
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell. Infect. Microbiol. , 15 July 2020
Sec. Parasite and Host
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00345
This article is part of the Research Topic Leishmaniasis: Control and Elimination View all 20 articles
Himachal Pradesh in India is a newer endemic state with co-existence of cutaneous and visceral leishmaniasis. The cutaneous leishmaniasis cases are on an increase in the region and reported to be unusually caused by Leishmania donovani with limited molecular validation. In order to molecularly characterize the causative parasite of the cutaneous disease, parasite specific Internal-Transcribed Spacer 1 (ITS1) PCR RFLP and sequence analysis was performed on skin lesional biopsies from cutaneous leishmaniasis patients. Interestingly, we found the presence of Leptomonas seymouri in 38.5% (22/57) of the patients along with L. donovani detected in all the samples. L. seymouri is a monoxenous insect trypanosomatid, generally incapable of infecting humans. In recent years, the parasite is also reported to co-infect humans with L. donovani in visceral and post kala-azar dermal leishmaniasis (PKDL) cases prevalent in northeastern India. The finding of L. seymouri-L. donovani co-infection in unusual cutaneous cases from Himachal Pradesh is the first ever to our knowledge and imply a newer disease paradigm. There is an urgent need to understand the biology of Leptomonas co-infection with L. donovani and its possible role in visceral and/or dermotropic disease outcome. Importantly, L. seymouri co-infection in cutaneous cases and previously reported visceral and PKDL cases needs to be recognized as a newer phenomenon by the leishmaniasis surveillance program in India.
Leishmaniasis is a disease complex caused by Leishmania parasite with a digenetic life cycle in the sandfly vector and the mammalian host. In India, visceral leishmaniasis (VL) caused by L. donovani predominates in the northeast belt in the state of Bihar, West Bengal, Uttar Pradesh, and Jharkhand with fewer cutaneous leishmaniasis (CL) cases by L. tropica in the hot arid western region of the Thar Desert in Rajasthan (Thakur et al., 2018). More recently, newer endemic pockets in the state of Kerala and Himachal Pradesh (HP) in India are exhibiting unusual disease presentation with L. donovani causing cutaneous leishmaniasis (Sharma et al., 2005; Kumar et al., 2015; Thakur et al., 2018). In recent years, the hilly state of Himachal Pradesh in India is coming up with increased number of CL cases caused by L. donovani in the previously non-endemic zones (Sharma et al., 2005; Kumari et al., 2017). Lack of in-depth study on the involvement of L. donovani in CL cases from HP made us perform a comprehensive molecular analysis of the parasite in the patients' skin lesional specimens. In our study, we report for the first time the presence of L. seymouri co-infection in the unusual CL cases in Himachal Pradesh (HP) caused by L. donovani variants (unpublished data). Classically, Leptomonas spp. comprise insect parasite with a monoxenous life cycle and are considered non-pathogenic to humans. Lately, adaption of the parasite to dixenous life cycle has been reported under specific conditions such that L. seymouri can exist as a co-infectant with other pathogens in immunocompromised subjects (Dedet and Pratlong, 2000; Kraeva et al., 2015; Selvapandiyan et al., 2015; Kaufer et al., 2017). In this context, our finding re-affirms the limited but significant emerging evidence on the newer parasitic capability of Leptomonas sp. as an opportunistic human pathogen in CL cases from HP. This is in line with the previous reports on L. seymouri as a co-infectant in clinical isolates as well as direct clinical specimens from VL and PKDL cases from northeast India (Srivastava et al., 2010; Ghosh et al., 2012; Singh et al., 2013).
Sixty CL patients, indigenous to Sutluj river belt in Himachal Pradesh were included in the study over the period from 2014 to 2018. Lesional skin biopsies were collected from the patients at Department of Dermatology, Indira Gandhi Medical College (IGMC), Shimla and Mahatma Gandhi Medical Services Complex (MGMSC) Khaneri, Rampur, Shimla at the time of diagnosis. Written informed consent was obtained from all the patients. The study design was approved by the Institutional Ethics Committee IGMC, Shimla, Himachal Pradesh, Approval no. HFW(MS)G-5(Ethics)/2014-10886 and Central University of Punjab, Approval no. CUPB/IEC/2016/034.
Lesional biopsy samples were processed for parasite detection using Giemsa stained touch smears, Hematoxylin and Eosin (H & E) stained paraffin-embedded tissue sections as per standard protocol (Elder et al., 2001; Bain et al., 2016). A part of the lesional biopsy from the CL cases was processed for examining CL specific histopathological changes. The samples were processed in 10% NBF, embedded in paraffin and processed to 4–5 μm thick tissue section. Tissue sections were stained with H&E and examined for histopathological changes specific to cutaneous lesions.
