- 1Newborn Research Zurich, Department of Neonatology, University Hospital Zurich and University of Zurich, Zurich, Switzerland
- 2Division of Transplantation Surgery, Department of Surgery, University of Colorado Hospital, Aurora, CO, United States
- 3Division of Pediatric Gastroenterology, Hepatology and Nutrition, Children’s Hospital Lucerne, Lucerne, Switzerland
- 4Department of Pediatric Surgery, University Children’s Hospital of Basel, Basel, Switzerland
- 5Institute of Physiology, University of Zurich, Zurich, Switzerland
The development of the neonatal gastrointestinal tract microbiota remains a poorly understood process. The interplay between neonatal (gestational age, genetic background), maternal (mode of delivery, nutritional status) and environmental factors (antibiotic exposure, available nutrition) are thought to influence microbial colonization, however, the exact mechanisms are unclear. Derangements in this process likely contribute to various gastrointestinal diseases including necrotizing enterocolitis and inflammatory bowel disease. As such, enhanced understanding of microbiota development may hold the key to significantly reduce the burden of gastrointestinal disease in the pediatric population. The most debatable topics during microbial seeding and possible future treatment approaches will be highlighted in this review.
Introduction
Humans are “holobionts”, which means that they host an assembly of their own human eukaryotic cells and all of the microorganisms living in/on them (Meyer-Abich, 1943; Margulis and Fester, 1991). It is well established that the human microbiota comprises a wide array of microorganisms including bacteria, archaea, fungi, and protozoa. The entirety of their corresponding genes are referred by the term “microbiome” as reviewed by Lynch et al. (Lynch and Pedersen, 2016). Because viruses are hosted in eukaryotic cells, bacteria or archaea, they are included under the umbrella of the microbiome as well (Virgin, 2014). Metagenomic data and new bioinformatic tools help to detect these hidden viral nucleotide sequences which may influence host phenotype (Angly et al., 2005; Virgin and Todd, 2011). The largest microbiota of the human body is found in the gastrointestinal tract (GIT) with about 1013–1014 microorganisms (Sender et al., 2016). As a nutritional inflow source, the GIT represents a fertile ground for microbial colonization. However, what types of microorganisms persist and in what quantity they do so, relies upon the methods by which microorganisms extract energy and provide commensal benefit to the GIT. The challenge of the host immune system is to both accept these commensal bacteria and defend against pathogens (Round and Mazmanian, 2009; Kim and Claud, 2019). Not only do resident microbiota extract energy for their survival, but they can also support the GIT in its function including pathogen defense (Freter, 1955; Abt and Pamer, 2014), strengthening the intestinal barrier function (Rakoff-Nahoum et al., 2004; Hayes et al., 2018) and promoting the immune development (O’Mahony et al., 2006; Round and Mazmanian, 2009). Additionally, the GIT microbiota helps to digests nutrients and improves gut motility (Abrams and Bishop, 1967; Dimidi et al., 2017) while supporting the synthesis of essential fatty acids (Høverstad and Midtvedt, 1986), amino acids (Jimenez et al., 2005), vitamins (Gustafsson et al., 1962) and hormones (Yano et al., 2015; Martin et al., 2019).
Existing literature suggests that children who are vaginally delivered at term without any instrumental assistance and are fed with maternal breast milk have the best chance to develop a healthy gastrointestinal microbiota which prevents dysbiosis (Levin et al., 2016; Martin et al., 2016). Dysbiosis refers to a phenomenon of microbiota “imbalance” or degeneracy in the microorganism make-up, which is thought to be associated with a wide range of metabolic/GIT diseases including obesity and metabolic syndrome (Turnbaugh et al., 2009), type 1 diabetes (Kostic et al., 2015), atopic conditions (Kalliomaki et al., 2001), inflammatory bowel disease (IBD) (Gevers et al., 2014), and necrotizing enterocolitis NEC (Fundora et al., 2020). As such, further understanding of dysbiosis is the first step to not only potentially prevent disease but also to offer hope for therapy.
This review summarizes the current evidence on the development of microbial colonization with a focus on factors which have been associated with dysbiosis including gestational age, mode of delivery, nutrition and antibiotic therapy.
First Microbial Colonization
The initiation of microbial colonization remains a controversial topic in developmental biology. The theory of “sterile womb” purports that the healthy fetus develops in a sterile environment in utero (Th and Bettelheim, 1988) and that microbial colonization starts after birth with the exception of intra-uterine infections during pregnancy (Küstner, 1877; Tissier, 1900). This theory has been challenged when microbial components have been detected in the placenta (Aagaard et al., 2014; Collado et al., 2016) amniotic fluid (Collado et al., 2016), umbilical cord blood (Jimenez et al., 2005), meconium (Jimenez et al., 2008; Chu et al., 2017; Tapiainen et al., 2018), and fetal membranes (Steel et al., 2005), even after uncomplicated pregnancies with healthy term born newborns (Perez-Munoz et al., 2017; Stinson et al., 2019; Patton and Neu, 2020). These microbial particles have typically been detected by sensitive polymerase chain reaction (PCR) methods. 16S ribosomal RNA is derived from the prokaryotic ribosome and is used to attribute detected RNA to respective bacterial strains (Woese and Fox, 1977). In all these studies, PCR mean copy numbers were low. Lauder et al. reported 5.72 × 102 gene copies for the maternal side and 1.2 × 102 for the fetal side in samples which were extracted from 0.1–0.5 g placental tissue (Lauder et al., 2016). When Rackaityte et al. aimed to control for procedural and environmental contamination, they found only 23.5 operational taxonomic units (OTUs) with ≥5 sequence read counts per meconium sample. Additionally, they analyzed the intestines of early terminated pregnancies (20 ± 2.2 weeks of gestation) and detected bacterial structures on electron scans (Rackaityte et al., 2020). Several sources of microbial fetal encounters have been proposed including ascension from the genitourinary tract (Zervomanolakis et al., 2007) or passage via mucosal membranes such as the oral cavity or the GIT (Han et al., 2004) of pregnant women (Baker et al., 2018). The analysis of potential bacterial seeding in utero is heavily complicated by intraamniotic infection. This infection occurs with an incidence of 3.9% of all women giving birth (Woodd et al., 2019). It may initially appear clinically silent but increases one’s risk of preterm birth (Hillier et al., 1988). Joint diagnosis of histological chorioamnionitis and bacterial growth in amnion cultures was found to be as low as 27.7% (Queiros da Mota et al., 2013), that is why detection of placental microbial particles could also represent clinically inapparent infections.
Defenders of the sterile womb hypothesis attribute the detected microbial particles to contamination (Olomu et al., 2020), because there was no evidence of viability of the detected bacterial structures (Rackaityte et al., 2020). Lim et al. found neither microbial nor viral communities in their amnion fluid samples from healthy term pregnancies (Lim et al., 2018; Lim et al., 2019). Correspondingly, in healthy pregnancies, the attempts to cultivate viable bacteria from placental specimen has thus far failed (Kuperman et al., 2020). Additionally, recent placental analyses of more than 500 placental tissue specimen assessed both with 16S- and metagenomic analyses revealed that besides pathogens (B streptococci), no placental microbiome has been detectable (de Goffau et al., 2019).
An interesting theory, which may help to join the two conflicting observations is that particles derived from bacteria, fungi or viruses can be transported via the placenta to various fetal sites and thereby contribute to the priming of the fetal immune system (Wilcox and Jones, 2018). Microbial structures might then occasionally be detected depending on the sensitivity of the method.
It has been demonstrated that bacteria, as part of the maternal microbiota can be absorbed by immune cells (Rescigno et al., 2001). In theory, they could be transported via the blood stream or the lymphatic system into the placenta (Funkhouser and Bordenstein, 2013). Taking into account the immunological challenge at the maternal-fetal interface of the placenta (Ander et al., 2019), we suspect that there is also the possibility, that dead bacterial components are expressed on placental dendritic cells and may be taken over to the fetal side to prime the fetal immune system as suspected for allergens (Szepfalusi et al., 2000).
However, the number of microbial agents which have been described in placental tissue remains low. Tenericutes, Firmicutes (Lactobacillus), Actinobacteria (Bifidobacterium, Propionibacterium, Rhodococcus, Streptomyces), Bacteroidetes (Bacteroides, Prevotella), Proteobacteria (E. coli, Neisseria, Enterobacteria), and Fusobacteria have been found in the placenta of healthy newborns at term (Aagaard et al., 2014; Parnell et al., 2017). Most taxa presented as “placental microbiome” correspond to the taxa found in the maternal oral microbiome (Fardini et al., 2010; Aagaard et al., 2014). Furthermore, it has been suggested that oral infections such as periodontitis are linked to complicated pregnancies and may contribute to prematurity or neonatal sepsis as reviewed by Zi et al. (2014).
With regard to umbilical cord blood of healthy term newborns Actinobacteria (Bifidobacterium, Propionibacterium), Proteobacteria (Escherichia), Firmicutes (Enterococcus, Staphylococcus, Streptococcus), and Bacteroidetes (Bacteroides) have been detected (Jimenez et al., 2005). Similarities between the microbiota of meconium, placenta, and amnion fluid of healthy infants either suggest a certain prenatal microbial antigen transfer or a common source of contamination. However, Chu et al. describe different bacteria on the newborn skin, mouth and nose depending on the mode of delivery (Chu et al., 2017). Contrastingly, they found similar bacteria in the newborn meconium with many samples harboring highly abundant Escherichia and Klebsiella (abundance 14.3% and 6.4%, respectively), not detectable in any other body site, speculating for a different microbial source prior to birth. It is tempting to assume that immunological priming with microbial particles starts to shape the fetal immune system prior to birth (Chu et al., 2017). Despite numerous papers published on this field, the concerns of contamination remain unsolved. In the newest study investigating this issue, evidences were put forward for contamination as the origin of bacteria found in human placenta samples (Gschwind et al., 2020).
Influence of Gestational Age
It has been well demonstrated that prematurity (birth before the completion of 37 weeks of pregnancy) may be triggered by intrauterine infections. Inflamed leaky or ruptured membranes facilitate the ascension of bacteria from the genitourinary tract (Hillier et al., 1995; Leitich et al., 2003). The gastrointestinal tract of premature infants is also known to have leaky barrier properties with a higher transepithelial and -mucosal permeability (Weaver et al., 1984a; Weaver et al., 1984b), impaired motility (Berseth, 1996), less active digestive enzymes (Demers-Mathieu et al., 2018) and lower absorption of nutrients (Neu and Koldovsky, 1996). Compared to their term counterparts, the immune system of premature infants displays fewer amounts of leukocytes, less proinflammatory cytokines, and less antibacterial peptides (Strunk et al., 2011; Melville and Moss, 2013).
