
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 16 September 2020
Sec. Molecular Bacterial Pathogenesis
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00483
This article is part of the Research Topic RNA-mediated Regulation of Bacterial Pathogenesis View all 9 articles
 Guang Yang1,2†
Guang Yang1,2† Boan Li2†
Boan Li2† Leili Jia1†
Leili Jia1† Huaiyu Qiu3†
Huaiyu Qiu3† Mingjuan Yang1†
Mingjuan Yang1† Binghua Zhu4†
Binghua Zhu4† Jing Xie1
Jing Xie1 Shaofu Qiu1
Shaofu Qiu1 Peng Li1
Peng Li1 Hui Ma5*
Hui Ma5* Hongbin Song1*
Hongbin Song1* Ligui Wang1*
Ligui Wang1*Regulation of the environmental stress response and virulence of Shigella flexneri may involve multiple signaling pathways; however, these mechanisms are not well-defined. In bacteria, small regulatory RNAs (sRNAs) regulate bacterial growth, metabolism, virulence, and environmental stress response. Therefore, identifying novel functional sRNAs in S. flexneri could help elucidate pathogenic adaptations to host micro-environmental stresses and associated virulence. The aim of this study was to confirm the presence of an sRNA, Ssr54, in S. flexneri and to determine its functions and possible mechanism of action. Ssr54 was found to regulate tolerance and virulence under hyperosmotic pressure. Its expression was verified by qRT-PCR and Northern blotting, and its genomic position was confirmed by 5′-rapid amplification of cDNA ends. Ssr54 expression was significantly decreased (~ 80%) under hyperosmotic conditions (680 mM NaCl), and the survival rate of the Ssr54 deletion strain increased by 20% under these conditions. This suggested that Ssr54 has been selected to promote host survival under hyperosmotic conditions. Additionally, virulence assessment, including guinea pig Sereny test and competitive invasion assays in mouse lungs, revealed that Ssr54 deletion significantly decreased S. flexneri virulence. Two-dimensional gel analyses suggest that Ssr54 may modulate the expression of tolC, ompA, and treF genes, which may affect the virulence and survival of S. flexneri under osmotic pressures. Furthermore, treF expression has been shown to improve the survival of S. flexneri under osmotic pressures. These results suggest that Ssr54 has a broad range of action in S. flexneri response to hyperosmotic environmental stresses and in controlling its virulence to adapt to environmental stresses encountered during host infection.
Shigella is a genus of gram-negative enteric bacteria, accounting for some of the most prominent pathogens causing bacterial diarrhea via fecal-oral route. Shigella infect colonic epithelial cells, leading to typical bacterial diarrheal symptoms. S. flexneri is the most prevalent endemic strain in developing countries (Kotloff et al., 1999; Wang et al., 2006). When S. flexneri enters the host body, it faces various host gastrointestinal environmental stresses characterized by fluctuating temperature, nutrients, bile salts, and pH conditions. Adaptations to these environmental stresses are important for S. flexneri survival and subsequent invasion and colonization of the intestinal tract, which are responsible for the pathogenic effects of the infection. To adapt, the bacteria require coordinated regulation of gene expression for rapid responses to and tolerance of these stresses. For instance, in S. flexneri, acid resistance is dependent on pH and controlled by RpoS under stress conditions (Small et al., 1994). Meanwhile, the two efflux pumps, AcrAB and MdtJI, of Shigella are reportedly associated with resistance to bile salts and the extrusion of toxic compounds (Leuzzi et al., 2015; Nickerson et al., 2017). Hence, developing strategies for tolerating changes in environmental osmolarity, is critical for pathogenic bacteria to not only permit survival during environmental osmotic stress, but also to establish infection (Mahmoud et al., 2017). Specifically, maintaining a stable osmotic balance between the cell cytoplasm and outer environment is an important challenge for all cell types, particularly bacteria.
Different bacteria respond to environmental stresses via different strategies, including the use of small regulatory RNAs (sRNAs). Most sRNAs are located in intergenic regions that range from 40 to 500 nt. They affect mRNA activity by base pairin with the 5'-untranslated regions (5′-UTRs) and/or the ribosome binding site (RBS) of these targets (Gottesman and Storz, 2011; Vanderpool et al., 2011). Antisense sRNAs, transcribed on the DNA strand opposite their target gene, are one type of sRNAs that act via extensive base pairing (Wagner and Romby, 2015). sRNAs have been identified in many bacteria, including Escherichia coli, Staphylococcus aureus, and Vibrio cholerae, in which they play pivotal roles in bacterial adaptation to environmental stress and virulence (Lenz et al., 2004; Mangold et al., 2004; Pichon and Felden, 2005; Bobrovskyy and Vanderpool, 2013; De Lay et al., 2013; Murina and Nikulin, 2015). For example, E. coli DsrA can bind to RpoS mRNA, which encodes the alternative sigma factor S, to repress formation of a closed secondary structure in the RBS of rpos transcripts and promote translation (Massé and Gottesman, 2002). Moreover, E. coli sRNA RyhB regulates intracellular Fe2+ utilization and storage, as well as iron binding proteins (Massé et al., 2005). sRNA also affects the pathogenesis of gram-positive Enterococcus faecalis by regulating its growth and survival under various environmental stresses, including iron stress (Michaux et al., 2014). In fact, studies have suggested that ~10–30% of all bacterial genes are subjected to sRNA regulation (Lewis et al., 2005).
Various studies have also reported on sRNA function in Shigella. For instance, in S. dysenteriae, the sRNA RyfA1 negatively impacts the virulence-associated process of cell-to-cell transfer, as well as expression of ompC, a gene that is important for the pathogenesis of Shigella (Fris et al., 2017). Additionally, CsrB/CsrC is regulated in response to carbon availability, RyhB is regulated in response to iron availability, while RnaG, as well as the activity of the shuA RNA thermometer, is regulated in response to temperature (Fris and Murphy, 2016). RnaG (450 nt), a small non-coding RNA encoded on the virulence plasmid pINV of Shigella flexneri, is able to directly interact with icsA mRNA, thereby promoting premature termination of transcription (Giangrossi et al., 2010) Additionally, VirF, which generally assists the RNA–RNA interaction, has been shown to directly activate the transcription of the small non-coding RnaG and icsA mRNA, which encodes two essential proteins that are involved in the pathogenicity process by binding their promoter regions (Giangrossi et al., 2017). RyhB (90 nt) modulates and inactivates ferritin-related genes to regulate iron metabolism and can inhibit the expression of virulence genes such as virB, a regulator of the T3SS secretory protein gene (Murphy and Payne, 2007). Meanwhile, in our previous study, Ssr1 (150 nt) was shown to help bacteria adapt to an acidic environment by regulating acid resistance genes and to regulate bacterial virulence by controlling the expression of T3SS genes (Wang L. et al., 2016). However, the functions of many other predicted sRNAs in Shigella have not yet been elucidated. The identification of novel S. flexneri sRNAs is pivotal to understanding Shigella virulence and adaptation to host microenvironment. Therefore, the aim of the present study was to confirm the presence of an sRNA, Ssr54, in S. flexneri that was predicted using a comparative genomics bioinformatics screening method (Wang L. et al., 2016), as well as determine the functions and possible mechanisms of action of this sRNA in S. flexneri.
