- 1Department of Epidemiology, University of Michigan, Ann Arbor, MI, United States
- 2Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI, United States
- 3Department of Biology and Center for Life in Extreme Environments, Portland State University, Portland, OR, United States
- 4Department of Biology, University of Texas at San Antonio, San Antonio, TX, United States
Regulatory RNAs contribute to gene expression control in bacteria. Antisense RNAs (asRNA) are a class of regulatory RNAs that are transcribed from opposite strands of their target genes. Typically, these untranslated transcripts bind to cognate mRNAs and rapidly regulate gene expression at the post-transcriptional level. In this article, we review asRNAs that modulate bacterial fitness and increase virulence. We chose examples that underscore the variety observed in nature including, plasmid- and chromosome-encoded asRNAs, a riboswitch-regulated asRNA, and asRNAs that require other RNAs or RNA-binding proteins for stability and activity. We explore how asRNAs improve bacterial fitness and virulence by modulating plasmid acquisition and maintenance, regulating transposon mobility, increasing resistance against bacteriophages, controlling flagellar production, and regulating nutrient acquisition. We conclude with a brief discussion on how this knowledge is helping to inform current efforts to develop new therapeutics.
Introduction
A major breakthrough in biology was the discovery of non-coding RNAs (ncRNAs) that regulate gene expression instead of coding for proteins. ncRNAs play important regulatory roles in all domains of life (Katayama et al., 2005; Beiter et al., 2009; Georg and Hess, 2011; Lybecker et al., 2014). In bacteria, ncRNAs regulate gene expression at the post-transcriptional level by binding to messenger RNAs (mRNAs) to control several processes, including pathogenesis (Gripenland et al., 2010; Gottesman and Storz, 2011; Kacharia et al., 2017). Typically, ncRNAs that are encoded on the opposite strands of target genes (complementary to sense transcript) are known as cis-acting antisense RNAs (asRNAs), while an ncRNA that is encoded in a separate part of the genome in relation to its target mRNAs is called a trans-acting small RNA (sRNA) (Waters and Storz, 2009; Thomason and Storz, 2010; Georg and Hess, 2011). Regulatory RNAs generally have an advantage over regulatory proteins because their synthesis require lower energy and they act rapidly. In addition, their co-degradation along with target mRNAs allow precise control of regulatory circuits, which is key for bacteria to quickly adapt to host immune response (Storz et al., 2011; Updegrove et al., 2015). asRNAs are particularly useful for rapid gene regulation because they bind to target mRNAs with perfect complementarity, whereas sRNAs form imperfect complementarity with target mRNAs and often require chaperone proteins such as Hfq and ProQ for stability and function (Georg and Hess, 2011; Saberi et al., 2016; Hoynes-O’Connor and Moon, 2016; Dutcher and Raghavan, 2018).
Initially, asRNAs were thought to be rare in bacteria, and the pervasive antisense transcription observed in microarray-based studies were assumed to be experimental artifacts (Johnson et al., 2005; Perocchi et al., 2007; Georg and Hess, 2011). Even with the advent of high-throughput sequencing, it was initially difficult to differentiate between bona fide asRNAs and transcriptional noise because of low sequence coverage (Georg et al., 2009; Yamaguchi et al., 2011; Georg and Hess, 2018). With the increase in sequencing resolution, recent studies have confirmed the presence of abundant asRNAs in bacteria and have revealed it to be a genome-wide phenomenon (Dornenburg et al., 2010; Georg and Hess, 2011; Raghavan et al., 2012; Thomason et al., 2015).
asRNAs have been shown to modulate bacterial pathogenicity by either regulating the expression of virulence genes (Giangrossi et al., 2010) or by controlling biochemical processes that improve bacterial fitness, which in turn boosts virulence (Gripenland et al., 2010; Lejars et al., 2019). In this mini review, we focus on the latter. In particular, we cover examples where we generally understand the mechanism of action and where the genome locations of asRNAs have been determined. We also chose examples that underscore the variety observed in nature, including short asRNAs and long asRNAs, those found in plasmids and those encoded on genomes, asRNAs that require binding stability from other RNAs or proteins, and asRNAs that work in concert with riboswitches. These examples are presented in several sections based on the main roles asRNAs play in pathogenesis: 1) acquisition and regulation of virulence plasmids, 2) modulation of transposon mobility, 3) increasing resistance against bacteriophages, 4) controlling flagellar production, and 5) regulating nutrient acquisition.
Modes of Antisense RNA-Based Improvement in Fitness and Virulence
Acquisition and Maintenance of Virulence Plasmids
A major avenue through which bacteria acquire new virulence factors is by obtaining new plasmids via conjugation, a process that involves asRNAs. An example of this is F-like plasmids, which are part of a large group of conjugative plasmids frequently found in Escherichia coli and throughout Enterobacteriaceae (Jerome et al., 1999). These plasmids frequently harbor accessory genes, including antibiotic resistance genes, enterotoxins, and other virulence genes (Koraimann, 2018). Conjugation is encoded by the Tra-operon, with initiation requiring TraJ. Initiation is regulated by FinOP, which consists of the RNA binding protein FinO and FinP, an asRNA. FinP attaches to the ribosome binding site of traJ, inhibiting its translation and promoting mRNA degradation (Figure 1A). FinO contributes by helping promote FinP binding to traJ, as well as protecting FinP from RNase E cleavage (Arthur et al., 2003). FinP levels are controlled by RNase E digestion, preventing binding to traJ. These processes play out temporally during conjugation, starting with initial high level of traJ expression, followed by dampening and repression, maintaining bacterial fitness by reducing the metabolic burden of the plasmid (Glover et al., 2015).
