- School of Medicine, Medical Sciences and Nutrition, Institute of Medical Sciences, University of Aberdeen, Aberdeen, United Kingdom
Candida species are known to differ in their ability to cause infection and have been shown to display varied susceptibilities to antifungal drugs. Treatment with the echinocandin, caspofungin, leads to compensatory alterations in the fungal cell wall. This study was performed to compare the structure and composition of the cell walls of different Candida species alone and in response to caspofungin treatment, and to evaluate how changes at the fungal cell surface affects interactions with macrophages. We demonstrated that the length of the outer fibrillar layer varied between Candida species and that, in most cases, reduced fibril length correlated with increased exposure of β-1,3-glucan on the cell surface. Candida glabrata and Candida guilliermondii, which had naturally more β-1,3-glucan exposed on the cell surface, were phagocytosed significantly more efficiently by J774 macrophages. Treatment with caspofungin resulted in increased exposure of chitin and β-1,3-glucan on the surface of the majority of Candida species isolates that were tested, with the exception of C. glabrata and Candida parapsilosis isolates. This increase in exposure of the inner cell wall polysaccharides, in most cases, correlated with reduced uptake by macrophages and in turn, a decrease in production of TNFα. Here we show that differences in the exposure of cell wall carbohydrates and variations in the repertoire of covalently attached surface proteins of different Candida species contributes to their recognition by immune cells.
Introduction
Candida species differ in their ability to cause infection. Candida albicans is the most common cause of Candida bloodstream infections (40%), followed by Candida glabrata (29%), Candida parapsilosis (11%), Candida tropicalis (4%), Candida dubliniensis (2%), and Candida lusitaniae (<1%) (Data captured from England; Health Protection Report, 2018). Candida species also have varied susceptibilities to antifungal drugs. The echinocandins act by specifically inhibiting the synthesis of β-1,3-glucan in the fungal cell wall. The inhibition of β-1,3-glucan synthesis occurs predominantly through inhibition of the catalytic Fks glucan synthase subunits (Kurtz and Douglas, 1997). Caspofungin is one of the most widely used of the echinocandins in the clinic and has fungicidal activity against the majority of Candida species. C. lusitaniae, C. parapsilosis, and Candida guilliermondii are known to have relatively reduced susceptibility compared to C. albicans and in recent years the incidence of clinical isolates of C. glabrata, which have acquired resistance to the echinocandins has increased (Garcia-Effron et al., 2010; Pfaller et al., 2013). Alarmingly echinocandin-resistant C. glabrata isolates (up to 38%) were also cross-resistant to fluconazole (Pfaller et al., 2012, 2013). Acquired resistance is predominantly mediated by point mutations within hotspot regions in the FKS genes (Park et al., 2005; Balashov et al., 2006; Garcia-Effron et al., 2010; Alexander et al., 2013; Pham et al., 2014; Marti-Carrizosa et al., 2015).
The fungal cell wall determines cell shape, maintains cell wall integrity and is recognized by the innate immune system. The cell walls of Candida spp. in general are composed of an inner core of chitin and β-1,3-glucan, which is covered by an outer layer of cell wall proteins, the majority of which are covalently linked to β-1,6-glucan by modified glycosylphosphatidylinositol (GPI) anchors (Gow et al., 2017). The cell wall is a dynamic structure which alters its composition in response to cell wall stress by upregulating genes involved in cell wall synthesis, in an attempt to restore the robustness of the cell wall (Walker et al., 2008). Treatment of C. albicans with caspofungin has been shown to lead to a compensatory increase in chitin content, in vitro and in vivo (Walker et al., 2008; Lee et al., 2012). This compensatory increase in chitin is not specific to C. albicans as C. tropicalis, C. parapsilosis, C. guilliermondii, and isolates of C. krusei also demonstrated an elevation in chitin content in response to caspofungin treatment (Walker et al., 2013). In addition, isolates of C. albicans, C. krusei, C. parapsilosis, and C. guilliermondii, which have increased chitin content are less susceptible to caspofungin (Walker et al., 2008, 2013). C. albicans cells with elevated chitin contents have also been shown to be less susceptible to caspofungin in a murine model of systemic infection (Lee et al., 2012).
Putative GPI-modified cell wall proteins have been implicated in susceptibility to caspofungin as deletion of specific proteins leads to alterations in cell wall composition and subsequently to differences in susceptibility to caspofungin (Plaine et al., 2008). As a result of the cell wall remodeling that occurs in response to caspofungin treatment, chitin and β-1,3-glucan also become more exposed on the cell surface (Wheeler and Fink, 2006; Wheeler et al., 2008; Mora-Montes et al., 2011).
The fungal cell wall plays an important role in immune recognition as it is the first point of contact between the host and pathogen. The main innate immune cells that are involved in the recognition of invading pathogens are neutrophils, monocytes and macrophages (Netea et al., 2008). Components of the cell wall act as pathogen associated molecular patterns (PAMPs), which are recognized by pattern recognition receptors (PRRs) on host cells (Brown and Gordon, 2001; Porcaro et al., 2003; Kohatsu et al., 2006; McGreal et al., 2006; Netea et al., 2006, 2008). The two main classes of PRRs are the Toll-like receptors (TLRs) and the C-type lectin receptors (CLRs). The TLRs recognize phospholipomannan and O-linked mannan, whereas the C-type lectin receptors recognize β-1,3-glucan and mannan (Stahl et al., 1978; Wileman et al., 1986; Jouault et al., 2003; McGreal et al., 2006; Netea et al., 2006, 2008; Sato et al., 2006; Gow et al., 2007; Taylor et al., 2007). The increased exposure of chitin and β-1,3-glucan on the cell surface of C. albicans, in response to caspofungin treatment, results in altered cytokine production by immune cells, indicating that remodeling of the cell wall influences interactions with host cells (Wheeler and Fink, 2006; Wheeler et al., 2008; Mora-Montes et al., 2011; Baltch et al., 2012a,b; Fidan et al., 2014). Caspofungin treatment of C. albicans cells, followed by UV inactivation led to increased recognition of fungal cells by the C-type lectin, Dectin-1, which in turn increased cytokine production (Wheeler and Fink, 2006; Wheeler et al., 2008). In contrast, increased exposure of chitin on the surface of C. albicans has been shown to result in reduced cytokine production (Mora-Montes et al., 2011). Naturally occurring variations in the cell walls of different C. albicans isolates influences the dependency on dectin-1 for recognition and clearance of fungal cells (Marakalala et al., 2013). In addition the role of dectin-1 in recognition of four different Candida species, with differences in their cell wall carbohydrate composition, also varies (Thompson et al., 2019).
Because different Candida species vary in their ability to cause infection and vary in their cell wall composition, we aimed to perform a detailed comparison of the cell wall architectures of seven most prevalent Candida spp. and examine how their surface carbohydrates and proteomes change in response to caspofungin (CSF) treatment. In addition, the effect of caspofungin-induced cell wall changes of the different Candida species on the response of host immune cells was investigated.
Materials and Methods
Strains, Media, and Growth Conditions
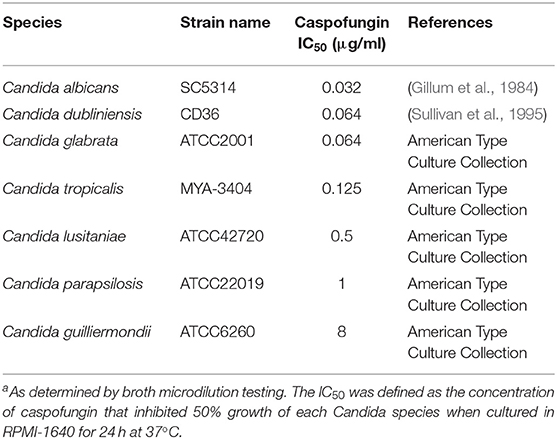
The strains of each Candida species that were used in this study had been typed and their genomes sequenced, these strains are listed in Table 1. Strains were maintained on solid YPD medium [1% (w/v) yeast extract, 2% (w/v) mycological peptone, 2% (w/v) glucose, 2% (w/v) agar]. RPMI-1640 medium (Gibco, Paisley, UK) was used for drug susceptibility testing and for growing cultures for HPLC and proteomic analysis.
Preparation of FITC-Stained Candida spp
Staining of Candida spp. yeast cells with fluorescein isothiocyanate (FITC) was performed as previously described (Graham et al., 2006). Briefly, Candida spp. were grown in YPD in the presence or absence of an IC50 concentration of caspofungin (Walker et al., 2013) in YPD at 30°C for 6 h. Candida spp. cells were washed twice with PBS and stained for 10 min at room temperature in the dark with 1 mg/ml fluorescein isothiocyanate (FITC) (Sigma, Dorset, United Kingdom) in 0.05 M carbonate-bicarbonate buffer (pH 9.6) (BDH Chemicals, VWR International, Leicestershire, United Kingdom). Cells were then washed three times in 1x PBS to remove residual FITC and were resuspended in 1x PBS. FITC-stained 6x105 Candida spp. cells that had been pre-grown with or without IC50 caspofungin were then added to the macrophages at a multiplicity of infection (MOI) of 3 Candida spp. cells per macrophage, and incubated at 37°C for 4 h before visualization.
