- Department of Food Hygiene and Environmental Health, Faculty of Veterinary Medicine, University of Helsinki, Helsinki, Finland
Yersinia pseudotuberculosis is a bacterium that not only survives, but also thrives, proliferates, and remains infective at cold-storage temperatures, making it an adept foodborne pathogen. We analyzed the differences in gene expression between Y. pseudotuberculosis IP32953 grown at 3 and 28°C to investigate which genes were significantly more expressed at low temperature at different phases of growth. We isolated and sequenced the RNA from six distinct corresponding growth points at both temperatures to also outline the expression patterns of the differentially expressed genes. Genes involved in motility, chemotaxis, phosphotransferase systems (PTS), and ATP-binding cassette (ABC) transporters of different nutrients such as fructose and mannose showed higher levels of transcripts at 3°C. At the beginning of growth, especially genes involved in securing nutrients, glycolysis, transcription, and translation were upregulated at 3°C. To thrive as well as it does at low temperature, Y. pseudotuberculosis seems to require certain cold shock proteins, especially those encoded by yptb3585, yptb3586, yptb2414, yptb2950, and yptb1423, and transcription factors, like Rho, IF-1, and RbfA, to maintain its protein synthesis. We also found that genes encoding RNA-helicases CsdA (yptb0468), RhlE (yptb1214), and DbpA (yptb1652), which unwind frozen secondary structures of nucleic acids with cold shock proteins, were significantly more expressed at 3°C, indicating that these RNA-helicases are important or even necessary during cold. Genes involved in excreting poisonous spermidine and acquiring compatible solute glycine betaine, by either uptake or biosynthesis, showed higher levels of transcripts at low temperatures. This is the first finding of a strong connection between the aforementioned genes and the cold adaptation of Y. pseudotuberculosis. Understanding the mechanisms behind the cold adaptation of Y. pseudotuberculosis is crucial for controlling its growth during cold storage of food, and will also shed light on microbial cold adaptation in general.
Introduction
Yersinia pseudotuberculosis is an enteropathogenic bacterium that causes the foodborne infection yersiniosis. Although it has an optimum growth temperature of around 28°C, Y. pseudotuberculosis can grow and multiply at cold-storage temperatures, even as low as 0°C (Keto-Timonen et al., 2018). Long storage at low temperature favors the growth of psychrotrophs like Y. pseudotuberculosis, as they can proliferate without much competition. Outbreaks of Y. pseudotuberculosis have been associated with vegetables, raw milk, and drinking untreated water (Sato and Komazawa, 1991; Nuorti et al., 2004; Rimhanen-Finne et al., 2009; Pärn et al., 2015). In order to survive the various ecological niches through which it could enter the food chain, Y. pseudotuberculosis carries many tools in its genome, such as operons for the transport of substrates more abundant in plants and soil than animal tissue, as well as type VI secretion systems (Jaakkola et al., 2015). The bacterium has been found in the intestines of many animals like domestic pigs, goats, sheep, wild lagomorphs, birds, rodents, and shrews (Niskanen et al., 2003; Laukkanen et al., 2008; Giannitti et al., 2014; Le Guern et al., 2016; Joutsen et al., 2017). It can also thrive in soil as well as in certain protozoans and nematodes (Buzoleva and Somov, 2003; Gengler et al., 2015; Santos-Montañez et al., 2015).
Adaptation to cold and long-term growth at low temperatures poses many challenges to bacteria. Low temperature decreases the fluidity of cell membranes, thereby interfering with normal membrane protein function. It also slows down protein folding, ribosomes, and translation as well as excessively stabilizes nucleic acid structures (Palonen et al., 2010). Cold also induces radical oxygen species production by both slowing metabolism and increasing oxygen solubility (Chattopadhyay et al., 2011). Psychrotrophic bacteria have many methods to deal with these problems. For example, bacteria can increase membrane fluidity by introducing unsaturated lipids that no longer fit as snugly together (Suutari and Laakso, 1994). Unsaturated fatty acids dominate the fatty acid composition of Y. pseudotuberculosis at low temperatures (Bakholdina et al., 2004).
Cold shock proteins (Csp) are small proteins whose mRNA carries a cold shock domain that enables its translation at low temperatures. Csps, along with helicases and translation factors exhibiting similar stabilizing secondary structures, unwind mRNA and support the translational apparatus at low temperatures. Y. pseudotuberculosis has nine csp genes, homologous to those of Escherichia coli, five of which are induced at low temperature in E. coli (Keto-Timonen et al., 2016). The bacterium also has five helicases with a conserved DEAD-box motif: CsdA (yptb0486), RhlE (yptb1214), RhlB (yptb0165), DbpA (yptb1652), and SrmA (yptb2900).
The main goal of this study was to determine how Y. pseudotuberculosis manages to thrive at refrigerator temperature. We identified which genes showed significantly more transcripts at 3°C when compared to 28°C, at each growth phase, mainly focusing on the beginning of growth and logarithmic phase. Of all cold shock proteins, those encoded by yptb1423, yptb3585, yptb3586, and especially yptb2414 and yptb2950, showed significantly more transcripts at low temperature, seemingly forming the backbone of cold acclimation of Y. pseudotuberculosis. Furthermore, we found that, in addition to CsdA, helicases RhlE, and DbpA were significantly upregulated at low temperature, which speaks to their importance in surviving low temperatures. The bacterium also seems to accumulate glycine betaine by uptake and biosynthesis, as the corresponding genes were upregulated at low temperature. Transcription termination factor Rho, along with IF-1 and RbfA, both acting on ribosomes, were also upregulated, which would seem to suggest that they play an important role. None of these genes and proteins have, to our knowledge, been linked to cold acclimation of Y. pseudotuberculosis before.
Materials and Methods
Bacterial Strain and Growth Conditions
Single Y. pseudotuberculosis IP32953 colonies grown on blood agar plates at 28°C were inoculated and grown separately in LB broth (Luria-Bertani; Sigma-Aldrich, St. Louis, MO, USA) at 28°C with shaking overnight. Overnight broths were diluted (1:100) in LB broth and divided into two groups so that four cultures (biological replicates) were grown with shaking both at 3°C and at 28°C. Biological replicates were used to better model true biological variability and improve the accuracy of statistical methods. Samples for total RNA extraction were collected at six corresponding points at different phases of growth across both temperatures (Figure 1). For total RNA extraction 1.25 ml of bacterial culture was mixed with 250 μl cold phenol-ethanol mixture (1:10) and kept on ice for 30 min. After incubation, samples were centrifuged at 4°C at 13,200 rpm for 2 min and the resulting cell pellets were stored at −70°C until RNA isolation.
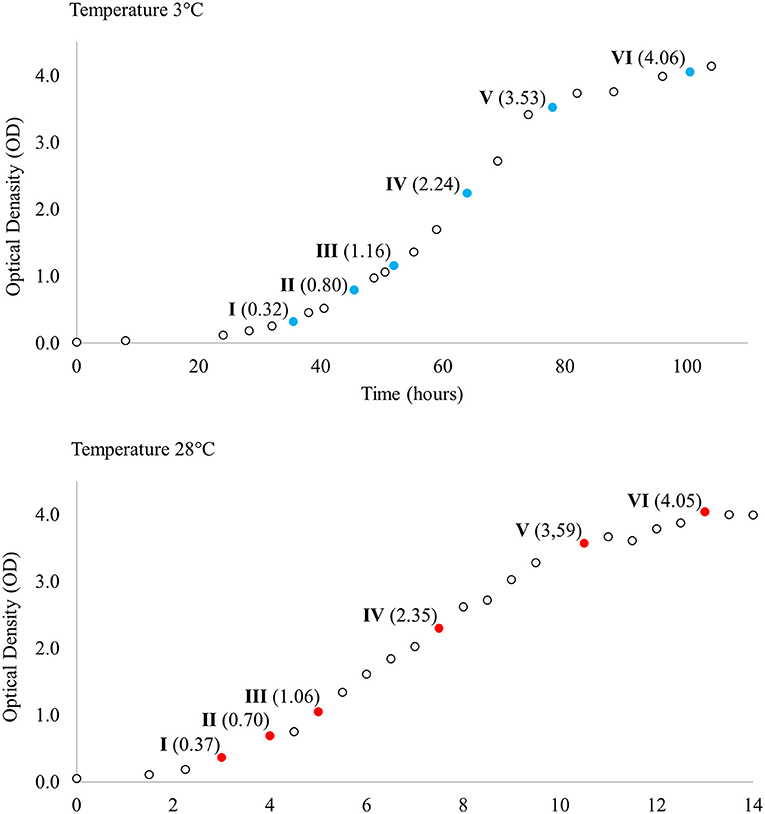
Figure 1. Growth curves of Yersinia pseudotuberculosis IP32953 grown at 3 and 28°C. Samples for total RNA extraction and sequencing were collected at both temperatures at six corresponding growth points I–VI selected by optical density (OD).
RNA Isolation
Total RNA was isolated using GeneJET RNA Purification Kit (Thermo Fisher Scientific, Waltham, MA, USA) and treated with DNA-Free DNA Removal Kit (Ambion, Life Technologies, Carlsbad, CA, USA) according to manufacturers' instructions. The quantity and quality of RNA was examined with a Nanodrop ND-1000 Spectrophotometer (Thermo Fisher Scientific) and Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). RNA was stored at −70°C until RNA-seq library preparation.
Library Preparation and Sequencing
Ribosomal RNA was depleted from total RNA using Ribo-Zero rRNA Removal Kit for Bacteria (Epicenter, Madison, WI, USA) following the manufacturer's protocol. cDNA libraries were prepared using Script-Seq v2 RNA-Seq Library Preparation Kit (Epicenter) following the manufacturer's instructions. The libraries were amplified twice by PCR and barcoded. PCR was performed using Phusion High-Fidelity DNA Polymerase Kit (Thermo Fisher Scientific). The libraries were purified using AMPure XP System (Beckman Coulter, Brea CA, USA) after both PCR procedures. The libraries were sequenced (75 bp single read) using the Illumina NextSeq500 platform at the Institute of Biotechnology, University of Helsinki, yielding four sets of sequence reads (biological replicates) per growth point at both 3 and 28°C. Raw sequences were deposited in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP144570.
CDNA Synthesis and RT-qPCR
To confirm the differences of transcript levels identified in the RNA-sequence expression analysis, RT-qPCR validation was performed for selected genes (betB, csdA, fabF, infA, mdtI, proX, rhlE, and rho) at growth points I and III at both temperatures (3 and 28°C). These genes or operons they represent are discussed in detail in this paper. A total of 500 ng of each RNA sample from three biological replicates per growth point was reverse-transcribed into cDNA in duplicate by using Maxima cDNA Synthesis Kit (Thermo Fisher Scientific) according to manufacturer's instructions. Primers for RT-qPCR were designed using Primer-BLAST software ((Ye et al., 2012); Table 1). Two replicate qPCR reactions for each cDNA sample were performed using the Dynamo Flash SYBR Green qPCR Kit (Thermo Scientific). Each reaction consisted of 1x Master Mix, 0,5 μM of forward and reverse primer and 4 μl of 1:20 (gene of interest) or 1:100,000 (16S rrn) diluted cDNA in a total volume of 20 μl. Rotor-Gene Q thermal cycler (Qiagen GmbH, Hilden Germany) was used in PCR runs with the cycling protocol consisting of initial heating step at 95°C for 7 min, followed by 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 15 s, extension at 72°C for 20 s, and a final extension at 60°C for 1 min. After each run a melt curve analysis was done to confirm specificity. Amplification reaction efficiencies for each primer pair were obtained from RT-qPCR standard curves prepared from serial dilutions of pooled cDNA samples. The duplicate Cq values for PCR replicates were averaged. The relative quantification of gene of interest transcript levels at 3°C, normalized to reference gene (16S rrn) transcript levels and calibrated to the samples taken at the same growth point at 28°C, were calculated using the Pfaffl method (Pfaffl, 2001). Given near equal primer efficiencies, the logFC values of Pfaffl gene expression ratios between 3 and 28°C should be proportional to the logFC values acquired by RNA-Seq. A linear regression analysis was performed on corresponding logFC values of RNA-Seq and RT-qPCR.
