- 1School of Medical Science, Griffith University, Southport, QLD, Australia
- 2Menzies Health Institute Queensland, Griffith University, Southport, QLD, Australia
- 3Institute for Glycomics, Griffith University, Southport, QLD, Australia
- 4School of Chemistry and Molecular Biosciences, University of Queensland, St. Lucia, QLD, Australia
Flagella are expressed on the surface of a wide range of bacteria, conferring motility and contributing to virulence and innate immune stimulation. Host-pathogen interaction studies of the roles of flagella in infection, including due to uropathogenic Escherichia coli (UPEC), have used various methods to purify and examine the biology of the major flagella subunit protein, FliC. These studies have offered insight into the ways in which flagella proteins interact with host cells. However, previous methods used to extract and purify FliC, such as mechanical shearing, ultracentrifugation, heterologous expression in laboratory E. coli strains, and precipitation-inducing chemical treatments have various limitations; as a result, there are few observations based on highly purified, non-denatured FliC in the literature. This is especially relevant to host-pathogen interaction studies such as immune assays that are designed to parallel, as closely as possible, naturally-occurring interactions between host cells and flagella. In this study, we sought to establish a new, carefully optimized method to extract and purify non-denatured, native FliC from the reference UPEC strain CFT073 to be suitable for immune assays. To achieve purification of FliC to homogeneity, we used a mutant CFT073 strain containing deletions in four major chaperone-usher fimbriae operons (type 1, F1C and two P fimbrial gene clusters; CFT073Δ4). A sequential flagella extraction method based on mechanical shearing, ultracentrifugation, size exclusion chromatography, protein concentration and endotoxin removal was applied to CFT073Δ4. Protein purity and integrity was assessed using SDS-PAGE, Western blots with anti-flagellin antisera, and native-PAGE. We also generated a fliC-deficient strain, CFT073Δ4ΔfliC, to enable the concurrent preparation of a suitable carrier control to be applied in downstream assays. Innate immune stimulation was examined by exposing J774A.1 macrophages to 0.05-1 μg of purified FliC for 5 h; the supernatants were analyzed for cytokines known to be induced by flagella, including TNF-α, IL-6, and IL-12; the results were assessed in the context of prior literature. Macrophage responses to purified FliC encompassed significant levels of several cytokines consistent with prior literature reports. The purification method described here establishes a new approach to examine highly purified FliC in the context of host-pathogen interaction model systems.
Introduction
Flagella are complex motility organelles expressed on the surface of a wide range of bacteria. The major structural component of flagella is the filament that affords helical propeller properties to bacterial cells and is the principle component recognized by the immune system, as reviewed elsewhere (Chaban et al., 2015; Hajam et al., 2017). The filament is a polymerized product of more than 20,000 protein monomers termed flagellin or FliC, encoded by the gene termed fliC (Zhou et al., 2015), as reviewed elsewhere (Iino et al., 1988). Collectively, more than fifty genes are required for flagellar biosynthesis, which are divided into 17 or more operons (Chilcott and Hughes, 2000). To enable the complex process of flagellar biogenesis, bacteria use hierarchical regulatory networks that involve transcriptional and post-translational mechanisms to control an ordered expression of flagellar structural components. So-called “early genes” are transcribed from a class 1 promoter in the flhDC operon, which is sensitive to environmental and cell state sensors (Silverman and Simon, 1974). As an early class operon, flhDC is termed the master operon reflecting its essentiality for the transcription of all the genes required for flagellar biosynthesis (Kutsukake et al., 1980). In contrast, “late genes” such as fliC are not engaged translationally until the latter stages of flagella biogenesis (Chilcott and Hughes, 2000). In addition, some E. coli isolates can carry two or more fliC genes (Ratiner, 1998), and sequencing of such alleles in pathogenic E. coli strains has been used to infer evolutionary relationships (Reid et al., 1999). Multiple types of flagellin in a pathogenic bacterium may be related to immune evasion or niche versatility, as discussed elsewhere (Mcquiston et al., 2008; Rossez et al., 2015). Finally, flagellar assembly is also affected by the growth-rate of bacteria, and flagellar abundance correlates with growth rate, whereby faster growing cells produce more flagella (Sim et al., 2017).
As an organelle, a flagellum comprises over 30 unique proteins that range in relative abundance from a few to tens of thousands of copies (Terashima et al., 2008; Chaban et al., 2015; Minamino and Imada, 2015). The structure of the flagellin monomer FliC was originally described in the context of supercoiling and different packing interactions (Samatey et al., 2001); FliC comprises four linearly connected domains; two core (D0 and D1) with alpha-helical structures in lateral N- and C- terminals, and two hypervariable (D3 and D4) that are exposed as folded beta-sheets in the central region. On the basis of the flagellar filament structure, 56 serogroups of E. coli are defined, termed H1 to H56 (Orskov and Orskov, 1992; Wang et al., 2003). H1-type flagella are produced by the commonly studied uropathogenic E. coli (UPEC) reference strain, CFT073; whereas multidrug resistant and globally disseminated ST131 strains of UPEC produce H4 flagella, and the UPEC reference cystitis strain, UTI89, produces H7 flagella. Studies examining the biology of UPEC flagella have contributed a great deal to our understanding of its roles in urinary tract infection and disease pathogenesis (Lane et al., 2005, 2007; Wright et al., 2005; Pichon et al., 2009; Hung et al., 2013; Kakkanat et al., 2015).
In addition to providing motility, flagella contribute to bacterial virulence and host-pathogen interactions via adhesive properties and by triggering immune responses, as reviewed elsewhere (Duan et al., 2013; Haiko and Westerlund-Wikstrom, 2013; Rossez et al., 2015). For example, strains of E. coli associated with meningitis are attenuated for adherence to brain microvascular endothelial cells when the bacteria lack flagella (Parthasarathy et al., 2007). H6 and H7 flagella of enterohemorrhagic and enteropathogenic E. coli exhibit adhesive properties (Giron et al., 2002; Erdem et al., 2007; Mahajan et al., 2009), and H48 flagella from enterotoxigenic E. coli adheres to human Caco-2 cells (Roy et al., 2009). Together, these observations signify a role for flagella in host colonization. In UPEC, flagella-mediated motility has been associated with the ascension of bacteria from the bladder to the kidneys, where host-pathogen interactions leading to inflammation can prompt pyelonephritis (Lane et al., 2007). Other studies have also reported that UPEC flagella can promote urinary tract colonization and invasion of host cells (Wright et al., 2005; Pichon et al., 2009) as well as biofilm formation (Duan et al., 2013; Hung et al., 2013).
There is also evidence supporting more nuanced roles for flagella in E. coli disease pathogenesis, including findings of no major contribution of flagella-mediated motility in urinary tract colonization (Lane et al., 2005), and no role for avian pathogenic or shiga toxin-producing E. coli flagellin in adhesion to Hep-2 cells (La Ragione et al., 2000) or epithelial cells (Rogers et al., 2012). Taken together, these observations are reflective of the highly diverse biological properties of the predicted thousands of distinct types of flagella in bacteria (Pallen and Matzke, 2006) and highlight the importance of careful examination of the roles of flagella and flagellin in experimental systems. Finally, the nature of bacterial flagella as a potent immune activator of innate and adaptive immunity via the Toll-Like Receptor (TLR) host protein, TLR5 has been described, as reviewed elsewhere (Ramos et al., 2004; Gewirtz, 2006; Miao et al., 2007; Hajam et al., 2017). In this context, innate immune responses to flagella can direct the development of flagellin-specific adaptive immune responses and these can modulate the production of flagella in the gut microbiome to help maintain mucosal barrier integrity (Cullender et al., 2013). Thus, there remains a need for improvement in our current understanding of flagella biology, including, in particular, advances in the tools used to examine the interactions between flagella and cells of the immune system.
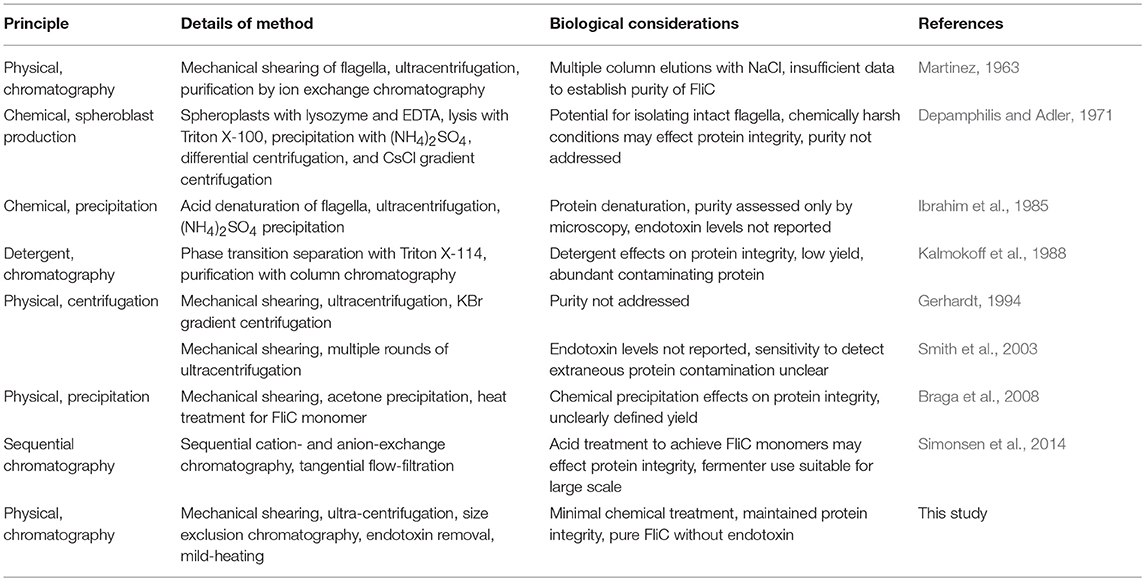
In studying FliC as the principle component of flagella, various methods for extraction and purification of the protein have been described in diverse biological systems, and these are summarized in Table 1. Methods described to date include assorted combinations of mechanical shearing, ultracentrifugation, heterologous expression in laboratory E. coli strains, and precipitation-inducing chemical treatments to isolate and concentrate FliC in order to explore how it interacts at the host cell interface. However, these purification methods have provided little insight into the biology of highly purified, native forms of FliC because of inherent limitations in the methods used, which can adversely affect protein integrity and/or lead to extraneous protein or endotoxin contamination, which can effect downstream (especially immune) assays (Petsch and Anspach, 2000; Gorbet and Sefton, 2005; Schwarz et al., 2014). Additionally, there have been few methodological advances to improve extraction and purification methods for FliC in recent years. A single method that yields pure FliC, free from endotoxin contamination and limiting processes of purification such as protein denaturation would be valuable for studies of the activities of FliC in host-pathogen interaction systems.