Phagocytosis and Cytokine Assays
Macrophage Cell Culture
J774.1 murine macrophages (European Collection of Cell Culture) were cultured in Dulbecco's modified Eagle's medium (DMEM; Lonza Group, Ltd., Braine-l'Alleud, Belgium), supplemented with 10% (vol/vol) fetal calf serum, 2% (wt/vol) penicillin and streptomycin antibiotics (Invitrogen, Ltd., Paisley, United Kingdom) in tissue culture flasks (Nalge Nunc, International, Hereford, United Kingdom) at 37°C and 5% (vol/vol) CO2. For phagocytosis assays, 2 × 105 J774.1 macrophages in 2 ml supplemented DMEM medium were seeded onto glass-based imaging dishes [Imaging dish CG 1.0; MACS Miltenyi Biotec (130-098-282), Surrey, UK] and incubated at 37°C with 5% CO2 for at least 2 h to allow for macrophage adherence to the dish. Immediately prior to experiments, DMEM medium was replaced with 2 ml pre-warmed supplemented CO2-independent medium (Gibco, Invitrogen, Paisley, UK) and cultures used immediately for phagocytosis assays.
After 4 h of co-incubation 50 μl of supernatant was removed from phagocytosis assays to allow determination of TNFα production from J774 macrophages as a result of incubation with each Candida species. TNFα concentrations were determined using enzyme-linked immunosorbent assays (R&D Systems) according to the manufacturer's instructions.
Cell Wall Staining of Exposed Chitin and Glucan
Candida spp. yeast cells were grown in YPD in the presence or absence of an IC50 concentration of caspofungin in YPD at 30°C for 6 h. Cells were stained with 5 μg/ml Fc:Dectin1 protein to visualize exposed β-1,3-glucan, as previously described (52). Briefly, 1 × 106 cells/sample were blocked with FACS block (0.5% BSA, 5% HI-rabbit serum, 5 mM ETDA, 2 mM NaAzide in PBS) for 30 min. Cell pellets were harvested and washed 3 × with 1 ml FACS wash (0.5% BSA, 5 mM EDTA, 2 mM NaAzide in PBS) at 4°C. After washing, the cell pellet was resuspended in 100 μl of 5 μg/ml Fc:Dectin1 protein (diluted from stock in FACS block) and incubated for 1 h on ice. Cells were then washed 3 times with 1 ml FACS wash and resuspended in 200 μl FACS block plus 1/200 anti-human Fc+Alexa-488 and incubated for 45 min on ice. After incubation cells were washed three times with 1 ml FACS wash and fixed with 200 μl 1% formaldehyde. After fixing cells were also stained with 25 μg/ml Wheat Germ Agglutinin conjugated to Texas Red (WGA-TR). All samples were examined by DIC and fluorescence microscopy using a Zeiss Axioplan 2 microscope. Images were recorded digitally using the Openlab system (Openlab v 4.04, Improvision, Coventry, UK) and a Hamamatsu C4742- 95 digital camera (Hamamatsu Photonics, Hamamatsu, Hertfordshire, UK). In all experiments the exposure time for a series of fluorescence images was fixed so the intensity of fluorescence relative to a control was used as standard. Chitin and β-1,3-glucan exposure were measured by quantifying WGA-TR and Fc:Dectin1-488 fluorescence, respectively, of individual yeast cells (Walker et al., 2008). Mean fluorescence intensities were calculated for 50 individual cells of each Candida species, for each condition.
High-Pressure Freezing (HPF)-Transmission Electron Microscopy (TEM)
Candida spp. yeast cells were grown in YPD at 30°C for 6 h and HPF was carried out as described previously (Walker et al., 2010) with the following modifications. Briefly, samples were prepared by high-pressure freezing with an EMPACT2 high-pressure freezer and rapid transport system (Leica Microsystems Ltd., Milton Keynes, United Kingdom). After freezing, cells were freeze-substituted in substitution reagent (1% [wt/vol] OsO4 in acetone) with a Leica EMAFS2. Samples were then embedded in Epoxy resin and additional infiltration was provided under a vacuum at 60°C before embedding in Leica FSP specimen containers and polymerizing at 60°C for 48 h. Semithin survey sections, 0.5 μm thick, were stained with 1% toluidine blue to identify areas containing cells. Ultrathin sections (60 nm) were prepared with a Diatome diamond knife on a Leica UC6 ultramicrotome and stained with uranyl acetate and lead citrate for examination with a Philips CM10 transmission microscope (FEI UK Ltd., Cambridge, United Kingdom) and imaging with a Gatan Bioscan 792 (Gatan United Kingdom, Abingdon, United Kingdom). Image J was used to measure the thickness of the inner (chitin and glucan) and outer cell wall by averaging measurements for 30 cells, for each condition.
Antifungal Susceptibility Testing
CSF (Merck Research Laboratories, New Jersey, USA) minimum inhibitory concentrations were determined by broth micro-dilution testing using the CLSI guidelines M27-A3. Drug concentrations ranged from 0.016 to 16 μg/ml CSF. Exponentially grown cultures were diluted to 2 × 106 cells/ml in 2x RPMI-1640 and 100 μl of culture was added to each well. Plates were incubated for 24 h at 37°C. After incubation, optical densities were read in a VERSAmax tunable microplate reader (Molecular Devices, California, USA) at 405 nm.
HPLC Analysis of Cell Wall Composition
Cell walls were extracted as described previously (Mora-Montes et al., 2011). Briefly, each Candida species was grown in RPMI-1640 for 3 h at 37°C with shaking at 200 rpm. Each Candida species was then subsequently treated with a sub-MIC concentration of CSF (as determined from Table 1) for an additional 2 h, or grown without addition of drug for a further 2 h, as an untreated control. Cells were collected by centrifugation at 4,000 g for 5 min, washed once with chilled deionized water, resuspended in deionized water, and physically fractured with glass beads in a FastPrep machine (Qbiogene). The lysed cells were collected by centrifugation at 4,000 g for 5 min. The pellet containing the cell debris and walls was washed five times with 1 M NaCl, resuspended in extraction buffer (500 mM Tris-HCl buffer, pH 7.5, 2% [wt/vol] SDS, 0.3 M β-mercaptoethanol, and 1 mM EDTA), boiled at 100°C for 10 min, and freeze-dried. To quantify glucan, mannan, and chitin the cell walls were acid hydrolyzed as previously described (Lee et al., 2012). The hydrolyzed samples were analyzed by high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) in a carbohydrate analyzer system from Dionex (Surrey, United Kingdom) as described previously (Plaine et al., 2008). The total concentration of each cell wall component was expressed as μg per mg of dried cell wall which was determined by calibration from the standard curves of glucosamine, glucose, and mannose monomers, and converted to a percentage of the total cell wall.
Proteomic Analysis
Analysis of cell wall proteins was as described previously (Dutton et al., 2014). Candida species were grown with or without CSF as described for the HPLC analysis above. Briefly, 2 mg of freeze dried cell wall was mixed with 0.5 M ammonium bicarbonate in water containing 3 mM dithiothreitol and heated at 60°C for 20 min. Iodoacetamide (30 μl of 55 mM stock solution) was then added, and the suspension was incubated in the dark, at 25°C for 10 min. Following addition of 30 μl of 20 mg/ml trypsin, the suspension was incubated at 37°C for 14 h and centrifuged at 14,000 × g for 10 min. The supernatant was freeze-dried and extracted with 10% formic acid, and peptides were purified using ZipTip mC18 pipette tips (Millipore) and dissolved in 0.1% formic acid.
Samples (3 μl) were injected into an LC-MS system which comprised an UltiMate 3000 LC instrument (Dionex Ltd., United Kingdom) fitted with a PepSwift monolithic poly(styrene-codivinylbenzene) (PS-DVB) column (200 μm inside diameter [i.d.] by 5 cm; Dionex) coupled to an HCTultra ion trap mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany) fitted with a low-flow nebulizer in the electrospray ionization (ESI) source and controlled by HyStar software (version 4.0; Bruker Daltonik). Peptides were separated at a flow rate of 2 μl/min using a linear gradient of 0–40% acetonitrile-water-formic acid (80:20:0.04) (solvent B) in water-acetonitrile-formic acid (97:3:0.05) (solvent A) over 40 min, followed by a 1 min column wash in 90% solvent B and a 12-min equilibration step in solvent A. MS/MS data (scan range, m/z 100–2,200; averages 2) were acquired in positive data-dependent AutoMS(n) mode using the esquireControl software program (version 6.2; Bruker Daltonik). Up to three precursor ions were selected from the MS scan (range, m/z 300–1,500; averages 3) in each AutoMS(n) cycle. Precursors were actively excluded after being selected twice within a 1-min window, and singly charged ions were also excluded. Peptide peaks were detected (maximum of 9,999 compounds above an intensity threshold of 50,000) and deconvoluted automatically using Data Analysis software (version 3.4; Bruker Daltonik). Mass lists in the form of Mascot Generic Format (*. mgf) files were created automatically and used as inputs to Mascot MS/MS ion searches via a local Mascot server (version 2.2; Matrix Science, London, United Kingdom) with a database built from sequence files available from Candida Genome Database (32). The Search parameters used were the following: enzyme = trypsin; fixed modifications = carbamidomethyl (C); variable modifications = oxidation (M); mass values = monoisotopic; peptide mass tolerance = 1.5 Da; fragment mass tolerance = 0.5 Da; max missed cleavages = 1; instrument type = ESI-TRAP. Search results were displayed by Mascot after selection of the following parameters: standard scoring; require bold red; ion score or expect cutoff = 0.05.
