
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 21 April 2020
Sec. Clinical Microbiology
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00145
This article is part of the Research Topic Immunoserological Diagnosis of Zoonoses and Emerging Infectious Diseases View all 6 articles
 Xin Yang1†
Xin Yang1† Zuoping He1,2†
Zuoping He1,2† Guoxia Zhang3†
Guoxia Zhang3† Jinhui Lu1
Jinhui Lu1 Hui Zhang4
Hui Zhang4 Hui Ren1
Hui Ren1 Yanjun Tian3
Yanjun Tian3 Heng Yang1,5
Heng Yang1,5 Chuangfu Chen4
Chuangfu Chen4 Linhai Li1,6
Linhai Li1,6 Yongshui Fu7
Yongshui Fu7 Jean-Pierre Allain1,8
Jean-Pierre Allain1,8 Chengyao Li1*
Chengyao Li1* Wenjing Wang1*
Wenjing Wang1*Brucellosis is a serious zoonosis occurring mainly in developing countries, and its diagnosis is largely dependent on serologic detection and bacterial culture. In this study, we developed the murine monoclonal antibodies (mAbs) against a conserved and major outer membrane protein 25 (Omp25) of Brucella species (B. spp.) for use in clinical diagnosis. The mAbs to Omp25 were produced by hybridoma technique, which were utilized for developing various immunoassays for detection of Brucellae, including Western blot (WB), enzyme-linked immunosorbent assay (ELISA), immunochemical staining (ICS), immunofluorescence staining (IFS), and flow cytometry assay (FCM). A number of five mAbs (2B10, 4A12, 4F10, 6C12, and 8F3) specific to Omp25 were selected, including 2 IgG1, 2 IgG2a, and 1 IgG2b. Among them, mAbs 6C12, 8F3, and 4A12 reacted highly with B. melitensis (M5-90), B. abortus (S19, 104M, and 2308), and B. suis strain (S2). No cross-reactivity with Yersinia enterocolitica O:9, Salmonella spp., and Escherichia coli was found. By mapping Omp25 epitopes, mAb 6C12 was found as reacting with a semi-conformational epitope, and mAbs 4A12 and 8F3 as recognizing a different linear epitope, respectively. The paired mAbs were tested for detecting Brucella species, suggesting that 8F3 was suitable for solid phase capture and 6C12 or 4A12 was suitable for conjugation with HRP for detection of Brucella Omp25 in ELISA. The FCM was established by mAb 6C12 for detecting intracellular Brucellae-infected peripheral blood mononuclear cells (PBMCs) from brucellosis patients. In conclusion, mAbs against Omp25 are precious reagents for detection of Brucellae in clinical samples with various immunoassays. mAb 6C12-based FCM could be potentially used for the monitoring of therapeutic efficacy for brucellosis in clinical practice.
Brucellosis is a severe zoonotic disease distributed worldwide, especially in the developing world (Wang et al., 2015). Sheep, goats, cattle, and pigs infected by Brucella species are the main sources of human brucellosis (Ducrotoy et al., 2018). In the past 10 years, human brucellosis has increased rapidly in China. There were 37,947 new cases of human brucellosis nationwide (morbidity 2.7318/100,000) in 2018 as reported by the Chinese CDC (http://www.nhc.gov.cn/jkj/s3578/201904/050427ff32704a5db64f4ae1f6d57c6c.shtml). New cases of human brucellosis were reported in non-endemic areas such as the Guangdong province (Chen et al., 2014), which is located in the south of China and far away from the high prevalence areas such as Inner Mongolia, Heilongjiang, Xinjiang, and Shanxi in the north or west of China (Wang et al., 2015).
At present, diagnosis of brucellosis mainly depends on serological methods used to detect antibodies against Brucellae in infected animals or humans (Araj, 2010). Diagnosing brucellosis by means of Brucellae cultures themselves can take at least 10 days. Once a Brucella infection becomes chronic, patients are likely to carry the bacteria for their whole lives. Anti-Brucellae methods are the most important approach for treatment of brucellosis. However, currently there are no rapid, simple, and quantitative methods for the evaluation of therapeutic efficacy of brucellosis during hospitalization. Brucella outer membrane proteins (Omps) are excellent candidates for the serologic diagnosis of a Brucella infection and potential antigens for recombinant subunit vaccines against brucellosis (Ahmed et al., 2015; Yousefi et al., 2016). Omp25 is one major Omps of the Brucella species and is considered to be closely related to virulence of Brucellae (Salhi et al., 2003; Goel and Bhatnagar, 2012). Brucella species without Omp25 survive for a shorter period of time than wild-type strains in mice (Edmonds et al., 2001, 2002). As a structural protein, Omp25 is highly conserved in various types and subtypes of Brucellae and induces a strong immune response (Cloeckaert et al., 2002; Goel et al., 2013; Ma et al., 2015). Therefore, it might be a useful diagnostic target for brucellosis. In a previous study, an antibody to Omp25 was used to identify rough Brucella isolates by means of a latex coagglutination test (Bowden et al., 1997). In this study, we aimed to generate novel monoclonal antibodies (mAbs) to Omp25 and to develop new immunoassays for diagnosis of brucellosis or evaluation of therapeutic efficacy of brucellosis in clinical practice.
Inactivated Brucella strains, including B. melitensis (M5-90), B. abortus (S19, 104M and 2308), and B. suis (S2), were provided from Shihezi University, Xinjiang, China. Yersinia enterocolitica O:9, Salmonella spp., and Escherichia coli (ATCC 23922) were provided from the Department of Microbiology, Southern Medical University (SMU), Guangzhou, China.
Recombinant proteins of Omp31, Omp19, Omp16, and periplasmic protein 26 (BP26) were produced from B. melitensis strain in the laboratory (Qiu et al., 2012; He et al., 2016; Li et al., 2017). Omp25 gene (642 bp) from B. melitensis M5-90 genomic DNA was cloned into the pET30a expression vector (Zhang et al., 2014; Yousefi et al., 2016), and designated as pET30a-Omp25. Recombinant Omp25 (rOmp25) was expressed as an inclusion body in Escherichia coli (DH5α) by induction with 1 mM IPTG. The rOmp 25 extract was denatured by 8 M urea and purified by Ni-NTA Agarose (GE Healthcare, Milwaukee, Wisconsin, USA), and then refolded by dialysis against 50 mM Tris-HCl buffer containing a declining gradient urea from 6, 4, 2, to 0 M (Qiu et al., 2012; Yousefi et al., 2016). Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was utilized to analyze rOmp25 (Yousefi et al., 2016). The purified soluble rOmp25 was used for mouse immunization and development of serologic tests.
