- 1Division of Molecular Parasitology, Proteo-Science Center, Ehime University, Toon, Japan
- 2Division of Malaria Research, Proteo-Science Center, Ehime University, Matsuyama, Japan
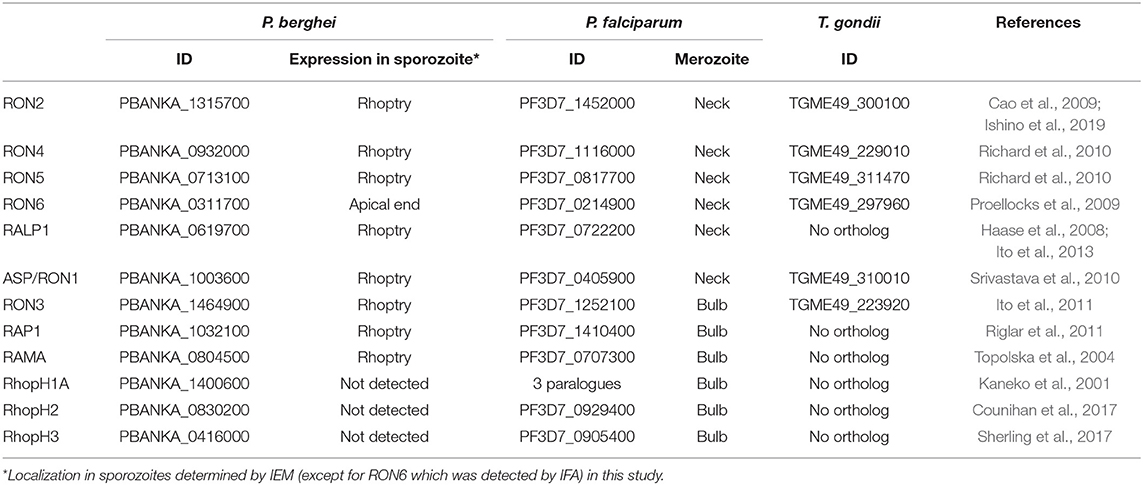
In the Plasmodium lifecycle two infectious stages of parasites, merozoites, and sporozoites, efficiently infect mammalian host cells, erythrocytes, and hepatocytes, respectively. The apical structure of merozoites and sporozoites contains rhoptry and microneme secretory organelles, which are conserved with other infective forms of apicomplexan parasites. During merozoite invasion of erythrocytes, some rhoptry proteins are secreted to form a tight junction between the parasite and target cell, while others are discharged to maintain subsequent infection inside the parasitophorous vacuole. It has been questioned whether the invasion mechanisms mediated by rhoptry proteins are also involved in sporozoite invasion of two distinct target cells, mosquito salivary glands and mammalian hepatocytes. Recently we demonstrated that rhoptry neck protein 2 (RON2), which is crucial for tight junction formation in merozoites, is also important for sporozoite invasion of both target cells. With the aim of comprehensively describing the mechanisms of sporozoite invasion, the expression and localization profiles of rhoptry proteins were investigated in Plasmodium berghei sporozoites. Of 12 genes representing merozoite rhoptry molecules, nine are transcribed in oocyst-derived sporozoites at a similar or higher level compared to those in blood-stage schizonts. Immuno-electron microscopy demonstrates that eight proteins, namely RON2, RON4, RON5, ASP/RON1, RALP1, RON3, RAP1, and RAMA, localize to rhoptries in sporozoites. It is noteworthy that most rhoptry neck proteins in merozoites are localized throughout rhoptries in sporozoites. This study demonstrates that most rhoptry proteins, except components of the high-molecular mass rhoptry protein complex, are commonly expressed in merozoites and sporozoites in Plasmodium spp., which suggests that components of the invasion mechanisms are basically conserved between infective forms independently of their target cells. Combined with sporozoite-stage specific gene silencing strategies, the contribution of rhoptry proteins in invasion mechanisms can be described.
Introduction
During the malaria lifecycle, three invasive forms, ookinetes, sporozoites, and merozoites, invade different types of cells in mosquito vectors and mammalian hosts. Among them, sporozoites, which transmit malaria disease from mosquitoes to mammalian hosts, firstly invade mosquito salivary glands prior to be injected into the mammalian skin during a blood meal. Sporozoites actively migrate through the skin to enter blood vessels and finally infect hepatocytes, where they develop into several thousand merozoites within a parasitophorous vacuole membrane (PVM). Merozoites invade erythrocytes by similarly forming a PVM, while ookinetes, the invasive form which develops in the midgut lumen after fertilization, simply traverse midgut epithelial cells without PVM formation.
The apical structures of invasive stage parasites in the phylum Apicomplexa are well-conserved as having microneme and rhoptry secretory organelles, which store proteins discharged prior to or post-invasion. Micronemes in the highly motile sporozoite and ookinete stages contain proteins involved in motility and cell traversal (Sultan et al., 1997; Yuda and Ishino, 2004; Kariu et al., 2006). In the case of the non-motile merozoite stage parasites micronemal proteins, such as merozoite surface proteins (MSPs), are involved in attachment to erythrocytes (Kadekoppala and Holder, 2010). Rhoptries are present only in infective stage parasites, such as Plasmodium merozoites and sporozoites which proliferate inside the PVM, and are absent in ookinetes, raising the possibility that rhoptry secretory proteins are involved in cell infection (reviewed in Baum et al., 2008; Frenal et al., 2017).
Rhoptry protein profiling has been conducted and characterized mainly in the related apicomplexans Toxoplasma tachyzoites and Plasmodium merozoites, and are classified into two protein groups based on their localization in rhoptry neck or rhoptry bulb (Bradley et al., 2005; Counihan et al., 2013). Rhoptry neck protein 2 (RON2), RON4, and RON5 are discharged as a complex prior to invasion and inserted into the target cellular membrane. The complex then interacts with apical merozoite protein 1 (AMA1) on the parasite plasma membrane to form a tight junction between the parasite and its target cell, a step which is essential for Plasmodium merozoite and Toxoplasma tachyzoite invasion of target cells (Alexander et al., 2005; Lebrun et al., 2005; Besteiro et al., 2009; Cao et al., 2009). Rhoptry bulb proteins are discharged subsequent to rhoptry neck proteins, to develop inside the parasitophorous vacuole. Many rhoptry blub proteins are species specific, in contrast to rhoptry neck proteins which are largely conserved between Plasmodium and Toxoplasma, such as components for the RON complex which is crucial for invasion by both parasites (Boothroyd and Dubremetz, 2008; Zuccala et al., 2012; Counihan et al., 2013; Kemp et al., 2013).
It has been studied whether tight junction formation mechanisms are critical for Plasmodium sporozoite invasion of mosquito salivary gland and mammalian hepatocyte target cells. This hypothesis is supported by the finding that the components of the RON complex, RON2, RON4, and RON5, are also expressed in sporozoites (Tufet-Bayona et al., 2009; Mutungi et al., 2014; Risco-Castillo et al., 2014). Moreover, a peptide inhibiting the interaction between AMA1 and RON2 reduced the P. falciparum sporozoite infection ability of cultured hepatocytes (Yang et al., 2017), and a conditional knockdown of RON2 or RON4 resulted in a reduction in sporozoite invasion ability (Giovannini et al., 2011; Ishino et al., 2019).
A comprehensive analysis of rhoptry proteins during sporozoite invasion using the sporozoite-stage specific knockdown system first requires the detailed profiling of rhoptry proteins in sporozoites. To date, about thirty rhoptry proteins have been classified in P. falciparum (reviewed in Counihan et al., 2013). In the present study, 12 proteins were selected as merozoite rhoptry proteins commonly expressed among Plasmodium spp.—i.e., expressed in both human and rodent malaria parasites—to examine their expression and localization in sporozoites. To achieve this goal, transgenic parasites were generated in P. berghei expressing target rhoptry proteins tagged with c-Myc tag at their C-terminus. Immuno-electron microscopy revealed that eight out of 12 candidate proteins are also localized to sporozoite rhoptries.
Materials and Methods
Parasites and Mosquitoes
A transgenic Plasmodium berghei ANKA parasite line was used in this study which constitutively expresses GFP under the control of the elongation factor 1A (ef1α) promoter without any drug resistance gene (Franke-Fayard et al., 2004), kindly given by Dr. Janse. Cryopreserved P. berghei ANKA infected erythrocytes were intraperitoneally injected into female ICR mice (4–6 weeks old, CLEA Japan, Tokyo, Japan) to obtain asexual stage parasites. To harvest mature schizonts, infected mouse erythrocytes with 0.5–1% parasitemia were cultured for 16 h and purified using Nycoprep 1.077 solution (Axis-Shield Diagnostics, Dundee, UK; Janse et al., 2006b). Anopheles stephensi SDA strain (An. stephensi) mosquitoes were maintained on a 5% sucrose solution during adult stages at 25°C. After feeding on P. berghei infected ICR mice, fully engorged mosquitoes were selected and kept at 20°C until dissection under a 12 h-light/12 h-dark cycle. All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Ehime University and the experiments were conducted according to the Ethical Guidelines for Animal Experiments of Ehime University.
