- 1Shenzhen Public Platform for Screening and Application of Marine Microbial Resources, The Graduate School at Shenzhen, Tsinghua University, Beijing, China
- 2The Department of Earth Science, Tsinghua University, Beijing, China
- 3Biology, Shenzhen Polytechnic, Shenzhen, China
The worldwide increase in antibiotic-resistant pathogens means that identification of alternative antibacterial drug targets and the subsequent development of new treatment strategies are urgently required. One such new target is the quorum sensing (QS) system. Coral microbial consortia harbor an enormous diversity of microbes, and are thus rich sources for isolating novel bioactive and pharmacologically valuable natural products. However, to date, the versatility of their bioactive compounds has not been broadly explored. In this study, about two hundred bacterial colonies were isolated from a coral species (Pocillopora damicornis) and screened for their ability to inhibit QS using the bioreporter strain Chromobacterium violaceum ATCC 12472. Approximately 15% (30 isolates) exhibited anti-QS activity, against the indicator strain. Among them, a typical Gram-positive bacterium, D11 (Staphylococcus hominis) was identified and its anti-QS activity was investigated. Confocal microscopy observations showed that the bacterial extract inhibited the biofilm formation of clinical isolates of wild-type P. aeruginosa PAO1 in a dose-dependent pattern. Chromatographic separation led to the isolation of a potent QS inhibitor that was identified by high-performance liquid chromatography-mass spectrometry (HPLC-MS) and nuclear magnetic resonance (NMR) spectroscopy as DL-homocysteine thiolactone. Gene expression analyses using RT-PCR showed that strain D11 led to a significant down-regulation of QS regulatory genes (lasI, lasR, rhlI, and rhlR), as well as a virulence-related gene (lasB). From the chemical structure, the target compound (DL-homocysteine thiolactone) is an analog of the acyl-homoserine lactones (AHLs), and we presume that DL-homocysteine thiolactone outcompetes AHL in occupying the receptor and thereby inhibiting QS. Whole-genome sequence analysis of S. hominis D11 revealed the presence of predicted genes involved in the biosynthesis of homocysteine thiolactone. This study indicates that coral microbes are a resource bank for developing QS inhibitors and they will facilitate the discovery of new biotechnologically relevant compounds that could be used instead of traditional antibiotics.
Introduction
The rising problem of microbial resistance to current antibiotics and high spreading rate of resistant bacterial species has become a major public health concern. Multidrug-resistance is the biggest challenge facing the healthcare sector field (Adonizio et al., 2008). Biofilm formation is one of the mechanisms used by bacteria for developing such resistance (Vuotto et al., 2014; Arendrup and Patterson, 2017). Biofilms can act as protective membranes and are difficult to eliminate, leading to both therapy failure and disease recurrence. In recent years, it has become apparent that improved strategies and new antimicrobials are urgently needed to control infectious diseases.
Biofilm formation is controlled by cellular signals, widely known as quorum sensing (QS). Inhibition of QS is one of the many different strategies deployed to control biofilm-forming microbes without causing drug resistance (Singh et al., 2013, 2016). Some opportunistic pathogens, such as Serratia marcescens and Pseudomonas aeruginosa, control production of their virulence factors including biofilm formation by using QS systems. For example, more than 6% of the genes in the genome of P. aeruginosa are regulated by QS and are involved in the control of pathogenesis (Schuster et al., 2003; Wagner et al., 2003). Therefore, much work has focused on targeting microbial pathogenesis by inhibiting QS or biofilm formation. This paradigm is neither bactericidal (it does not kill bacteria) nor bacteriostatic (it does not inhibit bacterial growth). It appears to be a particularly attractive alternative to other methods because it does not impose a strong selective pressure, and thus bacterial resistance is less likely to develop (Sommer et al., 2013). For this reason, the identification of compounds that interfere with QS systems is of considerable interest in an effort to develop treatments against biofilm-associated pathogens (Christensen et al., 2007). For this reason, an approach known as QS inhibition has been developed when an efficient screening for anti-QS agents is required.
In recent years, several anti-QS compounds were reported from plants and microbes (Choo et al., 2006; Ni et al., 2009; Kalia and Purohit, 2011; Kalia, 2012). A lot of bacteria and metabolites isolated from terrestrial environments have shown anti-QS properties that can decrease the expression of virulence factors produced by some pathogens (Okuda, 2005; Adonizio et al., 2008; Tolmacheva et al., 2014). Numerous reports are emerging that provide evidence demonstrating anti-QS activity from various land sources including plants, animal extracts, fungi, and host-associated bacteria (Jiang and Li, 2013; Defoirdt, 2017; Singh et al., 2017).
Interestingly, the ocean contains a rich microbial biodiversity in which plenty of bioactive compounds are produced by various aquatic microbes, indicating that the marine environment can serve as an important resource in the search for novel anti-QS substances (Dobretsov et al., 2009; Teasdale et al., 2011; Yaniv et al., 2017). Taking coral as an example, it contains an enormous diversity of microorganisms, which render the coral microbiota ideally suited to the search for new ecological functions and bioactive metabolic compounds (Pham et al., 2016). In previous studies, the bacterial species Oceanobacillus profundus was isolated from the octocoral Antillogorgia elisabethae and was reported for its anti-QS activity by yielded compounds tyrosol and tyrosol acetate (Martínez-Matamoros et al., 2016). In addition, Marinobacter sp. and a Proteobacteria associated with corals have also been reported to inhibit the QS-dependent virulence factors in an environmental isolate of S. marcescens, which further augmented our interest in exploring coral-associated bacterial isolates (Kvennefors et al., 2012). The likelihood of finding novel bioactive compounds from coral ecosystems seems high since many such symbiotic microorganisms in this ecosystem have not been well-characterized. With this milieu, the coral ecosystem, a hitherto under-explored reserve for novel bacteria, was screened for anti-QS producers. These bacteria were then evaluated for their anti-biofilm activity, with the hope that biomolecules from such novel bacteria will be of a new and unique type.
It is worth noting that despite the abundance of active compounds from marine environments, to date the discovery and isolation of anti-QS compounds from these sources has been slow compared with the synthetic chemistry approach or terrestrial counterparts (Dobretsov et al., 2011; Yaniv et al., 2017). More importantly, a detailed identification of compounds has still not been performed (Bakkiyaraj et al., 2012, 2013). The present study stresses the importance of the coral-associated bacteria as a potential model for naturally occurring products with anti-QS properties. More specifically, given the limited knowledge available on the production of these cues by coral bacteria, the purpose of this study was to gain a clearer understanding of the ecological role of the anti-QS substances secreted by coral-symbiotic microbes.
In this study, we take the coral Pocillopora damicornis as the material to screen for QS-inhibiting bacteria, and one isolated bacterium was further explored for anti-QS potential. The active compounds from this bacteria were identified, expression of regulatory key genes was analyzed, and a possible mechanism of action was inferred.
Materials and Methods
Bacterial Strains and Culture Conditions, and Coral Samples
Chromobacterium violaceum ATCC®. 12472™ and Pseudomonas aeruginosa PAO1 were used in this study. Both strains were cultured in lysogeny broth (LB) medium containing 1% peptone, 0.5% yeast extract, and 0.5% NaCl, either in liquid form or solidified using 1.5% agar as necessary.
Coral (Pocillopora damicornis) samples were collected from Xishan Islands (located at 3°57.058′E, 36°8.532′S) in the South China Sea. The samples were collected from six sites (three from Heilong Island and three from Daming Island) at a depth of 5–6 m. The salinity was around 33.1‰ (33.1 per thousand) and the seawater temperature was 29.7°C. At each site, five coral samples were collected. Samples were washed with sterile seawater, homogenized by grinding and agitation, and serially diluted in sterile seawater. Next, 50 μl of dilutions from 10−4 to 10−7 were surface-plated on marine agar 2216 (Difco, USA) and incubated at 30°C for 3–5 days. A quantity of pre-test colonies, chosen on the basis of their different colonial morphology, were collected by sterile toothpick and incubated in the conditions described above.
Screening and Identifying Anti-QS Bacteria
A disc diffusion assay (Bauer et al., 1966) was performed with biosensor strain C. violaceum ATCC 12472 to detect anti-QS activity (Busetti et al., 2014). Briefly, 5 ml overnight reporter strain culture is poured into 45 ml LB media containing 0.75% agar until the temperature of the media is about 45°C. The mixture is then plated and allowed to solidify before sterile filter paper circles (5 mm diameter) are placed on the LB surface at regular intervals. The screened single colony isolates are cultured overnight in LB medium at 30°C in 1.5 ml Eppendorf tubes with constant shaking at 150 rpm. The cultured individual as the test strains (OD600 near 0.1) and bacterial suspension (3 μl) are pipetted onto the filter paper. 2,5-Dimethyl-4-hydroxy-3[2H]-furanone (CAS No. 3658-77-3, Sigma-Aldrich, USA) dissolved in dimethyl sulfoxide (DMSO, 1 μl), DMSO solvent, and LB broth are used as positive, negative and blank controls, respectively, in this plate-based bioassay. After incubation for 24 h at 30°C, inhibition of pigment production around the disc (a colorless ring) is checked. Positive anti-QS activity will be recorded as visible colorless haloes like furanone. The bacterial isolates showing promising positive anti-QS activities are selected for further study. To ensure reliability of the experiment, the anti-QS activities of the selected isolates are repeated three times independently.
