- Department of Pathology, Michigan Medicine, University of Michigan, Ann Arbor, MI, United States
Klebsiella pneumoniae is a Gram-negative pathogen that has a large accessory genome of plasmids and chromosomal gene loci. This accessory genome divides K. pneumoniae strains into opportunistic, hypervirulent, and multidrug-resistant groups and separates K. pneumoniae from two closely related species, Klebsiella variicola and Klebsiella quasipneumoniae. Some strains of K. pneumoniae act as opportunistic pathogens, infecting critically ill and immunocompromised patients. These K. pneumoniae are a common cause of health-care associated infections including pneumonia, urinary tract infections (UTIs), and bloodstream infections. K. variicola and K. quasipneumoniae are often clinically indistinguishable from opportunistic K. pneumoniae. Other strains of K. pneumoniae are hypervirulent, infecting healthy people in community settings and causing severe infections including pyogenic liver abscess, endophthalmitis, and meningitis. A third group of K. pneumoniae encode carbapenemases, making them highly antibiotic-resistant. These strains act as opportunists but are exceedingly difficult to treat. All of these groups of K. pneumoniae and related species can colonize the gastrointestinal tract, and the accessory genome may determine if a colonizing strain remains asymptomatic or progresses to cause disease. This review will explore the associations between colonization and infection with opportunistic, antibiotic-resistant, and hypervirulent K. pneumoniae strains and the role of the accessory genome in distinguishing these groups and related species. As K. pneumoniae infections become progressively more difficult to treat in the face of antibiotic resistance and hypervirulent strains, an increased understanding of the epidemiology and pathogenesis of these bacteria is vital.
Introduction
Klebsiella pneumoniae was first described by Carl Friedlander in 1882 as a bacterium isolated from the lungs of patients who had died from pneumonia (Friedlaender, 1882). Klebsiella species are found ubiquitously in nature, including in plants, animals, and humans. They are the causative agent of several types of infections in humans, including respiratory tract infections, urinary tract infections (UTIs), and bloodstream infections (Podschun and Ullmann, 1998). Classically, these infections occur in hospitalized or otherwise immunocompromised patients and are routinely treated with β-lactams and other antibiotics effective against Enterobacteriaceae (Mandell, 2005). However, antibiotic-resistant K. pneumoniae and hypervirulent K. pneumoniae strains have emerged separately across the world. Furthermore, recent advances in molecular capabilities have shown that a portion of clinical isolates identified as K. pneumoniae are in fact other Klebsiella species (Brisse and Verhoef, 2001; Brisse et al., 2004; Maatallah et al., 2014; Berry et al., 2015; Long et al., 2017a). This review will focus on the epidemiology of endemic opportunistic, epidemic antibiotic-resistant, and emerging hypervirulent strains of K. pneumoniae, and the role of the accessory genome in each pathotype. Understanding how these emerging strains and species are similar and how they differ from one another, as well as the genetic factors contributing to their epidemiology, is necessary to successfully combat these infections.
Klebsiella pneumoniae: An Opportunistic, Hospital-Acquired Infection
Klebsiella pneumoniae is a Gram-negative pathogenic bacterium. On agar media, it has a mucoid phenotype that is conferred by the polysaccharide capsule attached to the bacterial outer membrane, and ferments lactose. K. pneumoniae is part of the Enterobacteriaceae family, which is comprised of other familiar pathogens such as Escherichia coli, Yersinia species, Salmonella species, and Shigella species. K. pneumoniae, a leading cause of hospital-acquired infections (HAIs) in the United States (Magill et al., 2014), has classically been considered an opportunistic pathogen, since it typically causes infections in hospitalized or otherwise immunocompromised individuals (Podschun and Ullmann, 1998). As the virulence of these bacteria and the demographic features of the patients they infect begin to shift, understanding how K. pneumoniae is transmitted and the factors responsible for pathogenicity is important in treating infected patients.
Klebsiella species have been identified as the third leading cause of HAIs in the United States (9.9%) behind Clostridium difficile and Staphylococcus aureus (Magill et al., 2014). K. pneumoniae causes serious infections including pneumonia, UTIs, and bloodstream infections (Magill et al., 2014). In fact, Klebsiella species have been identified as the number three cause of HA pneumonia in the United States, defined as a pneumonia occurring ≥48 h after hospital admission (Magill et al., 2014). Klebsiella species are also a leading cause of ventilator-associated pneumonia (VAP) among patients in intensive care units (ICUs) (Kalanuria et al., 2014; Selina et al., 2014), and VAP is responsible for 83% of hospital-acquired (HA) pneumonias (Richards et al., 2000). Mortality rates in K. pneumoniae pneumonia have been reported as high as 50% (Podschun and Ullmann, 1998).
K. pneumoniae are the second leading cause of bloodstream infections (BSI) caused by Gram-negative bacteria, behind only E. coli (Podschun and Ullmann, 1998; Magill et al., 2014). Cancer is the primary underlying disease associated with hospital-acquired BSI, while liver disease and diabetes mellitus had the highest association among community-acquired (CA) K. pneumoniae BSI (Kang et al., 2006). BSI can be a primary infection with no identifiable source. However, BSI is often a secondary infection that results from dissemination into the bloodstream from a known source. Common sources of secondary BSI include the urinary tract, the gastrointestinal tract, intravenous or urinary catheters, and respiratory sites (Montgomerie and Ota, 1980). The case mortality rate of BSI due to K. pneumoniae is ~20–30%, and the population mortality rate is estimated at 1.3 per 100,000 people (Podschun and Ullmann, 1998; Meatherall et al., 2009).
The urinary tract is the most common site of infection by K. pneumoniae (Podschun and Ullmann, 1998). As with other infections, UTI due to K. pneumoniae are associated with diabetes mellitus (Lye et al., 1992). Catheter-associated UTIs (CAUTIs) are another infection caused by K. pneumoniae. It is thought that these are facilitated by the ability for form biofilms and adhere to catheters (Schroll et al., 2010). Klebsiella are also responsible for wound/surgical site infections. This site represents ~13% of all infections caused by Klebsiella (Podschun and Ullmann, 1998; Magill et al., 2014). Together, K. pneumoniae infections at each of these body sites represent an endemic opportunistic pathogen that is a substantial burden for healthcare.
Klebsiella pneumoniae Commonly Colonize Human Mucosal Surfaces
The environment likely acts as a reservoir for human acquisition of K. pneumoniae, either as colonization or an infection. K. pneumoniae is frequently found in water, sewage, soil, and plant surfaces (Bagley, 1985; Podschun et al., 2001). Several studies have shown that K. pneumoniae in the environment are very similar to their clinical counterparts in biochemical patterns, virulence and pathogenicity, and bacteriocin susceptibility patterns (Matsen et al., 1974; Podschun et al., 1992; Podschun and Ullmann, 1996; Struve and Krogfelt, 2004), though capsule type representation differs between clinical/fecal and environmental sources (Podschun, 1990). However, environmental K. pneumoniae are significantly more susceptible to antibiotics than clinical K. pneumoniae (Matsen et al., 1974), suggesting that selective pressure exists in a clinical setting. Interestingly, both environmental temperature and dewpoint are positively predictive of increased bloodstream infection caused by K. pneumoniae, suggesting that acquisition from the environment may vary with season and climate (Anderson et al., 2008). In the hospital, there are several potential sources of transmission of K. pneumoniae. One source of transmission is person-to-person contact between healthcare workers and patients, with healthcare workers' hands being a significant source (Casewell and Phillips, 1977; Jarvis et al., 1985). Contaminated surfaces and instrumentation have also been identified as sources of transmission (Jarvis et al., 1985).
Once acquired, K. pneumoniae colonizes the mucosal surfaces in humans, including the nasopharynx and the gastrointestinal tract (Podschun and Ullmann, 1998). These bacteria can be found on skin, but are considered transient at this site rather than colonizing (Kloos and Musselwhite, 1975). Colonization rates differ by body site and whether K. pneumoniae are community- or hospital-acquired. Specifically, rates of CA colonization of the nasopharynx have been reported from 3 to 15% (Davis and Matsen, 1974; Wolf et al., 2001; Farida et al., 2013; Dao et al., 2014), and are typically higher in adults than children (Wolf et al., 2001; Farida et al., 2013). Nasopharyngeal colonization has also been associated with alcohol consumption (Fuxench-Lopez and Ramirez-Ronda, 1978; Dao et al., 2014). HA nasopharyngeal colonization rates are slightly higher, up to 19% (Pollack et al., 1972). In contrast to the nasopharynx, rates of CA gastrointestinal colonization vary but can reach as high as 35% (Davis and Matsen, 1974). Furthermore, rates of gastrointestinal colonization increase among hospitalized patients and have been reported as high as 77% (Podschun and Ullmann, 1998), though recent studies demonstrate rates around 20% (Martin et al., 2016; Gorrie et al., 2017). The wide variation in colonization rates may be due to differences in the patient populations sampled several decades ago (Rose and Schreier, 1968; Thom, 1970; Selden et al., 1971; Pollack et al., 1972). Colonization rates also increase following antibiotic treatment (Rose and Schreier, 1968). Among body sites, gastrointestinal colonization is likely a common and significant reservoir in terms of risk of transmission and infection (Dorman and Short, 2017). Published colonization rates are based on detectable colonization by nasopharyngeal, rectal, or fecal sampling. It is possible that many more people are truly colonized, but that colonization is only detected by culture when the density exceeds a certain threshold.
Progression from Colonization to Infection
Decades ago, serotyping based on the Klebsiella capsule indicated that gastrointestinal colonization is an important reservoir for strains causing healthcare-associated infections (Selden et al., 1971; Montgomerie, 1979). However, serotyping does not provide sufficient resolution to distinguish closely related strains. In seeking to understand how infection with K. pneumoniae progresses, two recent studies have investigated the link between colonization with K. pneumoniae and subsequent infection (Martin et al., 2016; Gorrie et al., 2017). Gastrointestinal carriage of K. pneumoniae was significantly associated with subsequent infections in hospitalized patients (Odds ratio > 4), even after adjusting for other risk factors for infection, and ~5% of colonized patients went on to get an infection. Both studies used genomic methods to explore if infections are caused by patients' colonizing strains and each found ~80% concordance between infecting and colonizing isolates of K. pneumoniae within infected patients. Understanding colonization as an important step in progression to infection provides the rationale for identifying colonized patients and potentially establishing intervention protocols to prevent subsequent infection.
If colonization is a step in progression to infection, then understanding the mechanisms of that progression is important. A 1990 study noted that the capsule serotypes of Klebsiella clinical isolates were more similar to fecal sample serotypes than those of environmental samples, suggesting that colonization and infection may be linked (Podschun, 1990). Several studies have suggested that the intestinal tract is a reservoir for hospital-acquired pathogens and have further suggested mechanisms for dissemination, however most have focused on pathogens other than K. pneumoniae (Donskey, 2004). Though the mechanism of progression from intestinal K. pneumoniae colonization to infection is not clearly understood, there are some apparent risk factors. Intestinal domination by Proteobacteria, which taxonomically includes K. pneumoniae, leads to a five-fold increase in the risk of bacteremia in allogenic hematopoietic stem cell transplant patients (Taur et al., 2012), suggesting that bacterial density of colonizing strains plays a role in progression to disease. Procedures such as endoscopy are a potential further source of endogenous infection (Spach et al., 1993). Several underlying diseases have also been identified as risk factors for infection, since they weaken host defenses and therefore increase susceptibility to infection. Specifically, cancer, diabetes mellitus, and alcoholism are associated with both CA and HA K. pneumoniae infection (Tsay et al., 2002; Happel and Nelson, 2005; Tsai et al., 2010), though whether they are associated with progression from colonization or only with infection is unclear. Compared to intestinal colonization, fluid and electrolyte disorders, neurologic disorders, and prior hospital admissions have been identified as significantly and independently associated with infection in hospitalized patients (Martin et al., 2016). While various risk factors for infection with K. pneumoniae have been identified, risk factors specifically associated with progression from colonization to infection have not been elucidated. Further studies are needed to understand how this progression occurs and what risk factors may be involved. In addition to epidemiological risk factors, variations in the accessory genome may modulate the risk for progression to disease.
