- 1Institute of Medical Microbiology and Hygiene, University of Ulm, Ulm, Germany
- 2Department of Microbiology and Immunology, Faculty of Pharmacy, Suez Canal University, Ismailia, Egypt
Group B streptococcus (GBS) is a leading cause of neonatal mortality and morbidity in the United States and Europe. It is part of the vaginal microbiota in up to 30% of pregnant women and can be passed on to the newborn through perinatal transmission. GBS has the ability to survive in multiple different host niches. The pathophysiology of this bacterium reveals an outstanding ability to withstand varying pH fluctuations of the surrounding environments inside the human host. GBS host pathogen interations include colonization of the acidic vaginal mucosa, invasion of the neutral human blood or amniotic fluid, breaching of the blood brain barrier as well as survival within the acidic phagolysosomal compartment of macrophages. However, investigations on GBS responses to acid stress are limited. Technologies, such as whole genome sequencing, genome-wide transcription and proteome mapping facilitate large scale identification of genes and proteins. Mechanisms enabling GBS to cope with acid stress have mainly been studied through these techniques and are summarized in the current review
Introduction
Streptococcus agalactiae or group B streptococcus (GBS) is an opportunistic pathogen which colonizes up to 30% of the genitourinary and gastrointestinal tracts of healthy women. At the same time GBS is a leading cause of life-threatening neonatal infections, such as meningitis, sepsis and pneumonia (Verani et al., 2010; Le Doare and Heath, 2013). A primary risk factor for GBS transmission to newborns is maternal colonization at birth, where GBS may spread either in utero by ascending infection or intrapartum through the aspiration of contaminated vaginal or amniotic fluids (Maisey et al., 2008a; Melin, 2011; Le Doare and Heath, 2013). Every tenth neonate may acquire GBS, which has also been proposed as a normal element of the neonatal microbiome (Landwehr-Kenzel and Henneke, 2014). Although GBS is harmless as a colonizer of healthy women, it can cause serious infections in pregnancy. In addition, GBS has been increasingly reported as being responsible for invasive disease in elderly and immunocompromised patients (Farley, 2001; Maisey et al., 2008a; Melin, 2011; Le Doare and Heath, 2013).
Assessment of GBS pathophysiology reveals that it has the capability to survive in various environments within the human host. It typically colonizes the vaginal mucosa, but also causes different types of invasive infections. GBS have thus successfully adapted to varying pH levels ranging from the acidic environment of the vagina or intracellular compartments to the almost neutral pH-values of amniotic fluid, the respiratory tract and human blood. These changes are most likely achieved by modifying the transcription of pathogen-host interaction related genes. The typical host environments and their respective pH-values that GBS can colonize and infect are depicted in Figure 1.
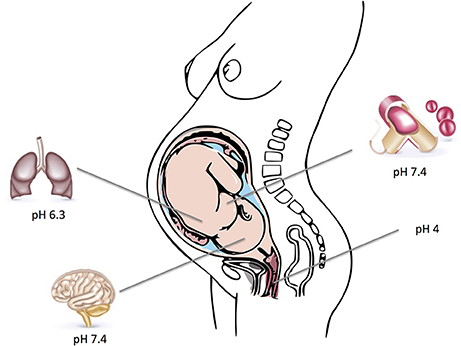
Figure 1. Depicted are typical host environments and their respective pH values that GBS can colonize and infect. Parts of the figure designed by Freepik.
In contrast to other Streptococci (Cotter and Hill, 2003; Kajfasz and Quivey, 2011), investigations on GBS responses to acid stress are limited. Whole GBS genomes sequencing projects (Glaser et al., 2002; Tettelin et al., 2002, 2005) lead to the identification of several genes with high similarities to streptococcal systems known to be involved in acid stress resistance. Several large scale genome-wide investigations employing technologies, such as DNA microarrays and proteomic analysis (two-dimensional polyacrylamide gel electrophoresis 2D-PAGE in combination with mass spectrometry) focused on pH-responsive GBS genes and facilitated the identification of new targets that are induced under acid stress (Cotter and Hill, 2003; Martin-Galiano et al., 2005; Bore et al., 2007; Gong et al., 2009; Martinez et al., 2010).