Potential anti-QS strains were grown overnight in LB broth at 30°C, and then 200 μl from each culture was transferred into a clean 1.5 ml Eppendorf tube and centrifuged at 7,000 g for 1 min (Chang et al., 2017). The flow-through in the tube was discarded, 100 μl TE buffer was added, and the sample was mixed gently, and then boiled for 10 min. The resulting supernatant contained the crude DNA extract (OD260/OD230 was more than 1.7, and OD260/OD280 was between 1.8 and 2.0). The 16S rRNA gene, which is approximately 1500 bp, was amplified by PCR using the forward primer 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and the reverse primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Lane, 1991), and sequenced at BGI-Shenzhen (BGI China, Mainland). The sequences obtained were assembled, analyzed, and manually edited using the CAP3 software package. The resulting sequences were compared against those from the NCBI database (http://www.ncbi.nlm.nih.gov) using BLAST analysis and the RDP online service (https://rdp.cme.msu.edu).
Determination of Growth and Violacein Production
The effect of the potential anti-QS bacterial extract on the growth of C. violaceum ATCC 12472 was determined by the colony count on plate method (Choo et al., 2006). Cultures of C. violaceum ATCC 12472 were serially diluted and 100 μl aliquots were spread on LB plates. The plates were incubated at 30°C for 24 h, and bacterial counts were compared with the control. For quantification of violacein production, 1 ml of culture was centrifuged at 13,000 rpm for 10 min to precipitate insoluble violacein. The culture supernatant was discarded and 1 ml DMSO was added to the pellet. The solution was vortexed vigorously for 30 s to completely solubilize violacein and was then centrifuged at 13,000 rpm for 10 min to remove cells (Choo et al., 2006). Two-hundred microliters of the violacein-containing supernatants were added to 96-well flat-bottomed microplates, three wells per sample, and the absorbance was read with a spectrophotometer (Infinite® 200 PRO, Tecan, Austria) at a wavelength of 585 nm (Blosser and Gray, 2000).
Extracting the Anti-QS Active Components
The possible anti-QS strains were incubated for 48 h in LB broth at 30°C with shaking at 200 rpm. Samples were then centrifuged at 6,000 g, 4°C for 20 min to remove bacterial cells, and the resulting supernatants were collected and extracted using an equal volume of ethyl acetate, with vigorous shaking for 15–20 min. The extraction was repeated twice and the aqueous extract fractions were discarded. The organic extract fractions (obtained by ethyl acetate extraction) were combined and evaporated in a rotary evaporator at 45°C. The organic residues were dissolved in methanol (Nithya et al., 2010c) and concentrated using nitrogen flow. All resulting extracts were sterilized using 0.22 μm filters.
A second-round of testing for anti-QS activity was carried out using the sterilized extracts and following the above-mentioned methods (see “Screening and identifying anti-QS bacteria”). The putative extracts (from the anti-QS strains identified in the preliminary screen) were pipetted onto the filter paper, and the QS inhibition activity was calculated by measuring the diameter of colorless haloes relative to equivalent furanone. Finally, the positive extracts were stored at −20°C and used for biofilm inhibition experiments at a range of concentrations.
Influence of Anti-QS Extract on P. aeruginosa PAO1 Biomass and Cellular Growth
The effects of the extract from the anti-QS positive strain on the biomass of biofilms produced by P. aeruginosa PAO1 were determined using the crystal violet (CV) method (Huber et al., 2003; Choo et al., 2006). Briefly, freshly cultured P. aeruginosa PAO1 was added to 96-well polystyrene plates (100 μl per well) and incubated in LB medium (Hinsa, 2006). The bacterial extracts (for example D11 strain) were added at 1, 2.5, 5, and 10 μg/ml (w/v). The mixtures were incubated at 30°C for 48 h. Planktonic cells and spent medium were removed from each culture. The remaining adherent cells were gently rinsed twice using deionized water. One-hundred microliters of 1% (w/v) CV solution was added to each well for 30 min at room temperature. The excess dye was discarded, and the plates were washed gently but thoroughly using deionized water. The CV-stained cells were solubilized in DMSO and the absorbance at 600 nm was determined using a microplate reader (Infinite® 200 PRO, Tecan). P. aeruginosa PAO1 cultures incubated in the absence of extract and lose QSI ability extract (ultrasonic method to destroy the chemical structure of the extract) served as negative controls. Pure water was used as a blank control. Experiments were performed with 12 replicates (12 replicate wells in 96-well plates) for each treatment. When absorbance was determined, three readings were recorded for each well.
To determine the effects of the extract on the growth of P. aeruginosa PAO1, a growth curve assay was conducted. P. aeruginosa PAO1 was cultured in LB broth in the presence or absence of extract from strain D11 (10 μg/ml, w/v). Cultures were incubated at 30°C for 48 h. After 0, 3, 6, 9, 12, 18, 27, 36, and 48 h, the optical density at 600 nm was determined using a microplate spectrophotometer (Infinite® 200 PRO, Tecan). Bacterial abundance was measured using a flow cytometer (BD Biosciences, USA). Briefly, samples (1 ml each) were fixed with glutaraldehyde (0.5% final concentration), then stained with SYBR green I solution (Molecular Probes) (at a 1000-fold dilution of the stock solution) at room temperature in the dark for 15 min (Gasol and del Giorgio, 2000). Fluorescent 1-μm latex beads (105 beads per ml) were added to the samples as an internal standard. Bacterial number (cells/ml) were calculated by their signatures in a side-scatter-vs.-green-fluorescence plot, as described by Pinder et al. (1990) and Gasol and del Giorgio (2000).
Separation and Identification of Anti-QS Active Compounds
Extract compounds were separated by preparative high-performance liquid chromatography (HPLC) (Agilent 1200, USA). Samples were kept at 4°C until injection, and 100 μl extract sample was injected onto a reverse-phase C18 core-shell column (50 × 2.1 mm, Waters, CA, USA) via an auto-sampler (ThermoFisher Scientific, USA). The mobile phase was obtained using 83% methanol and 17% water at a flow rate of 0.5 ml/min at 30°C. Samples, separated every 30 s, were collected with the fraction collector. At the end of the separation process, every peak sample was taken and concentrated by nitrogen-flow method. Every purified sample was then re-tested to confirm the anti-QS activity using the procedures described above (see “Screening and identifying anti-QS bacteria”).
The collected anti-QS active peaks were further purified on HPLC (Waters Delta Prep 4000, USA) using a C18 column and a linear water/acetonitrile gradient containing 0.1% trifluoroacetic acid. The residue was dissolved in 1 ml acetonitrile/water (1:1, v/v) to determine the molecular weight by mass spectrometry (MS) on a LTQ XL Orbitrap using a static nanospray (Thermo-Fisher, CA, USA) in positive-/negative-ion mode. To determine the active molecular structure, nuclear magnetic resonance spectroscopy (NMR) was performed on the purified anti-QS active sample to get a heteronuclear single quantum coherence (HSQC) and heteronuclear multiple bond coherence (HMBC) spectrogram.
Inhibition of Biofilm
Pre-sterilized glass microscope slides were used to observe biofilms by confocal laser scanning microscopy (CLSM) as described in the previous study (Ortlepp et al., 2007; Tolker-Nielsen and Sternberg, 2014). Briefly, P. aeruginosa PAO1 was grown in LB medium overnight and diluted with fresh medium to an OD600 of about 0.02. Then, 2 ml dilutions were incubated under static conditions with or without anti-QS extract (10 μg/mL, w/v) in 12-well plates with a glass microscope slide in each well. After 12 and 36 h, the glass slides were gently lifted out and rinsed with deionized water to remove loosely attached cells. The biofilms on one side were stained with 5 μM SYTO9 dye (Sigma, USA) in the dark, and those on the other side were wiped off. After 15 min, the slides were washed, and observed by CLSM (Zeiss, Germany) with a × 60 objective lens to visualize the biofilms. The 488 nm excitation and 520 nm emission filter settings were used for detection of SYTO9. Quantification of biofilm parameters was processed with the COMSTAT software using the CLSM images (Heydorn et al., 2000). Of the available parameters, we selected the three factors of total biomass, average thickness, and roughness coefficient to evaluate the biofilms (Hentzer et al., 2001). 3D transmission-fluorescence photos of the P. aeruginosa PAO1 biofilms were produced using FV10-ASW2.0 Viewer (Olympus, Japan). The optical sections were 5 μm apart on the Z-axis and taken at 640 × 640 pixels with a 12-bit intensity resolution (Chang et al., 2017). Digital images were processed using Leica Confocal Software Lite (Leica Microsystems, Germany).
