
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 15 May 2019
Sec. Molecular Bacterial Pathogenesis
Volume 9 - 2019 | https://doi.org/10.3389/fcimb.2019.00160
In pathogens that produce lipooligosaccharide (LOS), sugar residues within the surface-exposed LOS outer core mediate interactions with components of the host immune system, promoting bacterial infection. Many LOS structures are controlled by phase variation mediated by random slipped-strand base mispairing, which can reversibly switch gene expression on or off. Phase variation diversifies the LOS, however its adaptive role is not well-understood. Nontypeable Haemophilus influenzae (NTHi) is an important pathogen that causes a range of illnesses in the upper and lower respiratory tract. In NTHi a phase variable galactosyltransferase encoded by lic2A initiates galactose chain extension of the LOS outer core. The donor substrate for Lic2A, UDP-galactose, is generated from UDP-glucose by UDP-galactose epimerase encoded by galE. Our previous fitness profiling of H. influenzae mutants in a murine lung model showed that the galE mutant had a severe survival defect, while the lic2A mutant's defect was modest, leading us to postulate that unidentified factors act as suppressors of potential defects in a lic2A mutant. Herein we conducted a genome-wide genetic interaction screen to identify genes epistatic on lic2A for survival in the murine lung. An unexpected finding was that galE mutants exhibited restored virulence properties in a lic2A mutant background. We identified an alternative antibody epitope generated by Lic2A in the galE mutant that increased sensitivity to classical complement mediated killing in human serum. Deletion of lic2A or restoration of UDP-galactose synthesis alleviated the galE mutant's virulence defects. These studies indicate that when deprived of its galactosyl substrate, Lic2A acquires an alternative activity leading to increased recognition of NTHi by IgM and decreased survival in the lung model. Biofilm formation was increased by deletion of galE and by increased availability of UDP-GlcNAc precursors that can compete with UDP-galactose production. NTHi's ability to reversibly inactivate lic2A by phase-variation may influence survival in niches of infection in which UDP-Galactose levels are limiting.
Lipooligosaccharide (LOS), a short chain form of lipopolysaccharide (LPS) that lacks the long repetitive polysaccharide O-antigen of LPS, is the principal component of the outer-membrane of bacteria that produce it, mediating interactions with host immune defenses that determine the outcome of infection. Haemophilus influenzae, a Gram-negative bacterium that colonizes the human respiratory tract, causes a range of illnesses including otitis media, sinusitis, and exacerbation of chronic obstructive pulmonary disease (Klein, 1997; Murphy et al., 2004; Wiertsema et al., 2011; Ahearn et al., 2017). In H. influenzae, LOS structure varies between strains, yet several features are conserved such as lipid A, an inner core usually composed of a single 3-deoxy-D-manno-octulosonic acid linked to three heptose residues, and an outer core containing a short heteropolymer of glucose and galactose residues in different configurations extending from the heptosyl residues (Moxon and Murphy, 2000). Glucose and galactose extensions of the outer core often terminate in host-like structures including sialic acid, phosphorylcholine (PC), and N-acetylgalactosamine (Hood et al., 1999; Risberg et al., 1999) mediating numerous aspects of immune evasion. Sialylated LOS is critical for colonization of the middle ear and nasopharynx in a chinchilla model of otitis media (Bouchet et al., 2003) as well as in a gerbil middle ear infection model and a rat pulmonary model (Swords et al., 2004). PC modification of LOS promotes persistence in the respiratory tract in an infant rat model of infection (Weiser et al., 1998), and mutants deficient in the N-acetylgalactosamine structure exhibit reduced lethality in a mouse model of bacteremia (Hood et al., 1996).
In the H. influenzae Rd strain and NTHi strain 375, the lic2A encoded galactosyltransferase initiates galactose chain extension by addition of a β-galactose in a 1,4-linkage to a glucose appending from heptose III of the inner core (Hood et al., 1999, 2001). UDP-glucose 4-epimerase encoded by galE interconverts endogenous UDP-glucose (UDP-Glc) to UDP-galactose (UDP-Gal) (Frey, 1996), which then serves as donor of galactosyl units for biosynthesis of LOS outer core structures of H. influenzae, including the lic2A dependent extension (Maskell et al., 1992). From the glucose on Hep I, extensions containing several alternative “cryptic” LOS structures have also been identified that vary between strains and environmental conditions (Hood et al., 2004b; Figure 1). In our previous studies we utilized genome-wide mutant fitness analyses of H. influenzae Rd in the murine lung, a model of bacterial clearance that detects genes required for immune evasion and other determinants of host-pathogen interactions. Our approach, termed HITS (High-throughput Insertion Tracking by deep-Sequencing), revealed that mutations in galE conferred a severe survival defect relative to other potential virulence determinants. Surprisingly, mutations in lic2A conferred modest or insignificant defects (Gawronski et al., 2009; Wong et al., 2013). Because structural analysis of the major LOS species in Rd indicates that lic2A is required for addition of the initial galactose unit to a glucose extension in the outer core (Risberg et al., 1999; Hood et al., 2001), a similar LOS structure might be predicted for both the galE and lic2A mutants, in contrast to their very different in vivo survival levels identified by HITS in the murine model. It is possible that other LOS genes are functionally redundant with the lic2A gene during infection. For example, some cryptic LOS structures contain galactose, and therefore may contribute to the galE phenotype. As the cryptic structures vary under different environmental conditions and possess several alternative forms (Hood et al., 2004b), it is unclear which, if any, of them relate to these observations. We postulated that unidentified factors such as other complementary LOS genes or other genetic interactions act as suppressors of potential virulence defects in the lic2A mutant.
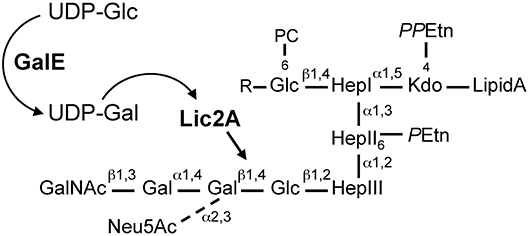
Figure 1. H. influenzae Rd LOS. GalE generates UDP-Gal for galactose extension by Lic2A. Diagram is based on structural information as described (Risberg et al., 1999; Hood et al., 2001, 2004b). R, tetrasaccharide extensions, Neu5Ac-α2,3-Gal-β1,4-GlcNAc-β1,3-Gal or PEtn-6-GalNAc-α1,6-Gal-β1,4-GlcNAc-β1,3-Gal. Glc, glucose; Gal, galactose; UDP-Glc, UDP-glucose; UDP-Gal, UDP-galactose; UDP, uridine diphosphate; Neu5Ac, N-acetylneuraminic acid; GalNAc, N-acetylgalactosamine; PC, phosphorylcholine; Hep, heptose; Kdo, 2-keto-3-deoxyoctulosonic acid; PPEtn, pyrophosphoethanolamine; PEtn, phosphoethanolamine.
In this report, we undertook a functional genomics approach using a genetic interaction screen to identify genes epistatic on lic2A that can account for the difference in the survival profiles between the galE and lic2A mutants. Specifically, this approach identifies secondary mutations that increase or decrease virulence phenotypes associated with deletion of lic2A. Our results implicated a diversity of genes in virulence when lic2A is deleted, including genes that influence the synthesis of cryptic LOS glycoforms, in addition to an unexpected genetic interaction in which deletion of lic2A specifically alleviated much of the requirement for galE in survival of NTHi in the lung. The results support a model in which an alternative LOS configuration controlled by precursor production levels modulates virulence properties of NTHi.
The non-encapsulated Rd derivative of H. influenzae type d (RdAW) (Akerley et al., 2002), and nontypeable H. influenzae (NTHi) clinical isolates 375 (NTHi375) (Bouchet et al., 2003) and NT127 (Wong et al., 2013) were grown at 35°C ± 1.5°C and 5% CO2 in Brain Heart Infusion supplemented with 10 μg/ml NAD and 10 μg/ml hemin (sBHI) on agar plates or in sBHI broth. Strains were also grown in a chemically defined medium (MIc) or on MIc agar as described (Barcak et al., 1991). DNA was transformed into naturally competent H. influenzae as described (Barcak et al., 1991). Kanamycin (Km), gentamicin (Gm), erythromycin (Erm), tetracycline (Tet), 3,4-cyclohexenoesculetin-β-D-galactopyranoside (S-Gal), and D-xylose were added to sBHI at 20 μg/ml, 15 μg/ml, 10 μg/ml, 3 μg/ml, 300 μg/L, and 1 mM, respectively. S-Gal is a chromogenic substrate for β-galactosidase for detection of bacterial colonies with the lac+ phenotype; D-xylose is used for induction of the lacZ gene encoding β-galactosidase under the xylA promoter at the xyl locus (Wong et al., 2007). MIc agar contains sialic acid (SA) at 25 μg/ml supplemented (MIcSA) with or without 0.5 or 1% galactose.
Standard methods were used for strain and plasmid construction (Ausubel et al., 1995). Strains are listed in Table 1, and primer sequences in Table S1. Replacing the coding region of interest with the appropriate antibiotic resistance cassette via transformation with PCR products containing each gene knockout generated nonpolar deletion mutants in H. influenzae. Complementing strains were created by cloning the gene of interest for complementation into allelic exchange vector pXT10 for integration at xyl of the desired mutant recipients (Wong and Akerley, 2003). Detailed description of strain construction and other strains/plasmids is provided in Supplementary Text S1.
Absorption of sera was performed as described (Loeb, 1984) with modifications (Supplementary Text S1), using strains RdgalElic2A/lic2 and RdgalElic2A grown on MIc agar containing sialic acid (25 μg/ml) (MIcSA).
Bacteria grown on MIcSA overnight were resuspended at ~0.4 OD600 units in Hank's Balanced Salt Solution with calcium and magnesium chloride (HBSS++) and NuPAGE LDS Sample buffer containing lithium dodecyl sulfate (ThermoFisher). Whole-cell lysates treated with or without 1 mg/ml proteinase K were separated on Invitrogen NuPAGE 4–12% Bis-Tris Protein Gels (ThermoFisher) using 2-(N-morpholino) ethanesulfonic acid (MES) running buffer, pH 7, transferred to PVDF membranes (Millipore) and blocked with TBS-0.1% Tween 20 containing 1% dry milk for 30 min at 24°C. Membranes were incubated with serum pre-absorbed with strains RdgalElic2A/lic2 (GalE−, Lic2A+) or RdgalElic2AV (GalE−, Lic2A−) diluted 1:120 in TBS-0.1% Tween 20 for 17 h at 4°C on a rotary shaker. Membrane-bound IgM and IgG were detected using alkaline phosphatase-conjugated anti-human IgM and IgG (Sigma) at a 1:1,000 dilution in TBS-0.1% Tween 20 and 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NPT) purple liquid substrate system (Sigma). Coomassie Blue staining of undigested samples was performed with a Colloidal Blue Staining Kit (ThermoFisher). Size markers were SeeBlue Plus2 pre-stained protein standards.
Bacteria were harvested after overnight growth on MIcSA with or without 0.5 or 1% galactose and resuspended in HBSS++ for serum bactericidal assays as previously described (Wong et al., 2011), with minor modifications (Supplementary Text S1). In designated experiments, 10 mM EGTA and 10 mM magnesium chloride (MgCl2) were added to NHS to selectively block the classical and lectin, but not the alternative complement pathway (Platts-Mills and Ishizaka, 1974; Des Prez et al., 1975). Bactericidal assays using strain specific absorbed sera were performed as described above except IgG/IgM antibody depleted human complement (HC) pooled serum (Pel-Freez Biologicals) was used as a source of active complement. Percent survival is calculated as indicated in the figure legends.
Rd and NTHi 375 strains were grown and harvested as described for the serum bactericidal assays above. Binding of serum antibodies to bacteria incubated with either NHSΔi or absorbed NHSΔi was detected with anti-human IgM and IgG conjugated to fluorescein isothiocyanate (FITC) (SouthernBiotech) on a NovoCyte flow cytometer with analysis using NovoExpress software (ACEA Biosciences, Inc., San Diego, CA). The reported median fluorescence intensities (MFI) of bacterial populations in replicate samples were obtained after subtraction of the background FITC-conjugated antibody only fluorescence values obtained for each strain.
Biofilm formation was assessed by the microtiter plate assay (O'toole, 2011). NTHi 375 strains grown to log phase in sBHI were diluted to OD600 of 0.01 in sBHI and seeded at 200 μl per well in replicates in polystyrene 96-well flat bottom microtiter plates (Corning Incorporated, Kennebunk, ME) in the presence of 0.1% galactose, 25 μg/ml sialic acid, 0.1% N-acetylglucosamine or in combinations of the three reagents for 24 hr at 36°C with 5% CO2. After assessing growth by measuring optical density at 600 nm, cultures were decanted, and biofilms quantified by crystal violet staining detected as absorbance at 595 nm, as previously described (O'toole, 2011).