The wild-type strain used in this study was S. flexneri 2a 301. This strain was cultured on Congo red solid culture medium at 37°C in 5% CO2 overnight. A single colony was selected, inoculated in liquid Luria-Bertani (LB) medium, and incubated at 37°C on an orbital shaker at 150 rpm for 6–8 h. When necessary, kanamycin, ampicillin, or chloramphenicol was added to the culture medium to obtain a final concentration of 50, 100, or 30 μg/mL, respectively. In the stress test, a bacterial culture with an OD600 of 0.6 was pelleted, resuspended in PBS, and inoculated into the following culture media: a hypoosmotic LB culture medium not containing NaCl, hyperosmotic LB culture media containing 340 or 680 mM NaCl, and LB culture medium with 160 mM NaCl as the negative control. The bacteria were cultured under these conditions for 30 min. Thereafter, we assessed sRNA expression by qRT-PCR and performed the survival assay under hyperosmotic conditions.
For the northern blot analysis, wild-type, Ssr54 mutant, and Ssr54 complement strain cells were harvested during the logarithmic growth phase (OD600 = 0.6). When required, the cultures were subjected to hypo/hyperosmotic stress conditions as described above. Non-stressed exponentially growing cultures were used as controls.
Total RNA was isolated with TRIzol (Invitrogen, Carlsbad, CA, USA, 15596108) from S. flexneri cultured to the middle logarithmic growth phase (OD600 = 0.6), and remaining DNA was digested with DNase (Promega, Madison, WI, USA, M6101). ODs were determined at 260 nm and 280 nm using a UV-Vis spectrophotometer to assess RNA concentration. DNase-treated total RNA (not more than 10 μg) was reverse-transcribed into cDNA using a reverse transcription kit (Promega, Madison, WI, USA, K1005S) in accordance with the manufacturer's instructions. cDNA was synthesized using 2 μg of total RNA and a Promega Reverse Transcription System. It was then diluted (1:5) in nuclease-free water. Primers were designed based on the Ssr54 sequence (Supplementary Table 1 for further details) and used the cDNA as PCR template. PCR was performed using ExTaq Mix (Takara Biochemicals, Beijing, China, RR001Q) under the following thermal cycling conditions: 10 min at 95°C for pre-incubation, followed by 30 cycles of amplification (30 s at 95°C, 30 s at 55°C, and 30 s at 72°C) (Absin Bioscience, Foster City, CA, USA). RNA preparation and qRT-PCR analysis were repeated at least three times.
Ssr54 size and expression levels were assessed by northern blotting with total RNA from the wild-type, Ssr54 mutant, and complement strains in normal and hyperosmotic conditions. Total RNA (15–20 μg from each strain) was separated via denaturing polyacrylamide gel electrophoresis (PAGE) (10% resolving gel) and electroblotted onto a nylon membrane where it was cross-linked under a 1,200-mJ UV light. The Ssr54 probe sequence was 5′-ATTCTTCAGAGATCACAAACTGGTTA-3′. The sequence of 5S rRNA probe, which was used as the internal control, was 5′-GTTTCACTTCTGAGTTCGGCATGGGGTCAGGTGGG-3′. These probes were synthesized by Shanghai Invitrogen Life Technology Service (Shanghai, China). The RNA was then hybridized with a biotin-labeled Ssr54 probe overnight, and the hybridization signal was detected using a North2South® Chemiluminescent Hybridization and Detection Kit from Thermo Fisher Scientific (Carlsbad, CA, USA, 17097) with an exposure time of 1–5 min.
The precise size of Ssr54 was determined by RACE using the 5′-Full RACE Kit (Takara Biochemicals, Beijing, China, D315) in accordance with the manufacturer's instructions. Total RNA was isolated from S. flexneri 2a 301 during the exponential phase (OD = 0.6). The Ssr54 RACE outer primer and inner primer sequences are presented in Supplementary Table 1. The resulting purified sRNA-derived PCR product was cloned into the pMD18T vector and transformed into competent E. coli DH5α cells. The inserts were sequenced to identify the TSSs of the individual sRNAs.
qRT-PCR was performed to assess Ssr54 sRNA expression in S. flexneri under different osmotic pressures. cDNA was synthesized using 2 μg of total RNA and a Promega Reverse Transcription System and then diluted (1:5) in nuclease-free water. With 3 μL of cDNA as the template and 16S rRNA as the internal control, Ssr54 was amplified by PCR using the 2 × SYBR Green I Master Mix protocol (Invitrogen, Carlsbad, CA, USA). The primers are presented in Supplementary Table 1. RT-PCR was performed using the ABI 7500 system (Applied Biosystems) with the following cycling conditions: 5 min denaturation step at 42°C, followed by 40 amplification cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C. Ssr54 gene transcript levels under various stress conditions were compared with those of the wild-type strain grown in normal LB medium at 37°C. Relative expression levels were calculated using the 2−ΔΔCT method (Gong et al., 2011). All experiments were conducted in triplicate.
The Ssr54 mutant strains were generated through λ-Red homologous recombination (Park et al., 2001). Genome segments 500–600 bp upstream and downstream of the homologous arm fragments in the Ssr54 sRNA sequence were amplified. The kanamycin resistance gene from the pUC19-kan plasmid was amplified, and the fusion-PCR method (with three fragments at a ratio of 1:1:1) was employed to generate a linear target DNA fragment. This linear fragment was transformed into a S. flexneri strain containing the pKD46 plasmid, which encoded the λ-Red recombination system, and the strain was cultured overnight on an antibiotic selection plate containing ampicillin and kanamycin to screen for the Ssr54-deficient mutant strain. This strain was confirmed using primers listed in Supplementary Table 1.
To construct the complementing plasmid pSsr54, DNA 200 bp upstream of the Ssr54, transcription start site (TSS), and its coding region was amplified from S. flexneri 2a 301. These primers targeted unique restriction enzyme sites, XbaI and SphI, such that when the PCR product was digested with these restriction enzymes, it could be ligated into a similarly digested plasmid, recombination pACYC184 (CmRTcR), in an orientation-specific manner. Recombinant DNA products were verified by sequencing. The resulting plasmid was used to transform the Ssr54 mutant strain via electroporation, followed by selection using chloromycetin. The final complement strain was dubbed ΔSsr54 + pSsr54. pACYC184 was electroporated into these strains to generate vector-only (no sRNA) controls.
Overnight cultures grown in LB medium or LB medium containing kanamycin were diluted 1:100 in LB medium with the hyperosmotic supplement and then grown at 37°C with shaking at 200 rpm/min. At time points of 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, and 24 h after inoculation, bacterial growth was monitored by measuring the OD600. Each sample was analyzed in triplicate and the assays were repeated at least three times. The average OD600 values were used to generate growth curves.
The wild-type and Ssr54 mutant strains were cultured at 37°C with agitation at 160 rpm until the early logarithmic phase of growth (OD600 of 0.6). Bacterial cultures were centrifuged for 5 min at 4°C; then, pellets were washed three times in PBS, resuspended in hyperosmotic LB culture medium containing 680 mM NaCl, and cultured for 30 min at 37°C (Gong et al., 2011). Final bacterial cultures were serially diluted and plated onto LB plates for overnight incubation to determine viability. The number of colony-forming units per milliliter (CFU/mL) was then calculated for each of the conditions, with each bacterial strain cultured in LB culture medium at pH 7.0 (containing 160 mM NaCl) serving as the control. Results are expressed as mean percentage ± SD from experiments performed in triplicate.
