell. Mol. Life Sci. 63, 865–876. doi: 10.1007/s00018-005-5454-z
Emlet C., Ruffin M., Lamendella R. (2020). Enteric Virome and Carcinogenesis in the Gut. Digest Dis. Sci. 65, 852–864. doi: 10.1007/s10620-020-06126-4
Enam S., Del Valle L., Lara C., Gan D. D., Ortiz-Hidalgo C., Palazzo J. P., et al. (2002). Association of human polyomavirus JCV with colon cancer: Evidence for interaction of viral T-antigen and β-catenin. Cancer Res. 62, 7093–7101.
Farrell P. J. (2019). Epstein–Barr Virus and Cancer. Annu. Rev. Pathol.: Mech. Dis. 14, 29–53. doi: 10.1146/annurev-pathmechdis-012418-013023
Fernandes Q., Gupta I., Vranic S., Al Moustafa A. E. (2020). Human papillomaviruses and epstein–barr virus interactions in colorectal cancer: A brief review. Pathogens 9, 1–20. doi: 10.3390/pathogens9040300
Flores C. O., Meyer J. R., Valverde S., Farr L., Weitz J. S. (2011). Statistical structure of host-phage interactions. Proc. Natl. Acad. Sci. U S A 108, 288–297. doi: 10.1073/pnas.1101595108
Flynn K. J., Baxter N. T., Schloss P. D. (2016). Metabolic and Community Synergy of Oral Bacteria in Colorectal Cancer. mSphere 1, 1–6. doi: 10.1128/msphere.00102-16
Fuchs C., Mitchell E. P., Hoff P. M. (2006). Irinotecan in the treatment of colorectal cancer. Cancer Treat Rev. 32, 491–503. doi: 10.1016/j.ctrv.2006.07.001
Fung K. Y., Cosgrove L., Lockett T., Head R., Topping D. L. (2012). A review of the potential mechanisms for the lowering of colorectal oncogenesis by butyrate. Br. J. Nutr. 108, 820–831. doi: 10.1017/S0007114512001948
Gal-Ben-Ari S., Barrera I., Ehrlich M., Rosenblum K. (2019). PKR: A Kinase to Remember. Front. Mol. Neurosci. 11:480. doi: 10.3389/fnmol.2018.00480
Ganeshan S. D., Hosseinidoust Z. (2019). Phage therapy with a focus on the human microbiota. Antibiotics 8, 131. doi: 10.3390/antibiotics8030131
Giuliani L., Ronci C., Bonifacio D., Di Bonito L., Favalli C., Perno C. F., et al. (2008). Detection of oncogenic DNA viruses in colorectal cancer. Anticancer Res. 28, 1405–1410.
Gock M., Kordt M., Matschos S., Mullins C. S., Linnebacher M. (2020). Patient-individual cancer cell lines and tissue analysis delivers no evidence of sequences from DNA viruses in colorectal cancer cells. BMC. Gastroenterology 20, 1–10. doi: 10.1186/s12876-020-01404-x
Goel A., Li M. S., Nagasaka T., Shin S. K., Fuerst F., Ricciardiello L., et al. (2006). Association of JC Virus T-Antigen Expression With the Methylator Phenotype in Sporadic Colorectal Cancers. Gastroenterology 130, 1950–1961. doi: 10.1053/j.gastro.2006.02.061
Goel S., Ocean A. J., Parakrama R. Y., Ghalib M. H., Chaudhary I., Shah U., et al. (2020). Elucidation of Pelareorep Pharmacodynamics in A Phase I Trial in Patients with KRAS-Mutated Colorectal Cancer. Mol. Cancer Ther. 19, 1148–1156. doi: 10.1158/1535-7163.MCT-19-1117
Guo Z.-S. (2020). Oncolytic immunotherapy for metastatic cancer: lessons and future strategies. Ann. Trans. Med. 8, 1113–1113. doi: 10.21037/atm.2020.04.42
Hannigan G. D., Duhaime M. B., Ruffin M. T., Koumpouras C. C., Schloss P. D. (2018). Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome. mBio 9, 1–13. doi: 10.1128/mBio.02248-18
Hansen R. M., Libnoch J. A. (1978). Remission of Chronic Lymphocytic Leukemia After Smallpox Vaccination. Arch. Internal Med. 138:1137. doi: 10.1001/archinte.1978.03630320073024
