- 1Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Laboratory for Marine Fisheries Science and Food Production Processes, National Laboratory for Marine Science and Technology (Qingdao), Key Laboratory of Maricultural Organism Disease Control, Ministry of Agriculture and Rural Affairs, Qingdao Key Laboratory of Mariculture Epidemiology and Biosecurity, Qingdao, China
- 2Shandong Provincial Key Laboratory of Animal Biotechnology and Disease Control and Prevention, Shandong Agricultural University, Tai'an, China
- 3Department of Pathology, Shanghai Tenth People's Hospital, Tongji University School of Medicine, Shanghai, China
Acute hepatopancreatic necrosis disease (AHPND) has caused sharp declines in aquaculture industries of whiteleg shrimp Penaeus vannamei in Asia and the Americas since 2010. Vibrio parahaemolyticus, V. campbellii, V. owensii, and V. punensis have been proved to cause AHPND. However, the mechanisms underlying the burgeoning number of Vibrio species that cause AHPND is not known. All of AHPND-causing Vibrio bacteria (VAHPND) harbor a highly homologous plasmid (designated as pVA1-type) carrying pirABvp toxin genes. In this study, we demonstrate conclusively that the pVA1-type plasmid can be transferred from VAHPND to non-pathogenic bacteria. We constructed a pVPGX1-Cmr plasmid (a pVA1-type plasmid) by adding a chloramphenicol resistance gene as a marker in a donor AHPND-causing V. parahaemolyticus 20130629002S01 (Vp2S01). Horizontal transfer of this plasmid was successfully performed from the AHPND-Vp2S01 to a non-pathogenic strain of V. campbellii at the transfer efficiency of 2.6×10−8 transconjugant/recipient, and DNase I treatment did not eliminate the transfer. The recipient V. campbellii acquired the pVA1-type plasmid and was shown to produce pirABvp RNA and proteins. Challenge studies using the transconjugant caused 100% mortality in exposed groups of P. vannamei. The challenged shrimp, infected with the transconjugant bacteria, showed typical gross signs and histological lesions of AHPND. These results demonstrated the conjugative transfer of an AHPND pVA1-type plasmid. It provides timely information for explaining the increased species of AHPND-causing Vibrio bacteria and will be useful in the development of management strategies leading to the prevention and control of AHPND.
Introduction
Acute hepatopancreatic necrosis disease (AHPND, also known as early mortality syndrome, EMS) affects marine shrimp Penaeus vannamei and P. monodon. Outbreaks of AHPND have been reported in shrimp farms in Asia and the Americas (Zhang et al., 2012; Tran et al., 2013; Gomez-Gil et al., 2014; Gomez-Jimenez et al., 2014; Kondo et al., 2014; Nunan et al., 2014; de la Pena et al., 2015; Lee et al., 2015), where this disease has resulted in substantial production and economic losses, especially in whiteleg shrimp P. vannamei. The disease progresses rapidly, usually within 30–35 days, after stocking postlarvae in growth out ponds. The disease emerged in 2010 and is estimated to cause economic losses of several billions of dollars each year (Shinn et al., 2018). This has provided an incentive to determine the causes of the disease.
The causative agent of AHPND was initially determined to be strains of Vibrio parahaemolyticus (Zhang et al., 2012; Tran et al., 2013). The virulent strains were found to contain a ~70 kb plasmid (pVA1) carrying genes pirABvp that encode homologs of the Photorhabdus insect-related (Pir) toxins (Han et al., 2015; Lee et al., 2015). The role of PirABvp in causing AHPND was demonstrated in laboratory studies using deletion and insertion mutants (Lee et al., 2015). Recent studies found that AHPND could be caused by strains of other Vibrio species, such as V. harveyi-like, V. campbellii, V. owensii, and V. punensis (Kondo et al., 2015; Liu et al., 2015, 2018; Dong et al., 2017b; Han et al., 2017; Restrepo et al., 2018). Strains of these Vibrio species, collectively abbreviated as VAHPND, that contain the pVA1-type plasmids as well as secreting both PirAvp and PirBvp proteins, can cause AHPND.
The pVA1 plasmid carries a set of genes related to transfer, suggesting that it has the ability to mobilize and transfer from one Vibrio cell to another. This set of genes, including conjugative transfer genes, mobilization genes and pndA PSK system, was first found in the pVA1 plasmid of AHPND-causing V. parahaemolyticus (VpAHPND) strain 3HP (Lee et al., 2015). Later, these genes were found in the pVA1-type plasmids from AHPND-causing strains of V. parahaemolyticus, V. harveyi-like (VhAHPND), V. owensii (VoAHPND), V. campbellii (VcAHPND), and V. punensis (VpuAHPND) (Kondo et al., 2015; Liu et al., 2015; Dong et al., 2017a,c; Xiao et al., 2017; Restrepo et al., 2018). We reported a high homology between the pVA1-type plasmids of strains of VpAHPND and VcAHPND isolated from the same AHPND-affected pond (Dong et al., 2017a), providing indirect evidence that strains of related Vibrio species can become virulent through the transfer of the pVA1-type plasmids. In association with our recent challenge studies, we found in vivo horizontal transfer of a pVA1-type plasmid from VcAHPND to non-AHPND-causing V. owensii (Dong et al., 2019).
In this work, we demonstrate conclusively, through conjugation experiments, that pVA1-type plasmid can be transferred from AHPND-V. parahaemolyticus to V. campbellii. This study provides insight into the genetic processes underlying the diversity found among VAHPND bacteria.
Materials and Methods
Bacterial Strains, Plasmids and Culture Conditions
Bacterial strains and plasmids used are shown in Table 1. Vp2S01 was isolated from AHPND-affected shrimp in Guangxi, China in 2013 (Dong et al., 2017a). Vp2S01 consists of two chromosomes (Genbank no. CP020034, CP020035) and two plasmids (CP020036, CP020037). V. campbellii LMB29 (VcLMB29) was isolated from a skin ulcer sample of red drum in Shenzhen, China (Liu J. et al., 2017). VcLMB29 consists of two chromosomes (CP019293, CP019294) and four plasmids (CP019295-CP019298). Vibrio strains were cultured in marine 2216E (5 g/L tryptone, 1 g/L yeast extract, FePO4 0.01g, 1 L filtered seawater, pH adjusted to 7.6) broth or agar, tryptic soy broth with 2% NaCl (TSB+), or Luria Bertani broth (LB) at 28°C.
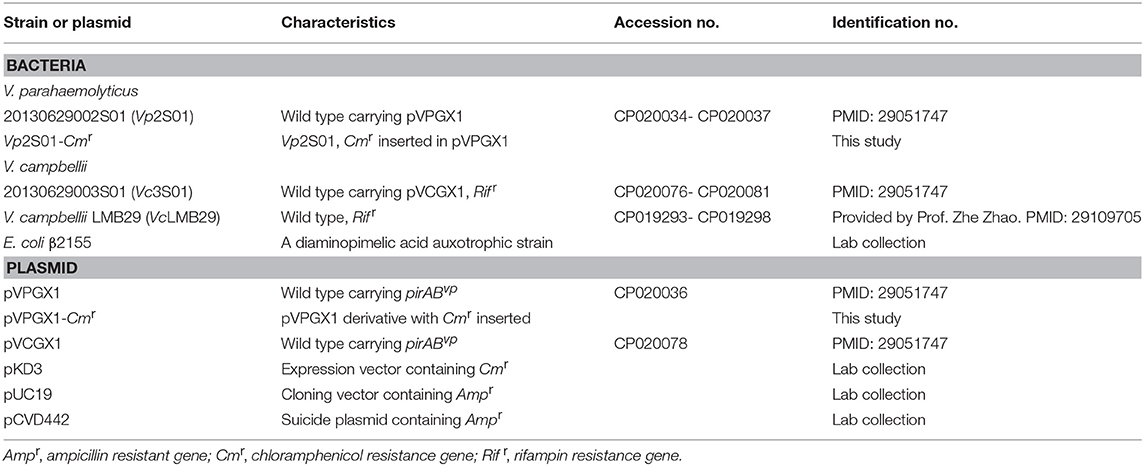
Table 1. Information of Vibrio parahaemolyticus and Vibrio campbellii strains and plasmids used in this study.