Alignment and Annotation of RNA-Seq Data
We aligned, annotated, and analyzed the sequence reads with Bioconductor (Huber et al., 2015). The complete reference genome sequence and genomic features of Y. pseudotuberculosis IP32953 (Johnson et al., 2015) were acquired from the Pathosystems Resource Integration Center, PATRIC (Wattam et al., 2014). The sequence reads were both aligned to the reference genome, allowing a maximum of 10 hits, and annotated with Bioconductor package QuasR (Gaidatzis et al., 2015).
Differential Gene Expression Analysis
One of the replicates of growth point IV at 28°C was discarded for its low alignment quality. The remaining annotated transcript counts were analyzed using Bioconductor package baySeq (Hardcastle and Kelly, 2010). Low count reads were filtered out. The replicate counts for each growth point at 3°C were compared to the counts of the respective growth point at 28°C. The prior distributions were acquired using a negative binomial distribution whose parameters were estimated by quasi-maximum-likelihood methods with a sample size of 10,000. The posterior likelihoods were established using 10 iterations to re-estimate the priors. Results were normalized by library size and results with FDR >0.05 were discarded. Log2 fold changes (logFC) across the two temperatures were calculated using transcript count averages of replicates, and genes with logFC ≥ 2 were considered significantly expressed at 3°C. For ease of viewing, genes were further grouped into operons retrieved from ProOpDB (Taboada et al., 2012). Amino acid sequence similarities were derived from multiple sequence alignment by Clustal Omega (Sievers et al., 2011). Upregulated genes sharing similar functions were classified into functional units (modules), acquired from the KEGG database (Kanehisa and Goto, 2000), using Bioconductor package clusterProfiler (Yu et al., 2012). P-values were adjusted for multiple comparisons by controlling the false discovery rate, using the Benjamini & Hochberg method implemented in clusterProfiler (Benjamini and Hochberg, 1995).
Clustering
Expression profiles of genes that were significantly more expressed at low temperature, at least at one growth point, were clustered using Pearson correlation distance. Count data was normalized for visualization by using median ratio normalization implemented in Bioconductor package DESeq2 (Love et al., 2014). Averages of replicate counts were used instead of individual counts. Normalized data for each gene (rows) across all growth points (columns) was then plotted in a heatmap (Figure 2).
Figure 2. Gene expression profile clustering of Yersinia pseudotuberculosis strain IP32953 genes that showed significantly more transcripts (logFC ≥ 2, FDR ≤ 0.05) at 3°C than 28°C at least once during growth. The tree has been cut to five subclusters (A–E). Subclusters (B–E) are of most interest because they comprise genes that express highly at the beginning and logarithmic phase of growth. Subcluster A consists of genes whose expression mainly peaks at stationary phase. Flagellar assembly (Figure S1) and chemotaxis genes (Figure S2) have been left out as Y. pseudotuberculosis is not motile at 28°C. In addition, tRNA-genes (Figure S3) have been filtered out, so that the final N = 482. The darker the color in the heatmap, the more transcripts the gene showed at that growth point.
Results
Expression profiles of Y. pseudotuberculosis IP32953 grown at 3 and 28°C were compared at corresponding phases of the growth based on growth curves determined by the optical density. We found 570 genes in total that showed significantly more transcripts at 3°C than 28°C at least at one of the growth points I–VI (Table 2). Motility and chemotaxis genes were at the top throughout growth since Y. pseudotuberculosis is non-motile at 28°C. The total number of upregulated genes was 482 when motility, chemotaxis, and tRNA genes were filtered out. Growth point VI held the fewest significantly expressed genes (N = 125) whereas growth points II and IV held the most (N = 162; Table 2). The difference in expression was largest for genes involved in motility, with a maximum logFC of 7.42.
Table 2. The number of Yersinia pseudotuberculosis strain IP32953 genes expressed significantly more at 3°C than at 28°C by growth point.
A gene that was expressed both differentially and in great numbers at 3°C probably plays an important role in cold growth. We clustered expression profiles of genes that at least once during growth showed more transcripts at 3°C than at 28°C. Five subclusters (A–E) could be identified in the expression profile heatmap (Figure 2). The genes of most interest in the context of cold growth express highly at the beginning and logarithmic phases of growth, clustered into subclusters B–E (see details in Supplementary Material). Subcluster A mostly held genes that were expressed highly at the stationary phase.
Validation of RNA Sequencing Results With RT-qPCR
The expression profiles of eight differentially expressed genes at two different growth points were evaluated by RT-qPCR and compared with the RNA-seq analysis results. In linear regression analysis between the RNA-seq and RT-qPCR logFC values (Figure 3), a 0.93 Pearson correlation coefficient value (R2 = 0.85) was observed, which confirmed the reproducibility and reliability of the RNA-seq method (Figure 3).
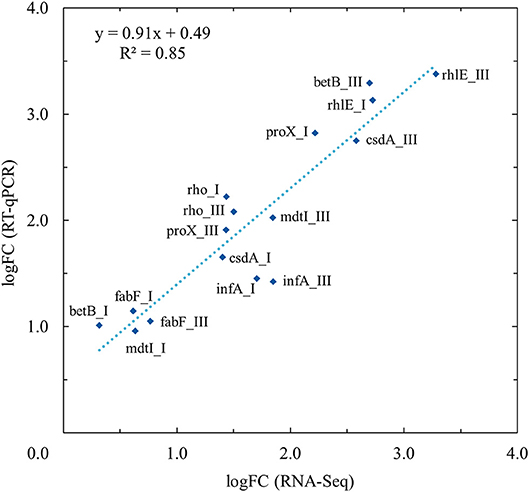
Figure 3. Validation of RNA-Seq results with quantitative real-time reverse-transcription PCR (RT-qPCR) using linear regression analysis. RT-qPCR validation was performed for selected genes (betB, csdA, fabF, infA, mdtI, proX, rhlE, and rho) at growth points I and III at both temperatures (3 and 28°C). Linear regression analysis showed an R2 coefficient of determination value of 0.85 between the RNA-Seq and RT-qPCR log2 fold changes (logFC).
Differentially Expressed Genes by Growth Point
At growth point I, genes involved in acquiring compatible solutes and various nutrients showed significantly more transcripts at 3°C, and this pattern continued until stationary phase (Figure 4). For example, a significant increase in transcripts at 3°C was displayed in the following genes: genes encoding a glycine betaine transporter (yptb2959–61); phosphotransferase systems (PTS) to import fructose (yptb1329–31; Figure 5), N-acetylglucosamine (yptb1120, yptb3075–82), and L-ascorbate (yptb2600–2; Figure 5); and ATP-binding cassette (ABC) transporters of maltose (yptb2521, yptb3095–102) and aldopentoses (yptb3591–6). A significant portion of upregulated genes at this growth point were associated with different PTSs and glycine betaine transport (Figure 4). As expected, genes encoding chaperone molecules such as helicase RhlE (yptb1214) and Csps (yptb2950, yptb2414, yptb3585, and yptb3586), which destabilize nucleic acid secondary structures, showed more transcripts at 3°C.
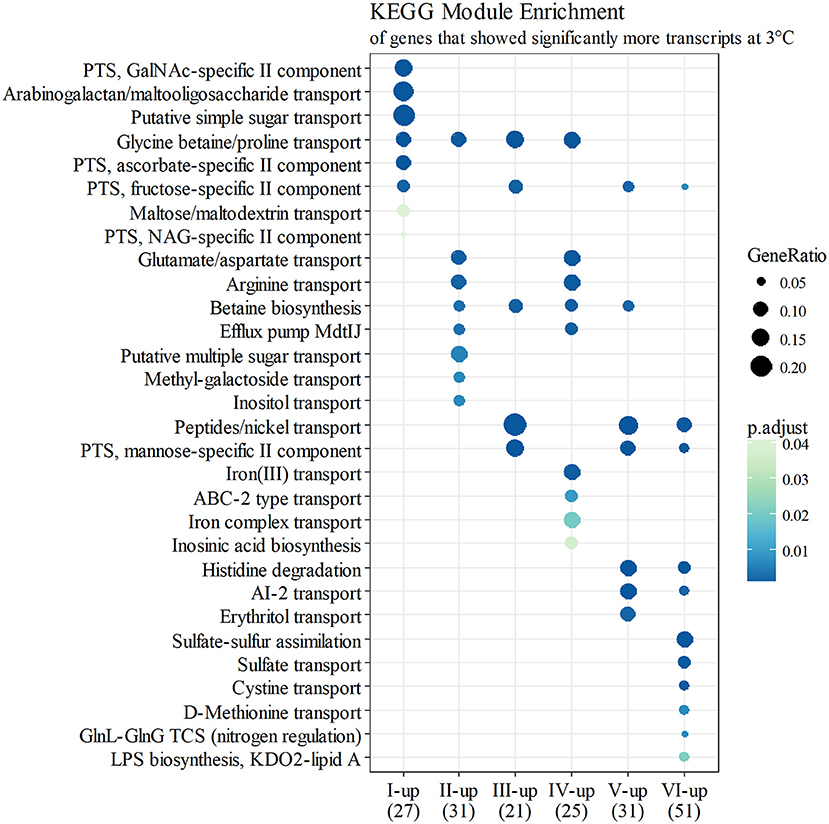
Figure 4. KEGG module enrichment (BH adjusted p ≤ 0.05) of genes that showed significantly more transcripts at 3°C (logFC ≥ 2 for at least one operon member, FDR ≤ 0.05). A gene can belong to multiple modules, or none. Some module names have been shortened for clarity.
