
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 21 December 2020
Sec. Microbiome in Health and Disease
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.587284
 Lei Xiang1†
Lei Xiang1† Yanfeng Lou2†
Yanfeng Lou2† Lingyu Liu3†
Lingyu Liu3† Yuanling Liu4
Yuanling Liu4 Weizheng Zhang5
Weizheng Zhang5 Jianxin Deng6
Jianxin Deng6 Yubin Guan7
Yubin Guan7 Miaoqin She8
Miaoqin She8 Xinchao You9
Xinchao You9 Minqi Liu7
Minqi Liu7 Hongwei Li10
Hongwei Li10 Xiaosong Xu7*
Xiaosong Xu7* Fang Liu2*
Fang Liu2* Xiangsheng Cai7,11*
Xiangsheng Cai7,11*Increasing evidence suggests that features of the gut microbiota correlate with ischemic stroke. However, the specific characteristics of the gut microbiota in patients suffering different types of ischemic stroke, or recovering from such strokes, have rarely been studied, and potential microbiotic predictors of different types of stroke have seldom been analyzed. We subjected fecal specimens from patients with lacunar or non-lacunar acute ischemic infarctions, and those recovering from such strokes, to bacterial 16S rRNA sequencing and compared the results to those of healthy volunteers. We identified microbial markers of different types of ischemic stroke and verified that these were of diagnostic utility. Patients with two types of ischemic stroke, and those recovering from ischemic stroke, exhibited significant shifts in microbiotic diversities compared to healthy subjects. Cluster of Orthologous Groups of Proteins (COG) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses revealed reduced metabolic and transport-related pathway activities in ischemic stroke patients. We performed fivefold cross-validation using a Random Forest model to identify two optimal bacterial species (operational taxonomic units; OTUs) serving as markers of lacunar infarction; these were Lachnospiraceae (OTU_45) and Bacteroides (OTU_4), and the areas under the receiver operating characteristic curves (AUCs under the ROCs) were 0.881 and 0.872 respectively. In terms of non-lacunar acute ischemic infarction detection, the two optimal species were Bilophila (OTU_330) and Lachnospiraceae (OTU_338); the AUCs under the ROCs were 0.985 and 0.929 respectively. In post-ischemic stroke patients, the three optimal species were Pseudomonas (OTU_35), Sphingomonadaceae (OTU_303), and Akkermansia (OTU_9); the AUCs under the ROCs were 1, 0.897, and 0.846 respectively. Notably, the gut microbial markers were of considerable value for utility when diagnosing lacunar infarction, non-lacunar acute ischemic infarction, and post-ischemic stroke. This study is the first to characterize the gut microbiotic profiles of patients with lacunar or non-lacunar, acute ischemic strokes, and those recovering from stroke, and to identify microbiotic predictors of such strokes.
Ischemic stroke is a major cause of death and severe neurological disability; global post-stroke mortality is 39% (Wang et al., 2017). Unhealthy living habits, excessive stress, and aging have increased the global incidence of cerebral ischemia. Acute ischemic infarction is the most common type of stroke, associated with high rates of death and disability (Paciaroni et al., 2008). Acute ischemic infarction subtypes were classified based on clinical data using the Oxfordshire Community Stroke Project (OCSP) classification scheme as total anterior circulation infarcts [TACIs], partial anterior circulation infarcts [PACIs], posterior circulation infarcts, and lacunar infarcts [LACIs]) (Sandset et al., 2015). Lacunar infarctions are small (<2-cm-diameter) infarctions that account for approximately 25% of all strokes; LIs differ pathologically from large artery strokes, including TACI, PACI, and POCI (Das et al., 2019). Thus, some studies divide acute ischemic infarctions into lacunar and non-lacunar acute ischemic infarctions (Regenhardt et al., 2019; Zhou et al., 2020). Post-ischemic stroke patients have experienced acute ischemic stroke and often deal with severe sequelae including cognitive and anxio-depressive disorders, fatigue, and restricted participation in daily life (Zhang et al., 2018). Though the progress were made in the treatments of stroke, the prognosis is poor, most ischemic stroke patients are diagnosed at advanced stages causing high risks of disability and mortality. Therefore, early diagnostic measures of ischemic stroke need to be explored so as to greatly improve the management of the disease.
Gut microbiotic alterations are newly identified risk factors for stroke; patients exhibit significant dysbiosis in terms of both microbial diversity and composition (Yin et al., 2015). A recent study found that ischemic stroke altered the gut microbiome, reduced microbiome diversity, and increased the immune response (Singh and Roth, 2016). Ischemic stroke triggered maladjustment of the mouse cecal microbiota within 72 h (Houlden et al., 2016). Thus, the gut microbiota was presumed to play roles in stroke initiation and development (Zhang et al., 2015; Langdon et al., 2016). Moreover, many neurological disorders (Alzheimer’s and Parkinson’s diseases, depression, and multiple sclerosis) are commonly accompanied by gastrointestinal symptoms (McDonald and Cervenka, 2018). Therefore, the microbiota-brain axis is proposed and is thought to constitute a bidirectional informational network linking the gut microbiota and the brain (Wang and Wang, 2016). Despite extensive analysis of the link between the gut microbiome and stroke, the microbiotic features of different types of stroke have rarely been studied. Moreover, it remains unclear whether the overall gut microbiota, or only specific bacterial species, affect stroke outcomes; no report has described microbiotic or bacterial predictors of stroke. Here, we describe changes in gut microbiotic species in patients with lacunar or non-lacunar, acute ischemic strokes, and those recovering from acute ischemic strokes, and we identify potential bacterial predictors of such strokes.
This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangdong Pharmaceutical University. All participants signed written informed consent prior to enrolment. Fecal samples were collected from 16 healthy volunteers (N group, n = 16); 20 patients with ischemic stroke: lacunar infarction (LI group, n = 10), non-lacunar acute ischemic infarction (AI group, n = 10); and 10 post-ischemic stroke patients who had undergone 15 days of treatment after acute ischemic stroke (PI group, n = 10). All of age, sex and the body mass index were comparable among the groups (Table 1; all P>0.05). The enrolled stroke patients were satisfied the clinical diagnostic criteria and were confirmed by magnetic resonance imaging (MRI) or brain computed tomography (CT). The exclusion criteria included respiratory or renal failure, a recent cardiac event, an immune system condition, intestinal disease or severe liver dysfunction, or the use of probiotics or antibiotics within 1 month prior to admission. The healthy volunteers who had not taken antibiotics, probiotics, or possibly confounding drugs within the prior 1 month were enrolled.
Fresh fecal samples were collected into Eppendorf tubes and frozen at −80°C. Bacterial genomic DNA was extracted using QIAamp DNA Stool Mini Kits according to the manufacturer’s instructions. The V3 variable regions of 16S rDNA genes were subjected to PCR amplification as described previously (Muyzer et al., 1993; Fan et al., 2016): predenaturation at 94°C for 5 min; 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 72°C for 30 s; and a final extension at 72°C for 10 min. Fragments were purified via agarose gel electrophoresis and re-electrophoresed to determine DNA concentrations using the Qubit 2 method. The mixed products were sent to our Sequencing Institute.
The data were filtered by Mothur software and clustered into operational taxonomic units (OTUs; species) at a similarity level of 97% using Quantitative Insights into Microbial Ecology (QIIMEv) software ver. 1.80 (Houlden et al., 2016). All OTUs in the discovery and validation sets were recorded. Gut microbiota alpha diversity indices (the Observed Species, Chao, ACE, Shannon, and Simpson indices) were calculated by Mothur ver. 1.31.2 software and dilution curves and box diagrams created with the aid of R software ver. 3.1.1. Beta diversity analysis was used to compare microbiotic species diversities. Principal component analysis (PCA) was performed with the aid of the Ade4 R package; this revealed similarities among relative microbial abundances. To compare microbial compositions among samples, principal coordinate analysis (PCoA) and nonmetric multidimensional scaling (NMDS) were applied, and the Bray-Curtis distances were used to generate two-dimensional plots.