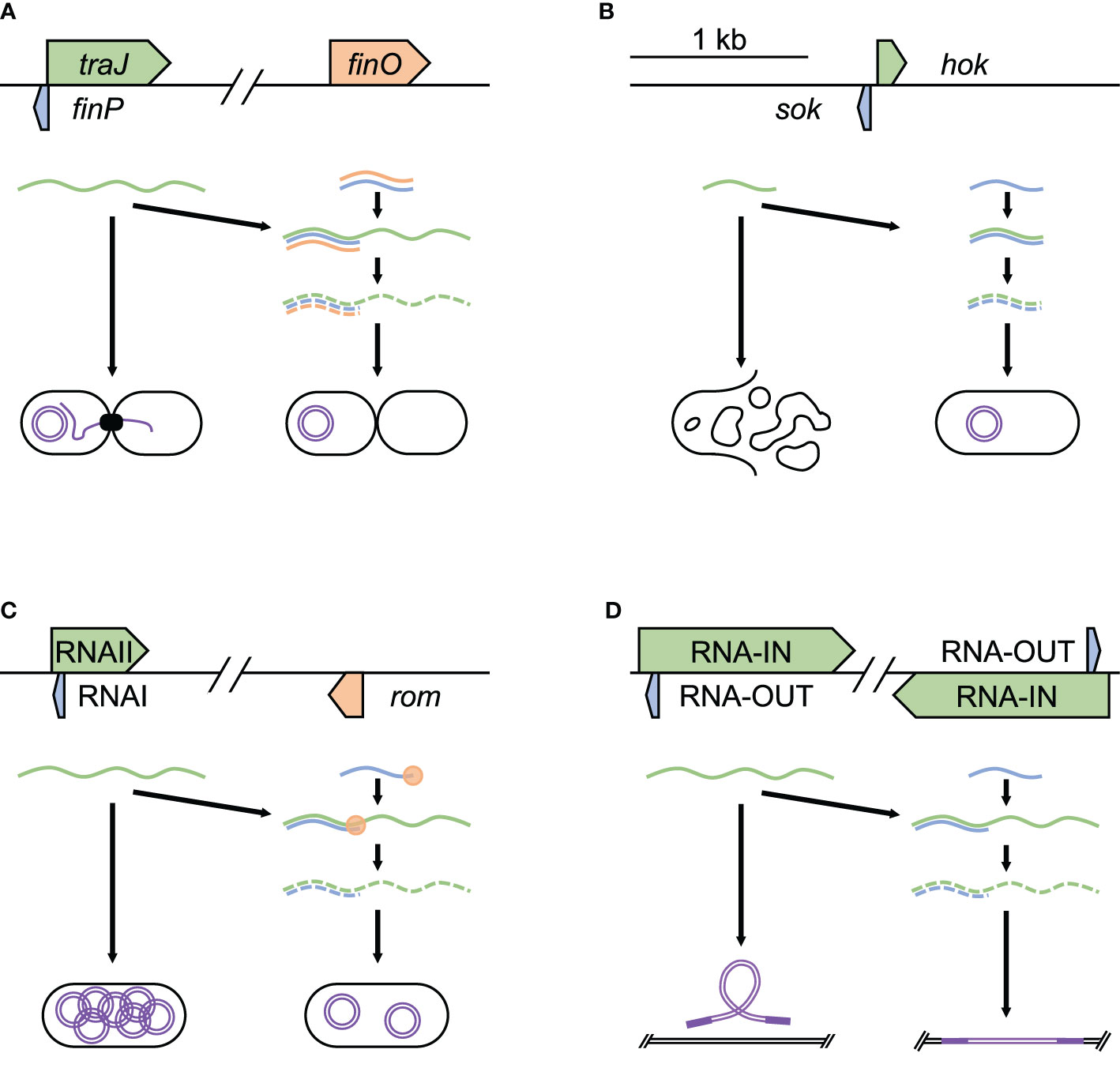
Figure 1 Acquisition and regulation of virulence plasmids and modulation of transposons. Examples of asRNA post-transcriptional regulation of acquisition and regulation of virulence plasmids and modulation of transposons. (A) finP control of plasmid acquisition through conjugation in E. coli F plasmid (NC_002483.1), (B) hok/sok toxin-antitoxin to maintain plasmids in S. flexneri R100 plasmid (NC_002134.1), (C) RNAI control of plasmid copy number in E. coli pColK plasmid (NC_006881.1), and (D) RNA-OUT regulation of transposase expression, controlling transposon movement in S. flexneri R100 plasmid (NC_002134.1). Each panel shows the relative position and size of the asRNA (blue), the target it regulates (green), and any factors required for stable binding (orange). In these examples, the successful binding of asRNA to its target promotes degradation.
Once bacteria acquire advantageous virulence factors through plasmids, some plasmids are retained through toxin/antitoxin systems (Gerdes and Wagner, 2007). These systems function by encoding a toxin and a paired strong antitoxin — many of which function as asRNAs, on the plasmid. During cell division, loss of the plasmid in a daughter cell results in loss of the strong antitoxin, leading to the death of cells without a plasmid copy. A well-studied system is the hok/sok system of R1 plasmids in E. coli and R100 in Shigella flexneri, known for harboring various antibiotic resistance genes (Ogata and Levine, 1980; Cox and Schildbach, 2017). This system encodes the Hok (host killing) toxin, which leads to cell death by depolarization of the cell membrane (Pecota et al., 2003), and Sok (suppression of killing), an asRNA antitoxin, which degrades very quickly (Gerdes and Wagner, 2007). Sok acts by binding to hok mRNA to block translation of the toxin (Figure 1B). Within the E. coli chromosome, sok gene has a very weak promoter, resulting in the production of small amounts of antitoxin that are degraded quickly and are unable to keep up with the Hok toxin, leading to cell death. On the R1 plasmid, the sok gene has a strong promoter, producing many times more of Sok than Hok (Gerdes et al., 1990). Hence, if the cell contains an R1 plasmid, excess Sok continues to bind all of Hok and prevent cell death. This ensures that after cell division, E. coli daughter cells will survive only if they maintain the plasmid. Thus, the Sok asRNA maintains bacterial fitness by promoting the retention of the R1/R100 plasmid, which has been found to improve bacterial stress response and growth in growth-limiting conditions (Chukwudi and Good, 2015).