In this study, we sought to establish an optimized protocol for extraction and purification of FliC from UPEC to homogeneity, with particular reference to its application to downstream immune assays. We developed and validated a new protocol based on a combination of physical extraction methods and fast protein liquid chromatography (FPLC), followed by protein concentration and endotoxin removal to generate highly pure FliC from UPEC CFT073. Finally, we applied purified FliC to macrophages in vitro, and measured proinflammatory cytokine responses to profile its immune stimulatory properties in the context of previous literature.
Materials and Methods
Bacterial Strains, Plasmids, and Primers
The UPEC reference strain, CFT073 was used in conjunction with several gene-deficient derivatives of the wild-type (Wt) parent, and the laboratory E. coli strain MC4100. fliC-deficient UPEC CFT073 (devoid of flagellar filament) was generated using lambda red recombinase with kanamycin resistance for selection, as previously described (Datsenko and Wanner, 2000). E. coli MC4100 deficient in flhDC (for flagella biosynthesis) was also used. Wt CFT073 and MC4100 strains containing IPTG-inducible pMG600 carrying flhDC were used as hyperflagellated strains. A summary of the bacterial strains used in this study is listed in Table 2.
Additionally, a multiple gene-deficient derivative of Wt CFT073 carrying combined mutations in the 4 genes that encode type 1 fimbriae (fim), F1C/S fimbriae (foc), and pyelonephritis-associated pili (pap1 and pap2), designated CFT073Δ4 (Wurpel et al., 2014), was used to generate a fliC-deficient derivative strain (GU2642; also devoid of fim, foc, pap1, and pap2). All gene deletions were confirmed by PCR and sequencing. The primers used for the construction of mutants are listed in Table 3. Bacteria were grown at 37°C in lysogeny broth (LB) and on LB agar (1.5% agar unless otherwise stated) with antibiotic selection (kanamycin, 50 μg/mL) and IPTG induction (20 mM), as needed.
Growth Conditions and Physical Extraction of Flagella
Initial protein preparations enriched for flagella were isolated from overnight E. coli cultures using ultracentrifugation techniques, essentially as described elsewhere (Gerhardt, 1994) but with minor modifications. Briefly, bacterial cultures were grown overnight in 500 mL LB at 37°C with slow shaking (60 rpm) and were harvested and washed twice in PBS (500 mL, 50 mL, 8,000 × g for 10 min at 4°C); bacterial suspensions were aliquoted into 1.8 mL volumes for flagella isolation. To physically shear the flagella, the bacteria were agitated in 2.0 mL Safe-Lock Microcentrifuge Tubes (Eppendorf) containing two stainless steel ball bearings (5 mm) using a Tissue Lyzer II (Qiagen, Netherlands) (5 × cycles of 30 s at 30 Hz/30 s on ice). The sheared bacterial suspensions were centrifuged at 12,000 × g for 10 min at 4°C to pellet the bacteria, and the supernatants, containing the sheared flagella, were passed through a 0.45 μM nitrocellulose filters (Millipore) to remove any remaining bacteria. In initial assays, we used protease inhibitor (Complete Ultra, EDTA free) at the concentration recommended by the manufacturer (Roche) to assess any protein degradation. Bacteria-free solutions containing the sheared flagella were centrifuged at 100,000 × g for 90 min (Beckman coulter L-90K) at 4°C. The pellets were resuspended in 2 mL PBS, frozen, and quantitated (Thermo Scientific Pierce BCA Protein Assay Kit #23227, USA).
In subsequent experiments, we used the protocol described by Tahoun et al. (2015) with minor modifications to achieve a higher degree of FliC purity. Briefly, following overnight growth of 250 mL LB cultures, cells were harvested at 4,100 × g for 30 min at 4°C. Mechanical shearing of flagella was achieved as described above, and pooled solutions were centrifuged again, and supernatant was collected and re-centrifuged twice. Any residual bacterial cells were removed by centrifugation at 15,000 × g for 10 min at 4°C. The supernatant was transferred into polycarbonate ultracentrifuge tubes and centrifuged at 135,000 × g for 90 min at 4°C. The translucent brown gelatinous pellet, representing flagella, was resuspended in 250 μl PBS and stored at −20°C for subsequent quantification and use. We also applied the protocol for purification of bacterial flagellin as reported by Smith et al. (2003) to compare protein yield and purity with that generated based on the protocol described in the current study.
Characterization of Protein Preparations by SDS-PAGE and Mass Spectrometry
Purity of FliC preparations was assessed by SDS-PAGE, Coomassie staining and western blots. Protein samples (between 1.0 and 12.5 μg per lane, unless otherwise stated and depending on the assay), representing whole cell lysates or flagella extract, were separated in 12% SDS-PAGE gels run at 200 V for 40 min. For Coomassie staining, gels were stained with brilliant blue solution for 1 h and de-stained (1% acetic acid) overnight for visualization using a Chemidoc XRS (Bio-Rad). For western blot, the gels were transferred to 0.45 μm nitrocellulose membranes (Bio-Rad #162-0115, USA) for 1 h at 100 V with cooling, and blocked with 5% skim milk in PBS-T. The membranes were incubated with a 1/100 dilution of polyclonal rabbit anti-flagella H-pool-E (H48+others) or pool-A (H1+others) antibody (Staten Serum Institut, Denmark) for 1 h and washed three times in PBS-T for 5 min. The secondary antibody was a 1:500 dilution of goat anti-rabbit IgG HRP-conjugate (Santa Cruz Biotech #sc-2030, USA) or Goat anti-rabbit IgG-AP (1:10,000; Santa Cruz Biotechnology) for 1 h, subsequently washed four times in PBS-T (5 min). Blots were developed using 3,3′-Diaminobenzidine substrate (Sigma #D4418, USA) or 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium (NBT) (Sigma). The reactions were stopped by addition of water prior to image capture using a flatbed scanner (Epson, Japan) or Chemidoc XRS.
Mass spectrophotometry (MS) was performed to identify several extraneous unknown proteins found to be present in initial FliC preparations. The MS was carried out using protein samples derived from CFT073Δ4 or its fliC-deficient derivative. Proteins (2.5 μg) were resolved by SDS-PAGE, stained with Coomassie blue and de-stained (1% acetic acid) overnight. Proteins bands were isolated in 1% acetic acid and were analyzed at the Translational Research Institute (University of Queensland), Proteomics Core Facility, Brisbane.
Fast Protein Liquid Chromatography (FPLC)
Post-purification of FliC extracts was undertaken by size exclusion chromatography on an ÄKTA Pure protein purification system (GE Lifesciences). We used the Superdex 200 Increase 10/300 GL column with a 24 mL bed volume (GE Lifesciences) equilibrated with 1.5 column volume (CV) of PBS buffer at 0.4 mL.min−1. Prior to application, the FliC extracts were resuspended in 250 μL of PBS, heated at 60°C (10 min) to generate monomers, cooled on ice for 2 min, and applied to the column using a 500 μL sample loop pre-filled with PBS. The 1.2 CV elution flow through was collected in 1 mL fractions using a Frac F9-R fraction collector (GE Lifesciences). The fractions were monitored for protein content by measuring the UV absorbance at both 215 and 280 nm throughout the elution. The protein containing fraction(s) were subsequently concentrated approximately 8-fold using Amicon Ultra-4 10K Centrifugal Filters (Merck Millipore) (for example, 3 mL pool of fraction 13 samples from three preparations concentrated to 400 μL). The samples were stored at 80°C or were used directly in procedures, including endotoxin removal, protein estimation, SDS-PAGE, or in vitro stimulation assays. All FLPC procedures were undertaken at 4°C. A scheme of the protocol used for FliC extraction and purification is illustrated in Figure 1.