Furthermore, preterm delivery is often linked to complicated pregnancies with a higher rate of caesarian sections and the use of prenatal antibiotics (Hill et al., 2017; Salvatore et al., 2019). The less mature newborns are, the longer time they have to spend in the neonatal intensive care unit (NICU) (Maier et al., 2018). They receive parenteral nutrition and/or enteral nutrition via nasogastric tubes (Viswanathan and Jadcherla, 2019). Additionally, these infants often need respiratory support (Shi et al., 2020). As a result of these well described, common consequences of prematurity, preterm infants often have a delayed development of their gastrointestinal bacterial microbiota, a lower bacterial load (Chernikova et al., 2018), fewer commensals, and obligate anaerobic bacteria and a higher number of pathogens such as Klebsiella pneumoniae and Clostridium difficile and facultative anaerobic bacteria (Dahl et al., 2018). The dominating taxa consist of Firmicutes (Staphylococcus, Enterococcus), Proteobacteria (Enterobacteriaceae, Escherichia, Klebsiella), Actinobacteria, and Bacteroidetes (Bacteroides) (Patel et al., 2016; Yuan et al., 2019). Their term counterparts are colonized with predominantly Actinobacteria (Bifidobacterium) (Penders et al., 2006) and Firmicutes (Staphylococcus, Streptococcus) (Palmer et al., 2007).
These microbial changes in the preterm infant may be associated with feeding intolerance (Ford et al., 2019; Salvatore et al., 2019), NEC (Baranowski and Claud, 2019), late-onset sepsis (LOS) (Stewart et al., 2017), and inferior long-term neurological outcomes (Niemarkt et al., 2019). Even if premature infants meet the «optimal microbial conditions» including vaginal delivery, nutrition with breast milk and no antibiotic therapy, the premature microbiome differs from the microbiome of term neonates (Leitich et al., 2003; Penders et al., 2006; Palmer et al., 2007). However, currently, there is no consensus concerning the exact time point when the microbiota of preterm and term infants align, with different studies reporting a time range between 4 months and 4 years (Dahl et al., 2018; Fouhy et al., 2019). A recent study of 5–11-year old children including 51 former preterm children (≤ 32 weeks of gestational age) could still find an inflammatory gut profile in the preterm group. The differences were attributed to a reduced gut phage richness (Jayasinghe et al., 2020).
Influence of Mode of Delivery
The mode of delivery influences the newborn gastrointestinal microbiota. During vaginal delivery, the infant’s GIT is colonized with vaginal (Dominguez-Bello et al., 2010) and intestinal (Makino et al., 2011; Makino et al., 2013) bacteria from the mother. Consequently, the infant’s gastrointestinal microbiota is dominated by Actinobacteria (Bifidobacterium, Atobium) (Dominguez-Bello et al., 2010; Reyman et al., 2019; Shao et al., 2019; Yang et al., 2019), Firmicutes (Lactobacillus, Megamonas) (Dominguez-Bello et al., 2010; Kuang et al., 2016), Bacteroidetes (Prevotella, Bacteroides, Parabacteroides) (Dominguez-Bello et al., 2010; Wampach et al., 2018), Fusobacteria (Sneathia) (Dominguez-Bello et al., 2010), and Proteobacteria (Shigella, Escherichia) (Kuang et al., 2016; Wampach et al., 2018; Shao et al., 2019; Yang et al., 2019). Most of these bacteria produce short chain fatty acids (SCFA), which lower the luminal pH and thereby inhibit the colonization of pathogens (Nagpal and Yamashiro, 2018). It is suggested that compared to children delivered via caesarean section, children after vaginal delivery display a higher diversity (Akagawa et al., 2019), fewer Staphylococci (Wampach et al., 2018) and C. difficile in their microbiota (Adlerberth and Wold, 2009).
In contrast, birth by caesarean (C-) section is theorized to interrupt the microbial transmission from the mother to child that occurs during vaginal birth (Backhed et al., 2015; Hill et al., 2017). Consequently, the GIT will first be colonized with bacteria present on the maternal skin (Akagawa et al., 2019) or in the direct neonatal environment (Dominguez-Bello et al., 2010). The intestinal microbiota after C-section is characterized by delayed bacterial colonization (Martin et al., 2016) and reduced number/diversity (Azad et al., 2013), but an increased number of opportunistic pathogens related to the hospital environment (Toscano et al., 2017a; Shao et al., 2019). After C-section the neonatal microbiome of the GIT is dominated by Firmicutes (Enterococcus, Staphylococcus, Streptococcus, Clostridium, Veillonella) (Azad et al., 2013; Martin et al., 2016; Kuang et al., 2016; Shao et al., 2019), and Proteobacteria (Klebsiella, Enterobacter, Haemophilus) (Shao et al., 2019). In relation to vaginal delivery, there are fewer Bacteroides, Bifidobacteria and Lactobacillus as well as SCFA (Nagpal and Yamashiro, 2018), and there is a general imbalance of the gut microbiome (Hoang et al., 2020). These findings correlate with a higher intraluminal pH and lower inhibition of pathogens (Nagpal and Yamashiro, 2018). There is some debate whether or not contractions might help to increase the microbial transfer to the baby (Levin et al., 2016; Shao et al., 2019) and how heavily the data is biased by antibiotic use as recommended prior to skin incision (Gholitabar et al., 2011). Mothers after C-section often additionally display a lower breastfeeding rate (Hobbs et al., 2016). In summary, C-section might be a contributing factor in the development of dysbiosis. However, the treatment of newborns with gauze swabs full of vaginal microbiota «vaginal seeding» did not show any benefit on long term outcomes but harbors the risk of pathogen transfer such as herpes, group B streptococci, Chlamydia trachomatis, and Neisseria gonorrhoeae (Cunnington et al., 2016; Haahr et al., 2018).
Influence of Nutrition
At the beginning of the 20th century, people realized that increasing alarming mortality rates of newborns and infants were associated with reduced rates of breastfeeding (Wolf, 2003). In the modern era, it is well understood that maternal breastfeeding indeed significantly reduces newborn and infant mortality and morbidity and contributes to maternal health (Ip et al., 2007; Zhao et al., 2020). However, the reasons underlying this association are more complex than initial theories related to breast milk providing nutrients and reducing pathogen transfer. Consequently, the composition of breastmilk and its influence on microbial composition is a growing area of research (Hennet and Borsig, 2016), not only due to the billion-dollar market linked to formula.
Important drivers for microbial seeding in the infant gut are pre- and probiotics in human breast milk (Sanders et al., 2019). Prebiotics are food components, which are not digested by human enzymes, but can be metabolized by certain bacteria, promote their growth and contribute to the health benefits of the host (Gibson and Roberfroid, 1995; Gibson, 1998; Gibson et al., 2004). Human milk oligosaccharides (HMO) are prebiotics and the third most common component of breast milk after lactose and lipids (Urashima et al., 2012). The first HMO has been described in 1954 as “bifidus factor” (Gyorgy et al., 1954a; Gyorgy et al., 1954b; Gauhe et al., 1954). These HMOs are not digested by pancreatic enzymes, but reach the colon intact, where they promote the growth of Bifidobacteria, Bacteroides and Lactobacillus (Marcobal et al., 2010; Thongaram et al., 2017). The digestion of HMO produces SCFA (such as acetate, propionate and butyrate), which can be used as energy source and lower the luminal pH, which inhibits the colonization of pathogens (Yu et al., 2013; David et al., 2014). Interestingly, the amount of specific fucosyl-oligosaccharides secreted into the milk seems to depend on the genetic background of the mother and whether it is preterm- or term breastmilk (Gabrielli et al., 2011). In addition to lipids and carbohydrates, human breast milk harbors proteins (immunoglobulins, enzymes) as well as hormones, growth factors, nucleotides, leukocytes, cytokines, lysozyme, and lactoferrin as reviewed by Hennet and Borsig (2016). Breast-fed children have an intestinal microbiota mainly dominated by Bifidobacteria and Lactobacilli (Cooke et al., 2005; Backhed et al., 2015), Bacteroides (which can digest HMO) (Wang et al., 2015) as well as Staphylococcus (Stewart et al., 2018). In contrast, children drinking formula tend to have a higher bacterial diversity and in addition to Bifidobacteriaceae, Clostridia, Enterococcus, and Enterobacteriaceae are detected (Harmsen et al., 2000; Li et al., 2014; Timmerman et al., 2017). However, studies are inconsistent (Adlerberth and Wold, 2009), possibly because they use different analytical approaches and infant nutrition and environmental influences are difficult to control in a large infant cohort.
In addition to the benefits named above, nutrition based on human milk is associated with a higher feeding tolerance (Schanler et al., 1999), lower risk of NEC (Miller et al., 2018), obesity (Ma et al., 2020), and atopic diseases (Lodge et al., 2015). However, the studies analyzing newborn and infant nutrition differ considerably concerning the duration and the amount of human milk provided as well as whether children were exclusively breastfed or human milk was provided with the bottle.
Prebiotic supplements such as galacto- and fructo-oligosaccharides added to infant formula shall mimic the effect of natural HMOs (Lodge et al., 2015), but to date, have not been demonstrated to lead to a complete approximation of the newborn microbiota (Bakker-Zierikzee et al., 2005; Haarman and Knol, 2005). In fact, intestinal microbiota of formula fed infants had more potential pathogens (Benno et al., 1984; Bezirtzoglou et al., 2011) as compared to breastfed children; dominated by Firmicutes (Staphylococcus, Streptococcus, Enterococcus, Lactobacillus, Clostridium), Bacteroidetes (Bacteroides), Proteobacteria (Enterobacteria), and Actinobacteria (Atopobium) (Fallani et al., 2010; Stewart et al., 2018). Probiotics are substances that contain vital microorganisms, which confer health benefits on their host (Food and Argiculture Organization of the United Nations, 2002; Hill et al., 2014). These microorganisms may change the microbial composition (Frese et al., 2017). They are believed to improve the barrier function of the intestinal epithelia, modify the immune response and protect against pathogens due to competition for nutrients and colonization with potential pathogens (Servin, 2004; Athalye-Jape et al., 2018). It is hypothesized that the supplementation of probiotics reduces the time to complete enteral feeding (Samanta et al., 2009; Braga et al., 2011), the duration of hospitalization (Romeo et al., 2011), and morbidity and mortality (Barrington, 2011). The best evidence available in this regard involves the combination of Bifidobacteria and Lactobacilli (Chang et al., 2017). A recent Cochrane review on this topic found 24 trials including 5,529 infants, all assessing probiotic treatment of preterm infants <37 weeks gestational age or <2.500 g birth weight. This meta-analysis found a significantly reduced incidence of severe NEC (Bell stage II or more) relative risk (RR) of 0.43 (95% confidence interval (CI) 0.33–0.56) and a reduced mortality RR 0.65 (CI 0.52–0.81) (Hobbs et al., 2016). However, the timing and composition of the probiotic treatment seems to be very important, because both the combination of Lactobacillus rhamnosus and Lactobacillus helveticus L (Freedman et al., 2018) and the supplementation of Lactobacillus rhamnosus alone were not sufficient to improve the outcome in children with gastroenteritis (Schnadower et al., 2018). Experts therefore suggest a personalized approach (Zmora et al., 2018).
