- 1Department of Clinical Sciences and Stomatology, Marche Polytechnic University, Ancona, Italy
- 2Department of Clinical and Experimental Medicine, University of Foggia, Foggia, Italy
- 3Department of Biomedical and Neuromuscular Sciences, University of Bologna, Bologna, Italy
- 4Dental Clinic, National Institute of Health and Science of Aging, IRCCS INRCA, Ancona, Italy
An altered oral microbiota has been linked with the development of several oral diseases, such as dental caries, periodontal disease, and oral stomatitis. Moreover, poor oral health has been linked to head and neck cancer, particularly oral cancer. In recent years a growing number of studies indicate that oral microbiota could be involved in the development of primary tumours outside of head and neck region. The aim of this article is to review the recent studies based on high-throughput technology to present evidences of a relationship between oral microbiota and “non-head and neck tumours.” Oral dysbiosis seem to be more pronounced in patients with tumours of gastrointestinal tract, in particular oesophageal, gastric, pancreatic, and colorectal cancers, paving the way for developing specific oral microbiota test to allow early cancer detection. Regarding other tumour types, the results are promising but highly preliminary and still debated. Currently, there are several factors that limit the generalization of the results, such as the small sample size, the lack of adequate clinical information about patients, the different sequencing techniques used, and biological sample heterogeneity. Although only at the beginning, the analysis of oral microbiota could be the next step in the evolution of cancer therapy and will help clinicians to develop individualised approaches to cancer prevention and treatment.
Introduction
The term “oral microbiota” refers to the heterogeneous group of microbial species colonizing all the oral cavity surfaces (Le Bars et al., 2017). In particular, about 700 bacterial species have been identified in oral cavity and, more importantly, 35% of them have not been cultured (Farrell et al., 2012). Indeed, the main limitation of microbial culture is the inability to identify the actual diversity of oral microbiota. To determine bacterial ecosystem as a whole, high-throughput sequencing methods have been developed, representing a leap toward characterization of oral microbial community (Benn et al., 2018). Currently, the most common approach to analyse oral microbiota is genome sequencing, through the analysis of 16S rDNA gene sequences. Indeed, the identification of this conserved DNA sequence has provided a universal system for the identification of bacteria. Recently, results from studies based on genome sequencing have started shedding light on the existence of site-specific microbial patterns that might be considered as “healthy/normal oral microbiota” (García-Castillo et al., 2016). The main obstacle to this definition is the highly variable composition of microbial community due to multiple factors, such as diet, age, and site (Arweiler and Netuschil, 2016). Nevertheless, ecological imbalance of microbial community, called dysbiosis, has been extensively studied both in human and animal models. It is characterised by: loss of beneficial microbes, expansion of pathogenic microbes, and general loss of microbial diversity (Meurman and Uittamo, 2008; Petersen and Round, 2014). It is becoming evident that the dysbiosis can lead to cancer development (Schwabe and Jobin, 2013). Three paradigms were proposed to describe the pathogenetic processes involving the microbiota in cancer development: trigger of chronic inflammation and immune responses that promote tumorigenesis and tumour growth; altered metabolic activity leading to increased production of toxic metabolites; abrogation of virus latency leading to malignancies (Plottel and Blaser, 2011).
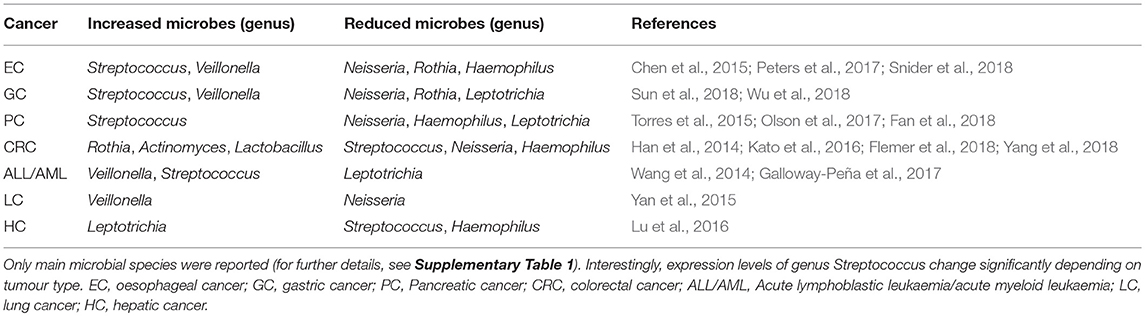
An increasing number of studies have shown that oral microbiota plays a role in the development of oral diseases, such as dental caries, periodontal disease, and oral stomatitis (Santarelli et al., 2015; Zhang et al., 2018). Poor oral hygiene and periodontal disease have been linked to oral cancer, and increasing evidences suggest that oral microbiota have a role in oral cancer development (García-Castillo et al., 2016). More broadly, is becoming evident that oral dysbiosis is associated with head and neck cancer development. Head and neck tumours originate from several anatomical sites, each associated with its own microbiota. Therefore, is possible that the crosstalk between microbial populations, combined with known risk factors, could drive head and neck carcinogenesis (Hayes et al., 2018). Oral dysbiosis has been also involved in the pathogenesis of systemic diseases. Indeed, recent studies indicate that oral microbiota seems to be involved in the tumours of distant organs, in particular “non-head and neck tumours” (Table 1; Klimesova et al., 2018).