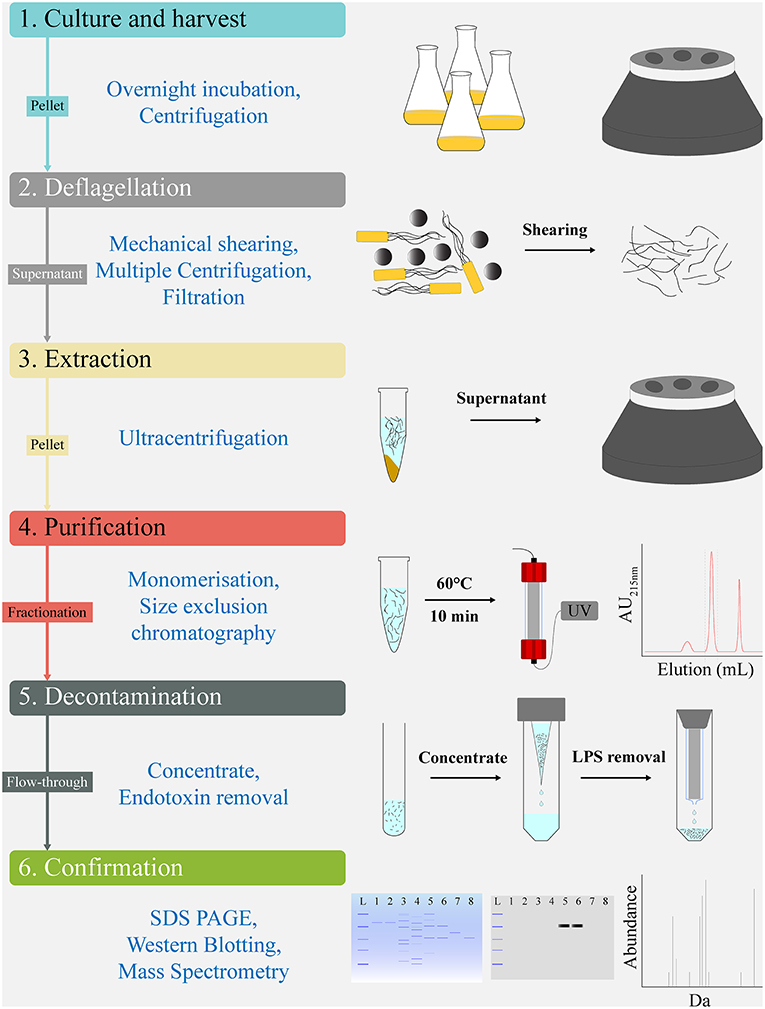
Figure 1. Work flow schematic for extraction and chromatographical purification of FliC from UPEC CFT073. The protocol's sequential steps of Culture and harvest (1), Deflagellation (2), Extraction (3), Purification (4), Decontamination (5), and Confirmation (6) are shown alongside schematics of the major elements comprising each step. Analytic tools used for quality control and validation of the FliC extracts, including MS are shown below the protocols sequential steps.
Endotoxin Removal and Measurement
FliC extracts were treated to remove residual endotoxin using High Capacity Endotoxin Removal Resin Columns (Pierce, 88274), according to the manufacturer's instructions (Thermo Scientific). Briefly, the columns were regenerated with 3.5 mL 0.2N NaOH overnight and after washes with 2M NaCl, ultrapure water and endotoxin free buffer, the FliC extracts were added and incubated at 4°C for 16 h. The proteins were recovered by centrifugation (500 × g for 1 min). Endotoxin levels in purified protein samples were measured using the ToxinSensor Chromogenic LAL Assay (Genscript), according to the manufacturer's instructions. Briefly, 100 μl volumes of samples were applied to endotoxin-free tubes after pH adjustment (pH 6–8) and were mixed with 100 μl of LAL reagent. The tubes were incubated for 15 min at 4°C in the dark, the substrate and color stabilizers were added, and absorbance at 545 nm was measured. Endotoxin concentration is reported in EU.μg−1.
Heat-Induced Monomerization of Flagellar Filament
De-polymerization of flagellar filaments into FliC monomers was assessed by heating proteins (4.5 μg) at temperatures ranging between 30 and 90°C for 10–15 min. Following heat treatment, proteins were mixed with native gel loading buffer containing 1M Tris (pH 6.8), glycerol and bromophenol blue, and were separated in 10% native gels run at 100 V for 1.5 h. The native gels were stained with Coomassie blue and de-stained (1% acetic acid) overnight. Gels from three independent experiments were imaged using a Chemidoc XRS and relative quantitation of protein bands was achieved using ImageJ software (1.6.0_24). Data are reported as mean arbitrary densitometric units (ADU) ±SEM. The tendency of FliC monomers to remain stable (or self-re-polymerize) following heat-induced monomerization was tested by incubating monomers at temperatures between 4 and 37°C for 2, 24, and 48 h; the proteins were then subsequently examined and compared with untreated FliC monomers using native gels.
Cell Culture and Immune Stimulation Assay
Mouse J774A.1 macrophages (ATCC#TIB-67) were grown at 37°C with 5% CO2 in complete RPMI (cRPMI) media, consisting of RPMI1640 (Life Technologies, USA) with 25 mM HEPES, 2 mM L-glutamine, 10% heat-inactivated fetal bovine serum, 100 mM non-essential amino acids, 1 mM sodium pyruvate, 100 U mL−1 penicillin, and 100 mg mL−1 streptomycin. In some experiments, human 5637 uroepithelial (ATCC#HTB-9) and U937 monocyte (ATCC#CRL-1593.2) cell lines were used for comparison. Approximately 1 x 105 host cells in 200 μl cRPMI were seeded into the wells of a 96-well tissue-culture treated microtiter plate, and stimulated with up to 1 μg of purified FliC (50 μl challenge volume consisting of protein [in PBS elution buffer] diluted with cRPMI) for 5 h. Control groups were treated with the equivalent volume of PBS elution buffer prepared from the corresponding FPLC fraction generated using fliC-deficient E. coli.
Additional comparisons were made of cells treated with purified FliC at the pre- and post-endotoxin removal stages of the protocol to determine the effect of the endotoxin removal. We also compared the responses of macrophages to amounts of FliC ranging between 0.05-1 μg. Cell culture supernatants from quadruplicate wells were collected, clarified at 1,000 × g for 10 min at 4°C, and stored at −80°C for subsequent cytokine assay. Experiments were performed in four independent assays. The cytokine concentrations in cell culture supernatants were measured using a multi-target Bio-Plex Assay (Bio-Rad) that included TNF-α, IL-1β, IL-6, mouse keratinocyte-derived chemokine (KC; chosen as a functional equivalent of human IL-8), IL-12(p40), and IL-12(p70), which was performed according to the manufacturer's instructions.
Statistics
Numbers of c.f.u. in suspension cultures are reported as mean ± SEM and were compared using students t-test. Kruskal-Wallis ANOVA and Dunn's multiple comparisons were used to analyze the cytokine levels in macrophage stimulation assays because some data did not satisfy Gaussian distribution and/or normality assumptions. The statistical analyses were performed using Graph Pad Prism v8.0 and SPSS Statistical software package v21. Statistical significance was accepted at p ≤ 0.05.
Results
FliC in Protein Preparations From UPEC CFT073 and MC4100
We initially assessed the yield and relative purity of FliC in extracts from whole cell lysates and flagella-enriched protein preparations generated by physical shearing, filtration and ultracentrifugation. Western blot analysis of whole cell lysates prepared from UPEC CFT073 and MC4100 (grown in liquid media) using anti-flagellin antisera showed no distinct bands for either CFT073 or MC4100 (which has inoperative flhDC) (Barembruch and Hengge, 2007). Introduction of pMG600 (containing the flhDC genes from Serratia) into MC4100 resulted in a band for FliC (~40 kDa) demonstrating reconstitution of flagella expression in this strain (Figure 2A).

Figure 2. Detection of FliC in whole cell lysate of UPEC CFT073 and MC4100 E. coli. (A) Western blot for FliC using whole cell lysate of CFT073 Wt (lane 1), MC4100 (lane 2), and MC4100+pflhDC (lane 3; band ~40 kDa). Cell lysates were derived from bacterial liquid cultures and reacted with anti-flagellin pool H-type antisera. Flagella overexpression in MC4100+pflhDC results in FliC detection. (B) Western blot for FliC using protein preparations that were enriched for flagella using physical methods of shearing, filtration and ultracentrifugation. Shown: flagella-enriched preparations of CFT073 Wt (lane 1–2; 1.0, and 2.5 μg protein, respectively, bands ~60 kDa), MC4100 (lane 3; no band), MC4100+pflhDC (4; bands ~40, ~60 kDa), CFT073ΔfliC (5; no band), and MC4100+pflhDC whole cell lysate prepared from 0.25% soft agar cultures (6; band ~40 kDa). (C) Coomassie stained SDS-PAGE gel of the protein samples shown used for Western blot. M, Marker.
Subsequent analysis of protein preparations that were enriched for flagella by physical methods showed a band for CFT073 (~60 kDa) and a similar reconstitution of flagella expression in MC4100 through the introduction of pflhDC in trans; in addition to a FliC band of expected size (~40 kDa) in MC4100 pflhDC flagella-enrichment revealed a band of equivalent size compared to CFT073 (~60 kDa) along with minor bands (Figure 2B). No bands were detected in protein preparations enriched for flagella derived from CFT073ΔfliC (Figure 2B).
Whole cell lysates prepared using 0.25% soft agar increased the expression of flagella and revealed a strong band for FliC but notable protein contaminants (Figures 2B,C). The addition of protease inhibitor had negligible effects in terms of proteolysis, as observed in a previous study (Lu et al., 2013). The agitation process in microcentrifuge tubes containing ball bearings had no detectable effect on the viability of E. coli, with 3 independent experiments comparing “pre-” and “post-”agitation cultures exhibiting equivalent numbers of c.f.u. (mean values ± SEM; “pre-” = 2.28 ± 0.1 × 108 c.f.u. mL−1 vs. “post-” 2.24 ± 0.1 × 108 mL−1). Taken together, these data show (i) FliC expression is conferred to MC4100 by flhDC supplied in trans, (ii) flagella-enrichment produces a predominant population of ~60 kDa (CFT073/pflhDC); and ~40 kDa (MC4100/pflhDC), and (iii) liquid culture is superior to soft agar culture to enrich FliC due to higher purity, but provides lower overall yield due to less flagella expression (even under culture conditions incorporating IPTG to induce flhDC expression in CFT073/pflhDC).
Purification of FliC From CFT073Δ4 and Identification of Co-purified Proteins
The methods described above achieved a maximum relative purity of FliC of approximately 85% based on densitometric intensities of the major band (FliC) compared to minor contaminating bands observed within Coomassie-stained SDS-PAGE gels and Western blots (data not shown). For FliC to be suitable for immunological assays we sought to attain a higher degree of purity and undertook a genetic approach to further minimize extraneous protein contaminants. To achieve this, we used a multiple mutant strain of CFT073, designated CFT073Δ4 (Wurpel et al., 2014), that contains deletions in genes encoding major surface fimbriae, including type 1, F1C and P fimbriae; this was used for FliC extraction alongside its derivative CFT073Δ4ΔfliC to generate carrier control for further assays.