Single strain colonization and in vivo competition assays were essentially as described previously (Gawronski et al., 2009; Wong et al., 2013; Supplementary Text S1). This model measures survival of H. influenzae in the otherwise healthy mammalian lung, resulting in transient weight loss and morbidity with clearance by approximately day 4. Attenuated mutants in this model are more rapidly eliminated, indicating loss of potential virulence genes. For in vivo selection of the H. influenzae Rd transposon insertion libraries in strains Δrep (parent) and Δreplic2A (isogenic lic2A deletion mutant), genomic DNA representing 105 mutants (Gawronski et al., 2009) was used to transform these two strains to KmR or KmRGmR to generate mutant banks termed parent library and lic2A library, respectively. We used the Δrep strain containing licA that is phase-on locked for LOS PC epitope to prevent phase variation at this locus, which has been shown previously to influence phenotypes of lic2A mutants (Langereis and Weiser, 2014). Forty microliter containing 107 CFU of each transposon insertion library was inoculated into the nares of five mice per strain. The remaining portion of the inoculum was processed for genomic DNA representing the input libraries. Output libraries were recovered from lungs at ~24 h post-infection, grown on sBHI agar plates and collected for genomic DNA isolation for transposon insertion mapping analysis. Experiments were conducted with approval and in accordance with guidelines of the Institutional Animal Care and Use Committee at the University of Mississippi Medical Center (Jackson, MS).
Genomic DNA from parent and lic2A libraries was isolated pre- and post-selection and sequenced using true single molecule DNA sequencing (tSMS) technology developed by SeqLL (Woburn, MA), formerly Helicos BioSciences (Cambridge, MA) (Thompson et al., 2012). The tSMS reads were aligned to the H. influenzae Rd KW20 genome sequence, GenBank: L42023.1 (National Center for Biotechnology Information, NCBI) using HeliSphere software (SeqLL) with the “best-only” setting and other parameters taken as defaults, assigned to genes using a revised version of NCBI protein table NC_000907 and filtered to remove reads corresponding to more than one location on the genome as described previously (Wong et al., 2013). The mariner transposon Himar1 inserts specifically into TA dinucleotides in target DNA (Lampe et al., 1996; Rubin et al., 1999). In the input parent and lic2A mutant banks, 64,348 (~50%) and 55,028 (~42%) of the 131,955 total possible chromosomal TA target sites were found to have sustained unique insertions, respectively. The number of sequencing reads mapping to the genome resulted in averages of 27-fold and 12-fold coverage of each TA insertion site within the parent and lic2A mutant libraries, respectively. Of the 1,724 predicted or annotated genes in the genome, we excluded 443 (from parent library) and 481 (from lic2A mutant library) loci essential for growth in vitro from our analysis as well as duplicated genes, genes with ≤ to 5 reads and those with only one TA insertion within the first 95% of their coding regions. Additional detailed analyses including classification of genes based on their fitness profiles, assigning gene essentiality and threshold requirements for genes considered to contribute to fitness during lung infection is provided in Supplementary Text S1.
A possibility to account for the difference in survival phenotypes of lic2A and galE mutants in the lung model would be the presence of other genes that have functions redundant with lic2A. In order to identify gene(s) in this category, we used HITS to examine the fitness of a large bank of mariner transposon insertion mutants (~75,000) in a lic2A deletion background vs. the control parental strain pre- and post-selection in the mouse lung model. This approach can unmask gene(s) that become required in the absence of lic2A during infection, potentially accounting for the difference in the survival profiles between the galE and lic2A mutants we observed previously. Moreover, the lic2A gene is subject to reversible inactivation by phase variation mediated by 5′-CAAT-3′ repeats in its coding region (High et al., 1996), and this approach can reveal genes that may be required during natural infection in which lic2A can reversibly switch off by this mechanism.
After excluding duplicated genes and genes with few insertions including those that are essential in vitro, the remaining 1,236 and 1,197 genes in the parent and lic2A mutant libraries, respectively, were further analyzed for roles in fitness in the mouse lung model (Figure 2A) (Materials and Methods and Supplementary Text S1). Fitness is expressed as a survival index (s.i.) representing the ratio of insertions detected after infection (output) to those detected in the in vitro grown inoculum (input). Values of s.i. <0.2 and p < 0.001 were set as requirements for genes to be considered to contribute to fitness during infection, resulting in 71 genes identified as required for infection in the parent, and 91 in the lic2A mutant bank dataset, of which 51 were required in both. A further cutoff of a fold difference of s.i. >3 between the two mutant banks was set as the criterion for a gene to be considered required for survival in one library but not the other, resulting in 3 and 29 genes that are considered uniquely required in the lung model by either the parent or lic2A mutant, respectively (Figures 2B,C). S.i. values for galE (0.04) and lic2A (0.8) in the parent library were consistent with our previous s.i. results for galE (0.025, 0.03) and lic2A (0.4, 1) in the mouse lung model (Gawronski et al., 2009; Wong et al., 2013). HITS data for all protein coding genes are listed in Table S2 and in vitro essential genes identified using the EL-ARTIST pipeline (Pritchard et al., 2014) are listed in Table S3. As expected, and further validating the method, 90% of genes within the common core set of 51 required for both parent and in the lic2A mutant background (Table S4) were found in our previous HITS results for H. influenzae infection in the murine lung model and have been discussed previously (Gawronski et al., 2009; Wong et al., 2013).
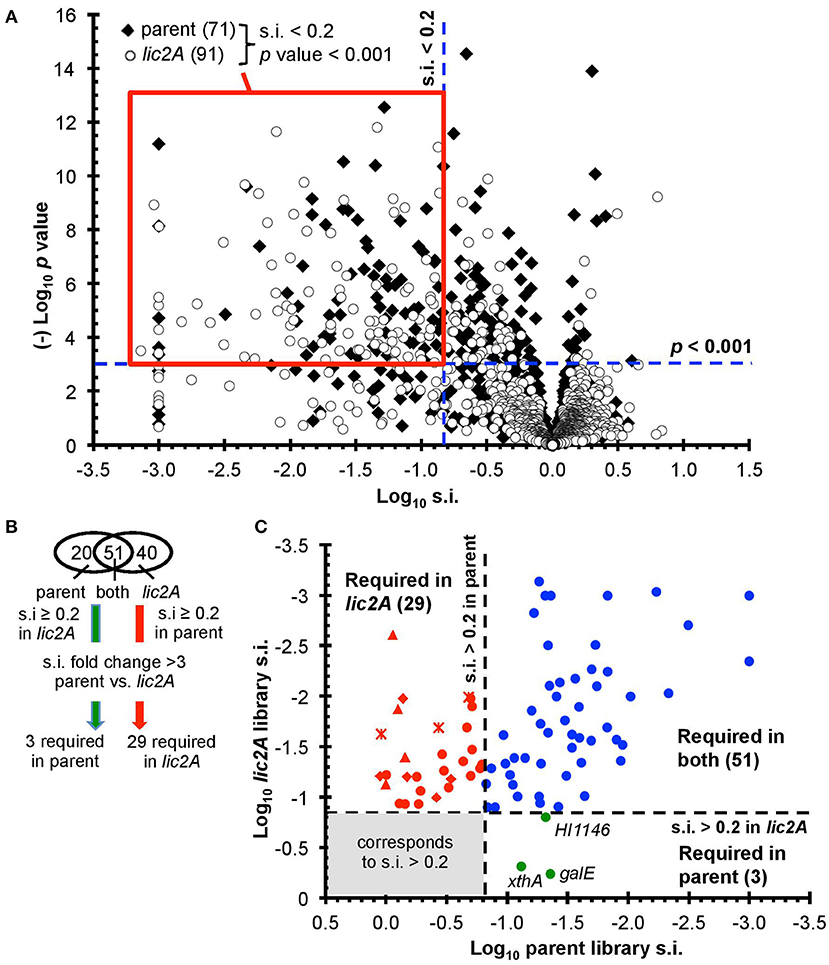
Figure 2. Genome-wide genetic interaction screen via HITS evaluating fitness of H. influenzae mutants in a lic2A deletion background in a mouse lung model. (A) Mutant fitness (survival index, s.i.) of candidate virulence genes in parent and lic2A deletion with no observed effects on fitness in vitro was plotted against significance (p-value). Red box demarcates the genes of interest meeting the requirements for further analysis (s.i. <0.2, p-value <0.001). (B) Venn diagram displaying these genes of interest sorted into categories. Further criteria were imposed to identify candidate genes required for infection in parent, lic2A only or in both. (C) Fitness of the transposon mutants attenuated in the parent or lic2A or in both was plotted against each other, highlighting their differential s.i. phenotypes. The 3 genes required for parent alone (green) or 29 in lic2A alone (red) are displayed (lsg, crosses; lic1, triangles; yrb, diamonds) (Table 2). The 51 genes required in both (blue) in vivo conditions are listed in Table S4.
Importantly, deletion of lic2A changed the genes required in the lung and yielded two categories of interest: mutations that resulted in survival defects only in the lic2A mutant background, and mutations that enhanced survival in this mutant but caused attenuation in the parent (Table 2). Several genes that were required only in the lic2A deletion background have roles in influencing the cell envelope integrity or changes in LOS structure: e.g., dacA encoding penicillin-binding protein 5 involved in peptidoglycan biosynthesis (Sauvage et al., 2008), the lic1 locus (licABCD) required for phosphorylcholine modification of the LOS in H. influenzae (Weiser et al., 1997), the lsgCDE genes involved in carbohydrate extension of the cryptic LOS structures (Hood et al., 2004b; Johansen et al., 2010) and the vacJ/yrbCDEF genes, encoding an ABC transport system that functions in maintenance of outer membrane lipid asymmetry (Malinverni and Silhavy, 2009) and influences the spatial arrangement of LOS in NTHi (Nakamura et al., 2011). Other genes in this category had annotated functions in DNA recombination/repair (recG, xerC), DNA recombination with regulatory roles (fis, ihfA), protein turnover/translation (lon, deaD), energy metabolism (aspA, pgi), L-tryptophan biosynthesis (trpG), transport of phosphate (pstA) and iron (hitC), and regulatory roles (sspA, phoB). Three genes were required in the parental strain background, but not required in the lic2A deletion background, indicating an unexpected suppression of virulence defects by the lic2A mutation. These genes have known or proposed functions in DNA repair (xthA), RNA degradation (HI1146, putative rapZ-like homolog) and LOS biosynthesis (galE).
Several genes of the lsg locus encoding glycosyltransferases were implicated in our genetic screen, and may have redundant functions with lic2A that could be responsible for suppressing in vivo defects of lic2A mutants. The HITS data indicate that mutations in these lsg genes in the lic2A mutant background gave rise to low survival indices suggesting their requirement in the absence of lic2A. As complement evasion is an important defense mechanism of H. influenzae, bactericidal assays with normal human serum (NHS) were used to evaluate whether these genes confer complement resistance in this context. Results of the serum bactericidal assays with the double lsg lic2A mutants showed that the lsgE, lsgD, and lsgC deletions in the lic2A mutant background do not increase serum bactericidal killing at statistically significant levels compared to the lic2A single mutant (Figure 3) indicating that the apparent attenuation of the double lic2A lsg mutants in vivo is likely effected by host defense mechanism(s) other than complement. Alternatively, lack of significant differential serum killing between the lic2A mutant and lsg lic2A double mutants could also result from a lack (or low levels) of expression or activity of the lsg gene products in vitro. These genes will require further studies to fully evaluate their interactions with lic2A.
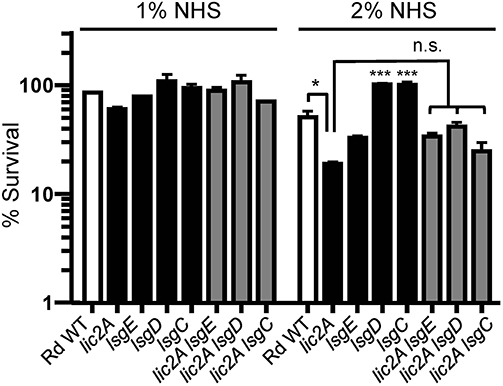
Figure 3. Deletion of lsg genes in parent or the lic2A mutant backgrounds does not significantly increase serum sensitivity. Viability of Rd wild-type (WT) and isogenic deletion mutants lic2A, lsgE, lsgD, lsgC, and double deletion mutants lic2A lsgE, lic2A lsgD, and lic2A lsgC grown in sBHI medium was assayed following incubation with 1 and 2% normal human serum (NHS) at 37°C for 30 min. Percent survival is the ratio of CFU recovered from serum treated samples at 30 min to CFU recovered from heat inactivated serum (NHSΔI) treated samples. The mean of two independent samples is shown. The lsgD and lsgC mutants each exhibited statistically significant increase in survival compared to all other samples at 2% NHS. Statistical significance is evaluated by one-way ANOVA with Bonferroni's multiple comparison test (***p < 0.001; *p < 0.05).