Real Time Reverse Transcription (RT)-PCR Analysis
Purified schizonts and infected-mosquito tissues (midguts and salivary glands) were collected in RNAlater (Thermo Fisher Scientific, San Jose, CA, USA) and stored at 4°C until RNA isolation. Total RNA was extracted using an RNeasy kit (Qiagen, GmbH, Hilden, Germany) and treated with DNaseI (Qiagen). Reverse transcription was conducted using a PrimeScript RT reagent Kit (Takara Bio, Otsu, Japan) with gDNA Eraser. Real-time RT-PCR reactions were performed using SYBR Premix Ex Taq (Takara Bio). The primer sequences used are listed in Supplementary Table S1. Real time PCR was performed using a TaKaRa PCR Thermal Cycler Dice (Takara Bio). Relative gene expressions were normalized by ef1α (PBANKA_1133300) mRNA levels and were compared using the delta, delta-Ct method (Pfaffl, 2001; Ishino et al., 2019).
Generation of c-Myc-Tagged Rhoptry Protein Expressing Transgenic Parasites
To generate transgenic parasites expressing a rhoptry protein tagged with c-Myc at its C-terminus, the native locus of the targeted rhoptry molecule in the WT-GFP genome was replaced by single crossover homologous recombination with an expression cassette of the C-terminus of the rhoptry protein fused with a c-Myc tag, similar to the generation of RON2-c-Myc expressing parasites (Ishino et al., 2019). Schematic representation of the transgenic vector construction is shown in Supplementary Figure S1. Approximately 1,000–2,000 base pair of DNA fragments including the C-terminus of each rhoptry protein were amplified with specific primers (sequences of primers used in this study were listed in Supplementary Table S1) by PCR from genomic DNA of WT-GFP. Amplified PCR fragments of RAP1, RhopH1A, RhopH2, and RhopH3 were inserted into the pL0033 plasmid (BEI Resources, Manassas, VA, USA) at SacII and NcoI sites just before the c-Myc tag coding region, and these plasmids were then linearized at endogenous HpaI, SpeI, and XbaI sites, respectively (see Supplementary Figure S1A). RON5 and RALP1 fragments were inserted into an NdeI site disrupted pL0033 vector, which was linearized at an endogenous NdeI site (see Supplementary Figure S1B). To introduce XbaI recognition sites for linearization into the PCR fragments of RON3 and RON4, site directed mutagenesis was performed to introduce mutations without amino acid substitution as follows: RON3, 5716A > T and 5717G > C; and RON4, 1711T > C. Using the same strategy, the endogenous NcoI site in the amplified RON6 fragment was disrupted, to avoid interference with ligation into the SacII and NcoI sites of pL0033 (1878C > A). These DNA fragments were inserted into the pL0033 plasmid at SacII and NcoI sites, which were linearized at an introduced XbaI site for RON3 and RON4, and at an endogenous BamHI site for RON6 (see Supplementary Figure S1C). Electroporation of 10–15 μg linearized DNA into schizont-enriched WT-GFP and selection of transgenic parasites were performed as described (Janse et al., 2006a). DNA integration occurs at the target locus in the WT-GFP genome by single crossover homologous recombination as illustrated in Supplementary Figure S2. DNA integration into the target locus was confirmed by PCR genotyping and transgenic parasites were cloned by limiting dilution.
Antibody Production
DNA fragments encoding amino acids 25–694 of ASP/RON1 and 899–1,072 of RON3 were amplified from P. berghei schizont cDNA by PCR and inserted into pEU-E01-GST-(TEV)-N1 (CellFree Sciences, Matsuyama, Japan) at EcoRV and BamHI sites to produce recombinant ASP/RON1 proteins and at XhoI and BamHI sites to produce recombinant RON3 with GST tag at their N-terminus. The GST-tagged recombinant proteins were produced using the wheat germ cell-free protein expression system (CellFree Sciences) and purified using a glutathione-Sepharose 4B column (GE Healthcare UK, Buckinghamshire, UK; Tsuboi et al., 2008). Purified GST-tagged recombinant proteins were used for immunization of Japanese white rabbits with Freund's adjuvant. Immunizations were done three times at 3-week intervals with 250 μg recombinant protein and the antisera were collected 14 days after the last immunization (Kitayama Labes, Ina, Japan). Anti-RON2 antibodies and anti-RAMA antibodies used in the study were prepared previously (Ishino et al., 2019).
Western Blotting Analysis
Purified schizonts were treated with 0.08% saponin for 15 min on ice and the schizont pellets were resuspended in sample buffer solution for SDS-PAGE (nacalai tesque, Kyoto, Japan) containing 5% 2-mercaptoethanol. Sporozoites collected from midguts of infected mosquitoes at days 24–26 post-feeding were purified by density gradient centrifugation using 17% Accudenz solution (Accurate Chemical & Scientific Corporation, NY, United States; Kennedy et al., 2012). Sporozoite pellets were resuspended in sample buffer containing 5% 2-mercaptoethanol. Proteins were separated by SDS-PAGE using 5–20% gradient acrylamide gels (ATTO, Tokyo, Japan) and electroblotted onto polyvinylidene difluoride (PVDF) membranes. The PVDF membranes were blocked with Blocking One (nacalai tesque) overnight at 4°C and then incubated with primary antibodies (1:100 anti-c-Myc rabbit antibodies (A-14), Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1:2,500, anti-ASP/RON1, RON3, or RAMA rabbit antibodies) diluted in PBS containing 0.01% Tween-20 (PBST) for 2 h at room temperature. After washing with PBST, the membranes were incubated with secondary antibodies conjugated to horseradish peroxidase (HRP; 1:30,000, Biosource, Camarillo, CA, USA) for 30 min at room temperature. Chemiluminescence detection was performed by adding Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore, Darmstadt, Germany), and the signal was detected using ImageQuant LAS 4000 (GE Healthcare UK).
Indirect Immunofluorescence Assay
Thin smears of purified schizonts were prepared on glass slides and fixed with cold acetone for 3 min. Sporozoites collected from midgut and salivary gland at day 24–26 post-feeding were seeded on 8-well multi-well slides, then air-dried and fixed with cold acetone for 3 min. The slides of schizonts and sporozoites were blocked with PBS containing 10% fetal calf serum at 37°C for 30 min and incubated with primary antibodies (1:50 for anti-c-Myc mouse monoclonal antibodies (9E10), 1:100 for anti-c-Myc rabbit antibodies (A-14), and 1:200 for anti RON2, ASP/RON1, and RAMA rabbit polyclonal antibodies) in blocking solution at 37°C for 2 h, followed by Alexa Fluor 488–conjugated goat anti-rabbit IgG and Alexa Fluor 568-conjugated goat anti-mouse IgG (Thermo Fisher Scientific) at 37°C for 30 min. Nuclei were stained with 1 mg/ml of 4′,6-diamidino-2-phenylindole (DAPI). The samples were mounted in ProLong Gold antifade reagent (Thermo Fisher Scientific) and observed with an inverted fluorescence microscope (Axio Observer Z1, Carl Zeiss, Oberkochen, Germany).
Immuno-Transmission Electron Microscopy
Cultivated schizonts of WT-GFP or transgenic parasites expressing c-Myc tagged rhoptry proteins were purified by density gradient centrifugation. Infected midguts (day 17 or 21 post-feeding) or salivary glands (day 24 or 26 post-feeding) were dissected. The samples were fixed in 1% paraformaldehyde, 0.2% glutaraldehyde and embedded in LR-White resin (Polyscience, PA, USA). Ultrathin sections were blocked in PBS containing 5% non-fat dry milk and 0.01% Tween 20 (PBS-MT), then incubated at 4°C overnight with anti-c-Myc antibody (Santa Cruz biotechnology) or specific antibodies against ASP/RON1 (1:25), RAMA (1:100), or RON3 (1:200). The sections were washed with PBS containing 0.4% Block Ace (Yukijirushi, Tokyo, Japan) and 0.01% Tween 20 (PBS-BT), and the grids were incubated for 1 h at 37°C with goat anti-rabbit IgG conjugated with 15 nm gold particles (GE Healthcare) diluted 1:20 in PBS-MT, rinsed with PBS-BT, and fixed in 2% glutaraldehyde for 10 min at 4°C to stabilize the gold particles. The sections were then stained with 2% uranyl acetate in 50% methanol and lead citrate. Samples were examined using a transmission electron microscope (JEM-1230; JEOL, Tokyo, Japan).
Gene IDs
The sequence information of genes in this article can be found in the PlasmoDB database (PlasmoDB.org) under the following gene ID numbers: ron2, PBANKA_1315700; ron4, PBANKA_0932000; ron5, PBANKA_0713100; ron6, PBANKA_0311700; ralp1, PBANKA_0619700; asp/ron1, PBANKA_1003600; rap1, PBANKA_1032100; ron3, PBANKA_1464900; rama, PBANKA_0804500; rhoph1a, PBANKA_1400600; rhoph2, PBANKA_0830200; and rhoph3, PBANKA_0416000.
Results
Selection of Plasmodium Rhoptry Proteins
In this study a rodent malaria parasite line, P. berghei ANKA strain expressing GFP under the control of ef1α promoter (WT-GFP; Janse et al., 2006a), was used to characterize rhoptry proteins in both merozoites and sporozoites. From the catalog of known P. falciparum rhoptry proteins (Counihan et al., 2013), 12 molecules whose orthologous genes exist in P. berghei were selected to compare their expression between merozoites and sporozoites. RON2, a sporozoite rhoptry protein demonstrated as transcribed predominantly in oocyst-derived sporozoites (Ishino et al., 2019), was included as a positive control. Among 12 selected proteins, six are conserved across the Apicomplexa phylum and another six are specific to Plasmodium spp. Gene IDs in P. berghei together with those of the orthologous genes in P. falciparum and in Toxoplasma gondii are listed in Table 1 (Aurrecoechea et al., 2009).