BALB/c mice were obtained from the Animal Experimental Center of Southern Medical University (SMU), Guangzhou, China. BALB/c mice were immunized with purified rOmp25. The hybridoma cells, secreting mAbs to rOmp25, were generated and selected according to a previously reported method (Qiu et al., 2012; Patra et al., 2014; Li et al., 2017). To prepare the ascitis fluid, BALB/c mice sensitized by the liquid paraffin were injected subcutaneously with 106/ml hybridoma cells. The mouse ascites fluid was collected and purified by Recombinant Protein G NUPharose Fast Flow, rProtein G NUPharose FF (Nuptec, Hangzhou, China) (Divya et al., 2018).
All animal experimentations were approved by Southern Medical University (SMU) Animal Care and Use Committee (permit numbers: NFYY-2008-043 and NFYY-2010-076). All mouse surgery was performed under anesthesia, and all efforts were made to minimize suffering of animals.
The isotype of mAbs was determined by IsoQuick Strips (a mouse mAb isotyping kit, Sigma-Aldrich, St Louis, Missouri, USA).
Plasma, or peripheral blood mononuclear cells (PBMCs), were isolated from healthy blood donors in Guangzhou and Harbin blood centers. Blood samples from brucellosis patients were collected at the General Hospital of Agricultural Reclamation Bureau, Harbin, Heilongjiang, China. All blood donor or patient samples were confirmed for negativity or positivity to Brucella infection by Standard Agglutination Test (SAT) and Polymerase Chain Reaction (PCR) (Mohammadi and Golchin, 2018). The PBMCs were prepared according to manufacturer's instructions (Ficoll Pague PLUS, GE Healthcare Life Sciences, USA).
Peptides spanning 213 amino acids of Omp25 were synthesized as P1–P10 (Table S1) by a commercial company (Chinese Yuantai Company, Nanjing, China). A peptide of Omp31 (EP24: EYLYTDLGKRNLVDVD) was used as a negative control. The purity of all peptides was more than 90% of total weight.
The concentration of purified mAbs was determined by NanoDrop 2000 (Thermo Fisher Scientific, USA) as 1.14 mg/ml for 4F10, 2.1 mg/ml for 2B10, 7.4 mg/ml for 6C12, 3.8 mg/ml for 8F3, and 1.9 mg/ml for 4A12, respectively. The titer of mAbs was measured by indirect ELISA (Qiu et al., 2012; Divya et al., 2018). The Immuno MicroWell plate was coated with 5 μg/ml rOmp25 in 0.1 M carbonate buffer (CBS, pH 9.6) overnight at 4°C and blocked in 1% bovine serum albumin (BSA) for 1 h at 37°C. The purified mAbs as primary antibody were serially diluted from 1:100 to 1:1,968,300, and then added to microwell plate for 1 h at 37°C. ELISA plates were incubated for 1 h at 37°C with 1:10,000 diluted goat anti-mouse IgG and IgM HRP-conjugate antibodies (Rockland Immunochemicals, Inc, USA). The optical densities (OD) of reactions were detected with Epoch Microplate Spectophotometer (BioTek Instruments, Inc. Winooski, USA). mAb titer was finally defined as the dilution fold at OD450 value of 1 by indirect ELISA.
mAb's relative affinity was determined according to reactivity in different concentration of ammonium thiocyanate (NH4SCN) solutions ranging between 0 and 4 M/L for 30 min at room temperature (Pullen et al., 1986; Macdonald et al., 1988). The appropriate concentration of primary mAbs was determined on the titration curve, which corresponded to the optical density observed near the top of the curve's linear portion (Macdonald et al., 1988). All experiments were carried out in triplicate and the mean of OD450 values obtained from three independent experiments was used to calculate the relative affinity.
The infectious recombinant lentivirus (LV-HAGE-Omp25) mediated Omp25 expression in 293FT cell as described previously, mimicking Brucella Omp25 antigen in the infected mammalian cells (Zhang et al., 2012; Li et al., 2017).
The supernatant of sonicated proteins (SSPs) from B. melitensis (M5-90), B. abortus (S19, 104M, and 2308), and B. suis (S2) were considered as the native Omp25 lysates, which were prepared by breaking Brucella spp. stains with ultrasonication (Ultrasonic Apparatus XO-650, Xianou, Nanjing, China). The native membrane protein extracts (NMPs) was prepared by Membrane Protein Extraction Kit (Bestbio, China). The rOmp25 and native Omp25 were electrophoresed on SDS-PAGE and transferred to Polyvinylidene Fluoride membranes (PVDF membranes, Millipore, Billerica, Massachusetts, USA). The blotted membrane was incubated in 1:1,000 diluted mAb for 2 h at room temperature. The strip was washed in TBS containing 0.05% Tween®20 (TBST) and incubated with 1:5,000 dilution of goat anti-mouse IgG and IgM HRP-conjugate antibodies. The blot was visualized by adding immuno-chemiluminescence reagent (ECL, Millipore, Billerica, Massachusetts, USA). An HCV rNS3 mAb was used as unrelated negative control (Qiu et al., 2012). The SSPs from E. coli (ATCC23922) strain was used as negative control.
In indirect ELISA, rOmp25 or native Omp25 was used as a coating antigen. Purified mAbs were used as the primary antibody at optimum working concentration. The microwell plate was incubated with a 1:10,000 dilution of goat anti-mouse IgG and IgM HRP-conjugate antibodies as reported previously (Tiwari et al., 2013; Ahmed et al., 2015). In peptide-ELISA, peptides P1–P10 were used to identify antigenic epitopes recognized by each mAb (Qiu et al., 2012; Li et al., 2017). The double-antibody sandwich ELISA (DAS-ELISA) was used to detect Brucella Omp25 by cross-matching mAbs in pair with a capture mAb coated on the microwell plate and a detection mAb conjugated with HRP (Luo et al., 2012).
293FT cells were transduced with recombinant lentivirus expressing Omp25 (LV-HAGE-Omp25) at multiplicities of infection (MOI = 5) for 8 h. The transduced 293FT cells were detected by immunofluorescent staining (IFS) with Omp25 mAbs as same as with Omp31 or BP26 described previously (Li et al., 2017). PBMCs were collected from two brucellosis patients (Guangzhou Center for Disease Prevention and Control) and were tested by IFS with mAb 6C12. PBMCs from a healthy blood donor were used as the negative control.
Intact Brucella were detected by using ICS (Li et al., 2017). Briefly, after pretreatment with 3% H2O2 and washing with PBS three times, the Brucella smears on the glass-slide reacted with Omp25 mAbs and saturated with goat anti-mouse IgG and IgM HRP conjugate (Rockland Immunochemicals, Inc, USA). Bacteria were visualized with diaminobenzidine (DAB) substrate for color development.