Statistical Analyses
Results from independent replicate experiments are expressed as means ± SD. One-way ANOVA with a Dunnett's post-hoc test was used for statistical analysis. Significance was determined as p < 0.05.
Results
IC50 of Candida spp to Caspofungin
The susceptibility of each of the sequenced Candida spp. strains, used in this study, to CSF was determined following the CLSI-M27-A3 guidelines. C. albicans, C. dublinensis, C. glabrata, and C. tropicalis were the most susceptible to CSF (Table 1). The IC50 range for these susceptible species was between 0.032 and 0.064 μg/ml CSF (Table 1). C. lusitaniae and C. parapsilosis had CSF IC50 measurements of 0.5 and 1 μg/ml, respectively, which was classified as intermediate susceptibility and C. guilliermondii was resistant to CSF (Table 1). Therefore, the order of susceptibility to CSF from most susceptible to most resistant of the isolates of the Candida spp. tested here was: C. albicans > C. dubliniensis > C. glabrata > C. tropicalis > C. lusitaniae > C. parapsilosis > C. guilliermondii.
Candida Species Have Differences in the Ultrastructure of Their Cell Wall
Ultrastructural differences in the cell wall of each Candida species were investigated using high pressure freezing transmission electron microscopy (TEM) which conserves the fungal cell wall architecture. TEM analysis revealed that the inner core of the cell wall of C. glabrata (Figure 1A, v) and C. tropicalis (Figure 1A, ii) isolates was significantly thicker than the inner core of C. albicans isolate (Figure 1A, i; Figure 1B). In contrast to this the inner core of the C. guilliermondii cell wall (Figure 1A, vii) was significantly thinner, in comparison to C. albicans (Figure 1A, i; Figure 1B). Overall, the largest difference in the cell wall between the Candida species was in the outer fibrillar layer (Figure 1B). The variations in length of the outer fibrillar layer were classified as either long (>65 nm), intermediate (30–65 nm), or short (<30 nm) (Figure 1B). C. albicans and C. tropicalis had the longest outer fibrillar layers, followed by C. dubliniensis and C. lusitaniae which were classified as having intermediate fibril length (Figure 1B). C. glabrata, C. parapsilosis, and C. guilliermondii isolates had short outer fibrils on the cell surface, in comparison to the C. albicans isolate (Figure 1B).
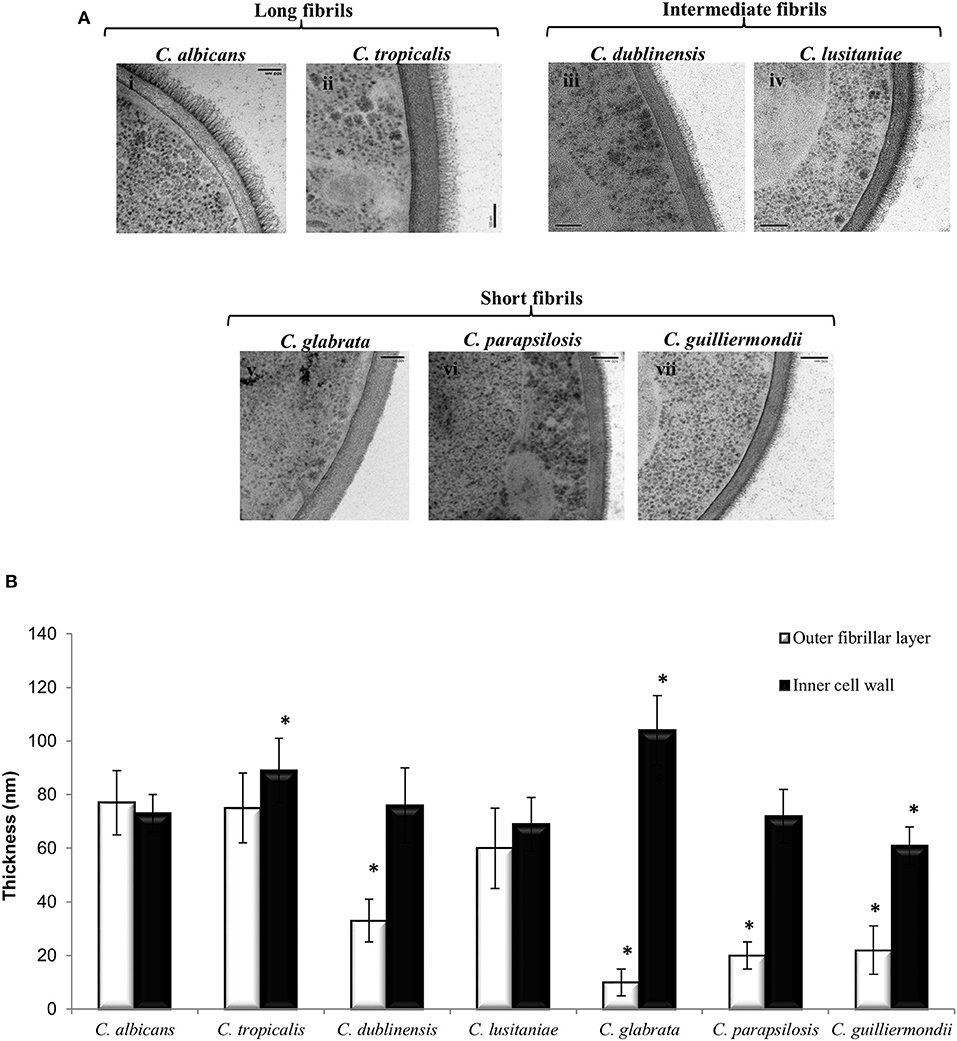
Figure 1. The ultrastructure of the cell wall varies between Candida species. (A) TEMs of the cell wall of different Candida species: C. albicans (i), C. tropicalis (ii), C. dublinensis (iii), C. lusitaniae (iv), C. glabrata (v), C. parapsilosis (vi) and C. guilliermondii (vii). Scale bars are 100 nm. (B) The thickness of the inner cell wall (glucan and chitin) and outer fibrillar layer was determined from TEM pictures. Measurements from 30 cells that were sectioned so that cell wall thickness was even all around the cell periphery. Significant differences (*p < 0.05) compared to C. albicans. Error bars are SD (n = 30).
Candida Species Have Species Specific GPI-Anchored Proteins Present on Their Cell Surface
To determine the differences in the cell wall proteome between the different Candida species, LC-MS/MS was used to identify which covalently attached, predicted GPI-modified proteins were present on the surface of each Candida species when they were cultured in RPMI-1640 medium (Table 2). Here we have annotated the proteins according to their C. albicans orthologs based on sequence similarity according to Candida Genome Database (Skrzypek et al., 2016). A core set of predicted cell wall proteins were identified in all species (Table 2). The hyphal specific protein, Hyr1, was uniquely detected in the cell wall of C. albicans, commensurate with it being a C. albicans-specific hyphal-associated protein and C. albicans will undergo filamentation in RPMI-1640 medium. GPI-anchored proteins that were identified specifically in the cell wall of C. dubliniensis included two Als proteins involved in adhesion and the secreted aspartyl protease, Sap9 (Table 2). A fungal-specific phospholipase and Pga59 orthologs were detected only on the cell surface of C. lusitaniae (Table 2). The cell wall of C. parapsilosis was found to contain orthologs of the yeast wall protein, Ywp1/Pga24, and Pga30, a protein of unknown function (Table 2).
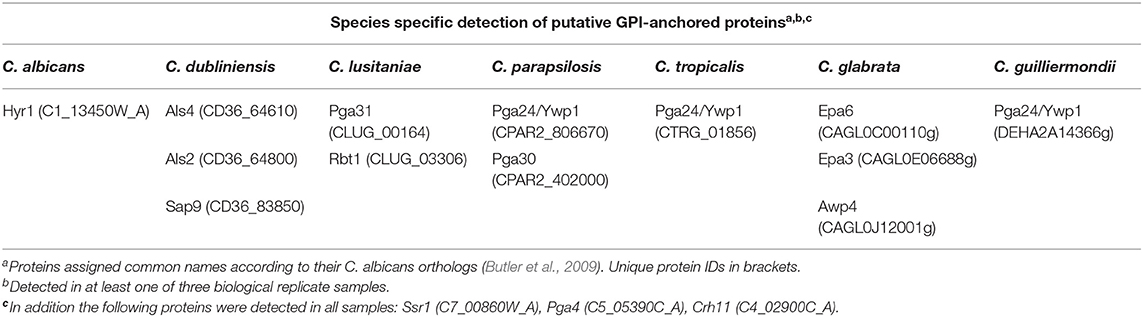
Table 2. Candida species-specific detection of cell wall GPI-anchored proteins by LC/MS/MS proteomics.
