- 1Department of Microbiology and Immunology, Emory University, Atlanta, GA, United States
- 2Research Service, Atlanta VA Healthcare System, Decatur, GA, United States
Multidrug resistant Acinetobacter baumannii is a serious healthcare threat. In fact, the Center for Disease Control recently reported that carbapenem-resistant A. baumannii is responsible for more than 8,500 infections, 700 deaths, and $281 million in healthcare costs annually in the United States with few, if any, treatment options available, leading to its designation as a pathogen of urgent concern and a priority for novel antimicrobial development. It is hypothesized that biofilms are, at least in part, responsible for the high prevalence of A. baumannii nosocomial and recurrent infections because they frequently contaminate hospital surfaces and patient indwelling devices; therefore, there has been a recent push for mechanistic understanding of biofilm formation, maturation and dispersal. However, most research has focused on A. baumannii pneumonia and bloodstream infections, despite a recent retrospective study showing that 17.1% of A. baumannii isolates compiled from clinical studies over the last two decades were obtained from urinary samples. This highlights that A. baumannii is an underappreciated uropathogen. The following minireview will examine our current understanding of A. baumannii biofilm formation and how this influences urinary tract colonization and pathogenesis.
Introduction
Acinetobacter baumannii is a public health menace recently rising to prominence due to the rapid increase in antibiotic resistance and infection rates. Infections caused by A. baumannii account for ~2% of all healthcare-associated infections in the United States and Europe (Sievert et al., 2013; Magill et al., 2014; Lob et al., 2016) and this rate is nearly doubled in Asia and the Middle East (Lob et al., 2016). Globally, it is estimated that nearly 45% of all A. baumannii isolates are multidrug-resistant (MDR; resistant to ≥3 antibiotics) with rates as eclipsing 70% in Latin America and the Middle East (Giammanco et al., 2017). We have reached a critical tipping point where antibiotic discovery cannot keep up with the rapidly evolving antibiotic resistance of A. baumannii without some type of intervention. Hence, the World Health Organization (WHO) and Centers for Disease Control (CDC) have signified A. baumannii as a pathogen of critical importance for the discovery of novel antimicrobials (WHO, 2017; CDC, 2019).
Acinetobacter baumannii primarily causes infections of the lung or bloodstream (Peleg et al., 2008). However, it was recently reported that up to one-fifth of all A. baumannii isolates are obtained from urinary sources, implying that this organism is an underappreciated uropathogen (Di Venanzio et al., 2019). Catheter-acquired urinary tract infections (CAUTI) are one of the most common hospital-acquired infections accounting for an estimated 100,000 infections annually in the United States (Zarb et al., 2012; Magill et al., 2014). It is hypothesized that bacterial biofilm formation along the catheter surface is the most important factor in the establishment of bacteriuria (Stickler, 2008). Acinetobacter baumannii's increasing prevalence in CAUTIs is due to its adept ability to form biofilms, with an estimated >75% of all isolates capable of forming a biofilm (Abdi-Ali et al., 2014; Azizi et al., 2016; Thummeepak et al., 2016). Therefore, understanding the mechanisms responsible for A. baumannii biofilm biogenesis and maturation are critical for elucidating the basis for uropathogenesis and may help with the development of future CAUTI anti-biofilm therapies. The following minireview examines existing data focused on the genetic regulation of A. baumannii biofilm lifestyle and its contribution to uropathogenesis as well as identifies current knowledge gaps to be addressed moving forward.
Biofilm Formation
Bacterial Cell Adherence
The initial step involved in the shift from planktonic to biofilm formation is surface contact and irreversible attachment (reviewed in Petrova and Sauer, 2012; Armbruster and Parsek, 2018). Acinetobacter baumannii has the ability to form biofilms on a wide range of surfaces including abiotic surfaces, like stainless steel and polypropylene, as well as host epithelial cells (Greene et al., 2016). Many virulence factors have been implicated in bacterial cell adherence, however the plasticity observed in A. baumannii genomes leads to significant strain-specific variations in biofilm formation. Investigation into the presence of known biofilm-associated genes in A. baumannii clinical isolates across several publications (Loehfelm et al., 2008; Badmasti et al., 2015; Zeighami et al., 2019) has shown that the most highly conserved genes were CsuE, the proposed tip subunit of the chaperone-usher pili (Csu), and OmpA (reported 81–100% detection). For the biofilm-associated protein (Bap) and class A extended β-lactamase blaPER-1 enzyme, detection was variable ranging from 30–66% to 2–64% of isolates, respectively. The Csu assembly system is composed of pilin subunits CsuA/B, CsuA, CsuB, and CsuE and transport proteins CsuC and CsuD, is highly conserved in biofilm-forming isolates and critical for adherence to abiotic surfaces, but not host surfaces (Tomaras et al., 2003; de Breij et al., 2009). Outer membrane protein A (OmpA) is a prominent porin that contributes to drug resistance, adhesion to epithelial cells and biofilm formation on plastic surfaces (C.H. Choi et al., 2008; Gaddy et al., 2009). Anti-OmpA serum and antibodies blocked A. baumannii's adherence and subsequent invasion of host cells (Schweppe et al., 2015). Biofilm-associated protein (Bap) is a surface-exposed, highly divergent protein that is required for adherence to bronchial cells and structural integrity and water channel formation within the biofilm (Loehfelm et al., 2008; Brossard and Campagnari, 2012; De Gregorio et al., 2015). One study found that disruption of the Bap gene led to significant reductions in biofilm thickness and volume, interbacterial cell adhesion and ability to form higher order structures on medically relevant abiotic surfaces (Loehfelm et al., 2008). Another recent study found that the variation in the bap coding sequence across A. baumannii lineages results in differential functions during biofilm development with some versions displaying better adherence properties and others forming more complex biofilms (Skerniskyte et al., 2019). β-lactamase blaPER-1-expressing strains displayed significantly increased cell adhesiveness and biofilm formation compared to strains lacking the β-lactamase (H.W. Lee et al., 2008). However, additional publications report no or limited correlation between blaPER-1 expression and biofilm formation (Sechi et al., 2004; Rao et al., 2008); thus, more research is required to elucidate its role.