The Accessory Genome
Within a bacterial species, there is typically a set of genes that is conserved amongst all members. This set of genes is considered the core genome. In K. pneumoniae the core genome, defined as present in >95% of isolates, is currently estimated to be comprised of ~2,000 genes (Holt et al., 2015). Genes that vary between isolates are referred to as the accessory genome. This includes chromosomally encoded genes and genes on plasmids. Since K. pneumoniae genomes are typically between 5,000 and 6,000 genes, this means that the majority of the genome is comprised of accessory genes. Genes in the accessory genome can aid in specific processes, such as nitrogen fixation (Fouts et al., 2008). They can also encode specific virulence factors, as discussed below. The accessory genome also carries genes encoding various antibiotic-resistant enzymes and mechanisms (Bi et al., 2015). Accessory genes can be acquired due to horizontal gene transfer among bacterial species, as evidenced by the presence of genomic islands and mobile genetic elements in many isolates. The genes encoded in genomic islands can help isolates adapt to specific sites of infection or colonization (Chen et al., 2010; Zhang et al., 2011; van Aartsen et al., 2012). Elements of the accessory genome can be identified or predicted using various in silico applications, including computation of GC content and comparative genomic analysis (Ou et al., 2007; Zhang et al., 2014). A recent study of 328 Klebsiella isolates identified almost 30,000 unique protein-coding sequences (Holt et al., 2015), estimating the current known Klebsiella “pangenome.” The authors further demonstrated that the pangenome is open, indicating that there are more accessory genes yet to be identified and characterized.
Not all K. pneumoniae strains cause disease in animal models of infection (Fouts et al., 2008; Fodah et al., 2014). Similarly, not all colonizing strains go on to cause disease in humans (Martin et al., 2016). In fact, these bacteria are traditionally considered commensals and opportunistic pathogens (Lau et al., 2008). Of those that do cause disease, certain genes, operons, and the high pathogenicity island encoding yersiniabactin (discussed below) have been identified in K. pneumoniae that are associated with virulence (Lau et al., 2007; Lawlor et al., 2007; Fodah et al., 2014; Holt et al., 2015). This indicates that progression to disease is not only reliant on host immunocompetence, but also on genes the bacteria possess.
The Accessory Genome in Progression to Disease and Virulence
K. pneumoniae virulence factors are encoded by genes in both the core and accessory genomes. Established virulence factors in K. pneumoniae include capsule, lipopolysaccharide, siderophores, and pili (Podschun and Ullmann, 1998). Allantoin utilization, other iron uptake systems, efflux pumps, and a type VI secretion system have been identified as virulence factors more recently. Allantoin utilization will be discussed with hypervirulent strains. These genes may determine the severity of an infection, and therefore the virulence of the infecting strain. They may also determine the ability of a colonizing strain to progress to infection, defining the pathogenic potential of a given strain.
The polysaccharide capsule is one of the most important virulence factors used by K. pneumoniae. It is primarily used to assist in evading the immune system during infection, by protecting bacteria from opsonophagocytosis (Domenico et al., 1994) and serum killing (Merino et al., 1992). The capsule is generated by the capsular polysaccharide synthesis (cps) locus in K. pneumoniae, and is a structure that lies on the outside of the bacterial cell attached to the outer membrane. It is composed of repeating subunits of four to six sugars, as well as uronic acids (Podschun and Ullmann, 1998). Based on serological testing, 77 capsular types have been identified (Ørskov and Ørskov, 1984). Recent advances in molecular techniques have led to further discrimination between K. pneumoniae types as well as complete sequencing of the cps locus in all serotypes (Pan et al., 2015). An example of increased discrimination is seen in wzi typing, which identifies types based on allelic differences in the wzi gene of the cps locus (Brisse et al., 2013). This method initially identified 135 distinct wzi types that correspond with traditional serological K types. Many of these genes are conserved, but other cps genes and gene alleles are present or absent depending on the capsule type. Thus, capsular synthesis is carried out by a combination of core and accessory genes.
Lipopolysaccharide (LPS), also termed endotoxin, is a major component decorating the outer membrane of Gram-negative bacteria. LPS is widely recognized as the most powerful mediator of septic shock caused by bacteria. Host sensing of LPS via Toll-like receptor 4 (TLR4) leads to an inflammatory cascade (Roger et al., 2009). It is this host response, rather than LPS itself, that leads to the devastating pathogenesis of sepsis and septic shock. LPS molecules are comprised of lipid A, a core domain, and the O-antigen. Variations in O-antigen structures provide various O-serotypes. In K. pneumoniae there are nine main O-serotypes. Three of these, O1, O2, and O3, are responsible for almost 80% of all Klebsiella infections (Trautmann et al., 1997; Hansen et al., 1999; Follador et al., 2016). K. pneumoniae isolates with shortened or absent O-antigen (rough LPS) are sensitive to serum complement, while isolates with full-length O-antigen (smooth LPS) are resistant to serum complement (Merino et al., 1992). Variations in LPS can also play a role in protecting bacteria from antimicrobial peptides, including polymyxin antibiotics (Papo and Shai, 2005; Cheng et al., 2015). As a primary component of the outer membrane of K. pneumoniae, LPS is considered part of the core genome.
Siderophores are high affinity, low-molecular weight iron-chelating molecules secreted by various bacteria to aid in iron acquisition (Griffiths, 1988). K. pneumoniae secrete multiple types of siderophores (Holden and Bachman, 2015). One of the common catecholates secreted by K. pneumoniae is enterobactin (Ent), which is encoded in the K. pneumoniae core genome (O'Brien and Gibson, 1970; Pollack and Neilands, 1970). Since Ent is a common siderophore, the innate immune system has developed a way to bind Ent, preventing bacteria from acquiring iron (Smith, 2007). Bacteria have therefore developed other siderophores, which are encoded in the accessory genome, to counteract this. A second catecholate siderophore is a glucosylated Ent-derivative, salmochelin (Sal). Sal, is a mechanism to evade the innate immune system (Hantke et al., 2003). Mixed-type siderophores such as yersiniabactin (Ybt) and aerobactin (Aer) are also commonly secreted by K. pneumoniae. The various siderophores are associated with different sites and severity of infection, and their pathogenic functions extend beyond simply iron acquisition. Ybt was first identified in Yersinia species, and the genes encoding the biosynthesis, transport, and regulation of this molecule are located on a transposable chromosomal fragment termed a “high pathogenicity island” (Carniel, 2001). Secretion and utilization of Ybt is associated with respiratory tract infections in patients and is sufficient to promote pneumonia in a murine model (Bachman et al., 2011). World-wide, Ybt is the most common virulence factor associated with human K. pneumoniae infections (Holt et al., 2015). Aer has also been identified as a major virulence factor in K. pneumoniae infections. Aer is typically plasmid-encoded (Nassif and Sansonetti, 1986). In addition to providing iron for K. pneumoniae replication, secretion of siderophores by K. pneumoniae isolates also induces inflammation and bacterial dissemination. Siderophore-dependent progression from pneumonia to bacteremia requires the host transcriptional regulatory protein HIF-1α that is activated by iron chelation (Holden et al., 2016). Siderophore systems are frequently dependent on TonB, a protein that regulates the transport of iron across the bacterial outer membrane (Moeck and Coulton, 1998). In fact, TonB itself is considered a virulence factor in K. pneumoniae (Hsieh et al., 2008). TonB is encoded by a gene on the bacterial chromosome. It is conserved among K. pneumoniae, and is used as one of the housekeeping genes for multilocus sequence typing in these bacteria (Diancourt et al., 2005). Whereas Ent and TonB are part of the core genome, Sal, Ybt and Aer are part of the accessory genome.
A critical step in progression to infection is for bacteria to adhere to host surfaces. In K. pneumoniae, this is frequently achieved using pili (fimbriae). Pili are filamentous structures extending from the surface of bacteria. They can be as long as 10 μm and between 1-11 nm in diameter (Ofek and Doyle, 1994). There are two common types of pili found on K. pneumoniae: type 1 (fim) pili and type 3 (mrk) pili. Type 1 pili are thought to aid virulence through their ability to adhere to human mucosal or epithelial surfaces. Type 3 pili similarly adhere to cell surfaces, but importantly have been identified as strong promoters of biofilm formation (Schroll et al., 2010). Both fim and mrk pili are considered part of the core genome (Andrade et al., 2014; Lery et al., 2014). It is thought that both types of pili play a role in colonization of urinary catheters, leading to catheter-associated UTIs (Murphy et al., 2013). In addition to fim and mrk pili, a number of additional usher-type pili have been identified in K. pneumoniae, with an average of ~8 pili clusters per strain (Wu et al., 2010; Khater et al., 2015). Based on varying gene frequencies, some of these appear to be part of the accessory genome.
Efflux pumps have frequently been associated with antibiotic resistance in K. pneumoniae due to their ability to export antibiotics from bacterial cells (Filgona et al., 2015). Interestingly, the efflux pump AcrAB contributes to virulence in murine respiratory infections caused by K. pneumoniae (Padilla et al., 2010). In other bacteria, efflux pumps have been demonstrated to mediate resistance against host-derived antimicrobial peptides, which are an important aspect of the host innate immune system (Shafer et al., 1998; Bengoechea and Skurnik, 2000; Tzeng et al., 2005; Pamp et al., 2008). It is possible that this mechanism plays a similar role in the virulence of K. pneumoniae.
The type VI secretion system (T6SS), first identified in V. cholerae (Pukatzki et al., 2006), is a syringe-like apparatus anchored within the bacterial cell membrane that serves to inject various effector molecules and toxins into other cells (Journet and Cascales, 2016). Present in several Gram-negative species, the T6SS has been shown to target both other bacteria as well as eukaryotic cells, suggesting a dual role in both competition and pathogenesis. Bioinformatic analysis of K. pneumoniae genomes has identified the presence of putative T6SS gene clusters, with up to 3 loci per strain, although the number and gene content vary (Sarris et al., 2011). More recent studies have begun to characterize T6SS effectors in K. pneumoniae. In hypervirulent strain Kp52.145, the phospholipase PLD1 is required for virulence in a pneumonia model, and in the carbapenemase producing strain HS11286 the phospholipase Tle1KP promotes inter- and intra-species killing and antibiotics induce its secretion (Lery et al., 2014; Liu et al., 2017). T6SS secretion systems appear to be present in both the core and accessory genome, promote competition with other bacteria in sites of colonization, and enhance fitness in sites of infection.
Emergence of Antibiotic Resistance in Klebsiella pneumoniae
The Centers for Disease Control and Prevention estimates that more than two million people contract infections due to antibiotic-resistant microorganisms each year in the United States. Of those infected, ~23,000 die (CDC, 2014). There are multiple factors believed to contribute to the spread of antibiotic resistance, including inappropriate antibiotic use in healthcare and agriculture, and lack of new antimicrobial therapeutics (CDC, 2014). K. pneumoniae are one of several bacteria that have experienced a dramatic increase in antibiotic resistance in the past decades. Several mechanisms of antibiotic resistance are found in K. pneumoniae, with resistance to β-lactams having the greatest impact on effective treatment. Colonization with antibiotic-resistant K. pneumoniae has been associated with subsequent infection with antibiotic-resistant K. pneumoniae in hospitalized patients, although the progression from colonization to infection is incompletely understood. The accessory genome is central to antibiotic resistance in K. pneumoniae, with plasmid based resistance genes rapidly reducing the armamentarium of effective antibiotics against this pathogen.
β-Lactamase-Producing Klebsiella pneumoniae
Resistance of bacteria to β-lactam antibiotics emerged before penicillin was widely used to treat infections. Alexander Fleming was the first to note that E. coli and other bacteria were not inhibited by penicillin (Fleming, 1929), a resistance which was later attributed to an enzyme produced by these bacteria (Abraham and Chain, 1940). Resistance to β-lactam antibiotics is achieved through hydrolysis of the antibiotic β-lactam ring by β-lactamases. In K. pneumoniae, resistance to some β-lactams is intrinsic since the enzyme is encoded in the core genome of the species. For example, SHV is consistently found in the chromosome, and corresponding ampicillin resistance is a hallmark of the species (Babini and Livermore, 2000; Bialek-Davenet et al., 2014). Other β-lactamases are part of the accessory genome. In the 1960s the first plasmid-mediated β-lactamase, TEM-1, was discovered in E. coli (Datta and Kontomichalou, 1965). K. pneumoniae are also known to harbor plasmid-mediated β-lactamases, such as AmpC enzymes which confer resistance to most penicillin antibiotics (Jacoby, 2009). β-lactamase enzymes are thought to have evolved from penicillin binding proteins due to selective pressure in the environment (Medeiros, 1984; Kelly et al., 1986; Massova and Mobashery, 1998; Meroueh et al., 2003).
