
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 22 November 2018
Sec. Molecular Bacterial Pathogenesis
Volume 8 - 2018 | https://doi.org/10.3389/fcimb.2018.00404
This article is part of the Research Topic The Pathogenic Yersiniae – Advances in the Understanding of Physiology and Virulence, Volume II View all 15 articles
 Jessica M. Morgan1
Jessica M. Morgan1 Hanh N. Lam2
Hanh N. Lam2 Jocelyn Delgado2
Jocelyn Delgado2 Justin Luu2
Justin Luu2 Sina Mohammadi3†
Sina Mohammadi3† Ralph R. Isberg3
Ralph R. Isberg3 Helen Wang4
Helen Wang4 Victoria Auerbuch2*
Victoria Auerbuch2*Dozens of Gram negative pathogens use one or more type III secretion systems (T3SS) to disarm host defenses or occupy a beneficial niche during infection of a host organism. While the T3SS represents an attractive drug target and dozens of compounds with T3SS inhibitory activity have been identified, few T3SS inhibitors have been validated and mode of action determined. One issue is the lack of standardized orthogonal assays following high throughput screening. Using a training set of commercially available compounds previously shown to possess T3SS inhibitory activity, we demonstrate the utility of an experiment pipeline comprised of six distinct assays to assess the stages of type III secretion impacted: T3SS gene copy number, T3SS gene expression, T3SS basal body and needle assembly, secretion of cargo through the T3SS, and translocation of T3SS effector proteins into host cells. We used enteropathogenic Yersinia as the workhorse T3SS-expressing model organisms for this experimental pipeline, as Yersinia is sensitive to all T3SS inhibitors we tested, including those active against other T3SS-expressing pathogens. We find that this experimental pipeline is capable of rapidly distinguishing between T3SS inhibitors that interrupt the process of type III secretion at different points in T3SS assembly and function. For example, our data suggests that Compound 3, a malic diamide, blocks either activity of the assembled T3SS or alters the structure of the T3SS in a way that blocks T3SS cargo secretion but not antibody recognition of the T3SS needle. In contrast, our data predicts that Compound 4, a haloid-containing sulfonamidobenzamide, disrupts T3SS needle subunit secretion or assembly. Furthermore, we suggest that misregulation of copy number control of the pYV virulence plasmid, which encodes the Yersinia T3SS, should be considered as a possible mode of action for compounds with T3SS inhibitory activity against Yersinia.
The type III secretion system (T3SS) is a macromolecular nanosyringe used by dozens of Gram negative pathogens, including Yersinia, Shigella, Salmonella, Chlamydia, and Pseudomonas, to inject effector proteins into target host cells (Deng et al., 2017). The core structure of the T3SS consists of a basal body that anchors the entire complex in the bacterial membrane and assembles first. The basal body is composed of three proteins, which oligomerize into rings. SctD and SctJ form two rings in the bacterial inner membrane (IM) and SctC, a member of the secretin family of proteins, forms a ring in the bacterial outer membrane (OM) (Bergeron et al., 2013). These three rings are connected in the center by a hollow inner rod and the IM rings are closely associated with the SctN ATPase complex whose integrity is essential to the T3SS (Diepold et al., 2010). In addition to ATP, the proton motive force is also important for T3SS activity (Wilharm et al., 2004), although how the T3SS harnesses the proton motive force remains unclear. Once active, the basal body secretes the SctF needle protein, which polymerizes into a straight, hollow tube protruding into the extracellular space. In Yersinia species pathogenic to mammals, the fully assembled Ysc T3SS needle is composed of ~140 SctF subunits, is 65 nm in length, and harbors a tip complex composed of a pentamer of the hydrophilic LcrV translocator protein (Broz et al., 2007). Upon host cell contact, two additional hydrophobic translocator proteins, YopD and YopB, are secreted through the Ysc needle to form a translocon complex that leads to pore formation in the host membrane, facilitating the translocation of effector proteins to the host cytoplasm (Büttner and Bonas, 2002).
The Yersinia Ysc T3SS is highly regulated at the transcriptional, translational, and post-translational levels (Francis et al., 2002; Heroven et al., 2012). The transcription factor LcrF directs transcription of genes encoding the T3SS structural, regulatory, and effector proteins, all of which are encoded on the 70 kb pYV virulence plasmid (Schwiesow et al., 2015). Several factors govern regulation of LcrF expression, including temperature and the transcription factor IscR (Schwiesow et al., 2015). Importantly, pYV copy number increases during active type III secretion, and this is important for Yersinia virulence (Wang et al., 2016). In addition, the T3SS functions on a positive feedback loop in which active secretion leads to upregulated transcription of T3SS genes (Cornelis et al., 1987; Francis et al., 2002), although the mechanism behind this remains unclear.
A number of pathogens require one or more T3SSs for virulence, as genetic ablation causes attenuation in animal models and clinical isolates harbor plasmids or pathogenicity islands that encode T3SS genes (Coburn et al., 2007). An antibody against the Pseudomonas aeruginosa T3SS needle tip protein PcrV is part of a current Phase II clinical trial to treat nosocomial ventilator-associated pneumonia (NCT02696902), indicating that antibodies targeting the T3SS may be used as therapeutics. However, antibodies have low oral bioavailability and must be administered by injection; small molecules with high oral bioavailability are more attractive as therapeutic agents. A number of putative small molecule T3SS inhibitors have been identified in the past 15 years (Duncan et al., 2012; Marshall and Finlay, 2014; Anantharajah et al., 2017), yet only one class of compounds can be considered validated. Many published T3SS inhibitors have off target effects that may underlie their T3SS disruption. For example, the best studied class of T3SS inhibitors, the salicylidene acylhydrazides, are thought to cause deregulation of T3SS genes through an unknown mechanism, yet the activity of some salicylidene acylhydrazides is dependent on iron chelation (Beckham and Roe, 2014). The phenoxyacetamides represent the only class of compounds that inhibit the T3SS in a physiologically relevant cellular context, protect against a bacterial infection (P. aeruginosa abscess formation in mice), and have a validated molecular target, the SctF needle subunit (Bowlin et al., 2014; Berube et al., 2017).
We have developed an experimental pipeline that can be employed to determine initial mode of action for compounds with T3SS inhibitory activity. We chose to use the enteropathogens Yersinia pseudotuberculosis and Yersinia enterocolitica as the workhorses for this assay pipeline because Yersinia is susceptible to the majority of T3SS inhibitors described and because of the wealth of genetic and biochemical tools available. In addition, Yersinia are extracellular pathogens that use their T3SS to prevent phagocytosis, negating the need for a T3SS inhibitor to cross the mammalian cell membrane to inhibit the T3SS-host cell interaction. Importantly, the assays selected for the experimental pipeline had to be amenable to miniaturization, as compound availability is often limiting. Since interfering with T3SS gene expression, basal body assembly, needle assembly, and host cell effector protein translocation could all lead to inhibition of T3SS activity, we designed our pipeline to consist of distinct assays that each measure a specific stage of type III secretion. Inhibitors of LcrF and its homolog ExsA from P. aeruginosa have been described (Marsden et al., 2015). In addition, it is possible that a small molecule with T3SS inhibitory activity could exert its effect by inhibiting secretion-associated increase of pYV copy number, leading to a decrease in T3SS gene expression. Therefore, two pipeline assays measure pYV copy number and T3SS gene expression. Once T3SS genes are expressed, the T3SS basal body assembles followed by the T3SS needle. Two additional pipeline assays measure SctD (YscD in Yersinia) localization to the IM and SctF (YscF) needle assembly. The last two pipeline assays monitor efficiency of T3SS effector protein (Yop) secretion in vitro or Yop translocation into target host cells. Each individual assay on its own provides a limited snapshot of a compound's T3SS inhibitory activity. However, when performed as a pipeline of assays, the resulting data can be used to predict the mode of action of a T3SS inhibitor. Furthermore, the training set of T3SS inhibitors we use to validate this pipeline are commercially available, and therefore can serve as controls to compare the activity of other T3SS inhibitors.
Compound 3 (CAS# 443329-02-0), compound 4 (CAS# 138323-28-1), and INP0007 (CAS# 300668-15-9) were obtained from Chembridge. INP0010 (CAS# 68639-26-9) was obtained from ChemDiv. Compound 20 (CAS# 489402-27-9) was obtained from TimTek. The anti-YscF antibody was raised in rabbits against the Y. pseudotuberculosis YscF peptide KDKPDNPALLADLQH (Morgan et al., 2017). DMSO concentration did not exceed 0.2%, except where indicated.
Bacterial strains used in this paper are listed in Table 1. Y. pseudotuberculosis and Y. enterocolitica were grown, unless otherwise specified, in 2xYT (yeast extract-tryptone) at 26°C shaking overnight. In order to induce the T3SS, overnight cultures were diluted into low calcium medium (2xYT plus 20 mM sodium oxalate and 20 mM MgCl2) to an optical density (OD600) of 0.2 and grown for 1.5 h at 26°C shaking followed by 2–3 h at 37°C to induce Yop synthesis, depending on the assay, as previously described (Auerbuch et al., 2009).
An expression cassette containing the yopH promoter and FLAG (5′ terminus)- and ssrA (3′ terminus)-tagged mCherry was generated using SOEing PCR and cloned into pMMB67EH (Horton et al., 1990; Pettersson et al., 1996; Karzai et al., 2000). The yopH promoter was amplified from pHYopT (gift from J. Bliska) using oligonucleotides SMP425 and SMP426 (Table 2). mCherry (gift from R. Tsien) was amplified using oligonucleotides SMP427 and SMP431. FLAG and ssrA sequences were incorporated into oligonucleotides SMP426/SMP427 and 431, respectively. Oligonucleotides SMP425 and SMP431 were used to generate the pyopH-FLAG-mCherry-ssrA cassette. PCR product was digested with BamHI and EcoRI and cloned into similarly digested pMMB67EH. Site directed mutagenesis using oligonucleotides SMP437 and SMP438 was used to generate the AAV variant of the ssrA tag.