Molecular identification of the parasite was performed using skin lesional specimens from the clinically confirmed CL patients. Genomic DNA (gDNA) isolation from patient samples and laboratory-grown L. donovani (Ld1S2D, LdBob) promastigote culture was done using standard protocol (Salotra et al., 2001). gDNA from 57 samples were used for species-specific ribosomal Internal-Transcribed Spacer 1 (ITS1) region PCR RFLP assay as described previously (El Tai et al., 2000). ITS1 specific PCR amplification was done with the primer set LITSR (5′-CTGGATCATTTTCCGATG-3′) and L5.8S (5′-TGATACCACTTATCGCACTT-3′). Briefly 50–100 ng of gDNA was used as template and amplified with 10 pmol of each primer using Go Taq Green Master mix, 1X (Promega, Cat # M7122) with an initial denaturation at 95°C for 2 min, 34 cycles of denaturation at 95°C for 20 s, annealing at 53°C for 30 s and extension at 72°C for 1 min with the final extension at 72°C for 6 min. The PCR product of ~ 320 bp size was subjected to HaeIII RFLP with overnight HaeIII digestion at 37°C and run on 2.5% agarose gel. The amplification product of ~320 bp and ~400 bp were eluted using Qiagen kit and submitted for Sanger sequencing for parasite identification.
Sequences corresponding to the ~320 bp and ~400 bp amplification products were retrieved for the representative samples with their accession numbers deposited in Genbank (Table 1). Each of the sequences was analyzed using BLAST with default parameters to identify the parasite in the cutaneous lesions. ITS1 nucleotide query sequences were aligned with relevant ITS1 reference sequences of standard WHO and region specific Leishmania spp., Leptomonas spp. and Trypanosoma spp. isolates using multiple alignment software, MUSCLE using default parameters (Table 1) (Edgar, 2004). The sequence alignment was done using Jalview multiple alignment editor version 2.10.4b1 (Clamp et al., 2004). The maximum likelihood tree from the aligned sequences was obtained with 1000 bootstraps with default parameters using the dnaml program of the phylip package (Felsenstein, 2004). The final tree was plotted using FigTree software (version 1.4.3).
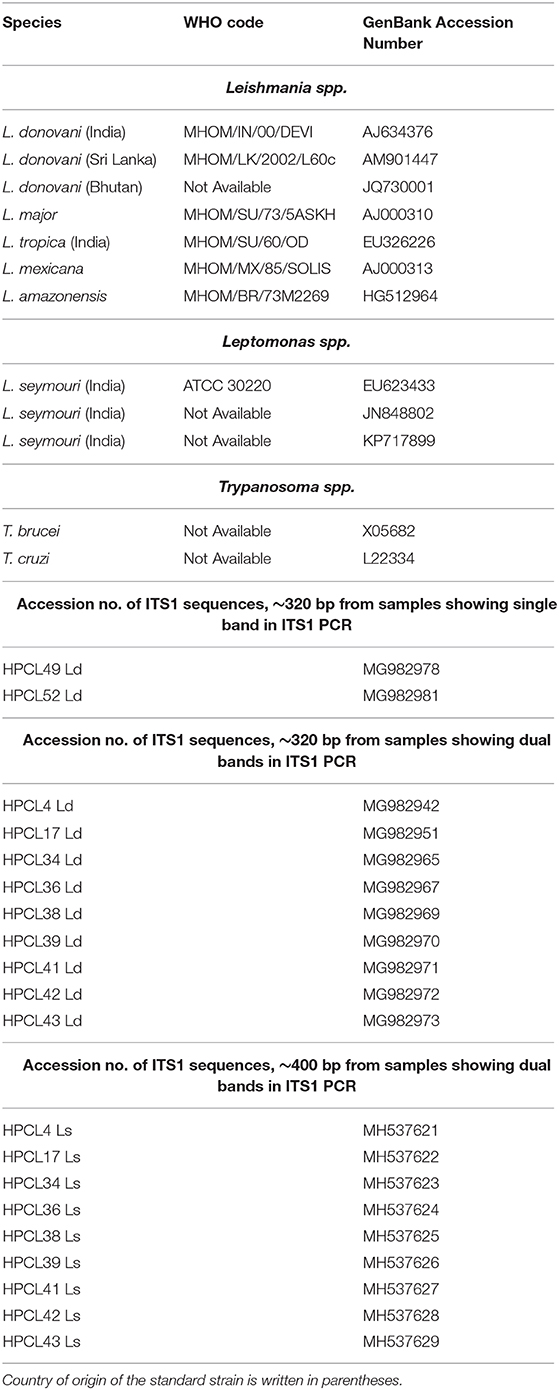
Table 1. GenBank Accession Numbers of ITS1 sequences of standard Leishmania, Leptomonas, and Trypanosoma spp. and ITS1 test sequences from CL patients used in phylogenetic analysis.