Analysis of keratoconjunctivitis induced in guinea pigs is a standard method to assess S. flexneri virulence, and it allows for direct evaluation of S. flexneri pathogenesis (Sereny, 1955). To verify the effect of Ssr54 sRNA on S. flexneri virulence, we investigated the effect of the wild-type, Ssr54 mutant, and complement strains on the corneas of guinea pigs. Single colonies of the wild-type, Ssr54 mutant, and Ssr54 complement strains were selected from TSA Congo red plates and inoculated in LB culture medium containing the appropriate antibiotics for incubation at 37°C overnight. Thereafter, 1% of each culture was inoculated into 5 mL of LB culture medium and incubated until it reached an OD600 of 1.0 (106 CFU/mL). After centrifugation for 5 min, we harvested 1 mL of each bacterial culture and resuspended it in 20 μL of PBS, which was then inoculated into the corneas of healthy guinea pigs with normal cornea and conjunctiva. Inoculated corneas were observed 24, 48, and 72 h after inoculation. Corneas inoculated with PBS served as controls. The degree of keratoconjunctivitis was graded as follows: – no keratoconjunctivitis; + mild conjunctival congestion; ++ palpebral edema, conjunctival congestion, corneal opacification, and no purulent discharge; +++ palpebral edema, corneal opacification, conjunctival congestion, and purulent discharge; ++++ palpebral edema, corneal gray patch, conjunctival congestion, and purulent discharge. The experiment was conducted in triplicate. Each group contained six guinea pigs.
The wild-type, mutant Ssr54, and complement strains were grown to the logarithmic growth phase, and then the concentrations of cultures were adjusted to 108 CFU/mL. Three aliquots (1 mL each) containing the same number of bacteria were pelleted by centrifugation for 5 min at 4°C and resuspended in 1 mL of PBS. Equal amounts of the wild-type and mutant strains were mixed, and an aliquot (20 μL) of the resulting mixture (2 × 106 CFU/mL), along with a similar aliquot of the mixture of equal amounts of wild-type and complement strains, were placed in the nasal cavities of BALB/c mice, under diethyl ether sedation, to infect lung tissues. After 24 h, the mice were euthanized via cervical dislocation, and lungs were collected from all and washed in RPMI (Roswell Park Memorial Institute) 1640 with 50 μg/mL gentamicin. Mouse lungs were harvested and incubated in PBS containing 0.1% Triton X-100 to release the bacteria. The extracts were then spread on either antibiotic-free or kanamycin antibiotic-containing LB plates for overnight cultivation at 37°C, and then the numbers of colonies were determined. The competitive index (CI) was defined as the ratio of mutant strain CFU after invasion to that of the wild-type strain CFU after invasion, divided by the ratio of the mutant strain CFU before invasion to that of the wild-type strain CFU before invasion. This experiment was performed in triplicate.
To further verify the influence of Ssr54 on virulence in S. flexneri, a competitive invasion assay was performed using HeLa cells. S. flexneri strain 301, Ssr54 mutant and complement strains were grown in LB medium to the exponential phase (OD600 of 0.6), adjusted to the same concentration, and mixed at a 1:1 (v/v) ratio. The two strains were added to DMEM cell culture medium (Gibco, Carlsbad, CA, USA), and the cultures formed were then added to HeLa cells at a multiplicity of infection of 100:1. After incubation for 4 h, infected HeLa cells were washed thrice with PBS and thrice with cell culture medium, following which cell culture medium containing 50 μg/mL gentamicin was added (Wang Y. et al., 2011). Gentamicin kills extracellular bacteria, while intracellular bacteria are protected from it. Following incubation at 37°C for 24 h, cell cultures were washed with PBS; the cultures were then harvested and lysed, and their cellular contents were cultured on LB and LB plates containing kanamycin. The CI was calculated, which represents the ratio of the CFUs of the recovered Ssr54 mutant and the wild-type, complement strains and the wild-type (normalized for the input ratio), and Student's t-test was used to determine the p-value (Christiansen et al., 2004). This assay was repeated three times, and the mean was reported as the final result.
Proteins from the wild-type and Ssr54 mutant strains were extracted and purified. The protein samples (total of 120 μg, as assessed by silver staining) were subjected to one-dimensional isoelectric focusing using IPG immobiline dry strip gels at pH 4–7 for 30 min at 300 V, and then 30 min at 700 V, 1.5 h at 1,500 V, and 8 h at 9,000 V until the voltage-time product reached 64 kVh. Thereafter, isoelectric focusing was discontinued, and the samples were separated in the other direction by sodium dodecyl sulfate (SDS)-PAGE (12.5% resolving gel). The gels were then stained, scanned, and subjected to image analysis. The samples underwent protein mass spectroscopy pre-treatment processes, including determination of restriction enzyme digestion sites, enzymatic digestion, peptide extraction, identification of target proteins, and mass spectroscopy measurement.
The animals were obtained from the Laboratory Animal Center (Academy of Military Medical Sciences). All methods were carried out in accordance with the approved guidelines of the Academy of Military Medical Sciences. The experimental protocols were approved by the Ethics Committee for Animal Experimentation of the Academy of Military Medical Sciences.
All standard biosecurity and institutional safety procedures were adhered to in all experimental procedures presented in this article.
In our previous study, we predicted 57 sRNAs using a transcription unit-based method and confirmed four of these sRNAs (Ssr1, Ssr9, Ssr44, and Ssr54) by reverse transcription (RT)-PCR and northern blotting assays (Wang L. et al., 2016). In the current study, we focused on determining the characteristics and functions of Ssr54. sRNA Ssr54 expression was detected by RT-PCR and verified by northern blotting. A biotin-labeled Ssr54 probe was hybridized with a band of ~100 bp in a wild-type S. flexneri strain and Ssr54 complement strain; no band was seen in the Ssr54 deletion strain (Figure 1A). In this experiment, 5S rRNA was used as the positive control. By 5′-RACE, the Ssr54 TSS was at 3635111 and located in a non-coding intergenic region (Figure 1B).