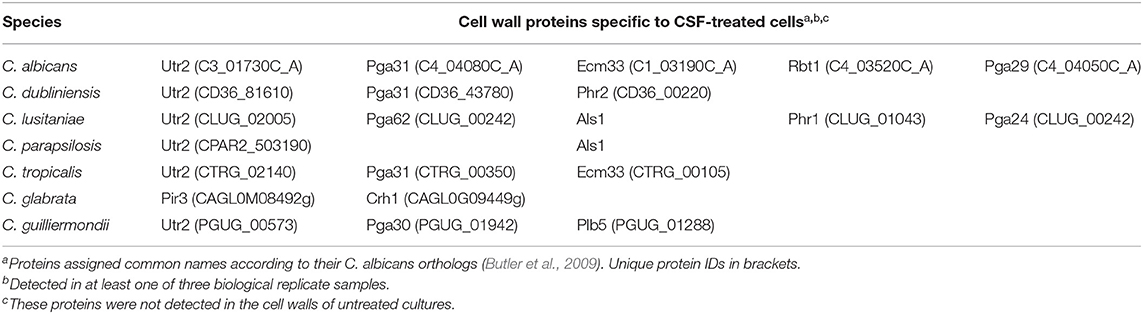
Caspofungin treatment has been previously shown to alter cell wall architecture and increases chitin production in C. albicans, C. tropicalis, C. krusei, C. parapsilosis, and C. guilliermondii (9). The effect of CSF treatment on the cell wall proteome of each Candida species was also investigated by treating each Candida spp. with their IC50 concentration of caspofungin (Table 3). Certain proteins were detected only at the cell surface in response to CSF treatment in addition to the proteins listed that are found on the surface of untreated cells. The proteins expressed in response to caspofungin varied between the different Candida spp. Orthologs of the transglycosidase, Utr2, were detected on the cell surface of the majority of Candida species in response to CSF treatment with the exception of C. glabrata (Table 3). Orthologs of Pga31, a GPI-anchored protein of unknown function was detected on the surface of the species that were most sensitive to CSF, such as C. albicans, C. dublinensis, and C. tropicalis in response to CSF treatment (Table 3). A protein related to Pga30, a member of the same family as Pga31 was detected in the cell wall of C. guilliermondii treated with CSF. C. lusitaniae and C. parapsilosis, which have intermediate susceptibility to CSF, were found to have Als-like proteins on their cell surface in response to CSF treatment (Table 3).
Cell Wall Composition of Different Candida Species in the Presence or Absence of CSF
The carbohydrate composition of the cell walls of the different Candida spp., in the presence or absence of an IC50 concentration of caspofungin, was determined by acid hydrolysis of isolated cell walls and analysis by high-performance liquid chromatography. C. tropicalis had substantially higher chitin content than C. albicans, whereas C. glabrata and C. guilliermondii had significantly less chitin compared to C. albicans (Table 4). The majority of Candida spp. had comparable levels of glucan in the cell wall, with the exception of C. tropicalis, which had significantly reduced glucan compared to C. albicans and C. glabrata, which had a higher glucan content (Table 4). The quantity of mannan in the cell wall was significantly reduced in C. glabrata, C. parapsilosis, and C. guilliermondii compared to C. albicans (Table 4) in agreement with the TEM analysis which suggested these species have shorter mannan fibrils on their outer surfaces. In most cases treatment with CSF resulted in an increase in chitin content, with the exception of C. glabrata and C. parapsilosis (Table 4). As expected all Candida spp. isolates tested had a reduction in β-1,3-glucan in response to CSF treatment. Exposure to an IC50 concentration of CSF led to a significant increase in mannan content in all Candida spp., with the exception of C. tropicalis and C. dublinensis isolates (Table 4).
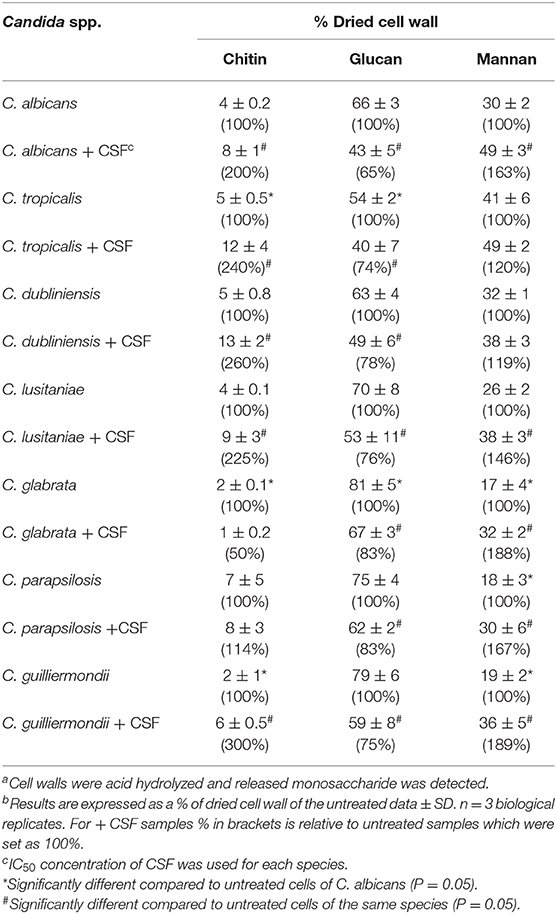
Table 4. Quantification of the components of the cell wall of Candida species, with and without caspofungin treatment a,b,c,d,e.
Candida Species Have Differences in Surface Exposure of Chitin and Glucan in Response to CSF Treatment
Exposure of chitin and glucan on the cell surface is known to influence the immune recognition of fungal cells (Wheeler and Fink, 2006; Wheeler et al., 2008; Mora-Montes et al., 2011). Fluorescent staining was used to determine whether the differences in outer fibril length between the Candida species isolates correlated with variations in exposure of chitin and β-1,3-glucan on the cell surface. The exposure of chitin was determined using 25 μg/ml Wheat Germ Agglutinin-Texas Red (WGA-TR) and exposed β-1,3-glucan was stained with 5 μg/ml Fc:Dectin1- Alexa 488 (Table 5, Supplementary Figure 1). The exposure of chitin and β-1,3-glucan, after treatment with an IC50 concentration of CSF for 6 h, was determined for each Candida species isolate. Exposure of β-1,3-glucan was significantly higher on the surface of untreated C. glabrata and C. guilliermondii compared to C. albicans (Table 5). Treatment with CSF, at the IC50, resulted in increased exposure of β-1,3-glucan on the surface of C. albicans, C. tropicalis, C. dubliniensis, C. lusitaniae, and C. guilliermondii (Table 5). However, CSF treatment had no effect on the exposure of β-1,3-glucan of C. glabrata and C. parapsilosis (Table 5). Measurement of exposed chitin on the cell surface determined that the C. glabrata isolate had less chitin exposed on the outer surface compared to C. albicans isolate (Table 5). Treatment of C. albicans, C. tropicalis, C. dubliniensis, C. lusitaniae, and C. guilliermondii with CSF resulted in increased detection of chitin on the cell surface (Table 5). The exception to this was C. glabrata and C. parapsilosis which had no change in exposure of chitin, in response to CSF treatment (Table 5).
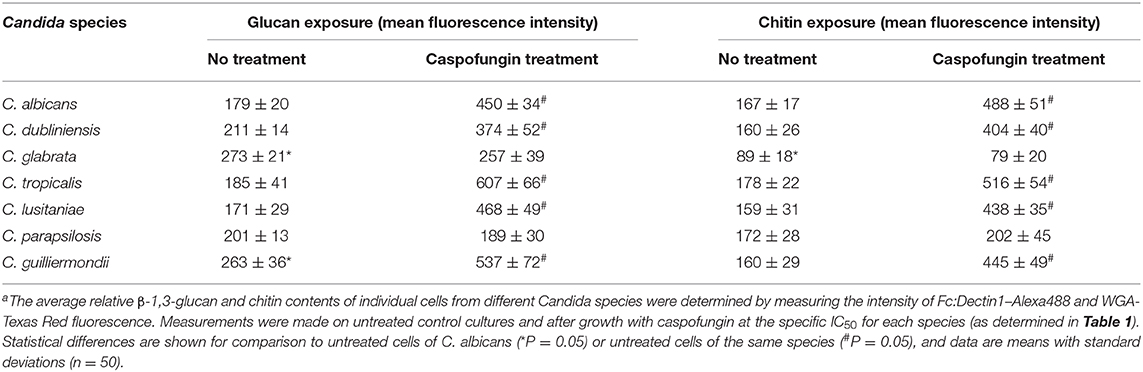
Table 5. Quantification of the exposure of β-1,3-glucan and chitin of Candida species, with and without caspofungin treatmenta.