While cultivated bacteria from breast milk samples have been attributed to contamination (Dudgeon and Jewesbury, 1924; Wright, 1947), living non-pathogenic bacteria below a density of 105 colony forming units/ml are now considered to be within normal range (Weaver et al., 2019), and potentially beneficial to newborn health (Toscano et al., 2017b). Several analyses have detected living bacteria such as Firmicutes (Staphylococcus, Streptococcus, Peptostreptococcus, Enterococcus, Clostridia, Lactobacillus), Actinobacteria (Bifidobacterium, Corynebacterium), Bacteroidetes (Bacteroides), and Proteobacteria (Escherichia, Serratia, Pseudomonas) in human breast milk. With the possibilities of large scale metagenomic analyses, it has now become possible to track the potential transfer of mobile genetic elements and antibiotic resistance genes via breast milk (Parnanen et al., 2018). Additionally, viable fungi have been cultured from breast milk samples recently at a density of ≥103/ml with the highest rate of Malassezia, Candida, and Saccharomyces taxae (Boix-Amoros et al., 2017). Vertical viral transmission from the mother to her newborn is evident for cytomegalovirus (CMV) (Bardanzellu et al., 2019), human immunodeficiency virus (HIV) (Van de Perre et al., 2012), and human-T-lymphotrope virus (HTLV). However, in general, breast-feeding has been associated with a lower risk of viral infections (Arifeen et al., 2001; Bahl et al., 2005). This viral reduction parallels with the finding, that prophages are also more abundant in formula-fed infants (Liang et al., 2020). There is a currently contentious debate in this regard to where microbial particles found in the human breast milk derive from and why microbial loads are reported to be divers (Biagi et al., 2017). Potential sources include the adjacent skin and areola of the breast and bacteria, fungi or viral particles located in the newborn nasopharyngeal tract, deriving from their direct environment. Some bacterial transfer can also be explained by reverse flow from the larger milk ducts near the nipple to smaller collecting ducts and ductules (Ramsay et al., 2004). However, Urbaniak et al. also found bacterial particles in breast samples which were taken from non-lactating women during an operation (Urbaniak et al., 2016). Some studies have suggested the origin of the human breast milk microbiota is the maternal GIT, because the transfer of obligate anaerobes such as Bifidobacterium breve which has been detected in breast milk samples is not possible via skin contact (Jost et al., 2014). If this was the case, bacteria in the intestinal lumen of the maternal GIT could be taken up by immune cells, transferred via the blood and/or lymphatic system and then secreted into the breast milk. Such a connection between the maternal GIT microbiota and breast milk production could be the key to new treatment opportunities in lactating mothers.
In 2019, Togo et al. reported the successful cultivation of methanogenic archaea from breast milk samples (Togo et al., 2019). The amount of DNA was low with 2 log 10 copies DNA/ml, but they are still suggested to be important commensals due to their H2 reducing properties (Hansen et al., 2011; Bang and Schmitz, 2015). Another driving force for human microbial diversity which has been nearly overlooked for a long time is the human phageome (Manrique et al., 2016). This highly dynamic system creates a high predation pressure, may be introduced with microbes in breast milk and shapes the human microbiome (Rodriguez-Valera et al., 2009). There is growing evidence that the maternal intestinal microbiota is an important driver of breast milk composition, suggesting that new interventions to optimize infant health could already start prior or during pregnancy.
After the introduction of solid foods, the differences between breast milk and formula fed infants become smaller and the microbiota starts to resemble the adult microbiota (Backhed et al., 2015). Some authors suggest that not the introduction of solid food but rather the cessation of breast milk leads to the alignment with the adult microbiota (Backhed et al., 2015; Levin et al., 2016). Given the fact that the human diet seem so decisively to influence microbial properties, the microbiota is potentially ripe for therapeutic intervention (Ku et al., 2020), especially during the newborn period.
Influence of Antibiotics
Unfortunately, globally the incidence of infections remains high in the newborn period (Fanos et al., 2007), and several diseases affecting newborns necessitate the administration of broad spectrum antibiotics (Isaacs, 2000; Gordon and Jeffery, 2005; Clark et al., 2006). Pre- as well as peri- and postnatal antibiotic therapy negatively influences the neonatal microbiota (Gibson et al., 2015; Tapiainen et al., 2019) and consequently the development of the infant’s immune system (Zeissig and Blumberg, 2014). Antibiotic therapy is associated with a lower number of commensal bacteria with delayed colonization with Bifidobacteria and Bacteroidetes (Coker et al., 2020; Eck et al., 2020) and a higher amount of potential pathogens (Aloisio et al., 2016). However, attempts to restore a healthy microbiota with probiotic treatment after antibiotic use even led to prolonged dysbiosis in healthy volunteers (Suez et al., 2018). In a cohort of infants aged 2–36 months, Yassour et al. observed that antibiotic treatment significantly reduced the strain diversity inducing a less stable microbiota. Moreover, bacteria increasingly acquired antibiotic resistance genes (Yassour et al., 2016). As such, the use of antibiotics increase the probability of fungal overgrowth (Kligman, 1952). Moreover, antibiotics may impact on long-term health outcomes such as modified nutrient absorption (Krajmalnik-Brown et al., 2012), lower vitamin production (LeBlanc et al., 2013), higher incidence of obesity (Dawson-Hahn and Rhee, 2019) and atopic diseases (Baron et al., 2020). The aim is to protect this delicate balance of bacterial and probably fungal interactions which support the healthy GIT-microbiota (Peleg et al., 2010). Thereby, ending antibiotic therapy expeditiously and narrowing antibiotic therapy should be encouraged for infants that do require antibiotics.
Development of the Microbiota in Early Childhood
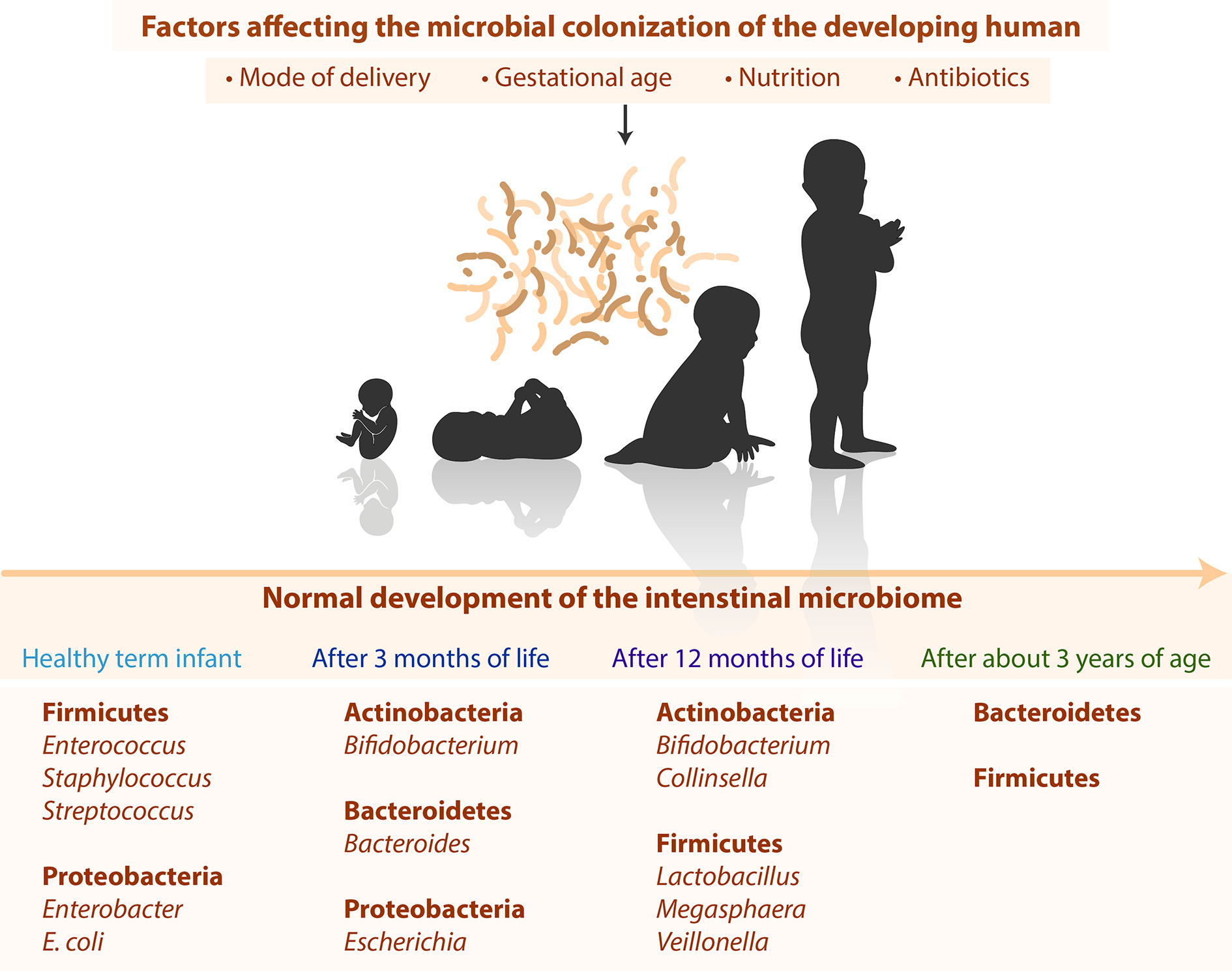
Despite huge individual differences (Eckburg et al., 2005; Ley et al., 2006; Turnbaugh et al., 2009) the development of the microbiota still follows typical timely changes as shown in Figure 1. Directly after birth, healthy term infants have a primarily aerobic GIT, which promotes the appearance of facultative anaerobe bacteria such as Firmicutes (Enterococcus, Staphylococcus, Streptococcus) and Proteobacteria (Enterobacter, Escherichia coli) (Palmer et al., 2007; Del Chierico et al., 2015). These bacteria reduce the oxygen content in the intestine and facilitate the occurrence of obligate anaerobic bacteria such as Actinobacteria (Bifidobacterium), Bacteroidetes (Bacteroides), and Firmicutes (Clostridium, Lactobacillus, Ruminococcus) (Koenig et al., 2011; Del Chierico et al., 2015). After three months of life, Actinobacteria (Bifidobacterium), Bacteroidetes (Bacteroides), and Proteobacteria (Escherichia) dominate the intestinal tract (Fallani et al., 2011; Hill et al., 2017). After 12 months of life, the infant’s GIT is dominated by Actinobacteria (Bifidobacterium, Collinsella) and Firmicutes (Lactobacillus, Megasphaera, Veillonella) (Penders et al., 2006). After 2–3 years of age, children display a higher diversity and lower inter-individual differences (Yatsunenko et al., 2012). The microbiota stabilizes after about 3 years and then resembles the adult microbiota in terms of diversity and complexity with high abundance of species from the firmicutes and bacteroidetes phyla (Arumugam et al., 2011).