Expression Profiling of Rhoptry Molecules in Schizonts and Developing Sporozoites
Firstly, the mRNA expression levels of the selected genes during sporozoite development in mosquito bodies were compared by real-time RT-PCR analysis with transcript levels in schizonts. Sporozoites, formed within oocysts on midguts of mosquitoes, are released into hemolymph and then invade salivary glands prior to being inoculated into mammalian skin with saliva. Since P. berghei sporozoite formation inside oocysts starts at days 10–14 post-feeding (Thathy et al., 2002; Ferguson et al., 2014), parasite-infected midguts were collected at days 9, 13, and 17 post-feeding. In addition, salivary glands of infected mosquitoes were collected at day 17 post-feeding. As a control, schizont-rich infected erythrocytes were purified and harvested. Relative mRNA amounts of selected genes were examined by real-time RT-PCR, normalized by ef1α mRNA expression.
Six molecules categorized as encoding rhoptry neck proteins in merozoites (ron2, ron4, ron5, ron6, rhoptry-associated leucine zipper-like protein 1 (ralp1), and apical sushi protein (asp)/ron1) are also transcribed in sporozoites (Figure 1A). Among six genes encoding rhoptry proteins localized to the bulb region in merozoites, rhoptry-associated protein 1 (rap1), ron3, and rhoptry associated membrane antigen (rama) are also transcribed in sporozoites (Figure 1B); while the other three, encoding the components of the high-molecular mass rhoptry protein complex (RhopH complex; Kaneko et al., 2001, 2005; Ling et al., 2003, 2004; Vincensini et al., 2008; Comeaux et al., 2011; Nguitragool et al., 2011; Counihan et al., 2017; Ito et al., 2017; Sherling et al., 2017), are transcribed far less in sporozoites than in schizonts (Figure 1C). This data raises the possibility that RhopH1A, RhopH2, and RhopH3 may play roles predominantly in merozoites. In contrast, ron5, ron6, asp/ron1, and ron3 are predominantly transcribed in sporozoites vs. schizonts. The transcripts of rhoptry molecules expressed in sporozoites increase during sporozoite maturation in oocysts. After sporozoite invasion of salivary glands, the transcript amounts of ron2, ron4, rap1, ron3, and rama are significantly decreased, while transcripts of ron5, ron6, ralp1, and asp1/ron1 remain high or increase. In salivary gland sporozoites, ron5 and ron6 are the highest transcribed among rhoptry molecules; however, their amounts remain ~200-fold less than that of a micronemal molecule, sporozoite protein essential for cell traversal 2 (spect2; Ishino et al., 2005), whose transcription is strongly enhanced after sporozoites invade salivary glands (Figure 1D). These results demonstrate that most merozoite rhoptry molecules, except for those encoding RhopH complex components, are expressed in both infective stages, merozoites and sporozoites.
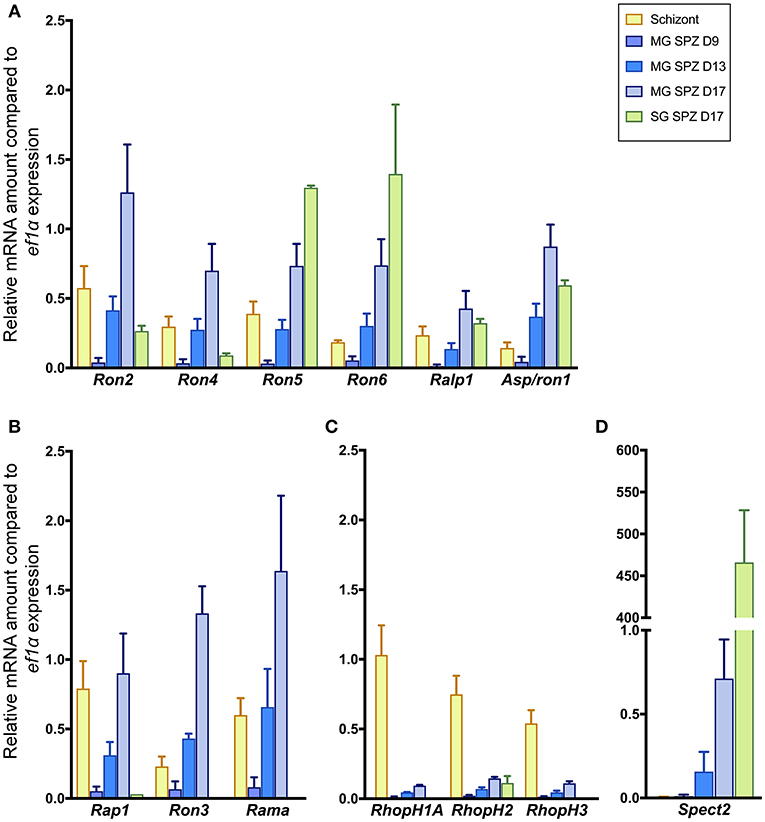
Figure 1. Transcriptional analyses of rhoptry genes in sporozoites and schizonts. Total RNA was extracted from parasite-infected mosquito midguts at days 9, 13, and 17 post-feeding (MG SPZ D9, MG SPZ D13, and MG SPZ D17); and salivary glands at day 17 post-feeding (SG SPZ D17). Total RNA was also extracted from schizont-enriched infected erythrocytes (Schizont). The mean values of relative mRNA amounts of each molecule, normalized by ef1α mRNA expression, are plotted as bar graphs with standard deviations from three independent experiments. (A) A group of molecules categorized as rhoptry neck proteins in merozoites. All examined molecules are transcribed in both merozoites and sporozoites. (B) A group of molecules categorized as rhoptry bulb proteins in merozoites whose transcriptions are detected in both merozoites and sporozoites. (C) A group of molecules categorized as rhoptry bulb proteins in merozoites whose transcriptions occur dominantly in merozoites. (D) Transcription profile of a typical micronemal protein, SPECT2, required for sporozoite migration toward hepatocytes after inoculation in the skin. Its transcript level drastically increases after sporozoite invasion of salivary glands.
Expression of Rhoptry Proteins in Sporozoites
To comprehensively examine protein expression patterns of rhoptry proteins in merozoites and sporozoites, transgenic parasite lines were generated by single-crossover homologous recombination to express each rhoptry protein fused with a C-terminal c-Myc tag (see Material and methods). Ten transgenic parasite lines were successfully isolated. Specific antibodies against recombinant protein were prepared for ASP/RON1 and RAMA, since they are predicted to have C-terminal glycosylphosphatidylinositol (GPI) anchored domains (Gilson et al., 2006) and therefore the modification of their C-terminal structure might disrupt their function (see Supplementary Figure S3). Specific antibodies against the middle region of RON3 were also prepared, because it was demonstrated that a 40 kDa fragment of C-terminal RON3 is cleaved during schizont maturation in P. falciparum (Ito et al., 2011).
Protein lysates of 1.5 × 105 schizonts and oocyst-derived sporozoites of each transgenic parasite line expressing c-Myc tagged rhoptry protein or WT-GFP were analyzed by western blotting using anti-c-Myc antibodies or specific antibodies against ASP/RON1, RAMA, and RON3. In schizonts all examined c-Myc tagged rhoptry proteins, except for RON3, were detected at the expected size of full-length (indicated by closed arrowheads, Figures 2A,B), demonstrating that c-Myc fused rhoptry proteins are successfully expressed. In addition, the processed forms of RON4 and RALP1 were detected at ~60 and 40 kDa (indicated by open arrowheads). In the case of RON3, anti-c-Myc antibodies detected ~40 kDa fragment as reported in P. falciparum, while anti-RON3 antibodies recognized two bands, near 250 kDa, corresponding to the full-length and processed RON3. It was confirmed that a roughly 40 kDa fragment of the C-terminal region in RON3 is cleaved in P. berghei mature schizonts as well as in P. falciparum. Antibodies against ASP/RON1 and RAMA recognized corresponding proteins at the size of expected full- and processed-proteins, confirming the specificity of these antibodies.