Visualization of Yersinia T3SS cargo secreted in broth culture by Coomassie staining of SDS-PAGE separated proteins was performed as previously described with some modifications (Auerbuch et al., 2009). After low calcium medium cultures were grown for 1.5 h at 26°C, compounds or DMSO were added and the cultures shifted to 37°C for another 2 h. Post-incubation cultures were spun down at 15,000 × g for 10 min at room temperature. Supernatants were transferred to a new eppendorf tube. Ten percent final trichloroacetic acid (TCA) was added and the mixture was vortexed vigorously. Samples were incubated on ice for 20 min and then spun down at 15,000 × g for 15 min at 4°C. The pellet was resuspended in final sample buffer (FSB) with 20% DTT. Samples were boiled for 15 min prior to running on a 12.5% SDS-PAGE gel. Sample loading was normalized for bacterial culture density (OD600) measured prior to centrifugation. Densitometric quantification of the bands was done using Image Lab software (Bio-Rad), setting the relevant DMSO-treated YopE band to 1.00.
The miniaturized secretion assay was done by using Y. pseudotuberculosis expressing a YopM-β-lactamase (YopM-Bla) reporter and the chromogenic β-lactamase substrate nitrocefin to detect secreted YopM-Bla (O'Callaghan et al., 1972; Lee et al., 2007; Green et al., 2016). Y. pseudotuberculosis Δyop6 pYopM-Bla and ΔyscNU pYopM-Bla (negative control) were grown overnight at 26°C with 250 rpm shaking in 2xYT supplemented with chloramphenicol (25 μg/ml). The bacterial culture was diluted in low calcium media to OD600 0.2, incubated at 26°C with 250 rpm shaking for 1.5 h, and 25 μl was distributed into a 384-well plate (NuncTM, Thermo Fisher). Fifty micromolars of compounds or equivalent volume of DMSO was added to each well and the plate incubated at 37°C with 250 rpm shaking for 2 h. The bacterial plate was spun down for 5 min and 10 μl of supernatant transferred to a new plate containing the same volume of nitrocefin (500 μg/ml). Absorbance at 490 nm was measured 30 min after addition of nitrocefin. Three technical replicates for each sample were carried out for each independent experiment.
Indicated concentrations of compounds, or the equivalent volumes of DMSO, were added to 26°C-grown low calcium medium cultures and treated cultures were shifted to 37°C for 3 h. RNA was isolated using an RNeasy Plus Micro Kit (Qiagen) according to the manufacturer's instructions and 2 μg RNA was used to make cDNA, as previously described (Auerbuch et al., 2009; Miller et al., 2014). SYBR Green PCR master mix (Applied Biosystems) was used for qPCR reactions according to the manufacturer's instructions and a 60°C annealing temperature, using the 16s rRNA gene as a reference for each sample. Three technical replicates were averaged for each sample/primer pair per independent experiment. Primers used are listed in Table 3. Results were analyzed using the Bio-Rad CFX software.
Indicated concentrations of compounds, or the equivalent volumes of DMSO, were added to 26°C-grown low calcium medium cultures and treated cultures were shifted to 37°C for 3 h. Two-hundred microliters of cultures were spun down at 3,000 × g for 5 min, resuspended in 200 μl 1X PBS, and mCherry fluorescence and optical density measured in black, clear bottom 96 well plates (Costar®, Corning Inc.) on a Perkin Elmer Victor X3 plate reader. Two technical replicates were averaged for each sample per independent experiment.
Quantification of YscF staining on the bacterial surface was carried out as described previously (Morgan et al., 2017). Indicated concentrations of compounds, or the equivalent volumes of DMSO, were added to 26°C-grown low calcium medium cultures and treated cultures were shifted to 37°C for 3 h. Bacteria were fixed by the addition of a mix of 800 μl 4% paraformaldehyde, 1 μl 25% glutaraldehyde, and 40 μl 0.5 M NaPO4 pH 7.4 to 500 μl of bacterial culture for 15 min at room temperature followed by 30 min or longer on ice. Fixed bacteria were gently sedimented (5,000 × g for 5 min), washed four times with PBS and stored in 250 mM glucose, 10 mM Tris-HCl pH 7.5, and 1 mM EDTA. Eight microliters of the fixed cells were added to coverslips and allowed to set until just dried. Coverslips were blocked overnight with PBST with 3% BSA (PBST/BSA) at 4°C. Blocking solution was removed and anti-YscF primary antibody was added at 1:10,000 in PBST/BSA and rocked at 4°C for 4 h. Coverslips were carefully rinsed in ice cold PBST with 0.1% Tween-20 several times and incubated with Alexa fluor 594 or 488 anti-rabbit secondary antibody (Invitrogen) at 1:10,000 in PBST/BSA and rocked at 4°C for 3 h followed by rinsing again in ice cold PBST with 0.1% Tween-20 several times. Coverslips were then stained for nuclear material with Hoechst 33342 (Thermo Scientific) at 1:10,000 in PBST/BSA and left in the dark at room temperature for 30 min. Coverslips were washed in ice cold PBST with 0.1% Tween-20 several times, allowed to dry briefly, mounted onto glass coverslips with Prolong Gold (Thermo Scientific), and sealed with clear nail polish. Images were taken with the Zeiss Axioimager Z2 widefield microscope under 63X/1.4 oil immersion using Zen software, pseudocolored, and merged in FIJI. YscF puncta were counted using IMARIS software.
Y. enterocolitica expressing EGFP-YscD were grown overnight in brain heart infusion (BHI) broth containing nalidixic acid (35 μg/ml) and diaminopimelic acid (80 μg/ml) (Diepold et al., 2010). Overnight cultures were diluted to an OD600 of 0.2 in M9 minimal medium supplemented with casamino acids. Cultures were grown for 1.5 h at 26°C. Compounds were added at indicated concentrations or the equivalent volumes of DMSO were added and the cultures switched to 37°C for another 3 h. Two microliters of culture were layered on a patch of 1% agarose in water (Skinner et al., 2013) supplemented with 80 μg/ml diaminopimelic acid, 5 mM EDTA, 10 mM MgCl2, and either compounds at indicated concentrations or the equivalent volume of DMSO (Diepold et al., 2010). Images were taken with a Zeiss Axioimager Z2 widefield microscope under 63X/1.4 oil immersion using Zen software, pseudocolored, and merged in FIJI. YscD puncta were counted using IMARIS software.
Y. pseudotuberculosis YpIII/pIBXN = 1 and YpIII/pIBX were grown overnight in 2xYT media containing kanamycin (30 μg/ml). Overnight cultures were diluted to an OD600 of 0.2 in M9 supplemented with casamino acids. Cultures were grown for 1.5 h at 26°C. Compounds were added at indicated concentrations or the equivalent volumes of DMSO were added and the cultures switched to 37°C for another 3 h. Two hundred microliters of each culture was added to clear bottom, white 96 well plates (Costar®, Corning Inc.) and optical density and luminescence were measured using a Perkin Elmer Victor X3 plate reader. Two technical replicates were averaged for each sample per independent experiment.
Measuring translocation of the YopM-β-lactamase (YopM-Bla) reporter effector protein was carried out as previously described (Duncan et al., 2014). A total of 6 × 103 CHO-K1 cells were plated in each well of a 384-well plate (CorningTM FalconTM) in 50 μl of F-12K medium plus 10% FBS and 1% glutamine, and incubated overnight. The following day, indicated concentrations of compounds, or the equivalent volumes of DMSO, were added to 26°C-grown low calcium medium cultures of Y. pseudotuberculosis expressing YopM-Bla and treated cultures were shifted to 37°C for 3 h. Immediately prior to infection of CHO-K1 cells, the F-12K media was removed and replaced with RPMI plus 10% FBS and compounds or DMSO were added. Y. pseudotuberculosis was added to the CHO-K1 plate at an MOI of 7. Five minutes after this transfer, the plate was centrifuged at 290 × g for 5 min to initiate bacterium-host cell contact and incubated for 1 h at 37°C and 5% CO2. Thirty minutes prior to the end of the infection, CCF2-AM (Invitrogen) was added to each well, and the plate covered in foil and incubated at room temperature. At the end of the infection, the medium was aspirated and fresh 4% paraformaldehyde added to each well for 20 min to fix the cells. The paraformaldehyde was then aspirated and the DNA dye DRAQ5 (Cell Signaling Technology) in PBS was added to each well. The monolayers were incubated at room temperature for 10 min, washed once with PBS, and visualized using an ImageXpressMICRO automated microscope and MetaXpress analysis software (Molecular Devices). The number of YopM-Bla-positive cells was calculated by dividing the number of blue (CCF2-cleaved) cells by the number of green (total CCF2) cells. Three technical replicates were averaged for each sample per independent experiment.
Overnight cultures of Y. pseudotuberculosis were diluted to an OD600 of 0.1 in 2xYT and 200 μl was added to each well of a 96-well plate (CorningTM FalconTM). Compounds were added at 50 μM and kanamycin at 50 μg/ml, plates incubated at 26°C, and the OD600 of the cultures measured every hour for 13 h using a VersaMax Tunable Microplate Reader (Molecular Devices). The 96-well plates were intermittently shaken throughout the experiment. One technical replicate was used for each sample per independent experiment.
Following overnight growth, 1 μl of Y. pseudotuberculosis overnight culture was spotted onto motility medium containing either 1% tryptone/0.25% agar or 1% tryptone/0.25% agar supplemented with 5 mM EGTA and 20 mM MgCl2 in six-well plates. Each well contained either 0.3% DMSO or 50 μM INP0007 or INP0010. The plates were incubated at 26°C for 24 h before the diameter of swimming motility was measured. One technical replicate was used for each sample per independent experiment.