In the present study, 60 patients with typical cutaneous lesions presented as plaques, nodules and/or papules were included. Most of the patients exhibited typical localized cutaneous lesions, often with raised borders, serous crusting and ulceration. The lesion number and size range in males and females is given in Supplementary Table 1 separately for L. donovani-L. seymouri co-infection cases and L. donovani alone infected cases with no major difference in the two groups. It is important to mention that all the CL cases analyzed were indigenous in nature with no VL history or any visit to VL endemic area. In both Leptomonas co-infected cases and L. donovani alone infected CL cases, males and females were almost equally represented with maximum disease frequency in the age group of 21–40 years, summarized in Supplementary Table 1.
All the patients with characteristic CL lesions were clinically confirmed for the presence of amastigotes in Giemsa stained lesional touch smears and Haematoxylin/Eosin stained paraffin embedded tissue sections (Supplementary Figures 1A,B). In Giemsa stained tissue smears, 35% of the samples were amastigote positive from the L. donovani-L. seymouri co-infected cases while ~65% samples were positive for amastogotes in L. donovani alone infected cases. Interestingly, ~ 40% of the Haematoxylin/Eosin stained biopsy samples were parasite positive for both the groups (Supplementary Table 1).
The CL lesion-specific histopathological analysis of biopsy samples from both the co-infected and L. donovani alone infected cases, showed characteristic CL lesion-specific epidermal changes with acanthosis and papillomatosis along with granulomatous inflammation (Supplementary Figures 2A,B). No apparent differences in the histopathological features were observed in the two groups with level of histopathological manifestations varying from sample to sample.
gDNA samples from lesional skin biopsies of 57 CL patients were used for parasite species specific identification by PCR RFLP analysis of ITS1 region along with gDNA from standard L. donovani (1S2D, LdBob) culture used as a positive control. HaeIII restriction enzyme digestion of the amplified ITS1 region between the small subunit (ssu) rRNA and 5.8S rRNA DNA provides a Leishmania spp. specific RFLP pattern (Dávila and Momen, 2000; El Tai et al., 2000). ITS1 specific PCR amplification gave the expected ~320 bp product in all the 57 CL samples similar to the positive control while 38.5 % (22/57) samples gave a dual band pattern with a unique ~400 bp extra band (Figures 1A,B, Left Panels). The ~320 bp band in all the samples resolved into HaeIII RFLP pattern identical to the L. donovani control sample with 3 discrete fragments of ~190 bp, ~80 bp, and ~50 bp (Figures 1A,B, Right Panels). The extra ~400 bp band, specific to 22 samples remained undigested as depicted in Figure 1B, Right Panel. Our result suggests possible co-infection of non-Leishmania trypanosomatid along with L. donovani in the test samples, based on existing literature. Ghosh et al reported a similar ITS1 PCR RFLP pattern in 4/29 of VL and 2/7 of PKDL specimens with the extra ~400 bp band corresponding to L. seymouri (Ghosh et al., 2012).
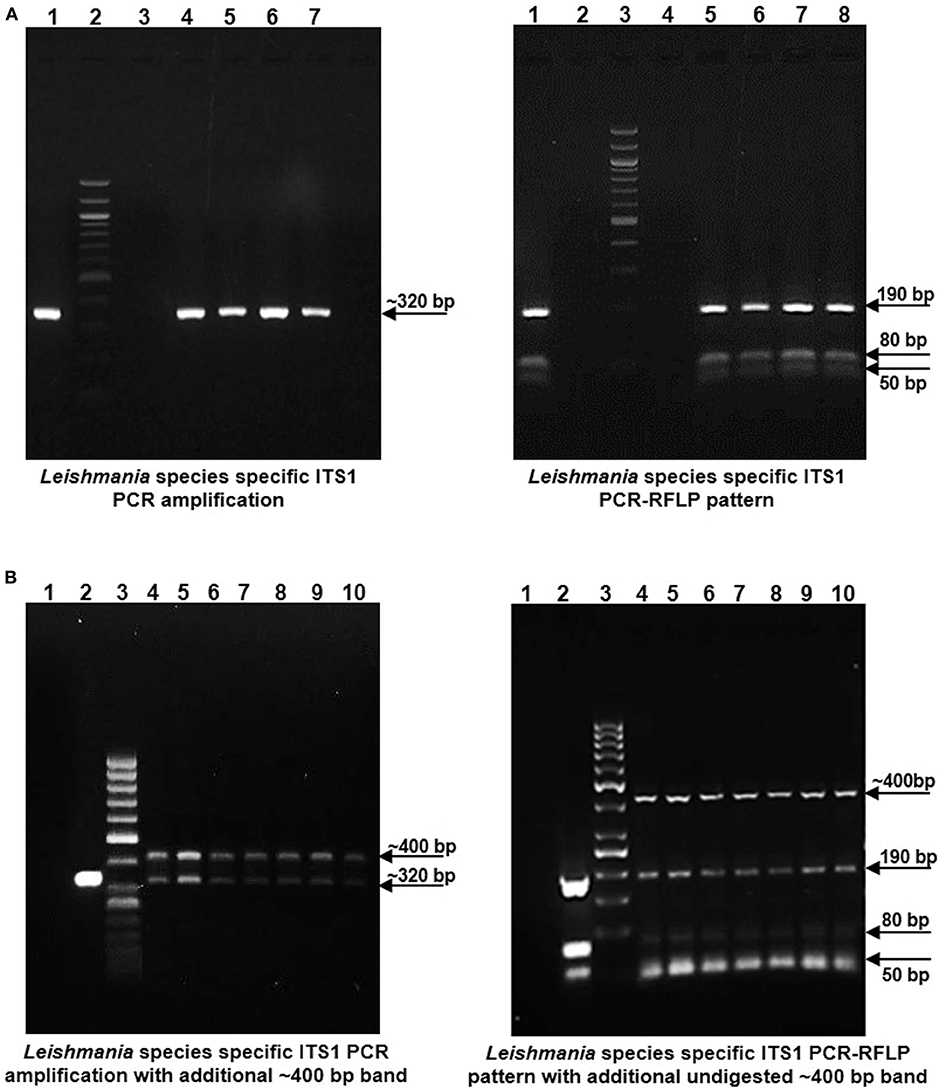
