
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Cell. Infect. Microbiol. , 27 June 2019
Sec. Clinical Microbiology
Volume 9 - 2019 | https://doi.org/10.3389/fcimb.2019.00226
This article is part of the Research Topic Novel Methods for Rapid Identification and Susceptibility Testing of Bacteria View all 19 articles
Fast dissemination of the mobilized colistin resistance (mcr) gene mcr-1 in Enterobacteriaceae causes a huge threat to the treatment of severe infection. In the current report, a multiple cross displacement amplification (MCDA) coupled with the detection of amplified products by gold nanoparticles-based lateral flow biosensor (LFB) assay (MCDA-LFB) was established to identify the mcr-1 gene with simpleness, rapidity, specificity, and sensitivity. The MCDA-LFB assay was performed at a isothermal temperature (63°C) for only 30 min during the amplification stage, and the reaction products were directly identified by using LFB which obtained the result within 2 min. The entire process of experiments, from templates extraction to result judging, was accomplished in <60 min. For the analytical specificity of this method, all of the 16 mcr-1-producing strains were positive, and all of the non-mcr-1 isolates produced the negative results. The sensitivity of mcr-1-MCDA-LFB assay was as little as 600 fg of plasmid DNA per reaction in pure culture, and approximately 4.5 × 103 CFU/mL (~4.5 CFU/reaction) in spiked fecal samples. Therefore, this technique established in the present study is suitable for the surveillance of mcr-1 gene in clinic and livestock industry.
The rapid increase of carbapenem-resistant Enterobacteriaceae (CRE) expressing Klebsiella pneumoniae carbapenemase (KPC), New Delhi metallo-blactamase (NDM) and oxacillinase (OXA) OXA-48 has risen serious concerns in clinic. Colistin, a “last resort” antibiotic, has a crucial role for treating the infection caused by those species (Nation and Li, 2009). Therefore, the number of colistin consumptions increasing along with the global augment of CRE will bring about the risk of emerging resistance (Gelband et al., 2015).
Resistance to colistin was linked with chromosomal resistance mechanisms in varieties of strains in the past (Olaitan et al., 2014). Since a new mobilized colistin resistance gene, mcr-1, carried by plasmid in an Escherichia coli was first reported in China in 2015 (Liu et al., 2016), which has been identified in numerous countries. China, Germany and Vietnam carry an important proportion of positive samples (Wang et al., 2018a). The mcr-1-positive bacterial species include Salmonella enterica, E. coli, Escherichia fergusonii, Enterobacter aerogenes, K. pneumoniae, Citrobacter braaki, and Klebsiella aerogenes (Doumith et al., 2016; Li et al., 2016; Stoesser et al., 2016; Zeng et al., 2016; Sennati et al., 2017; Wang et al., 2017a, 2018a). Besides discovered in Clinical samples, mcr-1 is also detected from environmental settings: meat and vegetable products purchased from markets, Animal feces collected from farms, fecal samples of pets gathered from pet hospital, river water, and seawater (Chen et al., 2017). The wide dissemination of mcr-1 across diversified species is benefited from many types of mcr-1-bearing plasmids covering IncHI2, IncI2, IncFI, IncX4, and IncX1-X2 hybrid type (McGann et al., 2016; Sun et al., 2016, 2017; Yang et al., 2016; Guo et al., 2017). Similarly, the genetic environments of mcr-1 gene also impact its transmission. A global data set of roughly 500 isolates producing mcr-1 analyzed by whole-genome sequencing (WGS) has revealed that an initial mobilized event of mcr-1 is mediated by an ISApl1-mcr-1-orf-ISApl1 transposon around 2006 (Wang et al., 2018a). The horizontal transfer of mcr-1 gene causing inflation of colistin-resistant isolates will lead to the shortage of effective measures for treating infections with multidrug-resistant bacteria. Therefore, a rapid, sensitive and specific diagnostic assay for mcr-1 detection is imperative to devise.
Currently, several categories of molecular diagnostic methodologies including conventional polymerase chain reaction (PCR) and real-time PCR methods have been devised to identify mcr-1 gene (Bontron et al., 2016). Nevertheless, the requirements of highly sophisticated devices, strictly experimental environments and well-trained personnel restrict those techniques to apply in resource-challenged areas and “on-site” detection (Niu et al., 2018). Recently, multiple cross displacement amplification (MCDA), a novel nucleic acid amplification technique, has been utilized in detection of bacterial agents, such as Listeria monocytogenes and methicillin-resistant Staphylococcus aureus (MRSA) (Wang et al., 2017b, 2018c). The approach was based on isothermal strand-displacement polymerization reaction, five pairs of primers were designed to amplify the corresponding regions of target sequence. Without the denaturing step, the primers of high concentration could bind to the template at a isothermal temperature. Several single stem-loop DNA structures and single-stranded DNAs were yielded with the primers binding and the reaction chain extending. Then, those DNA products were exponentially amplified as the reaction progressed (Wang et al., 2015). With the advantages of rapidity, specificity and sensitivity, MCDA operated in a simple heater can yield amplifcons from a few colonies (Wang et al., 2017b, 2018b), the amplicons are identified by a gold nanoparticles-based lateral flow biosensor (LFB) subsequently.
In this study, a MCDA-LFB assay for the rapid detection of mcr-1 was established, and the sensitivity and specificity of above method in pure culture and in spiked fecal samples were analyzed.
