- 1Structural Biology Unit, Center for Cooperative Research CIC bioGUNE, Derio, Spain
- 2Laboratorio de Virología Molecular, Centro de Investigaciones Nucleares, Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay
- 3Laboratorio de Inmunovirología, Institut Pasteur de Montevideo, Montevideo, Uruguay
Picornaviruses constitute one of the most relevant viral groups according to their impact on human and animal health. Etiologic agents of a broad spectrum of illnesses with a clinical presentation that ranges from asymptomatic to fatal disease, they have been the cause of uncountable epidemics throughout history. Picornaviruses are small naked RNA-positive single-stranded viruses that include some of the most important pillars in the development of virology, comprising poliovirus, rhinovirus, and hepatitis A virus. Picornavirus infectious particles use the fecal–oral or respiratory routes as primary modes of transmission. In this regard, successful viral spread relies on the capability of viral capsids to (i) shelter the viral genome, (ii) display molecular determinants for cell receptor recognition, (iii) facilitate efficient genome delivery, and (iv) escape from the immune system. Importantly, picornaviruses display a substantial amount of genetic variability driven by both mutation and recombination. Therefore, the outcome of their replication results in the emergence of a genetically diverse cloud of individuals presenting phenotypic variance. The host humoral response against the capsid protein represents the most active immune pressure and primary weapon to control the infection. Since the preservation of the capsid function is deeply rooted in the virus evolutionary dynamics, here we review the current structural evidence focused on capsid antibody evasion mechanisms from that perspective.
Picornavirus Historical Relevance
Picornaviruses have been pivotal in the foundations of virology. Original research on “ultra-filterable infectious agents” such as foot-and-mouth disease virus (FMDV) and poliovirus (PV) began the era of animal virology (Loeffler and Frosch, 1898; Eggers, 1999). The development of cell cultures for PV replication led to Salk's inactivated and Sabin's attenuated vaccines (Enders et al., 1949). The first animal virus engineered into an infectious clone (Racaniello and Baltimore, 1981) and the first virus synthesized outside the cell was PV (Molla et al., 1991).
Although vast knowledge has been gained, picornaviruses still challenge our understanding. The still open fundamental questions and public health challenges picornaviruses pose reflect that we are far from a conclusive comprehension (Holm-Hansen et al., 2016; Li et al., 2017; Zarocostas, 2018). In the following review, we examine how these agents evade host antibodies (Abs) based on their biological and evolutionary properties, with the spotlight on human picornaviruses.
Classification and Clinical Impact on Human Health
Picornaviridae is a large family of vertebrate viruses that produce both clinically asymptomatic infections but often mild and fatal disease. Their current classification includes more than 30 genera and 75 species (Zell et al., 2017), including several human picornaviruses. The genus Enterovirus comprises seven species infecting humans (enterovirus A-to-D and rhinovirus A-to-C). This genus contains poliovirus (PV), coxsackieviruses A/B (CVA/B), enteroviruses (EV), echoviruses (E), and rhinoviruses (RV). Further serological classification results in hundreds of serotypes. Hepatitis A virus (HAV) is the sole human-virus species in the genus Hepatovirus. Other human picornaviruses include members of the genera Cardiovirus (Saffold virus—SAFV), Cosavirus (CoSV), Parechovirus (Ljungan virus—LV), Kubovirus (Aichi virus—AiV), and Salivirus (Salivirus A—SaVA) (Nielsen et al., 2013).
RVs are airborne pathogens, while other enteroviruses and HAV use the fecal–oral route (Yin-Murphy and Almond, 1996). RVs cause the common cold, the most prevalent infectious disease worldwide, resulting in uncountable lost days from school and work. Epidemics occur yearly with outbreaks in the winter and spring (Drysdale et al., 2017). PV infection targets the central nervous system, destroying nerves and motor neurons, resulting in paralytic poliomyelitis. Until the PV worldwide eradication program based on global vaccination, polio epidemics have been the cause of high morbimortality (Minor, 2014). The so-called “non-polio enteroviruses” (CVs, EVs, and Es) cause several diseases with high morbimortality including meningitis, myocarditis, poliomyelitis-like syndrome, pancreatitis, and possibly the onset of diabetes. Outbreaks are common and have been considered to have pandemic potential (Zhang et al., 2015; Pons-Salort et al., 2018). Hepatitis produced by HAV is a mild disease producing liver damage usually leading to total recovery, but rare, severe cases are fatal in older age individuals. HAV produces large outbreaks probably due to its long 2 to 3 week incubation time (Jacobsen and Wiersma, 2010).
Genome Organization
Picornaviruses have single-stranded RNA positive-sense genomes (~7–9 kb) that serve as mRNA for viral protein synthesis (Baltimore, 1971) (Figure 1A). Their RNA holds a single ORF encoding a polyprotein precursor for all viral proteins. Importantly, the 5′-end of the genome is covalently bound to the Viral-Protein-genome (VPg) (Crawford and Baltimore, 1983), the primer for viral RNA synthesis (Nomoto et al., 1977). Two untranslated regions (UTR), 5′UTR and 3′UTR, flank the ORF and contain virus-specific RNA secondary structural elements implicated in replication and providing host specificity (Kloc et al., 2018). The 5′UTR bears an internal ribosomal entry site (IRES) and polypyrimidine tract (PPT) that elicit host ribosomes and PPT-binding protein for translation (Martinez-Salas et al., 2018). The 3′UTR finishes in a poly(A) tail that mimics host mRNA tail conferring genome stability.
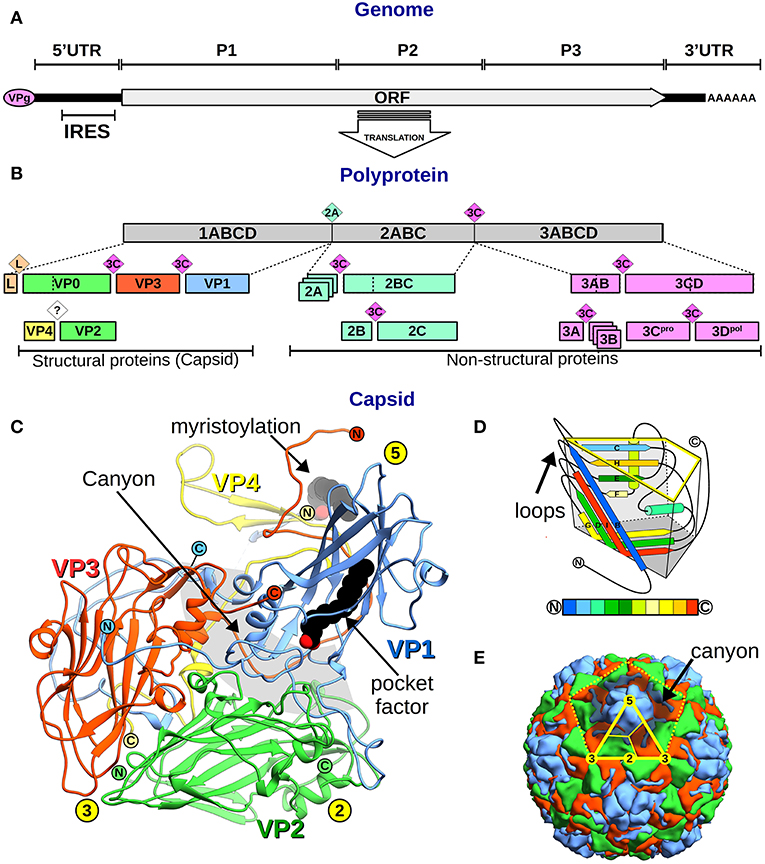
Figure 1. Picornavirus genome, proteins, and capsid organization. (A) Representation of the picornavirus genome, the VPg, and the polyA tail, showing the single ORF location. The position of the P1–3 regions, the flanking 5′ and 3′UTR, and the IRES are indicated. (B) A bar diagram showing the polyprotein (gray box) and the proteolytic cascade that leads to all picornaviral proteins (colored boxes). Boxes include the protein names following the genome-ORF regions' nomenclature (number–letters) or the VP1–4 nomenclature for the structural proteins. Colored rhombi indicate cleavage points and are labeled with the corresponding protease name. (C) Overall view of the canonical picornavirus protomer with the proteins VP1 (blue), VP2 (green), VP3 (red), and VP4 (yellow). The protein N- and C-termini are indicated as encircled N and C letters, and yellow circles show the 5-,−3, 2-fold symmetry axes positions. Lipid components as the VP4 myristoylation and the “pocket factor” are depicted as black spheres. The “canyon” region is shown as a gray circular segment shadow. (D) Schematics of the “jelly roll” fold of VP1–3 proteins inscribed in a trapezoidal prism where the yellow highlighted face corresponds to the external capsid surface, and the dark gray base faces the inner capsid. The secondary structure elements are colored from N- to C-terminus according to the color code bar below. External loops and N- and C-terminus are indicated. (E) Overall view of the picornavirus capsid showing the outer surface of VP1 (blue), VP2 (green), and VP3 (red). The yellow dotted line indicates the boundaries of one pentamer. The solid yellow line marks the icosahedral asymmetric subunit and thinner lines separate proteins following the trapezoidal schematics shown in (D). Symmetry 5-, 3-, 2-fold symmetry axes are indicated in yellow circles.