In general, streptococci possess an array of different defense mechanisms to cope with low pH. Proton pumps represent the most direct approach by transporting protons outside the cell to keep a proper level of intracellular pH. Inducing a buffering effect through increasing the concentration of intracellular alkaline compounds is another approach used to counteract cytoplasmic acidification. Additional mechanisms include repair or prevention of acid damage in macromolecules and modifying proton permeability of the cellular membrane. Regulation and control of these mechanisms is exerted through Two-component systems (TCSs), transcriptional regulators and sigma factors which respond to acid stress by modifying gene expression. Moreover, metal ion homeostasis, osmoregulation and oxidative stress response have been increasingly reported to contribute to acid adaptation mechanisms.
Low pH environments are often encountered by GBS inside the human host and mechanisms that enable GBS to cope with acid stress must therefore be essential for colonization as well as infection. However, our understanding of the acid stress response in GBS is incomplete. While the relevant mechanisms have often been studied at a functional level in other streptococci or gram positive pathogens studies in GBS largely rely on genomic and transcriptomic investigations. With this review, we try to summarize our current knowledge about mechanism permitting GBS survival at low pH and draw comparisons to other gram positive bacteria, especially streptococci. Figure 2 represents a simplified graph explaining different acid stress responses in GBS which will be discussed below.
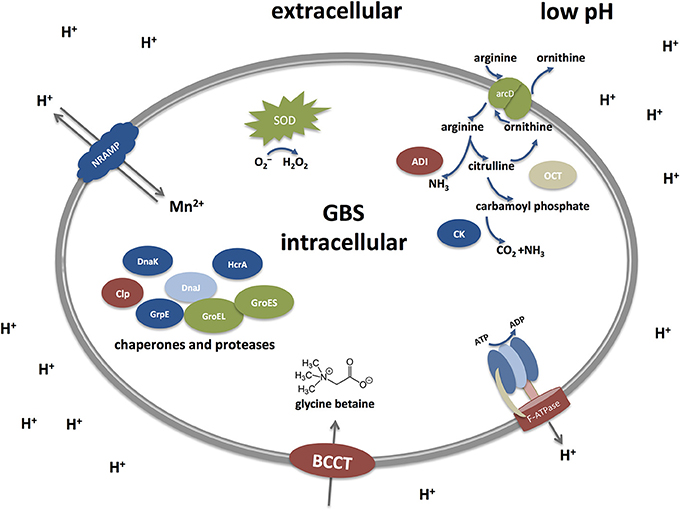
Figure 2. Acid stress responses in Streptococcus agalactiae (Group B streptococci, GBS) under low pH. GBS possess different defense mechanisms to cope with low pH. They include the Arginine deiminase system (ADI), an F-ATPase transporter, transporter of the BCCT family, chaperones and proteases, SodA, and a NRAMP-type transporter. The Arginine deiminase system (ADI) comprises three units: an arginine deiminase (AD), ornithine carbamoyltransferase (OTC) and carbamate kinase (CK). Arginine is taken up from the extracellular environment and cleaved by AD into citrulline and ammonia. Citrulline is further cleaved to yield ornithine and carbamoyl phosphate by the action of OTC. Finally, CK cleaves carbamoyl phosphate into carbon dioxide and ammonia, thereby generating an alkaline microenvironment. Proton pumps like the F-ATPase represent the most direct approach to counteract acid stress by transporting protons outside the cell to keep a proper level of intracellular pH. Under low pH, the F-ATPase system is induced to pump out protons extracellularly in order to maintain the alkalinity of the intracellular cytoplasm. Additional mechanisms include repair or prevention of acid damage in macromolecules by chaperones and proteases, such as Dnak, GroES, and CLp. Metal ion homeostasis also takes part in the acid response as the NRAMP metal ion symporter takes up Mn2+ and expels protons out of the cell. The osmotic stress is tightly controlled as well through the up-regulation of the glycine betaine osmoregulation system where choline and glycine betaine (a powerful osmoprotectant) are taken up by transporters of the BCCT (Betaine/Carnitine/Choline Transporter) family. Defense mechanisms to avoid the damaging effects of superoxide species generated during acid stress are mainly exerted through the activity of the streptococcal superoxide dismutase (SOD).
