- 1National Leading Research Laboratory of Drug Resistance Proteomics, Department of Biological Sciences, Myongji University, Yongin, South Korea
- 2Biotechnology Program, North Shore Community College, Danvers, MA, United States
- 3Department of Systems Biotechnology, College of Biotechnology and Natural Resources, Chung-Ang University, Anseong, South Korea
Klebsiella pneumoniae is one of the most clinically relevant species in immunocompromised individuals responsible for community-acquired and nosocomial infections, including pneumonias, urinary tract infections, bacteremias, and liver abscesses. Since the mid-1980s, hypervirulent K. pneumoniae, generally associated with the hypermucoviscosity phenotype, has emerged as a clinically significant pathogen responsible for serious disseminated infections, such as pyogenic liver abscesses, osteomyelitis, and endophthalmitis, in a generally younger and healthier population. Hypervirulent K. pneumoniae infections were primarily found in East Asia and now are increasingly being reported worldwide. Although most hypervirulent K. pneumoniae isolates are antibiotic-susceptible, some isolates with combined virulence and resistance, such as the carbapenem-resistant hypervirulent K. pneumoniae isolates, are increasingly being detected. The combination of multidrug resistance and enhanced virulence has the potential to cause the next clinical crisis. To better understand the basic biology of hypervirulent K. pneumoniae, this review will provide a summarization and discussion focused on epidemiology, hypervirulence-associated factors, and antibiotic resistance mechanisms of such hypervirulent strains. Epidemiological analysis of recent clinical isolates in China warns the global dissemination of hypervirulent K. pneumoniae strains with extensive antibiotic resistance in the near future. Therefore, an immediate response to recognize the global dissemination of this hypervirulent strain with resistance determinants is an urgent priority.
Introduction
Klebsiella pneumoniae is an opportunistic pathogen in immunocompromised individuals that causes a wide range of infections, including pneumonia, urinary tract infection, bacteremia, and meningitis, such as people suffering from diabetes or malignancies (Paczosa and Mecsas, 2016). K. pneumoniae has a polysaccharide capsule that is important for its pathogenesis and its ability to avoid phagocytosis (Cortés et al., 2002). Over the past two decades, unlike the “classic” K. pneumoniae (cKP), a new “hypervirulent” K. pneumoniae (hvKP) with hypermucoviscosity has emerged as a clinically significant pathogen causing highly invasive infections, such as liver abscesses, in both healthy and immunocompromised individuals (Prokesch et al., 2016). Remarkably, these infections are often complicated by devastating disseminated infections, including endophthalmitis and meningitis (Shon et al., 2013). This ability to disseminate is a common feature of certain Gram-positive pathogens, such as Staphylococcus aureus, but is generally uncommon for enteric Gram-negative bacilli, including K. pneumoniae, in the absence of host susceptibility factors (Cheng et al., 1991; Shon et al., 2013). In addition, unlike cKP infections, approximately half of all hvKP infections occur in young, healthy individuals (Shon et al., 2013; Struve et al., 2015; Paczosa and Mecsas, 2016). The hvKP strain is hypermucoviscous typically because of overproduction of its polysaccharide capsule (Paczosa and Mecsas, 2016). Unlike the typical nosocomial infections of cKP, hvKP can cause serious community-acquired infections in healthy individuals (Patel et al., 2014).
hvKP was reported initially in Taiwan and Southeast Asia in the mid-1980s and 1990s, but more than 30 cases have been reported worldwide (Shon et al., 2013). Although hvKP can cause pneumonias or lung abscesses, it is primarily associated with pyogenic liver abscesses. hvKP is now the most common cause of pyogenic liver abscesses in Asia (Siu et al., 2012; Wang et al., 2013). hvKP is rarely resistant to commonly used antimicrobial agents, except for an intrinsic resistance to ampicillin (Zhang et al., 2016). However, along with the global dissemination of mobile genetic elements conferring antibiotic resistance, antibiotic-resistant hvKP isolates are increasingly being reported (Yao et al., 2015; Zhang et al., 2015b; Zhan et al., 2017). Because carbapenem-resistant hvKP strains may cause severe, untreatable infections in healthy individuals, the emergence of these strains poses a great threat to public health. In this review, we summarize the epidemiology of hvKP isolates and discuss the resistance mechanisms involved.
Epidemiology of hvKP
The hypermucoviscosity/hypermucoviscous phenotype of hvKP is typically due to the increased production of capsular polysaccharide and the presence of specific virulence genes, such as rmpA (Siu et al., 2011; Paczosa and Mecsas, 2016). The hypermucoviscosity phenotype is generally determined by “the string test.” The string test is positive when a viscous string >5 mm in length is formed by stretching bacterial colonies on an agar plate (Fang et al., 2004). Because there are K. pneumoniae strains that are hypervirulent, but not hypermucoviscous (Catalán-Nájera et al., 2017), reports about non-hypermucoviscous hvKP types are included in our review (Table 1 and Table S1).
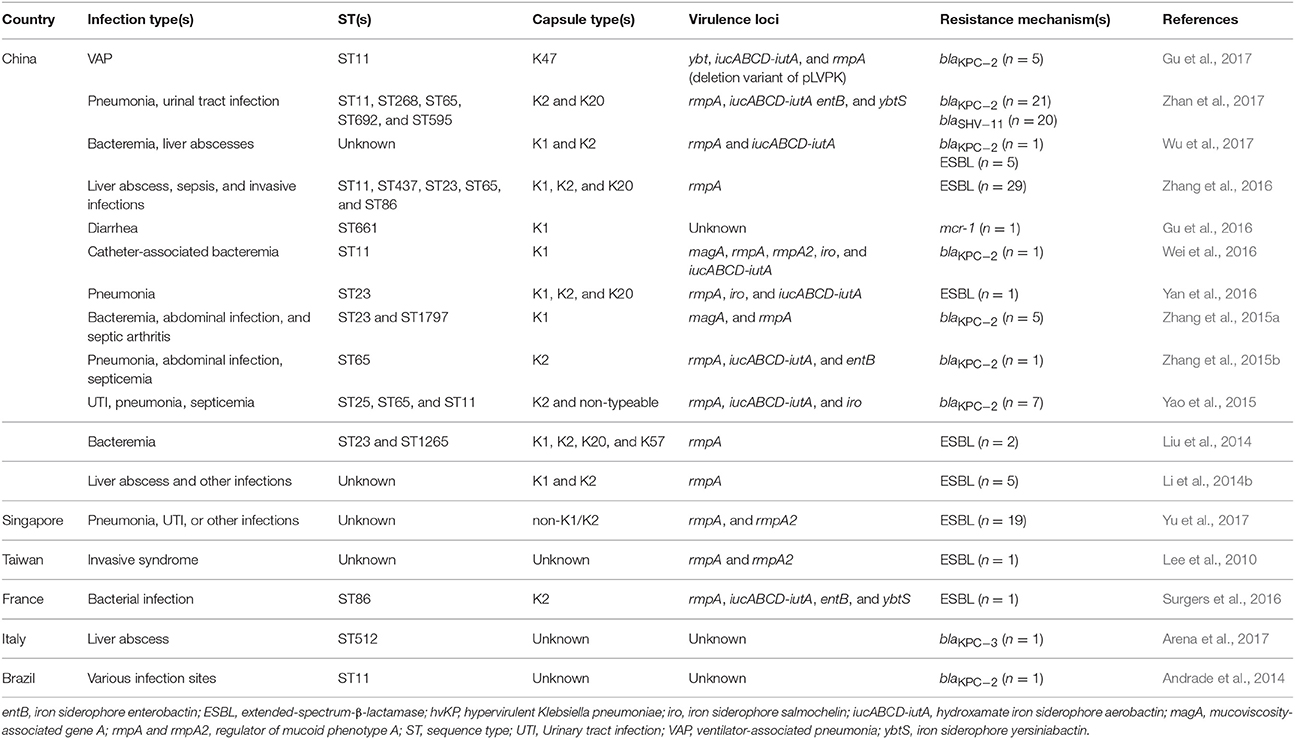
In the 1980s, case reports from Taiwan described community-acquired liver abscesses caused by hvKP in healthy patients with serious concomitant end-organ manifestations, such as meningitis and endophthalmitis (Liu et al., 1986; Chiu et al., 1988). Since the first report in Taiwan, the sporadic spread of hvKP has been observed in many Asian, European, and American countries (Figure 1). Although several cases of hvKP were reported in Europe and the Americas, the epidemic spread of hvKP occurred primarily in Asian countries, including Taiwan, China, South Korea, and Iran (Table S1). For example, in China, 22.8% (84/369) of K. pneumoniae clinical isolates associated with various types of invasive infections were identified as hvKP (Guo et al., 2017). Another report showed that 90.9% of the pathogens causing pyogenic liver abscesses were hvKP (Ye et al., 2016). In South Korea, 42.2% of K. pneumoniae strains isolated from patients with bacteremia were of the hypermucoviscosity phenotype (Jung et al., 2013). In Taiwan, 88.8% of K. pneumoniae isolates collected from patients with community-acquired extrahepatic abscesses had the hypermucoviscosity phenotype (Ku et al., 2008) and 41.5% of community-acquired K. pneumoniae bacteremia were caused by hvKP (Lee et al., 2006). Therefore, K. pneumoniae liver abscess is now considered as an endemic disease in Taiwan. The frequency of hvKP-associated diseases in these countries has continued to increase. Between 1996 and 2004, an almost 60% increase in the annual incidence of liver abscesses caused by hvKP were observed in Taiwan (Tsai et al., 2008). In South Korea, the rate of liver abscesses caused by K. pneumoniae rose 3.3% in the 1970s to 78.2% in the mid-2000s (Chung et al., 2007). A study in Iran showed that 60.95% of K. pneumoniae isolates collected from various patients were hypermucoviscosity-positive (Zamani et al., 2013). Although the reasoning for the hvKP predominance in Asia is unclear (Shon et al., 2013), some reports analyzing the hvKP mechanistic properties have provided clues. A study in 2002 analyzed the phenotypic and genotypic characteristics of K. pneumoniae isolates collected worldwide (Ko et al., 2002). Interestingly, a distinctive form of K. pneumoniae, often causing liver abscess, was identified almost exclusively from Taiwan (Ko et al., 2002), indicating that the spread of a specific hvKP clone may be the major factor of high predominance in Asia. Another analysis showed that hvKP infections in Western countries most commonly occur in Asians (Lederman and Crum, 2005; Keynan et al., 2007; Nadasy et al., 2007; Pastagia and Arumugam, 2008; Frazee et al., 2009; Gunnarsson et al., 2009; McCabe et al., 2010; Sobirk et al., 2010; Decré et al., 2011; Vila et al., 2011; Pomakova et al., 2012; Shon et al., 2013), suggesting that Asians may be more susceptible to hvKP infections than other ethnic groups. Because Asians in Western countries may have traveled to Asia or been exposed to individuals who recently traveled to Asia, these results do not conclusively establish a genetic risk of Asians for hvKP infections.