Discussion and Conclusion
This review demonstrates that there is still a large knowledge gap in regard to the microbial colonization of newborns. Neither the authors of the “sterile womb hypothesis” nor those defending the “in-utero colonization hypothesis” (Perez-Munoz et al., 2017) are able to completely explain the signaling mechanisms at the materno-fetal interface. Because the fetal intestinal immune system develops as early as 16 weeks of pregnancy (Stras et al., 2019) and fetal genetic particles have been found in the maternal blood (Lo et al., 1990; Lo et al., 1997), we assume that microbial particles derived from the mother are transported to the fetal side as well. This “microbial priming” (Ganal-Vonarburg et al., 2017) may help to prepare the offspring for microbial contact after birth. It could also be triggered via the transfer of bacterial, viral, archaeal or fungal components through the blood, the interstitium, or immune cells. Materno-fetal protein-transport and antigen-presentation has been previously described, for example via the placental Fc-receptors (Malek et al., 1998; Wilcox and Jones, 2018). Although exosomes become increasingly attentive (Czernek and Duchler, 2020), it is unclear, whether microbial particles might also be transferred via exosomes in healthy pregnancies.
The microbial seeding during the first days of life makes the newborn highly susceptible to microbial perturbations (Bokulich et al., 2016). The most important factors affecting microbial seeding are gestational age, mode of delivery, nutrition, and antibiotic therapy (Azad et al., 2016; Levin et al., 2016; Martin et al., 2016). Optimizing nutrition and medical treatment could potentially improve newborn growth, prevent NEC and support favorable long-term outcomes. However, the molecular mechanisms remain unclear. Additionally, most have used human feces as a surrogate to study the intestinal microbiota, although it is unknown to which extent the bacteria found in the feces represent the microbiota of the GIT and whether the luminal (transient) bacteria correlate with mucosal (resident) bacteria, which might differ depending on the gastrointestinal region (Sundin et al., 2020).
In summary, we are convinced that a deeper understanding of the development of the newborn and infant microbiota will help to discover further potentially modifying factors to improve long-term health and quality of life.
Author Contribution
VS and TR conceptualized the draft and wrote the manuscript. FS provided the figure and revised the paper. DB, RC, FR-G, and RV critically reviewed and improved the manuscript. All authors finally approved this final version. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Thierry Hennet (Insitute of Physiology, University of Zurich, Zurich Switzerland) for scientific support.
References
Aagaard, K., Ma, J., Antony, K. M., Ganu, R., Petrosino, J., Versalovic, J., et al. (2014). The placenta harbors a unique microbiome. Sci. Transl. Med. 6 (237), 237ra65. doi: 10.1126/scitranslmed.3008599
Abrams, G. D., Bishop, J. E. (1967). Effect of the normal microbial flora on gastrointestinal motility. Proc. Soc. Exp. Biol. Med. 126 (1), 301–304. doi: 10.3181/00379727-126-32430
Abt, M. C., Pamer, E. G. (2014). Commensal bacteria mediated defenses against pathogens. Curr. Opin. Immunol. 29, 16–22. doi: 10.1016/j.coi.2014.03.003
Adlerberth, I., Wold, A. E. (2009). Establishment of the gut microbiota in Western infants. Acta Paediatr. 98 (2), 229–238. doi: 10.1111/j.1651-2227.2008.01060.x
Akagawa, S., Tsuji, S., Onuma, C., Akagawa, Y., Yamaguchi, T., Yamagishi, M., et al. (2019). Effect of Delivery Mode and Nutrition on Gut Microbiota in Neonates. Ann. Nutr. Metab. 74 (2), 132–139. doi: 10.1159/000496427
Aloisio, I., Quagliariello, A., De Fanti, S., Luiselli, D., De Filippo, C., Albanese, D., et al. (2016). Evaluation of the effects of intrapartum antibiotic prophylaxis on newborn intestinal microbiota using a sequencing approach targeted to multi hypervariable 16S rDNA regions. Appl. Microbiol. Biotechnol. 100 (12), 5537–5546. doi: 10.1007/s00253-016-7410-2
Ander, S. E., Diamond, M. S., Coyne, C. B. (2019). Immune responses at the maternal-fetal interface. Sci. Immunol. 4 (31), 1–10. doi: 10.1126/sciimmunol.aat6114
Angly, F., Rodriguez-Brito, B., Bangor, D., McNairnie, P., Breitbart, M., Salamon, P., et al. (2005). PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinf. 6, 41. doi: 10.1186/1471-2105-6-41
Arifeen, S., Black, R. E., Antelman, G., Baqui, A., Caulfield, L., Becker, S., et al. (2001). Exclusive breastfeeding reduces acute respiratory infection and diarrhea deaths among infants in Dhaka slums. Pediatrics 108 (4), E67. doi: 10.1542/peds.108.4.e67
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473 (7346), 174–180. doi: 10.1038/nature09944
Athalye-Jape, G., Rao, S., Patole, S. (2018). Effects of probiotics on experimental necrotizing enterocolitis: a systematic review and meta-analysis. Pediatr. Res. 83 (1-1), 16–22. doi: 10.1038/pr.2017.218
Azad, M. B., Konya, T., Maughan, H., Guttman, D. S., Field, C. J., Chari, R. S., et al. (2013). Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ 185 (5), 385–394. doi: 10.1503/cmaj.121189
Azad, M. B., Konya, T., Persaud, R. R., Guttman, D. S., Chari, R. S., Field, C. J., et al. (2016). Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG 123 (6), 983–993. doi: 10.1111/1471-0528.13601
Backhed, F., Roswall, J., Peng, Y., Feng, Q., Jia, H., Kovatcheva-Datchary, P., et al. (2015). Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host. Microbe 17 (5), 690–703. doi: 10.1016/j.chom.2015.04.004
Bahl, R., Frost, C., Kirkwood, B. R., Edmond, K., Martines, J., Bhandari, N., et al. (2005). Infant feeding patterns and risks of death and hospitalization in the first half of infancy: multicentre cohort study. Bull. World Health Organ 83 (6), 418–426. doi: S0042-96862005000600009
Baker, J. M., Chase, D. M., Herbst-Kralovetz, M. M. (2018). Uterine Microbiota: Residents, Tourists, or Invaders? Front. Immunol. 9, 208. doi: 10.3389/fimmu.2018.00208
Bakker-Zierikzee, A. M., Alles, M. S., Knol, J., Kok, F. J., Tolboom, J. J., Bindels, J. G., et al. (2005). Effects of infant formula containing a mixture of galacto- and fructo-oligosaccharides or viable Bifidobacterium animalis on the intestinal microflora during the first 4 months of life. Br. J. Nutr. 94 (5), 783–790. doi: 10.1079/BJN20051451
Bang, C., Schmitz, R. A. (2015). Archaea associated with human surfaces: not to be underestimated. FEMS Microbiol. Rev. 39 (5), 631–648. doi: 10.1093/femsre/fuv010
Baranowski, J. R., Claud, E. C. (2019). Necrotizing Enterocolitis and the Preterm Infant Microbiome. Adv. Exp. Med. Biol. 1125, 25–36. doi: 10.1007/5584_2018_313
Bardanzellu, F., Fanos, V., Reali, A. (2019). Human Breast Milk-acquired Cytomegalovirus Infection: Certainties, Doubts and Perspectives. Curr. Pediatr. Rev. 15 (1), 30–41. doi: 10.2174/1573396315666181126105812
Baron, R., Taye, M., der Vaart, I. B., Ujcic-Voortman, J., Szajewska, H., Seidell, J. C., et al. (2020). The relationship of prenatal antibiotic exposure and infant antibiotic administration with childhood allergies: a systematic review. BMC Pediatr. 20 (1), 312. doi: 10.1186/s12887-020-02042-8
Barrington, K. J. (2011). Review: probiotics prevented necrotising enterocolitis and reduced mortality in preterm neonates. Arch. Dis. Child Educ. Pract. Ed. 96 (5), 199. doi: 10.1136/adc.2011.214569
Benno, Y., Sawada, K., Mitsuoka, T. (1984). The intestinal microflora of infants: composition of fecal flora in breast-fed and bottle-fed infants. Microbiol. Immunol. 28 (9), 975–986. doi: 10.1111/j.1348-0421.1984.tb00754.x
Berseth, C. L. (1996). Gastrointestinal motility in the neonate. Clin. Perinatol. 23 (2), 179–190. doi: 10.1016/S0095-5108(18)30237-9
Bezirtzoglou, E., Tsiotsias, A., Welling, G. W. (2011). Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 17 (6), 478–482. doi: 10.1016/j.anaerobe.2011.03.009
Biagi, E., Quercia, S., Aceti, A., Beghetti, I., Rampelli, S., Turroni, S., et al. (2017). The Bacterial Ecosystem of Mother’s Milk and Infant’s Mouth and Gut. Front. Microbiol. 8, 1214. doi: 10.3389/fmicb.2017.01214
Boix-Amoros, A., Martinez-Costa, C., Querol, A., Collado, M. C., Mira, A. (2017). Multiple Approaches Detect the Presence of Fungi in Human Breastmilk Samples from Healthy Mothers. Sci. Rep. 7 (1), 13016. doi: 10.1038/s41598-017-13270-x
Bokulich, N. A., Chung, J., Battaglia, T., Henderson, N., Jay, M., Li, H., et al. (2016). Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8 (343), 343ra82. doi: 10.1126/scitranslmed.aad7121
Braga, T. D., da Silva, G. A., de Lira, P. I., de Carvalho Lima, M. (2011). Efficacy of Bifidobacterium breve and Lactobacillus casei oral supplementation on necrotizing enterocolitis in very-low-birth-weight preterm infants: a double-blind, randomized, controlled trial. Am. J. Clin. Nutr. 93 (1), 81–86. doi: 10.3945/ajcn.2010.29799
Chang, H. Y., Chen, J. H., Chang, J. H., Lin, H. C., Lin, C. Y., Peng, C. C. (2017). Multiple strains probiotics appear to be the most effective probiotics in the prevention of necrotizing enterocolitis and mortality: An updated meta-analysis. PloS One 12 (2), e0171579. doi: 10.1371/journal.pone.0171579
Chernikova, D. A., Madan, J. C., Housman, M. L., Zain-Ul-Abideen, M., Lundgren, S. N., Morrison, H. G., et al. (2018). The premature infant gut microbiome during the first 6 weeks of life differs based on gestational maturity at birth. Pediatr. Res. 84 (1), 71–79. doi: 10.1038/s41390-018-0022-z
Chu, D. M., Ma, J., Prince, A. l., Antony, K. M., Seferovic, M. D., Aagaard, K. M. (2017). Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23 (3), 314–326. doi: 10.1038/nm.4272
Clark, R. H., Bloom, B. T., Spitzer, A. R., Gerstmann, D. R. (2006). Reported medication use in the neonatal intensive care unit: data from a large national data set. Pediatrics 117 (6), 1979–1987. doi: 10.1542/peds.2005-1707
Coker, M. O., Hoen, A. G., Dade, E., Lundgren, S., Li, Z., Wong, A. D., et al. (2020). Specific class of intrapartum antibiotics relates to maturation of the infant gut microbiota: a prospective cohort study. BJOG 127 (2), 217–227. doi: 10.1111/1471-0528.15799
Collado, M. C., Rautava, S., Aakko, J., Isolauri, E., Salminen, S. (2016). Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 6, 23129. doi: 10.1038/srep23129
Cooke, G., Behan, J., Clarke, N., Gorman, W., Costello, M. (2005). Comparing the gut flora of Irish breastfed and formula-fed neonates aged between birth and 6 weeks old. Microbial. Ecol. Health Dis. 17 (3), 163–168. doi: 10.3402/mehd.v17i3.7778
Cunnington, A. J., Sim, K., Deierl, A., Kroll, K. S., Brannigan, E., Darby, J. (2016). “Vaginal seeding” of infants born by caesarean section. BMJ 352, i227. doi: 10.1136/bmj.i227
Czernek, L., Duchler, M. (2020). Exosomes as Messengers Between Mother and Fetus in Pregnancy. Int. J. Mol. Sci. 21 (12), 1–17. doi: 10.3390/ijms21124264