Unexpectedly, our genetic screen revealed that the galE mutant was restored to virulence in the lic2A mutant background. To investigate the mechanism of this restoration, we performed bactericidal assays with NHS to determine the sensitivity profile of Rd wild-type compared to isogenic deletion mutants of galE, lic2A, and a galE lic2A double deletion mutant following growth in sBHI. Results showed that the galE mutant is more sensitive than the lic2A mutant to killing at 0.5, 1, and 2% NHS. Interestingly, relative to the galE mutant, the galE lic2A double deletion mutant exhibited significantly increased resistance to serum killing (Figure 4A). A hypothesis to account for serum sensitivity of the galE mutant is the proposed addition of an alternative structure to the LOS by Lic2A in the absence of UDP-Gal (Figure 4B). That this alternative lic2A dependent epitope is likely a glucose residue is supported by previous structural studies indicating the presence of an unexpected glucose residue at this position in an H. influenzae galE mutant (Masoud et al., 2008) (further described in Discussion).
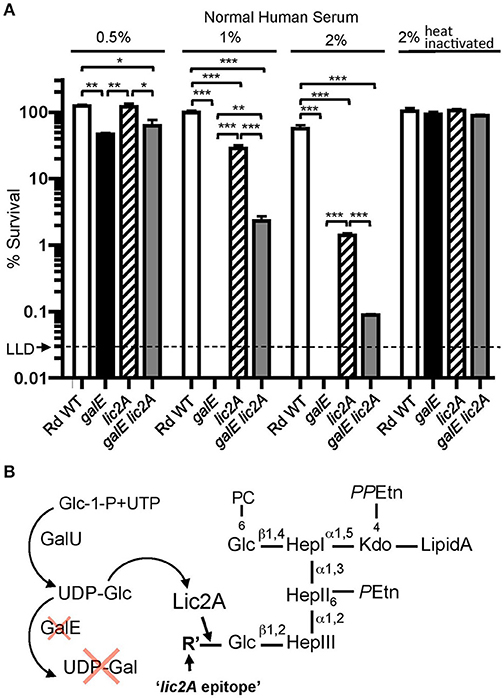
Figure 4. Deletion of lic2A increases serum resistance of the galE mutant. (A) Survival of Rd wild-type (WT) and isogenic single deletion mutants galE, lic2A and double deletion mutant galE lic2A grown in sBHI medium was assayed following incubation with 0.5, 1, and 2% NHS at 37°C for 30 min. Percent survival is the ratio of CFU recovered from samples treated with either NHS or NHSΔi at 30 min to CFU recovered from untreated samples. Log-transformed survival ratios were evaluated by one-way ANOVA with Bonferroni's multiple comparison test (*p < 0.05; **p < 0.01; ***p < 0.001). The mean of triplicate samples is shown. LLD, lower limit of detection. (B) Proposed model to account for complement sensitivity of the galE mutant resulting from addition of a lic2A dependent alternative epitope in the absence of galactose. GalU, glucosephosphate uridylyltransferase, R' is inferred to be Glc (see Results and Discussion).
Serum bactericidal assays typically have used H. influenzae grown in sBHI, an undefined rich medium. It has been noted that this medium contains trace amounts of sialic acid that could increase serum resistance in H. influenzae via sialylation of the LOS (Hood et al., 1999). Similarly, we found that wild-type NTHi strain NT127 was ~8-fold more resistant to 2% NHS compared to an isogenic siaB mutant following growth in sBHI in the absence of supplemental sialic acid (Figure S1). The siaB gene encodes CMP-Neu5Ac synthetase, which catalyzes the conversion of sialic acid to CMP-Neu5Ac, the activated nucleotide sugar donor essential for LOS sialylation. Therefore, these results indicate that wild-type NTHi are able to scavenge sialic acid from sBHI for LOS modification. To further refine our analysis of the bactericidal effect of NHS on the galE, lic2A, and galE lic2A mutants, we used a chemically defined medium, MIc, to control the levels of sialic acid in our assays, exploiting the fact that H. influenzae lacks the complete biosynthetic pathway for sialic acid synthesis and must acquire it from exogenous sources (Fleischmann et al., 1995; Vimr et al., 2000; Severi et al., 2007).
Serum bactericidal assays with Rd and mutants grown on MIc agar in either the presence or absence of supplemental sialic acid prior to treatment with 0.5 and 1% NHS confirmed sensitivity of the galE mutant to NHS killing (Figure 5). Because lic2A is subject to phase variation, we created strain RdgalE lic2A/lic2, a galE lic2A double deletion mutant containing lic2A complemented with a copy of lic2A that has been phase-on locked by deletion of CAAT repeats (Supplementary Text S1). This strain displayed a serum sensitivity phenotype comparable to that of the galE mutant, consistent with sequencing data indicating that our galE mutant is phase-on for lic2A (Supplementary Text S1). As expected, complementation of galE rescued serum resistance, and deletion of lic2A in the galE mutant background was also confirmed to rescue serum resistance similar to the results seen in Figure 4A. Because sialic acid is abundant on mucosal surfaces, we grew H. influenzae in MIc containing sialic acid, MIcSA for subsequent serum bactericidal assays and IgM binding studies to more closely resemble the host environment.
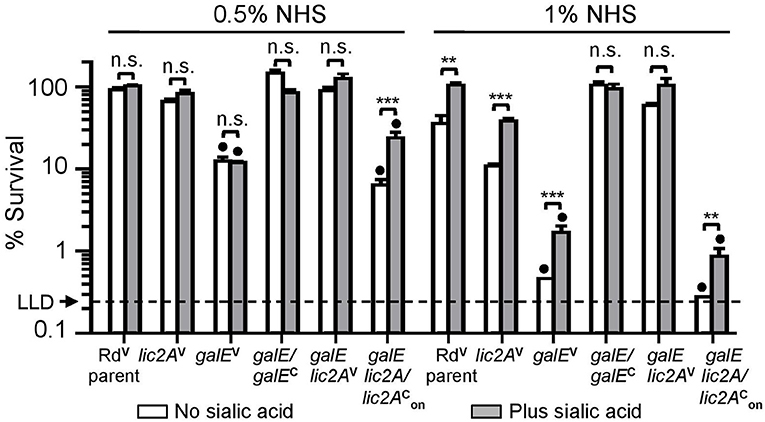
Figure 5. Deletion of lic2A increases serum resistance of the galE mutant when grown in the absence or presence of sialic acid on MIc medium. Viability of Rd containing empty cloning vector (V) (RdVparent), lic2A mutant containing V (lic2AV), galE mutant containing V (galEV), galE mutant with complementing copy of galE in trans (galE/galEC), galE lic2A mutant containing V (galE lic2AV), galE lic2A mutant with complementing copy of phase-on locked lic2A in trans, (galE lic2A/lic2) following incubation with 0.5% and 1% NHS at 37°C for 30 min. Percent survival is the ratio of CFU recovered at 30 min from serum treated samples to CFU recovered from samples treated with NHSΔi. Statistical significance of log-transformed survival ratios between each strain ± sialic acid is indicated (one-way ANOVA with Bonferroni's multiple comparison test; (**p < 0.01; ***p < 0.001; n.s., not statistically significant). galEV and galE lic2A/lic2 mutants significantly differed (denoted with black circles, p < 0.001) from all other strains except each other in −/+ sialic acid in both NHS concentration. The mean of triplicate samples is shown. LLD, lower limit of detection is approximately 0.25% survival.
The galE mutant is unable to generate UDP-Gal via the endogenous Leloir pathway, in which UDP-glucose pyrophosphorylase converts Glc-1-P to UDP-Glc, which the GalE epimerase then converts to UDP-Gal (Frey, 1996). If defects in LOS and serum resistance are caused by a deficiency in UDP-Gal, then resistance should be restored by provision of the galE mutant with an exogenous source of galactose, which can be converted to UDP-Gal via the exogenous Leloir pathway. The exogenous Leloir pathway requires galK, encoding galactokinase, which produces the phosphorylated intermediate galactose-1-phosphate, which is then converted to UDP-Gal by galactose-1-phosphate uridylyltransferase encoded by galT (Frey, 1996). Results of serum bactericidal assays showed that exogenous galactose restored serum resistance to galE mutants in both Rd and NTHi 375 strain backgrounds. A slight growth inhibition of the galE mutants by galactose under these conditions was observed (data not shown), however the rescued serum resistance of the galE mutants by galactose overcame this defect. The galE mutants of both of these strains grown in the absence of exogenous galactose were sensitive to NHS, and deletion of lic2A in their respective galE mutant backgrounds restored a significant level of resistance (Figures 6A,B).
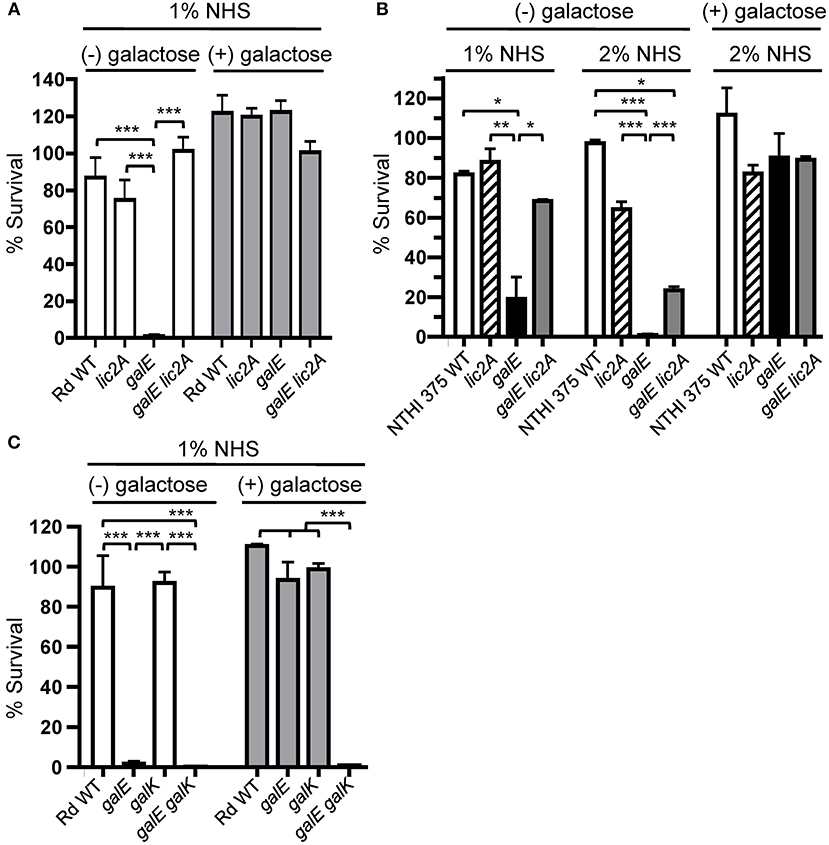
Figure 6. Deletion of lic2A or growth on galactose alleviates sensitivity of the galE mutant to bactericidal effects of NHS and blocking galactose catabolism in the galE mutant confers serum sensitivity. Viability of H. influenzae wild-type (WT) Rd (A,C) and NTHi strain 375 (B) and isogenic single and double deletion mutants in their respective strain backgrounds grown on MIc medium containing sialic acid (MIcSA) in the absence (-) or presence (+) of 1 or 0.5% galactose for Rd and NTHi 375, respectively, following incubation with the percent of NHS indicated at 37°C for 30 min. Percent survival is the ratio of CFU recovered at 30 min from serum treated samples to CFU recovered from NHSΔi treated samples. Log-transformed survival ratios were evaluated by one-way ANOVA with Bonferroni's multiple comparison test (*p < 0.05, **p < 0.01,***p < 0.001). The mean of duplicate assays is shown. Lower limit of detection is approximately 0.25% survival.
If catabolism of exogenous galactose is blocked in the galE mutant by deletion of galK, thus depriving cells of a source of activated galactose precursor for lic2A, then expression of the alternative lic2A structure is predicted to result in serum sensitivity. Serum bactericidal assays with the galE galK double deletion mutant confirmed that the exogenous galactose utilization pathway is required for serum resistance of the galE mutant (Figure 6C). Both galE and galE galK mutants were sensitive to serum killing when grown in the absence of galactose. As expected, galactose alleviated the serum sensitivity of the galE mutant, and the galK deletion mutant does not exhibit serum sensitivity regardless of exogenous galactose. However, the galE galK double mutant remained sensitive to serum killing even when supplied with exogenous galactose, confirming that blocking exogenous galactose catabolism in the galE mutant confers serum sensitivity.