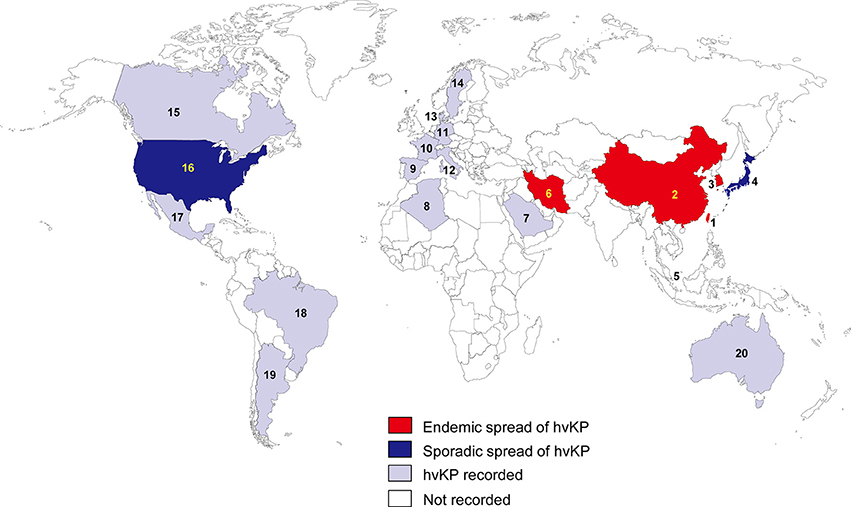
Figure 1. Epidemiological features of hvKP. The endemic spread of hvKP means that multiple outbreaks of hvKP were reported in an indicated region. The sporadic spread of hvKP means that only case studies (no outbreak) were reported in an indicated region. 1, Taiwan; 2, China; 3, South Korea; 4, Japan; 5, Singapore; 6, Iran; 7, Saudi Arabia; 8, Algeria; 9, Spain; 10, France; 11, Germany; 12, Italy; 13, Denmark; 14, Sweden; 15, Canada; 16, United States; 17, Mexico; 18, Brazil; 19, Argentina; 20, Australia.
In other regions of the world, the prevalence of hvKP is not high (Table S1). Studies from other regions are mostly case reports, but there are several studies analyzing various hvKP isolates. For example, a study in Spain showed that 5.4% of K. pneumoniae isolates collected from various infection sites had the hypermucoviscosity phenotype (Cubero et al., 2016). This rate is relatively low, compared with the high frequency (up to 90%) of hvKP in Asia. Similar results were obtained in Canada, Brazil, and Algeria (El Fertas-Aissani et al., 2013; Peirano et al., 2013; Pereira and Vanetti, 2015). In Canada, the hypermucoviscosity phenotype was present in 8.2% of K. pneumoniae isolates collected from patients with community-acquired bacteremia (Peirano et al., 2013). In Brazil, the hypermucoviscosity phenotype was observed in 6.7% of clinical K. pneumoniae isolates (Pereira and Vanetti, 2015). A study in Algeria showed that 9.2% of K. pneumoniae isolates obtained from various patients were hypermucoviscous-positive (El Fertas-Aissani et al., 2013). These results support the theory that the endemic spread of hvKP is mostly in Asian countries.
Factors Associated with Hypervirulence of hvKP
A variety of hypervirulence-associated factors are important in hvKP strains, including capsular serotypes, sequence types, a virulence plasmid, a pathogenicity island, and several virulence factors.
K1 and K2 Capsular Serotypes
The capsule is an important virulence factor of encapsulated pathogens, including K. pneumoniae. Capsular polysaccharides of K. pneumoniae are divided into at least 78 serotypes (Pan et al., 2008; Wyres et al., 2016). Notably, many reports have shown that K1 and K2 serotypes are strongly associated with hvKP (Paczosa and Mecsas, 2016; Catalán-Nájera et al., 2017) (Table 1 and Table S1). In China, among the hypermucoviscous K. pneumoniae clinical isolates associated with various types of invasive infections, including pyogenic liver abscess, 42.9% were the K2 serotype and 23.8% were the K1 serotype (Guo et al., 2017). Another study in China also showed that 68.75% of the hypermucoviscosity-positive K. pneumoniae isolates have the K2 serotype (Zhao et al., 2016). A South Korea study searching K. pneumoniae strains isolated from the urine of hospitalized patients showed that K1 and K2 capsular serotypes were more common in hypermucoviscosity-positive strains than in hypermucoviscosity-negative strains (Kim et al., 2017). Most hypermucoviscous K. pneumoniae isolates from patients with bacteremia also had the K1 or K2 serotype (78%) (Jung et al., 2013). Many hvKP isolates reported in Europe and America also had the K1 or K2 serotype (Table S1).
Why are the K1 and K2 serotypes so prevalent in hvKP? Two studies tried to resolve this question (Yeh et al., 2007; Lin et al., 2014). Serotype K1 and K2 K. pneumoniae isolates demonstrated significantly more virulence in the mouse model than did non-K1/K2 isolates (survival rates of 0 vs. 79.2%, respectively) (Yeh et al., 2007). Typically, K1 and K2 strains are more resistant to phagocytosis and intracellular killing by macrophages and neutrophils than strains with other serotypes, regardless of their hypermucoviscosities (Kabha et al., 1995; Lin et al., 2004; Lee et al., 2014; Paczosa and Mecsas, 2016). Several reasons were suggested for enhanced virulence of the K1 and K2 strains, relative to other strains. First, K1 and K2 strains lack the specific mannose residue repeats that are recognized by the host factors, such as the mannose-binding receptor on macrophage and the lung-secreted surfactant protein A (SP-A) (Kabha et al., 1995; Sahly et al., 2008). Second, K1 and K2 strains have a host-specific monosaccharide sialic acid on their surfaces (Lee et al., 2014), that is known to mimic host cells, thereby allowing evasion of the host immune cells. Third, K1 and K2 strains may induce a smaller release of reactive oxygen species by neutrophils than strains with other serotypes, allowing better survival in human tissues (Sahly et al., 2004; Paczosa and Mecsas, 2016). Fourth, K1 and K2 strains have more diverse O serotypes, compared with strains with other K serotypes (Follador et al., 2016), which may help K1 and K2 strains evade host immune systems. The O serotyping is based on the recognition of distinct variations of O-antigens, the outermost part of the lipopolysaccharide (LPS) (Trautmann et al., 1997).
Although the K1 and K2 serotypes are typically found in hvKP, several strains of cKP also possess these serotypes. At the same time, some hvKP strains have a non-K1 or -K2 serotype (Fang et al., 2007; Shon et al., 2013; Guo et al., 2017). In addition to K1 and K2 capsular serotypes, other capsular serotypes, including K5, K16, K20, K28, K54, K57, K63, and KN1, have also been found in hvKP (Table 1 and Table S1). A study compared the prevalence and importance of virulence factors for K. pneumoniae liver abscesses between isolates with capsular K1/K2 serotypes and isolates with non-K1/K2 serotypes (Yu et al., 2008). When injected into mice intraperitoneally, K. pneumoniae isolates with the hypermucoviscosity phenotype as well as presence of rmpA and iucABCD-iutA genes exhibited high virulence for mouse lethality, regardless of capsular type (Yu et al., 2008). However, in other reports, K1/K2 cKP strains were not significantly less virulent than K1/K2 hvKP strains (Lin et al., 2004), and serotype K1 and K2 isolates demonstrated significantly more phagocytic resistance and virulence than non-K1/K2 strains, regardless of the presence of the rmpA gene (Yeh et al., 2007), suggesting that the presence of the K1/K2 serotypes themselves is responsible for increased virulence. Therefore, these studies indicate that the presence of the K1/K2 serotypes themselves is partially responsible for the hypervirulence of the hvKP strains and the combination with other hypervirulence-associated factors may enhance virulence of K. pneumoniae.
ST23 and CC23 Sequence Types
Recent studies using a 694-gene core genome multilocus sequence typing (MLST) analysis or whole genome-sequencing revealed that K1 hvKP clonal complex 23 (CC23) isolates form a unique clonal lineage, whereas K2 hvKP isolates are genetically more diverse (Bialek-Davenet et al., 2014; Struve et al., 2015). These results indicate that the clonal lineage CC23 has a specific genetic background conferring hypervirulence and fitness. Another study has also revealed that the sequence type 23 (ST23) clone is strongly associated with the K1 capsular serotype in hvKP strains that were isolated specifically from liver abscesses (Siu et al., 2011). Among 47 serotype K1 isolates, 85.1% belonged to ST23, one isolate belonged to ST163 (a single-locus variant of ST23), and two isolates were ST249 (a three-locus variant of ST23) (Siu et al., 2011). In China, ST23 was found to be the most prevalent ST among 69 hypermucoviscous K. pneumoniae isolates and was found only among K1 isolates (Guo et al., 2017). Another study in China showed that 57.8% of K. pneumoniae strains from liver abscess patients belonged to ST23, and 96.2% of ST23 K. pneumoniae isolates were the K1 serotype (Qu et al., 2015). In South Korea, ST23 strains accounted for 97.3% of hvKP strains with the K1 serotype collected from patients with community-acquired liver abscesses (Chung et al., 2008). Similar results were found in many Western countries (Table S1). Reasons for the association of the K1 serotype with ST23 are unclear. Meanwhile, hvKP strains of the K2 serotype are genetically diverse (Shon et al., 2013). A recent study analyzing serotype K2 isolates from three different regions in Asia showed that eight different sequence types were found among serotype K2 isolates (ST65, ST66, ST86, ST373, ST374, ST375, ST380, and ST434) (Lin et al., 2014). There were two major groups, ST65-like (42%) and ST86-like (46%) (Lin et al., 2014). Notably, ST23 hvKP clones with the K1 serotype were associated with pyogenic liver abscess, whereas ST65 hvKP clones with the K2 serotype were correlated with various invasive infections (Guo et al., 2017).