Physical extraction of flagella from CFT073Δ4 improved the relative purity of the FliC extracts to ~95% according to densitometric analysis of SDS-PAGE gels (Figure 3A). It was vital in these assays to analyze higher amounts of proteins in the gels (up to 12.5 μg) than would typically be analyzed in order to achieve a higher level of sensitivity for the detection of trace protein contamination. We gel-purified the remaining contaminating bands and identified these proteins using mass spectrometry (MS) to assess their potential importance in modifying host-pathogen interactions in downstream assays. MS analysis identified these proteins as major outer membrane proteins OmpA and OmpC, and surface-localized fimbrillin and fimbrial protein (Table 4); these proteins have prominent roles in host-pathogen interactions, including binding to phagocyte scavenger receptors and acting as major immunogens (Jeannin et al., 2005; Liu et al., 2012). Comparing with the protocol for purification of bacterial flagellin described by Smith et al. (2003) we observed a higher yield of protein and contamination with fimbrillin but less extraneous higher molecular weight species using our protocol (Figure 3B). Therefore, we sought additional methods to separate these from FliC, post-purification.
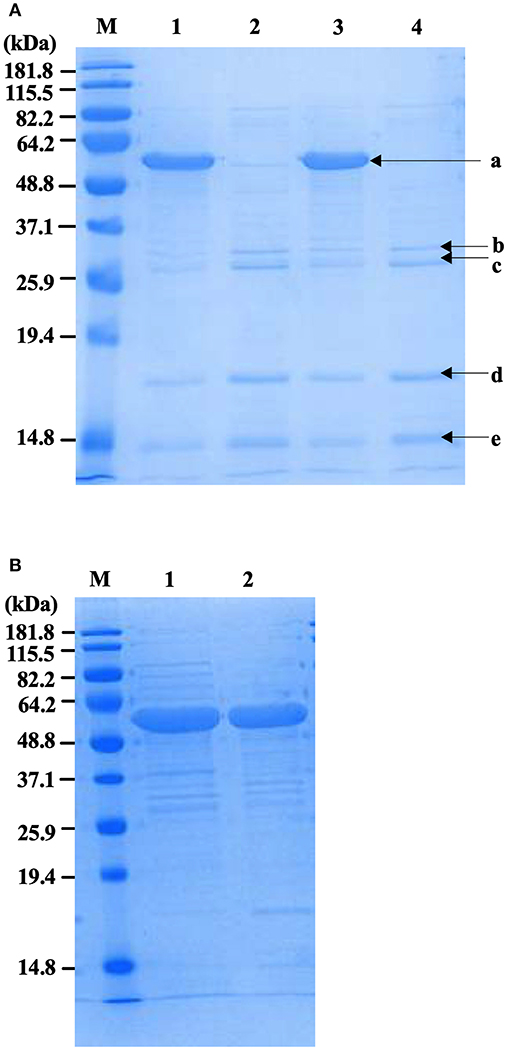
Figure 3. Protein profiles of fliC-enriched extracts from CFT073Δ4 (Δfim Δfoc Δpap1 Δpap2) and its fliC-deficient derivative. (A) Coomassie stained SDS-PAGE gel of FliC protein preparations (5 μg) enriched for flagella from CFT073/pflhDC (1), CFT073ΔfliC/pflhDC (2), CFT073Δ4/pflhDC (3), and CFT073Δ4ΔfliC/pflhDC (4); bands labeled a-e were gel extracted and subsequently analyzed by mass spectrophotometry to identify proteins and are listed in Table 3. (B) Coomassie stained SDS-PAGE gel of protein samples (10 μg) prepared using the protocol for purification of bacterial flagellin described by (Smith et al., 2003) (1) compared to the protocol used in the current study (2). Differences in relative amounts of fimbrillin and extraneous higher molecular weight species are shown in the gel. M, Marker.
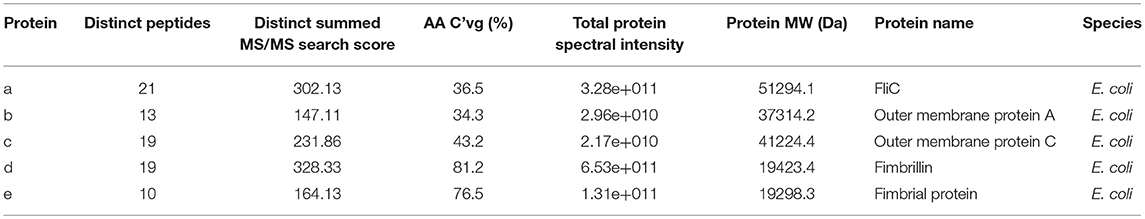
Table 4. Identities of co-purified proteins isolated with FliC from CFT073Δ4 and its fliC-deficient derivative using physical processes of shearing, filtration and ultracentrifugation.
Post-purification of FliC to Homogeneity Using FPLC and Endotoxin Removal
To generate highly pure FliC, protein extracts purified as above using physical processes of shearing, filtration and ultracentrifugation were subsequently separated by size exclusion chromatography using an ÄKTA Pure protein purification system with a Superdex 200 Increase 10/300 GL column. The fractions were analyzed by UV absorbance at 215 and 280 nm during elution, and were collected and protein containing fraction(s) were subsequently concentrated using Ultra Spin columns and analyzed by SDS-PAGE. FliC eluted in fraction 13 with a single peak, as illustrated in Figure 4A. Standard UV absorbance settings of 280 nm (used for monitoring proteins in eluted fractions) did not detect FliC due to an absence of tryptophan (and few tyrosine residues) in the protein, necessitating the application of the alternative 215 nm wavelength to monitor FliC.
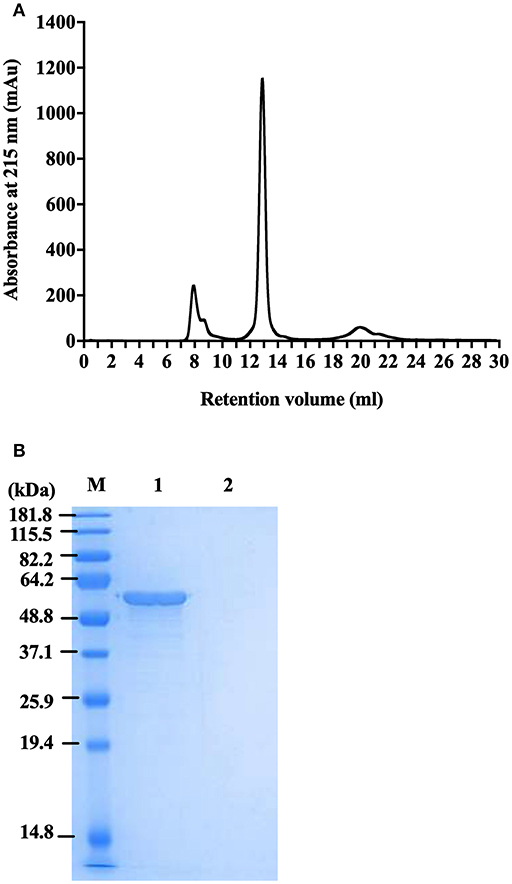
Figure 4. Protein profile of FliC-enriched extract from CFT073Δ4 (Δfim Δfoc Δpap1 Δpap2). (A) Chromatogram of FliC from extract of CFT073Δ4. (B) SDS-PAGE gel of protein (5 μg) in fraction 13 generated from CFT073Δ4/pflhDC (1) and CFT073Δ4ΔfliC/pflhDC (2) following protein concentration by centrifugal filters. M, Marker.
Importantly, these experiments demonstrated that FLPC achieved essentially complete removal of all extraneous protein contaminants from the CFT073Δ4 FliC extracts (Figure 4B). The post-purification process was concluded by applying the FliC preparations to endotoxin removal columns, which resulted in average levels of endotoxin in the final preparations of 0.005 ± 0.002 EU.μg−1; well-below the levels reported in previous immune stimulation studies of purified FliC (max. 0.125 EU.μg−1; Braga et al., 2008, 0.025 EU.μg−1 Metcalfe et al., 2010) and FliA (0.011 EU.μg−1; Schulke et al., 2010). Without performing this endotoxin removal step the FliC preparations routinely exhibited concentrations of contaminating endotoxin above 1 EU.μg−1. Together, these data combined with our findings on physical extraction methods establish FLPC-based post-purification and endotoxin removal as an excellent method for the post-purification of non-denatured UPEC FliC to homogeneity, ideally suited for downstream immunological assays.
Characterization of the Heat Stability of FliC Homopolymers
The flagellar filament is made up of 11 protofilaments, each comprised of FliC monomers stabilized as homopolymers by subunit hydrophobic interactions (Yonekura et al., 2003). Variation in the stability of flagella filaments between species (Yoon and Mekalanos, 2008) prompted us to characterize the heat stability of CFT073 FliC homopolymers with a goal of generating FliC monomers while minimizing protein denaturation so the purified proteins would be ideally suitable for host-pathogen interaction assays. Experiments performed with temperature gradients demonstrated that a treatment of 60°C for 10 min generated FliC monomers at an efficiency approaching 90% (Figure 5). Temperatures above 60°C resulted in minor increases in efficiency of monomerization whereas at temperatures below 50°C there was minimal monomerization. Therefore, 60°C was chosen as optimal to enable efficient generation of FliC monomers but limit the effects of heat denaturation. We then examined the tendency for monomerized FliC to self-polymerize following this heat treatment; for this, flagellar filaments were incubated at room temperature (RT), 60°C or 90°C; and the proteins were then stored at 4°C, RT or 37°C for 2, 24, or 48 h. Native gel analysis of proteins treated in this manner showed no appreciable re-polymerisation of FliC monomers into filaments after monomers were incubated and stored at 4°C. In contrast, FliC stored at RT or 37°C for 24 h or more, exhibited some re-polymerisation (Figure 6). We conclude from these data that UPEC FliC monomers are relatively stable as monomers for at least 2 h following de-polymerization at 60°C for 10 min but incubation of monomers at higher temperatures and/or for longer periods of time leads to some degree of self-re-polymerisation.