Antibodies, including IgM and to a lesser extent IgG, are potent activators of the classical complement pathway (CP), which is required for maximal bactericidal activity of complement against NTHi (Williams et al., 2001; Wong et al., 2011; Rosadini et al., 2014). Therefore, we investigated whether a functional lic2A gene in the galE mutant confers increased antibody binding leading to serum sensitivity. Increased IgM antibody binding of the galE mutant containing lic2A phase-on locked and the Rd galE mutant (also phase-on for lic2A) (Figures 7A,B) correlated with their serum sensitivity when grown without exogenous galactose (Figures 5, 6A). Also consistent with serum sensitivity results, deletion of lic2A in the galE mutant background and complementation of the galE mutant both reduced IgM binding to the levels seen in wild type. No differential IgG binding between Rd wild-type and the galE mutant was detected (Figure 7C). As expected, no significant differential binding of IgM was detected between Rd wild-type and the galE mutant when grown in the presence of galactose (Figure S2). Similarly, increased IgM binding of the NTHi 375 galE mutant (Figures 8A,B) correlated with increased serum sensitivity of this galE mutant grown in the absence, but not in the presence of exogenous galactose and deletion of lic2A in the galE mutant abrogated this increased binding correlating with decreased serum sensitivity (Figure 6B). As with Rd, there was no statistically significant differential binding of IgG between NTHi 375 wild-type and its isogenic galE mutant grown under either condition (Figure 8C).
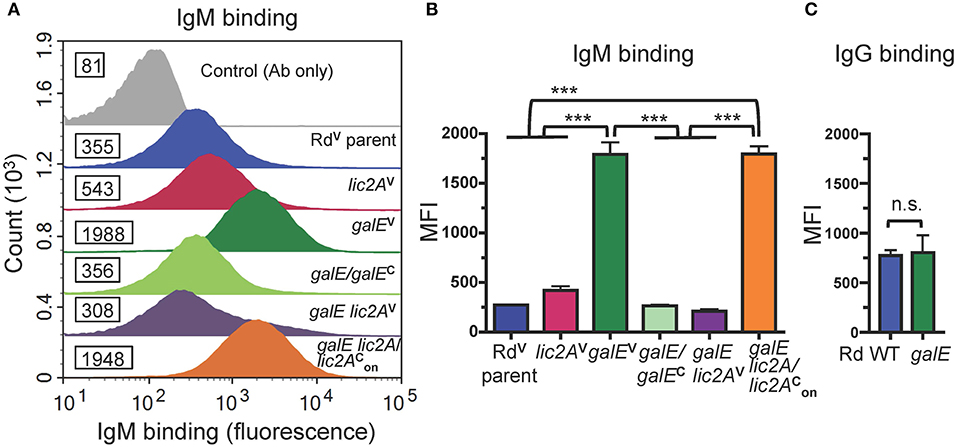
Figure 7. Increased binding of IgM but not IgG to the galE mutant and galE lic2A containing lic2A phase-on locked. (A) Rd strains grown in MIcSA were incubated with NHSΔi at 20% final concentration at 37°C for 30 min followed by detection via flow cytometry with anti-human IgM conjugated to FITC. X-axis, fluorescence; Y-axis, counts. Numbers alongside histograms indicate the median fluorescence intensity (MFI) of the bacterial population. Control antibody (Ab) only fluorescence values are similar for all strains and one representative is shown (RdV parent). One representative experiment of two reproducible repeats is shown. (B) IgM and (C) IgG binding for the mean of replicate samples of which one is shown in (A) for IgM. Statistical significance was evaluated by one-way ANOVA with Bonferroni's multiple comparison test (***p < 0.001; n.s., not statistically significant).
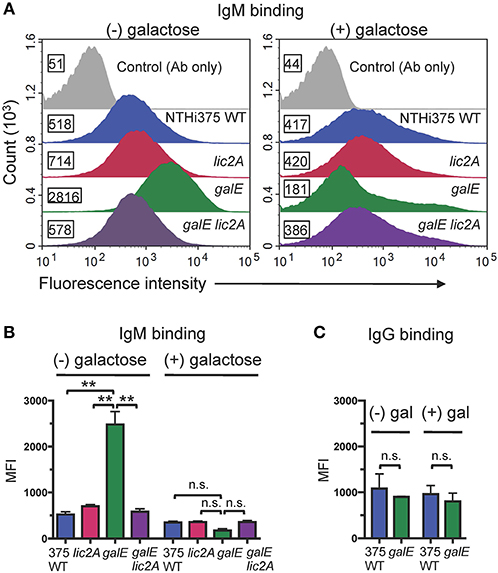
Figure 8. Increased binding of IgM but not IgG to the NTHi 375 galE mutant. (A) IgM binding to NTHi 375 wild-type (WT) and isogenic mutants grown in the absence or presence of 0.5% galactose in MIcSA then incubated with 20% NHSΔi followed by detection with anti-human IgM conjugated to FITC via flow cytometry. X-axis, fluorescence; Y-axis, counts. Numbers alongside the histograms indicate the MFI of the bacterial population. Control Ab only fluorescence values are similar for all strains and only one representative is shown (NTHi 375 WT). One representative experiment of two reproducible repeats is shown. (B) IgM and (C) IgG binding for the mean of replicate samples of which one is shown in (A) for IgM. Statistical significance was evaluated by one-way ANOVA with Bonferroni's multiple comparison test (**p < 0.01; n.s., not statistically significant).
To confirm the in vivo survival phenotype of galE mutants in the lic2A mutant background as identified in HITS data, survival of Rd, NTHi 375 and their respective isogenic mutants (lic2A, galE, galE lic2A) was evaluated in the mouse lung model. Supporting the HITS data, galE mutants exhibited the most severe attenuation in survival relative to wild-type strains in both backgrounds. The NTHi 375 lic2A mutant was moderately attenuated while the Rd lic2A mutant had no significant defect. Deletion of lic2A in the galE mutants of Rd and NTHi 375 partially rescued the persistence defect of the galE mutants in both strain backgrounds (Figures 9A,B). Moreover, locking lic2A in the phase-on state in the Rd galE mutant (galE lic2A/lic2) decreased persistence similar to that seen with the galE mutant, and complementation (galE/galEc) restored survival of the galE mutant in the lung (Figure 9A).
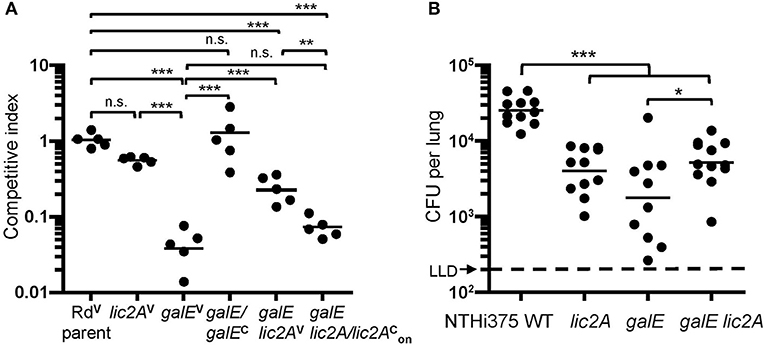
Figure 9. Deletion of lic2A in the galE mutant background partially rescues attenuation of the galE mutant for lung colonization in both Rd and NTHi strains. H. Influenzae Rd or NTHi 375 and isogenic mutants in their respective background strains recovered from lungs of C57BL/6 mice 24 h after intranasal inoculation. (A) H. influenzae Rd strains as described in Table 1 were co-inoculated with an equal amount of an H. influenzae reference strain expressing lacZ (RdLacZ) and recovered from mice (n = 5). Ratio of CFU of the experimental strains (LacZ–) to competitor strain (LacZ+) is reported as the competitive index (CI). Bars represent the geometric mean ratio of CFU of each designated strain relative to the LacZ+ reference strain. CI fold differences of parent vs. each of the mutants: lic2AV (1.9x), galEV (24x), galE lic2AV (4.3x), and galE lic2A/lic2 (14x) and between galE lic2AV vs. each of the mutants: galEV (5.5x), galE lic2A/lic2 (3.3x). (B) NTHi 375 wild-type (WT) and isogenic deletion mutants recovered from mice (n = 10–12). Bars represent the geometric mean CFU per lung. LLD, lower limit of detection. Fold differences in mean CFU recovered for WT vs. each of the mutants: lic2A (5.5x), galE (6.9x), and galE lic2A (4.3x) and between galE vs. galE lic2A (1.6x). Differences were evaluated via one-way ANOVA with Bonferroni's multiple comparison test of log-transformed values (*p < 0.05; **p < 0.01; ***p < 0.001; n.s., not statistically significant).
Increased IgM binding to the galE mutants (GalE−, Lic2A+) (Figures 7, 8) implicated the classical pathway of complement in their increased sensitivity to serum killing relative to the parental strains. To evaluate the role of the CP in differential killing, we performed bactericidal assays with NHS containing EGTA and MgCl2, which selectively blocks the classical and lectin pathways but not the alternative pathway. Chelation of calcium by EGTA blocks the calcium ion dependent assembly of the C1 complex of the classical pathway (Lepow et al., 1963). Calcium ions are also required for binding by C-type animal lectins (Iobst and Drickamer, 1994) and for activation of the lectin pathway by mannose-associated serine proteases (Wallis and Dodd, 2000).
Results in Figure 10A show that the NTHi 375 galE mutant was highly sensitive to 2% NHS as expected, but its resistance to killing is equivalent to wild type in 2% NHS with EGTA-MgCl2. Moreover, in NHS with EGTA-MgCl2 at 50%, the galE mutant approaches the survival levels of the wild type, retaining ~60% survival. We note, however, that at 50% EGTA-MgCl2, some killing of the galE mutant may occur by the alternative pathway, however this difference was not statistically significant. That the differential killing in NHS requires antibody mediated activation of the classical pathway was confirmed in bactericidal assays with NHS depleted of IgM and IgG as a source of active human complement (HC), with or without supplementation with heat inactivated NHS (NHSΔi) as a source of antibody (Figure 10B). Without a source of antibody from NHSΔi, there was no differential killing between strains, however in the presence of serum antibody, the galE mutant was killed 8-fold more compared to wild type, and deletion of lic2A in the galE mutant restored resistance to wild-type levels.
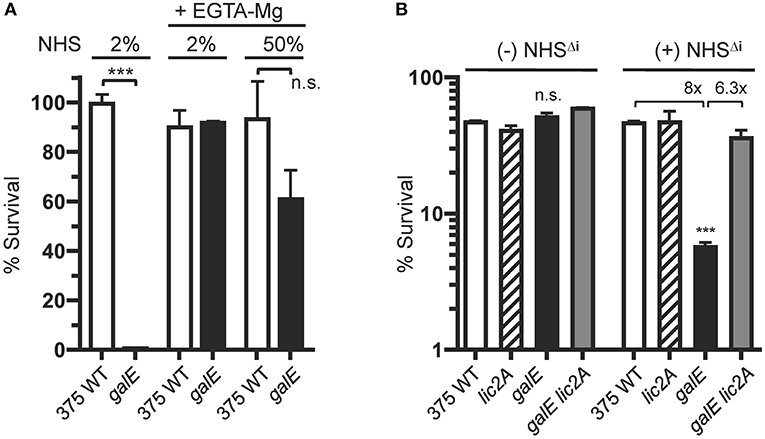
Figure 10. Killing of the NTHi 375 galE mutant by complement requires serum antibody. (A) Survival of NTHi 375 wild-type (WT) and isogenic galE mutant grown on MIcSA medium assayed following incubation with the indicated percentage of NHS in the absence or presence of EGTA and MgCl2 at 37°C for 30 min. Percent survival is the ratio of CFU recovered from serum treated samples (±EGTA and MgCl2) at 30 min to CFU recovered from NHSΔi treated samples (±EGTA and MgCl2). (B) Antibody-dependent killing of the galE mutant by NHS. Viability of NTHi 375 wild-type (WT) and isogenic deletion mutants grown on MIcSA following incubation with or without 10% unabsorbed NHSΔi followed by incubation with 3% IgG/IgM antibody depleted human complement active serum (HC) at 37°C for 30 min. Percent survival is the ratio of the number of CFU recovered at 30 min. relative to the input CFU at T = 0 min. of incubation. Log transformed survival ratios were evaluated by one-way ANOVA with Bonferroni's multiple comparison test (***p < 0.001; n.s., not statistically significant). In (B), the galE mutant differed significantly from the other three strains within the NHSΔi treatment group. The mean of duplicate samples is shown. Lower limit of detection is ~0.25% survival.