Dahl, C., Stigum, H., Valeur, J., Iszatt, N., Lenters, V., Peddada, S., et al. (2018). Preterm infants have distinct microbiomes not explained by mode of delivery, breastfeeding duration or antibiotic exposure. Int. J. Epidemiol. 47 (5), 1658–1669. doi: 10.1093/ije/dyy064
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505 (7484), 559–563. doi: 10.1038/nature12820
Dawson-Hahn, E. E., Rhee, K. E. (2019). The association between antibiotics in the first year of life and child growth trajectory. BMC Pediatr. 19 (1), 23. doi: 10.1186/s12887-018-1363-9
de Goffau, M. C., Lager, S., Sovio, U., Gaccioli, F., Cook, E., Peacock, S. J., et al. (2019). Human placenta has no microbiome but can contain potential pathogens. Nature 572 (7769), 329–334. doi: 10.1038/s41586-019-1451-5
Del Chierico, F., Vernocchi, P., Petrucca, A., Paci, P., Fuentes, S., Pratico, G., et al. (2015). Phylogenetic and Metabolic Tracking of Gut Microbiota during Perinatal Development. PloS One 10 (9), e0137347. doi: 10.1371/journal.pone.0137347
Demers-Mathieu, V., Qu, Y., Underwood, M. A, Borghese, R., Dallas, D. C. (2018). Premature Infants have Lower Gastric Digestion Capacity for Human Milk Proteins than Term Infants. J. Pediatr. Gastroenterol. Nutr. 66 (5), 816–821. doi: 10.1097/MPG.0000000000001835
Dimidi, E., Christodoulides, S., Scott, S. M., Whelan, K. (2017). Mechanisms of Action of Probiotics and the Gastrointestinal Microbiota on Gut Motility and Constipation. Adv. Nutr. 8 (3), 484–494. doi: 10.3945/an.116.014407
Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N., et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U. S. A. 107 (26), 11971–11975. doi: 10.1073/pnas.1002601107
Dudgeon, L. S., Jewesbury, R. C. (1924). The Bacteriology of Human Milk. J. Hyg. (Lond) 23 (1), 64–76. doi: 10.1017/S0022172400008470
Eck, A., Rutten, N., Singendonk, M. M. J., Rijkers, G. T., Savelkoul, P. H. M., Meijssen, C. B., et al. (2020). Neonatal microbiota development and the effect of early life antibiotics are determined by two distinct settler types. PloS One 15 (2), e0228133. doi: 10.1371/journal.pone.0228133
Eckburg, P. B., Bik, E. M., Bernstein, C. M., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308 (5728), 1635–1638. doi: 10.1126/science.1110591
Fallani, M., Young, D., Scott, J., Norin, E., Amarri, S., Adam, R., et al. (2010). Intestinal microbiota of 6-week-old infants across Europe: geographic influence beyond delivery mode, breast-feeding, and antibiotics. J. Pediatr. Gastroenterol. Nutr. 51 (1), 77–84. doi: 10.1097/MPG.0b013e3181d1b11e
Fallani, M., Amarri, S., Uusijarvi, A., Adam, R., Khanna, S., Aguilera, M., et al. (2011). Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology 157 (Pt 5), 1385–1392. doi: 10.1099/mic.0.042143-0
Fanos, V., Cuzzolin, L., Atzei, A., Testa, M. (2007). Antibiotics and antifungals in neonatal intensive care units: a review. J. Chemother. 19 (1), 5–20. doi: 10.1179/joc.2007.19.1.5
Fardini, Y., Chung, P., Dumm, R., Joshi, N., Han, Y. W. (2010). Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect. Immun. 78 (4), 1789–1796. doi: 10.1128/IAI.01395-09
Food and Argiculture Organization of the United Nations (2002). W.h.o., Guidelines for evaluating probiotics in food. Available at: www.who.int›probiotic_guidelines
Ford, S. L., Lohmann, P., Preidis, G. A., Gordon, P. S., O'Donnell, A., Hagan, J., et al. (2019). Improved feeding tolerance and growth are linked to increased gut microbial community diversity in very-low-birth-weight infants fed mother’s own milk compared with donor breast milk. Am. J. Clin. Nutr. 109 (4), 1088–1097. doi: 10.1093/ajcn/nqz006
Fouhy, F., Watkins, C., Hill, C. J., O'Shea, C. A., Nagle, B., Dempsey, E. M., et al. (2019). Perinatal factors affect the gut microbiota up to four years after birth. Nat. Commun. 10 (1), 1517. doi: 10.1038/s41467-019-09252-4
Freedman, S. B., Williamson-Urquhart, S., Farion, K. J., Gouin, S., Willan, A. R., Poonai, N., et al. (2018). Multicenter Trial of a Combination Probiotic for Children with Gastroenteritis. N. Engl. J. Med. 379 (21), 2015–2026. doi: 10.1056/NEJMoa1802597
Frese, S. A., Hutton, A. A., Contreras, L. N., Shaw, C. A., Palumbo, M. C., Casaburi, G., et al. (2017). Persistence of Supplemented Bifidobacterium longum subsinfantis EVC001 in Breastfed Infants. mSphere 2 (6), e00501–e00517. doi: 10.1128/mSphere.00501-17
Freter, R. (1955). The fatal enteric cholera infection in the guinea pig, achieved by inhibition of normal enteric flora. J. Infect. Dis. 97 (1), 57–65. doi: 10.1093/infdis/97.1.57
Fundora, J. B., Guha, P., Shores, D. R., Pammi, M., Maheshwari, A. (2020). Intestinal dysbiosis and necrotizing enterocolitis: assessment for causality using Bradford Hill criteria. Pediatr. Res. 87 (2), 235–248. doi: 10.1038/s41390-019-0482-9
Funkhouser, L. J., Bordenstein, S. R. (2013). Mom knows best: the universality of maternal microbial transmission. PloS Biol. 11 (8), e1001631. doi: 10.1371/journal.pbio.1001631
Gabrielli, O., Zampini, L., Galeazzi, T., Padella, L., Santoro, L., Peila, C., et al. (2011). Preterm milk oligosaccharides during the first month of lactation. Pediatrics 128 (6), e1520–e1531. doi: 10.1542/peds.2011-1206
Ganal-Vonarburg, S. C., Fuhrer, T., Gomez de Aguero, M. (2017). Maternal microbiota and antibodies as advocates of neonatal health. Gut. Microbes 8 (5), 479–485. doi: 10.1080/19490976.2017.1299847
Gauhe, A., Gyorgy, P., Hoover, J. R., Kuhn, R., Rose, C. S., Ruelius, H. W., et al. (1954). Bifidus factor. IV. Preparations obtained from human milk. Arch. Biochem. Biophys. 48 (1), 214–224. doi: 10.1016/0003-9861(54)90326-4
Gevers, D., Kugathasan, S., Denson, L. A., Vazquez-Baeza, Y., Van Treuren, W., Ren, B., et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host. Microbe 15 (3), 382–392. doi: 10.1016/j.chom.2014.02.005
Gholitabar, M., Ullman, R., James, D., Griffiths, M., Guideline Development Group of the National Institute for H, Clinical E. (2011). Caesarean section: summary of updated NICE guidance. BMJ 343, d7108. doi: 10.1136/bmj.d7108
Gibson, G. R., Roberfroid, M. B. (1995). Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125 (6), 1401–1412. doi: 10.1093/jn/125.6.1401
Gibson, G. R., Probert, H. M., Loo, J. V., Rastall, R. A., Roberfroid, M. B. (2004). Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr. Res. Rev. 17 (2), 259–275. doi: 10.1079/NRR200479
Gibson, M. K., Crofts, T. S., Dantas, G. (2015). Antibiotics and the developing infant gut microbiota and resistome. Curr. Opin. Microbiol. 27, 51–56. doi: 10.1016/j.mib.2015.07.007
Gibson, G. R. (1998). Dietary modulation of the human gut microflora using prebiotics. Br. J. Nutr. 80 (4), S209–S212. doi: 10.1017/S0007114500006048
Gordon, A., Jeffery, H. E. (2005). Antibiotic regimens for suspected late onset sepsis in newborn infants. Cochrane Database Syst. Rev. 3, CD004501. doi: 10.1002/14651858.CD004501.pub2
Gschwind, R., Fournier, T., Kennedy, S., Tsatsaris, V., Cordier, A. G., Barbut, F., et al. (2020). Evidence for contamination as the origin for bacteria found in human placenta rather than a microbiota. PloS One 15 (8), e0237232. doi: 10.1371/journal.pone.0237232
Gustafsson, B. E., Daft, F. S., McDaniel, E. G., Smith, J. C., Fitzgerald, R. J. (1962). Effects of vitamin K-active compounds and intestinal microorganisms in vitamin K-deficient germfree rats. J. Nutr. 78 (4), 461–468. doi: 10.1093/jn/78.4.461
Gyorgy, P., Norris, R. F., Rose, C. S. (1954a). Bifidus factor. I. A variant of Lactobacillus bifidus requiring a special growth factor. Arch. Biochem. Biophys. 48 (1), 193–201. doi: 10.1016/0003-9861(54)90323-9
Gyorgy, P., Kuhn, R., Rose, C. S., Zilliken, F. (1954b). Bifidus factor. II. Its occurrence in milk from different species and in other natural products. Arch. Biochem. Biophys. 48 (1), 202–208. doi: 10.1016/0003-9861(54)90324-0
Haahr, T., Glavind, J., Axelsson, P., Bistrup Fischer, M., Bjurstrom, J. (2018). Vaginal seeding or vaginal microbial transfer from the mother to the caesarean-born neonate: a commentary regarding clinical management. BJOG 125 (5), 533–536. doi: 10.1111/1471-0528.15050
Haarman, M., Knol, J. (2005). Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl. Environ. Microbiol. 71 (5), 2318–2324. doi: 10.1128/AEM.71.5.2318-2324.2005
Han, Y. W., Redline, R. W., Li, M., Yin, L., Hill, G. B., McCormick, T. S. (2004). Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect. Immun. 72 (4), 2272–2279. doi: 10.1128/IAI.72.4.2272-2279.2004
Hansen, E. E., Lozupone, C. A., Rey, F. E., Wu, M., Guruge, J. L., Narra, A., et al. (2011). Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc. Natl. Acad. Sci. U. S. A. 108 (Suppl 1), 4599–4606. doi: 10.1073/pnas.1000071108
Harmsen, H. J., Wildeboer-Veloo, A. C., Raangs, G. C., Wagendorp, A. A., Klijn, N., Bindels, J. G., et al. (2000). Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30 (1), 61–67. doi: 10.1097/00005176-200001000-00019
Hayes, C. L., Dong, J., Galipeau, H. J., Jury, J., McCarville, J., Huang, X., et al. (2018). Commensal microbiota induces colonic barrier structure and functions that contribute to homeostasis. Sci. Rep. 8 (1), 14184. doi: 10.1038/s41598-018-32366-6
Hennet, T., Borsig, L. (2016). Breastfed at Tiffany’s. Trends Biochem. Sci. 41 (6), 508–518. doi: 10.1016/j.tibs.2016.02.008
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11 (8), 506–514. doi: 10.1038/nrgastro.2014.66
Hill, C. J., Lynch, D. B., Murphy, K., Ulaszewska, M., Jeffery, I. B., O'Shea, C. A., et al. (2017). Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome 5 (1), 4. doi: 10.1186/s40168-016-0213-y
Hillier, S. L., Martius, J., Krohn, M., Kiviat, N., Holmes, K. K., Eschenbach, D. A. (1988). A case-control study of chorioamnionic infection and histologic chorioamnionitis in prematurity. N. Engl. J. Med. 319 (15), 972–978. doi: 10.1056/NEJM198810133191503
Hillier, S. L., Nugent, R. P., Eschenbach, D. A., Krohn, M. A., Gibbs, R. S., Martin, D. H., et al. (1995). Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. The Vaginal Infections and Prematurity Study Grou. N. Engl. J. Med. 333 (26), 1737–1742. doi: 10.1056/NEJM199512283332604
Hoang, D. M., Levy, E. I., Vandenplas, Y. (2020). The impact of Caesarean section on the infant gut microbiome. Acta Paediatr. 00, 1–8. doi: 10.1111/apa.15501
Hobbs, A. J., Mannion, C. A., McDonald, S. W., Brockway, M., Tough, S. C. (2016). The impact of caesarean section on breastfeeding initiation, duration and difficulties in the first four months postpartum. BMC Pregnancy Childbirth 16, 90. doi: 10.1186/s12884-016-0876-1
Høverstad, T., Midtvedt, T. (1986). Short-chain fatty acids in germfree mice and rats. J. Nutr. 116 (9), 1772–1776. doi: 10.1093/jn/116.9.1772
Ip, S., Chung, M., Raman, G., Chew, P., Magula, N., DeVine, D., et al. (2007). Breastfeeding and maternal and infant health outcomes in developed countries. Evid. Rep. Technol. Assess. (Full Rep) (153), 1–186.