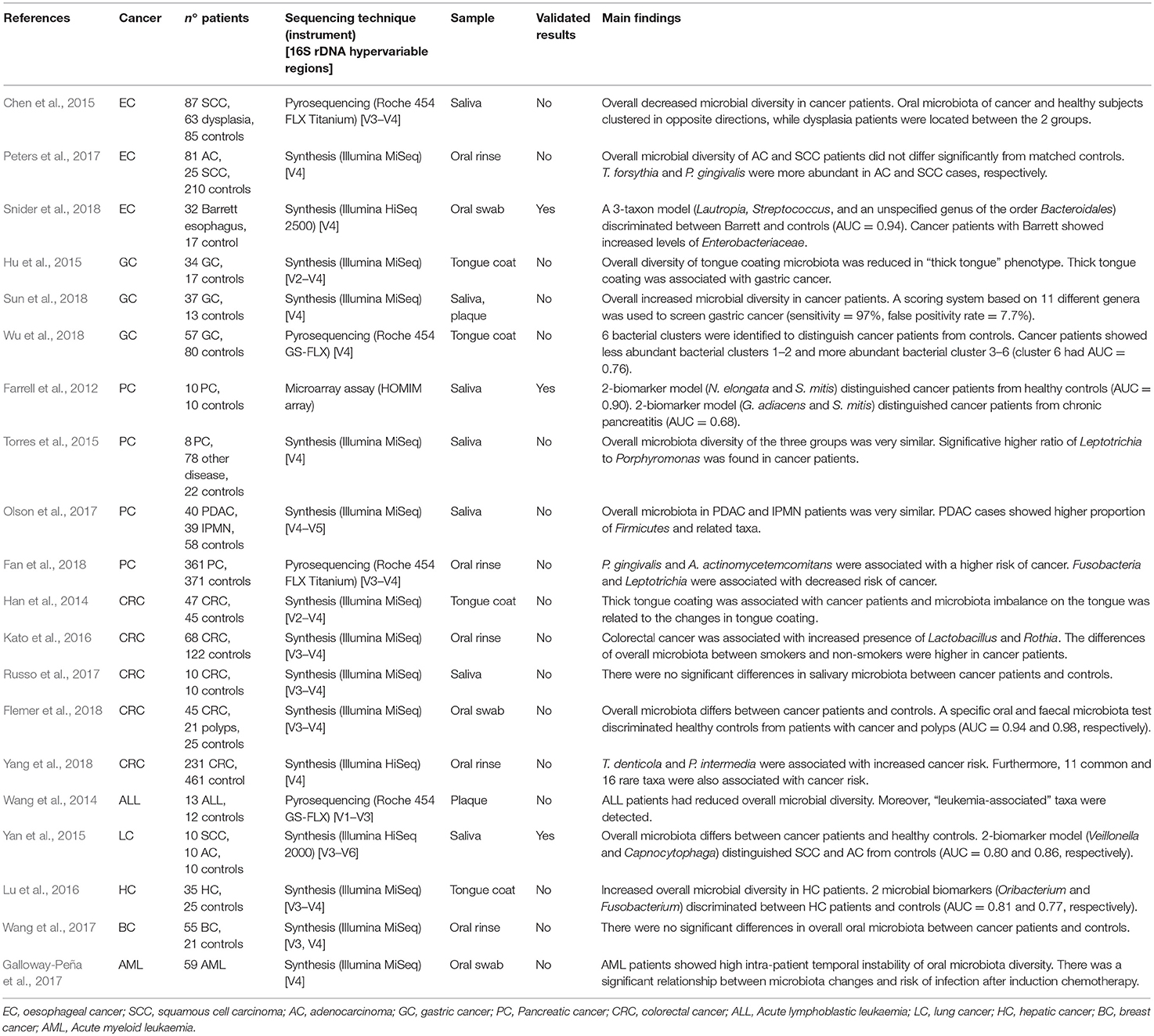
Table 1. Published studies on the relationship between oral microbiota and tumours of distant organs, showing principal results.
The aim of this article is to review the recent studies based on genome sequencing to present evidences of a relationship between oral microbiota and tumours of distant organs.
Pancreatic Cancer
Pancreatic cancer (PC) is a malignant tumour with a 5-year survival rate of 2–9% and a median survival time of 6 months (Mcguigan et al., 2018). Although localised PCs have better survival rate, these patients show absence or non-specific clinical manifestations. The diagnostic delay leads to a fatal outcome, with 80% of PCs that are unresectable at the time of presentation (Vincent et al., 2011). Despite the study of thousands of blood biomarkers for early detection of PC, none of them have been validated for clinical use, suggesting a continuous change of proteins expression over the course of disease evolution (Yachida et al., 2010).
