- Ribogenetics Biochemistry Lab, Department of Life Sciences and Chemistry, Jacobs University Bremen, Bremen, Germany
Viral infection had not been observed for amoebae, until the Acanthamoeba polyphaga mimivirus (APMV) was discovered in 2003. APMV belongs to the nucleocytoplasmatic large DNA virus (NCLDV) family and infects not only A. polyphaga, but also other professional phagocytes. Here, we review the Megavirales to give an overview of the current members of the Mimi- and Marseilleviridae families and their structural features during amoebal infection. We summarize the different steps of their infection cycle in A. polyphaga and Acanthamoeba castellani. Furthermore, we dive into the emerging field of virophages, which parasitize upon viral factories of the Megavirales family. The discovery of virophages in 2008 and research in recent years revealed an increasingly complex network of interactions between cell, giant virus, and virophage. Virophages seem to be highly abundant in the environment and occupy the same niches as the Mimiviridae and their hosts. Establishment of metagenomic and co-culture approaches rapidly increased the number of detected virophages over the recent years. Genetic interaction of cell and virophage might constitute a potent defense machinery against giant viruses and seems to be important for survival of the infected cell during mimivirus infections. Nonetheless, the molecular events during co-infection and the interactions of cell, giant virus, and virophage have not been elucidated, yet. However, the genetic interactions of these three, suggest an intricate, multilayered network during amoebal (co-)infections. Understanding these interactions could elucidate molecular events essential for proper viral factory activity and could implicate new ways of treating viruses that form viral factories.
Introduction to Giant Viruses
The discovery of giant viruses in the early 2000s led to a mind shift in the field of virology with respect to the potential origins of viruses (La Scola et al., 2003; Raoult et al., 2004). Originally, viruses were thought of as submicroscopic particles with a self-evident denial that viruses might exist, whose size would be large enough to be resolved with a simple light microscope (Lwoff, 1957; Raoult, 2013). Due to this mindset, the large, gram-positive particles in an Acanthamoeba polyphaga population were at first erroneously classified as bacteria (Birtles et al., 1997; La Scola et al., 2003; Raoult et al., 2007). Only the absence of ribosomal DNA in the presumed bacterium, led to the discovery and definition of the A. polyphaga mimivirus (APMV) in 2003 (La Scola et al., 2003). The acronym mimivirus (for mimicking microbe) reflects the resemblance to bacteria upon gram staining. At the same time, the discovery of APMV was the first ever report of a virus infecting amoebae. Amongst other features that are detailed below, APMV is unusual as it contains a large genome of 1.14 Mbp, thereby even surpassing the genome size of some bacterial species (Raoult et al., 2004). APMV particles are characterized by an up to 700 nm large capsid (Figure 1A), which is well above the resolution of a simple light microscope. Once it was established that giant DNA viruses of amoebae exist, many more such viruses, belonging to the nucleoplasmatic large DNA viruses (NCLDV) were found in the environment, as well as within a wide range of host organisms from humans, monkeys, and oysters (Boughalmi et al., 2013a; Dornas et al., 2014; Andrade et al., 2015). Ex vivo studies of human cell lines revealed that APMV is capable of infecting myeloid and mononuclear blood cells and interferes with the type I Interferon system (Silva et al., 2014). In addition, a distantly APMV-related NCLDV family member has been shown to productively infect T-lymphocytes under laboratory conditions (Popgeorgiev et al., 2013). In 2008, a small particle called Sputnik 1 (La Scola et al., 2008) was discovered in A. polyphaga, which parasitizes viral factories of giant viruses. Due to the functional similarity to bacteriophages in mediating lateral gene transfer, Sputnik was classified as a virophage (La Scola et al., 2008). Here, we will review the expanding family of virophages and discuss the implications for giant virus reproduction inside amoebae.
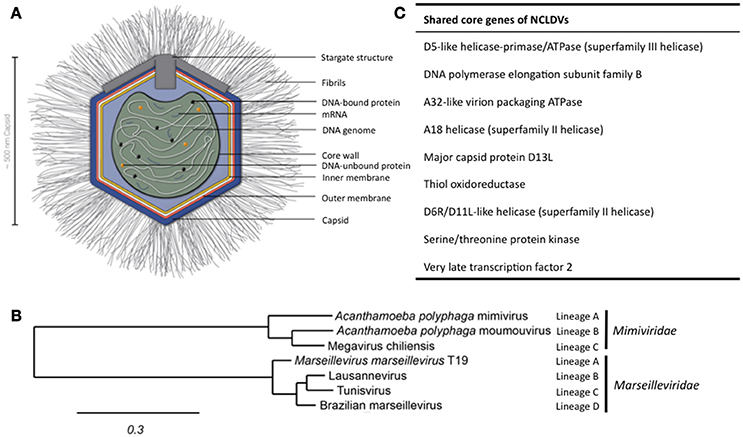
Figure 1. Structure of APMV and the core genes and relationship of giant viruses. (A) Viral particles of APMV feature a viral core with the genome, mRNAs, and prefabricated proteins. This core is surrounded by the indicated membranes and the capsid structure that contains a pentagonal, star-shaped structure termed “stargate,” which is involved in the release of the viral core into the host cell's cytosol upon phagocytosis. The capsid is decorated with a compact layer of fibrils. For details see main text. (B) Cladogram displaying the relationships of the different lineages of the Mimiviridae and Marseilleviridae. Since the discovery of APMV, over 100 new mimivirus strains have been characterized using samples of various origins in amoebal co-culture methods (Pagnier et al., 2013; Khalil et al., 2016a,b). All Mimiviridae share a capsid size between 370 and 600 nm and a 1.02–1.26 Mb AT-rich genome which encodes about 1.000 putative proteins (Colson et al., 2017). Based on sequence homology, the Mimiviridae can be divided into three distinct lineages: lineage A with APMV as prototype and a total of 18 members, as reviewed recently (Colson et al., 2017), lineage B with the moumouvirus as prototype and four additional members (Yoosuf et al., 2012; Colson et al., 2017), and lineage C with Megavirus chiliensis as prototype and a total of 12 members (Arslan et al., 2011; Colson et al., 2017). The tree was created using the sequences of the D13 major capsid proteins of the indicated prototype viruses using Phylogeny.fr, with the relative evolutionary distance indicated (Dereeper et al., 2008, 2010). (C) List of nine genes conserved throughout all NCLDV families.