The microbial compositions after grouping were compared at the phylum, class, and genus levels. Beta diversity heatmaps were plotted using the NMF package of R. The Bray-Curtis distances lay between 0 and 1; higher values indicated greater between-sample differences. To identify bacteria that differed significantly among groups, the taxon summaries were reformatted and linear discriminant analysis effect sizes (LEfSes) determined with the aid of the Huttenhower Lab Galaxy Server. In this setting, the Kruskal-Wallis sum-rank test (α = 0.05) was initially used to detect taxa exhibiting significantly different abundances. The false discovery rate (FDR) was the corrected P-value; an FDR<0.05 was considered significant. Biological consistency was then investigated via pairwise testing among subclasses using the Wilcoxon rank-sum test. Finally, linear discriminant analysis (LDA) was employed to estimate the effect sizes of differentially abundant gut microbiota. The logarithmic LDA threshold for discrimination was 2.0. The Clusters of Orthologous Group (COG) annotations were determined using PICRUSt2 software. The affected Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathways were identified using the KEGG Automatic Annotation Server (KAAS).
The Wilcoxon rank-sum test (two groups) or Kruskal-Wallis test (multiple groups) of R ver. 3.0.3 software were used for inter-group comparisons. The t- test, chi-squared test, and the Spearman rank correlation analysis of SPSS ver. 19.0 were employed as appropriate. A P-value <0.05 was considered to reflect statistical significance.
A species-based classifier was trained using the Random Forest package of R. The model was employed for fivefold cross-validation of the relative species abundance profile. Case probabilities were calculated by drawing receiver operating characteristic (ROC) curves.
The OTUs annotated for subsequent analyses included 22 phyla, 116 families, and 217 genera of gut microbes inferred via V4 amplicon sequences (39 to 297 base pairs); the among-sample similarity was 97% (Figure 1A). The total OTUs of the LI, PI, and AI groups (at the 97% similarity level) were 1,094, 1,528, and 1,650 respectively, and 2,053 for the control group. OTUs shared by the stroke and control groups numbered 360, as revealed by a Venn diagram (Figure 1B).
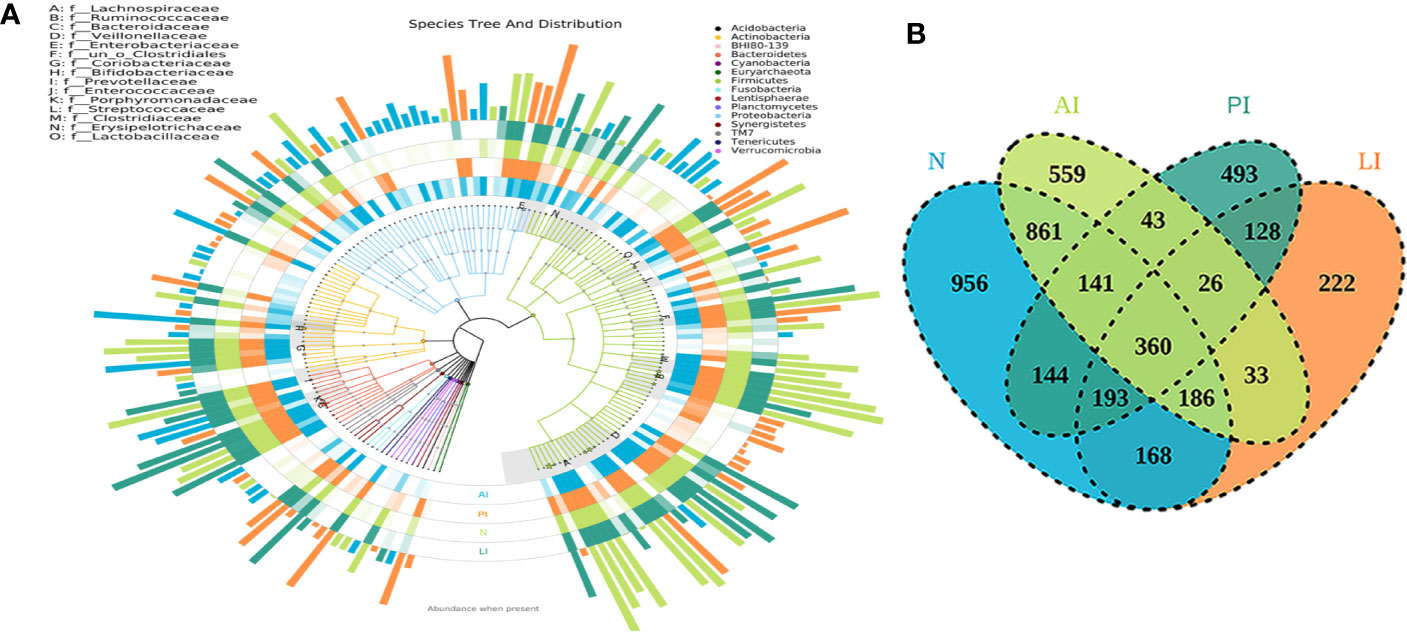
Figure 1 (A) The species trees and distributions of gut microbial communities. (B) A Venn diagram showing OTU similarities and differences among the groups.
In terms of alpha diversity, the chao1 richness index of the AI group differed significantly from those of the LI, PI, and control groups (61.3 P = 0.0; 51.9 P = 0.0002; and 28.6375 P = 0.0272). The Observed_species index of the AI group differed significantly from those of the LI and PI groups (52.75 P = 0.005; 45.95 P = 0.0015). The Shannon and Simpson indices of the PI group differed significantly from those of the AI and control groups (Shannon 35.8 P = 0.0369, 36.3375 P = 0.0369; Simpson 35 P = 0.0386 40.15 P = 0.0216) (Figure 2A). The ACE index of the AI group differed significantly from those of the LI, PI, and control groups (60.4 P = 0.0; 51.6 P = 0.0002; 26.8125 P = 0.0381). PCA and PCoA indicated that the AI microbiome differed significantly from those of the LI and PI groups (which were clustered), and was comparable to that of healthy controls (Figures 2B, C). Analysis of similarities (ANOSIM) indicated that the gut microbiotic structure differed significantly among the groups (ANOSIM, r = 0.371, P = 0.001) (Figure 2D). NMDS analysis based on the Bray-Curtis distances between microbial genera revealed significant differences between AI patients and the other three groups (Figure 2E).