Several reports have demonstrated an association between pathogenic microorganisms and PC, suggesting a role as diagnostic biomarkers. Furthermore, is possible that bacterial dysbiosis could contribute to the formation and progression of PC (Wang and Li, 2015). In addition to the well-known risk factors for PC, several studies have reported a positive association between PC and oral health status. In particular, the relationship between periodontal disease and tooth loss and an increased risk of PC was found, suggesting that toxins exposure and increase of systemic inflammation could promote tumour development (Hujoel et al., 2003; Stolzenberg-Solomon et al., 2003; Michaud et al., 2007). These findings led to the first studies on oral microbiota describing the possible link between oral dysbiosis and non-oropharyngeal cancers (Tables 1, 2).
In 2012 was conducted the first microbiota profiling of salivary samples of patients affected by PC (Farrell et al., 2012). A marked reduction of N. elongata and S. mitis and a significant increase of G. adiacens, suggesting a link between variations in oral microbiota and PC. In another study, saliva from 108 subjects was evaluated to determine microbiota profiles (Torres et al., 2015). Although the overall microbiota diversity of the groups was similar, a significative higher ratio of Leptotrichia to Porphyromonas (LP ratio) was found in PC patients. Despite the small sample size, salivary LP ratio was suggested as a simple biomarker for PC. Moreover, this study confirmed previous results showing that patients with high levels of plasma antibodies to P. gingivalis were at higher risk of PC (Ahn et al., 2012b; Michaud et al., 2013). Oral microbiota of patients with intraductal papillary mucinous neoplasms (IPMN) and pancreatic ductal adenocarcinoma (PDAC) was recently studied (Olson et al., 2017). The results found similar overall microbiota profile in saliva of PDAC and IPMN patients, but the mean relative proportion of taxa among IPMN patients was between that for PDAC patients and the controls. Recently, Fan et al. conducted the first prospective case-control study with the aim to evaluate microbiota profile in mouthwash samples taken from PC patients and healthy controls (Fan et al., 2018). Although no significant differences in overall microbiota composition were found, P. gingivalis and A. actinomycetemcomitans were associated with a higher risk of PC, while Fusobacteria and Leptotrichia were associated with decreased tumour risk.
Upper Gastrointestinal Tract Cancer
Upper gastrointestinal cancers are characterised by non-specific clinical manifestations, absence of effective treatments, and changing epidemiological trends. Oesophageal cancer is the sixth most common cause of cancer death, with a 5-year survival rate of 15–25%. Histologically, the two main types are oesophageal squamous cell carcinoma (SCC) and adenocarcinoma (AC) (Domper Arnal et al., 2015). Microbiota seems to play a role in oesophageal cancer aetiology, probably through the regulation of the barrier function, the modulation of inflammation, or the direct metabolism of carcinogens (Peters et al., 2017). Furthermore, periodontal disease seems to be associated with increased cancer risk, suggesting a role of oral microbiota (Michaud et al., 2007).
Salivary microbiota of oesophageal SCC, oesophageal dysplasia, and healthy subjects were compared, showing reduction of several genera in cancer patients (Chen et al., 2015). Overall oral microbiota of oesophageal SCC and controls clustered in opposite directions, while dysplasia patients were located between the 2 groups. Oral microbiota profiling was prospectively studied in oesophageal SCC and AC, using pre-diagnostic oral wash samples from cancer and healthy cohorts, but lack of significant diversity between cancer patients and matched controls was found (Peters et al., 2017). Nevertheless, two periodontal pathogens, T. forsythia and P. gingivalis, were more abundant in oesophageal AC and SCC cases, respectively. A recent work evaluated the feasibility of early detection of Barrett's oesophagus through the assessment of oral microbiota (Snider et al., 2018). A 3-taxon model including Lautropia, Streptococcus, and a genus of the order Bacteroidales distinguished Barrett's oesophagus from controls with high sensitivity (96.9%) and specificity (88.2%) (Tables 1, 2).
Gastric cancer (GC) is the third most common cause of cancer death, with a 5-year survival rate of 30%. Interestingly, 5-year survival rate in Japan is 70% for early stages of GC, suggesting the feasibility of mass screening programs (Karimi et al., 2014). The well-established role of H. pylori in GC development has given rise to the hypothesis that oral microbial dysbiosis could promote GC development (Ahn et al., 2012a). Several epidemiological studies corroborated such hypothesis showing the association between periodontal disease and tooth loss and an increased risk of GC (Stolzenberg-Solomon et al., 2003; Michaud et al., 2017). These findings led to the search for oral microbial biomarkers that allow an early and non-invasive diagnosis of GC (Table 1).