The Diverse Families of Giant Viruses that Infect Amoebae
The discovery of APMV sparked the interest in giant viruses and spawned a contemporary research field of its own (La Scola et al., 2003). Up until today, two giant virus families belonging to the NCLDV have been described that primarily infect amoebae: the Mimiviridae and the Marseilleviridae (Figure 1B). The latter has the A. polyphaga marseillevirus (APMaV) as founding member, which was discovered in 2009 (Boyer et al., 2009; Colson et al., 2013). In the last decade, nine additional viruses have been associated with the Marseilleviridae group (Colson et al., 2017). The Acanthamoeba castellani lausannevirus (ACLaV) was discovered by incubating water from the Seine river in France with A. castellani, a close relative of A. polyphaga (Thomas et al., 2011). ACLaV is the first known giant virus to encode histone-like proteins, which could point towards a DNA packaging mechanism similar to eukaryotes (Thomas et al., 2011). The Cannes 8 virus (Ca8V) (La Scola et al., 2010) and the Senegal virus (SNGV) (Lagier et al., 2012) have been isolated using similar co-culture methods and are grouped with the Marseilleviridae. The icosahedral capsid of the Marseilleviridae is between 190 and 250 nm in diameter (Colson et al., 2013). Like the genome of the Mimiviridae, the 370,000 bp dsDNA genome is encased in a lipid bilayer and encodes about 450 proteins (Boyer et al., 2009; Scola et al., 2010; Thomas et al., 2011; Lagier et al., 2012). Both, Mimiviridae and Marseilleviridae, share only nine core genes with all NCLDVs (Figure 1C) and 180 genes are shared with at least two of the NCLDV families (Yutin et al., 2009; Yutin and Koonin, 2012). Based on the discovery of APMV and its complex genome, it was suggested to incorporate viruses into the tree of life by defining them as capsid-encoding organisms contrary to the ribosome-encoding organisms, which are represented by eukarya, bacteria, and archaea (Raoult and Forterre, 2008).
APMV—The Best Studied Giant Virus of Amoebae
APMV was the first giant virus to be discovered (La Scola et al., 2008) and confronted the scientific community with features never observed in a virus before. Its capsid size and genetic complexity with many genes usually found in eukaryotic and prokaryotic cells challenged the Lwoff's characteristics of a virus (Raoult et al., 2004; Raoult and Forterre, 2008). The AT-rich 1.14 Mbp APMV genome features an impressive number of 979 protein-encoding genes in a dense arrangement (Raoult et al., 2004; Legendre et al., 2011). Several of its genes are only found in giant viruses of amoebae and code for virus-atypical proteins involved in DNA repair, protein folding, tRNA synthesis and translation, and more (Raoult et al., 2004). In addition, the APMV genome displays some plasticity and encodes self-splicing introns, inteins, and a specific set of mobile genetic elements called transpovirons (Desnues et al., 2012). Furthermore, the genome contains many genes likely acquired via horizontal gene transfer, paralogous genes, and so called ORFans, genes that encode proteins with unknown function (Suhre, 2005; Filée et al., 2007; Moreira and Brochier-Armanet, 2008; Forterre, 2010). Many of these genes are shared with the poxviruses, phycodnaviruses, and other NCLDVs (Filée et al., 2007). ORFans represent roughly 50% of genes and about 40% of the APMV proteome, which results in a high number of factors with unknown functions that might act during viral replication and morphogenesis (Renesto et al., 2006). Alike “classical” viruses, APMV genes are partly under the control of early and late stage-specific promoters (Raoult et al., 2004; Suhre et al., 2005).
The APMV particles possess remarkable structural features, separating them from the classical structures of viruses (Figure 1A). In its center, the viral DNA, mRNAs and proteins are packed into the core compartment (Xiao et al., 2009; Kuznetsov et al., 2013) and enclosed by a lipid membrane. Among the pre-packed proteins are 12 enzymes involved in transcription, five in DNA repair, two in RNA modification, and five in protein modification (Renesto et al., 2006). The central compartment is surrounded by an approximately 340 nm-large lipid bilayer and a secondary bilayer directly underneath an icosahedral capsid. This is comprised of major capsid proteins and features a five-branch proteinaceous structure, the “stargate,” at one vortex (Kuznetsov et al., 2013). The capsid itself is covered by a compact layer of about 120–140 nm long, heavily glycosylated fibrils, which potentially facilitate the attachment of APMV to its host cells (Rodrigues et al., 2015).
As of now, only four fiber associated proteins (FAP1-4) have been functionally associated with either fibril biosynthesis or as components of the fibrils (Sobhy et al., 2015). FAP1 is an aryl alcohol oxidase, which catalyzes the degradation of lignin or lignin derivatives. This suggests that APMV might also be able to infect lignin-containing algae (Klose et al., 2015; Rodrigues et al., 2015). However, the fibrils and associated proteins are not essential for the productive infection of amoebae: during long-term intraamoebal culture (150 generations), the responsible genes are lost (Boyer et al., 2011; Rodrigues et al., 2015). This indicates that the genomic complexity of APMV might be maintained to allow for a broad host range. If so, only a subset of its diverse molecular tools would come in use to enter and infect individual hosts.
Infection Cycles of Giant Viruses in Amoebae
Even though the replication cycle of most giant viruses differ in aspects like nuclear involvement, duration, assembly, and release of the viral progeny, key steps in the infection appear to be conserved, as summarized recently (Colson et al., 2017). For example, all known giant viruses enter the host cell by phagocytosis and release their DNA into the cytosol in a similar manner (Ghigo et al., 2008). Furthermore, viral replication takes place in specialized endoplasmatic reticulum (ER)-derived compartments that are found in the cytosol and are called viral factories (Xiao et al., 2009; Mutsafi et al., 2010; Kuznetsov et al., 2013).