Antibiotic Susceptibility Test
Antibiotic susceptibility was determined by minimal inhibitory concentration (MIC) using the broth microdilution method (Wiegand et al., 2008). Briefly, two-fold serial dilutions of each antibiotic, including ampicillin, cefazolin, ceftriaxone, chloramphenicol, gentamicin, rifampin, streptomycin, and tetracycline, were added into four 96-well microtiter plates, one row for each antibiotic with up to 10 dilutions. A volume of 90 μl bacterial suspension (106 CFU/mL) of each bacterial culture (i.e., Vp2S01, Vp2S01-Cmr, Vc3S01, or VcLMB29) was inoculated into one of the 96-well plates (column 1–11). After adding the antibiotic solution, the final bacterial concentration was 5 × 105 CFU/mL. A volume of 90 μL of broth was added to the control wells of column 12. All plates were incubated at 28°C for 24 h. Breakpoints were defined by the guidelines (M45-A2, 2010 and M100-S24, 2014) of the Clinical and Laboratory Standards Institute (CLSI)1.
Construction of pVPGX1-Cmr and Vp2S01-Cmr
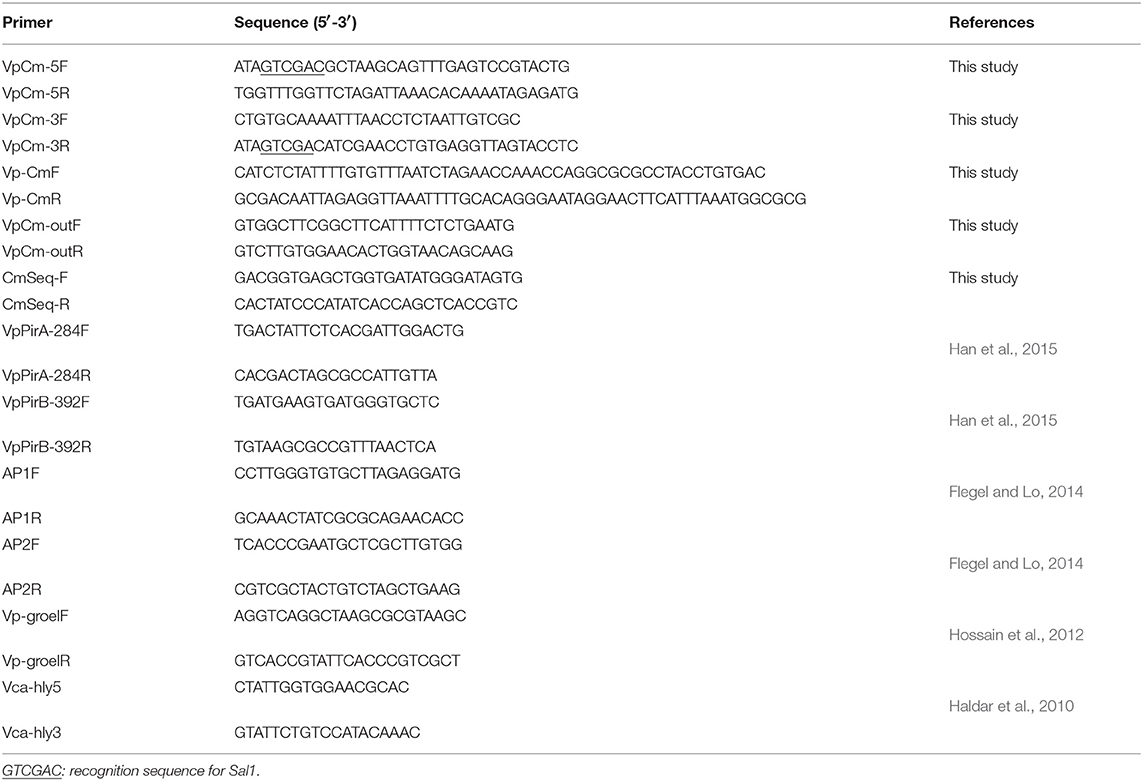
To insert the chloramphenicol resistance gene (Cmr) to the plasmid pVPGX1 by homologous recombination, we amplified the homologous flanking sequences and the Cmr fragment (660 bp) by PCR from pVPGX1 and pKD3 plasmids, respectively, using the primer pairs VpCm-5F/5R, VpCm-3F/3R, and Vp-CmF/R (Table 2). The target fragment was amplified using the primer pair VpCm-5F/3R; the PCR products were digested with SalI and ligated with pUC19. After confirming the expression of chloramphenicol in pUC19 by plating the transformed E. coli on LB agar plates containing 100 μg/mL of ampicillin and 17 μg/mL of chloramphenicol, the target fragment was then subcloned into pCVD442. The resulting plasmid pCVD442-Cmr was introduced into E. coli β2155 by electrotransformation. Then, the E. coli β2155 carrying pCVD442-Cmr was mated with Vp2S01 by conjugation. We validated the insertion of Cmr in the resulting Vp2S01-Cmr by PCR and sequencing using the primer pairs VpCm-outF/R and CmSeq-F/R (Table 2).
Conjugation Experiments
Conjugation experiments were carried out using a protocol described by Wiesner et al. (2013). Briefly, Vp2S01-Cmr (donor) was cultured in 2216E with chloramphenicol (Cm, 20 μg/mL); VcLMB29 (recipient) was cultured in 2216E with rifampin (Rif, 15 μg/mL). Equal amounts (50 mL each) of secondary inocula of donor and recipient cells were mixed, then washed and resuspended twice with 4 mL fresh 2216E to remove residual antibiotics. The mixture was resuspended with 100 μL fresh 2216E and dropped on 0.22 μm Millipore filter membranes placed on a 2216E agar plate and incubated at 28°C for 12 h. The conjugation mix was then resuspended in 2 mL of 2216E broth, diluted to 10−1, 10−2, 10−3, and spread on 2216E plates which contained Cm (60 μg/mL) and Rif (20 μg/mL) as transconjugants selection antibiotics. The mixture was further diluted to 10−7, 10−8, 10−9, then spread on 2216E plates containing Rif for determining the number of recipient cells. Transfer efficiency was determined by the ratio of transconjugants' counts to those of recipient strain. Transconjugants were analyzed for the presence of pirABvp genes and backbone of pVA1 plasmid with 4 pairs of PCR primers: VpPirA-284F/R, VpPirB-392F/R, AP1F/R, AP2F/R (Table 2). VpPirA-284F/R and VpPirB-392F/R primers target pirAvp and pirBvp gene, respectively (Han et al., 2015). AP1F/R and AP2F/R primers targeted the backbone of pVA1 plasmid (Flegel and Lo, 2014).
To determine which of the bacterial colonies grown on Cm and Rif plates were V. campbellii, all were analyzed with 2 pairs of PCR primers Vca-hly5/3 (amplicon length: 328 bp) and Vp-groelF/R (510 bp) (Table 2), which can be used to distinguish V. parahaemolyticus and V. campbellii (Haldar et al., 2010; Hossain et al., 2012).