Bacterial genomes extraction kits were obtained from Beijing ComWin Biotech Co., Ltd. (Beijing, China). QIAGEN plasmid kits and QIAamp fast DNA stool mini kits were purchased from Qiagen Co., Ltd. (Beijing, China). Isothermal amplification kits (including reaction buffer and Bst DNA polymerase 2.0), colorimetric indicator (Malachite Green), and disposable LFB were provided by BeiJing-HaiTaiZhengYuan Technology Co., Ltd. (Beijing, China). The heating thermostat (MTH-100) was purchased from Hangzhou MiU Instruments Co., Ltd. (Hangzhou, China). The UV transilluminator (UVsolo touch) was obtained from Analytik Jena (Jena, Germany). Nanodrop instrument (ND-2000) was purchased from Thermo Fisher Scientific Co., Ltd. (Massachusetts, America).
A total of 59 organisms consisting of 16 mcr-1-postive isolates and 43 non-mcr-1 bacteria were used in this study (Table 1). The confirmation of all genes used conventional PCR and sequencing. The mcr-1-postive bacterial species included 5 E. fergusonii, 10 E. coli, and 1 Salmonella enteritidis. The mcr-1-negative isolates involved KPC-2-postive stains (K. pneumoniae and Pseudomonas aeruginosa), NDM-1-postive isolates (E. coli, Enterobacter cloacae, and K. pneumoniae), NDM-5-producing E. coli, IMP-4-producing E. coli, and mcr-1/carbapenemase-negative species (Acinetobacter baumannii, P. aeruginosa, Serratia marcescens, E. coli, S. aureus, Enterococcus faecalis, and Enterococcus faecium). According to the handling instruction, the genomic DNA of all strains was extracted by bacterial genomes extraction kits, the plasmid DNA of mcr-1-producing E. fergusonii (ICDC-ZG2016M34-3) acted as a representative sample for optimization of reaction condition and sensitivity detection was acquired by QIAGEN Plasmid Kits, and quantified by a Nanodrop ND-2000 instrument.
A set of five pairs of primers, including 2 displacement primers (F1 and F2), 2 cross primers (CP1 and CP2), and 3 pairs of amplification primers (C1, C2, D1, D2, R1, and R2), targeted 10 distinct regions more than 150 bp on mcr-1 gene (Wang et al., 2015). Two softwares named Primer Premier 6.0 and PrimerExplorer V4 were used to design the 10 MCDA primers based on mcr-1 gene (GenBank accession number: KX886345). The dimer and hairpin structures of all primers were detected by Integrated DNA Technologies design tools (Wang et al., 2016), and the specificity of which was analyzed by using Basic Local Alignment Search Tool (Blast). The relevant information of primers pairs regarding positions and sequences was displayed in Figure 1 and Table 2. Furthermore, The FITC (Fluorescein isothiocyanate) and biotin labeled at 5′end of the C1 and D1 primers, respectively, and the labeled primers were named as C1* and D1*. All of the primers were synthesized and purified by Sangon Biotechnology Co., Ltd. (Shanghai, China) at HPLC purification grade.
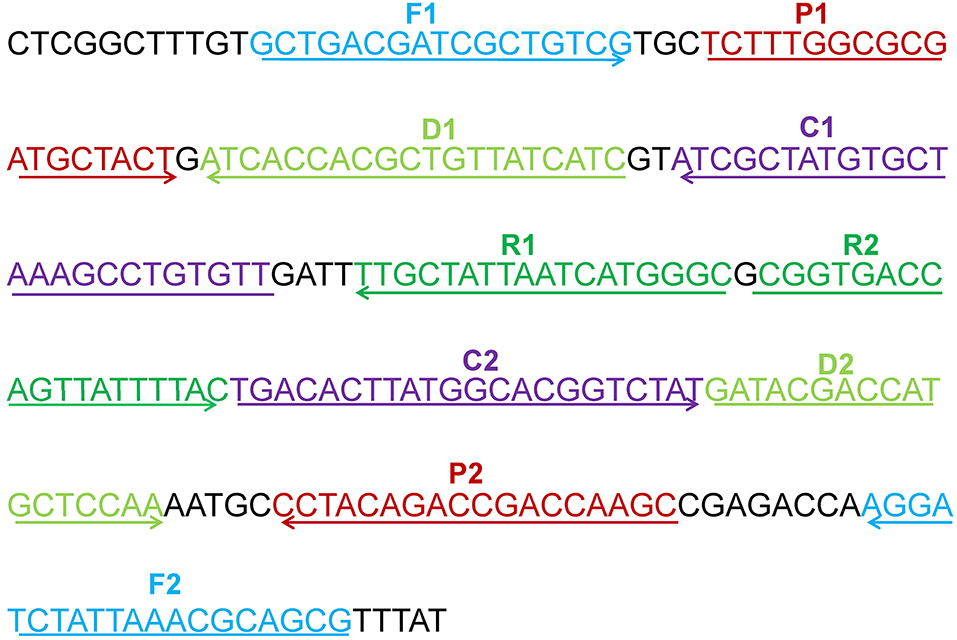
Figure 1. The location of the five primers pairs on the partial mcr-1 gene sequence. Different pairs of primers were represented by various of colors. The arrowed lines showed the amplification direction of primers.