To determine whether antibody dependent killing results from recognition of an epitope unique to the galE mutant, we performed bactericidal assays in HC supplemented with NHSΔi depleted of specific antibody by absorption. Sera were absorbed with a galE strain RdgalElic2A/lic2 (GalE−, Lic2A+) expressing lic2A locked phase-on or a galE lic2A double deletion strain RdgalElic2AV (GalE−, Lic2A−) predicted to lack the epitope. NTHi 375 and isogenic lic2A, galE and galE lic2A mutants were evaluated in serum killing assays with HC supplemented with depleted sera or control unabsorbed NHSΔI (Figure 11A). Results showed that the galE mutant was less sensitive to serum killing when treated with the serum pre-absorbed with strain RdgalElic2A/lic2. In contrast, sera pre-absorbed with strain RdgalElic2AV or control unabsorbed NHSΔi retained increased bactericidal activity against the galE mutant relative to wild type, lic2A and galE lic2A strains. Similar results were obtained with the same set of mutants in the Rd background (Figure S3). The decrease in serum sensitivity of the galE mutant treated with RdgalElic2A/lic2 pre-absorbed serum correlated with a parallel decrease in IgM binding by the galE mutant treated with this absorbed serum relative to unabsorbed serum or serum absorbed with the lic2A deficient strain, RdgalElic2AV (Figure 11B).
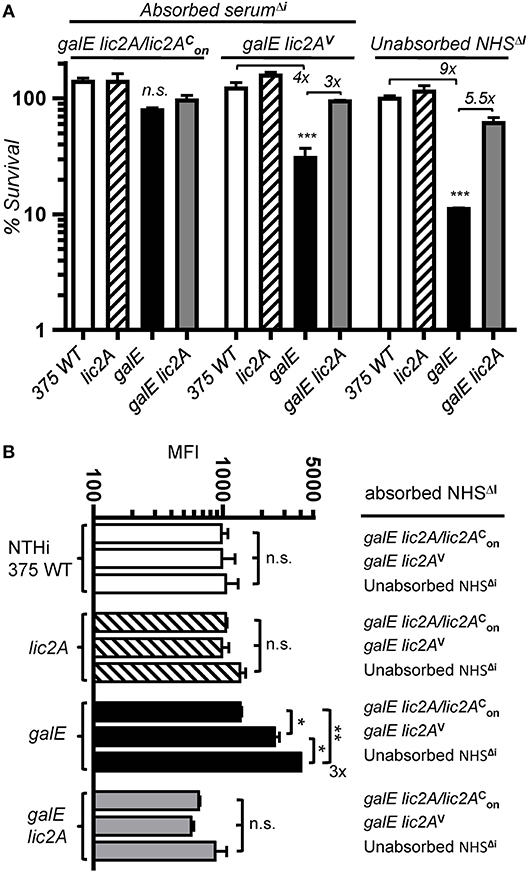
Figure 11. Absorption experiments indicating a lic2A dependent epitope in the galE mutant that confers increased killing by serum and IgM binding. (A) Serum pre-absorbed of the antibody that targets the lic2A dependent epitope in the galE mutant promotes decreased killing of the galE mutant. Viability of the indicated strains grown on MIcSA following incubation with 10% NHSΔi pre-absorbed with strains RdgalElic2A/lic2 (GalE−, Lic2A+) or RdgalElic2AV (GalE−, Lic2A−) vs. control unabsorbed NHSΔi, followed by incubation with 3% HC at 37°C for 30 min. Percent survival is the ratio of CFU recovered from absorbed or unabsorbed serum treated samples after HC incubation to CFU recovered from non-serum treated (buffer only) samples after HC incubation. Lower limit of detection is approximately 0.25% survival. Differences between the galE mutant vs. the other three strains within a treatment group were statistically significant by ANOVA, ***p < 0.001. (B) Serum pre-absorbed of the antibody targeting the lic2A dependent epitope in the galE mutant contains decreased IgM specific to the galE mutant. Strains grown on MIcSA were incubated with 10% NHSΔI pre-absorbed with RdgalElic2A/lic2 or RdgalElic2AV or control unabsorbed NHSΔI at 37°C for 30 min followed by detection via flow cytometry with anti-human IgM conjugated to FITC. IgM binding from the mean of replicate samples. Log-transformed survival ratios and MFI values were evaluated by one-way ANOVA with Bonferroni's multiple comparison test (*p < 0.05; **p < 0.01; n.s., not statistically significant).
Additional evidence for a lic2A dependent epitope recognized by IgM on the LOS of the galE mutant was obtained with immunoblots containing protease treated bacterial lysates probed with the two absorbed sera (Figure 12). IgM in the serum absorbed with the galE lic2A double deletion mutant bound to an ~5 kDa band, the expected size of the LOS. This IgM binding band was present in the galE mutant lysate, but was completely absent in lysates of NTHi 375 wild-type, lic2A, and galE lic2A deletion strains. A 15 kDa band, likely representing a protease resistant protein, bound equal amounts of IgM in all strains with both absorbed sera. Reactivity of IgM with the ~5 kDa band was completely eliminated with serum absorbed with the galE deletion strain. Consistent with a role for IgM but not IgG in complement sensitivity of the galE mutant, IgG binding to the galE mutant was not increased relative to other strains, but rather was undetectable in each mutant strain, while wild type exhibited substantial IgG reactivity with both absorbed sera despite its greater degree of complement resistance.
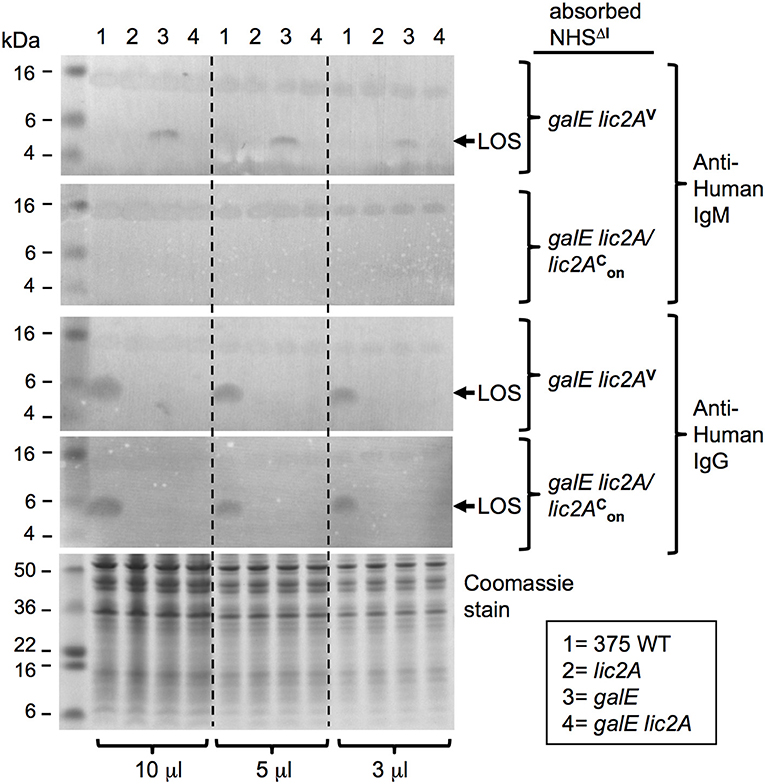
Figure 12. IgM binding to a lic2A dependent LOS epitope in the galE mutant. Whole-cell lysates from NTHi 375 and the indicated isogenic mutants grown on MIcSA were digested with proteinase K and separated by NuPAGE 4–12% Bis-Tris gels followed by western immunoblotting. Membranes were incubated with serum pre-absorbed with strains RdgalElic2A/lic2 (GalE−, Lic2A+) or RdgalElic2AV (GalE−, Lic2A−) followed by incubation with alkaline phosphatase conjugated anti-human IgM or IgG. Equal sample concentration was verified by Coomassie Blue staining of undigested samples. Each sample was electrophoresed at three different concentrations. Arrows indicate ~5kDa LOS; pre-stained protein standards in left lane.
Although the lic2A gene contributes positively to serum resistance under some conditions, deletion of lic2A restores serum resistance and improves lung colonization of galE mutants. Conditions in which galE activity is absent or low in wild-type NTHi have not been studied extensively, however the NTHi galE mutant exhibits enhanced production of biofilm suggesting that biofilm formation may involve decreased GalE activity (Greiner et al., 2004). We noticed in our HITS data that several genes required in the lung model in the lic2A mutant but not in the parent have been implicated in promoting biofilm formation, including genes of phosphate homeostasis (phoB, pstA) (Monds et al., 2001; Hsieh and Wanner, 2010; Xu et al., 2012); stress adaptation (lon, sspA, deaD, aspA) (Iost and Dreyfus, 2006; Marr et al., 2007; Van Melderen and Aertsen, 2009; Nijland and Burgess, 2010; Bernier et al., 2011; Giaouris et al., 2013; Karatan and Michael, 2013; Vakulskas et al., 2014; Rogers et al., 2016; Xie et al., 2016; Ching et al., 2018), and DNA replication, recombination, and repair (ihfA, xerC, fis) (Barre et al., 2001; Sheikh et al., 2001; Goodman et al., 2011; Moor et al., 2014; Atwood et al., 2016; Leite et al., 2017; Devaraj et al., 2018; Lv et al., 2018; Supplementary Text S1). Together, these observations suggest that during NTHi pathogenesis, biofilm formation may involve a switch to decreased reliance on LOS production. Such a switch could be induced by metabolites that NTHi scavenge from the host. Sialic acid has been shown to enhance NTHi biofilm production and both sialic acid and possibly N-acetylglucosamine (GlcNAc) are present in biofilm matrix (Greiner et al., 2004; Swords et al., 2004). Both GlcNAc and sialic acid are precursors of UDP-GlcNAc (Figure 13A), substrate donor for synthesis of poly-N-acetyl-glucosamine, a potential component of biofilm matrix that has been detected on the NTHi cell surface (Cywes-Bentley et al., 2013). Because likely orthologs of GalE of NTHi in other species can utilize UDP-GlcNAc as an alternative substrate (see Discussion), we postulate that exogenous GlcNAc/sialic acid in the environment could shift NTHi from UDP-Gal production toward synthesis of UDP-GlcNAc, leading to increased biofilm production. To begin to evaluate this hypothesis we conducted biofilm assays with NTHi 375 and isogenic galE and siaB mutants in the presence of these metabolites. The results showed that addition of exogenous GlcNAc and sialic acid fueled biofilm production in the wild type and in the galE mutant (Figure 13B). As previously shown, the asialylated siaB mutant exhibited reduced biofilm formation (Greiner et al., 2004; Swords et al., 2004). We note that exogenous galactose bi-passes the role of GalE, as enhanced biofilm production induced by sialic acid or GlcNAc is abrogated in the galE mutant by exogenous galactose. However, exogenous galactose did not influence biofilm production in the presence of both GlcNAc and sialic acid in the wild type, which synthesizes galactose endogenously.
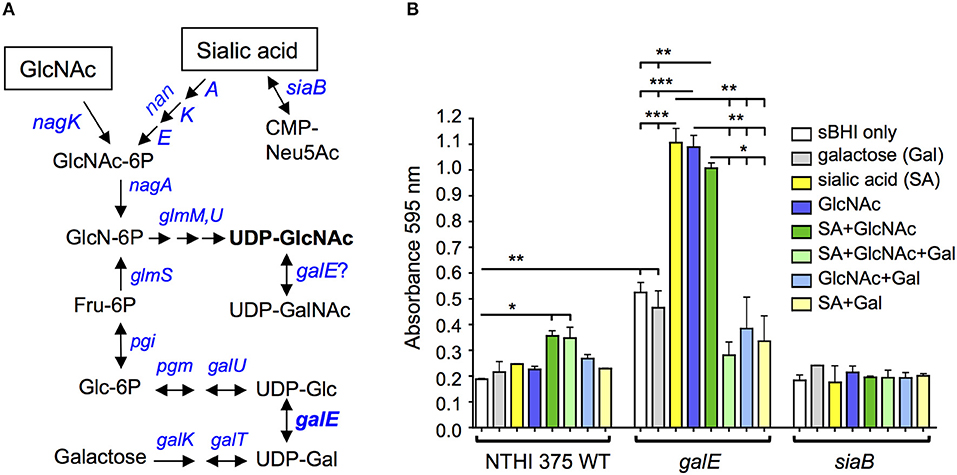
Figure 13. Exogenous UDP-GlcNAc precursors fuel biofilm production. (A) Amino sugar and nucleotide sugar metabolic pathway for generation of UDP-GlcNAc in H. influenzae (adapted from KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway database (https://www.genome.jp/kegg/pathway.html). (B) NTHi 375 wild-type (WT) with isogenic mutants galE and siaB were grown in sBHI for 24 hr in the presence of galactose, sialic acid, N-acetylglucosamine (GlcNAc) or combinations of the three in microtiter plates and biofilm formation was quantified by crystal violet staining. Log-transformed values were evaluated by one-way ANOVA with Bonferroni's multiple comparison test (*p < 0.05; **p < 0.01; ***p < 0.001). GlcNAc-6P, N-acetylglucosamine 6-phosphate; GlcN-6P, D-glucosamine 6-phosphate; UDP-GlcNAc, UDP-N-acetylglucosamine; UDP-GalNAc, UDP-N-acetylgalactosamine; Fru-6P, β-D-fructose 6-phosphate; Glc-6P, α-D-glucose 6-phosphate; CMP-Neu5Ac, cytidine monophosphate sialic acid; UDP-Glc, UDP-glucose; UDP-Gal, UDP-galactose; UDP, uridine diphosphate.