The relationship between tongue coating appearance and tongue microbiota profiling was investigated in GC patients and healthy controls (Hu et al., 2015). Tongue coating was significantly ticker in GC patients and was associated with reduced overall microbial diversity. Another study explored the alterations of tongue coating microbiota in patients with newly diagnosed GC and healthy controls, demonstrating a significative lower ratio of Bacteroidetes to Firmicutes in GC, especially in non-cardia GC patients (Wu et al., 2018). A recent case-control study was conducted with the aim to investigate the microbial community of both salivary and subgingival plaque in GC patients (Sun et al., 2018). Although saliva and plaque showed two different microbiotas, the overall microbial diversity was significantly increased in GC patients, and a screening system based on 11 different genera demonstrated high sensitivity (97%) (Table 2).
Lower Gastrointestinal Tract Cancer
Colorectal cancer (CRC) is one of the most diagnosed malignancy and the second most common cause of cancer death (Marley and Nan, 2016). A possible association between gut dysbiosis and CRC have been suggested. Indeed, both the whole colon microbiota and specific pathogens seems to contribute to the formation and progression of CRC (Klimesova et al., 2018). Findings of colonization of CRC tissues by typical oral bacteria raised the hypothesis of a possible involvement of oral microbiota in CRC development. The possible link between oral microbiota and CRC has been suggested by the fact that each day 1011 oral bacteria reach colorectal mucosa, influencing local microbiota (Klimesova et al., 2018). Furthermore, several epidemiological investigations suggested an association between periodontal disease and CRC (Hu et al., 2018). Although the interactions between CRC and the oral-gut microbiota axis are poorly understood, several studies have been conducted to investigate the role of oral bacteria as diagnostic biomarkers for CRC (Table 1).
Han et al. evaluated the relationship between tongue coating appearance and microbiota profiling in patients with CRC (Han et al., 2014). The results were similar to those obtained by the same research group in GC (Hu et al., 2015), showing a significant association between “thick tongue” phenotype and overall microbiota imbalance, especially in CRC patients. Another study investigated the association of smoking and CRC history with oral microbiota profile (Kato et al., 2016). The results failed to support earlier observations about the association between F. nucleatum and CRC, although increased presence of Lactobacillus and Rothia genera was found in cancer patients. Furthermore, smoking status was associated with a decrease in Betaproteobacteria class and Veilonellaceae family, especially in CRC patients. Microbial communities in a small group of CRC patients were investigated by comparing microbiota profiles from saliva, faeces and tissue samples (Russo et al., 2017). Although an altered microbial composition in faecal samples of CRC patients was noted, the differences in oral microbiota between cancer patients and healthy controls were inconsistent. A recent study compared microbiotas from faecal samples and colonic mucosa with oral microbiota in a larger cohort of patients (45 with CRC, 21 with colorectal polyps, and 25 healthy controls) (Flemer et al., 2018). Overall microbiota differs significantly between CRC patients and controls, suggesting that the colonic establishment of a putatively pathogenic oral-like community is more frequent in patients with CRC or polyps. Moreover, a specific oral and faecal microbiota test discriminated healthy controls from CRC patients with high sensitivity (76%) and specificity (94%). Recently, a large prospective case-control study investigated the association of specific periodontal pathogens and the overall oral microbiota with subsequent risk of CRC (Yang et al., 2018). The results showed a significant relationship between CRC risk and higher levels of T. denticola and P. intermedia. Furthermore, 11 common and 16 rare taxa seemed to be associated with cancer risk, especially among African-American and females.
Other Tumours
The promising results obtained by oral microbiota studies in gastrointestinal cancers led several research groups to investigate the diagnostic/prognostic applications of oral microbiota in other tumours. Although promising, these results are highly preliminary and far less clear than those reported in the aforementioned studies (Tables 1, 2).
Among the blood cancers, leukaemia is frequently associated with oral manifestations, like periodontitis and candidiasis. Therefore, imbalance of commensal oral microflora could be related to higher risk of both oral and systemic infections (Păunică et al., 2009). Oral microbiota profiling was evaluated in paediatric patients affected by acute lymphoblastic leukaemia (Wang et al., 2014). In particular, a genome analysis was conducted on supragingival plaque microbiota in 13 ALL patients, showing a reduced overall microbial diversity and detecting a specific pool of leukaemia-associated taxa. A longitudinal study was conducted on patients with acute myeloid leukaemia, with the aim to evaluate the variability of oral and gut microbiota during induction chemotherapy (Galloway-Peña et al., 2017). The results highlighted a high intra-patient temporal variability of oral microbiota and a correlation with higher risk of infection after induction chemotherapy.
Among solid malignancies, oral microbiota profile was investigated in lung cancer (LC) patients (Yan et al., 2015). Salivary samples from patients with SCC and AC showed overall microbiota difference between cancer patients and healthy controls. Furthermore, scoring system based on 2 genera, Veillonella and Capnocytophaga, distinguished healthy controls from SCC and AC. Another high-throughput sequencing-based study investigated tongue coating microbiota in cirrhotic patients with hepatic cancer (HC), finding an increased overall microbial diversity (Lu et al., 2016). Furthermore, 2 genera, Oribacterium and Fusobacterium, discriminated between HC patients and healthy controls. Finally, Wang et al. compared oral and urinary microbiota from 57 patients with breast cancer (BC) and 21 healthy subjects, but lack of significant relationship between oral microbiota and the presence of BC was found (Wang et al., 2017).