Figure 1. Leishmania species specific ITS1 PCR on DNA isolated from lesion biopsy samples from CL patients and HaeIII PCR RFLP analysis of ITS1 region of test samples and L. donovani culture as a positive control. (A) Left Panel: Representative patient samples with a single, Leishmania spp specific ~ 320 bp amplification product. Lane 1, L. donovani; Lane 2, 100 bp DNA marker; Lane 3, water control; Lanes 4–7, CL test samples. Right Panel: HaeIII ITS1 PCR-RFLP pattern for identification of Leishmania species. Lane 1, L. donovani; Lane 3, 100 bp marker; Lane 4, water control; Lanes 5–8, CL test samples. (B) Left Panel: Representative patient samples with dual amplification bands (~ 320 bp and ~ 400 bp). Lane 1; Water control, Lane 2; L. donovani positive control, Lane 3; 50 bp DNA marker, Lanes 4–10, CL test samples. Right Panel: HaeIII ITS1 PCR-RFLP pattern of samples with dual bands. Lane 1; Water control, Lane 2; L. donovani positive control, Lane 3; 50 bp DNA marker, Lanes 4–10, CL test samples.
Sequence based identification of the ~320 bp and ~400 bp amplicons was performed for the representative CL samples (2 samples with ~320 bp band alone and 9 samples with dual band pattern). All the sequences corresponding to ~320 bp band and ~400 bp band were deposited with their accession numbers in Genbank (Table 1). ITS1 sequences corresponding to ~320 bp suggested all the samples to be closest with L. donovani with maximum identity to L. donovani isolate from Bhutan (JQ730001) using BLAST analysis while the sequences corresponding to ~400 bp band, showed maximum identity with the standard L. seymouri ITS1 sequences with accession numbers KP717899, EU623433, and JN84880 (Table 1) (Ghosh et al., 2012; Yangzom et al., 2012; and unpublished data). A similar finding with the unique ~400 bp band representing L. seymouri specific ITS1 sequence and ~320 bp band specific to L. donovani ITS1 sequence has been demonstrated earlier in the VL and/or PKDL patients (Ghosh et al., 2012).
The multiple sequence alignment of the test sequences (~320 bp and ~400 bp) from the CL samples and the ITS1 sequences representing standard Leishmania, Leptomonas and Trypanosoma isolates from GenBank was performed and used for phylogenetic analysis using maximum likelihood method (Table 1). Phylogenetic analysis grouped all the HPCL isolates into two discrete clusters corresponding to Leishmania and Leptomonas with respect to the ~320 bp and ~400 bp ITS1 sequences respectively (Figure 2). All the 11 CL test samples clustered closely with the standard L. donovani isolates from India, Sri Lanka and Bhutan and L. infantum with respect to the ITS1 specific ~320 bp sequences represented by HPCL_L. donovani series (Figure 2, Table 1). Sequence analysis using BLAST and phylogenetic classification, suggest that the CL in HP is caused by L. donovani isolates closest to the Bhutan L. donovani isolate and distinct from the Indian and Sri Lankan L. donovani isolates (unpublished data). Nine out of 11 CL test samples also clustered closely with the standard L. seymouri isolates from India with respect to the ~400 bp sequence represented by HPCL_L. seymouri series (Figure 2, Table 1). Interestingly, both the HPCL derived parasite clusters representing ITS1 sequences corresponding to L. donovani and L. seymouri exhibited considerable heterogeneity signifying sequence variation among multiple isolates analyzed. The standard isolates representing Leishmania spp. known to cause CL in old and new world and Trypanosoma spp clustered independently as outgroups.