Effect of Anti-QS Extract on the Expression of QS Genes
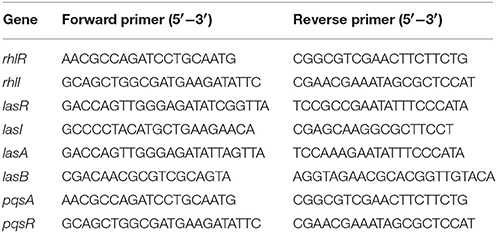
P. aeruginosa PAO1 was grown in 10 ml LB liquid medium to an OD600 of approximately 0.1. At this time point, treated groups had approximately 10 μl extract added (extract concentration was 10 μg/ml) to the P. aeruginosa PAO1 culture medium. The extract was dissolved in methanol and the final methanol concentration in the experimental system was 0.1% (v/v). Solvent control groups had 10 μl methanol only added. After 24–36 h, total RNA was extracted from control and treated groups using RNAiso Plus Reagent (Takara, China), and reverse-transcribed into cDNA with PrimeScript RT reagent kit (Takara) according to the manufacturer's protocol. Before performing the quantitative real-time PCR (qRT-PCR), RNA quality was determined (by measuring A260/A280 and A260/A230, and by gel electrophoresis). Eight reported functional genes coding for QS regulation activity were chosen for PCR analyses. Primers were designed using Primer Express 3.0 (Applied Biosystems) and are listed in Table 1. Thirty-two PCR cycles were run with denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and extension at 60°C for 45 s. The 16S rRNA gene was used as a control for standardization. A melt curve analysis was also done for the validation of specificity of the qRT-PCR. The relative transcription level of each gene was defined as the ratio of its transcript of biofilms grown in the indicated concentration of compounds over that in LB medium with methanol, using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Whole-Genome Sequencing of Strain D11
Genomic DNA of strain D11 was extracted using a GenEluteTMkit (Sigma-Aldrich, USA) and converted into a next-generation sequencing library using Next-era XT (Illumina, CA, USA) according to the manufacturer's instructions. Whole-genome sequencing was performed using the MiSeq at BGI Company (Shenzhen, China). SMRT Analysis 2.3.0 was used to filter low-quality reads and the sequences were assembled using Spades v2.5 (default setting) (Bankevich et al., 2012). The generated contigs were scaffolded and gap-closed using SSPACE and GAPFiller, respectively (Boetzer et al., 2011; Boetzer and Pirovano, 2012). Genome annotation was performed using Prokka and InterProScan5 (Jones et al., 2014; Seemann, 2014).
The software tRNAscan-SE v.1.23 and RNAmmer v.1.2 were used to identify presence of tRNA and rRNA, respectively (Lagesen et al., 2007). Gene prediction was performed by GeneMarkS with an integrated model that combined the GeneMarkS generated (native) and heuristic model parameters (Besemer et al., 2001). A whole-genome BLAST search (E-value less than 1 × 10−5), minimal alignment length percentage larger than 40%, was performed against the main databases, including KEGG (Kyoto Encyclopedia of genes and genomes), COG (Clusters of Orthologous Groups), and Swiss-Prot. The annotation predictions were manually evaluated and only genes predicted with consensus from two or more annotation pipelines were trusted in order to provide gene identification with high confidence.
Statistical Analysis
Differences in various data were determined using analysis of variance (ANOVA) at the P < 0.05 significance level. All analyses were performed using the SPSS software package 13.0 (NY, USA).
Results
Isolation and Identification of Anti-QS Coral Bacteria
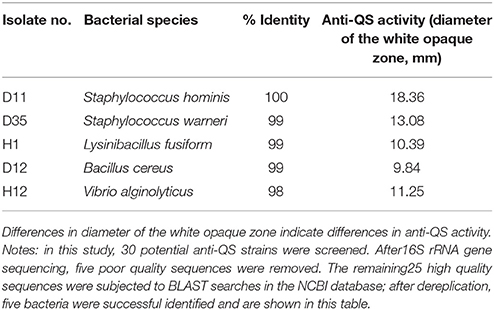
The possible anti-QS bacteria were screened using C. violaceum ATCC 12472 as an indicator strain since it produces the purple pigment violacein unless its QS system is interrupted. Using this technique, a lack of pigmentation from the indicator organism in the vicinity of the test organism indicates a potential anti-QS result (do Valle Gomes and Nitschke, 2012). A total of 200 culturable bacteria were isolated from the Pocillopora damicornis symbiotic environment and screened for anti-QS ability. About 15% (30 isolates) were positive in the screen for color reduction in C. violaceum ATCC 12472, with representative results shown in Figure 1. Some isolates showed promising anti-QS activity and a distinct white opaque zone of inhibition was observed in the biosensor plate containing reference strain C. violaceum ATCC 12472. The activity of positive isolates was recorded as either strong, medium or weak, based on the diameter of visible colorless haloes by the biosensor (Table 2). The isolate D11 caused the most significant reduction (the diameter of visible colorless haloes is 18.36 mm), in which the purple pigment of C. violaceum ATCC 12472 was completely eliminated (Figure 1). In comparison, the zone of inhibition was not detected with the negative control (DMSO solvent) or blank control (LB medium only).
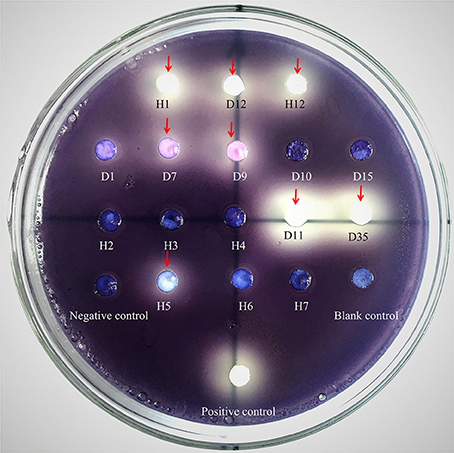
Figure 1. Screening of anti-QS strains on biosensor plates containing reference strain C. violaceum ATCC 12472 and filter paper for sample detection. Water, LB medium and furanone (diluted 10 times with DMSO) were used as blank, negative and positive controls, respectively. The absence of purple or formation of a pigment inhibition was considered to indicate a potential QS inhibitor. The red arrows refer to the positive anti-QS strains and the pigment inhibition can be observed on a clear background on the plate. Notes: the number indicated the test isolate strains. H samples come from Heilong Island, and D samples come from Daming Island.
The 16S rRNA gene sequences of these 30 positive isolates were aligned to the NCBI database using BLAST. Most of the representative isolates shared 99% sequence similarity with their respective reference strains. After filtering low-quality sequences and dereplication analyses, five representative strains were chosen from the candidates for anti-QS active substance studies. These five bacterial strains were Staphylococcus hominis, Lysinibacillus fusiform, Bacillus cereus, Staphylococcus warneri, and Vibrio alginolyticus. The 16S rRNA gene sequences for these five strains have been submitted to the GenBank database under the accession numbers MG761744–MG761748. Among the five strains, isolate D11 revealed a 100% sequence similarity to Staphylococcus hominis and has been tentatively named S. hominis D11 (GenBank accession number is MG761745). In the following experiment, we chose S. hominis D11, which has the most anti-QS activity, as the research object.
Effect of Anti-QS Extract on Growth and Violacein Production of C. violaceum ATCC 12472
The results of the colony count performed on LB plates at 24 h incubation showed no significant difference in the number of colony forming units (CFU) (Figure 2A). This indicates that the tested strains (D11, S. hominis; D35, L. fusiform; H1, B. cereus; D12, S. warneri; and H12, V. alginolyticus) have no effect on the growth of C. violaceum ATCC 12472. The five bacterial isolates showed a significant drop in violacein content, especially isolate D11 where violacein production was reduced by 92.3% (Figure 2B). Therefore, reduced production of violacein by bacterial culture was not due to the reduction of the “quorum,” but due to the interruption of the “sensing.”
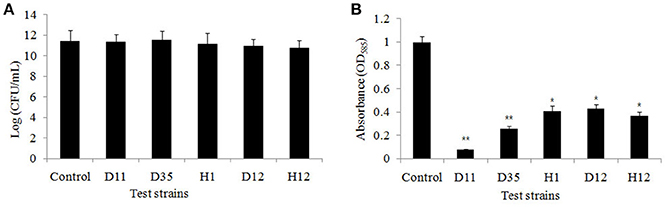
Figure 2. (A) Bacterial cell count of the flask incubation assay. The five test isolates were D11 (Staphylococcus hominis), D35 (Staphylococcus warneri), H1 (Lysinibacillus fusiform), D12 (Bacillus cereus), and H12 (Vibrio alginolyticus). C. violaceum ATCC 12472 was incubated for 16 h, and 100 μl of the bacteria, adjusted to OD600nm of 0.1 (approximately 1 × 108 CFU/ml), were spread on LB plates. The growth inhibition were compared with control. Data are presented as the logarithm of mean CFU ± SD. (B) Inhibition of violacein production by test strains. Violacein production was measured spectrophotometrically as described in the Materials and Methods. Data are presented as mean ± SD of absorbance at 585 nm. Asterisks indicate a statistically difference between experimental groups and control groups (*P < 0.05; **P < 0.01).