We selected a training set of commercially-available compounds to validate our assay pipeline (Table 4). All compounds selected met two strict requirements: demonstrated inhibitory activity on the Yersinia T3SS and absence of bactericidal activity within 6 h to accommodate the timeframe of the pipeline assays (Figures 1A–C).
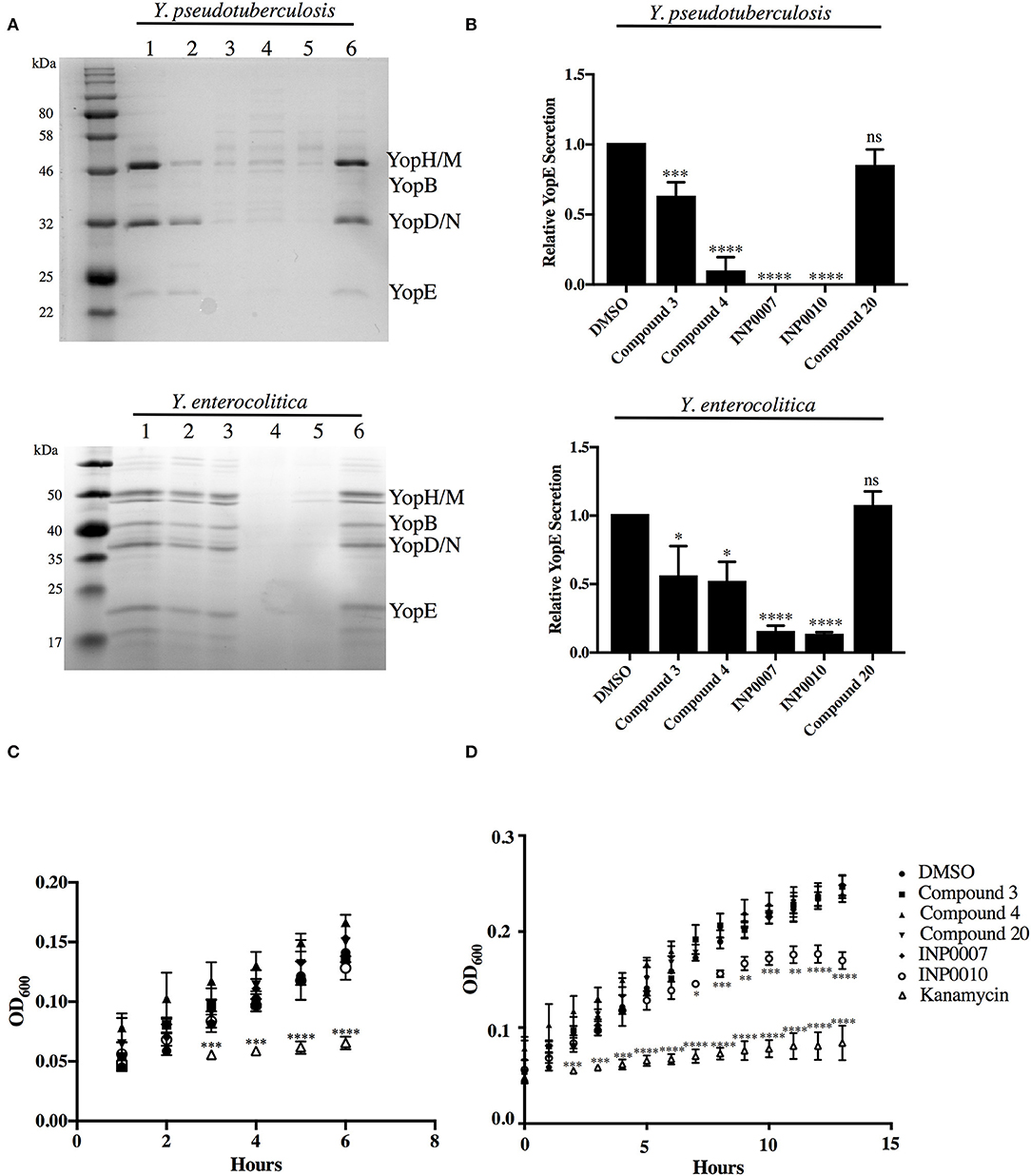
Figure 1. Efficiency of T3SS effector protein secretion and bacterial growth in the presence of T3SS inhibitors. (A,B) The relative efficiency of effector protein secretion into the culture supernatant was analyzed following bacterial growth for 2 h under T3SS-inducing conditions in the presence of either 50 μM compound or equivalent volume of DMSO. (A) The secretome of Y. pseudotuberculosis IP2666 and Y. enterocolitica 8081 was precipitated with trichloroacetic acid, separated by SDS-PAGE, and visualized by staining with Coomassie blue. Samples were normalized to culture optical density. (1) DMSO, (2) Compound 3, (3) Compound 4, (4) INP0007, (5) INP0010, (6) Compound 20. (B) Quantification of the YopE protein band by densitometry relative to the DMSO control. The average of 3 (Y. pseudotuberculosis) or 4 (Y. enterocolitica) biological replicates ± standard deviation is shown. (C,D) Y. pseudotuberculosis IP2666 growth at 26°C in the presence of 50 μM compound or DMSO was tracked by measuring optical density. The average of three biological replicates ± standard deviation is shown and statistical significance is represented comparing compounds relative to the DMSO control. *P < 0.03; **P < 0.004; ****P < 0.0001; ***P < 0.0007 (one way ANOVA with Dunnett's post-hoc test).
Compound 4 (C4), a haloid-containing sulfonamidobenzamide, was identified through a luciferase-based T3SS gene promoter fusion screen as a potent inhibitor of T3SS gene expression (Kauppi et al., 2003). We chose this compound because it was shown, albeit modestly, to inhibit expression of the LcrF master regulator of the T3SS in Yersinia (Kauppi et al., 2003) and we therefore expected that it would have a broad impact on all stages of type III secretion downstream of T3SS expression in our assay pipeline. From the same high throughput screening strategy that identified C4, two members of the salicylidene acylhydrazide class of inhibitors, INP0007 (Kauppi et al., 2003) and INP0010 (Nordfelth et al., 2005), were identified. Despite a large number of studies, the mechanism of action of these compounds remains unclear and there is evidence of off target effects on global virulence gene expression by at least some salicylidene acylhydrazides (Tree et al., 2009). In fact, INP0010 decreased Yersinia growth starting 7 h after initiation of treatment (Figure 1D), consistent with previous reports (Veenendaal et al., 2009). Compound 3 (C3), a malic diamide, was shown to inhibit ExoS secretion in P. aeruginosa and YopE secretion in Yersinia pestis (Aiello et al., 2010). This compound is structurally related to another class of T3SS inhibitors, the phenoxyacetamides, which were proposed to target the PscF needle subunit in Pseudomonas (Bowlin et al., 2014). Based on this structural relatedness, C3 was expected to disrupt T3SS needle assembly without impacting T3SS gene expression. Compound 20 (C20) was identified in a T3SS effector protein-β-lactamase reporter translocation screen, but had no significant effect on the ability of the bacteria to secrete T3SS effectors in the absence of host cells, pointing to the possibility that this compound specifically blocks the bacteria–host cell interaction (Harmon et al., 2010). Therefore, C20 was expected to inhibit the translocation of effector proteins into host cells but not impact T3SS gene expression and assembly.
An advantage of using Ysc-expressing Yersinia for our assay pipeline is that removing calcium from the culture medium enables T3SS effector proteins to be secreted into the supernatant in the absence host cells (Yother and Goguen, 1985; Perry et al., 1986; Straley and Bowmer, 1986; Sample et al., 1987; Forsberg and Wolf-Watz, 1988; Mehigh et al., 1989), providing a useful method of monitoring efficiency of T3SS inhibition in order to validate use of a given inhibitor in our assay pipeline. As expected, all test compounds except C20 exhibited significant inhibition of effector Yop secretion (Figures 1A,B). Importantly, this secretion assay was used routinely to ensure the efficacy of each batch of compound purchased and after 1 month in storage. Therefore, even when a compound had no effect in a particular assay, we could be confident that it was still active as a T3SS inhibitor.
We reasoned that inhibition of the pYV copy number upregulation that occurs during normal induction of type III secretion in Yersinia may impact T3SS gene expression, and therefore affect overall T3SS activity. To test this, we employed a Y. pseudotuberculosis strain with a luciferase reporter gene cluster integrated onto the pYV virulence plasmid (YpIII/pIBX) (Fahlgren et al., 2014). As a negative control, we used a strain in which a replication deficient copy of the virulence plasmid as well as the luciferase gene cluster was integrated into the chromosome (referred to as YpIII/pIBXN = 1) (Wang et al., 2016). In this YpIII/pIBXN = 1 strain, the ratio of T3SS and luciferase reporter genes vs. chromosomal genes remains constant even upon induction of type III secretion (Wang et al., 2016). As expected, expression of lcrF, the effector protein yopE, the integral IM ring protein yscD, and the needle subunit yscF were significantly decreased in the YpIII/pIBXN = 1 strain compared to the YpIII/pIBX strain (Figure 2A). While these data demonstrate that inhibition of pYV gene copy number could impact pYV gene expression, four of the five compounds in our training set did not significantly affect virulence plasmid copy number as measured by luciferase activity, while INP0007 significantly increased copy number (p = 0.0054; Figure 2B) for reasons that are unclear but that may reflect off target effects. In contrast, luciferase expression could be inhibited by blocking general transcription or translation using rifampicin or chloramphenicol (Figure 2B). Therefore, while inhibiting pYV copy number in Yersinia causes a decrease in T3SS activity, we conclude that none of the compounds in our training set act by inhibiting upregulation of pYV copy number.