Isaacs, D. (2000). Rationing antibiotic use in neonatal units. Arch. Dis. Child Fetal Neonatal Ed. 82 (1), F1–F2. doi: 10.1136/fn.82.1.F1
Jayasinghe, T. N., Vatanen, T., Chiavaroli, V., Jayan, S., McKenzie, E. J., Adriaenssens, E., et al. (2020). Differences in Compositions of Gut Bacterial Populations and Bacteriophages in 5-11 Year-Olds Born Preterm Compared to Full Term. Front. Cell Infect. Microbiol. 10, 276. doi: 10.3389/fcimb.2020.00276
Jimenez, E., Fernandez, L., Marin, M. L., Martin, R., Odriozola, J. M., Nueno-Palop, C., et al. (2005). Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 51 (4), 270–274. doi: 10.1007/s00284-005-0020-3
Jimenez, E., Marin, M. L., Martin, R., Odriozola, J. M., Olivares, M., Xaus, J., et al. (2008). Is meconium from healthy newborns actually sterile? Res. Microbiol. 159 (3), 187–193. doi: 10.1016/j.resmic.2007.12.007
Jost, T., Lacroix, C., Braegger, C. P., Rochat, F., Chassard, C. (2014). Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 16 (9), 2891–2904. doi: 10.1111/1462-2920.12238
Kalliomaki, M., Kirjavainen, P., Eerola, E., Kero, P., Salminen, S., Isolauri, E. (2001). Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J. Allergy Clin. Immunol. 107 (1), 129–134. doi: 10.1067/mai.2001.111237
Kim, C. S., Claud, E. C. (2019). Necrotizing Enterocolitis Pathophysiology: How Microbiome Data Alter Our Understanding. Clin. Perinatol. 46 (1), 29–38. doi: 10.1016/j.clp.2018.10.003
Kligman, A. M. (1952). Are Fungus infections increasing as a result of antibiotic therapy? J. Am. Med. Assoc. 149 (11), 979–983. doi: 10.1001/jama.1952.02930280001001
Koenig, J. E., Spor, A., Scalfone, N., Fricker, A. D., Stombaugh, J., Knight, R., et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U. S. A. 108 (Suppl 1), 4578–4585. doi: 10.1073/pnas.1000081107
Kostic, A. D., Gevers, D., Siljander, H., Vatanen, T., Hyotylainen, T., Hamalainen, A. M., et al. (2015). The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host. Microbe 17 (2), 260–273. doi: 10.1016/j.chom.2015.01.001
Krajmalnik-Brown, R., Ilhan, Z. E., Kang, D. W., DiBaise, J. K. (2012). Effects of gut microbes on nutrient absorption and energy regulation. Nutr. Clin. Pract. 27 (2), 201–214. doi: 10.1177/0884533611436116
Ku, H. J., Kim, Y. T., Lee, J. H. (2020). Microbiome Study of Initial Gut Microbiota from Newborn Infants to Children Reveals that Diet Determines Its Compositional Development. J. Microbiol. Biotechnol. 30 (7), 1067–1071. doi: 10.4014/jmb.2002.02042
Kuang, Y. S., Li, S. H., Guo, Y., Lu, J. H., He, J. R., Luo, B. J., et al. (2016). Composition of gut microbiota in infants in China and global comparison. Sci. Rep. 6:36666. doi: 10.1038/srep36666
Kuperman, A. A., Zimmerman, A., Hamadia, S., Ziv, O., Gurevich, V., Fichtman, B., et al. (2020). Deep microbial analysis of multiple placentas shows no evidence for a placental microbiome. BJOG 127 (2), 159–169. doi: 10.1111/1471-0528.15896
Küstner, O. (1877). Beitrag zur Lehre von der puerperalen Infection der Neugeborenen. Archiv. Für Gynäkol. 11 (2), 256–263. doi: 10.1007/BF01845161
Lauder, A. P., Roche, A. M., Sherrill-Mix, S., Bailey, A., Laughlin, A. L., Bittinger, K., et al. (2016). Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 4 (1), 29. doi: 10.1186/s40168-016-0172-3
LeBlanc, J. G., Milani, C., de Giori, G. S., Sesma, F., van Sinderen, D., Ventura, M. (2013). Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr. Opin. Biotechnol. 24 (2), 160–168. doi: 10.1016/j.copbio.2012.08.005
Leitich, H., Bodner-Adler, B., Brunbauer, M., Kaider, A., Egarter, C., Husslein, P. (2003). Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Am. J. Obstet. Gynecol. 189 (1), 139–147. doi: 10.1067/mob.2003.339
Levin, A. M., Sitarik, A. R., Havstad, S. L., Fujimura, K. E., Wegienka, G., Cassidy-Bushrow, A. E., et al. (2016). Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci. Rep. 6, 31775. doi: 10.1038/srep31775
Ley, R. E., Turnbaugh, P. J., Klein, S., Gordon, J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444 (7122), 1022–1023. doi: 10.1038/4441022a
Li, M., Wang, M., Donovan, S. M. (2014). Early development of the gut microbiome and immune-mediated childhood disorders. Semin. Reprod. Med. 32 (1), 74–86. doi: 10.1055/s-0033-1361825
Liang, G., Zhao, C., Zhang, H., Mattei, L., Sherrill-Mix, S., Bittinger, K., et al. (2020). The stepwise assembly of the neonatal virome is modulated by breastfeeding. Nature 581 (7809), 470–474. doi: 10.1038/s41586-020-2192-1
Lim, E. S., Rodriguez, C., Holtz, L. R. (2018). Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome 6 (1), 87. doi: 10.1186/s40168-018-0475-7
Lim, E. S., Rodriguez, C., Holtz, L. R. (2019). Correction to: Amniotic fluid from healthy term pregnancies does not harbor a detectable microbial community. Microbiome 7 (1), 22. doi: 10.1186/s40168-019-0641-6
Lo, Y. M., Patel, P., Sampietro, M., Gillmer, M. D., Fleming, K. A., Wainscoat, J. S. (1990). Detection of single-copy fetal DNA sequence from maternal blood. Lancet 335 (8703), 1463–1464. doi: 10.1016/0140-6736(90)91491-R
Lo, Y. M., Corbetta, N., Chamberlain, P. F., Rai, V., Sargent, I. L., Redman, C. W., et al. (1997). Presence of fetal DNA in maternal plasma and serum. Lancet 350 (9076), 485–487. doi: 10.1016/S0140-6736(97)02174-0
Lodge, C. J., Tan, D. J., Lau, M. X., Dai, X., Tham, R., Lowe, A. J., et al. (2015). Breastfeeding and asthma and allergies: a systematic review and meta-analysis. Acta Paediatr. 104 (467), 38–53. doi: 10.1111/apa.13132
Lynch, S. V., Pedersen, O. (2016). The Human Intestinal Microbiome in Health and Disease. N. Engl. J. Med. 375 (24), 2369–2379. doi: 10.1056/NEJMra1600266
Ma, J., Qiao, Y., Zhao, P., Li, W., Katzmarzyk, P. T., Chaput, J. P., et al. (2020). Breastfeeding and childhood obesity: A 12-country study. Matern. Child Nutr. 16 (3), e12984. doi: 10.1111/mcn.12984
Maier, R. F., Blondel, B., Piedvache, A., Misselwitz, B., Petrou, S., Van Reempts, P., et al. (2018). Duration and Time Trends in Hospital Stay for Very Preterm Infants Differ Across European Regions. Pediatr. Crit. Care Med. 19 (12), 1153–1161. doi: 10.1097/PCC.0000000000001756
Makino, H., Kushiro, A., Ishikawa, E., Muylaert, D., Kubota, H., Sakai, T., et al. (2011). Transmission of intestinal Bifidobacterium longum subslongum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl. Environ. Microbiol. 77 (19), 6788–6793. doi: 10.1128/AEM.05346-11
Makino, H., Kushiro, A., Ishikawa, E., Kubota, H., Gawad, A., Sakai, T., et al. (2013). Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant’s microbiota. PloS One 8 (11), e78331. doi: 10.1371/journal.pone.0078331
Malek, A., Sager, R., Schneider, H. (1998). Transport of proteins across the human placenta. Am. J. Reprod. Immunol. 40 (5), 347–351. doi: 10.1111/j.1600-0897.1998.tb00064.x
Manrique, P., Bolduc, B., Walk, S. T., van der Oost, J., de Vos, W. M., Young, M. J. (2016). Healthy human gut phageome. Proc. Natl. Acad. Sci. U. S. A. 113 (37), 10400–10405. doi: 10.1073/pnas.1601060113
Marcobal, A., Barboza, M., Froehlich, J. W., Block, D. E., German, J. M., Lebrilla, C. B., et al. (2010). Consumption of human milk oligosaccharides by gut-related microbes. J. Agric. Food Chem. 58 (9), 5334–5340. doi: 10.1021/jf9044205
Margulis, L., Fester, R. (1991). “Bellagio conference and book. Symbiosis as Source of Evolutionary Innovation,” in Speciation and Morphogenesis, Bellagio Conference Center, Italy. Symbiosis, Balaban Publishers, Conference–June 25-30, 1989, Vol. 11. (Balaban Publishers), 93–101.