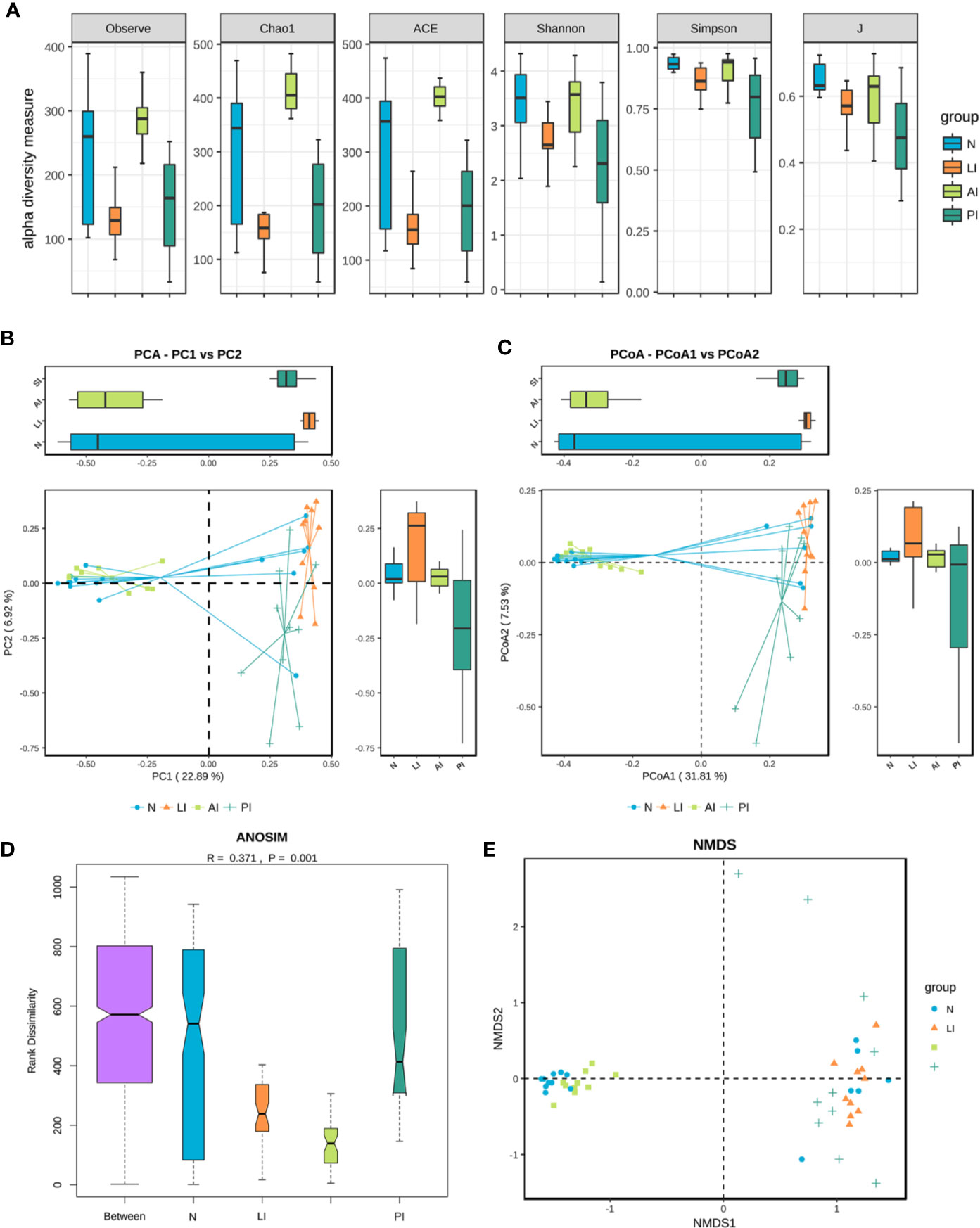
Figure 2 (A) Gut microbiotic alpha and beta diversity indices of ischemic stroke patients. The Observed_species, Chao1, ACE, Shannon, Simpson and J index values. (B) PCA scores based on the relative abundances of OTUs (at the 97% similarity level). (C) PCoA analysis based on weighted Unifrac distances. (D) Analysis of similarities. (E) Non-metric multi-dimensional scaling. Each dot represents a sample; the group is identified in the legend. In the graphs, closer samples are more similar.
The gut microbiota differed among the groups at the phylum, family, and genus level. At the phylum level, Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria were the most common phyla of the four groups, comprising 99.73% of all gut bacteria in the AI group; the figures for the PI, LI, and control groups were 87.97, 97.57, and 99.3% respectively (Figures 3A, B). All stroke patients exhibited fewer Firmicutes than controls (P<0.05). LI group contained more Cyanobacteria and Fusobacteria than the other groups. AI group exhibited markedly higher Actinobacteria levels (P<0.05) than the other groups. Verrucomicrobia, Synergistetes, and Proteobacteria were significantly enriched in the PI compared to the other groups (P<0.05). The ratio of Firmicutes : Bacteroidetes was lower in AI and LI groups but higher in PI group compared to healthy controls (1.30 in AI group, 3.47 in PI group, 1.47 in LI group, and 2.86 in control group).
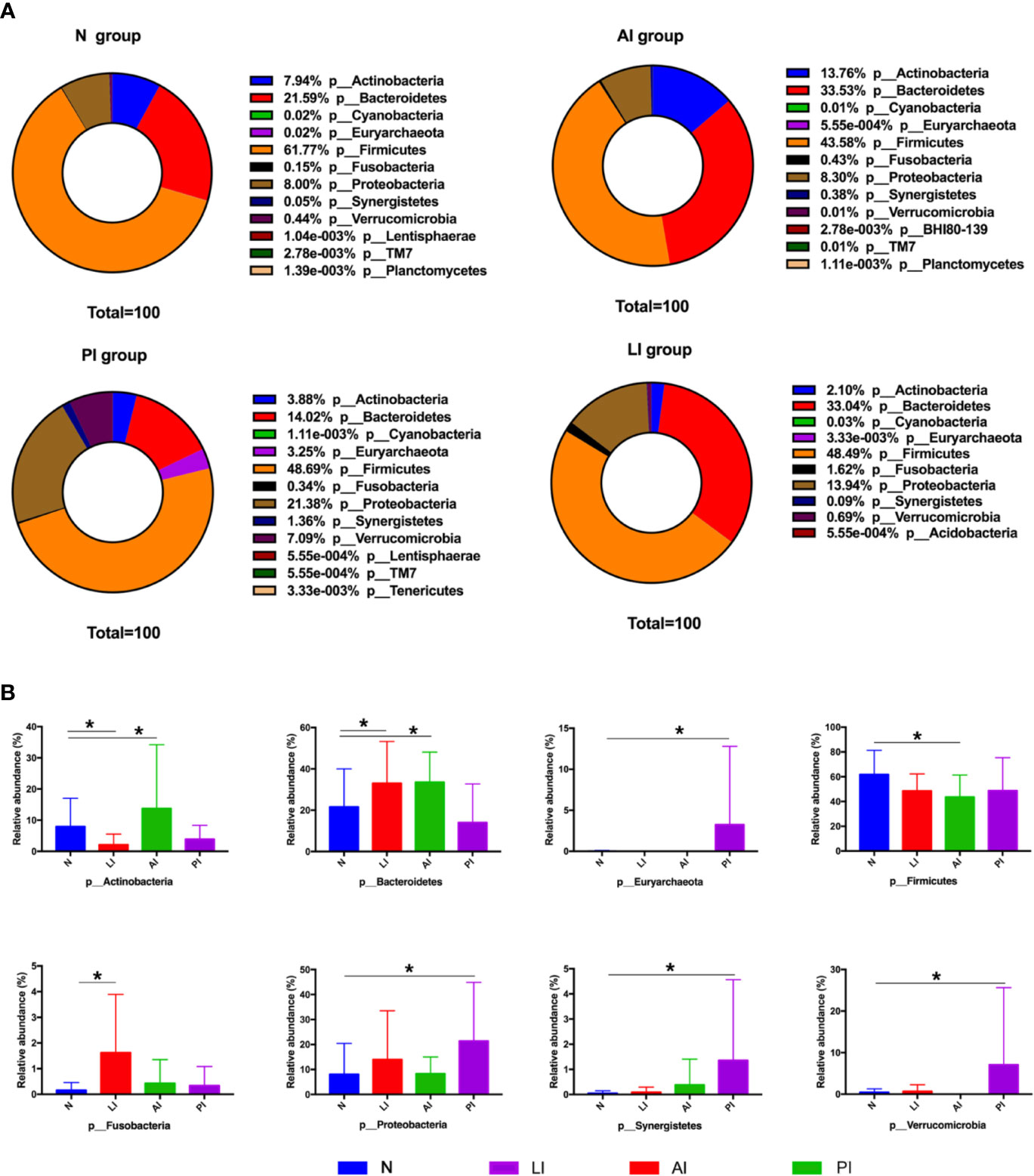
Figure 3 (A) Taxonomic profiling assigned most OTUs to Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Verrucomicrobia, and Fusobacteria at the phylum level. (B) Among-group differences at the phylum level. *P < 0.05.
At the family level, individual variations were more marked; the dominant taxa differed among the groups (Figure 4A). Of all families identified, 74, 60, 51, and 75 of the dominant families were present in AI, PI, LI, and control groups respectively. Bacteroidaceae, Lachnospiraceae, Ruminococcaceae, Bifidobacteriaceae, Enterobacteriaceae, Coriobacteriaceae, Veillonellaceae, and Prevotellaceae were the eight most abundant microbiotic components in all groups. Bacteroidaceae levels were significantly higher in AI and LI groups (P<0.05) compared to the other groups. Bifidobacteriaceae and Coriobacteriaceae were most abundant in AI group (P<0.05). Veillonellaceae levels were highest in the LI group (P<0.05). Enterobacteriaceae, Enterococcaceae, Lactobacillaceae, and Verrucomicrobiaceae levels were highest in PI group (P<0.05). Lachnospiraceae, Ruminococcaceae, and Prevotellaceae levels were maximal in control group (P<0.05).