Low pH Environments Encountered within the Human Host
The Human Vagina
A low pH of 4 ± 0.5 is crucial to maintain a healthy vaginal environment. These pH-values mediate a microbicidal impact, which has been demonstrated to be effective against various microbial pathogens of sexually transmitted diseases (STD), including HIV (Aldunate et al., 2013). Vaginal acidity develops during puberty due to elevated estrogen levels which promote the accumulation of glycogen in epithelial cells. The latter is then converted into acetic and lactic acids by fermentation through epithelial cells and lactobacilli of the vaginal microbiota (Boskey et al., 2001).
Lactobacilli are the dominant microbiota in the vagina of healthy women. They are responsible for maintaining vaginal acidity (Boskey et al., 1999, 2001) which inhibits colonization by bacterial pathogens and restores the vaginal ecosystem (Juarez Tomas et al., 2003). Any lactobacilli decline or eradication events render the host more prone to bacterial vaginosis (BV), genital tract infections (GTIs) by Neisseria gonorrhoeae or Trichomonas vaginalis, vulvovaginal candidiasis (VVC), and urinary tract infections (UTIs) (Falagas et al., 2007; Ruiz et al., 2009). Key findings in BV, for instance, are the overgrowth of anaerobic bacteria, diminished lactobacilli levels and most importantly an elevated vaginal pH above 4.5 (Brabin et al., 2005; Falagas et al., 2007). Lactobacilli have even been proposed as an alternative strategy for controlling GBS infections (Acikgoz et al., 2005; Ruiz et al., 2012). In addition to vaginal acidification, lactobacilli can block pathogens of the genitourinary tract by mechanisms, such as releasing adhesion-inhibiting bio-surfactants, antimicrobial substances and hydrogen peroxide as well as auto-aggregation, co-aggregating with other bacterial species and surface hydrophobicity (Ruiz et al., 2009). GBS need to be able to counteract or evade all these strategies in order to successfully colonize the vaginal mucosa.
The Phagosome
Phagocytic cells as part of the innate immune system are recruited to sites of infections in order to eliminate invading pathogens. Once recognized, microbial pathogens are phagocytosed, and incorporated into a phagosome. Phagosomal maturation then occurs leading to the creation of phagolysosomes (Flannagan et al., 2009; Russell et al., 2009).
Maturation involves alteration of the phagosomal membrane which results in an extremely acidic, oxidative and degradative environment. Early phagosome possesses a moderately acidic (pH 6.1–6.5) intraluminal pH. As maturation proceeds to later stages, the lumen becomes more acidic (pH 5.5–6.0) and enriched in proteases and lysosomal-associated membrane proteins (lAMPs). Terminal phagolysosomal compartments are strictly acidic, possessing intraluminal pH-values as low as 4.5, and contain reactive oxygen species (ROS), reactive nitrogen species (RNS), and antimicrobial peptides. Thus, during the maturation process, phagosomes become fully armed with destructive antimicrobial features (Ohkuma and Poole, 1978; Yates et al., 2007; Flannagan et al., 2009).
Despite considering GBS mainly as an extracellular pathogen, it has the ability to persist and survive within macrophages. Prolonged GBS survival within the phagolysosome has previously been shown (Valenti-Weigand et al., 1996; Cornacchione et al., 1998; Cumley et al., 2012). Surprisingly, GBS was found to be > 10-fold less prone to hydrogen peroxide killing than catalase-producing Staphylococcus aureus (Wilson and Weaver, 1985). Furthermore, it was recently reported that the acidification of phagosomes is essential for prolonged intracellular survival of GBS (Cumley et al., 2012) and that the pilus protein which is required for antimicrobial peptides resistance is up-regulated under acidic conditions (Maisey et al., 2008b). In line with these observations, the two-component regulator system CovS/CovR, which mediates GBS tolerance to acid stress, has been shown to be critical for survival within the phagosome (Cumley et al., 2012).