From 5′-to-3′, the ORF comprises three regions: (i) P1, encoding the structural capsid viral proteins (VP4–VP2–VP3–VP1), while, in some picornaviruses, also codifying a short leader L-protein; (ii) P2, encoding the viral non-structural proteins 2A−2B−2C; and (iii) P3 encoding the viral non-structural proteins 3A−3B−3C−3D (Palmenberg, 1990) (Figure 1B). The non-structural proteins' central role is replication, translation, and hijacking host-cell machinery (Figure 2). In particular, 3D is the RNA-dependent RNA polymerase (RdRp or 3Dpol) that synthesizes the virus genome and 3B (which is VPg) acts as its primer, being the only non-structural protein in the virion (Palmenberg, 1990).
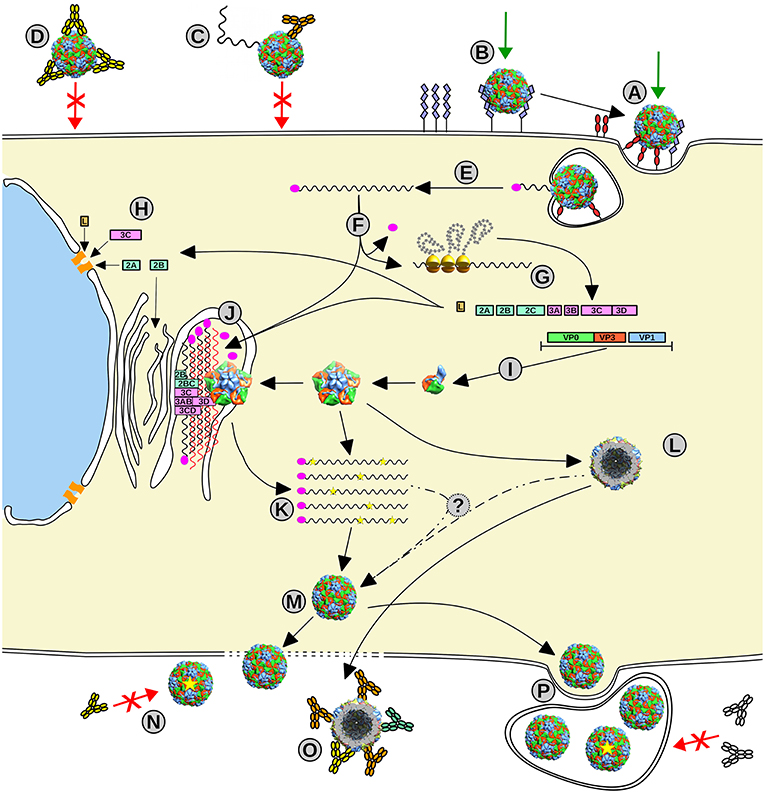
Figure 2. Picornavirus life cycle. (A, B) Picornavirus uses different receptors to enter the cell, some implicated in the signaling internalization (A), meanwhile others can act as carriers that transport the viral particle to meet the primary receptor (B). (C, D) This infection event can be impeded by the action of specific neutralizing antibodies that can destabilize the viral particle (C) or opsonize or stabilize the particle to impair receptor binding or conformational changes required for infection (D). (E) Once the virus enters the cell, the viral RNA delivery mechanism is triggered, and the viral genome (black wavy line) is released into the cytoplasm. (F) Upon removal of VPg (magenta oval), the genome starts the IRES-driven translation leading to the production of the viral polyprotein. (G) The proteolytic cascade produces all viral proteins, structural and non-structural. (H) Some proteins act by hijacking the host cellular systems such as the nuclear pore, the cell translation machinery, and the apoptotic systems and initiate the remodeling of the internal cell membranes. (I) The structural proteins assemble into the capsid intermediates, the protomer and the pentamer, and also procapsids (L). (J) The formed replication complex assembled from non-structural proteins and modified internal membranes firing the picornaviral genome replication by the 3D polymerase via RNA complementary (red wavy lines) and using VPg as a primer. (K) The new progeny genomes including eventual mutations (yellow stars). (M) Mature virions assemble from pentamers that surround and package the new viral genomes. Viral particles escape from the cell by cell lysis or budding within membranes that can protect the viral progeny (P). (N) Some progeny virus with mutations in their capsids (yellow star) may escape from to the action of specific NAbs. (O) Empty capsids can act as molecular decoys for Abs to protect the infecting particles from neutralization.
Picornavirus Capsid Anatomy
Picornaviruses were the first human viruses to be structurally defined at the atomic level (Rossmann et al., 1985). To date, several structures of human picornaviruses have been unveiled including HRV, PV, HAV, CVB, E, CVA, EV, SAFV, and AiV (Hogle et al., 1985; Rossmann et al., 1985; Filman et al., 1989, 1998; Muckelbauer et al., 1995; Zhao et al., 1996; Lentz et al., 1997; Hendry et al., 1999; Verdaguer et al., 2000, 2003; Stuart et al., 2002; Zhang et al., 2004; Venkataraman et al., 2008; Plevka et al., 2010, 2012; Zocher et al., 2014; Liu et al., 2015, 2016; Ren et al., 2015; Zhu et al., 2015; Mullapudi et al., 2016). Moreover, the structure of FMDV has been disclosed (Logan et al., 1993; Lea et al., 1994). All picornaviruses have a naked 30 nm icosahedral capsid composed of 60 identical tightly packed protomers (Figure 1E). Early in particle morphogenesis, immature protomers contain VP1 and VP3 together with VP0, the precursor of VP4 and VP2 (Jiang et al., 2014) (Figure 1B). Virus assembly likely goes through a dodecahedral pathway (Li et al., 2012), by the association of pentamers formed by five immature protomers leading to the icosahedral particle, defining 5-, 3-, and 2-fold symmetry axes. Upon genome encapsidation, VP0 is generally auto-catalytically cleaved into VP2 and VP4 generating the mature capsid (except for Parechovirus and Kubovirus) (Figure 2).
The larger proteins (VP1–3) form the external and internal capsid surface. These proteins have a common fold, the “jelly-roll,” formed by two 4-strand anti-parallel β-sheets and two helices (Figure 1D). Conversely, the small VP4 is located inside the capsid only and usually appears myristoylated at its N-terminus (Paul and Schultz, 1987; Belsham et al., 1991) (Figure 1C). The capsid external surface displays a rugged topography. Main surface features include (i) a principal protrusion built by the interaction of copies of VP1 forming a star-shaped 5-fold vertex, (ii) a 5-fold surrounding valley called the “canyon,” (iii) a VP2 loop protuberance or the “puff,” (iv) a VP3 loop rise or the “knob,” and (v) a large 2-fold depression (Muckelbauer et al., 1995). Loop differences result in distinctive surface traits between picornaviruses. Finally, some picornaviruses exhibit a lipid molecule, the “pocket factor,” bound in a cavity located inside the VP1 jelly-roll, which has been observed to play a role in particle stability (Figure 1C).
Receptors and Tropism
Picornavirus cell infection starts with its attachment to cell receptors (Figure 2). Therefore, virus-receptor usage is critical for tropism and its evolution can change virus targets at the level of cells to host ranges. The capsid binding sites of picornavirus receptors can be used to classify them into canyon binders and non-canyon binders.
Canyon binders are members of the immunoglobulin superfamily including: (i) ICAM-1 used by HRV and CVA (Greve et al., 1989; Staunton et al., 1989; Tomassini et al., 1989; Kolatkar et al., 1999; Xiao et al., 2001; Baggen et al., 2018), (ii) the PV receptor (PVR) (Mendelsohn et al., 1989; He et al., 2000; Strauss et al., 2015), (iii) the coxsackie and adenovirus receptor (CAR) used by CVBs (Bergelson, 1997; He et al., 2001; Organtini et al., 2014; Lee et al., 2016), (iv) αvβ3 and αvβ6 integrin used by some CVAs (Roivainen et al., 1994; Williams et al., 2004; Shakeel et al., 2013), and (v) the α2 subunit of VLA-2 used by E1 and E8 (Bergelson et al., 1992; Xing et al., 2004). Canyon binders' apical domain engages in the binding into the canyon, triggering conformational changes essential for infection, leading to the altered-particle (A-particle) conformational state (Greve et al., 1989; Xing et al., 2004; Xiao et al., 2005; Shakeel et al., 2013; Organtini et al., 2014; Strauss et al., 2015). These changes have been observed to depend on the number of binding events that stimulate the viral particle (Lee et al., 2016). Engagement of several receptors is known to bring the viral 5-fold vertex close to the cell membrane (Bubeck et al., 2005).