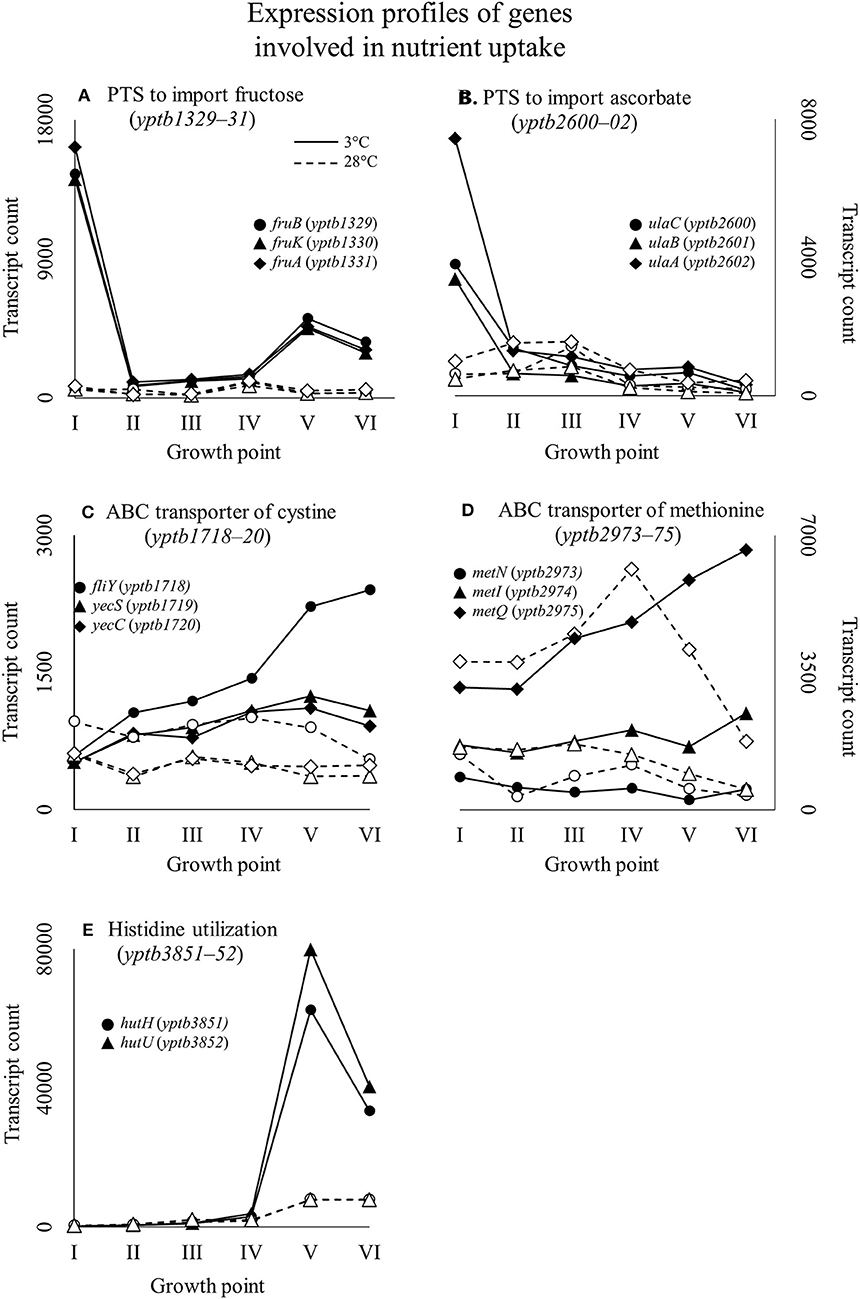
Figure 5. Expression profiles of example genes involved in nutrient uptake: fru-operon encoding a phosphotransferase system (PTS) to import fructose (A); ula-operon encoding a PTS to import ascorbate (B); ATP binding cassette (ABC) transporter of cystine, dimer form of cysteine (C); ABC transporter of methionine (D); and part of the histidine utilization system (E) of Yersinia pseudotuberculosis IP32953, that was grown at 3 and 28°C and sampled at corresponding growth points I–VI. Median ratio normalized counts were used. A large significant difference in expression of sugar importers between 3 and 28°C can be observed at the beginning of growth. Toward the end of growth at 3°C, amino acid utilization takes precedence when sugar importer expression tapers off.
At growth point II, genes involved in further processing of compatible solutes, spermidine efflux, synthesizing desaturated membrane lipids, biosynthesis of ribosomes, securing translation under cold stress, and posttranscriptional modification of RNA molecules showed significantly more transcripts at 3°C. Modules of different amino acid transporters and betaine synthesis were overexpressed (Figure 4). Genes encoding translation factors IF-1 (yptb1395) and Rho factor (yptb0167) peaked at this growth point. New additions to differentially expressed chaperone genes were helicase gene dbpA (yptb1652) and a new Csp gene (yptb1423).
At growth point III, a urease operon (yptb2938–44) and PTSs to import fructose (Figure 5), N-acetylglucosamine, and mannose showed significantly more transcripts at 3°C than 28°C. The gene encoding superoxide dismutase (yptb3925) also showed more transcripts at 3°C at this growth point. At growth point IV, it seems the nutrient strategy shifted as an operon involved in sulfur metabolism (yptb2309–14) was expressed more at 3°C than 28°C. In addition to genes encoding IF-1 and Rho factor, a new translation factor gene rbfA (yptb0481) showed more transcripts at 3°C at this point of growth.
At growth points V and VI, the nutrient strategy continued shifting as operons involved in the metabolism of histidine (yptb1965–9, yptb3851–2; Figure 5), cystine (yptb1717–20; Figure 5), methionine (yptb2973–5; Figure 5), and nitrogen compounds (yptb0022–3) showed more transcripts at 3°C. At the same time, Csp gene expression levels and differences tapered off. The gene encoding helicase CsdA (yptb0486) was significantly more expressed at 3°C at growth point III, but at 28°C at growth points V and VI. An operon encoding a system that utilizes autoinducer AI-2 showed significantly more transcripts at growth point V.
Expression of Cold Shock Protein Genes
Of all the Csp genes, yptb2414 showed most transcripts at 3°C from the beginning of growth throughout the logarithmic growth phase (Figure 6). Its difference in expression between 3 and 28°C was significant at all growth points and the largest of all Csp genes. Gene yptb1624 was expressed significantly more at 28°C (Figure 6). Gene yptb2950 showed many more transcripts at 3°C at growth points I–IV, with a peak logFC of over 7 (Figure 6). Its transcription counts dipped after growth point I and decreased toward stationary phase. Gene yptb1423 was expressed at lower levels, but showed significantly more transcripts at 3°C throughout growth points II–IV (Figure 6).
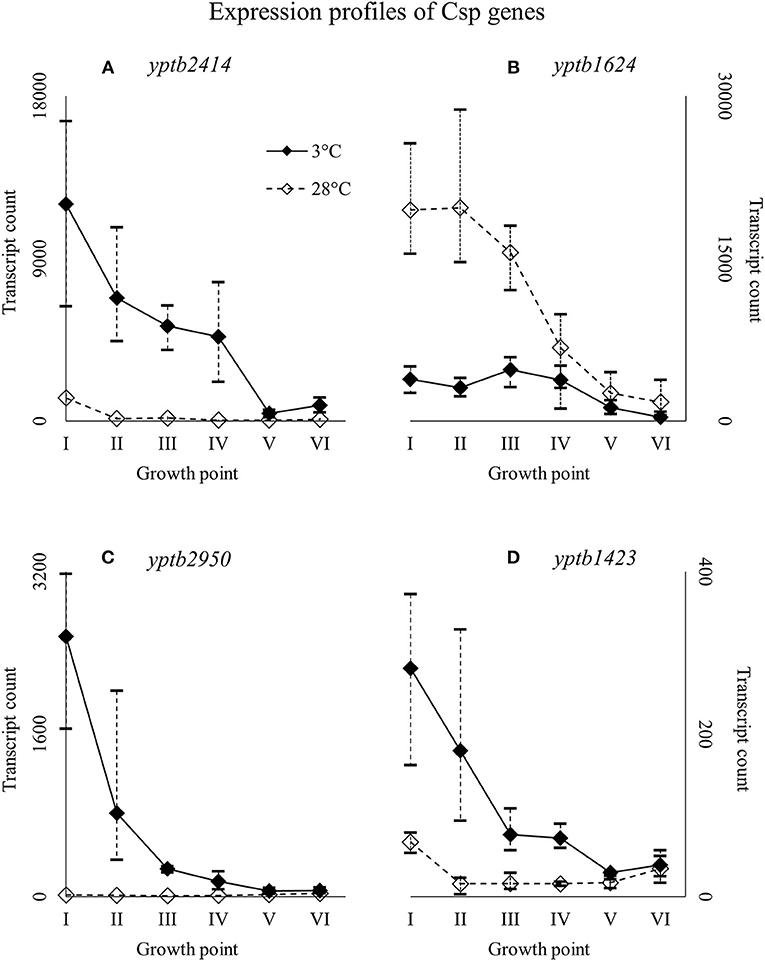
Figure 6. Expression profiles of cold shock protein genes yptb2414 (A), yptb1624 (B), yptb2950 (C), and yptb1423 (D) of Yersinia pseudotuberculosis IP32953, that was grown at 3 and 28°C and sampled at corresponding growth points I–VI. Median ratio normalized counts were used and the variation between replicates is shown by vertical lines. The expression curves are similar, but yptb2414, yptb2950, and yptb1423 showed significantly more transcripts at 3°C and yptb1624 at 28°C at the beginning and logarithmic phase of growth.
Nearly identical genes, yptb3585 and yptb3586, were expressed highly at growth point I, but their expression levels dipped right after (Figure 7). However, the differences in their expression between 3 and 28°C consistently favored cold at significant levels. Expression levels of yptb3587 and yptb1088 rose toward the stationary phase at 3°C, but the difference between 3 and 28°C was not significant. After starting growth, yptb3587 appeared slightly downregulated (Figure 7). Gene yptb1392 was expressed much in the same way, but showed significantly more transcripts at 28°C at the stationary phase of growth (Figure 7).
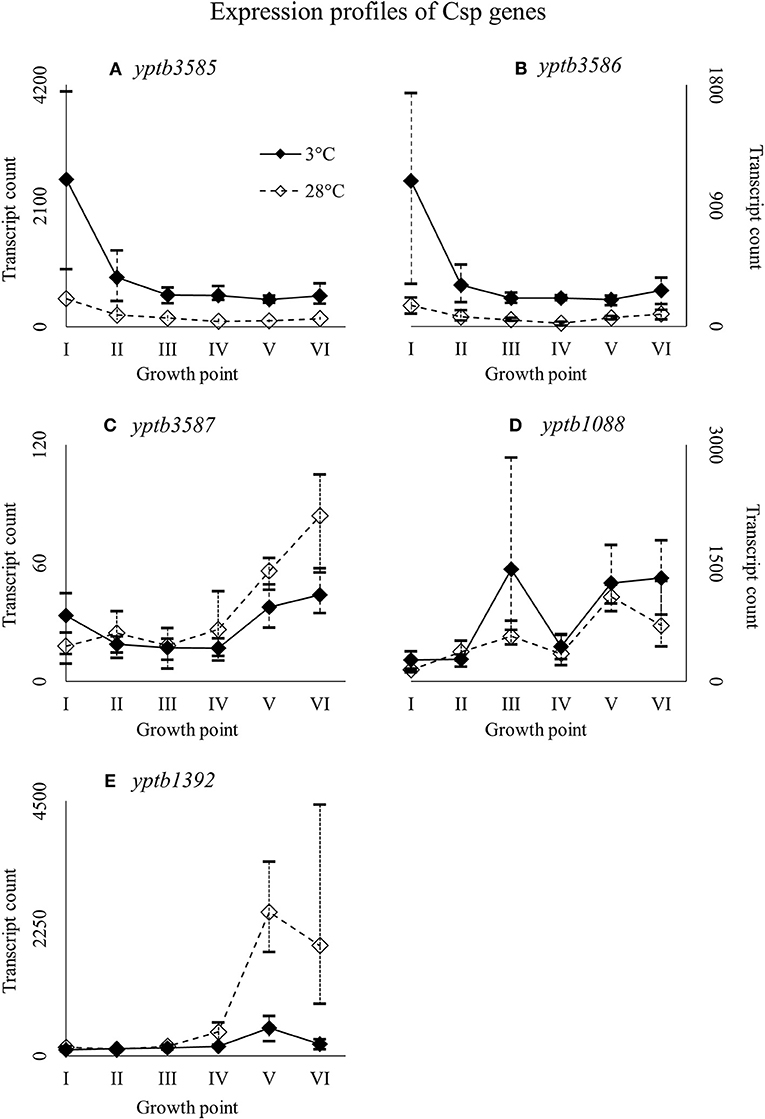
Figure 7. Expression profiles of cold shock protein genes yptb3585 (A), yptb3586 (B), yptb3587 (C), yptb1088 (D), and yptb1392 (E) of Yersinia pseudotuberculosis IP32953, that was grown at 3 and 28°C and sampled at corresponding growth points I–VI. Median ratio normalized counts were used and the variation between replicates is shown by vertical lines. Expression of yptb3585–86 decreased toward stationary phase whereas that of yptb3687, yptb1088, and yptb1392 increased.