We conducted a genome-wide genetic interaction screen to compare fitness of transposon mutants in a lic2A deletion background to that of the same mutants in the parental strain background, and thereby identify mutants with virulence phenotypes that are exacerbated by deletion of lic2A. This approach can identify genes that become required in natural phase variants in the lic2A off-phase, and also address the previously observed discrepancy between the virulence phenotypes of galE and lic2A mutants. A large number of mutations conferring virulence defects in the lic2A deletion background were identified in the screen, including mutations in known or putative LOS glycosyltransferase genes, a primary class we had anticipated finding. Surprisingly, the galE mutant exhibited a prominent phenotype in that its survival defect in the lung model was suppressed by deletion of lic2A. A major defect in serum resistance in the galE mutant was shown to result from an alternative epitope dependent on the presence of a functional lic2A gene but produced only when galE is inactivated. We found that serum IgM targets this epitope, leading to killing of NTHi by the classical pathway of complement. Therefore, a major virulence function of galE is to prevent alternative substrate utilization by the glycosyltransferase encoded by lic2A, which leads to generation of a novel IgM target and complement sensitivity. Nevertheless, the lic2A gene can also contribute positively to pathogenesis, as highlighted by survival defects in the lung model detected with transposon insertion mutations in a diversity of other genes in the lic2A deletion background. These genes are likely to be important in lic2A phase variants. We also cannot rule out a potential virulence role for this alternative Lic2A activity in some aspect of infection not reflected in the lung model. Finally, a question remains as to when UDP-Gal production by GalE might be decreased under natural conditions, a situation that could account for the evolution of phase-variation by lic2A as a means of preventing inappropriate epitope production. In this report, we suggest that the latter may occur in biofilms, as galE mutants exhibit increased biofilm formation, and host metabolites that increase biofilm production are also likely modulators of substrate utilization by GalE.
Deletion of lic2A generated or increased the requirement for numerous genes involved in host-pathogen interactions. Almost all of the vacJ/mla (yrb) homologs that function in phospholipid trafficking to maintain lipid asymmetry in the outer membrane were implicated (Malinverni and Silhavy, 2009). In NTHi, vacJ, yrbE, yrbB, and yrbD mutants are very sensitive to NHS (Nakamura et al., 2011), which may magnify sensitivity in the lic2A deletion background to killing by complement. We also observed the requirement for dacA (pbp5), which is involved in peptidoglycan biosynthesis and HI1658 (yraP), a predicted outer membrane lipoprotein that is a component of the divisome (Tsang et al., 2017) and contributes to maintaining the outer membrane barrier (Morris et al., 2018). The requirement for lic1, responsible for LOS modification with phosphorylcholine, is likely due to increased IgM binding of the lpsA dependent LOS structure exposed in a lic2A lic1 double mutant that was correlated with increased neutrophil-mediated killing in a previous study (Langereis and Weiser, 2014). We anticipated identifying LOS glycosyltransferase candidates such as the genes (hmg) required for the cryptic LOS glycoforms or genes (lsg) that associate with this structure (Hood et al., 2004b). Although, we did not find a requirement for hmg in the lic2A mutant background for in vivo survival, the lsg glycosyltransferase genes did appear to be functionally redundant with lic2A in vivo. But since the lsg mutations in the lic2A deletion background did not lead to increased killing compared to the lic2A mutation alone in the serum bactericidal assays, functional involvement of lsg genes with lic2A in pathogenesis is currently unclear, and may involve phenotypes other than complement resistance.
The most striking finding from our genetic screen is that galE, which supplies the precursor sugar for LOS incorporation by Lic2A, was not required for survival in the lung in the lic2A deletion strain. We identified a lic2A dependent immune target leading to serum sensitivity of the galE mutant and its attenuation in vivo. We hypothesize that the epitope added by lic2A in the absence of galE likely results from addition of a second glucose onto that added by lpsA on HepIII of the LOS. Structural analyses of LOS in H. influenzae type b strain Eagan indicate a galactose residue is normally attached by lic2A in a β1,4 linkage to a diglucoside on the middle HepII residue to form a Gal-Glc-Glc trisaccharide (Masoud et al., 1997; Hood et al., 2001, 2004a). However, both Eagan galE and double galE galK mutants display a novel triglucoside chain extension off HepII not normally found in H. influenzae LOS (Masoud et al., 2008), which is consistent with addition of Glc by Lic2A in the absence of UDP-Gal. In fact, it has been shown that there is some flexibility in the donor specificity of galactosyltransferases. For example, bovine β1,4-galactosyltransferase does not have an absolute requirement for UDP-Gal as donor, but at a lower efficiency can use other donors, such as UDP-Glc (Palcic and Hindsgaul, 1991). The amino acid sequence of the Rd Lic2A has 96 and 98% identities to Hib and NTHi 375 Lic2A, respectively, and they likely function similarly amongst diverse strains. Interestingly, reports in Neisseria meningitides indicated that a galE mutant has the expected major LOS species lacking galactose, but also displays a second LOS species (5–10% of the total) containing an extra glucose linked to the proximal glucose of the outer core (Lee et al., 1995; Wakarchuk et al., 1996). This extra glucose is dependent on the lgtE gene, whose product normally functions as a galactosyltransferase.
We speculate that a critical role of GalE in H. influenzae is to prevent Lic2A from adding an immune target, presumably a glucose extension, in the absence of a galactose source. Because the lic2A gene is subject to phase variation and can randomly and reversibly switch to the phase-off state, GalE activity in this scenario could decrease without triggering an adverse immune response, and adapting to niches that cause low GalE activity may be one of the roles of phase variation of lic2A. While such conditions have not been identified for NTHi in vivo, several of our results point to the biofilm mode of growth as a potential modulator of GalE, as discussed below. Biofilm formation by NTHi is known to occur in persistent infections in the airways (Swords, 2012) and has been found in the middle ear mucosa in a chinchilla experimental model of otitis media (OM) (Ehrlich et al., 2002) and in middle ear effusions in children with OM (Idicula et al., 2016).
We note that several genes required in the lic2A deletion background (Table 2) have potential orthologs in other bacterial species that participate in biofilm formation, such as phoB and pstA of the phosphate regulon; stress response/adaptation genes, lon, sspA, deaD, and aspA, and DNA metabolism genes, ihfA, xerC, and fis. In NTHi, the DNA binding protein IHF was found to be important in maintaining structural integrity of the extracellular DNA component of biofilm and treatment with anti-IHF could destabilize NTHi biofilms in vitro and in vivo in chinchilla middle ears (Goodman et al., 2011; Devaraj et al., 2018). Additional description of these genes is provided in Supplementary Text S1.
It is intriguing to speculate that a consequence of lic2A phasing off is an alleviated requirement for galE expression as part of adaptation within a biofilm. Because mutants lacking galE produce enhanced biofilm, it is possible that reduced UDP-Gal production by GalE may occur in biofilms. While sialic acid and at least one glycosyltransferase have been implicated in biofilm production (Pang et al., 2018), the exact composition of the carbohydrates in the matrix is not known. Nevertheless, cell surface poly-N-acetylglucosamine (PNAG), is a major polysaccharide component of biofilm matrix in other bacteria (Roux et al., 2015), and the precursor for PNAG, UDP-GlcNAc, has been proposed as a precursor of biofilm matrix material in NTHi (Greiner et al., 2004). Moreover, PNAG was recently detected on the NTHi cell surface (Cywes-Bentley et al., 2013), although the genes responsible for PNAG biosynthesis in NTHi have not been identified.
To begin to evaluate the hypothesis that biofilm formation may involve metabolic flux away from UDP-Gal production and toward higher UDP-GlcNAc levels, we conducted biofilm assays with precursors of this sugar nucleotide. Our biofilm assay results showed a statistically significant ~2-fold increase in biofilm formation in the NTHi 375 wild-type and galE mutant when grown in the presence of both GlcNAc and sialic acid supplied together (dark green bars) vs. sBHI alone (white bars) (Figure 13B), consistent with a potential role in biofilms for UDP-GlcNAc, which is generated from both of these substrates (Mengin-Lecreulx and Van Heijenoort, 1993, 1996; Figure 13A). The ~2-fold increase in biofilm formation in the galE mutant vs. wild type in sBHI alone is likely due to traces of sialic acid in sBHI as sialic acid is known to induce biofilm formation (Greiner et al., 2004; Swords et al., 2004).
In the NTHi 375 wild-type, GlcNAc or sialic acid alone did not enhance biofilm formation as much as it did in the galE mutant, and biofilm formation overall was higher in the galE mutant. These results could be accounted for if GalE in the wild-type strain has the capacity to interconvert UDP-GlcNAc to UDP-GalNAc (Figure 13A), providing GalNAc for LOS biosynthesis and thereby reducing levels UDP-GlcNAc available for biofilm formation, potentially involving PNAG production. GalE is a candidate for mediating this conversion as H. influenzae LOS contains GalNAc residues (Risberg et al., 1999; Hood et al., 2004b), yet there is no annotated UDP-GlcNAc 4-epimerase gene (gne) in its genome. In bacteria that do not possess gne homologs, such as Campylobacter jejuni and Neisseria spp, UDP-GlcNAc is synthesized by a bi-functional GalE epimerase (Bernatchez et al., 2005; Bartley et al., 2017). A single amino acid in GalE determines substrate specificity for UDP-Gal/Glc (mono-functional) vs. both UDP-Gal and UDP-GalNAc/GlcNAc (bi-functional) (Thoden et al., 2002; Bartley et al., 2017). Interestingly, a BLAST database search of GalE of 62 complete genome sequences of H. influenzae strains examined indicates that all possess a cysteine, C299, corresponding to the critical substrate specificity site, Y299, of E. coli GalE, in which a Y299C mutation converts it from a mono-functional to a bi-functional enzyme (Thoden et al., 2002). It is intriguing to speculate that GalE in H. influenzae may possess bi-functional activity and that this activity could potentially be modulated by UDP-GlcNAc precursors affecting biofilm production.
Aside from galE, the genetic screen also identified xthA and HI1146 as dispensable in the lic2A mutant but required in the parent. XthA (exodeoxyribonuclease III) participates in repair of oxidative damage to DNA (Lovett, 2011), resistance to oxidative stress (Demple et al., 1983) and resistance to hydrostatic pressure during stationary phase (Charoenwong et al., 2011). It is not clear why deletion of lic2A would alleviate these mutant phenotypes in H. influenzae. HI1146 encodes a putative RapZ (or YhbJ) homolog (RNase adaptor protein) that functions in a small RNA mediated regulatory feedback pathway, inhibiting expression of glucosamine-6-phosphate synthase (GlmS) in response to its product, glucosamine 6-phosphate (GlcN-6P), a key intermediate for the biosynthesis of UDP-GlcNAc and UDP-GalNAc (Figure 13A; Kalamorz et al., 2007; Göpel et al., 2013). Therefore, deletion of the putative rapZ homolog HI1146, is predicted to increase intracellular GlcN-6P, which may shift cellular production away from UDP-Gal and toward UDP-GlcNAc, leading to a Lic2A dependent virulence defect similar to that seen in a galE deletion mutant.
Our findings indicate that when deprived of its usual galactose donor substrate provided by GalE or the exogenous pathway, the lic2A encoded galactosyltransferase generates an alternative epitope that is an IgM antibody immune target leading to increased serum sensitivity and attenuation in the lung. Because virulence in a wide range of bacterial pathogens depends upon GalE, its role for NTHi in controlling alternative glycosyltransferase phenotypes identified herein has broad implications toward understanding its role in other bacterial pathogens of high clinical significance.
Transposon insertion site sequencing (HITS) read data have been deposited in the NCBI short read archive (SRA) with the Bioproject ID number: PRJNA516677.
Experiments were conducted with approval and in accordance with guidelines of the Institutional Animal Care and Use Committee at the University of Mississippi Medical Center (Jackson, MS).
SW and BA designed experiments, analyzed data and wrote the manuscript. SW performed all experiments with MJ contributing to studies of IgM binding, immunoblotting, serum bactericidal assays, and serum absorption studies. All authors read and approved the manuscript.
This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) R01AI095740 (to BA).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Sanjay Ram for critical reading of the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00160/full#supplementary-material
Ahearn, C. P., Gallo, M. C., and Murphy, T. F. (2017). Insights on persistent airway infection by non-typeable Haemophilus influenzae in chronic obstructive pulmonary disease. Pathog Dis. 75:ftx042. doi: 10.1093/femspd/ftx042
Akerley, B. J., Rubin, E. J., Novick, V. L., Amaya, K., Judson, N., and Mekalanos, J. J. (2002). A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. U.S.A. 99, 966–971. doi: 10.1073/pnas.012602299
Atwood, D. N., Beenken, K. E., Loughran, A. J., Meeker, D. G., Lantz, T. L., Graham, J. W., et al. (2016). XerC contributes to diverse forms of Staphylococcus aureus infection via agr-dependent and agr-independent pathways. Infect. Immun. 84, 1214–1225. doi: 10.1128/IAI.01462-15
Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. E., Seidman, J. G., Smith, J. A., and Struhl, K. (eds.). (1995). Current Protocols in Molecular Biology. New York, NY: John Wiley & Sons, Inc.