Conclusions
Overall, the results presented here showed a significant shift in oral microbiota composition between cancer patients and healthy controls. For example, Firmicutes and Actinobacteria were the most predominant phyla in cancer patients, while Proteobacteria, Fusobacteria, and Bacteroides were significantly more abundant in healthy controls (Table 2). Recently, the concept of resilience has been applied to the oral microbiota, defined as the capacity of this microbial ecosystem to deal with perturbation without shifting to potentially harmful states (Rosier et al., 2018). Strong and persistent perturbations to the oral environments can disrupt resilience, leading to a “regime shift” toward oral dysbiosis and, ultimately, to an increased risk of oral and systemic disease (Marsh, 2003).
Unfortunately, these results are heterogeneous among the studies, especially regarding specific cancer types, and do not allow drawing general conclusions. In particular, oral microbiota alterations seem to be more pronounced in patients with gastrointestinal tumours, paving the way for developing specific tests to allow early detection of cancer (Farrell et al., 2012; Torres et al., 2015; Flemer et al., 2018; Snider et al., 2018). The overlap between the microbiotas of oral cavity and gastrointestinal tract, probably due to colonization of gut mucosa by oral bacteria, could partially explain these results (Flynn et al., 2016).
There are several factors that currently limit the generalization of these results. First of all, the small sample size, mainly due to the high sequencing cost, limits the statistical power of the studies. Furthermore, lack of adequate information about patients (e.g., oral health status, active smoking, and antibiotic use) is a source of confounding factors (Snider et al., 2018). Most studies are conducted cross-sectionally, a design that is unable to infer causality between oral dysbiosis and tumour development. This aspect is particularly important, because the alternative hypothesis is the so-called “reverse causality,” namely that modifications in the normal oral microbiota are the result of tumour progression, secondary to systemic processes (Fan et al., 2018). Indeed, although different pathogenic mechanisms have been proposed, direct evidence is not yet available and the reverse causality cannot be excluded. In particular, oral microbiota could be involved in cancer development, but specific tissue tropism for microbial translocation and molecular mechanisms of microbiota-driven carcinogenesis at distal sites are yet to be proven (Chen et al., 2017).
Regarding methodological aspects, different types of oral samples were evaluated making the comparison of these microbial communities difficult. Bacteria colonize all sites within the oral cavity, forming different habitats, each with different non-overlapping microbial populations (Jia et al., 2018). Therefore, it is important to develop a method that can access all oral sites and the microbial diversity inherent to them (Lim et al., 2017). Saliva is the most commonly studied sample, due to several features that make it ideal for research purposes. Thanks to its role in “ecological stability” of oral cavity, saliva allows to early detect an overall dysbiotic trend of oral microbiota (Rosier et al., 2018; Belibasakis et al., 2019). Although different collection methods can alter salivary microbiota (e.g., muscle movements from spitting action may provide larger microbial coverage compared with passive drooling), this seems to not significantly affect the overall microbial composition (Lim et al., 2017). The studies reported here used different sequencing techniques and analysed different 16S rDNA hypervariable regions, with an increased risk for ambiguous data for some rare bacterial species (Table 1; Verma et al., 2018). Furthermore, sequencing depth of these studies could not be sensitive enough to adequately describe differences in oral microbiota in patients with and without tumour (Teng et al., 2018).
Nevertheless, the evidences linking oral microbiota with different tumours are promising. Furthermore, metatranscriptomic and metabolomic analyses are making a significant contribution to the understanding of the relationship between oral microbiota metabolism and human health (García-Castillo et al., 2016). The demonstration of a direct causality between oral dysbiosis and risk of tumour development could lead to new prevention strategies. This could also lead to a broader definition of oral health, including also the presence of a stable microbial homeostasis. Furthermore, causal link between oral microbiota and cancer could contribute to the search for new biomarkers (e.g., markers of resilience) and new therapeutic anticancer strategies, such as the use of pre- and pro-biotics (Rosier et al., 2018).
Although only at the beginning, oral microbiota analysis could be the next step in the evolution of cancer therapy and will help clinicians to develop tailored prevention and treatment strategies.