Extended-Spectrum β-Lactamases
Extended-spectrum β-lactamases (ESBL) are plasmid based resistance mechanisms that are part of the accessory genome. ESBL-producing K. pneumoniae were first identified in Europe in 1983 (Knothe et al., 1983) and in the United States in 1989 (Quinn et al., 1989). ESBLs are able to hydrolyze oxyimino-cephalosporins, such as third-generation cephalosporins and aztreonam, but are inhibited by clavulanic acid (Bush et al., 1995). Frequently plasmids encoding β-lactamases also possess resistance genes to other antibiotics as well as heavy metals (Jacoby and Sutton, 1991). Therefore, ESBLs are tightly linked to other accessory genes that could improve fitness of the strain it is in. And plasmid-based resistance mechanisms are associated with certain lineages that may have a characteristic accessory genome. For instance, a recent study identified that K. pneumoniae clonal group 307 (CG307) is associated with ESBL infections (Long et al., 2017b). Carbapenems have typically been the drug of choice to treat severe infections caused by ESBL-producing bacteria.
Carbapenem-Resistant Klebsiella pneumoniae (CR-Kp)
Perhaps due to the selective pressure of treating ESBL infections with carbapenems, carbapenem resistance has emerged and K. pneumoniae is the most common carbapenem-resistant Enterobacteriaceae (CRE). In 2013 the CDC declared CRE an urgent threat to public health in the United States (CDC, 2014). Of the ~9,000 infections due to CRE, Klebsiella species are responsible for ~80% of infections (CDC, 2014).
Carbapenem resistance is primarily driven by the accessory genome, sometimes in combination with mutations in the core genome. Carbapenem resistance in K. pneumoniae can be mediated in part through up-regulation of efflux pumps (Filgona et al., 2015) and alteration of outer membrane porins in the core genome (Kaczmarek et al., 2006), and hyperproduction of ESBL enzymes or AmpC β-lactamases in the accessory genome (Bush and Jacoby, 2010). For instance, hyperproduction of an ESBL or AmpC enzyme combined with a porin mutation can lead to a resistance phenotype, particularly to ertapenem (Bradford et al., 1997; García-Fernández et al., 2010). The most worrisome mechanism of carbapenem resistance is through plasmid-mediated carbapenemases. K. pneumoniae carbapenemase (KPC) β-lactamases are class A serine carbapenemases and the most frequent carbapenemases found in K. pneumoniae. KPC carbapenemases are associated with clonal group 258 (CG258) (Samuelsen et al., 2009; Breurec et al., 2013). Prominent sequence types within CG258 include ST258 that is found in Europe and North and South America, and ST11 in Asia (Baraniak et al., 2009; Kitchel et al., 2009; García-Fernández et al., 2012; Liu et al., 2012; Nicoletti et al., 2012). As of 2015, 22 KPC variants have been identified worldwide according to the Lahey Clinic website (http://www.lahey.org/studies/other.asp#table1). The association between the KPC gene and certain clonal groups suggests that these resistance plasmids have become stable members of the accessory genome in certain K. pneumoniae lineages.
Additional types of carbapenemases have also emerged in the K. pneumoniae accessory genome. The New Delhi metallo-β-lactamase-1 (NDM-1) is a plasmid-encoded class B metallo-β-lactamase (MBL). MBLs are characterized by a requirement for zinc at their active site and infections with MBL-producing strains are frequently associated with travel to and hospitalization in endemic regions (van der Bij and Pitout, 2012). For example, NDM-1 was discovered in a K. pneumoniae clinical urinary culture isolate from a Swedish patient who had recently traveled to India (Yong et al., 2009). Since then, acquisition of NDM-1 isolates has been strongly associated with individuals of Indian descent who travel to or live in the Indian subcontinent (Yong et al., 2009; Kumarasamy et al., 2010; Chen et al., 2017). K. pneumoniae carrying the Verona-integron encoded metallo-β-lactamase carbapenemases (VIM) were first detected in the United States in 2010 (Centers for Disease and Prevention, 2010). VIM-encoding isolates are endemic to Greece and Italy, and infections in the United States are associated with travel to and been hospitalization in these countries (Lauretti et al., 1999; van der Bij and Pitout, 2012; Lascols et al., 2013). VIM variants (blaVIM) are carried on integrons that can be integrated into either the chromosome or carried on plasmids (Lauretti et al., 1999; Pournaras et al., 2005). Imipenemase (IMP) type MBLs are endemic in Japan (Osano et al., 1994), but have been detected worldwide (Limbago et al., 2011). Similar to blaVIM, genes encoding IMP variants (blaIMP) are carried on integrons and can be encoded on either chromosomes or are plasmid-mediated (Arakawa et al., 1995; Docquier et al., 2003). OXA carbapenemases are class D enzymes are characterized by their ability to hydrolyze cloxacillin or oxacillin (Bush and Jacoby, 2010). The plasmid encoded OXA-48 (blaOXA-48) is found in K. pneumoniae and confers a high level of resistance to imipenem (Poirel et al., 2004).
Colistin Resistance in Klebsiella pneumoniae
Colistin resistance in K. pneumoniae is commonly caused by mutations in the core genome, but transmissible resistance genes in the accessory genome are a grave concern. Colistin is among the polymyxin class of antibiotics, used to treat Gram-negative infections in the 1960s and 1970s. Their use was discontinued due to renal- and neurotoxicity (Jerke et al., 2016). However, the recent emergence of CRE has made it necessary to return to colistin as a drug of last resort. Colistin resistance in K. pneumoniae typically occurs through mutations in regulatory genes such as mgrB that regulate modification of bacterial lipid A, the target of polymyxin antibiotics, decreasing the ability of polymyxins to interact (Cannatelli et al., 2013; Jayol et al., 2014, 2015; Olaitan et al., 2014; Poirel et al., 2015; Wright et al., 2015). In 2015, plasmid-mediated resistance to colistin was discovered in an E. coli isolate in China (Liu et al., 2016), conferred by the mcr-1 gene. This discovery heralds the potential for easily transmissible genes leading to pan-resistance. The prevalence of mcr-1 in K. pneumoniae BSI isolates in China is rare, and found more often in E. coli (Quan et al., 2017). The first reported incidence of mcr-1 in the United States occurred in 2016 in E. coli (McGann et al., 2016). In September of 2016 a pan-resistant isolate of K. pneumoniae was isolated (Chen et al., 2017), but colistin resistance in this isolate was not mediated by mcr-1.
Colonization as a Reservoir of Antibiotic-Resistant K. pneumoniae in Hospitals
Similar to recent findings across K. pneumoniae in general, intestinal colonization by antibiotic-resistant K. pneumoniae can precede infection with the same strain (Selden et al., 1971). A 1971 prospective study determined that patients colonized with antibiotic-resistant K. pneumoniae after hospital admission developed infection with antibiotic-resistant K. pneumoniae at a higher percentage within 21 days compared to those who did not become intestinal carriers. They further compared serotypes of colonization isolates and subsequent infecting isolates, finding that 14 of 31 (45.2%) patients colonized after admission were infected with the same serotype, indicating that hospitalized patients are often colonized with the same isolate they become infected with. There were, however, two predominant serotypes circulating and causing infection [serotype 30 (n = 11) and serotype 63 (n = 8)], confounding the significance of these findings. Serotype 30 was also the most common serotype identified in both rectal (n = 23/34) and antibiotic-resistant infecting (n = 18/20) isolates from a previous collection within the same facility (Selden et al., 1971). In a cohort study of ESBL-KP colonized patients, 22% progressed to a positive clinical culture, with a median time of 2.7 days from a positive rectal swab (Harris et al., 2007). For CR-Kp, colonization can persist and spread silently for years in long-term care facilities (Viau et al., 2012). CR-Kp colonization can also trigger a clonal outbreak and newly colonized patients can develop fatal infections (Snitkin et al., 2012).
Risk Factors for Infections Caused by Antibiotic-Resistant Klebsiella pneumoniae
Risk factors for colonization and infection with antibiotic-resistant K. pneumoniae are often considered together, so the risk factors for infection specifically are unclear (Selden et al., 1971; Pollack et al., 1972; Asensio et al., 2000). Risk factors associated with ESBL colonization and infection include prior treatment with antibiotics, prolonged hospitalization, prolonged ICU stay, and mechanical ventilation (Jacoby, 1998; Lautenbach et al., 2001; Nathisuwan et al., 2001). Intestinal colonization with ESBL bacteria has also been associated with ESBL infection. Risk factors associated with carbapenem-resistant K. pneumoniae colonization and infection include prior antibiotic treatment, renal dysfunction, older age, surgical procedures, and ICU admission (Kofteridis et al., 2014; Jiao et al., 2015). As with endemic strains of K. pneumoniae, hospitalization seems to be a key factor associated with infection. Though antibiotic-resistant strains can infect an array of body sites similar to endemic strains, they frequently cause UTIs (Selden et al., 1971; Lautenbach et al., 2001). This may represent inoculation of the urinary tract with K. pneumoniae from the gastrointestinal tract across the perineum.
Community-Acquired Hypervirulent Strains
In the 1980s and 1990s, reports began to emerge from the Asian Pacific Rim detailing severe infections due to K. pneumoniae (Liu et al., 1986; Cheng et al., 1991; Wang et al., 1998). These infections were unique in that they were community-acquired (CA), a departure from the classic presentation of K. pneumoniae infections in hospitalized patients. Common infections due to these hypervirulent K. pneumoniae (hvKP) include pyogenic liver abscess (PLA); endophthalmitis, an infection inside the eye; meningitis; and bloodstream infections (Fang et al., 2007). Symptoms of PLA vary between individuals and are frequently non-specific. Diagnosis requires radiographic imaging (Johannsen et al., 2000). The incidence of PLA rose sharply in Taiwan, from 11.1 to 17.6 per 100,000 people from 1996 to 2004 (Fung et al., 2012). Approximately 3–11% of patients with PLA will go on to develop endophthalmitis (Sng et al., 2008; Sheu et al., 2011). Patient risk factors for severe infection with hvKP include being aged 55–60 years, male, and having diabetes mellitus (Lee et al., 2008; Siu et al., 2012; Shon et al., 2013). Throughout China there is a high prevalence of hvKP strains among K. pneumoniae isolates that cause infection (31–37.8%), though the rate varies based on geographic location within China (Liu et al., 2014; Zhang et al., 2016). Reported mortality rates (14–30 days) vary among patients in Asia who are infected with hvKP (4.5%-31%) (Wang et al., 1998; Ko et al., 2002; Liu et al., 2014). Since hvKP strains emerged from the Asian Pacific Rim and are overrepresented in individuals of Asian descent (Wang et al., 1998), it has also been suggested that infection with these strains is associated with an individual's ethnicity, or that it may be a geographically-specific pathogen, though it is unclear if this is a significant association (Shon et al., 2013). Alarmingly, these strains have begun to emerge worldwide, including in the United States (Lederman and Crum, 2005; Nadasy et al., 2007; Pastagia and Arumugam, 2008; Frazee et al., 2009; McCabe et al., 2010; Fierer et al., 2011). However, individuals from Western countries who become infected with hvKP are frequently either of Asian descent and/or have recently traveled to or been in contact with someone from an Asian country (Lederman and Crum, 2005; Keynan et al., 2007; Frazee et al., 2009; Gunnarsson et al., 2009; McCabe et al., 2010; Decré et al., 2011; Pomakova et al., 2012). Fortunately, hvKP isolates are typically highly susceptible to most antibiotics (Fang et al., 2007). Recently, however, an outbreak due to a carbapenem-resistant ST11 hvKP isolate occurred in China, heralding the potential for dual-risk isolates that both cause severe infections and are increasingly difficult to treat or are fatal due to antibiotic resistance (Gu et al., 2017).