Extract From Strain D11 Inhibits Biofilm Formation
The anti-biofilm activity of the D11 extract was tested against the widely used biofilm-forming clinical isolate P. aeruginosa PAO1. Figure 3A presents quantitative analysis of P. aeruginosa PAO1 biofilm inhibition. Addition of S. hominis D11 extract (1, 2.5, 5, and 10 μg/ml) to P. aeruginosa PAO1 reduced biofilm formation by 18.2, 30.3, 46.7, and 62.1%, respectively, indicating that the inhibition occurred in a dose-dependent manner. The possibility of an inhibitory effect of the D11 extract on the growth of P. aeruginosa PAO1 was also analyzed. However, no significant effect on growth of P. aeruginosa PAO1 was observed in the presence of 10 μg/ml bacterial extracts (Figure 3B).
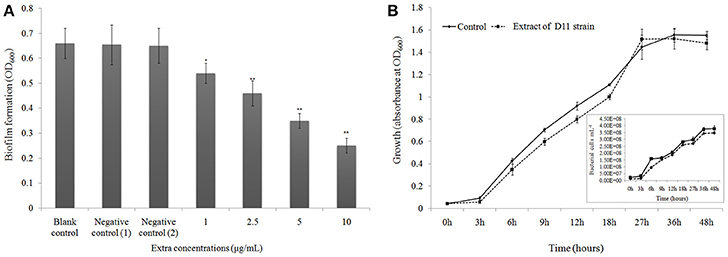
Figure 3. (A) Biofilm dispersal activity (crystal violet assay) of extract from isolate Staphylococcus hominis D11. Different concentrations of bacterial extract (1–10 μg/ml) were tested against the widely used biofilm-forming reference strain P. aeruginosa PAO1. Experiments with “extract + DMSO,” “lose QSI ability extract + DMSO,” “DMSO only,” and “no extract + no DMSO” were considered as test control, negative control (1), negative control (2), and blank control, respectively. Asterisks indicate a statistically significant difference (*P < 0.05; **P < 0.01) between experimental groups and control groups. Data are presented as mean ± SD (n = 3). (B) Effect of anti-QS compounds on growth of P. aeruginosa PAO1. Bacteria were grown in LB media with (dotted line) and without (solid line) D11 strain extract (10 μg/ml). The extract did not affect specific bacterial growth rate or bacterial abundance. Flow cytometry results (inset picture) show the count of bacterial cells. Data are presented as mean ± SD (n = 3).
Visualization of biofilms by microscopy analysis enabled precise evaluation of the biofilm 3D-structure. The topology of the biofilm developed by P. aeruginosa PAO1 and the effect of the D11 extract on it was analyzed by CLSM. A well-grown biofilm along with adhering bacterial cells was observed in control samples (normal biofilm developed by P. aeruginosa PAO1) at 12 and 36 h (Figures 4A,C), whereas dispersed bacterial cells were observed in treated samples (Figures 4B,D). Extremely thick biofilms (more cells and polysaccharides) were formed in the control relative to the experimental group. Also, the COMSTAT analysis clearly showed the disrupted surface topology and height distribution profile of the biofilm developed in the presence of the D11 extract compared to the control biofilm (taking36 h as the example) (Figure 5). In control groups, P. aeruginosa PAO1 developed a thick, dense biofilm, whereas on a surface coated with the D11 active crude extract, biofilm formation and bacterial adherence were prevented. Quantitative analysis showed that the D11 crude extract surface coating inhibited biofilm total biomass and average thickness by 43.9 and 58.7%, respectively (Figures 5A,B).
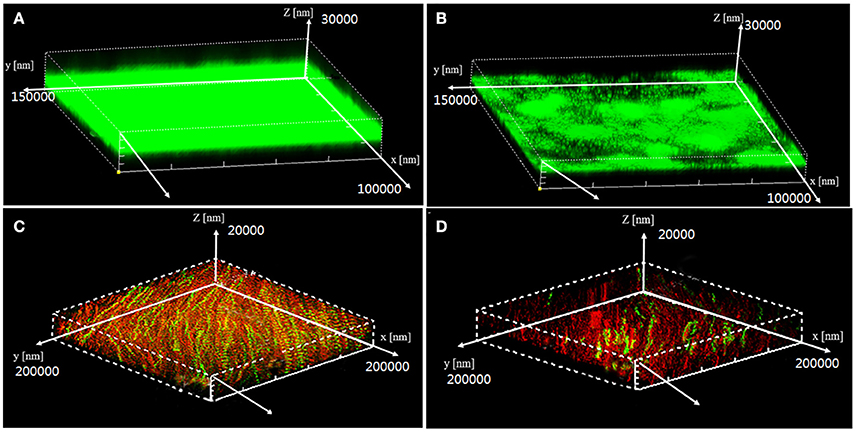
Figure 4. Confocal scanning laser microscopy (CLSM) z-stack 3-D images of P. aeruginosa PAO1 biofilm architecture in the presence (10 μg/ml)or absence (0 μg/ml) of D11 extract in media with 2% glucose. Data shown are early stage (12 h) biofilm structure of P. aeruginosa PAO1 in control group (A) and treatment group (B), and the post-stage (36 h) biofilm structure of P. aeruginosa PAO1 in control group (C) and experimental group (D). In these images, live bacterial cells produced green fluorescence, whereas dead cell sproduced red fluorescence.
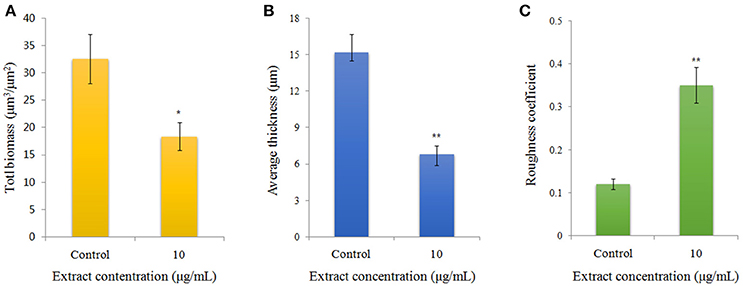
Figure 5. Quantification of biofilm formation of P. aeruginosa PAO1 (taking 36 h as example) using COMSTAT software, including (A) bio-volume, (B) average thickness, and (C) roughness coefficient. Error bars indicate SD (n = 3). Asterisks indicate a statistically significant difference (*P < 0.05; **P < 0.01) between experimental groups and control groups.
Identification of Anti-QS Compounds
Pre-HPLC analysis was applied to separate the crude extracts, with fractions collected every 30 s. The chromatogram from the liquid chromatography mass spectrometer (ThermoFisher Scientific™ TSQ Altis™, USA) showed that five main peaks exist (Figure 6A). The five fractions were collected and anti-QS activity was individually retested for each fraction using the biosensor plate containing C. violaceum ATCC 12472. Fraction peak 2 showed a maximum zone of QS inhibition; therefore, this fraction was selected for further characterization. Fraction peak 2 was subjected to HPLC and gas chromatography-mass spectrometry (GC-MS) analysis, and a main mass spectral peak, detected at m/z 118.03, was considered the corresponding experimental mass of the active fraction (Figure 6B). The detected mass spectra showed some resemblance to homocysteine thiolactone in the GC-MS library. The calculated (theoretical) or expected molecular mass of compound homocysteine thiolactone is 118. The molecular mass of the active fraction was further confirmed by NMR (C13 and H1) (Figures 6C,D).
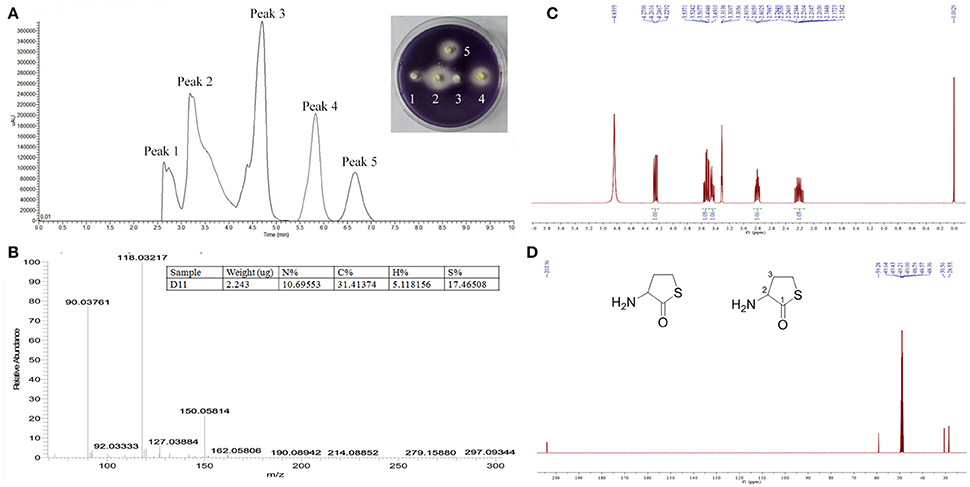
Figure 6. Analysis of active fraction showing anti-QS activity. (A) Pre-HPLC analysis of S. hominis D11 extract. The chromatogram shows the five main active peaks from S. hominis D11. The insert picture in (A) is the re-test of the anti-QS activity of the five peak compounds. (B) GC chromatograms and ESI-MS/MS of the active fraction (peak 2) of S. hominis D11 extract. Peaks are a function of intensity measured in milli-absorption units over time in minutes. (C) 1H-NMR spectrum of compound in Methanol-d4 at 400 MHz. (D) 13C-NMR spectrum of compound in Methanol-d4 at 100 MHz. 1H-NMR (Methanol-d4, 400 MHz) δ:4.25 (1H, dd, J = 12.9, 7.0 Hz, H-2), 3.53 (1H, td, J = 11.6, 5.2 Hz, H-4), 3.45 (1H, ddd, J = 11.6, 7.2, 1.1 Hz, H-4), 2.80 (1H, dddd, J = 12.2, 6.7, 5.2, 1.4 Hz, H-3), 2.21 (1H, m, H-3). 13C-NMR (Methanol-d4, 100 MHz) δ:204.0 (C-1), 59.3 (C-2), 30.6 (C-3), 28.6 (C-4). The insert picture in (D) is the structure of DL-homocysteine thiolactone (redrawn by ChemBioDraw Ultra 12.0).