Whole genome-sequencing of hvKP isolates from different geographic origins revealed the epidemiology of the hvKP CC23 expansion. Only minimal geographical grouping of the isolates was observed in the CC23 phylogeny, suggesting that CC23 may spread worldwide through multiple international transmission events rather than by local expansions (Struve et al., 2015). Indeed, infections of hvKP linked to recent travels to Asia were reported in several cases from Western countries (Fang et al., 2005; Gunnarsson et al., 2009; Rivero et al., 2010). However, because there were many cases not linked to travel or close contact with individuals from epidemic regions (Nadasy et al., 2007; Doud et al., 2009; Frazee et al., 2009; Pinsky et al., 2009; Decré et al., 2011; Vila et al., 2011; Merlet et al., 2012; Pomakova et al., 2012; Bilal et al., 2014; Coutinho et al., 2014; Gundestrup et al., 2014; Holmås et al., 2014; Melot et al., 2016; Surgers et al., 2016), an independent local establishment of the CC23 lineage outside of Asia is also possible.
The Virulence Plasmid pLVPK and KPHP1208 Pathogenicity Island
Notably, a large virulence plasmid (pLVPK, a pK2044-like plasmid) was detected in all whole genome-sequenced hvKP clonal lineages (Struve et al., 2015). This plasmid encodes aerobactin, salmochelin, and RmpA, which were found to be restricted to hvKP isolates (Struve et al., 2015). A CC23 strain lacking pLVPK was found to have significantly reduced virulence (Lin et al., 2011, 2012), indicating an important role of this plasmid in the hypervirulence of K. pneumoniae.
Additionally, a genomic comparison revealed a distinct variant of a pathogenicity island (KPHP1208) specifically associated with CC23 (Struve et al., 2015). Most CC23 isolates contain genes encoding yersiniabactin, colibactin, and microcin E492, in the KPHP1208 pathogenicity island (Struve et al., 2015). Colibactin is a polyketide-peptide genotoxin that causes DNA damage in eukaryotic cells (Lai et al., 2014). A recent study using a meningitis mouse model showed that colibactin was necessary for the meningeal tropism of K1 CC23 K. pneumoniae (Lu et al., 2017), indicating the importance of the KPHP1208 genomic island in the hypervirulence of K. pneumoniae.
RmpA and MagA
RmpA (regulator of the mucoid phenotype A gene) activates capsule production, resulting in the hypermucoviscosity phenotype and increase in virulence (Cheng et al., 2010). Three rmpA genes were reported in hvKP strains: two large plasmid-carried genes (p-rmpA and p-rmpA2) located in a virulence plasmid (pLVPK) and one chromosomal gene (c-rmpA) (Wacharotayankun et al., 1993; Hsu et al., 2011). In K. pneumoniae CG43, both p-rmpA and p-rmpA2 contributed to the enhancement of capsular production (Lai et al., 2003; Cheng et al., 2010), whereas in NTUH-K2044 (ST23, K1 serotype), the first genome-sequenced hvKP, only p-rmpA, but not p-rmpA2 or c-rmpA, contributed to the increased expression of capsule genes (Hsu et al., 2011). Therefore, additional studies in other strains are needed to clarify the role of each RmpA in hypervirulence.
Many results have revealed a strong association between RmpA and hypervirulence (Table 1 and Table S1). Whole-genome sequencing of 30 hvKP isolates from different geographic origins revealed that all hvKP were rmpA positive (Struve et al., 2015). Several studies from China also showed that 92–100% of hvKP isolates were positive for rmpA (Liu et al., 2014; Sun et al., 2016; Wu et al., 2017). Regardless of the investigated infection site, most hvKP isolates appear to have the rmpA gene (Ku et al., 2008; Yu et al., 2008; Lee et al., 2010; Decré et al., 2011; Qu et al., 2015; Guo et al., 2016; Zhang et al., 2016; Wu et al., 2017; Zhan et al., 2017). Despite the strong association between hvKP and RmpA, some rmpA-positive isolates did not exhibit the hypermucoviscosity phenotype (Yu et al., 2006). However, recent studies have shown that the lack of hypermucoviscosity and low virulence in some rmpA-positive K. pneumoniae strains were caused by a concurrent mutation of rmpA and rmpA2 genes in the absence of c-rmpA (Yu et al., 2015a,b).
The magA gene was discovered in 2004 as a gene required for the hypercapsular phenotype from K. pneumoniae strains isolated from invasive liver abscesses (Fang et al., 2004). Subsequent works using bioinformatic analysis and genetic experiments determined that magA is the serotype K1 allele of the polymerase gene wzy in the cps gene cluster (Fang et al., 2007; Yeh et al., 2010). Consequentially, magA was renamed wzy_K1 (Fang et al., 2010). A study in Taiwan showed that 83% of K. pneumoniae strains from patients with pyogenic liver abscess were magA positive, whereas only 3% of strains isolated from patients with other invasive infections were magA positive (Chuang et al., 2006). Additionally, all 36 magA-positive strains were identified as serotype K1, whereas the 38 magA-negative strains were not serotype K1 (Chuang et al., 2006). Another study also reported the association of magA with the capsule serotype K1 (Yeh et al., 2006). A recent study showed that magA was found more prevalently in hvKP strains with the K1 serotype than in non-K1 strains (Guo et al., 2017). These results suggest that the magA gene is responsible for the K. pneumoniae capsular serotype K1.
Aerobactin
Iron is a metal that is essential for bacterial growth and plays a crucial role in the progression of infection, including K. pneumoniae infection (Russo et al., 2014). K. pneumoniae can secrete various siderophores (aerobactin, salmochelin, enterobactin, and yersiniabactin) that acquire iron in iron-depleted environments, such as in a human host (Hsieh et al., 2008). The hvKP strains have a 6- to 10-fold increased siderophore activity compared with cKP strains (Russo et al., 2011, 2014). Enterobactin is a siderophore that is produced by more than 90% of examined enterobacterial isolates (Raymond et al., 2003), whereas genes encoding aerobactin and salmochelin are located on a large virulence plasmid (pLVPK) that is not present in most cKP strains (Nassif and Sansonetti, 1986; Struve et al., 2015) and the yersiniabactin locus located within the pathogenicity island of Yersinia species is believed to be acquired via horizontal gene transfer (Bach et al., 2000). Therefore, aerobactin and salmochelin were rarely expressed in cKP strains, but it was present in most hvKP isolates (Paczosa and Mecsas, 2016). Interestingly, although hvKP secretes four different siderophores, a study showed that aerobactin accounts for >90% of the siderophore activity (Russo et al., 2014). Furthermore, a recent study revealed that aerobactin, but not salmochelin, enterobactin, or yersiniabactin, plays a crucial role in the growth and survival of hvKP in human ascites fluid or serum and in the in vivo mouse infection models (Russo et al., 2015), suggesting that aerobactin is a crucial virulence factor of hvKP. Aerobactin is encoded by the iucABCD operon and its cognate receptor is encoded by the iutA gene (Russo et al., 2014).
Many reports showed that an iucABCD-iutA gene was more common in hvKP strains than in cKP strains (Table 1 and Table S1). In China, many studies have reported the high prevalence (69–96%) of aerobactin in hvKP isolates (Guo et al., 2016, 2017; Sun et al., 2016; Yan et al., 2016; Ye et al., 2016; Zhang et al., 2016; Zhao et al., 2016; Wu et al., 2017; Zhan et al., 2017). A study in Taiwan also showed that the prevalence of aerobactin was 100% in capsular K1 and K2 isolates and 86% in non-K1/K2 isolates. Furthermore, in this study, all K. pneumoniae isolates with rmpA and iucABCD-iutA genes in the presence of the hypermucoviscosity phenotype exhibited high virulence for mouse lethality, regardless of capsular type (Yu et al., 2008). When serotype K1 K. pneumoniae isolates from Hong Kong, Singapore, and Taiwan were analyzed, all hypervirulent isolates with a 50% lethal dose of <102 colony-forming units were identified as ST23 and carried both virulence-associated genes, rmpA and iucABCD-iutA (Siu et al., 2011). These results imply the importance of iucABCD-iutA and rmpA genes for hypervirulence. Notably, the expression of rmpA is regulated by the availability of iron and is negatively controlled by Fur, the iron-responsive global regulator (Cheng et al., 2010), implying that the availability of iron regulates the expression of rmpA, which could affect virulence of K. pneumoniae. Therefore, additional studies are needed to clarify the relationship between aerobactin and RmpA in hypervirulence.
Antibiotic Resistance of hvKP
The prevalence of antibiotic resistance in hvKP isolates is rare compared with the high prevalence of antibiotic-resistant cKP isolates (Lee et al., 2016; Paczosa and Mecsas, 2016). The reason for the low prevalence of antibiotic resistance in hvKP strains remains unclear; however, reports of antibiotic-resistant hvKP isolates are increasingly being reported worldwide, mostly in countries with an epidemic dissemination of hvKP (Table 1). Therefore, we will summarize and discuss the epidemiology and resistance mechanisms of antibiotic-resistant hvKP strains.
Antibiotic-Resistant hvKP in Asia
Except for an intrinsic resistance to ampicillin, most hvKP strains are rarely resistant to antibiotics and are generally susceptible to commonly used antimicrobial drugs (Paczosa and Mecsas, 2016; Zhang et al., 2016). However, with the global dissemination of mobile genetic elements harboring various antibiotic resistance genes, including the K. pneumoniae carbapenemase (KPC), NDM, and oxacillinases-48 (OXA-48) types of carbapenemases (Lee et al., 2016), antibiotic-resistant hvKP isolates have begun to emerge in the past few years (Zhang et al., 2015b). In 2010, a study from Taiwan analyzed various features of isolates with and without the hypermucoviscosity phenotype (Lee et al., 2010). The prevalence rates of extended-spectrum β-lactamase (ESBL) were significantly higher in K. pneumoniae isolates without the hypermucoviscosity phenotype (34%) than in isolates with the hypermucoviscosity phenotype (4%), and only one ESBL-producing strain among the 24 isolates with the hypermucoviscosity phenotype was detected (Lee et al., 2010). Several ESBL-producing hvKP strains causing bloodstream infections were found in China (Liu et al., 2014; Yan et al., 2016). Another report in China showed that 17% of hvKP strains expressed ESBL and several hvKP isolates showed resistance to all the tested antimicrobials, except carbapenems and amikacin (Li et al., 2014b). Furthermore, antimicrobial resistance in hvKP strains increased over time (Li et al., 2014b). Like the report in Taiwan, the resistance to antibiotics in China was found to be significantly greater in cKP strains than in hvKP strains (Li et al., 2014b). Similarly, a report in 2016 showed that 12.6% of hvKP isolates from several invasive infections produced ESBLs, and most of them carried blaCTX−M genes (Zhang et al., 2016).
