- 1Institutes of Biomedical Sciences, Shanxi University, Taiyuan, China
- 2Department of Microbiology, Changzhi Medical College, Changzhi, China
- 3Shanxi Academy of Advanced Research and Innovation, Taiyuan, China
The ADP ribosylation factor (ARF) GTPase activation protein ASAP1 possesses multiple biological functions, including regulation of cytoskeletal dynamics, small GTP-binding protein receptor recycling, and intracellular vesicle trafficking. Recently, ASAP1 polymorphisms have been reported to be associated with human susceptibility to tuberculosis (TB) according to a large-scale genome-wide association study (GWAS); ASAP1 expression affects dendritic cell migration, which may be involved in TB predisposition. However, it remains unclear whether ASAP1 affects TB in vivo. To address this issue, we used zebrafish as a model system to examine the effects of Asap1 against Mycobacterium marinum, an organism closely related to Mycobacterium tuberculosis. Two zebrafish asap1 homologs (asap1a and asap1b) were identified and characterized. By morpholino knockdown of asap1a and asap1b as a whole, we found that the asap1 morphants showed a higher mycobacterial load than the controls, which was almost rescued by injecting asap1 mRNA that confers resistance to mycobacterial infection. These Asap1-depleted zebrafish also exhibited decreased macrophage migration in response to tail injury or upon infection with M. marinum in the hindbrain ventricle, which was also proved in THP1-derived macrophages of knockdown ASAP1. Together, these findings represent a new perspective on the role of Asap1 in resistance to mycobacterial infection.
Introduction
Despite advances in diagnosis and treatment, tuberculosis (TB) remains a major global health concern, causing millions of deaths annually (Lewinsohn and Lewinsohn, 2019). It has been estimated that approximately one-third of the world's population has been infected with the pathogen Mycobacterium tuberculosis (Mtb), with nearly 5–10% of infected individuals progressing into active TB, although most remain in a latent state (Marino et al., 2004; Nguyen-Chi et al., 2015). The World Health Organization (WHO) set a goal to end TB by 2030, but with the emergence of antibiotic-resistant or multi-antibiotic-resistant strains, this is a huge challenge (WHO, 2019). Besides, as the specific pathogenesis of TB is still unclear, it is difficult to effectively treat patients with TB to completely eradicate the infection.
ASAP1 (ArfGAP with SH3 domain, ankyrin repeat, and PH domain 1), a member of the ArfGAP (ArfGTPase-activating protein) family, plays a major regulatory role in cell membrane remodeling, cytoskeletal organization, and tumor invasion and metastasis (Randazzo et al., 2000, 2007; Liu et al., 2002; Tien et al., 2014; Schreiber et al., 2019). Recently, the function of ASAP1 has also been explored in the field of infectious diseases, especially with regard to TB susceptibility. A recent genome-wide association study (GWAS) reported that single-nucleotide polymorphisms in the ASAP1 intron were associated with the risk of TB in a Russian population (Curtis et al., 2015). Subsequently, two genetic polymorphism sites in ASAP1 were also found to be related to TB susceptibility in a Chinese Xinjiang Muslim population (Wang et al., 2018). Moreover, an allele of rs4733781 in the ASAP1 intron has been associated with a decreased occurrence of TB in the Han Chinese population (Chen et al., 2019). Nevertheless, the detailed regulatory effects of ASAP1 during host infection with Mycobacterium remain to be elucidated.
The interaction between the host immune system and Mycobacterium is very complex, with the most focused site of interplay being the tuberculosis granuloma, an organized structure consisting of macrophages, T and B lymphocytes, and various other immune cells (Davis and Ramakrishnan, 2009; Silva Miranda et al., 2012; Huang et al., 2017). Among these cells, macrophages serve as important phagocytic and primary constituent cells, and play a significant role in the host innate immune response against mycobacterial infection, including from the primary infection to bacillary dissemination, and from latency to activation of TB (Das et al., 2013; Korb et al., 2016; Nathan, 2016). Notably, a recent study showed that the loss of ASAP1 impaired dendritic cell migration, which may affect the host adaptive immune response to TB infection (Curtis et al., 2015). However, the effect of ASAP1 expression level on macrophage migration has not been reported.
Progress toward fully understanding the occurrence and development of TB has also been hindered by the lack of an appropriate and tractable infectious animal model that can suitably account for the complexity of host genetic factors and pathogen interactions (Guirado et al., 2015). As an alternative, the model of zebrafish infected with M. marinum (Mm), a close genetic relative of Mtb, has proven very effective for understanding TB pathogenesis, given the ability of Mm to induce granuloma-like disease (Helguera-Repetto et al., 2004; Cronan and Tobin, 2014). This model also allows the evaluation of host innate immune reaction during the first week of life, as the zebrafish adaptive immunity is not yet mature at the early larval developmental stage (Tobin and Ramakrishnan, 2008; van der Vaart et al., 2013; Tien et al., 2014; Moule and Cirillo, 2020)
In this study, we explored the role of ASAP1 in macrophage migration during Mtb infection, taking advantage of the optical transparency of the zebrafish larvae infection model. In particular, we explored the function of Asap1 in the process of zebrafish infection with Mm within the first 10 days of development, when host and pathogen interact in the sole context of innate immunity in vivo. These analyses will expand current understanding on the role of ASAP1 in anti-mycobacterial infection, and explore a preliminary mechanism for ASAP1-mediated TB susceptibility.
Materials and Methods
Bacterial Culture
The pTEC27 plasmid (Takaki et al., 2013) expressing the tdTomato protein, a kind gift from Lalita Ramakrishnan (Department of Medicine, University of Cambridge, UK), was used to transform Mm by electroporation. Mm was a kind gift from Professor Chen Niu (School of Basic Medical Sciences, Fudan University, China). tdTomato-Mm were cultured at 28°C in 7H9 broth supplemented with 0.2% glycerol, 10% albumin–dextrose–catalase (BINDER, Qingdao, China), 0.05% Tween 80, and hygromycin (50 ng/mL) or kanamycin (50 ng/mL). Mtb strain H37Ra was a kind gift from Professor Yujiong Wang (School of Life Science, Ningxia University, China) and cultured in 7H9 broth with the same enrichment albeit at 37°C. All bacteria were washed with phosphate buffered saline (PBS), and bacterial clumps were destroyed by passing through syringes prior to use for injecting into zebrafish or infecting macrophages.
Zebrafish Lines and Ethics Statement
All zebrafish experiments were carried out according to the China guidelines for the handling of laboratory animals. The ethics committee of Shanxi University approved this study (SXULL2019004). Wild-type zebrafish (AB) were purchased from the China Zebrafish Resource Center (CZRC) and maintained in a recirculating aquatic system at a constant temperature (28°C) with a 10 h dark/14 h light cycle.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA of zebrafish embryos or larvae at different developmental stages were isolated using TRIzol reagent (TaKaRa, Shiga, Japan) according to the manufacturer's protocol, and 1,000 ng RNA was used for reverse transcription to cDNA using the PrimeScript™RT Master Mix kit (TaKaRa). qRT-PCR was performed on a LightCycler 480 system (Roche, Basel, Switzerland) using SYBR-Green mix (Bio-Rad, Hercules, CA, USA) under the following conditions: initial denaturation for 5 min at 95°C, 45 cycles of amplification including denaturation step: 15 s at 95°C, annealing stage: 45 s at 58°C, and extension step: 1 min at 72°C. PCR amplification of the zebrafish β-actin gene was concomitantly performed as a reference. Data were normalized to β-actin using the 2−ΔΔCt method. The primers used are shown in Table 1.