Effect of Low pH on GBS Adherence and Biofilm Formation
Adherence to host tissues is a fundamental step preceding GBS colonization and ensuing invasion. GBS is capable to adhere and invade various host cells, including epithelial cells of the vagina and the lung, endothelial cells and micro-vascular endothelial cells of the blood brain barrier (Maisey et al., 2008a). Following the initial attachment to certain proteins, GBS interacts with integrins of the host-cell surface and often enters into these cells. Numerous proteins have been reported as adhesins contributing to GBS binding to host cells and extracellular matrix (ECM) proteins (Nobbs et al., 2009; Park et al., 2012).
As a commensal of the female genital tract, GBS colonization preferentially takes place at low vaginal pH-values. Early reports demonstrated enhanced GBS adherence at low pH to vaginal and respiratory epithelial cells (Zawaneh et al., 1979; Tamura et al., 1994). Microarray analysis (Santi et al., 2009) detected the up-regulation of the dpsA gene under acidic conditions, which is essential for the attachment of GBS to eukaryotic cells, at low pH (Samen et al., 2004). Furthermore, the glutamine transport gene glnQ (Tamura et al., 2002) required for both adherence to fibronectin in vitro and for virulence in vivo was found to be up-regulated under decreasing pH-values (Santi et al., 2009). Comparative expression analysis between GBS strains with different clinical virulence resulted in the identification of a novel adhesin designated HvgA which is a specific signature for the hypervirulent GBS ST17 strain. HvgA promotes colonization, adhesion and host invasion. It was found to be regulated by the acid-sensing two-component regulator CovS/CovR and was reported to be crucial for GBS intestinal colonization (Tazi et al., 2010).
Low pH also promotes biofilm production in GBS (Rosini and Margarit, 2015). Bacteria in biofilms form high density populations that are more resistant to stress (Nobbs et al., 2009) and contribute to the acid resistance of Gram-positive bacteria (Cotter and Hill, 2003). GBS is able to form biofilms with a maximum biofilm formation time of 48 h (Yang et al., 2012), which is influenced by environmental pH changes. Increased biofilm formation by GBS is observed at acidic pH (Ho et al., 2013; D'Urzo et al., 2014). It is noteworthy that also elevated expression levels of pilus components are observed at acidic pH (Santi et al., 2009). Pilus proteins have previously been reported to be involved in GBS epithelial adherence and biofilm formation (Konto-Ghiorghi et al., 2009; Rinaudo et al., 2010) leading to the hypothesis that a higher pilus expression in the acidic environment of the vagina may favor colonization. D'Urzo et al. (2014) investigated 389 GBS isolates for their ability to produce biofilms. They reported clones belonging to the hypervirulent ST17 serotype III GBS lineage to be the best biofilm producers forming stronger biofilms under acidic pH. The hypervirulent ST17 harbors genes encoding pili and the HvgA adhesin (Tazi et al., 2010; D'Urzo et al., 2014; Teatero et al., 2016; Perichon et al., 2017) It is the most frequent cause for neonatal meningitis relative to other GBS clones (D'Urzo et al., 2014; Landwehr-Kenzel and Henneke, 2014; Teatero et al., 2016). Furthermore, the global virulence regulator CovS/CovR has been shown to control GBS adherence and biofilm production in response to environmental pH changes (Santi et al., 2009; Park et al., 2012). According to Santi et al. (2009), CovRS mediates the upregulation of virulence determinants and controls GBS transition from colonizing to invasive state upon translocation from the acidic vagina to neutral host environments. Consistently, Patras et al. (2013) demonstrated CovRS critical in modulating the host innate immune response by restricting virulence factors expression to promote colonization and maintain the GBS commensal phenotype in the vagina (Patras et al., 2013).