Non-canyon binders attach to the virus surface elsewhere outside the canyon, tethering the virus to the cell surface eventually signaling for virus internalization. Importantly, they are diverse in molecular characteristics. These receptors include (i) the LDL-receptor used by HRV-C (Verdaguer et al., 2004); (ii) the decay-accelerating factor (DAF), receptor of many echoviruses and CVBs (Bergelson et al., 1994, 1995; Pettigrew et al., 2006; Plevka et al., 2010; Pan et al., 2011; Yoder et al., 2012); (iii) P-selectin glycoprotein ligand-1 (PSGL-1) used by EV71 (Nishimura et al., 2009); and (iv) scavenger receptor B2 (ScaRB2), a receptor of EV71 (Yamayoshi et al., 2009; Zhou et al., 2018). This group possibly includes heparan sulfate used by several enteroviruses (Escribano-Romero, 2004; Zautner et al., 2006; McLeish et al., 2012; Tan et al., 2013; Nishimura et al., 2015) and cadherin-related family member 3 (CDHR3) used by HRV-C (Watters and Palmenberg, 2018). Non-canyon binders rarely induce substantial conformational changes due to their interaction, and some may not be essential for infection since few mutations allow or limit their usage (Pan et al., 2011; McLeish et al., 2012; Lee et al., 2013). This alternative receptor usage can modify the infection mechanism, as seen in the case of CVB3 binding to DAF which signals the trafficking of the attached virus to tight junctions where the virus can meet CAR (Coyne and Bergelson, 2006).
Population Dynamics and Genetic Variability
Picornaviruses display a great potential for adaptation and evolution, which is primarily dictated by their high mutation rate. Viral progenies are huge in population size and they have short generation times. Thus, the RNA virus population is a dynamic cloud of mutants where the average-consensus sequence of all variants represents the “genotype.” The mutation rate is the number of genetic changes, such as point mutations, insertions, or deletions introduced during viral replication. The first mutation rate measurement on RNA viruses was reported for CVA, disclosing a value of 1 mutation every 10,000 nucleotides copied (Eggers and Tamm, 1965).
Natural selection may have shaped picornaviral mutation rates in response to extremely dynamic ecosystems (Elena and Sanjuán, 2005). Therefore, this natural low replicative fidelity results in populations that quickly adapt to unexpected changes in the environment, such as immune pressure (Andino and Domingo, 2015). These observations have been conceptualized in the light of quasispecies evolution (Domingo et al., 2012). This adaptive capacity can be impaired by altering viral mutation rates (Vignuzzi et al., 2006). Indeed, the first support of the role of replicative fidelity in viral pathogenesis was observed in picornaviruses. Two groups isolated the first antimutator variant of an RNA virus, by serially passaging PV in the presence of ribavirin (Pfeiffer and Kirkegaard, 2003; Vignuzzi et al., 2006). The resistant variant contained a 3Dpol single point mutant (G64S) relatively resistant to lethal mutagenesis leading to (i) populations with lower mutation rates, (ii) reduced genetic diversity, and (iii) attenuated phenotype in mice (Vignuzzi et al., 2006). Recently, it has been suggested that this attenuation could be partly an outcome of a decrease in the replication speed (Fitzsimmons et al., 2018). Moreover, genetic engineering of viral polymerases has also identified several low-fidelity variants, called mutator variants (Thompson et al., 2007; Gong and Peersen, 2010; Gnädig et al., 2012; Rozen-Gagnon et al., 2014). For instance, using FMDV as a model, low-fidelity variants were found to increase mutation frequencies and render these viruses more susceptible to mutagenesis (Xie et al., 2014). Moreover, the same residue in the 3Dpol of FMDV is responsible to increase or decrease fidelity (Rai et al., 2017). Thus, picornaviral mutator variants were proven to increase mutation frequencies, decrease viral fitness, and also display an attenuated phenotype. Ostensibly, picornavirus mutation rates have been tuned to be near an upper limit (Crotty et al., 2001) yet evading population extinction by the accumulation of deleterious mutations by harmonizing (i) genetic integrity, (ii) genetic diversity, and (iii) replicative speed.
In addition to the classical view of single virus infectious unit of picornaviruses, structures containing many viral genomes support the existence of collective infectious units (Sanjuán, 2017). Lipid vesicles have been observed in HAV and EV infections (Feng et al., 2013; Chen et al., 2015; Kirkegaard, 2017). This current evidence incorporates the vesicle release and transmission to the standard lytic release and transmission of free virions, opening the debate on the “social evolution” of picornaviruses. Social evolution has been proposed to reduce detrimental mutations and negative interactions among the individuals within the population with direct implications for viral evolution, genetic diversity, and viral fitness (Bordería et al., 2015).
Recombination
During infection, RNA virus genomes can interchange nucleotide sequences resulting in genetic variation by recombination resulting in unpredictable advantages (Simon-Loriere and Holmes, 2011). This phenomenon was discovered in cells co-infected with PV escape mutants, resistant to antisera and guanidine, resulting in recombinant infectious PV (Ledinko, 1963). Recombination is widespread at intra-typic and inter-typic levels (Lukashev, 2005, 2010), often preceding the emergence of novel evolutionary lineages of picornaviruses (McWilliam Leitch et al., 2009, 2010; Meijer et al., 2012). For instance, there is evidence about recombinants of Sabin-related polioviruses harboring homologous sequences of other species of enteroviruses (Arita et al., 2005; Combelas et al., 2011; Bessaud et al., 2016).
Interestingly, recombination-deficient variants of PV have been identified. These viruses carry amino acid changes in the in 3Dpol that reduce recombination without conferring other detectable replication deficiencies. These non-recombining viruses accumulate a higher number of detrimental mutations, presumably by an inability to purge deleterious mutations, and fewer beneficial mutations (Xiao et al., 2016, 2017). Lately, the combined approach of mathematical modeling and experimental evolution experiments have predicted the frequency of recombination of picornaviruses such as PV and EV71 (Stern et al., 2017; Woodman et al., 2018).
Antibody Response Against Picornavirus Infection
Innate immune response detects foreign RNA using sensing proteins such as RIG-I, MDA-5, and Toll-like Receptor-3. These mediators act in the early control of picornaviral infection (Slater et al., 2010). Nevertheless, the adaptive response mediated by Abs plays the definitive role in the resolution of the infection. Several pieces of evidence support this view as (i) patients with agammaglobulinemia develop chronic infections (Wilfert et al., 1977; McKinney et al., 1987; Kainulainen et al., 2010; Bucciol et al., 2018), (ii) humoral response mediates the protective effect of picornavirus vaccines (Grant et al., 2017; Sun et al., 2017), (iii) mice lacking B-cells cannot clear enteroviral infections (Mena et al., 1999), and (iv) passive immunization with Abs is an efficient treatment of HAV infection (Stapleton, 1992), although their effectiveness for enteroviral neonatal infections is disputed (Yen et al., 2015). Therefore, it appears to be clear that an effective humoral immune response represents the final host weapon to shortcut the viral infection.
Antibody Escape Mutants
Picornaviruses' high mutation rates permit the rapid escape from the immune system. The change of residues in the exterior capsid surface overcomes the intense pressure exerted by host Abs. These properties were used to locate capsid antigenic sites by testing neutralizing Abs (nAbs) escape mutants in vitro (Minor et al., 1986; Sherry et al., 1986; Stapleton and Lemon, 1987). Successful progeny virus rely on capsid functionality. Therefore, the preservation of the architecture and receptor binding restrict viable mutations. The exposed jelly-roll loops can accommodate mutations easier than secondary structure elements and protein–protein interacting surfaces (Murray et al., 1988; Usherwood and Nash, 1995). Several structural studies by cryoEM revealed the way nAbs bind to solvent-exposed loops of VP1–3 by interacting with critical residues. Complexes of viruses with Fab-Abs fragments can display 1-Fab:1-protomer ratio following the icosahedral symmetry. Nevertheless, when epitopes are close to the symmetry axis, lower binding ratios are observed due to Fab–Fab steric hindrance (Lee et al., 2013). Three mechanisms of neutralization are interpreted from these structures: (i) destabilization of the virion by triggering conformational changes upon Fab binding, which is accompanied by the “pocket factor” release when present (Smith et al., 1996; Plevka et al., 2014; Dong et al., 2017; Zheng et al., 2019), (ii) stabilization of virions by cross-linking of protomers to prevent conformational changes for infection (Ye et al., 2016), (iii) virus aggregation by antibody cross-linking particles (Mosser et al., 1989), and (iv) opsonization that can interfere with virus-cell attachment and receptor binding (Lee et al., 2013; Wang et al., 2017).
The Canyon Hypothesis
Hiding the receptor-interacting surface from Abs surveillance is the hypothetical function of the canyon (Rossmann, 1989). Therefore, the canyon is the result of an evolutionary process to preserve and protect the critical residues required for host-cell receptor recognition. Conversely, accessible areas can mutate to disguise the virus from the humoral immune response. Nevertheless, some observations have challenged this view, proposing the receptor-binding site topology as an uncoating mechanism that dictates receptor binding to trigger the uncoating event (Smith et al., 1996). These views are not strictly incompatible; hence, both pictures contribute to the paradigm of picornavirus capsid evolution. Here, capsid topology arose out of and continues to be shaped by the interplay of host environmental pressure, random genome mutations, and fixation of mutations when beneficial.