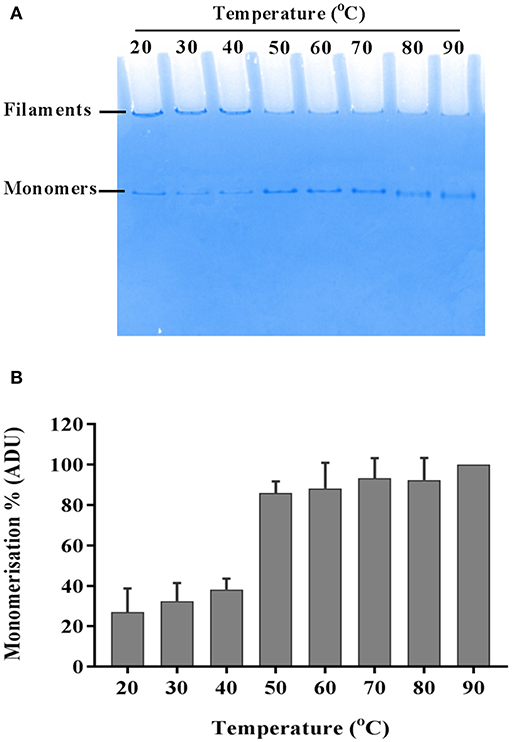
Figure 5. Heat-induced momerisation of purified FliC. Native PAGE gel showing dissociation of FliC from polymeric filaments to FliC monomers (A) and relative densitometric quantification of monomers at each temperature tested (B).
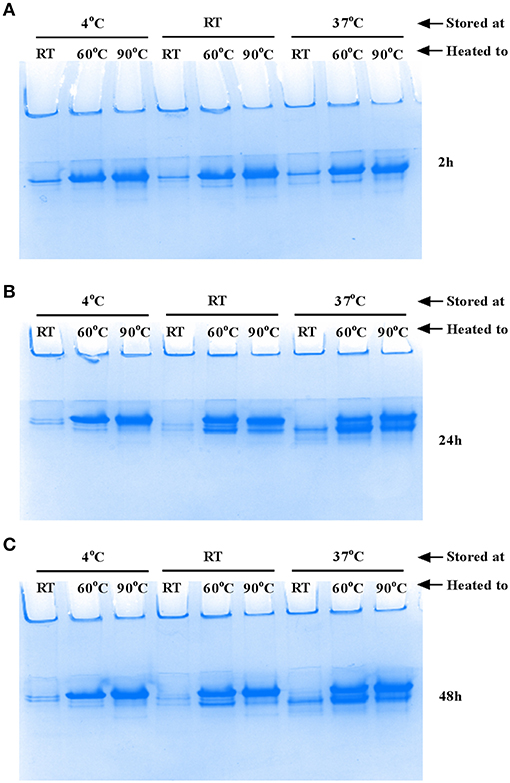
Figure 6. Stability of FliC monomers. Approximately 5 μg FliC (10 μl), diluted in native gel buffer were incubated at room temperature (RT), 60°C or 90°C; the proteins were then stored at 4°C, RT or 37°C for 2 h (A), 24 h (B) and 48 h (C) and subsequently resolved in native gels. The images show no appreciable re-polymerisation into flagella filaments of FliC that was incubated and stored at 4°C. In contrast, FliC stored at RT or 37°C for 24 h or more, exhibited some re-polymerisation apparent as two major bands (instead of a single band) in the images (B) and (C).
Cytokine Response of Macrophages to Highly Purified FliC
To assess the immunological activity of FliC purified to homogeneity, we next applied pure FliC to macrophage stimulation assays in vitro. Exposure of murine J774A.1 macrophages to 1 μg of FliC for 5 h led to significantly increased production of TNF-α, IL-1β, IL-6, KC, IL-12(p40) and IL-12(p70) (Figure 7), We generated a suitable carrier control for these experiments by preparing extracts from a fliC derivative of CFT073Δ4 (GU2642), which were processed in an identical manner to the FliC preparation above. In comparison to stimulation with FliC, there were no significant levels of any of these cytokines in cultures of macrophages that were exposed to either the carrier control (i.e., extracts from GU2642) or culture media only control (Figure 7). We did not observe significant levels of other cytokines that have not previously been associated with flagella, such as G-CSF and GM-CSF (data not shown) and also did not detect significant levels of IFN-γ that has previously been associated with cellular responses to flagella (Wyant et al., 1999). Investigation of the effect of endotoxin removal on macrophage responses demonstrated higher levels of several cytokines in cultures that were stimulated with purified FliC that was not treated for endotoxin removal (“pre-”) compared to FliC that was treated for endotoxin removal (“post-”) (Figure 8). Comparison of the responses of macrophages to different amounts of FliC showed that amounts of FliC <1 μg also triggered significant production of several cytokines; for example, 0.05 μg FliC induced significant production of TNF-α compared to control cultures that were treated with carrier alone (Figure 8). In separate assays using human U937 monocytes and 5,637 epithelial cells we observed similar significant responses for TNF-α, IL-1β, and IL-6 but no significant responses for IL-8 or IL-12(p70) (data not shown). Taken together, these findings show that macrophage proinflammatory cytokine responses as observed in this study are directly attributable to FliC and are consistent with prior reports of immune stimulation properties of flagella.
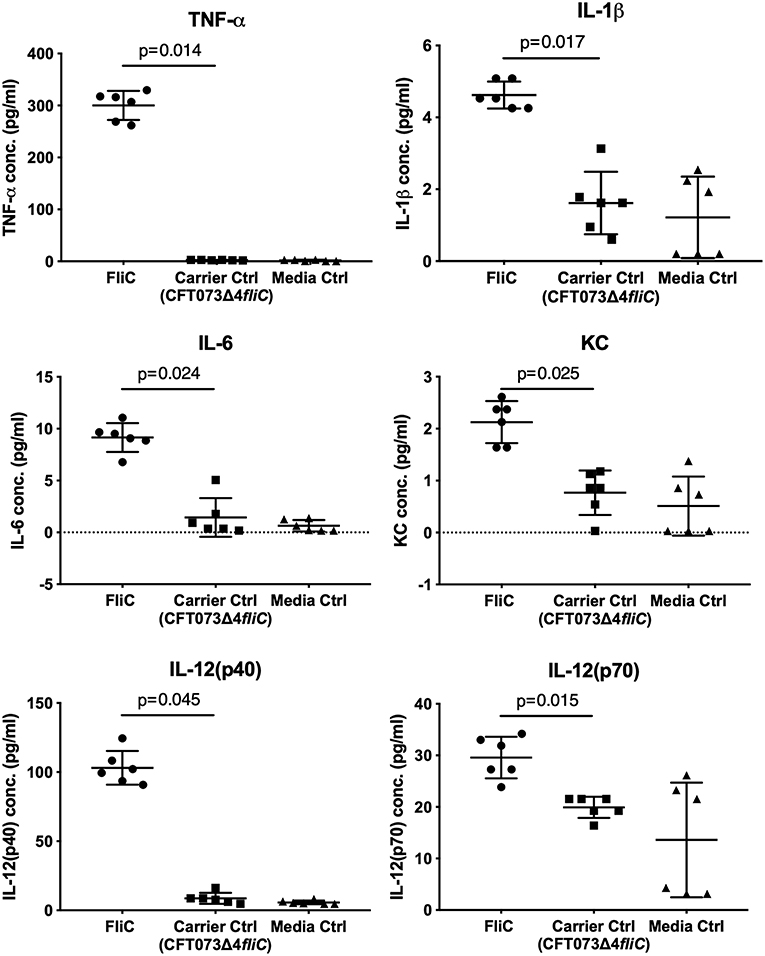
Figure 7. Cytokine response of macrophages to FliC purified to homogeneity from UPEC CFT073Δ4. J774A.1 macrophages were stimulated with 1 μg of FliC for 5 h and supernatants were used to measure the levels of TNF-α, IL-1β, IL-6, KC (chosen as a functional equivalent of human IL-8), IL-12(p40), and IL-12(p70). FliC induced higher concentrations of these cytokines compared to Carrier control (prepared from GU2642 using identical purification procedures) or Media only control.
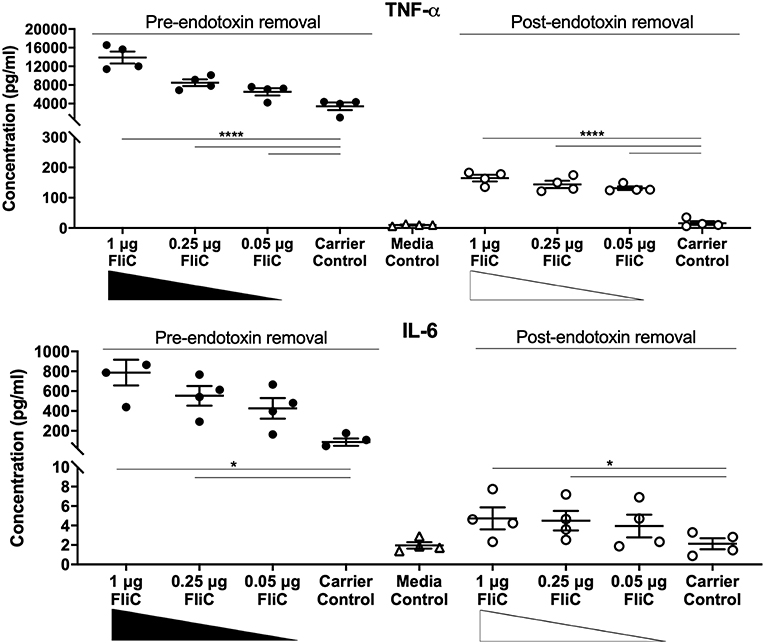
Figure 8. Cytokine response of macrophages to FliC purified to stages of the protocol designated pre- and post-endotoxin removal. J774A.1 macrophages were stimulated with 0.05-1 μg of FliC that had not been treated for endotoxin removal (“pre-“) vs. treated for endotoxin removal (“post-“) for 5 h and supernatants were used to measure the levels of TNF-α and IL-6. *P < 0.05; ****P < 0.0001.