Relationship Between Phagocytosis and Exposure of Chitin and Glucan
To determine the effect of exposure of chitin and glucan on phagocytosis, the uptake of isolates of each Candida species by J774 macrophages was determined. The yeasts were cultured with and without sub-MIC CSF treatment and phagocytosis correlated to the exposure of chitin and β-1,3-glucan on the cell surface (Figure 2, Supplementary Table 1). The majority of Candida species had low levels of chitin and β-1,3-glucan exposed naturally on the cell surface, under the growth conditions used (Figure 2A). The exception was C. glabrata and C. guilliermondii which had significantly more β-1,3-glucan exposed on the cell surface, compared to C. albicans, which correlated with these Candida species having the highest phagocytosis by J774 macrophages (Figure 2A). To determine the effect of altered exposure of β-1,3-glucan and chitin, the phagocytosis of CSF-treated cells was also measured (Figure 2B). In all cases the combined increase in exposure of chitin and β-1,3-glucan, in CSF-treated cells, resulted in a significant decrease in phagocytosis by macrophages (Figure 2B). Treatment of C. glabrata and C. parapsilosis with CSF had little effect on the exposure of the outer cell wall components and consequently there was no significant change in their phagocytosis by macrophages (Figure 2B). Pearson Correlation analysis demonstrated that there was a positive correlation between glucan exposure and chitin exposure and caspofungin treatment [r(12) = 0.703, p = 0.005; r(12) = 0.699, p = 0.005 respectively] but a negative correlation between % phagocytosis and caspofungin treatment [r(12) = −0.763, p = 0.002].
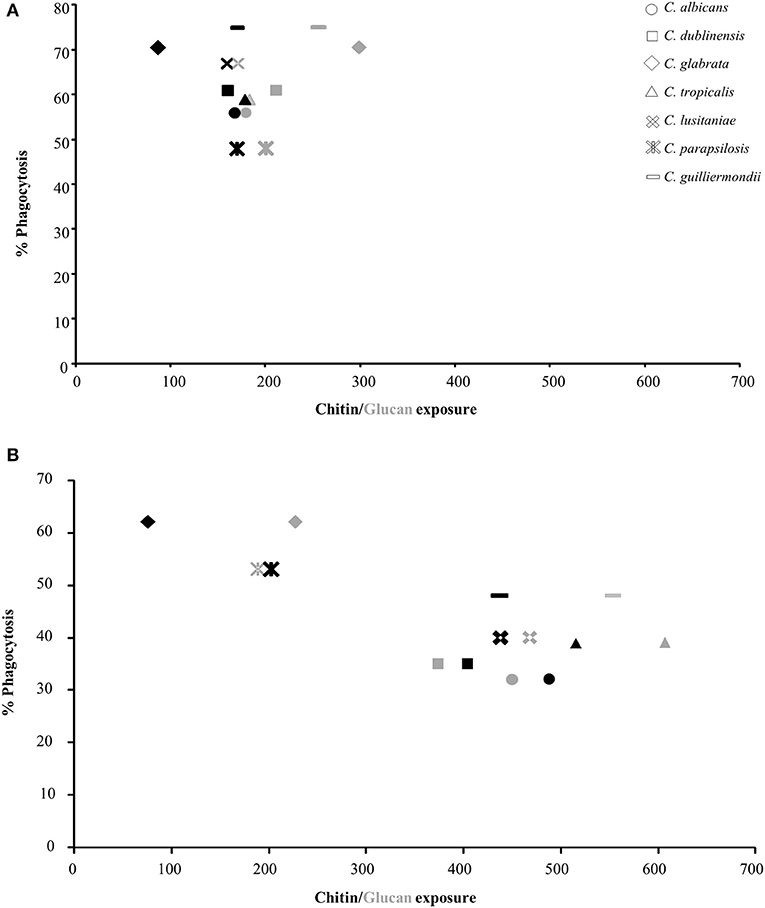
Figure 2. Correlation between exposure of chitin and glucan on the cell surface of Candida species and phagocytosis by J774 macrophages. The values for exposure of chitin and β-1,3-glucan were taken from Table 5. Phagocytosis of Candida spp. by macrophages was determined by fluorescence microscopy. All cells were stained with 10 μg/ml FITC prior to exposure to macrophages. FITC-stained cells were exposed to J774 macrophages for 1 h, after which non-phagocytosed cells were stained with 25 μg/ml CFW. The number of macrophages which had phagocytosed Candida cells were then counted (n = 300). (A) Untreated Candida spp. cells. (B) CSF treated Candida cells. Black shapes = chitin (WGA-TR), gray shapes = glucan (Fc:dectin1-alexa488).
Cytokine Production Varies Between Candida Species and Is Reduced in Response to CSF Treatment
Increased exposure of β-1,3-glucan has previously been shown to stimulate cytokine production whereas increased exposure of chitin has been shown to result in a decrease in cytokine production (Wheeler and Fink, 2006; Wheeler et al., 2008; Mora-Montes et al., 2011). To determine the cytokine production stimulated by the different Candida species, TNFα production was measured when untreated live cells of each Candida species were exposed to J774 macrophages for 4 h (Figure 3). Live cells of C. albicans and C. dubliniensis stimulated the highest levels of TNFα production (Figure 3). The other Candida species all stimulated very low levels of TNFα (Figure 3), which remained unchanged even after 24 h (data not shown). Treatment with an IC50 concentration of CSF, specific to each Candida species, led to a significant decrease in TNFα production in C. albicans and C. dubliniensis, compared to untreated cells (Figure 3). Treatment with CSF had no effect on TNFα production in the other Candida species isolates, with the exception of the C. tropicalis isolate which had a significant increase in TNFα production compared to untreated cells (Figure 3). C. tropicalis also had the highest level of surface-exposed β(1,3)-glucan when treated with CSF.
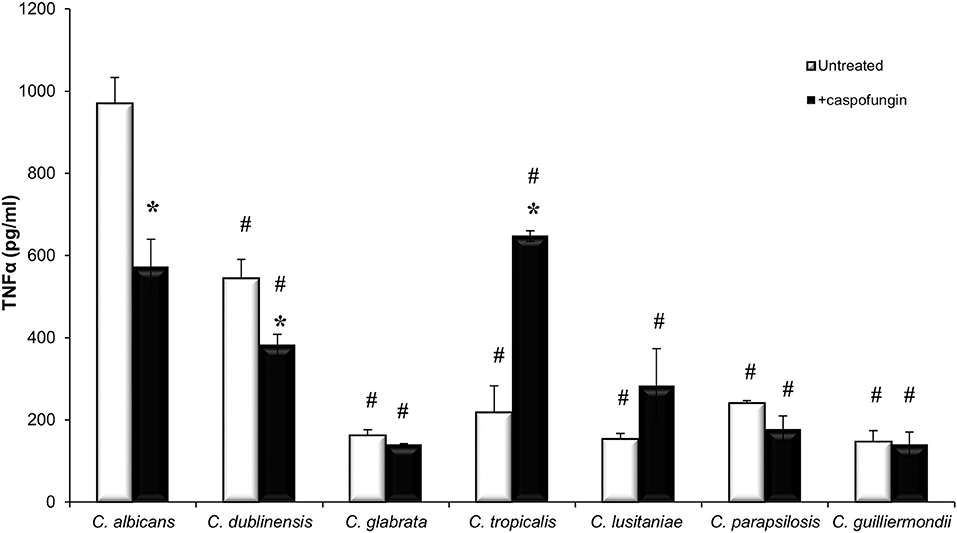
Figure 3. Candida species stimulate different immune responses. Live cells of each Candida species, which had been grown with or without an IC50 concentration of caspofungin for 6 h, were exposed to J774 macrophages for 4 h. After 4 h of co-incubation, supernatants were collected and cytokine production was measured. Significant differences (#p < 0.05) compared to C. albicans. Significant differences (*p < 0.05) compared to untreated cells of the same species. Error bars are SD (n = 3).
Discussion
This study aimed to directly compare the structure and composition of the cell walls of different Candida species, examine how the cell wall was altered in response to caspofungin treatment and assess how this impacted on fungal: murine macrophage interactions. We demonstrated that the length of the outer fibrillar layer varied between the species isolates tested and that, in most cases, reduced fibril length correlated with increased exposure of β-1,3-glucan on the cell surface. C. glabrata and C. guilliermondii, which had naturally more β-1,3-glucan exposed on the cell surface, were phagocytosed more efficiently by J774 macrophages. This is in agreement with a previous study that showed that in a mixed population of C. albicans and C. glabrata cells, C. glabrata is preferentially phagocytosed over C. albicans (Keppler-Ross et al., 2010). Similar to our observations, Estrada-Mata et al. (2016) also demonstrated that C. parapsilosis had less mannan content but more β-1,3-glucan exposed on the surface compared to C. albicans which correlated with an increase in cytokine production by human PBMCs (Estrada-Mata et al., 2016). Dectin-1 has been shown to play an important role in recognition of C. parapsilosis by human PBMCs (Toth et al., 2013). In our study C. parapsilosis induced little cytokine production from J774 macrophages at 4 h, but differences in the source of immune cells and the time that Candida cells were exposed to immune cells before performing the ELISAs may account for these differences.