To explore the capacity of mAb to recognize intracellular Brucellae in PBMCs of patients with brucellosis, the purified mAb 6C12 was labeled with phycoerythrin (PE) by PE/R-Phycoerythrin Conjugation Kit according to the manufacturer's instructions (Abcam, Cambridge, United Kingdom). FITC-Anti-CD14 antibody (M5E2 clone; BD, Bioscience, USA) was used to recognize the lineage of mononuclear cells. PBMCs collected from 28 brucellosis patients and 55 non-Brucella-infected blood donors were stained for intracellular Brucellae by FACSCalibur flow cytometer with FITC-Anti-CD14 antibody (M5E2) and PE-6C12 specific to FCM Omp25 (Delaporte et al., 2008; Okumura et al., 2015). Briefly, PBMCs were treated with 5 μl FITC-M5E2 for 30 min at room temperature and cells were washed twice with sample buffer [PBS containing 1% bovine serum albumin (BSA; Sigma-Aldrich, St Louis, Missouri, USA)]. After fixation and permeabilization with Intracellular Fixation & Permeabilization Buffer (eBioscience, California, USA), PBMCs were incubated with PE-6C12 for 45 min at 4°C and washed three times. Finally, 300 μl of resuspended PBMCs were tested by FCM. All steps were performed in the dark. The data were analyzed by FlowJo software version v10.0. The cutoff for non-Brucella-infected PBMCs from healthy blood donors was established as 1% by FCM.
Computer software (SPSS, Version 20.0, SPSS, Inc., Chicago, IL) was used for statistical analysis. All experiments were repeated at least three times independently. The results were presented as the mean ± SD.
Soluble rOmp25 was purified as an immunogen by Ni-NTA Agarose column. The purity of rOmp25 was about 95% of total proteins (Figure 1A). Five mAbs (2B10, 6C12, 8F3, 4A12, and 4F10) were selected for reacting with rOmp25 and NMPs of B. melitensis by ELISA, respectively (Figure 1B). Western blot analysis showed all five mAbs were highly reactive with denatured rOmp25, while among them three mAbs (6C12, 8F3, and 4A12) strongly reacted with the denatured NMPs of B. melitensis and two (2B10 and 4F10) reacted weakly (Figure 1C). In addition, only mAb 6C12 reacted with intracellular Brucella Omp25 expressed in 293FT cells (Figure 1C and Table 1). No mAbs reacted with other recombinant proteins of Brucella (Omp31, BP26, Omp19, and Omp16).
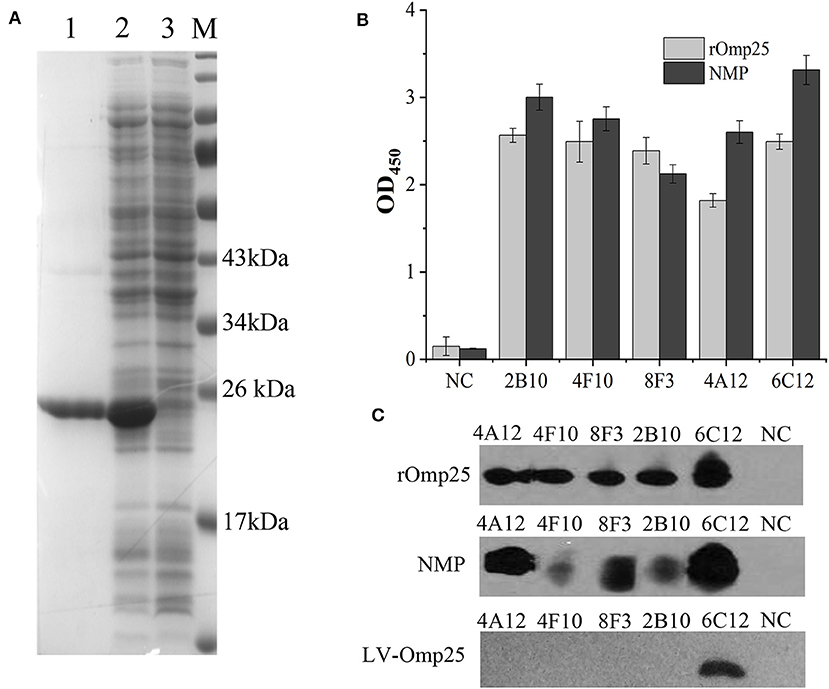
Figure 1. Purification and identification of recombinant Omp25. (A) The purified rOMP25 was analyzed by SDS-PAGE. Lane 1, purified Omp25 with 25 kD molecular weight; lane 2, cell lysate of pET30a-Omp25 transformed E. coli (DH5α) after IPTG induction; lane 3, cell lysate of pET30a-Omp25 transformed E. coli before IPTG induction. (B) Reactivity of five mAbs with rOMP25 or NMP of B. melitensis (M5-90 strain) in ELISA. (C) Reactivity of five mAbs with the denatured rOMP25 (1st panel) or NMP (2nd panel) of B. melitensis in WB. The third panel showed the denatured lentivirus expressed Omp25 in 293FT cells by WB. NC, negative control (recombinant NS3 to HCV).
mAbs 2B10, 6C12, 8F3, 4A12, and 4F10 were identified as 2 IgG1, 2 IgG2a and 1 IgG2b, respectively (Table 1). Antibody titers were determined by ELISA at value of 1 at OD450. The titer of mAbs 6C12, 8F3, and 4A12 was measured up to 1:200,000, 2B10 up to 1:50,000, and 4F10 up to 1:1,000, respectively (Figure 2A). Correspondingly, the optimum working concentration of these five mAbs was 1.14 μg/ml (4F10), 0.042 μg/ml (2B10), 0.037 μg/ml (6C12), 0.019 μg/ml (8F3), and 0.01 μg/ml (4A12), respectively. An appropriate dilution of 1:2,700 for 2B10, 1:900 for 4F10, 1:8,100 for 6C12 and 8F3, and 1:2,700 for 4A12 was used as initial concentration of mAb to determine its relative affinity. The relative affinity of mAb was estimated by thiocyanate elution assay, calculated at 50% reduction from initial antibody reactivity reached by increasing molarity of NH4SCN in the elution curves (Figure 2B). Relative affinity of mAbs was around 1.0 (Table 1), among them the affinity of 2B10 and 4F10 ranged from 1.0 to 1.5, 4A12 and 6C12 from 1.0 to 1.5, and 8F3 from 1.5 to 2.0, respectively.