Expression of DEAD-Box RNA Helicase Genes
Y. pseudotuberculosis has five helicases with a conserved DEAD-box motif: CsdA (yptb0486), RhlE (yptb1214), RhlB (yptb0165), DbpA (yptb1652), and SrmA (yptb2900). All the helicase genes were expressed more at 3°C throughout growth (Figure 8), but the difference was significant for csdA (yptb0486) at growth point III, rhlE (yptb1214) at growth points I–IV, and dbpA (yptb1652) at growth points II and IV.
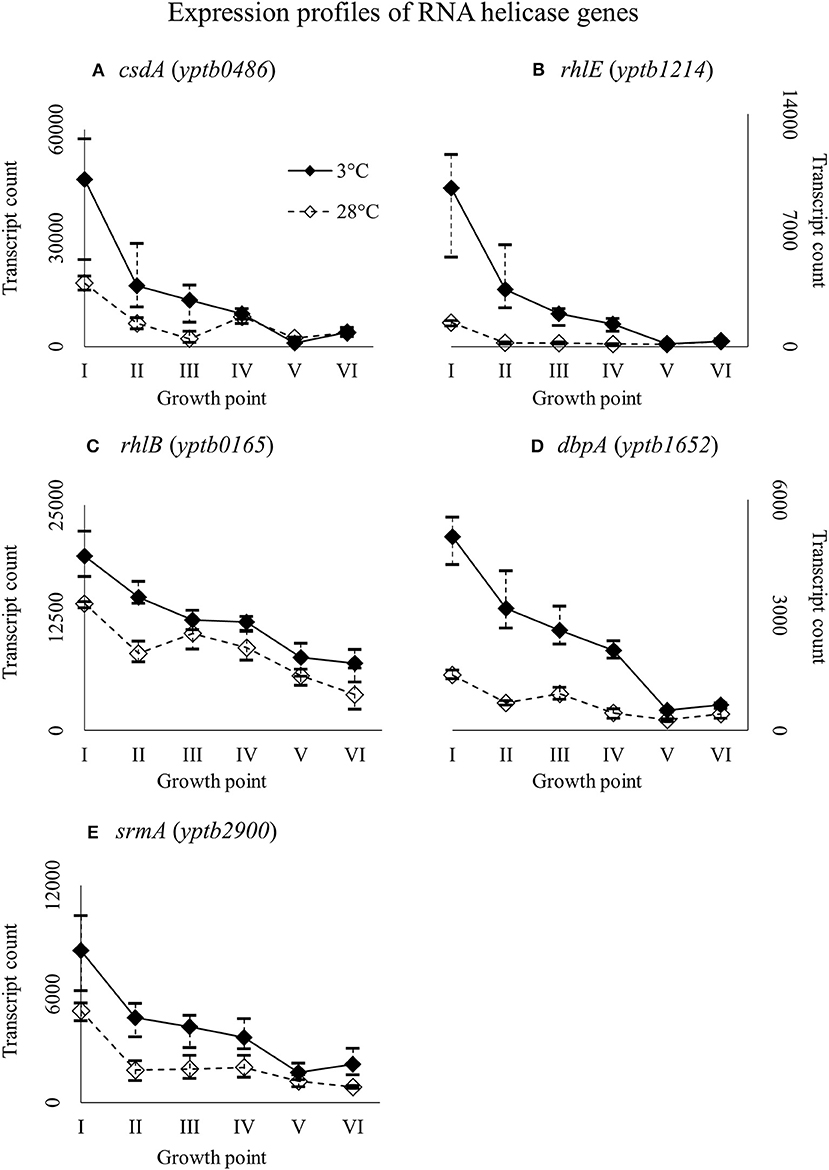
Figure 8. Expression profiles of DEAD-box RNA helicase genes csdA (A), rhlE (B), rhlB (C), dbpA (D), and srmA (E) of Yersinia pseudotuberculosis IP32953, that was grown at 3 and 28°C and sampled at corresponding growth points I–VI. Median ratio normalized counts were used and the variation between replicates is shown by vertical lines. Gene csdA showed significantly more transcripts at growth point III, rhlE at growth points I–IV, and dbpA at growth points II and IV, although all the genes expressed more at 3°C at least from the beginning through logarithmic phase.
Expression of Genes Handling Compatible Solutes
Genes yptb2959–61 form the proU operon that encodes a transport system for glycine betaine and proline; genes yptb1195–98 form the betIBA-betT divergent operon that is involved in glycine betaine biosynthesis; and genes yptb2052 and yptb2051 form the mdtIJ operon that encodes a spermidine efflux pump. Operon proU showed significantly more transcripts at 3°C at growth points I–IV (Figure 9), operon betIBA-betT at growth points II–IV (Figure 10), and operon mdtIJ at II and IV (Figure 11). Corresponding functional modules were also overexpressed at growth points I–IV for operon proU, points II–V for operon betIBA-betT, and points II and IV for operon mdtIJ (Figure 4). Another glycine betaine and L-proline transporter encoding gene proP (yptb0608) showed more transcripts at 3°C throughout the growth, but not quite at significant levels (Figure 9).
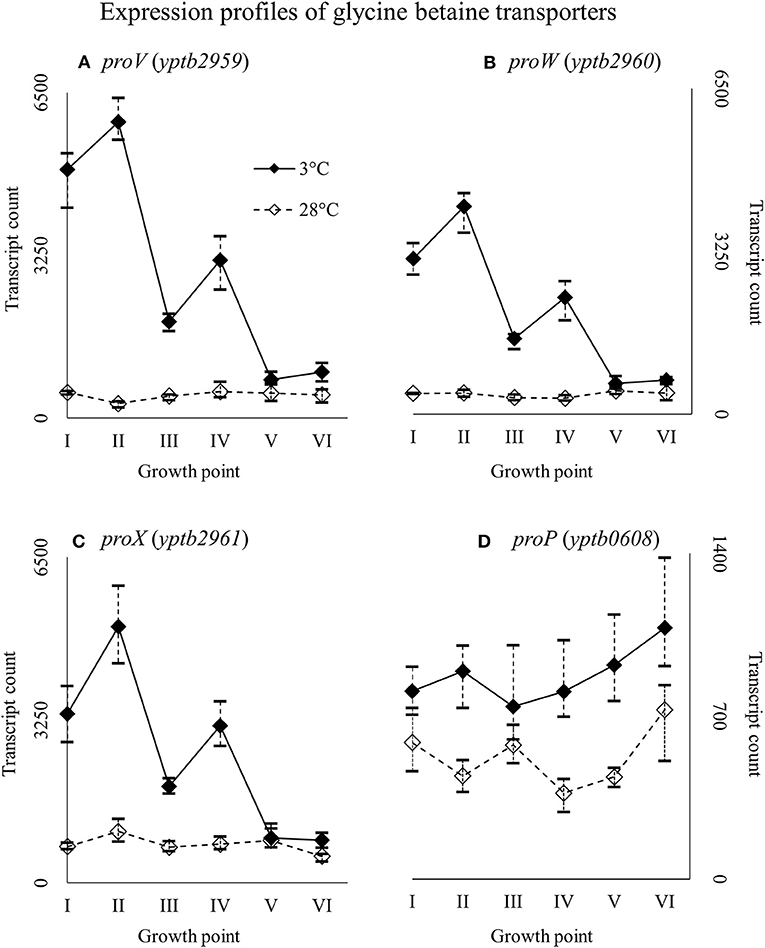
Figure 9. Expression profiles of genes encoding glycine betaine transporters, the proU operon (proVWX, A–C), and proP (D) of Yersinia pseudotuberculosis IP32953, that was grown at 3 and 28°C and sampled at corresponding growth points I–VI. Median ratio normalized counts were used and the variation between replicates is shown by vertical lines. The proU operon is hardly expressed at all at 28°C and proP also showed more transcripts at 3°C.
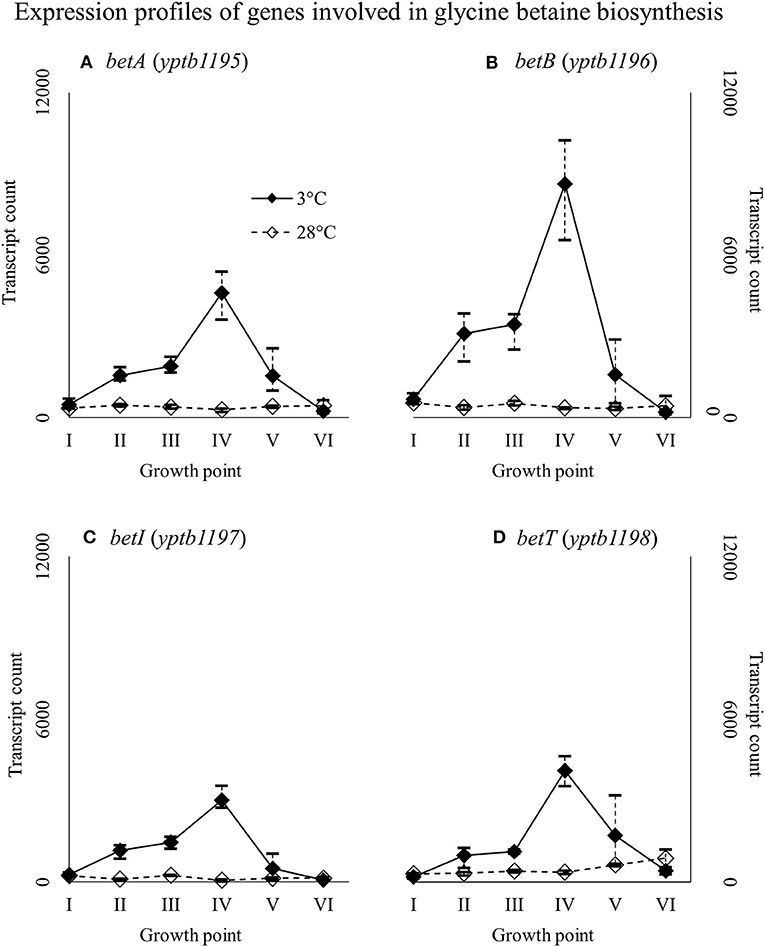
Figure 10. Expression profiles of genes involved in glycine betaine biosynthesis, the betIBA-betT divergent operon (A–D), of Yersinia pseudotuberculosis IP32953, that was grown at 3 and 28°C and sampled at corresponding growth points I–VI. Median ratio normalized counts were used and the variation between replicates is shown by vertical lines. The expression of the operon is all but diminished at 28°C.