Replication control is another asRNA-based mechanism used by bacteria to maintain plasmids. The presence of too many copies of a plasmid can increase the metabolic burden of the cell, lowering fitness through reduced growth rate and weakened competitiveness (Baltrus, 2013; Vogwill and MacLean, 2015). However, too few plasmid copies could result in the loss of a potentially useful plasmid in subsequent generations (Millan and MacLean, 2017; Pluta and Espinosa, 2018). Bacteria encode plasmid copy number control systems in order to maintain optimal number of plasmids. One that has been widely studied is found in ColE1-related plasmids (Lacatena and Cesareni, 1981) present in E. coli. The plasmid is named for containing the gene that encodes Colicin E1, the product of which is active against E. coli, as well as containing a gene for conferring immunity to Colicin E1. Under stressful conditions such as nutrient depletion, overcrowding, or antibiotics E. coli express Colicin E1, which promotes bacterial proliferation in mixed microbe niches such as the intestinal tract (Spangler et al., 1985; Riley and Gordon, 1999). To replicate the plasmid, RNAII (a pre-primer) attaches to DNA at origin of replication. RNAII is then trimmed into a primer, which initializes plasmid replication. The 5′ region of RNAII contains the asRNA RNAI, which inhibits ColE1 plasmid replication (Figure 1C). RNAI inhibits plasmid replication with the help of the Rom protein by binding to RNAII, preventing RNAII from binding to the plasmid origin of replication. As the copy number of ColE1 plasmid increases, so does the concentration of RNAI, resulting in a balance of copy number through negative control (del Solar and Espinosa, 2000). This ensures that there are enough copies of the virulence plasmid to pass on to daughter cells, while maintaining fitness by reducing the metabolic burden of what are often large — sometimes hundreds of kilobases long, plasmids (Sengupta and Austin, 2011).
Regulation of Transposon Mobility
Another role for asRNAs in maintaining bacterial fitness and virulence is by controlling the movement of transposons, which are genetic elements that move from one position to another within a genome (Bourque et al., 2018). Insertion of transposons in virulence associated genes could reduce a bacterium’s pathogenicity or increase its susceptibility to antibiotics (Murray et al., 2009; Murray et al., 2015; Kalindamar et al., 2019). In addition, transposable elements could modulate virulence by affecting biofilm formation (Arciola et al., 2004; Kiem et al., 2004; Perez et al., 2015) and reduce fitness by interrupting metabolic genes (Lapierre et al., 2002; Christie-Oleza et al., 2008; Moffatt et al., 2011). Bacteria defend against this by controlling transposases, the enzymes required for transposons’ mobility. An example of inhibition of bacterial transposase can be seen in the Tn10 transposable element, which is found in S. flexneri and other Enterobacteriaceae (Ma and Simons, 1990). Tn10 contains a number of tetracycline resistance genes and a pair of IS10 insertion sequences that each encode transposases that promote transposon mobility. IS10s also encode antisense RNA (RNA-OUT), which is found in the 5′-most segment of the transposase mRNA (RNA‐IN). RNA-OUT inhibits transposase translation by binding RNA‐IN and blocking ribosome binding site (Figure 1D). As the Tn10 copy number increases, asRNA increases to suppress the transposition of the transposon (Ellis et al., 2015); thus, bacteria are able to maintain fitness by reducing the chance of mobile elements disrupting essential genes or virulence factors.
Modulation of Bacteriophages
During infection, pathogenic bacteria have to outcompete other bacteria that share their niche and defend against both internal and external threats. One of the major dangers that bacteria face is from bacteriophages, many of which have lysogenic and lytic growth cycles (Echols, 1972). In the lysogenic cycle, phage DNA is integrated into the bacterial chromosome, allowing replication of the phage to occur more passively along with that of the bacterium. In lytic reproduction, the phage actively creates a large number of progeny and quickly lyse the bacterial cell to continue its lifecycle. Temperate phages include both cycles and are found in about half of microbial genomes currently sequenced (Touchon et al., 2016). While attempting to block infection by phages with lytic growth cycles often reduces virulence and fitness, allowing temperate phages to stay in the lysogenic cycle benefit bacteria by delaying eventual cell lysis (Seed et al., 2014; León and Bastías, 2015; Harrison and Brockhurst, 2017; Howard-Varona et al., 2017). An example of this can be seen in Salmonella’s maintenance of lysogeny in P22 phages (Liao et al., 1987). In P22, lytic growth is inhibited by the regulatory protein C2, which blocks the transcription of proteins needed for the development of lytic cycle. The switch to lytic growth is brought on by the anti-repressor protein Ant, which blocks C2 binding to the OR and OL operators of the P22 phage. Repression of this progression into the lytic replication cycle can be accomplished through Sar, an asRNA in the intergenic region of arc-ant mRNA (Schaefer and McClure, 1997). Sar blocks the ant ribosome binding site, which results in a failure to produce Ant and thus increasing bacterial fitness by preventing the escape of the prophage from the lysogenic state (Figure 2A).