Discussion
Flagellin has been studied as a trigger of immune defense in a various hosts, including humans, animals and plants (Wyant et al., 1999; Gomez-Gomez and Boller, 2000; Eaves-Pyles et al., 2001). In the context of host-pathogen interaction experiments, FliC applied to in vivo or in vitro models ideally should be highly purified by techniques that avoid protein denaturation or degradation and endotoxin contamination that can make interpreting experimental data difficult or impossible. It is likely that many studies have analyzed FliC preparations that are partially degraded, misfolded or contaminated with endotoxin or extraneous proteins as a result of limitations of extraction and purification methods. This is especially important for immune assays where experimental models are intended to provide insights into natural infection and therefore must parallel the natural interactive processes and native forms of protein as closely as possible. Despite notable increases in the numbers of studies of FliC as an immune modulator in recent years, few advances in the methods used for FliC purification have been reported in four decades (Ibrahim et al., 1985). In this study, we establish a method to purify FliC from UPEC which offers advantages of rapid time to purification (1-day), a high yield for purification of bacterial flagellin and generation of a FliC purified to homogeneity suited to immune assay.
Our initial approach of extracting flagellar filaments from UPEC by physical shearing methods followed by filtration and ultracentrifugation yielded FliC as the predominant protein. Several extraneous minor proteins were also present in these FliC extracts and identification of these as having potential to impact host-pathogen interaction studies dictated a further purification approach. Our approach was to further-purify FliC using FPLC-mediated size exclusion chromatography, followed by concentration of the protein and subsequent removal of residual endotoxin. Prior studies have applied chromatographic approaches for isolation of flagellin, such as ion exchange (Martinez, 1963) and sequential cation/anion exchange (Simonsen et al., 2014). However, to our knowledge, the current study is the first to exploit size exclusion (gel filtration) chromatography to purify bacterial flagellin. In the context of protein purification for downstream immune assays, size exclusion offers benefits including a lack of covalent binding of the protein to the exchanger, and no requirement for organic solvents or pH changes for elution; treatments such as these adversely affect protein integrity (Chang et al., 2013) and were therefore avoided in this study. Several studies from the 1970's demonstrate that chemically modified flagellin exhibits altered immunogenic properties (Parish, 1971a,b; Venning, 1975). Chromatographically purified proteins are typically highly purified and can be free from endotoxin. It is notable, however, that in our study, endotoxin removal subsequent to FPLC was essential because appreciable endotoxin was eluted with protein fractions and caused the production of much higher levels of multiple cytokines (e.g., TNF-α, IL-6) in macrophages compared to cytokine responses of macrophages exposed to FliC in the absence of contaminating endotoxin. This stage in the protocol led to substantial loss of protein; however, effective removal of endotoxin to levels considered acceptable for immunological assays circumvented the responses to endotoxin and enabled the study of native highly purified FliC. Comparing to prior literature, we consider the FliC applied in the current study to be among the most highly purified and useful reported to date in the context of immune study.
To our knowledge, the current study is the first study to describe the heat stability of flagellar filaments purified from UPEC. In avoiding acid and other chemical denaturation steps for FliC purification, this study identifies 60°C as an ideal temperature with which to monomerize UPEC flagellar filament. This treatment would avoid denaturation due to acid (Chang et al., 2013) and higher temperatures that can cause irreversible denaturation (Matsuura et al., 2015). Our finding that UPEC FliC monomers are stable as monomers for at least 2 h following de-polymerization is important to validate the use of FliC as monomers in downstream cell stimulation assays; in such assays, time periods of <2 h are realistic in a logistic workflow sense. The finding that incubation of UPEC FliC monomers at higher temperatures and for longer periods of time leads to some degree of self-re-polymerisation will require future work to elucidate the nature of this polymerization. Comparing to other bacteria, the filaments of Salmonella serovar Typhimurium and Vibrio cholerae are completely disassociated into monomers by heating at 80°C for 15 min; the former but not the latter is disassociated by lower temperatures down to 50°C (Yoon and Mekalanos, 2008). In another study, Salmonella flagella were depolymerized at temperatures as low as 37°C with mild acid treatment, whereas Lactobacillus agilis flagella were reported to require 57°C and stronger acid treatment to achieve monomers. It is also possible that fliC allelic variants may react differently in these kinds of dissociation assays. Jointly, however, these data can be used to infer that flagellar filaments of some bacteria are more resistant to thermal and/or acidic conditions than others (Kajikawa et al., 2016).
In many previous studies, bacterial flagella filaments are typically depolymerized into monomers either by heating at 60°C for 10 min or by acid treatment (pH < 2.5). Self-polymerization through the addition of short filaments as seeds can occur and lead to re-polymerization of filament; however, precipitation, such as with ammonium sulfate has been used by many studies to achieve efficient isolation of flagella filaments (Asakura, 1970). Analysis of some heat treatment protocols have led to suggestions that acid treatment for monomerization may be preferred for the maintenance of structural identity (Simonsen et al., 2014); however, acidic conditions (pH 3.6–4.4) as reported for the generation of FliC monomers (Yoon and Mekalanos, 2008) can also cause protein denaturation (Fink et al., 1994). We sought to identify the most mild heat treatment at which UPEC flagella filaments could be dissociated into monomers for immune assay given that TLR5 recognizes FliC in the context of monomers but not flagellar filament (Smith et al., 2003); the requirement for dissociation of flagellar filament into FliC monomers for cellular recognition might involve phagocytic acidification of the local cellular environment and/or cellular translocation of whole bacteria predicted to lead to depolymerization and thus afford recognition by TLR5.
In applying highly purified CFT073 FliC to a host-pathogen interaction assay, we exposed mouse macrophages as well as human monocytes and epithelial cells, to the protein. Several proinflammatory cytokines, including TNF-α, IL-1β, IL-6, KC, and IL-12 were induced in response to the purified FliC in a manner consistent with previous literature (Wyant et al., 1999; Eaves-Pyles et al., 2001; Hayashi et al., 2001; Moors et al., 2001; Kinnebrew et al., 2012). We also observed significant responses to lower doses of FliC (e.g., 0.05 μg), a finding consistent with prior studies that have reported activation of innate immune responses, including NF-κβ, TLR5, and host antimicrobial peptides following exposure to this level of FliC (Smith et al., 2003; Faber et al., 2018; Mowbray et al., 2018). We note that our use of a fliC mutant in these assays reflects a crucial control to conclude that the proinflammatory responses induced by FliC are not an artifact secondary to LPS contamination, which is an important consideration in studies of this type (Eaves-Pyles et al., 2001). We also point out that our expression system is based on Serratia flhDC, which may be a limitation to our study given a recent observation that heterologous expression of regulators that control flagella expression systems may not behave in exactly the same manner, even in systems that show significant synteny (Albanna et al., 2018). In the broader content of bacterial pathogenesis, Salmonella flagella, which occur with two antigenically distinct forms of flagellin, FliC and FljB (Eom et al., 2013), induce proinflammatory responses that encompass TNF-α, IL-1β, and IL-6 in human cells and mice (Ciacci-Woolwine et al., 1997; Mcdermott et al., 2000; Moors et al., 2001) and that appear to be independent of different flagellin variants (Horstmann et al., 2017). Listeria flagella also induce TNF-α in mouse macrophages and systemic IL-6 in vivo (Hayashi et al., 2001). These cytokines are key parts of disease pathogenesis during infection and our observations of UPEC FliC are fundamentally consistent with previous reports of innate immune responses to flagella. Mechanistically, it is well-established that FliC is a TLR5 agonist, and through recognition by this receptor, FliC activates varied immune responses; this mechanism is the basis of novel approaches to disease treatment and prevention such as a potential target for vaccine adjuvant and anti-tumor strategies (Ciacci-Woolwine et al., 1997; Wyant et al., 1999; Mcdermott et al., 2000; Hayashi et al., 2001; Hajam et al., 2017). A recent study identified flagellin functioning via the TLR5/NFkappaB pathway as a key UPEC virulence factor responsible for increased production of host-defense peptides, such as BD2, which mediate protection of urogenital tissues from infection (Ali et al., 2017).
In summary, we report a new method for physical extraction and chromatographical purification of flagella filament from UPEC. The purification of flagellin using this method will be particularly suited for immunological assays. Future studies of UPEC FliC purified to homogeneity using the methods described herein will provide new insight into the biology of this bacterial structure and will be especially useful to give a more complete understanding of the immunological implications of exposure to UPEC flagella in the context of infection and disease.
Author Contributions
DA, MJS, TE and BD performed the experiments. DA, MJS, and BD contributed to drafting of the manuscript and figures. MAS and GU provided expert input, analyzed the data, and contributed to the drafting of the manuscript. All authors contributed to final editing and revisions.
Funding
This work was funded by the National Medical Research Council (NHMRC; Australia) under grant number APP1084889.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dean Gosling and Lahiru Katupitiya for excellent technical assistance.
References
Albanna, A., Sim, M., Hoskisson, P. A., Gillespie, C., Rao, C. V., and Aldridge, P. D. (2018). Driving the expression of the Salmonella enterica sv Typhimurium flagellum using flhDC from Escherichia coli results in key regulatory and cellular differences. Sci. Rep. 8:16705. doi: 10.1038/s41598-018-35005-2
Ali, A. S. M., Mowbray, C., Lanz, M., Stanton, A., Bowen, S., Varley, C. L., et al. (2017). Targeting deficiencies in the TLR5 mediated vaginal response to treat female recurrent urinary tract infection. Sci. Rep. 7:11039. doi: 10.1038/s41598-017-10445-4
Asakura, S. (1970). Polymerization of flagellin and polymorphism of flagella. Adv. Biophys. 1, 99–155.