In order to study the stability of pVA1-type plasmid in transconjugants, one transconjugant colony (named as VcLMB29-pVPGX1) was passed 5 times on 2216E plates which containing Cm (60 μg/mL) and Rif (20 μg/mL) and analyzed for the presence of pirABvp genes with 4 pairs of PCR primers: VpPirA-284F/R VpPirB-392F/R, AP1F/R, and AP2F/R (Table 2).
Conjugation Experiments With DNase I
To determine the effect of DNase in plasmid transfer, this enzyme was added to mating mixture in conjugation experiments. The conjugation were carried out as described above. 2.5 μL of DNase I (5 unit; NEB) was added to the mixture of donor and recipient cells and washed with 4 mL 2216E. The mixture was resuspended with 90 μL fresh 2216E and 10 μL of DNase I (20 unit; NEB), then, dropped on a 0.22 μm Millipore filter membrane placed on the 2216E agar plate and incubated at 28°C for 12 h. The transfer efficiency was determined as described above.
Expression of pirABvp Genes in pVA1-Type Plasmid From a Transconjugant
For determination of the expression of pirABvp genes, total RNA of VcLMB29-pVPGX1, VcLMB29, and Vp2S01-Cmr were extracted using RNAprep pure Cell/Bacteria Kit (TIANGEN Biotech, China) and treated with DNase I. RT-PCR were performed to analyze the mRNA expression of pirAvp and pirBvp genes using 2 pairs of PCR primers VpPirA-284F/R and VpPirB-392F/R. The cDNA was generated with Random 6 mers at 42°C for 45 min, 95°C for 5 min using the TaKaRa PrimeScriptTM II 1st Strand cDNA Synthesis Kit (Takara Bio, Japan). The PCR were initiated at 94°C for 7 min, followed by 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, ending with 72°C for 7 min. Following PCR, 5 μL of the reaction mixture was analyzed in a 2% agarose gel containing GeneFinder (Bio-V, China).
To determine the production of PirAvp and PirBvp in the transconjugant, individual colonies of VcLMB29-pVPGX1, VcLMB29 (the recipient non-AHPND strain as a negative control), and Vp2S01 (an VpAHPND strain as a positive control) were cultured in 2216E broth at 28°C for 10 h. Fifty milliliter of each culture was centrifuged at 4,280 g for 10 min, each supernatant was concentrated into 2 mL by using Amicon 10 kDa filter tubes. All samples were analyzed using 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli, 1970) and then proteins in the target bands were identified by mass spectrometry (MS) analysis (Wang et al., 2011). All data were integrated and analyzed by the use of GPS Explorer V3.6 software with the MASCOT V2.3 search engine (Matrix Science Ltd., UK) with the following parameters: NCBI nr, trypsin digest with one missing cleavage, MS tolerance at 200 ppm, and MS/MS tolerance of 0.8 Da. Proteins were identified based on 95% or higher confidence interval of their scores.
Sequencing and Assembly of pVA1-type Plasmid Sequence From the Transconjugant
The sequence of pVCON1 from VcLMB29-pVPGX1 was determined after sequencing the bacteria with an Illumina HiSeq X Ten Sequencer (PE150 mode) and filtering out the bacterial chromosomal sequence (Liu J. et al., 2017). Raw sequencing data was generated by Illumina base calling software CASAVA v1.8.2 (http://support.illumina.com/sequencing/sequencing_software/casava.ilmn). Contamination reads were identified by Trimmomatic (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic) with default parameters. Clean data obtained by above quality control processes were used for further analysis.
We used ABySS (http://www.bcgsc.ca/platform/bioinfo/software/abyss) to perform a sequence assembly with multiple-Kmer parameters (Jackman et al., 2017). GapCloser software (https://sourceforge.net/projects/soapdenovo2/files/GapCloser/) was subsequently applied to fill up the remaining local inner gaps and correct the single nucleotide polymorphism for the final assembly results.
Comparative pVA1-type Plasmid Sequences Analysis
The reference sequences of pVA1-type plasmids downloaded from NCBI were used for comparative sequences analysis. These sequences included pVA1 (KP324996), pVPGX1 (CP020036), pVCGX1 (CP020078), and pLA16-2 (CP021148). We used the BLAST Ring Image Generator (BRIG) to generate the circle map. The innermost ring shows the reference sequence of pVA1. The second innermost ring shows the sequence of pVPGX1.The third innermost ring shows the sequence of pVCON1. The forth innermost ring shows the sequence of pVCGX1. The fifth innermost ring shows the sequence of pLA16-2. The sixth innermost ring shows the ORFs of the forward chain of pVA1. The seventh innermost ring shows the ORFs of the complementary chain of pVA1. The outermost ring highlights the gene clusters of pVA1.
Challenge Bioassays and pirABvp RT-PCR
To determine AHPND pathogenicity of the transconjugant VcLMB29-pVPGX1, a challenge study was undertaken. We used three Vibrio strains in the study, including Vp2S01-Cmr as the positive control, VcLMB29-pVPGX1 as the target strain, and VcLMB29 as the negative control. A blank control without Vibrio bacteria was also included. Each Vibrio strains was cultured in TSB+ at 28°C until its OD600nm reached 0.8–0.9 (~8–12 h) and added into 4 L seawater to make bacterial suspension at a concentration of 1 × 108 CFU/mL. Each group of 45–56 healthy P. vannamei (mean weight: 1 g) were immersed in each of the bacterial suspensions for 15 min. For each group, the shrimp and bacterial suspension were then divided into three parallels of 30 L tanks filled with seawater to give a final bacterial concentration of 1 × 106 CFU/mL. The shrimp in the blank control group were immersed in marine 2216E broth. The mortality of shrimp in each tank was monitored and recorded every 8 h. Moribund shrimp were fixed for histopathological examination with Davidson's alcohol-formalin-acetic acid (Bell and Lightner, 1988).
In order to analyze the expression of pirABvp toxin genes in shrimp, tissues of hepatopancreas and stomach from moribund shrimp were sampled to extract total RNA using RNAprep Pure Tissue Kit (TIANGEN Biotech, China) along with treatment with 0.3 U/μL DNase I at 25°C for 15 min. RT-PCR were performed to determine the mRNA expression of pirABvp genes as described above.
Nucleotide Sequence Accession Number
Complete plasmid sequence of pVCON1 from strain VcLMB29-pVPGX1 has been deposited in GenBank under the accession MH890610.