Hypervirulent K. pneumoniae Possess Unique Virulence Factors
The most striking aspect of hvKP isolates is their ability to cause severe infections in otherwise healthy patients. This severity is attributed to virulence factors encoded by the accessory genome. HvKP have a hypermucoviscous phenotype characterized by a positive “string” test: attempting to pick a colony with a loop results in a strand of bacteria that clings to the agar media. This phenotype has been determined to be conferred by two proteins: RmpA, which regulates capsule production (Hsu et al., 2011; Shon et al., 2013), and MagA, which is associated with the hypermucoviscous phenotype (Yu et al., 2006). Genes encoding RmpA and MagA are highly associated with hvKP, especially in Asia, and are considered virulence factors. Capsule types K1 and K2 are also highly associated with hvKP and may play a more important role in virulence than rmpA and magA (Yeh et al., 2007). Frequently, hvKP strains are K1 or K2 capsule type (Fung et al., 2002; Chung et al., 2007; Yu et al., 2008; Shon et al., 2013). They are also often sequence type 23 (ST23) a phylogroup strongly associated with the K1 capsule type (Liu et al., 2014). The siderophore Ybt has also been associated with hypervirulent strains (Holt et al., 2015), although it should be noted that Ybt is also found in many non-hypervirulent strains and is considered a general virulence factor in K. pneumoniae, as discussed above (Lawlor et al., 2007). Furthermore, the siderophore Aer has been distinguished as the most common siderophore secreted by hypervirulent K. pneumoniae (Russo et al., 2015).
Allantoin has been identified as a source of nitrogen in various bacterial species and as both a nitrogen source and a carbon source in K. pneumoniae (Cusa et al., 1999; Chou et al., 2004; Navone et al., 2014). Allantoin is a metabolic intermediate of purine degradation by various organisms including microbes (Vogels and Van der Drift, 1976). An allantoin utilization operon has been associated with hypervirulent K. pneumoniae strains that cause pyogenic liver abscesses (Chou et al., 2004). Deletion of a regulator gene in the operon corresponds to decreased virulence in a mouse model, indicating that the ability to use as a nitrogen source increases virulence in K. pneumoniae at certain sites of infection. Allantoin metabolism genes are variably encoded chromosomally in K. pneumoniae, and part of the accessory genome (Chou et al., 2004).
Colonization as a Reservoir for hvKP
Colonization appears to be a reservoir for hvKP that cause PLA and other severe infections. PFGE typing of liver and fecal isolates from patients with PLA demonstrates little or no differences within patients but discernable differences between patients. But each of these strains may have colonized many people, as a random sampling of fecal isolates from asymptomatic patients revealed some of the same PFGE types seen in PLA isolates (Fung et al., 2012). These findings suggest that, similar to opportunistic K. pneumoniae, hvKP infections are caused by endogenous colonizing strains.
If colonization is a potential reservoir for infection with hvKP strains, then understanding the community carriage rates is important. With that in mind, a recent study sought to identify the fecal carriage rate of K1 K. pneumoniae in healthy Koreans in order to identify this potential reservoir (Chung et al., 2012). They determined that 4.9% of individuals tested carry K1 K. pneumoniae. Of these, 94.7% were found to be ST23, which is strongly associated with PLA. The overall K. pneumoniae colonization rate was 21.1%. They further determined that the K1 carriage rate was higher in foreigners who lived in Korea compared to those of Korean descent who lived outside of Korea (24.1 vs. 5.6%, P = 0.024), suggesting that exposure to the Korean peninsula plays a role in K1 colonization. A similar study was undertaken where stool isolates from healthy Chinese adults living in various Asian countries were collected over 6 years and tested for the presence of K. pneumoniae with a K1/K2 serotype, both of which are associated with PLA (Lin et al., 2012). It was determined that 9.8% of K. pneumoniae isolates recovered were K1/K2. Rates were similar for Chinese living in all countries tested except Thailand and Vietnam. Strikingly, the overall K. pneumoniae colonization rate was also significantly higher than that seen in Western counties, with rates of >50% in cohorts from Taiwan and China. These findings establish that hvKP can be a significant proportion of K. pneumoniae colonization, and with high overall colonization rates, many people may be colonized with potentially deadly strains.
Convergence of Hypervirulence and Carbapenemase Accessory Genes
The most obvious concern about K. pneumoniae and its flexible accessory genome is a dual-risk isolate, one that is both antibiotic-resistant and hypervirulent. With treatment options already limited, this result could be devastating. These isolates will cause severe disease but will be difficult or impossible to treat. Genomic analyses of strains worldwide have already detected the convergence of hypervirulence and carbapenemase genes in a number of isolates (Bialek-Davenet et al., 2014; Holt et al., 2015) As proof of the legitimacy of these concerns, a recent study identified a fatal outbreak of VAP due to a hypervirulent carbapenem-resistant K. pneumoniae ST11 isolate in China (Gu et al., 2017). These isolates had acquired portions of the hypervirulent virulence plasmid pLVPK, and contained several virulence factors associated with hypervirulent strains including genes encoding the siderophore Aer and the mucoid phenotype regulator gene rmpA. Alarmingly, no antibiotics were effective in treating the infections caused by these hv CRE K. pneumoniae strains. The ST11 strains identified in this study also included genes encoding the siderophore Ybt. Interestingly, Ybt is also found in a subset of ST11 across Asia (Holt et al., 2015). These CRE K. pneumoniae isolates already encode a critical virulence factor in Ybt and can acquire more through acquisition of hypervirulence plasmids.
Klebsiella Species Vary in their Accessory Genome Content
Improved molecular epidemiology and sequencing capabilities have recently demonstrated that isolates frequently identified as K. pneumoniae can be divided into three distinct Klebsiella species (Brisse and Verhoef, 2001; Brisse et al., 2004, 2014; Maatallah et al., 2014; Berry et al., 2015). Although initially distinguished by variations in their core genome, these species can also be separated by the content of their accessory genome (Holt et al., 2015). Phylogroup KpI represents ~80% of opportunistic isolates and is the species K. pneumoniae (sensu stricto). Phylogroup KpII is identified as Klebsiella quasipneumoniae and KpIII as Klebsiella variicola. As three distinct species, these groups may vary in their epidemiology of colonization and infection.
Not much is known about the least frequently isolated of the three Kp phylogroups, K. quasipneumoniae. Approximately 94% of K. quasipneumoniae isolates have been recovered from humans, with over 50% of human isolates being associated with intestinal carriage rather than infection (compared to 24 and 39% intestinal carriage in KpI and KpIII, respectively; Holt et al., 2015). The most common sites of K. quasipneumoniae infections are the urinary and respiratory tracts, and some isolates are ESBL-producers (Holt et al., 2015).
Klebsiella variicola was proposed as a new, distinct species based on both genetic and biochemical differences from K. pneumoniae (Rosenblueth et al., 2004; Brisse et al., 2014). A consistent trait of K. variicola is the ability of this species to fix nitrogen, which explains their endophytic relationship with several plants such as maize, banana, sugarcane, and wheat. Genes responsible for nitrogen fixation, such as those in the nif operon, can be considered part of the core genome of K. variicola, but can also be found in related Klebsiella species as part of the inter-species accessory genome (Fouts et al., 2008). While colonization of plants is common, colonization rates in humans is unknown. Though this species is associated with environmental sources, the clinical importance of K. variicola is becoming more apparent. In fact, a recent study determined that patients with bloodstream infections due to K. variicola demonstrate a higher 30-day mortality rate (29.4%) compared to K. pneumoniae (13.5%) and K. quasipneumoniae (11.1%) species (Maatallah et al., 2014). This increased mortality was not due to any known virulence factors and was significant after controlling for patient co-morbidities, suggesting that K. variicola harbors yet undiscovered and potentially clinically relevant genes.
A comparison of the antimicrobial resistance patterns of clinical isolates of the three Kp phylogroups (n = 420) to 10 antimicrobial agents showed that K. pneumoniae (KpI) has the highest resistance levels, K. quasipneumoniae (KpII) has intermediate resistance levels, and K. variicola (KpIII) has the lowest resistance, with KpI having a resistance rate of two- to three-fold higher than KpIII (Brisse et al., 2004). A more recent study of isolates from the three phylogroups collected worldwide (n = 328) determined that ~50% of KpII isolates display an ESBL phenotype (Holt et al., 2015).
Since it is only relatively recently that these species have been differentiated, there is a paucity of information in the literature regarding colonization rates and virulence factors in K. quasipneumoniae and K. variicola. Along with K. pneumoniae, these three distinct but related species vary in their epidemiology of infection and antimicrobial resistance, likely due to the variation in genes each species possesses, indicating that exploration of the Klebsiella accessory genome is imperative for better understanding the nuances of how these bacteria cause disease. Indeed, genetic exchange across these three species can occur (Holt et al., 2015; Long et al., 2017a).
Summary and Gaps in Knowledge
Though classically considered an opportunistic, hospital-acquired pathogen that infects only immunocompromised hosts, two additional types of K. pneumoniae have emerged: carbapenem-resistant and hypervirulent. Across these three types, intestinal colonization rates are significant and serve as a reservoir for isolates capable of causing infection. For hospital-onset infections, the association between colonization and subsequent infection is established and strong. For hvKP and CR-KP the association between colonization and subsequent infection is unclear. And for all types of K. pneumoniae, the risk factors for progression to infection in colonized patients are poorly understood. Understanding colonization and infection as two distinct stages with potentially varying risk factors will further aid in understanding the pathogenesis of K. pneumoniae.
The accessory genome is likely critical in determining the differences in infection risk and outcomes of endemic, antibiotic-resistant, and hypervirulent K. pneumoniae. Several K. pneumoniae virulence factors identified to date are based on association with hypervirulent phenotypes, such as PLA. Furthermore, most studies involving K. pneumoniae tend to focus on one clonal group, such as carbapenem-resistant or hypervirulent isolates. While understanding of pathogenesis for each distinct type is important, a broad understanding of how K. pneumoniae causes the common infections of pneumonia, bacteremia, and UTIs could better aid in targeting common factors for diagnosis and treatment. Finally, it is only recently that we have begun to recognize that ~20% of nosocomial isolates identified as K. pneumoniae are actually K. variicola and K. quasipneumoniae. These species may have distinct epidemiological and resistance profiles based on the composition of their accessory genome. However, it is becoming evident that there is a reservoir of genes that are exchanged and assembled to create strains with varying infectious and antibiotic-resistant potential (Figure 1). Therefore, it is increasingly clear that Klebsiella can assemble a large accessory genome from a larger pool of available genes to determine their ability to colonize, infect, and resist antibiotics in humans.
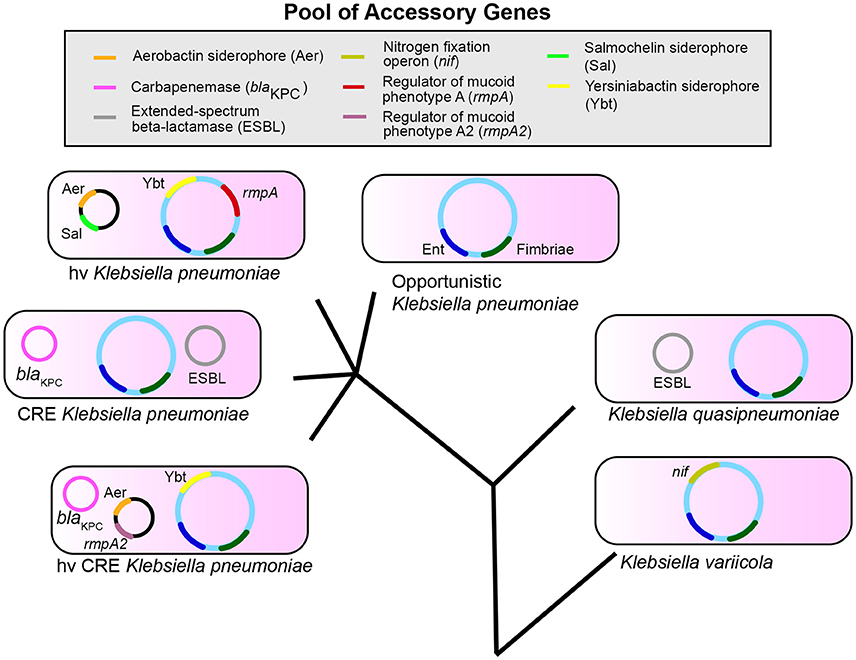
Figure 1. Klebsiella pneumoniae, variicola, and quasipneumoniae are three species that share a pool of accessory genes. The combination of genes in the accessory genome differs between species and pathotypes within K. pneumoniae such as opportunistic, carbapenem-resistant (CRE), and hypervirulent (hv) strains. These accessory genes can also combine to form new pathotypes (hvCRE) and can be shared across species. Enterobactin (Ent, blue) and Fimbriae (dark green) represent conserved genes; accessory genes are shown as examples and are not a definitive list. The evolutionary tree is not drawn to scale.