In order to confirm the QS inhibitory activity produced by strain D11 can be attributed to homocysteine thiolactone, the commercial product (DL-homocysteine thiolactone, CAS No. 6038-19-3) was purchased from the Macklin Biochemical Co., Ltd (Shanghai, China). The anti-QS activity of this commercial product was tested according to the above-mentioned methods. The inhibitory activity of DL-homocysteine thiolactone against bacterial QS was determined using violacein production by C. violaceum ATCC 12472. From Figure 7A, a concentration-dependent inhibitory activity was observed, with the tested concentrations (0.0625, 0.125, 0.25, 0.5, and 1.0 μg/ml) of DL-homocysteine thiolactone showing a significant inhibition in violacein content (ranged from 62.5 to 98.1%). A varying degree of white opaque zone of inhibition was also observed in the biosensor plate containing reference strain C. violaceum ATCC 12472 (Figure 7B).
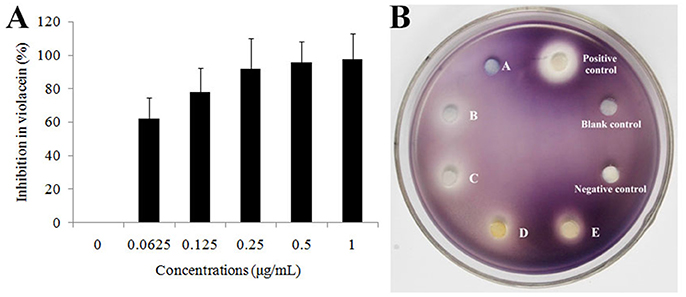
Figure 7. Effect of DL-homocysteine thiolactone on violacein production by C. violaceum ATCC 12472. (A) Percentage of inhibition of the violacein pigment after incubation with different concentrations of DL-homocysteine thiolactone. A dose-response effect in the production of violacein was observed in a series of concentrations (0.0625, 0.125, 0.25, 0.5, and 1.0 μg/mL) of DL-homocysteine thiolactone. Data show the mean (±SD) of three independent experiments. (B) Anti-QS activity of pure DL-homocysteine thiolactone at different concentrations. A, 0.0625 μg/ml; B, 0.125 μg/ml; C, 0.25 μg/ml; D, 0.5 μg/ml; and E, 1.0 μg/ml. LB medium, DMSO only and furanone (dissolved in DMSO, 1.0 μg/ml) were used as blank, negative and positive controls, respectively.
Expression Analysis by qRT-PCR
The transcriptional level of eight specific genes (lasI, lasR, lasA, lasB, rhlI, rhlR, pqsA, and pqsR) encoding putative biofilm-forming and QS factors was determined by RT-PCR in 24 h-old P. aeruginosa PAO1 cultures with extract and P. aeruginosa PAO1 cultures with methanol only as control. Approximately 2.5- to 5.1-fold down-regulation of the genes lasI, lasR, lasA, and lasB [responsible for acyl-homoserine lactone (AHL)-based biofilm formation] were observed in P. aeruginosa PAO1 cultured with the extract (P < 0.05 or P < 0.01) (Figure 8). Two virulence-related genes (rhlI and rhlR) also showed a significant decrease (72.3 and 88.5%, respectively) in expression level (P < 0.01). These results indicated that the general trend in expression for specific genes was similar between RT-PCR and biofilm state. As for the Pseudomonas quinolone signal (PQS) system in P. aeruginosa PAO1, there were no obvious differences between the experimental groups and the control groups (Figure 8).
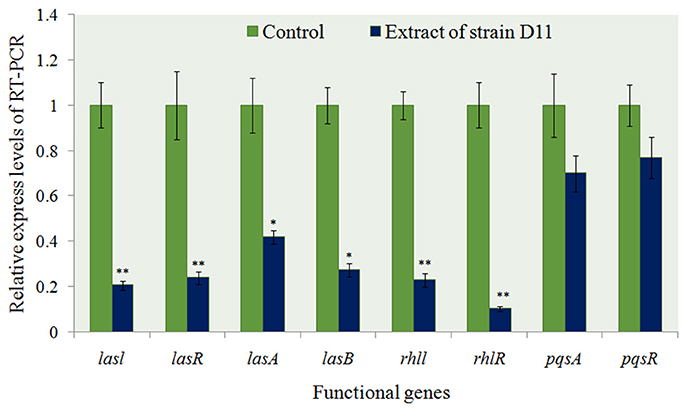
Figure 8. Expression profiling of some anti-QS regulatory genes from P. aeruginosa PAO1 with D11 extract measured by real-time PCR. The mRNA expression of these genes in the absence of extract served as a control. Results are based on three independent experiments and error bars represent means ± SD (n = 3). Asterisks indicate a statistically significant difference (*P < 0.05; **P < 0.01) between experimental groups and control groups.
Bio-Information From the Whole Genome of Strain D11
The whole genome of strain D11 comprised 5,392,014 nucleotides and the G+C content was 44.69%. It contains 71 contigs with an N50 contig length of 126,438 bp. The whole genome encodes 76 tRNA and 17 rRNA genes. The genome predicted a total of 4522 genes with 3738 protein-coding genes (Supplementary Table 1). Based on functional categories of COG (http://www.ncbi.nlm.nih.gov/COG/), a total of 593 genes were annotated to be participating in carbohydrate and amino acid metabolism, another 1018 genes were predicted to have general functions (Supplementary Figures 1, 2). In addition, 285 genes were predicted to encode signal transduction molecules.
In addition, we analyzed the candidate genes related to homocysteine thiolactone production. Homocysteine, an intermediate compound in the methionine metabolic cycle, is an amino acid that includes a thiol group. The homocysteine thiolactone forms adducts through irreversible reactions with epsilon-NH2 groups of lysine residues. We found several methionine-related genes (metI, metC, metF, metE, and mdh) (Supplementary Figure 3) located in the upstream position (contig 1), these genes showed relatively high sequence identity to another species of the same genus, Staphylococcus aureus (GenBank accession numbers SACOL0431-ACOL0427) (Schoenfelder et al., 2013). Our analysis predicts the presence of these genes identified in isolate D11 might be involved in methionine (or its intermediate product homocysteine thiolactone) biosynthesis. However, the corresponding mutants need to be constructed in the future in order to confirm this assumption.
Discussion
Among the marine environment, microorganisms and their metabolic products are a crucial source for the discovery of novel anti-QS compounds (Dong and Zhang, 2005; Choo et al., 2006; Dobretsov et al., 2006). Most of studies published on the production of QS inhibitors by marine bacteria have focused on bacteria that were collected from various niches, like surfaces, biofilms, and sediments (Teasdale et al., 2009, 2011). Indeed, many of the known anti-QS compounds have been discovered in sessile marine organisms such as sponges and microalgae that interact closely with bacteria (Stowe et al., 2011; Golberg et al., 2013). In the coral surface, Skindersoe et al. (2008) demonstrated that a large number of symbiotic microbes along the Great Barrier Reef corals possess anti-QS abilities. In addition, many studies were recently published indicating that QS inhibitors may be a frequently occurring feature in coral culturable bacteria such as Bacillus sp. and Vibrio sp. (Kanagasabhapathy et al., 2009; Thenmozhi et al., 2009; Nithya and Pandian, 2010a; Romero et al., 2012). These examples indicate that coral-derived bacteria may be potential sources of anti-QS compounds. In this work, approximately 15% of isolates from the hard coral species exhibited anti-QS activity. Among them, five strains (including S. hominis D11) were found to have significant anti-QS activity, supporting the hypothesis of Certner and Vollmer (2018), i.e., coral microbiota is a vast natural reservoir for developing new anti-QS substances.
Among the screened anti-QS bacteria, S. hominis D11 presented the most apparent opaque halo (growth of reporter strain with pigment inhibition) surrounding the isolate (Figure 1). The correlating results of violacein production (Figure 2B) further proved that the screened strains possess anti-QS activity against C. violaceum ATCC 12472. This inhibition activity seems similar to that of halogenated furanone, which inhibit QS function by interfering with luxS- or AI-2 system (Ren et al., 2002; Huang, 2009). Previously, some studies demonstrated that anti-QS activity and antimicrobial activity may co-occur (Busetti et al., 2014; Abudoleh and Mahasneh, 2017). In order to rule out the possibility that the inhibitory effect on the production of purple pigment was due to an antimicrobial effect, growth experiments with different test strains were performed, and no significant difference was observed among the experimental and control groups (Figure 2A). These results indicated that absence of violacein is mainly caused by QS disruption.