Bacterial Mechanisms to Cope with the Acid Stress
Production of Alkali: Arginine Deiminase System (ADI)
One of the common pathways to counteract acid stress is the release of buffering substances. The release of alkaline molecules, such as ammonia seems to be a universal approach of acid resistance in lactic acid bacteria where arginine is a well-known ammonia precursor. A classic enzymatic pathway for ammonia release is the Arginine deiminase system (ADI), it comprises an arginine deiminase, ornithine carbamoyltransferase (also referred as ornithine transcarbamylase), and carbamate kinase. These three enzymes are encoded by the arcA, arcB and arcC genes, respectively. ADI provides two moles of ammonia and one ATP unit per each molecule of arginine. To maintain pH homeostasis, ATP can be used to expel protons across the membrane while a by-product like ornithine is secreted out of the cell in place of arginine in an energy independent manner under the action of an ornithine/arginine antiporter, encoded by arcD (van de Guchte et al., 2002; Cotter and Hill, 2003; Kajfasz and Quivey, 2011).
Arginine deiminase system (ADI) has been characterized in oral streptococci (Burne and Marquis, 2000; Dong et al., 2002; Griswold et al., 2004; Gruening et al., 2006), S. pyogenes (Hering et al., 2013), and S. pneumoniae (Schulz et al., 2014). Complete genome projects show that GBS possess an ADI (Glaser et al., 2002; Tettelin et al., 2002, 2005) which, although not yet characterized, appears to resemble homologs in other streptococci (Griswold et al., 2004). Transcription analysis in GBS reported genes encoding a carbamate kinase and an ornithine carbamoyltransferase to be extremely up-regulated at low pH (Santi et al., 2009). Similar findings were reported during a global transcript profiling of growth phase regulated genes in GBS. The authors indicated 55–150 fold up-regulation of ADI genes during the stationary growth phase (Sitkiewicz and Musser, 2009) which can be considered as an acid stress phase for GBS as it produces lactic acid as a side-product of carbohydrate fermentation.
However, ADI function in GBS seems not to be restricted to acid tolerance. A recent proteomic study reported the up-regulation of arginine deiminase ArcA in response to human serum (Yang et al., 2011). Transcriptome analysis revealed 5–15 fold induction of arcA following incubation with human blood (Mereghetti et al., 2008) and about 2–3 fold increase at stationary phase growth in amniotic fluid (Sitkiewicz et al., 2009). According to Cotter and Hill (2003) environmental pH itself is not a major factor that triggers ADI expression in streptococci. Direct involvement of ADI in acid tolerance is not mandatory. Rather than low pH, ADI expression could be also driven by intracellular arginine, lack of energy, catabolite suppression or oxygenation (van de Guchte et al., 2002).
Proton Pumps: F0F1-ATPase
The F1F0-ATPase is a multi-subunit enzyme which translocates protons at the expense of ATP or conversely synthesizes ATP using protons. It is composed of a membrane-embedded F0 complex, which consists of the three subunits a, b, and c and a cytoplasmic-bound F1 complex. F0 translocates protons as a membrane-bound proton specific channel. The F1 domain comprises five subunits α, β, γ, δ, and ε. It promotes ATP synthesis during protons movement from the extracellular compartments into the cytoplasm, or cleaves ATP when protons are expelled out of the cell. The net result is a more alkaline cytoplasm in comparison to an extrinsic acidic environment (van de Guchte et al., 2002; Cotter and Hill, 2003).