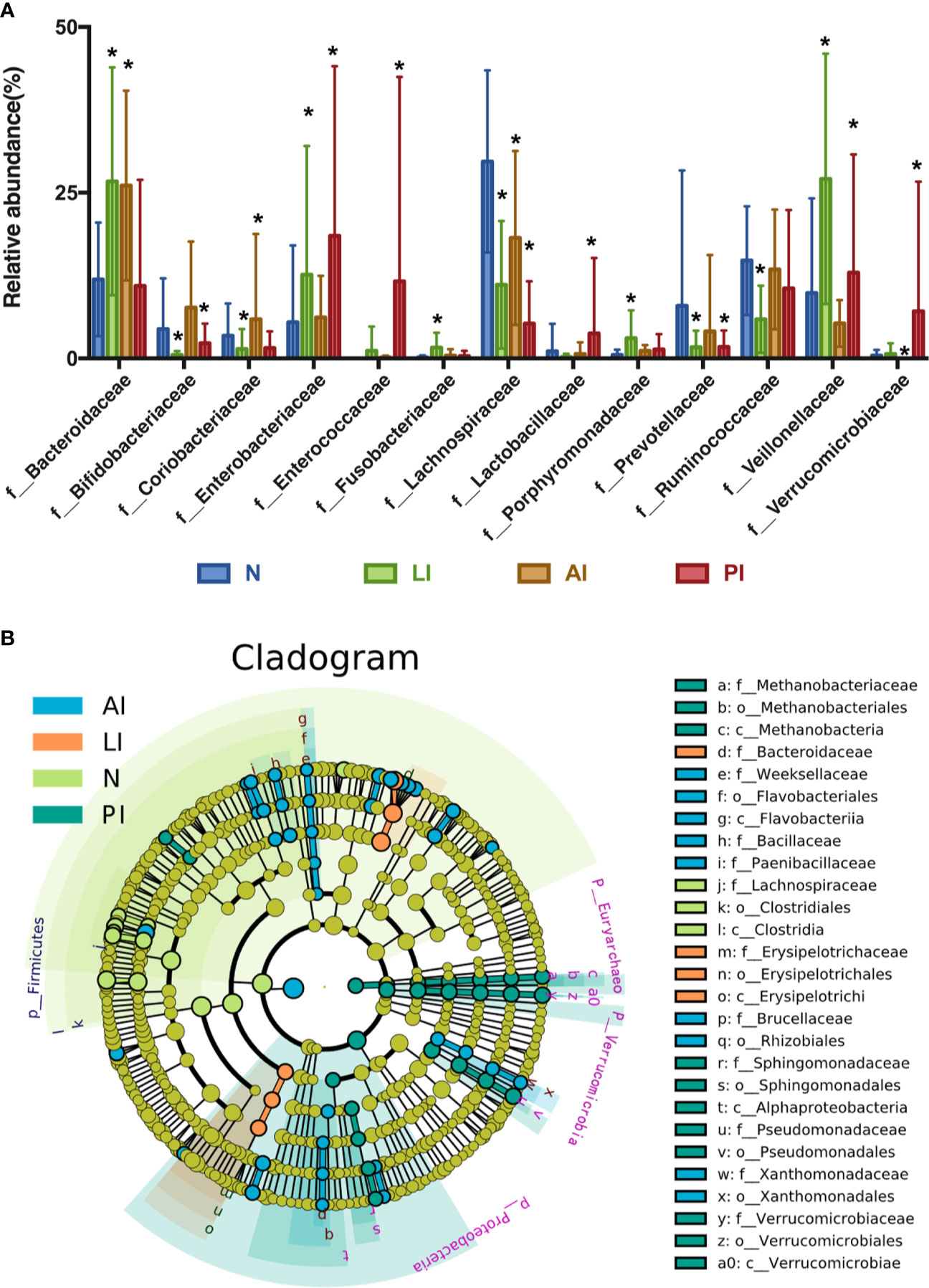
Figure 4 (A) The most abundant microbiotic components (at the family level) of each group. *P < 0.05. (B) LEfSE analysis reveals bacterial taxa, the levels of which differ significantly among groups.
LEfSE analysis was used to identify bacterial taxa, the levels of which differed significantly among groups (Figure 4B). Weeksellaceae, Bacillaceae, Paenibaciiaceae, Brucellaceae, and Xanthomnadaceae were significantly more abundant in AI group. Bacteroidaceae and Erysipelotrichaceae levels were highest in LI group. Methanobacteriaceae, Sphingomonadaceae, Pseudomonadaceae, and Verrucomicrobiaceae levels were maximal in PI group. Lactobacillaceae was most abundant in control group.
The genus-level compositions were more complex; the levels of many genera differed significantly among the groups (Figures 5A, B). In control group, Lachnospiraceae, Bacteroides, Ruminococcaceae, Prevotella, Blautia, Enterobacteriaceae, Bifidobacterium, and Ruminococcus, were the eight principal genera (of 132). In LI group, the eight predominated genera (of 100) were Bacteroides, Enterobacteriaceae, Phascolarctobacterium, Megasphaera, Lachnospiraceae, Acidaminococcus, Lachnospira, and Parabacteroides. In AI group, the eight common genera (of 136) were Bacteroides, Ruminococcaceae, Bifidobacterium, Blautia, Enterobacteriaceae, Lachnospiraceae, Coriobacteriaceae, and Prevotella. In PI group, Enterobacteriaceae, Enterococcus, Bacteroides, Veillonella, Ruminococcaceae, Akkermansia, Lactobacillus, and Lachnospiraceae were eight predominated genera (of 121).
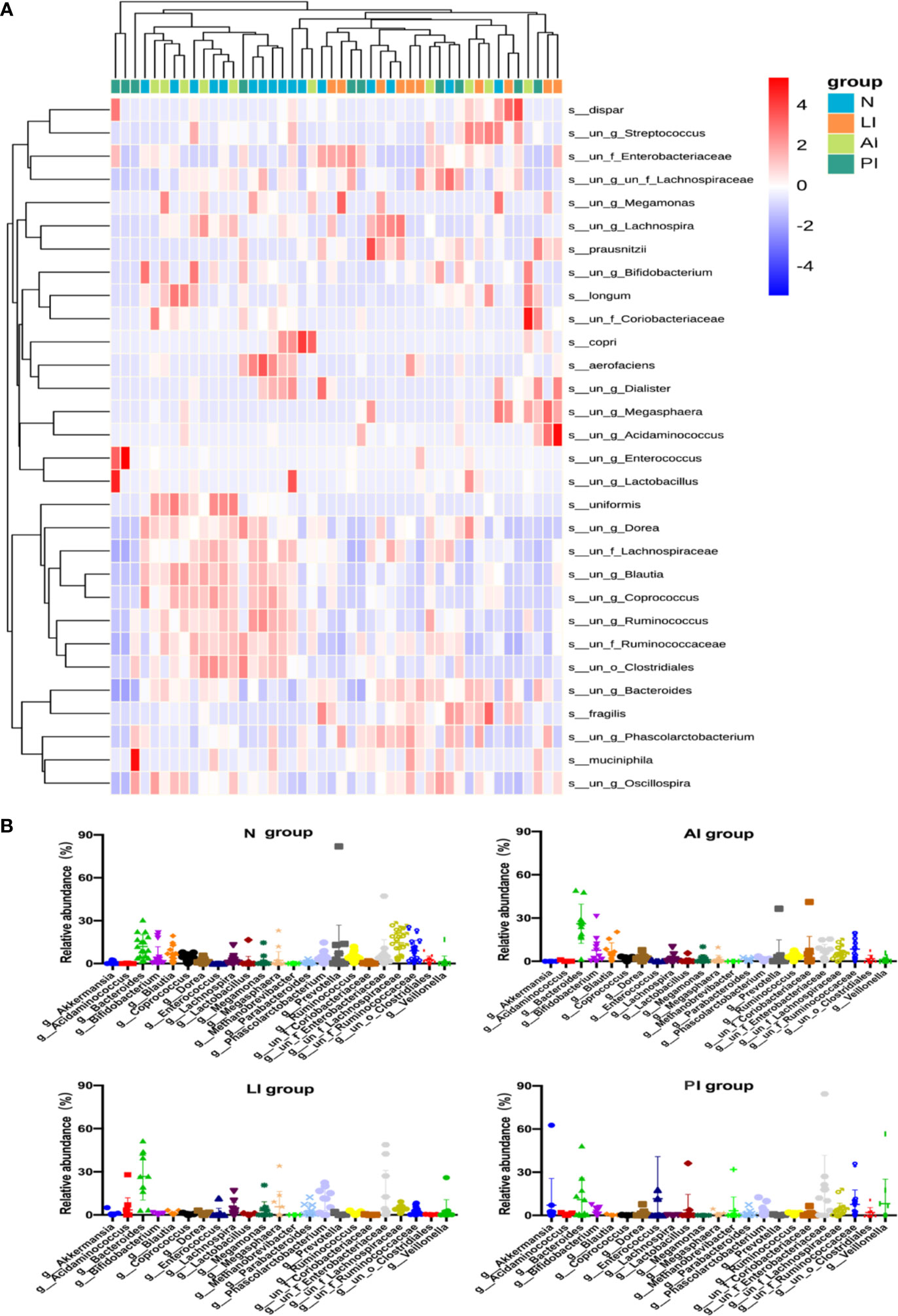
Figure 5 (A) A heat map showing the relative total abundances of the top 30 species, and the relative abundances of all species, at the genus level. (B) A taxonomic summary of the gut microbiota of each group at the genus level. Starring (*) indicates a significant difference between two groups.