Author Contributions
RM and MB: conceived design of the review, drafted, and critically revised the work, and provided final approval for publication.
Funding
RM and MB received support from the National Institutes of Health R01AI125307 to MB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Thank you to Nicholas W. Lukacs, Harry L. T. Mobley, Gabriel Núñez, and Evan S. Snitkin for reading an early draft of the review.
References
Abraham, E. P., and Chain, E. (1940). An enzyme from bacteria able to destroy penicillin. Nature 146, 837–837. doi: 10.1038/146837a0
Anderson, D. J., Hervé, R., Chen, L. F., Spelman, D. W. Y., Hung, J., Raoult, D., et al. (2008). Seasonal variation in Klebsiella pneumoniae bloodstream infection on 4 continents. J. Infect. Dis. 197, 752–756. doi: 10.1086/527486
Andrade, B. G. N., de Veiga Ramos, N., Marin, M. F. A., Fonseca, E. L., and Vicente, A. C. P. (2014). The genome of a clinical Klebsiella variicola strain reveals virulence-associated traits and a pl9-like plasmid. FEMS Microbiol. Lett. 360, 13–16. doi: 10.1111/1574-6968.12583
Arakawa, Y., Murakami, M., Suzuki, K., Ito, H., Wacharotayankun, R., Ohsuka, S., et al. (1995). A novel integron-like element carrying the metallo-beta-lactamase gene blaIMP. Antimicrob. Agents Chemother. 39, 1612–1615.
Asensio, A., Oliver, A., Gonzalez-Diego, P., Baquero, F., Perez-Diaz, J. C., Canton, R., et al. (2000). Outbreak of a multiresistant Klebsiella pneumoniae strain in an intensive care unit: antibiotic use as risk factor for colonization and infection. Clin. Infect. Dis. 30, 55–60. doi: 10.1086/313590
Babini, G. S., and Livermore, D. M. (2000). Are SHV β-lactamases universal in Klebsiella pneumoniae? Antimicrob. Agents Chemother. 44:2230. doi: 10.1128/AAC.44.8.2230-2230.2000
Bachman, M. A., Oyler, J. E., Burns, S. H., Caza, M., Lepine, F., Weiser, J. N., et al. (2011). Klebsiella pneumoniae yersiniabactin promotes respiratory tract infection through evasion of lipocalin 2. Infect. Immun. 79, 3309–3316. doi: 10.1128/IAI.05114-11
Bagley, S. T. (1985). Habitat association of Klebsiella species. Infect. Control 6, 52–58. doi: 10.1017/S0195941700062603
Baraniak, A., Izdebski, R., Herda, M., Fiett, J., Hryniewicz, W., Gniadkowski, M., et al. (2009). Emergence of Klebsiella pneumoniae ST258 with KPC-2 in Poland. Antimicrob. Agents Chemother. 53, 4565–4567. doi: 10.1128/AAC.00436-09
Bengoechea, J. A., and Skurnik, M. (2000). Temperature-regulated efflux pump/potassium antiporter system mediates resistance to cationic antimicrobial peptides in Yersinia. Mol. Microbiol. 37, 67–80. doi: 10.1046/j.1365-2958.2000.01956.x
Berry, G. J., Loeffelholz, M. J., and Williams-Bouyer, N. (2015). An investigation into laboratory misidentification of a bloodstream Klebsiella variicola infection. J. Clin. Microbiol. 53, 2793–2794. doi: 10.1128/JCM.00841-15
Bi, D., Jiang, X., Sheng, Z. K., Ngmenterebo, D., Tai, C., Wang, M., et al. (2015). Mapping the resistance-associated mobilome of a carbapenem-resistant Klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a “resistance-disarmed” model organism. J. Antimicrob. Chemother. 70, 2770–2774. doi: 10.1093/jac/dkv204
Bialek-Davenet, S., Criscuolo, A., Ailloud, F., Passet, V., Jones, L., Delannoy-Vieillard, A., et al. (2014). Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 20, 1812–1820. doi: 10.3201/eid2011.140206
Bradford, P. A., Urban, C., Mariano, N., Projan, S. J., Rahal, J. J., and Bush, K. (1997). Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC beta-lactamase, and the foss of an outer membrane protein. Antimicrob. Agents Chemother. 41, 563–569.
Breurec, S., Guessennd, N., Timinouni, M., Le, T. A., Cao, V., Ngandjio, A., et al. (2013). Klebsiella pneumoniae resistant to third-generation cephalosporins in five African and two Vietnamese major towns: multiclonal population structure with two major international clonal groups, CG15 and CG258. Clin. Microbiol. Infect. 19, 349–355. doi: 10.1111/j.1469-0691.2012.03805.x
Brisse, S., Passet, V., and Grimont, P. A. (2014). Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int. J. Syst. Evol. Microbiol. 64(Pt 9), 3146–3152. doi: 10.1099/ijs.0.062737-0
Brisse, S., Passet, V., Haugaard, A. B., Babosan, A., Kassis-Chikhani, N., Decré, D., et al. (2013). wzi gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J. Clin. Microbiol. 51, 4073–4078. doi: 10.1128/JCM.01924-13
Brisse, S., van Himbergen, T., Kusters, K., and Verhoef, J. (2004). Development of a rapid identification method for Klebsiella pneumoniae phylogenetic groups and analysis of 420 clinical isolates. Clin. Microbiol. Infect. 10, 942–945. doi: 10.1111/j.1469-0691.2004.00973.x
Brisse, S., and Verhoef, J. (2001). Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int. J. Syst. Evol. Microbiol. 51(Pt 3), 915–924. doi: 10.1099/00207713-51-3-915
Bush, K., and Jacoby, G. A. (2010). Updated functional classification of β-lactamases. Antimicrob. Agents Chemother. 54, 969–976. doi: 10.1128/AAC.01009-09
Bush, K., Jacoby, G. A., and Medeiros, A. A. (1995). A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39, 1211–1233. doi: 10.1128/AAC.39.6.1211
Cannatelli, A., D'Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Carniel, E. (2001). The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3, 561–569. doi: 10.1016/S1286-4579(01)01412-5
Casewell, M., and Phillips, I. (1977). Hands as route of transmission for Klebsiella species. Br. Med. J. 2, 1315–1317. doi: 10.1136/bmj.2.6098.1315
CDC (2014). Antibiotic Resistance Threats in the United States, 2013. Atlanta, GA: U.S. Department of Health and Human Services, Centers for Disease Control and Prevention.
Centers for Disease and Prevention (2010). Update: detection of a verona integron-encoded metallo-beta-lactamase in Klebsiella pneumoniae — United States, 2010. Morb. Mortal. Wkly. Rep. 59:1212.
Chen, L., Todd, R., Kiehlbauch, J., Walters, M., and Kallen, A. (2017). Notes from the field: pan-resistant New Delhi metallo-beta-lactamase-producing Klebsiella pneumoniae- Washoe County, Nevada, 2016. Morb. Mortal. Wkly. Rep. 66:33. doi: 10.15585/mmwr.mm6601a7
Chen, N., Ou, H. Y., van Aartsen, J. J., Jiang, X., Li, M., Yang, Z., et al. (2010). The pheV Phenylalanine tRNA Gene in Klebsiella pneumoniae clinical isolates is an integration hotspot for possible niche-adaptation genomic islands. Curr. Microbiol. 60, 210–216. doi: 10.1007/s00284-009-9526-4
Cheng, D. L., Liu, Y. C., Yen, M. Y., Liu, C. Y., and Wang, R. S. (1991). Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch. Intern. Med. 151, 1557–1559. doi: 10.1001/archinte.1991.00400080059010
Cheng, Y. H., Lin, T. L., Pan, Y. J., Wang, Y. P., Lin, Y. T., and Wang, J. T. (2015). Colistin resistance mechanisms in Klebsiella pneumoniae strains from Taiwan. Antimicrob. Agents Chemother. 59, 2909–2913. doi: 10.1128/AAC.04763-14
Chou, H. C., Lee, C. Z., Ma, L. C., Fang, C. T., Chang, S. C., and Wang, J. T. (2004). Isolation of a chromosomal region of Klebsiella pneumoniae associated with allantoin metabolism and liver infection. Infect. Immun. 72, 3783–3792. doi: 10.1128/IAI.72.7.3783-3792.2004
Chung, D. R., Lee, H., Park, M. H., Jung, S. I., Chang, H. H., Kim, Y. S., et al. (2012). Fecal carriage of serotype K1 Klebsiella pneumoniae ST23 strains closely related to liver abscess isolates in Koreans living in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 31, 481–486. doi: 10.1007/s10096-011-1334-7
Chung, D. R., Lee, S. S., Lee, H. R., Kim, H. B., Choi, H. J., Eom, J. S., et al. (2007). Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J. Infect. 54, 578–583. doi: 10.1016/j.jinf.2006.11.008
Cusa, E., Obradors, N., Baldomà, L., Badía, J., and Aguilar, J. (1999). Genetic analysis of a chromosomal region containing genes required for assimilation of allantoin nitrogen and linked glyoxylate metabolism in Escherichia coli. J. Bacteriol. 181, 7479–7484.
Dao, T. T., Liebenthal, D., Tran, T. K., Ngoc Thi Vu, B., Ngoc Thi Nguyen, D., Thi Tran, H., et al. (2014). Klebsiella pneumoniae oropharyngeal carriage in rural and urban Vietnam and the effect of alcohol consumption. PLoS ONE 9:e91999. doi: 10.1371/journal.pone.0091999
Datta, N., and Kontomichalou, P. (1965). Penicillinase synthesis controlled by infectious R factors in Enterobacteriaceae. Nature 208, 239–241. doi: 10.1038/208239a0
Davis, T. J., and Matsen, J. M. (1974). Prevalence and characteristics of Klebsiella species: relation to association with a hospital environment. J. Infect. Dis. 130, 402–405. doi: 10.1093/infdis/130.4.402
Decré, D., Verdet, C., Emirian, A., Le Gourrierec, T., Petit, J. C., Offenstadt, G., et al. (2011). Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J. Clin. Microbiol. 49, 3012–3014. doi: 10.1128/JCM.00676-11
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A. D., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
Docquier, J. D., Riccio, M. L., Mugnaioli, C., Luzzaro, F., Endimiani, A., Toniolo, A., et al. (2003). IMP-12, a new plasmid-encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob. Agents Chemother. 47, 1522–1528. doi: 10.1128/AAC.47.5.1522-1528.2003
Domenico, P., Salo, R. J., Cross, A. S., and Cunha, B. A. (1994). Polysaccharide capsule-mediated resistance to opsonophagocytosis in Klebsiella pneumoniae. Infect. Immun. 62, 4495–4499.
Donskey, C. J. (2004). The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin. Infect. Dis. 39, 219–226. doi: 10.1086/422002
Dorman, M. J., and Short, F. L. (2017). Klebsiella pneumoniae: when a colonizer turns bad. Nat. Rev. Microbiol. 15:384. doi: 10.1038/nrmicro.2017.64
Fang, C. T., Lai, S. Y., Yi, W. C., Hsueh, P. R., Liu, K. L., and Chang, S. C. (2007). Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin. Infect. Dis. 45, 284–293. doi: 10.1086/519262
Farida, H., Severin, J. A., Gasem, M. H., Keuter, M., van den Broek, P., Verbrugh, H. A. et al. (2013). Nasopharyngeal carriage of Klebsiella pneumoniae and other gram-negative Bacilli in pneumonia-prone age groups in Semarang, Indonesia. J. Clin. Microbiol. 51, 1614–1616. doi: 10.1128/JCM.00589-13
Fierer, J., Walls, L., and Chu, P. (2011). Recurring Klebsiella pneumoniae pyogenic liver abscesses in a resident of San Diego, California, due to a K1 strain carrying the virulence plasmid. J. Clin. Microbiol. 49, 4371–4373. doi: 10.1128/JCM.05658-11
Filgona, J., Banerjee, T., and Anupurba, S. (2015). Role of efflux pumps inhibitor in decreasing antibiotic resistance of Klebsiella pneumoniae in a tertiary hospital in North India. J. Infect. Dev. Ctries. 9, 815–820. doi: 10.3855/jidc.6216
Fleming, A. (1929). On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzæ. Br. J. Exp. Pathol. 10, 226–236.