Whole Mount in situ Hybridization
asap1a and asap1b probe template genes were amplified from 3 days post-fertilization (dpf) zebrafish by reverse transcription-polymerase chain reaction (RT-PCR) using the probe primers (Table 1) and ligated with pGEM-T vectors. Digoxigenin (DIG)-labeled sense and anti-sense RNA probes were transcribed according to the kit instructions (Promega, Madison, WI, USA) from recombinant pGEM-T-asap1a and pGEM-T-asap1b probe plasmids. The sense probe served as negative control. The detailed process of in situ hybridizations on whole mount embryos was previously described (Nguyen-Chi et al., 2015; Sanderson et al., 2015). Briefly, embryos from different periods were collected and the chorions were gently removed by hand. In particular, 0.0045% 1-phenyl-2-thiourea (PTU; Sigma Aldrich, St. Louis, MO, USA) solution was used to prevent the formation of melanin pigment in 24 h post-fertilization (hpf) embryos. Next, dechorionated embryos were fixed in 4% paraformaldehyde in PBS overnight at 4°C and dehydrated in 100% methanol for at least 2 h at −20°C. Then, the embryos were subjected to rehydration, prehybridization, and hybridization with antisense DIG-labeled RNA (Roche), blocked for unspecific binding sites in 1% bovine serum albumin (Solarbio, Beijing, China) and 1% sheep serum (Solarbio), and incubated with anti-DIG antibody (1:10,000; Roche). Finally, anti-DIG alkaline phosphatase (Roche) and chromogenic substrate NBT/BCIP (Roche) were used to stain the embryos. Images were obtained using light microscopy (SZX16, Olympus, Tokyo, Japan) with a 6.3 × objective.
Morpholino
Morpholino oligonucleotide (MO) (Gene Tools, Philomath, OR, USA) were diluted to 0.5 mM in 1 × Danieau buffer [58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca (NO3)2, and 5.0 mM HEPES; pH 7.6] containing 10% phenol red (Sigma Aldrich). 1 nL MO or MO mixture (MOs) were injected into 1-cell stage embryos using an injector (PV830; WPI, Sarasota, FL, USA) as described (Clay et al., 2008; Yuan and Sun, 2009; Roca and Ramakrishnan, 2013). asap1a (ENSDART00000144870.3) ATG-MO (5′-GAG GAC CTC AAG GCT CTG TAG TCA C-3′) plus asap1b (ENSDART00000145466.3) ATG-MO (5′-TAA GAC GAG AGG ATG ATG AC-3′) were used as the MO-asap1 mixture, asap1a control MO (5′-GAG CAG CTC ATC GCT CTC TAC TCA C-3′) plus asap1b control MO (5′-TAA CAC CAG ACG ATC ATC ACC TCA T-3′) were used as the MO-NC mixture.
Western Blotting of Zebrafish
Approximately 100 embryos or larvae at each stage of 1, 5, and 10 dpf were prepared for protein extraction. One dpf embryos were particularly removed from chorions in batches by using 1 mg/mL pronase (Sigma Aldrich) and swirling occasionally at 28°C for 17 min until complete dechorionation. The floating chorions were decanted after rinsing three times in cold Dulbecco's PBS (DPBS) solution. All the dechorionated embryos or larvae were gently blown repeatedly in DPBS solution and lightly centrifugated at 24 × g for 10 min at 4°C, the supernatant (yolk) was removed. Dechorionated and deyolked embryos or larvae were transferred to cold lysis buffer [20 mM Tris-HCl, pH 7.4, 10% glycerol, 187 mM NaCl, 2 mM ethylenediaminetetraacetic acid, 1% Triton X-100, and protein inhibitor cocktail (Sigma Aldrich)], lysed on ice for 30 min and pipetted every 10 min, followed by centrifugation at 10,000 × g for 10 min at 4°C. In particular, 5 and 10 dpf larvae were homogenized with ultrasonic disruption at 500 W until uniform in consistency and then centrifugated. The supernatant (zebrafish protein) was collected, resolved by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to immunoblot polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). The membrane was blocked using 5% non-fat milk with 0.5% Tween-20 for 1 h at room temperature and washed three times with Tris-Buffered Saline Tween-20 (each 10 min). The membrane was then incubated using the primary antibody against ASAP1 with a peptide from mouse as immunogen (1:2,500; Abcam11011, Cambridge, UK) and β-actin (1:5,000; Proteintech, Rosemont, IL, USA) at room temperature for 1 h, and then washed three times with Tris-Buffered Saline Tween-20. Next, the membrane was incubated using horseradish peroxidase-labeled secondary goat anti-rabbit (1:2,000; absin, Shanghai, China) or goat anti-mouse (1:2,000; absin) antibody for 1 h at room temperature, followed by three washes with Tris-Buffered Saline Tween-20. The membrane was visualized using a commercial ECL kit (GE Healthcare, Chicago, IL, USA).
Injection of Mm Into Zebrafish
Embryos were collected and kept in E3 medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, 10% methylene blue). tdTomato-Mm were prepared and 50 colony-forming units (CFU) microinjected into 16–1,000 cell stage or 100 CFU into the caudal vein of 28–30 hpf embryos according to published procedures (Takaki et al., 2013). Previously, PTU was added to the E3 medium to prevent melanization when the embryos were ~24 hpf. Subsequently, confocal images (LSM710, Carl Zeiss, Germany) were acquired from zebrafish larvae at 5 days post- infection (dpi) under anesthesia with 0.016% ethyl 3-aminobenzoate (Tricaine, Sigma Aldrich) in 2.5% methyl cellulose (Sigma Aldrich) drop. Finally, the overall bacterial burden of whole larvae was quantified by fluorescence quantification of the images using image J software (National Institutes of Health, Bethesda, MD, USA).
Phenotype Rescue Experiments in Zebrafish
The full-length coding gene sequence of asap1a or asap1b were amplified by using specific primers (Table 1) from the cDNA of 3 dpf zebrafish and subcloned into pCS2+ vectors using the Gibson Assembly enzyme-reagent mixture (NEB, Frankfurt am Main, Germany). The recombinant pCS2+-asap1a or pCS2+-asap1b plasmids were linearized using the Not I restriction enzyme (NEB). asap1a or asap1b mRNA were prepared by in vitro transcription using mMESSAGE mMACHINE™ SP6 Transcription Kit (Ambion, Austin, TX, USA) from pCS2+-asap1a or asap1b plasmids. The synthetic mRNA, at the concentration of 200 ng/μL, were co-injected into 1-cell stage embryos with asap1 morpholino mixture for rescue experiments according to the previous method (Rosen et al., 2009).