There were also notable differences in the cell wall proteins that reside on the surface of the different Candida species, for example, the GPI-anchored proteins, Epa3, Epa6, and Awp4, were identified specifically on the cell wall of C. glabrata under the growth conditions tested. These are known to be adhesion proteins in C. glabrata which are associated with biofilm formation (de Groot et al., 2008; Kraneveld et al., 2011). The presence of cell wall proteins on the cell surface of C. glabrata, C. parapsilosis, and C. tropicalis has been shown to be dependent on the growth conditions tested (Karkowska-Kuleta et al., 2019).
Treatment with CSF is known to result in a compensatory increase in cell wall chitin levels and result in increased exposure of chitin and β-1,3-glucan on the surface of C. albicans cells (Wheeler and Fink, 2006; Walker et al., 2008, 2013; Wheeler et al., 2008; Mora-Montes et al., 2011). All of the Candida species isolates tested, with the exception of C. glabrata and C. parapsilosis isolates, demonstrated a compensatory increase in cell wall chitin content in response to CSF treatment. Likewise, treatment with CSF also resulted in an increase in exposure of chitin and β-1,3-glucan on the cell surface for the majority of species tested. Notable exceptions were C. glabrata and C. parapsilosis with no changes in glucan exposure in response to caspofungin treatment. This increase in exposure of the inner cell wall polysaccharides, did not increase the number of macrophages that had engulfed the fungal cells in fact caspofungin treatment resulted in a reduction in % phagocytosis. The majority of live Candida spp. cells stimulated low levels of TNFα production. In comparison the C. albicans and C. dublinensis isolates stimulated elevated levels of TNFα but these levels were significantly reduced when macrophages were exposed to the same isolates after caspofungin treatment. A notable exception to this trend was CSF-treated C. tropicalis isolate which stimulated significantly elevated TNFα production compared to untreated cells attaining a level comparable to C. albicans and C. dublinensis. C. tropicalis had the highest level of Fc-dectin1-Alex488 staining with and without caspofungin treatment. These findings suggested that the immune stimulatory properties of the different Candida species is not solely dependent upon β-1,3-glucan exposure and recognition by Dectin-1 and other factors may come into play (Wagener et al., 2014). Recently, treatment with either fragments of chitin or β-1,3-glucan from Aspergillus fumigatus were shown to stimulate production of TNFα. In addition, combined treatment with both chitin and β-1,3-glucan resulted in a synergistic increase in TNFα production (Dubey et al., 2014). This is in contrast with the findings here where other changes at the cell surface including increased chitin exposure appeared to override any stimulation of the immune response elicited by glucan exposure. The differences in stimulation of the immune response may be due to variations in size of the cell wall polysaccharides and the way the polymer is presented to immune cells. Small particles of chitin are known to illicit a greater cytokine response compared to larger chitin fragments (Da Silva et al., 2009). Likewise, differences in polysaccharide structure may influence the immune response. The chitin microfibrils of A. fumigatus and C. albicans may have a different architecture, which could influence the immune response (Gow and Gooday, 1983; Lenardon et al., 2007). Recently, hyphal β-1,3-glucan of C. albicans was found to have a unique cyclical structure that is absent in β-1,3-glucan from the yeast form (Lowman et al., 2014). Compared to β-1,3-glucan from yeast cells, hyphal glucan was shown to induce an immune response in macrophages and human peripheral blood mononuclear cells through a Dectin-1-dependent mechanism. In addition CSF treatment has also been shown to not only effect β-1,3-glucan exposure but also its nano-structure with the density of dectin-1 binding sites increasing leading to increased phagocytosis by dendritic cells (Lin et al., 2016). To dissect further whether glucan and/or chitin plays the main role in caspofungin-mediated dampening of the immune response, it would be interesting to evaluate the effect of blocking receptors of both polysaccharides on interaction with macrophages. Another consideration is that the change in GPI-modified proteins on the cell surface, in response to CSF treatment, may also influence the interaction with immune cells. For example, the transglycosidase, Utr2, was detected on the surface of the majority of Candida species in response to CSF treatment. Utr2 is involved in crosslinking chitin and β-1,3-glucan and forms part of a compensatory mechanism activated in response to cell weakening induced by echinocandin treatment (Pardini et al., 2006). A C. albicans utr2Δ mutant was found to have the same susceptibility to micafungin as the wild type (Alberti-Segui et al., 2004). Deletion of UTR2 in C. albicans resulted in attenuated virulence in a mouse model of systemic candidiasis. Despite this attenuation in virulence the fungal burdens from the kidneys of mice infected with the utr2Δ mutant were comparable to that of mice infected with wild-type C. albicans. This suggests that in the absence of UTR2, cells do not elicit the same inflammatory response as wild-type cells and enables the utr2Δ mutant to be tolerated by the host, leading to survival of the infected mice (Pardini et al., 2006). Therefore, the increased abundance of Utr2 on the cell surface in response to caspofungin treatment may also contribute to altered interactions with host cells.
Candida guilliermondii and C. parapsilosis are known to be intrinsically less sensitive to the echinocandins. Both species had thin walls and their cell wall proteomes differed from CSF-susceptible species. Pga24/Ywp1 orthologs were detected on their cell surfaces when grown on RPMI-1640 medium. In C. albicans Pga24 is associated with the yeast cell wall (Granger et al., 2005) and here was not detected when C. albicans was grown on RPMI-1640 medium. Mutation of PGA24 did not alter echinocandin susceptibility of C. albicans (Plaine et al., 2008). The role of this surface protein in echinocandin susceptibility of C. guilliermondii and C. parapsilosis remains to be determined.
The ability of live cells of each isolate of the different Candida species to stimulate cytokine production varied significantly (Figure 3). C. albicans and C. dubliniensis isolates, tested here, elicited the highest production of TNFα. In contrast to this the other isolates representing the different Candida species stimulated negligible levels of TNFα when exposed to J774 macrophages for 4 h. Previous work has demonstrated that C. parapsilosis, C. tropicalis, C. lusitaniae, and C. guilliermondii in addition to C. albicans and C. dubliniensis, stimulated production of TNFα from murine peritoneal macrophages and human PBMCs after 24 h (Aybay and Imir, 1996; Navarro-Arias et al., 2019). The most likely reason for the difference observed at the 4 h time point is that C. albicans and C. dubliniensis are the only two species which form true hyphae and were capable of germinating within macrophages, and consequently escaping from macrophages, faster than any of the other Candida species. For example, C. glabrata yeast cells have been shown to replicate within macrophages for up to 3 days before causing lysis of the macrophage (Seider et al., 2011). Therefore, the low levels of cytokine production that have been observed as a result of exposure of macrophages to live C. glabrata is thought to be due to C. glabrata evading host defenses by hiding within immune cells (Seider et al., 2011, 2014; Brunke et al., 2014). However, it is possible for C. glabrata to filament under certain conditions. Prolonged exposure of C. glabrata to macrophages led to filamentous growth which was a result of a point mutation in CHS2 (Lewis et al., 2013). Treatment of C. albicans and C. dubliniensis with CSF resulted in a decrease in production of TNFα and reduced phagocytosis by macrophages. Exposure of C. albicans to caspofungin has previously been shown to reduce cytokine production in both live and heat killed C. albicans cells, as a result of increased exposure of chitin on the cell surface (Mora-Montes et al., 2011).
Here we examined caspofungin induced changes at the cell surface of representative isolates of different Candida species. We detected drug-induced changes in the repertoire of covalently attached surface proteins and alterations in the exposure of cell wall carbohydrates of contributing to their recognition by J774 macrophages. The cell surface is highly variable both between and within species and so these investigations should be expanded to a wider range of isolates of the different species to determine the extent of cell surface variability and how it impacts on host interactions and drug susceptibility.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
LW and CM contributed to acquisition, analysis, and interpretation of data for the work. All authors contributed to the design of the work and approved the submitted and final version.