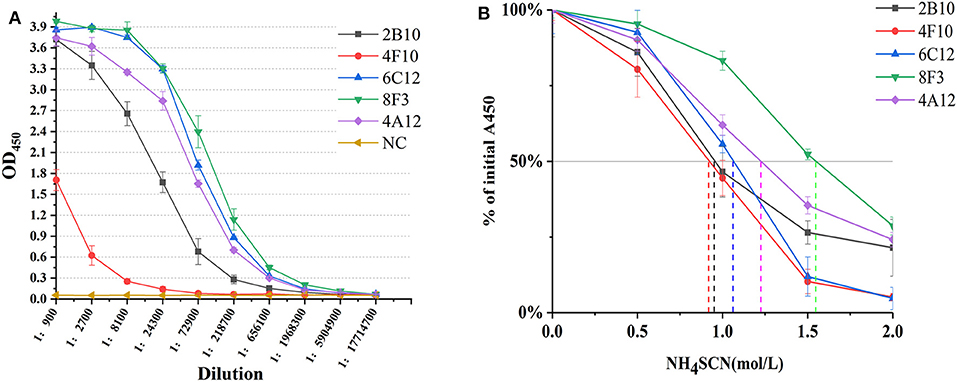
Figure 2. Determination of mAb titer and affinity. (A) Titration of mAb by an indirect ELISA. The mAb was serially diluted in 1:3. The optimum working concentration was determined for a midpoint of the steep portion of the curve. (B) The measurement of antibody relative affinity by thiocyanate elution assay. The affinity index was estimated by the molarity of NH4SCN causing 50% reduction from initial absorbance in the elution curves. All experiments were carried out in triplicate and the results were calculated from three independent experiments.
The ability of mAbs to Omp25 from different strains of Brucella, the SSPs extracted from B. melitensis (M5-90), B. abortus (S19, 104M, and 2308), and B. suis (S2) were tested by ELISA and WB. mAbs 4A12, 6C12, and 8F3 reacted with all three of Brucella species by ELISA (Figure 3A). The reactivity of 4A12 and 6C12 reached a relatively higher level than 8F3, but 2B10 and 4F10 reacted at low level or did not react. Western blot analysis showed these mAbs had a reaction pattern similar to ELISA (Figure 3B). Specifically, mAb 6C12 presented two reactive bands with Brucella strains 2308 (B. abortus), S19 (B. abortus), S2 (B. suis), and M5-90 (B. melitensis), corresponding to Omp25 (the lower) and Omp25d (the upper), respectively. The other four mAbs reacted only with Omp25 (Figure 3B). No mAbs reacted with lysates of Yersinia enterocolitica O:9, Salmonella spp., and Escherichia coli.
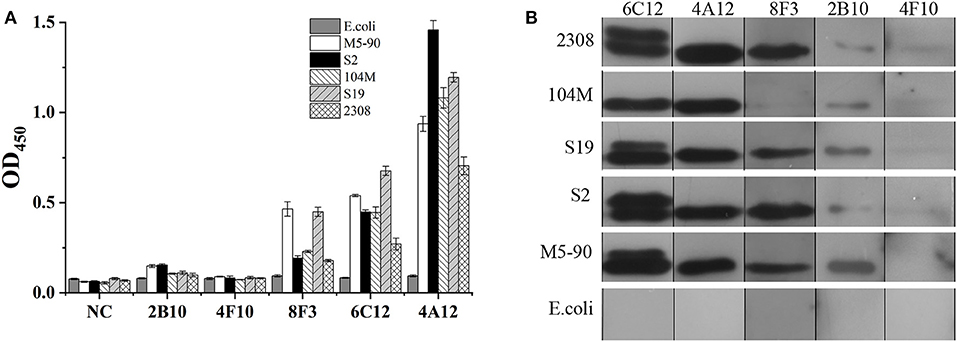
Figure 3. Detection of Brucella strains by mAbs to Omp25 in ELISA and WB. (A) The SSPs of M5-90, S19, 104M, 2308 and S2 strains were detected by an indirect ELISA. (B) Western blot analysis for identifying Brucella strains and non-Brucella stain with mAbs to Omp25. Brucella 2308 is a wild strain of B. abortus, 104M and S19 are vaccine strains of B. abortus, and S2 is a vaccine strain of B. suis, M5-90 is a vaccine strains of B. melitensis, respectively. E. coli (ATCC 23922) is used as a negative control.
In order to identify the Omp25 antigenic epitopes, all mAbs were tested in ELISA with 10 peptides derived from the 213 amino acid (aa) sequence of B. melitensis Omp25. Four mAbs (4A12, 8F3, 4F10, and 2B10) reacted with three different peptides (Figure 4A) identified as recognition of linear epitopes. mAb 6C12 did not react with any peptides but with denatured native Omp25 (Figure 3), corresponding to the recognition of a semi-conformational epitope (Table 1). The amino acid sequences of the three linear epitopes of Omp25 are presented in Figure 4B (19–20 mers peptides P2, P4, and P7).
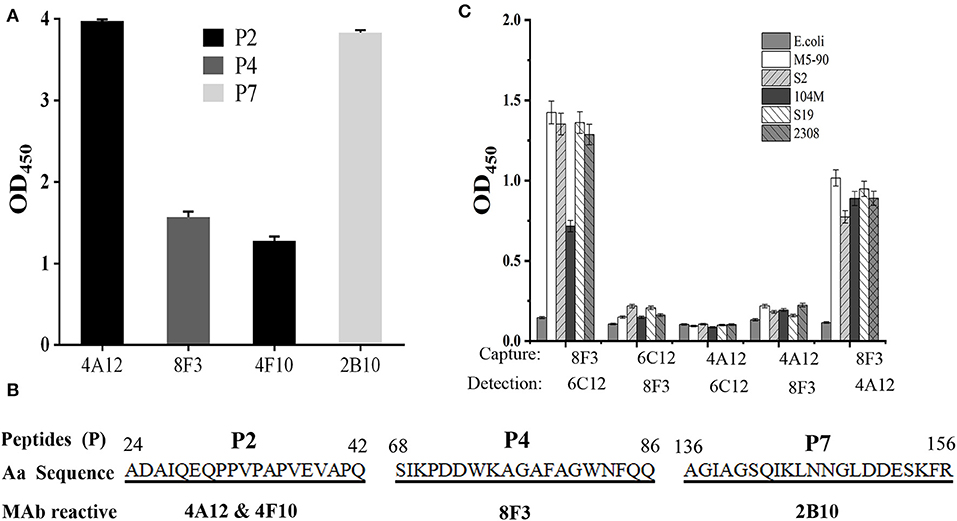
Figure 4. Epitope mapping of Omp25. (A) mAb reacted with peptides derived from Omp25 in ELISA. (B) mAb recognized linear epitopes within Omp25. Aa position is indicated at the beginning and the end of peptide sequence. mAb ID is indicated under the reactive peptide sequence. (C) Detection of Brucellae with cross-matching mAbs. The SSPs from B. abortus (S19, 104M, 2308), B. suis (S2) and B. menlitensis (M5-90) were used as antigens for cross-matching the mAb pair in DAS-ELISA. The capture mAb was used to coat microplates for Omp25 capture, and as detection mAb conjugated with HRP for detecting the captured Omp25 from various Brucella strains. E. coli was used as a negative control.
