
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 20 May 2020
Sec. Clinical Microbiology
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00228
This article is part of the Research Topic Bridging the Gap between Discovery and Clinical Development for Antimicrobial Peptides View all 8 articles
Host defense peptides (HDPs), also known as antimicrobial peptides, are naturally occurring polypeptides (~12–50 residues) composed of cationic and hydrophobic amino acids that adopt an amphipathic conformation upon folding usually after contact with membranes. HDPs have a variety of biological activities including immunomodulatory, anti-inflammatory, anti-bacterial, and anti-biofilm functions. Although HDPs have the potential to address the global threat of antibiotic resistance and to treat immune and inflammatory disorders, they have yet to achieve this promise. Indeed, there are several challenges associated with bringing peptide-based drug candidates from the lab bench to clinical practice, including identifying appropriate indications, stability, toxicity, and cost. These challenges can be addressed in part by the development of innate defense regulator (IDR) peptides and peptidomimetics, which are synthetic derivatives of HDPs with similar or better efficacy, increased stability, and reduced toxicity and cost of the original HDP. However, one of the largest gaps between basic research and clinical application is the validity and translatability of conventional model systems, such as cell lines and animal models, for screening HDPs and their derivatives as potential drug therapies. Indeed, such translation has often relied on animal models, which have only limited validity. Here we discuss the recent development of human organoids for disease modeling and drug screening, assisted by the use of omics analyses. Organoids, developed from primary cells, cell lines, or human pluripotent stem cells, are three-dimensional, self-organizing structures that closely resemble their corresponding in vivo organs with regards to immune responses, tissue organization, and physiological properties; thus, organoids represent a reliable method for studying efficacy, formulation, toxicity and to some extent drug stability and pharmacodynamics. The use of patient-derived organoids enables the study of patient-specific efficacy, toxicogenomics and drug response predictions. We outline how organoids and omics data analysis can be leveraged to aid in the clinical translation of IDR peptides.
The development of antibiotics is one of the greatest advances in modern medicine. However, excessive and improper uses of antibiotics have led to the rapid increase in antimicrobial resistant (AMR) organisms. Given the dearth of new antibiotics, AMR infections have become one of the most serious global health issues. AMR organisms are associated with high morbidity and mortality, and contribute to high economic burden (Zhen et al., 2019). According to the Centers for Disease Control and Prevention (CDC), at least 2.8 million Americans are infected by AMR organisms annually, with 35,000 people succumb to the infection (www.cdc.gov/drugresistance/index.html), although this situation may in fact be far worse since ~210,000 die of sepsis annually due to the failure of the front line therapy, antibiotics. In Canada, the Council of Canadian Academies (CCA) reported that AMR organisms mediated infection have cost the health care system >$2 billion, and claimed 5,400 Canadian lives in 2018 (cca-reports.ca/antimicrobial-resistance-poses-significant-risk-to-people-the-economy/). The urgent need for the development of new therapies against AMR organisms, was indicated by the World Health Organization (WHO) publication of a global action plan containing a global priority list of 12 families of AMR organisms that pose the greatest threat to human health, to guide research and development of new antibiotics (World Health Organization, 2015, 2017). The CDC has also stated that “without urgent action now, we are on the verge of returning to a pre-antibiotic era, where common infections and injury can become fatal, and surgeries and chemotherapy will become impossible” (www.cidrap.umn.edu/news-perspective/2016/10/cdc-chief-antibiotic-resistance-scary-threat-modern-medicine). It is thus evident that there is an urgent need to develop new strategies to fight against AMR organisms.
Recent research has demonstrated that host defense peptides (HDPs) and its analogs innate defense regulator (IDR) peptides and peptidomimetics have strong potential to be developed into alternative/adjunctive therapies to fight against AMR organisms. They also address an urgent need for new ways of treating inflammation that underlies the pathology of almost every human disease. However, one of the biggest hurdles is the validity and translatability of current in vitro and animal model systems used for drug screening. In this review, we examined the use of human organoid systems focusing on skin, lung, and intestinal organoids for disease modeling and drug screening. Together with omics analyses, we will discuss the prospect of using organoid systems to aid in clinical translation of HDP research (Figure 1).
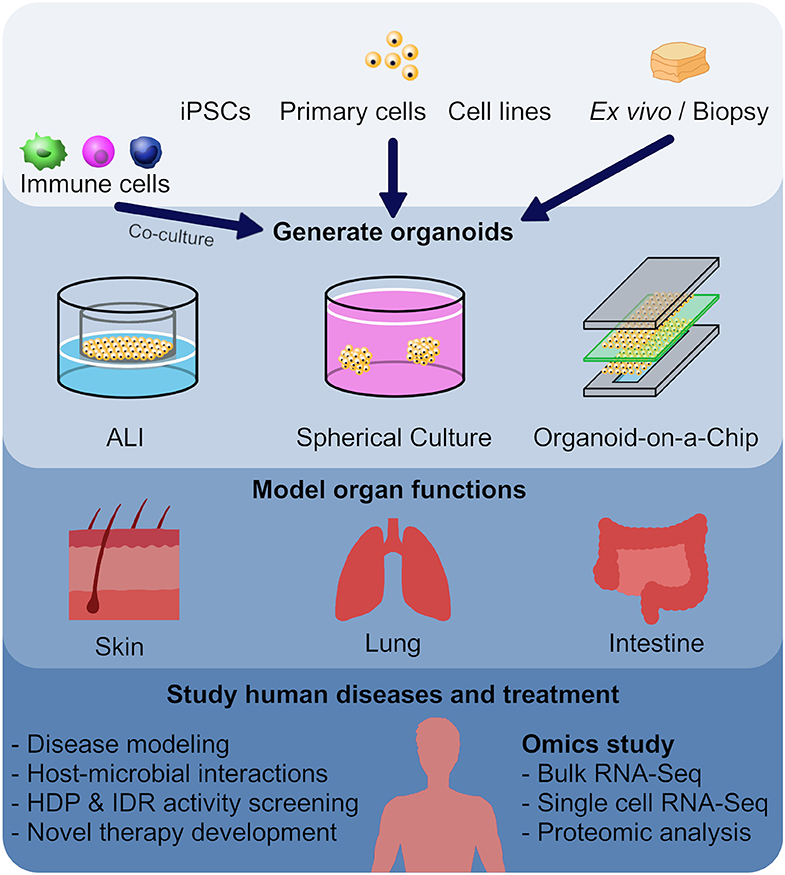
Figure 1. Utilizing organoid models as a screening method in the development of new host defense peptides. Human or animal induced pluripotent stem cells (iPSCs), embryonic stem cells, neonatal tissue stem cells, or ex vivo adult progenitors can all serve as starting materials to generate various organoids. In this review, we focused on skin, lung and intestinal organoids, which recapitulate the architecture, functions and multi-cellular components present in the tissue of origin. In general, there are three main forms of organoids: air-liquid interface (ALI) constructs, spheroids, and organ-on-a-chip models. These different forms of organoids, together with omics characterization, have provided mechanistic insights to diseases and host-microbial interactions, and provide novel tools for HDP and IDR screening.
HDPs, also known as antimicrobial peptides (AMPs), are naturally occurring cationic amphipathic polypeptides found ubiquitously in most species of life and play essential roles in providing protection against pathogens and modulating immunity (Hancock and Lehrer, 1998). To date, there are >3,000 HDPs described from the six kingdoms (animals, fungi, plants, and protists, with related molecules in bacteria and archaea): http://aps.unmc.edu/AP/main.php (Wang et al., 2016). These peptides tend to be relatively short (composed of ~12–50 amino acids), amphipathic, and have a net positive charge of +2 to +9 at physiological pH (Hancock and Sahl, 2006; Choi and Mookherjee, 2012). HDPs are an important component of the host immune system, participating in both innate and adaptive immunity (Hancock et al., 2016). They possess multifaceted biological functions in modulating host immune responses, including mediating immune cell recruitment and functions in part by regulating the production of cytokines and chemokines, suppression of inflammatory responses, enhancement of angiogenesis, and wound healing, etc. (Hancock et al., 2016). These host responses contribute to the resolution of infection and inflammation, which suggests that related synthetic IDR peptides may be excellent therapeutic candidates to treat infection and inflammatory diseases.
HDPs have broad-spectrum direct antimicrobial activities against Gram-positive and Gram-negative bacteria, viruses, fungi, and parasites (Ganz, 2003; Powers and Hancock, 2003; Straus and Hancock, 2006; De Zoysa et al., 2015). Several modes of actions had been proposed to explain antimicrobial effects of HDPs. Some of these mechanisms are directly targeting microorganisms to cause bactericidal effects, such as mediating damages to microbial cell membrane, inducing microbial DNA/RNA damages, and interacting with fungal mitochondria to cause cell lysis. While other mechanisms, such as inhibiting the synthesis of macromolecules and inhibiting enzyme activities leading to inhibition of bacterial cell growth, or mediate immune modulations of the hosts, contribute to bacteriostatic effects (Moravej et al., 2018; Haney et al., 2019; Lei et al., 2019). Many anti-biofilm HDP derivatives can target conserved stringent stress response leading to the degradation of the stringent response secondary-messengers guanosine pentaphosphate or tetraphosphate, which results in biofilm eradication and reduction in bacterial abscess formation (de la Fuente-Nunez et al., 2014; Mansour et al., 2016). These peptides can also work synergistically with conventional antibiotics (Pletzer et al., 2018). To date there are no HDP that have navigated through the clinical trial process to approval status, although peptides are clearly appropriate as medicines (Seo et al., 2012; Sachdeva et al., 2016; Mishra et al., 2017). Clear examples exist of HDP-like compounds (modified cationic amphipathic peptides) including the naturally-produced bacterial products polymyxin B, polymyxin E/colistin, and gramicidin S, that are used as over the counter medicines in triple antibiotic ointments, and for treatment of serious Gram-negative infections, wound infections, and genital ulcers, respectively (Costa et al., 2019). There are several peptides related to HDP in advanced clinical trials and most have been limited to topical application (Hancock et al., 2016; Costa et al., 2019). The reason why no HDP peptides have been approved to date is likely complex but may reflect issues such as selection of clinical targets, toxicity, low peptide stability, low bioavailability, and relatively high production costs (Seo et al., 2012; Mansour et al., 2014; Mishra et al., 2017). Some of these drawbacks have been addressed by creating synthetic analogs known as innate defense regulator (IDR) peptides and peptidomimetics using natural HDPs as templates. These synthetic analogs mimic the modes of action of the natural HDPs, but can be designed with enhanced stability, lower host toxicity, and lower cost of production (Hancock et al., 2016). Furthermore, advances in delivery systems and formulations, such as liposomes, polymeric nanoparticles, nanogels, cream, ointment, and wafers, can enhance the stability and bioavailability of HDPs (Piotrowska et al., 2017). To overcome peptide stability and high production costs, innovative chemistry and biotechnology allows the scale-up production of HDP and its analogs with lower synthetic costs (Bommarius et al., 2010; Ishida et al., 2016). For example, Vogel et al. successfully used calmodulin as a fusion partner to enhance the production of several HDP (e.g., melittin, fowlicidin-1, tritrpticin, indolicidin, lactoferrampin B, and human β-defensin 3) in Escherichia coli (Ishida et al., 2016).
With increasing interest in developing HDPs as alternative antimicrobial agents against AMR infections, there are researches addressing the potential for development of resistance to the antibacterial action of host defense peptides (Nizet, 2006; Cole and Nizet, 2016; Phoenix et al., 2016; Bechinger and Gorr, 2017). These have tended to show that for most peptides, resistance is very slow (over few 100 generations) to develop as compared to most conventional antibiotics (as little as one generation) (Cole and Nizet, 2016; Phoenix et al., 2016). Another consideration is that the direct killing effects of HDPs are drastically reduced in the presence of physiological concentrations of divalent cations and polyanions like the glycosaminoglycan heparin (Mansour et al., 2014). However, the ability to modulate host immunity to fight against infection without directly targeting the microbe gives HDPs an advantage in that this action occurs readily under physiological (tissue culture) conditions, and since it involves activation of host mechanisms does not induce bacterial resistance (Hancock and Sahl, 2006). This was exemplified by a recent study which showed that developing resistance to HDP in Staphylococcus aureus, evolved in vitro in the presence of HDPs, did not provide survival advantages to the bacteria in a host environment (El Shazely et al., 2019). In addition, there is increasing evidence that HDPs and their synthetic analogs are synergistic with conventional antibiotics, indicating that these peptides can also be use as adjunctive therapies (Rudilla et al., 2016; Wu et al., 2017). These unique characteristics have further shown that HDPs and their synthetic analogs are excellent candidates to be developed as alternative therapies against antibiotic resistant bacteria.
Currently, one of the biggest hurdles in drug development is the translation of research breakthroughs from basic science to clinical trials. In general, antimicrobial drug development often begins by identifying and screening possible drug candidates using efficacy assays with diverse bacteria (including in the context of host cells in the case of intracellular pathogens), toxicity assays with specific cells or cell lines in vitro, followed by efficacy and toxicity tests in in vivo animal models, before proceeding to clinical trials. For anti-inflammatory agents, initial screens of both efficacy and toxicity would involve specific cell lines and/or primary cells. Each of the above steps are essential, but also include certain limitations, the most important of which are in vitro assays with isolated cells do not capture the complexity of the infectious/inflammatory milieu in the body, while animal models often are incomplete mimics of human diseases. Here, we briefly discuss some of the limitations of using immortalized cell lines and primary cells for drug screening.
Utilizing immortalized cell lines to study complex biological systems have been invaluable to expand our understanding of disease biology. The advantages of cell lines lie in their consistency, reproducibility, and unlimited supply (Kaur and Dufour, 2012). However, many disadvantages also plague cell lines. The process of immortalization often freezes the cell in an immature state, such that they have to be further activated, often in a specific and/or non-physiological manner. For example, human monocytic cell lines like THP-1, UA37, and HL-60 require activation with phorbol 12-myristate-13-acetate and/or 1 α, 25-dihydroxyvitamin D3 for macrophage maturation (Daigneault et al., 2010). In the case of studying the facultative intracellular pathogen Salmonella enteric sv. typhimurium, THP-1 differentiation was largely dependent on phorbol 12-myristate-13-acetate dosage and time administered to mirror the infection rates found in human monocyte derived macrophages (Starr et al., 2018).
The major disadvantage is that immortalized cells require transformation to enable them to continuously replicate, and thus their underlying biology is by definition different from the cells from which they are derived. For example, deviations in transcriptomic and proteomic expression are observed when comparing established hepatocyte cell lines to its related primary cell (Olsavsky et al., 2007; Pan et al., 2009). In fact, important features can be lost resulting in divergent cell lines. It is also well-established that genetic differences persist between immortalized cell lines and their equivalent primary tissues which hints at a lack of environmental complexity. Other confounding features of cell lines include genetic drift over the course of serial passages, the potential for contamination by other cell types and/or microorganisms and the misidentification of cell lines (Drexler and Uphoff, 2002; Capes-Davis et al., 2010). Mycoplasma contamination in particular has incidence rates reported to be as high as 35% for immortal cell lines (Drexler and Uphoff, 2002). Awareness of these differences is thus important when establishing host influences in drug screening, and in monitoring host-pathogen interactions that can affect, hinder or alter the potential for clinical applications.
To overcome these obstacles, primary cells can be used, as many of the morphological and functional features of host tissues are conserved. Although the process of isolating and differentiating of primary cells such as monocyte-derived macrophages (MDM) are well-established, there are some limitations in using primary cells. Compared to immortalized cell lines, obtaining pure populations of primary cells on a regular basis from different donors (or even repeatedly obtaining cells from the same donor) is relatively-speaking more difficult, especially when low abundance cells or cells from patients with rare diseases are required. In the case of tissue macrophages, circulating tumor cells, and vascular epithelial cells, appreciable yields are difficult to obtain (Gudjonsson et al., 2002). Other primary cells, such as neutrophils, have relatively short half-lives that limit their use in many assays (Hirz and Dumontet, 2016). Meanwhile, high biological variability amongst donors, low self-renewal ability, and the relative difficulty of genetic manipulation also pose obstacles and can limit some experimental outcomes.
Animal models are an invaluable part of drug development and have helped in the advance of innumerable discoveries in human biology and diseases (Brom, 2002; Festing, 2004). Animal models provide a more complex system that allows us to address a variety of scientific questions, from exploration of physiological functions and systemic interactions between organs and the immune system, to development and assessment of toxicity and efficacy of vaccines or therapies (Barre-Sinoussi and Montagutelli, 2015). Despite their immense impact, the use of animal models also has certain limitations, and results obtained from animal research are not always confirmed in human studies.
In particular, there are genetic and physiological variations between species and within the same species. For example, laboratory mice and rats, two of the most common animals used in the scientific world, are often inbred and have highly homogeneous genetic backgrounds within each strain (Mouse Genome Sequencing et al., 2002; Coors et al., 2010; Smith et al., 2014). The inherent differences between each strain contribute to their relative susceptibility or resistance to both induced and spontaneously occurring diseases. For example, depending on the genetic background of the mice, some strains are completely resistant to Ebola virus, while others are highly susceptible to the infection (Rasmussen et al., 2014). Another example showed that certain strains of mice with host resistance factor SLC11A1 (NRAMP1) are more resistance to infections by intracellular pathogens, such as Mycobacterium avium subsp. Avium, and Salmonella (Cunrath and Bumann, 2019). Secondly, impacts of biological pathways in animals and humans can be different (Muzio et al., 2000; Heinz et al., 2003). For example, mice deficient in galactose-1-phosphate uridyltransferase did not show any adverse effects or mimic human clinical symptoms when placed on a high galactose diet or after injections of galactose, even though these mice mimic galactosemia, a disorder that results from enzymatic deficiency of enzymes in galactose metabolism (Leslie et al., 1996; Elsea and Lucas, 2002). Ning and colleagues suggested that the lack of toxicity in galactose-1-phosphate uridyltransferase deficient mice was due to differences in galactose transport across the blood brain barrier between humans and mice (Ning et al., 2001). Similarly, the immune systems of mice and men vary substantially (Zschaler et al., 2014). For example, the critical chemokine that attracts neutrophils in man, IL-8, does not exist in mouse. Thirdly, the toxicity or the lack of certain molecules or chemicals in animals is not always reflected in humans. Analyses done by Baily and colleagues comparing drug responses between preclinical species (dog, rat, mouse, rabbit, and non-human primates) and man showed that the absence of toxicity in animal species was not predictive of an absence of adverse drug reactions in humans (Bailey et al., 2014, 2015; Bailey and Balls, 2019). For instance, vancomycin, often used as a last resort antibiotic against multi-drug-resistant Gram-positive bacteria, can cause hypersensitivity reactions such as the Red Man Syndrome and anaphylaxis, but these are only observed in man (Sivagnanam and Deleu, 2003). In addition, some toxicity and adverse effects only appear after long-term usage. For example, Vioxx (rofecoxib), a non-steroidal anti-inflammatory drug approved in 1999 by the US Food and Drug Administration for the prevention of recurrent colorectal polyps in colorectal adenomas patients, was found to induce an increased risk of cardiovascular morbidity and mortality in patients who took the drug for more than 18 months (Juni et al., 2004). Withdrawal of the drug was announced by Merck in September 2004, after an estimated 88,000 people suffered heart attacks, and 38,000 died as the result of taking Vioxx (Juni et al., 2004).