Barcak, G. J., Chandler, M. S., Redfield, R. J., and Tomb, J. F. (1991). Genetic systems in Haemophilus influenzae. Meth. Enzymol. 204, 321–342. doi: 10.1016/0076-6879(91)04016-H
Barre, F. X., Søballe, B., Michel, B., Aroyo, M., Robertson, M., and Sherratt, D. (2001). Circles: the replication-recombination-chromosome segregation connection. Proc. Natl. Acad. Sci. U.S.A. 98, 8189–8195. doi: 10.1073/pnas.111008998
Bartley, S. N., Mowlaboccus, S., Mullally, C. A., Stubbs, K. A., Vrielink, A., Maiden, M. C., et al. (2017). Acquisition of the capsule locus by horizontal gene transfer in Neisseria meningitidis is often accompanied by the loss of UDP-GalNAc synthesis. Sci. Rep. 7:44442. doi: 10.1038/srep44442
Bernatchez, S., Szymanski, C. M., Ishiyama, N., Li, J., Jarrell, H. C., Lau, P. C., et al. (2005). A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J. Biol. Chem. 280, 4792–4802. doi: 10.1074/jbc.M407767200
Bernier, S. P., Létoffé, S., Delepierre, M., and Ghigo, J. M. (2011). Biogenic ammonia modifies antibiotic resistance at a distance in physically separated bacteria. Mol. Microbiol. 81, 705–716. doi: 10.1111/j.1365-2958.2011.07724.x
Bouchet, V., Hood, D. W., Li, J., Brisson, J. R., Randle, G. A., Martin, A., et al. (2003). Host-derived sialic acid is incorporated into Haemophilus influenzae lipopolysaccharide and is a major virulence factor in experimental otitis media. Proc. Natl. Acad. Sci. U.S.A. 100, 8898–8903. doi: 10.1073/pnas.1432026100
Charoenwong, D., Andrews, S., and Mackey, B. (2011). Role of rpoS in the development of cell envelope resilience and pressure resistance in stationary-phase Escherichia coli. Appl. Environ. Microbiol. 77, 5220–5229. doi: 10.1128/AEM.00648-11
Ching, C., Yang, B., Onwubueke, C., Lazinski, D., Camilli, A., and Godoy, V. G. (2018). Lon protease has multifaceted biological functions in Acinetobacter baumannii. J. Bacteriol. 201:e00536–18. doi: 10.1128/JB.00536-18
Cywes-Bentley, C., Skurnik, D., Zaidi, T., Roux, D., Deoliveira, R. B., Garrett, W. S., et al. (2013). Antibody to a conserved antigenic target is protective against diverse prokaryotic and eukaryotic pathogens. Proc. Natl. Acad. Sci. U.S.A. 110, E2209–E2218. doi: 10.1073/pnas.1303573110
Demple, B., Halbrook, J., and Linn, S. (1983). Escherichia coli xth mutants are hypersensitive to hydrogen peroxide. J. Bacteriol. 153, 1079–1082.
Des Prez, R. M., Bryan, C. S., Hawiger, J., and Colley, D. G. (1975). Function of the classical and alternate pathways of human complement in serum treated with ethylene glycol tetraacetic acid and MgCl2-ethylene glycol tetraacetic acid. Infect. Immun. 11, 1235–1243.
Devaraj, A., Buzzo, J., Rocco, C. J., Bakaletz, L. O., and Goodman, S. D. (2018). The DNABII family of proteins is comprised of the only nucleoid associated proteins required for nontypeable Haemophilus influenzae biofilm structure. Microbiologyopen 7:e00563. doi: 10.1002/mbo3.563
Ehrlich, G. D., Veeh, R., Wang, X., Costerton, J. W., Hayes, J. D., Hu, F. Z., et al. (2002). Mucosal biofilm formation on middle-ear mucosa in the chinchilla model of otitis media. JAMA 287, 1710–1715. doi: 10.1001/jama.287.13.1710
Fleischmann, R. D., Adams, M. D., White, O., Clayton, R. A., Kirkness, E. F., Kerlavage, A. R., et al. (1995). Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512. doi: 10.1126/science.7542800
Frey, P. A. (1996). The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 10, 461–470. doi: 10.1096/fasebj.10.4.8647345
Gawronski, J. D., Wong, S. M., Giannoukos, G., Ward, D. V., and Akerley, B. J. (2009). Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. U.S.A. 106, 16422–16427. doi: 10.1073/pnas.0906627106
Giaouris, E., Samoilis, G., Chorianopoulos, N., Ercolini, D., and Nychas, G. J. (2013). Differential protein expression patterns between planktonic and biofilm cells of Salmonella enterica serovar Enteritidis PT4 on stainless steel surface. Int. J. Food Microbiol. 162, 105–113. doi: 10.1016/j.ijfoodmicro.2012.12.023
Goodman, S. D., Obergfell, K. P., Jurcisek, J. A., Novotny, L. A., Downey, J. S., Ayala, E. A., et al. (2011). Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 4, 625–637. doi: 10.1038/mi.2011.27
Göpel, Y., Papenfort, K., Reichenbach, B., Vogel, J., and Görke, B. (2013). Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev. 27, 552–564. doi: 10.1101/gad.210112.112
Greiner, L. L., Watanabe, H., Phillips, N. J., Shao, J., Morgan, A., Zaleski, A., et al. (2004). Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect. Immun. 72, 4249–4260. doi: 10.1128/IAI.72.7.4249-4260.2004
High, N. J., Jennings, M. P., and Moxon, E. R. (1996). Tandem repeats of the tetramer 5'-CAAT-3' present in lic2A are required for phase variation but not lipopolysaccharide biosynthesis in Haemophilus influenzae. Mol. Microbiol. 20, 165–174. doi: 10.1111/j.1365-2958.1996.tb02498.x
Hood, D. W., Cox, A. D., Wakarchuk, W. W., Schur, M., Schweda, E. K., Walsh, S. L., et al. (2001). Genetic basis for expression of the major globotetraose-containing lipopolysaccharide from H. influenzae strain Rd (RM118). Glycobiology 11, 957–967. doi: 10.1093/glycob/11.11.957
Hood, D. W., Deadman, M. E., Allen, T., Masoud, H., Martin, A., Brisson, J. R., et al. (1996). Use of the complete genome sequence information of Haemophilus influenzae strain Rd to investigate lipopolysaccharide biosynthesis. Mol. Microbiol. 22, 951–965. doi: 10.1046/j.1365-2958.1996.01545.x
Hood, D. W., Deadman, M. E., Cox, A. D., Makepeace, K., Martin, A., and Richards, J. C. (2004a). Three genes, lgtF, lic2C and lpsA, have a primary role in determining the pattern of oligosaccharide extension from the inner core of Haemophilus influenzae LPS. Microbiology 150, 2089–2097. doi: 10.1099/mic.0.26912-0
Hood, D. W., Makepeace, K., Deadman, M. E., Rest, R. F., Thibault, P., Martin, A., et al. (1999). Sialic acid in the lipopolysaccharide of Haemophilus influenzae: strain distribution, influence on serum resistance and structural characterization. Mol. Microbiol. 33, 679–692. doi: 10.1046/j.1365-2958.1999.01509.x
Hood, D. W., Randle, G., Cox, A. D., Makepeace, K., Li, J., Schweda, E. K., et al. (2004b). Biosynthesis of cryptic lipopolysaccharide glycoforms in Haemophilus influenzae involves a mechanism similar to that required for O-antigen synthesis. J. Bacteriol. 186, 7429–7439. doi: 10.1128/JB.186.21.7429-7439.2004
Hsieh, Y. J., and Wanner, B. L. (2010). Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13, 198–203. doi: 10.1016/j.mib.2010.01.014
Idicula, W. K., Jurcisek, J. A., Cass, N. D., Ali, S., Goodman, S. D., Elmaraghy, C. A., et al. (2016). Identification of biofilms in post-tympanostomy tube otorrhea. Laryngoscope 126, 1946–1951. doi: 10.1002/lary.25826
Iobst, S. T., and Drickamer, K. (1994). Binding of sugar ligands to Ca(2+)-dependent animal lectins. II. Generation of high-affinity galactose binding by site-directed mutagenesis. J. Biol. Chem. 269, 15512–15519.
Iost, I., and Dreyfus, M. (2006). DEAD-box RNA helicases in Escherichia coli. Nucleic Acids Res. 34, 4189–4197. doi: 10.1093/nar/gkl500
Johansen, E. B., Szoka, F. C., Zaleski, A., Apicella, M. A., and Gibson, B. W. (2010). Utilizing the O-antigen lipopolysaccharide biosynthesis pathway in Escherichia coli to interrogate the substrate specificities of exogenous glycosyltransferase genes in a combinatorial approach. Glycobiology 20, 763–774. doi: 10.1093/glycob/cwq033
Johnston, D. M., and Cannon, J. G. (1999). Construction of mutant strains of Neisseria gonorrhoeae lacking new antibiotic resistance markers using a two gene cassette with positive and negative selection. Gene 236, 179–184.
Kalamorz, F., Reichenbach, B., März, W., Rak, B., and Görke, B. (2007). Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 65, 1518–1533. doi: 10.1111/j.1365-2958.2007.05888.x
Karatan, E., and Michael, A. J. (2013). A wider role for polyamines in biofilm formation. Biotechnol. Lett. 35, 1715–1717. doi: 10.1007/s10529-013-1286-3
Klein, J. O. (1997). Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr. Infect. Dis. J. 16, S5–S8. doi: 10.1097/00006454-199702001-00002
Lampe, D. J., Churchill, M. E., and Robertson, H. M. (1996). A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15, 5470–5479. doi: 10.1002/j.1460-2075.1996.tb00930.x
Langereis, J. D., and Weiser, J. N. (2014). Shielding of a lipooligosaccharide IgM epitope allows evasion of neutrophil-mediated killing of an invasive strain of nontypeable Haemophilus influenzae. MBio 5, e01478–e01414. doi: 10.1128/mBio.01478-14
Lee, F. K., Stephens, D. S., Gibson, B. W., Engstrom, J. J., Zhou, D., and Apicella, M. A. (1995). Microheterogeneity of Neisseria lipooligosaccharide: analysis of a UDP-glucose 4-epimerase mutant of Neisseria meningitidis NMB. Infect. Immun. 63, 2508–2515.
Leite, B., Werle, C. H., Carmo, C. P. D., Nóbrega, D. B., Milanez, G. P., Culler, H. F., et al. (2017). Integration host factor is important for biofilm formation by Salmonella enterica Enteritidis. Pathog. Dis. 75:femspd/ftx074. doi: 10.1093/femspd/ftx074
Lepow, I. H., Naff, G. B., Todd, E. W., Pensky, J., and Hinz, C. F. (1963). Chromatographic resolution of the first component of human complement into three activities. J. Exp. Med. 117, 983–1008. doi: 10.1084/jem.117.6.983
Loeb, M. R. (1984). Immunoblot method for identifying surface components, determining their cross-reactivity, and investigating cell topology: results with Haemophilus influenzae type b. Anal. Biochem. 143, 196–204. doi: 10.1016/0003-2697(84)90576-1
Lovett, S. T. (2011). The DNA exonucleases of Escherichia coli. EcoSal Plus 4:ecosalplus.4.4.7. doi: 10.1128/ecosal.4.4.7
Lv, M., Chen, Y., Liao, L., Liang, Z., Shi, Z., Tang, Y., et al. (2018). Fis is a global regulator critical for modulation of virulence factor production and pathogenicity of Dickeya zeae. Sci. Rep. 8:341. doi: 10.1038/s41598-017-18578-2
Malinverni, J. C., and Silhavy, T. J. (2009). An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc. Natl. Acad. Sci. U.S.A. 106, 8009–8014. doi: 10.1073/pnas.0903229106
Marr, A. K., Overhage, J., Bains, M., and Hancock, R. E. (2007). The Lon protease of Pseudomonas aeruginosa is induced by aminoglycosides and is involved in biofilm formation and motility. Microbiology 153, 474–482. doi: 10.1099/mic.0.2006/002519-0
Maskell, D. J., Szabo, M. J., Deadman, M. E., and Moxon, E. R. (1992). The gal locus from Haemophilus influenzae: cloning, sequencing and the use of gal mutants to study lipopolysaccharide. Mol. Microbiol. 6, 3051–3063. doi: 10.1111/j.1365-2958.1992.tb01763.x
Masoud, H., Moxon, E. R., Martin, A., Krajcarski, D., and Richards, J. C. (1997). Structure of the variable and conserved lipopolysaccharide oligosaccharide epitopes expressed by Haemophilus influenzae serotype b strain Eagan. Biochemistry 36, 2091–2103. doi: 10.1021/bi961989y
Masoud, H., Uhrin, D., Moxon, E. R., and Richards, J. C. (2008). Identification of a novel structural motif in the lipopolysaccharide of the galE/galK double mutant of Haemophilus influenzae strain Eagan. Carbohydr. Res. 343, 2763–2770. doi: 10.1016/j.carres.2008.04.027
Mengin-Lecreulx, D., and Van Heijenoort, J. (1993). Identification of the glmU gene encoding N-acetylglucosamine-1-phosphate uridyltransferase in Escherichia coli. J. Bacteriol. 175, 6150–6157. doi: 10.1128/jb.175.19.6150-6157.1993
Mengin-Lecreulx, D., and Van Heijenoort, J. (1996). Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J. Biol. Chem. 271, 32–39. doi: 10.1074/jbc.271.1.32
Monds, R. D., Silby, M. W., and Mahanty, H. K. (2001). Expression of the Pho regulon negatively regulates biofilm formation by Pseudomonas aureofaciens PA147-2. Mol. Microbiol. 42, 415–426. doi: 10.1046/j.1365-2958.2001.02641.x
Moor, H., Teppo, A., Lahesaare, A., Kivisaar, M., and Teras, R. (2014). Fis overexpression enhances Pseudomonas putida biofilm formation by regulating the ratio of LapA and LapF. Microbiology 160, 2681–2693. doi: 10.1099/mic.0.082503-0
Morris, F. C., Wells, T. J., Bryant, J. A., Schager, A. E., Sevastsyanovich, Y. R., Squire, D. J. P., et al. (2018). YraP contributes to cell envelope integrity and virulence of Salmonella enterica serovar typhimurium. Infect. Immun. 86:e00829–17. doi: 10.1128/IAI.00829-17
Moxon, E. R., and Murphy, T. F. (2000). “Haemophilus influenzae, p. 2369-2378,” in Mandell, Douglas, and Bennett's Principles and Practices of Infectious Diseases, eds G. L. Mandell, J.R. Bennett, and R. Dolin (New York, NY: Churchill Livingstone).