After uptake, the virus resides in a de-novo phagosome. Subsequently, the phagosomal and viral membranes fuse, which allows the release of the viral core, that contains the genome, proteins, and mRNAs into the cytosol (Zauberman et al., 2008; Mutsafi et al., 2010). Alike the well-described poxvirus (Broyles, 2003), the structural integrity of the viral core seems to be retained until viral factories arise (Claverie et al., 2009; Mutsafi et al., 2010). Intriguingly, recent experiments suggest that viral transcription might be initiated already before the release of the viral core (Mutsafi et al., 2014). Once in the cytosol, replication of the viral genome begins immediately and the expression of early stage genes leads to the formation of early viral factories (Suzan-Monti et al., 2007; Mutsafi et al., 2013, 2014). The replication cycle is confined to the cytosol, again a trait shared with the poxvirus (La Scola et al., 2003; Claverie et al., 2009). This also suggests that giant viruses (like the poxvirus) must carry transcription complexes to initiate transcription immediately after infection (Resch et al., 2007; Claverie et al., 2009). In later stages of infection, these viral factories merge into one large cytosolic compartment for replication and capsid assembly (Suzan-Monti et al., 2007; Mutsafi et al., 2014). It should be noted that viral factories are not chaotic, but rather appear to feature distinct assembly lines for their progeny. The viral factory is made up of functional regions playing discrete roles in replication, capsid assembly, DNA packaging, and attachment of fibrils (Suzan-Monti et al., 2007; Mutsafi et al., 2014). In the outermost layer of the viral factory, the internal membrane layers of APMV are assembled from host-derived membrane vesicles, which are thought to rupture, thereby forming open single-layer membrane sheets (Mutsafi et al., 2013). Capsid assembly occurs around these membrane sheets and is scaffolded by the major capsid protein L425 (Mutsafi et al., 2013). Upon capsid formation, the genome is deposited into the empty viral particle through a transient interstice distal from the “stargate” structure (Zauberman et al., 2008). There is little evidence for a nuclear stage of giant viruses. However, the nuclei of A. polyphaga and A. castellani exhibit transient changes in their morphology during the early stages of infection with members of the Marseilleviridae family (Arantes et al., 2016). This indicates that nuclear host factors might play a role in the APMV replication, a notion that is supported by a two-fold decrease of the nuclear size in infected A. polyphaga cells (Colson et al., 2017). This might be due to a substantial redistribution of nuclear factors for viral replication, transcription or other processes (Colson et al., 2017). Albeit indirectly, this scenario is supported by data on the cytoplasmic replication of the Vaccinia virus (a poxvirus), to which mimivirus replication bears similarities (Mutsafi et al., 2010) and for which the involvement of nuclear enzymes has been demonstrated (Oh and Broyles, 2005).
Virophages as Parasites of the Megavirales
The description of Megavirales infection of amoebae was followed by the discovery of the fascinating virophage Sputnik in 2008 (La Scola et al., 2008). Sputnik was found infecting the viral factories of the mamavirus, a close relative of APMV (La Scola et al., 2008). Replication of the Sputnik virophages inside APMV-infected A. castellani cells is deleterious to APMV replication and results in abortive DNA replication and disruption of capsid biogenesis (La Scola et al., 2008). There is an ongoing discussion on the classification of virophages, that are denoted in several articles as satellite viruses (Krupovic and Cvirkaite-Krupovic, 2011; Blanc et al., 2015; Koonin and Krupovic, 2017). Satellite viruses are characterized by their dependency on factors of a helper virus. However, the Sputnik genomes itself encodes factors involved in viral replication (La Scola et al., 2008), suggesting that Sputnik can be classified as a virus, rather than a defective viral particle or sub-viral agent (Fischer, 2011; Desnues and Raoult, 2012).
All known members of the virophage family parasitizing on giant viruses are categorized into the large virus-dependent or -associated (Lavida-)viridae family that is divided into the Sputnikvirus and Mavirus genera (Krupovic et al., 2016). At the species level, the Sputnikvirus genus can be differentiated into the APMV-dependent Sputnik virophage and the APMV-dependent Zamilon virophage (Table 1), while Mavirus genus contains only the Cafeteria roenbergensis virus (CroV)-dependent mavirus (Krupovic et al., 2016).
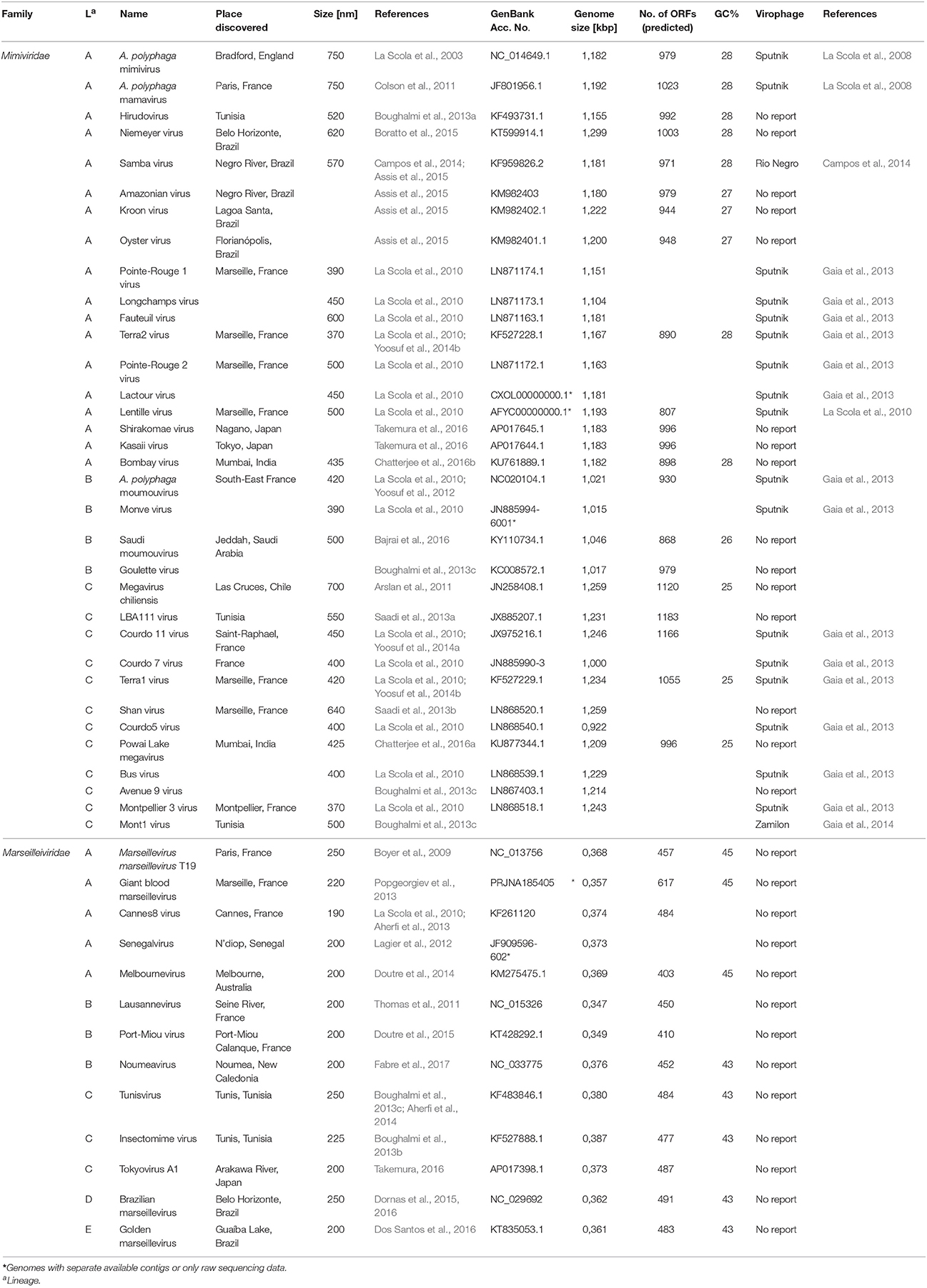
Table 1. Overview of the Mimiviridae and Marseilleviridae families and experimentally shown virophage infections.