Another major consideration is in the accuracy of animal models in predicting efficacy. Animal models can provide immense amounts of information, that is presumed to be more indicative than in vitro assays of efficacy in man, but there is no single animal model that perfectly mimics a given human disease (Hainsworth et al., 2017). For example, animal models of infection are often short term, with animals dying within 24 h from the major AMR pathogens, while human infections usually occur over much longer periods of time. Similarly, it is difficult to precisely replicate polygenic genetic diseases such as asthma, inflammatory bowel disease, etc., and long-term environmental influences in human disease are almost impossible to replicate. In addition, overall animal models are very costly, time-consuming, and labor intensive, while posing ethical concerns (Rollin, 2003; Ideland, 2009; Coors et al., 2010). The most appropriate and informative animal models must be carefully selected considering the principles of the three R's (Reduction, Replacement, and Refinement) whenever possible (Hubrecht and Carter, 2019).
In recent years, there has been an increased push for the development of alternative strategies to reduce or replace animal experiments (Adler et al., 2011; Hay et al., 2014; Swaminathan et al., 2019). To emphasize the need for alternative models to be developed, a directive was signed in the USA in 2019 to prioritize efforts to reduce animal studies by 30% by 2025, and eliminate all mammal studies by 2035 (Agency, 2019). The recent surge in the development of human pluripotent stem cell and organoid systems can address some of the limitations mentioned above and bring us a step closer to the development of more translatable models for drug discovery, transplantation, and personalized medicine.
In 2006, Takahashi and Yamanaka used a set of four transcription factors, Oct3/4, Sox2, c-Myc, and Klf4 to revert murine fibroblasts to a stem cell-like state, termed induced pluripotent stem cells (iPSC) (Takahashi and Yamanaka, 2006). With the same four transcription factors, this methodology was successfully applied to human dermal fibroblasts resulting in human iPSC (Takahashi et al., 2007). This discovery has spurred many groups to further refine the transcription factors needed to promote pluripotency in humans and murine somatic cells (Kim et al., 2008; Zhu et al., 2010; Wang L. et al., 2017). Chemical alternatives are also being interrogated to replace or enhance the stem cell-like qualities of adult mammalian somatic cells (Feng et al., 2009).
Macrophages, one of the main effectors of the immune system, have the ability to adopt different functional states depending on stimulus exposure. Originally described by Nobel Prize winner Elie Metchnikoff in 1908, macrophages were reported as phagocytic cells that engulf foreign invaders, leading him to name the process as phagocytosis (Kaufmann, 2008). Phagocytosis is a principal effector mechanism of innate immunity and is highly involved in the host's protective mechanisms against harmful pathogens. Metchnikoff's initial findings on the host defense and effector functions of macrophages have provided focus for much of the subsequent research on these cells. Monocyte functionality is diverse and encompasses features such as homeostasis, immune defense, and tissue repair (Ziegler-Heitbrock et al., 2010). The classification of macrophages is parallel to T-helper lymphocyte nomenclature with Th1 and Th2 lymphocytes, corresponding to classically activated (M1) and alternatively activated (M2) macrophages, although most authors consider that there is a broad spectrum of macrophage phenotypes spanning these two categories. The nomenclature is based on environmental stimuli, cellular phenotype, and the resulting physiology that separates macrophages into these broad categories. M1 are predominantly microbicidal macrophages with increased pro-inflammatory mediator levels, while M2 are macrophages with stunted inflammatory capacity and increased phagocytic activity and wound repair capabilities (Mosser and Edwards, 2008). Additional research has since further divided the M2 phenotype into three subsets: M2a, M2b, M2c (Tarique et al., 2015). Research from our lab has suggested that endotoxin tolerant macrophages (treated twice with endotoxin) are reprogrammed into an amnesic state that fails to recognize and respond to bacterial signatures, has characteristics similar to M2 macrophages and is predictive of the development of severe sepsis (Pena et al., 2011, 2014).
With the production of stem cell-like characteristics from adult somatic cells, many protocols have been established to differentiate these cells to specific cells of interest. Pluripotent iPSCs were generated with varying degrees of success depending on starting material and their associated conversion rates. In the case of human induced pluripotent stem cell derived macrophages (iPSDM), they have a strong genetic, functional, and transcriptomic similarity to monocyte derived macrophages (Hale et al., 2015). Furthermore, the generation of iPSDMs is scalable and therefore provides a source of unlimited genotype-specific cells that can potentially reflect the disease state from which the iPSCs are derived.
The continued characterization of myeloid development reports genetic distinctions between two cellular lineages. Each lineage originates from either the Myb-independent yolk-sac (YS) or Myb-dependent hematopoietic stem cells. The transcription factor Myb is required for YS-derived tissue resident macrophages (Schulz et al., 2012). Additional findings have reported that tissue resident macrophages like microglia and Kupffer cells are capable of self-renewal with minimal replenishment from hematopoietic stem cells (Hashimoto et al., 2013). Though there are broad similarities between iPSDMs and monocyte-derived macrophages (MDMs), iPSDMs develop in a Myb-independent manner and therefore may be more closely related to tissue resident macrophages when compared to macrophages derived from hematopoietic stem cells (Buchrieser et al., 2017).
Macrophages exhibit the features of plasticity and are famously known to respond to polarization factors that cause them to differentiate and exhibit a wide range of immune responses. Though many methods have been developed to generate macrophages from human iPSCs, the majority utilize similar differentiation steps. Dome-like iPSCs are maintained on a monolayer of irradiated murine feeder cells. Human iPSCs can then be converted into a three-germ-layer embryoid body (EB). Differentiation of the blastocyst-like EB can then be induced, upon the removal of recombinant human FGF2 plus the supplementation with IL-3 and M-CSF, to shed myeloid precursor cells into the surrounding medium. Enhanced concentrations of M-CSF result in the terminal differentiation of iPSDMs. These macrophages, established from iPSCs express a variety of macrophage markers that include CD11b, CD14, EMR1, and CD68 without or with limited SSEA-4 and OCT3/4 markers associated with undifferentiated stem cells (Hale et al., 2015). It has been established that iPSDMs can be polarized and express myeloid cell surface markers and secrete similar proteins compared to MDMs (Cao et al., 2019). Not only do macrophages generated from iPSCs have a strong genetic and functional similarities to host macrophages to better interrogate diverse disease states and infections, but their scalability enhances their use as potential drug screening tools (Hale et al., 2015).
Macrophages derived from human iPSCs established by Hale et al. have been harnessed to generate both wild-type and mutant varieties (Hale et al., 2015; Yeung et al., 2017, 2019). The generated iPSDMs were compared to blood monocyte derived macrophages and found to comparably phagocytize Chlamydia and generate very similar gene expression responses (Hale et al., 2015). Interestingly, iPSDMs had increased rates of infection with the knockout of IRF5 and IL10RA (Yeung et al., 2017), indicating that stimulation of these factors might counteract infection. iPSDMs also can serve as a genetic and drug screening tool by using guide RNA (gRNA) library methods. Yeung et al. utilized a mutant library of Cas9-expressing THP-1 cells and screened these for their inability to take up Salmonella. She consequently identified 183 loci that are required for Salmonella invasion of macrophages, each of which represented a potential target for host-directed therapy (Yeung et al., 2019). Such a strategy is easily translatable to iPSDMs since iPSCs are very genetically malleable. Overall, iPSDMs have the ability to mimic many of the features of MDMs and also provide insights into the host mechanisms that drive and promote C. trachomatis infections.
It is obvious that one cannot ethically perform whole organism infection model studies in man. Thus, it is of interest to create appropriate surrogate models for screening. One important approach utilizes organoids, which are self-organized, three-dimensional (3D) multi-cellular structures that resemble a miniature form of an organ. An organoid can be derived from human or animal iPSCs, embryonic stem cells (ESCs), neonatal tissue stem cells, or ex vivo adult progenitors (Clevers, 2016). Organoids recapitulate the crucial aspects of the architecture, functions and multi-cellular components present in the tissue of origin. In addition, organoids can be expanded and maintained over a long period of time while retaining genomic stability (Clevers, 2016). In general, there are three main forms of organoids: air-liquid interface (ALI) constructs, spheroids, and organ-on-a-chip models (Figure 1). Each model has their advantages and disadvantages, which will be discussed further below (Table 1).
Originally, organoids were used in basic research to understand fetal development, cell assembly, and neoplasia. Driven by rapid developments in stem cell research, the availability of human progenitor cells, and the desire to refine, reduce or replace the use of research animals, there is increasing interest in the field of biological and medical research to utilize organoids to study toxicology, oncology, microbiology, regenerative medicine, and drug development (Engelhart et al., 2005; Davies, 2018). To date, a wide variety of organoids have been developed to represent different organs of the body, including skin, brain, lung, heart, stomach, liver, and intestine. Organoids can also be used to model various diseases, including cancer and infectious disease (Clevers, 2016; Heo et al., 2018). In addition, the pathogenic processes of chronic diseases related to genetic defects, such as cystic fibrosis (CF), can be recapitulated using organoid models (McCauley et al., 2017). Here, we will focus on examining the use of human skin, lung, and intestinal epithelial organoids, and current knowledge regarding the use of omics analyses with organoids.
Human skin, as the largest organ of the body, forms a protective barrier that provides a first line of immune defense. The intensive crosstalk among epithelial cells, stromal cells, immune cells, and the skin commensal bacteria are essential for maintaining immune hemostasis in the skin (Pasparakis et al., 2014; Kabashima et al., 2019). Although animal models are widely used to study human immunological responses and to test the activities of novel therapeutics, there is concern about whether animal models can truly reflect human immunology (Bracken, 2009). Human and mouse skin have differences in their structure and cellular components. For example, human skin has a thicker dermis and less hair follicle density compared to that of the mouse skin (Pasparakis et al., 2014). In addition, Langerhans cells and CD8+ T cells are the major immune cell types in the epidermis of human skin, as compared to dendritic epidermal T cells which are more predominant in mouse epidermis (Mestas and Hughes, 2004; Pasparakis et al., 2014). Therefore, there is a great need for human systems to study human diseases. Three-dimensional skin organoid models are emerging tools implemented in many aspects of skin research such as the study of barrier properties (El Ghalbzouri et al., 2008; Danso et al., 2015), wound healing (El Ghalbzouri et al., 2004; Egles et al., 2010; van Kilsdonk et al., 2013), bacterial infection, biofilm formation, and screening of antimicrobial peptides (Holland et al., 2009; Shepherd et al., 2009; de Breij et al., 2012, 2018; Boekema et al., 2013; Haisma et al., 2013, 2014, 2016; van Drongelen et al., 2014; den Reijer et al., 2016), and inflammatory cutaneous diseases (van den Bogaard et al., 2014; Honzke et al., 2016; Hubaux et al., 2018).
Three main types of skin organoid models recapitulate the characteristics of human skin with fully stratified layers and a competent skin barrier, namely, in vitro generated ALI skin models, the ex vivo skin model and “skin-on-chip” technology. In ALI models, cells are grown on a permeable filter. Once the cells are confluent, media is removed from the apical side of the filter, allowing the cells to become exposed to air and initiate differentiation. ALI models can be built from human primary cells isolated from skin explants (de Breij et al., 2012; den Reijer et al., 2016; Haisma et al., 2016), cell lines [e.g., N/TERT cells (van Drongelen et al., 2014; Reijnders et al., 2015; Smits et al., 2017) or HaCaT cells (Springer et al., 2003; Engelhart et al., 2005)], keratinocytes or fibroblasts differentiated from iPSCs or ESCs (Guenou et al., 2009; Itoh et al., 2013), or primary cells nucleofected with siRNA targeting a gene of interest (Mildner et al., 2010; Honzke et al., 2016; Wallmeyer et al., 2017). N/TERT cells, for example, are immortalized by ectopic expression of the human telomerase reverse transcriptase gene while also losing the pRB/p16INK4a cell cycle control mechanism (Dickson et al., 2000). The epidermal skin model contains about 8–10 epidermal cell layers with fully stratified layers (e.g., stratum corneum, stratum granulosum, stratum spinosum, stratum basale) expressing structural proteins (e.g., Keratin 10 and Keratin 16), tight junction proteins (e.g., claudin-1, claudin-4 and occludin), antimicrobial peptides (e.g., hBD-2, hBD-3, and RNAse7), and cytokines and chemokines (e.g., IL-8, IL-1α, IL-1β) (de Breij et al., 2012; Basler et al., 2017). In contrast, the full-thickness human skin equivalent has, in addition of the epidermis, a continuous basement membrane and fibroblast-seeded collagen or fibrin matrix (Holland et al., 2008; Haisma et al., 2013, 2014). Alternatively, epidermis can be established on top of a human decellularized dermal scaffold referred to as a de-epidermized dermis (Tjabringa et al., 2008; Shepherd et al., 2009; van Kilsdonk et al., 2013). The in vitro models mimic the origin and the genetic make-up of the epidermis and dermis in human skin. Immune cells such as Langerhans cells (Regnier et al., 1997; Facy et al., 2004; Ouwehand et al., 2011), dermal-type macrophages (Bechetoille et al., 2011), melanocytes (Gibbs et al., 2000), and CD4+ T cells (Engelhart et al., 2005; van den Bogaard et al., 2014; Wallmeyer et al., 2017), as well as endothelial cells (Ponec et al., 2004) and hair follicles (Hoeller et al., 2001) have been incorporated allowing the in-depth investigation of cellular responses.
The ex vivo human skin model is established by ALI culturing of human surplus skin samples (e.g., breast or abdominal skin) obtained from surgery with ethical approval and donors' consent (Ng et al., 2009; Danso et al., 2015; de Breij et al., 2018). Ex vivo skin can be trimmed into various thicknesses (typically 300 μm-3 mm) and maintained under laboratory conditions for about 2 weeks. This model provides normal skin structures and immune cells (e.g., CD14+ or CD1c+ monocytes/macrophages, CD11c+ DCs, CD56+CD3− NK cells, CD4+ T cells, CD8+ T cells, and CD19+ B cells) (He et al., 2016), which reflect local immune responses and donor variability, but requires donor access and is difficult to genetically manipulate. Skin organoids can be thermally or physically wounded (Egles et al., 2010; Haisma et al., 2013, 2016) or injured by UV radiation (Bernerd and Asselineau, 2008) to create damaged skin models, which help the study of skin physiology and pathology with impaired skin barrier functions.
One major limitation of conventional ALI models is that these static culture systems make it difficult to study the dynamic processes such as nutrient exchange and immune cell migration. The microfluidic skin-on-chip technology helps to fill this gap since the skin is constructed in the presence of fluid flow under controlled microenvironments that closely mimics the mechanical force and biochemical gradients encountered by natural human skin (O'Neill et al., 2008; Wufuer et al., 2016; Lee et al., 2017). Monocytes (U937) have been co-cultured with HaCat keratinocyte-based epidermis in a bi-channel microfluidic setting for up to 17 days (Ramadan and Ting, 2016). Many skin-on-chip platforms contribute to drug development as biosensors can be implemented to provide non-invasive real-time tissue response readouts (e.g., membrane permeability and skin metabolism), which help in determining drug efficacy and toxicity (Wang Y. I. et al., 2017; Zhang et al., 2017; Alexander et al., 2018).
Skin models for cutaneous diseases such as atopic dermatitis and psoriasis have been employed to understand the initiation and progression of inflammatory skin diseases. Psoriasis and atopic dermatitis are prevalent chronic inflammatory skin diseases considered to be the paradigm of Th1/Th17 and Th2/Th22 types of disorders, respectively. One common approach for modeling disease symptoms is by stimulating skin equivalents with cytokines. For example, Tjabringa et al. discovered that skin equivalents stimulated with psoriasis-associated cytokines (IL-1α, TNF-α, and IL-6) displayed the molecular characteristics of psoriatic epidermis such as increased expression of skin-derived anti-leukoprotease (SKALP) and hBD-2 (Tjabringa et al., 2008). Smits and colleagues established psoriasis and atopic dermatitis models using N/TERT epidermal skin by adding Th1/Th17 (TNF-α, IL-6, IL-1α, IL-17, and IL-22) cytokines and Th2 (IL-4 and IL-13) cytokines, respectively, during the final stage of the skin maturation (Smits et al., 2017). In the psoriasis model, Th1 and Th17 cytokines caused thickening of the stratum corneum, which was accompanied by decreases in the mRNA expression of epidermal differentiation proteins filaggrin (FLG) and loricrin (LOR), and upregulation in hBD-2 and SKALP protein expression (Smits et al., 2017). Supplementing skin equivalents with Th2 cytokines triggered signs of spongiosis in the superbasal layers of skin and apoptosis of basal cells, downregulated FLG, involucrin (IVL) and LOR, and upregulated the expression of the atopic dermatitis-related genes CCL26 and CA2 (Smits et al., 2017). Rouaud-Tinguely et al. demonstrated that supplementing epidermal skin equivalents with Poly I:C, TNF-α, IL-4, and IL-13 in the last 2 days of skin reconstruction, led to reproduction of the features of atopic dermatitis, including altered epidermal reconstruction and differentiation, enhanced thymic stromal lymphopoietin (TSLP) and IL-8 secretions (Rouaud-Tinguely et al., 2015). Transcriptomic analysis by microarray showed that comparing skin supplemented with the inflammatory cocktail to control skin, 3,816 genes were differentially expressed, which were enriched in 4 relevant pathways including cell adhesion, cell migration, inflammation, and epidermal cell differentiation (Rouaud-Tinguely et al., 2015).
An alternative approach for skin disorders modeling is by incorporation of immune cells. Engelhart et al. developed an eczematous dermatitis model by integrating activated CD45RO+ T cells into skin equivalents (Engelhart et al., 2005). This model reproduced several clinical hallmarks of eczematous dermatitis including T cell-induced keratinocyte apoptosis and impaired epidermal barriers, upregulation of intercellular adhesion molecule-1 and neurotrophin-4, and increases in the production of proinflammatory cytokines (IL-1α and IL-6) and chemokines (IL-8, IP-10, TARC, MCP-1, RANTES, and eotaxin) (Engelhart et al., 2005). In a study of atopic dermatitis pathophysiology, Wallmeyer et al. added activated human CD4+ T cells beneath the dermis of ALI skin culture. CD4+ T cells were able to exclusively migrate upwards in the skin equivalents generated from keratinocytes with FLG knockdown (FLG−/−), and this process was directly stimulated by TSLP (Wallmeyer et al., 2017). Exposure of both FLG+ and FLG−/−skin equivalents to CD4+ T cells enhanced TSLP levels, which in turn shifted the polarization profile of activated CD4+ T cells from Th1/Th17 to Th2/Th22. This study highlights the critical role of TSLP in T cell migration as well as in the pathogenesis of atopic dermatitis (Wallmeyer et al., 2017). CD4+ T cell incorporation and migration in the skin model has also been used for studying psoriasis. Van den Bogaard et al. reported that skin equivalents populated with activated CD4+ T cells or in vitro polarized Th1 and Th17 cells induced the expression of psoriasis-associated marker genes DEFB4, LCE3A, PI3, and KRT16 and elicited psoriasis-like epidermal inflammation (van den Bogaard et al., 2014). These cutaneous disease models provide excellent platforms for studying the immunomodulatory aspects of HDPs and to aid in their therapeutic development.