Fodah, R. A., Scott, J. B., Tam, H. H., Yan, P., Pfeffer, T. L., Bundschuh, R., et al. (2014). Correlation of Klebsiella pneumoniae comparative genetic analyses with virulence profiles in a murine respiratory disease model. PLoS ONE 9:e107394. doi: 10.1371/journal.pone.0107394
Follador, R., Heinz, E., Wyres, K. L., Ellington, M. J., Kowarik, M., Holt, K. E., et al. (2016). The diversity of Klebsiella pneumoniae surface polysaccharides. Microbial Genomics 2:e000073. doi: 10.1099/mgen.0.000073
Fouts, D. E., Tyler, H. L., DeBoy, R. T., Daugherty, S., Ren, Q., Badger, J. H., et al. (2008). Complete genome sequence of the N2-fixing broad host range endophyte Klebsiella pneumoniae 342 and virulence predictions verified in mice. PLoS Genet. 4:e1000141. doi: 10.1371/journal.pgen.1000141
Frazee, B. W., Hansen, S., and Lambert, L. (2009). Invasive infection with hypermucoviscous Klebsiella pneumoniae: multiple cases presenting to a single emergency department in the United States. Ann. Emerg. Med. 53, 639–642. doi: 10.1016/j.annemergmed.2008.11.007
Friedlaender, C. (1882). Ueber die Schizomyceten bei der acuten fibrösen Pneumonie. Archiv Patholog. Anat. Physiol. Klinische Med. 87, 319–324. doi: 10.1007/BF01880516
Fung, C. P., Chang, F. Y., Lee, S. C., Hu, B. S., Kuo, B. I., Liu, C. Y., et al. (2002). A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 50, 420–424. doi: 10.1136/gut.50.3.420
Fung, C. P., Lin, Y. T., Lin, J. C., Chen, T. L., Yeh, K. M., Chang, F. Y., et al. (2012). Klebsiella pneumoniae in gastrointestinal tract and pyogenic liver abscess. Emerg. Infect. Dis. 18, 1322–1325. doi: 10.3201/eid1808.111053
Fuxench-Lopez, Z., and Ramirez-Ronda, C. H. (1978). Pharyngeal flora in ambulatory alcoholic patients: prevalence of gram-negative bacilli. Arch. Intern. Med. 138, 1815–1816. doi: 10.1001/archinte.1978.03630370033017
García-Fernández, A., Miriagou, V., Papagiannitsis, C. C., Giordano, A., Venditti, M., Mancini, C., et al. (2010). An ertapenem-resistant extended-spectrum-β-lactamase-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob. Agents Chemother. 54, 4178–4184. doi: 10.1128/AAC.01301-09
García-Fernández, A., Villa, L., Carta, C., Venditti, C., Giordano, A., Venditti, M., et al. (2012). Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob. Agents Chemother. 56, 2143–2145. doi: 10.1128/AAC.05308-11
Gorrie, C. L., Mirčeta, M., Wick, R. R., Edwards, D. J., Thomson, N. R., Strugnell, R. A., et al. (2017). Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208–215. doi: 10.1093/cid/cix270
Griffiths, E. (1988). “High-affinity iron uptake systems and bacterial virulence,” in Virulence Mechanisms of Bacterial Pathogens, ed J. A. Roth (Washington, DC: American Society for Microbiology), 121–137.
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2017). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37–46. doi: 10.1016/S1473-3099(17)30489-9
Gunnarsson, G. L., Brandt, P. B., Gad, D., Struve, C., and Justesen, U. S. (2009). Monomicrobial necrotizing fasciitis in a white male caused by hypermucoviscous Klebsiella pneumoniae. J. Med. Microbiol. 58, 1519–1521. doi: 10.1099/jmm.0.011064-0
Hansen, D. S., Mestre, F., Albertí, S., Hernández-Allés, S., Álvarez, D., Doménech-Sánchez, A., et al. (1999). Klebsiella pneumoniae lipopolysaccharide o typing: revision of prototype strains and o-group distribution among clinical isolates from different sources and countries. J. Clin. Microbiol. 37, 56–62.
Hantke, K., Nicholson, G., Rabsch, W., and Winkelmann, G. (2003). Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U.S.A. 100, 3677–3682. doi: 10.1073/pnas.0737682100
Happel, K. I., and Nelson, S. (2005). Alcohol, immunosuppression, and the lung. Proc. Am. Thorac. Soc. 2, 428–432. doi: 10.1513/pats.200507-065JS
Harris, A. D., Perencevich, E. N., Johnson, J. K., Paterson, D. L., Morris, J. G., Strauss, S. M., et al. (2007). Patient-to-patient transmission is important in extended-spectrum β-lactamase–producing Klebsiella pneumoniae acquisition. Clin. Infect. Dis. 45, 1347–1350. doi: 10.1086/522657
Holden, V. I., and Bachman, M. A. (2015). Diverging roles of bacterial siderophores during infection. Metallomics 7, 986–995. doi: 10.1039/C4MT00333K
Holden, V. I., Breen, P., Houle, S., Dozois, C. M., and Bachman, M. A. (2016). Klebsiella pneumoniae siderophores induce inflammation, bacterial dissemination, and HIF-1alpha stabilization during pneumonia. MBio 7:e01397-16. doi: 10.1128/mBio.01397-16
Holt, K. E., Wertheim, H., Zadoks, R. N., Baker, S., Whitehouse, C. A., Dance, D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci.U.S.A. 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Hsieh, P. F., Lin, T. L., Lee, C. Z., Tsai, S. F., and Wang, J. T. (2008). Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 197, 1717–1727. doi: 10.1086/588383
Hsu, C. R., Lin, T. L., Chen, Y. C., Chou, H. C., and Wang, J. T. (2011). The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 157(Pt 12), 3446–3457. doi: 10.1099/mic.0.050336-0
Jacoby, G. A. (1998). Editorial response: epidemiology of extended-spectrum β-Lactamases. Clin. Infect. Dis. 27, 81–83. doi: 10.1086/514644
Jacoby, G. A. (2009). AmpC β-lactamases. Clin. Microbiol. Rev. 22, 161–182. doi: 10.1128/CMR.00036-08
Jacoby, G. A., and Sutton, L. (1991). Properties of plasmids responsible for production of extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 35, 164–169. doi: 10.1128/AAC.35.1.164
Jarvis, W. R., Munn, V. P., Highsmith, A. K., Culver, D. H., and Hughes, J. M. (1985). The epidemiology of nosocomial infections caused by Klebsiella pneumoniae. Infect. Control 6, 68–74. doi: 10.1017/S0195941700062639
Jayol, A., Nordmann, P., Brink, A., and Poirel, L. (2015). Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 59, 2780–2784. doi: 10.1128/AAC.05055-14
Jayol, A., Poirel, L., Brink, A. M., Villegas, V., Yilmaz, M., and Nordmann, P. (2014). Resistance to colistin associated with a single amino acid change in protein PmrB among Klebsiella pneumoniae isolates of worldwide origin. Antimicrob. Agents Chemother. 58, 4762–4766. doi: 10.1128/AAC.00084-14
Jerke, K. H., Lee, M. J., and Humphries, R. M. (2016). Polymyxin susceptibility testing: a cold case reopened. Clin. Microbiol. Newslett. 38, 69–77. doi: 10.1016/j.clinmicnews.2016.04.003
Jiao, Y., Qin, Y., Liu, J., Li, Q., Dong, Y., Shang, Y., et al. (2015). Risk factors for carbapenem-resistant Klebsiella pneumoniae infection/colonization and predictors of mortality: a retrospective study. Pathog. Glob. Health 109, 68–74. doi: 10.1179/2047773215Y.0000000004
Johannsen, E. C., Sifri, C. D., and Madoff, L. C. (2000). Pyogenic liver abscesses. Infect Dis. Clin. North Am. 14, 547–563. doi: 10.1016/S0891-5520(05)70120-3
Journet, L., and Cascales, E. (2016). The type VI secretion system in Escherichia coli and related species. EcoSal Plus. doi: 10.1128/ecosalplus.ESP-0009-2015
Kaczmarek, F. M., Dib-Hajj, F., Shang, W., and Gootz, T. D. (2006). High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 β-lactamase production, Porin OmpK35/36 Insertional Inactivation, and Down-Regulation of the phosphate transport Porin pho. Antimicrob. Agents Chemother. 50, 3396–3406. doi: 10.1128/AAC.00285-06
Kalanuria, A. A., Zai, W., and Mirski, M. (2014). Ventilator-associated pneumonia in the ICU. Crit. Care 18:208. doi: 10.1186/cc13775
Kang, C. I., Kim, S. H., Bang, J. W., Kim, H. B., Kim, N. J., Kim, E. C., et al. (2006). Community-acquired versus nosocomial Klebsiella pneumoniae Bacteremia: clinical features, treatment outcomes, and clinical implication of antimicrobial resistance. J. Korean Med. Sci. 21, 816–822. doi: 10.3346/jkms.2006.21.5.816
Kelly, J. A., Dideberg, O., Charlier, P., Wery, J. P., Libert, M., Moews, P. C., et al. (1986). On the origin of bacterial resistance to penicillin: comparison of a b-lactamase and a penicillin target. Science 231, 1429–1431. doi: 10.1126/science.3082007
Keynan, Y., Karlowsky, J. A., Walus, T., and Rubinstein, E. (2007). Pyogenic liver abscess caused by hypermucoviscous Klebsiella pneumoniae. Scand. J. Infect. Dis. 39, 828–830. doi: 10.1080/00365540701266763
Khater, F., Balestrino, D., Charbonnel, N., Dufayard, J. F., Brisse, S., and Forestier, C. (2015). In silico analysis of usher encoding genes in Klebsiella pneumoniae and characterization of their role in adhesion and colonization. PLoS ONE 10:e0116215–e0116224. doi: 10.1371/journal.pone.0116215
Kitchel, B., Rasheed, J. K., Patel, J. B., Srinivasan, A., Navon-Venezia, S., Carmeli, Y., et al. (2009). Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob. Agents Chemother. 53, 3365–3370. doi: 10.1128/AAC.00126-09
Kloos, W. E., and Musselwhite, M. S. (1975). Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl. Microbiol. 30, 381–385.
Knothe, H., Shah, P., Krcmery, V., Antal, M., and Mitsuhashi, S. (1983). Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11, 315–317. doi: 10.1007/BF01641355
Ko, W. C., Paterson, D. L., Sagnimeni, A. J., Hansen, D. S., Von Gottberg, A., Mohapatra, S., et al. (2002). Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg. Infect. Dis. 8, 160–166. doi: 10.3201/eid0802.010025
Kofteridis, D. P., Valachis, A., Dimopoulou, D., Maraki, S., Christidou, A., Mantadakis, E., et al. (2014). Risk factors for carbapenem-resistant Klebsiella pneumoniae infection/colonization: a case-case-control study. J. Infect. Chemother. 20, 293–297. doi: 10.1016/j.jiac.2013.11.007
Kumarasamy, K. K., Toleman, M. A., Walsh, T. R., Bagaria, J., Butt, F., Balakrishnan, R., et al. (2010). Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect. Dis. 10, 597–602. doi: 10.1016/S1473-3099(10)70143-2
Lascols, C., Peirano, G., Hackel, M., Laupland, K. B., and Pitout, J. D. D. (2013). Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-Like enzymes in North America. Antimicrob. Agents Chemother. 57, 130–136. doi: 10.1128/AAC.01686-12
Lau, H. Y., Clegg, S., and Moore, T. A. (2007). Identification of Klebsiella pneumoniae genes uniquely expressed in a strain virulent using a murine model of bacterial pneumonia. Microb. Pathog. 42, 148–155. doi: 10.1016/j.micpath.2007.01.001
Lau, H. Y., Huffnagle, G. B., and Moore, T. A. (2008). Host and microbiota factors that control Klebsiella pneumoniae mucosal colonization in mice. Microbes Infect. 10, 1283–1290. doi: 10.1016/j.micinf.2008.07.040
Lauretti, L., Riccio, M. L., Mazzariol, A., Cornaglia, G., Amicosante, G., Fontana, R., et al. (1999). Cloning and characterization of bla(VIM), a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents chemother. 43, 1584–1590.