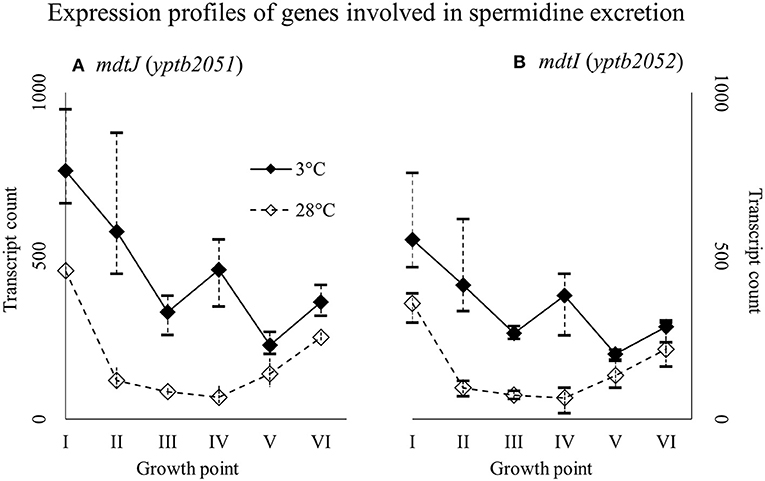
Figure 11. Expression profiles of genes encoding a spermidine efflux pump MdtIJ (A,B) of Yersinia pseudotuberculosis IP32953, that was grown at 3 and 28°C and sampled at corresponding growth points I–VI. Median ratio normalized counts were used and the variation between replicates is shown by vertical lines. The genes were expressed more at 3°C throughout the growth with a new expression peak at growth point IV.
Expression of Genes Encoding Rho Factor, IF-1, and RbfA
Genes rho (yptb0167), infA (yptb1395), and rbfA (yptb0481) encode the homologs of Rho factor, the translation initiation factor IF-1, and ribosome binding factor RbfA, respectively. The three factor genes were expressed highly from the beginning of growth to the end of the logarithmic phase (Figure 12). Their difference in expression between 3 and 28°C was significant at growth points II and IV, over 16-fold at the largest for yptb1395, but the three genes showed more transcripts at 3°C consistently throughout the growth.
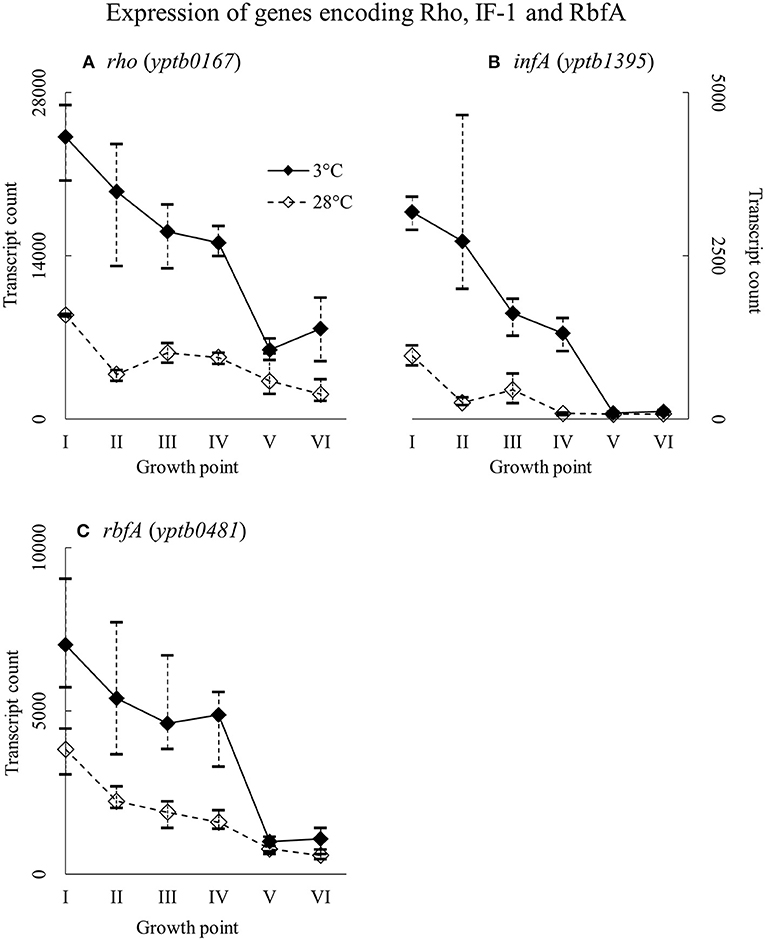
Figure 12. Expression profiles of genes encoding transcription termination factor Rho (A), translation initiation factor IF-1 (B), and ribosome binding factor RbfA (C) of Yersinia pseudotuberculosis IP32953, that was grown at 3 and 28°C and sampled at corresponding growth points I–VI. Median ratio normalized counts were used and the variation between replicates is shown by vertical lines. All three factors showed more transcripts at 3°C at least to the end of logarithmic phase.
Expression of Genes Involved in Modifying Membrane Lipid Composition
Genes yptb1450 and yptb2469, encoding FabA and FabF, were expressed highly at the beginning of growth whereas gene yptb2426 encoding FabB at stationary phase (Figure 13). Gene yptb2469 showed significantly more transcripts at 3°C at growth point II, although the difference favored cold during most of the growth.
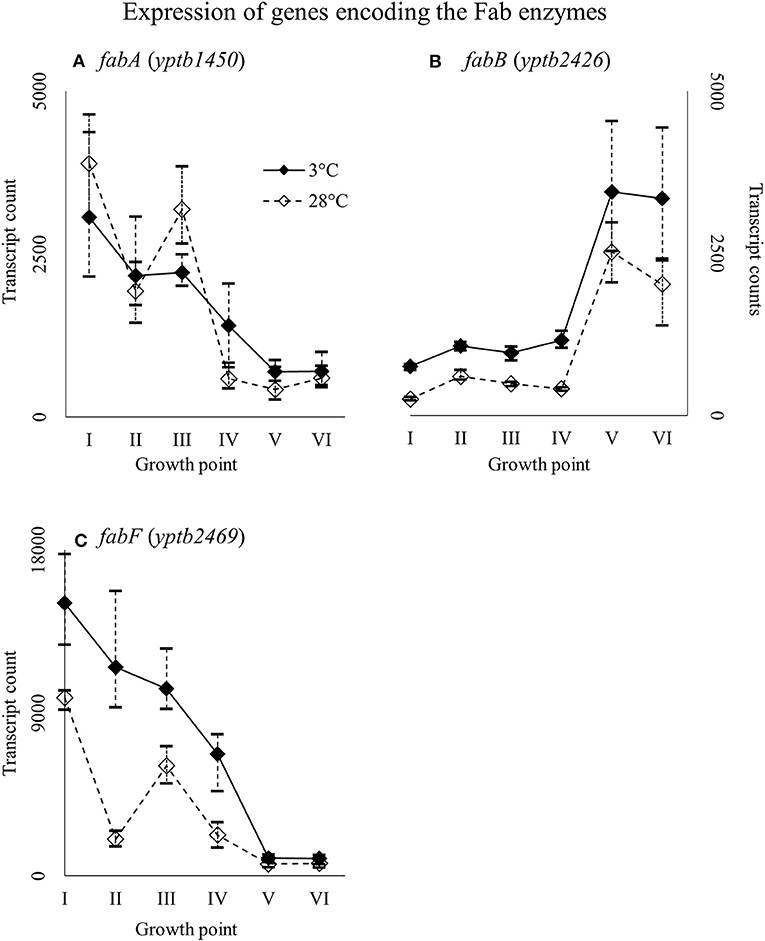
Figure 13. Expression profiles of genes encoding the Fab enzymes fabA (A), fabB (B), and fabF (C) of Yersinia pseudotuberculosis IP32953, that was grown at 3 and 28°C and sampled at corresponding growth points I–VI. Median ratio normalized counts were used and the variation between replicates is shown by vertical lines. Gene fabB and fabF showed more transcripts at 3°C during most of the growth.
Discussion
We identified several upregulated genes that may be important to cold adaptation of Y. pseudotuberculosis. We also analyzed gene expression profiles to better understand what happens at different phases of growth in cold. To tackle problems posed by refrigerator temperatures, Y. pseudotuberculosis has an extensive toolbox that includes Csps, DEAD-box RNA-helicases, compatible solutes, transcription factors, and fatty acid saturases.
Nutrient Acquisition
Y. pseudotuberculosis expresses many genes involved in securing nutrients significantly more at 3°C compared to 28°C (Figures 4,5), as successful growth in low temperatures requires more energy. In our results, a shift in nutrient utilization over time is observed. At the beginning of growth, transporters of fructose, ascorbate, maltooligosaccharides, and maltose were upregulated at 3°C. At the beginning of logarithmic phase, transporters of glutamate, aspartate, and arginine were upregulated. Toward the stationary phase, in addition to an operon turning histidine to glutamate, transporters of cystine, methionine, and sulfates were upregulated.
PTSs allow quick acquisition of carbohydrates from the environment (Postma et al., 1993). PTSs have also been linked to regulatory functions during cold stress (Wouters et al., 2000; Monedero et al., 2007). Y. pseudotuberculosis expresses several PTSs especially at the beginning of growth significantly more at 3°C than 28°C, as it is in dire need of carbon to sustain its laborious growth. Another large peak in expression of a fructose PTS can be observed at growth point V at 3°C, whereas the peak is located at growth point IV at 28°C. However, this is probably due to slower regulatory processes at 3°C. In Y. pestis, the malMBKEFG-operon, which is involved in maltose intake, was highly upregulated after cold shock (Han et al., 2005). Maltose can act as an cryoprotectant in addition to being a carbon source (Jain and Roy, 2009). In our results, parts of the mal-operon were upregulated especially at the beginning of growth.
E. coli has been shown to accumulate certain amino acids, including aspartic acid, glutamic acid, and methionine, when subjected to suboptimal temperatures (Jozefczuk et al., 2010). Accumulation has been theorized to result from protein degradation. In our results, ABC transporters for cystine and methionine were upregulated in cold at the stationary phase.
Cold Shock Proteins
Y. pseudotuberculosis seems to express Csps encoded by yptb2414 and yptb2950 almost exclusively at low temperature, i.e., with the highest significant difference of all Csps, and at high levels. Additionally, yptb3585, yptb3586, and yptb1423 were upregulated throughout the growth. Csps encoded by yptb2950 and yptb3585/6 are comparable to CspG and CspB of E. coli, respectively (Keto-Timonen et al., 2016). Csp encoded by yptb2414 is 80% similar to CspC and CspE of E. coli K-12 W3110.
Csp encoded by yptb1088, which is almost identical to CspE of E. coli (Keto-Timonen et al., 2016), showed more transcripts at 3°C, but not at significant levels. However, its transcript count rose toward stationary phase like has been shown to happen in E. coli (Czapski and Trun, 2014). In E. coli, CspE was also the most abundant of all csps after cold shock and present in the bacterium at all times. The expression of yptb1392, similar to CspD of E. coli, ramped up at stationary phase in 28°C and a similar but less dramatic bump can be seen in expression profile in 3°C. In E. coli, CspD is not cold-induced and is expressed mainly at stationary phase (Yamanaka et al., 2001). It is involved in persister cell and biofilm formation (Kim and Wood, 2010).