Other virulence factors that have been implicated in adherence and biofilm formation include Pap, Prp, Cup, and Type IV pili systems as well as Acinetobacter trimeric autotransporter (Ata) (reviewed in Gaddy and Actis, 2009; Eijkelkamp et al., 2014; Longo et al., 2014; Harding et al., 2018). The pap operon encodes proteins homologous to P pili in E. coli, which has been found to be important for migration of bacteria from the bladder to the kidney (Wullt et al., 2000). The prpABCD operon encodes a photoregulated pilus associated with light-regulated motility and biofilm formation in ATCC 17978 (Wood et al., 2018). In addition, this operon is conserved in several other A. baumannii strains, including the hyper-biofilm forming MAR002, which displayed a 25-fold increase in the prpD homolog in sessile cells (Alvarez-Fraga et al., 2016). CUP2 pili were recently discovered as a prp operon homolog in UTI pathogen UPAB1, which when deleted resulted in reduced adhesion to both the catheter surface and bladder lumen in a CAUTI murine model (Di Venanzio et al., 2019). Type IV pili, encoded by the pil operon, have been shown to play a role in adhesion to cells and stainless steel (Ronish et al., 2019). Ata is a surface-exposed protein that has been shown to play an important role in biofilm formation as well as adherence to host cells and various host extracellular components (Bentancor et al., 2012; Weidensdorfer et al., 2019).
Biofilm Formation Cues and Detection
Following adhesion to a surface, the bacterial cells are now primed to continue the shift to the biofilm state. The next step in biofilm formation involves environmental signal sensing and signal transduction, which will lead to downstream cellular responses. Many signals and signaling components that have been implicated in the control of biofilm formation and virulence factor production in A. baumannii are described below.
Acinetobacter baumannii and its close relative Acinetobacter nosocomialis have one quorum sensing (QS) system, which plays an integral role in regulating virulence factors, biofilm formation and surface motility (Niu et al., 2008; Clemmer et al., 2011; Bhargava et al., 2015; Subhadra et al., 2019). AbaI is the autoinducer synthase that generates the QS molecule N-(3-hydroxydodecanoyl)-L-HSL (AHL), which at high enough density interacts with the cognate receptor AbaR leading to downstream cellular responses. Several publications have found that AbaI and AbaR gene disruption leads to reduced biofilm formation (Niu et al., 2008; Anbazhagan et al., 2012; Guo and Xiang, 2017). Furthermore, cells cultured in the presence of AHL exhibited increased expression of Csu pili and stimulation of biofilm formation (Luo et al., 2015). Additionally, the activity of AbaI and biofilm production are regulated by iron in a dose-dependent manner (Modarresi et al., 2015), suggesting that iron is a possible environmental signal for nutrient limitation and the shift to survival mechanisms.
Several two component systems (TCS) have been shown to play a critical role in biofilm formation. BfmRS is predicted to contribute to the enhanced biofilm formation on abiotic surfaces since a knockout mutant of bfmS displayed drastic reduction in biofilm formation, adherence to eukaryotic cells and serum killing resistance compared to the wildtype strain (Liou et al., 2014). Furthermore, the csu operon is regulated by BfmRS, suggesting that the TCS plays an integral role in the initial adhesion step of biofilm formation (Tomaras et al., 2008; Shin et al., 2009). AdeRS is another TCS implicated in biofilm formation because an adeS deletion mutant resulted in decreased biofilm formation (Richmond et al., 2016). GacSA TCS was initially discovered for its role in citrate metabolism (Dorsey et al., 2002). However, further characterization of a gacS deletion mutant revealed its involvement in the control of pili synthesis, motility, biofilm formation, resistance against human serum, and metabolism of aromatic compounds by the paa operon (Cerqueira et al., 2014). Finally, A1S_2811 is a recently characterized hybrid sensor kinase expressed in an operon with pilGHIJ genes, suggesting a potential link to adhesion. Further, the A1S_2811 deletion mutant displayed a significant reduction in surface motility, pellicle formation and abaI protein (Chen et al., 2017), suggesting a second putative control mechanism associated with QS.
Many other signals and sensing systems have been recently implicated in biofilm formation by A. baumannii. One study showed that cyclic di-GMP may play a role in A. baumannii biofilm formation since small molecule inhibitors of diguanylate cyclase enzymes (DGC) significantly reduced biofilm density (Sambanthamoorthy et al., 2014). Furthermore, another publication identified 2 DGCs that control biofilm and pellicle formation (Ahmad et al., 2020). When these enzymes are overexpressed, it drives early poly-N-acetyl-β-(1-6)-glucosamine (PNAG) production, which is an important biofilm extracellular matrix component. Temperature influences biofilm robustness since 26°C biofilms displayed significantly increased biofilm mass compared to 30 and 37°C (Eze and El Zowalaty, 2019). Mussi et al. showed that A. baumannii senses and responds to blue light as motility and biofilm formation were only observed in cultures grown in darkness, with the responsiveness level influenced by temperature (Mussi et al., 2010). The predicted photoreceptor protein is conserved in other A. baumannii strains, suggesting that light sensing is a potential widespread cue in Acinetobacter species. Deletion of A1S_0114 displayed an increase in csuAB expression as well as a decrease in other pilin proteins and ompA (Rumbo-Feal et al., 2017). Further, this mutant was unable to form complex 3D biofilm structures on abiotic surfaces and reduced airway epithelial adhesion. Recently, a Zur-regulated lipoprotein ZrlA was described to be involved in biofilm formation and motility through BfmRS signaling and subsequent control of csu expression (E.K. Lee et al., 2020).