Lautenbach, E., Patel, J. B., Bilker, W. B., Edelstein, P. H., and Fishman, N. O. (2001). Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin. Infect. Dis. 32, 1162–1171. doi: 10.1086/319757
Lawlor, M. S., O'Connor, C., and Miller, V. L. (2007). Yersiniabactin is a virulence factor for Klebsiella pneumoniae during pulmonary infection. Infect. Immun. 75, 1463–1472. doi: 10.1128/IAI.00372-06
Lederman, E. R., and Crum, N. F. (2005). Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am. J. Gastroenterol. 100, 322–331. doi: 10.1111/j.1572-0241.2005.40310.x
Lee, S. S., Chen, Y. S., Tsai, H. C., Wann, S. R., Lin, H. H., Huang, C. K., et al. (2008). Predictors of septic metastatic infection and mortality among patients with Klebsiella pneumoniae liver abscess. Clin. Infect. Dis. 47, 642–650. doi: 10.1086/590932
Lery, L. M., Frangeul, L., Tomas, A., Passet, V., Almeida, A. S., Bialek-Davenet, S., et al. (2014). Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 12:41. doi: 10.1186/1741-7007-12-41
Limbago, B. M., Rasheed, J. K., Anderson, K. F., Zhu, W., Kitchel, B., Watz, N., et al. (2011). IMP-producing carbapenem-resistant Klebsiella pneumoniae in the United States. J. Clin. Microbiol. 49, 4239–4245. doi: 10.1128/JCM.05297-11
Lin, Y. T., Siu, L. K., Lin, J. C., Chen, T. L., Tseng, C. P., Yeh, K. M., et al. (2012). Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy chinese and overseas chinese adults in Asian countries. BMC Microbiol. 12:13. doi: 10.1186/1471-2180-12-13
Liu, L., Ye, M., Li, X., Li, J., Deng, Z., Yao, Y. F., et al. (2017). Identification and characterization of an antibacterial type VI secretion system in the carbapenem-resistant strain Klebsiella pneumoniae HS11286. Front. Cell. Infect. Microbiol. 7:442. doi: 10.3389/fcimb.2017.00442
Liu, P., Li, P., Jiang, X., Bi, D., Xie, Y., Tai, C., et al. (2012). Complete genome sequence of Klebsiella pneumoniae subsp. pneumoniae HS11286, a multidrug-resistant strain isolated from human sputum. J. Bacteriol. 194, 1841–1842. doi: 10.1128/JB.00043-12
Liu, Y. C., Cheng, D. L., and Lin, C. L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146, 1913–1916. doi: 10.1001/archinte.1986.00360220057011
Liu, Y. M., Li, B. B., Zhang, Y. Y., Zhang, W., Shen, H., Li, H., et al. (2014). Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob. Agents Chemother. 58, 5379–5385. doi: 10.1128/AAC.02523-14
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Shen, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Long, S. W., Linson, S. E., Ojeda Saavedra, M., Cantu, C., Davis, J. J., Brettin, T., et al. (2017a). Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2:e00290–17. doi: 10.1128/mSphereDirect.00290-17
Long, S. W., Olsen, R. J., Eagar, T. N., Beres, S. B., Zhao, P., Davis, J. J., et al. (2017b). Population genomic analysis of 1,777 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates, Houston, Texas: unexpected abundance of clonal group 307. MBio 8:e00489-17. doi: 10.1128/mBio.00489-17
Lye, W. C., Chan, R. K. T., Lee, E. J. C., and Kumarasinghe, G. (1992). Urinary tract infections in patients with diabetes mellitus. J. Infect. 24, 169–174. doi: 10.1016/0163-4453(92)92876-K
Maatallah, M., Vading, M., Kabir, M. H., Bakhrouf, A., Kalin, M., Naucler, P., et al. (2014). Klebsiella variicola is a frequent cause of bloodstream infection in the Stockholm area, and associated with higher mortality compared to K. pneumoniae. PLoS ONE 9:e113539. doi: 10.1371/journal.pone.0113539
Magill, S. S., Edwards, J. R., Bamberg, W., Beldavs, Z. G., Dumyati, G., Kainer, M. A., et al. (2014). Multistate point-prevalence survey of health care–associated infections. N. Engl. J. Med. 370, 1198–1208. doi: 10.1056/NEJMoa1306801
Mandell, G. L. (2005). Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. Philadelphia, PA: Elsevier.
Martin, R. M., Cao, J., Brisse, S., Passet, V., Wu, W., Zhao, L., et al. (2016). Molecular epidemiology of colonizing and infecting isolates of Klebsiella pneumoniae. mSphere 1:e00261-16. doi: 10.1128/mSphere.00261-16
Massova, I., and Mobashery, S. (1998). Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob. Agents Chemother. 42, 1–17.
Matsen, J. M., Spindler, J. A., and Blosser, R. O. (1974). Characterization of Klebsiella isolates from natural receiving waters and comparison with human isolates. Appl. Microbiol. 28, 672–678.
McCabe, R., Lambert, L., and Frazee, B. (2010). Invasive Klebsiella pneumoniae infections, California, USA. Emerging Infect. Dis. 16, 1490–1491. doi: 10.3201/eid1609.100386
McGann, P., Snesrud, E., Maybank, R., Corey, B., Ong, A. C., Clifford, R., et al. (2016). Escherichia coli harboring mcr-1 and blaCTX-M on a novel IncF plasmid: first report of mcr-1 in the United States. Antimicrob. Agents Chemother. 60, 4420–4421. doi: 10.1128/AAC.01103-16
Meatherall, B. L., Gregson, D., Ross, T., Pitout, J. D. D., and Laupland, K. B. (2009). Incidence risk factors, and outcomes of Klebsiella pneumoniae bacteremia. Am. J. Med. 122, 866–873. doi: 10.1016/j.amjmed.2009.03.034
Medeiros, A. A. (1984). β-Lactamases. Br. Med. Bull. 40, 18–27. doi: 10.1093/oxfordjournals.bmb.a071942
Merino, S., Camprubí, S., Albertí, S., Benedí, V. J., and Tomás, J. M. (1992). Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect. Immun. 60, 2529–2535.
Meroueh, S. O., Minasov, G., Lee, W., Shoichet, B. K., and Mobashery, S. (2003). Structural aspects for evolution of β-lactamases from penicillin-binding proteins. J. Am. Chem. Soc. 125, 9612–9618. doi: 10.1021/ja034861u
Moeck, G. S., and Coulton, J. W. (1998). TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport†. Mol. Microbiol. 28, 675–681. doi: 10.1046/j.1365-2958.1998.00817.x
Montgomerie, J. Z. (1979). Epidemiology of Klebsiella and hospital-associated infections. Rev. Infect. Dis. 1, 736–753. doi: 10.1093/clinids/1.5.736
Montgomerie, J. Z., and Ota, J. K. (1980). Klebsiella bacteremia. Arch. Intern. Med. 140, 525–527. doi: 10.1001/archinte.1980.00330160085032
Murphy, C. N., Mortensen, M. S., Krogfelt, K. A., and Clegg, S. (2013). Role of Klebsiella pneumoniae type 1 and type 3 fimbriae in colonizing silicone tubes implanted into the bladders of mice as a model of catheter-associated urinary tract infections. Infect. Immun. 81, 3009–3017. doi: 10.1128/IAI.00348-13
Nadasy, K. A., Domiati-Saad, R., and Tribble, M. A. (2007). Invasive Klebsiella pneumoniae syndrome in North America. Clin. Infect. Dis. 45, e25–e28. doi: 10.1086/519424
Nassif, X., and Sansonetti, P. J. (1986). Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54, 603–608.
Nathisuwan, S., Burgess, D. S., and Lewis, J. S. II. (2001). Extended-spectrum beta-lactamases: epidemiology, detection, and treatment. Pharmacotherapy 21, 920–928. doi: 10.1592/phco.21.11.920.34529
Navone, L., Casati, P., Licona-Cassani, C., Marcellin, E., Nielsen, L. K., Rodriguez, E., et al. (2014). Allantoin catabolism influences the production of antibiotics in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 98, 351–360. doi: 10.1007/s00253-013-5372-1
Nicoletti, A. G., Fehlberg, L. C. C., Picão, R. C., Machado, O. A., and Gales, A. C. (2012). Clonal complex 258, the most frequently found multilocus sequence type complex in KPC-2-producing Klebsiella pneumoniae isolated in Brazilian hospitals. Antimicrob. Agents Chemother. 56, 4563–4564. doi: 10.1128/AAC.00219-12
O'Brien, I. G., and Gibson, F. (1970). The structure of enterochelin and related 2, 3-dihydroxy-N-benzoylserine conjugates from Escherichia coli. Biochim. Biophys. Acta 215, 393–402. doi: 10.1016/0304-4165(70)90038-3
Olaitan, A. O., Diene, S. M., Kempf, M., Berrazeg, M., Bakour, S., Gupta, S. K., et al. (2014). Worldwide emergence of colistin resistance in Klebsiella pneumoniae from healthy humans and patients in Lao PDR, Thailand, Israel, Nigeria and France owing to inactivation of the PhoP/PhoQ regulator mgrB: an epidemiological and molecular study. Int. J. Antimicrob. Agents 44, 500–507. doi: 10.1016/j.ijantimicag.2014.07.020
Ørskov, I., and Ørskov, F. (1984). 4 serotyping of Klebsiella. Methods Microbiol. 14, 143–164. doi: 10.1016/S0580-9517(08)70449-5
Osano, E., Arakawa, Y., Wacharotayankun, R., Ohta, M., Horii, T., Ito, H., et al. (1994). Molecular characterization of an enterobacterial metallo beta-lactamase found in a clinical isolate of Serratia marcescens that shows imipenem resistance. Antimicrob. Agents Chemother. 38, 71–78. doi: 10.1128/AAC.38.1.71
Ou, H. Y., He, X., Harrison, E. M., Kulasekara, B. R., Thani, A. B., Kadioglu, A., et al. (2007). MobilomeFINDER: web-based tools for in silico and experimental discovery of bacterial genomic islands. Nucleic Acids Res. 35, W97–W104. doi: 10.1093/nar/gkm380
Padilla, E., Llobet, E., Doménech-Sánchez, A., Martínez-Martínez, L., Bengoechea, J. A., and Albertí, S. (2010). Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob. Agents Chemother. 54, 177–183. doi: 10.1128/AAC.00715-09
Pamp, S. J., Gjermansen, M., Johansen, H. K., and Tolker-Nielsen, T. (2008). Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68, 223–240. doi: 10.1111/j.1365-2958.2008.06152.x
Pan, Y. J., Lin, T. L., Chen, C. T., Chen, Y. Y., Hsieh, P. F., Hsu, C. R., et al. (2015). Genetic analysis of capsular polysaccharide synthesis gene clusters in 79 capsular types of Klebsiella spp. Sci. Rep. 5:15573. doi: 10.1038/srep15573
Papo, N., and Shai, Y. (2005). A molecular mechanism for lipopolysaccharide protection of Gram-negative bacteria from antimicrobial peptides. J. Biol. Chem. 280, 10378–10387. doi: 10.1074/jbc.M412865200
Pastagia, M., and Arumugam, V. (2008). Klebsiella pneumoniae liver abscesses in a public hospital in Queens, New York. Travel Med. Infect. Dis. 6, 228–233. doi: 10.1016/j.tmaid.2008.02.005
Podschun, R. (1990). Phenotypic properties of Klebsiella pneumoniae and K. oxytoca isolated from different sources. Zentralbl. Hyg. Umweltmed. 189, 527–535.
Podschun, R., Fischer, A., and Ullmann, U. (1992). Siderophore production of Klebsiella species isolated from different sources. Zentralbl. Bakteriol. 276, 481–486. doi: 10.1016/S0934-8840(11)80673-0
Podschun, R., Pietsch, S., Höller, C., and Ullmann, U. (2001). Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl. Environ. Microbiol. 67, 3325–3327. doi: 10.1128/AEM.67.7.3325-3327.2001
Podschun, R., and Ullmann, U. (1996). Bacteriocin typing of Klebsiella spp. isolated from different sources. Zentralbl. Hyg. Umweltmed. 198, 258–264.