Results
Comparative Sequence Analysis of pVA1-type Plasmids in AHPND-Causing Vibrio Strains
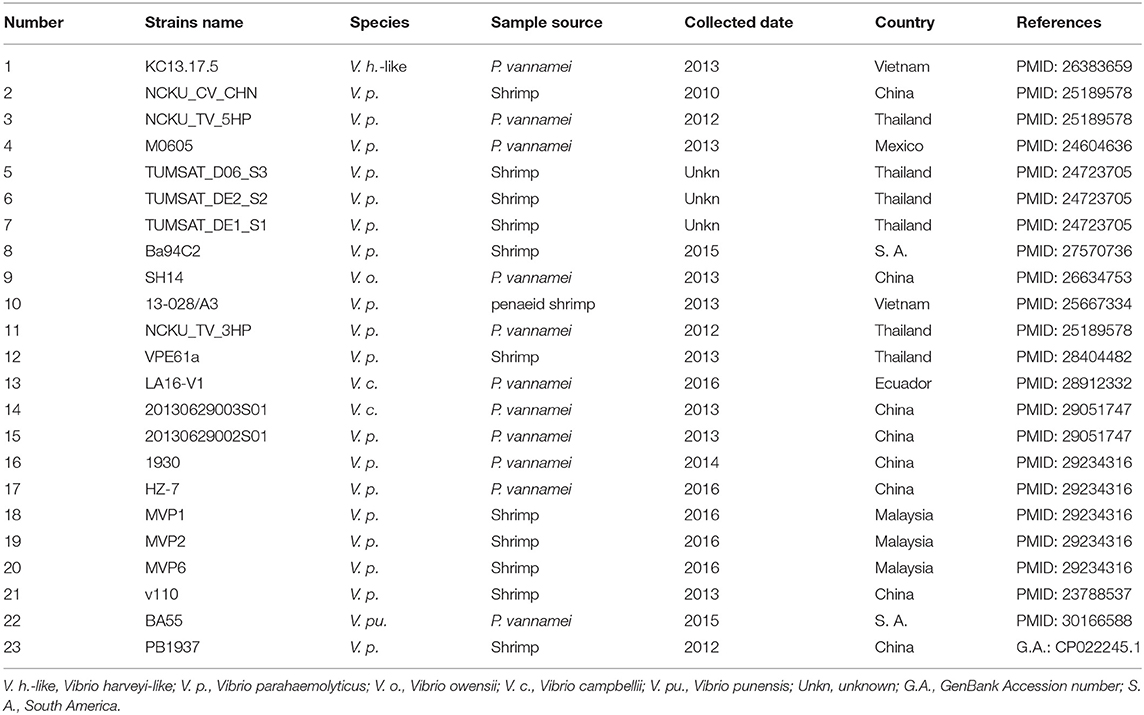
We retrieved 23, complete or draft, sequences of pVA1-type plasmids harbored in VAHPND bacteria from the NCBI RefSeq database (Table 3). There were 9 completely sequenced pVA1 plasmids, ranging in size from 69 to 73 kb. These plasmids have 99.92–100.00% identity in nucleotide sequence. We compared, with regard to genes known to be involved in conjugation, the genes of these plasmids, and found that all of the plasmid genomes contained two gene clusters related to conjugation (Figure 1). In one 9.5 kb gene cluster, we found a T4SS gene cluster with 11 genes (CDSs: pVA1074 to PVA1084, based on the reference plasmid pVA1) that, with the exception of pVA1080, code for conjugative transfer genes (trbC, trbD, trbF, trbJ, trbB, trbL, trbF, trbG, trbH, trbI). In the second gene cluster, pVA1010 and pVA1011 were found to be related to conjugation, the CDSs of which have high homology to T4SS-related proteins TraF and TrbN. The pVA1023 is a conjugative gene predicted to encode molybdopterin-guanine dinucleotide biosynthesis protein MobB (Figure 1).
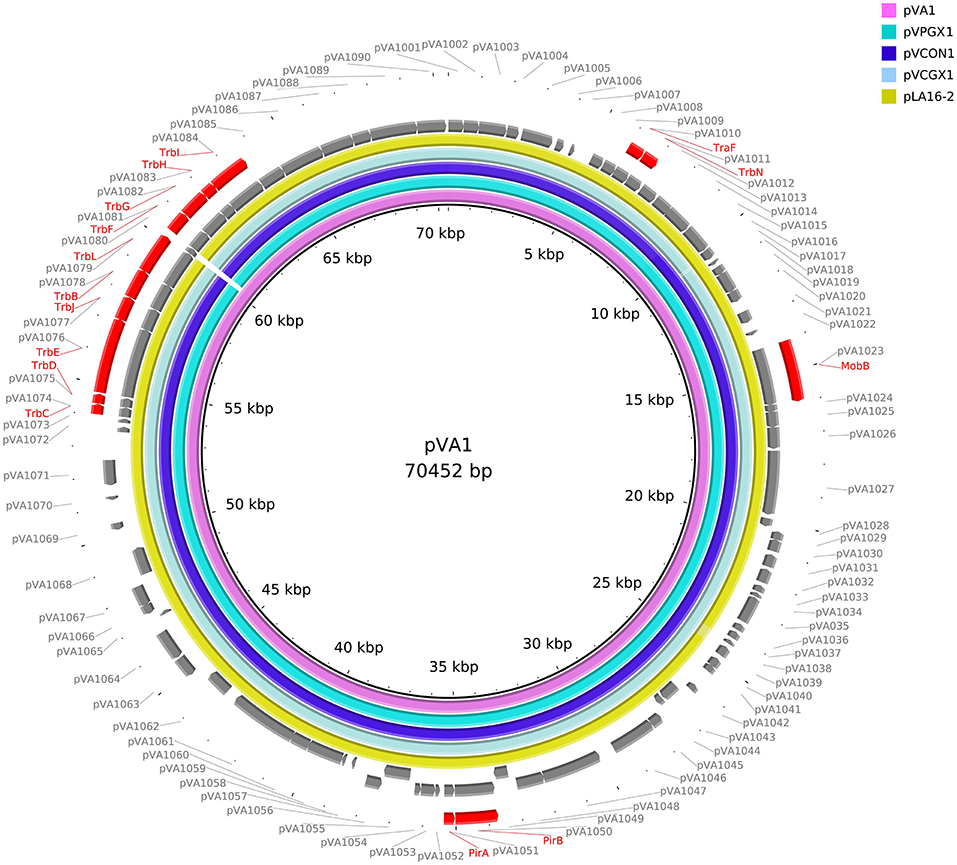
Figure 1. Comparative sequences analysis of pVA1-type plasmids. The reference sequences of pVA1-type plasmids downloaded from NCBI were used for comparative plasmid sequences analysis. The tracks from inside to outside represent the pVA1-type plasmid sequences of pVA1 (KP324996), pVPGX1 (CP020036), pVCON1 (MH890610), pVCGX1 (CP020078), pLA16-2 (CP021148), ORF (+) of pVA1, ORF (–) of pVA1, and marked genes, ORF numbers.
Construction of Chloramphenicol Resistant Strain Vp2S01-Cmr and Antibiotic Resistance Patterns of Vibrio Bacteria
To determine if the pVA1-type plasmid can be horizontally transferred, we constructed a testing plasmid in the donor strain Vp2S01-Cmr by homologous recombination, incorporating a chloramphenicol resistance gene (Cmr) into its virulence plasmid pVPGX1 (69,227 bp). The Cmr was inserted between the ORF9 (Vp2S01_p10009) and ORF10 (Vp2S01_p10010) region, the size of pVPGX1-Cmr is 70,125 bp.
All 3 VAHPND strains (Vp2S01, Vp2S01-Cmr, Vc3S01) and a non-pathogenic strain (VcLMB29) were determined for their antibiotic resistance. The results of MIC tests showed that, of the four strains tested, only Vp2S01 was susceptible to chloramphenicol (Cm) (Table 4). Among the other 7 antibiotics analyzed, this strain was intermediate to both ceftriaxone (Cro) and rifampin (Rif). Strain Vp2S01-Cmr exhibited a similar pattern of antibiotic resistance to that of Vp2S01 except that it was resistant to Cm as expected. Strain Vc3S01 was resistant to 7 of the antibiotics and was only susceptible to Cm. The VcLMB29 exhibited intermediate to Cm, but resistant to Rif and susceptible to 6 other antibiotics. Thus, Cm and Rif were selected to screen transconjugants.
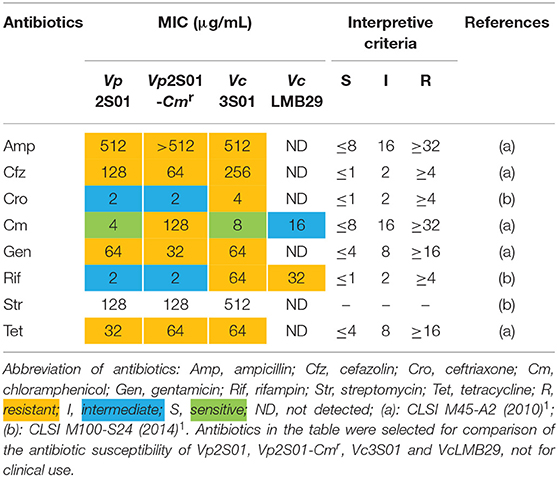
Table 4. Minimal inhibitory concentrations (MICs) of selected antibiotics on Vibrio parahaemolyticus and V. campbellii.