Accumulating data evidence that AHL-dependent QS is a key factor for formation of biofilms, indicating that anti-QS substances can inhibit biofilm development. Research by Adonizio et al. (2008) and Nithya et al. (2010b) suggest that P. aeruginosa PAO1 biofilm maturation can be inhibited by marine-derived bacterial species Callistemon viminalis and Bacillus pumilus S8-07, respectively. Teasdale et al. (2009) also found that the anti-QS properties exhibited by the marine bacterium Halobacillus salinus C42 were present in the solvent phase, in which the solvent was ethyl acetate. In this study, we found that addition of D11 organic extract resulted in significant reduction in the P. aeruginosa PAO1 biofilm (Figure 3A), indicating that active anti-QS extract may contain non-enzymatic compounds. In addition, inhibition of the AHL-dependent QS system by bacterial extracts was also observed. We speculated that the activity substance was associated with AHL analogs that acted as AHL-antagonists by competing with AHL for receptor binding and eventually inhibit biofilm formation of P. aeruginosa PAO1. In order to test the hypothesis and elucidate the possible mechanisms responsible for the inhibitory properties, related studies aimed at purifying and characterizing D11 extracts were carried out. After HPLC-MS-NMR analysis, we determined from the chemical structure that the activity substance was DL-homocysteine thiolactone (Figure 6D). Interestingly, this compound is very similar to homoserine lactone (HSL), produced from hydrolysis of AHLs, a common QS signal molecule (McInnis and Blackwell, 2011). This result supported our hypothesis along with further confirmation that AHL-based analogs have been extensively developed as QS modulators or anti-biofilm agents (Melvin et al., 2016). Interestingly, the violacein inhibition and anti-QS activities were confirmed by using the pure commercial DL-homocysteine thiolactone substance, which leads to the proposal that the QS inhibitory activity produced by strain D11 is homocysteine thiolactone. This is the first report of anti-biofilm activity of DL-homocysteine thiolactone on P. aeruginosa; further study is required to develop this substance as an anti-bacterial agent for treatment of the biofilm-forming pathogenic bacteria.
Quorum-sensing genes are key regulators of biofilm development, various extracellular virulence factors, luminescence and the antibiotic resistance of bacterial pathogens (Schuster and Greenberg, 2006; deKievit, 2009; Sharma et al., 2014). There are three well-characterized QS networks that have been identified in P. aeruginosa: las-, rhl-, and pqs-pathways. The three pathways utilize the corresponding AHLs: respectively, N-3-oxo-dodecanoyl homoserine-lactone (3OC12-HSL), N-butanoylhomoserine lactone (C4-HSL), and 2-heptyl-3-hydroxyl-4-quinolone (Pseudomonas quinolone signal, or PQS) (Zhang and Dong, 2004). In these systems, lasI and rhlI are involved in autoinducer synthesis, and lasR and rhlR code for transcriptional activators (Sharma et al., 2014). In our work, significantly reduced lasI, lasR, rhlI, and rhlR expression were observed (Figure 8), indicating that the D11 extract (DL-homocysteine thiolactone) has the ability to inhibit lasR and rhlR regulatory systems. It perhaps suggests that the mechanism for QS inhibition is via interaction with both las and rhl receptors. This result was also found by Vattem et al. (2007) who achieved the same result with extracts of Kigelia africana. In addition, unlike the chemically synthesized QS inhibitors, such as furanone, cyclopentanols and furanone derivatives (Givskov et al., 1996; Hentzer et al., 2002; Ishida et al., 2007; Geske et al., 2008; Kim et al., 2008), the natural compounds (for instance, DL-homocysteine thiolactone) have several advantages, including low toxicity and being environment-friendly. These features expand their potential utility in the biomedical field as natural QS inhibitors. In addition to anti-biofilm activity, strain D11 also inhibited the production of P. aeruginosa PAO1 virulence factors such as las genes (Figure 8). These results are similar to findings by Park et al. (2005), which showed that the Streptomyces strain M664 produces an AHL-degrading acylase enzyme that degrades AHL-regulated elastase and total and lasA proteases by 43–50%. Musthafa et al. (2011) also demonstrated that the marine-derived Bacillus sp. SS4 inhibited AHL-regulated production of P. aeruginosa virulence factors. In addition, our previous work found that the inhibition of elastase activity and siderophore production by Rhizobium sp. NAO1 occurs via interference with QS activity because these virulence factors are under the control of the las-coding gene systems (Chang et al., 2017).
For the whole-genome data of strain D11 (S. hominis), genome annotation on predicted genes was carried out by BLAST searches against anon-redundant protein sequence database and other databases available online, such as COG and KEGG. Based on the functional categories and gene annotation analysis (Supplementary Figure 2), 216 genes of strain D11 are involved in carbohydrate metabolism and 472 genes participate in nitrogen utilization and energy conversion, which allows this microorganism to adapt to coral-bacteria symbiosis. After gene annotation analysis, 377 genes were related to amino acids processing. Potentially, these genes are a key feature of strain D11 that enable it to biosynthesize all kinds of amino acids, including an intermediate compound (homocysteine) in methionine metabolism. For methionine or related by-products (such as homocysteine thiolactone), several metI/E/F-encoding genes were predicted to be located at contig1 (Supplementary Figure 3). These genes showed relatively high identity to another species of the same genus, Staphylococcus aureus (Grundy and Henkin, 1998). Our results further support the previous viewpoint, i.e., many microorganisms are able to synthesize methionine de novo and staphylococci employ the trans-sulfuration pathway to generate methionine (Rodionov et al., 2004). In this work, the whole-genome sequence of strain D11 provides deeper understanding of the molecular mechanism of the anti-QS ability of strain D11, and also may facilitate insights into the active product biosynthesis process.
Conclusions
In this work, we uncovered the anti-QS activity of a marine bacterial species isolated from the coral Pocillopora damicornis. The extract of strain D11 (S. hominis) was antagonistic to P. aeruginosa PAO1 QS and affected QS-regulated functional genes, including those involved in biofilm formation and virulence production. It is possible that the analog molecule DL-homocysteine thiolactone produced by strain D11 (S. hominis) competed with the auto-inducers produced by P. aeruginosa PAO1. Interestingly, DL-homocysteine thiolactone did not affect the growth of P. aeruginosa PAO1. These characteristics may accelerate development of QS inhibitors with broad-spectrum activity, and facilitate the discovery of novel drugs with greater efficacy to deal with bacterial infections in the current post-antibiotic era.
Author Contributions
Z-PM and JZ performed the experiments and drafting of the manuscript. YS and Z-HC acquired and analyzed data. Z-JL prepared figures and tables. YW and G-HL completed critical revision.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by NSFC (41741015), as well as the S&T Projects of Shenzhen Science and Technology Innovation Committee (JCYJ20170412171959157, JCYJ20150529164918736, JCYJ20150831192329178, and JCYJ20170412171947159).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00144/full#supplementary-material
References
Abudoleh, S. M., and Mahasneh, A. M. (2017). Anti-quorum sensing activity of substances isolated from wild berry associated bacteria. Avicenna J. Med. Biotechnol. 9, 23–30.