Zebrafish Macrophages Recruitment Assay
Neutral red staining was used to observe the macrophage, following published procedures (Herbomel et al., 2001; Davis and Ramakrishnan, 2009; Shiau et al., 2015). The 3 dpf zebrafish were wounded consistently by tailfin transection with a sterile scalpel. Subsequently, zebrafish were immersed in neutral red solution (2.5 μg/mL + 0.003% PTU) (Sigma Aldrich) at 28°C in the dark for 7–9 h. Furthermore, in order to observe the response of macrophages against mycobacterial infection, Mm were injected into the hindbrain of 30–32 hpf zebrafish embryos with 50 CFU according to the previous method (Gutzman and Sive, 2009). Zebrafish were then incubated in neutral red solution as already described. Anesthetized zebrafish were embedded into solidified agarose drops and viewed under light microscopy to acquire images (SteREO Discovery.V20; Carl Zeiss, Oberkochen, Germany).
siRNA Transfection
Human monocyte-like THP-1 cells were purchased from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) and cultured in RPMI-1640 supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA). A pool of three ASAP1-specific small-interfering RNA (siRNA) or a control siRNA (GenePharma, Shanghai, China) (ASAP1 siRNA: 5′-CAC CUU GGA UUC UUU GUU A-3′, 5′-GAC CUG AUA UCA CAU AAU A-3′, 5′-CUG CCC UAG ACA UAG CAA A-3′; control siRNA 5′-CAU GCA UCG UAG CUA UGC AUU-3′) was transfected into human monocyte-like THP-1 cells using Lipofectamine3000 (Thermo Fisher Scientific) following treatment using phorbol-12-myristate-13-acetate (PMA, Sigma Aldrich) at 100 ng/mL for 48 h. Total protein of 24 h post transfection was extracted from the THP-1 cells using lysis buffer (Solarbio). Western blotting was performed using an anti-ASAP1 antibody (1:1,000; sc374410, Santa Cruz Biotechnology, Dallas, TX, USA) and anti-β-actin antibody (1:5,000; Proteintech) to assess knockdown efficiency.
Transwell Migration Assay
THP-1 cells were infected with recombinant strains tdTomato-H37Ra according to a previously described protocol (Behar et al., 2011). Firstly, THP-1 cells were seeded at a density of 106 cells per well in RPMI-1640 medium supplemented with 10% fetal bovine serum for 48 h with PMA incubation and then infected with H37Ra at multiplicity of infection (MOI) of 5. After 12 h, the supernatant was collected for use as a chemoattractant in the subsequent Transwell assay. The cell migration assay was conducted in 24-well Transwell plates of 8.0-μm pore size (Corning Costar, Armonk, NY, USA), referring to the previous method (Smith et al., 2015). Prior to the assay, THP-1 cells with PMA incubation were starved overnight in RPMI-1640, and 2 × 104 cells suspended in serum-free RPMI-1640 were seeded onto each upper chamber, whereas 10% FBS RPMI-1640 medium and the supernatant obtained following cell infection with H37Ra were mixed in equal volumes to 1 mL, and added to the lower chambers. The Transwell plates were maintained at 37°C in a humidified atmosphere with 5% CO2. After 24 h, cells remaining on the upper surface of the membrane were completely removed with cotton swabs, whereas cells that migrated through the permeable membrane to the lower surface were fixed in 4% paraformaldehyde followed by staining with 0.1% crystal violet (Solarbio). Images of migrated cells were acquired using light microscopy (DMi1, Leica, Wetzlar, Germany).
Statistics
Results are presented as the mean ± SEM. All data were analyzed using Prism 7 software (GraphPad, La Jolla, CA, USA). Statistical significances were evaluated by the Kruskal-Wallis and the Dunn post-test among groups, the Mann-Whitney test or the Student's unpaired t-test between two groups. The significant value was set at P < 0.05.
Results
Characterization of the Zebrafish asap1 Gene
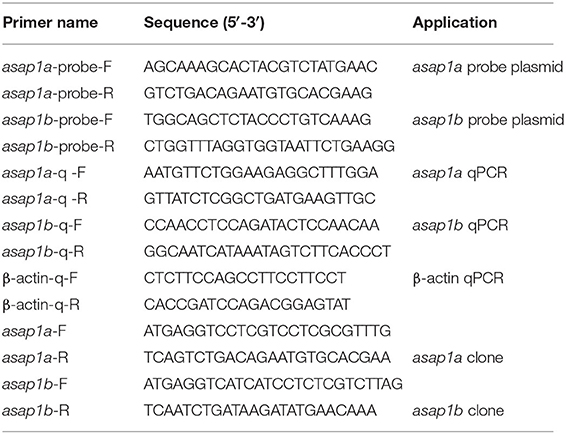
To identify the zebrafish asap1 genes, the human ASAP1 protein sequence was used as the reference to perform the p-n Blast against zebrafish genomes (GRCz11). In contrast to the single human ASAP1 gene, we found two hits (asap1a and asap1b) in the zebrafish genome. Localized on chromosome 2, asap1a contains 29 exons and encodes a protein of 1151 amino acids, while asap1b on chromosome 24 contains 29 exons and encodes a protein of 1140 amino acids. Primary structure analysis shows that both zebrafish Asap1a and Asap1b share identical functional domains with human ASAP1, including BAR (Bin, Amphiphysin and Rvs167 and Rvs161), ArfGAP (ADP ribosylation factor-GTPase activating protein), SH3 (Src Homolog 3), ANK (ankyrin) repeat, and PH (Pleckstrin Homology) domains. Zebrafish Asap1a and Asap1b share ~76 and 78% identity with human ASAP1, respectively (Figure 1). Syntenic and phylogenetic analysis reveals that zebrafish asap1a and asap1b are evolutionarily conserved in their chromosome linkage positions and evolutionary relationship (Supplementary Figure 1). Through temporal expression lapse, we found asap1a and asap1b mRNA major concentrated at 3 hpf, suggesting that asap1 may be under maternal control (Figure 2A). However, the mRNA of asap1a and asap1b were at a low level from 12 to 48 hpf, and subsequently increased from 72 hpf according to the qRT-PCR analysis. Whole mount in situ hybridization demonstrates that asap1a and asap1b transcripts are ubiquitously distributed at early stages of 4 and 10 hpf, but the expression of asap1a and asap1b are getting weaker at 18 and 24 hpf, especially in the tail muscle tissue at 24 hpf (Figure 2B).

Figure 1. Multiple sequence alignment of zebrafish Asap1a (ENSDART00000144870.3) and Asap1b (ENSDART00000145466.3) with human ASAP1 (ENST00000518721.6), rat ASAP1 (ENSRNOT00000079524.1), and mouse ASAP1 (ENSMUST00000177374.7). Five domains common between all ASAP1 are indicated by different color boxes.