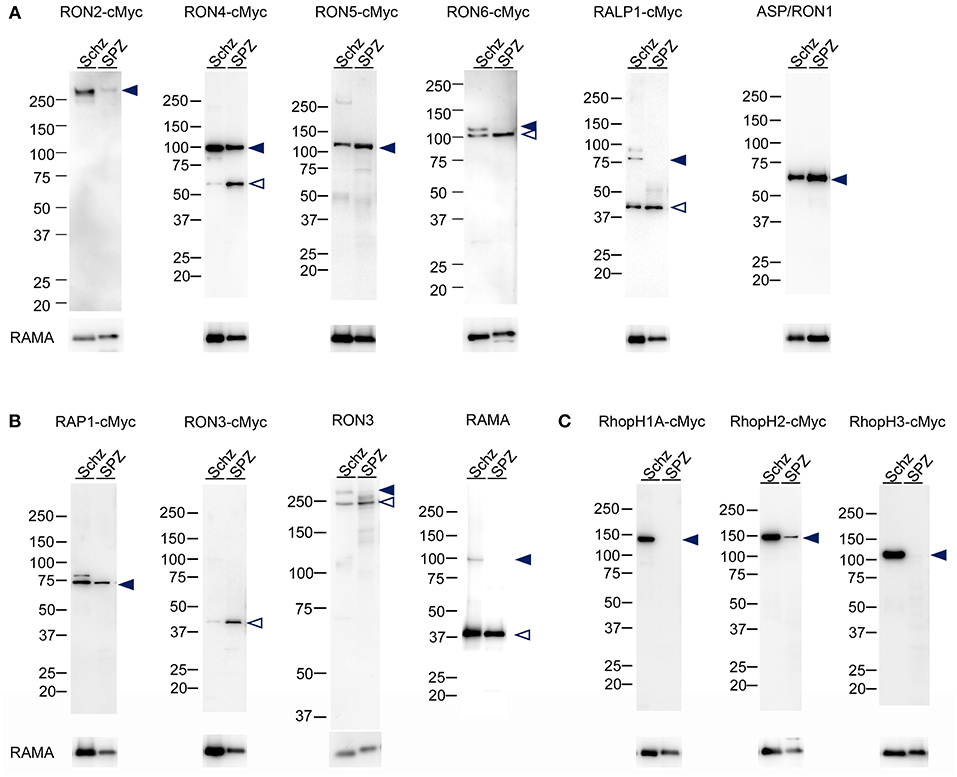
Figure 2. Western blot analyses of rhoptry proteins in schizonts and sporozoites. Proteins were extracted from schizonts purified after in vitro culture of parasite infected erythrocytes (Schz) or sporozoites purified from midguts at days 24–26 post-feeding (SPZ). Each lane contains proteins from 1.5 × 105 schizonts or sporozoites of transgenic parasites expressing c-Myc tagged rhoptry proteins or WT-GFP. Target proteins were detected by western blotting using anti c-Myc antibodies or specific antibodies against ASP/RON1, RON3, or RAMA. The transgenic parasite lines used as antigens are indicated above panels; for example, RON2-cMyc. When specific antibodies were used to detect target molecules in WT-GFP parasites, the name of the target molecule is instead indicated. Closed- and open- arrowheads demonstrate the expected full-length and cleaved target proteins, respectively. The sizes of protein markers (kDa) are indicated on the left of each image. (A) Expression patterns of proteins categorized as rhoptry neck proteins in merozoites. ASP/RON1 was detected using rabbit specific antibodies as it contains a C-terminal GPI-anchor domain and is likely refractory to C-terminal c-Myc integration. (B) Expression patterns of proteins which are categorized as rhoptry bulb proteins in merozoites and transcribed in both merozoites and sporozoites. In the case of RON3, anti-c-Myc antibodies only recognized a roughly 40 kDa protein (RON3-cMyc), demonstrating that the C-terminal region was cleaved in both merozoites and sporozoites. Specific anti-RON3 antibodies detected full-length and cleaved forms of RON3 (RON3). (C) Expression patterns of proteins which are categorized as rhoptry bulb proteins in merozoites and predominantly transcribed in schizonts. Consistent with transcription analyses, the protein amounts of RhopH1A, RhopH2, and RhopH3 are far less in sporozoites than those in merozoites, indicated by closed arrowheads.
RhopH1A and RhopH3 proteins were not detected in sporozoites, while RhopH2 was detected as a far weaker band compared to that in schizonts, which is in good agreement with the transcriptional data (Figure 2C). In addition, RON2 production in sporozoites was significantly less than in schizonts. Proteolysis patterns are conserved between schizonts and sporozoites, although the ratio of uncleaved protein is less in oocyst-derived sporozoites than in schizonts. Since it takes longer for sporozoite maturation in oocysts than merozoites in schizonts, proteolysis of rhoptry proteins might be enhanced in sporozoites.
Protein Localization of Rhoptry Molecules in Merozoites and Sporozoites
Rhoptry proteins in P. falciparum merozoites and T. gondii tachyzoites are categorized according to their detailed localization in rhoptries; specifically, rhoptry neck proteins (RON2, RON4, RON5, RON6, ASP/RON1, and RALP1) and rhoptry bulb proteins (RON3, RAP1, RAMA, RhopH1A, RhopH2, and RhopH3) (Counihan et al., 2013; Kemp et al., 2013). All rhoptry proteins examined in this study were confirmed to localize to the apical end region of P. berghei merozoites, similar to the RON2 marker signal, by immunofluorescent assay (IFA) using anti-c-Myc or specific antibodies (Figure 3, left columns). By comparison to RON2 localization, RON4, RON5, RON6, ASP/RON1, and RALP1 were suggested to be localized to the rhoptry neck region. This indicates that the C-terminal c-Myc tagging does not interfere with proper rhoptry protein localization. Additionally, their localization was examined in sporozoites collected from midguts and salivary glands (Figure 3, right columns). Specific signals corresponding to RhopH1A and RhopH3 were barely detected in sporozoites by IFA, confirming the western blotting data. The signal for RhopH2 was detected as a diffuse pattern in the cytoplasm of sporozoites. Taking into consideration the western blotting result, a small amount of RhopH2 is also expressed in sporozoites; however, it is not localized to rhoptries. Other examined proteins were detected at the apical end of sporozoites from both midguts and salivary glands, demonstrating that these proteins were transported to the apical region during sporozoite formation in oocysts and maintained even after sporozoite invasion of salivary glands.
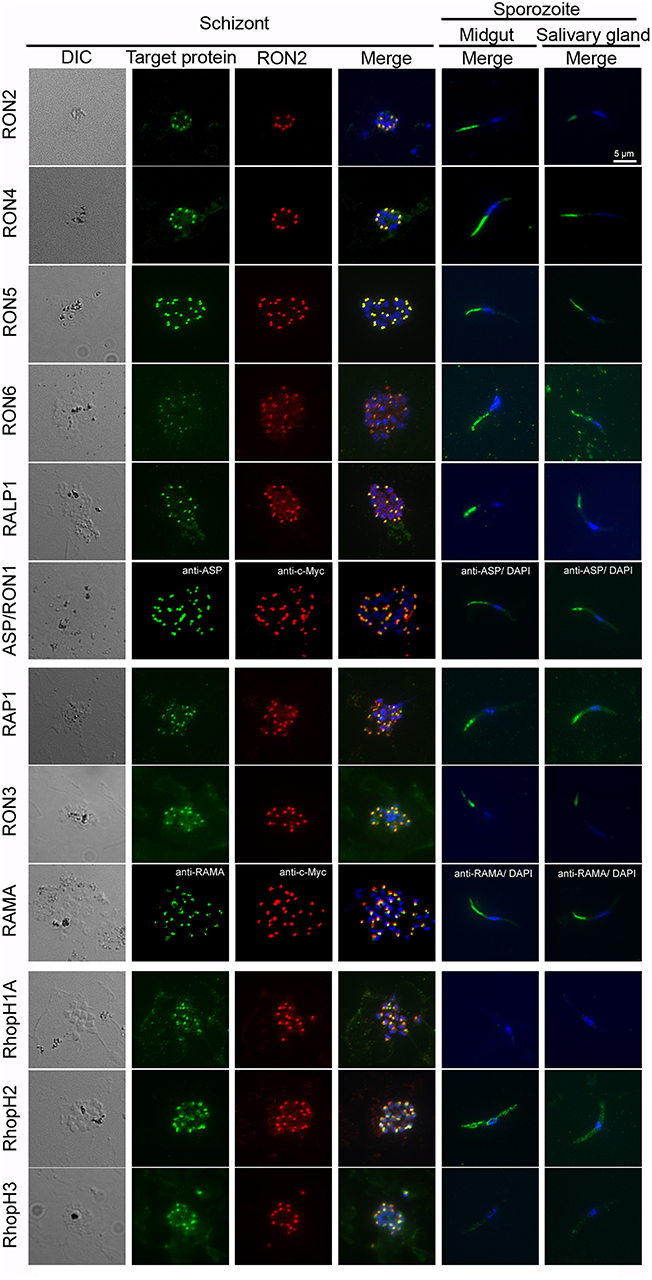
Figure 3. Expression pattern of rhoptry proteins in merozoites and sporozoites. Schizonts and sporozoites collected from midguts or salivary glands at day 24–26 post-feeding were fixed with acetone on glass slides. In schizonts, anti-c-Myc antibodies were used to detect target c-Myc fused rhoptry protein (shown in green) in each transgenic parasite, which is compared with the localization pattern of RON2 (shown in red) and nuclei (blue) stained by anti-RON2 antibodies and DAPI, respectively. To detect ASP/RON1 and RAMA, RON2-c-Myc transgenic parasites were used as antigens and target proteins and RON2 was detected by specific antibodies (green) and anti-c-Myc antibodies (red). Differential interference contrast (DIC) images are shown in the left panels. In sporozoites (right two panels), the localization of target proteins (green) and nuclei (blue) are shown in the merged images. Bar indicates 5 μm.
Eight Proteins Are Localized to Rhoptries in Sporozoites
To determine the precise localization of rhoptry proteins, immuno-electron microscopy (IEM) was performed using schizont stage merozoites and oocyst sporozoites. In P. berghei merozoites, RON2, RON4, RON5, RALP1, and ASP/RON1, which are categorized as rhoptry neck proteins in Pf merozoites, were confirmed to localize to the rhoptry neck region (Figure 4A). In addition, RAP1, RON3, RhopH1A, RhopH2, and RhopH3 are observed in the rhoptry bulb region, as reported for Pf merozoites (Figure 4B). RAMA is observed on the rhoptry membrane at the bulb region. RON6 could not be detected by anti-c-Myc antibodies, possibly because its protein amount in merozoites is not sufficient to be observed by IEM. This is the first comprehensive demonstration of rhoptry protein localization in Plasmodium merozoites.