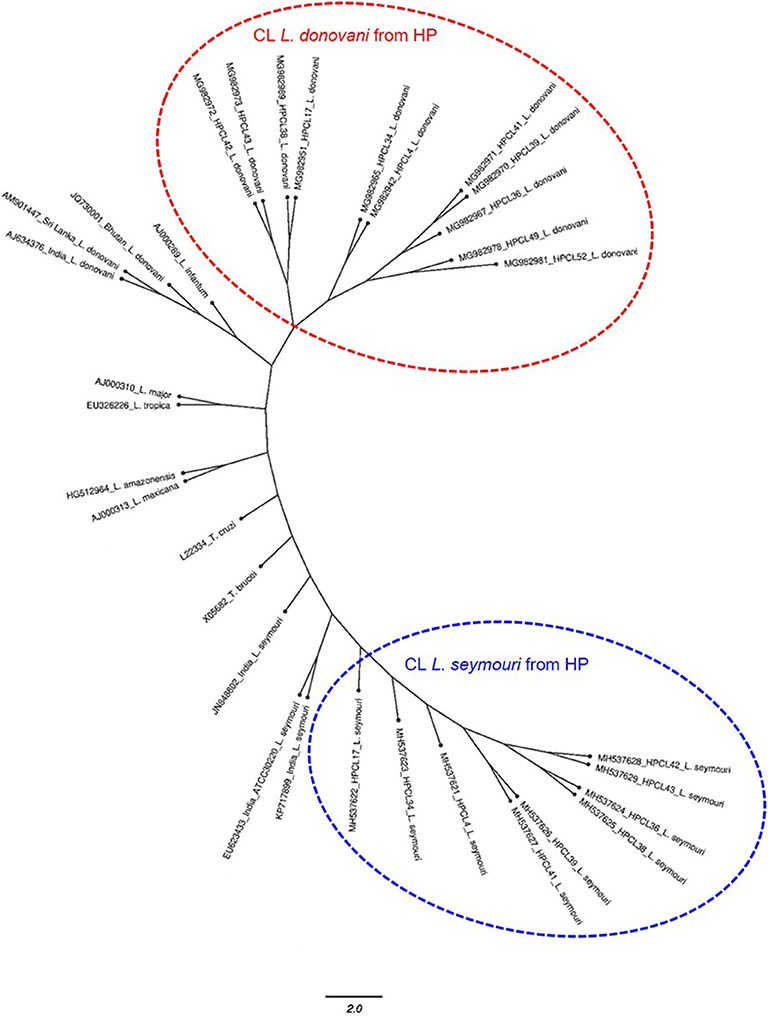
Figure 2. Phylogenetic tree of ITS1 gene sequences from CL test samples with single and dual-band amplification patterns (~320 bp and ~400 bp band size test samples, designated as HPCL_Ld series corresponding to ~320 bp band and HPCL_Ls series corresponding to ~400 bp band, numbered in order of their collection) and ITS1 sequences of standard Leishmania, Leptomonas and Trypanosoma isolates from Genbank was obtained using Maximum Likelihood method with 1,000 bootstraps using dnaml program of PHYLIP package.
ITS1 DNA region is highly conserved among different trypanosomatid parasite species (Dávila and Momen, 2000; Borghesan et al., 2013). Multiple sequence alignment of the sequences corresponding to the representative HP L. donovani, ~320 bp and HP L. seymouri, ~400 bp ITS1 sequences (MG982942, MG982978, MH537621) along with the ~320 bp ITS1 sequences of standard L. donovani isolates from India (AJ634376), Sri Lanka (AM901447), Bhutan (JQ730001) and ~400 bp ITS1 sequences of standard L. seymouri isolates from India (KP717899, EU623433, JN848802) exhibited distinct differences in ITS1 sequences specific to L. donovani and L. seymouri. Additional nucleotide patches unique to L. seymouri ITS1 sequence in comparison to L. donovani ITS1 region were apparent in the sequence alignment shown in Figure 3.

Figure 3. Multiple sequence alignment of ITS1 sequences representing Leishmania specific ~320 bp band (MG982942_HPCL4_L. donovani and MG982978_HPCL49_L. donovani) and Leptomonas specific ~400 bp band (MH537621_HPCL4_L. seymouri) along with standard L. donovani and L. seymouri specific ITS1 sequences retrieved from Genbank (AJ634376_India_ L.‘ donovani, JQ730001_Bhutan _L. donovani, AM901447_Sri Lanka_ L. donovani, KP717899_ India_L. seymouri, EU623433_India_ ATCC30220_L. seymouri and JN848802_India_L. seymouri). Sequences were aligned using the Jalview sequence alignment program. Differences in L. donovani and L. seymouri ITS1 sequences are highlighted in red.