Harris H. C., Edwards C. A., Morrison D. J. (2017). Impact of glycosidic bond configuration on short chain fatty acid production from model fermentable carbohydrates by the human gut microbiota. Nutrients 9, 26. doi: 10.3390/nu9010026
He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., Da Costa L. T., et al. (1998). Identification of c-MYC as a target of the APC pathway. Science 281, 1509–1512. doi: 10.1126/science.281.5382.1509
Heise C., Sampson-Johannes A., Williams A., Mccormick F., Von Hoff D. D., Kirn D. H. (1997). ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat. Med. 3, 639–645. doi: 10.1038/nm0697-639
Henderson B. R. (2000). Nuclear-cytoplasmic shuttling of APC regulates β-catenin subcellular localization and turnover. Nat. Cell Biol. 2, 653–660. doi: 10.1038/35023605
Hibberd A. A., Lyra A., Ouwehand A. C., Rolny P., Lindegren H., Cedgård L., et al. (2017). Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 4, e000145. doi: 10.1136/bmjgast-2017-000145
Hill A., Jugovic P., York L., Russ G., Bennink J., Yewdell J., et al. (1995). Herpes simplex virus turns off the TAP to evade host immunity. Nature 375, 411–415. doi: 10.1038/375411a0
Hori R., Murai Y., Tsuneyama K., Abdel-Aziz H. O., Nomoto K., Takahashi H., et al. (2005). Detection of JC virus DNA sequences in colorectal cancers in Japan. Virchows Archiv. 447, 723–730. doi: 10.1007/s00428-005-0014-3
Hosseinidoust Z., van de Ven T. G. M., Tufenkji N. (2013). Evolution of Pseudomonas aeruginosa virulence as a result of phage predation. Appl. Environ. Microbiol. 79, 6110–6116. doi: 10.1128/AEM.01421-13
Hoster H. A., Zanes R. P. Jr., von Haam E. (1949). Studies in Hodgkin’s syndrome; the association of viral hepatitis and Hodgkin’s disease; a preliminary report. Cancer Res. 9 (8), 473–480.
Hsu B. B., Gibson T. E., Yeliseyev V., Liu Q., Lyon L., Bry L., et al. (2019). Dynamic Modulation of the Gut Microbiota and Metabolome by Bacteriophages in a Mouse Model. Cell Host Microbe 25, 803–814. doi: 10.1016/j.chom.2019.05.001
Ijssennagger N., Belzer C., Hooiveld G. J., Dekker J., van Mil S. W. C., Müller M., et al. (2015). Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc. Natl. Acad. Sci. 112, 10038–10043. doi: 10.1073/pnas.1507645112
Jarzyński A., Zajac P., Żebrowski R., Boguszewska A., Polz-Dacewicz M. (2017). Occurrence of BK virus and human papilloma virus in colorectal cancer. Ann. Agric. Environ. Med. 24, 440–445. doi: 10.26444/aaem/74648
Jung W. T., Li M. S., Goel A., Boland C. R. (2008). JC virus T-antigen expression in sporadic adenomatous polyps of the colon. Cancer 112, 1028–1036. doi: 10.1002/cncr.23266
Kakasis A., Panitsa G. (2019). Bacteriophage therapy as an alternative treatment for human infections. A comprehensive review. Int. J. Antimicrobial. Agents 53, 16–21. doi: 10.1016/j.ijantimicag.2018.09.004
Kaufman H. L., Ruby C. E., Hughes T., Slingluff C. L. (2014). Current status of granulocyte-macrophage colony-stimulating factor in the immunotherapy of melanoma. J. ImmunoTher. Cancer 2, 11. doi: 10.1186/2051-1426-2-11
Kean J. M., Rao S., Wang M., Garcea R. L. (2009). Seroepidemiology of human polyomaviruses. PLoS Pathog. 5, e1000363. doi: 10.1371/journal.ppat.1000363
Keum N. N., Giovannucci E. (2019). Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 16, 713–732. doi: 10.1038/s41575-019-0189-8