Virophage replication has been extensively studied in particular for Sputnik, Zamilon and mavirus. Studies on amoebae infected with different mimiviruses revealed that Sputnik virophages can parasitize mimiviruses from all Mimiviridae lineages but apparently not the Marseilleviridae lineages (Gaia et al., 2013). Sputnik replicates inside mamavirus-infected A. castellani cells within the viral factories, nonetheless, with different kinetics as the mamavirus and at multiple hot spots inside the factory (La Scola et al., 2008). In APMVs viral factories, Sputnik infection results in the emergence of newly generated particles 6 h post infection with a concomitant decrease of infective APMV particles (Ogata and Claverie, 2008). The 18,343-kilobase circular dsDNA genome of Sputnik possesses 21 partly overlapping open-reading frames (ORFs) encoding for several factors involved in DNA replication (La Scola et al., 2008). Interestingly, four of the ORFs are strongly homologous to APMV-encoded genes (La Scola et al., 2008; Gaia et al., 2013). Since Sputnik virophages encodes a lambda-type integrase, the molecular tools for genomic integration are present (La Scola et al., 2008). Indeed, an integration of the Sputnik genome into the genome of the Lentille virus, a relative of APMV, could be observed experimentally (Desnues et al., 2012). There is no indication of Sputnik genome integration into the host cell genome, in line with the lack of indications for a nuclear phase.
The Zamilon virophage (belonging to the Sputnikvirus genus) was discovered together with the Mont1 mimivirus in soil samples from Tunisia (Boughalmi et al., 2013a; Gaia et al., 2014). The 60 nm-wide, spherical virophage carries a 17,276 bp dsDNA genome encoding 20 genes. Although Zamilon shares 76% of its genomic sequence with Sputnik, Zamilon can only infect lineages B and C (Gaia et al., 2014). Furthermore, the tv_L8 protein, encoded in the transpovirons of the Monve mimivirus, shares significant homology with the ORF8-encoded protein of Zamilon (Gaia et al., 2014). This suggests that an exchange of genetic material can in principle occur between the giant virus and the Zamilon virophage within co-infected amoebae, although this has not been observed experimentally so far.
The Maverick-related virus (mavirus), lonely member of the Mavirus genus, parasitizes the viral factories of CroV that infects the marine heterotrophic nanoflagellate C. roenbergensis (Fischer et al., 2010; Fischer and Suttle, 2011). Although this review is predominantly concerned with infection of amoebae, mavirus is included here for its unique features for a virophage. Its 19,063 bp circular genome possesses 20 ORFs including a retroviral integrase, an unsual, protein-primed DNA polymerase, plus four additional proteins, all of which are also found conserved in Maverick/Polinton (MP) retroelements (Fischer and Suttle, 2011; Krupovic et al., 2014, 2016). Additionally, the termini of the mavirus genome consist of long terminal repeats similar to those found in MP retroelements (Yutin et al., 2013; Krupovic et al., 2016). Both findings suggest that these retroelements might have originated from mavirus genome integration events in mavirus co-infected cells (Fischer and Suttle, 2011; Krupovic et al., 2016). Nonetheless, this hypothesis for the origins of MP retroelements remains to be tested experimentally. Fischer and Hackl (2016) succeeded to monitor the integration of mavirus into the C. roenbergensis genome by co-infection with a low multiplicity of infection of CroV. Intriguingly, genes in the mavirus genome possess promoter sequences similar to the late stage promoter of CroV (Fischer and Hackl, 2016). As a consequence, re-infection of C. roenbergensis carrying the integrated mavirus genome with CroV resulted in inhibition of CroV DNA replication, concomitantly with an increased survival of C. roenbergensis (Fischer and Hackl, 2016).
Other virophages have been discovered by metagenomic analysis of water samples [e.g., the Organic Lake virophage (Yau et al., 2011), the Yellowstone Lake virophages (Zhou et al., 2013, 2015)]. However, the viral and cellular host for these remain to be determined (Krupovic et al., 2016), unlike the situation of the Rio Negro virophage that has the Samba virus as viral host (Campos et al., 2014).
Outlook
Since the discovery of its first member APMV in 2003, new giant viruses are discovered continuously in samples from all over the world and added to the Megavirales family. The addition of virophages as parasites of giant viruses, their high abundance in the environment, and the genetic interactions between cell, giant virus, and virophage, suggest an intricate, multilayered network during amoebal co- and super-infections. Future studies of these dynamic interactions could elucidate the inner mechanics of viral factories.