Lung epithelium is the first point of contact with airborne pathogens, allergens, and pollutants. Airway lung epithelium is composed mainly of basal cells, ciliated cells, and secretory cells (club cells and goblet cells). Basal cells serve as progenitors for the ciliated and secretory cells (Shaykhiev, 2015). The ciliated cells contain beating cilia responsible for transporting foreign material trapped by the mucus out of the respiratory tract (Bustamante-Marin and Ostrowski, 2017). The secretory cells produce a thin layer of liquid containing mucin and glycoproteins, which entraps pathogens or allergens, and acts as lubricant to facilitate ciliary movement and moisten incoming air (Barkauskas et al., 2017; Bustamante-Marin and Ostrowski, 2017). The epithelium of the alveoli mainly consists of two cell types: alveolar type I cells (AEC1), which are responsible for gas exchange, and alveolar type II cells (AEC2) that secrete surfactants and proteins (Barkauskas et al., 2017). Under ideal conditions, where there are no airborne foreign materials, the turnover rate of lung epithelium is relatively low (Bowden, 1983; Kotton and Morrisey, 2014). However, in the presence of foreign materials or cellular injury, basal cells quickly proliferate and differentiate into one or more cell types to regenerate the epithelium and restore barrier function (Shaykhiev, 2015). Interestingly, when basal cells were selectively killed off via genetic modification, researchers have discovered that club cells were able to undergo reprogramming to become Krt5+Trp63+ basal cells and fill in the role (Tata et al., 2013; Pardo-Saganta et al., 2015). This robust flexibility and the ability to self-regenerate and repair the epithelium has sparked interest in the field of regenerative medicine and biological sciences, enabling a search for cell-based therapies or drug targets that can induce regeneration and repair damages induced by infection or chronic airway diseases. However, the relevance and validity of using traditional submerged in vitro cell lines (such as HBE16o−, Calu-3, A549) for toxicity and drug efficacy assessment is highly questionable (Frohlich and Salar-Behzadi, 2014; Clippinger et al., 2016). In addition, submerged in vitro models lack the ability to produce the physiological features of airway epithelium. Due to substantial differences in lung anatomy, physiology and molecular mechanisms between human and mice, results from in vivo animal model, especially toxicity studies, cannot be translated to man (Wright et al., 2008; Landsiedel et al., 2014). In addition, many animal lung disease models, such as CF, chronic obstructive pulmonary disease (COPD) and cancer, fail to sufficiently recapitulate the human symptoms of the diseases (Mortaz and Adcock, 2012; Kim et al., 2019). Furthermore, certain behaviors of laboratory animals, such as mice being obligate nose-breathers, can hinder the translatability of results to man (Benam et al., 2016). Thus, there is a need for an in vitro lung model that can recapitulate human lung physiology. The development of lung organoids has provided models that more closely recapitulate human lung physiology.
Lung organoids can be derived from both healthy and diseased primary lung cells (such as human bronchial epithelial cells, HBEC), ESCs or iPSCs (Barkauskas et al., 2017; Nikolic et al., 2017; Yamamoto et al., 2017; Miller et al., 2019). In order to form organoids, progenitors have to undergo stepwise directed differentiation, using culture medium with defined growth factor cocktails that activate and inhibit specific signaling pathways (Huang et al., 2015; Gkatzis et al., 2018). Briefly, human iPSCs are first differentiated into definitive endoderm by using high concentrations of activin A, followed induction of anterior foregut endoderm using Noggin to inhibit bone morphogenetic protein (BMP), as well as transforming growth factor-β (TGF-β) and activation of Wnt signaling (Huang et al., 2015; Mou et al., 2016). Subsequently, patterning into ventral anterior foregut fate by applying Wnt, BMP, fibroblast growth factor (FGF), retinoic acid and GSK3β antagonist CHIR9901, ultimately gives rise to lung and airway progenitors. Finally, these progenitor cells are subjected to maturation into airway and lung epithelial cells using Wnt, FGF, cAMP, and glucocorticoid agonism. The matured organoid consists of functional ciliated cells with beating cilia, basal cells (TRP63+KRT5+), and goblet cells (MUC5B+) (Huang et al., 2015; Hild and Jaffe, 2016). Using RNA-Seq, Dye and colleagues showed that the gene expression activity of the developed lung organoids resembles that of a developing human fetus. Chen and colleagues developed an improved technique to further induce the matured lung and airway progenitors into branching airway, which form alveolar-like structures in a matrigel 3D culture that expresses alveolar epithelial cell markers (Chen Y. W. et al., 2017). This model was further used to show that the organoids were able to recapitulate the in vivo characteristics of human lung development and enabled studies of respiratory syncytial virus infection and pulmonary fibrotic lung disease (Chen Y. W. et al., 2017). Recently, Sachs and colleagues demonstrated a method that allows long term expansion of pseudostratified airway organoids (Sachs et al., 2019). They used the model to assess a variety of pulmonary diseases: (1) organoids derived from CF patient progenitors cells were used to examine the function of cystic fibrosis transmembrane conductance regulator (CFTR); (2) Lung tumoroid were made to enable assessment of histopathology, gene mutation and drug screening; and (3) infected organoids were shown to recapitulate the central diseases features of respiratory syncytial virus infection, showing that neutrophil recruitment occurred during co-culture (Sachs et al., 2019). Yamamoto et al. performed long-term expansion of alveolar organoids containing alveolar epithelial type II (AEC2) derived from human iPSCs (Yamamoto et al., 2017). Using single-cell RNA-Seq, the authors showed that these alveolar stem cells have heterogeneous gene expression patterns that mimic lung cells from the late gestational phase rather than adult phase (Yamamoto et al., 2017). Furthermore, even though these alveolar organoids contained representative stem cell markers and mature lamellar bodies, they lacked MHC class II gene expression, indicating that they might require interaction with immune cells such as macrophages for further maturation (Yamamoto et al., 2017). However, transcriptomic analyses of these organoids treated with two drugs known to impact on AEC2 cells, namely amiodarone and GNE7915, provided putative mechanistic insights as to how these two drugs differentially induce toxicity (Yamamoto et al., 2017). In addition, several reports have shown that organoids derived from primary HBEC or patient specific iPSC maintain the patient's genetic and phenotypic signatures in vitro (Gras et al., 2012; Kim et al., 2019; Leibel et al., 2019). Thus overall, the spherical culture is a promising tool to study lung development and can be used to model a variety of chronic and infectious diseases. However, the interiorized lumen (facing inwards rather than outwards) makes stimulation and sample collection difficult. This limitation can be overcome with the use of an ALI model.
In ALI models, iPSC derived or primary lung epithelial cells are grown to confluence on a permeable filter (Upadhyay and Palmberg, 2018), and media then removed from the apical side of the filter. This exposes the cells to air and initiates differentiation into a matured, polarized pseudo-stratified epithelium, consisting of functional basal cells, ciliated cells, and secretory cells. The validity of the ALI lung model is supported by many studies that compare its structures, functions, and genetic profiles with nasal or bronchoscopically-obtained tracheal and bronchial brushings from human airways (Dvorak et al., 2011; Pezzulo et al., 2011; Fulcher and Randell, 2013). For example, using a combination of ALI model and mass spectrometry-based approaches, Kesimer et al. showed that human tracheobronchial epithelial cells grown in the ALI system shared 84/186 similar secretory host defense proteins with human tracheobronchial normal induced sputum (Kesimer et al., 2009). The ALI model has both apical (top) and basal (bottom) chambers, which allows for the convenient addition of any stimulus, as well as sample collection. The ALI model provides a convenient platform for determining epithelium integrity, by measuring the trans-epithelial electric resistance, mucociliary clearance, and cilia beat frequency (Gras et al., 2012; Gamez et al., 2015). To further assess the dynamic mechanical and biochemical microenvironment of the lung, the lung-on-a-chip model can be used. Lung-on-a-chip or airway-on-a-chip are microfluidic devices on microchips, consisting of a 3D cell culture system separated by a porous membrane, containing canals that allow continuous perfusion to mimic circulation in the body, and vacuum chambers to mimic breathing in human lungs (Konar et al., 2016). Various microsensors within the microchip enable the collection of real-time data, such as barrier function, surfactant production, protein production, fluid pressure, and cellular migration (Bhatia and Ingber, 2014). These organ-on-a-chip models are able to recapitulate in vivo-like environments and allow for the comparison of biological responses under normal and disease conditions (Benam et al., 2016). For example, Benam and colleagues used bronchiolar cells from normal or COPD patients to build airway-on-a-chip models to perform a matched study on electronic cigarette smoke-induced pathophysiology, such as oxidative stress and ciliary function (Benam et al., 2016). By combining omics analyses, they were able to identify 335 differentially expressed genes in normal smoking chips compared to non-smoking chips and 276 differentially expressed genes in COPD smoking chips compared to non-smoking chips, of which 147 were COPD specific and 129 were shared with smoke-exposed normal chips (Benam et al., 2016). This study showed the possibility of using a relatively “simple” organ-on-a-chip model to identify disease-specific responses and molecular signatures that might reveal novel potential therapeutic targets or diagnostic biomarkers.
The intestinal epithelium is composed of a layer of epithelial cells that serve to facilitate digestion and absorption of nutrients, and is in constant contact with dietary antigens and diverse microorganisms (Turner, 2009). Human intestinal epithelium is comprised of multiple cell types, such as enterocytes, goblet cells, Paneth cells, and neuroendocrine cells. Enterocytes are columnar cells that are responsible for absorbing nutrients and secreting immunoglobulins (Kong et al., 2018). Neuroendocrine cells acts as chemo-receptors that detect the presence of microorganisms or dietary antigens, and initiate the appropriate responses by releasing hormones or peptides into the blood stream (Cox, 2016). Goblet cells are responsible for secreting mucins, which provide lubrication for food passage and create a barrier to prevent bacterial invasion (Hansson and Johansson, 2010). Paneth cells can sense the presence of bacteria through Toll-like receptor (TLR) activation, synthesize antimicrobial peptides and proteins, and provide signals to support intestinal stem cell self-renewal (Bevins and Salzman, 2011). Microarray and proteomic studies showed that goblet or Paneth lineage-specific intestinal organoids recapitulate the functions and phenotypes of their in vivo counterparts (Yin et al., 2014; Luu et al., 2018). Furthermore, by comparing label-free quantitative proteomic data from goblet or Paneth cell-enriched organoids with the Human Protein Atlas, novel markers for goblet and Paneth cells were identified (Luu et al., 2018). Transcriptomic profiling of intestinal epithelial cells and enteroids also provided insights into the spatial compartmentalization of TLRs and unique TLR-induced responses in the intestinal epithelium (Price et al., 2018). Finally, single-cell RNA-Seq of intestinal organoids has identified a rare population of hormone-producing enteroendocrine cells that express Reg4 (Grun et al., 2015).
The intestinal epithelium possess a very high rate of self-renewal (every 4–5 days), where new intestinal epithelial cells arise in the crypt from intestinal stem cells (ISC) expressing leucine-rich-repeat-containing G-protein coupled receptor 5 (Lgr5), migrate up the crypt-villus axis, and differentiate into various mature epithelial cell subsets (Clevers, 2013). Due to the constant assaults from interacting with pathogens and foreign antigens, the maintenance of proper integrity, function, and immune responses of the intestinal epithelial is essential to prevent disease development. In order to understand the various intestinal diseases, host-microbe relationships, development and pathologies of the intestinal tract, researchers have utilized various in vitro, ex vivo, and in vivo models over the years.
Several intestinal submerged in vitro and ex vivo models have been developed using transformed carcinoma cell lines, and human fetal or germ-free rodent primary cells (George et al., 2019). However, the use of cell lines and primary cell models often encounters several limitations including: (1) lack of mature epithelial cell types and reduced genomic stability; (2) cell lines usually contain one type of cell that does not reflect the composition of intestinal epithelium, which is composed of multiple cell types; (3) cell lines do not produce mucin, which is an important host defense mechanism of the intestine; (4) cell lines tend to proliferate until sub-confluency, while in vivo intestinal epithelial cells proliferate to confluency essential for maintenance of epithelium integrity; (5) primary cells and ex vivo models are highly variable, expensive, and can only be maintain within a very short period of time; (6) species-specific issues of using non-human cell lines; and (7) cell line models lack the presence of microbiome. Studies showed that interaction between intestinal epithelium and commercial microflora in the gut is an essential aspect of health (Cornick et al., 2015; Jandhyala et al., 2015; Valdes et al., 2018; George et al., 2019). Alternatively, there are also many animal models that have been developed over the years to study intestine-related diseases, such as pathogen gastrointestinal infections and inflammatory bowel disease (IBD). Some examples of in vivo models include, but are not limited to ligated loops of animal intestines, chemically induced experimental intestinal injury and intestinal xenograft in immune-deficient mice. The in vivo models contain many of the essential cellular subsets, the microbiome and a more “complete” immune system, which allows the exploration of intestinal functions and diseases. Also, mouse and rat intestinal development is fairly similar to human intestine, while non-human primates have an even high degree of genetic (90%) and physiological similarity to human (Mouse Genome Sequencing et al., 2002; Coors et al., 2010; Smith et al., 2014). However, there are still many limitations in that: (1) non-human primates are expensive, invoke substantial ethical considerations, and have a potential hazard of contracting with zoonotic agents (Ideland, 2009; Coors et al., 2010); (2) harsh chemicals or genetic modification used in animal models, usually induce phenotypes similar, but not identical, to human intestinal diseases; and (3) differences in species-specific expression of Toll-like receptors affects the responses to various stimuli (Muzio et al., 2000; Heinz et al., 2003; Low et al., 2013). Due to these limitations, especially from a translational perspective, there is a need for human models that more closely reflect human intestinal diseases.
In this regard, intestinal organoids can provide a more representative culture system for the study of intestinal development, physiology, and diseases. Intestinal organoids, enteroids, or colonoids, can be formed from iPSCs or adult ISC (George et al., 2019). Enteroids contain various differentiated epithelial cell types that mimic small intestinal architecture, while colonoids consists of epithelial cell subsets similar to those found in large intestine or colon (Luu et al., 2018; Attili et al., 2019; In et al., 2019; Jones et al., 2019). The iPSC-derived organoids require meticulous maintenance, while adult ISC-derived organoids are established based on tissue of the intestinal crypts, obtained from surgery or endoscopic biopsies, but require relatively less maintenance (George et al., 2019). Briefly, the isolated Lgr5+ ISCs are embedded in Matrigel (a gelatinous protein mixture secreted by mouse sarcoma cells), with culture medium containing small molecules and growth factors (Wnt agonist R-spondin1, the bone morphogenetic protein (BMP) antagonist Noggin, and epidermal growth factor (EGF), Wnt3a, gastrin, progstaglandin E2, nicotinamide, the TGF-β receptor inhibitor A83-01 and p38 inhibitor SB202190) required for proliferation, differentiation and self-organize into a 3D organoid (Nanthakumar et al., 2000). Once formed, the structure of the organoid resembles normal intestinal epithelium, comprising of multiple crypt-like domains that contains villi, Paneth cells, goblet cells, and enterocytes (Sato et al., 2009; Yin et al., 2014). With proper maintenance, both types of intestinal organoids can retain the gene expression profile, function, and pathological characteristics of the normal intestine and have the ability to self-renewal, even after many passages (Sato et al., 2009; Middendorp et al., 2014; Suzuki et al., 2018). A 3D organoid can capture patient-specific in vivo-like complexity and architecture, making it an ideal model to study the mechanisms of tissue homeostasis and disease development for diseases such as ulcerative colitis or colorectal cancer (Otte et al., 2004). CRISPR/Cas9 technology can be used in conjunction with organoids to study many genetic gastroenterological disorders or to screen anticancer candidates in different stages of tumorigenesis (Cario and Podolsky, 2000; Otte et al., 2004). Pathogens, such as Vibrio cholera, enteropathogenic Escherichia coli and rotavirus, can be microinjected into the lumen of the organoid to study host-microbe interactions (Dotan et al., 2007; Cho, 2008; Spence et al., 2011; Dedhia et al., 2016). Furthermore, each organoid line can be expanded, frozen, and revived for multiple uses, which can potentially be use as biobanks (van de Wetering et al., 2015).
Although both the organoid and ALI have many advantages over conventional in vitro and in vivo models, both still lack other supporting cells, immune cells and physiological luminal and blood flow. In addition, spherical organoids have an enclosed lumen, which makes the process of stimulation and sample collection difficult (Bhatia and Ingber, 2014). To overcome these hurdles, researchers have developed a microfluidic Organ Chip model for the human intestine, also known as gut-on-a-chip, where microchannels are lined with human microvascular endothelium, immune cells or bacteria. In addition, culture medium or stimuli can be perfused through each microchannel with a syringe or peristaltic pump (Vickerman et al., 2008; Bhatia and Ingber, 2014).
Given that a number of severe human gut diseases, such as Crohn's disease or salmonellosis are associated with Paneth cell dysfunction, Jones et al. used the integration of various omics data from Paneth cell-enriched enteroids to investigate the role of autophagy in these pathologies (Jones et al., 2019). Specifically, previous genome-wide association studies identified a mutation in the key autophagy gene, ATG16L1, that prevent autophagy-mediated defense against bacterial pathogens. By comparing the quantitative proteome of Paneth cell-enriched enteroids from mutant ATG16L1 with that of wild-type cells, the authors showed an increase in protein abundance in Atg16l1-deficient cells due to autophagy impairment (Jones et al., 2019), as confirmed by protein-protein interaction network analysis. Furthermore, integration of transcriptomic and proteomic data, together with functional enrichment and network analysis showed that these autophagy impaired Atg16l1-deficient organoids had reduced exocytosis-mediated secretion of antimicrobial peptides (Jones et al., 2019).
Intestinal organoids also serve as an important model for characterizing host-microbe-interactions. For example, characterization of Salmonella enteric sv. typhimurium interaction with the intestinal epithelium has been challenging using whole mouse infection models due to rare epithelial interactions in the unligated intestine. Similarly using traditional immortalized intestinal cell lines is difficult as they lack the different cell types and architecture of a typical intestinal epithelium. However, microinjection of S. typhimurium into intestinal organoids generated from human iPSCs provided an opportunity to understand how Salmonella interactions with different intestinal cell types impact on its ability to spread. Transcriptomic analyses by RNA-Seq of intestinal organoids upon S. typhimurium infection identified the upregulation of genes, including those encoding interleukins and proinflammatory cytokines, in response to Salmonella infection (Forbester et al., 2015). Furthermore, goblet cell-associated genes such as GCNT3 and MUC2 were also upregulated indicating that multiple cell types in the enteroids can respond to Salmonella infection (Forbester et al., 2015). The direct modeling of wild-type S. typhimurium and the intestinal cells of enteroids was further confirmed by microscopy and required S. typhimurium invasion protein InvA (Forbester et al., 2015). These enteroids also were used to demonstrate the importance of alpha-defensins in restricting S. enteric sv. typhimurium (Wilson et al., 2015). Similarly, intestinal organoids can serve as a useful experimental system to characterize host transcriptional responses to microbiota-produced short-chain fatty acids (Lukovac et al., 2014).