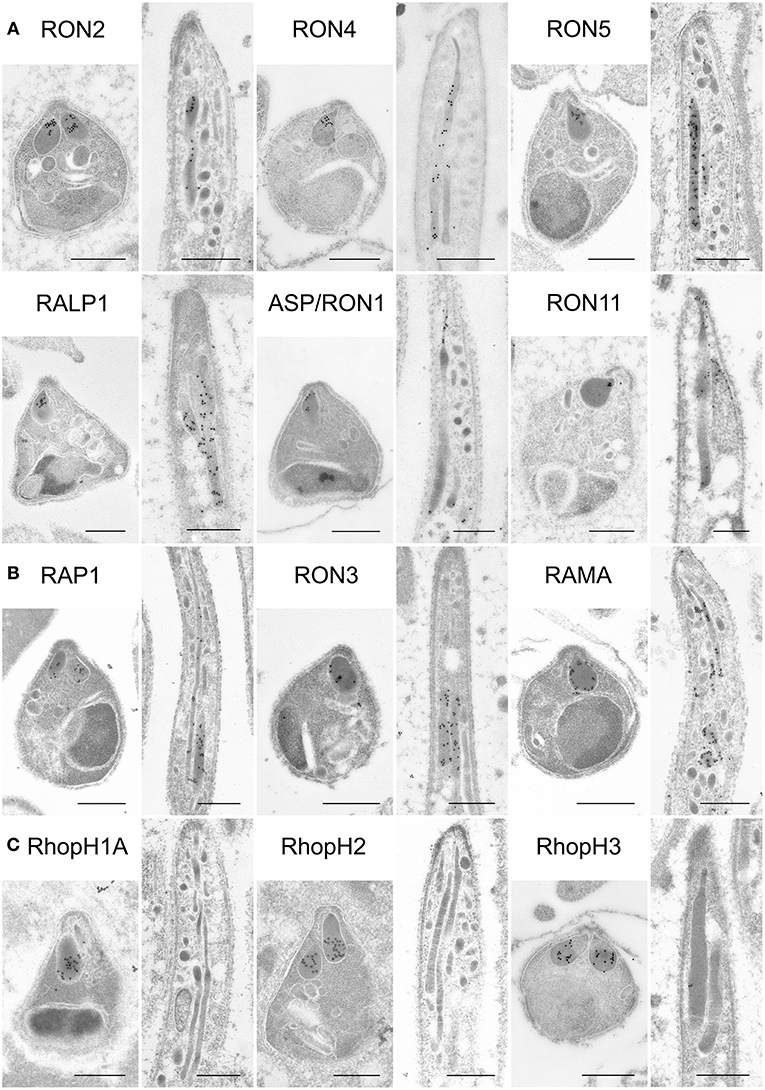
Figure 4. Localization analyses of rhoptry proteins in merozoites and sporozoites by immuno-electron microscopy. Longitudinally sectioned merozoites in cultivated schizonts and oocyst-derived sporozoites (at day 17 or 21 post-feeding) of each transgenic parasite line or WT-GFP were prepared for IEM analyses. Target proteins were detected by incubation with anti-c-Myc rabbit antibodies or specific antibodies against ASP/RON1 or RAMA, followed by secondary antibodies conjugated to gold particles. (A) A group of rhoptry proteins categorized as rhoptry neck proteins in merozoites. All examined proteins are confirmed to be localized to the rhoptry neck region in P. berghei merozoites. In contrast, most of them, except ASP/RON1, are distributed throughout rhoptries in sporozoites, as demonstrated previously regarding RON11. (B) A group of proteins expressed both in merozoites and sporozoites and categorized as rhoptry bulb proteins in merozoites. It is noteworthy that RAMA localizes to the rhoptry membrane in merozoites. In sporozoites, they are distributed in the rhoptry body region. (C) A group of proteins categorized as rhoptry bulb proteins in merozoites, which are produced predominantly in merozoites. IEM confirmed that none of them, RhopH1A, RhopH2, and RhopH3, are localized to rhoptries in sporozoites, despite their clear localization to the rhoptry bulb in merozoites. Bars, 500 nm.
In sporozoites formed inside oocysts, it was confirmed that three components for the RhopH complex do not accumulate in rhoptries, as expected from the observation of far less amounts of transcripts and proteins in sporozoites compared to merozoites (Figure 4C). Other than the RhopH complex, all proteins examined are localized to rhoptries in sporozoites as well as in merozoites. However, most proteins are distributed throughout rhoptries in sporozoites, despite their sub-localization in merozoites, suggesting that sub-compartmentation in rhoptries might be different between merozoites and sporozoites. This is consistent with the observation that the sub-localization of RON11 in rhoptries differs between merozoites and sporozoites (Figure 4A, Bantuchai et al., 2019). Only ASP/RON1 tends to accumulate in the thinner part in rhoptries near the tip of sporozoites. It is not clear whether RAMA localizes to the rhoptry membrane in sporozoites as in merozoites, as the maximum width of rhoptries is shorter in sporozoites than in merozoites. Matured sporozoites are released into the haemocoel followed by invasion of salivary glands. It was reported that, although morphologically similar, sporozoites in salivary glands show higher infectivity to the liver than sporozoites developed inside oocysts (Vanderberg, 1975); and accordingly transcription of some genes are upregulated or downregulated after sporozoite invasion of salivary glands (Kaiser et al., 2004; Mikolajczak et al., 2008; Tarun et al., 2008; Zhang et al., 2010). To determine rhoptry protein localization in liver infective sporozoites, salivary glands of transgenic parasites or WT-GFP infected mosquitoes were fixed for IEM analyses (Figure 5). RON2, RON4, RON5, RALP1, RAP1, and RAMA were detected in rhoptries of sporozoites residing in salivary glands, indicating that rhoptry proteins reside in rhoptries after sporozoite invasion of salivary glands, presumably available for subsequent invasion of hepatocytes in mammalian hosts. ASP/RON1 and RON3 could not be detected by anti-ASP/RON1 or anti-c-Myc antibodies, which might due to less target or c-Myc tagged protein amounts in salivary gland sporozoites.
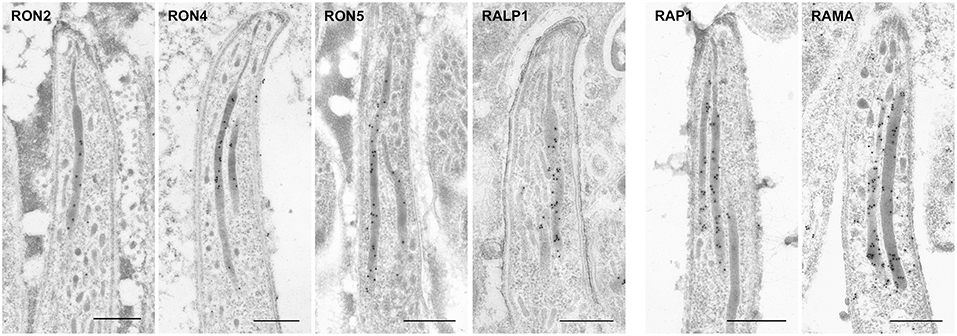
Figure 5. Localization of rhoptry proteins in sporozoites residing in mosquito salivary glands. Salivary glands of parasite infected mosquitoes were harvested at day 24 or 26 post-feeding and fixed for IEM. Target proteins were detected by anti-c-Myc antibodies or specific antibodies against ASP/RON1 or RAMA. All examined rhoptry proteins, despite their sub-localization in merozite rhoptries, are distributed throughout rhoptries in sporozoites residing in salivary glands, as well as oocyst-derived sporozoites. Bars, 500 nm.
Discussion
In this study it is demonstrated that most rhoptry proteins reported in merozoites are expressed in sporozoites, with the exception of components of the RhopH complex. Together with three reported proteins, RAP2/3, RON11, and RON12 (Tufet-Bayona et al., 2009; Bantuchai et al., 2019; Oda-Yokouchi et al., 2019), nine rhoptry proteins are demonstrated to be commonly expressed in the infective stages of P. berghei. Our results on mRNA expression in schizonts and sporozoites of rhoptry molecules are mostly confirmed by single cell RNA-seq analyses in P. berghei (Reid et al., 2018). The orthologs of these proteins in P. yoelii, another rodent malaria parasite, are transcribed in oocyst-derived sporozoites (Tarun et al., 2008) and their proteins, except for RALP1, were detected in salivary gland residing sporozoites (Lindner et al., 2013). In addition, the orthologous proteins in the human malaria parasite Plasmodium falciparum were detected in salivary gland residing sporozoites (Lindner et al., 2013). Taken together, our finding that the most rhoptry proteins, except for RhopH complex components, are expressed in both merozoites and sporozoites indicates conservation of expression among Plasmodium species.