Author Contributions
All authors designed the mini review. JD: generated the figures and drafted the text; MH and CH: wrote the manuscript.
Funding
This work is supported by a grant of the Tönjes-Vagt-Stiftung Bremen, Germany.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aherfi, S., Boughalmi, M., Pagnier, I., Fournous, G., La Scola, B., Raoult, D., et al. (2014). Complete genome sequence of Tunisvirus, a new member of the proposed family Marseilleviridae. Arch. Virol. 159, 2349–2358. doi: 10.1007/s00705-014-2023-5
Aherfi, S., Pagnier, I., Fournous, G., Raoult, D., La Scola, B., and Colson, P. (2013). Complete genome sequence of Cannes 8 virus, a new member of the proposed family “Marseilleviridae.” Virus Genes 47, 550–555. doi: 10.1007/s11262-013-0965-4
Andrade, K. R., Boratto, P. P. V. M., Rodrigues, F. P., Silva, L. C. F., Dornas, F. P., Pilotto, M. R., et al. (2015). Oysters as hot spots for mimivirus isolation. Arch. Virol. 160, 477–482. doi: 10.1007/s00705-014-2257-2
Arantes, T. S., Rodrigues, R. A. L., Dos Santos Silva, L. K., Oliveira, G. P., Souza, H. L. D., Khalil, J. Y., et al. (2016). The large Marseillevirus explores different entry pathways by forming giant infectious vesicles. J. Virol. 90, 5246–5255. doi: 10.1128/JVI.00177-16
Arslan, D., Legendre, M., Seltzer, V., Abergel, C., and Claverie, J.-M. (2011). Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc. Natl. Acad. Sci. U.S.A. 108, 17486–17491. doi: 10.1073/pnas.1110889108
Assis, F. L., Bajrai, L., Abrahao, J. S., Kroon, E. G., Dornas, F. P., Andrade, K. R., et al. (2015). Pan-genome analysis of Brazilian lineage A amoebal mimiviruses. Viruses 7, 3483–3499. doi: 10.3390/v7072782
Bajrai, L. H., de Assis, F. L., Azhar, E. I., Jardot, P., Robert, C., Abrahão, J., et al. (2016). Saudi Moumouvirus, the first group B mimivirus isolated from Asia. Front. Microbiol. 7:2029. doi: 10.3389/fmicb.2016.02029
Birtles, R. J., Rowbotham, T. J., Storey, C., Marrie, T. J., and Raoult, D. (1997). Chlamydia-like obligate parasite of free-living amoebae. Lancet 349, 925–926. doi: 10.1016/S0140-6736(05)62701-8
Blanc, G., Gallot-Lavallée, L., and Maumus, F. (2015). Provirophages in the Bigelowiella genome bear testimony to past encounters with giant viruses. Proc. Natl. Acad. Sci. U.S.A. 112, E5318–E5326. doi: 10.1073/pnas.1506469112
Boratto, P. V., Arantes, T. S., Silva, L. C. F., Assis, F. L., Kroon, E. G., La Scola, B., et al. (2015). Niemeyer virus: a new mimivirus group A isolate harboring a set of duplicated aminoacyl-tRNA synthetase genes. Front. Microbiol. 6:1256. doi: 10.3389/fmicb.2015.01256
Boughalmi, M., Pagnier, I., Aherfi, S., Colson, P., Raoult, D., and La Scola, B. (2013a). First isolation of a giant virus from wild Hirudo medicinalis leech: Mimiviridae isolation in Hirudo medicinalis. Viruses 5, 2920–2930. doi: 10.3390/v5122920
Boughalmi, M., Pagnier, I., Aherfi, S., Colson, P., Raoult, D., and La Scola, B. (2013b). First isolation of a Marseillevirus in the Diptera Syrphidae Eristalis tenax. Intervirology 56, 386–394. doi: 10.1159/000354560
Boughalmi, M., Saadi, H., Pagnier, I., Colson, P., Fournous, G., Raoult, D., et al. (2013c). High-throughput isolation of giant viruses of the Mimiviridae and Marseilleviridae families in the Tunisian environment. Environ. Microbiol. 15, 2000–2007. doi: 10.1111/1462-2920.12068
Boyer, M., Azza, S., Barrassi, L., Klose, T., Campocasso, A., Pagnier, I., et al. (2011). Mimivirus shows dramatic genome reduction after intraamoebal culture. Proc. Natl. Acad. Sci. U.S.A. 108, 10296–10301. doi: 10.1073/pnas.1101118108
Boyer, M., Yutin, N., Pagnier, I., Barrassi, L., Fournous, G., Espinosa, L., et al. (2009). Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. U.S.A. 106, 21848–21853. doi: 10.1073/pnas.0911354106
Broyles, S. S. (2003). Vaccinia virus transcription. J. Gen. Virol. 84, 2293–2303. doi: 10.1099/vir.0.18942-0
Campos, R. K., Boratto, P. V., Assis, F. L., Aguiar, E. R. G. R., Silva, L. C. F., Albarnaz, J. D., et al. (2014). Samba virus: a novel mimivirus from a giant rain forest, the Brazilian Amazon. Virol. J. 11:95. doi: 10.1186/1743-422X-11-95
Chatterjee, A., Ali, F., Bange, D., and Kondabagil, K. (2016a). Complete genome sequence of a new megavirus family member isolated from an Inland Water Lake for the First Time in India. Genome Announc. 4:e00402-16. doi: 10.1128/genomeA.00402-16
Chatterjee, A., Ali, F., Bange, D., and Kondabagil, K. (2016b). Isolation and complete genome sequencing of Mimivirus bombay, a Giant Virus in sewage of Mumbai, India. Genomics Data 9, 1–3. doi: 10.1016/j.gdata.2016.05.013
Claverie, J.-M., Grzela, R., Lartigue, A., Bernadac, A., Nitsche, S., Vacelet, J., et al. (2009). Mimivirus and Mimiviridae: giant viruses with an increasing number of potential hosts, including corals and sponges. J. Invertebr. Pathol. 101, 172–180. doi: 10.1016/j.jip.2009.03.011
Colson, P., La Scola, B., Levasseur, A., Caetano-Anollés, G., and Raoult, D. (2017). Mimivirus: leading the way in the discovery of giant viruses of amoebae. Nat. Rev. Microbiol. 15, 243–254. doi: 10.1038/nrmicro.2016.197