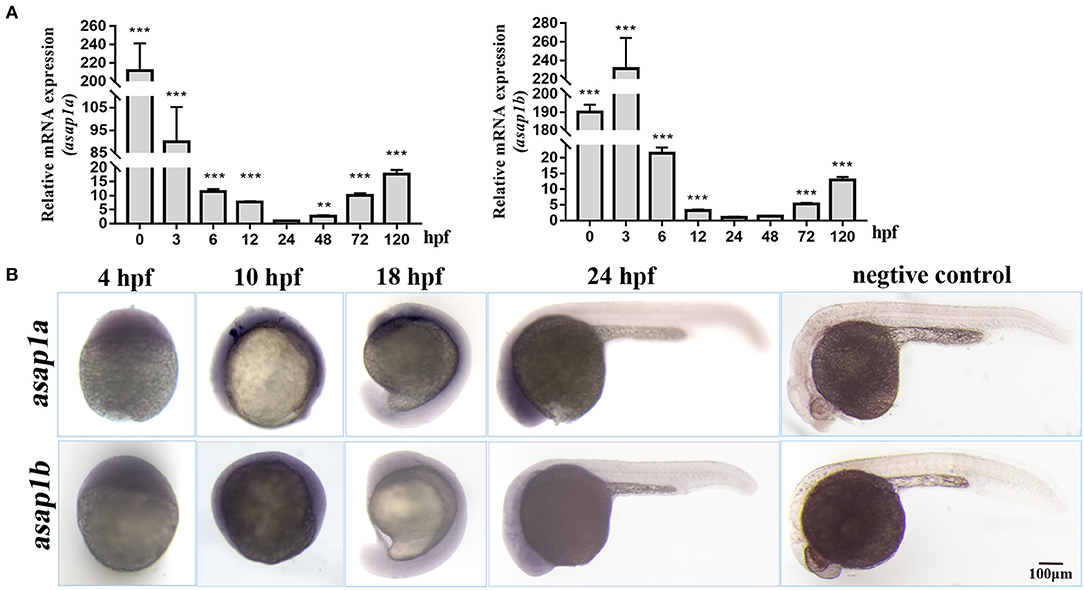
Figure 2. Expression pattern of zebrafish asap1 homologs during embryonic development. (A) Temporal expression profiles of asap1a and asap1b by qRT-PCR. **P < 0.01, ***P < 0.001 vs. 24 hpf control group using Student's unpaired t-test. Data shown are representative of 3 independent experiments. (B) Spatial expression of asap1a (upper panels) and asap1b (lower panels) by whole mount in situ hybridization in wild-type embryos, the sense probe served as negative control (Scale bar, 100 μm).
asap1 Morphants Are Susceptible to Mm Infection
To explore the function of Asap1 in zebrafish against Mm infection, firstly the function model of zebrafish asap1 loss was generated by morpholino mediated knockdown methods (Stainier et al., 2017). By injecting of specific anti-sense morpholinos targeting the exon 1 of premature asap1a and asap1b mRNA, we successfully detected high efficiency knockdown for asap1 until 10 dpf by qRT-PCR and western blotting (Figures 3A,B), which makes the asap1 morphants acceptable for follow-up studies. Notably, the Asap1 antibody used in this experiment was derived from a recombinant peptide of 20 amino acids corresponding to the mouse ASAP1 protein, and the antigen region was highly conserved in zebrafish Asap1 proteins (Figure 1). Thus, it was supposed to recognize both Asap1a and Asap1b. The result of western blotting revealed that we successfully detected the expression level of the zebrafish protein but failed to distinguish the Asap1 homologs, owing to the high similarity of the two Asap1 proteins. However, all morphants developed normally and didn't show any obvious morphological defects within 2 weeks. Next, to evaluate the roles of asap1 against Mm infection in vivo, Mm were firstly injected into the yolk of morphant embryos at the 16–1,000 cells stage (Figure 3C). We then followed existing protocol to detect tdTomato-Mm fluorescent expression in 5 dpi zebrafish (Benard et al., 2012; Takaki et al., 2013). Preliminary experimental results showed that Asap1 knockdown enhanced Mm infection efficiency by ~2-fold (Figures 3D,E), but a single knockdown of Asap1a or Asap1b did not cause increased susceptibility to Mm over controls (Supplementary Figures 2A–D). Intravenous injection with Mm is commonly used to evaluate the infection in zebrafish embryos (Figure 3F). Here, Mm was also intravenously injected into 28–32 hpf embryos after treatment with control or asap1 MOs. The results also showed that compared with the control group, asap1 morphants markedly increased the infection efficiency of Mm with 2.5-fold more bacteria (Figures 3G,H) but asap1a or asap1b morphants did not (Supplementary Figures 2E–H). Meanwhile, we also found that mortality rate did not significantly differ (no statistical difference) among Mm infected morphants no matter whether it was introduced through yolk (Supplementary Figure 3A) or intravenous injection (Supplementary Figure 3B). Additionally, to definitively demonstrate that the loss of Asap1 was causative for susceptibility phenotype, we performed a rescue experiment in asap1 morphants injecting with synthetic asap1a and asap1b mRNA. Expectedly, the re-expression of asap1a and asap1b in zebrafish, as evidenced by the qRT-PCR and western blotting (Supplementary Figure 4), can partially rescue the susceptibility of asap1 morphants to Mm infection and the survival of zebrafish post-infection (Figures 3D–H, Supplementary Figure 3A,B). Taken together, these results showed that knockdown of Asap1 as a whole weakened zebrafish resistance to Mm infection even if they didn't obviously affect the survival status within 10 d post-infection.
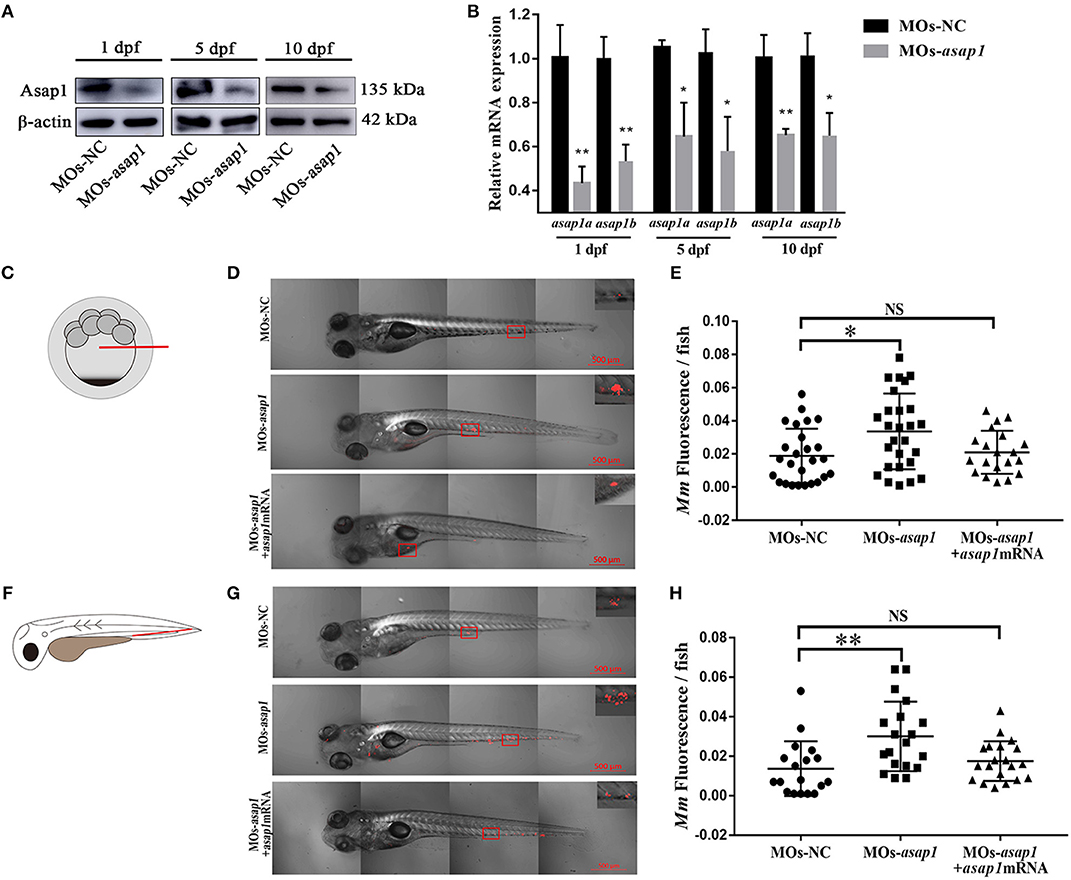
Figure 3. asap1 morphants exhibit altered susceptibility to Mm infection. (A,B) Western blotting (A) and qRT-PCR (B) analysis of the knockdown efficiency of asap1 morphants in 1, 5, and 10 dpf zebrafish. *P < 0.05, **P < 0.01 vs. control morphants using Student's unpaired t-test. The antibody was against both Asap1a and Asap1b. Data shown are representative of 3 independent experiments. (C) Schematic diagram of the yolk injection method. (D,E) Representative fluorescence images and quantification of bacterial burdens by fluorescence pixel counts of controls, asap1 morphants, or asap1 morphants treated with asap1 mRNA at 5 dpi with equivalent bacterial inocula by the yolk injection method (scale bar 500 μm); *P < 0.05, P-values calculated by Kruskal-Wallis with Dunn's post-test. n (MOs-NC group) = 27, n (MOs-asap1 group) = 27, n (MOs-asap1+asap1mRNA group) = 20. Mean ± SEM from four pooled independent experiments. (F) Schematic diagram of the intravenous injection method. (G,H) Representative fluorescence images and quantification of bacterial burdens by fluorescence pixel counts of controls, asap1 morphants or asap1 morphants treated with asap1 mRNA at 5 dpi with equivalent bacterial inocula by the intravenous injection method (scale bar, 500 μm); **P < 0.01, NS, not significant, P-values calculated by Kruskal-Wallis with Dunn's post-test. n (MOs-NC group) = 18, n (MOs-asap1 group) = 19, n (MOs-asap1+asap1mRNA group) = 20. Each dot represents one larva. Mean ± SEM from three pooled independent experiments. MOs, morpholino oligonucleotide mixture. asap1 means asap1a and asap1b.