Recently, in China, the prevalence of hvKP in invasive infections was studied in several reports. In a study in 2016, 90.9% of the pathogens causing pyogenic liver abscess were hvKP (Ye et al., 2016). A study analyzing 369 K. pneumoniae isolates associated with various invasive infections revealed that 22.8% (84/369) of the isolates were identified as hvKP (Guo et al., 2017). With a national prevalence of hvKP, several studies in China have revealed the emergence of hvKP that have acquired extensive antibiotic resistances, including carbapenem resistance. In 2015, among 33 patients with carbapenem-resistant K. pneumoniae, carbapenem-resistant hvKP isolates were found in four patients (Yao et al., 2015). From these four patients, seven carbapenem-resistant hvKP isolates were identified and the blaKPC−2 gene was detected in six of the seven hvKP isolates (Yao et al., 2015). Notably, carbapenem-resistant hvKP strains belonged to three sequence types, [ST65 (five isolates), ST25 (one isolate), and ST11 (one isolate)], and two capsular serotypes, [K2 (six isolates) and non-typeable (one isolate)] (Yao et al., 2015). A retrospective study analyzing 28 cases of carbapenem-resistant K. pneumoniae infections from nine cities in China showed a similar result. Among the 28 carbapenem-resistant K. pneumoniae isolates, five carbapenem-resistant hvKP isolates were detected and three isolates had the blaKPC−2 gene (Zhang et al., 2015b). Interestingly, the sequence types of carbapenem-resistant hvKP strains were ST11 (three isolates), ST65 (one isolate), and ST1700 (one isolate). The serotypes were K2 (one isolate) and non-typeable (four isolates) (Zhang et al., 2015b). A recent study analyzed a total of 1,838 K. pneumoniae isolates collected from various patient specimens of patients between 2013 and 2015. Results were similar, identifying 21 carbapenem-resistant hvKP isolates, with all 21 isolates positive for blaKPC−2 (Zhan et al., 2017). Additionally, all the carbapenem-resistant hvKP isolates except one harbored blaSHV−11. Notably, the carbapenem-resistant hvKP strains belonged to five sequence types, [ST11 (16 isolates), ST65 (one isolate), ST268 (two isolates), ST595 (one isolate), and ST692 (one isolate)], and three capsular serotypes, [K20 (six isolates), K2 (one isolate), and non-typeable (14 isolates)] (Zhan et al., 2017). In a 2016 case report in China, the carbapenem-resistant ST11 hvKP strain was detected (Wei et al., 2016). This strain also harbored blaKPC−2. In conclusion, the prevalence of hvKP among carbapenem-resistant K. pneumoniae isolates in China have significantly been high, ranging from 7.4 to 15% (Zhang et al., 2015a; Zhan et al., 2017).
Carbapenem-resistant ST23 hvKP strains with the K1 serotype have not often been detected in China. Most of the identified carbapenem-resistant hvKP strains have the blaKPC−2 gene. However, a study in 2016 revealed the detection of carbapenem-resistant ST23 hvKP strains with the K1 serotype. A retrospective study in 2016 was conducted on 148 meropenem-resistant K. pneumoniae strains collected from two hospitals in Zhejiang Province in 2013, and among these isolates, seven K1, one K2, and five non-K1/K2 hvKP strains were identified by the loop string test (Zhang et al., 2015a). Among them, seven carbapenem-resistant hvKP strains with the K1 serotype belonged to only two sequence types, ST23 (four isolate) and ST1797 (three isolates); five strains harbored blaKPC−2 (Zhang et al., 2015a). In this study, the blaKPC−2 gene also was detected in most carbapenem-resistant hvKP strains (Zhang et al., 2015a). In China, the KPC types of carbapenemases are more prevalent than other global carbapenemases, such as NDM and OXA-48 types (Lee et al., 2016). KPC-2 was the most frequently identified carbapenemase in K. pneumoniae in many countries, including China (Gupta et al., 2011; Munoz-Price et al., 2013; Li et al., 2014a; Lee et al., 2016), and ST11, which is rarely found in hvKP strains (Liu et al., 2014; Zhang et al., 2016; Table 1), is the prevalent clone associated with the spread of KPC-producing K. pneumoniae in Asia (particularly in China and Taiwan) (Qi et al., 2011; Lee et al., 2016). Therefore, these epidemiological features of carbapenemase-producing K. pneumoniae in China imply the emergence of carbapenem-resistant hvKP strains by interaction between cKP and hvKP strains.
In China, the predominant clone of carbapenem-resistant K. pneumoniae is KPC-2-producing ST11 (Qi et al., 2011; Lee et al., 2016) and ST23 is one of the dominant clones of hvKP (Table 1). Among 36 carbapenem-resistant hvKPs identified in China at the present time (Yao et al., 2015; Zhang et al., 2015a,b; Wei et al., 2016; Wu et al., 2017; Zhan et al., 2017), the rates of KPC-2-producing ST11 and KPC-2-producing ST23 clones are 50% (18/36) and 8.3% (3/36), respectively. Notably, KPC-2 is found in 94.4% (34/36) of carbapenem-resistant hvKP isolates. These results imply that the exchange of mobile genetic elements between KPC-2-producing ST11 cKP and ST23 hvKP has occurred. Additionally, because the rate (50%) of KPC-2-producing ST11 hvKP isolates is far higher than that of KPC-2-producing ST23 hvKP (8.3%), the transmission of hypervirulence-related mobile genetic elements, such as the pLVPK plasmid, from ST23 hvKP strains to KPC-2-producing ST11 cKP strains seems to be a prevalent event. Of course, this assumption should be verified by many additional carbapenem-resistant clinical isolates of hvKP, because the number of currently identified isolates is too small (36 isolates). A study in 2014 showed the in vitro transmission and retainment of a plasmid carrying blaKPC−2 from a parental non-serotype ST258 K. pneumoniae to a recipient invasive virulent K2 ST65 K. pneumoniae strain (Siu et al., 2014). However, the in vitro transmission of plasmids with hypervirulence-related genes, such as pLVPK, from the parental serotype K1 ST23 K. pneumoniae to the recipient carbapenem-resistant ST11 K. pneumoniae strain has not been reported yet. Notably, a recent study in China showed that five carbapenem-resistant ST11 hvKP strains had the pLVPK-like plasmid (Gu et al., 2017). Genomic analyses showed that the emergence of these ST11 carbapenem-resistant hvKP strains was due to the acquisition of the pLVPK-like plasmid by ST11 carbapenem-resistant cKP strains (Gu et al., 2017). Therefore, additional studies are needed to verify the transmission of mobile genetic elements between cKP and hvKP strains.
94.4% of carbapenemases found in carbapenem-resistant hvKP isolates in China are KPC-2. KPC-2 was the most prevalent carbapenemase in K. pneumoniae in China (Li et al., 2014a; Lee et al., 2016). The global epidemiology of carbapenem-resistant K. pneumoniae showed that the most prevalent carbapenemase of each country varies (Lee et al., 2016). For example, the most prevalent carbapenemases of Pakistan and Turkey is NDM and OXA-48, respectively (Lee et al., 2016). If the most prevalent carbapenemase of carbapenem-resistant hvKPs in Pakistan and Turkey are identified as NDM and OXA-48, respectively, then these results suggest the necessity of investigating the possibility for the direct transmission of mobile genetic elements between cKP and hvKP. In any case, results from China caution researchers and clinicians of the potential for the emergence and global dissemination of hvKP strains with acquired carbapenem resistance.
Colistin (polymyxin E) is regarded as a key component for the combination antimicrobial therapy used for the treatment of severe carbapenem-resistant K. pneumoniae infections (Lee et al., 2016). However, many recent studies have shown that some strains acquire colistin resistance through LPS modification (Cannatelli et al., 2013, 2014), and the frequency of these colistin-resistant isolates is gradually increasing (Giani et al., 2015; Lee et al., 2016). Particularly, the emergence of a plasmid carrying the mcr-1 gene, which mediates colistin resistance by modifying LPS, was reported in China (Liu et al., 2016). A recent study in China reported the detection of an hvKP isolate harboring the mcr-1 gene (Gu et al., 2016). This isolate was identified from a stool specimen of an infant patient with diarrhea in China and was identified as the ST661 clone with the K1 serotype (Gu et al., 2016). This particular case indicates that hvKP can acquire various types of antibiotic resistances.
A recent study showed that the acquisition of colistin resistance of ST23 hvKP strains was accompanied by reduced capsule production, impaired virulence, and a significant fitness cost (Choi and Ko, 2015). Similar results regarding the effects of colistin resistance on virulence and fitness costs have been observed in other Gram-negative pathogens, including Acinetobacter baumannii and Salmonella enterica (Sun et al., 2009; López-Rojas et al., 2011; Hraiech et al., 2013; Beceiro et al., 2014). Although the effect of antibiotic resistance on virulence and fitness costs in hvKP strains has not been assessed for other antibiotics, including carbapenems, high fitness burdens in antibiotic-resistant hvKP strains may explain partially the low prevalence and restricted spread of antibiotic-resistant hvKP strains.
Antibiotic-Resistant hvKP in Europe and America
A few cases of antibiotic-resistant hvKP were reported in Europe and America (Table 1). In France, an ESBL (blaCTX−M−3)-producing hvKP strain was identified in a patient born in Algeria, who alternately resided in France and Algeria for several years without travel to any other countries (Surgers et al., 2016). The strain was the ST86 clone with the K2 serotype. In Brazil in 2014, a polymyxin B-resistant and KPC-2-producing hvKP was detected (Andrade et al., 2014); the strain was the ST11 clone. KPC-2 is the most prevalent carbapenemase in Brazil (Lee et al., 2016), and the correlation is consistent with cases in China. Recently, a study in Italy showed the detection of a KPC-3-producing hvKP isolate from a 52 year old Caucasian patient with liver abscess (Arena et al., 2017). The sequence type for this strain was ST512 (a single-locus variant of ST258). KPC-type enzymes were the most common carbapenemase in Italy (89.5% of carbapenemase producers), followed by VIM-1 (9.2%) and OXA-48 (1.3%) (Giani et al., 2013; Lee et al., 2016). These results imply the possibility for the direct transmission of mobile genetic elements between cKP and hvKP strains.