Author Contributions
MM, AS, and LL conceived the literature review. MM, LT, and GT described pancreatic studies. GT, VC, and LT described upper gastrointestinal tract studies. DG, LM, and AS described lower gastrointestinal tract studies. LL, MP, and LM wrote the concluding remarks. All authors discussed and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00232/full#supplementary-material
References
Ahn, J., Chen, C. Y., and Hayes, R. B. (2012a). Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control 23, 399–404. doi: 10.1007/s10552-011-9892-7
Ahn, J., Segers, S., and Hayes, R. B. (2012b). Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis 33, 1055–1058. doi: 10.1093/carcin/bgs112
Arweiler, N. B., and Netuschil, L. (2016). The oral microbiota. Adv. Exp. Med. Biol. 902, 45–60. doi: 10.1007/978-3-319-31248-4_4
Belibasakis, G. N., Bostanci, N., Marsh, P. D., and Zaura, E. (2019). Applications of the oral microbiome in personalized dentistry. Arch. Oral Biol. 104, 7–12. doi: 10.1016/j.archoralbio.2019.05.023
Benn, A., Heng, N., Broadbent, J. M., and Thomson, W. M. (2018). Studying the human oral microbiome: challenges and the evolution of solutions. Aust. Dent. J. 63, 14–24. doi: 10.1111/adj.12565
Chen, J., Domingue, J. C., and Sears, C. L. (2017). Microbiota dysbiosis in select human cancers: evidence of association and causality. Semin. Immunol. 32, 25–34. doi: 10.1016/j.smim.2017.08.001
Chen, X., Winckler, B., Lu, M., Cheng, H., Yuan, Z., Yang, Y., et al. (2015). Oral microbiota and risk for esophageal squamous cell carcinoma in a high-risk area of China. PLoS ONE 10:e0143603. doi: 10.1371/journal.pone.0143603
Domper Arnal, M. J., Ferrández Arenas, Á., and Lanas Arbeloa, Á. (2015). Esophageal cancer: risk factors, screening and endoscopic treatment in Western and Eastern countries. World J. Gastroenterol. 21, 7933–7943. doi: 10.3748/wjg.v21.i26.7933
Fan, X., Alekseyenko, A. V., Wu, J., Peters, B. A., Jacobs, E. J., Gapstur, S. M., et al. (2018). Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut 67, 120–127. doi: 10.1136/gutjnl-2016-312580
Farrell, J. J., Zhang, L., Zhou, H., Chia, D., Elashoff, D., Akin, D., et al. (2012). Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61, 582–588. doi: 10.1136/gutjnl-2011-300784
Flemer, B., Warren, R. D., Barrett, M. P., Cisek, K., Das, A., Jeffery, I. B., et al. (2018). The oral microbiota in colorectal cancer is distinctive and predictive. Gut 67, 1454–1463. doi: 10.1136/gutjnl-2017-314814
Flynn, K. J., Baxter, N. T., and Schloss, P. D. (2016). Metabolic and community synergy of oral bacteria in colorectal cancer. mSphere 1:e00102–16. doi: 10.1128/mSphere.00102-16
Galloway-Peña, J. R., Smith, D. P., Sahasrabhojane, P., Wadsworth, W. D., Fellman, B. M., Ajami, N. J., et al. (2017). Characterization of oral and gut microbiome temporal variability in hospitalized cancer patients. Genome Med. 9:21. doi: 10.1186/s13073-017-0409-1
García-Castillo, V., Sanhueza, E., Mcnerney, E., Onate, S. A., and García, A. (2016). Microbiota dysbiosis: a new piece in the understanding of the carcinogenesis puzzle. J. Med. Microbiol. 65, 1347–1362. doi: 10.1099/jmm.0.000371
Han, S., Chen, Y., Hu, J., and Ji, Z. (2014). Tongue images and tongue coating microbiome in patients with colorectal cancer. Microb. Pathog. 77, 1–6. doi: 10.1016/j.micpath.2014.10.003
Hayes, R. B., Ahn, J., Fan, X., Peters, B. A., Ma, Y., Yang, L., et al. (2018). Association of oral microbiome with risk for incident head and neck squamous cell cancer. JAMA Oncol. 4, 358–365. doi: 10.1001/jamaoncol.2017.4777
Hu, J., Han, S., Chen, Y., and Ji, Z. (2015). Variations of tongue coating microbiota in patients with gastric cancer. Biomed Res. Int. 2015:173729. doi: 10.1155/2015/173729
Hu, J. M., Shen, C. J., Chou, Y. C., Hung, C. F., Tian, Y. F., You, S. L., et al. (2018). Risk of colorectal cancer in patients with periodontal disease severity: a nationwide, population-based cohort study. Int. J. Colorectal Dis. 33, 349–352. doi: 10.1007/s00384-018-2965-2
Hujoel, P. P., Drangsholt, M., Spiekerman, C., and Weiss, N. S. (2003). An exploration of the periodontitis-cancer association. Ann. Epidemiol. 13, 312–316. doi: 10.1016/S1047-2797(02)00425-8