Picornavirus Antibody Decoy Particles
Picornaviruses are known to produce a significant amount of procapsids during the infectious cell cycle, which appears as an inefficient way to replicate (Shingler et al., 2015). This wastefulness looks aggravated considering each polyprotein translation event would lead to a single protomer. Finally, the so-called procapsid may be an off-pathway particle (Cifuente et al., 2013). Although counterintuitive in appearance, the function of the procapsid could be to act as an immune decoy to enhance the infectivity of mature virions providing an evolutive advantage (Shingler et al., 2015; Liu et al., 2016). In this regard, empty Dane particles also have been proposed to be decoy particles for the hepatitis B virus (Rydell et al., 2017).
Several picornavirus procapsids are larger particles compared to the mature virion and similar in shape to the A-particle. Procapsids have some viral epitopes more accessible and consequently can bind nAbs more efficiently. This phenomenon has been observed for the procapsid and mature virion of EV71, revealing that they are antigenically distinct (Shingler et al., 2015). Moreover, a non-nAb has been structurally solved in complex with procapsids but not disclosed for the mature virion (Hewat and Blaas, 2006), which suggests that procapsid may also lead to effects in the modulation of non-nAbs immune response.
Vaccines
Inactivated vaccines for PV, HAV, and FMDV are shown to prevent associated illnesses by inducing specific antibody defenses (Salk, 1957). Nevertheless, it was the live-attenuated oral PV vaccine (OPV) responsible for most of the success in controlling polio epidemics. PV attenuation was obtained by serial passage in cell lines from non-human hosts (Sabin, 1965). Rare cases of vaccination-derived paralytic disease can occur as well as vaccines shedding of virulent poliovirus revertant (Platt et al., 2014).
New ideas for picornavirus vaccines, currently under development, exploit evolutionary concepts including (i) codon usage, (ii) mutational robustness, (iii) modification of translational efficiency, and (iv) RdRp fidelity manipulation (Burns et al., 2006; Mueller et al., 2006; Coleman et al., 2008; Le Nouën et al., 2014; Tulloch et al., 2014; McDonald et al., 2016). In this regard, synonymous codon deoptimization was first applied for PV (Burns et al., 2006; Mueller et al., 2006) and other picornaviruses such as FMDV (Diaz-San Segundo et al., 2016). Interestingly, increasing the CpG and UpA dinucleotide frequencies using synonymous codon substitutions leads to increased activation of the immune system (Tulloch et al., 2014). The maintenance of phenotype despite changing the genotype (mutations), or mutational robustness, can be altered by codon substitutions, correlating less robust viruses with attenuation in mice (Lauring et al., 2012). This approach was further extended using synonymous codons that upon mutation became stop codons (Moratorio et al., 2017). The increase in stop codon mutations in codon-engineered CVB3 during replication led to a loss of infectivity in vitro and attenuation in vivo. These new methods can improve the safety of already existing live-attenuated vaccines and can be broadly applied by re-coding any viral genome (Jorge et al., 2015).
Further elucidation of the mechanisms underlying these phenotypes could be used for rational codon rewiring in combination with increasing CpG and UpA frequencies to activate innate host immunity (Kumagai et al., 2008; Tulloch et al., 2014).
Final Remarks
The control and eradication of pathogenic picornaviruses is an ongoing problem. Picornavirus evolution, ruled by high mutation rates and recombination, has made this viral group genetically and antigenically highly variable. Moreover, changes in virulence represent an unpredictable threat. Important lessons drawn from the polio eradication battle indicate the necessity of a new generation of vaccines (Agol et al., 2016). A holistic approach based on big data and mathematical modeling combining views from structural and evolutionary biology, cellular, and molecular immunology will allow a better understanding of picornavirus–host interactions. This knowledge could have the potential to foresee possible outbreaks and changes on viral virulence. Furthermore, these models may redefine the way new vaccines and antiviral therapies will be designed.
Author Contributions
JC and GM conceived the review project, designed figures, wrote the manuscirpt, and approved it for publication. They both contributed equally to the work.
Funding
This work was supported by PEDECIBA, Uruguay, and Comisión Sectorial de Investigación Científica (CSIC), UdelaR, Uruguay.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We especially thank James Weger-Lucarelli for critical reading.
References
Agol, V., Cello, J., Chumakov, K., Ehrenfeld, E., and Wimmer, E. (2016). Eradicating polio: a balancing act. Science 351:348. doi: 10.1126/science.351.6271.348-b
Andino, R., and Domingo, E. (2015). Viral quasispecies. Virology 479–480, 46–51. doi: 10.1016/j.virol.2015.03.022
Arita, M., Zhu, S.-L., Yoshida, H., Yoneyama, T., Miyamura, T., and Shimizu, H. (2005). A sabin 3-derived poliovirus recombinant contained a sequence homologous with indigenous human enterovirus species C in the viral polymerase coding region. J. Virol. 79, 12650–12657. doi: 10.1128/JVI.79.20.12650-12657.2005
Baggen, J., Hurdiss, D. L., Zocher, G., Mistry, N., Roberts, R. W., Slager, J. J., et al. (2018). Role of enhanced receptor engagement in the evolution of a pandemic acute hemorrhagic conjunctivitis virus. Proc. Natl. Acad. Sci. U.S.A. 115, 397–402. doi: 10.1073/pnas.1713284115
Belsham, G. J., Abrams, C. C., King, A. M., Roosien, J., and Vlak, J. M. (1991). Myristoylation of foot-and-mouth disease virus capsid protein precursors is independent of other viral proteins and occurs in both mammalian and insect cells. J. Gen. Virol. 72, 747–751. doi: 10.1099/0022-1317-72-3-747
Bergelson, J. M. (1997). Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275, 1320–1323. doi: 10.1126/science.275.5304.1320
Bergelson, J. M., Chan, M., Solomon, K. R., St John, N. F., Lin, H., and Finberg, R. W. (1994). Decay-accelerating factor (CD55), a glycosylphosphatidylinositol-anchored complement regulatory protein, is a receptor for several echoviruses. Proc. Natl. Acad. Sci. U.S.A. 91, 6245–6248.
Bergelson, J. M., Mohanty, J. G., Crowell, R. L., St John, N. F., Lublin, D. M., and Finberg, R. W. (1995). Coxsackievirus B3 adapted to growth in RD cells binds to decay-accelerating factor (CD55). J. Virol. 69, 1903–1906.
Bergelson, J. M., Shepley, M. P., Chan, B. M., Hemler, M. E., and Finberg, R. W. (1992). Identification of the integrin VLA-2 as a receptor for echovirus 1. Science 255, 1718–1720.
Bessaud, M., Joffret, M.-L., Blondel, B., and Delpeyroux, F. (2016). Exchanges of genomic domains between poliovirus and other cocirculating species C enteroviruses reveal a high degree of plasticity. Sci. Rep. 6:38831. doi: 10.1038/srep38831
Bordería, A. V., Isakov, O., Moratorio, G., Henningsson, R., Agüera-González, S., Organtini, L., et al. (2015). Group selection and contribution of minority variants during virus adaptation determines virus fitness and phenotype. PLoS Pathog. 11:e1004838. doi: 10.1371/journal.ppat.1004838
Bubeck, D., Filman, D. J., Cheng, N., Steven, A. C., Hogle, J. M., and Belnap, D. M. (2005). The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79, 7745–7755. doi: 10.1128/JVI.79.12.7745-7755.2005
Bucciol, G., Moens, L., Payne, K., Wollants, E., Mekahli, D., Levtchenko, E., et al. (2018). Chronic Aichi virus infection in a patient with x-linked agammaglobulinemia. J. Clin. Immunol. 38, 748–752. doi: 10.1007/s10875-018-0558-z
Burns, C. C., Shaw, J., Campagnoli, R., Jorba, J., Vincent, A., Quay, J., et al. (2006). Modulation of poliovirus replicative fitness in HeLa cells by deoptimization of synonymous codon usage in the capsid region. J. Virol. 80, 3259–3272. doi: 10.1128/JVI.80.7.3259-3272.2006
Chen, Y. H., Du, W., Hagemeijer, M. C., Takvorian, P. M., Pau, C., Cali, A., et al. (2015). Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160, 619–630. doi: 10.1016/j.cell.2015.01.032
Cifuente, J. O., Lee, H., Yoder, J. D., Shingler, K. L., Carnegie, M. S., Yoder, J. L., et al. (2013). Structures of the procapsid and mature virion of enterovirus 71 strain 1095. J. Virol. 87, 7637–7645. doi: 10.1128/JVI.03519-12
Coleman, J. R., Papamichail, D., Skiena, S., Futcher, B., Wimmer, E., and Mueller, S. (2008). Virus attenuation by genome-scale changes in codon pair bias. Science 320, 1784–1787. doi: 10.1126/science.1155761
Combelas, N., Holmblat, B., Joffret, M.-L., Colbère-Garapin, F., and Delpeyroux, F. (2011). Recombination between poliovirus and coxsackie A viruses of species C: a model of viral genetic plasticity and emergence. Viruses 3, 1460–1484. doi: 10.3390/v3081460
Coyne, C. B., and Bergelson, J. M. (2006). Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124, 119–131. doi: 10.1016/j.cell.2005.10.035
Crawford, N. M., and Baltimore, D. (1983). Genome-linked protein VPg of poliovirus is present as free VPg and VPg-PUpU in poliovirus-infected cells. Proc. Natl. Acad. Sci. U.S.A. 80, 7452–7455.