Conjugation, Stability of pVA1-type Plasmid and Expression of pirABvp Genes
In conjugation experiments, filter mating protocol was used to transfer the pVpGX1-Cmr plasmid from VpAHPND strain Vp2S01-Cmr (donor) to V. campbellii strain VcLMB29 (recipient). The transconjugants were obtained by selecting for colonies that were resistant to both Cm and Rif and were further verified to be V. campbellii by species-specific PCR showing that a 328 bp fragment was generated with Vca-hly5/3 primers. We found positive transconjugants and the transfer efficiency was 2.6 × 10−8 transconjugant/recipient at 12 h of conjugation. A similar transfer efficiency, 6.0 × 10−9, was obtained when DNase I was added to the conjugation mixture. One transconjugant VcLMB29-pVPGX1 was passed for 5 times, and all of the subcultures had positive PCR reactions with primers VpPirA-284F/R VpPirB-392F/R, AP1F/R and AP2F/R, indicating that pVA1-type plasmid was stable in this transconjugant.
The strain VcLMB29-pVPGX1 grown to a mid-logarithmic phase was pelleted. Total RNA was extracted and analyzed for the expression of pirABvp using RT-PCR. The result showed that bands of pirAvp and pirBvp products appeared at expected sizes of 284 and 392 bp in the transconjugant (Figure 2A, lanes 2 and 6), not seen in the recipient bacteria (Figure 2A, lanes 1 and 5). There were no amplified products when the RT step was omitted. Furthermore, the extracellular extracts from the cultured bacteria were analyzed for the presence of PirAvp and PirBvp proteins in an SDS-PAGE. The result showed that 2 bands, 17 kDa (predicted size of PirAvp) and 50 kDa (predicted size of PirBvp), were observed in the sample from strain VcLMB29-pVPGX1 (Figure 2B, lane 2) and in the VpAHPND strain Vp2S01 (as the positive control, Figure 2B, lane 3). The 17 and 50 kDa bands from VcLMB29-pVPGX1 (Figure 2B, lane 2) were excised and analyzed by mass spectrometry (LC-MS/MS) and MASCOT, confirming the presence of PirAvp and PirBvp proteins. These two bands were not present in the sample of the recipient bacteria VcLMB29 (Figure 2B, lane 1).
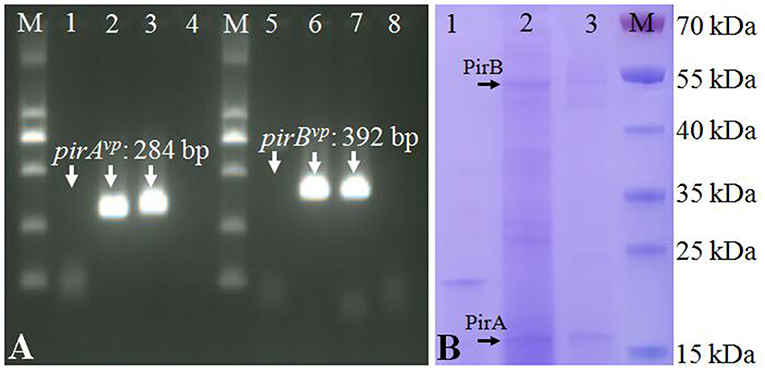
Figure 2. Expression of pirABvp genes in Vibrio campbellii and V. parahaemolyticus. (A) RT-PCR detection of pirAvp and pirBvp expression. Lanes 1–4: RT-PCR amplification of pirAvp in RNA samples from strain VcLMB29, VcLMB29-pVPGX1, Vp2S01-Cmr, and water (blank control), respectively. Lanes 5–8: RT-PCR amplification of pirBvp in RNA samples from strain VcLMB29, VcLMB29-pVPGX1, Vp2S01-Cmr, and water (blank control), respectively. M: 2 kb plus DNA ladder. (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of PirAvp and PirBvp. Lane 1: VcLMB29. Lane 2: VcLMB29-pVPGX1. Lane 3: Vp2S01. M: the PageRuler Prestained Protein Ladder.
Sequence Analysis of pVA1-type Plasmids From Donor and Transconjugant Cells
To determine if strain VcLMB29-pVPGX1 acquired the pVPGX1-Cmr, the bacterial genome was sequenced and compared with that of recipient strain VcLMB29. The result showed that the VcLMB29-pVPGX1 contains a large (70,053 bp, named as pVCON1) plasmid. This transconjugant was determined to be the recipient bacterium based on that its chromosomal sequence was 99.9% identical to that of VcLMB29. The pVCON1 displayed a 100% identity to pVPGX1-Cmr from donor Vp2S01-Cmr. Further comparative sequence analysis among the pVA1-type plasmids, including pVA1, pVPGX1, pVCON1, pVCGX1 and pLA16-2, revealed that pVCON1 sequence shared high (>99%) homologies with those in other pVA1-type plasmid (Figure 1). Interestingly, the smaller plasmid pVPGX2 (38 kb) from Vp2S01-Cmr was not found in VcLMB29-pVPGX1.
Ability of VcLMB29-pVPGX1 to cause AHPND in P. vannamei
From the laboratory bioassays, P. vannamei exposed to VcLMB29-pVPGX1 or Vp2S01-Cmr showed typical gross signs of AHPND within 6 h, displaying a pale and atrophied hepatopancreas (HP), and an empty stomach (ST) and midgut (MG) (Figure 3A); all the exposed shrimp died within 24 h (Figure 3B). The shrimp in the negative control and blank control groups had a normal size HP, dark orange in color, and a full ST and MG.
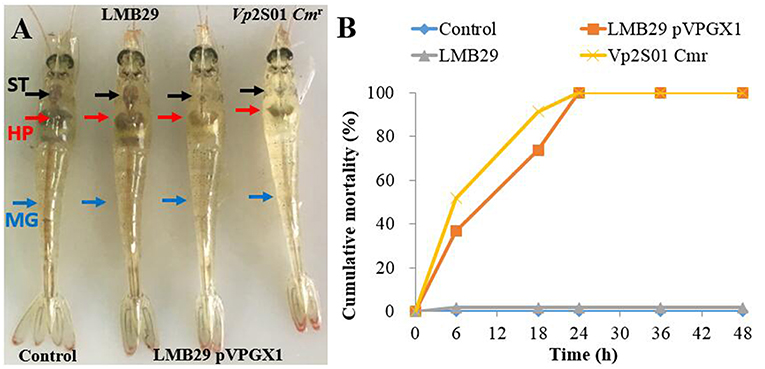
Figure 3. Gross signs and mortality of Penaeus vannamei exposed to AHPND-bacteria. (A) Gross signs of AHPND-affected shrimp. Normal shrimp in the blank control and the group infected with VcLMB29: a normal size hepatopancreas (HP) with dark orange color and a full stomach (ST) and midgut (MG). AHPND-affected shrimp from the group infected with VcLMB29-pVPGX1 and the group infected with Vp2S01-Cmr: pale, atrophied HP, and an empty stomach (ST) and midgut (MG). (B) Cumulative mortality of shrimp infected with VcLMB29-pVPGX1, shrimp were exposed to Virbio bacteria through immersion infection.