Lack of Treatment Options for Carbapenem-Resistant hvKP
Although most hvKP strains are susceptible to antimicrobial drugs, except for ampicillin (Shon et al., 2013), and a fair number of patients with hvKP infection are young with no other illnesses, mortality rates of hvKP infections are high, ranging from 3 to 42% (Han, 1995; Wang et al., 1998; Fang et al., 2000, 2007; Ko et al., 2002; Ku et al., 2008; Decré et al., 2011; Shon et al., 2013). Cases of community-acquired pneumonia with bacteremia and necrotizing fasciitis have presented high mortality rates of 55 and 47%, respectively (Lin et al., 2010; Cheng et al., 2012). Furthermore, survivors of hvKP infections have often suffered catastrophic morbidity such as loss of vision or neurologic sequelae (Liu et al., 1986; Cheng et al., 1991, 2012; Han, 1995; Fang et al., 2007; Shon et al., 2013).
Management of hvKP infections was made more difficult by the emergence of carbapenem-resistant hvKP strains. In cases of 34 patients with carbapenem-resistant hvKPs (Andrade et al., 2014; Yao et al., 2015; Zhang et al., 2015a; Arena et al., 2017; Zhan et al., 2017), 10 patients died and all treatment of 11 patients ceased because of lack of effective antibiotics. Difficulties managing carbapenem-resistant hvKPs could make this strain the next worldwide “superbug” in waiting.
Conclusions
This review summarizes the epidemiology and antibiotic resistance mechanisms of hvKPs. Invasive hvKPs infections have several dangerous features that are uncommon for cKP. First, approximately half of the patients with hvKP infections are young or do not have co-morbidities. Second, most hvKP infections are community-acquired infections that are unusual for cKP, which are generally nosocomial infections in patients with immunosuppression. Third, hvKP infections are associated with a mortality rate, ranging from 3 to 42%. Fourth, hvKP infections are often complicated by devastating disseminated infections, including endophthalmitis and meningitis, which are uncommon for enteric Gram-negative bacilli, including K. pneumoniae in the absence of host susceptibility factors. These pathogenic features of hvKP may be primarily involved in its hypervirulence, but the exact mechanisms remain unclear. Other features of hvKP are its epidemic spread in Asia and the low prevalence of antibiotic resistance. Although hvKP infections have increasingly been reported in Western countries, most cases of hvKP infections were reported along the Asian Pacific Rim, including Taiwan, China, South Korea, Singapore, and Japan (Figure 1). The reasons for the prevalence of hvKP infections in Asia remain unclear. Despite the global prevalence of antibiotic-resistant cKP strains, the rate of antibiotic resistance in hvKP strains is relatively low and the exact reasons for this also remain unclear.
The exact definition of hvKP is a controversial issue. Although the string test is generally accepted as a method for determining hvKP, many cases of string test-positive K. pneumoniae isolates do not exhibit hypervirulence. Several virulence factors, such as the K1/K2 serotypes, RmpA, and aerobactin, seem to be associated strongly with the hypervirulence of hvKP. Therefore, the exact phenotypic and genotypic definitions of hvKP need to be established firmly. The predominant hvKP clone is ST23, and most ST23 clones have the K1 serotype. The reason for this also remains unclear. It is interesting that the predominant clone of carbapenem-resistant cKP, ST258, is not the predominant clone of hvKP. Additional studies analyzing epidemiological and pathogenic characteristics of each clone are required to understand the exact relationship between the specific clones and specific phenotypes of K. pneumoniae, such as hypervirulence or antibiotic resistance.
The threat of hvKP acquiring carbapenem resistance is becoming a reality in certain countries, such as China, where both carbapenem-resistant cKP and carbapenem-susceptible hvKP strains are prevalent. The prevalence of hvKP among carbapenem-resistant K. pneumoniae isolates in China is high, ranging from 7.4% to 15%. The epidemiology of carbapenem-resistant hvKP in China implies the possibility of the transmission of mobile genetic elements between hvKP and cKP strains. Until now, ST11 and ST23 have been identified as the major clones of carbapenem-resistant hvKP strains in China. In China, ST11 and ST23 are the major clones of cKP and hvKP strains, respectively. Additionally, KPC-2 is found in 94.4% of carbapenem-resistant hvKP isolates in China. KPC-2 is the most prevalent carbapenemase in China. These results also imply the transfer of mobile genetic elements between hvKP and cKP strains. A recent study using genomic analyses showed that ST11 carbapenem-resistant cKP strains could become carbapenem-resistant hvKP strains through the acquisition of the pLVPK-like plasmid. Therefore, an immediate response to the global emergence of hvKP with resistance determinants, especially against carbapenems, is required.
In conclusion, great knowledge gaps exist for hvKP, despite a great deal of clinical, physiological, and pathogenic studies of hvKP over the past 30-years. The definition of hvKP remains unclear. Hypervirulence-associated determinants of hvKP require further study. These hypervirulence-associated factors may become a good target for the development of novel therapeutic agents. Epidemiological analysis of recent hvKP clinical isolates in China indicates a potential for the global dissemination of hvKP strains with extensive antibiotic resistances in the near future. The development of effective diagnostic tools for identification of hvKP is required for the effective control of hvKP in the clinical field. The development of novel antimicrobial agents also is absolutely required.
Author Contributions
C-RL, JL, KP, and SL contributed to the conception and the design of the review and C-RL, JL, KP, JJ, YK, C-JC, BJ, and SL researched and wrote the review.
Funding
This review was supported by research grants from the Bio Medical Technology Development Program of the NRF funded by the MSIT (numbers NRF-2017M3A9E4078009, NRF-2017M3A9E4078014 and NRF-2017M3A9E4078017); the Research of Korea Centers for Disease Control and Prevention (number 2017N-ER5404-00); the NRF funded by the Ministry of Science and ICT (number NRF-2017R1A2B4002315).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2017.00483/full#supplementary-material
References
Andrade, L. N., Vitali, L., Gaspar, G. G., Bellissimo-Rodrigues, F., Martinez, R., and Darini, A. L. (2014). Expansion and evolution of a virulent, extensively drug-resistant (polymyxin B-resistant), QnrS1-, CTX-M-2-, and KPC-2-producing Klebsiella pneumoniae ST11 international high-risk clone. J. Clin. Microbiol. 52, 2530–2535. doi: 10.1128/JCM.00088-14
Arena, F., Henrici De Angelis, L., D'Andrea, M. M., Cannatelli, A., Fossati, L., Di Pilato, V., et al. (2017). Infections caused by carbapenem-resistant Klebsiella pneumoniae with hypermucoviscous phenotype: a case report and literature review. Virulence. doi: 10.1080/21505594.2017.1286439. [Epub ahead of print].
Bach, S., de Almeida, A., and Carniel, E. (2000). The Yersinia high-pathogenicity island is present in different members of the family Enterobacteriaceae. FEMS Microbiol. Lett. 183, 289–294. doi: 10.1111/j.1574-6968.2000.tb08973.x
Beceiro, A., Moreno, A., Fernández, N., Vallejo, J. A., Aranda, J., Adler, B., et al. (2014). Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob. Agents Chemother. 58, 518–526. doi: 10.1128/AAC.01597-13
Bialek-Davenet, S., Criscuolo, A., Ailloud, F., Passet, V., Jones, L., Delannoy-Vieillard, A. S., et al. (2014). Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 20, 1812–1820. doi: 10.3201/eid2011.140206
Bilal, S., Volz, M. S., Fiedler, T., Podschun, R., and Schneider, T. (2014). Klebsiella pneumoniae-induced liver abscesses, Germany. Emerg. Infect. Dis. 20, 1939–1940. doi: 10.3201/eid2011.140149
Cannatelli, A., D'Andrea, M. M., Giani, T., Di Pilato, V., Arena, F., Ambretti, S., et al. (2013). In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob. Agents Chemother. 57, 5521–5526. doi: 10.1128/AAC.01480-13
Cannatelli, A., Giani, T., D'Andrea, M. M., Di Pilato, V., Arena, F., Conte, V., et al. (2014). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. doi: 10.1128/AAC.03110-14
Catalán-Nájera, J. C., Garza-Ramos, U., and Barrios-Camacho, H. (2017). Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp. phenotypes? Virulence. doi: 10.1080/21505594.2017.1317412. [Epub ahead of print].