Detection of the ITS1 sequences corresponding to the two bands in clinical specimens clearly exhibited the presence of L. seymouri and L. donovani co-infection in CL patients from HP. We report L. seymouri as a co-infecting parasite in almost 38.5% (22/57) of the CL cases caused by L. donovani. This is the first-ever report of L. seymouri co-infection in CL patients, typed with unusual cutaneous manifestation by L. donovani with no visceral features, detailed in a recent communication from our laboratory (unpublished data).
Our study concludes another instance of L. seymouri as a L. donovani co-infectant in leishmaniasis patients from HP, a newer endemic region in India in line with earlier reports of L. seymouri co-infection in VL and PKDL patients from the northeastern VL zone (Srivastava et al., 2010; Ghosh et al., 2012; Singh et al., 2013). Importantly, the CL patients diagnosed in our study with L. seymouri, exhibit cutaneous manifestation caused by L. donovani (Sharma et al., 2005; and unpublished data). A possible role of Leptomonas co-infection in disease pathogenesis and phenotypic outcome with viscerotropic and/or dermotropic manifestation in VL vs. PKDL cases is not much explored. In addition to L. seymouri co-infection detected in cutaneous L. donovani disease in our study, the parasite has been reported from PKDL cases and HIV patients with a diffuse cutaneous phenotype (Dedet et al., 1995; Ghosh et al., 2012). Earlier notion of Leptomonas spp incapable of causing infection in vertebrates is getting blur with increasing evidence of its dixenous existence as a real co-infecting partner rather than a culture contaminant (Srivastava et al., 2010; Ghosh et al., 2012; Singh et al., 2013; Kraeva et al., 2015; Selvapandiyan et al., 2015). This is evident from the detection of L. seymouri in 22/57 clinical specimens directly processed for the molecular detection of the parasite with a rare chance of interim contamination.
L. seymouri is currently understood as an opportunistic parasite in immuno-compromised hosts such as HIV and Leishmaniasis cases (Kraeva et al., 2015; Selvapandiyan et al., 2015). Human cases of Leptomonas co-infection have been underestimated due to its morphological, antigenic and genomic similarity with Leishmania such that many of the DNA sequences specific to L. seymouri have been assigned to L. donovani (Nasereddin et al., 2008; Ghosh et al., 2012; Kraeva et al., 2015; Selvapandiyan et al., 2015). In this regard a rapid diagnostic method to detect and differentiate Leishmania and Leptomonas in clinical samples needs to be adopted for reliable experimentation and data inter-presentation along with specifically understanding the clinical correlates of Leptomonas co-infection (Ahuja et al., 2020). Interestingly, the genetic similarity of Leptomonas with Leishmania along with few nucleotide variations in ITS1 sequences among the two clusters retrieved in our study raises the possibility of the existence of genetic hybrids with heterogeneous Leishmania as well as Leptomonas genotypes albeit with limited evidence of natural L. seymouri infection in P. argentipe known for L. donovani transmission in India.
Importantly, Leptomonas like parasite has been demonstrated to naturally co-infect P. argentipes along with L. donovani in VL endemic region of Nepal (Bhattarai et al., 2009). Also, L. seymouri and L. donovani have been shown to co-persist in P. argentipes experimental infection (Kraeva et al., 2015). With the possibility of P. longiductus mediated L. seymouri-L. donovani co-infection in HP, natural co-infection of P. longipalpis by Leptomonas and L. donovani has been demonstrated in a focus of kala-azar (Deane and Deane, 1954; Sharma et al., 2009). Additionally, P. longiductus has been identified as one of the vector in Bhutan and China (World Health Organization, 2010; Yangzom et al., 2012). Thus, the identity of sandfly vector capable of co-transmitting Leptomonas and Leishmania parasite needs to be further ascertained in different regions with reports of Leptomonas co-infection in humans. The similarity in antigenicity of the two organisms further warranties a comparative immune-profiling of the VL and CL patients with and without L. seymouri co-infection relevant in immune response studies like deciphering L. donovani mediated immune-suppression that possibly makes infected patients vulnerable to Leptomonas co-infection and for vaccine development studies (Selvapandiyan et al., 2015).