Histopathological examination of moribund shrimp infected with VcLMB29-pVPGX1 or Vp2S01-Cmr revealed typical AHPND lesions in the HP compare to that in the negative control and blank control groups (Figures 4A,B). These lesions were characterized by acute sloughing of epithelium cells in the HP tubules, followed by extensive necrosis in most tubules (Figures 4C,D). The PCR detection of Vibrio bacteria showed that V. parahaemolyticus could be detected in all groups and V. campbellii could be only detected in LMB29-infected shrimp and LMB29 pVPGX1-infected shrimp (Supplementary Figure 1). In addition, we investigated the expression of pirAvp and pirBvp in shrimp by RT-PCR using primers VpPirA-284F/R and VpPirB-392F/R. The result showed that bands of both amplificated products appeared at expected sizes of 284 bp from pirAvp and 392 bp from pirBvp in the sample of the shrimp infected with VcLMB29-pVPGX1 (Figure 5, lanes 4 and 9). There were no amplification products from the healthy shrimp and the shrimp infected with non-AHPND-causing V. campbellii strain VcLMB29 (Figure 5, lanes 2, 3 and 6, 7).
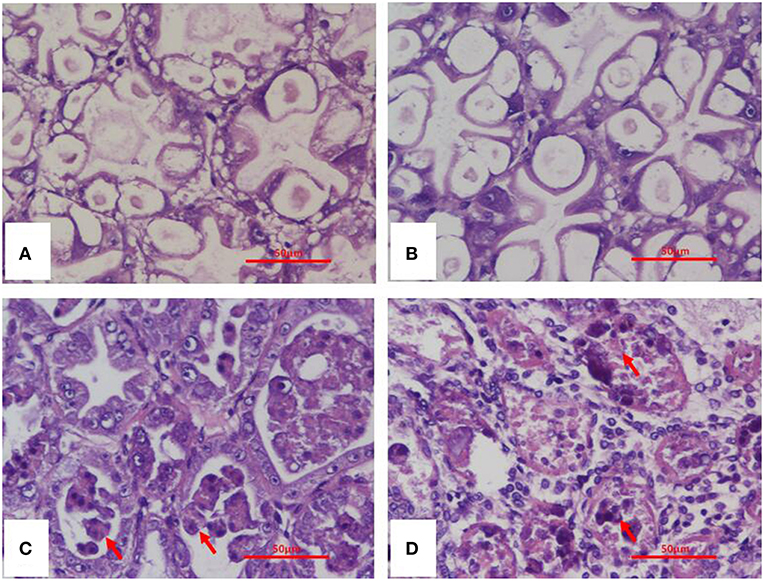
Figure 4. H&E stained histological sections of the hepatopancreas of Penaeus vannamei from AHPND-Vibrio bacteria challenge studies. (A) Healthy (blank-control) shrimp; (B) shrimp infected with VcLMB29; (C) shrimp infected with VcLMB29-pVPGX1; (D) shrimp infected with Vp2S01-Cmr. (A,B) normal histology of hepatopancreas; (C,D) AHPND histopathology characterized by sloughing epithelial cells of hepatopancreatic tubule (arrows). Scale bars = 50 μm.

Figure 5. RT-PCR detection of pirAvp and pirBvp mRNA. Total RNA extracted from tissues in cephalothorax containing hepatopancreas and stomach were analyzed by RT-PCR using primers VpPirA-284F/R (lanes 1–5) and primers VpPirB-392F/R (lanes 6–10). Lanes 1 and 6: non-template control, lanes 2 and 7: healthy shrimp, lanes 3 and 8: shrimp infected with VcLMB29, lanes 4 and 9: shrimp infected with VcLMB29-pVPGX1; lanes 5 and 10: shrimp infected with Vp2S01-Cmr. M: 2 kb plus DNA ladder.
Discussion
Horizontal gene transfer is a driver for diversification of pathogenic bacteria via mobile genetic elements (MGEs) including plasmids, transposons, insertion sequences (ISs), prophages, integrons and associated gene cassettes, integrative conjugative elements (ICEs), and genomic islands (GIs) (Antonenka et al., 2005; Bi, 2015). However, little is known in this regard in relation to pathogenic bacteria of crustaceans. This is the first study to definitively show conjugative transfer of the pVA1-type plasmid which exists in AHPND-Vibrio bacteria.
Initially, it was thought that the virulence of VpAHPND may be due to lysogenic infection of the V. parahaemolyticus genome with a bacteriophage (FAO, 2013), as lysogenic phages frequently harbor bacterial toxin genes. For example, V. cholera toxin is encoded by ctxAB genes residing on the CTXϕ phage (Waldor and Mekalanos, 1996). However, our study have found lysogenic phages in only two of five VpAHPND isolates (Wang et al., 2016), indicating that the AHPND toxin genes do not reside on a phage. Our previous study showed that pVA1-type plasmids from VpAHPND (Vp2S01) and VcAHPND (Vc3S01), isolated from a diseased pond, had a sequence similarity of 99.9% (Dong et al., 2017a). Then, in a subsequent study, we found that VoAHPND appeared in a tank where shrimp were being challenged with VcAHPND (Dong et al., 2019). Furthermore, pVA1-type plasmids can be found in all published sequences of AHPND-causing Vibrio strains (Table 3). All of these results suggested the possible horizontal transfer of pVA1-type plasmids from AHPND-Vibrio to non-pathogenic bacteria. In the current study, the transfer of the pVA1-type plasmid from Vp2S01-Cmr to V. campbellii was demonstrated through conjugation experiments.
The demonstration of the transfer of a virulent plasmid to non-pathogenic strains of Vibrio has practical relevance to the development of protocols for the diagnosis of AHPND and for strategies for prevention and control of the disease. For example, some shrimp hatcheries provide a quality analysis of postlarvae based on the number of V. parahaemolyticus and related bacteria. The procedure involves culturing Vibrio bacteria from hepatopancreas tissue homogenates on TCBS agar plates, in these tests, colonies of V. parahaemolyticus are green. The CFU of a particular sample is usually determined only by the counts of green colonies; however, this would miss virulent strains of other Vibrio species present, such as V. owensii, potentially reducing the effectiveness of this metric. In addition, some shrimp farms routinely use probiotics, many of which include or are contaminated with species of Vibrio, to control the disease and to improve water quality; however, increasing levels of Vibrio in the pond water could also increase the risk of the virulent plasmid being transferred and could subsequently increase the risk of AHPND. Also, antibiotics, antimicrobial peptides, and phages have been considered in aquaculture farms and hatcheries to control the bacterial diseases as prophylactic and therapeutic agents (Jun et al., 2016, 2018). If pVA1 transfers to other bacteria in the shrimp or pond environment, the new strains may not be susceptible to these treatments.
This study showed that pVA1-type plasmid could be conjugatively transferred with an efficiency of 10−8 in a 12 h period. Thus, such transfer events may be very frequent in diseased shrimp ponds, where numerous Vibrio spp. are present. The conjugation process can bring a high risk of developing the diversity and complexity for AHPND bacteria.
There are over 100 species of bacteria in the genus Vibrio. Of these, only 4 have been reported to contain pVA1-type plasmids. Three of the four species are closely related and in the harveyi-clade, and the fourth, V. punensis (Restrepo et al., 2018), is a more distant relative within the genus. This suggests that the number of potential recipient strains, those capable of receiving AHPND toxin genes and attaining virulence through horizontal transfer of a pVA1-type plasmid, is relatively small. This is not unexpected as, in general, the horizontal exchange of genetic material through conjugation occurs only between closely related individuals (Bernstein et al., 2018). Therefore, management of pond microbial ecology to reduce the prevalence of these potential recipient strains, such as through the use of probiotics to effect competitive exclusion, could prove to be a useful strategy to reduce the risks of AHPND. It would be useful to determine the range of bacterial species that are capable of being recipients for the pVA1-type plasmids. This can be studied through the use of established pVA1 plasmid transfer systems, both in vitro (described here) and in vivo (Dong et al., 2019). Recent studies have shown that the conjugation efficiency is affected by both duration and temperature (Wiesner et al., 2013; Liu y. et al., 2017). In order to effectively assess the risk, more information is needed on factors affecting transfer efficiency of pVA1-type plasmids in pond environments.