Crotty, S., Cameron, C. E., and Andino, R. (2001). RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. U.S.A. 98, 6895–6900. doi: 10.1073/pnas.111085598
Diaz-San Segundo, F. D.-S., Medina, G. N., Ramirez-Medina, E., Velazquez-Salinas, L., Koster, M., Grubman, M. J., et al. (2016). Synonymous deoptimization of foot-and-mouth disease virus causes attenuation in vivo while inducing a strong neutralizing antibody response. J. Virol. 90, 1298–1310. doi: 10.1128/JVI.02167-15
Domingo, E., Sheldon, J., and Perales, C. (2012). Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 76, 159–216. doi: 10.1128/MMBR.05023-11
Dong, Y., Liu, Y., Jiang, W., Smith, T. J., Xu, Z., and Rossmann, M. G. (2017). Antibody-induced uncoating of human rhinovirus B14. Proc. Natl. Acad. Sci. U.S.A. 114, 8017–8022. doi: 10.1073/pnas.1707369114
Drysdale, S. B., Mejias, A., and Ramilo, O. (2017). Rhinovirus—not just the common cold. J. Infect. 74(Suppl. 1), S41–S46. doi: 10.1016/S0163-4453(17)30190-1
Eggers, H. J. (1999). Milestones in early poliomyelitis research (1840 to 1949). J. Virol. 73, 4533–4535.
Eggers, H. J., and Tamm, I. (1965). Coxsackie A9 virus: mutation from drug dependence to drug independence. Science 148, 97–98. doi: 10.1126/science.148.3666.97
Elena, S. F., and Sanjuán, R. (2005). Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 79, 11555–11558. doi: 10.1128/JVI.79.18.11555-11558.2005
Enders, J. F., Weller, T. H., and Robbins, F. C. (1949). Cultivation of the Lansing strain of poliomyelitis virus in cultures of various human embryonic tissues. Science 109, 85–87. doi: 10.1126/science.109.2822.85
Escribano-Romero, E. (2004). Heparan sulphate mediates swine vesicular disease virus attachment to the host cell. J. Gen. Virol. 85, 653–663. doi: 10.1099/vir.0.19603-0
Feng, Z., Hensley, L., McKnight, K. L., Hu, F., Madden, V., Ping, L., et al. (2013). A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371. doi: 10.1038/nature12029
Filman, D. J., Syed, R., Chow, M., Macadam, A. J., Minor, P. D., and Hogle, J. M. (1989). Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8, 1567–1579.
Filman, D. J., Wien, M. W., Cunningham, J. A., Bergelson, J. M., and Hogle, J. M. (1998). Structure determination of echovirus 1. Acta Crystallogr. D Biol. Crystallogr. 54, 1261–1272. doi: 10.1107/S0907444998002790
Fitzsimmons, W. J., Woods, R. J., McCrone, J. T., Woodman, A., Arnold, J. J., and Yennawar, M. (2018). A speed–fidelity trade-off determines the mutation rate and virulence of an RNA virus. PLoS Biol. 16:2006459. doi: 10.1371/journal.pbio.2006459
Gnädig, N. F., Beaucourt, S., Campagnola, G., Bordería, A. V., Sanz-Ramos, M., Gong, P., et al. (2012). Coxsackievirus B3 mutator strains are attenuated in vivo. Proc. Natl. Acad. Sci. U.S.A. 109, E2294–E2303. doi: 10.1073/pnas.1204022109
Gong, P., and Peersen, O. B. (2010). Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 107, 22505–22510. doi: 10.1073/pnas.1007626107
Grant, C. F. J., Carr, B. V., Kotecha, A., van den Born, E., Stuart, D. I., Hammond, J. A., et al. (2017). The B cell response to foot-and-mouth disease virus in cattle following sequential vaccination with multiple serotypes. J. Virol. 91, 10–1128. doi: 10.1128/JVI.02157-16
Greve, J. M., Davis, G., Meyer, A. M., Forte, C. P., Yost, S. C., Marlor, C. W., et al. (1989). The major human rhinovirus receptor is ICAM-1. Cell 56, 839–847.
He, Y., Bowman, V. D., Mueller, S., Bator, C. M., Bella, J., Peng, X., et al. (2000). Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. U.S.A. 97, 79–84. doi: 10.1073/pnas.97.1.79
He, Y., Chipman, P. R., Howitt, J., Bator, C. M., Whitt, M. A., Baker, T. S., et al. (2001). Interaction of coxsackievirus B3 with the full length coxsackievirus–adenovirus receptor. Nat. Struct. Biol. 8, 874–878. doi: 10.1038/nsb1001-874
Hendry, E., Hatanaka, H., Fry, E., Smyth, M., Tate, J., Stanway, G., et al. (1999). The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7, 1527–1538. doi: 10.1016/S0969-2126(00)88343-4
Hewat, E. A., and Blaas, D. (2006). Nonneutralizing human rhinovirus serotype 2-specific monoclonal antibody 2G2 attaches to the region that undergoes the most dramatic changes upon release of the viral RNA. J. Virol. 80, 12398–12401. doi: 10.1128/JVI.01399-06
Hogle, J. M., Chow, M., and Filman, D. J. (1985). Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229, 1358–1365. doi: 10.1126/SCIENCE.2994218
Holm-Hansen, C. C., Midgley, S. E., and Fischer, T. K. (2016). Global emergence of enterovirus D68: a systematic review. Lancet Infect. Dis. 16, 64–75. doi: 10.1016/S1473-3099(15)00543-5
Jacobsen, K. H., and Wiersma, S. T. (2010). Hepatitis A virus seroprevalence by age and world region, 1990 and 2005. Vaccine 28, 6653–6657. doi: 10.1016/j.vaccine.2010.08.037
Jiang, P., Liu, Y., Ma, H.-C., Paul, A. V., and Wimmer, E. (2014). Picornavirus morphogenesis. Microbiol. Mol. Biol. Rev. 78, 418–437. doi: 10.1128/MMBR.00012-14
Jorge, D. M., Mills, R. E., and Lauring, A. S. (2015). CodonShuffle: a tool for generating and analyzing synonymously mutated sequences. Virus Evol. 1:vev012. doi: 10.1093/ve/vev012
Kainulainen, L., Vuorinen, T., Rantakokko-Jalava, K., Osterback, R., and Ruuskanen, O. (2010). Recurrent and persistent respiratory tract viral infections in patients with primary hypogammaglobulinemia. J. Allergy Clin. Immunol. 126, 120–126. doi: 10.1016/j.jaci.2010.04.016
Kirkegaard, K. (2017). Unconventional secretion of hepatitis A virus. Proc. Natl. Acad. Sci. U. S.A. 114, 6653–6655. doi: 10.1073/pnas.1707142114
Kloc, A., Rai, D. K., and Rieder, E. (2018). The roles of picornavirus untranslated regions in infection and innate immunity. Front. Microbiol. 9:485. doi: 10.3389/fmicb.2018.00485
Kolatkar, P. R., Bella, J., Olson, N. H., Bator, C. M., Baker, T. S., and Rossmann, M. G. (1999). Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18, 6249–6259. doi: 10.1093/emboj/18.22.6249
Kumagai, Y., Takeuchi, O., and Akira, S. (2008). TLR9 as a key receptor for the recognition of DNA. Adv. Drug Deliv. Rev. 60, 795–804. doi: 10.1016/j.addr.2007.12.004
Lauring, A. S., Acevedo, A., Cooper, S. B., and Andino, R. (2012). Codon usage determines the mutational robustness, evolutionary capacity, and virulence of an RNA virus. Cell Host Microbe 12, 623–632. doi: 10.1016/j.chom.2012.10.008
Le Nouën, C., Brock, L. G., Luongo, C., McCarty, T., Yang, L., and Mehedi, M. (2014). Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc. Natl. Acad. Sci. U.S.A. 111, 13169–13174. doi: 10.1073/pnas.1411290111
Lea, S., Hernández, J., Blakemore, W., Brocchi, E., Curry, S., Domingo, E., et al. (1994). The structure and antigenicity of a type C foot-and-mouth disease virus. Structure 2, 123–139.