Kilcher S., Loessner M. J. (2019). Engineering Bacteriophages as Versatile Biologics. Trends Microbiol. 27, 355–367. doi: 10.1016/j.tim.2018.09.006
Kim J. H., Oh J. Y., Park B. H., Lee D. E., Kim J. S., Park H. E., et al. (2006). Systemic armed oncolytic and immunologic therapy for cancer with JX-594, a targeted poxvirus expressing GM-CSF. Mol. Ther. J. Am. Soc. Gene Ther. 14, 361–370. doi: 10.1016/j.ymthe.2006.05.008
Kirk M. D., Pires S. M., Black R. E., Caipo M., Crump J. A., Devleesschauwer B., et al. (2015). World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases 2010: A Data Synthesis. PLoS Med. 12, 1–21. doi: 10.1371/journal.pmed.1001921
Knecht L. E., Veljkovic M., Fieseler L. (2020). Diversity and Function of Phage Encoded Depolymerases. Front. Microbiol. 10, 1–16. doi: 10.3389/fmicb.2019
Kohlhapp F. J., Kaufman H. L. (2016). Molecular pathways: Mechanism of action for talimogene laherparepvec, a new oncolytic virus immunotherapy. Clin. Cancer Res. 22, 1048–1054. doi: 10.1158/1078-0432.CCR-15-2667
Kostic A. D., Gevers D., Pedamallu C. S., Michaud M., Duke F., Earl A. M., et al. (2012). Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 22, 292–298. doi: 10.1101/gr.126573.111
Kostic A. D., Chun E., Robertson L., Glickman J. N., Gallini C. A., Michaud M., et al. (2013). Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe 14, 207–215. doi: 10.1016/j.chom.2013.07.007
Kuhn I., Harden P., Bauzon M., Chartier C., Nye J., Thorne S., et al. (2008). Directed evolution generates a novel oncolytic virus for the treatment of colon cancer. PLoS One 3, 1–11. doi: 10.1371/journal.pone.0002409
Kutateladze M. (2015). Experience of the Eliava Institute in bacteriophage therapy. Virol. Sin. 30, 80–81. doi: 10.1007/s12250-014-3557-0
Laghi L., Randolph A. E., Chauhan D. P., Marra G., Major E. O., Neel J. V., et al. (1999). JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc. Natl. Acad. Sci. U S A 96, 7484–7489. doi: 10.1073/pnas.96.13.7484
Lange K., Buerger M., Stallmach A., Bruns T. (2016). Effects of Antibiotics on Gut Microbiota. Digest Dis. 34, 260–268. doi: 10.1159/000443360
Lange J., Fraune S., Bosch T. C., Lachnit T. (2019). The neglected part of the microbiome: Prophage TJ1 regulates the bacterial community of the metaorganism Hydra. bioRxiv. doi: 10.1101/607325
Lee H. L., Shen H., Hwang I. Y., Ling H., Yew W. S., Lee Y. S., et al. (2018). Targeted Approaches for In Situ Gut Microbiome Manipulation. Genes 9, 351. doi: 10.3390/genes9070351
Lehti T. A., Pajunen M. II, Skog M. S., Finne J. (2017). Internalization of a polysialic acid-binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat. Commun. 8, 1915. doi: 10.1038/s41467-017-02057-3
Levy M., Kolodziejczyk A. A., Thaiss C. A., Elinav E. (2017). Dysbiosis and the immune system. Nat. Rev. Immunol. 17, 219–232. doi: 10.1038/nri.2017.7
Li J., Zhao F., Wang Y., Chen J., Tao J., Tian G., et al. (2017). Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 5, 1–19. doi: 10.1186/s40168-016-0222-x
Lin M. V., King L. Y., Chung R. T. (2015). Hepatitis C Virus–Associated Cancer. Annu. Rev. Pathol.: Mech. Dis. 10, 345–370. doi: 10.1146/annurev-pathol-012414-040323
Link A., Shin S. K., Nagasaka T., Balaguer F., Koi M., Jung B., et al. (2009). JC virus mediates invasion and migration in colorectal metastasis. PLoS One 4, e8146. doi: 10.1371/journal.pone.0008146