In conclusion, the presence of L. seymouri with L. donovani in VL, PKDL and CL cases imply a real yet unappreciated phenomenon with the possible implication in disease outcome. We emphasize the emerging L. seymouri- L. donovani co-infected CL cases in the newer Sutluj river belt in HP in addition to co-infected VL/PKDL cases, reported in the northeast belt of India. The finding needs to be taken as a new challenge for the leishmaniasis surveillance and elimination program operational in India.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by Institutional Ethics Committee IGMC, Shimla, Himachal Pradesh, Approval no. HFW(MS)G-5(Ethics)/2014-10886 and Central University of Punjab, Approval no. CUPB/IEC/2016/034. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
LT collected the samples, performed the experiments, and analyzed the data. HK constructed the phylogenetic tree. AN provided the patient samples. AJ and MJ contributed in concept and design of study, data analysis, drafting of article, critical revision, and final approval of manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by Research Seed Money Grant (RSM Grant, GP# 25) Central University of Punjab, Bathinda, Punjab, India. LT was funded with a doctoral fellowship, Central University of Punjab, Bathinda.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We acknowledge Emeritus Prof. Rentala Madhubala, JNU, Delhi, India for generously gifting L. donovani standard culture.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00345/full#supplementary-material
Ahuja, K., Vats, A., Beg, M. A., Kariyawasam, K., Chaudhury, A., Chatterjee, M., et al. (2020). High resolution melting based method for rapid discriminatory diagnosis of co-infecting Leptomonas seymouri in Leishmania donovani-induced leishmaniasis. Parasitol. Int 75:102047. doi: 10.1016/j.parint.2019.102047
Bain, B. J., Bates, I., and Laffan, M. A. (2016). Dacie and Lewis Practical Haematology E-Book: Elsevier Health Sciences. Available online at: https://www.elsevier.com/books/dacie-and-lewis-practicalhaematology/bain/978-0-7020-6696-2 (accessed May 27, 2020).
Bhattarai, N. R., Das, M. L., Rijal, S., van der Auwera, G., Picado, A., Khanal, B., et al. (2009). Natural infection of Phlebotomus argentipes with Leishmania and other trypanosomatids in a visceral leishmaniasis endemic region of Nepal. Trans. R. Soc. Trop. Med. Hyg. 103, 1087–1092. doi: 10.1016/j.trstmh.2009.03.008
Borghesan, T. C., Ferreira, R. C., Takata, C. S., Campaner, M., Borda, C. C., Paiva, F., et al. (2013). Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist 164, 129–152. doi: 10.1016/j.protis.2012.06.001
Clamp, M., Cuff, J., Searle, S., and Barton, G. (2004). The Jalview Java alignment editor. Bioinformatics 20, 426–427. doi: 10.1093/bioinformatics/btg430
Dávila, A., and Momen, H. (2000). Internal-transcribed-spacer (ITS) sequences used to explore phylogenetic relationships within Leishmania. Ann. Trop. Med. Parasitol. 94, 651–654. doi: 10.1080/00034983.2000.11813588
Deane, M., and Deane, L. (1954). Natural infection of Phlebotomus longipalpis by Leptomonas, probably Leishmania donovani, in a focus of kala-azar in Ceará. Hospital (Rio de Janeiro, Brazil) 45:697.
Dedet, J., Roche, B., Pratlong, F., Cales-Quist, D., Jouannelle, J., Benichou, J., et al. (1995). Diffuse cutaneous infection caused by a presumed monoxenous trypanosomatid in a patient infected with HIV. Trans. R. Soc. Trop. Med. Hyg. 89, 644–646. doi: 10.1016/0035-9203(95)90427-1
Dedet, J. P., and Pratlong, F. (2000). Leishmania, trypanosoma and monoxenous trypanosomatids as emerging opportunistic agents 1. J. Eukarytic Microbiol. 47, 37–39. doi: 10.1111/j.1550-7408.2000.tb00008.x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
El Tai, N., Osman, O., El Fari, M., Presber, W., and Schönian, G. (2000). Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans. R. Soc. Trop. Med. Hyg. 94, 575–579. doi: 10.1016/S0035-9203(00)90093-2
Elder, D., Elenitsas, R., Johnson Jr, B., Murphy, G., and Xu, G. (2001). Lever's histopathology of the skin, 2009. Philadelphia: Wolters Kluwer/Lipplncott Williams & Wilkins Cop. Guillen DR, Cockerell CJ. Cutaneous and subcutaneous sarcomas. Clin. Dermatol. 19, 262–268. doi: 10.1016/S0738-081X(01)00177-8
Felsenstein, J. (2004). PHYLIP (Phylogeny Inference Package) Version 3.6. Distributed by the Author. Available online at: http://evolution.genetics.washington.edu/phylip.html (accessed May 25, 2020)