asap1 Morphants Exhibit Delayed Macrophage Recruitment
A previous study showed that ASAP1 was involved in regulating dendritic cell migration (Curtis et al., 2015). In the zebrafish infectious model, neutrophils and macrophages are the primary cells to act against pathogens (Bernut et al., 2016; Rosowski et al., 2016). To dissect the mechanism of Asap1 in regulating zebrafish against infection, we further examined the effect of Asap1 on macrophage migration in zebrafish by tailfin click and Mm hindbrain injection. In tailfin transection assay, asap1 morphants exhibited significant defects of macrophage migration in response to injury (Figures 4A,B). The hindbrain ventricle is a suitable site to evaluate macrophage recruitment toward local infections (Benard et al., 2012; Pagan et al., 2015). When we further injected hindbrain ventricles of asap1 morphants with Mm, we noted that far fewer macrophages were recruited and crossed the brain-blood barrier than in control morphants (Figures 4C,D). Meanwhile, we confirmed that total macrophage numbers did not differ between asap1 and control morphants (Supplementary Figure 5), using a published method that quantified neutral red stained macrophages in whole zebrafish larvae (Tobin et al., 2010). These results suggest that impaired macrophage migration is not due to a reduction in total macrophage number.
Figure 4. asap1 morphant macrophages exhibit impaired migration. (A) Representative light microscopy images of neutral red stained macrophages of transgenic morphants migrated to tailfin injury at 9 h post injury. (B) Numbers of recruited neutral red stained macrophages in the tailfin injury at 9 h post injury (scale bar, 100 μm); *** P < 0.001, P-values calculated by Student's unpaired t-test. n (MOs-NC group) = 45, n (MOs-asap1 group) = 40. (C) Representative light microscopy images of neutral red stained macrophages following hindbrain injection with Mm (left panels) or mock (right panels) into morphants at 9 h post infection. (D) Numbers of recruited neutral red stained macrophages in the hindbrain at 9 h post infection (scale bar, 100 μm); ***P < 0.001, NS, not significant, P-values calculated between two groups by Student's unpaired t-test. n (MOs-NC group/Mm) = 77, n (MOs-asap1 group/Mm) = 81, n (MOs-NC group/mock) =34, n (MOs-asap1 group/mock) =29. Each dot represents one larva. Mean ± SEM from two pooled independent experiments. MOs, morpholino oligonucleotide mixture. asap1 means asap1a and asap1b.
Knockdown of ASAP1 Impaired THP-1 Cells Migration
To examine the role of ASAP1 in regulating macrophage motility, THP-1 cells were transfected with either siRNA against ASAP1 or control siRNA, and then subjected to the Transwell assay to determine cell migration ability. Western blotting analysis revealed that ASAP1 was knocked down successfully (Figure 5A). At 24 h post treatment with ASAP1 siRNA or control siRNA, the cells were transferred to the upper chamber of a Transwell culture plate. Then, the medium supernatant of THP-1 cells infected with H37Ra for 24 h was added to the lower chamber (Figure 5B). The results showed that ASAP1 knockdown THP-1 cells exhibited reduced migration ability compared to that of their control-transfected counterparts (Figures 5C,D).
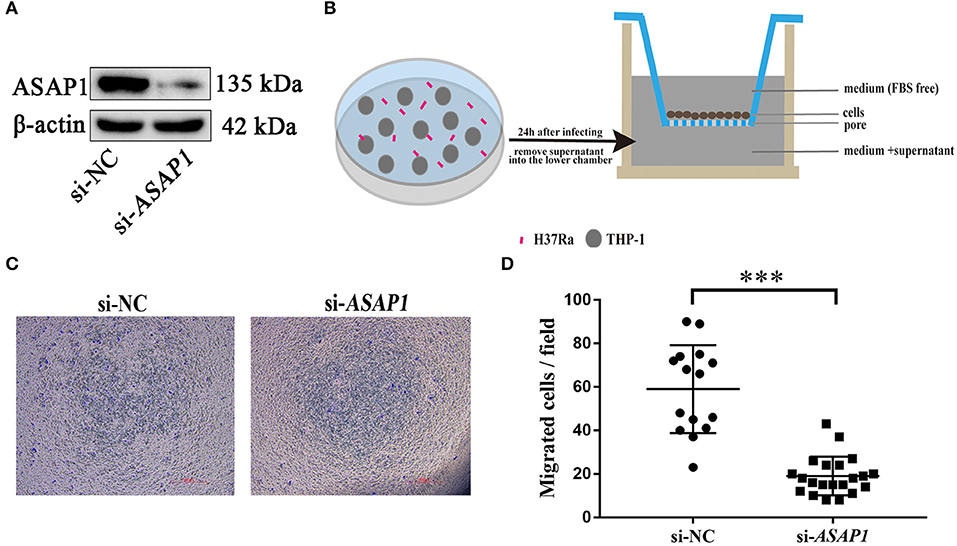
Figure 5. Migration of cells is impaired in THP-1 cells upon knockdown of ASAP1. (A) Western blotting analysis of the knockdown efficiency of ASAP1 when THP-1 cells were transfected with control or ASAP1 siRNA. (B) Schematic diagram of the Transwell protocol. (C,D) Transwell assay was applied to evaluate the effect of ASAP1 on cell migration (scale bar, 500 μm); ***P < 0.001, P-values calculated by the Mann-Whitney test. Each dot represents one scope. Mean ± SEM from two independent experiments.