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603.
Poirel, L., Heritier, C., Tolun, V., and Nordmann, P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48, 15–22. doi: 10.1128/AAC.48.1.15-22.2004
Poirel, L., Jayol, A., Bontron, S., Villegas, M. V., Ozdamar, M., Turkoglu, S., et al. (2015). The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J. Antimicrob. Chemother. 70, 75–80. doi: 10.1093/jac/dku323
Pollack, J. R., and Neilands, J. B. (1970). Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem. Biophys. Res. Commun. 38, 989–992. doi: 10.1016/0006-291X(70)90819-3
Pollack, M., Charache, P., Nieman, R. E., Jett, M. P., Reimhardt, J. A., and Hardy, P. H. Jr. (1972). Factors influencing colonisation and antibiotic-resistance patterns of gram-negative bacteria in hospital patients. Lancet 2, 668–671. doi: 10.1016/S0140-6736(72)92084-3
Pomakova, D. K., Hsiao, C. B., Beanan, J. M., Olson, R., MacDonald, U., Keynan, Y., et al. (2012). Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur. J. Clin. Microbiol. Infect. Dis. 31, 981–989. doi: 10.1007/s10096-011-1396-6
Pournaras, S., Ikonomidis, A., Tzouvelekis, L. S., Tokatlidou, D., Spanakis, N., Maniatis, A. N., et al. (2005). VIM-12, a novel plasmid-mediated metallo-β-lactamase from Klebsiella pneumoniae that resembles a VIM-1/VIM-2 hybrid. Antimicrob. Agents Chemother. 49, 5153–5156. doi: 10.1128/AAC.49.12.5153-5156.2005
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Quan, J., Li, X., Chen, Y., Jiang, Y., Zhou, Z., Zhang, H., et al. (2017). Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect. Dis. 17, 400–410. doi: 10.1016/S1473-3099(16)30528-X
Quinn, J. P., Miyashiro, D., Sahm, D., Flamm, R., and Bush, K. (1989). Novel plasmid-mediated beta-lactamase (TEM-10) conferring selective resistance to ceftazidime and aztreonam in clinical isolates of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 33, 1451–1456. doi: 10.1128/AAC.33.9.1451
Richards, M. J., Edwards, J. R., Culver, D. H., and Gaynes, R. P. (2000). Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 21, 510–515. doi: 10.1086/501795
Roger, T., Froidevaux, C., Le Roy, D., Reymond, M. K., Chanson, A. L., Mauri, D., et al. (2009). Protection from lethal gram-negative bacterial sepsis by targeting Toll-like receptor 4. Proc. Natl. Acad. Sci. U.S.A. 106, 2348–2352. doi: 10.1073/pnas.0808146106
Rose, H. D., and Schreier, J. (1968). The effect of hospitalization and antibiotic therapy on the gram-negative fecal flora. Am. J. Med. Sci. 255, 228–236. doi: 10.1097/00000441-196804000-00003
Rosenblueth, M., Martínez, L., Silva, J., and Martínez-Romero, E. (2004). Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst. Appl. Microbiol. 27, 27–35. doi: 10.1078/0723-2020-00261
Russo, T. A., Olson, R., MacDonald, U., Beanan, J., and Davidson, B. A. (2015). Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun. 83, 3325–3333. doi: 10.1128/IAI.00430-15
Samuelsen, O., Naseer, U., Tofteland, S., Skutlaberg, D. H., Onken, A., Hjetland, R., et al. (2009). Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J. Antimicrob. Chemother. 63, 654–658. doi: 10.1093/jac/dkp018
Sarris, P. F., Zoumadakis, C., Panopoulos, N. J., and Scoulica, E. V. (2011). Distribution of the putative type VI secretion system core genes in Klebsiella spp. Infect. Genet. Evol. 11, 157–166. doi: 10.1016/j.meegid.2010.09.006
Schroll, C., Barken, K. B., Krogfelt, K. A., and Struve, C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. doi: 10.1186/1471-2180-10-179
Selden, R., Lee, S., Wang, W. L., Bennett, J. V., and Eickhoff, T. C. (1971). Nosocomial klebsiella infections: intestinal colonization as a reservoir. Ann. Intern. Med. 74, 657–664. doi: 10.7326/0003-4819-74-5-657
Selina, F., Talha, K. A., Islam, A., Hasan, Z., Hyder, M., and Selvapandian, S. (2014). Organisms associated with ventilator associated pneumonia (VAP) in intensive care units (ICU). J. BSA 22, 72–77. doi: 10.3329/jbsa.v22i2.18146
Shafer, W. M., Qu, X. D., Waring, A. J., and Lehrer, R. I. (1998). Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. U.S.A. 95, 1829–1833. doi: 10.1073/pnas.95.4.1829
Sheu, S. J., Kung, Y. H., Wu, T. T., Chang, F. P., and Horng, Y. H. (2011). Risk factors for endogenous endophthalmitis secondary to Klebsiella pneumoniae liver abscess: 20-year experience in Southern Taiwan. Retina 31, 2026–2031. doi: 10.1097/IAE.0b013e31820d3f9e
Shon, A. S., Bajwa, R. P., and Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Siu, L. K., Yeh, K. M., Lin, J. C., Fung, C. P., and Chang, F. Y. (2012). Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 12, 881–887. doi: 10.1016/S1473-3099(12)70205-0
Smith, K. D. (2007). Iron metabolism at the host pathogen interface: lipocalin 2 and the pathogen-associated iroA gene cluster. Int. J. Biochem. Cell Biol. 39, 1776–1780. doi: 10.1016/j.biocel.2007.07.003
Sng, C. C., Jap, A., Chan, Y. H., and Chee, S. P. (2008). Risk factors for endogenous Klebsiella endophthalmitis in patients with Klebsiella bacteraemia: a case-control study. Br. J. Ophthalmol. 92, 673–677. doi: 10.1136/bjo.2007.132522
Snitkin, E. S., Zelazny, A. M., Thomas, P. J., Stock, F., Henderson, D. K., Palmore, T. N., et al. (2012). Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 4:148ra116. doi: 10.1126/scitranslmed.3004129
Spach, D. H., Silverstein, F. E., and Stamm, W. E. (1993). Transmission of infection by gastrointestinal endoscopy and bronchoscopy. Ann. Intern. Med. 118, 117–128. doi: 10.7326/0003-4819-118-2-199301150-00008
Struve, C., and Krogfelt, K. A. (2004). Pathogenic potential of environmental Klebsiella pneumoniae isolates. Environ. Microbiol. 6, 584–590. doi: 10.1111/j.1462-2920.2004.00590.x
Taur, Y., Xavier, J. B., Lipuma, L., Ubeda, C., Goldberg, J., Gobourne, A., et al. (2012). Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 55, 905–914. doi: 10.1093/cid/cis580
Trautmann, M., Ruhnke, M., Rukavina, T., Held, T. K., Cross, A. S., Marre, R., et al. (1997). O-antigen seroepidemiology of Klebsiella clinical isolates and implications for immunoprophylaxis of Klebsiella infections. Clin. Diagn. Lab. Immunol. 4, 550–555.
Tsai, S. S., Huang, J. C., Chen, S. T., Sun, J. H., Wang, C. C., Lin, S. F., et al. (2010). Characteristics of Klebsiella pneumoniae bacteremia in community-acquired and nosocomial infections in diabetic patients. Chang Gung Med. J. 33, 532–539.
Tsay, R., Siu, L. K., Fung, C., and Chang, F. (2002). Characteristics of bacteremia between community-acquired and nosocomial klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch. Intern. Med. 162, 1021–1027. doi: 10.1001/archinte.162.9.1021
Tzeng, Y. L., Ambrose, K. D., Zughaier, S., Zhou, X., Miller, Y. K., Shafer, W. M., et al. (2005). Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol. 187, 5387–5396. doi: 10.1128/JB.187.15.5387-5396.2005
van Aartsen, J. J., Stahlhut, S. G., Harrison, E. M., Crosatti, M. H., Ou, Y., Krogfelt, K., et al. (2012). Characterization of a novel chaperone/usher fimbrial operon present on KpGI-5, a methionine tRNA gene-associated genomic island in Klebsiella pneumoniae. BMC Microbiol. 12, 59–59. doi: 10.1186/1471-2180-12-59
van der Bij, A. K., and Pitout, J. D. D. (2012). The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J. Antimicrob. Chemother. 67, 2090–2100. doi: 10.1093/jac/dks214
Viau, R. A., Hujer, A. M., Marshall, S. H., Perez, F., Hujer, K. M., Briceño, D. F., et al. (2012). “Silent” dissemination of Klebsiella pneumoniae isolates bearing K. pneumoniae carbapenemase in a long-term care facility for children and young adults in Northeast Ohio. Clin. Infect. Dis. 54, 1314–1321. doi: 10.1093/cid/cis036
Vogels, G. D., and Van der Drift, C. (1976). Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 40, 403–468.
Wang, J. H., Liu, Y. C., Lee, S. S., Yen, M. Y., Chen, Y. S., Wang, J. H., et al. (1998). Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26, 1434–1438. doi: 10.1086/516369
Wolf, B., Rey, L. C., Moreira, L. B., Milatovic, D., Fleer, A., Verhoef, J., et al. (2001). Carriage of gram-negative bacilli in young Brazilian children with community-acquired pneumonia. Int. J. Infect. Dis. 5, 155–159. doi: 10.1016/S1201-9712(01)90091-8
Wright, M. S., Suzuki, Y., Jones, M. B., Marshall, S. H., Rudin, S. D., van Duin, D., et al. (2015). Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob. Agents Chemother. 59, 536–543. doi: 10.1128/AAC.04037-14
Wu, C. C., Huang, Y. J., Fung, C.-P., and Peng, H. L. (2010). Regulation of the Klebsiella pneumoniae Kpc fimbriae by the site-specific recombinase KpcI. Microbiology 156, 1983–1992. doi: 10.1099/mic.0.038158-0
Yeh, K. M., Kurup, A., Siu, L. K., Koh, Y. L., Fung, C. P., Lin, J. C., et al. (2007). Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J. Clin. Microbiol. 45, 466–471. doi: 10.1128/JCM.01150-06
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Yu, W. L., Ko, W. C., Cheng, K. C., Lee, C. C., Lai, C. C., and Chuang, Y. C. (2008). Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn. Microbiol. Infect. Dis. 62, 1–6. doi: 10.1016/j.diagmicrobio.2008.04.007
Yu, W. L., Ko, W. C., Cheng, K. C., Lee, H. C., Ke, D. S., Lee, C. C., et al. (2006). Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 42, 1351–1358. doi: 10.1086/503420
Zhang, J., van Aartsen, J. J., Jiang, X., Shao, Y., Tai, C., He, X., et al. (2011). Expansion of the known Klebsiella pneumoniae species gene pool by characterization of novel alien DNA islands integrated into tmRNA gene sites. J. Microbiol. Methods 84, 283–289. doi: 10.1016/j.mimet.2010.12.016
Zhang, R., Ou, H. Y., Gao, F., and Luo, H. (2014). Identification of horizontally-transferred genomic islands and genome segmentation points by using the GC profile method. Curr. Genomics 15, 113–121. doi: 10.2174/1389202915999140328163125
Zhang, Y., Zhao, C., Wang, Q., Wang, X., Chen, H., Li, H., et al. (2016). High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob. Agents Chemother. 60, 6115–6120. doi: 10.1128/AAC.01127-16
Keywords: Klebsiella pneumoniae, antibiotic resistance, carbapenem resistance, hypervirulent, colonization, hospital-acquired infection, CRE
Citation: Martin RM and Bachman MA (2018) Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front. Cell. Infect. Microbiol. 8:4. doi: 10.3389/fcimb.2018.00004
Received: 30 September 2017; Accepted: 05 January 2018;
Published: 22 January 2018.
Edited by:
Kathryn Elizabeth Holt, University of Melbourne, AustraliaReviewed by:
Haijian Zhou, National Institute for Communicable Disease Control and Prevention (China CDC), ChinaHong-Yu Ou, Shanghai Jiao Tong University, China
Sylvain Brisse, Institut Pasteur, France
Copyright © 2018 Martin and Bachman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael A. Bachman, bWlrZWJhY2hAbWVkLnVtaWNoLmVkdQ==
 Rebekah M. Martin
Rebekah M. Martin Michael A. Bachman
Michael A. Bachman