We compared the COG and KEGG pathways among the groups; we sought differences in microbiotic functions between stroke patients and controls. Figure 6A shows that nine functional KEGG pathways were highly enriched in both AI patients and healthy controls. These included the pathways of folate biosynthesis, photosynthesis, peroxisomal action, the citrate cycle (TCA cycle), galactose metabolism, phenyltransferase, other glycan degradation, amino sugar and nucleotide sugar metabolism, and fructose and mannose metabolism. Figure 6B shows that 12 COG categories were highly enriched in AI patients and healthy subjects, including transposase (and its inactivated derivatives), the predicted Rossmann fold nucleotide-binding protein, the ribonucleotide reductase alpha subunit, the L-asparaginase/archaeal Glu-tRNAGln amidotransferase subunit D, the Mn2+ and Fe2+ transporters of the NRAMP family, the ABC-type transport system involved in Fe-S assembly, a permease component, the site-specific recombinase XerC, an exopolyphosphatase, 7,8-dihydro-6-hydroxymethylpterin-pyrophosphokinase, a predicted membrane protein, an enzyme related to GTP cyclohydrolase I, and a peptidylarginine deaminase and related enzymes. The transposase (and inactivated derivatives), predicted Rossmann fold nucleotide-binding protein, ribonucleotide reductase alpha subunit, and L-asparaginase/archaeal Glu-tRNAGln amidotransferase subunit D were significantly more abundant in the AI than the LI or PI groups (all P<0.05).
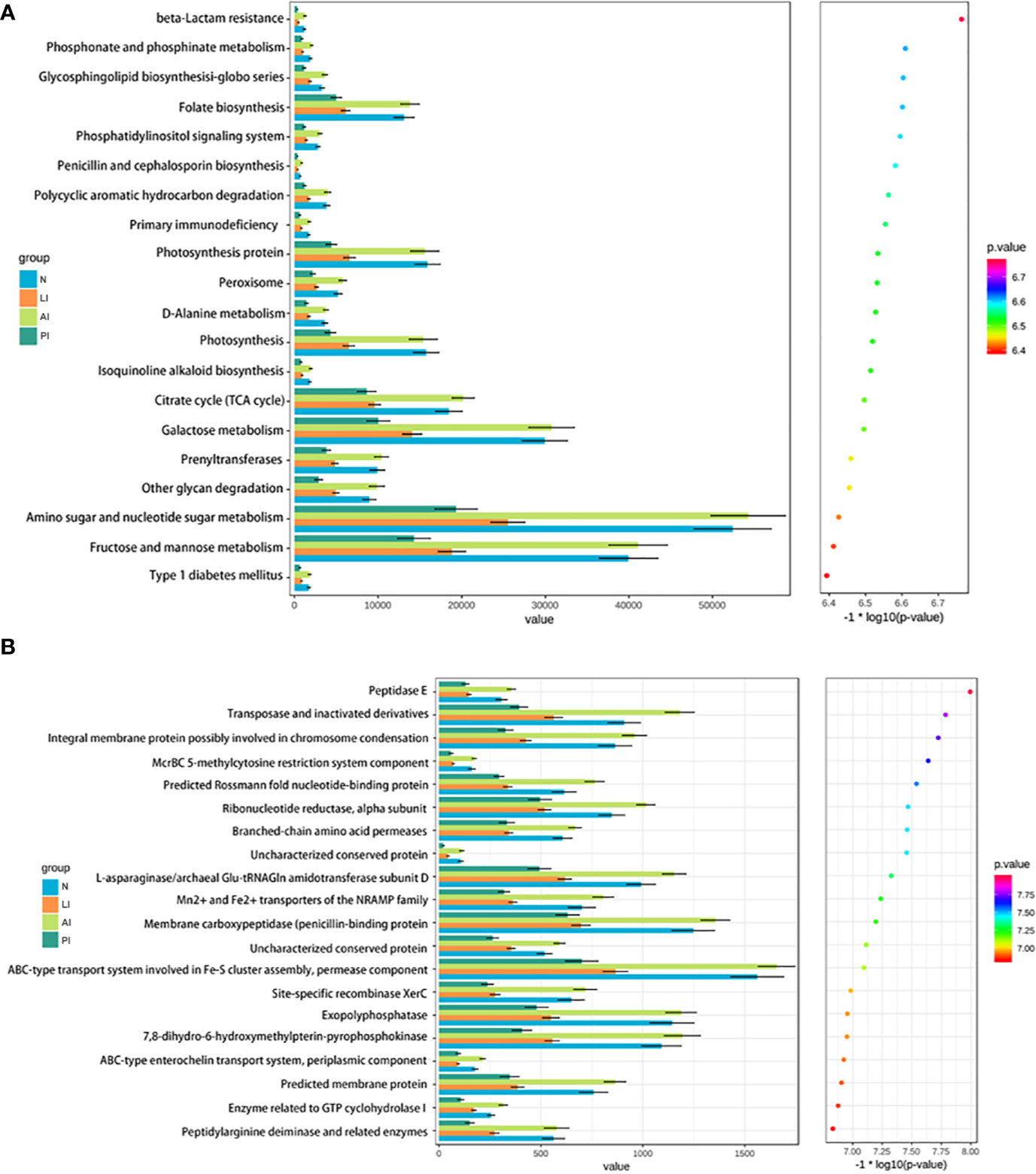
Figure 6 KEGG pathways (A) and COG categories (B) that was less frequent in stroke groups than healthy controls.
We used a Random Forest model to identify microbial profiles that optimally differentiated the ischemic stroke and healthy groups; the Random Forest is a robust machine-learning technique that can handle nonlinear relationships and dependencies among microbiotic features. We performed fivefold cross-validation five times using a validation set (Figures 7A–C). Two species afforded optimal LI detection: Lachnospiraceae (OTU_45) and Bacteroides (OTU_4); the areas under the receiver operating characteristic curves (AUCs under the ROCs) were 0.881 and 0.872 respectively (Figure 8A, Table 2). Two species optimally detected AI: Bilophila (OTU_330) and Lachnospiraceae (OTU_338); the AUCs under the ROCs were 0.985 and 0.929 respectively (Figure 8B, Table 2). Three species optimally detected PI: Pseudomonas (OTU_35), Sphingomonadaceae (OTU_303), and Akkermansia (OTU_9); the AUCs under the ROCs were 1, 0.897, and 0.846 respectively (Figure 8C, Table 2).