Barembruch, C., and Hengge, R. (2007). Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 65, 76–89. doi: 10.1111/j.1365-2958.2007.05770.x
Braga, C. J., Massis, L. M., Alencar, B. C., Rodrigues, M. M., Sbrogio-Almeida, M. E., and Ferreira, L. C. (2008). Cytotoxic T cell adjuvant effects of three Salmonella enterica flagellins. Brazil. J. Microbiol. 39, 44–49. doi: 10.1590/S1517-838220080001000011
Chaban, B., Hughes, H. V., and Beeby, M. (2015). The flagellum in bacterial pathogens: For motility and a whole lot more. Semin. Cell Dev. Biol. 46, 91–103. doi: 10.1016/j.semcdb.2015.10.032
Chang, K. C., Cheng, S. J., Chen, Y. C., Huang, H. R., and Liou, J. W. (2013). Nanoscopic analysis on pH induced morphological changes of flagella in Escherichia coli. J. Microbiol. Immunol. Infect. 46, 405–412. doi: 10.1016/j.jmii.2012.08.004
Chilcott, G. S., and Hughes, K. T. (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708. doi: 10.1128/MMBR.64.4.694-708.2000
Ciacci-Woolwine, F., Kucera, L. S., Richardson, S. H., Iyer, N. P., and Mizel, S. B. (1997). Salmonellae activate tumor necrosis factor alpha production in a human promonocytic cell line via a released polypeptide. Infect. Immunity 65, 4624–4633.
Cullender, T. C., Chassaing, B., Janzon, A., Kumar, K., Muller, C. E., Werner, J. J., et al. (2013). Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 14, 571–581. doi: 10.1016/j.chom.2013.10.009
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S. A. 97, 6640–6645. doi: 10.1073/pnas.120163297
Depamphilis, M. L., and Adler, J. (1971). Purification of intact flagella from Escherichia coli and Bacillus subtilis. J. Bacteriol. 105, 376–383.
Duan, Q., Zhou, M., Zhu, L., and Zhu, G. (2013). Flagella and bacterial pathogenicity. J. Basic Microbiol. 53, 1–8. doi: 10.1002/jobm.201100335
Eaves-Pyles, T., Murthy, K., Liaudet, L., Virag, L., Ross, G., Soriano, F. G., et al. (2001). Flagellin, a novel mediator of Salmonella-induced epithelial activation and systemic inflammation: I kappa B alpha degradation, induction of nitric oxide synthase, induction of proinflammatory mediators, and cardiovascular dysfunction. J. Immunol. 166, 1248–1260. doi: 10.4049/jimmunol.166.2.1248
Eom, J. S., Seok Kim, J., Im Jang, J., Kim, B. H., Young Yoo, S., Hyeon Choi, J., et al. (2013). Enhancement of host immune responses by oral vaccination to Salmonella enterica serovar Typhimurium harboring both FliC and FljB flagella. PLoS ONE 8:e74850. doi: 10.1371/journal.pone.0074850
Erdem, A. L., Avelino, F., Xicohtencatl-Cortes, J., and Giron, J. A. (2007). Host protein binding and adhesive properties of H6 and H7 flagella of attaching and effacing Escherichia coli. J. Bacteriol. 189, 7426–7435. doi: 10.1128/jb.00464-07
Faber, E., Tedin, K., Speidel, Y., Brinkmann, M. M., and Josenhans, C. (2018). Functional expression of TLR5 of different vertebrate species and diversification in intestinal pathogen recognition. Sci. Rep. 8:11287. doi: 10.1038/s41598-018-29371-0
Fink, A. L., Calciano, L. J., Goto, Y., Kurotsu, T., and Palleros, D. R. (1994). Classification of acid denaturation of proteins: intermediates and unfolded states. Biochemistry 33, 12504–12511.
Gerhardt, P. (1994). Methods for General and Molecular Bacteriology. Washington, DC: American Society for Microbiology.
Gewirtz, A. T. (2006). Flag in the crossroads: flagellin modulates innate and adaptive immunity. Curr. Opin. Gastroenterol. 22, 8–12. doi: 10.1097/01.mog.0000194791.59337.28
Giron, J. A., Torres, A. G., Freer, E., and Kaper, J. B. (2002). The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44, 361–379. doi: 10.1046/j.1365-2958.2002.02899.x
Givskov, M., Eberl, L., Christiansen, G., Benedik, M. J., and Molin, S. (1995). Induction of phospholipase- and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol. Microbiol. 15, 445–454.
Gomez-Gomez, L., and Boller, T. (2000). FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. doi: 10.1016/S1097-2765(00)80265-8
Gorbet, M. B., and Sefton, M. V. (2005). Endotoxin: the uninvited guest. Biomaterials 26, 6811–6817. doi: 10.1016/j.biomaterials.2005.04.063
Haiko, J., and Westerlund-Wikstrom, B. (2013). The role of the bacterial flagellum in adhesion and virulence. Biology 2, 1242–1267. doi: 10.3390/biology2041242
Hajam, I. A., Dar, P. A., Shahnawaz, I., Jaume, J. C., and Lee, J. H. (2017). Bacterial flagellin-a potent immunomodulatory agent. Exp. Mol. Med. 49:e373. doi: 10.1038/emm.2017.172
Hayashi, F., Smith, K. D., Ozinsky, A., Hawn, T. R., Yi, E. C., Goodlett, D. R., et al. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103. doi: 10.1038/35074106
Horstmann, J. A., Zschieschang, E., Truschel, T., De Diego, J., Lunelli, M., Rohde, M., et al. (2017). Flagellin phase-dependent swimming on epithelial cell surfaces contributes to productive Salmonella gut colonisation. Cell. Microbiol. 19:e12739. doi: 10.1111/cmi.12739
Hung, C., Zhou, Y., Pinkner, J. S., Dodson, K. W., Crowley, J. R., Heuser, J., et al. (2013). Escherichia coli biofilms have an organized and complex extracellular matrix structure. MBio 4, e00645–e00613. doi: 10.1128/mBio.00645-13
Ibrahim, G. F., Fleet, G. H., Lyons, M. J., and Walker, R. A. (1985). Method for the isolation of highly purified Salmonella flagellins. J. Clin. Microbiol. 22, 1040–1044.
Iino, T., Komeda, Y., Kutsukake, K., Macnab, R. M., Matsumura, P., Parkinson, J. S., et al. (1988). New unified nomenclature for the flagellar genes of Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 52, 533–535.
Jeannin, P., Bottazzi, B., Sironi, M., Doni, A., Rusnati, M., Presta, M., et al. (2005). Complexity and complementarity of outer membrane protein A recognition by cellular and humoral innate immunity receptors. Immunity 22, 551–560. doi: 10.1016/j.immuni.2005.03.008
Kajikawa, A., Midorikawa, E., Masuda, K., Kondo, K., Irisawa, T., Igimi, S., et al. (2016). Characterization of flagellins isolated from a highly motile strain of Lactobacillus agilis. BMC Microbiol. 16:49. doi: 10.1186/s12866-016-0667-x
Kakkanat, A., Totsika, M., Schaale, K., Duell, B. L., Lo, A. W., Phan, M. D., et al. (2015). The role of H4 flagella in Escherichia coli ST131 virulence. Sci. Rep. 5:16149. doi: 10.1038/srep16149
Kalmokoff, M. L., Jarrell, K. F., and Koval, S. F. (1988). Isolation of flagella from the archaebacterium Methanococcus voltae by phase separation with Triton X-114. J. Bacteriol. 170, 1752–1758.
Kinnebrew, M. A., Buffie, C. G., Diehl, G. E., Zenewicz, L. A., Leiner, I., Hohl, T. M., et al. (2012). Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity 36, 276–287. doi: 10.1016/j.immuni.2011.12.011
Kutsukake, K., Iino, T., Komeda, Y., and Yamaguchi, S. (1980). Functional homology of fla genes between Salmonella typhimurium and Escherichia coli. Mol. General Genet. 178, 59–67.
La Ragione, R. M., Sayers, A. R., and Woodward, M. J. (2000). The role of fimbriae and flagella in the colonization, invasion and persistence of Escherichia coli O78:K80 in the day-old-chick model. Epidemiol. Infect. 124, 351–363.
Lane, M. C., Alteri, C. J., Smith, S. N., and Mobley, H. L. (2007). Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc. Natl. Acad. Sci. U.S.A. 104, 16669–16674. doi: 10.1073/pnas.0607898104
Lane, M. C., Lockatell, V., Monterosso, G., Lamphier, D., Weinert, J., Hebel, J. R., et al. (2005). Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect. Immunity 73, 7644–7656. doi: 10.1128/IAI.73.11.7644-7656.2005
Liu, Y. F., Yan, J. J., Lei, H. Y., Teng, C. H., Wang, M. C., Tseng, C. C., et al. (2012). Loss of outer membrane protein C in Escherichia coli contributes to both antibiotic resistance and escaping antibody-dependent bactericidal activity. Infect. Immunity 80, 1815–1822. doi: 10.1128/IAI.06395-11
Lu, Y., Welsh, J. P., Chan, W., and Swartz, J. R. (2013). Escherichia coli-based cell free production of flagellin and ordered flagellin display on virus-like particles. Biotechnol. Bioeng. 110, 2073–2085. doi: 10.1002/bit.24903
Mahajan, A., Currie, C. G., Mackie, S., Tree, J., Mcateer, S., Mckendrick, I., et al. (2009). An investigation of the expression and adhesin function of H7 flagella in the interaction of Escherichia coli O157: H7 with bovine intestinal epithelium. Cell. Microbiol. 11, 121–137. doi: 10.1111/j.1462-5822.2008.01244.x
Martinez, R. J. (1963). A method for the purification of bacterial flagella by ion exchange chromatography. J. General Microbiol. 33, 115–120. doi: 10.1099/00221287-33-1-115
Matsuura, Y., Takehira, M., Joti, Y., Ogasahara, K., Tanaka, T., Ono, N., et al. (2015). Thermodynamics of protein denaturation at temperatures over 100 degrees C: CutA1 mutant proteins substituted with hydrophobic and charged residues. Sci. Rep. 5:15545. doi: 10.1038/srep15545
Mcdermott, P. F., Ciacci-Woolwine, F., Snipes, J. A., and Mizel, S. B. (2000). High-affinity interaction between gram-negative flagellin and a cell surface polypeptide results in human monocyte activation. Infect. Immunity 68, 5525–5529. doi: 10.1128/IAI.68.10.5525-5529.2000
Mcquiston, J. R., Fields, P. I., Tauxe, R. V., and Logsdon, J. M. Jr. (2008). Do Salmonella carry spare tyres? Trends Microbiol. 16, 142–148. doi: 10.1016/j.tim.2008.01.009
Metcalfe, H. J., Best, A., Kanellos, T., La Ragione, R. M., and Werling, D. (2010). Flagellin expression enhances Salmonella accumulation in TLR5-positive macrophages. Dev. Compar. Immunol. 34, 797–804. doi: 10.1016/j.dci.2010.02.008
Miao, E. A., Andersen-Nissen, E., Warren, S. E., and Aderem, A. (2007). TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin. Immunopathol. 29, 275–288. doi: 10.1007/s00281-007-0078-z
Minamino, T., and Imada, K. (2015). The bacterial flagellar motor and its structural diversity. Trends Microbiol. 23, 267–274. doi: 10.1016/j.tim.2014.12.011
Mobley, H. L., Green, D. M., Trifillis, A. L., Johnson, D. E., Chippendale, G. R., Lockatell, C. V., et al. (1990). Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immunity 58, 1281–1289.