Five mAbs were tested for reactivity with Omp25 by cross-matching pairs with capture and detection antibodies by DAS-ELISA. Five pairs of mAbs (4A12/6C12-HRP, 4A12/8F3-HRP, 6C12/8F3-HRP, 8F3/4A12-HRP, and 8F3/6C12-HRP) reacted with rOmp25 (Table S2). These five pairs were tested for reactivity with the SSPs of 2308, 104M, S19, S2, and M5-90 strains from B. melitensis, B. abortus, or B. suis. Two mAb pairs, 8F3/6C12-HRP and 8F3/4A12-HRP, were found the be more reactive for detection of various Brucella species (Figure 4C).
To determine the ability of Omp25 mAbs to detect intracellular Brucellae or intact Brucella strains by microscopy, infected cells or intact bacteria were stained by IFS or ICS, respectively. mAbs 6C12, 2B10, and 4F10 showed a strong fluorescent reactivity with the lentivirus-Omp25 transduced 293FT cells by IFS (Figure S1A). In addition, by IFS with mAb 6C12 from two patients with brucellosis, visible fluorescent intracellular Brucellae infected PBMCs were observed, but not seen with controls from healthy blood donors (Figure S1B).
By ICS, the cultured Brucella could be seen by microscopy after staining with HRP-conjugated 6C12, 8F3, or 4A12 on a glass slide (Figure S2). The overall results are presented in Table 1.
On the basis of the above analysis, mAb 6C12 was considered the most functional antibody for Brucella detection. Hence, mAb 6C12 was selected for labeling with PE to detect intracellular Brucellae-infected PBMCs by FCM (Figure 5). The mononuclear cells were sorted from freshly isolated PBMCs by FITC-Anti-CD14 antibody (M5E2), and the ratio of Brucella-infected monocytes was calculated by staining with PE-6C12. Stained monocytes from healthy blood donor controls over brucellosis patients were stratified as the cutoff or threshold between two levels of intracellular Brucellae: <1.0%, indicating a lower level or negative intracellular Brucellae-infected cells (Figure 5A), and ≥1.0%, indicating a higher level of intracellular Brucellae-infected cells (Figure 5B). A total of 28 brucellosis patients and 20 blood donors were tested by FCM with both mAbs M5E2 and 6C12, showing that 46.4% (13/28) of brucellosis patients but none of blood donors carried intra-PBMCs Brucellae (≥1.0%) (Table 2). Only one patient (No. 8) had 1.13% of stained monocytes with unclear boundary by FCM. These results suggested that FCM with mAb 6C12 might be a practical assay to determine the frequency of intracellular Brucellae-infected PBMCs in individual brucellosis patients with clinical diagnosis.
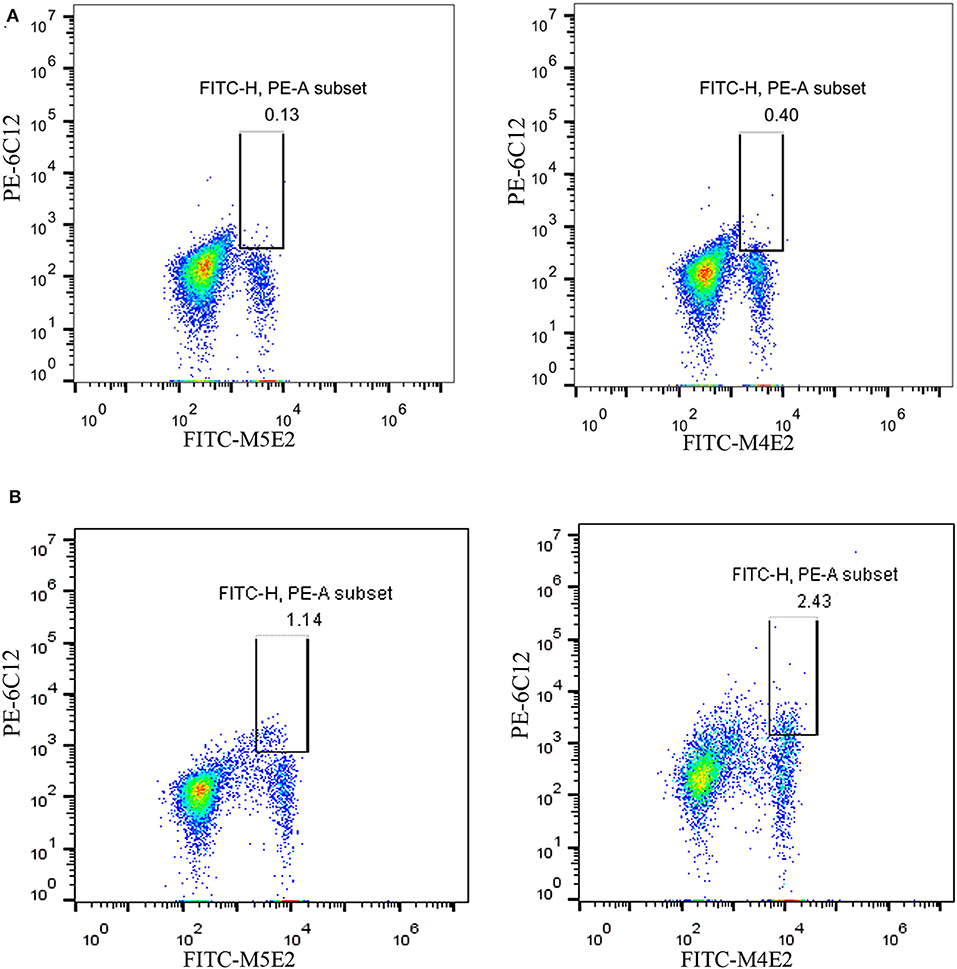
Figure 5. Detection of intracellular Brucellae-infected PBMCs from brucellosis patients by FCM. Monocytes were sorted by staining with mAb to CD14 (FITC-M5E2), and were further calculated by mAb to Brucella Omp25 (PE-6C12) as a percentage of total monocytes. (A) Detection of PBMCs from Healthy blood donors who were used as negative control. (B) Detection of PBMCs from brucellosis patients who were confirmed positive by SAT and PCR at administration to hospital.