Colson, P., Pagnier, I., Yoosuf, N., Fournous, G., La Scola, B., and Raoult, D. (2013). “Marseilleviridae,” a new family of giant viruses infecting amoebae. Arch. Virol. 158, 915–920. doi: 10.1007/s00705-012-1537-y
Colson, P., Yutin, N., Shabalina, S. A., Robert, C., Fournous, G., La Scola, B., et al. (2011). Viruses with More Than 1,000 Genes: Mamavirus, a New Acanthamoeba polyphaga mimivirus Strain, and Reannotation of Mimivirus Genes. Genome Biol. Evol. 3, 737–742. doi: 10.1093/gbe/evr048
Dereeper, A., Audic, S., Claverie, J.-M., and Blanc, G. (2010). BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 10:8. doi: 10.1186/1471-2148-10-8
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Desnues, C., La Scola, B., Yutin, N., Fournous, G., Robert, C., Azza, S., et al. (2012). Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc. Natl. Acad. Sci. U.S.A. 109, 18078–18083. doi: 10.1073/pnas.1208835109
Desnues, C., and Raoult, D. (2012). Virophages question the existence of satellites. Nat. Rev. Microbiol. 10:234. doi: 10.1038/nrmicro2676-c3
Dornas, F. P., Assis, F. L., Aherfi, S., Arantes, T., Abrahão, J. S., Colson, P., et al. (2016). A Brazilian Marseillevirus is the founding member of a lineage in family Marseilleviridae. Viruses 8:76. doi: 10.3390/v8030076
Dornas, F. P., Khalil, J. Y. B., Pagnier, I., Raoult, D., Abrahão, J., and La Scola, B. (2015). Isolation of new Brazilian giant viruses from environmental samples using a panel of protozoa. Front. Microbiol. 6:1086. doi: 10.3389/fmicb.2015.01086
Dornas, F. P., Rodrigues, F. P., Boratto, P. V. M., Silva, L. C. F., Ferreira, P. C. P., Bonjardim, C. A., et al. (2014). Mimivirus circulation among wild and domestic mammals, Amazon Region, Brazil. Emerging Infect. Dis. 20, 469–472. doi: 10.3201/eid2003.131050
Dos Santos, R. N., Campos, F. S., Medeiros De Albuquerque, N. R., Finoketti, F., Côrrea, R. A., Cano-Ortiz, L., et al. (2016). A new marseillevirus isolated in Southern Brazil from Limnoperna fortunei. Sci. Rep. 6:35237. doi: 10.1038/srep35237
Doutre, G., Arfib, B., Rochette, P., Claverie, J.-M., Bonin, P., and Abergel, C. (2015). Complete genome sequence of a new member of the Marseilleviridae recovered from the Brackish submarine spring in the Cassis Port-Miou Calanque, France. Genome Announc. 3:e01148-15. doi: 10.1128/genomeA.01148-15
Doutre, G., Philippe, N., Abergel, C., and Claverie, J.-M. (2014). Genome analysis of the first Marseilleviridae representative from Australia indicates that most of its genes contribute to virus fitness. J. Virol. 88, 14340–14349. doi: 10.1128/JVI.02414-14
Fabre, E., Jeudy, S., Santini, S., Legendre, M., Trauchessec, M., Couté, Y., et al. (2017). Noumeavirus replication relies on a transient remote control of the host nucleus. Nat. Commun. 8:15087. doi: 10.1038/ncomms15087
Filée, J., Siguier, P., and Chandler, M. (2007). I am what I eat and I eat what I am: acquisition of bacterial genes by giant viruses. Trends Genet. 23, 10–15. doi: 10.1016/j.tig.2006.11.002
Fischer, M. G. (2011). Sputnik and Mavirus: more than just satellite viruses. Nat. Rev. Microbiol. 10:78. doi: 10.1038/nrmicro2676-c1
Fischer, M. G., Allen, M. J., Wilson, W. H., and Suttle, C. A. (2010). Giant virus with a remarkable complement of genes infects marine zooplankton. Proc. Natl. Acad. Sci. U.S.A. 107, 19508–19513. doi: 10.1073/pnas.1007615107
Fischer, M. G., and Hackl, T. (2016). Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature 540:288. doi: 10.1038/nature20593
Fischer, M. G., and Suttle, C. A. (2011). A virophage at the origin of large DNA transposons. Science 332:231. doi: 10.1126/science.1199412
Forterre, P. (2010). Giant viruses: conflicts in revisiting the virus concept. Intervirology 53, 362–378. doi: 10.1159/000312921
Gaia, M., Benamar, S., Boughalmi, M., Pagnier, I., Croce, O., Colson, P., et al. (2014). Zamilon, a novel virophage with Mimiviridae host specificity. PLoS ONE 9:e94923. doi: 10.1371/journal.pone.0094923
Gaia, M., Pagnier, I., Campocasso, A., Fournous, G., Raoult, D., and La Scola, B. (2013). Broad spectrum of Mimiviridae virophage allows its isolation using a mimivirus reporter. PLoS ONE 8:e61912. doi: 10.1371/journal.pone.0061912
Ghigo, E., Kartenbeck, J., Lien, P., Pelkmans, L., Capo, C., Mege, J.-L., et al. (2008). Ameobal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathog. 4:e1000087. doi: 10.1371/journal.ppat.1000087
Khalil, J. Y. B., Andreani, J., and La Scola, B. (2016a). Updating strategies for isolating and discovering giant viruses. Curr. Opin. Microbiol. 31, 80–87. doi: 10.1016/j.mib.2016.03.004
Khalil, J. Y. B., Robert, S., Reteno, D. G., Andreani, J., Raoult, D., and La Scola, B. (2016b). High-throughput isolation of giant viruses in liquid medium using automated flow cytometry and fluorescence staining. Front. Microbiol. 7:26. doi: 10.3389/fmicb.2016.00026
Klose, T., Herbst, D. A., Zhu, H., Max, J. P., Kenttämaa, H. I., and Rossmann, M. G. (2015). A mimivirus enzyme that participates in viral entry. Structure 23, 1058–1065. doi: 10.1016/j.str.2015.03.023