The MCDA reaction systems were performed according to the previous studies (Wang et al., 2016). Each reaction, the total volumes of 25 μL, included reaction buffer (12.5 μL), Bst DNA polymerase 2.0 (1 μL), colorimetric indicator (1 μL), cross primers (1.6 μM each), displacement primers (0.4 μM each), amplification primers (0.4 μM each), and 1 μL of DNA template. The NDM-1-positive E. coli (WHCDC-WH67) and KPC-2-producing K. pneumoniae (WHCDC-WH108) were regarded as the negative controls, and the distilled water was served as the blank control. To assess the optimal reaction temperature of mcr-1-MCDA, the MCDA amplification systems were executed with a constant temperature in the range of 60–67°C for 40 min.
The MCDA reaction products were analyzed by three detection methods including 2% agarose gel electrophoresis, colorimetric indicator, and LFB (Wang et al., 2017b). When employing gel electrophoresis, 3 μL of reaction mixtures were run at 110 volts for 60 min. A ladder of multiple bands could be observed in the positive reactions, but not in the negative and blank controls. Reaction products were detected by using colorimetric indicator, the color of amplified products remained unchanged. Nevertheless, the negative and blank controls reactions changed from blue to colorless. The material, theory and operation procedure of LFB were previously described by Wang et al. (2017b). 0.2 μL of amplicons followed by three drops of the running buffer consisting of 1% Tween 20 and 0.01 mol/L phosphate-buffered saline were added to the well of sample pad (Niu et al., 2018). After 1–2 min, two red lines named test line (TL) and control line (CL), respectively, could be visualized in positive products, while only the CL was observed for the negative and blank control.
The plasmid DNA of E. fergusonii ICDC-ZG2016M34-3 was serially diluted (6 ng, 60 pg, 600 fg, 60 fg, and 6 fg per μl) for sensitivity analysis of mcr-1-MCDA-LFB detection. The colorimetric indicator and agarose gel electrophoresis were carried out simultaneously. Each test was repeated three times. The specificity of mcr-1-MCDA-LFB assay was evaluated with the DNA templates of 16 mcr-1-producing strains and 43 non-mcr-1 strains (Table 1). The specificity evaluations were confirmed twice.
In order to screen the optimal time for the mcr-1-MCDA-LFB assay, The MCDA mixture was completed at the reaction temperature in the range from 10 to 40 min at 10 min intervals. Subsequently, the MCDA products were detected by LFB detection. Each amplification time was operated at two times.
The fecal samples were obtained from a healthy man in Wuhan, China. Mcr-1 gene was not detected in those samples according to the microbial culture and PCR identification. The volumes of 100 μL were taken out from the mcr-1-positive E. fergusonii ICDC-ZG2016M34-3 cultures when the optical density (OD) of that reached to 0.6. The suspensions were serially diluted (10−1−10−8), and the aliquots of 100 μL dilutions (10−3−10−6) were incubated on nutrient agar plates with three replicates, colony forming units (CFUs) were counted subsequently. One hundred microliter of diluted mcr-1-producing cultures (10−2−10−7) with known amounts (4.5 × 106−4.5 × 101 CFU/ mL) was, respectively, added to 0.2 g of fecal sample and mixed well. DNA templates were extracted with the manufacturer's protocol by using QIAamp fast DNA stool mini kits. The extracted genomic DNA was dissolved in 100 μl of elution buffer, and 1 μl of which was used for MCDA-LFB detection as templates. A non-spiked feces sample was tested as negative control. The products of MCDA were also detected by colorimetric indicator and 2% agarose gel electrophoresis. The evaluation assay for limit of detection in fecal samples was conducted triplicate.
Mcr-1 gene was amplified by conventional PCR, primers: mcr-1-F, 5′-CGG TCA GTC CGT TTG TTC-3′; mcr-1-R, 5′-CGG TCA GTC CGT TTG TTC-3′. And amplicons were subsequently sequenced (Liu et al., 2016). The LOD of mcr-1-MCDA-LFB and conventional PCR in pure culture and in fecal samples were compared, respectively.
The mcr-1 MCDA assays were performed at a constant temperature (63°C) for 40 min to verify the availability of MCDA primers. Positive reaction appeared with DNA from mcr-1-producing E. fergusonii (ICDC-ZG2016M34-3), but not with NDM-1-positive E. coli (WHCDC-WH67), KPC-2-producing K. pneumoniae (WHCDC-WH108), and the blank control (Figure 2). Therefore, the primers of mcr-1-MCDA was suitable for establishment of the MCDA-LFB assay to detect mcr-1 gene.