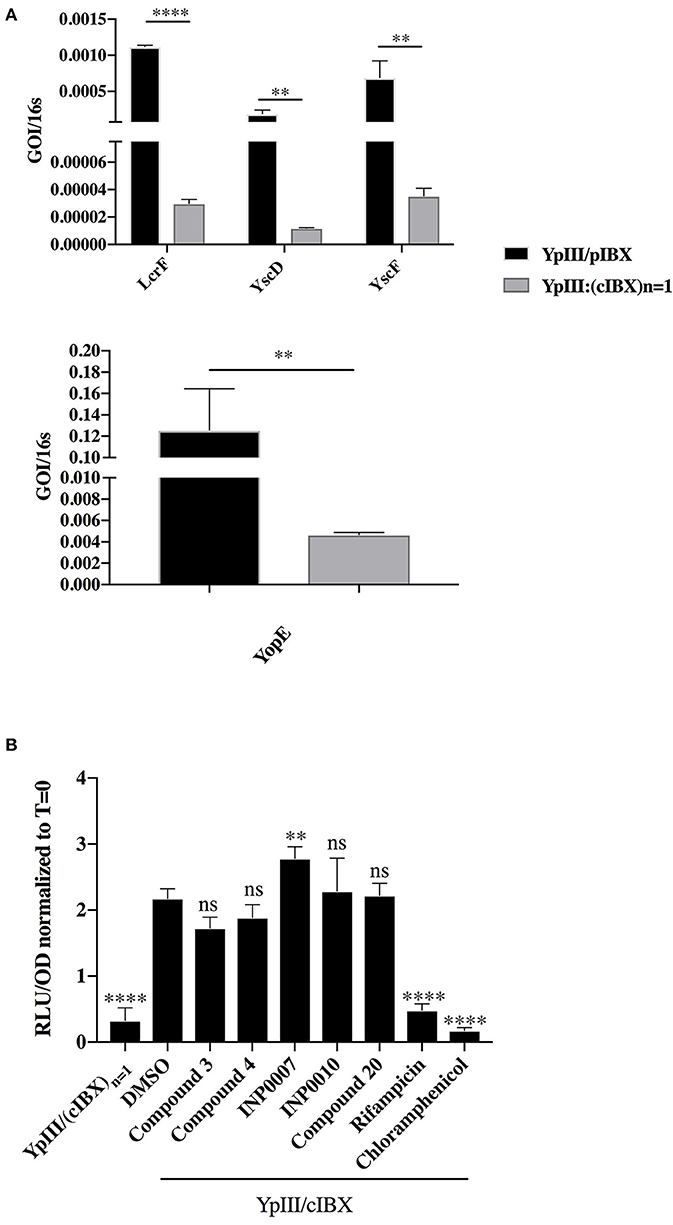
Figure 2. Correlation between pYV copy number and T3SS gene expression. (A) Y. pseudotuberculosis encoding luciferase genes on pYV (YPIII/pIBX) or a strain in which pYV-encoded T3SS and luciferase genes were incorporated into the chromosome in single copy [YPIII/(cIBX)n = 1] were grown for 3 h under T3SS-inducing conditions in 50 μM compound or equivalent volume of DMSO and T3SS gene expression evaluated by qPCR. GOI, gene of interest. Expression levels were normalized to 16s rRNA and then the fold change compared to DMSO calculated [(GOIcompound/16scompound)/(GOIDMSO/16sDMSO)]. The average of four biological replicates ± standard deviation is shown. ****P < 0.0001, **P < 0.008 (Student T-test). (B) Y. pseudotuberculosis YpIII/pIBX and YPIII/(cIBX)n = 1 were grown under T3SS-inducing conditions for 3 h in 50 μM compound or equivalent volume of DMSO and luminescence measured as a readout of pYV gene copy number. The average of four biological replicates ± standard deviation is shown and statistical significance is represented comparing compounds relative to the DMSO control. ****P < 0.0001; **P = 0.008 (one way ANOVA with Dunnett's post-hoc test).
In order to determine if the training set compounds negatively impacted T3SS gene expression, we used a Yersinia reporter strain containing an unstable YopH-mCherry-AAV transcriptional reporter (Andersen et al., 1998). C4, INP0007, and INP0010 inhibited YopH-mCherry-AAV expression when T3SS activity was induced by incubation in low calcium medium at 37°C (Figure 3A). In order to validate and extend these results, we measured transcript levels of T3SS and non-T3SS genes by qPCR. Consistent with the YopH-mCherry-AAV data, C3 and C20 had no significant impact on gene expression (Figure 4). However, C4 inhibited expression of all T3SS genes tested, but did not impact expression of two non-T3SS-associated genes: the iron sulfur cluster loading protein erpA and the small ribosomal subunit L9. Interestingly, INP0007 and INP0010 significantly inhibited transcript levels of lcrF, the needle tip subunit lcrV, and effector proteins yopH, yopE, and yopK, but did not inhibit expression of the ATPase yscN, yscF, and yscD under these conditions (Figure 4). To test whether inhibition of secretion via the positive feedback loop indirectly inhibits T3SS gene expression, we measured YopH-mCherry fluorescence and yscD mRNA levels via qPCR during growth at 37°C in high calcium (Figures 3B,C). Under these conditions, Yersinia can assemble the T3SS (Diepold et al., 2010), but no secretion of Yop effectors occurs (Forsberg and Wolf-Watz, 1988); therefore, the positive feedback loop is not active in high calcium. While overall expression levels of YscD and YopH are lower in high calcium compared to low calcium conditions, C4, INP0007, and INP0010 did not significantly decrease YopH-mCherry-AAV fluorescence in high calcium conditions (Figure 3B), suggesting an indirect effect of these compounds on T3SS gene expression through inhibition of secretion in low calcium conditions. Likewise, yscD levels were not decreased by C4 in high calcium (Figure 3C). Importantly, yscD expression was significantly lower in the ΔlcrF mutant compared to WT under high calcium conditions in the absence of compound. These data indicate that C4 is not likely to target LcrF activity, as yscD is predicted to be under LcrF control (Schwiesow et al., 2015).
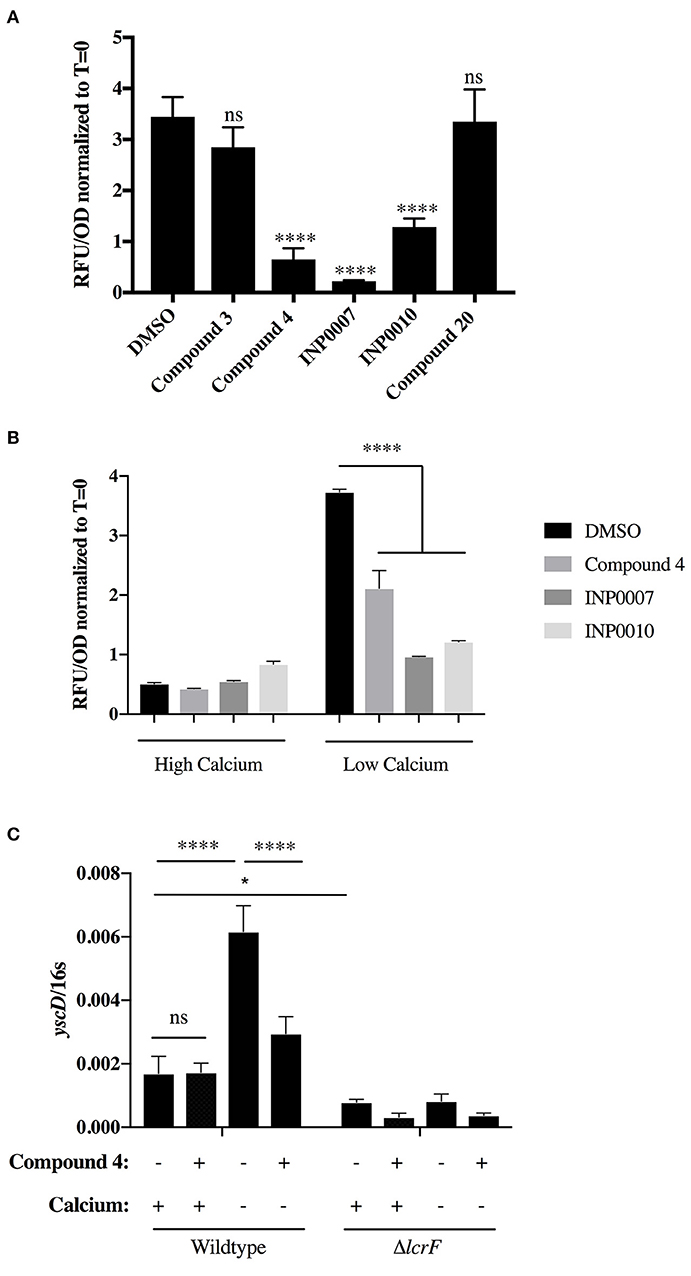
Figure 3. Analysis of yopH-mCherry fluorescence to assess T3SS gene expression. (A,B) Y. pseudotuberculosis pyopH FLAG mCherry was grown under T3SS-inducing (A,B, low calcium) or non-inducing (B, high calcium) conditions and relative mCherry fluorescence measured at 3 h after addition of 50 μM compound or equivalent volume of DMSO. The average of three biological replicates ± standard deviation is shown and statistical significance is represented comparing compounds relative to the DMSO control. ****P < 0.0001 (one way ANOVA with Dunnett's post-hoc test). (C) Y. pseudotuberculosis wildtype or ΔlcrF were grown in low or high calcium media for 3 h in 50 μM Compound 4 or equivalent volume of DMSO and yscD mRNA levels measured by qPCR. Expression levels were normalized to 16s rRNA. Statistical significance is represented comparing the indicated pairs of conditions. ****P < 0.0001, *P < 0.002 (Student t-test).