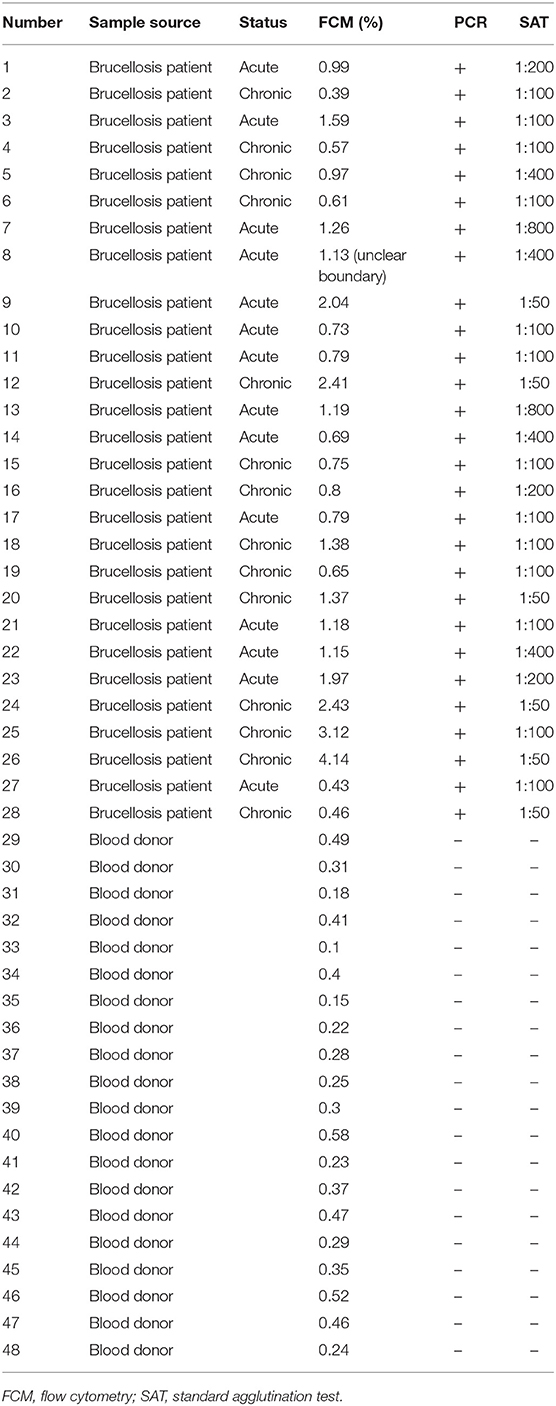
Table 2. Characterization of blood samples and detection results of Brucellae-infected PBMCs by FCM with mAb PE-6C12.
Several early studies focused on the immunogenicity and protective activity of Brucella Omp25 (Bowden et al., 1998). In this study, three non-overlapping linear epitopes (P2, P4, and P7) were recognized by mAbs 4A12 and 4F10, 8F3, and 2B10, respectively (Figure 4B). Previously mAb A59/05F01/C09 was reported to recognize a liner epitope of Omp25 (aa 24-43) (Salhi et al., 2003), which overlaps with the epitope P2 (aa24-42) recognized by mAbs 4A12 and 4F10 in this study. P1 peptide (aa1-30) has seven amino acids (24-ADAIQEQ-30) overlapping with P2 peptide (Table S1) but does not react with these mAbs, suggesting that mAbs 4A12 and 4F10 likely recognize the linear epitope PPVPAPVEVAPQ (aa 31-42) within Omp25. Another mAb A76/02C12/C11 to Omp25 was reported to strongly bind to rough Brucella isolates except B. ovis tested with a latex co-agglutination assay (Bowden et al., 1997), but the epitope was not clearly defined. In addition to the three different linear epitopes (P2, P4, and P7) revealed by mAbs 4A12 and 4F10, 8F3, and 2B10, a semi-conformational epitope within the denatured native Brucella Omp25 was identified by mAb 6C12 recognition. Interestingly, mAb 6C12 recognized two bands of 25 kD and 26 kD proteins from Brucella strains in WB (Figure 3B). The larger band (26 kD) was designated as Omp25d as previously described (Salhi et al., 2003), while the other four mAbs reacted only with the linear epitopes of 25 kD protein (Omp25). This might explain that 6C12 had better binding capacity in FCM than other mAbs for detecting Brucellae of PBMCs in comparison with mAb 5H3 to Omp31 or 5A5 to BP26 (Yang et al., 2019). These Omp25 mAbs had no cross-reactivity with Yersinia enterocolitica O:9, Salmonella spp., and Escherichia coli reported previously (Muñoz et al., 2015). However, previous studies found the high serological cross-reactivity between Brucella and other Alphaproteobacteria such as Ochrobactrum and Sinorhizobium (Cloeckaert et al., 1999; Velasco et al., 2000; Delpino et al., 2004). By aligning Brucella Omp25 aa sequence with Ochrobactrum anthropi and Rhizobiales 63-22 strains, the high homologous sequences are observed including the linear epitopes (P2, P4, and P7) within Brucella Omp25 recognized by mAbs 4A12/4F10, 8F3, and 2B10 (Figure S3), which indicate the cross-reactivity of these mAbs may exist in other Alphaproteobacteria.
B. melitensis, B. abortus, and B. suis are three major pathogenic strains for human brucellosis transmitted from infected sheep and goats, cattle, or pigs (Chen et al., 2014; Ducrotoy et al., 2018). Among five mAbs, 8F3, 4A12, and 6C12 had higher relative affinity (>1) reacted with Omp25 of five strains (M5-90, S19, 104M, 2308, and S2) from three Brucella in various immunoassays. mAb 8F3 presented a strong capacity as a capture antibody (Figure 4C), while 6C12 and 4A12 appeared to be the best detection antibody when conjugated with HRP or fluorescein (Figure 4C, Figures S1, S2). The best-matched antibody pairs were 8F3/6C12-HRP and 8F3/4A12-HRP for detection of Brucellae in DAS-ELISA, while mAb 6C12 was the most suitable for detection of intracellular Brucellae in PBMCs from patients by FCM (Figure 5).
Currently, there is no an optimal immunoassay for detecting intracellular Brucellae in terms of counting the infected monocytes for evaluation of anti-Brucella efficacy in clinical treatment of brucellosis. Conventional method of blood culture requires a biosafety level 3 laboratory (BSL-3) and takes 1–2 weeks (Pappas et al., 2006). In this study, we sought to develop an Omp25 mAb-based FCM assay to detect intracellular Brucellae-infected PBMCs from brucellosis patients, which might be helpful for evaluating therapeutic efficacy by determining the percentage of Brucella-infected monocytes before or after treatment of individual patients with brucellosis. By analyzing 28 patients with brucellosis, who were found positive upon admission to hospital by SAT and PCR, eight out of 28 (28.6%) and five out of 28 (17.8%) brucellosis patients had more than 1 or 2% intracellular Brucellae infected PBMCs, respectively. Overall, 46.4% (13 out of 28) patients carried a higher level (>1%) of intracellular Brucellae monocytes and 53.6% (15 out of 28) patients had a lower level (<1%) or no detectable intracellular Brucellae monocytes (Table 2). The ratios of mAb 6C12-stained monocytes from PBMCs were consistent with the amounts of intracellular Brucellae in brucellosis patients. In our previous study, we utilized IFS for detecting Brucella-infected PBMCs stained with Omp31 mAbs (Yang et al., 2019) and found 50–70% positive PBMCs in patients receiving zero to three treatment courses of anti-Brucella drugs. IFS is a qualitative test unsuitable to quantify the number of Brucella-infected cells in a patient's PBMCs. The selected mAb 6C12 has been shown effective for detection of intracellular Brucella by IFS (Figure S1). However, the established FCM with mAb 6C12 specific to Omp25 has an advantage over IFS for counting the frequency of intracellular Brucellae-infected monocytes and might be a potential assay for evaluation of therapeutic efficacy of brucellosis patients in clinical practice.