In E. coli, the genes cspA, cspB, cspE, and cspG all must be deleted to achieve a cold-sensitive phenotype, and the deletion of only one or two genes leads to overexpression of the rest (Xia et al., 2001). In addition, CspC and CspE seem to double as regulatory elements in stress responses (Phadtare and Inouye, 2001; Phadtare et al., 2006). The two proteins have been shown to be required for expression of fructose PTS at suboptimal temperatures (Phadtare et al., 2006). E. coli expresses cspA, cspB, and cspG even when its protein synthesis is inhibited by antimicrobials (Etchegaray and Inouye, 1999). In Clostridium botulinum, deletion of cspB, or cspC leads to a cold-sensitive phenotype, and cspC deletion even hinders growth at 37°C (Söderholm et al., 2011). Genes cspB and cspC also provide the bacterium resistance to NaCl, pH, and ethanol stress (Derman et al., 2015). In Staphylococcus aureus, cspB mutation renders the bacterium susceptible to cold and various antimicrobials (Duval et al., 2010). By contrast, CspC has been shown to induce more strongly in response to antimicrobials, hydrogen peroxide, and arsenate than cold (Chanda et al., 2009).
In our experiment, the expression of the genes yptb3585–86, comparable to cspB of E. coli (Keto-Timonen et al., 2016), dipped after the beginning of growth at 3°C but showed more transcripts at 3°C throughout the growth. Pathogenic Yersinia have been shown to hold a tandem gene duplication cspA1/A2 (Neuhaus et al., 1999), which is very nearly identical to genes yptb3585–87 and needs to be downregulated after the initial cold shock for growth to continue (Neuhaus et al., 2000). In Yersinia enterocolitica, cspA and cspB were the very first genes expressed after cold-shock with their transcript levels decreasing after the beginning (Bresolin et al., 2006). Similarly, cspA and cspB expression fluctuated accordingly with temperature cycling in E. coli (Ivancic et al., 2013). In light of our results, yptb3585–86 might play an important role in the initial regulation of cold-inducible genes also in Y. pseudotuberculosis. Genes yptb2414 and yptb2950 seem to be important at low temperature growth considering their differential expression and transcript levels. It is also probable that most Csps of Y. pseudotuberculosis are interchangeable. Csp encoded by yptb1423 is an odd one out, with a similarity of 54% at best to other Csps of Y. pseudotuberculosis (Keto-Timonen et al., 2016), with a peak similarity of 50% to the Csps of E. coli K-12 W3110.
DEAD-Box RNA Helicases
Y. pseudotuberculosis is unable to grow at 3°C without a functional RNA helicase gene csdA (yptb0486) (Palonen et al., 2012). Of other foodborne pathogens, Listeria monocytogenes and C. botulinum have been shown to require RNA-helicases, including a CsdA homolog, to grow at suboptimal temperatures (Markkula et al., 2012; Söderholm et al., 2015). In fact, CsdA was the most abundant protein extracted from the microbial mats of Lake Joyce in Antarctica (Koo et al., 2016). CsdA and SrmB of E. coli are directly involved in the biogenesis of large ribosomal subunits, presumably at subsequent steps with some functional overlap, and deletion of either gene stunts growth at low temperatures (Charollais et al., 2003, 2004). The deletion of dbpA in E. coli did not lead to similar accumulation of ribosome precursors and stunted growth at either 25 or 37°C (Peil et al., 2008).
Our results suggest that DbpA (encoded by yptb1652) may indeed be important at low temperatures, at least in Y. pseudotuberculosis. DbpA is missing completely from Pasteurellales, an order of symbionts and parasites growing at unchanging, high-temperature, nutrient-rich niches, which suggests that DbpA, like other DEAD-box helicases, has become specialized to operate in adverse growth conditions (Iost and Dreyfus, 2006). Amino acid similarities between corresponding helicases of Y. pseudotuberculosis IP32953 and E. coli K-12 W3110 range from 69 to 90%.
Gene yptb1214 coding RhlE had the largest and most consistent significant difference in expression in favor of 3°C. It has been suggested by Jain (2008) that RhlE may modulate the function of CsdA and SrmB in priming immature ribosomal RNA. This would also help explain the gene's prominence at low temperature in our results at the beginning and during logarithmic phase when ribosomal activity is at its highest. In E. coli, RhlE has been shown to complement the function of both CsdA and RhlB (Khemici et al., 2004; Awano et al., 2007), but deletion of rhlE did not affect growth at 37°C in E. coli (Phadtare, 2011).
Compatible Solutes
Our results show that Y. pseudotuberculosis expresses genes involved in glycine betaine intake and biosynthesis significantly more at 3°C during critical points of growth, i.e., the beginning and logarithmic phase. It has been shown that Y. enterocolitica, with almost identical proU and proP operons, accumulates glycine betaine at low temperature, but glycine betaine only protects it against osmotic stress (Park et al., 1995; Annamalai and Venkitanarayanan, 2009). However, glycine betaine has been shown to also grant cold protection to gram-positive L. monocytogenes, B. subtilis, and gram-negative Vibrio anguillarum (Ko et al., 1994; Hoffmann and Bremer, 2011; Ma et al., 2017). Furthermore, choline/carnitine/betaine transporter gene of C. botulinum was upregulated in cold shock (Dahlsten et al., 2014). The precise mechanism behind glycine betaine meditated cold protection is unclear, but the molecule has been suggested to hinder ice crystal formation and displace water around macromolecules, reducing aggregation and denaturation (Hoffmann and Bremer, 2011).
In our results, an operon encoding spermidine efflux pump MdtIJ was expressed significantly more at 3°C. Bacteria need spermidine for various purposes, but at low temperatures excess spermidine is detrimental (Higashi et al., 2008). Spermidine displaces magnesium-ions from ribosomes, thus deactivating them, but it also prolongs synthesis of Csps after cold shock (Limsuwun and Jones, 2000). It has been shown that prolonged synthesis of Csps after cold shock occupies all available ribosomes effectively stopping growth in Y. enterocolitica (Neuhaus et al., 2000). Our results suggest that also Y. pseudotuberculosis ejects excess spermidine to avoid these problems.
Rho Factor, IF-1, and RbfA
Translation factors like IF-1 and RbfA act directly on ribosomes, securing protein synthesis. Initiation factor IF-1 separates the overly stabilized large and small subunits of ribosomes so that they can fuse and begin translating again elsewhere (Giangrossi et al., 2007), while RbfA primes 16S rRNA for new small subunits (Xia et al., 2003). Both IF-1 and RbfA have been shown to be significantly more expressed during cold shock in E. coli (Xia et al., 2003; Giangrossi et al., 2007). E. coli IF-1 resembles the CspA homologs (Phadtare et al., 2007), whereas in Y. pseudotuberculosis they are only 42–50% similar in nucleic acid and 14–21% in amino acid sequence.
The rho gene is associated with bacteria that are subjected to various stresses in their environment (D'heygère et al., 2013). Rho factor is needed at low temperature to clear the bacterial DNA of frozen ribosomes and polymerases (D'heygère et al., 2013), and the structure of its RNA-binding subunit is similar to that of a cold shock domain (Briercheck et al., 1996). Rho factor also coordinates homeostasis of magnesium ions, which are needed by ribosomes, by controlling transporter genes (Kriner and Groisman, 2015). The Rho factor has been shown to be upregulated at low temperature in at least Acidithiobacillus ferrooxidans (Mykytczuk et al., 2011), Pseudomonas haloplanktis strain TAC125 (Piette et al., 2010), and Bacillus subtilis (Quirk et al., 1993). To our knowledge, none of these factors have been linked to cold growth in Yersinia genus before.
Y. pseudotuberculosis expressed the genes encoding IF-1, RbfA, and Rho all through the critical phases of growth at high levels but also significantly more at 3°C. Massive difference of IF-1 (yptb1395) expression in favor of cold during logarithmic phase suggests that the initiation factor is important for continued growth at low temperature. It is probable that IF-1 and RbfA of Y. pseudotuberculosis provide the bacterium with working ribosomes at low temperature. Y. pseudotuberculosis might need Rho factor not only to terminate inefficient transcription but also to provide ribosomes with much needed magnesium ions. The upregulation of mgt transporter genes at 3°C throughout the growth supports this theory.
Cell Membranes
Lipid synthesis and cell growth cease in cyanobacteria until an adequate membrane lipid composition has been achieved (Sinetova and Los, 2016). However, gram-negative cyanobacteria and gram-positive Bacillus subtilis possess a two-component system, DesK-DesR, that senses increasing membrane rigidity at low temperatures (Beranová et al., 2010; Sinetova and Los, 2016). Although a corresponding two-component system has not been identified in Y. pseudotuberculosis (Palonen et al., 2011), the ratio of unsaturated to saturated fatty acids in its membranes rises at low temperature (Bakholdina et al., 2004). In another gram-negative bacterium, E. coli, temperature drop directly affects activity of the FabF enzyme in its cytosol and this enzyme is involved in unsaturated lipid synthesis with FabA and FabB (Mansilla et al., 2004). Cold stress has also been shown to induce the transcription of fabF in Y. pestis (Han et al., 2005).
It is also possible that the csp encoded by yptb2414, a homolog of E. coli CspC and CspE, is involved in cold adaptation of membranes in Y. pseudotuberculosis. CspC and CspE bind specific uracil-rich segments of mRNA, and these segments often encode hydrophobic structures of membrane proteins (Benhalevy et al., 2015). It has been suggested that bacteria use these segments as addresses for directing membrane protein mRNA to where they are needed in the cell (Benhalevy et al., 2015).
Conclusions
Y. pseudotuberculosis has extensive tools to keep its protein synthesis running at low temperature. It is probable, that the functions of its Csps, RNA helicases and factors acting on ribosomes overlap greatly and thus form a robust network to protect nucleic acids from cold damage. Csps encoded by yptb1423, yptb2414, yptb2950, yptb3585–86, and RNA helicases CsdA, RhlE, and DbpA, seem to form the backbone of cold survival of Y. pseudotuberculosis with their regulatory and nucleic acid unwinding functions. IF-1, RbfA, and by extension, Rho, keep the ribosomes of Y. pseudotuberculosis running even at suboptimal temperatures. Rho factor also terminates frozen ribosomes and RNA polymerases, freeing scant resources under cold stress.
Increased expression of motility, chemotaxis, and nutrient uptake genes at low temperature suggests that Y. pseudotuberculosis actively tries to find and secure sufficient resources to grow in refrigerator temperatures. The nutrients acquired are diverse and the nutrient profile seems to shift as growth progresses. The bacterium also possibly changes its membrane lipid composition with Fab enzymes, much like E. coli, to battle increasing membrane rigidity.
The exact role of glycine betaine in cold resistance is uncertain, but Y. pseudotuberculosis seems to accumulate it during cold growth like many other bacteria. Genes involved in defense against foreign DNA as well as oxidative stress were upregulated at 3°C, which would seem to suggest that stress responses are linked in some way.
Author Contributions
RK-T and HK designed the study. NS and RK-T performed the experiments. J-PV and KJ performed the transcriptome analysis. J-PV, KJ, RK-T, and HK contributed to the data analysis and interpretation. J-PV drafted the manuscript. RK-T, NS, and HK contributed to manuscript revision. All authors have read and approved the final manuscript.