Adonizio, A., Kong, K. F., and Mathee, K. (2008). Inhibition of quorum sensing controlled virulence factor production in Pseudomonas aeruginosa by south Florida plant extracts. Antimicrob. Agents Chemother. 52, 198–203. doi: 10.1128/AAC.00612-07
Arendrup, M. C., and Patterson, T. F. (2017). Multidrug-resistant candida: epidemiology, molecular mechanisms, and treatment. J. Infect. Dis. 216(Suppl. 3), S445–S451. doi: 10.1093/infdis/jix131
Bakkiyaraj, D., Sivasankar, C., and Pandian, S. K. (2012). Inhibition of quorum sensing regulated biofilm formation in Serratia marcescens causing nosocomial infections. Bioorg. Med. Chem. Lett. 22, 3089–3094. doi: 10.1016/j.bmcl.2012.03.063
Bakkiyaraj, D., Sivasankar, C., and Pandian, S. K. (2013). Anti-pathogenic potential of coral associated bacteria isolated from Gulf of Mannar against Pseudomonas aeruginosa. Indian J. Microbiol. 53, 111–113. doi: 10.1007/s12088-012-0342-3
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bauer, A. W., Kirby, W. M., Sherris, J. C., and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. doi: 10.1093/ajcp/45.4ts.493
Besemer, J., Lomsadze, A., and Borodovsky, M. (2001). GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29, 2607–2618. doi: 10.1093/nar/29.12.2607
Blosser, R. S., and Gray, K. M. (2000). Extraction of violacein from Chromobacterium violaceum provides a new quantitative bioassay for N-acyl homoserine lactone autoinducers. J. Microbiol. Methods 40, 47–55. doi: 10.1016/S0167-7012(99)00136-0
Boetzer, M., Henkel, C. V., Jansen, H. J., Butler, D., and Pirovano, W. (2011). Scaffolding preassembled contigs using SSPACE. Bioinformatics 27, 578–579. doi: 10.1093/bioinformatics/btq683
Boetzer, M., and Pirovano, W. (2012). Toward almost closed genomes with GapFiller. Gen. Biol. 13:R56. doi: 10.1186/gb-2012-13-6-r56
Busetti, A., Shaw, G., Megaw, J., Gorman, S. P., Maggs, C. A., and Gilmore, B. F. (2014). Marine-derived quorum-sensing inhibitory activities enhance the antibacterial efficacy of tobramycin against Pseudomonas aeruginosa. Mar. Drugs. 13, 1–28. doi: 10.3390/md13010001
Certner, R. H., and Vollmer, S. V. (2018). Inhibiting bacterial quorum sensing arrests coral disease development and disease-associated microbes. Environ. Microbiol. 20, 645–657. doi: 10.1111/1462-2920.13991
Chang, H., Zhou, J., Zhu, X. S., Yu, S. C., Chen, L., Jin, H., et al. (2017). Strain identification and quorum sensing inhibition characterization of marine-derived Rhizobium sp. NAO1. R. Soc. Open. Sci. 4:170025. doi: 10.1098/rsos.170025
Choo, J. H., Rukayadi, Y., and Hwang, J. K. (2006). Inhibition of bacterial quorum sensing by vanilla extract. Lett. Appl. Microbiol. 42, 637–641. doi: 10.1111/j.1472-765X.2006.01928.x
Christensen, L. D., Moser, C., Jensen, P. Ø., Rasmussen, T. B., Christophersen, L., Kjelleberg, S., et al. (2007). Impact of Pseudomonas aeruginosa quorum sensing on biofilm persistence in an in vivo intraperitoneal foreign-body infection model. Microbiology 153, 2312–2320. doi: 10.1099/mic.0.2007/006122-0
Defoirdt, T. (2017). Quorum-sensing systems as targets for anti-virulence therapy. Trends. Microbiol. 26, 313–328. doi: 10.1016/j.tim.2017.10.005
deKievit, T. R. (2009). Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 11, 279–288. doi: 10.1111/j.1462-2920.2008.01792.x
Dobretsov, S., Dahms, H. U., and Qian, P. Y. (2006). Inhibition of biofouling by marine microorganisms and their metabolites. Biofouling 22, 43–54. doi: 10.1080/08927010500504784
Dobretsov, S., Teplitski, M., Bayer, M., Gunasekera, S., Proksch, P., and Paul, V. J. (2011). Inhibition of marine biofouling by bacterial quorum sensing inhibitors. Biofouling 27, 893–905. doi: 10.1080/08927014.2011.609616
Dobretsov, S., Teplitski, M., and Paul, V. (2009). Mini-review: quorum sensing in the marine environment and its relationship to biofouling. Biofouling 25, 413–427. doi: 10.1080/08927010902853516
Dong, Y. H., and Zhang, L. H. (2005). Quorum sensing and quorum-quenching enzymes. J. Microbiol. 43, 101–109.
do Valle Gomes, M. Z., and Nitschke, M. (2012). Evaluation of rhamnolipid and surfactin to reduce the adhesion and remove biofilms of individual and mixed cultures of food pathogenic bacteria. Food Control 25, 441–447. doi: 10.1016/j.foodcont.2011.11.025
Gasol, J., and del Giorgio, P. (2000). Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64, 197–224. doi: 10.3989/scimar.2000.64n2197
Geske, G. D., O'Neill, J. C., Miller, D. M., Wezeman, R. J., Mattmann, M. E., Lin, Q., et al. (2008). Comparative analyses of N-acylated homoserine lactones reveal unique structural features that dictate their ability to activate or inhibit quorum sensing. Chembiochem 9, 389–400. doi: 10.1002/cbic.200700551
Givskov, M., de Nys, R., Manefield, M., Gram, L., Maximilien, R., Eberl, L., et al. (1996). Eukaryotic interference with homoserine lactone-mediated prokaryotic signaling. J. Bacteriol. 178, 6618–6622. doi: 10.1128/jb.178.22.6618-6622.1996
Golberg, K., Pavlov, V., Marks, R. S., and Kushmaro, A. (2013). Coral-associated bacteria, quorum sensing disrupters, and the regulation of biofouling. Biofouling 29, 669–682. doi: 10.1080/08927014.2013.796939
Grundy, F. J., and Henkin, T. M. (1998). The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30, 737–749. doi: 10.1046/j.1365-2958.1998.01105.x
Hentzer, M., Riedel, K., Rasmussen, T. B., Heydorn, A., Andersen, J. B., Parsek, M. R., et al. (2002). Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology 148, 87–102. doi: 10.1099/00221287-148-1-87
Hentzer, M., Teitzel, G. M., Balzer, G. J., Heydorn, A., Molin, S., Givskov, M., et al. (2001). Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183, 5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001
Heydorn, A., Nielsen, A. T., Hentzer, M., Sternberg, C., Givskov, M., Ersbøll, B. K., et al. (2000). Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146, 2395–2407. doi: 10.1099/00221287-146-10-2395
Hinsa, S. M. (2006). Biofilm formation by Pseudomonas fluorescens WCS365: a role for LapD. Microbiology 52, 1375–1383. doi: 10.1099/mic.0.28696-0
Huang, Z., Meric, G., Liu, Z., Ma, R., Tang, Z., and Lejeune, P. (2009) luxS-based quorum-sensing signaling affects biofilm formation in Streptococcus mutans. J. Mol. Microbiol. Biotechnol. 17, 12–19. doi: 10.1159/000159193
Huber, B., Eberl, L., Feucht, W., and Polster, J. (2003). Influence of polyphenols on bacterial biofilm formation and quorum-sensing. Z. Naturforsch. C 58, 879–884. doi: 10.1515/znc-2003-11-1224
Ishida, T., Ikeda, T., Takiguchi, N., Kuroda, A., Ohtake, H., and Kato, J. (2007). Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl. Environ. Microbiol. 73, 3183–3188. doi: 10.1128/AEM.02233-06
Jiang, T., and Li, M. (2013). Quorum sensing inhibitors: a patent review. Expert. Opin. Ther. Pat. 23, 867–894. doi: 10.1517/13543776.2013.779674
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kalia, V. C. (2012). Quorum sensing inhibitors: an overview. Biotechnol. Adv. 31, 224–245. doi: 10.1016/j.biotechadv.2012.10.004
Kalia, V. C., and Purohit, H. J. (2011). Quenching the quorum sensing system: potential antibacterial drug targets. Crit. Rev. Microbiol. 37, 121–140. doi: 10.3109/1040841X.2010.532479
Kanagasabhapathy, M., Yamazaki, G., Ishida, A., Sasaki, H., and Nagata, S. (2009). Presence of quorum-sensing inhibitor-like compounds from bacteria isolated from the brown alga Colpomenia sinuosa. Lett. Appl. Microbiol. 49, 573–579. doi: 10.1111/j.1472-765X.2009.02712.x
Kim, C., Kim, J., Park, H. Y., Park, H. J., Lee, J. H., Kim, C. K., et al. (2008). Furanone derivatives as quorum-sensing antagonists of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 80, 37–47. doi: 10.1007/s00253-008-1474-6
Kvennefors, E. C., Sampayo, E., Kerr, C., Vieira, G., Roff, G., and Barnes, A. C. (2012). Regulation of bacterial communities through antimicrobial activity by the coral holobiont. Microb. Ecol. 63, 605–618. doi: 10.1007/s00248-011-9946-0
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt, W. Goodfellow (New York, NY: John Wiley & Sons, Inc.), 115–118.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Martínez-Matamoros, D., Fonseca, M. L., Duque, C., Ramos, F. A., and Castellanos, L. (2016). Screening of marine bacterial strains as source of quorum sensing inhibitors (QSI): first chemical study of Oceanobacillus profundus (RKHC-62B). Vitae 23, 30–47. doi: 10.17533/udea.vitae.v23n1a04