In conclusion, mAbs 8F3 was suitable as an immobilized capture antibody, and 6C12 or 4A12 was suitable as a conjugated detection antibody. These mAbs specific to Brucella Omp25 appear precious reagents in various immunoassays, such as WB, ELISA, ICS, IFS, and FCM assays for detection of different Brucella species in the diagnosis of brucellosis or evaluation of therapeutic efficacy of anti-Brucella treatment in clinical practice.
All datasets generated for this study are included in the article/Supplementary Material.
The animal study and studies involving human participants were reviewed and approved by Southern Medical University (SMU) Nan Fang Hospital Medical Ethics Committee (permit numbers: NFYY-2009-23). The patients/participants provided their written informed consent to participate in this study.
WW, CL, and XY participated in the study design, analysis of data, and writing of the manuscript. XY, GZ, CC, LL, and YF collected blood samples and interpreted the patient data regarding the brucellosis disease. XY, ZH, JL, YT, HZ, HR, and HY performed the laboratory examination. J-PA analyzed the data and revised the manuscript. All authors read and approved the final version of manuscript.
This study was funded by National Key Research and Development Program of China (No. 2017YFD0500300), National Natural Science Foundation of China (No. 31372443), Innovative R&D Team Introduction Program of Guangdong (No. 2014ZT05S123), Military Logistics Research Project (No. CWH17C017), and Research Initiative Project of SMU (CX2017N007). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Xia Rong at the Guangzhou Blood Centre for the collection of blood donor samples and the physicians in the Department of Infectious Disease, Heilongjiang General Hospital of Agriculture Bureau for their helpful clinical information analysis. We thank Dr. Yandong Tang in Harbin Veterinary Research Institute for his assistance in laboratory experiments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.00145/full#supplementary-material
Ahmed, I. M., Khairani-Bejo, S., Hassan, L., Bahaman, A. R., and Omar, A. R. (2015). Serological diagnostic potential of recombinant outer membrane proteins (rOMPs) from Brucella melitensis in mouse model using indirect enzyme-linked immunosorbent assay. BMC. Vet. Res. 11:275. doi: 10.1186/s12917-015-0587-2
Araj, G. F. (2010). Update on laboratory diagnosis of human brucellosis. Int. J. Antimicrob. Agents. 36(Suppl. 1), S12–S17. doi: 10.1016/j.ijantimicag.2010.06.014
Bowden, R. A., Cloeckaert, A., Zygmunt, M. S., and Dubray, G. (1998). Evaluation of immunogenicity and protective activity in BALB/c mice of the 25-kDa major outer-membrane protein of Brucella melitensis (Omp25) expressed in Escherichia coli. J. Med. Microbiol. 47, 39–48. doi: 10.1099/00222615-47-1-39
Bowden, R. A., Verger, J. M., Grayon, M., and Cloeckaert, A. (1997). Rapid identification of rough Brucella isolates by a latex coagglutination assay with the 25-kilodalton outer membrane protein and rough-lipopolysaccharide-specific monoclonal antibodies. Clin. Diagn. Lab. Immunol. 4, 611–614. doi: 10.1128/CDLI.4.5.611-614.1997
Chen, S., Zhang, H., Liu, X., Wang, W., Hou, S., Li, T., et al. (2014). Increasing threat of brucellosis to low-risk persons in urban settings. Emerg. Infect. Dis. 20, 126–130. doi: 10.3201/eid2001.130324
Cloeckaert, A., Tibor, A., and Zygmunt, M. S. (1999). Brucella outer membrane lipoproteins share antigenic determinants with bacteria of the family rhizobiaceae. Clin. Diagn. Lab. Immunol. 6, 627–629. doi: 10.1128/CDLI.6.4.627-629.1999
Cloeckaert, A., Vizcaino, N., Paquet, J. Y., Bowden, R. A., and Elzer, P. H. (2002). Major outer membrane proteins of Brucella spp.: past, present and future. Vet. Microbiol. 90, 229–247. doi: 10.1016/S0378-1135(02)00211-0
Delaporte, M., McKenna, P., Siah, A., and Berthe, F. C. J. (2008). Immunophenotyping of mya arenaria neoplastic hemocytes using propidium iodide and a specific monoclonal antibody by flow cytometry. J. Invertebr. Pathol. 99, 120–122. doi: 10.1016/j.jip.2008.04.008
Delpino, M. V., Fossati, C. A., and Baldi, P. C. (2004). Occurrence and potential diagnostic applications of serological cross-reactivities between Brucella and other alpha-proteobacteria. Clin. Diagn. Lab. Immunol. 11, 868–873. doi: 10.1128/CDLI.11.5.868-873.2004
Divya, G., Jayaprakash, N. S., and Venkataraman, K. (2018). Development and characterization of monoclonal antibodies against nitro-166 tyrosine of high-density lipoprotein: apolipoprotein A1. Monoclon. Antib. Immunodiagn. Immunother. 37, 167–174. doi: 10.1089/mab.2018.0018
Ducrotoy, M. J., Muñoz, P. M., Conde-Álvarez, R., Blasco, J. M., and Moriyón, I. (2018). A systematic review of current immunological tests for the diagnosis of cattle brucellosis. Prev. Vet. Med. 151, 57–72. doi: 10.1016/j.prevetmed.2018.01.005
Edmonds, M. D., Cloeckaert, A., Booth, N. J., Fulton, W. T., Hagius, S. D., Walker, J. V., et al. (2001). Attenuation of a Brucella abortus mutant lacking a major 25 kDa outer membrane protein in cattle. Am. J. Vet. Res. 62, 1461–1466. doi: 10.2460/ajvr.2001.62.1461
Edmonds, M. D., Cloeckaert, A., and Elzer, P. H. (2002). Brucella species lacking the major outer membrane protein Omp25 are attenuated in mice and protect against Brucella melitensis and Brucella ovis. Vet. Microbiol. 88, 205–221. doi: 10.1016/S0378-1135(02)00110-4
Goel, D., and Bhatnagar, R. (2012). Intradermal immunization with outer membrane protein 25 protects Balb/c mice from virulent B. abortus 544. Mol. Immunol. 51, 159–168. doi: 10.1016/j.molimm.2012.02.126
Goel, D., Rajendran, V., Ghosh, P. C., and Bhatnagar, R. (2013). Cell mediated immune response after challenge in Omp25 liposome immunized mice contributes to protection against virulent Brucella abortus 544. Vaccine 31, 1231–1237. doi: 10.1016/j.vaccine.2012.12.043
He, Z., Luo, P., Hu, F., Weng, Y., Wang, W., and Li, C. (2016). Expression and identification of eukaryotic expression vectors of Brucella melitensis lipoprotein OMP19. Chin. J. Cell Mol Immunol. 32, 470–473.