Funding
This work was funded by the Finnish Veterinary Foundation and the Walter Ehrström Foundation.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge Dr. Elisabeth Carniel, Institut Pasteur, Paris, France for providing the Y. pseudotuberculosis IP32953 strain. We thank Henna Niinivirta, Erika Pitkänen, and Kirsi Ristkari for technical assistance.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00416/full#supplementary-material
References
Annamalai, T., and Venkitanarayanan, K. (2009). Role of proP and proU in betaine uptake by Yersinia enterocolitica under cold and osmotic stress conditions. Appl. Environ. Microbiol. 75, 1471–1477. doi: 10.1128/AEM.01644-08
Awano, N., Xu, C., Ke, H., Inoue, K., Inouye, M., and Phadtare, S. (2007). Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J. Bacteriol. 189, 5808–5815. doi: 10.1128/JB.00655-07
Bakholdina, S. I., Sanina, N. M., Krasikova, I. N., Popova, O. B., and Solov'eva, T. F. (2004). The impact of abiotic factors (temperature and glucose) on physicochemical properties of lipids from Yersinia pseudotuberculosis. Biochimie 86, 875–881. doi: 10.1016/j.biochi.2004.10.011
Benhalevy, D., Bochkareva, E. S., Biran, I., and Bibi, E. (2015). Model uracil-rich RNAs and membrane protein mRNAs interact specifically with cold shock proteins in Escherichia coli. PLoS ONE 10:e0134413. doi: 10.1371/journal.pone.0134413
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300.
Beranová, J., Mansilla, M. C., De Mendoza, D., Elhottová, D., and Konopásek, I. (2010). Differences in cold adaptation of Bacillus subtilis under anaerobic and aerobic conditions. J. Bacteriol. 192, 4164–4171. doi: 10.1128/JB.00384-10
Bresolin, G., Neuhaus, K., Scherer, S., and Fuchs, T. M. (2006). Transcriptional analysis of long-term adaptation of Yersinia enterocolitica to low-temperature growth. J. Bacteriol. 188, 2945–2958. doi: 10.1128/JB.188.8.2945-2958.2006
Briercheck, D. M., Allison, T. J., Richardson, J. P., Ellena, J. F., Wood, T. C., and Rule, G. S. (1996). 1H, 15N and 13C resonance assignments and secondary structure determination of the RNA-binding domain of E. coli Rho protein. J. Biomol. NMR 8, 429–444. doi: 10.1007/BF00228145
Buzoleva, L. S., and Somov, G. P. (2003). Adaptation variability of Yersinia pseudotuberculosis during long-term persistence in soil. Bull. Exp. Biol. Med. 135, 456–459. doi: 10.1023/A:1024915409187
Chanda, P. K., Mondal, R., Sau, K., and Sau, S. (2009). Antibiotics, arsenate and H2O2 induce the promoter of Staphylococcus aureus cspC gene more strongly than cold. J. Basic Microbiol. 49, 205–211. doi: 10.1002/jobm.200800065
Charollais, J., Dreyfus, M., and Iost, I. (2004). CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32, 2751–2759. doi: 10.1093/nar/gkh603
Charollais, J., Pflieger, D., Vinh, J., Dreyfus, M., and Iost, I. (2003). The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 48, 1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x
Chattopadhyay, M. K., Raghu, G., Sharma, Y. V. R. K., Biju, A. R., Rajasekharan, M. V., and Shivaji, S. (2011). Increase in oxidative stress at low temperature in an antarctic bacterium. Curr. Microbiol. 62, 544–546. doi: 10.1007/s00284-010-9742-y
Czapski, T. R., and Trun, N. (2014). Expression of csp genes in E. coli K-12 in defined rich and defined minimal media during normal growth, and after cold-shock. Gene 547, 91–97. doi: 10.1016/j.gene.2014.06.033
Dahlsten, E., Isokallio, M., Somervuo, P., Lindström, M., and Korkeala, H. (2014). Transcriptomic analysis of (Group I) Clostridium botulinum ATCC 3502 cold shock response. PLoS ONE 9:e89958. doi: 10.1371/journal.pone.0089958
Derman, Y., Söderholm, H., Lindström, M., and Korkeala, H. (2015). Role of csp genes in NaCl, pH, and ethanol stress response and motility in Clostridium botulinum ATCC 3502. Food Microbiol. 46, 463–470. doi: 10.1016/j.fm.2014.09.004
D'heygère, F., Rabhi, M., and Boudvillain, M. (2013). Phyletic distribution and conservation of the bacterial transcription termination factor Rho. Microbiology 159, 1423–1436.
Duval, B. D., Mathew, A., Satola, S. W., and Shafer, W. M. (2010). Altered growth, pigmentation, and antimicrobial susceptibility properties of Staphylococcus aureus due to loss of the major cold shock gene cspB. Antimicrob. Agents Chemother. 54, 2283–2290. doi: 10.1128/AAC.01786-09
Etchegaray, J. P., and Inouye, M. (1999). CspA, CspB, and CspG, major cold shock proteins of Escherichia coli, are induced at low temperature under conditions that completely block protein synthesis. J. Bacteriol. 181, 1827–1830.
Gaidatzis, D., Lerch, A., Hahne, F., and Stadler, M. B. (2015). QuasR: quantification and annotation of short reads in R. Bioinformatics 31, 1130–1132. doi: 10.1093/bioinformatics/btu781
Gengler, S., Laudisoit, A., Batoko, H., and Wattiau, P. (2015). Long-Term Persistence of Yersinia pseudotuberculosis in entomopathogenic nematodes. PLoS ONE 10:e0116818. doi: 10.1371/journal.pone.0116818
Giangrossi, M., Brandi, A., Giuliodori, A. M., Gualerzi, C. O., and Pon, C. L. (2007). Cold-shock-induced de novo transcription and translation of infA and role of IF1 during cold adaptation. Mol. Microbiol. 64, 807–821. doi: 10.1111/j.1365-2958.2007.05699.x
Giannitti, F., Barr, B. C., Brito, B. P., Uzal, F. A., Villanueva, M., and Anderson, M. (2014). Yersinia pseudotuberculosis infections in goats and other animals diagnosed at the California animal health and food safety laboratory system: 1990–2012. J. Vet. Diagn. Invest. 26, 88–95. doi: 10.1177/1040638713516624
Han, Y., Zhou, D., Pang, X., Zhang, L., Song, Y., Tong, Z., et al. (2005). DNA microarray analysis of the heat- and cold-shock stimulons in Yersinia pestis. Microbes Infect. 7, 335–348. doi: 10.1016/j.micinf.2004.11.005
Hardcastle, T. J., and Kelly, K. A. (2010). baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinformatics 11:422. doi: 10.1186/1471-2105-11-422
Higashi, K., Ishigure, H., Demizu, R., Uemura, T., Nishino, K., Yamaguchi, A., et al. (2008). Identification of a spermidine excretion protein complex (MdtJI) in Escherichia coli. J. Bacteriol. 190, 872–878. doi: 10.1128/JB.01505-07
Hoffmann, T., and Bremer, E. (2011). Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J. Bacteriol. 193, 1552–1562. doi: 10.1128/JB.01319-10
Huber, W., Carey, V. J., Gentleman, R., Anders, S., Carlson, M., Carvalho, B. S., et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12:115. doi: 10.1038/nmeth.3252
Iost, I., and Dreyfus, M. (2006). DEAD-box RNA helicases in Escherichia coli. Nucl. Acids Res. 34, 4189–4197. doi: 10.1093/nar/gkl500
Ivancic, T., Jamnik, P., and Stopar, D. (2013). Cold shock CspA and CspB protein production during periodic temperature cycling in Escherichia coli. BMC Res. Notes 6:248. doi: 10.1186/1756-0500-6-248
Jaakkola, K., Somervuo, P., and Korkeala, H. (2015). Comparative genomic hybridization analysis of Yersinia enterocolitica and Yersinia pseudotuberculosis identifies genetic traits to elucidate their different ecologies. BioMed. Res. Int. 2015:760494. doi: 10.1155/2015/760494
Jain, C. (2008). The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA 14, 381–389. doi: 10.1261/rna.800308
Jain, N. K., and Roy, I. (2009). Effect of trehalose on protein structure. Protein Sci. 18, 24–36. doi: 10.1002/pro.3
Johnson, S. L., Daligault, H. E., Davenport, K. W., Jaissle, J., Frey, K. G., Ladner, J. T., et al. (2015). Thirty-two complete genome assemblies of nine Yersinia species, including Y. pestis, Y. pseudotuberculosis, and Y. enterocolitica. Genome Announc. 3:e00148–15. doi: 10.1128/genomeA.00148-15
Joutsen, S., Laukkanen-Ninios, R., Henttonen, H., Niemimaa, J., Voutilainen, L., Kallio, E. R., et al. (2017). Yersinia spp. in Wild Rodents and Shrews in Finland. Vector-Borne Zoonotic Dis. 17, 303–311. doi: 10.1089/vbz.2016.2025
Jozefczuk, S., Klie, S., Catchpole, G., Szymanski, J., Cuadros-Inostroza, A., Steinhauser, D., et al. (2010). Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 6:364. doi: 10.1038/msb.2010.18
Kanehisa, M., and Goto, S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Keto-Timonen, R., Hietala, N., Palonen, E., Hakakorpi, A., Lindström, M., and Korkeala, H. (2016). Cold shock proteins: a minireview with special emphasis on csp-family of enteropathogenic Yersinia. Front. Microbiol. 7:1151. doi: 10.3389/fmicb.2016.01151
Keto-Timonen, R., Pontinen, A., Aalto-Araneda, M., and Korkeala, H. (2018). Growth of Yersinia pseudotuberculosis strains at different temperatures, pH values, and NaCl and ethanol concentrations. J. Food Prot. 81, 142–149. doi: 10.4315/0362-028X.JFP-17-223
Khemici, V., Toesca, I., Poljak, L., Vanzo, N. F., and Carpousis, A. J. (2004). The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: functional replacement of RhlB by RhlE. Mol. Microbiol. 54, 1422–1430. doi: 10.1111/j.1365-2958.2004.04361.x
Kim, Y., and Wood, T. K. (2010). Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 391, 209–213. doi: 10.1016/j.bbrc.2009.11.033
Ko, R., Smith, L. T., and Smith, G. M. (1994). Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J. Bacteriol. 176, 426–431. doi: 10.1128/jb.176.2.426-431.1994
Koo, H., Hakim, J. A., Fisher, P. R. E., Grueneberg, A., Andersen, D. T., and Bej, A. K. (2016). Distribution of cold adaptation proteins in microbial mats in Lake Joyce, Antarctica: analysis of metagenomic data by using two bioinformatics tools. J. Microbiol. Methods 120, 23–28. doi: 10.1016/j.mimet.2015.11.008
Kriner, M. A., and Groisman, E. A. (2015). The bacterial transcription termination factor Rho coordinates Mg2+ homeostasis with translational signals. J. Mol. Biol. 427, 3834–3849. doi: 10.1016/j.jmb.2015.10.020
Laukkanen, R., Martínez, P. O., Siekkinen, K.-M., Ranta, J., Maijala, R., and Korkeala, H. (2008). Transmission of Yersinia pseudotuberculosis in the Pork Production Chain from Farm to Slaughterhouse. Appl. Environ. Microbiol. 74, 5444–5450. doi: 10.1128/AEM.02664-07
Le Guern, A.-S., Martin, L., Savin, C., and Carniel, E. (2016). Yersiniosis in France: overview and potential sources of infection. Int. J. Infect. Dis. 46, 1–7. doi: 10.1016/j.ijid.2016.03.008
Limsuwun, K., and Jones, P. G. (2000). Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli. J. Bacteriol. 182, 5373–5380. doi: 10.1128/JB.182.19.5373-5380.2000
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Ma, Y., Wang, Q., Gao, X., and Zhang, Y. (2017). Biosynthesis and uptake of glycine betaine as cold-stress response to low temperature in fish pathogen Vibrio anguillarum. J. Microbiol. 55, 44–55. doi: 10.1007/s12275-017-6370-2
Mansilla, M. C., Cybulski, L. E., Albanesi, D., and De Mendoza, D. (2004). Control of membrane lipid fluidity by molecular thermosensors. J. Bacteriol. 186, 6681–6688. doi: 10.1128/JB.186.20.6681-6688.2004
Markkula, A., Mattila, M., Lindström, M., and Korkeala, H. (2012). Genes encoding putative DEAD-box RNA helicases in Listeria monocytogenes EGD-e are needed for growth and motility at 3°C. Environ. Microbiol. 14, 2223–2232. doi: 10.1111/j.1462-2920.2012.02761.x
Monedero, V., Maze, A., Boel, G., Zuniga, M., Beaufils, S., Hartke, A., et al. (2007). The phosphotransferase system of Lactobacillus casei: regulation of carbon metabolism and connection to cold shock response. J. Mol. Microbiol. Biotechnol. 12, 20–32. doi: 10.1159/000096456
Mykytczuk, N. C. S., Trevors, J. T., Foote, S. J., Leduc, L. G., Ferroni, G. D., and Twine, S. M. (2011). Proteomic insights into cold adaptation of psychrotrophic and mesophilic Acidithiobacillus ferrooxidans strains. Antonie van Leeuwenhoek 100, 259–277. doi: 10.1007/s10482-011-9584-z
Neuhaus, K., Francis, K. P., Rapposch, S., Görg, A., and Scherer, S. (1999). Pathogenic Yersinia species carry a novel, cold-inducible major cold shock protein tandem gene duplication producing both bicistronic and monocistronic mRNA. J. Bacteriol. 181, 6449–6455.