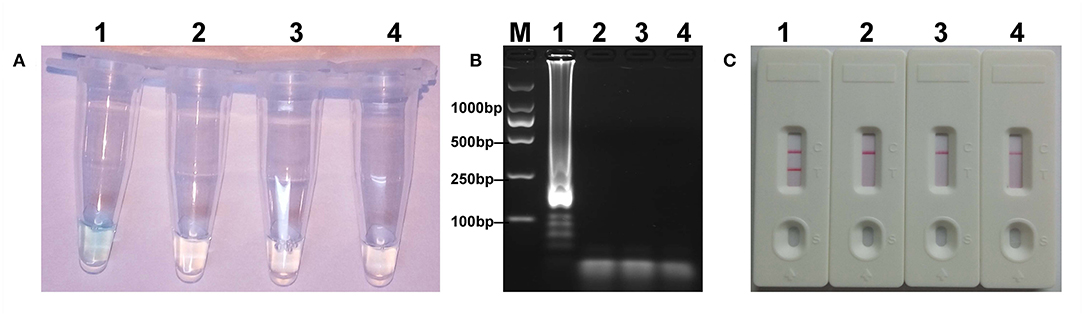
Figure 2. Confirmation of the mcr-1-MCDA-LFB assay. There were three detection methods to identify MCDA products: (A) colorimetric indicators, (B) agarose gel electrophoresis, (C) LFB. Four samples were tested: (1) mcr-1-positive strain of Escherichia fergusonii (ICDC-ZG2016M34-3); (2) NDM-1-positive Escherichia coli (WHCDC-WH67); (3) KPC-2-producing Klebsiella pneumoniae (WHCDC-WH108); (4) distilled water. Only the amplification with Escherichia fergusonii (ICDC-ZG2016M34-3) showed the positive.
To optimize the reaction temperature of MCDA-LFB assay during the amplification stage, the plasmid DNA of E. fergusonii (ICDC-ZG2016M34-3) at the level of 6 pg per reaction was used as the templates. A series of temperatures (ranging from 60 to 67°C, with 1°C intervals) was compared for amplifying efficiency of MCDA-LFB assay by employing 2% agarose gel electrophoresis. The result showed that 62 and 63°C were the better candidates for this method (Figure 3). Therefore, the reaction temperature of 63°C was performed for the subsequent MCDA-LFB experiments.
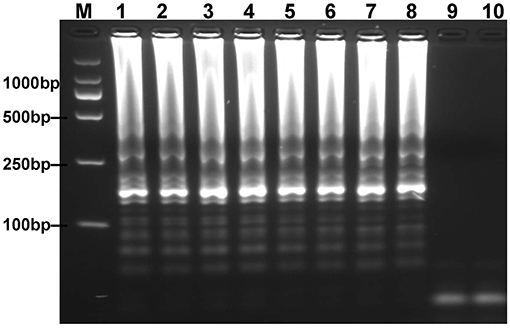
Figure 3. Optimal reaction temperature for mcr-1-MCDA assay was determined by using agarose gel electrophoresis. The plasmid DNA of Escherichia fergusonii (ICDC-ZG2016M34-3) (6 pg) was amplified in different temperatures. Lane 1–8 represented the amplification temperature in the range from 60 to 67°C (1°C intervals); lane 9, negative control (6 pg of Escherichia coli WHCDC-WH67 genomic DNA); lane 10, blank control (DW).
To acquire the detection limit of this assay, serial dilutions of E. fergusonii ICDC-ZG2016M34-3 plasmid DNA were used in mcr-1-MCDA-LFB assay. It indicated that the threshold was as little as 600 fg of plasmid DNA (Figure 4). The same results were observed by using colorimetric indicator and agarose gel electrophoresis analysis.
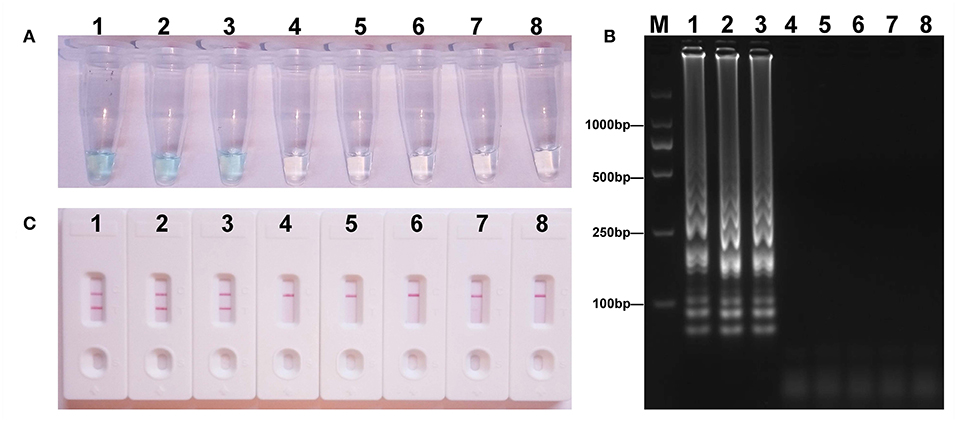
Figure 4. Sensitivity of the mcr-1-MCDA assay using serially diluted plasmid DNA with Escherichia fergusonii ICDC-ZG2016M34-3. Three detection techniques, including colorimetric indicators (A), gel electrophoresis (B), and LFB (C), were applied for testing MCDA products. Tubes (A)/lanes (B)/biosensors (C) 1–8 represented the plasmid DNA levels of 6 ng, 60 pg, 600 fg, 60 fg, and 6 fg per reaction, negative control (NDM-1-positive Escherichia coli WHCDC-WH67 genomic DNA, 6 pg per reaction), negative control (KPC-2-producing Klebsiella pneumoniae WHCDC-WH108 genomic DNA, 6 pg per reaction), and blank control (DW).