It is noteworthy that all examined commonly expressed rhoptry proteins localize to the neck region in merozoites, that is RON2, RON4, RON5, RALP1, ASP1/RON1, RON11, and RON12; while some rhoptry bulb proteins, RhopH1A, RhopH2, and RhopH3, are expressed predominantly in merozoites. The orthologs of proteins localized to the rhoptry neck are highly conserved across the infective stages of apicomplexan parasites, such as Plasmodium, Toxoplasma, Babesia, and Cryptosporidium; however, proteins localized to the rhoptry bulb tend to be species specific. Since it has been proposed that rhoptry neck proteins, such as RON2, RON4, and RON5, are discharged to form the tight junction between parasites and host cells, this invasion mechanism could be conserved across the Apicomplexa phylum. Recently it was reported that the Toxoplasma genome contains paralogous genes for RON2 and AMA1, selectively expressed in sporozoites, suggesting that Toxoplasma has developed stage-specific invasion mechanisms (Poukchanski et al., 2013). Since the Plasmodium genome contains only one set of genes encoding RON2 and AMA1, it would be interesting to examine whether the rhoptry neck protein complex is formed during sporozoite invasion of mosquito salivary glands and mammalian hepatocytes. In contrast, it has been revealed that many of the Toxoplasma rhoptry bulb proteins are involved in interaction with host signaling pathways (Lim et al., 2012) and interference with host immunity after invasion (Fentress et al., 2010; Ong et al., 2010; Steinfeldt et al., 2010; Yamamoto et al., 2011; Niedelman et al., 2012). These mechanisms are likely host cell-specific adaptations, and therefore the expression of rhoptry bulb proteins might be variable among parasites species and stages. As most rhoptry proteins are refractory to gene disruption in both Plasmodium and Toxoplasma, the sporozoite-specific gene knockdown system by promoter swapping is a powerful tool to reveal functions of rhoptry proteins in sporozoites (Ishino et al., 2019). To differentiate the conserved- or specific- mechanisms of invasion among species or infective stages would give clues to understand the comprehensive molecular basis of parasite infection.
The mRNA amounts of ASP/RON1 and RON3 are clearly greater in sporozoites than in schizonts. This suggests that ASP/RON1 and RON3 might mainly have roles or additional roles in sporozoites. Indeed, unlike other rhoptry proteins, ASP/RON1 is dispensable for parasite proliferation during the intraerythrocytic stage in P. berghei (Bushell et al., 2017). Understanding its function in sporozoites would reveal the different contributions of rhoptry proteins depending on infective stages or target host cells.
This is the first profiling data showing the expression and localization of rhoptry proteins in both merozoites and sporozoites by immuno-electron microscopy. By generation of transgenic parasite lines expressing c-Myc tagged rhoptry proteins, comprehensive localization analyses by immuno-electron microscopy could be performed using anti-c-Myc antibodies, thus overcoming the difficulty to obtain specific and high titer antibodies against Plasmodium proteins. The c-Myc tag, a peptide of 10 amino acids and ~1,200 Da, is relatively small and unlikely to interfere with protein folding and function following addition to the C-terminus of target proteins that do not possess a C-terminal GPI-anchor domain. All the transgenic parasite lines, in which native rhoptry proteins were replaced by c-Myc tagged rhoptry proteins, proliferate normally during the intra-erythrocytic stage. Considering that most rhoptry proteins are refractory to gene deletion due to their necessity for parasite proliferation in the blood stage (Tufet-Bayona et al., 2009; Giovannini et al., 2011; Counihan et al., 2013; Bushell et al., 2017), this indicates that c-Myc tagging at the C-terminus of each protein doesn't affect rhoptry protein localization and function. This c-Myc tagging application combined with IEM will facilitate the observation of protein trafficking and discharge; for example, by overcoming the difficulties in producing specific antibodies with high titer.
In merozoites of P. berghei ANKA, as well as in P. falciparum, RON2, RON4, RON5, RALP1, and ASP /RON1 are localized to the rhoptry neck, while RAP1, RON3, RAMA, RhopH1A, RhopH2, and RhopH3 are localized to the rhoptry bulb. It has been proposed that sub-localization of rhoptry proteins reflects the order of discharge during merozoite invasion; that is, rhoptry neck proteins are secreted prior to invasion and followed by release of rhoptry bulb proteins (Zuccala et al., 2012). In this study we demonstrate that rhoptry neck proteins are also expressed in sporozoites; however, their localization, except for ASP/RON1, is not restricted to the rhoptry neck. This observation is in good agreement with reports demonstrating that the localization of the rhoptry proteins RON11 and RON12 differ between merozoites and sporozoites (Bantuchai et al., 2019; Oda-Yokouchi et al., 2019). RON2 in sporozoites is required for salivary gland invasion as well as hepatocyte infection (Ishino et al., 2019), which is not the case for RON2 in merozoites involved in erythrocyte invasion, supporting that localization in sporozoite rhoptries might be different from that in merozoites. Further analyses will reveal whether the protein secretion mechanisms and/or timing is different between merozoites and sporozoites. In addition, the same strategy can be used to address protein secretion mechanisms, such as how proteins might be transported to but differentially localized within rhoptries in sporozoites vs. merozoites.
Data Availability
All datasets generated for this study are included in the manuscript/Supplementary Files.
Ethics Statement
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Ehime University and the experiments were conducted according to the Ethical Guidelines for Animal Experiments of Ehime University.
Author Contributions
NT, MTo, and TI conceived and designed the experiments. NT, MN, MTa, MB, KM, MTo, and TI performed the experiments and analyzed the data. MN, MTo, and TI wrote manuscript. NT, MTa, MB, KM, and TT critically reviewed it.
Funding
This work was supported in part by JSPS KAKENHI (JP22590379, JP23790459, JP24390101, and JP19H03459) and Takeda Science Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Chris J. Janse, Leiden University, for supplying the Plasmodium berghei ANKA parasites expressing GFP under the control of ef1α promoter. The following reagents were obtained through BEI Resources, NIAID, NIH: Plasmid pL0033, for generation of transgenic parasites expressing c-Myc tagged rhoptry proteins in Plasmodium berghei, and MRA-802, contributed by Dr. Andrew P. Waters. This study was supported by the Division of Analytical Bio-Medicine, the Advanced Research Support Center (ADRES), Ehime University. We also thank A. Konishi and S. Sadaoka for rearing mice and technical support. We would like to thank Dr. Thomas Templeton for critical reading of the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00316/full#supplementary-material
Abbreviations
AMA1, apical merozoite protein 1; An, Anopheles; ASP, apical sushi protein; ef1α, elongation factor 1A; GPI, glycosylphosphatidylinositol; IFA, immunofluorescent assay; IEM, immuno-electron microscopy; Pb, P. berghei; RAMA, rhoptry associated membrane antigen; PBS-MT, PBS containing 5% non-fat dry milk and 0.01% Tween 20; PBS-BT, PBS containing 0.4% Block Ace and 0.01% Tween 20; PVM, parasitophorous vacuole membrane; RALP1, rhoptry-associated leucine zipper-like protein 1; RAP1, rhoptry-associated protein 1; RhopH, high-molecular mass rhoptry protein; RON2, rhoptry neck protein; RT-PCR, reverse transcription-PCR; SPECT2, sporozoite protein essential for cell traversal 2.
References
Alexander, L., Mital, J., Ward, G. E., Bradley, P., and Boothroyd, J. C. (2005). Identification of the moving junction complex of Toxoplasma gondii: a collaboration between distinct secretory organelles. PLoS Pathog. 1:e17. doi: 10.1371/journal.ppat.0010017
Aurrecoechea, C., Brestelli, J., Brunk, B. P., Dommer, J., Fischer, S., Gajria, B., et al. (2009). PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–D543. doi: 10.1093/nar/gkn814
Bantuchai, S., Nozaki, M., Thongkukiatkul, A., Lorsuwannarat, N., Tachibana, M., Baba, M., et al. (2019). Rhoptry neck protein 11 has crucial roles during malaria parasite sporozoite invasion of salivary glands and hepatocytes. Int. J. Parasitol. 49, 725–735. doi: 10.1016/j.ijpara.2019.05.001