Currently, newly developed drug treatments for human diseases face limitations due to individual differences between patients, thus making it challenging to predict therapeutic outcomes and effectiveness. However, organoids based on a specific disease or a variety of individual scan potentially revolutionize precision therapy by improving disease modeling and drug screening to improve gene-drug associations (Bartfeld and Clevers, 2017; Takahashi, 2019). Below, we will discuss how various iPSC-derived immune cells and organoid models have been utilized in drug discovery and their potential use for screening HDPs.
Infections from the obligate intracellular bacterium C. trachomatis is the leading cause of bacterial sexually transmitted diseases (STDs) and preventable blindness, and are a global health burden (Pinsent and Gambhir, 2017; Rowley et al., 2019). Chlamydial infections cause chronic disease and often are asymptomatic. If left untreated, STD infections can progress to the upper genital tract and result in pelvic inflammatory disease (PID) and infertility (Moodley et al., 2002). The complexity of chlamydial infections is in part, due the biphasic lifestyle of the infecting bacterium. Infectious, non-growing extracellular elementary bodies infect cells leading to differentiations into non-infectious, replicating intracellular reticulate bodies and are able to reside within infected macrophages (Gracey et al., 2013; Elwell et al., 2016; Yeung et al., 2017). Studying the host-pathogen interactions of M0, M1 or M2 polarized murine macrophage infected with C. muridarum demonstrated the susceptibility of M0 and M2 macrophages, and resistance of M1 macrophages, to infection (Gracey et al., 2013), as also shown for C. trachomatis (Yeung et al., 2017). While internalized, Chlamydia spp. survival and proliferation is favored in M2 polarized macrophages (Buchacher et al., 2015). Alternatively, classically activated M1 macrophages secrete high levels of proinflammatory cytokines and exhibit antimicrobial effects. Since Chlamydia only replicates in human cells, iPSDM (and possibly organoid models) would appear to be ideal for studying the mechanisms of infection and screening for anti-Chlamydia therapeutics, including host directed therapies (Yeung et al., 2017).
Unlike many pathogenic bacteria, antibiotic resistance is a rare occurrence with Chlamydia spp. Like antibiotics, some HDPs can have robust antimicrobial effects. The 21-residue synthetic peptide, Pep1 demonstrated direct antimicrobial activity against intracellular Chlamydia. Application of Pep-1 to infected macrophages resulted in complete eradication of chlamydial inclusions while also blocking growth of infectious progeny (Park et al., 2009). The only human cathelicidin, a C-terminal 37 amino acid LL-37 also demonstrated both antimicrobial activity and immune modulation. C. trachomatis has a highly conserved cryptic plasmid which encodes eight open reading frames. Of the eight, virulence factor Pgp3 (pORF5) is the only protein secreted into chlamydial inclusions and the cell cytosol and then binds to Li et al. (2008) and inactivates (Hou et al., 2015) cathelicidin human HDP LL-37.
Two of the potential limitations in the use of HDPs as anti-infection or anti-inflammatory therapy are peptide instability and off-target toxicity (Johansson et al., 1998; Sieprawska-Lupa et al., 2004; Heilborn et al., 2005; Vandamme et al., 2012; Mishra et al., 2017; Zhou et al., 2019). To address these concerns, Zhou and colleagues proposed to use stem cell-based gene delivery therapy with the human HDP LL-37 to provide local protection against pulmonary infections (Zhou et al., 2019). The HDP peptide LL-37 is known to have a broad range of antimicrobial and immunomodulatory activities that provides protection against infections (Vandamme et al., 2012; van der Does et al., 2012). A transgenic mouse strain was constructed that constitutively expressed LL-37 (LL-37+/+) to confirm the in vivo protective effects of the peptide against P. aeruginosa PA01 infection (Zhou et al., 2019). Using RNA-Seq, they compared the transcriptomic profiles of wild-type and LL-37+/+ mice before and after PA01 infection (Figure 2). In contract to the wild-type mice, the transcriptomic profiles of LL-37+/+ mice before and after PA01 infection were very similar, indicating that LL-37+/+ mice were protected from PA01 challenge-induced alterations. In response to PA01 infection, WT mice revealed an inflammation driven protein-protein interaction network whereas a lung development-related molecular network was found in LL-37+/+ mice, indicating a protective role of LL-37 expression in mouse lung (Figure 2). Secondly, Zhou et al. introduced the LL-37 gene into mouse P63+/Krt5+ distal airway stem cells (DASC) by lentiviral transduction to create mDASC that constitutively expresses LL-37 (LL-37-mDASC). Using a 3D organoid system, they showed that the genetically engineered LL-37-mDASC were able to form alveolar-like sphere structures, consisting of differentiated cells expressing type I alveolar cell markers, and were able to secrete functional LL-37 that inhibited growth of both PA01 and Escherichia coli (Figure 2). In a lung injury mouse model, transplanted LL-37-mDASCs incorporated only into the injured lung tissues, but not healthy lung tissues, which suggested that LL-37-mDASC specifically targeted injured lung tissues. The regenerated LL-37 lung, containing LL-37-mDASC, enhanced bacterial clearance after intratracheal challenge with either PA01 or E. coli. By using a combination of genetically engineered stem cells, animal and organoid models and RNA-Seq analyses, Zhou and colleagues showed that genetically engineered DASCs, which constitutively express the human HDP LL-37, can provide protection against pulmonary infections and enhance the repair of lung injury (Zhou et al., 2019).
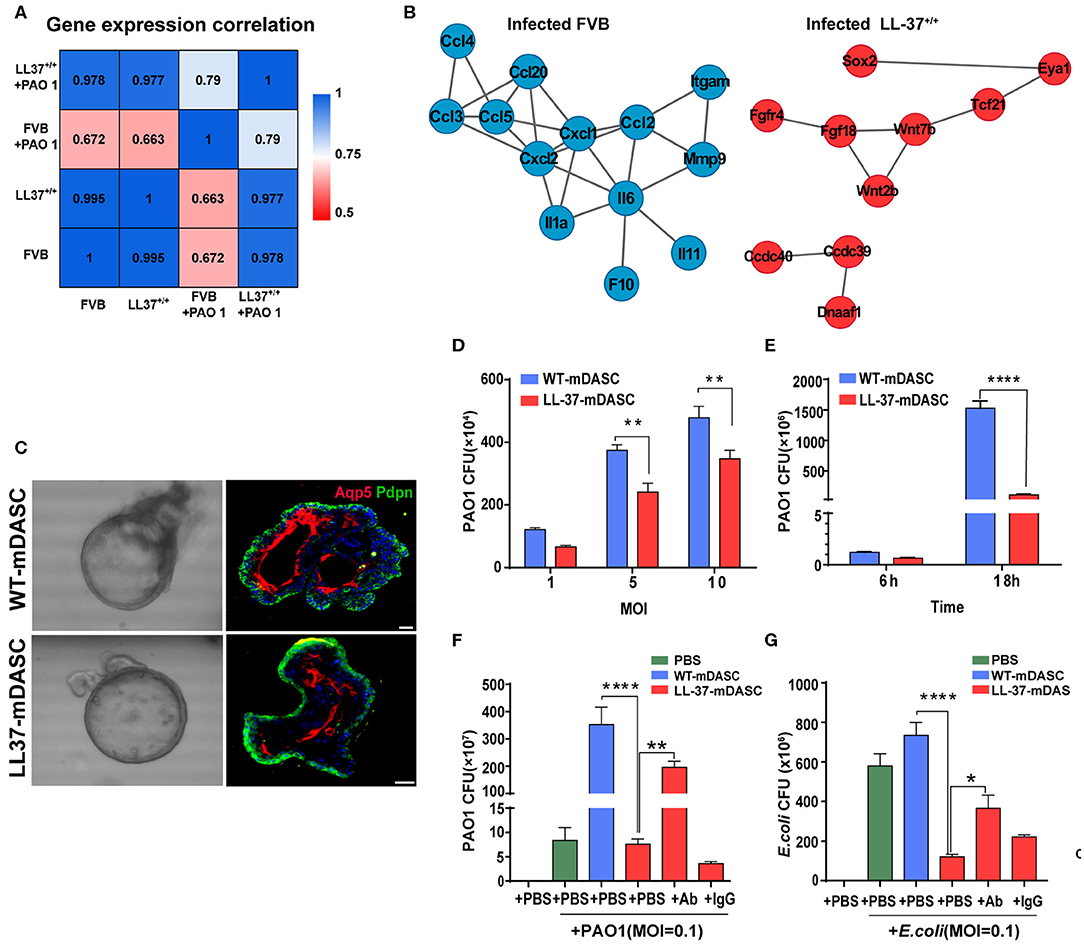
Figure 2. Genetically engineered distal airway stem cell transplantation protects mice from pulmonary infection. (A) Heatmap showing transcriptome profile correlation values of indicated lung tissue samples before and after PAO1 infection. (B) Protein-protein interaction network of selected genes with high expression level in PAO1-infected wild-type lung (blue) and PAO1-infected LL-37+/+ lung (red), respectively. (C) Representative 3D organoid culture of mDASCs with expression of type I alveolar cell markers (Aqp5 and Pdpn). Left panels, bright-field imaging of 3D organoids. Right panels, immunofluorescence of organoid sections. Scale bar, 20 μm. (D) Co-culture of bacteria with DASCs shows antimicrobial effects in dose-dependent manner. Initial additions of PAO1 were 0.1 ×, 0.5 × and 1 × 104 CFU, respectively. Co-culture duration, 6 h. n = 4. Error bars, SEM. MOI, multiplicity of infection. (E) Co-culture of bacteria with DASCs shows antimicrobial effects in time-dependent manner. Initial concentration of PAO1 was 1 × 104 CFU. MOI = 1. n = 3. Error bars, SEM. (F,G) Preincubation of cells with anti-LL-37 antibody, but not IgG control, significantly reduced anti-PAO1 (F) and anti-Escherichia coli (G) effects of LL-37-mDASCs. Initial dose of bacteria was 103 CFU. Co-culture duration, 18 h. n = 4 in (F) and n = 3 in (G). Error bars, SEM. Statistics for graphs: two-way ANOVA followed by Sidak's test (D,E) and one-way ANOVA followed by Tukey's test (F,G). *P < 0.05; **P < 0.01; ****P < 0.0001. These results were from Zhou et al. (2019), published under the terms of the CC by 4.0 license. Full terms at https://creativecommons.org/licenses/by/4.0/. This figure combined Figures 2, 3 from the original article with permission from the corresponding author.
Researchers have employed skin models to investigate host-microbial interactions including skin commensal organisms such as Staphylococcus epidermidis and Propionibacterium acnes (Holland et al., 2008, 2009), and pathogens that are the leading cause of skin and soft tissue infections including Staphylococcus aureus (Charles et al., 2009; Shepherd et al., 2009; Haisma et al., 2013, 2014, 2016; van Drongelen et al., 2014; den Reijer et al., 2016; de Breij et al., 2018) and P. aeruginosa (Charles et al., 2009; Shepherd et al., 2009; Boekema et al., 2013). In the study of antimicrobial and immunomodulatory activities of HDPs and IDR peptides, these skin infection models are helpful tools when used complementarily with conventional microtiter plate assays that help bridge the gap between HDP discovery and potential drug development. Indeed, it could be argued that given the immunomodulatory potential of HDPs/IDR peptides, models involving both host and microbes provide superior potential to demonstrate efficacy under in vivo-like circumstances. Haisma et al. studied a group of synthetic peptides derived from P60.4Ac, a peptide based on LL-37, using a full-thickness skin model that was thermally wounded and infected with different strains of S. aureus (Haisma et al., 2014). One P60.4Ac derivative, Peptide 10, was found to reduce MRSA LUH14616 and mupirocin-resistant MRSA LUH15051 burden without affecting the level of IL-8 produced by thermally wounded skin (Haisma et al., 2014). A human epidermal model was later used to test the efficacy of various P60.4Ac formulations (Haisma et al., 2016). In particular, P60.4Ac could be stably formulated in a hypromellose gel, which was highly effective (from as low as 0.1% wt/wt) in eradicating planktonic and biofilm-associated MRSA on epidermal skin models without causing cytotoxicity (up to 2%) (Haisma et al., 2016). De Breij et al. used an ex vivo wounded skin infection model to confirm the activity of a hypromellose gel-formulated peptide SAAP-148 that showed superior efficacy in biofilm inhibition and eradication assays in vitro (de Breij et al., 2018). Likewise, a single treatment with SAAP-148 ointment eradicated acute infections and pre-formed biofilms of MRSA LUH14616 and Acinetobacter baumannii RUH875 from the wounded human skin (Figure 3) and abraded murine skin (de Breij et al., 2018). Ventress et al. showed that synthetic peptides derived from the EC2 domain of the tetraspanin CD9 could block the adherence of S. aureus to wounded skin models (Ventress et al., 2016). A role for LL-37 in the pathogenesis of the skin inflammatory condition rosacea could be demonstrated by the application of the anti-parasitic ivermectin into keratinocytes, reconstructed human epidermis and ex vivo skin models. This led to an anti-inflammatory effect mediated by suppressing expression of LL-37 and the serine protease kallikrein-5 (Thibaut de Menonville et al., 2017).
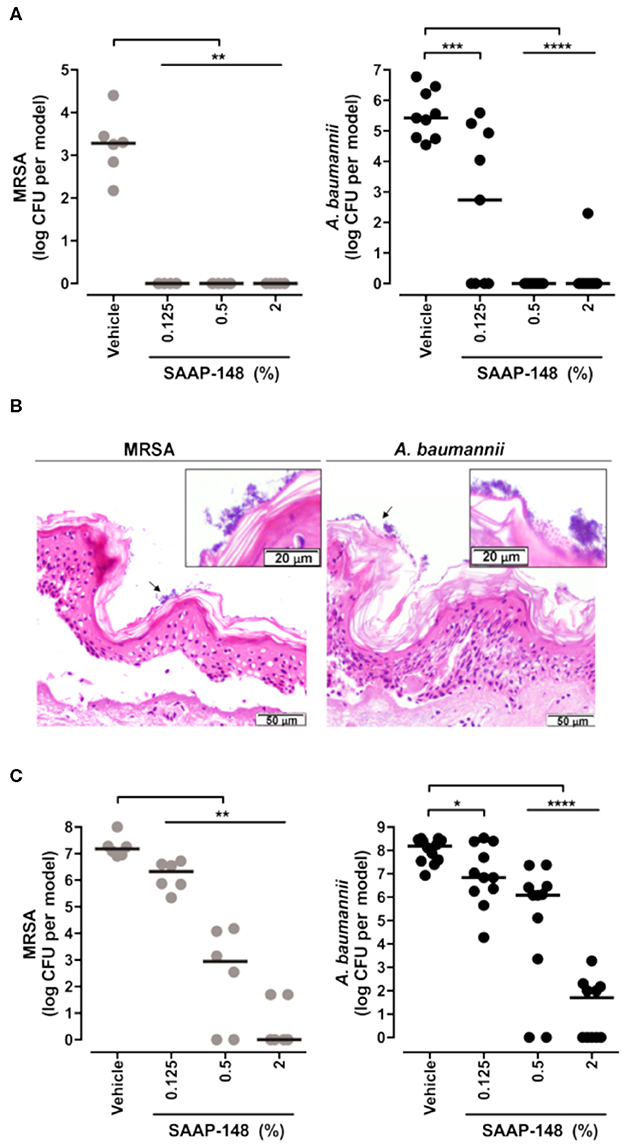
Figure 3. Topical application of SAAP-148 ointment eradicates acute and established infections of MRSA and A. baumannii from the skin. Use of an ex vivo human skin ALI model to demonstrate that topical application of peptide SAAP-148 ointment eradicates acute (A) and established (C) infections of methicillin resistant Staphylococcus aureus (MRSA) and Acinetobacter baumannii infections from the skin within 4 h of treatment. Results are expressed as the numbers of viable bacteria (in log10 CFU) per skin model of three to six donors. Each circle represents one skin sample, and bars indicate medians. *Significantly different (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001) as compared to the vehicle, as calculated using the Mann-Whitney rank sum test. (B) shows that these organisms form biofilms on the skin surface 24 h after inoculation. These results were excerpted from de Breij et al. (2018), with permission from AAAS.
The production of HDPs can be assessed in both lung and skin ALI models (Hertz et al., 2003; Beisswenger et al., 2006; Diamond et al., 2010). The comparison of HDP expressions between patients and healthy lung ALI cultures permit an understanding of the important roles of HDPs in respiratory diseases and can potentially identify key mechanisms or drug targets (Diamond et al., 2010; Zuyderduyn et al., 2011; Amatngalim et al., 2017). For example, Amatngalim et al. showed that the expression of non-typeable Haemophilus influenzae (NTHi)-induced human β-defensin 2 (DEFB4) and S110A7, but not LL37 (CAMP) or β-defensin 1 and 3 (hDB-1 and hBD-3), were reduced in ALI from COPD patients when compared to non-COPD cultures (Amatngalim et al., 2017). In addition, cigarette smoke attenuated NTHi-reduced gene expression of DEFB4, LCN2, S100A7, and CCL20, while increasing IL-8 and IL-6 gene expression (Amatngalim et al., 2017). This study utilized a lung ALI model to demonstrate that the imbalance of reduced expression of HDPs and increased expression of pro-inflammatory cytokines might have contributed to the susceptibility to microbial infections of smokers and COPD patients. It was suggested that exogenous application of HDPs might provide a potential route of therapy in COPD (Amatngalim et al., 2017). Harcourt et al. used an ALI Calu-3 model to show that the efficacy of LL-37 in reducing respiratory syncytial virus (RSV) infections differs when the peptide is given as a prophylactic or therapeutic (Harcourt et al., 2016). When used in a prophylactic (pretreatment) regimen, LL-37 significantly reduced the intracellular viral genome counts as well as the production of cytokines and chemokines mediated by RSV (Harcourt et al., 2016). An ALI mode derived from primary HBE cells was used to show that the frog skin-derived HDP Esc (1-21) and its synthetic derivative Esc (1-21)-1c were both able to preserve epithelial layer barrier integrity upon P. aeruginosa challenge (Chen C. et al., 2017). It was also demonstrated that Esc (1-21)-1c, created by substituting two amino acids and using the corresponding D-enantiomer of Esc (1-21), had significantly reduced peptide-induced cellular toxicity and enhanced biostability and antibiofilm activity (Cappiello et al., 2016). In order to predict acute local lung toxicity, Ritter et al. utilized an ALI model with an optimized aerosol delivery system to evaluate different combinations of HDPs and nanocarriers (Ritter et al., 2020). This study demonstrated that the ALI culture system is more sensitive when compared to the submerged culture systems. It was suggested that the immediate contact between the deposited aerosol and epithelial surfaces contributed to a larger, faster bioavailability and more realistic estimation of the lowest observable adverse effect levels (Ritter et al., 2020).