Cheng, D. L., Liu, Y. C., Yen, M. Y., Liu, C. Y., and Wang, R. S. (1991). Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch. Intern. Med. 151, 1557–1559. doi: 10.1001/archinte.1991.00400080059010
Cheng, H. Y., Chen, Y. S., Wu, C. Y., Chang, H. Y., Lai, Y. C., and Peng, H. L. (2010). RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J. Bacteriol. 192, 3144–3158. doi: 10.1128/JB.00031-10
Cheng, N. C., Yu, Y. C., Tai, H. C., Hsueh, P. R., Chang, S. C., Lai, S. Y., et al. (2012). Recent trend of necrotizing fasciitis in Taiwan: focus on monomicrobial Klebsiella pneumoniae necrotizing fasciitis. Clin. Infect. Dis. 55, 930–939. doi: 10.1093/cid/cis565
Chiu, C. T., Lin, D. Y., and Liaw, Y. F. (1988). Metastatic septic endophthalmitis in pyogenic liver abscess. J. Clin. Gastroenterol. 10, 524–527. doi: 10.1097/00004836-198810000-00009
Choi, M. J., and Ko, K. S. (2015). Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob. Agents Chemother. 59, 6763–6773. doi: 10.1128/AAC.00952-15
Chuang, Y. P., Fang, C. T., Lai, S. Y., Chang, S. C., and Wang, J. T. (2006). Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 193, 645–654. doi: 10.1086/499968
Chung, D. R., Lee, H. R., Lee, S. S., Kim, S. W., Chang, H. H., Jung, S. I., et al. (2008). Evidence for clonal dissemination of the serotype K1 Klebsiella pneumoniae strain causing invasive liver abscesses in Korea. J. Clin. Microbiol. 46, 4061–4063. doi: 10.1128/JCM.01577-08
Chung, D. R., Lee, S. S., Lee, H. R., Kim, H. B., Choi, H. J., Eom, J. S., et al. (2007). Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. J. Infect. 54, 578–583. doi: 10.1016/j.jinf.2006.11.008
Cortés, G., Borrell, N., de Astorza, B., Gómez, C., Sauleda, J., and Albertí, S. (2002). Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect. Immun. 70, 2583–2590. doi: 10.1128/IAI.70.5.2583-2590.2002
Coutinho, R. L., Visconde, M. F., Descio, F. J., Nicoletti, A. G., Pinto, F. C., Silva, A. C., et al. (2014). Community-acquired invasive liver abscess syndrome caused by a K1 serotype Klebsiella pneumoniae isolate in Brazil: a case report of hypervirulent ST23. Mem. Inst. Oswaldo Cruz. 109, 970–971. doi: 10.1590/0074-0276140196
Cubero, M., Grau, I., Tubau, F., Pallarés, R., Dominguez, M. A., Linares, J., et al. (2016). Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin. Microbiol. Infect. 22, 154–160. doi: 10.1016/j.cmi.2015.09.025
Decré, D., Verdet, C., Emirian, A., Le Gourrierec, T., Petit, J. C., Offenstadt, G., et al. (2011). Emerging severe and fatal infections due to Klebsiella pneumoniae in two university hospitals in France. J. Clin. Microbiol. 49, 3012–3014. doi: 10.1128/JCM.00676-11
Doud, M. S., Grimes-Zeppegno, R., Molina, E., Miller, N., Balachandar, D., Schneper, L., et al. (2009). A k2A-positive Klebsiella pneumoniae causes liver and brain abscess in a Saint Kitt's man. Int. J. Med. Sci. 6, 301–304. doi: 10.7150/ijms.6.301
El Fertas-Aissani, R., Messai, Y., Alouache, S., and Bakour, R. (2013). Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol. Biol. 61, 209–216. doi: 10.1016/j.patbio.2012.10.004
Fang, C. T., Chen, Y. C., Chang, S. C., Sau, W. Y., and Luh, K. T. (2000). Klebsiella pneumoniae meningitis: timing of antimicrobial therapy and prognosis. QJM 93, 45–53. doi: 10.1093/qjmed/93.1.45
Fang, C. T., Chuang, Y. P., Shun, C. T., Chang, S. C., and Wang, J. T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Fang, C. T., Lai, S. Y., Yi, W. C., Hsueh, P. R., and Liu, K. L. (2010). The function of wzy_K1 (magA), the serotype K1 polymerase gene in Klebsiella pneumoniae cps gene cluster. J. Infect. Dis. 201, 1268–1269. doi: 10.1086/652183
Fang, C. T., Lai, S. Y., Yi, W. C., Hsueh, P. R., Liu, K. L., and Chang, S. C. (2007). Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin. Infect. Dis. 45, 284–293. doi: 10.1086/519262
Fang, F. C., Sandler, N., and Libby, S. J. (2005). Liver abscess caused by magA+ Klebsiella pneumoniae in North America. J. Clin. Microbiol. 43, 991–992. doi: 10.1128/JCM.43.2.991-992.2005
Follador, R., Heinz, E., Wyres, K. L., Ellington, M. J., Kowarik, M., Holt, K. E., et al. (2016). The diversity of Klebsiella pneumoniae surface polysaccharides. Microb. Genom. 2:e000073. doi: 10.1099/mgen.0.000073
Frazee, B. W., Hansen, S., and Lambert, L. (2009). Invasive infection with hypermucoviscous Klebsiella pneumoniae: multiple cases presenting to a single emergency department in the United States. Ann. Emerg. Med. 53, 639–642. doi: 10.1016/j.annemergmed.2008.11.007
Giani, T., Arena, F., Vaggelli, G., Conte, V., Chiarelli, A., Henrici De Angelis, L., et al. (2015). Large nosocomial outbreak of colistin-resistant, carbapenemase-producing Klebsiella pneumoniae traced to clonal expansion of an mgrB deletion mutant. J. Clin. Microbiol. 53, 3341–3344. doi: 10.1128/JCM.01017-15
Giani, T., Pini, B., Arena, F., Conte, V., Bracco, S., Migliavacca, R., et al. (2013). Epidemic diffusion of KPC carbapenemase-producing Klebsiella pneumoniae in Italy: results of the first countrywide survey, 15 May to 30 June 2011. Euro. Surveill. 18:20489. doi: 10.2807/ese.18.22.20489-en
Gu, D., Dong, N., Zheng, Z., Lin, D., Huang, M., Wang, L., et al. (2017). A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. doi: 10.1016/S1473-3099(17)30489-9. [Epub ahead of print].
Gu, D. X., Huang, Y. L., Ma, J. H., Zhou, H. W., Fang, Y., Cai, J. C., et al. (2016). Detection of Colistin Resistance Gene mcr-1 in Hypervirulent Klebsiella pneumoniae and Escherichia coli Isolates from an Infant with Diarrhea in China. Antimicrob. Agents Chemother. 60, 5099–5100. doi: 10.1128/AAC.00476-16
Gundestrup, S., Struve, C., Stahlhut, S. G., and Hansen, D. S. (2014). First case of liver abscess in Scandinavia due to the international hypervirulent Klebsiella pneumoniae clone ST23. Open Microbiol. J. 8, 22–24. doi: 10.2174/1874285801408010022
Gunnarsson, G. L., Brandt, P. B., Gad, D., Struve, C., and Justesen, U. S. (2009). Monomicrobial necrotizing fasciitis in a white male caused by hypermucoviscous Klebsiella pneumoniae. J. Med. Microbiol. 58, 1519–1521. doi: 10.1099/jmm.0.011064-0
Guo, S., Xu, J., Wei, Y., Xu, J., Li, Y., and Xue, R. (2016). Clinical and molecular characteristics of Klebsiella pneumoniae ventilator-associated pneumonia in mainland China. BMC Infect. Dis. 16:608. doi: 10.1186/s12879-016-1942-z
Guo, Y., Wang, S., Zhan, L., Jin, Y., Duan, J., Hao, Z., et al. (2017). Microbiological and clinical characteristics of hypermucoviscous Klebsiella pneumoniae isolates associated with invasive infections in China. Front. Cell. Infect. Microbiol. 7:24. doi: 10.3389/fcimb.2017.00024
Gupta, N., Limbago, B. M., Patel, J. B., and Kallen, A. J. (2011). Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin. Infect. Dis. 53, 60–67. doi: 10.1093/cid/cir202
Han, S. H. (1995). Review of hepatic abscess from Klebsiella pneumoniae. an association with diabetes mellitus and septic endophthalmitis. West. J. Med. 162, 220–224.
Holmås, K., Fostervold, A., Stahlhut, S. G., Struve, C., and Holter, J. C. (2014). Emerging K1 serotype Klebsiella pneumoniae primary liver abscess: three cases presenting to a single university hospital in Norway. Clin. Case Rep. 2, 122–127. doi: 10.1002/ccr3.77
Hraiech, S., Roch, A., Lepidi, H., Atieh, T., Audoly, G., Rolain, J. M., et al. (2013). Impaired virulence and fitness of a colistin-resistant clinical isolate of Acinetobacter baumannii in a rat model of pneumonia. Antimicrob. Agents Chemother. 57, 5120–5121. doi: 10.1128/AAC.00700-13
Hsieh, P. F., Lin, T. L., Lee, C. Z., Tsai, S. F., and Wang, J. T. (2008). Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 197, 1717–1727. doi: 10.1086/588383
Hsu, C. R., Lin, T. L., Chen, Y. C., Chou, H. C., and Wang, J. T. (2011). The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 157, 3446–3457. doi: 10.1099/mic.0.050336-0
Jung, S. W., Chae, H. J., Park, Y. J., Yu, J. K., Kim, S. Y., Lee, H. K., et al. (2013). Microbiological and clinical characteristics of bacteraemia caused by the hypermucoviscosity phenotype of Klebsiella pneumoniae in Korea. Epidemiol. Infect. 141, 334–340. doi: 10.1017/S0950268812000933
Kabha, K., Nissimov, L., Athamna, A., Keisari, Y., Parolis, H., Parolis, L. A., et al. (1995). Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect. Immun. 63, 847–852.
Keynan, Y., Karlowsky, J. A., Walus, T., and Rubinstein, E. (2007). Pyogenic liver abscess caused by hypermucoviscous Klebsiella pneumoniae. Scand. J. Infect. Dis. 39, 828–830. doi: 10.1080/00365540701266763
Kim, Y. J., Kim, S. I., Kim, Y. R., Wie, S. H., Lee, H. K., Kim, S. Y., et al. (2017). Virulence factors and clinical patterns of hypermucoviscous Klebsiella pneumoniae isolated from urine. Infect. Dis. (Lond). 49, 178–184. doi: 10.1080/23744235.2016.1244611
Ko, W. C., Paterson, D. L., Sagnimeni, A. J., Hansen, D. S., Von Gottberg, A., Mohapatra, S., et al. (2002). Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerging Infect. Dis. 8, 160–166. doi: 10.3201/eid0802.010025
Ku, Y. H., Chuang, Y. C., and Yu, W. L. (2008). Clinical spectrum and molecular characteristics of Klebsiella pneumoniae causing community-acquired extrahepatic abscess. J. Microbiol. Immunol. Infect. 41, 311–317.