Moors, M. A., Li, L., and Mizel, S. B. (2001). Activation of interleukin-1 receptor-associated kinase by gram-negative flagellin. Infect. Immunity 69, 4424–4429. doi: 10.1128/IAI.69.7.4424-4429.2001
Mowbray, C. A., Shams, S., Chung, G., Stanton, A., Aldridge, P., Suchenko, A., et al. (2018). High molecular weight hyaluronic acid: a two-pronged protectant against infection of the urogenital tract? Clin. Transl. Immunol. 7:e1021. doi: 10.1002/cti2.1021
Orskov, F., and Orskov, I. (1992). Escherichia coli serotyping and disease in man and animals. Can. J. Microbiol. 38, 699–704.
Pallen, M. J., and Matzke, N. J. (2006). From The Origin of Species to the origin of bacterial flagella. Nat. Rev. Microbiol. 4, 784–790. doi: 10.1038/nrmicro1493
Parish, C. R. (1971a). Immune response to chemically modified flagellin. I. Induction of antibody tolerance to flagellin by acetoacetylated derivatives of the protein. J. Exp. Med. 134, 1–20.
Parish, C. R. (1971b). Immune response to chemically modified flagellin. II. Evidence for a fundamental relationship between humoral and cell-mediated immunity. J. Exp. Med. 134, 21–47.
Parthasarathy, G., Yao, Y., and Kim, K. S. (2007). Flagella promote Escherichia coli K1 association with and invasion of human brain microvascular endothelial cells. Infect. Immunity 75, 2937–2945. doi: 10.1128/IAI.01543-06
Peters, J. E., Thate, T. E., and Craig, N. L. (2003). Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 185, 2017–2021. doi: 10.1128/JB.185.6.2017-2021.2003
Petsch, D., and Anspach, F. B. (2000). Endotoxin removal from protein solutions. J. Biotechnol. 76, 97–119. doi: 10.1016/S0168-1656(99)00185-6
Pichon, C., Hechard, C., Du Merle, L., Chaudray, C., Bonne, I., Guadagnini, S., et al. (2009). Uropathogenic Escherichia coli AL511 requires flagellum to enter renal collecting duct cells. Cell. Microbiol. 11, 616–628. doi: 10.1111/j.1462-5822.2008.01278.x
Ramos, H. C., Rumbo, M., and Sirard, J. C. (2004). Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12, 509–517. doi: 10.1016/j.tim.2004.09.002
Ratiner, Y. A. (1998). New flagellin-specifying genes in some Escherichia coli strains. J. Bacteriol. 180, 979–984.
Reid, S. D., Selander, R. K., and Whittam, T. S. (1999). Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J. Bacteriol. 181, 153–160.
Rogers, T. J., Thorpe, C. M., Paton, A. W., and Paton, J. C. (2012). Role of lipid rafts and flagellin in invasion of colonic epithelial cells by Shiga-toxigenic Escherichia coli O113:H21. Infect. Immunity 80, 2858–2867. doi: 10.1128/IAI.00336-12
Rossez, Y., Wolfson, E. B., Holmes, A., Gally, D. L., and Holden, N. J. (2015). Bacterial flagella: twist and stick, or dodge across the kingdoms. PLoS Pathog. 11:e1004483. doi: 10.1371/journal.ppat.1004483
Roy, K., Hilliard, G. M., Hamilton, D. J., Luo, J., Ostmann, M. M., and Fleckenstein, J. M. (2009). Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature 457, 594–598. doi: 10.1038/nature07568
Samatey, F. A., Imada, K., Nagashima, S., Vonderviszt, F., Kumasaka, T., Yamamoto, M., et al. (2001). Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410, 331–337. doi: 10.1038/35066504
Schulke, S., Waibler, Z., Mende, M. S., Zoccatelli, G., Vieths, S., Toda, M., et al. (2010). Fusion protein of TLR5-ligand and allergen potentiates activation and IL-10 secretion in murine myeloid DC. Mol. Immunol. 48, 341–350. doi: 10.1016/j.molimm.2010.07.006
Schwarz, H., Schmittner, M., Duschl, A., and Horejs-Hoeck, J. (2014). Residual endotoxin contaminations in recombinant proteins are sufficient to activate human CD1c+ dendritic cells. PLoS ONE 9:e113840. doi: 10.1371/journal.pone.0113840
Silverman, M., and Simon, M. (1974). Characterization of Escherichia coli flagellar mutants that are insensitive to catabolite repression. J. Bacteriol. 120, 1196–1203.
Sim, M., Koirala, S., Picton, D., Strahl, H., Hoskisson, P. A., Rao, C. V., et al. (2017). Growth rate control of flagellar assembly in Escherichia coli strain RP437. Sci. Rep. 7:41189. doi: 10.1038/srep41189
Simonsen, K. A., Anderson-Berry, A. L., Delair, S. F., and Davies, H. D. (2014). Early-onset neonatal sepsis. Clin. Microbiol. Rev. 27, 21–47. doi: 10.1128/CMR.00031-13
Smith, K. D., Andersen-Nissen, E., Hayashi, F., Strobe, K., Bergman, M. A., Barrett, S. L., et al. (2003). Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4, 1247–1253. doi: 10.1038/ni1011
Tahoun, A., Jensen, K., Corripio-Miyar, Y., Mcateer, S. P., Corbishley, A., Mahajan, A., et al. (2015). Functional analysis of bovine TLR5 and association with IgA responses of cattle following systemic immunisation with H7 flagella. Vet. Res. 46:9. doi: 10.1186/s13567-014-0135-2
Terashima, H., Kojima, S., and Homma, M. (2008). Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270, 39–85. doi: 10.1016/s1937-6448(08)01402-0
Venning, M. M. (1975). The effect of chemical modification of flagellin on its subsequent immunogenicity. Immunochemistry 12, 365–372.
Wang, L., Rothemund, D., Curd, H., and Reeves, P. R. (2003). Species-wide variation in the Escherichia coli flagellin (H-antigen) gene. J. Bacteriol. 185, 2936–2943.
Wright, K. J., Seed, P. C., and Hultgren, S. J. (2005). Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immunity 73, 7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005
Wurpel, D. J., Totsika, M., Allsopp, L. P., Hartley-Tassell, L. E., Day, C. J., Peters, K. M., et al. (2014). F9 fimbriae of uropathogenic Escherichia coli are expressed at low temperature and recognise Galbeta1-3GlcNAc-containing glycans. PLoS ONE 9:e93177. doi: 10.1371/journal.pone.0093177
Wyant, T. L., Tanner, M. K., and Sztein, M. B. (1999). Salmonella typhi flagella are potent inducers of proinflammatory cytokine secretion by human monocytes. Infect. Immunity 67, 3619–3624.
Yonekura, K., Maki-Yonekura, S., and Namba, K. (2003). Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424, 643–650. doi: 10.1038/nature01830
Yoon, S. S., and Mekalanos, J. J. (2008). Decreased potency of the Vibrio cholerae sheathed flagellum to trigger host innate immunity. Infect. Immunity 76, 1282–1288. doi: 10.1128/IAI.00736-07
Keywords: uropathogenic Escherichia coli, UPEC, Flagella, FliC flagellin, fast protein liquid chromatography, FPLC, immune assay
Citation: Acharya D, Sullivan MJ, Duell BL, Eveno T, Schembri MA and Ulett GC (2019) Physical Extraction and Fast Protein Liquid Chromatography for Purifying Flagella Filament From Uropathogenic Escherichia coli for Immune Assay. Front. Cell. Infect. Microbiol. 9:118. doi: 10.3389/fcimb.2019.00118
Received: 24 December 2018; Accepted: 03 April 2019;
Published: 24 April 2019.
Edited by:
Jonathan Shaw, University of Sheffield, United KingdomReviewed by:
Phillip Aldridge, Newcastle University, United KingdomSusana Merino, University of Barcelona, Spain
Copyright © 2019 Acharya, Sullivan, Duell, Eveno, Schembri and Ulett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Glen C. Ulett, Zy51bGV0dEBncmlmZml0aC5lZHUuYXU=
†Present Address: Tanguy Eveno, Biotechnologies Co., France
Benjamin L. Duell, CanImGuide Therapeutics AB, Malmö, Sweden
 Dhruba Acharya1,2
Dhruba Acharya1,2 Matthew J. Sullivan
Matthew J. Sullivan Tanguy Eveno
Tanguy Eveno Mark A. Schembri
Mark A. Schembri Glen C. Ulett
Glen C. Ulett