Interestingly, the genetic organization of the encoding atp operon in streptococci, atpEBFHAGDC (Smith et al., 1996; Quivey et al., 2001; Cotter and Hill, 2003), is somewhat different from that described earlier for E. coli (Walker et al., 1984). The F0 gene order in streptococci consists of atpEBF. This is dissimilar to the atpBEF order identified in other bacteria. If this variant genetic organization is important in regard to function is however still under investigation. Generally, the F-ATPase is the primary mechanism pumping out protons in order to keep pH homeostasis of streptococci under acidic condition. The transcriptional machinery of F-ATPase appears to be similar for many streptococcal species. Transcription analysis of the F-ATPase systems demonstrated an upregulation of the encoding genes at low pH for S. mutans, S. suis, S. sanguinis, and S. pneumoniae (Martín-Galiano et al., 2001; Kuhnert et al., 2004; Gong et al., 2009; Wei et al., 2011). Genome sequencing projects have predicted GBS to possess the F-ATPase operon similar to other streptococci (Glaser et al., 2002; Tettelin et al., 2002, 2005), but a detailed functional analysis hat not been conducted in GBS. The available large scale transcriptome analysis in GBS indicated a steady or slightly increasing transcript level of the F1-ATP synthase subunits atpABEF genes over time independently of growth phase (Sitkiewicz and Musser, 2009). Likewise, a recent proteomic investigation reported the abundance of the F1-ATP synthease subunit gamma during mid-exponential growth phase of GBS (Yang et al., 2010). F-ATPase activity under acid stress in GBS may thus not only be restricted to the extrusion of protons but may also be needed for ATP synthesis during growth and maintenance. Being a lactic acid producing organism, a steady accumulation of acids is expected during GBS growth and may explain the elevated transcription levels of the F1-ATP system. Supporting this interpretation, previous investigations indicated the existence of a basal level of ATPases at alkaline pH, which was attributed to allow rapid resumption of growth upon pH decrease (Kobayashi et al., 1986; Kakinuma, 1998; Cotter and Hill, 2003).
Acid Tolerance Response (ATR)
Some bacterial species display elevated survival rates after exposure to lethal acidic pH provided that they are only briefly challenged with sub-lethal acidic levels. This phenomenon is recognized as the acid tolerance response (ATR) (Cotter and Hill, 2003). However, other bacteria possess additional systems to cope with acid stress at levels that are too acidic to permit growth (pH 2.5 and below). A response which is designated acid resistance (AR) or extreme acid resistance (XAR) (Lund et al., 2014). So ATR comprises mechanisms that keep intracellular pH homoeostasis, while the XAR or AR represents an extreme acid stress response that includes mechanisms to avoid the intracellular pH from dropping to life-threating levels (Cotter and Hill, 2003; Lund et al., 2014). It is a remarkable observation that F0F1-ATPase and not ADI has been frequently reported to play a role in the induction of ATR (Cotter and Hill, 2003; Lund et al., 2014). Both ATR and AR have been defined for E. coli (Goodson and Rowbury, 1989), Salmonella Typhimurium (Foster and Hall, 1990), Lactococcus lactis, and Lactobacilli (Lund et al., 2014) while ATR has been described in oral streptococci (Hamilton and Buckley, 1991; Nascimento et al., 2004; Papadimitriou et al., 2007; Martinez et al., 2010) and for S. pneumoniae (Martin-Galiano et al., 2005).
Neither ATR nor AR has been investigated or addressed in GBS. The only available study is that done by Yang et al. (2012). They observed that short term acid exposure does not drive an ATR in GBS when inoculating acid adapted cells into pH 5 and monitoring them for long term survival. The authors were not able to detect any improvement in GBS survival. Despite these findings, there remains a possibility that GBS is able to mount an ATR under different culture conditions. Nascimento et al. (2004) was able to detect ATR in S. sobrinus when using bacterial cells cultivated in a continuous chemostat culture in contrast to Svensäter et al. (1997) who were unable to distinguish any ATR when employing buffered media and batch-cultivated cells of S. sobrinus.
Sensing Acid Stress and Signaling
Modulation of gene expression as a consequence of varying extracellular environmental conditions is a fundamental adaptation response that is mandatory for bacteria to replicate and survive (Cotter and Hill, 2003). Alternative sigma factors, transcriptional regulators and two-component signal transduction systems (TCSs) have been demonstrated to control the coordinated gene expression in bacteria experiencing changing environmental conditions (Cotter and Hill, 2003).