Li, J., Hu, F., Chen, S., Luo, P., He, Z., Wang, W., et al. (2017). Characterization of novel Omp31 antigenic epitopes of Brucella melitensis by monoclonal antibodies. BMC Microbiol. 17:115. doi: 10.1186/s12866-017-1025-3
Luo, Y., Terkawi, M. A., Jia, H., Aboge, G. O., Goo, Y. K., Cao, S, et al. (2012). A double antibody sandwich enzyme-linked immunosorbent assay for detection of secreted antigen 1 of Babesia microti using hamster model. Exp.Parasitol. 130, 178–182. doi: 10.1016/j.exppara.2011.10.012
Ma, Q., Liu, A., Ma, X., Wang, Y., Hou, Y., and Wang, Z. (2015). Brucella outer membrane protein Omp25 induces microglial cells in vitro to secrete inflammatory cytokines and inhibit apoptosis. Int. J. Clin. Exp. Med. 8, 17530–17535.
Macdonald, R. A., Hosking, C. S., and Jones, C. L. (1988). The measurement of relative antibody affinity by ELISA using thiocyanate elution. J. Immunol. Methods. 106, 191–194. doi: 10.1016/0022-1759(88)90196-2
Mohammadi, E., and Golchin, M. (2018). Detection of Brucella abortus by immunofluorescence assay using anti outer membrane protein of 19 kDa antibody. Adv. Clin. Exp. Med. 27, 643–648. doi: 10.17219/acem/85081
Muñoz, P. M., Marín, C. M., Monreal, D., González, D., Garin-Bastuji, B., Díaz, R., et al. (2015). Efficacy of several serological tests and antigens for diagnosis of bovine brucellosis in the presence of false-positive serological results due to Yersinia enterocolitica O:9. Clin. Diagn. Lab. Immunol. 12, 141–151. doi: 10.1128/CDLI.12.1.141-151.2005
Okumura, T., Masuda, K., Watanabe, K., Miyadai, K., Nonaka, K., Yabuta, M., et al. (2015). Efficient enrichment of high-producing recombinant Chinese hamster ovary cells for monoclonal antibody by flow cytometry. J. Biosci. Bioeng. 120, 340–346. doi: 10.1016/j.jbiosc.2015.01.007
Pappas, G., Panagopoulou, P., Christou, L., and Akritidis, N. (2006). Brucella as a biological weapon. Cell Mol. Life. Sci. 63, 2229–2236. doi: 10.1007/s00018-006-6311-4
Patra, K. P., Saito, M., Atluri, V. L., Rolan, H. G., Young, B., Kerrinnes, T., et al. (2014). A protein-conjugate approach to develop a monoclonal antibody-based antigen detection test for the diagnosis of human brucellosis. PLoS Negl. Trop. Dis. 8:e29266. doi: 10.1371/journal.pntd.0002926
Pullen, G. R., Fitzgerald, M. G., and Hosking, C. S. (1986). Antibody avidity determination by ELISA using thiocyanate elution. J. Immunol. Methods. 86:83–87. doi: 10.1016/0022-1759(86)90268-1
Qiu, J., Wang, W., Wu, J., Wang, Y., Qiao, J., Chen, C., et al. (2012). Characterization of periplasmic protein BP26 epitopes of Brucella melitensis reacting with murine monoclonal and sheep antibodies. PLoS ONE 7:e34246. doi: 10.1371/journal.pone.0034246
Salhi, I., Boigegrain, R. A., Machold, J., Weise, C., Cloeckaert, A., and Rouot, B. (2003). Characterization of new members of the group 3 outer membrane protein family of Brucella spp. Infect. Immun. 71, 4326–4332. doi: 10.1128/IAI.71.8.4326-4332.2003
Tiwari, S., Kumar, A., Thavaselvam, D., Mangalgi, S., Rathod, V., Prakash, A., et al. (2013). Development and comparative evaluation of a plate enzyme-linked immunosorbent assay based on recombinant outer membrane antigens Omp28 and Omp31 for diagnosis of human brucellosis. Clin. Vaccine Immunol. 20, 1217–1222. doi: 10.1128/CVI.00111-13
Velasco, J., Bengoechea, J. A., Brandenburg, K., Lindner, B., Seydel, U., González, D., et al. (2000). Brucella abortus and its closest phylogenetic relative, Ochrobactrum spp., differ in outer membrane permeability and cationic peptide resistance. Infect. Immun. 68, 3210–3218. doi: 10.1128/IAI.68.6.3210-3218.2000
Wang, W., Liao, Q., Wu, X., Hou, S., Wang, Y., Wu, J., et al. (2015). Potential risk of blood transfusion-transmitted brucellosis in an endemic area of China. Transfusion 55, 586–592. doi: 10.1111/trf.12853
Yang, H., Zhang, G., Luo, P., He, Z., Hu, F., Li, L., et al. (2019). Detection of Brucellae in peripheral blood mononuclear cells for monitoring therapeutic efficacy of brucellosis infection. Antimicrob. Resist. Infect. Control. 8:154. doi: 10.1186/s13756-019-0607-2
Yousefi, S., Tahmoorespur, M., and Sekhavati, M. H. (2016). Cloning, expression and molecular analysis of Iranian Brucella melitensis Omp25 gene for designing a subunit vaccine. Res. Pharm. Sci. 11, 412–418. doi: 10.4103/1735-5362.192493
Zhang, J., Guo, F., Huang, X., Chen, C., Liu, R., Zhang, H., et al. (2014). A novel Omp25-binding peptide screened by phage display can inhibit Brucella abortus 2308 infection in vitro and in vivo. J. Med. Microbiol. 63, 780–787. doi: 10.1099/jmm.0.069559-0
Keywords: brucellosis, Brucella, Omp25, mAb, flow cytometry assay, diagnosis
Citation: Yang X, He Z, Zhang G, Lu J, Zhang H, Ren H, Tian Y, Yang H, Chen C, Li L, Fu Y, Allain J-P, Li C and Wang W (2020) Evaluation of Reactivity of Monoclonal Antibodies Against Omp25 of Brucella spp.. Front. Cell. Infect. Microbiol. 10:145. doi: 10.3389/fcimb.2020.00145
Received: 30 September 2019; Accepted: 19 March 2020;
Published: 21 April 2020.
Edited by:
Max Maurin, Université Grenoble Alpes, FranceReviewed by:
Jalal Abdolalizadeh, Tabriz University of Medical Sciences, IranCopyright © 2020 Yang, He, Zhang, Lu, Zhang, Ren, Tian, Yang, Chen, Li, Fu, Allain, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengyao Li, Y2hlbmd5YW9saUBob3RtYWlsLmNvbQ==; Wenjing Wang, d2VuamluZ0BzbXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.