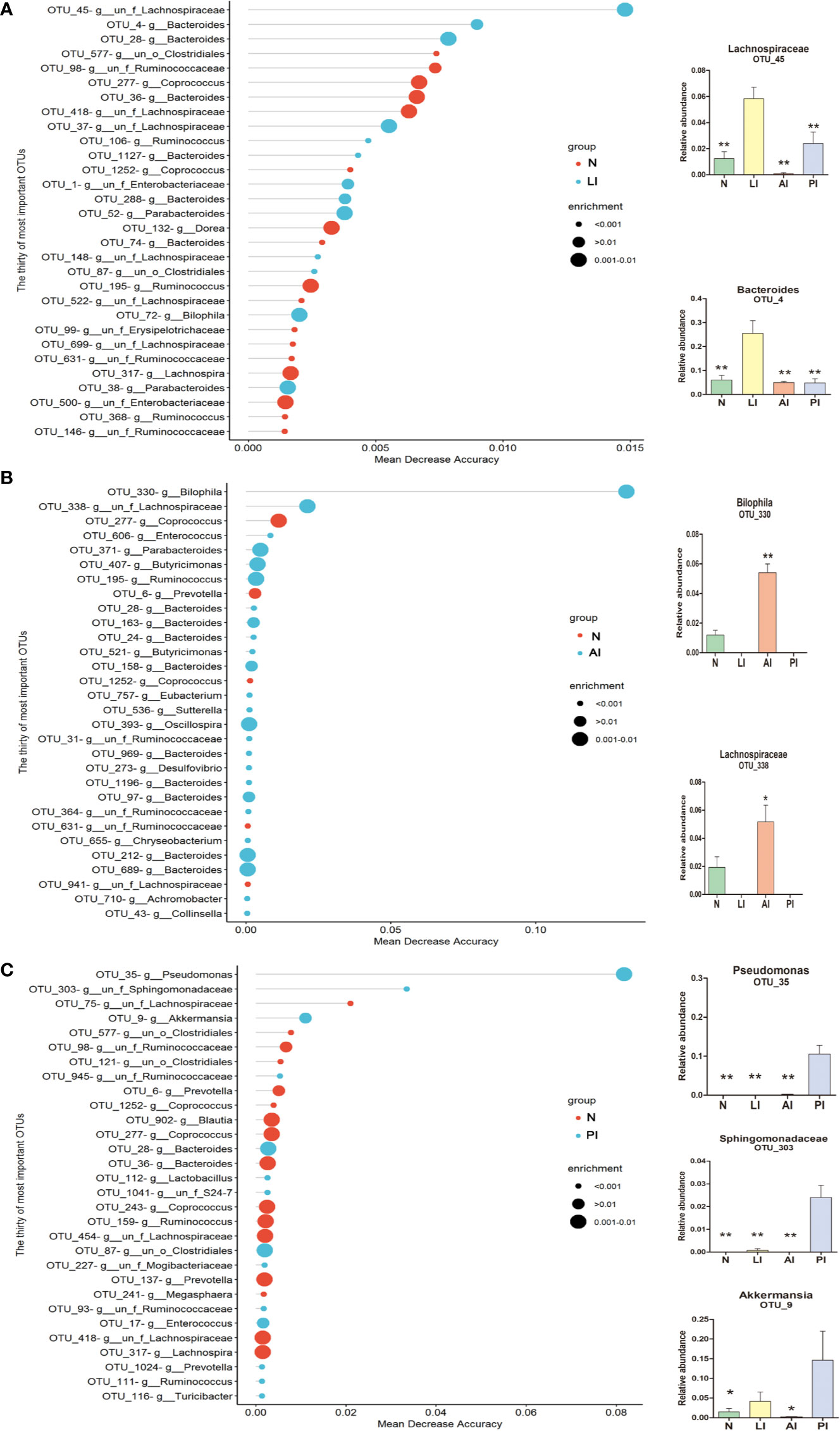
Figure 7 OTU-based markers of ischemic stroke identified by the Random Forest model. (A) lacunar infarction. **p < 0.01 compared with the LI group. (B) non-lacunar acute ischemic infarction. *p < 0.05 and **p < 0.01 compared with the AI group. (C) post-ischemic stroke. *p < 0.05 and **p < 0.01 compared with the PI group.
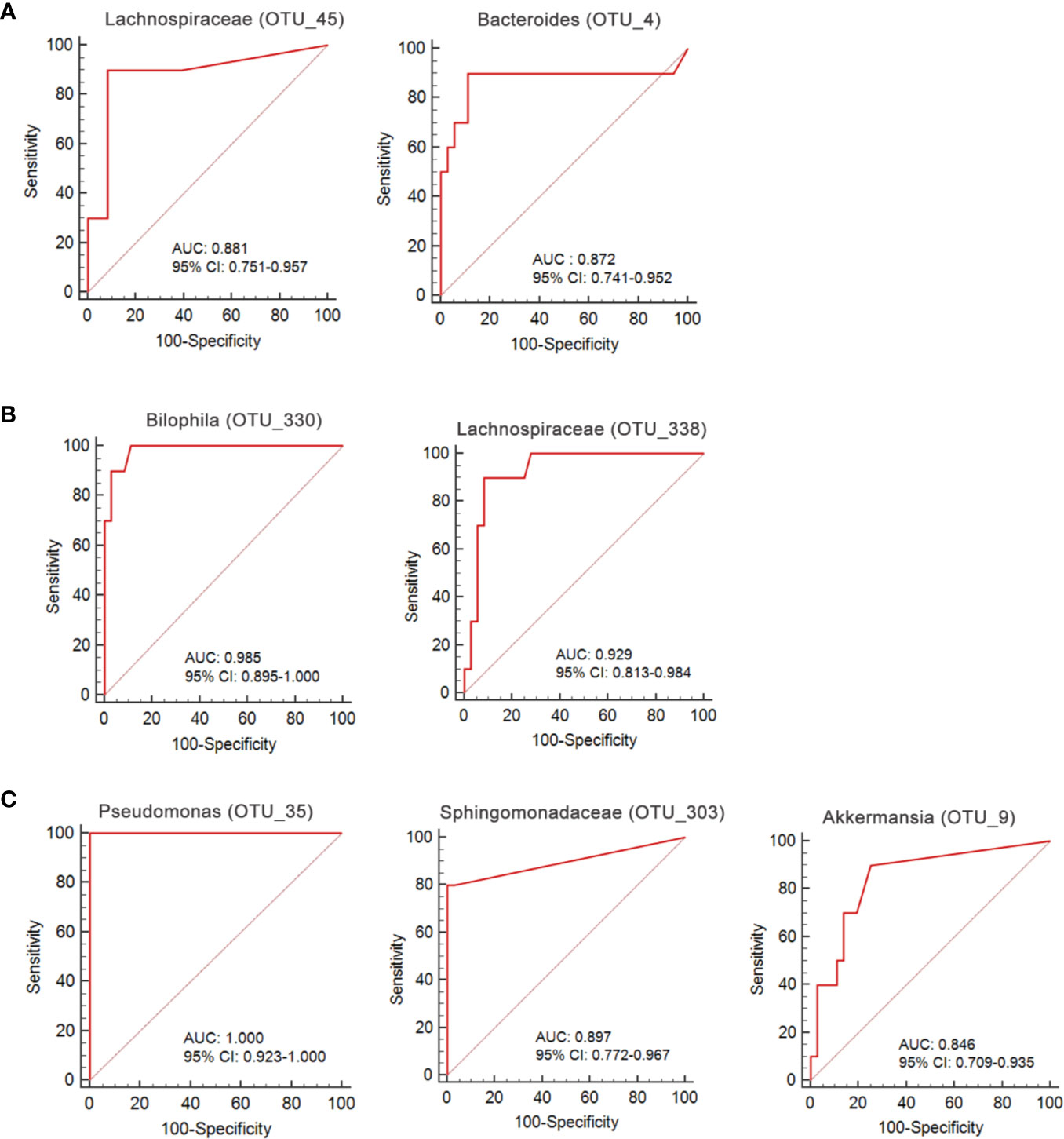
Figure 8 ROC curves for the OTU-based diagnostic biomarkers of ischemic stroke. (A) lacunar infarction; (B) non-lacunar acute ischemic infarction; (C) post-ischemic stroke.