Other than growing as spheroids, both enteroid and colonoid cells can also be grown as polarized epithelial monolayers on permeable tissue culture membranes, allowing easy access to the lumen for infectious studies and drug screening assays (In et al., 2019). A recent study demonstrated the use of enteroid monolayers to perform screening of 2,000 candidate drugs, which shows that organoid models can provide a high-throughput drug screening tool (Kozuka et al., 2017). In other examples, transgenic expression of α-defensins in enteroids led to inhibition of Salmonella growth, illustrating the anti-bacterial activities of α-defensins at physiologically relevant conditions (Wilson et al., 2015). Furthermore single-cell RNA-Seq was used to demonstrate that Paneth-cell enriched enteroids mimic their in vivo counterparts, and can express a set of antimicrobial peptide marker genes including Defa22, Defa21, and Ang4 (Mead et al., 2018). Intestinal organoids have also provided a model for the mechanistic characterization of the mode of action of bacitracin beyond its ability to inhibit cell wall synthesis in Gram-positive bacteria. Specifically, bacitracin can neutralize the Clostridium difficile TcdB toxin by preventing TcdB translocation into the cytosol of intestinal epithelial cells. Therefore reducing TcdB-induced glucosylation of Rac1 and destruction of F-Actin, which ultimately leads to the maintenance of epithelial integrity (Figure 4) (Zhu et al., 2019).
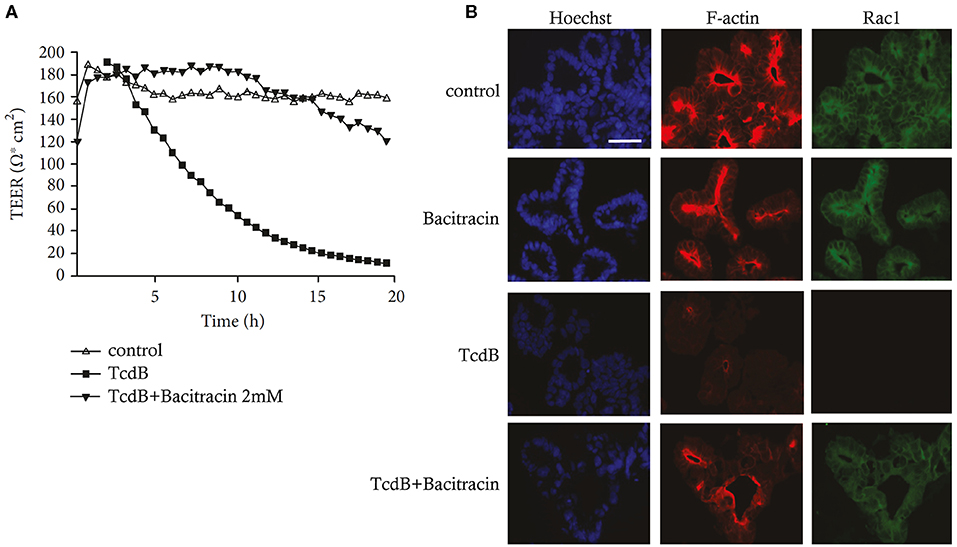
Figure 4. The antibiotic Bacitracin protected human gut epithelia and human intestinal organoids from Clostridium difficile Toxin TcdB. (A) Bacitracin preserves epithelial integrity of CaCo-2 monolayers from TcdB activity. Epithelial CaCo-2 monolayers grown on filters in a Transwell chamber were preincubated with or without Bacitracin (2 mM) for 30 min at 37°C and then stimulated apically with Clostridium difficile toxin B (TcdB, 6 ng/ml). The epithelial integrity was recorded by measuring the transepithelial electrical resistance (TEER) every 35 min over a time course of 20 h. (B) Bacitracin reduces the TcdB-induced glucosylation of Rac1 and destruction of F-actin in stem cell-derived human intestinal organoids. Intestinal organoids preincubated with or without Bacitracin (1 mM) for 30 min at 37°C were challenged with 60 ng/ml TcdB for 3 h. Nuclei, F-actin, and non-glucosylated Rac1 were specifically stained and visualized by confocal fluorescence microscopy. Bar = 50 μm. These results were from Zhu et al. (2019), published under the terms of the CC by 4.0 license. Full terms at https://creativecommons.org/licenses/by/4.0/. This figure combined Figures 3, 4 from the original article with permission from the corresponding author.
In addition, epithelial monolayers in a transwell plate provide researchers with the opportunity to co-culture the organoid with other immune cells (Hensel et al., 2014). Dekkers et al. generated intestinal organoids from rectal biopsies of cystic fibrosis (CF) patients and used a microscopic assay to functionally assess CFTR in these organoids (Dekkers et al., 2013). By using forskolin to raise the intracellular cyclic AMP levels and activate CFTR, ion uptake followed by fluid secretion into the lumen of the intestinal organoids was shown to lead to swelling of these organoids. However, intestinal organoids from CF patients showed reduced swelling compared to organoids from healthy individuals as a consequence of defective CFTR (Dekkers et al., 2013). Using these CF intestinal organoids and the forskolin assay, it was subsequently shown that the CFTR-modulating drug VX-770 (ivacaftor/kalydeco) can treat an uncharacterized rare CFTR genotype and improve clinical outcome for these patients (Dekkers et al., 2016). High-throughput screens have also been developed to identify drug sensitivities and putative protein kinase inhibitor using tumor organoids (Aboulkheyr Es et al., 2018; Phan et al., 2019).
Although the various organoid culture systems have several advantages over conventional submerged in vitro models and in vivo animal models, there are certain hurdles that need to be overcome. First, most current organoid models consists of only the epithelial layer, which lacks the interactions with vascular, immune, and nervous systems (Xu et al., 2018). Researchers are addressing this limitation with continuous development and improvements of organoid models. For example, a multi-organ-on-chips system is currently being developed and refined, whereby multiple organ-on-a-chip systems can be connected, allowing real-time monitoring of the pharmacokinetics and pharmacodynamics of multi-organ interactions (Zhao et al., 2019). Co-culture systems are also utilized to increase the complexity and functionality of organoid models. For example, Workman et al. used a tissue-engineering approach to co-culture human iPSC-derived neural crest cells and human intestinal organoids (Heinz et al., 2003), giving rise to functional enteric nervous system -containing intestinal organoids that were capable of responding to calcium stimulation, demonstrating neuronal activities and expressing neurochemical markers (Workman et al., 2017). Using RNA-Seq analysis, the authors revealed the effects of the enteric nervous system on HIO development, revealing differentially expressed genes associated with digestive-tract development (e.g., EGF signaling components and TGF-β signaling factors), and absorptive (e.g., FABP2, LCT, TREH, SI, and MGAM) and secretory (MUC2, Paneth cell markers, and WNT3), lineage markers when compared to HIO alone (Workman et al., 2017). Moreover, this co-culture system could be used to mechanistically study of Hirschsprung's disease, caused by a mutation in the PHOX2B gene in human (Workman et al., 2017). Another study used a co-culture system containing primary human macrophages with enteroids, and showed that macrophages enhanced the barrier functions and maturity of enteroids (Noel et al., 2017).
Second, most organoid models currently reflect only the fetal stages of organ development. However, recent studies showed that transplantation into immune-compromized hosts promotes organoid maturation and enhances its functionality. For example, Dye et al. showed that transplantation of ESC-derived lung organoids into immune deficient NSG mice enhanced epithelial organization, structure development and cellular differentiation (Dye et al., 2016). In addition, the structure and cellular diversities of the transplanted lung organoids resemble adult human airways to a greater extent than do in vitro grown lung organoids (Dye et al., 2016). Third, most organoid models are relatively expensive due to the requirement for reagents and growth factors/inhibitors, and their dependence on extracellular matrix (such as Matrigel) and long-term culture (Xu et al., 2018). Animal-derived extracellular matrices, such as Matrigel, are prone to batch-to-batch variability and have poorly defined protein and growth factor compositions. The use of various growth factors and inhibitors during the organoid formation has the potential to mask drug responses. In addition, the process of organoid production is time consuming and labor intensive. To address some of these limitations, bioengineers are making an effort to create a well-defined culture media and extracellular matrix systems, such as collagen I gels and fibrin hydrogels, and utilize bioprinting technologies (Sachs et al., 2017; Yui et al., 2018). Lastly, current organoid models do not accurately recapitulate the metabolism of their parental organs (Fan et al., 2018). Further efforts in the development and refinement of organoid model building will help to overcome some of these limitations.
Over the past few years, researchers had made extensive efforts to understand and improve the use of induced pluripotent stem cell-derived immune cells and organoid systems in the field of drug discovery (Figure 1). As discussed in some of the examples above, iPSC-derived epithelial, immune cell, and organoid models can be used as a powerful screening tool for precision therapy (Table 1). ALI cultures and organoids derived from primary cells or iPSC maintain the patient's genetic, functional and phenotypic signatures in vitro, allowing for accurate prediction of drug responses of individual patients (Gras et al., 2012; Bartfeld and Clevers, 2017; Oost et al., 2018; Kim et al., 2019; Leibel et al., 2019). In combination with genetic modification tools, such as lentiviruses or CRISPR/Cas9, organoids, and ALI cultures can be used to study the effects of specific genetic mutations in various diseases and identify potential drug targets and cell therapies (Schwank et al., 2013; Zhou et al., 2019). In addition, ALI and organoids can be expanded from small amounts of starting materials, and can be maintained over a long period of time to provide enough samples for a wide variety of laboratory analysis, such as efficacy, toxicology, multi-omics analyses (such as RNA-Seq, proteomic, and metabolomic). With regards to biofilm infections, which represent 65% of all human infections, are very resistant to antibiotics, and lack targeted therapies. ALI cultures and organoids offer a screening model for agents, including antimicrobial peptides to address such infections. We propose that future HDP and IDR peptide screens can be performed using both iPSC-derived and patient-specific ALI cultures/organoids with integrating multi-omics analysis, to not only identify new HDPs and IDR peptides, but also potentially determine which patients will benefit from a combination treatment using HDPs and antibiotics. Furthermore, ALI cultures and organoids can be used for toxicology testing to complement or replace animal testing.
K-YC, BW, AL, and BB wrote the manuscript. RH extensively edited the manuscript. All the authors reviewed and edited the final draft of the manuscript.
Our own peptide, organoid, and systems biology research was supported by grants to REWH from the Canadian Institutes for Health Research FDN-154287, and the Michael Smith Foundation for Health Research grant 17774. K-YC was supported by Michael Smith Foundation for Health Research and Lotte & John Hecht Memorial Foundation Research Trainee Award. AL is supported by the Simon Fraser University New Faculty Start-Up Grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Canadian Institutes for Health Research or Michael Smith Foundation for Health Research.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
RH is supported by the Canadian Institutes for Health Research and Michael Smith Foundation, and holds a Canada Research Chair in Health and Genomics and a University of British Columbia Killam Professorship.
Aboulkheyr Es, H., Montazeri, L., Aref, A. R., Vosough, M., and Baharvand, H. (2018). Personalized cancer medicine: an organoid approach. Trends Biotechnol. 36, 358–371. doi: 10.1016/j.tibtech.2017.12.005
Adler, S., Basketter, D., Creton, S., Pelkonen, O., van Benthem, J., Zuang, V., et al. (2011). Alternative (non-animal) methods for cosmetics testing: current status and future prospects-2010. Arch. Toxicol. 85, 367–485. doi: 10.1007/s00204-011-0693-2
Agency, U. S. E. P. (2019). Efforts to Reduce Animal Testing at EPA. Available online at: https://www.epa.gov/research/efforts-reduce-animal-testing-epa
Alexander, F. A., Eggert, S., and Wiest, J. (2018). Skin-on-a-chip: transepithelial electrical resistance and extracellular acidification measurements through an automated air-liquid interface. Genes 9:114. doi: 10.3390/genes9020114
Amatngalim, G. D., Schrumpf, J. A., Henic, A., Dronkers, E., Verhoosel, R. M., Ordonez, S. R., et al. (2017). Antibacterial defense of human airway epithelial cells from chronic obstructive pulmonary disease patients induced by acute exposure to nontypeable haemophilus influenzae: modulation by cigarette smoke. J. Innate Immun. 9, 359–374. doi: 10.1159/000455193
Attili, D., McClintock, S. D., Rizvi, A. H., Pandya, S., Rehman, H., Nadeem, D. M., et al. (2019). Calcium-induced differentiation in normal human colonoid cultures: Cell-cell / cell-matrix adhesion, barrier formation and tissue integrity. PLoS ONE 14:e0215122. doi: 10.1371/journal.pone.0215122
Bailey, J., and Balls, M. (2019). Recent efforts to elucidate the scientific validity of animal-based drug tests by the pharmaceutical industry, pro-testing lobby groups, and animal welfare organisations. BMC Med. Ethics 20:16. doi: 10.1186/s12910-019-0352-3
Bailey, J., Thew, M., and Balls, M. (2014). An analysis of the use of animal models in predicting human toxicology and drug safety. Altern. Lab. Anim. 42, 181–199. doi: 10.1177/026119291404200306
Bailey, J., Thew, M., and Balls, M. (2015). Predicting human drug toxicity and safety via animal tests: can any one species predict drug toxicity in any other, and do monkeys help? Altern. Lab. Anim. 43, 393–403. doi: 10.1177/026119291504300607
Barkauskas, C. E., Chung, M. I., Fioret, B., Gao, X., Katsura, H., and Hogan, B. L. (2017). Lung organoids: current uses and future promise. Development 144, 986–997. doi: 10.1242/dev.140103
Barre-Sinoussi, F., and Montagutelli, X. (2015). Animal models are essential to biological research: issues and perspectives. Future Sci. OA 1:FSO63. doi: 10.4155/fso.15.63
Bartfeld, S., and Clevers, H. (2017). Stem cell-derived organoids and their application for medical research and patient treatment. J. Mol. Med. 95, 729–738. doi: 10.1007/s00109-017-1531-7
Basler, K., Galliano, M. F., Bergmann, S., Rohde, H., Wladykowski, E., Viadal-Y-Sy, S., et al. (2017). Biphasic influence of Staphylococcus aureus on human epidermal tight junctions. Ann. N. Y. Acad. Sci. 1405, 53–70. doi: 10.1111/nyas.13418
Bechetoille, N., Vachon, H., Gaydon, A., Boher, A., Fontaine, T., Schaeffer, E., et al. (2011). A new organotypic model containing dermal-type macrophages. Exp. Dermatol. 20, 1035–1037. doi: 10.1111/j.1600-0625.2011.01383.x
Bechinger, B., and Gorr, S. U. (2017). Antimicrobial peptides: mechanisms of action and resistance. J. Dent. Res. 96, 254–260. doi: 10.1177/0022034516679973
Beisswenger, C., Kandler, K., Hess, C., Garn, H., Felgentreff, K., Wegmann, M., et al. (2006). Allergic airway inflammation inhibits pulmonary antibacterial host defense. J. Immunol. 177, 1833–1837. doi: 10.4049/jimmunol.177.3.1833
Benam, K. H., Novak, R., Nawroth, J., Hirano-Kobayashi, M., Ferrante, T. C., Choe, Y., et al. (2016). Matched-comparative modeling of normal and diseased human airway responses using a microengineered breathing lung chip. Cell Syst. 3, 456–466 e4. doi: 10.1016/j.cels.2016.10.003
Bernerd, F., and Asselineau, D. (2008). An organotypic model of skin to study photodamage and photoprotection in vitro. J. Am. Acad. Dermatol. 58(5 Suppl. 2), S155–S159. doi: 10.1016/j.jaad.2007.08.050
Bevins, C. L., and Salzman, N. H. (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368. doi: 10.1038/nrmicro2546
Bhatia, S. N., and Ingber, D. E. (2014). Microfluidic organs-on-chips. Nat. Biotechnol. 32, 760–772. doi: 10.1038/nbt.2989
Boekema, B. K., Pool, L., and Ulrich, M. M. (2013). The effect of a honey based gel and silver sulphadiazine on bacterial infections of in vitro burn wounds. Burns 39, 754–759. doi: 10.1016/j.burns.2012.09.008
Bommarius, B., Jenssen, H., Elliott, M., Kindrachuk, J., Pasupuleti, M., Gieren, H., et al. (2010). Cost-effective expression and purification of antimicrobial and host defense peptides in Escherichia coli. Peptides 31, 1957–1965. doi: 10.1016/j.peptides.2010.08.008
Bracken, M. B. (2009). Why animal studies are often poor predictors of human reactions to exposure. J. R. Soc. Med. 102, 120–122. doi: 10.1258/jrsm.2008.08k033
Brom, F. W. (2002). Science and society: different bioethical approaches towards animal experimentation. ALTEX 19, 78–82.