Koonin, E. V., and Krupovic, M. (2017). Polintons, virophages and transpovirons: a tangled web linking viruses, transposons and immunity. Curr. Opin. Virol. 25, 7–15. doi: 10.1016/j.coviro.2017.06.008
Krupovic, M., Bamford, D. H., and Koonin, E. V. (2014). Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses. Biol. Direct 9:6. doi: 10.1186/1745-6150-9-6
Krupovic, M., and Cvirkaite-Krupovic, V. (2011). Virophages or satellite viruses? Nat. Rev. Microbiol. 9:762. doi: 10.1038/nrmicro2676
Krupovic, M., Kuhn, J. H., and Fischer, M. G. (2016). A classification system for virophages and satellite viruses. Arch. Virol. 161, 233–247. doi: 10.1007/s00705-015-2622-9
Kuznetsov, Y. G., Klose, T., Rossmann, M., and Mcpherson, A. (2013). Morphogenesis of mimivirus and its viral factories: an atomic force microscopy study of infected cells. J. Virol. 87, 11200–11213. doi: 10.1128/JVI.01372-13
Lagier, J.-C., Armougom, F., Million, M., Hugon, P., Pagnier, I., Robert, C., et al. (2012). Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 18, 1185–1193. doi: 10.1111/1469-0691.12023
La Scola, B., Audic, S., Robert, C., Jungang, L., Lamballerie, X. D., Drancourt, M., et al. (2003). A giant virus in amoebae. Science 299:2033. doi: 10.1126/science.1081867
La Scola, B., Campocasso, A., N'dong, R., Fournous, G., Barrassi, L., Flaudrops, C., et al. (2010). Tentative characterization of new environmental giant viruses by MALDI-TOF mass spectrometry. Intervirology 53, 344–353. doi: 10.1159/000312919
La Scola, B., Desnues, C., Pagnier, I., Robert, C., Barrassi, L., Fournous, G., et al. (2008). The virophage as a unique parasite of the giant mimivirus. Nature 455, 100–104. doi: 10.1038/nature07218
Legendre, M., Santini, S., Rico, A., Abergel, C., and Claverie, J.-M. (2011). Breaking the 1000-gene barrier for Mimivirus using ultra-deep genome and transcriptome sequencing. Virol. J. 8:99. doi: 10.1186/1743-422X-8-99
Lwoff, A. (1957). The concept of virus. J. Gen. Microbiol. 17, 239–253. doi: 10.1099/00221287-17-2-239
Moreira, D., and Brochier-Armanet, C. (2008). Giant viruses, giant chimeras: the multiple evolutionary histories of Mimivirus genes. BMC Evol. Biol. 8:12. doi: 10.1186/1471-2148-8-12
Mutsafi, Y., Fridmann-Sirkis, Y., Milrot, E., Hevroni, L., and Minsky, A. (2014). Infection cycles of large DNA viruses: emerging themes and underlying questions. Virology 466–467, 3–14. doi: 10.1016/j.virol.2014.05.037
Mutsafi, Y., Shimoni, E., Shimon, A., and Minsky, A. (2013). Membrane assembly during the infection cycle of the giant Mimivirus. PLoS Pathog. 9:e1003367. doi: 10.1371/journal.ppat.1003367
Mutsafi, Y., Zauberman, N., Sabanay, I., and Minsky, A. (2010). Vaccinia-like cytoplasmic replication of the giant Mimivirus. Proc. Natl. Acad. Sci. U.S.A. 107, 5978–5982. doi: 10.1073/pnas.0912737107
Ogata, H., and Claverie, J.-M. (2008). How to infect a mimivirus. Science 321:1305. doi: 10.1126/science.1164839
Oh, J., and Broyles, S. S. (2005). Host cell nuclear proteins are recruited to cytoplasmic vaccinia virus replication complexes. J. Virol. 79, 12852–12860. doi: 10.1128/JVI.79.20.12852-12860.2005
Pagnier, I., Reteno, D.-G. I., Saadi, H., Boughalmi, M., Gaia, M., Slimani, M., et al. (2013). A decade of improvements in Mimiviridae and Marseilleviridae isolation from amoeba. Intervirology 56, 354–363. doi: 10.1159/000354556
Popgeorgiev, N., Boyer, M., Fancello, L., Monteil, S., Robert, C., Rivet, R., et al. (2013). Marseillevirus-like virus recovered from blood donated by asymptomatic humans. J. Infect. Dis. 208, 1042–1050. doi: 10.1093/infdis/jit292
Raoult, D. (2013). TRUC or the need for a new microbial classification. Intervirology 56, 349–353. doi: 10.1159/000354269
Raoult, D., Audic, S., Robert, C., Abergel, C., Renesto, P., Ogata, H., et al. (2004). The 1.2-megabase genome sequence of Mimivirus. Science 306, 1344–1350. doi: 10.1126/science.1101485
Raoult, D., and Forterre, P. (2008). Redefining viruses: lessons from Mimivirus. Nat. Rev. Microbiol. 6, 315–319. doi: 10.1038/nrmicro1858
Raoult, D., La Scola, B., and Birtles, R. (2007). The discovery and characterization of Mimivirus, the largest known virus and putative pneumonia agent. Clin. Infect. Dis. 45, 95–102. doi: 10.1086/518608
Renesto, P., Abergel, C., Decloquement, P., Moinier, D., Azza, S., Ogata, H., et al. (2006). Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J. Virol. 80, 11678–11685. doi: 10.1128/JVI.00940-06
Resch, W., Hixson, K. K., Moore, R. J., Lipton, M. S., and Moss, B. (2007). Protein composition of the vaccinia virus mature virion. Virology 358, 233–247. doi: 10.1016/j.virol.2006.08.025
Rodrigues, R. A. L., Dos Santos Silva, L. K., Dornas, F. P., Oliveira, D. B. D., Magalhães, T. F. F., Santos, D. A., et al. (2015). Mimivirus fibrils are important for viral attachment to the microbial world by a diverse glycoside interaction repertoire. J. Virol. 89, 11812–11819. doi: 10.1128/JVI.01976-15