Ledinko, N. (1963). Genetic recombination with poliovirus type 1. Virology 20, 107–119. doi: 10.1016/0042-6822(63)90145-4
Lee, H., Cifuente, J. O., Ashley, R. E., Conway, J. F., Makhov, A. M., Tano, Y., et al. (2013). A strain-specific epitope of enterovirus 71 identified by cryo-electron microscopy of the complex with Fab from neutralizing antibody. J. Virol. 87, 11363–11370. doi: 10.1128/JVI.01926-13
Lee, H., Shingler, K. L., Organtini, L. J., Ashley, R. E., Makhov, A. M., Conway, J. F., et al. (2016). The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs. Sci. Adv. 2:e1501929. doi: 10.1126/sciadv.1501929
Lentz, K. N., Smith, A. D., Geisler, S. C., Cox, S., Buontempo, P., Skelton, A., et al. (1997). Structure of poliovirus type 2 Lansing complexed with antiviral agent SCH48973: comparison of the structural and biological properties of three poliovirus serotypes. Structure 5, 961–978. doi: 10.1016/S0969-2126(97)00249-9
Li, C., Wang, J. C.-Y., Taylor, M. W., and Zlotnick, A. (2012). In vitro assembly of an empty picornavirus capsid follows a dodecahedral path. J. Virol. 86, 13062–13069. doi: 10.1128/JVI.01033-12
Li, J., Wang, J., Xu, C., Yin, Q., Hu, M., Sun, Z., et al. (2017). Hand, foot, and mouth disease in Mainland China before it was listed as category C disease. Lancet Infect. Dis. 17, 1017–1018. doi: 10.1016/S1473-3099(17)30471-1
Liu, Y., Hill, M. G., Klose, T., Chen, Z., Watters, K., Bochkov, Y. A., et al. (2016). Atomic structure of a rhinovirus C, a virus species linked to severe childhood asthma. Proc. Natl. Acad. Sci. U.S.A. 113, 8997–9002. doi: 10.1073/pnas.1606595113
Liu, Y., Sheng, J., Fokine, A., Meng, G., Shin, W.-H., Long, F., et al. (2015). Structure and inhibition of EV-D68, a virus that causes respiratory illness in children. Science 347, 71–74. doi: 10.1126/science.1261962
Loeffler, F., and Frosch, P. (1898). Report of the commission for research on the foot-and-mouth disease. Zent. Bakt. Parasitkde. Abt.I 23, 371–391.
Logan, D., Abu-Ghazaleh, R., Blakemore, W., Curry, S., Jackson, T., King, A., et al. (1993). Structure of a major immunogenic site on foot-and-mouth disease virus. Nature 362, 566–568. doi: 10.1038/362566a0
Lukashev, A. N. (2005). Recombination in circulating human enterovirus B: independent evolution of structural and non-structural genome regions. J. Gen. Virol. 86, 3281–3290. doi: 10.1099/vir.0.81264-0
Lukashev, A. N. (2010). Recombination among picornaviruses. Rev. Med. Virol. 20, 327–337. doi: 10.1002/rmv.660
Martinez-Salas, E., Francisco-Velilla, R., Fernandez-Chamorro, J., and Embarek, A. M. (2018). Insights into structural and mechanistic features of viral IRES elements. Front. Microbiol. 8:2629. doi: 10.3389/fmicb.2017.02629
McDonald, S., Block, A., Beaucourt, S., Moratorio, G., Vignuzzi, M., and Peersen, O. B. (2016). Design of a genetically stable high fidelity coxsackievirus B3 polymerase that attenuates virus growth in vivo. J. Biol. Chem. 291, 13999–14011. doi: 10.1074/jbc.M116.726596
McKinney, R. E., Katz, S. L., and Wilfert, C. M. (1987). Chronic enteroviral meningoencephalitis in agammaglobulinemic patients. Clin. Infect. Dis. 9, 334–356. doi: 10.1093/clinids/9.2.334
McLeish, N. J., Williams, C. H., Kaloudas, D., Roivainen, M. M., and Stanway, G. (2012). Symmetry-related clustering of positive charges is a common mechanism for heparan sulfate binding in enteroviruses. J. Virol. 86, 11163–11170. doi: 10.1128/JVI.00640-12
McWilliam Leitch, E. C., Bendig, J., Cabrerizo, M., Cardosa, J., Hyypiä, T., Ivanova, O. E., et al. (2009). Transmission networks and population turnover of echovirus 30. J. Virol. 83, 2109–2118. doi: 10.1128/JVI.02109-08
McWilliam Leitch, E. C., Cabrerizo, M., Cardosa, J., Harvala, H., Ivanova, O. E., Kroes, A. C. M., et al. (2010). Evolutionary dynamics and temporal/geographical correlates of recombination in the human enterovirus echovirus types 9, 11, and 30. J. Virol. 84, 9292–9300. doi: 10.1128/JVI.00783-10
Meijer, A., van der Sanden, S., Snijders, B. E. P., Jaramillo-Gutierrez, G., Bont, L., van der Ent, C. K., et al. (2012). Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology 423, 49–57. doi: 10.1016/j.virol.2011.11.021
Mena, I., Perry, C. M., Harkins, S., Rodriguez, F., Gebhard, J., and Whitton, J. L. (1999). The role of B lymphocytes in coxsackievirus B3 infection. Am. J. Pathol. 155, 1205–1215. doi: 10.1016/S0002-9440(10)65223-6
Mendelsohn, C. L., Wimmer, E., and Racaniello, V. R. (1989). Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56, 855–865.
Minor, P. (2014). The polio endgame. Hum. Vaccin. Immunother. 10, 2106–2108. doi: 10.4161/21645515.2014.981115
Minor, P. D., Ferguson, M., Evans, D. M., Almond, J. W., and Icenogle, J. P. (1986). Antigenic structure of polioviruses of serotypes 1, 2 and 3. J. Gen. Virol. 67(Pt 7), 1283–1291. doi: 10.1099/0022-1317-67-7-1283
Molla, A., Paul, A. V., and Wimmer, E. (1991). Cell-free, de novo synthesis of poliovirus. Science 254, 1647–1651.
Moratorio, G., Henningsson, R., Barbezange, C., Carrau, L., Bordería, A. V., and Blanc, H. (2017). Attenuation of RNA viruses by redirecting their evolution in sequence space. Nat. Microbiol. 2:17088. doi: 10.1038/nmicrobiol.2017.88
Mosser, A. G., Leippe, D. M., and Rueckert, R. R. (1989). “Neutralization of picornaviruses: support for the pentamer bridging hypothesis,” in Molecular Aspects of Picornavirus Infection and Detection, eds B. L. Semler and E. Ehrenfeld (Washington, DC: American Society for Microbiology (ASM)), 155–167.
Muckelbauer, J. K., Kremer, M., Minor, I., Diana, G., Dutko, F. J., Groarke, J., et al. (1995). The structure of coxsackievirus B3 at 3.5 å resolution. Structure 3, 653–667. doi: 10.1016/S0969-2126(01)00201-5
Mueller, S., Papamichail, D., Coleman, J. R., Skiena, S., and Wimmer, E. (2006). Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J. Virol. 80, 9687–9696. doi: 10.1128/JVI.00738-06
Mullapudi, E., Nováček, J., Pálková, L., Kulich, P., Lindberg, A. M., van Kuppeveld, F. J. M., et al. (2016). Structure and genome release mechanism of the human cardiovirus saffold virus 3. J. Virol. 90, 7628–7639. doi: 10.1128/JVI.00746-16
Murray, M. G., Bradley, J., Yang, X. F., Wimmer, E., Moss, E. G., and Racaniello, V. R. (1988). Poliovirus host range is determined by a short amino acid sequence in neutralization antigenic site I. Science 241, 213–215.
Nielsen, A. C., Gyhrs, M. L., Nielsen, L. P., Pedersen, C., and Böttiger, B. (2013). Gastroenteritis and the novel picornaviruses aichi virus, cosavirus, saffold virus, and salivirus in young children. J. Clin. Virol. 57, 239–242. doi: 10.1016/j.jcv.2013.03.015
Nishimura, Y., McLaughlin, N. P., Pan, J., Goldstein, S., Hafenstein, S., Shimizu, H., et al. (2015). The suramin derivative NF449 interacts with the 5-fold vertex of the enterovirus A71 capsid to prevent virus attachment to PSGL-1 and heparan sulfate. PLOS Pathog. 11:e1005184. doi: 10.1371/journal.ppat.1005184
Nishimura, Y., Shimojima, M., Tano, Y., Miyamura, T., Wakita, T., and Shimizu, H. (2009). Human P-selectin glycoprotein ligand-1 is a functional receptor for enterovirus 71. Nat. Med. 15, 794–797. doi: 10.1038/nm.1961
Nomoto, A., Detjen, B., Pozzatti, R., and Wimmer, E. (1977). The location of the polio genome protein in viral RNAs and its implication for RNA synthesis. Nature 268, 208–213.
Organtini, L. J., Makhov, A. M., Conway, J. F., Hafenstein, S., and Carson, S. D. (2014). Kinetic and structural analysis of coxsackievirus B3 receptor interactions and formation of the A-particle. J. Virol. 88, 5755–5765. doi: 10.1128/JVI.00299-14
Palmenberg, A. C. (1990). Proteolytic processing of picornaviral polyprotein. Annu. Rev. Microbiol. 44, 603–623. doi: 10.1146/annurev.mi.44.100190.003131
Pan, J., Narayanan, B., Shah, S., Yoder, J. D., Cifuente, J. O., Hafenstein, S., et al. (2011). Single amino acid changes in the virus capsid permit coxsackievirus B3 to bind decay-accelerating factor. J. Virol. 85, 7436–7443. doi: 10.1128/JVI.00503-11
Paul, A. V., and Schultz, A. (1987). Capsid protein VP4 of poliovirus is N-myristoylated. Proc. Natl. Acad. Sci. U.S.A. 84, 7827–7831.