Discussion
In our work, we initially characterized zebrafish asap1a and asap1b, both of encoding proteins with very high amino-acid sequence identity to mammalian ASAP1. We then found that Asap1 deficiency in zebrafish significantly enhanced susceptibility to Mm, which could be rescued by injecting with asap1 mRNA. To fully validate susceptibility phenotypes, we compared two different mycobacterial injection methods for evaluating the Asap1-mediated susceptibility to TB: yolk injection and intravenous injection. In the former method, Mm were injected into 16–1,000 cell embryos, which affords easy manipulation but confers a high mortality rate. In the latter method, Mm were intravenously injected into 28–32 h embryos, which represents the optimal method for researching Mm susceptibility in zebrafish owing to the formation of a rapid systemic infection and allowing precise fluorescence-based measurements of overall bacterial burden. Additionally, these two different zebrafish asap1 isoforms, asap1a and asap1b, did not alter the susceptibility of zebrafish to Mm when they were knocked down separately, indicating that both of these proteins contribute to susceptibility phenotypes and may have partially overlapping functions in zebrafish. Furthermore, asap1a and asap1b share 76% identity in protein sequence with each other (Figure 1), which makes it difficult to distinguish the different functions and the redundant degree between asap1a and asap1b. Therefore, it is still unclear whether the roles of asap1a and asap1b are overlapping or distinct. Here, we should point out that mortality rate in asap1 morphants was not significantly higher than that in the controls after an increase in bacterial load, unlike lta4h morphants and csf1r mutants (Tobin et al., 2010; Pagan et al., 2015). This outcome suggests that Asap1-induced elevation in bacterial load might not strongly influence infection lethality initiated via normal Mm inocula, at least in the zebrafish model. The lack of an effect on mortality rates may also be due to differences in the complex progressive disease of TB across hosts. Future studies should explore in greater detail for the association between bacterial load and TB pathological mechanisms when Asap1 is deleted in zebrafish. Despite the need for further elucidation of mechanisms and infection processes, our zebrafish model highlights the importance of ASAP1 in host resistance to mycobacterial entry.
ASAP1 polymorphisms are associated with TB susceptibility in many populations (Curtis et al., 2015; Wang et al., 2018; Chen et al., 2019). But we know little about how ASAP1 polymorphisms confer altered TB susceptibility or the underlying pathophysiological mechanisms. Moreover, currently recognized ASAP1 polymorphisms associated with TB susceptibility are all intronic, increasing the difficulty of analyzing the relationship between ASAP1 and TB susceptibility. Genetic defects in ASAP1 mediate susceptibility to Salmonella infections, but a similar connection has not been investigated for Mycobacterium. The ArfGEF (Arf Guanine-nucleotide exchange factor) and ArfGAP cycles ensure the alternation of Arf activation and deactivation, which jointly regulate pathogens entry into cells (Humphreys et al., 2013). Recent findings have illustrated that ArfGAP engages to modulate mycobacterial colonization by controlling the actin cytoskeleton (Song et al., 2018). It appears possible that bacteria might interfere with ArfGAP in order to sustain activation of Arf, which is required for rearrangements of the actin cytoskeleton and facilitates bacterial invasion via the cell membrane surface complex, such as through ruffles and podosomes (Bharti et al., 2007; Rafiq et al., 2017). Therefore, we examined the role of ASAP1 on the migration effect of phagocytes to Mm infection in the zebrafish model.
Macrophages phagocytose Mtb and then secrete a large number of inflammatory factors to recruit other macrophages or immune cells to form an organized granuloma structure, which constitutes the initial reaction in TB disease and benefits the host through the control of Mtb in the body and the limitations of the lesion. However, from another perspective, once macrophages parasitized by Mtb in the granuloma become necrotic, bacteria are released, and spread to the whole body, such that the macrophages and granuloma actually function as a bacterial multiplicative tool for expanding infection (Cambier et al., 2014; Guirado et al., 2015; Martin et al., 2016). Nevertheless, the ability of macrophages in immune response to kill Mtb is crucial in terms of fighting TB. A 2016 study reported that inhibition of macrophage migration in zebrafish delayed phagocytosis of Mm, thus potentially driving susceptibility to TB (Berg et al., 2016). In the present study, we found that the knockdown of Asap1 in zebrafish significantly decreased the migration of macrophages during Mm infection, moreover, similar result was obtained using THP-1 cells as determined with a Transwell assay, which together strongly indicated that ASAP1 regulated the migration of macrophages. We further suppose that such impaired migration of macrophages may result in a slow microbicidal response or increase the time required to initiate an effectively adaptive immune response for Mycobacterium, thereby inducing a weak innate and adaptive immune response to control mycobacterial proliferation in zebrafish larvae. In contrast, Torraca et al. (2015) and Sommer et al. (2020) reported that diminished macrophage migration caused by cxcr3 depletion might limit dissemination of mycobacterial infection and lead to an reduction in total bacterial burden in zebrafish. Basing on the above viewpoints, it will therefore be of great interest to investigate how macrophages interact with Mm in the context of ASAP1 deletion, using adult zebrafish with mature immune system. Together, Our work highlights the importance of ASAP1 in modulating the balance between innate immune control and mycobacterial infection and suggests a mechanistic explanation for the reported GWAS findings between ASAP1 and TB susceptibility. Moreover, our findings may provide an explanation regarding how a single genetic determinant might influence antimicrobial and immunological dysfunction of phagocytic cells. Considering a further finding that ArfGEFs and ArfGAPs might collaborate to control pathogen infection (Liu et al., 2005; Davidson et al., 2015), we speculate that ASAP1-mediated regulation of mycobacterial invasion occurs through various temporally and spatially adjusted mechanisms. Although herein we reveal only the basic morphology and give a preliminary description of the relationship between ASAP1 and Mycobacterium, the results of zebrafish Asap1 provide a new perspective on the debate regarding host susceptibility genes and Mycobacterium infection, which may facilitate the development of improved preventive and therapeutic strategies for TB. Toward this end, further detailed examination of the mechanisms underlying ASAP1 function in regulating mycobacterial invasion and persistence in macrophages is warranted.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The Ethics Committee of Shanxi University approved this study (SXULL2019004). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
ZZ and CW contributed to project conception and supervised the projects. JC, DW, and YW performed experiments. JC, GC, and ZZ performed data analysis and statistical tests and wrote the manuscript. All authors contributed to manuscript revision and approved the final submitted manuscript.