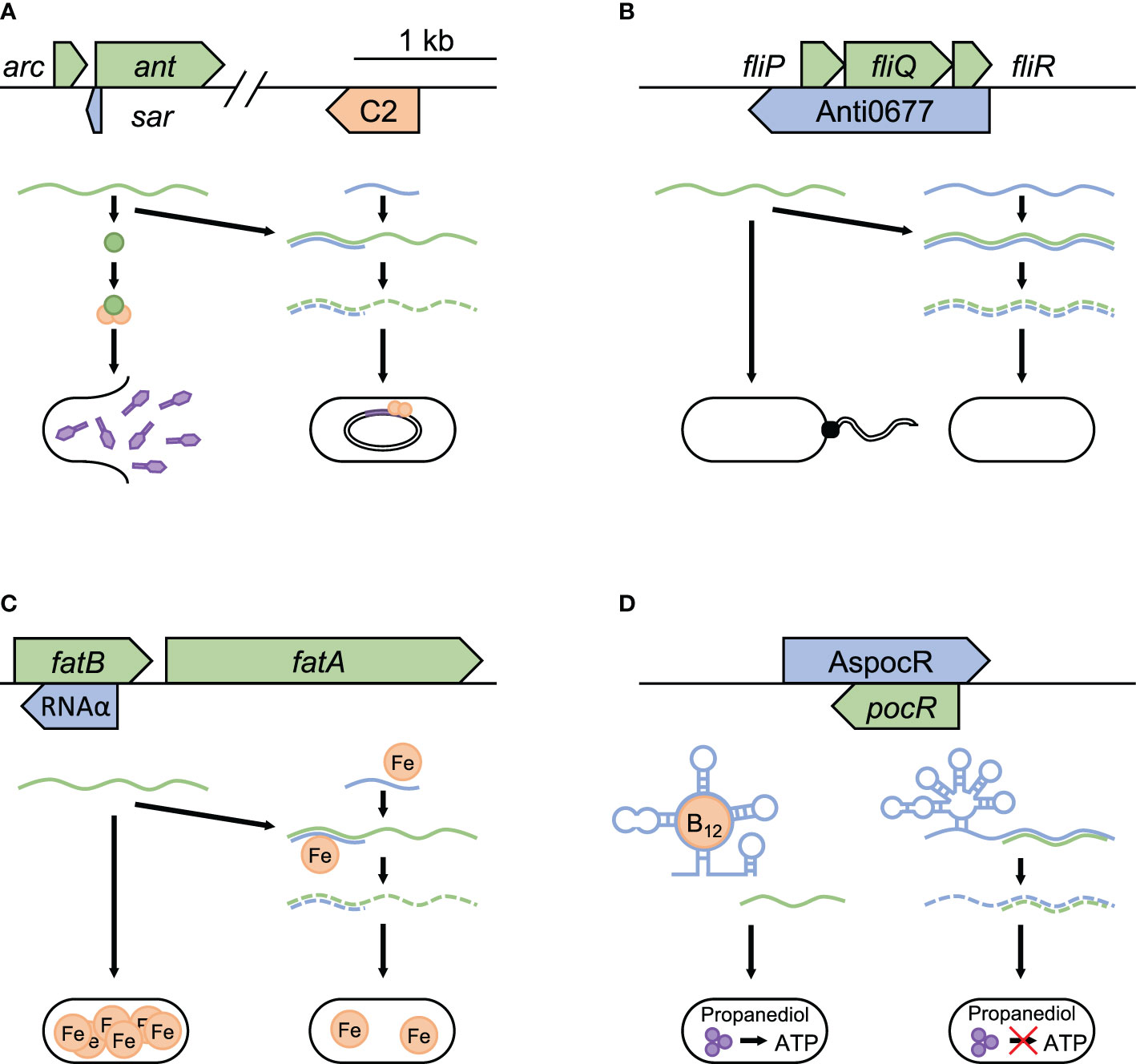
Figure 2 Modulation of bacteriophages, flagella production, and nutrient acquisition. Examples of asRNA post-transcriptional regulation in pathogenic bacteria to increase fitness during infection. Repression of temperate bacteriophages is seen in (A) sar repression of lytic growth in Enterobacteria P22 phage (NC_002371.2). sar blocks the anti-repressor ant from binding to C2 (orange), which prevents escape from lysogenic growth. Temperature dependent regulation in bacteria is represented by (B) Anti0677 control of flagella formation in response to temperature change in L. monocytogenes (NC_003210.1). Examples that highlight regulation of nutrient acquisition can be seen in (C) RNAα control of iron acquisition in V. anguillarum pJM1 plasmid (NC_005250.1) and (D) AspocR/B12 riboswitch control of opportunistic propanediol catabolism in L. monocytogenes (NC_003210.1). Each panel shows the relative position and size of the asRNA (blue), and the target it regulates (green). In (A–C), the successful binding of the asRNA to its target promotes mRNA degradation, with iron required in (C) for stable binding. In (D), when B12 is absent, a full-length version of the asRNA is transcribed, which binds to pocR mRNA and blocks propanediol fermentation. When B12 is present and binds to the riboswitch, transcription ends prematurely, resulting in the production of PocR, which promotes propanediol fermentation.
Control of Flagella Production
By modifying their outer structures bacteria evade immune response and improve persistence within hosts. An example of asRNA’s involvement in this process is observed in Listeria monocytogenes, where a long asRNA regulates flagella production in response to temperature (Toledo-Arana et al., 2009). At 30°C, L. monocytogenes expresses flagella on its surface and exhibits swimming motility. Producing flagella requires the expression of a number of genes, including the flagellum export apparatus genes (fliP, fliQ, and fliR). When the temperature rises to 37°C, the motility gene repressor mogR switches off flagella formation (Lebreton and Cossart, 2017). Overlapping fliP, fliQ, and fliR is a large asRNA, Anti0677, which negatively regulates their expression by promoting mRNA degradation by direct interaction (Figure 2B). Additionally, the end of Anti0677 both contains the coding sequence for and drives the expression of MogR. These two effects — the antisense component Anti0677 and increased expression of MogR — together suppress flagella formation within the host, possibly reducing the host inflammatory response attempting to lyse the invading bacteria (Hayashi et al., 2001). The term “excludon” has been proposed for transcripts such as Anti0677 that both code for proteins and regulate the expression of multiple genes or operons encoded divergently from them (Sesto et al., 2013).