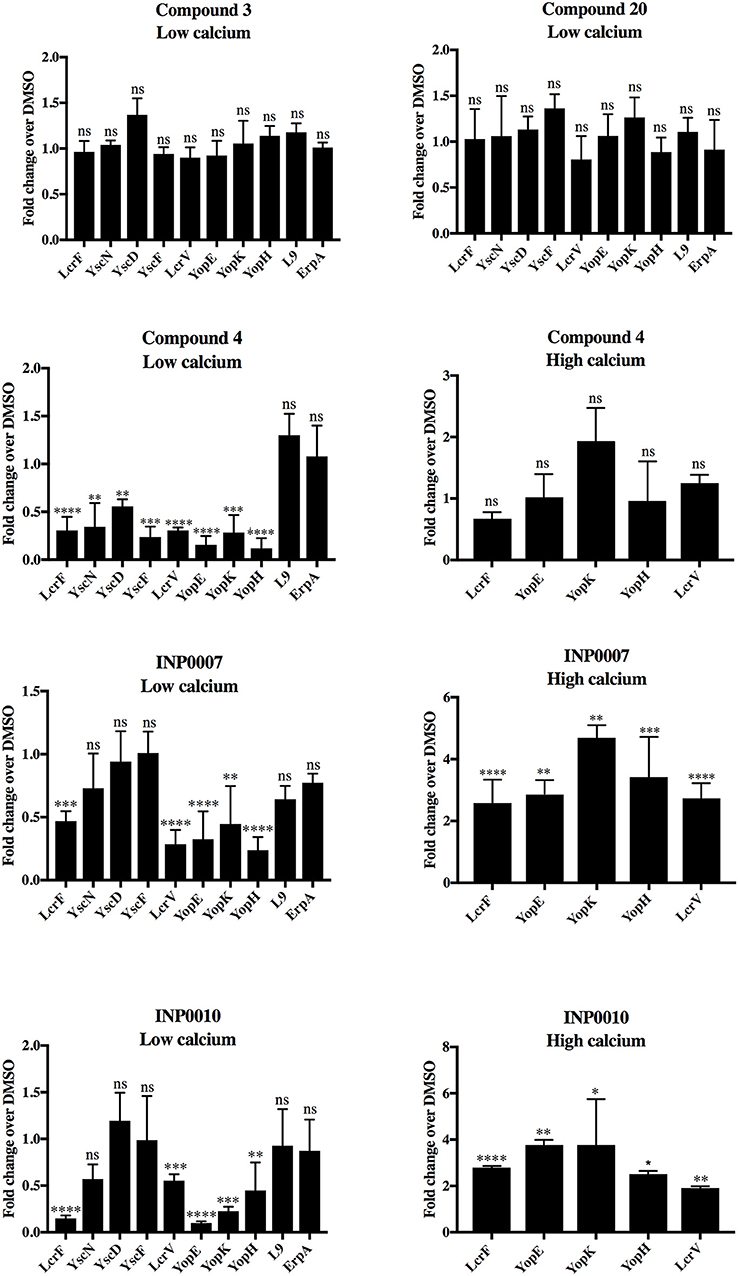
Figure 4. T3SS gene mRNA levels under low and high calcium conditions. Wildtype Y. pseudotuberculosis was grown under high or low calcium conditions in the presence of 50 μM compound or equivalent volume of DMSO and mRNA levels of T3SS (lcrF, yscN, yscD, yscF, lcrV, yopE, yopK, yopH) and non-T3SS genes (L9, erpA) assessed by qPCR. The average of four biological replicates ± standard deviation is shown and statistical significance is represented comparing compounds relative to the DMSO control for each gene. ****P < 0.0001, ***P < 0.0005, **P < 0.003; *P < 0.022 (one way ANOVA with Dunnett's post-hoc test). ns, not significant.
In order to monitor T3SS assembly, we used fluorescence microscopy to quantify YscD localization in Y. enterocolitica and YscF puncta formation in Y. pseudotuberculosis as a proxy for T3SS basal body and T3SS needle assembly, respectively, as previously described (Davis and Mecsas, 2007; Diepold et al., 2010; Morgan et al., 2017). Only INP0007 significantly impacted YscD-EGFP puncta formation (Figure 5). Importantly, the level of diffuse fluorescence in INP0007-treated Yersinia was greater than that of the non-T3SS-inducing condition (26°C-grown Yersinia; Figure 5A, inset), indicating that while YscD is expressed at 37°C in the presence of INP0007, it is not assembled into basal bodies in the presence of this compound. In contrast, while C3 and C20 had no significant effect on YscF puncta formation as measured with an anti-YscF antibody, C4, INP0007, and INP0010 significantly inhibited needle assembly (Figure 6).
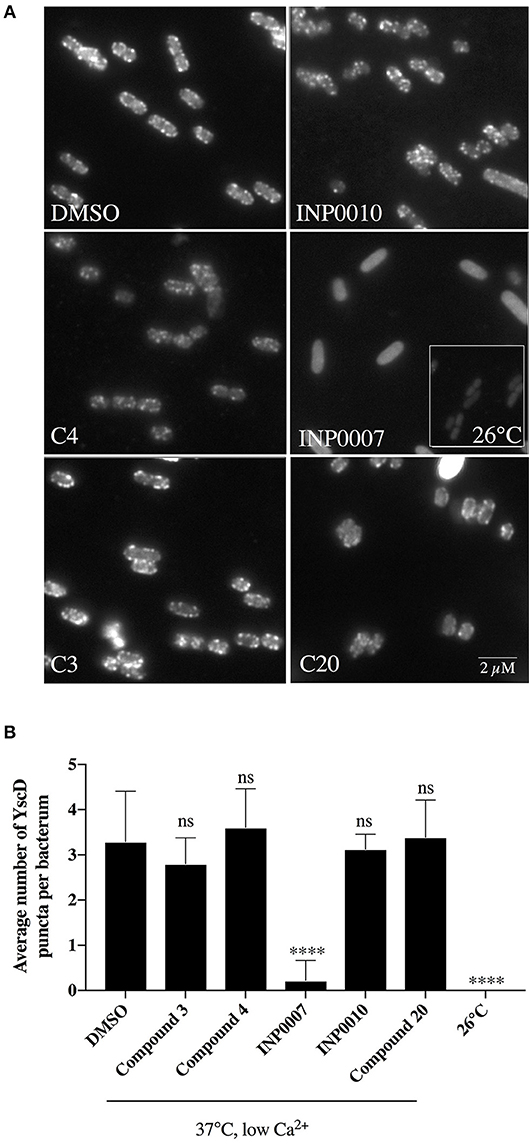
Figure 5. YscD-EGFP puncta formation in Y. enterocolitica. Y. enterocolitica pYV40-EGFP-yscD was grown under T3SS-inducing conditions for 3 h in the presence of 50 μM compound or equivalent volume of DMSO and fluorescent foci quantified using IMARIS software. Representative images (A) and the average number of foci ± standard deviation per cell (B) are shown from three biological replicates, with N > 2,000 cells quantified per condition. Statistical significance is represented comparing compounds relative to the DMSO control. ****P < 0.008 (one way ANOVA with Dunnett's post-hoc test).
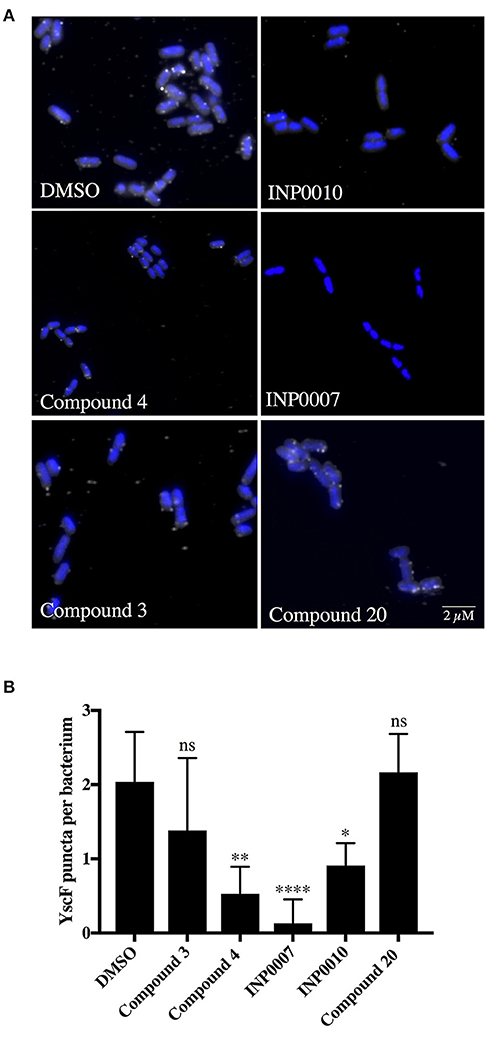
Figure 6. YscF puncta visualization in Y. pseudotuberculosis using immunofluorescence. Wildtype Y. pseudotuberculosis was grown under T3SS-inducing conditions for 3 h in 50 μM compound or equivalent volume of DMSO and an anti-YscF antibody used to visualize T3SS needles. (A) Representative confocal microscopy images and (B) the average number of YscF puncta per bacterium from three biological replicates ± the standard deviation are shown, with N > 1,500 bacteria quantified per condition. Statistical significance is represented comparing compounds relative to the DMSO control. ****P < 0.0001, **P < 0.002, *P < 0.04 (one way ANOVA with Dunnett's post-hoc test).