Funding
This study was supported by a sub-project of the National Major Research and Development Project of the Ministry of Science and Technology (No. 2017YFD0500303), the Shanxi Province Higher Education Technology Innovation Project (No. 2019L0035), and the Changzhi Medical College Spring sunshine Project (No. QDZ201906).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank professor Lalita Ramakrishnan (Department of Medicine, University of Cambridge, UK) for plasmid pTEC27, professor Chen Niu (School of Basic Medical Sciences, Fudan University, China) for Mm and professor Yujong Wang (College of Life Sciences, Ningxia University, China) for H37Ra.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.519503/full#supplementary-material
References
Behar, S. M., Martin, C. J., Booty, M. G., Nishimura, T., Zhao, X., Gan, H. X., et al. (2011). Apoptosis is an innate defense function of macrophages against Mycobacterium tuberculosis. Mucosal. Immunol. 4, 279–287. doi: 10.1038/mi.2011.3
Benard, E. L., Van Der Sar, A. M., Ellett, F., Lieschke, G. J., Spaink, H. P., and Meijer, A. H. (2012). Infection of zebrafish embryos with intracellular bacterial pathogens. J. Vis. Exp. 61:3781. doi: 10.3791/3781
Berg, R. D., Levitte, S., O'sullivan, M. P., O'leary, S. M., Cambier, C. J., Cameron, J., et al. (2016). Lysosomal disorders drive susceptibility to tuberculosis by compromising macrophage migration. Cell 165, 139–152. doi: 10.1016/j.cell.2016.02.034
Bernut, A., Nguyen-Chi, M., Halloum, I., Herrmann, J. L., Lutfalla, G., and Kremer, L. (2016). Mycobacterium abscessus-induced granuloma formation is strictly dependent on TNF signaling and neutrophil trafficking. PLoS Pathog. 12:e1005986. doi: 10.1371/journal.ppat.1005986
Bharti, S., Inoue, H., Bharti, K., Hirsch, D. S., Nie, Z., Yoon, H. Y., et al. (2007). Src-dependent phosphorylation of ASAP1 regulates podosomes. Mol. Cell. Biol. 27, 8271–8283. doi: 10.1128/MCB.01781-06
Cambier, C. J., Takaki, K. K., Larson, R. P., Hernandez, R. E., Tobin, D. M., Urdahl, K. B., et al. (2014). Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505, 218–222. doi: 10.1038/nature12799
Chen, C., Zhao, Q., Shao, Y., Li, Y., Song, H., Li, G., et al. (2019). A Common variant of ASAP1 is associated with tuberculosis susceptibility in the han chinese population. Dis. Markers 2019:7945429. doi: 10.1155/2019/7945429
Clay, H., Volkman, H. E., and Ramakrishnan, L. (2008). Tumor necrosis factor signaling mediates resistance to Mycobacteria by inhibiting bacterial growth and macrophage death. Immunity 29, 283–294. doi: 10.1016/j.immuni.2008.06.011
Cronan, M. R., and Tobin, D. M. (2014). Fit for consumption: zebrafish as a model for tuberculosis. Dis. Model Mech. 7, 777–784. doi: 10.1242/dmm.016089
Curtis, J., Luo, Y., Zenner, H. L., Cuchet-Lourenco, D., Wu, C., Lo, K., et al. (2015). Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration. Nat. Genet. 47, 523–527. doi: 10.1038/ng.3248
Das, R., Koo, M. S., Kim, B. H., Jacob, S. T., Subbian, S., Yao, J., et al. (2013). Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, E2997–3006. doi: 10.1073/pnas.1301128110
Davidson, A. C., Humphreys, D., Brooks, A. B., Hume, P. J., and Koronakis, V. (2015). The arf GTPase-activating protein family is exploited by Salmonella enterica serovar Typhimurium to invade nonphagocytic host cells. mBio. 6:14. doi: 10.1128/mBio.02253-14
Davis, J. M., and Ramakrishnan, L. (2009). The role of the granuloma in expansion and dissemination of early tuberculous infection. Cell 136, 37–49. doi: 10.1016/j.cell.2008.11.014
Guirado, E., Mbawuike, U., Keiser, T. L., Arcos, J., Azad, A. K., Wang, S. H., et al. (2015). Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. MBio 6, e02537–e02514. doi: 10.1128/mBio.02537-14
Gutzman, J. H., and Sive, H. (2009). Zebrafish brain ventricle injection. J. Vis. Exp. 26:1218. doi: 10.3791/1218
Helguera-Repetto, C., Cox, R. A., Munoz-Sanchez, J. L., and Gonzalez-Y-Merchand, J. A. (2004). The pathogen Mycobacterium marinum, a faster growing close relative of Mycobacterium tuberculosis, has a single rRNA operon per genome. FEMS Microbiol. Lett. 235, 281–288. doi: 10.1111/j.1574-6968.2004.tb09600.x
Herbomel, P., Thisse, B., and Thisse, C. (2001). Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 238, 274–288. doi: 10.1006/dbio.2001.0393
Huang, Z., Su, R., Deng, Z., Xu, J., Peng, Y., Luo, Q., et al. (2017). Identification of differentially expressed circular RNAs in human monocyte derived macrophages response to Mycobacterium tuberculosis infection. Sci. Rep. 7:13673. doi: 10.1038/s41598-017-13885-0
Humphreys, D., Davidson, A. C., Hume, P. J., Makin, L. E., and Koronakis, V. (2013). Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proc. Natl. Acad. Sci. U.S.A. 110, 16880–16885. doi: 10.1073/pnas.1311680110
Korb, V. C., Chuturgoon, A. A., and Moodley, D. (2016). Mycobacterium tuberculosis: manipulator of protective immunity. Int. J. Mol. Sci. 17:131. doi: 10.3390/ijms17030131
Lewinsohn, D. M., and Lewinsohn, D. A. (2019). New concepts in tuberculosis host defense. Clin. Chest. Med. 40, 703–719. doi: 10.1016/j.ccm.2019.07.002
Liu, Y., Loijens, J. C., Martin, K. H., Karginov, A. V., and Parsons, J. T. (2002). The association of ASAP1, an ADP ribosylation factor-GTPase activating protein, with focal adhesion kinase contributes to the process of focal adhesion assembly. Mol. Biol. Cell. 13, 2147–2156. doi: 10.1091/mbc.e02-01-0018
Liu, Y., Yerushalmi, G. M., Grigera, P. R., and Parsons, J. T. (2005). Mislocalization or reduced expression of Arf GTPase-activating protein ASAP1 inhibits cell spreading and migration by influencing Arf1 GTPase cycling. J. Biol. Chem. 280, 8884–8892. doi: 10.1074/jbc.M412200200
Marino, S., Pawar, S., Fuller, C. L., Reinhart, T. A., Flynn, J. L., and Kirschner, D. E. (2004). Dendritic cell trafficking and antigen presentation in the human immune response to Mycobacterium tuberculosis. J. Immunol. 173, 494–506. doi: 10.4049/jimmunol.173.1.494
Martin, C. J., Carey, A. F., and Fortune, S. M. (2016). A bug's life in the granuloma. Semin. Immunopathol. 38, 213–220. doi: 10.1007/s00281-015-0533-1
Moule, M. G., and Cirillo, J. D. (2020). Mycobacterium Tuberculosis dissemination plays a critical role in pathogenesis. Front. Cell Infect. Microbiol. 10:65. doi: 10.3389/fcimb.2020.00065