Buchacher, T., Ohradanova-Repic, A., Stockinger, H., Fischer, M. B., and Weber, V. (2015). M2 polarization of human macrophages favors survival of the intracellular pathogen Chlamydia pneumoniae. PLoS ONE 10:e0143593. doi: 10.1371/journal.pone.0143593
Buchrieser, J., James, W., and Moore, M. D. (2017). Human induced pluripotent stem cell-derived macrophages share ontogeny with MYB-independent tissue-resident macrophages. Stem Cell Rep. 8, 334–345. doi: 10.1016/j.stemcr.2016.12.020
Bustamante-Marin, X. M., and Ostrowski, L. E. (2017). Cilia and mucociliary clearance. Cold Spring Harb. Perspect. Biol. 9:a028241. doi: 10.1101/cshperspect.a028241
Cao, X., Yakala, G. K., F. E., van den Hil, Cochrane, A., Mummery, C. L., and Orlova, V. V. (2019). Differentiation and functional comparison of monocytes and macrophages from hiPSCs with peripheral blood derivatives. Stem Cell Rep. 12, 1282–1297. doi: 10.1016/j.stemcr.2019.05.003
Capes-Davis, A., Theodosopoulos, G., Atkin, I., Drexler, H. G., Kohara, A., MacLeod, R. A., et al. (2010). Check your cultures! A list of cross-contaminated or misidentified cell lines. Int. J. Cancer 127, 1–8. doi: 10.1002/ijc.25242
Cappiello, F., Di Grazia, A., Segev-Zarko, L. A., Scali, S., Ferrera, L., Galietta, L., et al. (2016). Esculentin-1a-derived peptides promote clearance of Pseudomonas aeruginosa internalized in bronchial cells of cystic fibrosis patients and lung cell migration: biochemical properties and a plausible mode of action. Antimicrob. Agents Chemother. 60, 7252–7262. doi: 10.1128/AAC.00904-16
Cario, E., and Podolsky, D. K. (2000). Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect. Immun. 68, 7010–7017. doi: 10.1128/IAI.68.12.7010-7017.2000
Charles, C. A., Ricotti, C. A., Davis, S. C., Mertz, P. M., and Kirsner, R. S. (2009). Use of tissue-engineered skin to study in vitro biofilm development. Dermatol. Surg. 35, 1334–1341. doi: 10.1111/j.1524-4725.2009.01238.x
Chen, C., Mangoni, M. L., and Di, Y. P. (2017). In vivo therapeutic efficacy of frog skin-derived peptides against Pseudomonas aeruginosa-induced pulmonary infection. Sci. Rep. 7:8548. doi: 10.1038/s41598-017-08361-8
Chen, Y. W., Huang, S. X., de Carvalho, A., Ho, S. H., Islam, M. N., Volpi, S., et al. (2017). A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 19, 542–549. doi: 10.1038/ncb3510
Cho, J. H. (2008). The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 8, 458–466. doi: 10.1038/nri2340
Choi, K. Y., and Mookherjee, N. (2012). Multiple immune-modulatory functions of cathelicidin host defense peptides. Front. Immunol. 3:149. doi: 10.3389/fimmu.2012.00149
Clevers, H. (2013). The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284. doi: 10.1016/j.cell.2013.07.004
Clevers, H. (2016). Modeling development and disease with organoids. Cell 165, 1586–1597. doi: 10.1016/j.cell.2016.05.082
Clippinger, A. J., Ahluwalia, A., Allen, D., Bonner, J. C., Casey, W., Castranova, V., et al. (2016). Expert consensus on an in vitro approach to assess pulmonary fibrogenic potential of aerosolized nanomaterials. Arch. Toxicol. 90, 1769–1783. doi: 10.1007/s00204-016-1717-8
Cole, J. N., and Nizet, V. (2016). Bacterial evasion of host antimicrobial peptide defenses. Microbiol. Spectr. 4. doi: 10.1128/microbiolspec.VMBF-0006-2015
Coors, M. E., Glover, J. J., Juengst, E. T., and Sikela, J. M. (2010). The ethics of using transgenic non-human primates to study what makes us human. Nat. Rev. Genet. 11, 658–662. doi: 10.1038/nrg2864
Cornick, S., Tawiah, A., and Chadee, K. (2015). Roles and regulation of the mucus barrier in the gut. Tissue Barriers 3:e982426. doi: 10.4161/21688370.2014.982426
Costa, F., Teixeira, C., Gomes, P., and Martins, M. C. L. (2019). Clinical application of AMPs. Adv. Exp. Med. Biol. 1117, 281–298. doi: 10.1007/978-981-13-3588-4_15
Cox, H. M. (2016). Neuroendocrine peptide mechanisms controlling intestinal epithelial function. Curr. Opin. Pharmacol. 31, 50–56. doi: 10.1016/j.coph.2016.08.010
Cunrath, O., and Bumann, D. (2019). Host resistance factor SLC11A1 restricts Salmonella growth through magnesium deprivation. Science 366, 995–999. doi: 10.1126/science.aax7898
Daigneault, M., Preston, J. A., Marriott, H. M., Whyte, M. K., and Dockrell, D. H. (2010). The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS ONE 5:e8668. doi: 10.1371/journal.pone.0008668
Danso, M. O., Berkers, T., Mieremet, A., Hausil, F., and Bouwstra, J. A. (2015). An ex vivo human skin model for studying skin barrier repair. Exp. Dermatol. 24, 48–54. doi: 10.1111/exd.12579
Davies, J. A. (2018). “Organoids and mini-organs,” in Organs and Organoids, eds J. A. Davies and M. L. Lawrence (Cambridge, MA: Academic Press). p. 3–23.
de Breij, A., Haisma, E. M., Rietveld, M., El Ghalbzouri, A., van den Broek, P. J., Dijkshoorn, L., et al. (2012). Three-dimensional human skin equivalent as a tool to study Acinetobacter baumannii colonization. Antimicrob. Agents Chemother. 56, 2459–2464. doi: 10.1128/AAC.05975-11
de Breij, A., Riool, M., Cordfunke, R. A., Malanovic, N., de Boer, L., Koning, R. I., et al. (2018). The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 10:eaan4044. doi: 10.1126/scitranslmed.aan4044
de la Fuente-Nunez, C., Reffuveille, F., Haney, E. F., Straus, S. K., and Hancock, R. E. (2014). Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLoS Pathog. 10:e1004152. doi: 10.1371/journal.ppat.1004152
De Zoysa, G. H., Cameron, A. J., Hegde, V. V., Raghothama, S., and Sarojini, V. (2015). Antimicrobial peptides with potential for biofilm eradication: synthesis and structure activity relationship studies of battacin peptides. J. Med. Chem. 58, 625–639. doi: 10.1021/jm501084q
Dedhia, P. H., Bertaux-Skeirik, N., Zavros, Y., and Spence, J. R. (2016). Organoid models of human gastrointestinal development and disease. Gastroenterology 150, 1098–1112. doi: 10.1053/j.gastro.2015.12.042
Dekkers, J. F., Berkers, G., Kruisselbrink, E., Vonk, A., de Jonge, H. R., Janssens, H. M., et al. (2016). Characterizing responses to CFTR-modulating drugs using rectal organoids derived from subjects with cystic fibrosis. Sci. Transl. Med. 8:344ra84. doi: 10.1126/scitranslmed.aad8278
Dekkers, J. F., Wiegerinck, C. L., de Jonge, H. R., Bronsveld, I., Janssens, H. M., de Winter-de Groot, K. M., et al. (2013). A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 19, 939–945. doi: 10.1038/nm.3201
den Reijer, P. M., Haisma, E. M., Lemmens-den Toom, N. A., Willemse, J., Koning, R. I., Demmers, J. A., et al. (2016). Detection of alpha-toxin and other virulence factors in biofilms of Staphylococcus aureus on polystyrene and a human epidermal model. PLoS ONE 11:e0145722. doi: 10.1371/journal.pone.0145722
Diamond, G., Yim, S., Rigo, I., and McMahon, L. (2010). Measuring antimicrobial peptide activity on epithelial surfaces in cell culture. Methods Mol. Biol. 618, 371–382. doi: 10.1007/978-1-60761-594-1_23
Dickson, M. A., Hahn, W. C., Ino, Y., Ronfard, V., Wu, J. Y., Weinberg, R. A., et al. (2000). Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell Biol. 20, 1436–1447. doi: 10.1128/MCB.20.4.1436-1447.2000
Dotan, I., Allez, M., Nakazawa, A., Brimnes, J., Schulder-Katz, M., and Mayer, L. (2007). Intestinal epithelial cells from inflammatory bowel disease patients preferentially stimulate CD4+ T cells to proliferate and secrete interferon-gamma. Am. J. Physiol. Gastrointest Liver Physiol. 292, G1630–G1640. doi: 10.1152/ajpgi.00294.2006
Drexler, H. G., and Uphoff, C. C. (2002). Mycoplasma contamination of cell cultures: incidence, sources, effects, detection, elimination, prevention. Cytotechnology 39, 75–90. doi: 10.1023/A:1022913015916
Dvorak, A., Tilley, A. E., Shaykhiev, R., Wang, R., and Crystal, R. G. (2011). Do airway epithelium air-liquid cultures represent the in vivo airway epithelium transcriptome? Am. J. Respir. Cell. Mol. Biol. 44, 465–473. doi: 10.1165/rcmb.2009-0453OC
Dye, B. R., Dedhia, P. H., Miller, A. J., Nagy, M. S., White, E. S., Shea, L. D., et al. (2016). A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. Elife 5:e19732. doi: 10.7554/eLife.19732.023
Egles, C., Garlick, J. A., and Shamis, Y. (2010). Three-dimensional human tissue models of wounded skin. Methods Mol. Biol. 585, 345–359. doi: 10.1007/978-1-60761-380-0_24
El Ghalbzouri, A., Hensbergen, P., Gibbs, S., Kempenaar, J., van der Schors, R., and Ponec, M. (2004). Fibroblasts facilitate re-epithelialization in wounded human skin equivalents. Lab. Invest. 84, 102–112. doi: 10.1038/labinvest.3700014
El Ghalbzouri, A., Siamari, R., Willemze, R., and Ponec, M. (2008). Leiden reconstructed human epidermal model as a tool for the evaluation of the skin corrosion and irritation potential according to the ECVAM guidelines. Toxicol. In Vitro 22, 1311–1320. doi: 10.1016/j.tiv.2008.03.012
El Shazely, B., Urbanski, A., Johnston, P. R., and Rolff, J. (2019). In vivo exposure of insect AMP resistant Staphylococcus aureus to an insect immune system. Insect Biochem. Mol. Biol. 110, 60–68. doi: 10.1016/j.ibmb.2019.04.017
Elsea, S. H., and Lucas, R. E. (2002). The mousetrap: what we can learn when the mouse model does not mimic the human disease. ILAR J. 43, 66–79. doi: 10.1093/ilar.43.2.66
Elwell, C., Mirrashidi, K., and Engel, J. (2016). Chlamydia cell biology and pathogenesis. Nat. Rev. Microbiol. 14, 385–400. doi: 10.1038/nrmicro.2016.30
Engelhart, K., El Hindi, T., Biesalski, H. K., and Pfitzner, I. (2005). In vitro reproduction of clinical hallmarks of eczematous dermatitis in organotypic skin models. Arch. Dermatol. Res. 297, 1–9. doi: 10.1007/s00403-005-0575-7
Facy, V., Flouret, V., Regnier, M., and Schmidt, R. (2004). Langerhans cells integrated into human reconstructed epidermis respond to known sensitizers and ultraviolet exposure. J. Invest. Dermatol. 122, 552–553. doi: 10.1046/j.0022-202X.2004.22209.x
Fan, T. W., El-Amouri, S. S., Macedo, J. K. A., Wang, Q. J., Song, H., Cassel, T., et al. (2018). Stable isotope-resolved metabolomics shows metabolic resistance to anti-cancer selenite in 3D spheroids versus 2D cell cultures. Metabolites 8:40. doi: 10.3390/metabo8030040
Feng, B., Ng, J. H., Heng, J. C., and Ng, H. H. (2009). Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 4, 301–312. doi: 10.1016/j.stem.2009.03.005
Festing, M. F. (2004). Is the use of animals in biomedical research still necessary in 2002? Unfortunately, “yes”. Altern. Lab. Anim. 32(Suppl. 1B), 733–739. doi: 10.1177/026119290403201s121
Forbester, J. L., Goulding, D., Vallier, L., Hannan, N., Hale, C., Pickard, D., et al. (2015). Interaction of Salmonella enterica Serovar typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect. Immun. 83, 2926–2934. doi: 10.1128/IAI.00161-15
Frohlich, E., and Salar-Behzadi, S. (2014). Toxicological assessment of inhaled nanoparticles: role of in vivo, ex vivo, in vitro, and in silico studies. Int. J. Mol. Sci. 15, 4795–4822. doi: 10.3390/ijms15034795
Fulcher, M. L., and Randell, S. H. (2013). Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol. Biol. 945, 109–121. doi: 10.1007/978-1-62703-125-7_8
Gamez, A. S., Gras, D., Petit, A., Knabe, L., Molinari, N., Vachier, I., et al. (2015). Supplementing defect in club cell secretory protein attenuates airway inflammation in COPD. Chest 147, 1467–1476. doi: 10.1378/chest.14-1174
Ganz, T. (2003). Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3, 710–720. doi: 10.1038/nri1180
George, M. M., Rahman, M., Connors, J., and Stadnyk, A. W. (2019). Opinion: are organoids the end of model evolution for studying host intestinal epithelium/microbe interactions? Microorganisms 7:406. doi: 10.3390/microorganisms7100406
Gibbs, S., Murli, S., De Boer, G., Mulder, A., Mommaas, A. M., and Ponec, M. (2000). Melanosome capping of keratinocytes in pigmented reconstructed epidermis–effect of ultraviolet radiation and 3-isobutyl-1-methyl-xanthine on melanogenesis. Pigment Cell Res. 13, 458–466. doi: 10.1034/j.1600-0749.2000.130608.x
Gkatzis, K., Taghizadeh, S., Huh, D., Stainier, D. Y. R., and Bellusci, S. (2018). Use of three-dimensional organoids and lung-on-a-chip methods to study lung development, regeneration and disease. Eur. Respir. J. 52:1800876. doi: 10.1183/13993003.00876-2018
Gracey, E., Lin, A., Akram, A., Chiu, B., and Inman, R. D. (2013). Intracellular survival and persistence of Chlamydia muridarum is determined by macrophage polarization. PLoS ONE 8:e69421. doi: 10.1371/journal.pone.0069421
Gras, D., Bourdin, A., Vachier, I., de Senneville, L., Bonnans, C., and Chanez, P. (2012). An ex vivo model of severe asthma using reconstituted human bronchial epithelium. J. Allergy Clin. Immunol. 129, 1259–1266 e1. doi: 10.1016/j.jaci.2012.01.073
Grun, D., Lyubimova, A., Kester, L., Wiebrands, K., Basak, O., Sasaki, N., et al. (2015). Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525, 251–255. doi: 10.1038/nature14966
Gudjonsson, T., Villadsen, R., Nielsen, H. L., Ronnov-Jessen, L., Bissell, M. J., and Petersen, O. W. (2002). Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev. 16, 693–706. doi: 10.1101/gad.952602
Guenou, H., Nissan, X., Larcher, F., Feteira, J., Lemaitre, G., Saidani, M., et al. (2009). Human embryonic stem-cell derivatives for full reconstruction of the pluristratified epidermis: a preclinical study. Lancet 374, 1745–1753. doi: 10.1016/S0140-6736(09)61496-3
Hainsworth, A. H., Allan, S. M., Boltze, J., Cunningham, C., Farris, C., Head, E., et al. (2017). Translational models for vascular cognitive impairment: a review including larger species. BMC Med. 15:16. doi: 10.1186/s12916-017-0793-9
Haisma, E. M., de Breij, A., Chan, H., van Dissel, J. T., Drijfhout, J. W., Hiemstra, P. S., et al. (2014). LL-37-derived peptides eradicate multidrug-resistant Staphylococcus aureus from thermally wounded human skin equivalents. Antimicrob. Agents Chemother. 58, 4411–4419. doi: 10.1128/AAC.02554-14
Haisma, E. M., Goblyos, A., Ravensbergen, B., Adriaans, A. E., Cordfunke, R. A., Schrumpf, J., et al. (2016). Antimicrobial peptide P60.4Ac-containing creams and gel for eradication of methicillin-resistant Staphylococcus aureus from cultured skin and airway epithelial surfaces. Antimicrob. Agents Chemother. 60, 4063–4072. doi: 10.1128/AAC.03001-15
Haisma, E. M., Rietveld, M. H., de Breij, A., van Dissel, J. T., El Ghalbzouri, A., and Nibbering, P. H. (2013). Inflammatory and antimicrobial responses to methicillin-resistant Staphylococcus aureus in an in vitro wound infection model. PLoS ONE 8:e82800. doi: 10.1371/journal.pone.0082800
Hale, C., Yeung, A., Goulding, D., Pickard, D., Alasoo, K., Powrie, F., et al. (2015). Induced pluripotent stem cell derived macrophages as a cellular system to study Salmonella and other pathogens. PLoS ONE 10:e0124307. doi: 10.1371/journal.pone.0124307
Hancock, R. E., Haney, E. F., and Gill, E. E. (2016). The immunology of host defence peptides: beyond antimicrobial activity. Nat. Rev. Immunol. 16, 321–334. doi: 10.1038/nri.2016.29
Hancock, R. E., and Lehrer, R. (1998). Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16, 82–88. doi: 10.1016/S0167-7799(97)01156-6
Hancock, R. E., and Sahl, H. G. (2006). Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24, 1551–1557. doi: 10.1038/nbt1267
Haney, E. F., Straus, S. K., and Hancock, R. E. W. (2019). Reassessing the host defense peptide landscape. Front. Chem. 7:43. doi: 10.3389/fchem.2019.00043
Hansson, G. C., and Johansson, M. E. (2010). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Gut Microbes 1, 51–54. doi: 10.4161/gmic.1.1.10470
Harcourt, J. L., McDonald, M., Svoboda, P., Pohl, J., Tatti, K., and Haynes, L. M. (2016). Human cathelicidin, LL-37, inhibits respiratory syncytial virus infection in polarized airway epithelial cells. BMC Res. Notes 9:11. doi: 10.1186/s13104-015-1836-y
Hashimoto, D., Chow, A., Noizat, C., Teo, P., Beasley, M. B., Leboeuf, M., et al. (2013). Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804. doi: 10.1016/j.immuni.2013.04.004
Hay, M., Thomas, D. W., Craighead, J. L., Economides, C., and Rosenthal, J. (2014). Clinical development success rates for investigational drugs. Nat. Biotechnol. 32, 40–51. doi: 10.1038/nbt.2786
He, X., de Oliveira, V. L., Keijsers, R., Joosten, I., and Koenen, H. J. (2016). Lymphocyte isolation from human skin for phenotypic analysis and ex vivo cell culture. J. Vis. Exp. 52564. doi: 10.3791/52564
Heilborn, J. D., Nilsson, M. F., Jimenez, C. I., Sandstedt, B., Borregaard, N., Tham, E., et al. (2005). Antimicrobial protein hCAP18/LL-37 is highly expressed in breast cancer and is a putative growth factor for epithelial cells. Int. J. Cancer 114, 713–719. doi: 10.1002/ijc.20795
Heinz, S., Haehnel, V., Karaghiosoff, M., Schwarzfischer, L., Muller, M., Krause, S. W., et al. (2003). Species-specific regulation of Toll-like receptor 3 genes in men and mice. J. Biol. Chem. 278, 21502–21509. doi: 10.1074/jbc.M301476200
Hensel, K. O., Boland, V., Postberg, J., Zilbauer, M., Heuschkel, R., Vogel, S., et al. (2014). Differential expression of mucosal trefoil factors and mucins in pediatric inflammatory bowel diseases. Sci. Rep. 4:7343. doi: 10.1038/srep07343