Transcriptomic and Proteomic Changes
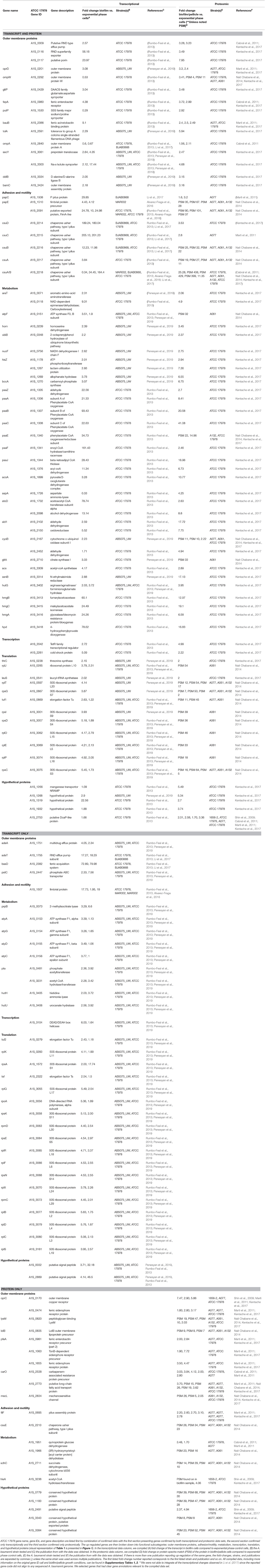
Several studies have compared the transcriptomic and proteomic profiles of A. baumannii grown in various growth conditions, including exponential, late stationary, pellicle and biofilm states, to elucidate the functional and metabolic differences between various bacterial lifestyles (Shin et al., 2009; Cabral et al., 2011; Marti et al., 2011; Chopra et al., 2013; Rumbo-Feal et al., 2013; Han et al., 2014; Kentache et al., 2017; Li et al., 2017; Penesyan et al., 2019). To gain more insight into the differential cellular response associated with biofilms, we compiled transcriptional and proteomic data reported from 9 publications, focusing specifically on up-regulated genes in biofilm/pellicle states compared to exponential growth (Table 1, Supplementary Tables 1, 2). Our efforts evaluated a total of 854 reported up-regulated genes (473 up-regulated transcripts and 381 up-regulated proteins) across 7 different A. baumannii strains (ATCC 17978, AB5075_UW, A077, A061, A132, 1656-2, BJAB0868), 3 of which were isolated from urinary sources (A077, A061, A132). Overall, we found 132 up-regulated genes to be corroborated between independent strains and/or separate publications (Table 1). Seventy-six genes were confirmed by both transcriptional and proteomic data with 43 of those genes validated across at least 2 different A. baumannii strains. Further, 35 and 21 genes were verified by at least two independent collections of transcriptional data and proteomic data, respectively. We further broke down these 132 up-regulated biofilm genes into basic biological function categories: Outer membrane proteins, Attachment/Motility, Metabolism, Transcription, Translation, and Hypothetical proteins (Table 1 and Figure 1). The largest represented categories were metabolism (49 genes), translation (30 genes), and outer membrane proteins (29 genes). This suggests that the transition and maintenance of the biofilm state involves significant changes to metabolic processes and outer membrane composition supported by translational machinery required to produce nascent proteins.
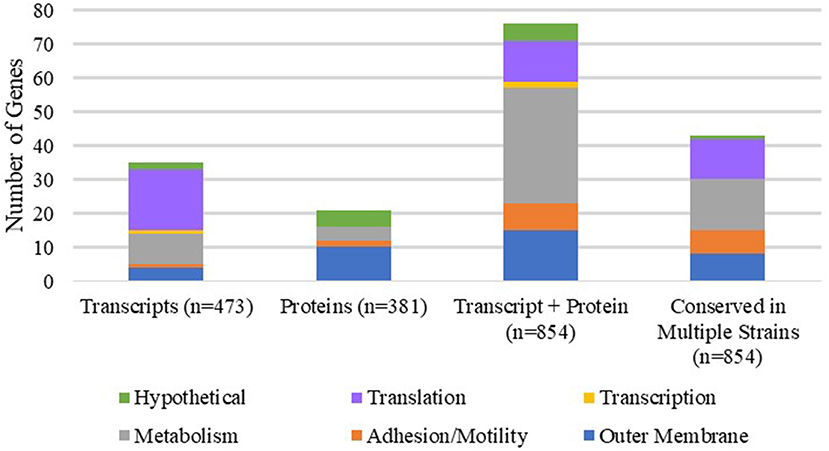
Figure 1. Functional categorization of corroborated genes up-regulated in biofilms. Each stacked bar represents the number of corroborated genes confirmed by transcriptional, proteomic, transcriptional and proteomic data sets (as listed in Table 1). The last stacked bar represents the number of genes confirmed in at least two different strains of A. baumannii regardless of data set. The total number of genes compared in each data set are listed after the bar title (i.e., 473 up-regulated transcripts compiled and compared, 381 up-regulated proteins compiled and compared, and so on). Each stacked bar is further broken down into functional subcategories: outer membrane proteins (blue), adhesion and motility (orange), metabolism (gray), transcription (yellow), translation (purple), and hypothetical proteins (green).
The most highly upregulated genes observed in biofilm associated cells were the csu operon (csuABCDE) exhibiting overexpression levels ranging from 11- to 205-fold increase over exponential phase cells. The other attachment/motility genes identified to be upregulated include pili genes filF, fimA, and papCE. In the metabolic category, we observed the significant upregulation of the phenylacetate degradation operon (paaZABCEFJ), which has been linked to neutrophil evasion and regulation by the GacS/GacA TCS (Cerqueira et al., 2014). Of the 30 translational genes up-regulated, 24 are components composing the small and large subunits of the ribosome, suggesting an overall increase in translational capacity within biofilm cells. In the outer membrane protein category, we observe significant increases in RND efflux pump proteins and iron acquisition systems, which are important for intrinsic antibiotic resistance and nutrient procurement.
Though not confirmed in our gene list, the pgaABCD operon encodes the enzymes that produce PNAG, an important structural component for biofilm formation (A.H. Choi et al., 2009). Further, it has been shown that expression of pgaB is positively correlated with biofilm formation capacity in clinical isolates from burn wound infections in Iran (Amin et al., 2019). It is clear that the A. baumannii growth state results in different transcriptional, proteomic, and metabolic profiles, which account for variable cellular responses.
Recent Developments
As mentioned previously, A. baumannii has only just begun to be recognized as an important uropathogen. A recent study discovered that a large conjugative plasmid (pAB5) in the MDR A. baumannii urinary isolate UPAB1 increases virulence in a first-of-its-kind CAUTI murine model of infection (Di Venanzio et al., 2019). Furthermore, UPAB1 grew better than ATCC 19606 in pooled human urine in vitro and co-localized with fibrinogen similar to previous observations in common UTI pathogens such as E. faecalis and MRSA (Walker et al., 2017; Xu et al., 2017). To identify adhesins involved in colonization of the bladder, Di Venanzio et al. identified two loci encoding putative CUP pili (CUP1 and CUP2). Deletion of these operons revealed loss of distinct surface appendages observed in the wildtype control and reduction in bacterial burden both on the catheter implant and within the bladder. Further, loss of pAB5 resulted in significantly reduced bacterial burden on the implant and within the bladder; however, the presence of pAB5 attenuated virulence and dissemination to other organs in an acute pneumonia murine model, which led the researchers to conclude that pAB5 confers niche specificity. To identify potential virulence factors differentially regulated by pAB5, researchers utilized proteomic and transcriptional approaches. Overall, their data indicated that pAB5 repressed type VI secretion system and differential regulation of PNAG biosynthesis and CUP1/2 pili are influenced by growth condition; thus, indicating that plasmid-encoded genes may influence biofilm formation and uropathogenesis by modulating the expression of chromosomal genes.