Sigma factors play an essential part in the bacterial response to low pH. Analysis of the GBS genome revealed the presence of three putative sigma factors, the major sigma factor σA, ComX and an ECF-type sigma factor (Glaser et al., 2002). The later has been reported in S. equi but not in S. pneumoniae, S. mutans, or S. pyogenes (Glaser et al., 2002). In addition, Glaser et al. (2002) identified numerous transcriptional regulators in GBS representing 5% of the predicted genes, several of which were upregulated at pH 5.5 (Santi et al., 2009). Similar findings were made for S. mutans another streptococcal species highly adapted to an acidic environment (Gong et al., 2009).
Two-component signal transduction systems (TCSs) are composed of a membrane-associated histidine kinase sensor and a cytoplasmic response regulator. They contribute to adaptation, virulence and survival through detecting environmental fluctuations and providing a proper response (Cotter and Hill, 2003). The S. mutans genome contains 14 TCS of which LiaRS, CiaRH, ComDE, and CovRS were found to be up-regulated upon acid adaptation (Gong et al., 2009). Besides, they were formerly reported to participate in acid stress and considered as crucial virulence elements in S. mutans (Li et al., 2002; Ahn et al., 2006; Lévesque et al., 2007; Gong et al., 2009; Kawada-Matsuo et al., 2009). The GBS genome was found to encode as many as 20 sensor histidine kinases and 21 response regulators (Glaser et al., 2002). Interestingly genome comparison revealed a much higher number of TCSs in GBS than those reported in related species, such as S. pyogenes, S. pneumoniae, S. mutans, and L. lactis suggesting a higher capacity of GBS to adapt to varying environmental conditions (Glaser et al., 2002). However, only few TCSs have been characterized in GBS to date (Klinzing et al., 2013). One of the most well-studied TCSs is the CovRS system (or CsrRS) (Faralla et al., 2014). It is best known as a major virulence regulator of pathogenic streptococci and its direct contribution to acid stress tolerance has previously been reported for GBS (Lamy et al., 2004; Santi et al., 2009; Firon et al., 2013; Faralla et al., 2014; Perichon et al., 2017). Genome-wide transcription analysis found 90% of the down-regulated genes and 60% of the up-regulated genes at pH 5.5 were CovRS dependent (Santi et al., 2009). Another TCS in GBS is the CiaR/H system. Consistent to S. mutans, reports on GBS demonstrated CiaR/H contribution to acid adaptation and enhancing intracellular survival in macrophages (Quach et al., 2009). The CiaR/H-dependent genes in GBS have shown significant homology to acid and multi-stress tolerant genes of L. lactis (Quach et al., 2009). Furthermore, recent microarray analysis proposed the involvement of CiaR/H in acid tolerance of S. suis (Wei et al., 2011).
Osmoregulation
Since most bacteria do not possess active water transport mechanisms to maintain cell turgor, alternative approaches have been developed to endure osmotic stress (Poolman et al., 2002). Shifts in osmotic pressure result in altered gene expression patterns of transporters or enzymes to sustain water equilibrium (Poolman et al., 2002). One of the well-known bacterial strategies in response to osmotic stress is to accumulate solutes, such as glycine betaine, proline, carnitine, and choline as osmoprotectants. These are soluble zwitterionic substances which can be retrieved from the surrounding milieu or biosynthesized intracellularly in high concentrations without interfering in critical physiological pathways (Kempf and Bremer, 1998).
Evidence for a putative connection between surviving acidic conditions and osmotic stress was reported by Santi et al. for GBS (Santi et al., 2009). They found an up-regulation of genes encoding components of the glycine betaine osmoregulation system upon shifting GBS from pH 7 to pH 5.5. The glycine betaine system has previously been reported to counteract osmotic pressure in Bacillus subtilis (Kempf and Bremer, 1995, 1998) and L. lactis (Obis et al., 1999). Consistently, in S. mutans it was observed that acid tolerance responses following an acid shock from pH 7.5 to pH 5.5 may protect against osmotic stress (Svensater et al., 2000). A potential connection between ATR and osmotic stress is supported by findings in S. pneumoniae that showed an increased expression level of a choline transporter under acid stress (Martin-Galiano et al., 2005). A choline transporter is comparable to the glycine betaine transporter of B. subtilis (Kappes et al., 1996) where an osmotically mediated choline uptake is also reported (Kappes et al., 1999). Choline is a necessary precursor for the biosynthesis of glycine betaine (Kappes et al., 1996, 1999).