Heo, I., Dutta, D., Schaefer, D. A., Iakobachvili, N., Artegiani, B., Sachs, N., et al. (2018). Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814–823. doi: 10.1038/s41564-018-0177-8
Hertz, C. J., Wu, Q., Porter, E. M., Zhang, Y. J., Weismuller, K. H., Godowski, P. J., et al. (2003). Activation of Toll-like receptor 2 on human tracheobronchial epithelial cells induces the antimicrobial peptide human beta defensin-2. J. Immunol. 171, 6820–6826. doi: 10.4049/jimmunol.171.12.6820
Hild, M., and Jaffe, A. B. (2016). Production of 3-D airway organoids from primary human airway basal cells and their use in high-throughput screening. Curr. Protoc. Stem Cell Biol. 37, IE 9 1–IE 9 15. doi: 10.1002/cpsc.1
Hirz, T., and Dumontet, C. (2016). Neutrophil isolation and analysis to determine their role in lymphoma cell sensitivity to therapeutic agents. J. Vis. Exp. e53846. doi: 10.3791/53846
Hoeller, D., Huppertz, B., Roos, T. C., Poblete Gutierrez, P., Merk, H. F., Frank, J., et al. (2001). An improved and rapid method to construct skin equivalents from human hair follicles and fibroblasts. Exp. Dermatol. 10, 264–271. doi: 10.1034/j.1600-0625.2001.100406.x
Holland, D. B., Bojar, R. A., Farrar, M. D., and Holland, K. T. (2009). Differential innate immune responses of a living skin equivalent model colonized by Staphylococcus epidermidis or Staphylococcus aureus. FEMS Microbiol. Lett. 290, 149–155. doi: 10.1111/j.1574-6968.2008.01402.x
Holland, D. B., Bojar, R. A., Jeremy, A. H., Ingham, E., and Holland, K. T. (2008). Microbial colonization of an in vitro model of a tissue engineered human skin equivalent–a novel approach. FEMS Microbiol. Lett. 279, 110–115. doi: 10.1111/j.1574-6968.2007.01021.x
Honzke, S., Wallmeyer, L., Ostrowski, A., Radbruch, M., Mundhenk, L., Schafer-Korting, M., et al. (2016). Influence of Th2 cytokines on the cornified envelope, tight junction proteins, and ss-defensins in filaggrin-deficient skin equivalents. J. Invest. Dermatol. 136, 631–639. doi: 10.1016/j.jid.2015.11.007
Hou, S., Dong, X., Yang, Z., Li, Z., Liu, Q., and Zhong, G. (2015). Chlamydial plasmid-encoded virulence factor Pgp3 neutralizes the antichlamydial activity of human cathelicidin LL-37. Infect. Immun. 83, 4701–4709. doi: 10.1128/IAI.00746-15
Huang, S. X., Green, M. D., de Carvalho, A. T., Mumau, M., Chen, Y. W., D'Souza, S. L., et al. (2015). The in vitro generation of lung and airway progenitor cells from human pluripotent stem cells. Nat. Protoc. 10, 413–425. doi: 10.1038/nprot.2015.023
Hubaux, R., Bastin, C., and Salmon, M. (2018). On the relevance of an in vitro reconstructed human epidermis model for drug screening in atopic dermatitis. Exp. Dermatol. 27, 1403–1407. doi: 10.1111/exd.13810
Hubrecht, R. C., and Carter, E. (2019). The 3Rs and humane experimental technique: implementing change. Animals 9:754. doi: 10.3390/ani9100754
Ideland, M. (2009). Different views on ethics: how animal ethics is situated in a committee culture. J. Med. Ethics 35, 258–261. doi: 10.1136/jme.2008.026989
In, J. G., Foulke-Abel, J., Clarke, E., and Kovbasnjuk, O. (2019). Human colonoid monolayers to study interactions between pathogens, commensals, and host intestinal epithelium. J. Vis. Exp. e59357. doi: 10.3791/59357
Ishida, H., Nguyen, L. T., Gopal, R., Aizawa, T., and Vogel, H. J. (2016). Overexpression of antimicrobial, anticancer, and transmembrane peptides in Escherichia coli through a calmodulin-peptide fusion system. J. Am. Chem. Soc. 138, 11318–11326. doi: 10.1021/jacs.6b06781
Itoh, M., Umegaki-Arao, N., Guo, Z., Liu, L., Higgins, C. A., and Christiano, A. M. (2013). Generation of 3D skin equivalents fully reconstituted from human induced pluripotent stem cells (iPSCs). PLoS ONE 8:e77673. doi: 10.1371/journal.pone.0077673
Jandhyala, S. M., Talukdar, R., Subramanyam, C., Vuyyuru, H., Sasikala, M., and Nageshwar Reddy, D. (2015). Role of the normal gut microbiota. World J. Gastroenterol. 21, 8787–8803. doi: 10.3748/wjg.v21.i29.8787
Johansson, J., Gudmundsson, G. H., Rottenberg, M. E., Berndt, K. D., and Agerberth, B. (1998). Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J. Biol. Chem. 273, 3718–3724. doi: 10.1074/jbc.273.6.3718
Jones, E. J., Matthews, Z. J., Gul, L., Sudhakar, P., Treveil, A., Divekar, D., et al. (2019). Integrative analysis of Paneth cell proteomic and transcriptomic data from intestinal organoids reveals functional processes dependent on autophagy. Dis. Model. Mech. 12:dmm037069. doi: 10.1242/dmm.037069
Juni, P., Nartey, L., Reichenbach, S., Sterchi, R., Dieppe, P. A., and Egger, M. (2004). Risk of cardiovascular events and rofecoxib: cumulative meta-analysis. Lancet 364, 2021–2029. doi: 10.1016/S0140-6736(04)17514-4
Kabashima, K., Honda, T., Ginhoux, F., and Egawa, G. (2019). The immunological anatomy of the skin. Nat. Rev. Immunol. 19, 19–30. doi: 10.1038/s41577-018-0084-5
Kaufmann, S. H. (2008). Immunology's foundation: the 100-year anniversary of the Nobel Prize to Paul Ehrlich and Elie Metchnikoff. Nat. Immunol. 9, 705–712. doi: 10.1038/ni0708-705
Kaur, G., and Dufour, J. M. (2012). Cell lines: Valuable tools or useless artifacts. Spermatogenesis 2, 1–5. doi: 10.4161/spmg.19885
Kesimer, M., Kirkham, S., Pickles, R. J., Henderson, A. G., Alexis, N. E., Demaria, G., et al. (2009). Tracheobronchial air-liquid interface cell culture: a model for innate mucosal defense of the upper airways? Am. J. Physiol. Lung Cell. Mol. Physiol. 296, L92–L100. doi: 10.1152/ajplung.90388.2008
Kim, J. B., Zaehres, H., Wu, G., Gentile, L., Ko, K., Sebastiano, V., et al. (2008). Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454, 646–650. doi: 10.1038/nature07061
Kim, M., Mun, H., Sung, C. O., Cho, E. J., Jeon, H. J., Chun, S. M., et al. (2019). Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 10:3991. doi: 10.1038/s41467-019-11867-6
Konar, D., Devarasetty, M., Yildiz, D. V., Atala, A., and Murphy, S. V. (2016). Lung-on-a-chip technologies for disease modeling and drug development. Biomed. Eng. Comput. Biol. 7, 17–27. doi: 10.4137/BECB.S34252
Kong, S., Zhang, Y. H., and Zhang, W. (2018). Regulation of intestinal epithelial cells properties and functions by amino acids. Biomed. Res. Int. 2018:2819154. doi: 10.1155/2018/2819154
Kotton, D. N., and Morrisey, E. E. (2014). Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat. Med. 20, 822–832. doi: 10.1038/nm.3642
Kozuka, K., He, Y., Koo-McCoy, S., Kumaraswamy, P., Nie, B., Shaw, K., et al. (2017). Development and characterization of a human and mouse intestinal epithelial cell monolayer platform. Stem Cell Rep. 9, 1976–1990. doi: 10.1016/j.stemcr.2017.10.013
Landsiedel, R., Ma-Hock, L., Hofmann, T., Wiemann, M., Strauss, V., Treumann, S., et al. (2014). Application of short-term inhalation studies to assess the inhalation toxicity of nanomaterials. Part Fibre Toxicol. 11:16. doi: 10.1186/1743-8977-11-16
Lee, S., Jin, S. P., Kim, Y. K., Sung, G. Y., Chung, J. H., and Sung, J. H. (2017). Construction of 3D multicellular microfluidic chip for an in vitro skin model. Biomed. Microdevices 19:22. doi: 10.1007/s10544-017-0156-5
Lei, J., Sun, L., Huang, S., Zhu, C., Li, P., He, J., et al. (2019). The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 11, 3919–3931.
Leibel, S. L., Winquist, A., Tseu, I., Wang, J., Luo, D., Shojaie, S., et al. (2019). Reversal of surfactant protein b deficiency in patient specific human induced pluripotent stem cell derived lung organoids by gene therapy. Sci. Rep. 9:13450. doi: 10.1038/s41598-019-49696-8
Leslie, N. D., Yager, K. L., McNamara, P. D., and Segal, S. (1996). A mouse model of galactose-1-phosphate uridyl transferase deficiency. Biochem. Mol. Med. 59, 7–12. doi: 10.1006/bmme.1996.0057
Li, Z., Chen, D., Zhong, Y., Wang, S., and Zhong, G. (2008). The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect. Immun. 76, 3415–3428. doi: 10.1128/IAI.01377-07
Low, D., Nguyen, D. D., and Mizoguchi, E. (2013). Animal models of ulcerative colitis and their application in drug research. Drug Des. Devel. Ther. 7, 1341–1357. doi: 10.2147/DDDT.S40107
Lukovac, S., Belzer, C., Pellis, L., Keijser, B. J., de Vos, W. M., Montijn, R. C., et al. (2014). Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. mBio 5:e01438-14. doi: 10.1128/mBio.01438-14
Luu, L., Matthews, Z. J., Armstrong, S. D., Powell, P. P., Wileman, T., Wastling, J. M., et al. (2018). Proteomic profiling of enteroid cultures skewed toward development of specific epithelial lineages. Proteomics 18:e1800132. doi: 10.1002/pmic.201800132
Mansour, S. C., Pena, O. M., and Hancock, R. E. (2014). Host defense peptides: front-line immunomodulators. Trends Immunol. 35, 443–450. doi: 10.1016/j.it.2014.07.004
Mansour, S. C., Pletzer, D., de la Fuente-Nunez, C., Kim, P., Cheung, G. Y. C., Joo, H.-S., et al. (2016). Bacterial abscess formation is controlled by the stringent stress response and can be targeted therapeutically. EBioMedicine 12, 219–226. doi: 10.1016/j.ebiom.2016.09.015
McCauley, K. B., Hawkins, F., Serra, M., Thomas, D. C., Jacob, A., and Kotton, D. N. (2017). Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of Wnt signaling. Cell Stem Cell 20, 844–857 e6. doi: 10.1016/j.stem.2017.03.001
Mead, B. E., Ordovas-Montanes, J., Braun, A. P., Levy, L. E., Bhargava, P., Szucs, M. J., et al. (2018). Harnessing single-cell genomics to improve the physiological fidelity of organoid-derived cell types. BMC Biol. 16:62. doi: 10.1186/s12915-018-0527-2
Mestas, J., and Hughes, C. C. (2004). Of mice and not men: differences between mouse and human immunology. J. Immunol. 172, 2731–2738. doi: 10.4049/jimmunol.172.5.2731
Middendorp, S., Schneeberger, K., Wiegerinck, C. L., Mokry, M., Akkerman, R. D., van Wijngaarden, S., et al. (2014). Adult stem cells in the small intestine are intrinsically programmed with their location-specific function. Stem Cells 32, 1083–1091. doi: 10.1002/stem.1655
Mildner, M., Jin, J., Eckhart, L., Kezic, S., Gruber, F., Barresi, C., et al. (2010). Knockdown of filaggrin impairs diffusion barrier function and increases UV sensitivity in a human skin model. J. Invest. Dermatol. 130, 2286–2294. doi: 10.1038/jid.2010.115
Miller, A. J., Dye, B. R., Ferrer-Torres, D., Hill, D. R., Overeem, A. W., Shea, L. D., et al. (2019). Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 14, 518–540. doi: 10.1038/s41596-018-0104-8
Mishra, B., Reiling, S., Zarena, D., and Wang, G. (2017). Host defense antimicrobial peptides as antibiotics: design and application strategies. Curr. Opin. Chem. Biol. 38, 87–96. doi: 10.1016/j.cbpa.2017.03.014
Moodley, P., Wilkinson, D., Connolly, C., Moodley, J., and Sturm, A. W. (2002). Trichomonas vaginalis is associated with pelvic inflammatory disease in women infected with human immunodeficiency virus. Clin. Infect. Dis. 34, 519–522. doi: 10.1086/338399
Moravej, H., Moravej, Z., Yazdanparast, M., Heiat, M., Mirhosseini, A., Moosazadeh Moghaddam, M., et al. (2018). Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb. Drug Resist. 24, 747–767. doi: 10.1089/mdr.2017.0392
Mortaz, E., and Adcock, I. A. (2012). Limitation of COPD studies in animal modeling. Tanaffos 11, 7–8.
Mosser, D. M., and Edwards, J. P. (2008). Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 8, 958–969. doi: 10.1038/nri2448
Mou, H., Vinarsky, V., Tata, P. R., Brazauskas, K., Choi, S. H., Crooke, A. K., et al. (2016). Dual SMAD signaling inhibition enables long-term expansion of diverse epithelial basal cells. Cell Stem Cell 19, 217–231. doi: 10.1016/j.stem.2016.05.012
Mouse Genome Sequencing, C., Waterston, R. H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J. F., et al. (2002). Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562. doi: 10.1038/nature01262
Muzio, M., Bosisio, D., Polentarutti, N., D'Amico, G., Stoppacciaro, A., Mancinelli, R., et al. (2000). Differential expression and regulation of toll-like receptors (TLR) in human leukocytes: selective expression of TLR3 in dendritic cells. J. Immunol. 164, 5998–6004. doi: 10.4049/jimmunol.164.11.5998
Nanthakumar, N. N., Fusunyan, R. D., Sanderson, I., and Walker, W. A. (2000). Inflammation in the developing human intestine: a possible pathophysiologic contribution to necrotizing enterocolitis. Proc. Natl. Acad. Sci. U.S.A. 97, 6043–6048. doi: 10.1073/pnas.97.11.6043
Ng, K. W., Pearton, M., Coulman, S., Anstey, A., Gateley, C., Morrissey, A., et al. (2009). Development of an ex vivo human skin model for intradermal vaccination: tissue viability and Langerhans cell behaviour. Vaccine 27, 5948–5955. doi: 10.1016/j.vaccine.2009.07.088
Nikolic, M. Z., Caritg, O., Jeng, Q., Johnson, J. A., Sun, D., Howell, K. J., et al. (2017). Human embryonic lung epithelial tips are multipotent progenitors that can be expanded in vitro as long-term self-renewing organoids. Elife 6:e26575. doi: 10.7554/eLife.26575
Ning, C., Reynolds, R., Chen, J., Yager, C., Berry, G. T., Leslie, N., et al. (2001). Galactose metabolism in mice with galactose-1-phosphate uridyltransferase deficiency: sucklings and 7-week-old animals fed a high-galactose diet. Mol. Genet. Metab. 72, 306–315. doi: 10.1006/mgme.2001.3152
Nizet, V. (2006). Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr. Issues Mol. Biol. 8, 11–26.
Noel, G., Baetz, N. W., Staab, J. F., Donowitz, M., Kovbasnjuk, O., Pasetti, M. F., et al. (2017). A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 7:45270. doi: 10.1038/srep46790
Olsavsky, K. M., Page, J. L., Johnson, M. C., Zarbl, H., Strom, S. C., and Omiecinski, C. J. (2007). Gene expression profiling and differentiation assessment in primary human hepatocyte cultures, established hepatoma cell lines, and human liver tissues. Toxicol. Appl. Pharmacol. 222, 42–56. doi: 10.1016/j.taap.2007.03.032
O'Neill, A. T., Monteiro-Riviere, N. A., and Walker, G. M. (2008). Characterization of microfluidic human epidermal keratinocyte culture. Cytotechnology 56, 197–207. doi: 10.1007/s10616-008-9149-9
Oost, K. C., van Voorthuijsen, L., Fumagalli, A., Lindeboom, R. G. H., Sprangers, J., Omerzu, M., et al. (2018). Specific labeling of stem cell activity in human colorectal organoids using an ASCL2-responsive minigene. Cell Rep. 22, 1600–1614. doi: 10.1016/j.celrep.2018.01.033
Otte, J. M., Cario, E., and Podolsky, D. K. (2004). Mechanisms of cross hyporesponsiveness to Toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology 126, 1054–1070. doi: 10.1053/j.gastro.2004.01.007
Ouwehand, K., Spiekstra, S. W., Waaijman, T., Scheper, R. J., de Gruijl, T. D., and Gibbs, S. (2011). Technical advance: langerhans cells derived from a human cell line in a full-thickness skin equivalent undergo allergen-induced maturation and migration. J. Leukoc. Biol. 90, 1027–1033. doi: 10.1189/jlb.0610374
Pan, C., Kumar, C., Bohl, S., Klingmueller, U., and Mann, M. (2009). Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol. Cell Proteomics, 8, 443–450. doi: 10.1074/mcp.M800258-MCP200
Pardo-Saganta, A., Law, B. M., Tata, P. R., Villoria, J., Saez, B., Mou, H., et al. (2015). Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell 16, 184–197. doi: 10.1016/j.stem.2015.01.002
Park, N., Yamanaka, K., Tran, D., Chandrangsu, P., Akers, J. C., de Leon, J. C., et al. (2009). The cell-penetrating peptide, Pep-1, has activity against intracellular chlamydial growth but not extracellular forms of Chlamydia trachomatis. J. Antimicrob. Chemother. 63, 115–123. doi: 10.1093/jac/dkn436
Pasparakis, M., Haase, I., and Nestle, F. O. (2014). Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 14, 289–301. doi: 10.1038/nri3646
Pena, O. M., Hancock, D. G., Lyle, N. H., Linder, A., Russell, J. A., Xia, J., et al. (2014). An endotoxin tolerance signature predicts sepsis and organ dysfunction at initial clinical presentation. EBioMedicine 1, 64–71. doi: 10.1016/j.ebiom.2014.10.003
Pena, O. M., Pistolic, J., Raj, D., Fjell, C. D., and Hancock, R. E. (2011). Endotoxin tolerance represents a distinctive state of alternative polarization (M2) in human mononuclear cells. J. Immunol. 186, 7243–7254. doi: 10.4049/jimmunol.1001952
Pezzulo, A. A., Starner, T. D., Scheetz, T. E., Traver, G. L., Tilley, A. E., Harvey, B. G., et al. (2011). The air-liquid interface and use of primary cell cultures are important to recapitulate the transcriptional profile of in vivo airway epithelia. Am. J. Physiol. Lung Cell. Mol. Physiol. 300, L25–31. doi: 10.1152/ajplung.00256.2010
Phan, N., Hong, J. J., Tofig, B., Mapua, M., Elashoff, D., Moatamed, N. A., et al. (2019). A simple high-throughput approach identifies actionable drug sensitivities in patient-derived tumor organoids. Commun. Biol. 2:78. doi: 10.1038/s42003-019-0305-x
Phoenix, D.A., Dennison, S. R., and Harris, F. (2016). “Bacterial resistance to host defence peptides,” in Host Defense Peptides and Their Potential as Therapeutic Agents, ed R. M. Epand (Cham: Springer International Publishing), 161–204.