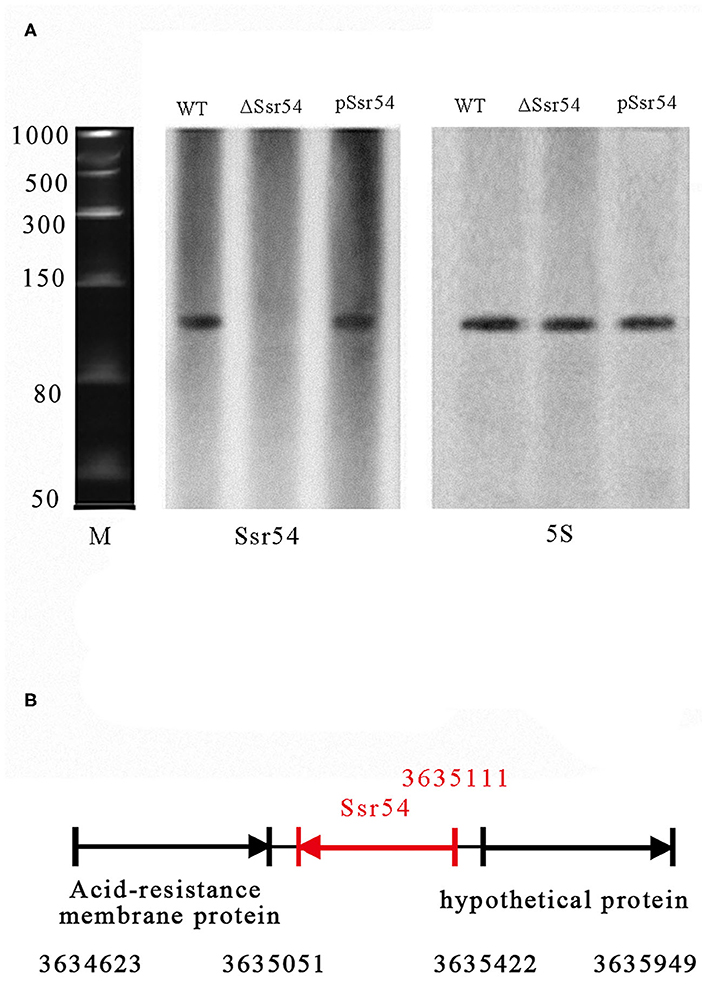
Figure 1. Experimental verification and expression of S. flexneri small RNAs (sRNAs). (A) Northern blot of the novel sRNA (Ssr54) and 5S rRNA in S. flexneri wild-type, Ssr54 mutant, and Ssr54 complement strains. Ssr54 expression in the S. flexneri strains grown to logarithmic growth phase was verified by Northern blotting; (B) Ssr54 genomic position in S. flexneri was assessed via 5′ rapid amplification of cDNA ends.
We then examined whether Ssr54 is required for S. flexneri to cope with osmotic stress. To this end, we first quantified its expression in osmotic conditions. Northern blot analysis was used to examine the levels of Ssr54 sRNA in S. flexneri grown under various hyperosmotic conditions supplemented with 340 mM and 680 mM NaCl (Figure 2A). At osmotic pressures of 340 mM NaCl and 680 mM NaCl, the expression of Ssr54 was predominantly low in the wild-type strain (Figure 2A). In this experiment, 5S rRNA was used as the positive control.
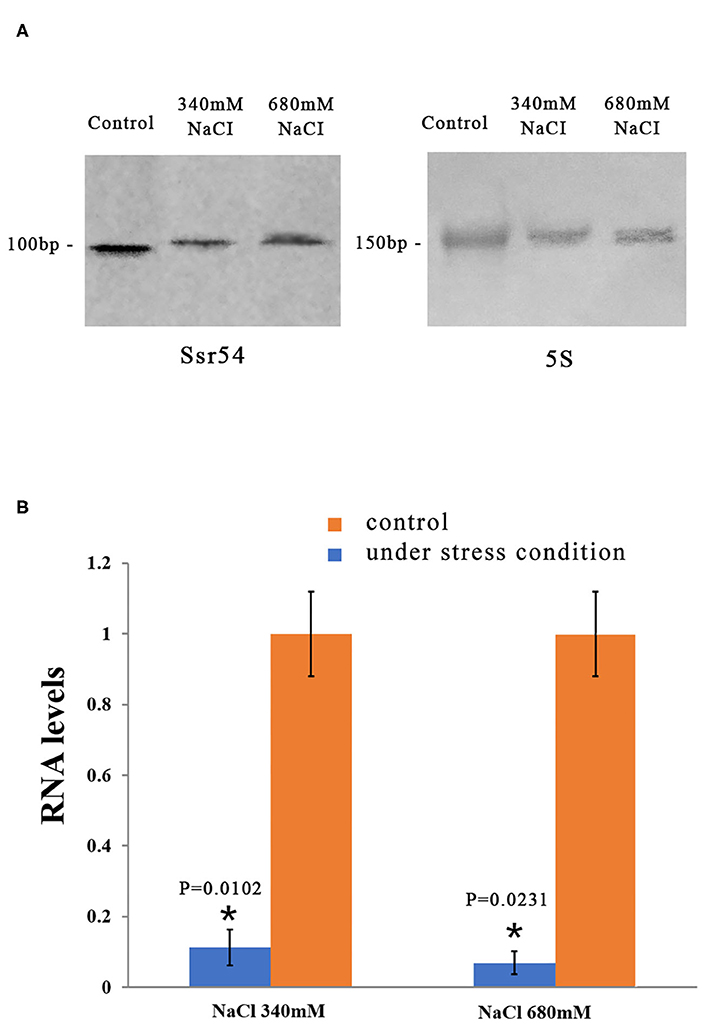
Figure 2. Levels of the small RNA Ssr54 in S. flexneri strain 301 when exposed to high osmotic conditions for 30 min. (A) Northern blot of Ssr54 and 5S rRNA in the S. flexneri wild-type strain under hyperosmotic conditions. Control cultured in 160 mM NaCl; (B) RNA levels of Ssr54, qRT-PCR analyses of the relative Ssr54 levels in S. flexneri under high osmotic conditions compared with that in 160 mM NaCl. Relative abundance was calculated using the 2−ΔΔCt method. Bars indicate fold changes calculated as the mean of triplicate experiments, representing the ratios of Ssr54 expression levels. Black bars represent the control transcript values; Red bars represent results under stress conditions. Error bars indicate standard deviations. Statistical analyses were performed using Student's t-test.
Reverse transcription quantitative PCR (RT-qPCR) was performed to further assess Ssr54 expression in S. flexneri under different osmotic pressures. Ssr54 was significantly downregulated under hyperosmotic pressures of 680 mM NaCl (>90% downregulation, P = 0.0231) and 340 mM NaCl (>80% downregulation, P = 0.0102) (Figure 2B). These results were consistent with the northern blot results, confirming changes in Ssr54 expression under different osmotic pressures.
To investigate the role of Ssr54 in adaptation of S. flexneri to hyperosmotic pressures, we constructed an Ssr54 mutant strain using λ-red homologous recombination and a low-copy plasmid Ssr54 complement strain. All mutant and complementary Ssr54 constructs were confirmed by PCR and agarose gel electrophoresis (Supplementary Figure 1). To determine whether Ssr54 plays a role in regulating growth under hyperosmotic conditions, we placed the wild-type, Ssr54 mutant, and complement strains in LB culture medium under different stress conditions at 37°C for 24 h. Optical densities at 600 nm (OD600) were measured every 2 h until the late stationary phase (24 h), and growth curves were plotted. A mutant carrying an empty vector served as a negative control. Under hypoosmotic conditions (0 mM NaCl), growth of the wild-type strain was not significantly different from that of the Ssr54 mutant and complement strains (Figure 3A). Under hyperosmotic conditions (680 mM NaCl), the OD600 values for the Ssr54 mutant strain were significantly higher than those for the wild-type and Ssr54 complement strains, and the Ssr54 mutant strain entered the stationary phase later than the wild-type, hence indicating that entry into the stationary phase was accelerated in the deletion strain (Figure 3B). This suggested that Ssr54 expression plays an important role in S. flexneri growth under hyperosmotic conditions.