Jia, G., Zhi, A., Lai, P. F. H., Wang, G., Xia, Y., Xiong, Z., et al. (2018). The oral microbiota - a mechanistic role for systemic diseases. Br. Dent. J. 224, 447–455. doi: 10.1038/sj.bdj.2018.217
Karimi, P., Islami, F., Anandasabapathy, S., Freedman, N. D., and Kamangar, F. (2014). Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol. Biomarkers Prev. 23, 700–713. doi: 10.1158/1055-9965.EPI-13-1057
Kato, I., Vasquez, A. A., Moyerbrailean, G., Land, S., Sun, J., Lin, H. S., et al. (2016). Oral microbiome and history of smoking and colorectal cancer. J. Epidemiol. Res. 2, 92–101. doi: 10.5430/jer.v2n2p92
Klimesova, K., Jiraskova Zakostelska, Z., and Tlaskalova-Hogenova, H. (2018). Oral bacterial and fungal microbiome impacts colorectal carcinogenesis. Front. Microbiol. 9:774. doi: 10.3389/fmicb.2018.00774
Le Bars, P., Matamoros, S., Montassier, E., Le Vacon, F., Potel, G., Soueidan, A., et al. (2017). The oral cavity microbiota: between health, oral disease, and cancers of the aerodigestive tract. Can. J. Microbiol. 63, 475–492. doi: 10.1139/cjm-2016-0603
Lim, Y., Totsika, M., Morrison, M., and Punyadeera, C. (2017). The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols. Sci. Rep. 7:8523. doi: 10.1038/s41598-017-07885-3
Lu, H., Ren, Z., Li, A., Zhang, H., Jiang, J., Xu, S., et al. (2016). Deep sequencing reveals microbiota dysbiosis of tongue coat in patients with liver carcinoma. Sci. Rep. 6:33142. doi: 10.1038/srep33142
Marley, A. R., and Nan, H. (2016). Epidemiology of colorectal cancer. Int. J. Mol. Epidemiol. Genet. 7, 105–114.
Marsh, P. D. (2003). Are dental diseases examples of ecological catastrophes? Microbiology 149, 279–294. doi: 10.1099/mic.0.26082-0
Mcguigan, A., Kelly, P., Turkington, R. C., Jones, C., Coleman, H. G., and Mccain, R. S. (2018). Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 24, 4846–4861. doi: 10.3748/wjg.v24.i43.4846
Meurman, J. H., and Uittamo, J. (2008). Oral micro-organisms in the etiology of cancer. Acta Odontol. Scand. 66, 321–326. doi: 10.1080/00016350802446527
Michaud, D. S., Fu, Z., Shi, J., and Chung, M. (2017). Periodontal disease, tooth loss, and cancer risk. Epidemiol. Rev. 39, 49–58. doi: 10.1093/epirev/mxx006
Michaud, D. S., Izard, J., Wilhelm-Benartzi, C. S., You, D. H., Grote, V. A., Tjønneland, A., et al. (2013). Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 62, 1764–1770. doi: 10.1136/gutjnl-2012-303006
Michaud, D. S., Joshipura, K., Giovannucci, E., and Fuchs, C. S. (2007). A prospective study of periodontal disease and pancreatic cancer in US male health professionals. J. Natl. Cancer Inst. 99, 171–175. doi: 10.1093/jnci/djk021
Olson, S. H., Satagopan, J., Xu, Y., Ling, L., Leong, S., Orlow, I., et al. (2017). The oral microbiota in patients with pancreatic cancer, patients with IPMNs, and controls: a pilot study. Cancer Causes Control 28, 959–969. doi: 10.1007/s10552-017-0933-8
Păunică, S. C., Dumitriu, A., Mogoş, M., Georgescu, O., and Mogoş, I. (2009). The evaluation of the periodontium in patients with leukemia using thermographic imaging. Hematology 14, 341–346. doi: 10.1179/102453309X12473408860505
Peters, B. A., Wu, J., Pei, Z., Yang, L., Purdue, M. P., Freedman, N. D., et al. (2017). Oral microbiome composition reflects prospective risk for esophageal cancers. Cancer Res. 77, 6777–6787. doi: 10.1158/0008-5472.CAN-17-1296
Petersen, C., and Round, J. L. (2014). Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 16, 1024–1033. doi: 10.1111/cmi.12308
Plottel, C. S., and Blaser, M. J. (2011). Microbiome and malignancy. Cell Host Microbe 10, 324–335. doi: 10.1016/j.chom.2011.10.003
Rosier, B. T., Marsh, P. D., and Mira, A. (2018). Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J. Dent. Res. 97, 371–380. doi: 10.1177/0022034517742139
Russo, E., Bacci, G., Chiellini, C., Fagorzi, C., Niccolai, E., Taddei, A., et al. (2017). Preliminary comparison of oral and intestinal human microbiota in patients with colorectal cancer: a pilot study. Front. Microbiol. 8:2699. doi: 10.3389/fmicb.2017.02699