Lai, Y. C., Lin, A. C., Chiang, M. K., Dai, Y. H., Hsu, C. C., Lu, M. C., et al. (2014). Genotoxic Klebsiella pneumoniae in Taiwan. PLoS ONE 9:e96292. doi: 10.1371/journal.pone.0096292
Lai, Y. C., Peng, H. L., and Chang, H. Y. (2003). RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J. Bacteriol. 185, 788–800. doi: 10.1128/JB.185.3.788-800.2003
Lederman, E. R., and Crum, N. F. (2005). Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: an emerging disease with unique clinical characteristics. Am. J. Gastroenterol. 100, 322–331. doi: 10.1111/j.1572-0241.2005.40310.x
Lee, C. H., Chang, C. C., Liu, J. W., Chen, R. F., and Yang, K. D. (2014). Sialic acid involved in hypermucoviscosity phenotype of Klebsiella pneumoniae and associated with resistance to neutrophil phagocytosis. Virulence 5, 673–679. doi: 10.4161/viru.32076
Lee, C. H., Liu, J. W., Su, L. H., Chien, C. C., Li, C. C., and Yang, K. D. (2010). Hypermucoviscosity associated with Klebsiella pneumoniae-mediated invasive syndrome: a prospective cross-sectional study in Taiwan. Int. J. Infect. Dis. 14, e688–e692. doi: 10.1016/j.ijid.2010.01.007
Lee, C. R., Lee, J. H., Park, K. S., Kim, Y. B., Jeong, B. C., and Lee, S. H. (2016). Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front. Microbiol. 7:895. doi: 10.3389/fmicb.2016.00895
Lee, H. C., Chuang, Y. C., Yu, W. L., Lee, N. Y., Chang, C. M., Ko, N. Y., et al. (2006). Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: association with invasive syndrome in patients with community-acquired bacteraemia. J. Intern. Med. 259, 606–614. doi: 10.1111/j.1365-2796.2006.01641.x
Li, H., Zhang, J., Liu, Y., Zheng, R., Chen, H., Wang, X., et al. (2014a). Molecular characteristics of carbapenemase-producing Enterobacteriaceae in China from 2008 to 2011: predominance of KPC-2 enzyme. Diagn. Microbiol. Infect. Dis. 78, 63–65. doi: 10.1016/j.diagmicrobio.2013.10.002
Li, W., Sun, G., Yu, Y., Li, N., Chen, M., Jin, R., et al. (2014b). Increasing occurrence of antimicrobial-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in China. Clin. Infect. Dis. 58, 225–232. doi: 10.1093/cid/cit675
Lin, A. C., Liao, T. L., Lin, Y. C., Lai, Y. C., Lu, M. C., and Chen, Y. T. (2012). Complete genome sequence of Klebsiella pneumoniae 1084, a hypermucoviscosity-negative K1 clinical strain. J. Bacteriol. 194:6316. doi: 10.1128/JB.01548-12
Lin, J. C., Chang, F. Y., Fung, C. P., Xu, J. Z., Cheng, H. P., Wang, J. J., et al. (2004). High prevalence of phagocytic-resistant capsular serotypes of Klebsiella pneumoniae in liver abscess. Microbes Infect. 6, 1191–1198. doi: 10.1016/j.micinf.2004.06.003
Lin, J. C., Koh, T. H., Lee, N., Fung, C. P., Chang, F. Y., Tsai, Y. K., et al. (2014). Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. 6:21. doi: 10.1186/1757-4749-6-21
Lin, Y. C., Lu, M. C., Tang, H. L., Liu, H. C., Chen, C. H., Liu, K. S., et al. (2011). Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol. 11:50. doi: 10.1186/1471-2180-11-50
Lin, Y. T., Jeng, Y. Y., Chen, T. L., and Fung, C. P. (2010). Bacteremic community-acquired pneumonia due to Klebsiella pneumoniae: clinical and microbiological characteristics in Taiwan, 2001-2008. BMC Infect. Dis. 10:307. doi: 10.1186/1471-2334-10-307
Liu, Y. C., Cheng, D. L., and Lin, C. L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146, 1913–1916. doi: 10.1001/archinte.1986.00360220057011
Liu, Y. M., Li, B. B., Zhang, Y. Y., Zhang, W., Shen, H., Li, H., et al. (2014). Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob. Agents Chemother. 58, 5379–5385. doi: 10.1128/AAC.02523-14
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
López-Rojas, R., Domínguez-Herrera, J., McConnell, M. J., Docobo-Peréz, F., Smani, Y., Fernández-Reyes, M., et al. (2011). Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J. Infect. Dis. 203, 545–548. doi: 10.1093/infdis/jiq086
Lu, M. C., Chen, Y. T., Chiang, M. K., Wang, Y. C., Hsiao, P. Y., Huang, Y. J., et al. (2017). Colibactin contributes to the hypervirulence of pks+ K1 CC23 Klebsiella pneumoniae in mouse meningitis infections. Front. Cell. Infect. Microbiol. 7:103. doi: 10.3389/fcimb.2017.00103
McCabe, R., Lambert, L., and Frazee, B. (2010). Invasive Klebsiella pneumoniae infections, California, USA. Emerging Infect. Dis. 16, 1490–1491. doi: 10.3201/eid1609.100386
Melot, B., Brisse, S., Breurec, S., Passet, V., Malpote, E., Lamaury, I., et al. (2016). Community-acquired meningitis caused by a CG86 hypervirulent Klebsiella pneumoniae strain: first case report in the Caribbean. BMC Infect. Dis. 16:736. doi: 10.1186/s12879-016-2065-2
Merlet, A., Cazanave, C., Dutronc, H., de Barbeyrac, B., Brisse, S., and Dupon, M. (2012). Primary liver abscess due to CC23-K1 virulent clone of Klebsiella pneumoniae in France. Clin. Microbiol. Infect. 18, E338–E339. doi: 10.1111/j.1469-0691.2012.03953.x
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Nadasy, K. A., Domiati-Saad, R., and Tribble, M. A. (2007). Invasive Klebsiella pneumoniae syndrome in North America. Clin. Infect. Dis. 45, e25–e28. doi: 10.1086/519424
Nassif, X., and Sansonetti, P. J. (1986). Correlation of the virulence of Klebsiella pneumoniae K1 and K2 with the presence of a plasmid encoding aerobactin. Infect. Immun. 54, 603–608.
Paczosa, M. K., and Mecsas, J. (2016). Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 80, 629–661. doi: 10.1128/MMBR.00078-15
Pan, Y. J., Fang, H. C., Yang, H. C., Lin, T. L., Hsieh, P. F., Tsai, F. C., et al. (2008). Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J. Clin. Microbiol. 46, 2231–2240. doi: 10.1128/JCM.01716-07
Pastagia, M., and Arumugam, V. (2008). Klebsiella pneumoniae liver abscesses in a public hospital in Queens, New York. Travel Med. Infect. Dis. 6, 228–233. doi: 10.1016/j.tmaid.2008.02.005
Patel, P. K., Russo, T. A., and Karchmer, A. W. (2014). Hypervirulent Klebsiella pneumoniae. Open Forum. Infect. Dis. 1:ofu028. doi: 10.1093/ofid/ofu028
Peirano, G., Pitout, J. D., Laupland, K. B., Meatherall, B., and Gregson, D. B. (2013). Population-based surveillance for hypermucoviscosity Klebsiella pneumoniae causing community-acquired bacteremia in Calgary, Alberta. Can. J. Infect. Dis. Med. Microbiol. 24, e61–64. doi: 10.1155/2013/828741
Pereira, S. C., and Vanetti, M. C. (2015). Potential virulence of Klebsiella sp. isolates from enteral diets. Braz. J. Med. Biol. Res. 48, 782–789. doi: 10.1590/1414-431X20154316
Pinsky, B. A., Baron, E. J., Janda, J. M., and Banaei, N. (2009). Bartholin's abscess caused by hypermucoviscous Klebsiella pneumoniae. J. Med. Microbiol. 58, 671–673. doi: 10.1099/jmm.0.006734-0
Pomakova, D. K., Hsiao, C. B., Beanan, J. M., Olson, R., MacDonald, U., Keynan, Y., et al. (2012). Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumoniae: an emerging and under-recognized pathogenic variant. Eur. J. Clin. Microbiol. Infect. Dis. 31, 981–989. doi: 10.1007/s10096-011-1396-6
Prokesch, B. C., TeKippe, M., Kim, J., Raj, P., TeKippe, E. M., and Greenberg, D. E. (2016). Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 16, e190–e195. doi: 10.1016/S1473-3099(16)30021-4
Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P., and Yu, Y. (2011). ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66, 307–312. doi: 10.1093/jac/dkq431
Qu, T. T., Zhou, J. C., Jiang, Y., Shi, K. R., Li, B., Shen, P., et al. (2015). Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect. Dis. 15:161. doi: 10.1186/s12879-015-0899-7
Raymond, K. N., Dertz, E. A., and Kim, S. S. (2003). Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U.S.A. 100, 3584–3588. doi: 10.1073/pnas.0630018100
Rivero, A., Gomez, E., Alland, D., Huang, D. B., and Chiang, T. (2010). K2 serotype Klebsiella pneumoniae causing a liver abscess associated with infective endocarditis. J. Clin. Microbiol. 48, 639–641. doi: 10.1128/JCM.01779-09
Russo, T. A., Olson, R., MacDonald, U., Beanan, J., and Davidson, B. A. (2015). Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect. Immun. 83, 3325–3333. doi: 10.1128/IAI.00430-15
Russo, T. A., Olson, R., Macdonald, U., Metzger, D., Maltese, L. M., Drake, E. J., et al. (2014). Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 82, 2356–2367. doi: 10.1128/IAI.01667-13
Russo, T. A., Shon, A. S., Beanan, J. M., Olson, R., MacDonald, U., Pomakov, A. O., et al. (2011). Hypervirulent K. pneumoniae secretes more and more active iron-acquisition molecules than “classical” K. pneumoniae thereby enhancing its virulence. PLoS ONE 6:e26734. doi: 10.1371/journal.pone.0026734
Sahly, H., Aucken, H., Benedi, V. J., Forestier, C., Fussing, V., Hansen, D. S., et al. (2004). Impairment of respiratory burst in polymorphonuclear leukocytes by extended-spectrum β-lactamase-producing strains of Klebsiella pneumoniae. Eur. J. Clin. Microbiol. Infect. Dis. 23, 20–26. doi: 10.1007/s10096-003-1047-7
Sahly, H., Keisari, Y., Crouch, E., Sharon, N., and Ofek, I. (2008). Recognition of bacterial surface polysaccharides by lectins of the innate immune system and its contribution to defense against infection: the case of pulmonary pathogens. Infect. Immun. 76, 1322–1332. doi: 10.1128/IAI.00910-07
Shon, A. S., Bajwa, R. P., and Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Siu, L. K., Fung, C. P., Chang, F. Y., Lee, N., Yeh, K. M., Koh, T. H., et al. (2011). Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J. Clin. Microbiol. 49, 3761–3765. doi: 10.1128/JCM.00977-11
Siu, L. K., Huang, D. B., and Chiang, T. (2014). Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae. BMC Infect. Dis. 14:176. doi: 10.1186/1471-2334-14-176
Siu, L. K., Yeh, K. M., Lin, J. C., Fung, C. P., and Chang, F. Y. (2012). Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 12, 881–887. doi: 10.1016/S1473-3099(12)70205-0
Sobirk, S. K., Struve, C., and Jacobsson, S. G. (2010). Primary Klebsiella pneumoniae liver abscess with metastatic spread to lung and eye, a North-European case report of an emerging syndrome. Open Microbiol. J. 4, 5–7. doi: 10.2174/1874285801004010005
Struve, C., Roe, C. C., Stegger, M., Stahlhut, S. G., Hansen, D. S., Engelthaler, D. M., et al. (2015). Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio. 6:e00630. doi: 10.1128/mBio.00630-15
Sun, S., Negrea, A., Rhen, M., and Andersson, D. I. (2009). Genetic analysis of colistin resistance in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 53, 2298–2305. doi: 10.1128/AAC.01016-08
Sun, Y., Wu, H., and Shen, D. (2016). Clinical and molecular analysis of Klebsiella pneumoniae causing liver abscess in China. J. Mol. Microbiol. Biotechnol. 26, 245–251. doi: 10.1159/000444367
Surgers, L., Boyd, A., Girard, P. M., Arlet, G., and Decré, D. (2016). ESBL-Producing Strain of Hypervirulent Klebsiella pneumoniae K2, France. Emerging Infect. Dis. 22, 1687–1688. doi: 10.3201/eid2209.160681
Trautmann, M., Ruhnke, M., Rukavina, T., Held, T. K., Cross, A. S., Marre, R., et al. (1997). O-antigen seroepidemiology of Klebsiella clinical isolates and implications for immunoprophylaxis of Klebsiella infections. Clin. Diagn. Lab. Immunol. 4, 550–555.