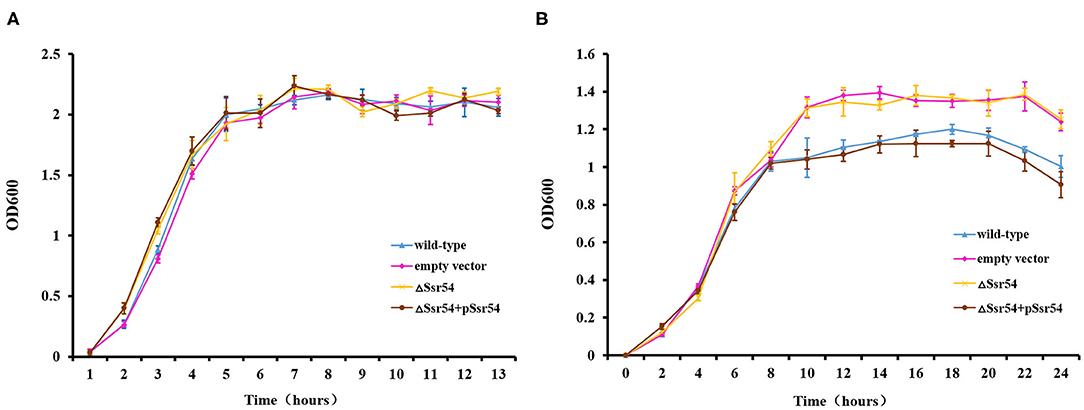
Figure 3. Osmotic stress of the Ssr54 mutant. Growth characteristics of the wild-type, Ssr54 mutant, and complement strains, as well as the mutant strain carrying the pACYC184 empty vector, in LB under different osmotic conditions: (A) 0 mM NaCl; (B) 680 mM NaCl. The error bars indicate standard deviations based on triplicate experiments. Statistical analyses were performed using Student's t-test.
In addition, we assessed survival of the wild-type and Ssr54 mutant strains for 30 min following hyperosmotic stimulation and found no significant difference in growth. This survival assay was used to verify survival of the wild-type and Ssr54 mutant strains under different osmolarities. After exposure to hyperosmotic pressure, survival of the Ssr54 mutant strain improved by ~20% compared with that of the wild-type strain (P = 0.0109) (Figure 4). There was no difference in survival between the Ssr54 complement strain and wild-type strain with the empty plasmid (P = 0.0701), indicating Ssr54 deletion enhanced S. flexneri tolerance to hyperosmotic pressure and survival. The empty vector mutant served as a negative control.
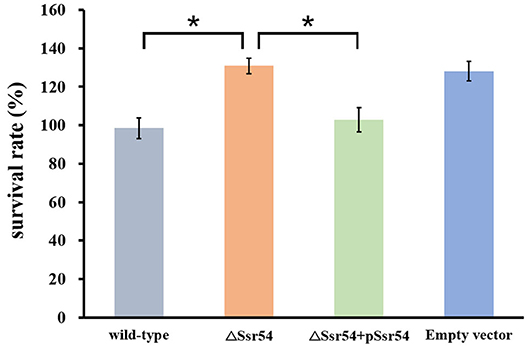
Figure 4. Survival of the sRNA Ssr54 mutant strain (ΔSsr54) relative to that of the wild-type S. flexneri 301 strain when exposed to high osmotic stress. Wild-type S. flexneri 301, ΔSsr54, the complement (ΔSsr54 + pSsr54) strain, and mutant strain carrying the pACYC184 empty vector were grown in Luria-Bertani medium (160 mM NaCl) up to the exponential phase and then subjected to high osmotic stress (680 mM NaCl). Viability was determined by counting bacteria plated in serial dilutions. Bars represent the mean percent survival compared with untreated controls. Statistical analyses were performed using Student's t-test. Data represent the average of biological triplicates, and errors bars indicate one standard deviation. *Denotes a significant difference with P < 0.01.
The wild-type, Ssr54 mutant, and complement strains were inoculated into the corneas of healthy guinea pigs (Figure 5). After 24 h of infection, mild conjunctival congestion was observed in the wild-type and complement strain-inoculated group, whereas no keratoconjunctivitis was detected in the Ssr54 mutant strain-inoculated group or the PBS (control) group. After 48 h of infection, the wild-type and complement strain-inoculated group presented palpebral edema and corneal opacification, and keratoconjunctivitis was still absent in the Ssr54 mutant strain-inoculated and PBS (control) groups. After 72 h of infection, purulent discharge was observed in the wild-type and complement strain-inoculated group, and keratoconjunctivitis still remained absent in the Ssr54 mutant strain-inoculated and PBS (control) groups (Table 1). These data indicate that there was significant inflammatory cell infiltration following inoculation with the wild-type strain; however, there was no inflammatory response following inoculation with the Ssr54 mutant strain or PBS (Supplementary Figure 3). These data, which show the induction of keratoconjunctivitis in guinea pigs, strongly suggest that the inflammatory response time and infectivity of the Ssr54 mutant strain were significantly less than those of the wild-type and complement strains and imply that deletion of Ssr54 attenuates S. flexneri invasiveness.
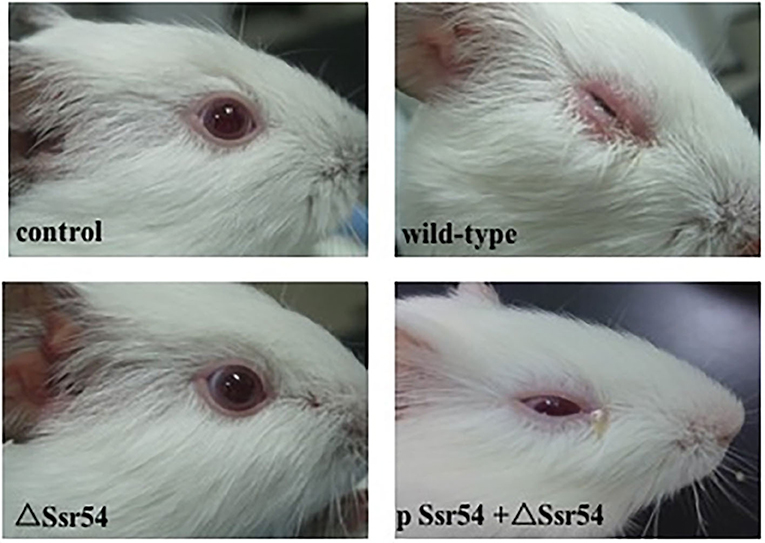
Figure 5. Phenotypes of wild-type S. flexneri and sRNA mutant ΔSsr54 in guinea pigs. More inflammatory cells were present in the corneas of guinea pigs inoculated with the wild-type and complement strains than those inoculated with the mutant ΔSsr54 strain, whereas the NaCl (control) group showed no inflammatory cells.
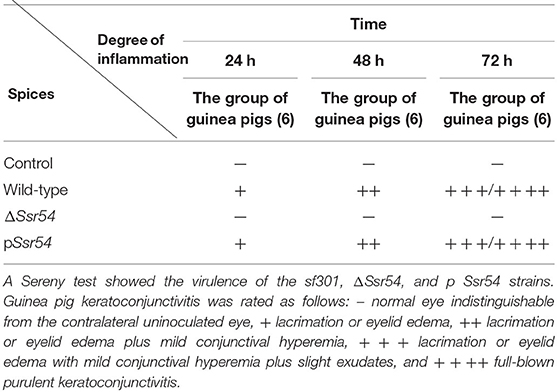
Table 1. Keratoconjunctivitis in guinea pigs inoculated with wild-type and ΔSsr54 Shigella flexneri strains or an NaCl control.