The analytical specificity of the mcr-1-MCDA-LFB assay was assessed with genomic DNA extracted from 16 mcr-1-producing strains and 43 non-mcr-1 isolates. As shown in Figure 5, all products derived from the strains carrying mcr-1 gene exhibited two red bands (TL and CL) in the LFB, but each sample from the mcr-1-negative organisms and blank control yielded only one red band. The results certified that the MCDA-LFB assay had a complete specificity for mcr-1 detection.
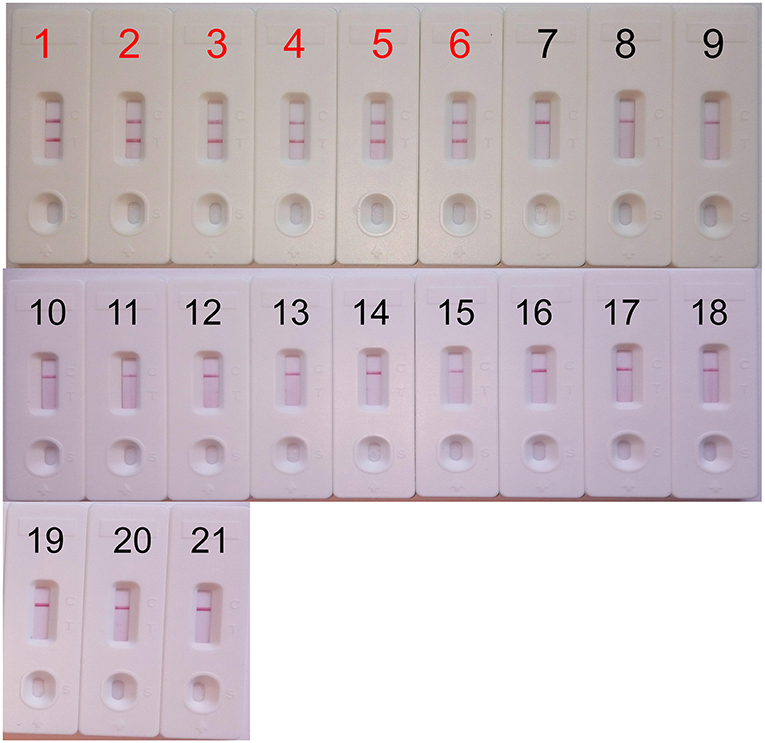
Figure 5. Specificity of the mcr-1-MCDA-LFB assay. Biosensor 1, mcr-1-positive Escherichia fergusonii ICDC-ZG2016M34-3. Biosensor 2, mcr-1-positive Escherichia fergusonii from ICDC. Biosensor 3, mcr-1-positive Escherichia fergusonii from WHCDC. Biosensor 4, mcr-1-producing Escherichia coli from ICDC. Biosensor 5, mcr-1-producing Escherichia coli from WHCDC. Biosensor 6, mcr-1-producing Salmonella enteritidis from ICDC. Biosensors 7–17, KPC-2-postive Klebsiella pneumoniae, KPC-2-postive Pseudomonas aeruginosa, NDM-1-postive Escherichia coli, NDM-1-postive Enterobacter cloacae, NDM-1-postive Klebsiella pneumoniae, NDM-5-postive Escherichia coli, IMP-4-postive Escherichia coli, Acinetobacter baumannii carring non-mcr-1/non-carbapenemase gene, Pseudomonas aeruginosa carring non-mcr-1/non carbapenemase gene, mcr-1/carbapenemase-negative Serratia marcescens, mcr-1/carbapenemase-negative Escherichia coli, all of the stains derived from WHCDC. Biosensor 18, blank control (DW). Biosensors 19–21, Staphylococcus aureus carring non-mcr-1/non-carbapenemase gene, Enterococcus faecalis carring non-mcr-1/non carbapenemase gene, mcr-1/carbapenemase-negative Enterococcus faecium, those stains were isolated from WHCDC.
To evaluate the optimum time, four reaction times (10–40 min at 10 min intervals) were tested for the mcr-1-MCDA-LFB assay during the amplification stage. The mcr-1-producing E. fergusonii (ICDC-ZG2016M34-3) plasmid DNA, 600 fg/μl (the LOD of the method), did not gain the positive results until the reaction had operated for 30 min (Figure 6). Hence, the amplification time of 30 min at 63°C was considered as an optimal reaction condition for the current assay.
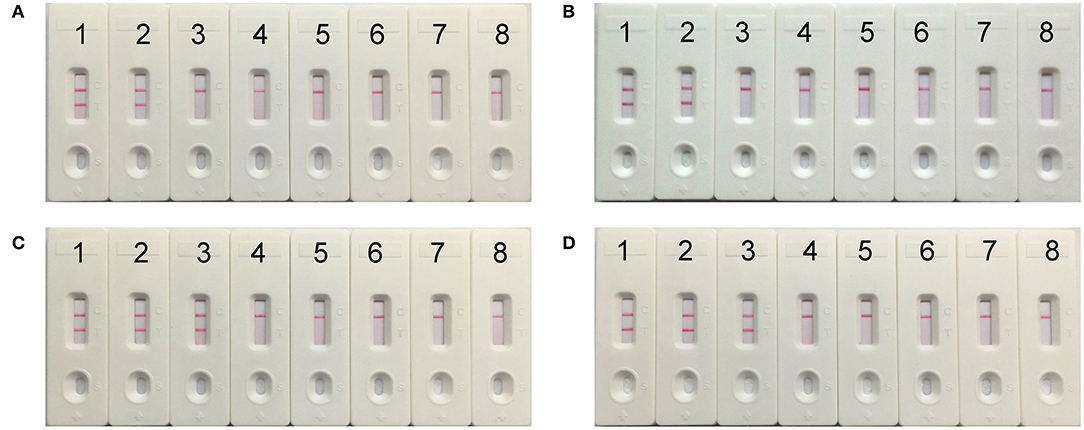
Figure 6. The optimal reaction time for mcr-1-MCDA-LFB assay. Four different amplification times, covering 10 min (A), 20 min (B), 30 min (C), and 40 min (D), were compared at 63°C. Biosensors 1–8 represented the plasmid DNA levels of 60 pg, 6 pg, 600 fg, 60 fg, and 6 fg of Escherichia fergusonii ICDC-ZG2016M34-3, negative control (6 pg of NDM-1-positive Escherichia coli WHCDC-WH67 genomic DNA), negative control (6 pg of KPC-2-producing Klebsiella pneumoniae WHCDC-WH108 genomic DNA) and blank control (DW).