McInnis, C. E., and Blackwell, H. E. (2011). Thiolactone modulators of quorum sensing revealed through library design and screening. Bioorg. Med. Chem. 19, 4820–4828. doi: 10.1016/j.bmc.2011.06.071
Melvin, J. A., Montelaro, R. C., and Bomberger, J. M. (2016). Clinical potential of engineered cationic antimicrobial peptides against drug resistant biofilms. Expert. Rev. Anti. Infect. Ther. 14, 989–991. doi: 10.1080/14787210.2016.1236687
Musthafa, K. S., Saroja, V., Pandian, S. K., and Ravi, A. V. (2011). Antipathogenic potential of marine Bacillus sp. SS4 on N-acyl-homoserine-lactone-mediated virulence factors production in Pseudomonas aeruginosa (PAO1). J. Biosci. 36, 55–67. doi: 10.1007/s12038-011-9011-7
Ni, N., Li, M., Wang, J., and Wang, B. (2009). Inhibitors and antagonists of bacterial quorum sensing. Med. Res. Rev. 29, 65–124. doi: 10.1002/med.20145
Nithya, C., Aravindraja, C., and Pandian, S. K. (2010b). Bacillus pumilus of Palk Bay origin inhibits quorum-sensing-mediated virulence factors in Gram-negative bacteria. Res. Microbiol. 161, 293–304. doi: 10.1016/j.resmic.2010.03.002
Nithya, C., Begum, M. F., and Pandian, S. K. (2010c). Marine bacterial isolates inhibit biofilm formation and disrupt mature biofilms of Pseudomonas aeruginosa PAO1. Appl. Microbiol. Biotechnol. 88, 341–358. doi: 10.1007/s00253-010-2777-y
Nithya, C., and Pandian, S. K. (2010a). The in vitro antibiofilm activity of selected marine bacterial culture supernatants against Vibrio spp. Arch. Microbiol. 192, 843–854. doi: 10.1007/s00203-010-0612-6
Okuda, T. (2005). Systematics and health effects of chemically distinct tannins in medicinal plants. Phytochem Rev. 66, 2012–2031. doi: 10.1016/j.phytochem.2005.04.023
Ortlepp, S., Sjögren, M., Dahlström, M., Weber, H., Ebel, R., Edrada, R., et al. (2007). Antifouling activity of bromotyrosine-derived sponge metabolites and synthetic analogues. Mar. Biotechnol. 9, 776–785. doi: 10.1007/s10126-007-9029-x
Park, S. Y., Kang, H. O., Jang, H. S., Lee, J. K., Koo, B. T., and Yum, D. Y. (2005). Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 71, 2632–2641. doi: 10.1128/AEM.71.5.2632-2641.2005
Pham, T. M., Wiese, J., Wenzel-Storjohann, A., and Imhoff, J. F. (2016). Diversity and antimicrobial potential of bacterial isolates associated with the soft coral Alcyoniumdigitatum from the Baltic Sea. Antonie Van Leeuwenhoek. 109, 105–119. doi: 10.1007/s10482-015-0613-1
Pinder, A. C., Purdy, P. W., Poulter, S. A., and Clark, D. C. (1990). Validation of flow cytometry for rapid enumeration of bacterial concentrations in pure cultures. J. Appl. Bacteriol. 69, 92–100. doi: 10.1111/j.1365-2672.1990.tb02916.x
Ren, D., Sims, J., and Wood, T. (2002). Inhibition of biofilm formation and swarming of Bacillus subtilis by (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2 (5H)-furanone. Lett. Appl. Microbiol. 34, 293–299. doi: 10.1046/j.1472-765x.2002.01087.x
Rodionov, D. A., Vitreschak, A. G., Mironov, A. A., and Gelfand, M. S. (2004). Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 32, 3340–3353. doi: 10.1093/nar/gkh659
Romero, M., Martin-Cuadrado, A. B., and Otero, A. (2012). Determination of whether quorum quenching is a common activity in marine bacteria by analysis of cultivable bacteria and metagenomic sequences. Appl. Environ. Microbiol. 78, 6345–6348. doi: 10.1128/AEM.01266-12
Schoenfelder, S. M., Marincola, G., Geiger, T., Goerke, C., Wolz, C., and Ziebuhr, W. (2013). Methionine biosynthesis in Staphylococcus aureus is tightly controlled by a hierarchical network involving an initiator tRNA-specific T-box riboswitch. PLoS Pathog. 9:e1003606. doi: 10.1371/journal.ppat.1003606
Schuster, M., and Greenberg, E. P. (2006). A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296, 73–81. doi: 10.1016/j.ijmm.2006.01.036
Schuster, M., Lohstroh, C. P., Ogi, T., and Greenberg, E. P. (2003). Identification, timing and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185, 2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sharma, I. M., Petchiappan, A., and Chatterji, D. (2014). Quorum sensing and biofilm formation in mycobacteria: role of c-di-GMP and methods to study this second messenger. IUBMB Life 66, 823–834. doi: 10.1002/iub.1339
Singh, V. K., Kavita, K., Prabhakaran, R., and Jha, B. (2013). Cis-9-octadecenoic acid from the rhizospheric bacterium Stenotrophomonas maltophilia BJ01 shows quorum quenching and anti-biofilm activities. Biofouling 29, 855–867. doi: 10.1080/08927014.2013.807914
Singh, V. K., Mishra, A., and Jha, B. (2016). “Marine bacterial extracellular polymeric substances: characteristics and applications,” in Marine Glycobiology: Principles and Applications, ed S. Kim (Boca Raton, FL: Taylor and Francis Group; CRC Press), 369–377.
Singh, V. K., Mishra, A., and Jha, B. (2017). Anti-quorum sensing and anti-biofilm activity of Delftia tsuruhatensis extract by attenuating the quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 7:337. doi: 10.3389/fcimb.2017.00337
Skindersoe, M. E., Ettinger-Epstein, P., Rasmussen, T. B., Bjarnsholt, T., de Nys, R., and Givskov, M. (2008). Quorum sensing antagonism from marine organisms. Mar. Biotechnol. 10, 56–63. doi: 10.1007/s10126-007-9036-y
Sommer, R., Joachim, I., Wagner, S., and Titz, A. (2013). New approaches to control infections: anti-biofilm strategies against gram-negative bacteria. Chimia 67, 286–290. doi: 10.2533/chimia.2013.286
Stowe, S. D., Richards, J. J., Tucker, A. T., Thompson, R., Melander, C., and Cavanagh, J. (2011). Anti-biofilm compounds derived from marine sponges. Mar. Drugs 9, 2010–2035. doi: 10.3390/md9102010
Teasdale, M. E., Donovan, K. A., Forschner-Dancause, S. R., and Rowley, D. C. (2011). Gram-positive marine bacteria as a potential resource for the discovery of quorum sensing inhibitors. Mar. Biotechnol. 13, 722–732. doi: 10.1007/s10126-010-9334-7
Teasdale, M. E., Liu, J., Wallace, J., Akhlaghi, F., and Rowley, D. C. (2009). Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in Gram-negative bacteria. Appl. Environ. Microbiol. 75, 567–572. doi: 10.1128/AEM.00632-08
Thenmozhi, R., Nithyanand, P., Rathna, J., and Karutha, P. S. (2009). Antibiofilm activity of coral-associated bacteria against different clinical M serotypes of Streptococcus pyogenes. FEMS Immunol. Med. Microbiol. 573, 284–294. doi: 10.1111/j.1574-695X.2009.00613.x
Tolker-Nielsen, T., and Sternberg, C. (2014). Methods for studying biofilm formation: flow cells and confocal laser scanning microscopy. Methods Mol. Biol. 1149, 615–629. doi: 10.1007/978-1-4939-0473-047
Tolmacheva, A. A., Rogozhin, E. A., and Deryabin, D. G. (2014). Antibacterial and quorum sensing regulatory activities of some traditional Eastern-European medicinal plants. Acta Pharm. 64, 173–186. doi: 10.2478/acph-2014-0019
Vattem, D. A., Mihalik, K., Crixell, S. H., and McLean, R. J. (2007). Dietary phytochemicals as quorum sensing inhibitors. Fitoterapia 78, 302–310. doi: 10.1016/j.fitote.2007.03.009
Vuotto, C., Longo, F., Balice, M. P., Donelli, G., and Varaldo, P. E. (2014). Antibiotic resistance related to biofilm formation in Klebsiella pneumoniae. Pathogens 3, 743–758. doi: 10.3390/pathogens3030743
Wagner, V. E., Bushnell, D., Passador, L., Brooks, A. I., and Iglewski, B. H. (2003). Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185, 2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003
Yaniv, K., Golberg, K., Kramarsky-Winter, E., Marks, R., Pushkarev, A., Béjà, O., et al. (2017). Functional marine metagenomic screening for anti-quorum sensing and anti-biofilm activity. Biofouling 33, 1–13. doi: 10.1080/08927014.2016.1253684
Keywords: anti-quorum sensing, coral microbes, S. hominis, HPLC-MS-NMR, marine drug
Citation: Ma Z-P, Song Y, Cai Z-H, Lin Z-J, Lin G-H, Wang Y and Zhou J (2018) Anti-quorum Sensing Activities of Selected Coral Symbiotic Bacterial Extracts From the South China Sea. Front. Cell. Infect. Microbiol. 8:144. doi: 10.3389/fcimb.2018.00144
Received: 29 January 2018; Accepted: 20 April 2018;
Published: 08 May 2018.
Edited by:
Maria Tomas, Complexo Hospitalario Universitario A Coruña, SpainReviewed by:
Naybi Muñoz, Colegio de Postgraduados (COLPOS), MexicoJ. Christopher Fenno, University of Michigan, United States
Copyright © 2018 Ma, Song, Cai, Lin, Lin, Wang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Wang, d2FuZ3lhbm1hcnlAc3pwdC5lZHUuY24=
Jin Zhou, emhvdS5qaW5Ac3oudHNpbmdodWEuZWR1LmNu
 Zhi-Ping Ma
Zhi-Ping Ma Yu Song2
Yu Song2 Jin Zhou
Jin Zhou