Virulence of the wild-type, Ssr54 mutant, and complement strains was also assessed using a competitive invasion model in mouse lungs. Following competitive lung invasion by the wild-type, Ssr54 mutant, and complement strains, total viability of the Ssr54 mutant strain (2.07 × 102) was found to be significantly lower than that of the wild-type strain (6.15 × 104) (P = 0.0068), whereas total viability of the complement strain (5.86 × 104) (P > 0.05) was not significantly different from that of the wild-type strain. The CI between the Ssr54 mutant and wild-type strains was 0.028 (P < 0.05), which indicates a significant difference, whereas that between the Ssr54 complement and wild-type strains was 0.958 (P > 0.05), which was not significantly different (Figure 6). This finding suggested that deletion of Ssr54 significantly attenuated S. flexneri virulence, which was consistent with the experimental data regarding keratoconjunctivitis induction in guinea pigs.
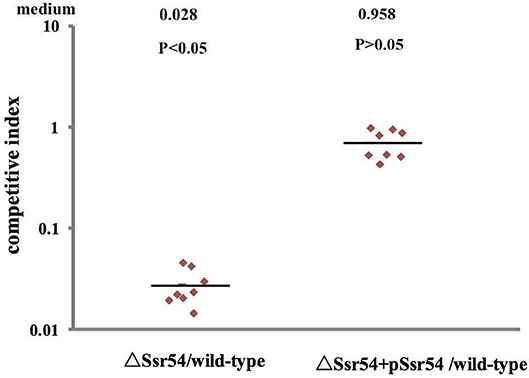
Figure 6. Mit4ce were infected with wild-type, mutant ΔSsr54, and complement strains. A competitive index (CI) of 1 represents recovery of equivalent amounts of wild-type and mutant bacteria. The CI of the Ssr54 mutant and wild-type was lower than 1 (P < 0.05); The CI of the Ssr54 complement strain and wild-type was ~ 1 (P > 0.05).
In addition, we assessed the virulence of the wild-type, Ssr54 mutant, and complement strains in an in vivo HeLa cell invasion assay. The CFU recovered for the S. flexneri wild-type strain had a mean value of 1.696 × 107 (6 replicates); the mean CFU of the wild-type was significantly higher than that of the Ssr54 mutant (1.3 × 106 CFU (6 replicates); P < 0.05). The calculated median value for the CI values between the Ssr54 mutant and wild-type strains was 0.0865 (Figure 7). This value being significantly < 1 (Student's t-test, P < 0.05) strongly suggests that Ssr54 deletion has an impact on the ability of S. flexneri to effectively invade epithelial cell in vivo. However, the total viability of the complement strain (1.7 × 107) (6 replicates) was not significantly different from that of the wild-type strain (P > 0.05). The CI between the Ssr54 complement and wild-type strains was 1.086 (P > 0.05), which was not significant (Figure 7).
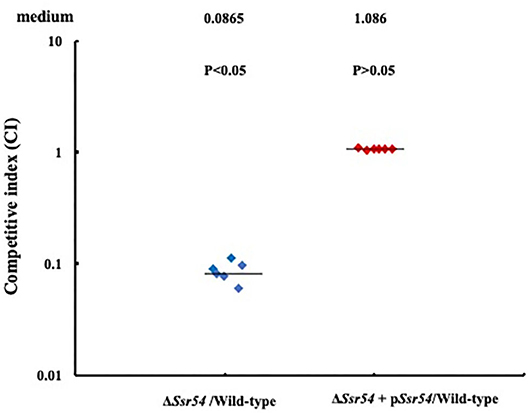
Figure 7. HeLa cell competitive invasion assays of wild-type, mutant ΔSsr54, and complement strains. The results show the competitive index (CI) of the Ssr54 mutant. A CI of 1 represents recovery of equivalent amounts of wild-type and mutant bacteria. The CI of the Ssr54 mutant and wild-type was significantly lower than 1 (P < 0.05); The CI of the Ssr54 complement strain and wild-type was ~1 (P > 0.05).
To determine the targets of Ssr54, whole bacterial protein extracts of the wild-type and Ssr54 mutant strains were prepared from logarithmic growth phase cultures, followed by two-dimensional PAGE. The same sample was tested in triplicate to confirm reproducibility of the two-dimensional electrophoresis maps. By comparing maps, we ascertained spots indicating highly differentially expressed proteins and conducted in-gel protease digestion, followed by mass spectroscopic analysis. Compared with the Ssr54 mutant strain, the wild-type strain had 137 differentially expressed protein spots. Protein spots with a difference in expression of at least three-fold were identified, subjected to robotic spot excision, trypsin digested, and subsequently analyzed using MALDI-TOF mass spectrometry. Protein identification was based on highly accurate masses and MS/MS sequence data from multiple tryptic peptides. Ssr54 deletion resulted in a total of 40 differentially expressed proteins, of which 25 were downregulated and 15 were upregulated (Supplementary Figure 2). Supplementary Figure 2A shows the wild-type strain as per our previous results (Wang L. et al., 2016); however, when compared to the Ssr54 deletion strain, the protein spots differ. Twenty-six of the proteins differentially expressed in the wild-type and Ssr54 mutant strains were cytoplasmic, one was periplasmic, two were on the cytoplasmic leaflet of the plasma membrane, six were on the extracellular leaflet of the plasma membrane, and four were unidentified (Table 2).
After Ssr54 deletion, mRNA of proteins with the most significantly altered expression levels (TreF, OmpA [C43], and TolC) and their roles in stress tolerance and virulence were analyzed by qRT-PCR. The tolC and ompA genes were significantly downregulated in the Ssr54 mutant strain compared to those in the wild-type strain, whereas treF was significantly upregulated in the Ssr54 mutant strain.
To determine whether hyperosmotic conditions affect the expression of these target genes, treF, tolC, and ompA expression levels were determined under hyperosmotic conditions using qRT-PCR (Figure 8). Hyperosmotic conditions activated treF expression (1.99-fold) (P = 0.0115) and downregulated tolC and ompA expression (approximately two-fold) (P = 0.0207 and 0.0218, respectively).
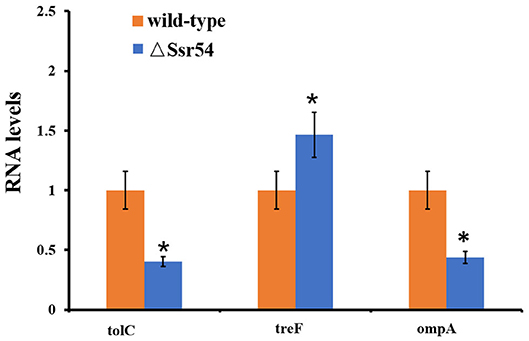
Figure 8. qRT-PCR for three mRNA targets of Ssr54. Level of three mRNA targets in the ΔSsr54 mutant strain under hyperosmotic conditions expressed relative to the level in wild-type. Statistical analyses were performed using Student's t-test. Data are representative of three independent experiments. Error bars indicate one standard deviation. *Denotes a significant difference with P < 0.01.