This is the first study to explore the characteristics of the gut microbiota in patients suffering from different types of acute ischemic stroke, or recovering from such strokes. The gastrointestinal tract is a major organ of the immune system, containing over 70% of all immune cells and the largest population of macrophages in the human body. The composition of the gut microbiota depends on many factors, including lifestyle, diet, metabolism, antibiotic use, and hygiene; all are also closely associated with ischemic stroke (Bibbò et al., 2016). Several reports have described intestinal dysfunction and gut dysbiosis after stroke, highlighting the delicate interplay between the brain, gut, and microbiome after acute brain injury (Benakis et al., 2016; Singh and Roth, 2016; Zhao et al., 2018). The “brain-gut axis” is considered to facilitate bidirectional communication between the central nervous system and the gastrointestinal tract (Khlevner et al., 2018). Although many studies have confirmed the relationship between stroke and gut microbiome, the microbiotic status of patients differing in terms of stroke status has not been explored.
In this study, the phylum Firmicutes was less abundant in three stroke groups than the control group. The genera of butyrate-producing bacteria (e.g., Lachnospiraceae) were less common in the stroke groups. Increasing evidence indicates that the short-chain fatty acid (SCFA) metabolites (especially butyric acid) of gut bacteria are key signaling molecules (Stilling et al., 2016). Butyric acid supplementation significantly reduced intestinal leakage in patients with cerebral ischemic strokes and reduced levels of butyrate-producing bacteria may trigger stroke (Singh and Roth, 2016). Transplantation of fecal bacteria that produce SCFAs, and butyric acid supplementation, may effectively treat cerebral ischemic stroke.
AI and LI groups exhibited lower Firmicutes: Bacteroidetes ratios, and a higher ratio in PI group, than healthy controls. A reduced ratio reflects dysbiotic gastrointestinal tract metabolism, associated with low levels of circulating SCFAs, compromising systemic immunity and triggering systemic inflammation. It may be possible to reduce the risk of stroke, and prevent stroke progression and complications, by manipulating the ratio (Eckburg et al., 2005).
Each stroke group exhibited unique microbiotic features. Fusobacteria and Cyanobacteria were most abundant in LI group. Notably, LEfSE analysis showed that Bacteroidaceae and Erysipelotrichaceae were more abundant in LI than other groups. To the best of our knowledge, this is the first such report. The opportunistic pathogens of the Veillonellaceae were more abundant in LI group, as previously reported by Zeng et al., (2019). Lacunar infarcts are thought to be primarily attributable to intracranial, small vessel disease. Endothelial dysfunction may play important roles in the pathogenesis and progression of such disease (Regenhardt et al., 2018). Ruminococcaceae levels were dramatically decreased in LI group, and beneficial bacteria such as Meganonas enriched. Emerging evidence indicates that microbiotic interventions may enhance the efficacy of therapy and ameliorate toxicity; the gut microbiome performs several vital functions including vitamin production and dietary metabolism, and protects against gut pathogen expansion and systemic infiltration.
AI group exhibited markedly higher levels of Actinobacteria than other groups. A mouse study found that the gut microbiota affected stroke progression by acting on the immune system (Benakis et al., 2016). The opportunistic pathogens Bacteroidaceae, and lactate-producing bacteria (e.g., Bifidobacterium), were enriched in AI group. The serum trimethylamine N-oxide (TMAO) concentration is positively associated with the first stroke in hypertensive patients. TMAO is produced when hepatic flavin mono-oxygenases act on trimethylamine, a waste product of gut microbes (Zeisel and Warrier, 2017). Ruminococcaceae, Bifidobacteriaceae, and Coriobacteriaceae were most abundant in the AI group. Notably, LEfSE analysis revealed that Xanthomonadaceae, Bacillaceae, Brucellaceae, Paenibacillaceae, and Weeksellaceae were most abundant in AI group. To the best of our knowledge, this is the first report to show that these bacteria were more common in acute ischemic stroke patients than controls. KEGG analysis revealed a cluster of affected metabolic modules in AI group, including folate biosynthesis, photosynthesis, the citrate cycle (TCA cycle), galactose metabolism, amino sugar and nucleotide sugar metabolism, and fructose and mannose metabolism. Stroke alters the gut microbiotic composition; conversely, microbiotic dysbiosis substantially affects stroke outcomes by modulating the immune response and the metabolic system. Intestinal microorganisms affect host metabolism and immune status, in turn modulating neuronal pathways of the enteric and central nervous systems. Intestinal probiotics may protect against stroke.
Verrucomicrobia, Synergistetes, and Proteobacteria levels were significantly higher in PI than the other groups. Lactobacillus levels were highest in PI group; these bacteria help to maintain good health and optimal immune function. Moreover, Lactobacillus exerted protective effects in rat models of cerebral ischemic stroke, inhibiting neural cell apoptosis, decreasing the cerebral infarction volume, reducing oxidative stress, and restoring neurobehavioral impairment (Chen et al., 2019). We thus speculate that Lactobacillus may aid recovery after stroke.
Together, the gut and brain control neurodevelopment, neurotransmitter production, and microglial function, thus modulating cerebral biochemistry and behavior (Moschopoulos and Kratimenos, 2018; Blacher et al., 2019). Clostridium butyricum alleviated ischemia stroke injury in diabetic mice by regulating the gut microbiota (Sun et al., 2016). Butyric acid significantly enhanced the alpha diversity of gut microbiota and reduced the levels of pathogenic bacteria such as Bacteroides. Conversely, brain lesions of various etiologies change the composition of the microbiota. Microbiotic manipulation may open new therapeutic approaches to various neurological diseases.
We go beyond a simple description of gut microbiotic changes in ischemic stroke patients; we propose that the gut microbiota contains non-invasive markers of early ischemic stroke. Thus, we performed fivefold cross-validation using a Random Forest model. Two species allowed of optimal LI detection: Lachnospiraceae (OTU_45), and Bacteroides (OTU_4); the AUCs under the ROCs were 0.881 and 0.872 respectively. Two species optimally detected AI: Bilophila (OTU_330) and Lachnospiraceae (OTU_338); the AUCs under the ROCs were 0.985 and 0.929 respectively. Three species optimally detected PI: Pseudomonas (OTU_35), Sphingomonadaceae (OTU_303), and Akkermansia (OTU_9); the AUCs under the ROCs were 1, 0.897, and 0.846 respectively. To the best of our knowledge, the microbiotic markers mentioned above have not previously been used to predict ischemic stroke.
The main limitation of the study is the small samples size, which is conduce to some potential bacteria serving as useful predictors of stroke may not be discovered. Moreover, patients with non-lacunar acute ischemic infarctions were not further subdivided into subpopulations because of the small sample size. In this condition, it is difficult to define some bacterial flora is associated with a particular subtypes of acute ischemic infarction.
We analyzed the gut microbiota of ischemic stroke patients. The microbiotas differed from those of healthy controls. Fecal microbial markers (OTUs) may be developed as useful utility when diagnosing various types of ischemic stroke, and may facilitate early appropriate therapy.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The studies involving human participants were reviewed and approved by the Ethics Committee of the First Affiliated Hospital of Guangdong Pharmaceutical University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
LX, YFL, and LL designed the study, collected samples, compiled the data, drafted the manuscript, and provided critical revision. YLL, WZ, JD,YG, MS, and XY compiled the data and performed the statistical analysis and data interpretation. ML collected samples and compiled the data. HL provided critical revision. XC, FL, and XX designed this study, collected samples, drafted the manuscript, and provided critical revision. All authors contributed to the article and approved the submitted version.
This research was supported by the National Natural Science Foundation of China (grant no. 81804030).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Benakis C., Brea D., Caballero S., Faraco G., Moore J., Murphy M., et al. (2016). Commensal microbiota affects ischemic stroke outcome by regulating intestinal γδ T cells. Nat. Med. 22 (5), 516–523. doi: 10.1038/nm.4068
Bibbò S., Ianiro G., Giorgio V., Scaldaferri F., Masucci L., Gasbarrini A., et al. (2016). The role of diet on gut microbiota composition. Eur. Rev. Med. Pharmacol. Sci. 20 (22), 4742–4749.