Pettigrew, D. M., Williams, D. T., Kerrigan, D., Evans, D. J., Lea, S. M., and Bhella, D. (2006). Structural and functional insights into the interaction of echoviruses and decay-accelerating factor. J. Biol. Chem. 281, 5169–5177. doi: 10.1074/jbc.M510362200
Pfeiffer, J. K., and Kirkegaard, K. (2003). A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. U.S.A. 100, 7289–7294. doi: 10.1073/pnas.1232294100
Platt, L. R., Estívariz, C. F., and Sutter, R. W. (2014). Vaccine-associated paralytic poliomyelitis: a review of the epidemiology and estimation of the global burden. J. Infect. Dis. 210, S380–S389. doi: 10.1093/infdis/jiu184
Plevka, P., Hafenstein, S., Harris, K. G., Cifuente, J. O., Zhang, Y., Bowman, V. D., et al. (2010). Interaction of decay-accelerating factor with echovirus 7. J. Virol. 84, 12665–12674. doi: 10.1128/JVI.00837-10
Plevka, P., Lim, P.-Y., Perera, R., Cardosa, J., Suksatu, A., Kuhn, R. J., et al. (2014). Neutralizing antibodies can initiate genome release from human enterovirus 71. Proc. Natl. Acad. Sci. U.S.A. 111, 2134–2139. doi: 10.1073/pnas.1320624111
Plevka, P., Perera, R., Cardosa, J., Kuhn, R. J., and Rossmann, M. G. (2012). Crystal structure of human enterovirus 71. Science 336:1274. doi: 10.1126/science.1218713
Pons-Salort, M., Oberste, M. S., Pallansch, M. A., Abedi, G. R., Takahashi, S., Grenfell, B. T., et al. (2018). The seasonality of nonpolio enteroviruses in the United States: patterns and drivers. Proc. Natl. Acad. Sci. U.S.A. 115, 3078–3083. doi: 10.1073/pnas.1721159115
Racaniello, V. R., and Baltimore, D. (1981). Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214, 916–919. doi: 10.1126/science.6272391
Rai, D. K., Diaz-San Segundo, F., Campagnola, G., Keith, A., Schafer, E. A., Kloc, A., et al. (2017). Attenuation of foot-and-mouth disease virus by engineered viral polymerase fidelity. J. Virol. 91:e00081-17. doi: 10.1128/JVI.00081-17
Ren, J., Wang, X., Zhu, L., Hu, Z., Gao, Q., Yang, P., et al. (2015). Structures of coxsackievirus A16 capsids with native antigenicity: implications for particle expansion, receptor binding, and immunogenicity. J. Virol. 89, 10500–10511. doi: 10.1128/JVI.01102-15
Roivainen, M., Piirainen, L., Hovi, T., Virtanen, I., Riikonen, T., Heino, J., et al. (1994). Entry of coxsackievirus A9 into host cells: specific interactions with αvβ3 integrin, the vitronectin receptor. Virology 203, 357–365. doi: 10.1006/viro.1994.1494
Rossmann, M. G. (1989). The canyon hypothesis. Hiding the host cell receptor attachment site on a viral surface from immune surveillance. J. Biol. Chem. 264, 14587–14590.
Rossmann, M. G., Arnold, E., Erickson, J. W., Frankenberger, E. A., Griffith, J. P., Hecht, H.-J., et al. (1985). Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317, 145–153. doi: 10.1038/317145a0
Rozen-Gagnon, K., Stapleford, K. A., Mongelli, V., Blanc, H., Failloux, A.-B., and Saleh, M.-C. (2014). Alphavirus mutator variants present host-specific defects and attenuation in mammalian and insect models. PLoS Pathog 10:e1003877. doi: 10.1371/journal.ppat.1003877
Rydell, G. E., Prakash, K., Norder, H., and Lindh, M. (2017). Hepatitis B surface antigen on subviral particles reduces the neutralizing effect of anti-HBs antibodies on hepatitis B viral particles in vitro. Virology 509, 67–70. doi: 10.1016/j.virol.2017.05.017
Sabin, A. B. (1965). Oral poliovirus vaccine. History of its development and prospects for eradication of poliomyelitis. JAMA 194, 872–876.
Salk, J. E. (1957). Poliomyelitis vaccination in the fall of 1956. Am. J. Public Health 47, 1–18. doi: 10.2105/ajph.47.1.1
Sanjuán, R. (2017). Collective infectious units in viruses. Trends Microbiol. 25, 402–412. doi: 10.1016/j.tim.2017.02.003
Shakeel, S., Seitsonen, J. J., Kajander, T., Laurinmäki, P., Hyypiä, T., Susi, P., et al. (2013). Structural and functional analysis of coxsackievirus A9 integrin αvβ6 binding and uncoating. J. Virol. 87, 3943–3951. doi: 10.1128/JVI.02989-12
Sherry, B., Mosser, A. G., Colonno, R. J., and Rueckert, R. R. (1986). Use of monoclonal antibodies to identify four neutralization immunogens on a common cold picornavirus, human rhinovirus 14. J. Virol. 57, 246–257.
Shingler, K. L., Cifuente, J. O., Ashley, R. E., Makhov, A. M., Conway, J. F., and Hafenstein, S. (2015). The enterovirus 71 procapsid binds neutralizing antibodies and rescues virus infection in vitro. J. Virol. 89, 1900–1908. doi: 10.1128/JVI.03098-14
Simon-Loriere, E., and Holmes, E. C. (2011). Why do RNA viruses recombine? Nat. Rev. Microbiol. 9, 617–626. doi: 10.1038/nrmicro2614
Slater, L., Bartlett, N. W., Haas, J. J., Zhu, J., Message, S. D., Walton, R. P., et al. (2010). Co-ordinated role of TLR3, RIG-I and MDA5 in the innate response to rhinovirus in bronchial epithelium. PLoS Pathogens 6:e1001178. doi: 10.1371/journal.ppat.1001178
Smith, T. J., Chase, E. S., Schmidt, T. J., Olson, N. H., and Baker, T. S. (1996). Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature 383, 350–354. doi: 10.1038/383350a0
Stapleton, J. T. (1992). Passive immunization against hepatitis A. Vaccine 10, S45–S47. doi: 10.1016/0264-410X(92)90541-Q
Stapleton, J. T., and Lemon, S. M. (1987). Neutralization escape mutants define a dominant immunogenic neutralization site on hepatitis A virus. J. Virol. 61, 491–498.
Staunton, D. E., Merluzzi, V. J., Rothlein, R., Barton, R., Marlin, S. D., and Springer, T. A. (1989). A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell 56, 849–853. doi: 10.1016/0092-8674(89)90689-2
Stern, A., Yeh, M. T., Zinger, T., Smith, M., Wright, C., Ling, G., et al. (2017). The evolutionary pathway to virulence of an RNA virus. Cell 169, 35–46.e19. doi: 10.1016/j.cell.2017.03.013
Strauss, M., Filman, D. J., Belnap, D. M., Cheng, N., Noel, R. T., and Hogle, J. M. (2015). Nectin-like interactions between poliovirus and its receptor trigger conformational changes associated with cell entry. J. Virol. 89, 4143–4157. doi: 10.1128/JVI.03101-14
Stuart, A. D., McKee, T. A., Williams, P. A., Harley, C., Shen, S., Stuart, D. I., et al. (2002). Determination of the structure of a decay accelerating factor-binding clinical isolate of echovirus 11 allows mapping of mutants with altered receptor requirements for infection. J. Virol. 76, 7694–7704. doi: 10.1128/JVI.76.15.7694-7704.2002
Sun, M., Li, C., Xu, W., Liao, G., Li, R., Zhou, J., et al. (2017). Immune serum from sabin inactivated poliovirus vaccine immunization neutralizes multiple individual wild and vaccine-derived polioviruses. Clin. Infect. Dis. 64, 1317–1325. doi: 10.1093/cid/cix110
Tan, C. W., Poh, C. L., Sam, I.-C., and Chan, Y. F. (2013). Enterovirus 71 uses cell surface heparan sulfate glycosaminoglycan as an attachment receptor. J. Virol. 87, 611–620. doi: 10.1128/JVI.02226-12
Thompson, A. A., Albertini, R. A., and Peersen, O. B. (2007). Stabilization of poliovirus polymerase by NTP binding and fingers–thumb interactions. J. Mol. Biol. 366, 1459–1474. doi: 10.1016/j.jmb.2006.11.070
Tomassini, J. E., Graham, D., DeWitt, C. M., Lineberger, D. W., Rodkey, J. A., and Colonno, R. J. (1989). CDNA cloning reveals that the major group rhinovirus receptor on HeLa cells is intercellular adhesion molecule 1. Proc. Natl. Acad. Sci. U.S.A. 86, 4907–4911.