Liu B. L., Robinson M., Han Z.-Q., Branston R. H., English C., Reay P., et al. (2003). ICP34.5 deleted herpes simplex virus with enhanced oncolytic, immune stimulating, and anti-tumour properties. Gene Ther. 10, 292–303. doi: 10.1038/sj.gt.3301885
Loc-Carrillo C., Abedon S. T. (2011). Pros and cons of phage therapy. Bacteriophage 1, 111–114. doi: 10.4161/bact.1.2.14590
Lynch S. V., Pedersen O. (2016). The Human Intestinal Microbiome in Health and Disease. New Engl. J. Med. 375, 2369–2379. doi: 10.1056/NEJMra1600266
Mar Rodríguez M., Pérez D., Javier Chaves F., Esteve E., Marin-Garcia P., Xifra G., et al. (2015). Obesity changes the human gut mycobiome. Sci. Rep. 5, 1–15. doi: 10.1038/srep14600
Mariggiò G., Koch S., Schulz T. F. (2017). Kaposi sarcoma herpesvirus pathogenesis. Philos. Trans. R. Soc. B.: Biol. Sci. 372, 20160275. doi: 10.1098/rstb.2016.0275
Melcher A., Parato K., Rooney C. M., Bell J. C. (2011). Thunder and lightning: Immunotherapy and oncolytic viruses collide. Mol. Ther. 19, 1008–1016. doi: 10.1038/mt.2011.65
Mima K., Nishihara R., Qian Z. R., Cao Y., Sukawa Y., Nowak J. A., et al. (2016). Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 65, 1973–1980. doi: 10.1136/gutjnl-2015-310101
Minot S., Sinha R., Chen J., Li H., Keilbaugh S. A., Wu G. D., et al. (2011). The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 21, 1616–1625. doi: 10.1101/gr.122705.111
Morin P. J., Sparks A. B., Korinek V., Barker N., Clevers H., Vogelstein B., et al. (1997). Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Sci. (N. Y. N. Y.) 275, 1787–1790. doi: 10.1126/science.275.5307.1787
Mullard A. (2015). Regulators approve the first cancer-killing virus. Nat. Rev. Drug Discov 14, 811–811. doi: 10.1038/nrd4805
Nakatsu G., Zhou H., Wu W. K. K., Wong S. H., Coker O. O., Dai Z., et al. (2018). Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology 155, 529–541. doi: 10.1053/j.gastro.2018.04.018
Nale J. Y., Spencer J., Hargreaves K. R., Buckley A. M., Trzepiński P., Douce G. R., et al. (2016). Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrobial Agents Chemother. 60, 968–981. doi: 10.1128/AAC.01774-15
Nale J. Y., Redgwell T. A., Millard A., Clokie M. R. J. (2018). Efficacy of an Optimised Bacteriophage Cocktail to Clear Clostridium difficile in a Batch Fermentation Model. Antibiot. (Basel Switzerland) 7, 13. doi: 10.3390/antibiotics7010013
Newcomb P. A., Bush A. C., Stoner G. L., Lampe J. W., Potter J. D., Bigler J. (2004). No evidence of an association of JC virus and colon neoplasia. Cancer Epidemiol. Biomarkers Prev. 13, 662–666.
Nguyen S., Baker K., Padman B. S., Patwa R., Dunstan R. A., Weston T. A., et al. (2017). Bacteriophage Transcytosis Provides a Mechanism To Cross Epithelial Cell Layers. mBio 8, 1–14. doi: 10.1128/mBio.01874-17
Niller H. H., Wolf H., Minarovits J. (2011). Viral hit and run-oncogenesis: Genetic and epigenetic scenarios. Cancer Lett. 305, 200–217. doi: 10.1016/j.canlet.2010.08.007
Norman J. M., Handley S. A., Baldridge M. T., Droit L., Liu C. Y., Keller B. C., et al. (2015). Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160, 447–460. doi: 10.1016/j.cell.2015.01.002
Nosho K., Shima K., Kure S., Irahara N., Baba Y., Chen L., et al. (2009). JC virus T-antigen in colorectal cancer is associated with p53 expression and chromosomal instability, independent of CpG island methylator phenotype. Neoplasia 11, 87–95. doi: 10.1593/neo.81188