Another recent publication supports the hypothesis of niche-specific plasmid acquisition. They found distinct genome expansions in strains isolated from the similar sites of infections whereas strains isolated from another site of infection maintained different plasmids (Yakkala et al., 2019).
Given the wide-ranging phenotypic changes observed during the transition from planktonic to biofilm growth, it is likely that there are many levels of regulation involved in coordinating the cellular response. In recent years, the role of small RNAs (sRNA) in transcriptional regulation networks have been increasingly recognized. To this end, Alvarez-Fraga et al. compared the expression of sRNAs in ATCC 17978 biofilm cells and found 60 sRNAs were differentially regulated compared to planktonic cells (Alvarez-Fraga et al., 2017). Additionally, they were able to show that sRNA 13573 is involved in the biofilm formation and attachment to eukaryotic cells, suggesting that biofilm biogenesis and adhesion properties in ATCC 17978 are coordinately regulated. Interestingly, another group found a distinct set of differentially expressed sRNAs in A. baumannii strain MTCC1425 compared to ATCC 17978, suggesting that the sRNAs involved in transcriptional control display some strain specificity (Sharma et al., 2014).
Mangas et al. compared nearly 2000 A. baumannii genomes. They observed that strains carrying CRISPR systems were enriched for biofilm-associated genes (>70 vs. <2% non-CRISPR strains), suggesting a link between CRISPR immunity and biofilm formation (Mangas et al., 2019). Previous research has shown that Cas3 endonuclease is involved in the control of biofilm formation in both gram-positive and gram-negative bacteria (Tang et al., 2019; Cui et al., 2020).
Perspectives
While investigations into the mechanisms behind A. baumannii biofilm formation and CAUTI-associated pathogenesis have expanded recently, there remains many questions left to be addressed in order to produce a fully developed model.
A general concern across all pathogenic organism studies is that in vitro assays have been important for identifying virulence factors responsible for pathogenesis. However, studies within animal models of these putative virulence factors have often lacked direct correlation with in vivo outcomes, including in A. baumannii studies (Wand et al., 2012; Giannouli et al., 2013; Zimbler et al., 2013; Beceiro et al., 2014; Lazaro-Diez et al., 2016). These results highlight the importance of the confirmation of virulence in vivo, especially in models reflecting human infection. The first CAUTI murine infection model was recently established and requires more investigation for validation (Di Venanzio et al., 2019), but is a good first step in addressing this concern.
Another major complication that is evident across the array of A. baumannii pathogenesis publications is that some of the biological roles associated with identified virulence factors seem to be strain specific. For example, Wood et al. described and characterized a light-regulated pilus system involved in ATCC 17978 biofilm formation; however, this operon displayed no changes in expression in the hyper-biofilm producing strain MAR002 (Alvarez-Fraga et al., 2016; Wood et al., 2018). Further, Eze and El Zowalaty observed significant strain variation in biofilm formation across strains tested under differing temperatures, nutrient levels and agitation conditions (Eze and El Zowalaty, 2019). Future work should investigate conservation and incorporate several different A. baumannii lineages to strengthen the original discovery.
One observation we encountered while compiling up-regulated genes involved in biofilm cell growth is the wide variation in methods used to measure biofilm formation (Shin et al., 2009; Cabral et al., 2011; Marti et al., 2011; Rumbo-Feal et al., 2013; Nait Chabane et al., 2014; Alvarez-Fraga et al., 2016; Kentache et al., 2017; Li et al., 2017; Penesyan et al., 2019). Publications reported using different incubation times (24–144 h), incubation temperatures (25–37°C), abiotic surfaces supplied (glass, polystyrene), and growth conditions (continuous flow, stationary). While we were able to identify a large set of genes up-regulated in biofilm cells despite differential growth conditions, we are concerned that many other genes may have been missed in these studies. For example, previously reported biofilm-associated genes, bap and the pga operon, were not reported to be up-regulated in any publication examined. Moving forward, transcriptional and proteomic profiling over time during biofilm formation and maturation will provide important information into the dynamic, rapidly transitioning cellular responses within sessile cells.
Recently, a novel, phase-variable colony opacity switch has been described in AB5075 and other A. baumannii clinical isolates, in which colonies interconvert at a high-frequency between opaque and translucent variants (Tipton et al., 2015). Further characterization of the two opacity forms showed significant differences in biofilm formation, virulence and transcriptional profiles (Chin et al., 2018). However, none of the publications discussed in this review mentioned focusing a specific phase variant, which likely means their results were generated from a mixed pool of cell types. This implies that transcriptional and proteomic data may be missing important differences since the average of the mixed population may match control even though one subpopulation could have the gene significantly up-regulated and the other subpopulation has the gene significantly down. This leads us to wonder what the individual contributions that each of the phase types have in biofilm formation and maturation. It is important to note that we have not observed colony opacity variation in ATCC 17978 and therefore, this phenotypic variation may not apply to studies using this strain.
Finally, there are nearly 2500 A. baumannii genome sequences publicly available comprising a core genome of ~2,200 genes and a collapsed pan-genome size of almost 20,000 genes (Chan et al., 2015; Mangas et al., 2019), showing the broad variation across this pathogen. Furthermore, 42% of the pan-genome is of unknown function displaying our superficial knowledge of the roles these genes play in A. baumannii growth, virulence and environmental adaptability. Overall, the accumulation of decades of research has revealed many genes that are involved in the transition from planktonic growth to biofilms in A. baumannii. Only recently has this organism begun to be appreciated as a uropathogen and research into this area has commenced. Many more studies are required to fully understand how biofilm-associated genes may contribute to urinary tract infection. As we gain more insight into the underlying mechanisms of biofilm formation and uropathogenesis, this work will lay the foundation for potential anti-infective targets to combat surmounting obstacle of MDR A. baumannii.
Author Contributions
JC conceived and wrote the majority of the manuscript. PR contributed to manuscript revision. Both authors read and approved the submitted version.