Pinsent, A., and Gambhir, M. (2017). Improving our forecasts for trachoma elimination: what else do we need to know? PLoS Negl. Trop. Dis. 11:e0005378. doi: 10.1371/journal.pntd.0005378
Piotrowska, U., Sobczak, M., and Oledzka, E. (2017). Current state of a dual behaviour of antimicrobial peptides-therapeutic agents and promising delivery vectors. Chem. Biol. Drug. Des. 90, 1079–1093. doi: 10.1111/cbdd.13031
Pletzer, D., Mansour, S. C., and Hancock, R. E. W. (2018). Synergy between conventional antibiotics and anti-biofilm peptides in a murine, sub-cutaneous abscess model caused by recalcitrant ESKAPE pathogens. PLoS Pathog. 14:e1007084. doi: 10.1371/journal.ppat.1007084
Ponec, M., El Ghalbzouri, A., Dijkman, R., Kempenaar, J., van der Pluijm, G., and Koolwijk, P. (2004). Endothelial network formed with human dermal microvascular endothelial cells in autologous multicellular skin substitutes. Angiogenesis 7, 295–305. doi: 10.1007/s10456-004-6315-3
Powers, J. P., and Hancock, R. E. (2003). The relationship between peptide structure and antibacterial activity. Peptides 24, 1681–1691. doi: 10.1016/j.peptides.2003.08.023
Price, A. E., Shamardani, K., Lugo, K. A., Deguine, J., Roberts, A. W., Lee, B. L., et al. (2018). A map of toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity 49, 560–575 e6. doi: 10.1016/j.immuni.2018.07.016
Ramadan, Q., and Ting, F. C. (2016). In vitro micro-physiological immune-competent model of the human skin. Lab. Chip 16, 1899–1908. doi: 10.1039/C6LC00229C
Rasmussen, A. L., Okumura, A., Ferris, M. T., Green, R., Feldmann, F., Kelly, S. M., et al. (2014). Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science 346, 987–991. doi: 10.1126/science.1259595
Regnier, M., Staquet, M. J., Schmitt, D., and Schmidt, R. (1997). Integration of Langerhans cells into a pigmented reconstructed human epidermis. J. Invest. Dermatol. 109, 510–512. doi: 10.1111/1523-1747.ep12336627
Reijnders, C. M., van Lier, A., Roffel, S., Kramer, D., Scheper, R. J., and Gibbs, S. (2015). Development of a full-thickness human skin equivalent in vitro model derived from TERT-immortalized keratinocytes and fibroblasts. Tissue Eng. Part A 21, 2448–2459. doi: 10.1089/ten.tea.2015.0139
Ritter, D., Knebel, J., Niehof, M., Loinaz, I., Marradi, M., Gracia, R., et al. (2020). In vitro inhalation cytotoxicity testing of therapeutic nanosystems for pulmonary infection. Toxicol. In Vitro 63:104714. doi: 10.1016/j.tiv.2019.104714
Rollin, B. E. (2003). Toxicology and new social ethics for animals. Toxicol. Pathol. 31(Suppl), 128–131. doi: 10.1080/01926230390175011
Rouaud-Tinguely, P., Boudier, D., Marchand, L., Barruche, V., Bordes, S., Coppin, H., et al. (2015). From the morphological to the transcriptomic characterization of a compromised three-dimensional in vitro model mimicking atopic dermatitis. Br. J. Dermatol. 173, 1006–1014. doi: 10.1111/bjd.14012
Rowley, J., Vander Hoorn, S., Korenromp, E., Low, N., Unemo, M., Abu-Raddad, L. J., et al. (2019). Chlamydia, gonorrhoea, trichomoniasis and syphilis: global prevalence and incidence estimates, 2016. Bull. World Health Organ. 97, 548–562. doi: 10.2471/BLT.18.228486
Rudilla, H., Fuste, E., Cajal, Y., Rabanal, F., Vinuesa, T., and Vinas, M. (2016). Synergistic antipseudomonal effects of synthetic peptide AMP38 and carbapenems. Molecules 21:1223. doi: 10.3390/molecules21091223
Sachdeva, S., Lobo, S., and Goswami, T. (2016). What is the future of noninvasive routes for protein- and peptide-based drugs? Ther. Deliv. 7, 355–357. doi: 10.4155/tde-2016-0031
Sachs, N., Papaspyropoulos, A., Zomer-van Ommen, D. D., Heo, I., Bottinger, L., Klay, D., et al. (2019). Long-term expanding human airway organoids for disease modeling. EMBO J. 38:e100300. doi: 10.15252/embj.2018100300
Sachs, N., Tsukamoto, Y., Kujala, P., Peters, P. J., and Clevers, H. (2017). Intestinal epithelial organoids fuse to form self-organizing tubes in floating collagen gels. Development 144, 1107–1112. doi: 10.1242/dev.143933
Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. doi: 10.1038/nature07935
Schulz, C., Gomez Perdiguero, E., Chorro, L., Szabo-Rogers, H., Cagnard, N., Kierdorf, K., et al. (2012). A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90. doi: 10.1126/science.1219179
Schwank, G., Koo, B. K., Sasselli, V., Dekkers, J. F., Heo, I., Demircan, T., et al. (2013). Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 13, 653–658. doi: 10.1016/j.stem.2013.11.002
Seo, M. D., Won, H. S., Kim, J. H., Mishig-Ochir, T., and Lee, B. J. (2012). Antimicrobial peptides for therapeutic applications: a review. Molecules 17, 12276–12286. doi: 10.3390/molecules171012276
Shaykhiev, R. (2015). Multitasking basal cells: combining stem cell and innate immune duties. Eur. Respir. J. 46, 894–897. doi: 10.1183/13993003.00521-2015
Shepherd, J., Douglas, I., Rimmer, S., Swanson, L., and MacNeil, S. (2009). Development of three-dimensional tissue-engineered models of bacterial infected human skin wounds. Tissue Eng. Part C Methods 15, 475–484. doi: 10.1089/ten.tec.2008.0614
Sieprawska-Lupa, M., Mydel, P., Krawczyk, K., Wojcik, K., Puklo, M., Lupa, B., et al. (2004). Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 48, 4673–4679. doi: 10.1128/AAC.48.12.4673-4679.2004
Smith, C. M., Finger, J. H., Hayamizu, T. F., McCright, I. J., Xu, J., Berghout, J., et al. (2014). The mouse Gene Expression Database (GXD): 2014 update. Nucl. Acids Res. 42, D818–D824. doi: 10.1093/nar/gkt954
Smits, J. P. H., Niehues, H., Rikken, G., van Vlijmen-Willems, I., van de Zande, G., Zeeuwen, P., et al. (2017). Immortalized N/TERT keratinocytes as an alternative cell source in 3D human epidermal models. Sci. Rep. 7:11838. doi: 10.1038/s41598-017-12041-y
Spence, J. R., Mayhew, C. N., Rankin, S. A., Kuhar, M. F., Vallance, J. E., Tolle, K., et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109. doi: 10.1038/nature09691
Springer, M., Engelhart, K., and Biesalski, H. K. (2003). Effects of 3-isobutyl-1-methylxanthine and kojic acid on cocultures and skin equivalents composed of HaCaT cells and human melanocytes. Arch. Dermatol. Res. 295, 88–91. doi: 10.1007/s00403-003-0401-z
Starr, T., Bauler, T. J., Malik-Kale, P., and Steele-Mortimer, O. (2018). The phorbol 12-myristate-13-acetate differentiation protocol is critical to the interaction of THP-1 macrophages with Salmonella typhimurium. PLoS ONE 13:e0193601. doi: 10.1371/journal.pone.0193601
Straus, S. K., and Hancock, R. E. (2006). Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim. Biophys. Acta 1758, 1215–1223. doi: 10.1016/j.bbamem.2006.02.009
Suzuki, K., Murano, T., Shimizu, H., Ito, G., Nakata, T., Fujii, S., et al. (2018). Single cell analysis of Crohn's disease patient-derived small intestinal organoids reveals disease activity-dependent modification of stem cell properties. J. Gastroenterol. 53, 1035–1047. doi: 10.1007/s00535-018-1437-3
Swaminathan, S., Kumar, V., and Kaul, R. (2019). Need for alternatives to animals in experimentation: an Indian perspective. Indian J. Med. Res. 149, 584–592. doi: 10.4103/ijmr.IJMR_2047_17
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., et al. (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. doi: 10.1016/j.cell.2007.11.019
Takahashi, K., and Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. doi: 10.1016/j.cell.2006.07.024
Takahashi, T. (2019). Organoids for drug discovery and personalized medicine. Annu. Rev. Pharmacol. Toxicol. 59, 447–462. doi: 10.1146/annurev-pharmtox-010818-021108
Tarique, A. A., Logan, J., Thomas, E., Holt, P. G., Sly, P. D., and Fantino, E. (2015). Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages. Am. J. Respir. Cell Mol. Biol. 53, 676–688. doi: 10.1165/rcmb.2015-0012OC
Tata, P. R., Mou, H., Pardo-Saganta, A., Zhao, R., Prabhu, M., Law, B. M., et al. (2013). Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature 503, 218–223. doi: 10.1038/nature12777
Thibaut de Menonville, S., Rosignoli, C., Soares, E., Roquet, M., Bertino, B., Chappuis, J. P., et al. (2017). Topical treatment of rosacea with ivermectin inhibits gene expression of cathelicidin innate immune mediators, LL-37 and KLK5, in reconstructed and ex vivo skin models. Dermatol. Ther. 7, 213–225. doi: 10.1007/s13555-017-0176-3
Tjabringa, G., Bergers, M., van Rens, D., de Boer, R., Lamme, E., and Schalkwijk, J. (2008). Development and validation of human psoriatic skin equivalents. Am. J. Pathol. 173, 815–823. doi: 10.2353/ajpath.2008.080173
Turner, J. R. (2009). Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 9, 799–809. doi: 10.1038/nri2653
Upadhyay, S., and Palmberg, L. (2018). Air-liquid interface: relevant in vitro models for investigating air pollutant-induced pulmonary toxicity. Toxicol. Sci. 164, 21–30. doi: 10.1093/toxsci/kfy053
Valdes, A. M., Walter, J., Segal, E., and Spector, T. D. (2018). Role of the gut microbiota in nutrition and health. BMJ 361:k2179. doi: 10.1136/bmj.k2179
van de Wetering, M., Francies, H. E., Francis, J. M., Bounova, G., Iorio, F., Pronk, A., et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945. doi: 10.1016/j.cell.2015.03.053
van den Bogaard, E. H., Tjabringa, G. S., Joosten, I., Vonk-Bergers, M., van Rijssen, E., Tijssen, H. J., et al. (2014). Crosstalk between keratinocytes and T cells in a 3D microenvironment: a model to study inflammatory skin diseases. J. Invest. Dermatol. 134, 719–727. doi: 10.1038/jid.2013.417
van der Does, A. M., Bergman, P., Agerberth, B., and Lindbom, L. (2012). Induction of the human cathelicidin LL-37 as a novel treatment against bacterial infections. J. Leukoc. Biol. 92, 735–742. doi: 10.1189/jlb.0412178
van Drongelen, V., Haisma, E. M., Out-Luiting, J. J., Nibbering, P. H., and El Ghalbzouri, A. (2014). Reduced filaggrin expression is accompanied by increased Staphylococcus aureus colonization of epidermal skin models. Clin. Exp. Allergy 44, 1515–1524. doi: 10.1111/cea.12443
van Kilsdonk, J. W., van den Bogaard, E. H., Jansen, P. A., Bos, C., Bergers, M., and Schalkwijk, J. (2013). An in vitro wound healing model for evaluation of dermal substitutes. Wound Repair. Regen. 21, 890–896. doi: 10.1111/wrr.12086
Vandamme, D., Landuyt, B., Luyten, W., and Schoofs, L. (2012). A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell Immunol. 280, 22–35. doi: 10.1016/j.cellimm.2012.11.009
Ventress, J. K., Partridge, L. J., Read, R. C., Cozens, D., MacNeil, S., and Monk, P. N. (2016). Peptides from Tetraspanin CD9 Are Potent Inhibitors of Staphylococcus aureus adherence to keratinocytes. PLoS ONE 11:e0160387. doi: 10.1371/journal.pone.0160387
Vickerman, V., Blundo, J., Chung, S., and Kamm, R. (2008). Design, fabrication and implementation of a novel multi-parameter control microfluidic platform for three-dimensional cell culture and real-time imaging. Lab Chip 8, 1468–1477. doi: 10.1039/b802395f
Wallmeyer, L., Dietert, K., Sochorova, M., Gruber, A. D., Kleuser, B., Vavrova, K., et al. (2017). TSLP is a direct trigger for T cell migration in filaggrin-deficient skin equivalents. Sci. Rep. 7:774. doi: 10.1038/s41598-017-00670-2
Wang, G., Li, X., and Wang, Z. (2016). APD3: the antimicrobial peptide database as a tool for research and education. Nucl. Acids Res. 44, D1087–D1093. doi: 10.1093/nar/gkv1278
Wang, L., Huang, D., Huang, C., Yin, Y., Vali, K., Zhang, M., et al. (2017). Enhanced human somatic cell reprogramming efficiency by fusion of the MYC transactivation domain and OCT4. Stem Cell Res. 25, 88–97. doi: 10.1016/j.scr.2017.10.014
Wang, Y. I., Oleaga, C., Long, C. J., Esch, M. B., McAleer, C. W., Miller, P. G., et al. (2017). Self-contained, low-cost Body-on-a-Chip systems for drug development. Exp. Biol. Med. 242, 1701–1713. doi: 10.1177/1535370217694101
Wilson, S. S., Tocchi, A., Holly, M. K., Parks, W. C., and Smith, J. G. (2015). A small intestinal organoid model of non-invasive enteric pathogen-epithelial cell interactions. Mucosal. Immunol. 8, 352–361. doi: 10.1038/mi.2014.72
Workman, M. J., Mahe, M. M., Trisno, S., Poling, H. M., Watson, C. L., Sundaram, N., et al. (2017). Engineered human pluripotent-stem-cell-derived intestinal tissues with a functional enteric nervous system. Nat. Med. 23, 49–59. doi: 10.1038/nm.4233
World Health Organization (2015). Global Priority List of Antibiotic Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva: World Health Organization
World Health Organization (2017). WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Geneva.
Wright, J. L., Cosio, M., and Churg, A. (2008). Animal models of chronic obstructive pulmonary disease. Am. J. Physiol. Lung Cell Mol. Physiol. 295, L1–15. doi: 10.1152/ajplung.90200.2008
Wu, X., Li, Z., Li, X., Tian, Y., Fan, Y., Yu, C., et al. (2017). Synergistic effects of antimicrobial peptide DP7 combined with antibiotics against multidrug-resistant bacteria. Drug Des. Devel. Ther. 11, 939–946. doi: 10.2147/DDDT.S107195
Wufuer, M., Lee, G., Hur, W., Jeon, B., Kim, B. J., Choi, T. H., et al. (2016). Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 6:37471. doi: 10.1038/srep37471
Xu, H., Jiao, Y., Qin, S., Zhao, W., Chu, Q., and Wu, K. (2018). Organoid technology in disease modelling, drug development, personalized treatment and regeneration medicine. Exp. Hematol. Oncol. 7:30. doi: 10.1186/s40164-018-0122-9
Yamamoto, Y., Gotoh, S., Korogi, Y., Seki, M., Konishi, S., Ikeo, S., et al. (2017). Long-term expansion of alveolar stem cells derived from human iPS cells in organoids. Nat. Methods 14, 1097–1106. doi: 10.1038/nmeth.4448
Yeung, A. T. Y., Choi, Y. H., Lee, A. H. Y., Hale, C., Ponstingl, H., Pickard, D., et al. (2019). A genome-wide knockout screen in human macrophages identified host factors modulating Salmonella infection. mBio 10:e02169-19. doi: 10.1128/mBio.02169-19
Yeung, A. T. Y., Hale, C., Lee, A. H., Gill, E. E., Bushell, W., Parry-Smith, D., et al. (2017). Exploiting induced pluripotent stem cell-derived macrophages to unravel host factors influencing Chlamydia trachomatis pathogenesis. Nat. Commun. 8:15013. doi: 10.1038/ncomms15013
Yin, X., Farin, H. F., van Es, J. H., Clevers, H., Langer, R., and Karp, J. M. (2014). Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods 11, 106–112. doi: 10.1038/nmeth.2737
Yui, S., Azzolin, L., Maimets, M., Pedersen, M. T., Fordham, R. P., Hansen, S. L., et al. (2018). YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22, 35–49 e7. doi: 10.1016/j.stem.2017.11.001
Zhang, Y. S., Aleman, J., Shin, S. R., Kilic, T., Kim, D., Mousavi Shaegh, S. A., et al. (2017). Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. U.S.A. 114, E2293–E2302. doi: 10.1073/pnas.1612906114
Zhao, Y., Kankala, R. K., Wang, S. B., and Chen, A. Z. (2019). Multi-organs-on-chips: towards long-term biomedical investigations. Molecules 24:675. doi: 10.3390/molecules24040675
Zhen, X., Lundborg, C. S., Sun, X., Hu, X., and Dong, H. (2019). Economic burden of antibiotic resistance in ESKAPE organisms: a systematic review. Antimicrob. Resist. Infect. Control. 8:137. doi: 10.1186/s13756-019-0590-7
Zhou, Y. Q., Shi, Y., Yang, L., Sun, Y. F., Han, Y. F., Zhao, Z. X., et al. (2019). Genetically engineered distal airway stem cell transplantation protects mice from pulmonary infection. EMBO Mol. Med. 12:e10233. doi: 10.15252/emmm.201810233
Zhu, S., Li, W., Zhou, H., Wei, W., Ambasudhan, R., Lin, T., et al. (2010). Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell 7, 651–655. doi: 10.1016/j.stem.2010.11.015
Zhu, Z., Schnell, L., Muller, B., Muller, M., Papatheodorou, P., and Barth, H. (2019). The antibiotic bacitracin protects human intestinal epithelial cells and stem cell-derived intestinal organoids from Clostridium difficile toxin TcdB. Stem Cells Int. 2019:4149762. doi: 10.1155/2019/4149762
Ziegler-Heitbrock, L., Ancuta, P., Crowe, S., Dalod, M., Grau, V., Hart, D. N., et al. (2010). Nomenclature of monocytes and dendritic cells in blood. Blood 116, e74–80. doi: 10.1182/blood-2010-02-258558
Zschaler, J., Schlorke, D., and Arnhold, J. (2014). Differences in innate immune response between man and mouse. Crit. Rev. Immunol. 34, 433–454. doi: 10.1615/CritRevImmunol.2014011600
Zuyderduyn, S., Ninaber, D. K., Schrumpf, J. A., van Sterkenburg, M. A., Verhoosel, R. M., Prins, F. A., et al. (2011). IL-4 and IL-13 exposure during mucociliary differentiation of bronchial epithelial cells increases antimicrobial activity and expression of antimicrobial peptides. Respir. Res. 12:59. doi: 10.1186/1465-9921-12-59
Keywords: host-defense peptide, organoid, air-liquid interface, disease model, drug screening, antimicrobial resistant organisms
Citation: Choi K-YG, Wu BC, Lee AH-Y, Baquir B and Hancock REW (2020) Utilizing Organoid and Air-Liquid Interface Models as a Screening Method in the Development of New Host Defense Peptides. Front. Cell. Infect. Microbiol. 10:228. doi: 10.3389/fcimb.2020.00228
Received: 10 February 2020; Accepted: 23 April 2020;
Published: 20 May 2020.
Edited by:
Florie Desriac, Université de Caen Normandie, FranceReviewed by:
Sadri Znaidi, Institut Pasteur de Tunis, TunisiaCopyright © 2020 Choi, Wu, Lee, Baquir and Hancock. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert E. W. Hancock, Ym9iQGhhbmNvY2tsYWIuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.