Neuhaus, K., Rapposch, S., Francis, K. P., and Scherer, S. (2000). Restart of exponential growth of cold-shocked Yersinia enterocolitica occurs after down-regulation of cspA1/A2 mRNA. J. Bacteriol. 182, 3285–3288. doi: 10.1128/JB.182.11.3285-3288.2000
Niskanen, T., Waldenström, J., Fredriksson-Ahomaa, M., Olsen, B., and Korkeala, H. (2003). virF-Positive Yersinia pseudotuberculosis and Yersinia enterocolitica found in migratory birds in Sweden. Appl. Environ. Microbiol. 69, 4670–4675. doi: 10.1128/AEM.69.8.4670-4675.2003
Nuorti, J. P., Niskanen, T., Hallanvuo, S., Mikkola, J., Kela, E., Hatakka, M., et al. (2004). A widespread outbreak of Yersinia pseudotuberculosis O:3 infection from iceberg lettuce. J. Infect. Dis. 189, 766–774. doi: 10.1086/381766
Palonen, E., Lindström, M., Karttunen, R., Somervuo, P., and Korkeala, H. (2011). Expression of signal transduction system encoding genes of Yersinia pseudotuberculosis IP32953 at 28°C and 3°C. PLoS ONE 6:e25063. doi: 10.1371/journal.pone.0025063
Palonen, E., Lindström, M., and Korkeala, H. (2010). Adaptation of enteropathogenic Yersinia to low growth temperature. Crit. Rev. Microbiol. 36, 54–67. doi: 10.3109/10408410903382581
Palonen, E., Lindström, M., Somervuo, P., Johansson, P., Björkroth, J., and Korkeala, H. (2012). Requirement for RNA Helicase CsdA for Growth of Yersinia pseudotuberculosis IP32953 at low temperatures. Appl. Environ. Microbiol. 78, 1298–1301. doi: 10.1128/AEM.07278-11
Park, S., Smith, L. T., and Smith, G. M. (1995). Role of glycine betaine and related osmolytes in osmotic stress adaptation in Yersinia enterocolitica ATCC 9610. Appl. Environ. Microbiol. 61, 4378–4381.
Pärn, T., Hallanvuo, S., Salmenlinna, S., Pihlajasaari, A., Heikkinen, S., Telkki-Nykänen, H., et al. (2015). Outbreak of Yersinia pseudotuberculosis O:1 infection associated with raw milk consumption, Finland, spring 2014. Eurosurveillance 20:30033. doi: 10.2807/1560-7917.ES.2015.20.40.30033
Peil, L., Virumäe, K., and Remme, J. (2008). Ribosome assembly in Escherichia coli strains lacking the RNA helicase DeaD/CsdA or DbpA. FEBS J. 275, 3772–3782. doi: 10.1111/j.1742-4658.2008.06523.x
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Phadtare, S. (2011). Unwinding activity of cold shock proteins and RNA metabolism. RNA Biol. 8, 394–397. doi: 10.4161/rna.8.3.14823
Phadtare, S., and Inouye, M. (2001). Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J. Bacteriol. 183, 1205–1214. doi: 10.1128/JB.183.4.1205-1214.2001
Phadtare, S., Kazakov, T., Bubunenko, M., Court, D. L., Pestova, T., and Severinov, K. (2007). Transcription antitermination by translation initiation factor IF1. J. Bacteriol. 189, 4087–4093. doi: 10.1128/JB.00188-07
Phadtare, S., Tadigotla, V., Shin, W.-H., Sengupta, A., and Severinov, K. (2006). Analysis of Escherichia coli global gene expression profiles in response to overexpression and deletion of CspC and CspE. J. Bacteriol. 188, 2521–2527. doi: 10.1128/JB.188.7.2521-2527.2006
Piette, F., D'amico, S., Struvay, C., Mazzucchelli, G., Renaut, J., Tutino, M. L., et al. (2010). Proteomics of life at low temperatures: trigger factor is the primary chaperone in the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Mol. Microbiol. 76, 120–132. doi: 10.1111/j.1365-2958.2010.07084.x
Postma, P. W., Lengeler, J. W., and Jacobson, G. R. (1993). Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57, 543–594.
Quirk, P. G., Dunkley, E. A., Lee, P., and Krulwich, T. A. (1993). Identification of a putative Bacillus subtilis rho gene. J. Bacteriol. 175, 647–654. doi: 10.1128/jb.175.3.647-654.1993
Rimhanen-Finne, R., Niskanen, T., Hallanvuo, S., Makary, P., Haukka, K., Pajunen, S., et al. (2009). Yersinia pseudotuberculosis causing a large outbreak associated with carrots in Finland, 2006. Epidemiol. Infect. 137, 342–347. doi: 10.1017/S0950268807000155
Santos-Montañez, J., Benavides-Montano, J. A., Hinz, A. K., and Vadyvaloo, V. (2015). Yersinia pseudotuberculosis IP32953 survives and replicates in trophozoites and persists in cysts of Acanthamoeba castellanii. FEMS Microbiol. Lett. 362:fnv091. doi: 10.1093/femsle/fnv091
Sato, K., and Komazawa, M. (1991). Yersinia pseudotuberculosis infection in children due to untreated drinking water. Contrib. Microbiol. Immunol. 12, 5–10.
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Sinetova, M. A., and Los, D. A. (2016). New insights in cyanobacterial cold stress responses: Genes, sensors, and molecular triggers. Biochim. Biophys. Acta 1860, 2391–2403. doi: 10.1016/j.bbagen.2016.07.006
Söderholm, H., Derman, Y., Lindström, M., and Korkeala, H. (2015). Functional csdA is needed for effective adaptation and initiation of growth of Clostridium botulinum ATCC 3502 at suboptimal temperature. Int. J. Food Microbiol. 208, 51–57. doi: 10.1016/j.ijfoodmicro.2015.05.013
Söderholm, H., Lindström, M., Somervuo, P., Heap, J., Minton, N., Lindén, J., et al. (2011). cspB encodes a major cold shock protein in Clostridium botulinum ATCC 3502. Int. J. Food Microbiol. 146, 23–30. doi: 10.1016/j.ijfoodmicro.2011.01.033
Suutari, M., and Laakso, S. (1994). Microbial Fatty Acids and Thermal Adaptation. Crit. Rev. Microbiol. 20, 285–328. doi: 10.3109/10408419409113560
Taboada, B., Ciria, R., Martinez-Guerrero, C. E., and Merino, E. (2012). ProOpDB: prokaryotic operon database. Nucleic Acids Res. 40, D627–D631. doi: 10.1093/nar/gkr1020
Wattam, A. R., Abraham, D., Dalay, O., Disz, T. L., Driscoll, T., Gabbard, J. L., et al. (2014). PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 42, D581–D591. doi: 10.1093/nar/gkt1099
Wouters, J. A., Kamphuis, H. H., Hugenholtz, J., Kuipers, O. P., De Vos, W. M., and Abee, T. (2000). Changes in glycolytic activity of Lactococcus lactis induced by low temperature. Appl. Environ. Microbiol. 66, 3686–3691. doi: 10.1128/AEM.66.9.3686-3691.2000
Xia, B., Ke, H., and Inouye, M. (2001). Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol. Microbiol. 40, 179–188. doi: 10.1046/j.1365-2958.2001.02372.x
Xia, B., Ke, H., Shinde, U., and Inouye, M. (2003). The role of RbfA in 16S rRNA processing and cell growth at low temperature in Escherichia coli. J. Mol. Biol. 332, 575–584. doi: 10.1016/S0022-2836(03)00953-7
Yamanaka, K., Zheng, W., Crooke, E., Wang, Y.-H., and Inouye, M. (2001). CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol. Microbiol. 39, 1572–1584. doi: 10.1046/j.1365-2958.2001.02345.x
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., and Madden, T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13:134. doi: 10.1186/1471-2105-13-134
Keywords: cold stress, stress tolerance, compatible solute, RNA helicase, cold shock protein, transcription factor
Citation: Virtanen J-P, Keto-Timonen R, Jaakkola K, Salin N and Korkeala H (2018) Changes in Transcriptome of Yersinia pseudotuberculosis IP32953 Grown at 3 and 28°C Detected by RNA Sequencing Shed Light on Cold Adaptation. Front. Cell. Infect. Microbiol. 8:416. doi: 10.3389/fcimb.2018.00416
Received: 07 May 2018; Accepted: 09 November 2018;
Published: 27 November 2018.
Edited by:
Victoria Auerbuch, University of California, Santa Cruz, United StatesReviewed by:
Karl M. Thompson, Howard University, United StatesMichael Marceau, Université Lille Nord de France, France
Copyright © 2018 Virtanen, Keto-Timonen, Jaakkola, Salin and Korkeala. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Riikka Keto-Timonen, cmlpa2thLmtldG8tdGltb25lbkBoZWxzaW5raS5maQ==
 Jussa-Pekka Virtanen
Jussa-Pekka Virtanen Riikka Keto-Timonen
Riikka Keto-Timonen Kaisa Jaakkola
Kaisa Jaakkola Noora Salin
Noora Salin Hannu Korkeala
Hannu Korkeala