Regulation of Nutrient Acquisition
Bacteria can optimize their growth rates by modulating nutrient acquisition. This capability bestows increased fitness to pathogens by allowing them to survive under nutrient-poor conditions such as infections (Parrow et al., 2013; Fonseca and Swanson, 2014). An example of this phenomenon is iron uptake suppression in the fish pathogen Vibrio anguillarum (Chen and Crosa, 1996; Sesto et al., 2013). Iron is an essential nutrient for most bacteria because it plays critical roles in numerous metabolic processes (León-Sicairos et al., 2015). V. anguillarum contains the pJM1 plasmid, which encodes most genes necessary for iron-anguibactin siderophore transport and biosynthesis (Naka et al., 2010). Among these are transport proteins FatA and FatB, which are encoded by genes fatA and fatB that are transcribed together as a polycistronic mRNA. An asRNA termed RNAα, which is encoded within fatB, is expressed in response to increasing iron levels. It binds to the fatB portion of the fatA-fatB mRNA and represses the translation of both genes (Figure 2C). Iron further stabilizes the binding of RNAα to fatA-fatB mRNA, leading to its degradation. This system helps to reduce the fitness cost associated with metabolic burden by synthesizing iron siderophores only when confronted with iron-poor conditions, thereby allowing the bacterium to optimize its resources to outcompete other bacteria.
Another example of improving metabolic fitness through regulating bacterial nutrient acquisition is the regulation of propanediol catabolism in L. monocytogenes (Mellin et al., 2013). Propanediol is a byproduct of the fermentation of rhamnose and fucose, and is often produced by commensal bacteria in host intestines (Bobik et al., 1997; Degnan et al., 2014). Propanediol fermentation is facilitated through a coenzyme B12-dependent process and can support bacterial growth by providing ATP (Toraya et al., 1979). Some studies suggest that propanediol catabolism gives bacteria a competitive advantage, with mutations in related genes resulting in a virulence defect (Conner et al., 1998). Within L. monocytogenes, the presence of propanediol activates the transcription factor PocR, which controls the expression of propanediol catabolism genes that require vitamin B12 as a cofactor. On the opposite strand of pocR gene, there is a vitamin B12 riboswitch-regulated asRNA, AspocR (Figure 2D). When vitamin B12 is absent, a full-length version of AspocR is transcribed, which inhibits pocR expression. When vitamin B12 is bound to the riboswitch, AspocR transcript ends prematurely, and hence cannot inhibit pocR. This leads to the production of PocR, which promotes the expression of propanediol catabolism genes. Thus, the riboswitch-regulated asRNA allows the expression of propanediol fermentation genes only when both propanediol and B12 are present, thereby reducing the fitness cost associated with unnecessary metabolic burden.
Conclusions
asRNAs are ubiquitous in bacteria and are involved in a multitude of pathogenesis-related mechanisms. The wide range of asRNA functions span the control of intra- and extra-chromosomal DNA, as well as adaption strategies to improve persistence under changing environments. Some asRNAs are only found in specific bacterial species, while others are found across bacteria. Because asRNAs play important roles in modulating the fitness of pathogenic bacteria, current research is focused not only on identifying new asRNAs, but also to use them to our advantage by developing novel asRNA-based therapeutics. For instance, bacterial antibiotic resistance genes can be targeted with synthetic asRNAs, resulting in antibiotic sensitive bacteria (Ji et al., 2004; Nikravesh et al., 2007). Other possible applications include using asRNA to silence bacterial metabolism or ribosomal protein coding genes (successfully shown in E. coli) and protection from bacteriophages in the production of live mucosal vaccines (Sturino and Klaenhammer, 2006; Alessandra et al., 2008; Suzukia et al., 2020). Applications of these techniques beyond in vitro studies have been limited due to difficulties in delivering asRNAs to the site of infection (Good and Stach, 2011; Saberi et al., 2016). As these impediments are addressed, the use of asRNAs in therapeutics will likely expand and contribute to the understanding of the rich landscape of bacterial control systems.
Author Contributions
JM drafted the initial manuscript and figures. JM and RR read and edited the initial manuscript. All authors contributed to the article and approved the submitted version.
Funding
JM is supported by a grant from the National Science Foundation GRFP (DGE‐1256260) and through the Rackham Merit Fellowship through the University of Michigan. RR is supported by NIH grants AI133023 and DE028409.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Carl F. Marrs for the original inspiration for this review and Kenetta Nunn and Ihudiya Finda Williams for feedback.