The LOD for strains expressing mcr-1 in fecal samples was determined to assess the practical application of the established assay. The detected threshold of mcr-1-positive bacteria was approximately 4.5 × 103 CFU/mL (~4.5 CFU/reaction) in 0.2 g fecal samples spiked with 100 μl of dilutions of strains (Figure 7). The results of other subjects including the lower suspensions concentrations, negative control, and blank control were negative. As the same to the aforementioned experiments, detection of the amplicons with three methods got an equivalent conclusion.
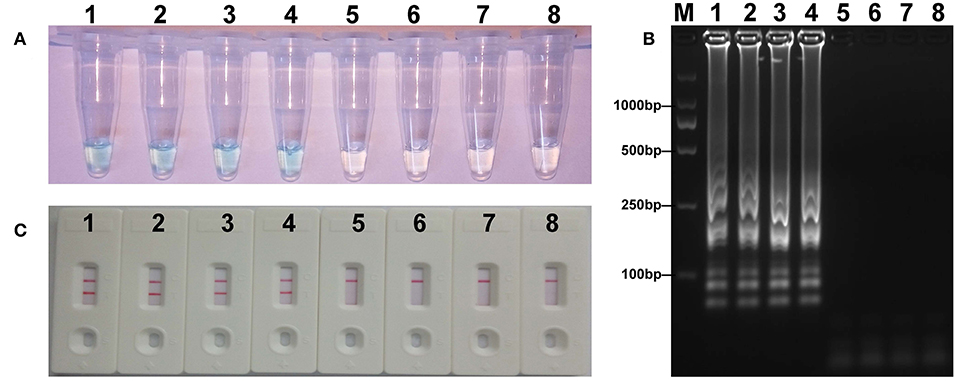
Figure 7. Sensitivity of mcr-1-MCDA-LFB assay in spiked feces samples. Three measurement techniques, including colorimetric indicators (A), gel electrophoresis (B), and LFB (C), were applied for testing MCDA products. Tubes (A)/lanes (B)/biosensors (C) 1–8 represented the DNA levels of 4.5 × 103, 4.5 × 102, 4.5 × 101, 4.5 × 100, 4.5 × 10−1, and 4.5 × 10−2 CFU per reaction, negative control (non-spiked feces sample) and blank control (DW).
The detection limit of the conventional PCR assay was 6 pg of plasmid DNA per microliter in pure culture and 4.5 × 104 CFU/mL (~45 CFU/reaction) in spiked fecal samples. Mcr-1-MCDA-LFB method was thus highly sensitive, 10-fold more so than that of PCR (Table 3).
Polymyxins (polymyxin B and colistin) have nearly become last-resort drugs for treating the severe infections caused by multidrug-resistant or pan-resistant Enterobacteriaceae (Wang et al., 2017a). The defensive line will be destroyed by the emergence of mcr-1-positive strains resisting to colistin. Moreover, the mcr-1 gene mediated by plasmids or transposons can transfer in different species freely. Thus, the isolates carrying mcr-1 gene will undoubtedly become a major issue for public health. Under the circumstances, a convenient and fast technique for detection of mcr-1 in various samples is of great importance. Here, an approach was reported to detect target gene by MCDA united with lateral flow biosensor (MCDA-LFB). In the MCDA-LFB assay, the high specificity was guaranteed, as a set of 10 primers was employed for specially amplifying the target sequence. The specificity of mcr-1-MCDA-LFB was successfully confirmed with the genomic templates extracted from mcr-1-producing strains and non-mcr-1 organisms, and the results were positive for all mcr-1-positive isolates, but negative for non-mcr-1 isolates and blank control. Therefore, the diagnostic test based on MCDA-LFB for the detection of mcr-1 in bacteria identifies target gene with high selectivity.
The MCDA products can be analyzed with LFB, colorimetric indicator and agarose gel electrophoresis, respectively. LFB, as a detection technique by observing the number of red lines on sensor bar, is more objective than colorimetric indicator, which reports the result through color change. Maybe the latter is in trouble when the concentration of target gene is very low (Wang et al., 2017c). Likewise, LFB is more rapid and convenient than gel electrophoresis, which requires the use of an additional operation procedure and complex equipment. Hence, LFB will be a better candidate for the results' judge of MCDA products.