Tsai, F. C., Huang, Y. T., Chang, L. Y., and Wang, J. T. (2008). Pyogenic liver abscess as endemic disease, Taiwan. Emerging Infect. Dis. 14, 1592–1600. doi: 10.3201/eid1410.071254
Vila, A., Cassata, A., Pagella, H., Amadio, C., Yeh, K. M., Chang, F. Y., et al. (2011). Appearance of Klebsiella pneumoniae liver abscess syndrome in Argentina: case report and review of molecular mechanisms of pathogenesis. Open Microbiol. J. 5, 107–113. doi: 10.2174/1874285801105010107
Wacharotayankun, R., Arakawa, Y., Ohta, M., Tanaka, K., Akashi, T., Mori, M., et al. (1993). Enhancement of extracapsular polysaccharide synthesis in Klebsiella pneumoniae by RmpA2, which shows homology to NtrC and FixJ. Infect. Immun. 61, 3164–3174.
Wang, J. H., Liu, Y. C., Lee, S. S., Yen, M. Y., Chen, Y. S., Wang, J. H., et al. (1998). Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26, 1434–1438. doi: 10.1086/516369
Wang, J., Yan, Y., Xue, X., Wang, K., and Shen, D. (2013). Comparison of pyogenic liver abscesses caused by hypermucoviscous Klebsiella pneumoniae and non-Klebsiella pneumoniae pathogens in Beijing: a retrospective analysis. J. Int. Med. Res. 41, 1088–1097. doi: 10.1177/0300060513487645
Wei, D. D., Wan, L. G., Deng, Q., and Liu, Y. (2016). Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in Mainland China. Diagn. Microbiol. Infect. Dis. 85, 192–194. doi: 10.1016/j.diagmicrobio.2015.03.012
Wu, H., Li, D., Zhou, H., Sun, Y., Guo, L., and Shen, D. (2017). Bacteremia and other body site infection caused by hypervirulent and classic Klebsiella pneumoniae. Microb. Pathog. 104, 254–262. doi: 10.1016/j.micpath.2017.01.049
Wyres, K. L., Wick, R. R., Gorrie, C., Jenney, A., Follador, R., Thomson, N. R., et al. (2016). Identification of Klebsiella capsule synthesis loci from whole genome data. Microb. Genom. 2:e000102. doi: 10.1099/mgen.0.000102
Yan, Q., Zhou, M., Zou, M., and Liu, W. E. (2016). Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur. J. Clin. Microbiol. Infect. Dis. 35, 387–396. doi: 10.1007/s10096-015-2551-2
Yao, B., Xiao, X., Wang, F., Zhou, L., Zhang, X., and Zhang, J. (2015). Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int. J. Infect. Dis. 37, 107–112. doi: 10.1016/j.ijid.2015.06.023
Ye, M., Tu, J., Jiang, J., Bi, Y., You, W., Zhang, Y., et al. (2016). Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front. Cell. Infect. Microbiol. 6:165. doi: 10.3389/fcimb.2016.00165
Yeh, K. M., Chang, F. Y., Fung, C. P., Lin, J. C., and Siu, L. K. (2006). magA is not a specific virulence gene for Klebsiella pneumoniae strains causing liver abscess but is part of the capsular polysaccharide gene cluster of K. pneumoniae serotype K1. J. Med. Microbiol. 55, 803–804. doi: 10.1099/jmm.0.46368-0
Yeh, K. M., Kurup, A., Siu, L. K., Koh, Y. L., Fung, C. P., Lin, J. C., et al. (2007). Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J. Clin. Microbiol. 45, 466–471. doi: 10.1128/JCM.01150-06
Yeh, K. M., Lin, J. C., Yin, F. Y., Fung, C. P., Hung, H. C., Siu, L. K., et al. (2010). Revisiting the importance of virulence determinant magA and its surrounding genes in Klebsiella pneumoniae causing pyogenic liver abscesses: exact role in serotype K1 capsule formation. J. Infect. Dis. 201, 1259–1267. doi: 10.1086/606010
Yu, W. L., Ko, W. C., Cheng, K. C., Lee, C. C., Lai, C. C., and Chuang, Y. C. (2008). Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn. Microbiol. Infect. Dis. 62, 1–6. doi: 10.1016/j.diagmicrobio.2008.04.007
Yu, W. L., Ko, W. C., Cheng, K. C., Lee, H. C., Ke, D. S., Lee, C. C., et al. (2006). Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 42, 1351–1358. doi: 10.1086/503420
Yu, W. L., Lee, M. F., Chang, M. C., and Chuang, Y. C. (2015a). Intrapersonal mutation of rmpA and rmpA2: a reason for negative hypermucoviscosity phenotype and low virulence of rmpA-positive Klebsiella pneumoniae isolates. J. Glob. Antimicrob. Resist. 3, 137–141. doi: 10.1016/j.jgar.2015.03.008
Yu, W. L., Lee, M. F., Chen, C. C., Tang, H. J., Ho, C. H., and Chuang, Y. C. (2017). Impacts of hypervirulence determinants on clinical features and outcomes of bacteremia caused by extended-spectrum β-lactamase-producing Klebsiella pneumoniae. Microb. Drug Resist. 23, 376–383. doi: 10.1089/mdr.2016.0018
Yu, W. L., Lee, M. F., Tang, H. J., Chang, M. C., and Chuang, Y. C. (2015b). Low prevalence of rmpA and high tendency of rmpA mutation correspond to low virulence of extended spectrum beta-lactamase-producing Klebsiella pneumoniae isolates. Virulence 6, 162–172. doi: 10.1080/21505594.2015.1016703
Zamani, A., Yousefi Mashouf, R., Ebrahimzadeh Namvar, A. M., and Alikhani, M. Y. (2013). Detection of magA gene in Klebsiella spp. isolated from clinical samples. Iran J. Basic Med. Sci. 16, 173–176. doi: 10.22038/ijbms.2013.298
Zhan, L., Wang, S., Guo, Y., Jin, Y., Duan, J., Hao, Z., et al. (2017). Outbreak by Hypermucoviscous Klebsiella pneumoniae ST11 Isolates with Carbapenem Resistance in a Tertiary Hospital in China. Front. Cell. Infect. Microbiol. 7:182. doi: 10.3389/fcimb.2017.00182
Zhang, R., Lin, D., Chan, E. W., Gu, D., Chen, G. X., and Chen, S. (2015a). Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in china. Antimicrob. Agents Chemother. 60, 709–711. doi: 10.1128/AAC.02173-15
Zhang, Y., Zeng, J., Liu, W., Zhao, F., Hu, Z., Zhao, C., et al. (2015b). Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J. Infect. 71, 553–560. doi: 10.1016/j.jinf.2015.07.010
Zhang, Y., Zhao, C., Wang, Q., Wang, X., Chen, H., Li, H., et al. (2016). High prevalence of hypervirulent klebsiella pneumoniae infection in china: geographic distribution, clinical characteristics, and antimicrobial resistance. Antimicrob. Agents Chemother. 60, 6115–6120. doi: 10.1128/AAC.01127-16
Zhao, J., Chen, J., Zhao, M., Qiu, X., Chen, X., Zhang, W., et al. (2016). Multilocus sequence types and virulence determinants of hypermucoviscosity-positive Klebsiella pneumoniae isolated from community-acquired infection cases in Harbin, North China. Jpn. J. Infect. Dis. 69, 357–360. doi: 10.7883/yoken.JJID.2015.321
Keywords: antimicrobial resistance, hypervirulent Klebsiella pneumoniae, epidemiology, resistance mechanism, serotype, sequence type
Citation: Lee C-R, Lee JH, Park KS, Jeon JH, Kim YB, Cha C-J, Jeong BC and Lee SH (2017) Antimicrobial Resistance of Hypervirulent Klebsiella pneumoniae: Epidemiology, Hypervirulence-Associated Determinants, and Resistance Mechanisms. Front. Cell. Infect. Microbiol. 7:483. doi: 10.3389/fcimb.2017.00483
Received: 19 July 2017; Accepted: 09 November 2017;
Published: 21 November 2017.
Edited by:
Kelly L. Wyres, University of Melbourne, AustraliaReviewed by:
Russel Lee, National University of Singapore, SingaporeXingmin Sun, University of South Florida, United States
Scott Wesley Long, Houston Methodist Hospital, United States
Copyright © 2017 Lee, Lee, Park, Jeon, Kim, Cha, Jeong and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang Hee Lee, c2FuZ2hlZWxlZUBtanUuYWMua3I=
†These authors have contributed equally to this work.
 Chang-Ro Lee
Chang-Ro Lee Jung Hun Lee
Jung Hun Lee Kwang Seung Park
Kwang Seung Park Jeong Ho Jeon
Jeong Ho Jeon Young Bae Kim
Young Bae Kim Chang-Jun Cha
Chang-Jun Cha Byeong Chul Jeong
Byeong Chul Jeong Sang Hee Lee
Sang Hee Lee