References
Alessandra S., Alessandro T., Flavio S., Alejandro H. (2008). Artificial antisense RNAs silence lacZ in E. coli by decreasing target mRNA concentration. BMB Rep. 41, 568–574. doi: 10.5483/bmbrep.2008.41.8.568
Arciola C. R., Campoccia D., Gamberini S., Rizzi S., Donati M. E., Baldassarri L., et al. (2004). Search for the insertion element IS256 within the ica locus of Staphylococcus epidermidis clinical isolates collected from biomaterial-associated infections. Biomaterials 25, 4117–4125. doi: 10.1016/j.biomaterials.2003.11.027
Arthur D. C., Ghetu A. F., Gubbins M. J., Edwards R. A., Frost L. S., Glover J. N. (2003). FinO is an RNA chaperone that facilitates sense-antisense RNA interactions. EMBO J. 22, 6346–6355. doi: 10.1093/emboj/cdg607
Baltrus D. A. (2013). Exploring the costs of horizontal gene transfer. Trends Ecol. Evol. 28, 489–495. doi: 10.1016/j.tree.2013.04.002
Beiter T., Reich E., Williams R. W., Simon P. (2009). Antisense transcription: A critical look in both directions. Cell Mol. Life Sci. 66, 94–112. doi: 10.1007/s00018-008-8381-y
Bobik T. A., Xu Y., Jeter R. M., Otto K. E., Roth J. R. (1997). Propanediol utilization genes (pdu) of Salmonella typhimurium: Three genes for the propanediol dehydratase. J. Bacteriol. 179, 6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997
Bourque G., Burns K. H., Gehring M., Gorbunova V., Seluanov A., Hammell M., et al. (2018). Ten things you should know about transposable elements. Genome Biol. 19, 199. doi: 10.1186/s13059-018-1577-z
Chen Q., Crosa J. H. (1996). Antisense RNA, fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J. Biol. Chem. 271, 18885–18891. doi: 10.1074/jbc.271.31.18885
Christie-Oleza J. A., Nogales B., Martín-Cardona C., Lanfranconi M. P., Albertí S., Lalucat J., et al. (2008). ISPst9, an ISL3-like insertion sequence from Pseudomonas stutzeri AN10 involved in catabolic gene inactivation. Int. Microbiol. 11, 101–110. doi: 10.2436/20.1501.01.49
Chukwudi C. U., Good L. (2015). The role of the hok/sok locus in bacterial response to stressful growth conditions. Microb. Pathog. 79, 70–79. doi: 10.1016/j.micpath.2015.01.009
Conner C. P., Heithoff D. M., Julio S. M., Sinsheimer R. L., Mahan M. J. (1998). Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl. Acad. Sci. 95, 4641–4645. doi: 10.1073/pnas.95.8.4641
Cox K. E. L., Schildbach J. F. (2017). Sequence of the R1 plasmid and comparison to F and R100. Plasmid 91, 53–60. doi: 10.1016/j.plasmid.2017.03.007
Degnan P. H., Taga M. E., Goodman A. L. (2014). Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 20, 769–778. doi: 10.1016/j.cmet.2014.10.002
del Solar G., Espinosa M. (2000). Plasmid copy number control: An ever-growing story. Mol. Microbiol. 37, 492–500. doi: 10.1046/j.1365-2958.2000.02005.x
Dornenburg J., DeVita A., Palumbo M., Wade J. (2010). Widespread antisense transcription in Escherichia coli. MBio 1, e00024–e00010. doi: 10.1128/mBio.00024-10
Dutcher H. A., Raghavan R. (2018). Origin, evolution, and loss of bacterial small RNAs. Microbiol. Spectr. 6, RWR–0004-2017. doi: 10.1128/microbiolspec.RWR-0004-2017
Echols H. (1972). Developmental pathways for the temperate phage: Lysis vs lysogeny. Annu. Rev. Genet. 6, 157–190. doi: 10.1146/annurev.ge.06.120172.001105
Ellis M. J., Trussler R. S., Haniford D. B. (2015). Hfq binds directly to the ribosome-binding site of IS10 transposase mRNA to inhibit translation. Mol. Microbiol. 96, 633–650. doi: 10.1111/mmi.12961
Fonseca M. V., Swanson M. S. (2014). Nutrient salvaging and metabolism by the intracellular pathogen Legionella pneumophila. Front. Cell. Infect. Microbiol. 4:12. doi: 10.3389/fcimb.2014.00012
Georg J., Hess W. R. (2011). cis-antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 75, 286–300. doi: 10.1128/MMBR.00032-10
Georg J., Hess W. R. (2018). Widespread antisense transcription in prokaryotes. Microbiol. Spectr. 6, RWR–0029. doi: 10.1128/microbiolspec.RWR-0029-2018
Georg J., Voss B., Scholz I., Mitschke J., Wilde A., Hess W. R. (2009). Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 5, 305. doi: 10.1038/msb.2009.63
Gerdes K., Wagner E. G. (2007). RNA antitoxins. Curr. Opin. Microbiol. 10, 117–124. doi: 10.1016/j.mib.2007.03.003
Gerdes K., Thisted T., Martinussen J. (1990). Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol. Microbiol. 4, 1807–1818. doi: 10.1111/j.1365-2958.1990.tb02029.x
Giangrossi M., Prosseda G., Tran C. N., Brandi A., Colonna B., Falconi M. (2010). A novel antisense RNA regulates at transcriptional level the virulence gene icsA of Shigella flexneri. Nucleic. Acids Res. 38, 3362–3375. doi: 10.1093/nar/gkq025
Glover J. N., Chaulk S. G., Edwards R. A., Arthur D., Lu J., Frost L. S. (2015). The FinO family of bacterial RNA chaperones. Plasmid 78, 79–87. doi: 10.1016/j.plasmid.2014.07.003
Good L., Stach J. E. (2011). Synthetic RNA silencing in bacteria - antimicrobial discovery and resistance breaking. Front. Microbiol. 2, 185. doi: 10.3389/fmicb.2011.00185
Gottesman S., Storz G. (2011). Bacterial small RNA regulators: Versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3, a003798. doi: 10.1101/cshperspect.a003798
Gripenland J., Netterling S., Loh E., Tiensuu T., Toledo-Arana A., Johansson J. (2010). RNAs: Regulators of bacterial virulence. Nat. Rev. Microbiol. 8, 857–866. doi: 10.1038/nrmicro2457
Harrison E., Brockhurst M. A. (2017). Ecological and evolutionary benefits of temperate phage: What does or doesn’t kill you makes you stronger. Bioessays 39:1700112. doi: 10.1002/bies.201700112
Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., et al. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103. doi: 10.1038/35074106
Howard-Varona C., Hargreaves K. R., Abedon S. T., Sullivan M. B. (2017). Lysogeny in nature: Mechanisms, impact and ecology of temperate phages. ISME J. 11, 1511–1520. doi: 10.1038/ismej.2017.16
Hoynes-O’Connor A., Moon T. S. (2016). Development of design rules for reliable antisense RNA behavior in E. coli. ACS Synth. Biol. 5, 1441–1454. doi: 10.1021/acssynbio.6b00036
Jerome L. J., van Biesen T., Frost L. S. (1999). Degradation of FinP antisense RNA from F-like plasmids: The RNA-binding protein, FinO, protects FinP from ribonuclease E. J. Mol. Biol. 285, 1457–1473. doi: 10.1006/jmbi.1998.2404
Ji Y., Yin D., Fox B., Holmes D. J., Payne D., Rosenberg M. (2004). Validation of antibacterial mechanism of action using regulated antisense RNA expression in Staphylococcus aureus. FEMS Microbiol. Lett. 231, 177–184. doi: 10.1016/S0378-1097(03)00931-5
Johnson J. M., Edwards S., Shoemaker D., Schadt E. E. (2005). Dark matter in the genome: Evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 21, 93–102. doi: 10.1016/j.tig.2004.12.009
Kacharia F. R., Millar J. A., Raghavan R. (2017). Emergence of new sRNAs in enteric bacteria is associated with low expression and rapid evolution. J. Mol. Evol. 84, 204–213. doi: 10.1007/s00239-017-9793-9
Kalindamar S., Lu J., Abdelhamed H., Tekedar H. C., Lawrence M. L., Karsi A. (2019). Transposon mutagenesis and identification of mutated genes in growth-delayed Edwardsiella ictaluri. BMC Microbiol. 19, 55. doi: 10.1186/s12866-019-1429-3
Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., et al. (2005). Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566. doi: 10.1126/science.1112009
Kiem S., Oh W. S., Peck K. R., Lee N. Y., Lee J., Song J., et al. (2004). Phase variation of biofilm formation in Staphylococcus aureus by IS256 insertion and its impact on the capacity adhering to polyurethane surface. J. Korean Med. Sci. 19, 779–782. doi: 10.3346/jkms.2004.19.6.779
Koraimann G. (2018). Spread and persistence of virulence and antibiotic resistance genes: A ride on the F plasmid conjugation module. EcoSal Plus 8. doi: 10.1128/ecosalplus.ESP-0003-2018
Lacatena R. M., Cesareni G. (1981). Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature 294, 623–626. doi: 10.1038/294623a0
Lapierre L., Mollet B., Germond J. (2002). Regulation and adaptive evolution of lactose operon expression in Lactobacillus delbrueckii. J. Bacteriol. 184, 928–935. doi: 10.1128/jb.184.4.928-935.2002
Lebreton A., Cossart P. (2017). RNA- and protein-mediated control of Listeria monocytogenes virulence gene expression. RNA Biol. 14, 460–470. doi: 10.1080/15476286.2016.1189069
Lejars M., Kobayashi A., Hajnsdorf E. (2019). Physiological roles of antisense RNAs in prokaryotes. Biochimie 164, 3–16. doi: 10.1016/j.biochi.2019.04.015
León M., Bastías R. (2015). Virulence reduction in bacteriophage resistant bacteria. Front. Microbiol. 6, 343. doi: 10.3389/fmicb.2015.00343
León-Sicairos N., Angulo-Zamudio U. A., de la Garza M., Velázquez-Román J., Flores-Villaseñor H. M., Canizalez-Román A. (2015). Strategies of Vibrio parahaemolyticus to acquire nutritional iron during host colonization. Front. Microbiol. 6:702. doi: 10.3389/fmicb.2015.00702
Liao S. M., Wu T. H., Chiang C. H., Susskind M. M., McClure W. R. (1987). Control of gene expression in bacteriophage P22 by a small antisense RNA. I. Characterization in vitro of the Psar promoter and the sar RNA transcript. Genes Dev. 1, 197–203. doi: 10.1101/gad.1.2.197
Lybecker M., Bilusic I., Raghavan R. (2014). Pervasive transcription: Detecting functional RNAs in bacteria. Transcription 5, e944039. doi: 10.4161/21541272.2014.944039
Ma C., Simons R. W. (1990). The IS10 antisense RNA blocks ribosome binding at the transposase translation initiation site. EMBO J. 9, 1267–1274. doi: 10.1002/j.1460-2075.1990.tb08235.x
Mellin J. R., Tiensuu T., Bécavin C., Gouin E., Johansson J., Cossart P. (2013). A riboswitch-regulated antisense RNA in Listeria monocytogenes. Proc. Natl. Acad. Sci. 110, 13132–13137. doi: 10.1073/pnas.1304795110
Millan A. S., MacLean R. C. (2017). Fitness costs of plasmids: A limit to plasmid transmission. Microbiol. Spectr. 5, MTBP–0016. doi: 10.1128/microbiolspec.MTBP-0016-2017
Moffatt J. H., Harper M., Adler B., Nation R. L., Li J., Boyce J. D. (2011). Insertion sequence ISAba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 3022–3024. doi: 10.1128/AAC.01732-10
Murray G. L., Morel V., Cerqueira G. M., Croda J., Srikram A., Henry R., et al. (2009). Genome-wide transposon mutagenesis in pathogenic Leptospira species. Infect. Immun. 77, 810–816. doi: 10.1128/IAI.01293-08
Murray J. L., Kwon T., Marcotte E. M., Whiteley M. (2015). Intrinsic antimicrobial resistance determinants in the superbug Pseudomonas aeruginosa. mBio 6, e01603–e01615. doi: 10.1128/mBio.01603-15
Naka H., López C. S., Crosa J. H. (2010). Role of the pJM1 plasmid-encoded transport proteins FatB, C and D in ferric anguibactin uptake in the fish pathogen Vibrio anguillarum. Environ. Microbiol. Rep. 2, 104–111. doi: 10.1111/j.1758-2229.2009.00110.x
Nikravesh A., Dryselius R., Faridani O. R., Goh S., Sadeghizadeh M., Behmanesh M., et al. (2007). Antisense PNA accumulates in Escherichia coli and mediates a long post-antibiotic effect. Mol. Ther. 15, 1537–1542. doi: 10.1038/sj.mt.6300209
Ogata R. T., Levine R. R. (1980). Characterization of complement resistance in Escherichia coli conferred by the antibiotic resistance plasmid R100. J. Immunol. 125, 1494–1498.