Murphy, T. F., Brauer, A. L., Schiffmacher, A. T., and Sethi, S. (2004). Persistent colonization by Haemophilus influenzae in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 170, 266–272. doi: 10.1164/rccm.200403-354OC
Nakamura, S., Shchepetov, M., Dalia, A. B., Clark, S. E., Murphy, T. F., Sethi, S., et al. (2011). Molecular basis of increased serum resistance among pulmonary isolates of non-typeable Haemophilus influenzae. PLoS Pathog. 7:e1001247. doi: 10.1371/journal.ppat.1001247
Nijland, R., and Burgess, J. G. (2010). Bacterial olfaction. Biotechnol. J. 5, 974–977. doi: 10.1002/biot.201000174
O'toole, G. A. (2011). Microtiter dish biofilm formation assay. J. Vis. Exp. 47:2437. doi: 10.3791/2437
Palcic, M. M., and Hindsgaul, O. (1991). Flexibility in the donor substrate specificity of beta 1,4-galactosyltransferase: application in the synthesis of complex carbohydrates. Glycobiology 1, 205–209. doi: 10.1093/glycob/1.2.205
Pang, B., Armbruster, C. E., Foster, G., Learman, B. S., Gandhi, U., and Swords, W. E. (2018). Autoinducer 2 (AI-2) production by nontypeable Haemophilus influenzae 86-028NP promotes expression of a predicted glycosyltransferase that is a determinant of biofilm maturation, prevention of dispersal, and persistence in vivo. Infect. Immun. 86:e00506–18 doi: 10.1128/IAI.00506-18
Platts-Mills, T. A., and Ishizaka, K. (1974). Activation of the alternate pathway of human complements by rabbit cells. J. Immunol. 113, 348–358.
Pritchard, J. R., Chao, M. C., Abel, S., Davis, B. M., Baranowski, C., Zhang, Y. J., et al. (2014). ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet. 10:e1004782. doi: 10.1371/journal.pgen.1004782
Risberg, A., Masoud, H., Martin, A., Richards, J. C., Moxon, E. R., and Schweda, E. K. (1999). Structural analysis of the lipopolysaccharide oligosaccharide epitopes expressed by a capsule-deficient strain of Haemophilus influenzae Rd. Eur. J. Biochem. 261, 171–180. doi: 10.1046/j.1432-1327.1999.00248.x
Rogers, A., Townsley, L., Gallego-Hernandez, A. L., Beyhan, S., Kwuan, L., and Yildiz, F. H. (2016). The LonA protease regulates biofilm formation, motility, virulence, and the type VI secretion system in Vibrio cholerae. J. Bacteriol. 198, 973–985. doi: 10.1128/JB.00741-15
Rosadini, C.V., Gawronski, J.D, Raimunda, D, Arguello, J.M, and Akerley, B.J. (2011). A novel zinc binding system, ZevAB, is critical for survival of nontypeable Haemophilus influenzae in a murine lung infection model. Infect. Immun. 79:3366–3376.
Rosadini, C. V., Ram, S., and Akerley, B. J. (2014). Outer membrane protein P5 is required for resistance of nontypeable Haemophilus influenzae to both the classical and alternative complement pathways. Infect. Immun. 82, 640–649. doi: 10.1128/IAI.01224-13
Roux, D., Cywes-Bentley, C., Zhang, Y. F., Pons, S., Konkol, M., Kearns, D. B., et al. (2015). Identification of poly-N-acetylglucosamine as a major polysaccharide component of the Bacillus subtilis biofilm matrix. J. Biol. Chem. 290, 19261–19272. doi: 10.1074/jbc.M115.648709
Rubin, E. J., Akerley, B. J., Novik, V. N., Lampe, D. J., Husson, R. N., and Mekalanos, J. J. (1999). In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 96, 1645–1650. doi: 10.1073/pnas.96.4.1645
Sauvage, E., Kerff, F., Terrak, M., Ayala, J. A., and Charlier, P. (2008). The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol. Rev. 32, 234–258. doi: 10.1111/j.1574-6976.2008.00105.x
Severi, E., Hood, D. W., and Thomas, G. H. (2007). Sialic acid utilization by bacterial pathogens. Microbiology 153, 2817–2822. doi: 10.1099/mic.0.2007/009480-0
Sheikh, J., Hicks, S., Dall'agnol, M., Phillips, A. D., and Nataro, J. P. (2001). Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol. 41, 983–997. doi: 10.1046/j.1365-2958.2001.02512.x
Swords, W. E. (2012). Nontypeable Haemophilus influenzae biofilms: role in chronic airway infections. Front. Cell. Infect. Microbiol. 2:97. doi: 10.3389/fcimb.2012.00097
Swords, W. E., Moore, M. L., Godzicki, L., Bukofzer, G., Mitten, M. J., and Voncannon, J. (2004). Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect. Immun. 72, 106–113. doi: 10.1128/IAI.72.1.106-113.2004
Thoden, J. B., Henderson, J. M., Fridovich-Keil, J. L., and Holden, H. M. (2002). Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks. J. Biol. Chem. 277, 27528–27534. doi: 10.1074/jbc.M204413200
Thompson, J. F., Reifenberger, J. G., Giladi, E., Kerouac, K., Gill, J., Hansen, E., et al. (2012). Single-step capture and sequencing of natural DNA for detection of BRCA1 mutations. Genome Res. 22, 340–345. doi: 10.1101/gr.122192.111
Tsang, M. J., Yakhnina, A. A., and Bernhardt, T. G. (2017). NlpD links cell wall remodeling and outer membrane invagination during cytokinesis in Escherichia coli. PLoS Genet. 13:e1006888. doi: 10.1371/journal.pgen.1006888
Vakulskas, C. A., Pannuri, A., Cortés-Selva, D., Zere, T. R., Ahmer, B. M., and Babitzke, P. (2014). Global effects of the DEAD-box RNA helicase DeaD (CsdA) on gene expression over a broad range of temperatures. Mol. Microbiol. 92, 945–958. doi: 10.1111/mmi.12606
Van Melderen, L., and Aertsen, A. (2009). Regulation and quality control by Lon-dependent proteolysis. Res. Microbiol. 160, 645–651. doi: 10.1016/j.resmic.2009.08.021
Vimr, E., Lichtensteiger, C., and Steenbergen, S. (2000). Sialic acid metabolism's dual function in Haemophilus influenzae. Mol. Microbiol. 36, 1113–1123. doi: 10.1046/j.1365-2958.2000.01925.x
Wakarchuk, W., Martin, A., Jennings, M. P., Moxon, E. R., and Richards, J. C. (1996). Functional relationships of the genetic locus encoding the glycosyltransferase enzymes involved in expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. J. Biol. Chem. 271, 19166–19173. doi: 10.1074/jbc.271.32.19166
Wallis, R., and Dodd, R. B. (2000). Interaction of mannose-binding protein with associated serine proteases: effects of naturally occurring mutations. J. Biol. Chem. 275, 30962–30969. doi: 10.1074/jbc.M004030200
Weiser, J. N., Pan, N., Mcgowan, K. L., Musher, D., Martin, A., and Richards, J. (1998). Phosphorylcholine on the lipopolysaccharide of Haemophilus influenzae contributes to persistence in the respiratory tract and sensitivity to serum killing mediated by C-reactive protein. J. Exp. Med. 187, 631–640. doi: 10.1084/jem.187.4.631
Weiser, J. N., Shchepetov, M., and Chong, S. T. (1997). Decoration of lipopolysaccharide with phosphorylcholine: a phase-variable characteristic of Haemophilus influenzae. Infect. Immun. 65, 943–950.
Wiertsema, S. P., Kirkham, L. A., Corscadden, K. J., Mowe, E. N., Bowman, J. M., Jacoby, P., et al. (2011). Predominance of nontypeable Haemophilus influenzae in children with otitis media following introduction of a 3+0 pneumococcal conjugate vaccine schedule. Vaccine 29:5163–5170. doi: 10.1016/j.vaccine.2011.05.035
Williams, B. J., Morlin, G., Valentine, N., and Smith, A. L. (2001). Serum resistance in an invasive, nontypeable Haemophilus influenzae strain. Infect. Immun. 69, 695–705. doi: 10.1128/IAI.69.2.695-705.2001
Wong, S.M., and Akerley, B.J. (2005). Environmental and genetic regulation of the phosphorylcholine epitope of Haemophilus influenzae lipooligosaccharide. Mol. Microbiol. 55:724–738. doi: 10.1111/j.1365-2958.2004.04439.x
Wong, S. M., and Akerley, B. J. (2003). Inducible expression system and marker-linked mutagenesis approach for functional genomics of Haemophilus influenzae. Gene 316, 177–186. doi: 10.1016/S0378-1119(03)00762-5
Wong, S. M., Alugupalli, K. R., Ram, S., and Akerley, B. J. (2007). The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol. Microbiol. 64, 1375–1390. doi: 10.1111/j.1365-2958.2007.05747.x
Wong, S. M., Bernui, M., Shen, H., and Akerley, B. J. (2013). Genome-wide fitness profiling reveals adaptations required by Haemophilus in coinfection with influenza A virus in the murine lung. Proc. Natl. Acad. Sci. U.S.A. 110, 15413–15418. doi: 10.1073/pnas.1311217110
Wong, S. M., St. Michael, F., Cox, A., Ram, S., and Akerley, B. J. (2011). ArcA-regulated glycosyltransferase lic2B promotes complement evasion and pathogenesis of nontypeable Haemophilus influenzae. Infect. Immun. 79, 1971–1983. doi: 10.1128/IAI.01269-10
Xie, F., Li, G., Zhang, Y., Zhou, L., Liu, S., Liu, S., and Wang, C. (2016). The Lon protease homologue LonA, not LonC, contributes to the stress tolerance and biofilm formation of Actinobacillus pleuropneumoniae. Microb. Pathog. 93, 38–43. doi: 10.1016/j.micpath.2016.01.009
Keywords: lipooligosaccharide (LOS), lung infection, Haemophilus influenzae, NTHi, Tn-seq, HITS, immune evasion
Citation: Wong SM, Jackson MD and Akerley BJ (2019) Suppression of Alternative Lipooligosaccharide Glycosyltransferase Activity by UDP-Galactose Epimerase Enhances Murine Lung Infection and Evasion of Serum IgM. Front. Cell. Infect. Microbiol. 9:160. doi: 10.3389/fcimb.2019.00160
Received: 08 March 2019; Accepted: 29 April 2019;
Published: 15 May 2019.
Edited by:
W. Edward Swords, University of Alabama at Birmingham, United StatesReviewed by:
Timothy Murphy, University at Buffalo, United StatesCopyright © 2019 Wong, Jackson and Akerley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian J. Akerley, YmFrZXJsZXlAdW1jLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.