Santarelli, A., Mascitti, M., Rubini, C., Bambini, F., Zizzi, A., Offidani, A., et al. (2015). Active inflammatory biomarkers in oral lichen planus. Int. J. Immunopathol. Pharmacol. 28, 562–568. doi: 10.1177/0394632015592101
Schwabe, R. F., and Jobin, C. (2013). The microbiome and cancer. Nat. Rev. Cancer 13, 800–812. doi: 10.1038/nrc3610
Snider, E. J., Compres, G., Freedberg, D. E., Giddins, M. J., Khiabanian, H., Lightdale, C. J., et al. (2018). Barrett's esophagus is associated with a distinct oral microbiome. Clin. Transl. Gastroenterol. 9:135. doi: 10.1038/s41424-018-0005-8
Stolzenberg-Solomon, R. Z., Dodd, K. W., Blaser, M. J., Virtamo, J., Taylor, P. R., and Albanes, D. (2003). Tooth loss, pancreatic cancer, and Helicobacter pylori. Am. J. Clin. Nutr. 78, 176–181. doi: 10.1093/ajcn/78.1.176
Sun, J. H., Li, X. L., Yin, J., Li, Y. H., Hou, B. X., and Zhang, Z. (2018). A screening method for gastric cancer by oral microbiome detection. Oncol. Rep. 39, 2217–2224. doi: 10.3892/or.2018.6286
Teng, F., Darveekaran Nair, S. S., Zhu, P., Li, S., Huang, S., Li, X., et al. (2018). Impact of DNA extraction method and targeted 16S-rRNA hypervariable region on oral microbiota profiling. Sci. Rep. 8:16321. doi: 10.1038/s41598-018-34294-x
Torres, P. J., Fletcher, E. M., Gibbons, S. M., Bouvet, M., Doran, K. S., and Kelley, S. T. (2015). Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ 3:e1373. doi: 10.7717/peerj.1373
Verma, D., Garg, P. K., and Dubey, A. K. (2018). Insights into the human oral microbiome. Arch. Microbiol. 200, 525–540. doi: 10.1007/s00203-018-1505-3
Vincent, A., Herman, J., Schulick, R., Hruban, R. H., and Goggins, M. (2011). Pancreatic cancer. Lancet 378, 607–620. doi: 10.1016/S0140-6736(10)62307-0
Wang, C., and Li, J. (2015). Pathogenic microorganisms and pancreatic cancer. Gastrointest. Tumors 2, 41–47. doi: 10.1159/000380896
Wang, H., Altemus, J., Niazi, F., Green, H., Calhoun, B. C., Sturgis, C., et al. (2017). Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 8, 88122–88138. doi: 10.18632/oncotarget.21490
Wang, Y., Xue, J., Zhou, X., You, M., Du, Q., Yang, X., et al. (2014). Oral microbiota distinguishes acute lymphoblastic leukemia pediatric hosts from healthy populations. PLoS ONE 9:e102116. doi: 10.1371/journal.pone.0102116
Wu, J., Xu, S., Xiang, C., Cao, Q., Li, Q., Huang, J., et al. (2018). Tongue coating microbiota community and risk effect on gastric cancer. J. Cancer 9, 4039–4048. doi: 10.7150/jca.25280
Yachida, S., Jones, S., Bozic, I., Antal, T., Leary, R., Fu, B., et al. (2010). Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 467, 1114–1117. doi: 10.1038/nature09515
Yan, X., Yang, M., Liu, J., Gao, R., Hu, J., Li, J., et al. (2015). Discovery and validation of potential bacterial biomarkers for lung cancer. Am. J. Cancer Res. 5, 3111–3122.
Yang, Y., Cai, Q., Shu, X. O., Steinwandel, M. D., Blot, W. J., Zheng, W., et al. (2018). Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int. J. Cancer. 77:4931. doi: 10.1158/1538-7445.AM2017-4931
Keywords: oral microbiota, oral microbiome, pancreatic cancer, gastrointestinal tract cancer, high-throughput sequencing
Citation: Mascitti M, Togni L, Troiano G, Caponio VCA, Gissi DB, Montebugnoli L, Procaccini M, Lo Muzio L and Santarelli A (2019) Beyond Head and Neck Cancer: The Relationship Between Oral Microbiota and Tumour Development in Distant Organs. Front. Cell. Infect. Microbiol. 9:232. doi: 10.3389/fcimb.2019.00232
Received: 15 April 2019; Accepted: 14 June 2019;
Published: 26 June 2019.
Edited by:
Frederic Antonio Carvalho, INSERM U1107 Douleur et Biophysique Neurosensorielle (Neuro-Dol), FranceReviewed by:
Nele Brusselaers, Karolinska Institute (KI), SwedenMohamed Helmy, University of Toronto, Canada
Copyright © 2019 Mascitti, Togni, Troiano, Caponio, Gissi, Montebugnoli, Procaccini, Lo Muzio and Santarelli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco Mascitti, bWFyY29tYXNjaXR0aTg2QGhvdG1haWwuaXQ=
 Marco Mascitti
Marco Mascitti Lucrezia Togni
Lucrezia Togni Giuseppe Troiano2
Giuseppe Troiano2 Vito Carlo Alberto Caponio
Vito Carlo Alberto Caponio Lorenzo Lo Muzio
Lorenzo Lo Muzio