Oechslin F., Piccardi P., Mancini S., Gabard J., Moreillon P., Entenza J. M., et al. (2016). Synergistic interaction between phage therapy and antibiotics clears Pseudomonas aeruginosa infection in endocarditis and reduces virulence. J. Infect. Dis. 215:jiw632. doi: 10.1093/infdis/jiw632
Olano-Martin E., Mountzouris K. C., Gibson G. R., Rastall R. A. (2000). In vitro fermentability of dextran, oligodextran and maltodextrin by human gut bacteria. Br. J. Nutr. 83, 247–255. doi: 10.1017/S0007114500000325
O’Cathail S. M., Davis S., Holmes J., Brown R., Fisher K., Seymour L., et al. (2020). A phase 1 trial of the safety, tolerability and biological effects of intravenous Enadenotucirev, a novel oncolytic virus, in combination with chemoradiotherapy in locally advanced rectal cancer (CEDAR). Radiat. Oncol. 15, 1–8. doi: 10.1186/s13014-020-01593-5
Orvedahl A., Alexander D., Tallóczy Z., Sun Q., Wei Y., Zhang W., et al. (2007). HSV-1 ICP34.5 Confers Neurovirulence by Targeting the Beclin 1 Autophagy Protein. Cell Host Microbe 1, 23–35. doi: 10.1016/j.chom.2006.12.001
Panagi M., Georgila K., Eliopoulos A. G., Apidianakis Y. (2019). Constructing personalized longitudinal holo’omes of colon cancer-prone humans and their modeling in flies and mice. Oncotarget 10, 4224–4246. doi: 10.18632/oncotarget.6463
Park S. H., Breitbach C. J., Lee J., Park J. O., Lim H. Y., Kang W. K., et al. (2015). Phase 1b Trial of Biweekly Intravenous Pexa-Vec (JX-594), an Oncolytic and Immunotherapeutic Vaccinia Virus in Colorectal. Cancer Mol. Ther. 23, 1532–1540. doi: 10.1038/mt.2015.109
Paule A., Frezza D., Edeas M. (2018). Microbiota and Phage Therapy: Future Challenges in Medicine. Med. Sci. 6:86. doi: 10.3390/medsci6040086
Piekarowicz A., Kłyż A., Adamczyk-Popławska M., Stein D. C. (2020). Association of host proteins with the broad host range filamentous phage NgoΦ6 of Neisseria gonorrhoeae. PLoS One 15, e0240579. doi: 10.1371/journal.pone.0240579
Powell S. M., Zilz N., Beazer-Barclay Y., Bryan T. M., Hamilton S. R., Thibodeau S. N., et al. (1992). APC mutations occur early during colorectal tumorigenesis. Nature 359, 235–237. doi: 10.1038/359235a0
Raman S. S., Hecht J. R., Chan E. (2019). Talimogene laherparepvec: review of its mechanism of action and clinical efficacy and safety. Immunotherapy 11, 705–723. doi: 10.2217/imt-2019-0033
Reid T., Galanis E., Abbruzzese J., Sze D., Andrews J., Romel L., et al. (2001). Intra-arterial administration of a replication-selective adenovirus (dl1520) in patients with colorectal carcinoma metastatic to the liver: a phase I trial. Gene Ther. 8, 1618–1626. doi: 10.1038/sj.gt.3301512
Reinhardt C., Bergentall M., Greiner T. U., Schaffner F., Östergren-Lundén G., Petersen L. C., et al. (2012). Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature 483, 627–631. doi: 10.1038/nature10893
Reyes A., Wu M., McNulty N. P., Rohwer F. L., Gordon J. II (2013). Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc. Natl. Acad. Sci. 110, 20236–20241. doi: 10.1073/pnas.1319470110
Ricciardiello L., Baglioni M., Giovannini C., Pariali M., Cenacchi G., Ripalti A., et al. (2003). Induction of Chromosomal Instability in Colonic Cells by the Human Polyomavirus JC Virus. Cancer Res. 63, 7256–7262.