The bacterial flagellar apparatus is composed of a basal body that is structurally related to the injectisome T3SS basal body and mediates secretion of the flagellar hook and filament proteins (Macnab, 2004). Therefore, it is possible that compounds with the ability to inhibit injectisome T3SS basal body assembly may inhibit flagellar assembly and therefore flagellar motility. INP0007 has been shown to inhibit motility in Yersinia (Kauppi et al., 2003), but neither INP0007 nor INP0010 inhibited motility in Salmonella (Negrea et al., 2007). In our standard Yersinia motility agar (Morgan et al., 2017), INP0007 significantly decreased flagellar motility in Y. pseudotuberculosis, while INP0010 decreased motility weakly albeit significantly (Figure 7). Addition of the chelating agent EGTA in the motility agar, as used in the Kauppi et al study (Kauppi et al., 2003), led to an even greater inhibitory effect on motility by INP0010 (Figure 7). The Y. pseudotuberculosis flhDCYpestis non-motile strain served as the negative control for the motility assay. Taken together, these data indicate that INP0007 and INP0010 impact both the flagellar and injectisome T3SS.
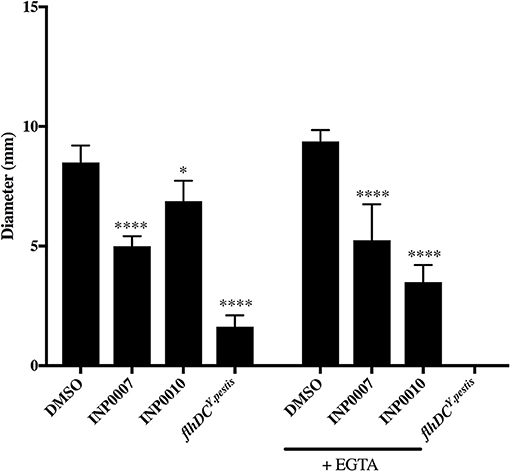
Figure 7. INP0007 and INP0010 inhibit Y. pseudotuberculosis motility. Y. pseudotuberculosis wildtype or the non-motile flhDCY.pestis mutant were spotted onto motility agar containing the 50 μM compounds or the equivalent volume of DMSO and allowed to grow for ~24 h. Average swimming diameter of the colony was measured from three biological replicates ± standard deviation and statistical significance is represented comparing compounds relative to the DMSO control. ****P < 0.0001, *P < 0.03 (one way ANOVA with Dunnett's post-hoc test).
While the Yop in vitro secretion assay shown in Figure 1 is critical for assessing the impact of a compound on overall Yop secretion, the method used (Coomassie staining of SDS-PAGE separated proteins precipitated from the culture supernatant) is not amenable to a high throughput format. In order to assess Yop secretion in vitro in microtiter plates, Y. pseudotuberculosis expressing a YopM-β-lactamase (YopM-Bla) reporter was used in conjunction with the chromogenic β-lactamase substrate nitrocefin (O'Callaghan et al., 1972; Lee et al., 2007; Green et al., 2016). Consistent with the block in Yop secretion observed in Figure 1, C3, C4, INP0007, and INP0010 significantly blocked YopM-Bla secretion into the culture supernatant while C20 did not (Figure 8). Surprisingly, INP0010 blocked YopM-Bla secretion by only 32% as measured using nitrocefin (Figure 8), yet appeared to have a much more significant effect on native YopM secretion as measured using Coomassie staining (Figure 1). The reason for this discrepancy is unclear; however, we note that INP0010 has cytotoxic effects on bacteria and eukaryotic cells (see below) and is likely to have the greatest off target effects of the compounds in our training set.
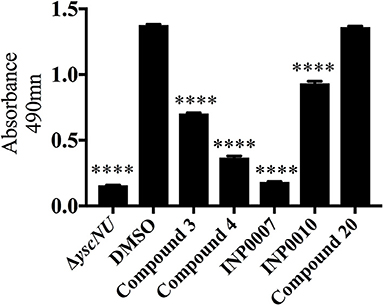
Figure 8. Secretion of YopM-Bla into the culture supernatant. Y. pseudotuberculosis Δyop6 expressing the Yop reporter YopM-Bla were grown under T3SS-inducing in vitro conditions for 2 h in the presence of 50 μM compound or equivalent volumes of DMSO and cleavage of the β-lactamase substrate nitrocefin used to measure the relative quantity of secreted YopM-Bla. The average of three independent experiments ± standard deviation are shown and statistical significance is represented comparing compounds relative to the DMSO control. ****P < 0.0001 (one way ANOVA with Dunnett's post-hoc test).
One of the most important requirements of a T3SS inhibitor is its ability to prevent translocation of effector proteins into target host cells. We employed a different type of YopM-Bla reporter assay to assess translocation of effector proteins into CHO K1 cells loaded with CCF2, a fluorescent β-lactamase substrate that can enter eukaryotic cells (Dewoody et al., 2011). While C3, C4, C20, INP0007, and INP0010 significantly blocked YopM-Bla translocation at 50 μM, INP0010 appeared to be cytotoxic at this concentration (Figure 9).
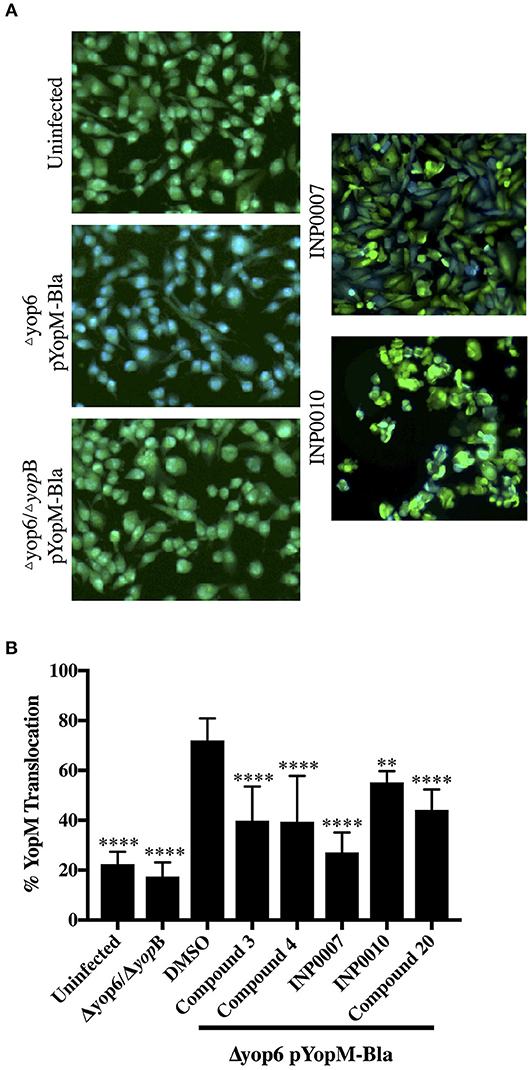
Figure 9. Translocation of YopM-Bla into mammalian cells in the presence of T3SS inhibitors. (A) Representative fluorescent micrographs of CCF2-AM loaded CHO K1 cells in the absence of bacteria (uninfected) or infected for 2 h with Y. pseudotuberculosis lacking YopHEMOJT but expressing the Yop reporter YopM-Bla (Δyop6 pYopM-Bla) at MOI 7 in the presence of 50 μM compound or equivalent volume of DMSO. A T3SS-defective mutant lacking the pore-forming protein YopB was used as a negative control (Δyop6/ΔyopB pYopM-Bla). (B) CCF2 green (uncleaved) and blue (cleaved) fluorescence was measured and the average from three biological replicates ± standard deviation is shown and statistical significance is represented relative to the DMSO control. ****P < 0.0001, **P < 0.007 (one way ANOVA with Dunnett's post-hoc test).
A methodical approach to primary characterization of inhibitors for the T3SS has been lacking in the field. Here, we describe a pipeline of miniaturized assays, using enteropathogenic Yersinia as the workhorse organism, that enable rapid, initial characterization of the stage of T3SS expression, assembly, or function targeted by compounds with T3SS inhibitory activity. Furthermore, we used a training set of compounds with previously-identified T3SS inhibitory activity to test the utility of this pipeline (Table 5). INP0007 blocked T3SS basal body and needle assembly and therefore inhibited Yop secretion and translocation. C4 and INP0010 did not prevent basal body assembly but blocked needle assembly, secretion, and translocation. However, INP0010 exhibited cytotoxicity, complicating interpretation of some assay results. C3 allowed T3SS assembly but prevented Yop secretion and translocation. Lastly, C20 allowed T3SS assembly and Yop secretion in vitro, but blocked translocation of Yops into target host cells. These results demonstrate the ability of our assay pipeline both to validate T3SS inhibitors and provide testable hypotheses on their mode of action.
C3 blocked the ability of the Yersinia T3SS to secrete T3SS effector proteins into low calcium culture supernatant and to translocate effector proteins into target host cells. Yet C3 did not block T3SS basal body and needle assembly, as determined by imaging YscD basal body and YscF needle puncta formation. C3 showed a modest inhibition of YopE secretion (28%), but notably inhibited secretion of the translocator protein YopD (Figure 1A). These data provide a possible explanation for why C3 strongly inhibited Yop translocation into host cells (40%) despite less potent inhibition of YopE secretion, as YopD is required for Yop delivery into host cells (Rosqvist et al., 1994) C3 has structural similarities with the phenoxyacetamides, which target the SctF needle subunit (Aiello et al., 2010; Bowlin et al., 2014). However, our data indicate that C3 blocks a specific activity of the assembled T3SS. Alternatively, the T3SS needle structure may be altered in the presence of C3, still allowing recognition by our anti-YscF antibody but impeding cargo egress.