Nathan, C. (2016). Macrophages' choice: take it in or keep it out. Immunity. 45, 710–711. doi: 10.1016/j.immuni.2016.10.002
Nguyen-Chi, M., Laplace-Builhe, B., Travnickova, J., Luz-Crawford, P., Tejedor, G., Phan, Q. T., et al. (2015). Identification of polarized macrophage subsets in zebrafish. Elife. 4:e07288. doi: 10.7554/eLife.07288
Pagan, A. J., Yang, C. T., Cameron, J., Swaim, L. E., Ellett, F., Lieschke, G. J., et al. (2015). Myeloid growth factors promote resistance to mycobacterial infection by curtailing granuloma necrosis through macrophage replenishment. Cell Host Microbe. 18, 15–26. doi: 10.1016/j.chom.2015.06.008
Rafiq, N. B., Lieu, Z. Z., Jiang, T., Yu, C. H., Matsudaira, P., Jones, G. E., et al. (2017). Podosome assembly is controlled by the GTPase ARF1 and its nucleotide exchange factor ARNO. J. Cell. Biol. 216, 181–197. doi: 10.1083/jcb.201605104
Randazzo, P. A., Andrade, J., Miura, K., Brown, M. T., Long, Y. Q., Stauffer, S., et al. (2000). The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc. Nat. Acad. Sci. U.S.A. 97, 4011–4016. doi: 10.1073/pnas.070552297
Randazzo, P. A., Inoue, H., and Bharti, S. (2007). Arf GAPs as regulators of the actin cytoskeleton. Biol. Cell. 99, 583–600. doi: 10.1042/BC20070034
Roca, F. J., and Ramakrishnan, L. (2013). TNF dually mediates resistance and susceptibility to mycobacteria via mitochondrial reactive oxygen species. Cell. 153, 521–534. doi: 10.1016/j.cell.2013.03.022
Rosen, J. N., Sweeney, M. F., and Mably, J. D. (2009). Microinjection of zebrafish embryos to analyze gene function. J. Vis. Exp. 2009:1115. doi: 10.3791/1115
Rosowski, E. E., Deng, Q., Keller, N. P., and Huttenlocher, A. (2016). Rac2 functions in both neutrophils and macrophages to mediate motility and host defense in larval zebrafish. J. Immunol. 197, 4780–4790. doi: 10.4049/jimmunol.1600928
Sanderson, L. E., Chien, A. T., Astin, J. W., Crosier, K. E., Crosier, P. S., and Hall, C. J. (2015). An inducible transgene reports activation of macrophages in live zebrafish larvae. Dev. Comp. Immunol. 53, 63–69. doi: 10.1016/j.dci.2015.06.013
Schreiber, C., Saraswati, S., Harkins, S., Gruber, A., Cremers, N., Thiele, W., et al. (2019). Loss of ASAP1 in mice impairs adipogenic and osteogenic differentiation of mesenchymal progenitor cells through dysregulation of FAK/Src and AKT signaling. PLoS Genet. 15:e1008216. doi: 10.1371/journal.pgen.1008216
Shiau, C. E., Kaufman, Z., Meireles, A. M., and Talbot, W. S. (2015). Differential requirement for irf8 in formation of embryonic and adult macrophages in zebrafish. PLoS ONE 10:e0117513. doi: 10.1371/journal.pone.0117513
Silva Miranda, M., Breiman, A., Allain, S., Deknuydt, F., and Altare, F. (2012). The tuberculous granuloma: an unsuccessful host defence mechanism providing a safety shelter for the bacteria? Clin. Dev. Immunol. 2012:139127. doi: 10.1155/2012/139127
Smith, M. P., Young, H., Hurlstone, A., and Wellbrock, C. (2015). Differentiation of THP1 cells into macrophages for transwell co-culture assay with melanoma cells. Bio. Protoc. 5:1638. doi: 10.21769/BioProtoc.1638
Sommer, F., Torraca, V., Kamel, S. M., Lombardi, A., and Meijer, A. H. (2020). Frontline science: antagonism between regular and atypical Cxcr3 receptors regulates macrophage migration during infection and injury in zebrafish. J. Leukoc. Biol. 107, 185–203. doi: 10.1002/JLB.2HI0119-006R
Song, O. R., Queval, C. J., Iantomasi, R., Delorme, V., Marion, S., Veyron-Churlet, R., et al. (2018). ArfGAP1 restricts Mycobacterium tuberculosis entry by controlling the actin cytoskeleton. EMBO Rep. 19, 29–42. doi: 10.15252/embr.201744371
Stainier, D. Y. R., Raz, E., Lawson, N. D., Ekker, S. C., Burdine, R. D., Eisen, J. S., et al. (2017). Guidelines for morpholino use in zebrafish. PLoS Genet. 13:e1007000. doi: 10.1371/journal.pgen.1007000
Takaki, K., Davis, J. M., Winglee, K., and Ramakrishnan, L. (2013). Evaluation of the pathogenesis and treatment of mycobacterium marinum infection in zebrafish. Nat. Protoc. 8, 1114–1124. doi: 10.1038/nprot.2013.068
Tien, D. N., Kishihata, M., Yoshikawa, A., Hashimoto, A., Sabe, H., Nishi, E., et al. (2014). AMAP1 as a negative-feedback regulator of nuclear factor-κB under inflammatory conditions. Sci. Rep. 4:5094. doi: 10.1038/srep05094
Tobin, D. M., and Ramakrishnan, L. (2008). Comparative pathogenesis of mycobacterium marinum and Mycobacterium tuberculosis. Cell Microbiol. 10, 1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x
Tobin, D. M., Vary, J. C. Jr., Ray, J. P., Walsh, G. S., Dunstan, S. J., Bang, N. D., et al. (2010). The lta4h locus modulates susceptibility to mycobacterial infection in zebrafish and humans. Cell. 140, 717–730. doi: 10.1016/j.cell.2010.02.013
Torraca, V., Cui, C., Boland, R., Bebelman, J. P., Van Der Sar, A. M., Smit, M. J., et al. (2015). The CXCR3-CXCL11 signaling axis mediates macrophage recruitment and dissemination of mycobacterial infection. Dis. Model Mech. 8, 253–269. doi: 10.1242/dmm.017756
van der Vaart, M., Van Soest, J. J., Spaink, H. P., and Meijer, A. H. (2013). Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system. Dis. Model Mech. 6, 841–854. doi: 10.1242/dmm.010843
Wang, X., Ma, A., Han, X., Litifu, A., and Xue, F. (2018). ASAP1 gene polymorphisms are associated with susceptibility to tuberculosis in a Chinese Xinjiang Muslim population. Exp. Ther. Med. 15, 3392–3398. doi: 10.3892/etm.2018.5800
WHO (2019). Global Tuberculosis Report 2019. Geneva: World Health Organization. Available online at: https://www.who.int/tb/en/
Keywords: ASAP1, Mycobacterium, migration, zebrafish, macrophage
Citation: Cui J, Chen G, Wen D, Wang Y, Zhao Z and Wu C (2020) Asap1 Affects the Susceptibility of Zebrafish to Mycobacterium by Regulating Macrophage Migration. Front. Cell. Infect. Microbiol. 10:519503. doi: 10.3389/fcimb.2020.519503
Received: 12 December 2019; Accepted: 11 September 2020;
Published: 29 October 2020.
Edited by:
Teresa Thurston, Imperial College London, United KingdomReviewed by:
Michiel Van Der Vaart, Leiden University, NetherlandsVincenzo Torraca, University of London, United Kingdom
Copyright © 2020 Cui, Chen, Wen, Wang, Zhao and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonghua Zhao, emh6aGFvQHN4dS5lZHUuY24=; Changxin Wu, Y3h3MjBAc3h1LmVkdS5jbg==
 Jia Cui
Jia Cui Guangxin Chen1
Guangxin Chen1 Zhonghua Zhao
Zhonghua Zhao Changxin Wu
Changxin Wu