Data Availability
The datasets generated for this study can be found in Complete plasmid sequence of pVCON1 from strain VcLMB29-pVPGX1 has been deposited in GenBank under the accession MH890610.
Author Contributions
The experiments, data analysis, and manuscript writing were performed by JS, XD, and JH. HW isolated and identified the bacterial strain 20130629003S01 and provide the Vibrio campbellii strain 20130629003S01, V. parahaemolyticus strain 20130629002S01 and revised the manuscript. JS, JC, WW, GW, and YR performed experiments. XW and KT discussed the results and revised the manuscript. DB provided vital guidance, technical support, and proofreading for the work. All authors read and approved the final manuscript.
Funding
This work was supported by the projects under the National Natural Science Foundation of China (31802342), Opening Fund of Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology (2017-4B02), Pilot National Laboratory for Marine Science and Technology (Qingdao) (QNLM201706), the Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. 2018SDKJ0502), and China Agriculture Research System (CARS-48).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Xiaohua Zhang for the strains of VcHHS18 and VcVIB285. We thank Prof. Zhe Zhao for the strain of VcLMB29. We thank Prof. Zhaolan Mo for her kind support.
Footnotes
1. ^https://clsi.org/standards/products/microbiology/documents
References
Antonenka, U., Nolting, C., Heesemann, J., and Rakin, A. (2005). Horizontal transfer of Yersinia high-pathogenicity island by the conjugative RP4 attB target-presenting shuttle plasmid. Mol. Microbiol. 57, 727–734. doi: 10.1111/j.1365-2958.2005.04722.x
Bell, T. A., and Lightner, D. V. (1988). A Handbook of Normal Penaeid Shrimp Histology. Baton Rouge, LA: World Aquaculture Society.
Bernstein, H., Bernstein, C., and Michod, R. E. (2018). Sex in microbial pathogens. Infect. Genet. Evol. 57, 8–25. doi: 10.1016/j.meegid.2017.10.024
Bi, D. (2015). Study on the mobile genome of multidrug-resistant Klebsiella pneumoniae [dissertation/doctor's thesis]. Shanghai Jiao Tong University, Shanghai, China.
de la Pena, L. D., Cabillon, N. A., Catedral, D. D., Amar, E. C., Usero, R. C., Monotilla, W. D., et al. (2015). Acute hepatopancreatic necrosis disease (AHPND) outbreaks in Penaeus vannamei and P. monodon cultured in the Philippines. Dis. Aquat. Organ. 116, 251–254. doi: 10.3354/dao02919
Dong, X., Bi, D., Wang, H., Zou, P., Xie, G., Wan, X., et al. (2017a). pirABvp -bearing Vibrio parahaemolyticus and Vibrio campbellii pathogens isolated from the same AHPND-affected pond possess highly similar pathogenic plasmids. Front. Microbiol. 8:1859. doi: 10.3389/fmicb.2017.01859
Dong, X., Chen, J., and Song, J. (2019). Evidence of the horizontal transfer of pVA1-type plasmid from AHPNDcausing V. campbellii to non-AHPND V. owensii. Aquaculture 503, 396–402. doi: 10.1016/j.aquaculture.2019.01.016
Dong, X., Wang, H., Xie, G., Zou, P., Guo, C., Liang, Y., et al. (2017b). An isolate of Vibrio campbellii carrying the pirVP gene causes acute hepatopancreatic necrosis disease. Emerg. Microbes Infect. 6:e2. doi: 10.1038/emi.2016.131
Dong, X., Wang, H., Zou, P., Chen, J., Liu, Z., Wang, X., et al. (2017c). Complete genome sequence of Vibrio campbellii strain 20130629003S01 isolated from shrimp with acute hepatopancreatic necrosis disease. Gut Pathog. 9:31. doi: 10.1186/s13099-017-0180-2
FAO (2013). Report of the FAO/MARD Technical Workshop on Early Mortality Syndrome (EMS) or Acute Hepatopancreatic Necrosis Syndrome (AHPNS) of Cultured Shrimp (Under TCP/VIE/3304). FAO Fisheries and Aquaculture Report No. 1053, Rome, 54.
Flegel, T. W., and Lo, C. F. (2014). Free Release of Primers for Specific Detection of Bacterial Isolates That Cause Acute Hepatopancreatic Necrosis Disease (AHPND). Network of Aquaculture Centres in Asia-Pacific (Bangkok).
Gomez-Gil, B., Soto-Rodriguez, S., Lozano, R., and Betancourt-Lozano, M. (2014). Draft genome sequence of Vibrio parahaemolyticus strain M0605, which causes severe mortalities of shrimps in Mexico. Genome Announc. 2:e00055-14. doi: 10.1128/genomeA.00055-14
Gomez-Jimenez, S., Noriega-Orozco, L., Sotelo-Mundo, R. R., Cantu-Robles, V. A., Cobian-Guemes, A. G., Cota-Verdugo, R. G., et al. (2014). High-quality draft genomes of two Vibrio parahaemolyticus strains aid in understanding acute hepatopancreatic necrosis disease of cultured shrimps in Mexico. Genome Announc. 2:e00800-14. doi: 10.1128/genomeA.00800-14
Haldar, S., Neogi, S. B., Kogure, K., Chatterjee, S., Chowdhury, N., Hinenoya, A., et al. (2010). Development of a haemolysin gene-based multiplex PCR for simultaneous detection of Vibrio campbellii, Vibrio harveyi and Vibrio parahaemolyticus. Lett. Appl. Microbiol. 50, 146–152. doi: 10.1111/j.1472-765X.2009.02769.x
Han, J. E., Tang, K. F., Tran, L. H., and Lightner, D. V. (2015). Photorhabdus insect-related (Pir) toxin-like genes in a plasmid of Vibrio parahaemolyticus, the causative agent of acute hepatopancreatic necrosis disease (AHPND) of shrimp. Dis. Aquat. Org. 113, 33–40. doi: 10.3354/dao02830
Han, J. E., Tang, K. F. J., Aranguren, L. F., and Piamsomboon, P. (2017). Characterization and pathogenicity of acute hepatopancreatic necrosis disease natural mutants, pirABvp (-) V. parahaemolyticus, and pirABvp (+) V. campbellii strains. Aquaculture 470, 84–90. doi: 10.1016/j.aquaculture.2016.12.022
Hossain, M. T., Kim, E. Y., Kim, Y. R., Kim, D. G., and Kong, I. S. (2012). Application of groEL gene for the species-specific detection of Vibrio parahaemolyticus by PCR. Lett. Appl. Microbiol. 54, 67–72. doi: 10.1111/j.1472-765X.2011.03174.x
Jackman, S. D., Vandervalk, B. P., Mohamadi, H., Chu, J., Yeo, S., Hammond, S. A., et al. (2017). ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 27, 768–777. doi: 10.1101/gr.214346.116
Jun, J. W., Han, J. E., Giri, S. S., Tang, K. F. J., Zhou, X., Aranguren, L. F., et al. (2018). Phage application for the protection from acute hepatopancreatic necrosis disease (AHPND) in Penaeus vannamei. Indian J. Microbiol. 58, 114–117. doi: 10.1007/s12088-017-0694-9