In contrast to C3, C20 specifically blocked translocation of a T3SS effector protein into host cells without blocking any other stage of type III secretion, including T3SS effector protein secretion into low calcium culture. As C20 was previously shown to inhibit only translocation of T3SS effector proteins into target host cells (Harmon et al., 2010), our verification of this finding further demonstrates the robustness of our experimental pipeline. Harmon et al. hypothesized that C20 inhibited the host cell-bacterial interaction, as C20 strongly inhibited adherence of Y. pseudotuberculosis to HEp-2 cells. Possible modes of action of compounds that inhibit Yop translocation but not any other stage of type III secretion include interruption of bacterial adhesins, host integrin receptors, or YopBD-mediated pore formation on the host membrane.
C4 was shown to inhibit promoter activity of lcrF, the Ysc T3SS master regulator (Kauppi et al., 2003). However, while we observed a C4-dependent decrease in mRNA levels of all T3SS genes tested under low calcium T3SS-inducing conditions, C4 treatment did not impact T3SS gene expression under high calcium conditions when T3SSs are assembled but no Yop secretion occurs. As a ΔlcrF mutant had significantly less T3SS gene expression under high calcium conditions compared to wildtype Yersinia, this argues against C4 inhibiting LcrF activity. C4 did not alter the number of YscD puncta observed but did decrease the number of YscF puncta per cell by greater than half. These data suggest that C4 does not impact YscD assembly in the plasma membrane but disrupts overall T3SS basal body formation in such a way that needle formation is compromised or interferes with YscF secretion or assembly.
The salicylidene acylhydrazides INP0007 and INP0010 inhibited expression of lcrF, lcrV, and yopKEH, but did not decrease expression of yscN, yscD, or yscF. While INP0007 and INP0010 are related structurally, INP0010 prevents Yersinia growth if the bacteria are exposed longer than 6 h, while INP0007 does not affect bacterial growth even up to 13 h of exposure. Surprisingly, while INP0010 did not inhibit YscD puncta formation, INP0007 caused a 10-fold decrease in YscD puncta. Both compounds significantly inhibited YscF puncta formation, Yop secretion into low calcium culture, and Yop translocation into host cells. However, measuring Yop translocation into host cells in the presence of INP0010 is complicated by the obvious toxic effects of the compound on host cells. Therefore, utilization of the Yop translocation assay of our experimental pipeline is limited to compounds without toxic effects on host cells. However, this also shows the benefit of using microscopy to observe CCF2 green-to-blue fluorescence conversion, as it allows observation of host cell morphology. The salicylidene acylhydrazide class of compounds have been suggested to have a broad impact on expression of horizontally acquired genes (Tree et al., 2009). However, we only observe a defect in T3SS gene expression in low calcium, but not high calcium medium, for INP0007. In addition, we observe a significant defect in flagellar motility by INP0007, suggesting a broader effect of the compound either on the flagellar T3SS, ATP synthesis, or the proton motive force. In total, our results argue that INP0007 inhibits formation of the injectisome T3SS and, either through a similar mechanism or by targeting multiple pathways, also disrupts flagellar T3SS activity. Therefore, we suggest that for T3SS inhibitors that block T3SS basal body assembly, a flagellar motility assay be performed to assess their breadth of action.
Given the previously-demonstrated positive feedback exerted by active type III secretion on T3SS gene transcription (Cornelis et al., 1987), we expected that C3 would inhibit T3SS gene expression in low calcium medium. C4 and INP0007 inhibited YopE secretion by >70–80% and this correlated with the ability to affect T3SS gene expression under low calcium conditions. C3 inhibited YopE secretion < 50% and this was insufficient to impact the feedback loop on T3SS gene expression. Similarly, the T3SS inhibitor piericidin A1 inhibits YopE secretion by ~40–45% and also does not repress transcription of T3SS genes (Morgan et al., 2017). These data suggest that only compounds that potently inhibit Yop secretion will affect the T3SS gene expression feedback loop. This indicates that screening strategies based on Yop gene expression as the readout for T3SS inhibition may miss less robust compounds that could be improved by structure-activity relationship analysis.
While none of the inhibitors we tested affected pYV copy number, our data shows that prevention of pYV plasmid copy number upregulation during active type III secretion leads to dramatically reduced T3SS gene mRNA steady-state levels. Therefore, when using Yersinia as a model organism for assessing T3SS inhibitor mechanism of action, it is important to consider pYV copy number. Furthermore, as chloramphenicol and rifampicin significantly inhibited luminescence as a readout of pYV copy number, this assay also sheds light on whether a compound affects general transcription or translocation. One caveat is that compounds that act as luminescence quenchers would decrease luminescence in this assay. In this case, analyzing expression of non-T3SS genes, such as the L9 and ErpA genes shown here, via qPCR can serve as a way to test this possibility.
In summary, we propose that the experimental pipeline described here can be used to rapidly bin T3SS inhibitors into categories depending on the stage of type III secretion they inhibit, providing testable hypotheses on mode of action. The commercially available training set shown here can be used to establish these assays in other labs and perhaps enable a more standardized approach to T3SS inhibitor research.
JM: experimental design, performing experiments, writing paper; HL: experimental design, performing experiments; JD and JL: performing experiments; SM, RI, HW: providing reagents; VA: experimental design, writing paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Benjamin Abrams of the UCSC Life Sciences Microscopy Center and Walter Bray of the UCSC Chemical Screening Center for technical support. We thank Andreas Diepold for providing the fluorescent YscD Y. enterocolitica strain. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Numbers R01AI106930 and R01AI119082. JM was supported by the National Human Genome Research Institute of the National Institutes of Health under Award Number 4R25HG006836-04. RI was supported by NIAID R01AI110684. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Aiello, D., Williams, J. D., Majgier-Baranowska, H., Patel, I., Peet, N. P., Huang, J., et al. (2010). Discovery and characterization of inhibitors of Pseudomonas aeruginosa type III secretion. Antimicrob. Agents Chemother. 54, 1988–1999. doi: 10.1128/AAC.01598-09
Anantharajah, A., Buyck, J. M., Sundin, C., Tulkens, P. M., Mingeot-Leclercq, M. P., and Van Bambeke, F. (2017). Salicylidene acylhydrazides and hydroxyquinolines act as inhibitors of type three secretion systems in Pseudomonas aeruginosa by distinct mechanisms. Antimicrob. Agents Chemother. 61:e02566-16. doi: 10.1128/AAC.02566-16
Andersen, J. B., Sternberg, C., Poulsen, L. K., Bjorn, S. P., Givskov, M., and Molin, S. (1998). New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64, 2240–2246.
Auerbuch, V., Golenbock, D. T., and Isberg, R. R. (2009). Innate immune recognition of Yersinia pseudotuberculosis type III secretion. PLoS Pathog. 5:e1000686. doi: 10.1371/journal.ppat.1000686
Beckham, K. S., and Roe, A. J. (2014). From screen to target: Insights and approaches for the development of anti-virulence compounds. Front. Cell. Infect. Microbiol. 4:139. doi: 10.3389/fcimb.2014.00139
Bergeron, J., Worrall, L., Sgourakis, N., DiMaio, F., Pfuetzner, R., Felise, H., et al. (2013). A refined model of the prototypical Salmonella spi-1 t3ss basal body reveals the molecular basis for its assembly. PLoS Pathog. 9:e1003307. doi: 10.1371/journal.ppat.1003307
Berube, B. J., Murphy, K. R., Torhan, M. C., Bowlin, N. O., Williams, J. D., Bowlin, T. L., et al. (2017). Impact of type III secretion effectors and of phenoxyacetamide inhibitors of type III secretion on abscess formation in a mouse model of Pseudomonas aeruginosa infection. Antimicrob. Agents Chemother. 61:e01202-17. doi: 10.1128/AAC.01202-17
Bliska, J. B., Guan, K. L., Dixon, J. E., and Falkow, S. (1991). Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc. Natl. Acad. Sci. USA. 88, 1187–1191.
Bowlin, N. O., Williams, J. D., Knoten, C. A., Torhan, M. C., Tashjian, T. F., Li, B., et al. (2014). Mutations in the Pseudomonas aeruginosa needle protein gene pscF confer resistance to phenoxyacetamide inhibitors of the type III secretion system. Antimicrob. Agents Chemother. 58, 2211–2220. doi: 10.1128/AAC.02795-13
Broz, P., Mueller, C. A., Müller, S. A., Philippsen, A., Sorg, I., Engel, A., et al. (2007). Function and molecular architecture of the Yersinia injectisome tip complex. Mol. Microbiol. 65, 1311–1320. doi: 10.1111/j.1365-2958.2007.05871.x
Büttner, D., and Bonas, U. (2002). Port of entry–the type III secretion translocon. Trends Microbiol. 10, 186–192. doi: 10.1016/S0966-842X(02)02331-4
Coburn, B., Sekirov, I., and Finlay, B. (2007). Type III secretion systems and disease. Clin. Microbiol. Rev. 20, 535–549. doi: 10.1128/CMR.00013-07
Cornelis, G., Vanootegem, J. C., and Sluiters, C. (1987). Transcription of the yop regulon from Y. enterocolitica requires trans acting pYV and chromosomal genes. Microb Pathog. 2, 367–379.