Ripple M. J., Struckhoff A. P., Trillo-Tinoco J., Li L., Margolin D. A., McGoey R., et al. (2014). Activation of c-Myc and cyclin D1 by JCV T-Antigen and β-Catenin in colon cancer. PLoS One 9, e106257. doi: 10.1371/journal.pone.0106257
Rossmann F. S., Racek T., Wobser D., Puchalka J., Rabener E. M., Reiger M., et al. (2015). Phage-mediated dispersal of biofilm and distribution of bacterial virulence genes is induced by quorum sensing. PLoS Pathog. 11, e1004653. doi: 10.1371/journal.ppat.1004653
Sarker S. A., Sultana S., Reuteler G., Moine D., Descombes P., Charton F., et al. (2016). Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine 4, 124–137. doi: 10.1016/j.ebiom.2015.12.023
Schooley R. T., Biswas B., Gill J. J., Hernandez-Morales A., Lancaster J., Lessor L., et al. (2017). Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails To Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrobial Agents Chemother. 61, e00954–17. doi: 10.1128/AAC.00954-17
Secor P. R., Sweere J. M., Michaels L. A., Malkovskiy A. V., Lazzareschi D., Katznelson E., et al. (2015). Filamentous Bacteriophage Promote Biofilm Assembly and Function. Cell Host Microbe 18, 549–559. doi: 10.1016/j.chom.2015.10.013
Sender R., Fuchs S., Milo R. (2016). Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 14, e1002533. doi: 10.1371/journal.pbio.1002533
Shkoporov A. N., Hill C. (2019). Bacteriophages of the Human Gut: The “Known Unknown” of the Microbiome. Cell Host Microbe 25, 195–209. doi: 10.1016/j.chom.2019.01.017
Shkoporov A. N., Clooney A. G., Sutton T. D., Ryan F. J., Daly K. M., Nolan J. A., et al. (2019). The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific. Cell Host Microbe 26, 527–541. doi: 10.1016/j.chom.2019.09.009
Siegel R. L., Torre L. A., Soerjomataram I., Hayes R. B., Bray F., Weber T. K., et al. (2019). Global patterns and trends in colorectal cancer incidence in young adults. Gut 68, 2179–2185. doi: 10.1136/gutjnl-2019-319511
Siegel R. L., Miller K. D., Goding Sauer A., Fedewa S. A., Butterly L. F., Anderson J. C., et al. (2020a). Colorectal cancer statistics 2020. CA. A. Cancer J. Clin. 70, 145–164. doi: 10.3322/caac.21601
Siegel R. L., Miller K. D., Jemal A. (2020b). Cancer statistics 2020. CA. A. Cancer J. Clin. 70, 7–30. doi: 10.3322/caac.21590
Spencer S. P., Fragiadakis G. K., Sonnenburg J. L. (2019). Pursuing Human-Relevant Gut Microbiota-Immune Interactions. Immunity 51, 225–239. doi: 10.1016/j.immuni.2019.08.002
Steenbergen L., Sellaro R., van Hemert S., Bosch J. A., Colzato L. S. (2015). A randomized controlled trial to test the effect of multispecies probiotics on cognitive reactivity to sad mood. Brain Behav. Immun. 48, 258–264. doi: 10.1016/j.bbi.2015.04.003
Su F.-H., Le T. N., Muo C.-H., Te S. A., Sung F.-C., Yeh C.-C. (2020). Chronic Hepatitis B Virus Infection Associated with Increased Colorectal Cancer Risk in Taiwanese Population. Viruses 12:97. doi: 10.3390/v12010097
Sulakvelidze A., Alavidze Z., Morris J. G. (2001). Bacteriophage therapy. Antimicrobial Agents Chemother. 45, 649–659. doi: 10.1128/AAC.45.3.649-659.2001
Sweere J. M., Van Belleghem J. D., Ishak H., Bach M. S., Popescu M., Sunkari V., et al. (2019). Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science (N. Y. N. Y.) 363, eaat9691. doi: 10.1126/science.aat9691
Tallóczy Z., Jiang W., Virgin IV H. W., Leib D. A., Scheuner D., Kaufman R. J., et al. (2002). Regulation of starvation- and virus-induced autophagy by the elF2α kinase signaling pathway. Proc. Natl. Acad. Sci. U S A 99, 190–195. doi: 10.1073/pnas.012485299
Taneja S., MacGregor J., Markus S., Ha S., Mohr I. (2001). Enhanced antitumor efficacy of a herpes simplex virus mutant isolated by genetic selection in cancer cells. Proc. Natl. Acad. Sci. U S A 98, 8804–8808. doi: 10.1073/pnas.161011798