Besides specificity, the superb sensitivity is also very important for the newly established assay. The mcr-1-MCDA-LFB method sufficed to detect as little as 600 fg of mcr-1-positive plasmid DNA per microliter in pure culture and 4.5 × 103 CFU/mL (~4.5 CFU/reaction) in fecal samples spiked with 100 μl of strains. This technique has the same sensitivity to mcr-1-LAMP described in previously report (Zou et al., 2017), and is 10 times more sensitive than that of conventional PCR. Moreover, in spite of the equivalent specificity and sensitivity in both isothermal amplification assays, the mcr-1-MCDA-LFB method which expends less reaction time than the other will provide faster detection for mcr-1. Likewise, although the Real-time PCR and MALDI-TOF MS-based method could also test the mcr-1 gene in low limit of detection and less time, respectively (Bontron et al., 2016; Dortet et al., 2018), they needed expensive apparatus and immaculately experimental condition that were not well-equipped in resource-challenged fields, especially in the livestock industry where a large quantity of stains carrying mcr gene were identified (Dortet et al., 2018).
The mcr-1-MCDA-LFB assay only required a constant reaction temperature at 63°C. The entire process of experiments, including sample processing (25 min), isothermal amplification (30 min), and detection (2 min), could be accomplished in <60 min. Herein, this assay economizes the test time and device, and is suitable for timely identification on the spot particularly.
In conclusion, we devised a reliable MCDA-LFB assay for the detection of mcr-1 with simplicity, rapidity, and low-cost facility. The LOD of this assay was only 600 fg per reaction with pure culture, and the specificity was 100% according to the trial results. From the above, the mcr-1-MCDA-LFB assay built in this study will greatly improve the detection efficiency for the monitor of target gene in practical application.
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
LG and JiL designed the experiments. LG and EL carried out the experiments. JuL and JC contributed the partial isolates. XL and HX provided the materials and reagents. LG analyzed the related data and wrote the paper.
We acknowledge the financial support (the research projects of clinical medicine in Wuhan, Grant Number: WG17Q02) from the Wuhan Municipal Health Commission, People's Republic of China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer LD declared a shared affiliation, with no collaboration, with several of the authors, JC and JuL, to the handling editor at time of review.
We would like to thank JuL and JC (Chinese Center for Disease Control and Prevention) for providing partial isolates.
Bontron, S., Poirel, L., and Nordmann, P. (2016). Real-time PCR for detection of plasmid-mediated polymyxin resistance (mcr-1) from cultured bacteria and stools. J. Antimicrob. Chemother. 71, 2318–2320. doi: 10.1093/jac/dkw139
Chen, K., Chan, E. W., Xie, M., Ye, L., Dong, N., and Chen, S. (2017). Widespread distribution of mcr-1-bearing bacteria in the ecosystem, 2015 to 2016. Euro. Surveill. 22:17-00206. doi: 10.2807/1560-7917.ES.2017.22.39.17-00206
Dortet, L., Bonnin, R. A., Pennisi, I., Gauthier, L., Jousset, A. B., Dabos, L., et al. (2018). Rapid detection and discrimination of chromosome- and MCR-plasmid- mediated resistance to polymyxins by MALDI-TOF MS in Escherichia coli: the MALDIxin test. J. Antimicrob. Chemother. 73, 3359–3367. doi: 10.1093/jac/dky330
Doumith, M., Godbole, G., Ashton, P., Larkin, L., Dallman, T., Day, M., et al. (2016). Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother. 71, 2300–2305. doi: 10.1093/jac/dkw093
Gelband, H., Miller-Petrie, M. K., Pant, S., Gandra, S., Levinson, J., Barter, D., et al. (2015). The state of the world's antibiotics 2015. Wound Heal. South. Afr. 8, 30–34. Available online at: https://hdl.handle.net/10520/EJC180082
Guo, Q., Su, J., McElheny, C. L., Stoesser, N., Doi, Y., and Wang, M. (2017). IncX2 and IncX1-X2 hybrid plasmids coexisting in fosA6-producing Escherichia coli. Antimicrob. Agents Chemother. 61, e00536–e00517. doi: 10.1128/AAC.00536-17
Li, Z., Tan, C., Lin, J., and Feng, Y. (2016). Diversified variants of the mcr-1-carrying plasmid reservoir in the swine lung microbiota. Sci. China Life Sci. 59, 971–973. doi: 10.1007/s11427-016-5111-9
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism mcr-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
McGann, P., Snesrud, E., Maybank, R., Corey, B., Ong, A. C., Clifford, R., et al. (2016). Escherichia coli harboring mcr-1 and blaCTX−M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob. Agents Chemother. 60, 4420–4421. doi: 10.1128/AAC.01103-16
Nation, R. L., and Li, J. (2009). Colistin in the 21st century. Curr. Opin. Infect. Dis. 22, 535–543. doi: 10.1097/QCO.0b013e328332e672