Davis, A., and Mecsas, J. (2007). Mutations in the Yersinia pseudotuberculosis type III secretion system needle protein, yscF, that specifically abrogate effector translocation into host cells. J. Bacteriol. 189, 83–97. doi: 10.1128/JB.01396-06
Deng, W., Marshall, N. C., Rowland, J. L., McCoy, J. M., Worrall, L. J., Santos, A. S., et al. (2017). Assembly, structure, function and regulation of type III secretion systems. Nat. Rev. Microbiol. 15, 323–337. doi: 10.1038/nrmicro.2017.20
Dewoody, R., Merritt, P. M., Houppert, A. S., and Marketon, M. M. (2011). YopK regulates the Yersinia pestis type III secretion system from within host cells. Mol. Microbiol. 79, 1445–1461. doi: 10.1111/j.1365-2958.2011.07534.x
Diepold, A., Amstutz, M., Abel, S., Sorg, I., Jenal, U., and Cornelis, G. R. (2010). Deciphering the assembly of the yersinia type III secretion injectisome. EMBO J. 29, 1928–1940. doi: 10.1038/emboj.2010.84
Duncan, M. C., Linington, R. G., and Auerbuch, V. (2012). Chemical inhibitors of the type three secretion system: disarming bacterial pathogens. Antimicrob. Agents Chemother.. 56(11):5433–5441. doi: 10.1128/AAC.00975-12
Duncan, M. C., Wong, W. R., Dupzyk, A. J., Bray, W. M., Linington, R. G., and Auerbuch, V. (2014). An nF-κβ-based high-throughput screen identifies piericidins as inhibitors of the Yersinia pseudotuberculosis type III secretion system. Antimicrob. Agents Chemother. 58, 1118–1126. doi: 10.1128/AAC.02025-13
Fahlgren, A., Avican, K., Westermark, L., Nordfelth, R., and Fällman, M. (2014). Colonization of cecum is important for development of persistent infection by Yersinia pseudotuberculosis. Infect. Immun. 82, 3471–3482. doi: 10.1128/IAI.01793-14
Forsberg, A., and Wolf-Watz, H. (1988). The virulence protein yop5 of Yersinia pseudotuberculosis is regulated at transcriptional level by plasmid-plb1 -encoded trans-acting elements controlled by temperature and calcium. Mol. Microbiol. 2, 121–133.
Francis, M. S., Wolf-Watz, H., and Forsberg, A. (2002). Regulation of type III secretion systems. Curr. Opin. Microbiol. 5, 166–172. doi: 10.1016/S1369-5274(02)00301-6
Green, E. R., Clark, S., Crimmins, G. T., Mack, M., Kumamoto, C. A., and Mecsas, J. (2016). Fis is essential for Yersinia pseudotuberculosis virulence and protects against reactive oxygen species produced by phagocytic cells during infection. PLoS Pathog. 12:e1005898. doi: 10.1371/journal.ppat.1005898
Harmon, D. E., Davis, A. J., Castillo, C., and Mecsas, J. (2010). Identification and characterization of small-molecule inhibitors of yop translocation in Yersinia pseudotuberculosis. Antimicrob. Agents Chemother. 54, 3241–3254. doi: 10.1128/AAC.00364-10
Heroven, A. K., Böhme, K., and Dersch, P. (2012). The Csr/Rsm system of Yersinia and related pathogens: a post-transcriptional strategy for managing virulence. RNA Biol. 9, 379–391. doi: 10.4161/rna.19333
Horton, R. M., Cai, Z. L., Ho, S. N., and Pease, L. R. (1990). Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques 8, 528–535.
Karzai, A. W., Roche, E. D., and Sauer, R. T. (2000). The ssrA-smpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7, 449–455. doi: 10.1038/75843
Kauppi, A. M., Nordfelth, R., Uvell, H., Wolf-Watz, H., and Elofsson, M. (2003). Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10, 241–249. doi: 10.1016/S1074-5521(03)00046-2
Lee, V. T., Pukatzki, S., Sato, H., Kikawada, E., Kazimirova, A. A., Huang, J., et al. (2007). Pseudolipasin a is a specific inhibitor for phospholipase A2 activity of Pseudomonas aeruginosa cytotoxin exoU. Infect. Immun. 75, 1089–1098. doi: 10.1128/IAI.01184-06
Macnab, R. M. (2004). Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta. 1694, 207–217. doi: 10.1016/j.bbamcr.2004.04.005
Marsden, A. E., King, J. M., Spies, M. A., Kim, O. K., and Yahr, T. L. (2015). Inhibition of Pseudomonas aeruginosa ExsA DNA binding activity by H-hydroxybenzimidazoles. Antimicro. Agents Chemother. 60, 766–776. doi: 10.1128/AAC.02242-15
Marshall, N. C., and Finlay, B. B. (2014). Targeting the type III secretion system to treat bacterial infections. Expert Opin. Ther. Targets 18, 137–152. doi: 10.1517/14728222.2014.855199
Mehigh, R. J., Sample, A. K., and Brubaker, R. R. (1989). Expression of the low calcium response in Yersinia pestis. Microb. Pathog. 6, 203–217. doi: 10.1016/0882-4010(89)90070-3
Merriam, J. J., Mathur, R., Maxfield-Boumil, R., and Isberg, R. R. (1997). Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65, 2497–2501.
Miller, H. K., Kwuan, L., Schwiesow, L., Bernick, D. L., Mettert, E., Ramirez, H. A., et al. (2014). IscR is essential for Yersinia pseudotuberculosis type III secretion and virulence. PLoS Pathog. 10:e1004194. doi: 10.1371/journal.ppat.1004194
Morgan, J. M., Duncan, M. C., Johnson, K. S., Diepold, A., Lam, H., Dupzyk, A. J., et al. (2017). Piericidin A1 blocks Yersinia ysc type III secretion system needle assembly. mSphere 2:e00030–17. doi: 10.1128/mSphere.00030-17
Negrea, A., Bjur, E., Ygberg, S. E., Elofsson, M., Wolf-Watz, H., and Rhen, M. (2007). Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enterica serovar typhimurium. Antimicrob. Agents Chemother. 51, 2867–2876. doi: 10.1128/AAC.00223-07
Nordfelth, R., Kauppi, A. M., Norberg, H. A., Wolf-Watz, H., and Elofsson, M. (2005). Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73, 3104–3114. doi: 10.1128/IAI.73.5.3104-3114.2005
O'Callaghan, C. H., Morris, A., Kirby, S. M., and Shingler, A. H. (1972). Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1, 283–288. doi: 10.1128/AAC.1.4.283
Perry, R. D., Harmon, P. A., Bowmer, W. S., and Straley, S. C. (1986). A low-Ca2+ response operon encodes the V antigen of Yersinia pestis. Infect. Immun. 54, 428–434.
Pettersson, J., Nordfelth, R., Dubinina, E., Bergman, T., Gustafsson, M., Magnusson, K. E., et al. (1996). Modulation of virulence factor expression by pathogen target cell contact. Science 273, 1231–1233.
Portnoy, D. A., Moseley, S. L., and Falkow, S. (1981). Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect. Immun. 31, 775–782.
Rosqvist, R., Magnusson, K. E., and Wolf-Watz, H. (1994). Target cell contact triggers expression and polarized transfer of Yersinia yopE cytotoxin into mammalian cells. EMBO J. 13, 964–972.
Sample, A. K., Fowler, J. M., and Brubaker, R. R. (1987). Modulation of the low-calcium response in Yersinia pestis via plasmid-plasmid interaction. Microb. Pathog. 2, 443–453. doi: 10.1016/0882-4010(87)90051-9
Schwiesow, L., Lam, H., Dersch, P., and Auerbuch, V. (2015). Yersinia type III secretion system master regulator LcrF. J. Bacteriol. 198, 604–614. doi: 10.1128/JB.00686-15
Skinner, S. O., Sepúlveda, L. A., Xu, H., and Golding, I. (2013). Measuring mRNA copy number in individual Escherichia coli cells using single-molecule fluorescent in situ hybridization. Nat. Protoc. 8, 1100–1113. doi: 10.1038/nprot.2013.066
Straley, S. C., and Bowmer, W. S. (1986). Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect. Immun. 51, 445–454.
Tree, J. J., Wang, D., McInally, C., Mahajan, A., Layton, A., Houghton, I., et al. (2009). Characterization of the effects of salicylidene acylhydrazide compounds on type III secretion in Escherichia coli o157:H7. Infect. Immun. 77, 4209–4220. doi: 10.1128/IAI.00562-09
Veenendaal, A. K., Sundin, C., and Blocker, A. J. (2009). Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol. 191, 563–570. doi: 10.1128/JB.01004-08
Wang, H., Avican, K., Fahlgren, A., Erttmann, S. F., Nuss, A. M., Dersch, P., et al. (2016). Increased plasmid copy number is essential for Yersinia T3SS function and virulence. Science 353, 492–495. doi: 10.1126/science.aaf7501
Wilharm, G., Lehmann, V., Krauss, K., Lehnert, B., Richter, S., Ruckdeschel, K., et al. (2004). Yersinia enterocolitica type III secretion depends on the proton motive force but not on the flagellar motor components motA and motB. Infect. Immun. 72, 4004–4009. doi: 10.1128/IAI.72.7.4004-4009.2004
Keywords: type III secretion system, T3SS, T3SS inhibitor, Yersinia, pYV
Citation: Morgan JM, Lam HN, Delgado J, Luu J, Mohammadi S, Isberg RR, Wang H and Auerbuch V (2018) An Experimental Pipeline for Initial Characterization of Bacterial Type III Secretion System Inhibitor Mode of Action Using Enteropathogenic Yersinia. Front. Cell. Infect. Microbiol. 8:404. doi: 10.3389/fcimb.2018.00404
Received: 30 July 2018; Accepted: 26 October 2018;
Published: 22 November 2018.
Edited by:
Matthew C. Wolfgang, University of North Carolina at Chapel Hill, United StatesReviewed by:
Deborah Anderson, University of Missouri, United StatesCopyright © 2018 Morgan, Lam, Delgado, Luu, Mohammadi, Isberg, Wang and Auerbuch. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Victoria Auerbuch, dmFzdG9uZUB1Y3NjLmVkdQ==
†Present Address: Sina Mohammadi, Merck Exploratory Science Center, Cambridge, MA, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.