Tetsu O., McCormick F. (1999). β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426. doi: 10.1038/18884
Theodoropoulos G., Panoussopoulos D., Papaconstantinou I., Gazouli M., Perdiki M., Bramis J., et al. (2005). Assessment of JC polyoma virus in colon neoplasms. Dis. Colon. Rectum 48, 86–91. doi: 10.1007/s10350-004-0737-2
Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. II (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Twumasi-Boateng K., Pettigrew J. L., Kwok Y. Y., Bell J. C., Nelson B. H. (2018). Oncolytic viruses as engineering platforms for combination immunotherapy. Nat. Rev. Cancer 18, 419–432. doi: 10.1038/s41568-018-0009-4
Van Belleghem J. D., Clement F., Merabishvili M., Lavigne R., Vaneechoutte M. (2017). Pro- and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci. Rep. 7, 8004. doi: 10.1038/s41598-017-08336-9
Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X., et al. (2012). Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 6, 320–329. doi: 10.1038/ismej.2011.109
Warren F. J., Fukuma N. M., Mikkelsen D., Flanagan B. M., Williams B. A., Lisle A. T., et al. (2018). Food Starch Structure Impacts Gut Microbiome Composition. mSphere 3, 1–13. doi: 10.1128/msphere.00086-18
Wigington C. H., Sonderegger D., Brussaard C. P., Buchan A., Finke J. F., Fuhrman J. A., et al. (2016). Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 1, 15024. doi: 10.1038/nmicrobiol.2015.24
Wright A., Hawkins C. H., Anggård E. E., Harper D. R. (2009). A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. Off. J. ENT-UK Off. J. Netherlands Soc. Oto-Rhino-Laryngol. Cervico-Facial Surg. 34, 349–357. doi: 10.1111/j.1749-4486.2009.01973.x
Wu S., Rhee K. J., Albesiano E., Rabizadeh S., Wu X., Yen H. R., et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15, 1016–1022. doi: 10.1038/nm.2015
Wu N., Yang X., Zhang R., Li J., Xiao X., Hu Y., et al. (2013). Dysbiosis Signature of Fecal Microbiota in Colorectal Cancer Patients. Microbial. Ecol. 66, 462–470. doi: 10.1007/s00248-013-0245-9
Wu J., Li Q., Fu X. (2019). Fusobacterium nucleatum Contributes to the Carcinogenesis of Colorectal Cancer by Inducing Inflammation and Suppressing Host Immunity. Trans. Oncol. 12, 846–851. doi: 10.1016/j.tranon.2019.03.003
Xie Y. (2017). Hepatitis B Virus-Associated Hepatocellular Carcinoma. Adv. Exp. Med. Biol. 1018, 11–21. doi: 10.1007/978-981-10-5765-6_2
Zhang L., Shay J. W. (2017). Multiple Roles of APC and its Therapeutic Implications in Colorectal Cancer. J. Natl. Cancer Institute 109, 1–10. doi: 10.1093/jnci/djw332
Zhang X., Zhang D., Jia H., Feng Q., Wang D., Liang D., et al. (2015). The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905. doi: 10.1038/nm.3914
Zheng D. W., Dong X., Pan P., Chen K. W., Fan J. X., Cheng S. X., et al. (2019). Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 3, 717–728. doi: 10.1038/s41551-019-0423-2
Zheng M., Huang J., Tong A., Yang H. (2019). Oncolytic Viruses for Cancer Therapy: Barriers and Recent Advances. Mol. Ther. Oncolyt. 15, 234–247. doi: 10.1016/j.omto.2019.10.007
Keywords: virus, bacteriophage, cancer, colorectal, disease dynamics, bacteriophage therapy, oncolysis, microbiome
Citation: Turkington CJR, Varadan AC, Grenier SF and Grasis JA (2021) The Viral Janus: Viruses as Aetiological Agents and Treatment Options in Colorectal Cancer. Front. Cell. Infect. Microbiol. 10:601573. doi: 10.3389/fcimb.2020.601573
Received: 01 September 2020; Accepted: 23 November 2020;
Published: 07 January 2021.
Edited by:
Andrey Shkoporov, University College Cork, IrelandReviewed by:
Yiorgos Apidianakis, University of Cyprus, CyprusCopyright © 2021 Turkington, Varadan, Grenier and Grasis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher J. R. Turkington, cturkington@ucmerced.edu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.