Funding
This work was supported by funding from the Department of Veterans Affairs I01 BX001725, IK6BX004470 and NIH R21AI142489 to PR.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge the other members of the Rather Lab, Drs. Sarah Anderson and Maria Pérez-Varela, and Aimee Tierney, for their insightful discussions while conceiving and writing this review.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00253/full#supplementary-material
References
Abdi-Ali, A., Hendiani, S., Mohammadi, P., and Gharavi, S. (2014). Assessment of biofilm formation and resistance to imipenem and ciprofloxacin among clinical isolates of Acinetobacter baumannii in Tehran. Jundishapur J. Microbiol. 7:e8606. doi: 10.5812/jjm.8606
Ahmad, I., Nygren, E., Khalid, F., Myint, S. L., and Uhlin, B. E. (2020). A cyclic-di-GMP signalling network regulates biofilm formation and surface associated motility of Acinetobacter baumannii 17978. Sci. Rep. 10:1991. doi: 10.1038/s41598-020-58522-5
Alvarez-Fraga, L., Perez, A., Rumbo-Feal, S., Merino, M., Vallejo, J. A., Ohneck, E. J., et al. (2016). Analysis of the role of the LH92_11085 gene of a biofilm hyper-producing Acinetobacter baumannii strain on biofilm formation and attachment to eukaryotic cells. Virulence 7, 443–455. doi: 10.1080/21505594.2016.1145335
Alvarez-Fraga, L., Rumbo-Feal, S., Perez, A., Gomez, M. J., Gayoso, C., Vallejo, J. A., et al. (2017). Global assessment of small RNAs reveals a non-coding transcript involved in biofilm formation and attachment in Acinetobacter baumannii ATCC 17978. PLoS ONE 12:e0182084. doi: 10.1371/journal.pone.0182084
Amin, M., Navidifar, T., Shooshtari, F. S., Rashno, M., Savari, M., Jahangirmehr, F., et al. (2019). Association between biofilm formation, structure, and the expression levels of genes related to biofilm formation and biofilm-specific resistance of Acinetobacter baumannii strains isolated from burn infection in Ahvaz, Iran. Infect. Drug Resist. 12, 3867–3881. doi: 10.2147/IDR.S228981
Anbazhagan, D., Mansor, M., Yan, G. O., Md Yusof, M. Y., Hassan, H., and Sekaran, S. D. (2012). Detection of quorum sensing signal molecules and identification of an autoinducer synthase gene among biofilm forming clinical isolates of Acinetobacter spp. PLoS ONE 7:e36696. doi: 10.1371/journal.pone.0036696
Armbruster, C. R., and Parsek, M. R. (2018). New insight into the early stages of biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 115, 4317–4319. doi: 10.1073/pnas.1804084115
Azizi, O., Shahcheraghi, F., Salimizand, H., Modarresi, F., Shakibaie, M. R., Sh Mansouri, Ramazanzadeh, R., et al. (2016). Molecular analysis and expression of bap gene in biofilm-forming multi-drug-resistant Acinetobacter baumannii. Rep. Biochem. Mol. Biol. 5, 62–72.
Badmasti, F., Siadat, S. D., Bouzari, S., Ajdary, S., and Shahcheraghi, F. (2015). Molecular detection of genes related to biofilm formation in multidrug-resistant Acinetobacter baumannii isolated from clinical settings. J. Med. Microbiol. 64 (Pt 5), 559–564. doi: 10.1099/jmm.0.000058
Beceiro, A., Moreno, A., Fernandez, N., Vallejo, J. A., Aranda, J., Adler, B., et al. (2014). Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 58, 518–526. doi: 10.1128/AAC.01597-13
Bentancor, L. V., Routray, A., Bozkurt-Guzel, C., Camacho-Peiro, A., Pier, G. B., and Maira-Litran, T. (2012). Evaluation of the trimeric autotransporter Ata as a vaccine candidate against Acinetobacter baumannii infections. Infect. Immun. 80, 3381–3388. doi: 10.1128/IAI.06096-11
Bhargava, N., Singh, S. P., Sharma, A., Sharma, P., and Capalash, N. (2015). Attenuation of quorum sensing-mediated virulence of Acinetobacter baumannii by Glycyrrhiza glabra flavonoids. Future Microbiol. 10, 1953–1968. doi: 10.2217/fmb.15.107
Brossard, K. A., and Campagnari, A. A. (2012). The Acinetobacter baumannii biofilm-associated protein plays a role in adherence to human epithelial cells. Infect. Immun. 80, 228–233. doi: 10.1128/IAI.05913-11
Cabral, M. P., Soares, N. C., Aranda, J., Parreira, J. R., Rumbo, C., Poza, M., et al. (2011). Proteomic and functional analyses reveal a unique lifestyle for Acinetobacter baumannii biofilms and a key role for histidine metabolism. J. Proteome Res. 10, 3399–3417. doi: 10.1021/pr101299j
CDC (2019). Antibiotic Resistance Threats in the United States Atlanta, GA: U.S. Department of Health and Human Services, CDC.