Saadi, H., Pagnier, I., Colson, P., Cherif, J. K., Beji, M., Boughalmi, M., et al. (2013a). First isolation of mimivirus in a patient with pneumonia. Clin. Infect. Dis. 57, e127–e134. doi: 10.1093/cid/cit354
Saadi, H., Reteno, D. G. I., Colson, P., Aherfi, S., Minodier, P., Pagnier, I., et al. (2013b). Shan virus: a new mimivirus isolated from the stool of a Tunisian Patient with pneumonia. Intervirology 56, 424–429. doi: 10.1159/000354564
Silva, L. C. F., Almeida, G. M. F., Oliveira, D. B., Dornas, F. P., Campos, R. K., La Scola, B., et al. (2014). A resourceful giant: APMV is able to interfere with the human type I interferon system. Microbes Infect. 16, 187–195. doi: 10.1016/j.micinf.2013.11.011
Sobhy, H., La Scola, B., Pagnier, I., Raoult, D., and Colson, P. (2015). Identification of giant Mimivirus protein functions using RNA interference. Front. Microbiol. 6:345. doi: 10.3389/fmicb.2015.00345
Suhre, K. (2005). Gene and genome duplication in Acanthamoeba polyphaga Mimivirus. J. Virol. 79, 14095–14101. doi: 10.1128/JVI.79.22.14095-14101.2005
Suhre, K., Audic, S., and Claverie, J.-M. (2005). Mimivirus gene promoters exhibit an unprecedented conservation among all eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 14689–14693. doi: 10.1073/pnas.0506465102
Suzan-Monti, M., La Scola, B., Barrassi, L., Espinosa, L., and Raoult, D. (2007). Ultrastructural characterization of the giant volcano-like virus factory of Acanthamoeba polyphaga Mimivirus. PLoS ONE 2:e328. doi: 10.1371/journal.pone.0000328
Takemura, M. (2016). Draft genome sequence of tokyovirus, a member of the family Marseilleviridae Isolated from the Arakawa River of Tokyo, Japan. Genome Announc. 4:e00429-16. doi: 10.1128/genomeA.00429-16
Takemura, M., Mikami, T., and Murono, S. (2016). Nearly complete genome sequences of two mimivirus strains isolated from a Japanese Freshwater Pond and River Mouth. Genome Announc. 4:e01378-16. doi: 10.1128/genomeA.01378-16
Thomas, V., Bertelli, C., Collyn, F., Casson, N., Telenti, A., Goesmann, A., et al. (2011). Lausannevirus, a giant amoebal virus encoding histone doublets. Environ. Microbiol. 13, 1454–1466. doi: 10.1111/j.1462-2920.2011.02446.x
Xiao, C., Kuznetsov, Y. G., Sun, S., Hafenstein, S. L., Kostyuchenko, V. A., Chipman, P. R., et al. (2009). Structural studies of the giant mimivirus. PLoS Biol. 7:e92. doi: 10.1371/journal.pbio.1000092
Yau, S., Lauro, F. M., Demaere, M. Z., Brown, M. V., Thomas, T., Raftery, M. J., et al. (2011). Virophage control of antarctic algal host–virus dynamics. Proc. Natl. Acad. Sci. U.S.A. 108, 6163–6168. doi: 10.1073/pnas.1018221108
Yoosuf, N., Pagnier, I., Fournous, G., Robert, C., La Scola, B., Raoult, D., et al. (2014a). Complete genome sequence of Courdo11 virus, a member of the family Mimiviridae. Virus Genes 48, 218–223. doi: 10.1007/s11262-013-1016-x
Yoosuf, N., Pagnier, I., Fournous, G., Robert, C., Raoult, D., La Scola, B., et al. (2014b). Draft genome sequences of Terra1 and Terra2 viruses, new members of the family Mimiviridae isolated from soil. Virology 452–453, 125–132. doi: 10.1016/j.virol.2013.12.032
Yoosuf, N., Yutin, N., Colson, P., Shabalina, S. A., Pagnier, I., Robert, C., et al. (2012). Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage. Genome Biol. Evol. 4, 1324–1330. doi: 10.1093/gbe/evs109
Yutin, N., and Koonin, E. V. (2012). Hidden evolutionary complexity of Nucleo-Cytoplasmic Large DNA viruses of eukaryotes. Virol. J. 9:161. doi: 10.1186/1743-422X-9-161
Yutin, N., Raoult, D., and Koonin, E. V. (2013). Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virol. J. 10:158. doi: 10.1186/1743-422X-10-158
Yutin, N., Wolf, Y. I., Raoult, D., and Koonin, E. V. (2009). Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virol. J. 6:223. doi: 10.1186/1743-422X-6-223
Zauberman, N., Mutsafi, Y., Halevy, D. B., Shimoni, E., Klein, E., Xiao, C., et al. (2008). Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol. 6:e114. doi: 10.1371/journal.pbio.0060114
Zhou, J., Sun, D., Childers, A., Mcdermott, T. R., Wang, Y., and Liles, M. R. (2015). Three novel virophage genomes discovered from Yellowstone Lake metagenomes. J. Virol. 89, 1278–1285. doi: 10.1128/JVI.03039-14
Keywords: Acanthamoeba polyphaga mimivirus (APMV), virophage, nucleocytoplasmatic large DNA virus (NCLDV), mimivirus, pathogen defense
Citation: Diesend J, Kruse J, Hagedorn M and Hammann C (2018) Amoebae, Giant Viruses, and Virophages Make Up a Complex, Multilayered Threesome. Front. Cell. Infect. Microbiol. 7:527. doi: 10.3389/fcimb.2017.00527
Received: 15 October 2017; Accepted: 13 December 2017;
Published: 11 January 2018.
Edited by:
Thierry Soldati, Université de Genève, SwitzerlandReviewed by:
Guruprasad R. Medigeshi, Translational Health Science and Technology Institute, IndiaBernard La Scola, Aix-Marseille University, France
Copyright © 2018 Diesend, Kruse, Hagedorn and Hammann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Hammann, Yy5oYW1tYW5uQGphY29icy11bml2ZXJzaXR5LmRl
 Jan Diesend
Jan Diesend Janis Kruse
Janis Kruse Monica Hagedorn
Monica Hagedorn Christian Hammann
Christian Hammann