Parrow N. L., Fleming R. E., Minnick M. F. (2013). Sequestration and scavenging of iron in infection. Infect. Immun. 81, 3503–3514. doi: 10.1128/IAI.00602-13
Pecota D. C., Osapay G., Selsted M. E., Wood T. K. (2003). Antimicrobial properties of the Escherichia coli R1 plasmid host killing peptide. J. Biotechnol. 100, 1–12. doi: 10.1016/s0168-1656(02)00240-7
Perez M., Calles-Enríquez M., del Rio B., Ladero V., Martín M. C., Fernández M., et al. (2015). IS256 abolishes gelatinase activity and biofilm formation in a mutant of the nosocomial pathogen Enterococcus faecalis V583. Can. J. Microbiol. 61, 517–519. doi: 10.1139/cjm-2015-0090
Perocchi F., Xu Z., Clauder-Münster S., Steinmetz L. M. (2007). Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 35, e128. doi: 10.1093/nar/gkm683
Pluta R., Espinosa M. (2018). Antisense and yet sensitive: Copy number control of rolling circle-replicating plasmids by small RNAs. Wiley Interdiscip. Rev. RNA 9, e1500. doi: 10.1002/wrna.1500
Raghavan R., Sloan D. B., Ochman H. (2012). Antisense transcription is pervasive but rarely conserved in enteric bacteria. MBio 3, e00156–e00112. doi: 10.1128/mBio.00156-12
Riley M. A., Gordon D. M. (1999). The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 7, 129–133. doi: 10.1016/s0966-842x(99)01459-6
Saberi F., Kamali M., Najafi A., Yazdanparast A., Moghaddam M. M. (2016). Natural antisense RNAs as mRNA regulatory elements in bacteria: A review on function and applications. Cell Mol. Biol. Lett. 21, 6. doi: 10.1186/s11658-016-0007-z
Schaefer K. L., McClure W. R. (1997). Antisense RNA control of gene expression in bacteriophage P22. I. Structures of sar RNA and its target, ant mRNA. RNA 3, 141–156.
Seed K. D., Yen M., Shapiro J., Hilaire I. J., Charles R. C., Teng J. E., et al. (2014). Evolutionary consequences of intra-patient phage predation on microbial populations. Elife 3, e03497. doi: 10.7554/eLife.03497
Sengupta M., Austin S. (2011). Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect. Immun. 79, 2502–2509. doi: 10.1128/IAI.00127-11
Sesto N., Wurtzel O., Archambaud C., Sorek R., Cossart P. (2013). The excludon: A new concept in bacterial antisense RNA-mediated gene regulation. Nat. Rev. Microbiol. 11, 75–82. doi: 10.1038/nrmicro2934
Spangler R., Zhang S. P., Krueger J., Zubay G. (1985). Colicin synthesis and cell death. J. Bacteriol. 163, 167–173. doi: 10.1128/JB.163.1.167-173.1985
Storz G., Vogel J., Wassarman K. M. (2011). Regulation by small RNAs in bacteria: Expanding frontiers. Mol. Cell. 43, 880–891. doi: 10.1016/j.pestbp.2011.02.012.Investigations
Sturino J. M., Klaenhammer T. R. (2006). Engineered bacteriophage-defence systems in bioprocessing. Nat. Rev. Microbiol. 4, 395–404. doi: 10.1038/nrmicro1393
Suzukia Y., Ishimoto T., Fujita S., Kiryu S., Wada M., Akatsuka T., et al. (2020). Antimicrobial antisense RNA delivery to F-pili producing multidrug-resistant bacteria via a genetically engineered bacteriophage. Biochem. Biophys. Res. Commun. 530, 533–540. doi: 10.1016/j.bbrc.2020.06.088
Thomason M., Storz G. (2010). Bacterial antisense RNAs: How many are there and what are they doing? Annu. Rev. Genet. 44, 167–188. doi: 10.1146/annurev-genet-102209-163523
Thomason M. K., Bischler T., Eisenbart S. K., Förstner K. U., Zhang A., Herbig A., et al. (2015). Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 197, 18–28. doi: 10.1128/JB.02096-14
Toledo-Arana A., Dussurget O., Nikitas G., Sesto N., Guet-Revillet H., Balestrino D., et al. (2009). The Listeria transcriptional landscape from saprophytism to virulence. Nature 459, 950–956. doi: 10.1038/nature08080
Toraya T., Honda S., Fukui S. (1979). Fermentation of 1,2-propanediol with 1,2-ethanediol by some genera of Enterobacteriaceae, involving coenzyme B12-dependent diol dehydratase. J. Bacteriol. 139, 39–47. doi: 10.1128/JB.139.1.39-47.1979
Touchon M., Bernheim A., Rocha E. P. (2016). Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 10, 2744–2754. doi: 10.1038/ismej.2016.47
Updegrove T. B., Shabalina S. A., Storz G. (2015). How do base-pairing small RNAs evolve? FEMS Microbiol. Rev. 39, 379–391. doi: 10.1093/femsre/fuv014
Vogwill T., MacLean R. C. (2015). The genetic basis of the fitness costs of antimicrobial resistance: A meta-analysis approach. Evol. Appl. 8, 284–295. doi: 10.1111/eva.12202
Waters L. S., Storz G. (2009). Regulatory RNAs in bacteria. Cell 136, 615–628. doi: 10.1016/j.cell.2009.01.043
Keywords: asRNA, anti-sense RNA, pathogenesis, virulence, bacteria, regulation, riboregulation
Citation: Millar JA and Raghavan R (2021) Modulation of Bacterial Fitness and Virulence Through Antisense RNAs. Front. Cell. Infect. Microbiol. 10:596277. doi: 10.3389/fcimb.2020.596277
Received: 18 August 2020; Accepted: 30 December 2020;
Published: 11 February 2021.
Edited by:
D. Scott Samuels, University of Montana, United StatesReviewed by:
Meghan Catherine Lybecker, University of Colorado Colorado Springs, United StatesClayton Caswell, Virginia Tech, United States
Copyright © 2021 Millar and Raghavan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jess A. Millar, amFtaWxsYXJAdW1pY2guZWR1
 Jess A. Millar
Jess A. Millar Rahul Raghavan
Rahul Raghavan