Baum, J., Gilberger, T. W., Frischknecht, F., and Meissner, M. (2008). Host-cell invasion by malaria parasites: insights from Plasmodium and Toxoplasma. Trends Parasitol. 24, 557–563. doi: 10.1016/j.pt.2008.08.006
Besteiro, S., Michelin, A., Poncet, J., Dubremetz, J. F., and Lebrun, M. (2009). Export of a Toxoplasma gondii rhoptry neck protein complex at the host cell membrane to form the moving junction during invasion. PLoS Pathog. 5:e1000309. doi: 10.1371/journal.ppat.1000309
Boothroyd, J. C., and Dubremetz, J. F. (2008). Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat. Rev. Microbiol. 6, 79–88. doi: 10.1038/nrmicro1800
Bradley, P. J., Ward, C., Cheng, S. J., Alexander, D. L., Coller, S., Coombs, G. H., et al. (2005). Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in Toxoplasma gondii. J. Biol. Chem. 280, 34245–34258. doi: 10.1074/jbc.M504158200
Bushell, E., Gomes, A. R., Sanderson, T., Anar, B., Girling, G., Herd, C., et al. (2017). Functional profiling of a Plasmodium genome reveals an abundance of essential genes. Cell 170, 260–272.e8. doi: 10.1016/j.cell.2017.06.030
Cao, J., Kaneko, O., Thongkukiatkul, A., Tachibana, M., Otsuki, H., Gao, Q., et al. (2009). Rhoptry neck protein RON2 forms a complex with microneme protein AMA1 in Plasmodium falciparum merozoites. Parasitol. Int. 58, 29–35. doi: 10.1016/j.parint.2008.09.005
Comeaux, C. A., Coleman, B. I., Bei, A. K., Whitehurst, N., and Duraisingh, M. T. (2011). Functional analysis of epigenetic regulation of tandem RhopH1/clag genes reveals a role in Plasmodium falciparum growth. Mol. Microbiol. 80, 378–390. doi: 10.1111/j.1365-2958.2011.07572.x
Counihan, N. A., Chisholm, S. A., Bullen, H. E., Srivastava, A., Sanders, P. R., Jonsdottir, T. K., et al. (2017). Plasmodium falciparum parasites deploy RhopH2 into the host erythrocyte to obtain nutrients, grow and replicate. Elife 6:e23217. doi: 10.7554/eLife.23217
Counihan, N. A., Kalanon, M., Coppel, R. L., and de Koning-Ward, T. F. (2013). Plasmodium rhoptry proteins: why order is important. Trends Parasitol. 29, 228–236. doi: 10.1016/j.pt.2013.03.003
Fentress, S. J., Behnke, M. S., Dunay, I. R., Mashayekhi, M., Rommereim, L. M., Fox, B. A., et al. (2010). Phosphorylation of immunity-related GTPases by a Toxoplasma gondii-secreted kinase promotes macrophage survival and virulence. Cell Host Microbe. 8, 484–495. doi: 10.1016/j.chom.2010.11.005
Ferguson, D. J., Balaban, A. E., Patzewitz, E. M., Wall, R. J., Hopp, C. S., Poulin, B., et al. (2014). The repeat region of the circumsporozoite protein is critical for sporozoite formation and maturation in Plasmodium. PLoS ONE 9:e113923. doi: 10.1371/journal.pone.0113923
Franke-Fayard, B., Trueman, H., Ramesar, J., Mendoza, J., van der Keur, M., van der Linden, R., et al. (2004). A Plasmodium berghei reference line that constitutively expresses GFP at a high level throughout the complete life cycle. Mol. Biochem. Parasitol. 137, 23–33. doi: 10.1016/j.molbiopara.2004.04.007
Frenal, K., Dubremetz, J. F., Lebrun, M., and Soldati-Favre, D. (2017). Gliding motility powers invasion and egress in Apicomplexa. Nat. Rev. Microbiol. 15, 645–660. doi: 10.1038/nrmicro.2017.86
Gilson, P. R., Nebl, T., Vukcevic, D., Moritz, R. L., Sargeant, T., Speed, T. P., et al. (2006). Identification and stoichiometry of glycosylphosphatidylinositol-anchored membrane proteins of the human malaria parasite Plasmodium falciparum. Mol. Cell Proteomics 5, 1286–1299. doi: 10.1074/mcp.M600035-MCP200
Giovannini, D., Spath, S., Lacroix, C., Perazzi, A., Bargieri, D., Lagal, V., et al. (2011). Independent roles of apical membrane antigen 1 and rhoptry neck proteins during host cell invasion by apicomplexa. Cell Host Microbe 10, 591–602. doi: 10.1016/j.chom.2011.10.012
Haase, S., Cabrera, A., Langer, C., Treeck, M., Struck, N., Herrmann, S., et al. (2008). Characterization of a conserved rhoptry-associated leucine zipper-like protein in the malaria parasite Plasmodium falciparum. Infect. Immun. 76, 879–887. doi: 10.1128/IAI.00144-07
Ishino, T., Chinzei, Y., and Yuda, M. (2005). A Plasmodium sporozoite protein with a membrane attack complex domain is required for breaching the liver sinusoidal cell layer prior to hepatocyte infection. Cell Microbiol. 7, 199–208. doi: 10.1111/j.1462-5822.2004.00447.x
Ishino, T., Murata, E., Tokunaga, N., Baba, M., Tachibana, M., Thongkukiatkul, A., et al. (2019). Rhoptry neck protein 2 expressed in Plasmodium sporozoites plays a crucial role during invasion of mosquito salivary glands. Cell Microbiol. 21:e12964. doi: 10.1111/cmi.12964
Ito, D., Han, E. T., Takeo, S., Thongkukiatkul, A., Otsuki, H., Torii, M., et al. (2011). Plasmodial ortholog of Toxoplasma gondii rhoptry neck protein 3 is localized to the rhoptry body. Parasitol. Int. 60, 132–138. doi: 10.1016/j.parint.2011.01.001
Ito, D., Hasegawa, T., Miura, K., Yamasaki, T., Arumugam, T. U., Thongkukiatkul, A., et al. (2013). RALP1 is a rhoptry neck erythrocyte-binding protein of Plasmodium falciparum merozoites and a potential blood-stage vaccine candidate antigen. Infect. Immun. 81, 4290–4298. doi: 10.1128/IAI.00690-13
Ito, D., Schureck, M. A., and Desai, S. A. (2017). An essential dual-function complex mediates erythrocyte invasion and channel-mediated nutrient uptake in malaria parasites. Elife 6:e23485. doi: 10.7554/eLife.23485
Janse, C. J., Franke-Fayard, B., and Waters, A. P. (2006a). Selection by flow-sorting of genetically transformed, GFP-expressing blood stages of the rodent malaria parasite, Plasmodium berghei. Nat. Protoc. 1, 614–623. doi: 10.1038/nprot.2006.88
Janse, C. J., Ramesar, J., and Waters, A. P. (2006b). High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 1, 346–356. doi: 10.1038/nprot.2006.53
Kadekoppala, M., and Holder, A. A. (2010). Merozoite surface proteins of the malaria parasite: the MSP1 complex and the MSP7 family. Int. J. Parasitol. 40, 1155–1161. doi: 10.1016/j.ijpara.2010.04.008
Kaiser, K., Matuschewski, K., Camargo, N., Ross, J., and Kappe, S. H. (2004). Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol. Microbiol. 51, 1221–1232. doi: 10.1046/j.1365-2958.2003.03909.x
Kaneko, O., Tsuboi, T., Ling, I. T., Howell, S., Shirano, M., Tachibana, M., et al. (2001). The high molecular mass rhoptry protein, RhopH1, is encoded by members of the clag multigene family in Plasmodium falciparum and Plasmodium yoelii. Mol. Biochem. Parasitol. 118, 223–231. doi: 10.1016/S0166-6851(01)00391-7
Kaneko, O., Yim Lim, B. Y., Iriko, H., Ling, I. T., Otsuki, H., Grainger, M., et al. (2005). Apical expression of three RhopH1/Clag proteins as components of the Plasmodium falciparum RhopH complex. Mol. Biochem. Parasitol. 143, 20–28. doi: 10.1016/j.molbiopara.2005.05.003
Kariu, T., Ishino, T., Yano, K., Chinzei, Y., and Yuda, M. (2006). CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol. Microbiol. 59, 1369–1379. doi: 10.1111/j.1365-2958.2005.05024.x
Kemp, L. E., Yamamoto, M., and Soldati-Favre, D. (2013). Subversion of host cellular functions by the apicomplexan parasites. FEMS Microbiol. Rev. 37, 607–631. doi: 10.1111/1574-6976.12013
Kennedy, M., Fishbaugher, M. E., Vaughan, A. M., Patrapuvich, R., Boonhok, R., Yimamnuaychok, N., et al. (2012). A rapid and scalable density gradient purification method for Plasmodium sporozoites. Malar J. 11:421. doi: 10.1186/1475-2875-11-421
Lebrun, M., Michelin, A., El Hajj, H., Poncet, J., Bradley, P. J., Vial, H., et al. (2005). The rhoptry neck protein RON4 re-localizes at the moving junction during Toxoplasma gondii invasion. Cell Microbiol. 7, 1823–1833. doi: 10.1111/j.1462-5822.2005.00646.x
Lim, D. C., Cooke, B. M., Doerig, C., and Saeij, J. P. (2012). Toxoplasma and Plasmodium protein kinases: roles in invasion and host cell remodelling. Int. J. Parasitol. 42, 21–32. doi: 10.1016/j.ijpara.2011.11.007
Lindner, S. E., Swearingen, K. E., Harupa, A., Vaughan, A. M., Sinnis, P., Moritz, R. L., et al. (2013). Total and putative surface proteomics of malaria parasite salivary gland sporozoites. Mol. Cell Proteomics 12, 1127–1143. doi: 10.1074/mcp.M112.024505
Ling, I. T., Florens, L., Dluzewski, A. R., Kaneko, O., Grainger, M., Yim Lim, B. Y., et al. (2004). The Plasmodium falciparum clag9 gene encodes a rhoptry protein that is transferred to the host erythrocyte upon invasion. Mol. Microbiol. 52, 107–118. doi: 10.1111/j.1365-2958.2003.03969.x
Ling, I. T., Kaneko, O., Narum, D. L., Tsuboi, T., Howell, S., Taylor, H. M., et al. (2003). Characterisation of the rhoph2 gene of Plasmodium falciparum and Plasmodium yoelii. Mol. Biochem. Parasitol. 127, 47–57. doi: 10.1016/S0166-6851(02)00302-X