Cerqueira, G. M., Kostoulias, X., Khoo, C., Aibinu, I., Qu, Y., Traven, A., et al. (2014). A global virulence regulator in Acinetobacter baumannii and its control of the phenylacetic acid catabolic pathway. J. Infect. Dis. 210, 46–55. doi: 10.1093/infdis/jiu024
Chan, A. P., Sutton, G., DePew, J., Krishnakumar, R., Choi, Y., Huang, X. Z., et al. (2015). A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol. 16:143. doi: 10.1186/s13059-015-0701-6
Chen, R., Lv, R., Xiao, L., Wang, M., Du, Z., Tan, Y., et al. (2017). A1S_2811, a CheA/Y-like hybrid two-component regulator from Acinetobacter baumannii ATCC17978, is involved in surface motility and biofilm formation in this bacterium. Microbiologyopen 6 :e00510. doi: 10.1002/mbo3.510
Chin, C. Y., Tipton, K. A., Farokhyfar, M., Burd, E. M., Weiss, D. S., and Rather, P. N. (2018). A high-frequency phenotypic switch links bacterial virulence and environmental survival in Acinetobacter baumannii. Nat. Microbiol. 3, 563–569. doi: 10.1038/s41564-018-0151-5
Choi, A. H., Slamti, L., Avci, F. Y., Pier, G. B., and Maira-Litran, T. (2009). The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-beta-1-6-N-acetylglucosamine, which is critical for biofilm formation. J. Bacteriol. 191, 5953–5963. doi: 10.1128/JB.00647-09
Choi, C. H., Lee, J. S., Lee, Y. C., Park, T. I., and Lee, J. C. (2008). Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol. 8:216. doi: 10.1186/1471-2180-8-216
Chopra, S., Ramkissoon, K., and Anderson, D. C. (2013). A systematic quantitative proteomic examination of multidrug resistance in Acinetobacter baumannii. J. Proteomics 84, 17–39. doi: 10.1016/j.jprot.2013.03.008
Clemmer, K. M., Bonomo, R. A., and Rather, P. N. (2011). Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157(Pt 9), 2534–2544. doi: 10.1099/mic.0.049791-0
Cui, L., Wang, X., Huang, D., Zhao, Y., Feng, J., Lu, Q., et al. (2020). CRISPR-cas3 of salmonella upregulates bacterial biofilm formation and virulence to host cells by targeting quorum-sensing systems. Pathogens 9:53. doi: 10.3390/pathogens9010053
de Breij, A., Gaddy, J. J., van der Meer, Koning, R., Koster, A. P., van den Broek, Actis, L., Nibbering, P., et al. (2009). CsuA/BABCDE-dependent pili are not involved in the adherence of Acinetobacter baumannii ATCC19606(T) to human airway epithelial cells and their inflammatory response. Res. Microbiol. 160, 213–218. doi: 10.1016/j.resmic.2009.01.002
De Gregorio, E., Del Franco, M., Martinucci, M., Roscetto, E., Zarrilli, R., Di Nocera, P. P., et al. (2015). Biofilm-associated proteins: news from Acinetobacter. BMC Genomics 16:933. doi: 10.1186/s12864-015-2136-6
Di Venanzio, G., Flores-Mireles, A. L., Calix, J. J., Haurat, M. F., Scott, N. E., Palmer, L. D., Potter, R. F., et al. (2019). Urinary tract colonization is enhanced by a plasmid that regulates uropathogenic Acinetobacter baumannii chromosomal genes. Nat. Commun. 10:2763. doi: 10.1038/s41467-019-10706-y
Dorsey, C. W., Tomaras, A. P., and Actis, L. A. (2002). Genetic and phenotypic analysis of Acinetobacter baumannii insertion derivatives generated with a transposome system. Appl. Environ. Microbiol. 68, 6353–6360. doi: 10.1128/AEM.68.12.6353-6360.2002
Eijkelkamp, B. A., Stroeher, U. H., Hassan, K. A., Paulsen, I. T., and Brown, M. H. (2014). Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genomics 15:1020. doi: 10.1186/1471-2164-15-1020
Eze, E. C., and El Zowalaty, M. E. (2019). Combined effects of low incubation temperature, minimal growth medium, and low hydrodynamics optimize Acinetobacter baumannii biofilm formation. Infect. Drug Resist. 12, 3523–3536. doi: 10.2147/IDR.S203919
Gaddy, J. A., and Actis, L. A. (2009). Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol. 4, 273–278. doi: 10.2217/fmb.09.5
Gaddy, J. A., Tomaras, A. P., and Actis, L. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77, 3150–3160. doi: 10.1128/IAI.00096-09
Giammanco, A., Cala, C., Fasciana, T., and Dowzicky, M. J. (2017). Global assessment of the activity of tigecycline against multidrug-resistant gram-negative pathogens between 2004 and 2014 as part of the tigecycline evaluation and surveillance trial. mSphere 2:e00310–16. doi: 10.1128/mSphere.00310-16
Giannouli, M., Antunes, L. C., Marchetti, V., Triassi, M., Visca, P., and Zarrilli, R. (2013). Virulence-related traits of epidemic Acinetobacter baumannii strains belonging to the international clonal lineages I-III and to the emerging genotypes ST25 and ST78. BMC Infect. Dis. 13:282. doi: 10.1186/1471-2334-13-282
Greene, C., Wu, J., Rickard, A. H., and Xi, C. (2016). Evaluation of the ability of Acinetobacter baumannii to form biofilms on six different biomedical relevant surfaces. Lett. Appl. Microbiol. 63, 233–239. doi: 10.1111/lam.12627
Guo, H. N., and Xiang, J. (2017). Influences of abaR gene on biofilm formation of Acinetobacter baumannii. Zhonghua Shao Shang Za Zhi 33, 200–205. doi: 10.3760/cma.j.issn.1009-2587.2017.04.003
Han, X., Li, Q., Shen, L., Hu, D., and Qu, Y. (2014). Correlation between the biofilm-forming ability, biofilm-related genes and antimicrobial resistance of Acinetobacter baumannii. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue 26, 639–643. doi: 10.3760/cma.j.issn.2095-4352.2014.09.007
Harding, C. M., Hennon, S. W., and Feldman, M. F. (2018). Uncovering the mechanisms of Acinetobacter baumannii virulence. Nat. Rev. Microbiol. 16, 91–102. doi: 10.1038/nrmicro.2017.148
Kentache, T., Ben Abdelkrim, A., Jouenne, T., De, E., and Hardouin, J. (2017). Global dynamic proteome study of a pellicle-forming Acinetobacter baumannii strain. Mol. Cell Proteomics 16, 100–112. doi: 10.1074/mcp.M116.061044
Lazaro-Diez, M., Navascues-Lejarza, T., Remuzgo-Martinez, S., Navas, J., Icardo, J. M., Acosta, F., et al. (2016). Acinetobacter baumannii and A. pittii clinical isolates lack adherence and cytotoxicity to lung epithelial cells in vitro. Microbes Infect. 18, 559–564. doi: 10.1016/j.micinf.2016.05.002
Lee, E. K., Choi, C. H., and Oh, M. H. (2020). Zur-regulated lipoprotein A contributes to the fitness of Acinetobacter baumannii. J. Microbiol. 58, 67–77. doi: 10.1007/s12275-020-9531-7
Lee, H. W., Koh, Y. M., Kim, J., Lee, J. C., Lee, Y. C., Seol, S. Y., et al. (2008). Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 14, 49–54. doi: 10.1111/j.1469-0691.2007.01842.x
Li, S., Li, H., Qi, T., Yan, X., Wang, B., Guan, J., et al. (2017). Comparative transcriptomics analyses of the different growth states of multidrug-resistant Acinetobacter baumannii. Biomed. Pharmacother. 85, 564–574. doi: 10.1016/j.biopha.2016.11.065
Liou, M. L., Soo, P. C., Ling, S. R., Kuo, H. Y., Tang, C. Y., and Chang, K. C. (2014). The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. J. Microbiol. Immunol. Infect. 47, 275–281. doi: 10.1016/j.jmii.2012.12.004
Lob, S. H., Hoban, D. J., Sahm, D. F., and Badal, R. E. (2016). Regional differences and trends in antimicrobial susceptibility of Acinetobacter baumannii. Int. J. Antimicrob. Agents 47, 317–323. doi: 10.1016/j.ijantimicag.2016.01.015
Loehfelm, T. W., Luke, N. R., and Campagnari, A. A. (2008). Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 190, 1036–1044. doi: 10.1128/JB.01416-07
Longo, F., Vuotto, C., and Donelli, G. (2014). Biofilm formation in Acinetobacter baumannii. New Microbiol. 37, 119–27.