Blacher E., Bashiardes S., Shapiro H., Rothschild D., Mor U., Dori-Bachash M., et al. (2019). Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 572 (7770), 474–480. doi: 10.1038/s41586-019-1443-5
Chen R., Xu Y., Wu P., Zhou H., Lasanajak Y., Fang Y., et al. (2019). Transplantation of fecal microbiota rich in short chain fatty acids and butyric acid treat cerebral ischemic stroke by regulating gut microbiota. Pharmacol. Res. 148, 104403. doi: 10.1016/j.phrs.2019.104403
Das A. S., Regenhardt R. W., Feske S. K., Gurol M. E. (2019). Treatment Approaches to Lacunar Stroke. J. Stroke Cerebrovasc. Dis. 28 (8), 2055–2078. doi: 10.1016/j.jstrokecerebrovasdis.2019.05.004
Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308 (5728), 1635–1638. doi: 10.1126/science.1110591
Fan W. T., Yan Y. M., Bie Y. L., Wang Q. (2016). [Diversity of intestinal microflora in patients with depression after stroke]. Nan Fang Yi Ke Da Xue Xue Bao 36 (10), 1305–1311.
Houlden A., Goldrick M., Brough D., Vizi E. S., Lénárt N., Martinecz B., et al. (2016). Brain injury induces specific changes in the caecal microbiota of mice via altered autonomic activity and mucoprotein production. Brain Behav. Immun. 57, 10–20. doi: 10.1016/j.bbi.2016.04.003
Khlevner J., Park Y., Margolis K. G. (2018). Brain-Gut Axis: Clinical Implications. Gastroenterol. Clin. North Am. 47 (4), 727–739. doi: 10.1016/j.gtc.2018.07.002
Langdon A., Crook N., Dantas G. (2016). The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 8 (1), 39. doi: 10.1186/s13073-016-0294-z
McDonald T. J. W., Cervenka M. C. (2018). Ketogenic Diets for Adult Neurological Disorders. Neurotherapeutics 15 (4), 1018–1031. doi: 10.1007/s13311-018-0666-8
Moschopoulos C., Kratimenos P. (2018). The Neurodevelopmental Perspective of Surgical Necrotizing Enterocolitis: The Role of the Gut-Brain Axis. Mediators Inflamm. 2018, 7456857. doi: 10.1155/2018/7456857
Muyzer G., de Waal E. C., Uitterlinden A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol 59 (3), 695–700. doi: 10.1128/AEM.59.3.695-700.1993
Paciaroni M., Agnelli G., Corea F., Ageno W., Alberti A., Lanari A., et al. (2008). Early hemorrhagic transformation of brain infarction: rate, predictive factors, and influence on clinical outcome: results of a prospective multicenter study. Stroke 39 (8), 2249–2256. doi: 10.1161/strokeaha.107.510321
Regenhardt R. W., Das A. S., Lo E. H., Caplan L. R. (2018). Advances in Understanding the Pathophysiology of Lacunar Stroke: A Review. JAMA Neurol. 75 (10), 1273–1281. doi: 10.1001/jamaneurol.2018.1073
Regenhardt R. W., Das A. S., Ohtomo R., Lo E. H., Ayata C., Gurol M. E. (2019). Pathophysiology of Lacunar Stroke: History’s Mysteries and Modern Interpretations. J. Stroke Cerebrovasc. Dis. 28 (8), 2079–2097. doi: 10.1016/j.jstrokecerebrovasdis.2019.05.006
Sandset E. C., Jusufovic M., Sandset P. M., Bath P. M., Berge E. (2015). Effects of blood pressure-lowering treatment in different subtypes of acute ischemic stroke. Stroke 46 (3), 877–879. doi: 10.1161/strokeaha.114.008512
Singh V., Roth S. (2016). Microbiota Dysbiosis Controls the Neuroinflammatory Response after Stroke. J. Neurosci. 36 (28), 7428–7440. doi: 10.1523/jneurosci.1114-16.2016
Stilling R. M., van de Wouw M., Clarke G., Stanton C., Dinan T. G., Cryan J. F. (2016). The neuropharmacology of butyrate: The bread and butter of the microbiota-gut-brain axis? Neurochem. Int. 99, 110–132. doi: 10.1016/j.neuint.2016.06.011
Sun J., Wang F., Ling Z., Yu X., Chen W., Li H., et al. (2016). Clostridium butyricum attenuates cerebral ischemia/reperfusion injury in diabetic mice via modulation of gut microbiota. Brain Res. 1642, 180–188. doi: 10.1016/j.brainres.2016.03.042
Wang H. X., Wang Y. P. (2016). Gut Microbiota-brain Axis. Chin Med. J. (Engl) 129 (19), 2373–2380. doi: 10.4103/0366-6999.190667
Wang W., Jiang B., Sun H., Ru X., Sun D., Wang L., et al. (2017). Prevalence, Incidence, and Mortality of Stroke in China: Results from a Nationwide Population-Based Survey of 480 687 Adults. Circulation 135 (8), 759–771. doi: 10.1161/circulationaha.116.025250
Yin J., Liao S. X., He Y., Wang S., Xia G. H., Liu F. T., et al. (2015). Dysbiosis of Gut Microbiota With Reduced Trimethylamine-N-Oxide Level in Patients With Large-Artery Atherosclerotic Stroke or Transient Ischemic Attack. J. Am. Heart Assoc. 4 (11), e002699. doi: 10.1161/jaha.115.002699
Zeisel S. H., Warrier M. (2017). Trimethylamine N-Oxide, the Microbiome, and Heart and Kidney Disease. Annu. Rev. Nutr. 37, 157–181. doi: 10.1146/annurev-nutr-071816-064732
Zeng X., Gao X., Peng Y., Wu Q., Zhu J., Tan C., et al. (2019). Higher Risk of Stroke Is Correlated With Increased Opportunistic Pathogen Load and Reduced Levels of Butyrate-Producing Bacteria in the Gut. Front. Cell Infect. Microbiol 9, 4. doi: 10.3389/fcimb.2019.00004
Zhang Y. J., Li S., Gan R. Y., Zhou T., Xu D. P., Li H. B. (2015). Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 16 (4), 7493–7519. doi: 10.3390/ijms16047493
Zhang P., Wang J., Xu Q., Song Z., Dai J., Wang J. (2018). Altered functional connectivity in post-ischemic stroke depression: A resting-state functional magnetic resonance imaging study. Eur. J. Radiol 100, 156–165. doi: 10.1016/j.ejrad.2018.01.003
Zhao L., Xiong Q., Stary C. M., Mahgoub O. K., Ye Y., Gu L., et al. (2018). Bidirectional gut-brain-microbiota axis as a potential link between inflammatory bowel disease and ischemic stroke. J. Neuroinflammation 15 (1), 339. doi: 10.1186/s12974-018-1382-3
Keywords: lacunar infarction, post-ischemic stroke, gut microbiota, acute ischemic infarction, non-lacunar acute ischemic infarction, random forest model
Citation: Xiang L, Lou Y, Liu L, Liu Y, Zhang W, Deng J, Guan Y, She M, You X, Liu M, Li H, Xu X, Liu F and Cai X (2020) Gut Microbiotic Features Aiding the Diagnosis of Acute Ischemic Stroke. Front. Cell. Infect. Microbiol. 10:587284. doi: 10.3389/fcimb.2020.587284
Received: 25 July 2020; Accepted: 18 November 2020;
Published: 21 December 2020.
Edited by:
Marloes Dekker Nitert, The University of Queensland, AustraliaReviewed by:
Chengwen Sun, North Dakota State University, United StatesCopyright © 2020 Xiang, Lou, Liu, Liu, Zhang, Deng, Guan, She, You, Liu, Li, Xu, Liu and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangsheng Cai, eGlhbmdzaGVuZ2NhaUBnZHB1LmVkdS5jbg==; Fang Liu, bGl1ZmFuZ25keXhAMTYzLmNvbQ==; Xiaosong Xu, eHV4aWFvc29uZ216MjAxNkAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.