Martin, R., Makino, H., Cetinyurek Yavuz, A., Ben-Amor, K., Roelofs, M. (2016). Early-Life Events, Including Mode of Delivery and Type of Feeding, Siblings and Gender, Shape the Developing Gut Microbiota. PloS One 11 (6), e0158498. doi: 10.1371/journal.pone.0158498
Martin, A. M., Sun, E. W., Rogers, G. B., Keating, D. J. (2019). The Influence of the Gut Microbiome on Host Metabolism Through the Regulation of Gut Hormone Release. Front. Physiol. 10, 428. doi: 10.3389/fphys.2019.00428
Melville, J. M., Moss, T. J. (2013). The immune consequences of preterm birth. Front. Neurosci. 7, 79. doi: 10.3389/fnins.2013.00079
Meyer-Abich, A. (1943). Beiträge zur Theorie der Evolution derOrganismen. I. Das typologische Grundgesetz und seine Folgerungenfür Phylogenie und Entwicklungsphysiologie [Contributions to theevolutionary theory of organisms: I. The basic typological law and itsimplications for phylogeny and developmental physiology]. ActaBiotheoretica 7, 1–80. doi: 10.1007/BF01603792
Miller, J., Tonkin, E., Damarell, R. A., McPhee, A. J., Suganuma, M., Suganuma, H., et al. (2018). A Systematic Review and Meta-Analysis of Human Milk Feeding and Morbidity in Very Low Birth Weight Infants. Nutrients 10 (6), 1–35. doi: 10.3390/nu10060707
Nagpal, R., Yamashiro, Y. (2018). Gut Microbiota Composition in Healthy Japanese Infants and Young Adults Born by C-Section. Ann. Nutr. Metab. 73 (Suppl 3), 4–11. doi: 10.1159/000490841
Neu, J., Koldovsky, O. (1996). Nutrient absorption in the preterm neonate. Clin. Perinatol. 23 (2), 229–243. doi: 10.1016/S0095-5108(18)30240-9
Niemarkt, H. J., De Meij, T. G., van Ganzewinkel, C. J., de Boer, N. K. H., Andriessen, P., Hutten, M. C., et al. (2019). Necrotizing Enterocolitis, Gut Microbiota, and Brain Development: Role of the Brain-Gut Axis. Neonatology 115 (4), 423–431. doi: 10.1159/000497420
Olomu, I. N., Pena-Cortes, L. C., Long, R. A., Vyas, A., Krichevskiy, O., Luellwitz, R., et al. (2020). Elimination of “kitome” and “splashome” contamination results in lack of detection of a unique placental microbiome. BMC Microbiol. 20 (1), 157. doi: 10.1186/s12866-020-01839-y
O’Mahony, L., O’Callaghan, L., McCarthy, J., Shilling, D., Scully, P., Sibartie, S., et al. (2006). Differential cytokine response from dendritic cells to commensal and pathogenic bacteria in different lymphoid compartments in humans. Am. J. Physiol. Gastrointest Liver Physiol. 290 (4), G839–G845. doi: 10.1152/ajpgi.00112.2005
Palmer, C., Bik, E. M., DiGiulio, D. B, Relman, D. A., Brown, P. O. (2007). Development of the human infant intestinal microbiota. PloS Biol. 5 (7), e177. doi: 10.1371/journal.pbio.0050177
Parnanen, K., Karkman, A., Hultman, J., Lyra, C, Bengtsson-Palme, J., Larsson, D. G. J., et al. (2018). Maternal gut and breast milk microbiota affect infant gut antibiotic resistome and mobile genetic elements. Nat. Commun. 9 (1), 3891. doi: 10.1038/s41467-018-06393-w
Parnell, L. A., Briggs, C. M., Cao, B., Delannoy-Bruno, O, Schrieffer, A. E., Mysorekar, I. U. (2017). Microbial communities in placentas from term normal pregnancy exhibit spatially variable profiles. Sci. Rep. 7 (1), 11200. doi: 10.1038/s41598-017-11514-4
Patel, A. L., Mutlu, E. A., Sun, Y., Koenig, L., Green, S., Jakubowicz, A., et al. (2016). Longitudinal Survey of Microbiota in Hospitalized Preterm Very-Low-Birth-Weight Infants. J. Pediatr. Gastroenterol. Nutr. 62 (2), 292–303. doi: 10.1097/MPG.0000000000000913
Patton, L., Neu, J. (2020). “Chapter 1 - Microbes and the womb: does it matter?,” in The Developing Microbiome. Ed. Claud, E. C. (Academic Press, Elsevier Inc.), 1–20. doi: 10.1016/B978-0-12-820602-7.00001-5
Peleg, A. Y., Hogan, D. A., Mylonakis, E. (2010). Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 8 (5), 340–349. doi: 10.1038/nrmicro2313
Penders, J., Thijs, C., Vink, C., Stelma, F. F., Snijders, B., Kummeling, I., et al. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118 (2), 511–521. doi: 10.1542/peds.2005-2824
Perez-Munoz, M. E., Arrieta, M. C., Ramer-Tait, A. E., Walter, J. (2017). A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5 (1), 48. doi: 10.1186/s40168-017-0268-4
Queiros da Mota, V., Prodhom, G., Yan, P., Hohlfheld, P., Greub, G., Rouleau, C. (2013). Correlation between placental bacterial culture results and histological chorioamnionitis: a prospective study on 376 placentas. J. Clin. Pathol. 66 (3), 243–248. doi: 10.1136/jclinpath-2012-201124
Rackaityte, E., Halkias, J., Fukui, E. M., Mendoza, V. F., Hayzelden, C., Crawford, E. D., et al. (2020). Viable bacterial colonization is highly limited in the human intestine in utero. Nat. Med. 26 (4), 599–607. doi: 10.1038/s41591-020-0761-3
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S., Medzhitov, R. (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118 (2), 229–241. doi: 10.1016/j.cell.2004.07.002
Ramsay, D. T., Kent, J. C., Owens, R. A., Hartmann, P. E. (2004). Ultrasound imaging of milk ejection in the breast of lactating women. Pediatrics 113 (2), 361–367. doi: 10.1542/peds.113.2.361
Rescigno, M., Urbano, M., Valzasina, B., Francolini, M., Rotta, G., Bonasio, R., et al. (2001). Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2 (4), 361–367. doi: 10.1038/86373
Reyman, M., van Houten, M. A., van Baarle, D., Bosch, A., Man, W. H., Chu, M., et al. (2019). Impact of delivery mode-associated gut microbiota dynamics on health in the first year of life. Nat. Commun. 10 (1), 4997. doi: 10.1038/s41467-019-13373-1
Rodriguez-Valera, F., Martin-Cuadrado, A. B., Rodriguez-Brito, B., Pasic, L., Thingstad, T. F., Rohwer, F., et al. (2009). Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 7 (11), 828–836. doi: 10.1038/nrmicro2235
Romeo, M. G., Romeo, D. M., Trovato, L., Oliveri, S., Palermo, F., Cota, F., et al. (2011). Role of probiotics in the prevention of the enteric colonization by Candida in preterm newborns: incidence of late-onset sepsis and neurological outcome. J. Perinatol. 31 (1), 63–69. doi: 10.1038/jp.2010.57
Round, J. L., Mazmanian, S. K. (2009). The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9 (5), 313–323. doi: 10.1038/nri2515
Salvatore, S., Baldassarre, M. E., Di Mauro, A., Laforgia, N., Tafuri, S., Bianchi, F. P., et al. (2019). Neonatal Antibiotics and Prematurity Are Associated with an Increased Risk of Functional Gastrointestinal Disorders in the First Year of Life. J. Pediatr. 212, 44–51. doi: 10.1016/j.jpeds.2019.04.061
Samanta, M., Sarkar, M., Ghosh, P., Ghosh, J., Sinha, M., Chatterjee, S. (2009). Prophylactic probiotics for prevention of necrotizing enterocolitis in very low birth weight newborns. J. Trop. Pediatr. 55 (2), 128–131. doi: 10.1093/tropej/fmn091
Sanders, M. E., Merenstein, D. J., Reid, G., Gibson, G. R., Rastall, R. A. (2019). Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16 (10), 605–616. doi: 10.1038/s41575-019-0173-3
Schanler, R. J., Shulman, R. J., Lau, C. (1999). Feeding strategies for premature infants: beneficial outcomes of feeding fortified human milk versus preterm formula. Pediatrics 103 (6 Pt 1), 1150–1157. doi: 10.1542/peds.103.6.1150
Schnadower, D., Tarr, P. I., Casper, T. C., Gorelick, M. H., Dean, J. M., O’Connell, K. J., et al. (2018). Lactobacillus rhamnosus GG versus Placebo for Acute Gastroenteritis in Children. N. Engl. J. Med. 379 (21), 2002–2014. doi: 10.1056/NEJMoa1802598
Sender, R., Fuchs, S., Milo, R. (2016). Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 164 (3), 337–340. doi: 10.1016/j.cell.2016.01.013
Servin, A. L. (2004). Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28 (4), 405–440. doi: 10.1016/j.femsre.2004.01.003
Shao, Y., Forster, S. C., Tsaliki, E., Vervier, K., Strang, A., Simpson, N., et al. (2019). Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574 (7776), 117–121. doi: 10.1038/s41586-019-1560-1
Shi, Y., Muniraman, H., Biniwale, M., Ramanathan, R. (2020). A Review on Non-invasive Respiratory Support for Management of Respiratory Distress in Extremely Preterm Infants. Front. Pediatr. 8, 270. doi: 10.3389/fped.2020.00270
Steel, J. H., Malatos, S., Kennea, N., Edwards, A. D., Miles, L., Duggan, P., et al. (2005). Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr. Res. 57 (3), 404–411. doi: 10.1203/01.PDR.0000153869.96337.90
Stewart, C. J., Embleton, N. D., Marrs, E. C. L., Smith, D. P., Fofanova, T., Nelson, A., et al. (2017). Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome 5 (1), 75. doi: 10.1186/s40168-017-0295-1
Stewart, C. J., Ajami, N. J., O’Brien, J. L., Hutchinson, D. S., Smith, D. P., Wong, M. C., et al. (2018). Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562 (7728), 583–588. doi: 10.1038/s41586-018-0617-x
Stinson, L. F., Boyce, M. C., Payne, M. S., Keelan, J. A. (2019). The Not-so-Sterile Womb: Evidence That the Human Fetus Is Exposed to Bacteria Prior to Birth. Front. Microbiol. 10, 1124. doi: 10.3389/fmicb.2019.01124
Stras, S. F., Werner, L., Toothaker, J. M., Olaloye, O. O., Oldham, A. L., McCourt, C. C., et al. (2019). Maturation of the Human Intestinal Immune System Occurs Early in Fetal Development. Dev. Cell 51 (3), 357–373.e5. doi: 10.1016/j.devcel.2019.09.008
Strunk, T., Currie, A., Richmond, P., Simmer, K., Burgner, D. (2011). Innate immunity in human newborn infants: prematurity means more than immaturity. J. Matern. Fetal Neonatal Med. 24 (1), 25–31. doi: 10.3109/14767058.2010.482605
Suez, J., Zmora, N., Zilberman-Schapira, G., Mor, U., Dori-Bachash, M., Bashiardes, S., et al. (2018). Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell 174 (6), 1406–1423.e16. doi: 10.1016/j.cell.2018.08.047
Sundin, J., Aziz, I., Nordlander, S., Polster, A., Hu, Y. O. O., Hugerth, L. W., et al. (2020). Evidence of altered mucosa-associated and fecal microbiota composition in patients with Irritable Bowel Syndrome. Sci. Rep. 10 (1), 593. doi: 10.1038/s41598-020-57468-y
Szepfalusi, Z., Loibichler, C., Pichler, J., Reisenberger, K., Ebner, C., Urbanek, R., et al. (2000). Direct evidence for transplacental allergen transfer. Pediatr. Res. 48 (3), 404–407. doi: 10.1203/00006450-200009000-00024
Tapiainen, T., Paalanne, N., Tejesvi, M. V., Koivusaari, P., Korpela, K., Pokka, T., et al. (2018). Maternal influence on the fetal microbiome in a population-based study of the first-pass meconium. Pediatr. Res. 84 (3), 371–379. doi: 10.1038/pr.2018.29
Tapiainen, T., Koivusaari, P., Brinkac, L., Lorenzi, H. A., Salo, J., Renko, M., et al. (2019). Impact of intrapartum and postnatal antibiotics on the gut microbiome and emergence of antimicrobial resistance in infants. Sci. Rep. 9 (1), 10635. doi: 10.1038/s41598-019-46964-5
Th, E., Bettelheim, K. S. (1988). The Intestinal Bacteria of the Neonate and Breast-Fed Infant. Rev. Infect. Dis. 10 (6), 1220–1225. doi: 10.1093/clinids/10.6.1220
Thongaram, T., Hoeflinger, J. L., Chow, J., Miller, M. J. (2017). Human milk oligosaccharide consumption by probiotic and human-associated bifidobacteria and lactobacilli. J. Dairy Sci. 100 (10), 7825–7833. doi: 10.3168/jds.2017-12753
Timmerman, H. M., Rutten, N., Boekhorst, J., Saulnier, D. M., Kortman, G. A. M., Contractor, N., et al. (2017). Intestinal colonisation patterns in breastfed and formula-fed infants during the first 12 weeks of life reveal sequential microbiota signatures. Sci. Rep. 7 (1), 8327. doi: 10.1038/s41598-017-08268-4
Tissier, H. (1900). Dissertation: Recherches sur la Flore Intestinale des Nourrissons (état Normal et Pathologique). Ed. Naud, G. C. E. C. (Paris).