Luo, L. M., Wu, L. J., Xiao, Y. L., Zhao, D., Chen, Z. X., Kang, M., et al. (2015). Enhancing pili assembly and biofilm formation in Acinetobacter baumannii ATCC19606 using non-native acyl-homoserine lactones. BMC Microbiol. 15:62. doi: 10.1186/s12866-015-0397-5
Magill, S. S., Edwards, J. R., Beldavs, Z. G., Dumyati, G., Janelle, S. J., Kainer, M. A., et al. (2014). Prevalence of antimicrobial use in US acute care hospitals, May-September 2011. JAMA 312, 1438–1446. doi: 10.1001/jama.2014.12923
Mangas, E. L., Rubio, A., Alvarez-Marin, R., Labrador-Herrera, G., Pachon, J., Pachon-Ibanez, M. E., et al. (2019). Pangenome of Acinetobacter baumannii uncovers two groups of genomes, one of them with genes involved in CRISPR/Cas defence systems associated with the absence of plasmids and exclusive genes for biofilm formation. Microb. Genom. 5:e000309. doi: 10.1099/mgen.0.000309
Marti, S., Nait Chabane, Y., Alexandre, S., Coquet, L., Vila, J., Jouenne, T., et al. (2011). Growth of Acinetobacter baumannii in pellicle enhanced the expression of potential virulence factors. PLoS ONE 6:e26030. doi: 10.1371/journal.pone.0026030
Modarresi, F., Azizi, O., Shakibaie, M. R., Motamedifar, M., Mosadegh, E., and Mansouri, S. (2015). Iron limitation enhances acyl homoserine lactone (AHL) production and biofilm formation in clinical isolates of Acinetobacter baumannii. Virulence 6, 152–161. doi: 10.1080/21505594.2014.1003001
Mussi, M. A., Gaddy, J. A., Cabruja, M., Arivett, B. A., Viale, A. M., Rasia, R., et al. (2010). The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J. Bacteriol. 192, 6336–6345. doi: 10.1128/JB.00917-10
Nait Chabane, Y., Marti, S., Rihouey, C., Alexandre, S., Hardouin, J., Lesouhaitier, O., et al. (2014). Characterisation of pellicles formed by Acinetobacter baumannii at the air-liquid interface. PLoS ONE 9:e111660. doi: 10.1371/journal.pone.0111660
Niu, C., Clemmer, K. M., Bonomo, R. A., and Rather, P. N. (2008). Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J. Bacteriol. 190, 3386–3392. doi: 10.1128/JB.01929-07
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/CMR.00058-07
Penesyan, A., Nagy, S. S., Kjelleberg, S., Gillings, M. R., and Paulsen, I. T. (2019). Rapid microevolution of biofilm cells in response to antibiotics. NPJ Biofilms Microbiomes 5:34. doi: 10.1038/s41522-019-0108-3
Petrova, O. E., and Sauer, K. (2012). Sticky situations: key components that control bacterial surface attachment. J. Bacteriol. 194, 2413–2425. doi: 10.1128/JB.00003-12
Rao, R. S., Karthika, R. U., Singh, S. P., Shashikala, P., Kanungo, R., Jayachandran, S., et al. (2008). Correlation between biofilm production and multiple drug resistance in imipenem resistant clinical isolates of Acinetobacter baumannii. Indian J. Med. Microbiol. 26, 333–337. doi: 10.4103/0255-0857.43566
Richmond, G. E., Evans, L. P., Anderson, M. J., Wand, M. E., Bonney, L. C., Ivens, A., et al. (2016). The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. mBio 7, e00430–16. doi: 10.1128/mBio.00430-16
Ronish, L. A., Lillehoj, E., Fields, J. K., Sundberg, E. J., and Piepenbrink, K. H. (2019). The structure of PilA from Acinetobacter baumannii AB5075 suggests a mechanism for functional specialization in Acinetobacter type IV pili. J. Biol. Chem. 294, 218–230. doi: 10.1074/jbc.RA118.005814
Rumbo-Feal, S., Gomez, M. J., Gayoso, C., Alvarez-Fraga, L., Cabral, M. P., Aransay, A. M., et al. (2013). Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS ONE 8:e72968. doi: 10.1371/journal.pone.0072968
Rumbo-Feal, S., Perez, A., Ramelot, T. A., Alvarez-Fraga, L., Vallejo, J. A., Beceiro, A., et al. (2017). Contribution of the A. baumannii A1S_0114 gene to the interaction with eukaryotic cells and virulence. Front. Cell Infect. Microbiol. 7:108. doi: 10.3389/fcimb.2017.00108
Sambanthamoorthy, K., Luo, C., Pattabiraman, N., Feng, X., Koestler, B., Waters, C. M., et al. (2014). Identification of small molecules inhibiting diguanylate cyclases to control bacterial biofilm development. Biofouling 30, 17–28. doi: 10.1080/08927014.2013.832224
Schweppe, D. K., Harding, C., Chavez, J. D., Wu, X., Ramage, E., Singh, P. K., et al. (2015). Host-microbe protein interactions during bacterial infection. Chem. Biol. 22, 1521–1530. doi: 10.1016/j.chembiol.2015.09.015
Sechi, L. A., Karadenizli, A., Deriu, A., Zanetti, S., Kolayli, F., Balikci, E., et al. (2004). PER-1 type beta-lactamase production in Acinetobacter baumannii is related to cell adhesion. Med. Sci. Monit. 10, BR180–4.