Jun, J. W., Han, J. E., Tang, K. F. J., Lightner, D. V., Kim, J., Seo, S. W., et al. (2016). Potential application of bacteriophage pVp-1: agent combating Vibrio parahaemolyticus strains associated with acute hepatopancreatic necrosis disease (AHPND) in shrimp. Aquaculture 457, 100–103. doi: 10.1016/j.aquaculture.2016.02.018
Kondo, H., Tinwongger, S., Proespraiwong, P., Mavichak, R., Unajak, S., Nozaki, R., et al. (2014). Draft genome sequences of six strains of Vibrio parahaemolyticus isolated from early mortality syndrome/acute hepatopancreatic necrosis disease shrimp in Thailand. Genome Announc. 2:e00221-14. doi: 10.1128/genomeA.00221-14
Kondo, H., Van, P. T., Dang, L. T., and Hirono, I. (2015). Draft genome sequence of non-Vibrio parahaemolyticus acute hepatopancreatic necrosis disease strain KC13.17.5, isolated from diseased shrimp in Vietnam. Genome Announc. 3:e00978-15. doi: 10.1128/genomeA.00978-15
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Lee, C. T., Chen, I. T., Yang, Y. T., Ko, T. P., Huang, Y. T., Huang, J. Y., et al. (2015). The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. U.S.A. 112, 10798–10803. doi: 10.1073/pnas.1503129112
Liu, J., Zhao, Z., Deng, Y., Shi, Y., Liu, Y., Wu, C., et al. (2017). Complete genome sequence of Vibrio campbellii LMB 29 isolated from red drum with four native megaplasmids. Front. Microbiol. 8:2035. doi: 10.3389/fmicb.2017.02035
Liu, L., Xiao, J., Xia, X., Pan, Y., Yan, S., and Wang, Y. (2015). Draft genome sequence of Vibrio owensii strain SH-14, which causes shrimp acute hepatopancreatic necrosis disease. Genome Announc. 3:e01395-15. doi: 10.1128/genomeA.01395-15
Liu, L. Y., Xiao, J. Z., Zhang, M. M., Zhu, W. Y., Xia, X. M., Dai, X. L., et al. (2018). A Vibrio owensii strain as the causative agent of AHPND in cultured shrimp, Litopenaeus vannamei. J. Invertebr. Pathol. 153, 156–164. doi: 10.1016/j.jip.2018.02.005
Liu, Y., Gao, Y. N., Liu, X. H., Liu, Q., Zhang, Y. X., Wang, Q. Y., et al. (2017). Transposon insertion sequencing reveals T4SS as the major genetic trait for conjugation transfer of multi-drug resistance pEIB202 from Edwardsiella. BMC Microbiol. 17:112. doi: 10.1186/s12866-017-1013-7
Nunan, L., Lightner, D., Pantoja, C., and Gomez-Jimenez, S. (2014). Detection of acute hepatopancreatic necrosis disease (AHPND) in Mexico. Dis. Aquat. Org. 111, 81–86. doi: 10.3354/dao02776
Restrepo, L., Bayot, B., Arciniegas, S., Bajana, L., Betancourt, I., Panchana, F., et al. (2018). pirVP genes causing AHPND identified in a new Vibrio species (Vibrio punensis) within the commensal Orientalis clade. Sci. Rep. 8:13080. doi: 10.1038/s41598-018-30903-x
Shinn, A. P., Pratoomyot, J., Griffiths, D., Trong, T. Q., Vu, N. T., Jiravanichpaisal, P., et al. (2018). Asian shrimp production and the economic costs of disease. Asian Fish. Sci. 31S, 29–58. Available online at: www.asianfisheriessociety.org/publication/archivedetails.php?id=152&q=1
Tran, L., Nunan, L., Redman, R. M., Mohney, L. L., Pantoja, C. R., Fitzsimmons, K., et al. (2013). Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Org. 105, 45–55. doi: 10.3354/dao02621
Waldor, M. K., and Mekalanos, J. J. (1996). Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272, 1910–1914. doi: 10.1126/science.272.5270.1910
Wang, J., Jiang, J., Zhang, H., Wang, J., Cai, H., Li, C., et al. (2011). Integrated transcriptional and proteomic analysis with in vitro biochemical assay reveal the important role of CYP3A46 in T-2 toxin hydroxylation in porcine primary hepatocytes. Mol. Cell. Proteomics 10:M111008748. doi: 10.1074/mcp.M111.008748
Wang, N., Wang, H. L., Bai, N., Huang, J., Xie, D. X., Xie, Q. L., et al. (2016). Isolation of lysogenic phage in Vibrio parahaemolyticus and its relationship with the pathogenicity of the host bacteria [in Chinese]. Prog. Fishery Sci. 37, 105–110. doi: 10.11758/yykxjz.20150417002
Wiegand, I., Hilpert, K., and Hancock, R. E. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Wiesner, M., Fernández-Mora, M., Cevallos, M. A., Zavala-Alvarado, C., Zaidi, M. B., Calva, E., et al. (2013). Conjugative transfer of an IncA/C plasmid-borne blaCMY−2 gene through genetic re-arrangements with an IncX1 plasmid. BMC Microbiol. 13:264. doi: 10.1186/1471-2180-13-264
Xiao, J. Z., Liu, L. Y., Ke, Y. Y., Li, X. F., Liu, Y. F., Pan, Y. J., et al. (2017). Shrimp AHPND-causing plasmids encoding the PirAB toxins as mediated by pirAB-Tn903 are prevalent in various Vibrio species. Sci. Rep. 7:42177. doi: 10.1038/srep42177
Zhang, B., Liu, F., Bian, H., Liu, J., Pan, L., and Huang, J. (2012). Isolation, identification, and pathogenicity analysis of a Vibrio parahaemolyticus strain from Litopenaeus vannamei [in Chinese]. Prog. Fishery Sci. 33, 56–62. Available online at: https://www.researchgate.net/publication/281080473_Isolation_identification_and_pathogenicity_analysis_of_a_Vibrio_parahaemolyticus_strain_from_Litopenaeus_vannamei_yizhufannabinduixiabingyuanjundefenlijiandingjiqizhibinglifenxi
Keywords: acute hepatopancreatic necrosis disease (AHPND), conjugative transfer, pVA1-type plasmid, pirABvp genes, bacterial diversity, AHPND-causing Vibrio bacteria
Citation: Dong X, Song J, Chen J, Bi D, Wang W, Ren Y, Wang H, Wang G, Tang KFJ, Wang X and Huang J (2019) Conjugative Transfer of the pVA1-Type Plasmid Carrying the pirABvp Genes Results in the Formation of New AHPND-Causing Vibrio. Front. Cell. Infect. Microbiol. 9:195. doi: 10.3389/fcimb.2019.00195
Received: 09 April 2019; Accepted: 21 May 2019;
Published: 07 June 2019.
Edited by:
Jyl S. Matson, University of Toledo, United StatesReviewed by:
Parin Chaivisuthangkura, Srinakharinwirot University, ThailandJinzhou Xiao, Shanghai Ocean University, China
Copyright © 2019 Dong, Song, Chen, Bi, Wang, Ren, Wang, Wang, Tang, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuan Dong, ZG9uZ3h1YW4mI3gwMDA0MDt5c2ZyaS5hYy5jbg==; Jie Huang, aHVhbmdqaWUmI3gwMDA0MDt5c2ZyaS5hYy5jbg==
†These authors have contributed equally to this work
 Xuan Dong
Xuan Dong Jipeng Song
Jipeng Song Jiayuan Chen1,2
Jiayuan Chen1,2 Dexi Bi
Dexi Bi Xuepeng Wang
Xuepeng Wang