Mikolajczak, S. A., Silva-Rivera, H., Peng, X., Tarun, A. S., Camargo, N., Jacobs-Lorena, V., et al. (2008). Distinct malaria parasite sporozoites reveal transcriptional changes that cause differential tissue infection competence in the mosquito vector and mammalian host. Mol. Cell. Biol. 28, 6196–6207. doi: 10.1128/MCB.00553-08
Mutungi, J. K., Yahata, K., Sakaguchi, M., and Kaneko, O. (2014). Expression and localization of rhoptry neck protein 5 in merozoites and sporozoites of Plasmodium yoelii. Parasitol. Int. 63, 794–801. doi: 10.1016/j.parint.2014.07.013
Nguitragool, W., Bokhari, A. A., Pillai, A. D., Rayavara, K., Sharma, P., Turpin, B., et al. (2011). Malaria parasite clag3 genes determine channel-mediated nutrient uptake by infected red blood cells. Cell 145, 665–677. doi: 10.1016/j.cell.2011.05.002
Niedelman, W., Gold, D. A., Rosowski, E. E., Sprokholt, J. K, Lim, D., Farid Arenas, A., et al. (2012). The rhoptry proteins ROP18 and ROP5 mediate Toxoplasma gondii evasion of the murine, but not the human, interferon-gamma response. PLoS Pathog. 8:e1002784. doi: 10.1371/journal.ppat.1002784
Oda-Yokouchi, Y., Tachibana, M., Iriko, H., Torii, M., Ishino, T., and Tsuboi, T. (2019). Plasmodium RON12 localizes to the rhoptry body in sporozoites. Parasitol. Int. 68, 17–23. doi: 10.1016/j.parint.2018.10.001
Ong, Y. C., Reese, M. L., and Boothroyd, J. C. (2010). Toxoplasma rhoptry protein 16 (ROP16) subverts host function by direct tyrosine phosphorylation of STAT6. J. Biol. Chem. 285, 28731–28740. doi: 10.1074/jbc.M110.112359
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Poukchanski, A., Fritz, H. M., Tonkin, M. L., Treeck, M., Boulanger, M. J., and Boothroyd, J. C. (2013). Toxoplasma gondii sporozoites invade host cells using two novel paralogues of RON2 and AMA1. PLoS ONE 8:e70637. doi: 10.1371/journal.pone.0070637
Proellocks, N. I., Kats, L. M., Sheffield, D. A., Hanssen, E., Black, C. G., Waller, K. L., et al. (2009). Characterisation of PfRON6, a Plasmodium falciparum rhoptry neck protein with a novel cysteine-rich domain. Int. J. Parasitol. 39, 683–692. doi: 10.1016/j.ijpara.2008.11.002
Reid, A. J., Talman, A. M., Bennett, H. M., Gomes, A. R., Sanders, M. J., Illingworth, C. J. R., et al. (2018). Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. Elife 7:e33105. doi: 10.7554/eLife.33105
Richard, D., MacRaild, C. A., Riglar, D. T., Chan, J. A., Foley, M., Baum, J., et al. (2010). Interaction between Plasmodium falciparum apical membrane antigen 1 and the rhoptry neck protein complex defines a key step in the erythrocyte invasion process of malaria parasites. J. Biol. Chem. 285, 14815–14822. doi: 10.1074/jbc.M109.080770
Riglar, D. T., Richard, D., Wilson, D. W., Boyle, M. J., Dekiwadia, C., Turnbull, L., et al. (2011). Super-resolution dissection of coordinated events during malaria parasite invasion of the human erythrocyte. Cell Host Microbe 9, 9–20. doi: 10.1016/j.chom.2010.12.003
Risco-Castillo, V., Topcu, S., Son, O., Briquet, S., Manzoni, G., and Silvie, O. (2014). CD81 is required for rhoptry discharge during host cell invasion by Plasmodium yoelii sporozoites. Cell Microbiol. 16, 1533–1548. doi: 10.1111/cmi.12309
Sherling, E. S., Knuepfer, E., Brzostowski, J. A., Miller, L. H., Blackman, M. J., and van Ooij, C. (2017). The Plasmodium falciparum rhoptry protein RhopH3 plays essential roles in host cell invasion and nutrient uptake. Elife 6:e23239. doi: 10.7554/eLife.23239
Srivastava, A., Singh, S., Dhawan, S., Mahmood Alam, M., Mohmmed, A., and Chitnis, C. E. (2010). Localization of apical sushi protein in Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 174, 66–69. doi: 10.1016/j.molbiopara.2010.06.003
Steinfeldt, T., Konen-Waisman, S., Tong, L., Pawlowski, N., Lamkemeyer, T., Sibley, L. D., et al. (2010). Phosphorylation of mouse immunity-related GTPase (IRG) resistance proteins is an evasion strategy for virulent Toxoplasma gondii. PLoS Biol. 8:e1000576. doi: 10.1371/journal.pbio.1000576
Sultan, A. A., Thathy, V., Frevert, U., Robson, K. J., Crisanti, A., Nussenzweig, V., et al. (1997). TRAP is necessary for gliding motility and infectivity of Plasmodium sporozoites. Cell 90, 511–522. doi: 10.1016/S0092-8674(00)80511-5
Tarun, A. S., Peng, X., Dumpit, R. F., Ogata, Y., Silva-Rivera, H., Camargo, N., et al. (2008). A combined transcriptome and proteome survey of malaria parasite liver stages. Proc. Natl. Acad. Sci. U. S. A. 105, 305–310. doi: 10.1073/pnas.0710780104
Thathy, V., Fujioka, H., Gantt, S., Nussenzweig, R., Nussenzweig, V., and Menard, R. (2002). Levels of circumsporozoite protein in the Plasmodium oocyst determine sporozoite morphology. EMBO J. 21, 1586–1596. doi: 10.1093/emboj/21.7.1586
Topolska, A. E., Lidgett, A., Truman, D., Fujioka, H., and Coppel, R. L. (2004). Characterization of a membrane-associated rhoptry protein of Plasmodium falciparum. J. Biol. Chem. 279, 4648–4656. doi: 10.1074/jbc.M307859200
Tsuboi, T., Takeo, S., Iriko, H., Jin, L., Tsuchimochi, M., Matsuda, S., et al. (2008). Wheat germ cell-free system-based production of malaria proteins for discovery of novel vaccine candidates. Infect. Immun. 76, 1702–1708. doi: 10.1128/IAI.01539-07
Tufet-Bayona, M., Janse, C. J., Khan, S. M., Waters, A. P., Sinden, R. E., and Franke-Fayard, B. (2009). Localisation and timing of expression of putative Plasmodium berghei rhoptry proteins in merozoites and sporozoites. Mol. Biochem. Parasitol. 166, 22–31. doi: 10.1016/j.molbiopara.2009.02.009
Vanderberg, J. P. (1975). Development of infectivity by the Plasmodium berghei sporozoite. J. Parasitol. 61, 43–50. doi: 10.2307/3279102
Vincensini, L., Fall, G., Berry, L., Blisnick, T., and Braun Breton, C. (2008). The RhopH complex is transferred to the host cell cytoplasm following red blood cell invasion by Plasmodium falciparum. Mol. Biochem. Parasitol. 160, 81–89. doi: 10.1016/j.molbiopara.2008.04.002
Yamamoto, M., Ma, J. S., Mueller, C., Kamiyama, N., Saiga, H., Kubo, E., et al. (2011). ATF6beta is a host cellular target of the Toxoplasma gondii virulence factor ROP18. J. Exp. Med. 208, 1533–1546. doi: 10.1084/jem.20101660
Yang, A. S. P., Lopaticki, S., O'Neill, M. T., Erickson, S. M., Douglas, D. N., Kneteman, N. M., et al. (2017). AMA1 and MAEBL are important for Plasmodium falciparum sporozoite infection of the liver. Cell Microbiol. 19:e12745 doi: 10.1111/cmi.12745
Yuda, M., and Ishino, T. (2004). Liver invasion by malarial parasites–how do malarial parasites break through the host barrier? Cell Microbiol. 6, 1119–1125. doi: 10.1111/j.1462-5822.2004.00474.x
Zhang, M., Fennell, C., Ranford-Cartwright, L., Sakthivel, R., Gueirard, P., Meister, S., et al. (2010). The Plasmodium eukaryotic initiation factor-2alpha kinase IK2 controls the latency of sporozoites in the mosquito salivary glands. J. Exp. Med. 207, 1465–1474. doi: 10.1084/jem.20091975
Zuccala, E. S., Gout, A. M., Dekiwadia, C., Marapana, D. S., Angrisano, F., Turnbull, L., et al. (2012). Subcompartmentalisation of proteins in the rhoptries correlates with ordered events of erythrocyte invasion by the blood stage malaria parasite. PLoS ONE 7:e46160. doi: 10.1371/journal.pone.0046160
Keywords: malaria, Plasmodium, merozoite, sporozoite, rhoptry, immuno-electron microscopy
Citation: Tokunaga N, Nozaki M, Tachibana M, Baba M, Matsuoka K, Tsuboi T, Torii M and Ishino T (2019) Expression and Localization Profiles of Rhoptry Proteins in Plasmodium berghei Sporozoites. Front. Cell. Infect. Microbiol. 9:316. doi: 10.3389/fcimb.2019.00316
Received: 25 June 2019; Accepted: 22 August 2019;
Published: 10 September 2019.
Edited by:
Rhoel Dinglasan, University of Florida, United StatesReviewed by:
Scott E. Lindner, Pennsylvania State University, United StatesFriedrich Frischknecht, Heidelberg University, Germany
Copyright © 2019 Tokunaga, Nozaki, Tachibana, Baba, Matsuoka, Tsuboi, Torii and Ishino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tomoko Ishino, dGlzaGlub0BtLmVoaW1lLXUuYWMuanA=
†These authors have contributed equally to this work
‡Present Address: Naohito Tokunaga, Division of Analytical Bio-Medicine, The Advanced Research Support Center (ADRES), Ehime University, Matsuyama, Japan
Mamoru Nozaki, Plant and Cell Engineering Laboratory, Graduate School of Engineering, Toyama Prefectural University, Imizu, Japan
Kazuhiro Matsuoka, Department of Infectious Diseases and Immunology, Clinical Research Center, National Hospital Organization Nagoya Medical Center, Nagoya, Japan
 Naohito Tokunaga
Naohito Tokunaga Mamoru Nozaki
Mamoru Nozaki Mayumi Tachibana
Mayumi Tachibana Minami Baba
Minami Baba Kazuhiro Matsuoka1‡
Kazuhiro Matsuoka1‡ Motomi Torii
Motomi Torii Tomoko Ishino
Tomoko Ishino