Tulloch, F., Atkinson, N. J., Evans, D. J., Ryan, M. D., and Simmonds, P. (2014). RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. Elife 3:e04531. doi: 10.7554/eLife.04531
Usherwood, E. J., and Nash, A. A. (1995). Lymphocyte recognition of picornaviruses. J. Gen. Virol. 76, 499–508. doi: 10.1099/0022-1317-76-3-499
Venkataraman, S., Reddy, S. P., Loo, J., Idamakanti, N., Hallenbeck, P. L., and Reddy, V. S. (2008). Structure of seneca valley virus-001: an oncolytic picornavirus representing a new genus. Structure 16, 1555–1561. doi: 10.1016/j.str.2008.07.013
Verdaguer, N., Blaas, D., and Fita, I. (2000). Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol. 300, 1179–1194. doi: 10.1006/JMBI.2000.3943
Verdaguer, N., Fita, I., Reithmayer, M., Moser, R., and Blaas, D. (2004). X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat. Struct. Mol. Biol. 11, 429–434. doi: 10.1038/nsmb753
Verdaguer, N., Jimenez-Clavero, M. A., Fita, I., and Ley, V. (2003). Structure of swine vesicular disease virus: mapping of changes occurring during adaptation of human coxsackie B5 virus to infect swine. J. Virol. 77, 9780–9789. doi: 10.1128/JVI.77.18.9780-9789.2003
Vignuzzi, M., Stone, J. K., Arnold, J. J., Cameron, C. E., and Andino, R. (2006). Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439, 344–348. doi: 10.1038/nature04388
Wang, X., Zhu, L., Dang, M., Hu, Z., Gao, Q., Yuan, S., et al. (2017). Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site. Proc. Natl. Acad. Sci. U.S.A. 114, 770–775. doi: 10.1073/pnas.1616502114
Watters, K., and Palmenberg, A. C. (2018). CDHR3 extracellular domains EC1-3 mediate rhinovirus C interaction with cells and as recombinant derivatives, are inhibitory to virus infection. PLOS Pathog. 14:1007477. doi: 10.1371/journal.ppat.1007477
Wilfert, C. M., Buckley, R. H., Mohanakumar, T., Griffith, J. F., Katz, S. L., Whisnant, J. K., et al. (1977). Persistent and fatal central-nervous-system ECHOvirus infections in patients with agammaglobulinemia. N. Engl. J. Med. 296, 1485–1489. doi: 10.1056/NEJM197706302962601
Williams, C. H., Kajander, T., Hyypiä, T., Jackson, T., Sheppard, D., and Stanway, G. (2004). Integrin alpha v beta 6 is an RGD-dependent receptor for coxsackievirus A9. J. Virol. 78, 6967–6973. doi: 10.1128/JVI.78.13.6967-6973.2004
Woodman, A., Lee, K.-M., Janissen, R., Gong, Y.-N., Dekker, N. H., Shih, S.-R., et al. (2018). Predicting intraserotypic recombination in enterovirus 71. J. Virol. 93:e02057-18. doi: 10.1128/JVI.02057-18
Xiao, C., Bator, C. M., Bowman, V. D., Rieder, E., He, Y., Hébert, B., et al. (2001). Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J. Virol. 75, 2444–2451. doi: 10.1128/JVI.75.5.2444-2451.2001
Xiao, C., Bator-Kelly, C. M., Rieder, E., Chipman, P. R., Craig, A., Kuhn, R. J., et al. (2005). The crystal structure of coxsackievirus A21 and its interaction with ICAM-1. Structure 13, 1019–1033. doi: 10.1016/j.str.2005.04.011
Xiao, Y., Dolan, P. T., Goldstein, E. F., Li, M., Farkov, M., Brodsky, L., et al. (2017). Poliovirus intrahost evolution is required to overcome tissue-specific innate immune responses. Nat. Commun. 8:375. doi: 10.1038/s41467-017-00354-5
Xiao, Y., Rouzine, I. M., Bianco, S., Acevedo, A., Goldstein, E. F., Farkov, M., et al. (2016). RNA recombination enhances adaptability and is required for virus spread and virulence. Cell Host Microbe 19, 493–503. doi: 10.1016/j.chom.2016.03.009
Xie, X., Wang, H., Zeng, J., Li, C., Zhou, G., Yang, D., et al. (2014). Foot-and-mouth disease virus low-fidelity polymerase mutants are attenuated. Arch. Virol. 159, 2641–2650. doi: 10.1007/s00705-014-2126-z
Xing, L., Huhtala, M., Pietiäinen, V., Käpylä, J., Vuorinen, K., Marjomäki, V., et al. (2004). Structural and functional analysis of integrin α2 I domain interaction with echovirus 1. J. Biol. Chem. 279, 11632–11638. doi: 10.1074/jbc.M312441200
Yamayoshi, S., Yamashita, Y., Li, J., Hanagata, N., Minowa, T., Takemura, T., et al. (2009). Scavenger receptor B2 is a cellular receptor for enterovirus 71. Nat. Med. 15, 798–801. doi: 10.1038/nm.1992
Ye, X., Fan, C., Ku, Z., Zuo, T., Kong, L., Zhang, C., et al. (2016). Structural basis for recognition of human enterovirus 71 by a bivalent broadly neutralizing monoclonal antibody. PLoS Pathog 12:e1005454. doi: 10.1371/journal.ppat.1005454
Yen, M.-H., Huang, Y.-C., Chen, M.-C., Liu, C.-C., Chiu, N.-C., Lien, R., et al. (2015). Effect of intravenous immunoglobulin for neonates with severe enteroviral infections with emphasis on the timing of administration. J. Clin. Virol. 64, 92–96. doi: 10.1016/j.jcv.2015.01.013
Yin-Murphy, M., and Almond, J. W. (1996). “Picornaviruses,” in Medical Microbiology, ed S. Baron (Galveston, TX: University of Texas Medical Branch).
Yoder, J. D., Cifuente, J. O., Pan, J., Bergelson, J. M., and Hafenstein, S. (2012). The crystal structure of a coxsackievirus B3-RD variant and a refined 9-angstrom cryo-electron microscopy reconstruction of the virus complexed with decay-accelerating factor (DAF) provide a new footprint of DAF on the virus surface. J. Virol. 86, 12571–12581. doi: 10.1128/JVI.01592-12
Zarocostas, J. (2018). WHO keeps polio on the international health emergency list. Lancet 392:2425. doi: 10.1016/S0140-6736(18)33115-5
Zautner, A. E., Jahn, B., Hammerschmidt, E., Wutzler, P., and Schmidtke, M. (2006). N- and 6-O-sulfated heparan sulfates mediate internalization of coxsackievirus B3 variant PD into CHO-K1 cells. J. Virol. 80, 6629–6636. doi: 10.1128/JVI.01988-05
Zell, R., Delwart, E., Gorbalenya, E., and Hovi, T. (2017). ICTV virus taxonomy profile. Picornaviridae. 98, 2421–2422. doi: 10.1099/jgv.0.000911
Zhang, C., Zhu, R., Yang, Y., Chi, Y., Yin, J., Tang, X., et al. (2015). Phylogenetic analysis of the major causative agents of hand, foot and mouth disease in Suzhou city, Jiangsu province, China, in 2012–2013. Emerg. Microbes Infect. 4, 1–10. doi: 10.1038/emi.2015.12
Zhang, Y., Simpson, A. A., Ledford, R. M., Bator, C. M., Chakravarty, S., Skochko, G. A., et al. (2004). Structural and virological studies of the stages of virus replication that are affected by antirhinovirus compounds. J. Virol. 78, 11061–11069. doi: 10.1128/JVI.78.20.11061-11069.2004
Zhao, R., Pevear, D. C., Kremer, M. J., Giranda, V. L., Kofron, J. A., Kuhn, R. J., et al. (1996). Human rhinovirus 3 at 3.0 Å resolution. Structure 4, 1205–1220.
Zheng, Q., Zhu, R., Xu, L., He, M., Yan, X., Liu, D., et al. (2019). Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization. Nat. Microbiol. 4, 124–133. doi: 10.1038/s41564-018-0275-7
Zhou, D., Zhao, Y., Kotecha, A., Fry, E. E., Kelly, J. T., Wang, X., et al. (2018). Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2. Nat. Microbiol. 4, 414–419. doi: 10.1038/s41564-018-0319-z
Zhu, L., Wang, X., Ren, J., Porta, C., Wenham, H., Ekström, J.-O., et al. (2015). Structure of Ljungan virus provides insight into genome packaging of this picornavirus. Nat. Commun. 6:8316. doi: 10.1038/ncomms9316
Keywords: picornavirus, capsid, antibody, genetic variability, structure, vaccine
Citation: Cifuente JO and Moratorio G (2019) Evolutionary and Structural Overview of Human Picornavirus Capsid Antibody Evasion. Front. Cell. Infect. Microbiol. 9:283. doi: 10.3389/fcimb.2019.00283
Received: 12 February 2019; Accepted: 24 July 2019;
Published: 20 August 2019.
Edited by:
Ricardo Martin Gomez, CONICET Institute of Biotechnology and Molecular Biology (IBBM), ArgentinaReviewed by:
Fayna Diaz San Segundo, Plum Island Animal Disease Center (ARS-USDA), United StatesElizabeth Rieder, Plum Island Animal Disease Center (ARS-USDA), United States
Copyright © 2019 Cifuente and Moratorio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Javier Orlando Cifuente, amNpZnVlbnRlQGNpY2Jpb2d1bmUuZXM=; Gonzalo Moratorio, Z29uemFsby5tb3JhdG9yaW9AcGFzdGV1ci5mcg==
†These authors have contributed equally to this work
 Javier Orlando Cifuente
Javier Orlando Cifuente Gonzalo Moratorio
Gonzalo Moratorio