Numerous recent studies have investigated bacterial sRNAs, with more than 900 novel sRNAs reported in the sRNA database RNAMap (Huang et al., 2009). Of these, 79 were found in E. coli. However, there have been very few functional studies of sRNAs, and only 57 sRNA targets have been identified as per the sRNATarBase database (Wang L. et al., 2016). Hence, investigations on the mechanism of action of novel sRNAs are warranted. Interestingly, the sequence of sRNA ArrS, identified in E. coli strain K-12, is identical to that of Ssr54 (Bianco et al., 2019). However, there are significant differences between their lengths and functions. The length of ArrS is 50 nt, which is less than that of Ssr54. ArrS regulates the response of E. coli to low pH, whereas Ssr54 demonstrates different functions in S. flexneri. In the present study, we confirmed the presence of a novel sRNA, Ssr54, in S. flexneri, observed its role in environmental stress tolerance and virulence, and identified possible targets to suggest mechanisms of action.
In pathogenic bacteria, response to osmotic stress is a critical mechanism for survival during environmental osmotic stress as well as for establishing infection. For instance, the ProU system functions to allow S. sonnei to tolerate hyperosmotic stress in vitro, while also facilitating the survival and proliferation of the bacteria within stressful intracellular niches (Mahmoud et al., 2017). Herein, we observed that when exposed to hyperosmotic stress, Ssr54 was downregulated and survival of the Ssr54 deletion strain increased. Generally, bacteria tolerate changes in osmolarity by solute efflux or gene regulation (Mahmoud et al., 2017), allowing them to adapt for survival. In this way, the downregulation of Ssr54 may serve as a protective mechanism for S. flexneri in the gastrointestinal tract. Furthermore, Ssr54 deletion was shown to upregulate expression of the target gene treF. TreF, the cytosolic trehalase, is weakly induced by high osmolarity of the medium and is partially dependent on RpoS, the stationary - phase sigma factor. Hence, treF is expressed primarily under hyperosmotic conditions (Horlacher et al., 1996; Weber et al., 2006); accordingly, Ssr54 expression seems to inhibit treF, with treF upregulated under high osmotic concentrations when Ssr54 is repressed. Ssr54 may, therefore, upregulate treF to enhance survival under osmotic stress. Thus, virulence genes in Shigella are expressed in response to increases in osmolarity.
Shigella guinea pig Sereny tests, lung invasion assays, and HeLa invasion assays were also performed to determine whether Ssr54 regulates S. flexneri virulence. Results suggest that the virulence of S. flexneri ΔSsr54 was significantly decreased as compared to that of the wild-type strain. TolC is an outer membrane channel protein that contributes to bacterial adhesion, antibiotic resistance, and invasion during disease pathogenesis (Stone and Miller, 1995; Sharff et al., 2001; Platz et al., 2010). Ssr54 deletion downregulated tolC, which may have attenuated the virulence of S. flexneri. Our study also showed that deletion of Ssr54 downregulated ompA. OmpA, the major outer membrane protein, plays important roles in the virulence of pathogens (Scribano et al., 2016). Some studies have suggested that micA and rseX sRNAs bind to the ompA translation initiation region and block ribosome binding to prevent expression of ompA in E. coli (Udekwu et al., 2005). Hence, we speculate that ompA was downregulated in the Ssr54 deletion strain, decreasing the virulence of S. flexneri. Overall, Ssr54 may affect S. flexneri virulence by regulating the expression of putative target genes tolC and ompA, either directly or indirectly.
Studies have suggested that hyperosmotic pressures affect virulence gene expression in S. flexneri (Porter and Dorman, 1994). Under hyperosmotic stress, the hyperosmotic regulator gene treF was upregulated, whereas the virulence regulator genes tolc and ompA were downregulated, indicating that Ssr54 regulates treF to increase S. flexneri tolerance to hyperosmotic conditions and regulates virulence genes tolc and ompA to reduce pathogenicity of this bacterium. These results have, therefore, established the role of Ssr54 in survival of Shigella under hyperosmotic environments; however, whether it is required in the context of infection remains unknown. Moreover, whether Ssr54 regulates virulence and hyperosmotic stress tolerance by regulating genes directly, or via other pathways, remains to be answered; we are currently investigating this mechanism.
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study.
The animal study was reviewed and approved by the Ethics Committee for Animal Experimentation of the Academy of Military Medical Sciences.
HS, LW, and HM designed the research, assessed and interpreted the results, and prepared the manuscript. GY, BL, LJ, HQ, MY, and BZ carried out the data analysis and designed the experiments. JX, SQ, and PL assisted in carrying out the experiments. All authors contributed to the article and approved the submitted version.
This study was supported by the PLA Youth Training Project for Medical Science (16QNP127) and Natural Science Foundation of China (31000589).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Editage (www.editage.cn) for providing English language editing service.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00483/full#supplementary-material
Bianco, C. M., Fröhlich, K. S., and Vanderpool, C. K. (2019). Bacterial cyclopropane fatty acid synthase mRNA is targeted by activating and repressing small RNAs. J. Bacteriol. 201:e00461-19. doi: 10.1128/JB.00461-19
Bobrovskyy, M., and Vanderpool, C. K. (2013). Regulation of bacterial metabolism by small RNAs using diverse mechanisms. Annu. Rev. Genet. 47, 209–232. doi: 10.1146/annurev-genet-111212-133445
Christiansen, J. K., Larsen, M. H., Ingmer, H., Søgaard-Andersen, L., and Kallipolitis, B. H. (2004). The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 186, 3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004
De Lay, N., Schu, D. J., and Gottesman, S. (2013). Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 288, 7996–8003. doi: 10.1074/jbc.R112.441386
Fris, M. E., Broach, W. H., Klim, S. E., Coschigano, P. W., Carroll, R. K., Caswell, C. C., et al. (2017). Sibling sRNA RyfA1 influences Shigella dysenteriae pathogenesis. Genes 8:50. doi: 10.3390/genes8020050
Fris, M. E., and Murphy, E. R. (2016). Riboregulators: fine-tuning virulence in Shigella. Front. Cell Infect. Microbiol. 6:2. doi: 10.3389/fcimb.2016.00002
Giangrossi, M., Giuliodori, A. M., Tran, C. N., Amici, A., Marchini, C., and Falconi, M. (2017). VirF relieves the transcriptional attenuation of the virulence gene icsA of Shigella flexneri affecting the icsA mRNA-RnaG complex formation. Front. Microbiol. 8:650. doi: 10.3389/fmicb.2017.00650
Giangrossi, M., Prosseda, G., Tran, C. N., Brandi, A., Colonna, B., and Falconi, M. (2010). A novel antisense RNA regulates at transcriptional level the virulence gene icsA of Shigella flexneri. Nucleic Acids Res. 38, 3362–3375. doi: 10.1093/nar/gkq025
Gong, H., Vu, G. P., Bai, Y., Chan, E., Wu, R., Yang, E., et al. (2011). A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog. 7:e1002120. doi: 10.1371/journal.ppat.1002120
Gottesman, S., and Storz, G. (2011). Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect. Biol. 3:a003798. doi: 10.1101/cshperspect.a003798
Horlacher, R., Uhland, K., Klein, W., Ehrmann, M., and Boos, W. (1996). Characterization of a cytoplasmic trehalase of Escherichia coli. J. Bacteriol. 178, 6250–6257. doi: 10.1128/JB.178.21.6250-6257.1996
Huang, H. Y., Chang, H. Y., Chou, C. H., Tseng, C. P., Ho, S. Y., Yang, C. D., et al. (2009). sRNAMap: genomic maps for small non-coding RNAs, their regulators and their targets in microbial genomes. Nucleic Acids Res. 37, D150–D154. doi: 10.1093/nar/gkn852
Kotloff, K. L., Winickoff, J. P., Ivanoff, B., Clemens, J. D., Swerdlow, D. L., Sansonetti, P. J., et al. (1999). Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. World Health Organ. 77, 651–666.