Niu, L., Zhao, F., Chen, J., Nong, J., Wang, C., Wang, J., et al. (2018). Isothermal amplification and rapid detection of Klebsiella pneumoniae based on the multiple cross displacement amplification (MCDA) and gold nanoparticle lateral flow biosensor (LFB). PLoS ONE 13:e0204332. doi: 10.1371/journal.pone.0204332
Olaitan, A. O., Morand, S., and Rolain, J. M. (2014). Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5:643. doi: 10.3389/fmicb.2014.00643
Sennati, S., Di Pilato, V., Riccobono, E., Di Maggio, T., Villagran, A. L., Pallecchi, L., et al. (2017). Citrobacter braakii carrying plasmid-borne mcr-1 colistin resistance gene from ready-to-eat food from a market in the Chaco region of Bolivia. J. Antimicrob. Chemother. 72, 2127–2129. doi: 10.1093/jac/dkx078
Stoesser, N., Mathers, A. J., Moore, C. E., Day, N. P., and Crook, D. W. (2016). Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect. Dis. 16, 285–286. doi: 10.1016/S1473-3099(16)00010-4
Sun, J., Fang, L. X., Wu, Z., Deng, H., Yang, R. S., Li, X. P., et al. (2017). Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci. Rep. 7:424. doi: 10.1038/s41598-017-00095-x
Sun, J., Li, X., Yang, R., Fang, L., Huo, W., Li, S., et al. (2016). Complete nucleotide sequence of an IncI2 plasmid coharboring blaCTX−M−55 and mcr-1. Antimicrob. Agents Chemother. 60, 5014–5017. doi: 10.1128/AAC.00774-16
Wang, Q., Sun, J., Li, J., Ding, Y., Li, X. P., Lin, J., et al. (2017a). Expanding landscapes of the diversified mcr-1-bearing plasmid reservoirs. Microbiome 5:70. doi: 10.1186/s40168-017-0288-0
Wang, R., Dorp, L. V., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018a). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. doi: 10.1038/s41467-018-03205-z
Wang, Y., Li, H., Li, D., Li, K., Wang, Y., Xu, J., et al. (2016). Multiple cross displacement amplification combined with gold nanoparticle-based lateral flow biosensor for detection of Vibrio parahaemolyticus. Front. Microbiol. 7:2047. doi: 10.3389/fmicb.2016.02047
Wang, Y., Li, H., Wang, Y., Li, H., Luo, L., Xu, J., et al. (2017b). Development of multiple cross displacement amplification label-based gold nanoparticles lateral flow biosensor for detection of Listeria monocytogenes. Int. J. Nanomed. 12, 473–486. doi: 10.2147/IJN.S123625
Wang, Y., Li, H., Wang, Y., Xu, H., Xu, J., and Ye, C. (2018b). Antarctic thermolabile uracil-DNA- glycosylase-supplemented multiple cross displacement amplification using a label-based nanoparticle lateral flow biosensor for the simultaneous detection of nucleic acid sequences and elimination of carryover contamination. Nano. Res. 11, 2632–2647. doi: 10.1007/s12274-017-1893-z
Wang, Y., Li, H., Wang, Y., Zhang, L., Zhang, J., Xu, J., et al. (2017c). Nanoparticle-based lateral flow biosensor combined with multiple cross displacement amplification for rapid, visual and sensitive detection of Vibrio cholerae. FEMS Microbiol. Lett. 15:364. doi: 10.1093/femsle/fnx234
Wang, Y., Wang, Y., Ma, A. J., Li, D. X., Luo, L. J., Liu, D. X., et al. (2015). Rapid and sensitive isothermal detection of nucleic-acid sequence by multiple cross displacement amplification. Sci. Rep. 5:11902. doi: 10.1038/srep11902
Wang, Y., Yan, W., Fu, S., Hu, S., Wang, Y., Xu, J., et al. (2018c). Multiple cross displacement amplification coupled with nanoparticles-based lateral flow biosensor for detection of Staphylococcus aureus and identification of methicillin-resistant S. aureus. Front. Microbiol. 9:907. doi: 10.3389/fmicb.2018.00907
Yang, R., Feng, Y., Lv, X., Duan, J., Chen, J., Fang, L., et al. (2016). Emergence of NDM-5 and MCR-1-producing Escherichia coli clone ST648 and ST156 from a single Muscovy duck (Cairina moschata). Antimicrob. Agents. Chemother. 60, 6899–6902. doi: 10.1128/AAC.01365-16
Zeng, K. J., Doi, Y., Patil, S., Huang, X., and Tian, G. B. (2016). Emergence of plasmid-mediated mcr-1 gene in colistin-resistant Enterobacter aerogenes and Enterobacter cloacae. Antimicrob. Agents. Chemother. 60, 3862–3863. doi: 10.1128/AAC.00345-16
Keywords: mcr-1, colistin resistance, multiple cross displacement amplification, lateral flow biosensor, detection assay
Citation: Gong L, Liu E, Che J, Li J, Liu X, Xu H and Liang J (2019) Multiple Cross Displacement Amplification Coupled With Gold Nanoparticles-Based Lateral Flow Biosensor for Detection of the Mobilized Colistin Resistance Gene mcr-1. Front. Cell. Infect. Microbiol. 9:226. doi: 10.3389/fcimb.2019.00226
Received: 03 April 2019; Accepted: 11 June 2019;
Published: 27 June 2019.
Edited by:
Bing Gu, Affiliated Hospital of Xuzhou Medical University, ChinaReviewed by:
Yi Wang, Beijing Children's Hospital, ChinaCopyright © 2019 Gong, Liu, Che, Li, Liu, Xu and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiansheng Liang, d2gtbGpzQHNvaHUuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.