Funding
We acknowledge funding from the British Society for Antimicrobial Chemotherapy, the Medical Research Council (G0400284) and Wellcome Trust Strategic Award for Medical Mycology and Fungal Immunology 097377.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Gillian Milne from the University of Aberdeen Microscopy and Histology facility for help with EM, Dr. David Stead (Aberdeen Proteomics) for proteomics analysis and Prof. Gordon Brown for Fc:Dectin1. We thank Dr. Judith Bain and Prof. Lars Erwig for advice on the macrophage assays.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00164/full#supplementary-material
References
Alberti-Segui, C., Morales, A. J., Xing, H., Kessler, M. M., Willins, D. A., Weinstock, K. G., et al. (2004). Identification of potential cell-surface proteins in Candida albicans and investigation of the role of a putative cell-surface glycosidase in adhesion and virulence. Yeast 21, 285–302. doi: 10.1002/yea.1061
Alexander, B. D., Johnson, M. D., Pfeiffer, C. D., Jimenez-Ortigosa, C., Catania, J., Booker, R., et al. (2013). Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 56, 1724–1732. doi: 10.1093/cid/cit136
Aybay, C., and Imir, T. (1996). Tumor necrosis factor (TNF) induction from monocyte/macrophages by Candida species. Immunobiology 196, 363–374. doi: 10.1016/S0171-2985(96)80059-3
Balashov, S. V., Park, S., and Perlin, D. S. (2006). Assessing resistance to the echinocandin antifungal drug caspofungin in Candida albicans by profiling mutations in FKS1. Antimicrob. Agents Chemother. 50, 2058–2063. doi: 10.1128/AAC.01653-05
Baltch, A. L., Lawrence, D., Ritz, W. J., Andersen, N., Bopp, L. H., Michelsen, P. B., et al. (2012b). Effects of anidulafungin and voriconazole, singly and in combination, on cytokine/chemokine production by human monocyte-derived macrophages infected with Candida glabrata or activated by lipopolysaccharide. Chemotherapy 58, 146–151. doi: 10.1159/000337076
Baltch, A. L., Lawrence, D. A., Ritz, W. J., Andersen, N. J., Bopp, L. H., Michelsen, P. B., et al. (2012a). Effects of echinocandins on cytokine/chemokine production by human monocytes activated by infection with Candida glabrata or by lipopolysaccharide. Diagn. Microbiol. Infect. Dis. 72, 226–233. doi: 10.1016/j.diagmicrobio.2011.11.004
Brown, G. D., and Gordon, S. (2001). Immune recognition. A new receptor for beta-glucans. Nature 413, 36–37. doi: 10.1038/35092620
Brunke, S., Seider, K., Fischer, D., Jacobsen, I. D., Kasper, L., Jablonowski, N., et al. (2014). One small step for a yeast–microevolution within macrophages renders Candida glabrata hypervirulent due to a single point mutation. PLoS Pathog. 10:e1004478. doi: 10.1371/journal.ppat.1004478
Butler, G., Rasmussen, M. D., Lin, M. F., Santos, M. A., Sakthikumar, S., Munro, C. A., et al. (2009). Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature. 459, 657–662. doi: 10.1038/nature08064
Da Silva, C. A., Chalouni, C., Williams, A., Hartl, D., Lee, C. G., and Elias, J. A. (2009). Chitin is a size-dependent regulator of macrophage TNF and IL-10 production. J. Immunol. 182, 3573–3582. doi: 10.4049/jimmunol.0802113
de Groot, P. W., Kraneveld, E. A., Yin, Q. Y., Dekker, H. L., Gross, U., Crielaard, W., et al. (2008). The cell wall of the human pathogen Candida glabrata: differential incorporation of novel adhesin-like wall proteins. Eukaryot. Cell 7, 1951–1964. doi: 10.1128/EC.00284-08
Dubey, L. K., Moeller, J. B., Schlosser, A., Sorensen, G. L., and Holmskov, U. (2014). Induction of innate immunity by Aspergillus fumigatus cell wall polysaccharides is enhanced by the composite presentation of chitin and beta-glucan. Immunobiology 219, 179–188. doi: 10.1016/j.imbio.2013.10.003
Dutton, L. C., Nobbs, A. H., Jepson, K., Jepson, M. A., Vickerman, M. M., Aqeel Alawfi, S., et al. (2014). O-mannosylation in Candida albicans enables development of interkingdom biofilm communities. MBio 5:e00911–14. doi: 10.1128/mBio.00911-14
Estrada-Mata, E., Navarro-Arias, M. J., Perez-Garcia, L. A., Mellado-Mojica, E., Lopez, M. G., Csonka, K., et al. (2016). Members of the Candida parapsilosis complex and Candida albicans are differentially recognized by human peripheral blood mononuclear cells. Front. Microbiol. 6:1527. doi: 10.3389/fmicb.2015.01527
Fidan, I., Yesilyurt, E., Kalkanci, A., Aslan, S. O., Sahin, N., Ogan, M. C., et al. (2014). Immunomodulatory effects of voriconazole and caspofungin on human peripheral blood mononuclear cells stimulated by Candida albicans and Candida krusei. Am. J. Med. Sci. 348, 219–223. doi: 10.1097/MAJ.0000000000000236
Garcia-Effron, G., Chua, D. J., Tomada, J. R., DiPersio, J., Perlin, D. S., Ghannoum, M., et al. (2010). Novel FKS mutations associated with echinocandin resistance in Candida species. Antimicrob. Agents Chemother. 54, 2225–2227. doi: 10.1128/AAC.00998-09
Gillum, A. M., Tsay, E. Y., and Kirsch, D. R. (1984). Isolation of the Candida albicans gene for orotidine-5'-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 198, 179–182. doi: 10.1007/bf00328721
Gow, N. A., Netea, M. G., Munro, C. A., Ferwerda, G., Bates, S., Mora-Montes, H. M., et al. (2007). Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 196, 1565–1571. doi: 10.1086/523110
Gow, N. A. R., and Gooday, G. W. (1983). Ultrastructure of chitin in hyphae of Candida albicans and other dimorphic and mycelial fungi. Protoplasma 115, 52–58. doi: 10.1007/BF01293580
Gow, N. A. R., Latge, J.-P., and Munro, C. A. (2017). The fungal cell wall: structure, biosynthesis, and function. Microbiol. Spectr. 5, 1–25. doi: 10.1128/microbiolspec.FUNK-0035-2016
Graham, L. M., Tsoni, S. V., Willment, J. A., Williams, D. L., Taylor, P. R., Gordon, S., et al. (2006). Soluble Dectin-1 as a tool to detect beta-glucans. J. Immunol. Methods 314, 164–169. doi: 10.1016/j.jim.2006.05.013
Granger, B. L., Flenniken, M. L., Davis, D. A., Mitchell, A. P., and Cutler, J. E. (2005). Yeast wall protein 1 of Candida albicans. Microbiology 151, 1631–1644. doi: 10.1099/mic.0.27663-0
Jouault, T., Ibata-Ombetta, S., Takeuchi, O., Trinel, P. A., Sacchetti, P., Lefebvre, P., et al. (2003). Candida albicans phospholipomannan is sensed through toll-like receptors. J. Infect. Dis. 188, 165–172. doi: 10.1086/375784
Karkowska-Kuleta, J., Satala, D., Bochenska, O., Rapala-Kozik, M., and Kozik, A. (2019). Moonlighting proteins are variably exposed at the cell surfaces of Candida glabrata, Candida parapsilosis and Candida tropicalis under certain growth conditions. BMC Microbiol. 19, 145–149. doi: 10.1186/s12866-019-1524-5
Keppler-Ross, S., Douglas, L., Konopka, J. B., and Dean, N. (2010). Recognition of yeast by murine macrophages requires mannan but not glucan. Eukaryot. Cell 9, 1776–1787. doi: 10.1128/EC.00156-10
Kohatsu, L., Hsu, D. K., Jegalian, A. G., Liu, F. T., and Baum, L. G. (2006). Galectin-3 induces death of Candida species expressing specific beta-1,2-linked mannans. J. Immunol. 177, 4718–4726. doi: 10.4049/jimmunol.177.7.4718
Kraneveld, E. A., de Soet, J. J., Deng, D. M., Dekker, H. L., de Koster, C. G., Klis, F. M., et al. (2011). Identification and differential gene expression of adhesin-like wall proteins in Candida glabrata biofilms. Mycopathologia 172, 415–427. doi: 10.1007/s11046-011-9446-2
Kurtz, M. B., and Douglas, C. M. (1997). Lipopeptide inhibitors of fungal glucan synthase. Med. Mycol. 35, 79–86. doi: 10.1080/02681219780000961
Lee, K. K., Maccallum, D. M., Jacobsen, M. D., Walker, L. A., Odds, F. C., Gow, N. A., et al. (2012). Elevated cell wall chitin in Candida albicans confers echinocandin resistance in vivo. Antimicrob. Agents Chemother. 56, 208–217. doi: 10.1128/AAC.00683-11
Lenardon, M. D., Whitton, R. K., Munro, C. A., Marshall, D., and Gow, N. A. (2007). Individual chitin synthase enzymes synthesize microfibrils of differing structure at specific locations in the Candida albicans cell wall. Mol. Microbiol. 66, 1164–1173. doi: 10.1111/j.1365-2958.2007.05990.x
Lewis, L. E., Bain, J. M., Okai, B., Gow, N. A., and Erwig, L. P. (2013). Live-cell video microscopy of fungal pathogen phagocytosis. J. Vis. Exp. 2013:50196. doi: 10.3791/50196
Lin, J., Wester, M. J., Graus, M. S., Lidke, K. A., and Neumann, A. K. (2016). Nanoscopic cell-wall architecture of an immunogenic ligand in Candida albicans during antifungal drug treatment. Mol. Biol. Cell 27, 1002–1014. doi: 10.1091/mbc.E15-06-0355