Lenz, D. H., Mok, K. C., Lilley, B. N., Kulkarni, R. V., Wingreen, N. S., and Bassler, B. L. (2004). The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118, 69–82. doi: 10.1016/j.cell.2004.06.009
Leuzzi, A., Di Martino, M. L., Campilongo, R., Falconi, M., Barbagallo, M., Marcocci, L., et al. (2015). Multifactor regulation of the MdtJI polyamine transporter in Shigella. PLoS ONE 10:e0136744. doi: 10.1371/journal.pone.0136744
Lewis, B. P., Burge, C. B., and Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20. doi: 10.1016/j.cell.2004.12.035
Mahmoud, R. Y., Li, W., Eldomany, R. A., Emara, M., and Yu, J. (2017). The Shigella ProU system is required for osmotic tolerance and virulence. Virulence 8, 362–374. doi: 10.1080/21505594.2016.1227906
Mangold, M., Siller, M., Roppenser, B., Vlaminckx, B. J., Penfound, T. A., Klein, R., et al. (2004). Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol. Microbiol. 53, 1515–1527. doi: 10.1111/j.1365-2958.2004.04222.x
Massé, E., and Gottesman, S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625. doi: 10.1073/pnas.032066599
Massé, E., Vanderpool, C. K., and Gottesman, S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005
Michaux, C., Hartke, A., Martini, C., Reiss, S., Albrecht, D., Budin-Verneuil, A., et al. (2014). Involvement of Enterococcus faecalis small RNAs in stress response and virulence. Infect. Immun. 82, 3599–3611. doi: 10.1128/IAI.01900-14
Murina, V. N., and Nikulin, A. D. (2015). Bacterial small regulatory RNAs and Hfq protein. Biochemistry 80, 1647–1654. doi: 10.1134/S0006297915130027
Murphy, E. R., and Payne, S. M. (2007). RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect. Immun. 75, 3470–3477. doi: 10.1128/IAI.00112-07
Nickerson, K. P., Chanin, R. B., Sistrunk, J. R., Rasko, D. A., Fink, P. J., Barry, E. M., et al. (2017). Analysis of Shigella flexneri resistance, biofilm formation, and transcriptional profile in response to bile salts. Infect. Immun. 85, e01067–e01016. doi: 10.1128/IAI.01067-16
Park, S. H., Oh, K. H., and Kim, C. K. (2001). Adaptive and cross-protective responses of Pseudomonas sp. DJ-12 to several aromatics and other stress shocks. Curr. Microbiol. 43, 176–181. doi: 10.1007/s002840010283
Pichon, C., and Felden, B. (2005). Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. U.S.A. 102, 14249–14254. doi: 10.1073/pnas.0503838102
Platz, G. J., Bublitz, D. C., Mena, P., Benach, J. L., Furie, M. B., and Thanassi, D. G. (2010). A tolC mutant of Francisella tularensis is hypercytotoxic compared to the wild type and elicits increased proinflammatory responses from host cells. Infect. Immun. 78, 1022–1031. doi: 10.1128/IAI.00992-09
Porter, M. E., and Dorman, C. J. (1994). A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J. Bacteriol. 176, 4187–4191. doi: 10.1128/JB.176.13.4187-4191.1994
Scribano, D., Damico, R., Ambrosi, C., Superti, F., Marazzato, M., Conte, M. P., et al. (2016). The Shigella flexneri OmpA amino acid residues 188 EVQ 190 are essential for the interaction with the virulence factor PhoN2. Biochem. Biophys. Rep. 8, 168–173. doi: 10.1016/j.bbrep.2016.08.010
Sereny, B. (1955). Experimental Shigella keratoconjunctivitis; a preliminary report. Acta. Microbiol. 2, 293–296.
Sharff, A., Fanutti, C., Shi, J., Calladine, C., and Luisi, B. (2001). The role of the TolC family in protein transport and multidrug efflux. From stereochemical certainty to mechanistic hypothesis. Eur. J. Biochem. 268, 5011–5026. doi: 10.1046/j.0014-2956.2001.02442.x
Small, P., Blankenhorn, D., Welty, D., Zinser, E., and Slonczewski, J. L. (1994). Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176, 1729–1737. doi: 10.1128/JB.176.6.1729-1737.1994
Stone, B. J., and Miller, V. L. (1995). Salmonella enteritidis has a homologue of tolC that is required for virulence in BALB/c mice. Mol. Microbiol. 17, 701–712. doi: 10.1111/j.1365-2958.1995.mmi_17040701.x
Udekwu, K. I., Darfeuille, F., Vogel, J., Reimegård, J., Holmqvist, E., and Wagner, E. G. (2005). Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 19, 2355–2366. doi: 10.1101/gad.354405
Vanderpool, C. K., Balasubramanian, D., and Lloyd, C. R. (2011). Dual-function RNA regulators in bacteria. Biochimie 93, 1943–1949. doi: 10.1016/j.biochi.2011.07.016
Wagner, E. G. H., and Romby, P. (2015). Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 90, 133–208. doi: 10.1016/bs.adgen.2015.05.001
Wang, L., Yang, G., Qi, L., Li, X., Jia, L., Xie, J., et al. (2016). A novel small RNA regulates tolerance and virulence in Shigella flexneri by responding to acidic environmental changes. Front. Cell. Infect. Microbiol. 6:24. doi: 10.3389/fcimb.2016.00024
Wang, X. Y., Tao, F., Xiao, D., Lee, H., Deen, J., Gong, J., et al. (2006). Trend and disease burden of bacillary dysentery in China (1991–2000). Bull. World Health Organ. 84, 561–568. doi: 10.2471/BLT.05.023853
Wang, Y., Bai, Y., Qu, Q., Xu, J., Chen, Y., Zhong, Z., et al. (2011). The 16MΔvjbR as an ideal live attenuated vaccine candidate for differentiation between Brucella vaccination and infection. Vet. Microbiol. 151, 354–362. doi: 10.1016/j.vetmic.2011.03.031
Keywords: Shigella flexneri, sRNA, environmental stress, virulence, Ssr54
Citation: Yang G, Li B, Jia L, Qiu H, Yang M, Zhu B, Xie J, Qiu S, Li P, Ma H, Song H and Wang L (2020) A Novel sRNA in Shigella flexneri That Regulates Tolerance and Virulence Under Hyperosmotic Pressure. Front. Cell. Infect. Microbiol. 10:483. doi: 10.3389/fcimb.2020.00483
Received: 17 April 2020; Accepted: 04 August 2020;
Published: 16 September 2020.
Edited by:
Rakhi Rajan, University of Oklahoma, United StatesReviewed by:
Christina S. Faherty, Massachusetts General Hospital and Harvard Medical School, United StatesCopyright © 2020 Yang, Li, Jia, Qiu, Yang, Zhu, Xie, Qiu, Li, Ma, Song and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ligui Wang, d2FuZ2xpZ3VpMTk4M0AxMjYuY29t; Hongbin Song, aG9uZ2JpbnNvbmdAMjYzLm5ldA==; Hui Ma, bWFodWktMjhAMTYzLmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.