Togo, A. H., Grine, G., Khelaifia, S., des Robert, C., Brevaut, V., Caputo, A., et al. (2019). Culture of Methanogenic Archaea from Human Colostrum and Milk. Sci. Rep. 9 (1), 18653. doi: 10.1038/s41598-019-54759-x
Toscano, M., De Grandi, R., Peroni, D. G., Grossi, E., Facchin, V., Comberiati, P., et al. (2017a). Impact of delivery mode on the colostrum microbiota composition. BMC Microbiol. 17 (1), 205. doi: 10.1186/s12866-017-1109-0
Toscano, M., De Grandi, R., Grossi, E., Drago, L. (2017b). Role of the Human Breast Milk-Associated Microbiota on the Newborns’ Immune System: A Mini Review. Front. Microbiol. 8, 2100. doi: 10.3389/fmicb.2017.02100
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457 (7228), 480–484. doi: 10.1038/nature07540
Urashima, T., Asakuma, S., Leo, F., Fukuda, K., Messer, M., Oftedal, O. T., et al. (2012). The predominance of type I oligosaccharides is a feature specific to human breast milk. Adv. Nutr. 3 (3), 473S–482S. doi: 10.3945/an.111.001412
Urbaniak, C., Burton, J. P., Reid, G. (2016). The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 82 (16), 5039–5048. doi: 10.1128/AEM.01235-16
Van de Perre, P., Rubbo, P. A., Viljoen, J., Nagot, N., Tylleskar, T., Lepage, P., et al. (2012). HIV-1 reservoirs in breast milk and challenges to elimination of breast-feeding transmission of HIV-1. Sci. Transl. Med. 4 (143), 143sr3. doi: 10.1126/scitranslmed.3003327
Virgin, H. W., Todd, J. A. (2011). Metagenomics and personalized medicine. Cell 147 (1), 44–56. doi: 10.1016/j.cell.2011.09.009
Virgin, H. W. (2014). The virome in mammalian physiology and disease. Cell 157 (1), 142–150. doi: 10.1016/j.cell.2014.02.032
Viswanathan, S., Jadcherla, S. (2019). Transitioning from gavage to full oral feeds in premature infants: When should we discontinue the nasogastric tube? J. Perinatol. 39 (9), 1257–1262. doi: 10.1038/s41372-019-0446-2
Wampach, L., Heintz-Buschart, A., Fritz, J. V., Ramiro-Garcia, J., Habier, J., Herold, M., et al. (2018). Birth mode is associated with earliest strain-conferred gut microbiome functions and immunostimulatory potential. Nat. Commun. 9 (1), 5091. doi: 10.1038/s41467-018-07631-x
Wang, M., Li, M., Wu, S., Lebrilla, C. B., Chapkin, R. S., Ivanov, I., et al. (2015). Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J. Pediatr. Gastroenterol. Nutr. 60 (6), 825–833. doi: 10.1097/MPG.0000000000000752
Weaver, L. T., Laker, M. F., Nelson, R. (1984a). Intestinal permeability in the newborn. Arch. Dis. Child 59 (3), 236–241. doi: 10.1136/adc.59.3.236
Weaver, L. T., Laker, M. F., Nelson, R. (1984b). Enhanced intestinal permeability in preterm babies with bloody stools. Arch. Dis. Child 59 (3), 280–281. doi: 10.1136/adc.59.3.280
Weaver, G., Bertino, E., Gebauer, C., Grovslien, A., Mileusnic-Milenovic, R., Arslanoglu, S., et al. (2019). Recommendations for the Establishment and Operation of Human Milk Banks in Europe: A Consensus Statement From the European Milk Bank Association (EMBA). Front. Pediatr. 7, 53. doi: 10.3389/fped.2019.00053
Wilcox, C. R., Jones, C. E. (2018). Beyond Passive Immunity: Is There Priming of the Fetal Immune System Following Vaccination in Pregnancy and What Are the Potential Clinical Implications? Front. Immunol. 9, 1548. doi: 10.3389/fimmu.2018.01548
Woese, C. R., Fox, G. E. (1977). Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. U. S. A. 74 (11), 5088–5090. doi: 10.1073/pnas.74.11.5088
Wolf, J. H. (2003). Low breastfeeding rates and public health in the United States. Am. J. Public Health 93 (12), 2000–2010. doi: 10.2105/AJPH.93.12.2000
Woodd, S. L., Montoya, A., Barreix, M., Pi, L., Calvert, C., Rehman, A. M., et al. (2019). Incidence of maternal peripartum infection: A systematic review and meta-analysis. PloS Med. 16 (12), e1002984. doi: 10.1371/journal.pmed.1002984
Wright, J. (1947). Bacteriology of the collection and preservation of human milk. Lancet 2 (6465), 121–124. doi: 10.1016/S0140-6736(47)90193-1
Yang, R., Gao, R., Cui, S., Zhong, H., Zhang, X., Chen, Y., et al. (2019). Dynamic signatures of gut microbiota and influences of delivery and feeding modes during the first 6 months of life. Physiol. Genomics 51 (8), 368–378. doi: 10.1152/physiolgenomics.00026.2019
Yano, J. M., Yu, K., Donaldson, G. P., Shastri, G. G., Ann, P., Ma, L., et al. (2015). Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161 (2), 264–276. doi: 10.1016/j.cell.2015.02.047
Yassour, M., Vatanen, T., Siljander, H., Hamalainen, A. M., Harkonen, T., Ryhanen, S. J., et al. (2016). Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8 (343), 343ra81. doi: 10.1126/scitranslmed.aad0917
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486 (7402), 222–227. doi: 10.1038/nature11053
Yu, Z. T., Chen, C., Newburg, D. S. (2013). Utilization of major fucosylated and sialylated human milk oligosaccharides by isolated human gut microbes. Glycobiology 23 (11), 1281–1292. doi: 10.1093/glycob/cwt065
Yuan, Z., Yan, J., Wen, H., Deng, X., Li, X., Su, S. (2019). Feeding intolerance alters the gut microbiota of preterm infants. PloS One 14 (1), e0210609. doi: 10.1371/journal.pone.0210609
Zeissig, S., Blumberg, R. S. (2014). Life at the beginning: perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat. Immunol. 15 (4), 307–310. doi: 10.1038/ni.2847
Zervomanolakis, I., Ott, H. W., Hadziomerovic, D., Mattle, V., Seeber, B. E., Virgolini, I., et al. (2007). Physiology of upward transport in the human female genital tract. Ann. N. Y. Acad. Sci. 1101, 1–20. doi: 10.1196/annals.1389.032
Zhao, M., Wu, H., Liang, Y., Liu, F., Bovet, P., Xi, B. (2020). Breastfeeding and Mortality Under 2 Years of Age in Sub-Saharan Africa. Pediatrics 145 (5):e20192209. doi: 10.1542/peds.2019-2209
Zi, M. Y., Longo, P. L., Bueno-Silva, B., Mayer, M. P. (2014). Mechanisms Involved in the Association between Periodontitis and Complications in Pregnancy. Front. Public Health 2, 290. doi: 10.3389/fpubh.2014.00290
Keywords: microbiome, microbiota, fetus, newborn, infant
Citation: Senn V, Bassler D, Choudhury R, Scholkmann F, Righini-Grunder F, Vuille-dit-Bille RN and Restin T (2020) Microbial Colonization From the Fetus to Early Childhood—A Comprehensive Review. Front. Cell. Infect. Microbiol. 10:573735. doi: 10.3389/fcimb.2020.573735
Received: 19 June 2020; Accepted: 28 September 2020;
Published: 30 October 2020.
Edited by:
Shai Bel, Bar-Ilan University, IsraelReviewed by:
Moran Yassour, The Hebrew University of Jerusalem, IsraelMelanie Lynn Conrad, Charité—Universitätsmedizin Berlin, Germany
Copyright © 2020 Senn, Bassler, Choudhury, Scholkmann, Righini-Grunder, Vuille-dit-Bille and Restin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tanja Restin, dGFuamEucmVzdGluQHV6aC5jaA==
 Viola Senn
Viola Senn Dirk Bassler
Dirk Bassler Rashikh Choudhury2
Rashikh Choudhury2 Tanja Restin
Tanja Restin