Lowman, D. W., Greene, R. R., Bearden, D. W., Kruppa, M. D., Pottier, M., Monteiro, M. A., et al. (2014). Novel structural features in Candida albicans hyphal glucan provide a basis for differential innate immune recognition of hyphae versus yeast. J. Biol. Chem. 289, 3432–3443. doi: 10.1074/jbc.M113.529131
Marakalala, M. J., Vautier, S., Potrykus, J., Walker, L. A., Shepardson, K. M., Hopke, A., et al. (2013). Differential adaptation of Candida albicans in vivo modulates immune recognition by dectin-1. PLoS Pathog. 9:e1003315. doi: 10.1371/journal.ppat.1003315
Marti-Carrizosa, M., Sanchez-Reus, F., March, F., Canton, E., and Coll, P. (2015). Implication of Candida parapsilosis FKS1 and FKS2 mutations in reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 59, 3570–3573. doi: 10.1128/AAC.04922-14
McGreal, E. P., Rosas, M., Brown, G. D., Zamze, S., Wong, S. Y., Gordon, S., et al. (2006). The carbohydrate-recognition domain of Dectin-2 is a C-type lectin with specificity for high mannose. Glycobiology 16, 422–430. doi: 10.1093/glycob/cwj077
Mora-Montes, H. M., Netea, M. G., Ferwerda, G., Lenardon, M. D., Brown, G. D., Mistry, A. R., et al. (2011). Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect. Immun. 79, 1961–1970. doi: 10.1128/IAI.01282-10
Navarro-Arias, M. J., Hernandez-Chavez, M. J., Garcia-Carnero, L. C., Amezcua-Hernandez, D. G., Lozoya-Perez, N. E., Estrada-Mata, E., et al. (2019). Differential recognition of Candida tropicalis, Candida guilliermondii, Candida krusei, and Candida auris by human innate immune cells. Infect. Drug Resist. 12, 783–794. doi: 10.2147/IDR.S197531
Netea, M. G., Brown, G. D., Kullberg, B. J., and Gow, N. A. (2008). An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. 6, 67–78. doi: 10.1038/nrmicro1815
Netea, M. G., Gow, N. A., Munro, C. A., Bates, S., Collins, C., Ferwerda, G., et al. (2006). Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Invest. 116, 1642–1650. doi: 10.1172/JCI27114
Pardini, G., De Groot, P. W., Coste, A. T., Karababa, M., Klis, F. M., de Koster, C. G., et al. (2006). The CRH family coding for cell wall glycosylphosphatidylinositol proteins with a predicted transglycosidase domain affects cell wall organization and virulence of Candida albicans. J. Biol. Chem. 281, 40399–40411. doi: 10.1074/jbc.M606361200
Park, S., Kelly, R., Kahn, J. N., Robles, J., Hsu, M. J., Register, E., et al. (2005). Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob. Agents Chemother. 49, 3264–3273. doi: 10.1128/AAC.49.8.3264-3273.2005
Pfaller, M., Neofytos, D., Diekema, D., Azie, N., Meier-Kriesche, H. U., Quan, S. P., et al. (2012). Epidemiology and outcomes of candidemia in 3648 patients: data from the Prospective Antifungal Therapy [PATH Alliance(R)] registry, 2004-2008. Diagn. Microbiol. Infect. Dis. 74, 323–331. doi: 10.1016/j.diagmicrobio.2012.10.003
Pfaller, M. A., Messer, S. A., Woosley, L. N., Jones, R. N., and Castanheira, M. (2013). Echinocandin and triazole antifungal susceptibility profiles for clinical opportunistic yeast and mold isolates collected from 2010 to 2011: application of new CLSI clinical breakpoints and epidemiological cutoff values for characterization of geographic. J. Clin. Microbiol. 51, 2571–2581. doi: 10.1128/JCM.00308-13
Pham, C. D., Iqbal, N., Bolden, C. B., Kuykendall, R. J., Harrison, L. H., Farley, M. M., et al. (2014). Role of FKS mutations in Candida glabrata: MIC values, echinocandin resistance, and multidrug resistance. Antimicrob. Agents Chemother. 58, 4690–4696. doi: 10.1128/AAC.03255-14
Plaine, A., Walker, L., Da Costa, G., Mora-Montes, H. M., McKinnon, A., Gow, N. A., et al. (2008). Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 45, 1404–1414. doi: 10.1016/j.fgb.2008.08.003
Porcaro, I., Vidal, M., Jouvert, S., Stahl, P. D., and Giaimis, J. (2003). Mannose receptor contribution to Candida albicans phagocytosis by murine E-clone J774 macrophages. J. Leukoc. Biol. 74, 206–215. doi: 10.1189/jlb.1202608
Sato, K., Yang, X. L., Yudate, T., Chung, J. S., Wu, J., Luby-Phelps, K., et al. (2006). Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor gamma chain to induce innate immune responses. J. Biol. Chem. 281, 38854–38866. doi: 10.1074/jbc.M606542200
Seider, K., Brunke, S., Schild, L., Jablonowski, N., Wilson, D., Majer, O., et al. (2011). The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J. Immunol. 187, 3072–3086. doi: 10.4049/jimmunol.1003730
Seider, K., Gerwien, F., Kasper, L., Allert, S., Brunke, S., Jablonowski, N., et al. (2014). Immune evasion, stress resistance, and efficient nutrient acquisition are crucial for intracellular survival of Candida glabrata within macrophages. Eukaryot. Cell 13, 170–183. doi: 10.1128/EC.00262-13
Skrzypek, M. S., Binkley, J., Binkley, G., Miyasato, S. R., Simison, M., and Sherlock, G. (2016). The Candida Genome Database (CGD): incorporation of Assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res. 45, D592–D596. doi: 10.1093/nar/gkw924
Stahl, P. D., Rodman, J. S., Miller, M. J., and Schlesinger, P. H. (1978). Evidence for receptor-mediated binding of glycoproteins, glycoconjugates, and lysosomal glycosidases by alveolar macrophages. Proc. Natl. Acad. Sci. U.S.A. 75, 1399–1403. doi: 10.1073/pnas.75.3.1399
Sullivan, D. J., Westerneng, T. J., Haynes, K. A., Bennett, D. E., and Coleman, D. C. (1995). Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141, 1507–1521. doi: 10.1099/13500872-141-7-1507
Taylor, P. R., Tsoni, S. V., Willment, J. A., Dennehy, K. M., Rosas, M., Findon, H., et al. (2007). Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol. 8, 31–38. doi: 10.1038/ni1408
Thompson, A., Griffiths, J. S., Walker, L., da Fonseca, D. M., Lee, K. K., Taylor, P. R., et al. (2019). Dependence on Dectin-1 Varies With Multiple Candida Species. Front. Microbiol. 10:1800. doi: 10.3389/fmicb.2019.01800
Toth, A., Csonka, K., Jacobs, C., Vagvolgyi, C., Nosanchuk, J. D., Netea, M. G., et al. (2013). Candida albicans and Candida parapsilosis induce different T-cell responses in human peripheral blood mononuclear cells. J. Infect. Dis. 208, 690–698. doi: 10.1093/infdis/jit188
Wagener, J., Malireddi, R. K., Lenardon, M. D., Koberle, M., Vautier, S., MacCallum, D. M., et al. (2014). Fungal chitin dampens inflammation through IL-10 induction mediated by NOD2 and TLR9 activation. PLoS Pathog. 10:e1004050. doi: 10.1371/journal.ppat.1004050
Walker, C. A., Gomez, B. L., Mora-Montes, H. M., Mackenzie, K. S., Munro, C. A., Brown, A. J., et al. (2010). Melanin externalization in Candida albicans depends on cell wall chitin structures. Eukaryot. Cell 9, 1329–1342. doi: 10.1128/EC.00051-10
Walker, L. A., Gow, N. A., and Munro, C. A. (2013). Elevated chitin content reduces the susceptibility of Candida species to caspofungin. Antimicrob. Agents Chemother. 57, 146–154. doi: 10.1128/AAC.01486-12
Walker, L. A., Munro, C. A., de Bruijn, I., Lenardon, M. D., McKinnon, A., and Gow, N. A. (2008). Stimulation of chitin synthesis rescues Candida albicans from echinocandins. PLoS Pathog. 4:e1000040. doi: 10.1371/journal.ppat.1000040
Wheeler, R. T., and Fink, G. R. (2006). A drug-sensitive genetic network masks fungi from the immune system. PLoS Pathog. 2:e35. doi: 10.1371/journal.ppat.0020035
Wheeler, R. T., Kombe, D., Agarwala, S. D., and Fink, G. R. (2008). Dynamic, morphotype-specific Candida albicans beta-glucan exposure during infection and drug treatment. PLoS Pathog. 4:e1000227. doi: 10.1371/journal.ppat.1000227
Keywords: chitin, β-1, 3-glucan, GPI-anchored proteins, fungal cell wall, echinocandin, macrophages, immune response
Citation: Walker LA and Munro CA (2020) Caspofungin Induced Cell Wall Changes of Candida Species Influences Macrophage Interactions. Front. Cell. Infect. Microbiol. 10:164. doi: 10.3389/fcimb.2020.00164
Received: 19 December 2019; Accepted: 27 March 2020;
Published: 12 May 2020.
Edited by:
James Bernard Konopka, Stony Brook University, United StatesReviewed by:
Jeniel E. Nett, University of Wisconsin-Madison, United StatesRobert T. Wheeler, University of Maine, United States
Copyright © 2020 Walker and Munro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Louise A. Walker, bG91aXNlLndhbGtlckBhYmRuLmFjLnVr
 Louise A. Walker
Louise A. Walker Carol A. Munro
Carol A. Munro