Sharma, R., Arya, S., Patil, S. D., Sharma, A., Jain, P. K., Navani, N. K., et al. (2014). Identification of novel regulatory small RNAs in Acinetobacter baumannii. PLoS ONE 9:e93833. doi: 10.1371/journal.pone.0093833
Shin, J. H., Lee, H. W., Kim, S. M., and Kim, J. (2009). Proteomic analysis of Acinetobacter baumannii in biofilm and planktonic growth mode. J. Microbiol. 47, 728–735. doi: 10.1007/s12275-009-0158-y
Sievert, D. M., Ricks, P., Edwards, J. R., Schneider, A., Patel, J., Srinivasan, A., et al. (2013). Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National healthcare safety network at the centers for disease control and prevention, 2009-2010. Infect. Control Hosp. Epidemiol. 34, 1–14. doi: 10.1086/668770
Skerniskyte, J., Karazijaite, E., Deschamps, J., Krasauskas, R., Armalyte, J., Briandet, R., et al. (2019). Blp1 protein shows virulence-associated features and elicits protective immunity to Acinetobacter baumannii infection. BMC Microbiol. 19:259. doi: 10.1186/s12866-019-1615-3
Stickler, D. J. (2008). Bacterial biofilms in patients with indwelling urinary catheters. Nat. Clin. Pract. Urol. 5, 598–608. doi: 10.1038/ncpuro1231
Subhadra, B., Surendran, S., Lim, B. R., Yim, J. S., Kim, D. H., Woo, K., et al. (2019). Complete genome sequence and phylogenetic analysis of nosocomial pathogen Acinetobacter nosocomialis strain NCTC 8102. Genes Genomics 41, 1063–1075. doi: 10.1007/s13258-019-00834-6
Tang, B., Gong, T., Zhou, X., Lu, M., Zeng, J., Peng, X., et al. (2019). Deletion of cas3 gene in Streptococcus mutans affects biofilm formation and increases fluoride sensitivity. Arch. Oral Biol. 99, 190–197. doi: 10.1016/j.archoralbio.2019.01.016
Thummeepak, R., Kongthai, P., Leungtongkam, U., and Sitthisak, S. (2016). Distribution of virulence genes involved in biofilm formation in multi-drug resistant Acinetobacter baumannii clinical isolates. Int. Microbiol. 19, 121–129. doi: 10.2436/20.1501.01.270
Tipton, K. A., Dimitrova, D., and Rather, P. N. (2015). Phase-variable control of multiple phenotypes in Acinetobacter baumannii strain AB5075. J. Bacteriol. 197, 2593–2599. doi: 10.1128/JB.00188-15
Tomaras, A. P., Dorsey, C. W., Edelmann, R. E., and Actis, L. A. (2003). Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149(Pt 12), 3473–3484. doi: 10.1099/mic.0.26541-0
Tomaras, A. P., Flagler, M. J., Dorsey, C. W., Gaddy, J. A., and Actis, L. A. (2008). Characterization of a two-component regulatory system from Acinetobacter baumannii that controls biofilm formation and cellular morphology. Microbiology 154(Pt 11), 3398–3409. doi: 10.1099/mic.0.2008/019471-0
Walker, J. N., Flores-Mireles, A. L., Pinkner, C. L., th Schreiber, H. L., Joens, M. S., Park, A. M., et al. (2017). Catheterization alters bladder ecology to potentiate Staphylococcus aureus infection of the urinary tract. Proc. Natl. Acad. Sci. U.S.A. 114, E8721–E8730. doi: 10.1073/pnas.1707572114
Wand, M. E., Bock, L. J., Turton, J. F., Nugent, P. G., and Sutton, J. M. (2012). Acinetobacter baumannii virulence is enhanced in Galleria mellonella following biofilm adaptation. J. Med. Microbiol. 61(Pt 4), 470–477. doi: 10.1099/jmm.0.037523-0
Weidensdorfer, M., Ishikawa, M., Hori, K., Linke, D., Djahanschiri, B., Iruegas, R., et al. (2019). The Acinetobacter trimeric autotransporter adhesin Ata controls key virulence traits of Acinetobacter baumannii. Virulence 10, 68–81. doi: 10.1080/21505594.2018.1558693
WHO (2017) Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2016–2017. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO.
Wood, C. R., Ohneck, E. J., Edelmann, R. E., and Actis, L. A. (2018). A light-regulated type I pilus contributes to Acinetobacter baumannii biofilm, motility, and virulence functions. Infect. Immun. 86:e00442–18. doi: 10.1128/IAI.00442-18
Wullt, B., Bergsten, G., Connell, H., Rollano, P., Gebretsadik, N., Hull, R., et al. (2000). P fimbriae enhance the early establishment of Escherichia coli in the human urinary tract. Mol. Microbiol. 38, 456–464. doi: 10.1046/j.1365-2958.2000.02165.x
Xu, W., Flores-Mireles, A. L., Cusumano, Z. T., Takagi, E., Hultgren, S. J., and Caparon, M. G. (2017). Host and bacterial proteases influence biofilm formation and virulence in a murine model of enterococcal catheter-associated urinary tract infection. NPJ Biofilms Microbiomes 3:28. doi: 10.1038/s41522-017-0036-z
Yakkala, H., Samantarrai, D., Gribskov, M., and Siddavattam, D. (2019). Comparative genome analysis reveals niche-specific genome expansion in Acinetobacter baumannii strains. PLoS ONE 14:e0218204. doi: 10.1371/journal.pone.0218204
Zarb, P., Coignard, B., Griskeviciene, J., Muller, A., Vankerckhoven, V., Weist, K., et al. (2012). The European Centre for Disease Prevention and Control (ECDC) pilot point prevalence survey of healthcare-associated infections and antimicrobial use. Euro. Surveill. 17:20316. doi: 10.2807/ese.17.46.20316-en
Zeighami, H., Valadkhani, F., Shapouri, R., Samadi, E., and Haghi, F. (2019). Virulence characteristics of multidrug resistant biofilm forming Acinetobacter baumannii isolated from intensive care unit patients. BMC Infect. Dis. 19:629. doi: 10.1186/s12879-019-4272-0
Keywords: Acinetobacter baumannii, bacterial biofilm, uropathogen, CAUTI, virulence, environmental sensing, gene expression
Citation: Colquhoun JM and Rather PN (2020) Insights Into Mechanisms of Biofilm Formation in Acinetobacter baumannii and Implications for Uropathogenesis. Front. Cell. Infect. Microbiol. 10:253. doi: 10.3389/fcimb.2020.00253
Received: 22 March 2020; Accepted: 30 April 2020;
Published: 29 May 2020.
Edited by:
Paola Scavone, Instituto de Investigaciones Biológicas Clemente Estable (IIBCE), UruguayReviewed by:
Mario Feldman, Washington University in St. Louis, United StatesMaría Dolores Alcántar-Curiel, National Autonomous University of Mexico, Mexico
Copyright © 2020 Colquhoun and Rather. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Philip N. Rather, cHJhdGhlckBlbW9yeS5lZHU=
 Jennifer M. Colquhoun
Jennifer M. Colquhoun Philip N. Rather
Philip N. Rather