Ghosh, S., Banerjee, P., Sarkar, A., Datta, S., and Chatterjee, M. (2012). Coinfection of Leptomonas seymouri and Leishmania donovani in Indian leishmaniasis. J. Clin. Microbiol. 50, 2774–2778. doi: 10.1128/JCM.00966-12
Kaufer, A., Ellis, J., Stark, D., and Barratt, J. (2017). The evolution of trypanosomatid taxonomy. Parasit. Vectors 10, 287. doi: 10.1186/s13071-017-2204-7
Kraeva, N., Butenko, A., Hlaváčová, J., Kostygov, A., Myškova, J., Grybchuk, D., et al. (2015). Leptomonas seymouri: adaptations to the dixenous life cycle analyzed by genome sequencing, transcriptome profiling and co-infection with Leishmania donovani. PLoS Pathog. 11:5127. doi: 10.1371/journal.ppat.1005127
Kumar, N. P., Srinivasan, R., Anish, T., Nandakumar, G., and Jambulingam, P. (2015). Cutaneous leishmaniasis caused by Leishmania donovani in the tribal population of the Agasthyamala Biosphere Reserve forest, Western Ghats, Kerala, India. J.Med. Microbiol. 64, 157–163. doi: 10.1099/jmm.0.076695-0
Kumari, S., Negi, A., Garg, A., Thakur, K., Lal, P., and Ahluwalia, A. K. (2017). Clinico-epidemiological trends of cutaneous leishmaniasis along the satluj valley of Himachal Pradesh-A new364 focus with emerging infection. J. Med. Sci. Clin. Res. 5, 24973–24978. doi: 10.18535/jmscr/v5i7.110
Nasereddin, A., Bensoussan-Hermano, E., Schönian, G., Baneth, G., and Jaffe, C. L. (2008). Molecular diagnosis of Old World cutaneous leishmaniasis and species identification by use of a reverse line blot hybridization assay. J. Clin. Microbiol. 46, 2848–2855. doi: 10.1128/JCM.00951-08
Salotra, P., Sreenivas, G., Pogue, G. P., Lee, N., Nakhasi, H. L., Ramesh, V., et al. (2001). Development of a species-specific PCR assay for detection of Leishmania donovani in clinical samples from patients with kala-azar and post-kala-azar dermal leishmaniasis. J. Clin. Microbiol. 39, 849–854. doi: 10.1128/JCM.39.3.849-854.2001
Selvapandiyan, A., Ahuja, K., Puri, N., and Krishnan, A. (2015). Implications of co-infection of Leptomonas in visceral leishmaniasis in India. Parasitology 142, 1657–1662. doi: 10.1017/S0031182015001389
Sharma, N. L., Mahajan, V. K., Kanga, A., Sood, A., Katoch, V. M., Mauricio, I., et al. (2005). Localized cutaneous leishmaniasis due to Leishmania donovani and Leishmania tropica: preliminary findings of the study of 161 new cases from a new endemic focus in Himachal Pradesh, India. Am. J. Trop. Med. Hyg 72, 819–824. doi: 10.4269/ajtmh.2005.72.819
Sharma, N. L., Mahajan, V. K., Ranjan, N., Verma, G. K., Negi, A. K., and Mehta, K. I. S. (2009). The sandflies of the Satluj river valley, Himachal Pradesh (India): some possible vectors of the parasite causing human cutaneous and visceral leishmaniases in this endemic focus. J. Vector Borne Dis 46:136.
Singh, N., Chikara, S., and Sundar, S. (2013). SOLiD™ sequencing of genomes of clinical isolates of Leishmania donovani from India confirm leptomonas co-infection and raise some key questions. PLoS ONE 8:e55738. doi: 10.1371/journal.pone.0055738
Srivastava, P., Prajapati, V. K., Vanaerschot, M., Van der Auwera, G., Dujardin, J. C., and Sundar, S. (2010). Detection of Leptomonas sp. parasites in clinical isolates of Kala-azar patients from India. Infect. Genet. Evol. 10, 1145–1150. doi: 10.1016/j.meegid.2010.07.009
Thakur, L., Singh, K. K., Shanker, V., Negi, A., Jain, A., Matlashewski, G., et al. (2018). Atypical leishmaniasis: A global perspective with emphasis on the Indian subcontinent. PLoS Negl. Trop. Dis. 12:659. doi: 10.1371/journal.pntd.0006659
World Health Organization (2010). Control of the Leishmaniases: Report of a Meeting of the WHO Expert Committee on the Control of Leishmaniases, Geneva, 22-26 March 2010. Available online at: https://apps.who.int/iris/handle/10665/44412 (accessed March 31, 2020).
Keywords: Leptomonas seymouri, cutaneous leishmaniasis, Leishmania donovani, Himachal Pradesh, India
Citation: Thakur L, Kushwaha HR, Negi A, Jain A and Jain M (2020) Leptomonas seymouri Co-infection in Cutaneous Leishmaniasis Cases Caused by Leishmania donovani From Himachal Pradesh, India. Front. Cell. Infect. Microbiol. 10:345. doi: 10.3389/fcimb.2020.00345
Received: 11 April 2020; Accepted: 04 June 2020;
Published: 15 July 2020.
Edited by:
Om Prakash Singh, Banaras Hindu University, IndiaReviewed by:
Angamuthu Selvapandiyan, Jamia Hamdard University, IndiaCopyright © 2020 Thakur, Kushwaha, Negi, Jain and Jain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manju Jain, bWFuanVqYWlubWRhQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.