Protection or Repair of Macromolecules: Chaperones
Rapid adaptive bacterial responses to abrupt environmental changes involve the release of proteases and chaperones in order to protect and repair macromolecules like DNA and proteins which are essential for optimal acid adaptation (Cotter and Hill, 2003). Chaperones provide protection against different environmental stresses by aiding in protein folding, renaturation, and eradication of damaged proteins (Cotter and Hill, 2003). The most common bacterial molecular chaperones include DnaK, DnaJ, GrpE and HrcA, which are encoded by the DnaK operon (hrcA-grpE-dnaK-dnaJ), GroEL and GroES, encoded by the GroE operon (groES-groEL), and Clp proteases (Jayaraman et al., 1997; Lemos et al., 2001; Nair et al., 2003; Henderson et al., 2006; Tomoyasu et al., 2012). DnaK is an ATP-dependent chaperone that works together with the co-chaperone DnaJ and the nucleotide exchange factor GrpE. It prevents protein aggregation and ensures proper folding by binding to hydrophobic peptides sequences during protein synthesis. GroEL possesses an interior cavity where substrates are protected by the union of GroES to GroEL in an ATP-dependent manner. Both DnaK and GroEL are negatively regulated by HrcA (Lemos et al., 2001; Wong and Houry, 2004). The Clp ATPases constitutes a huge family of highly conserved and universal proteins that are present in both prokaryotic and eukaryotic organisms. They act as molecular chaperones and contribute to protein assembly, folding, as well as proteolysis (Nair et al., 2003).
Chaperones and proteases are greatly conserved throughout different genomes implying their fundamental tasks for cellular life (Jordan et al., 2002). A clear linkage between acid stress and chaperones can be demonstrated in numerous Gram-positive bacteria. S. mutans grown in continuous chemostate culture displayed a marked increase in DnaK levels in response to acid shock (Jayaraman et al., 1997). Supporting these findings an up-regulated expression of DnaK, DnaJ and GroEL in response to acidic stimuli were reported for S. mutans in different studies (Lemos et al., 2001; Wilkins et al., 2002; Matsui and Cvitkovitch, 2010). Moreover, similar results were found for S. pneumoniae (Martin-Galiano et al., 2005) and S. sobrinus (Nascimento et al., 2004) under acid stress. Consistently, proteomic investigation in GBS identified GroES among the more abundantly expressed proteins in cells grown at pH 5 (Yang et al., 2010). Sitkiewicz and Musser (2009) could demonstrate the genes hrcA, grpE, dnaK to be significantly up-regulated in the more acidic stationary growth phase of GBS. However, a different scenario was presented by Santi et al. (2009) as genes belonging to the hrcA-grpE operon were detected to be down-regulated upon shifting from pH 7 to pH 5.5. Interestingly a down-regulated DnaK operon during acid shock was also found by Wei et al. (2011) in S. suis. In agreement, a S. intermedius dnaK mutant could not show significant acid sensitivity (Tomoyasu et al., 2012). One possible explanation for the contradictory results in GBS could be attributed to differences in growth conditions and most notably the brief exposure time to acid stress (30 min) utilized by Santi et al. (2009).
The Clp chaperones have also been linked to acid tolerance responses. Previous studies in S. mutans employing a ClpL-deficient mutant exhibited impaired viability of the mutant under acidic conditions in comparison to the parent strain (Lemos and Burne, 2002; Kajfasz et al., 2009). Likewise, Len et al. (2004) found ClpL protein levels to increase under acidic pH in S. mutans. The gene encoding ClpL protease was also up-regulated in response to acid in S. pneumoniae (Martin-Galiano et al., 2005) and reported to take part in ATR response where an early activation of ClpL (5 min after acid shock) could be detected. Genome analysis revealed (Glaser et al., 2002), that GBS encodes a bulk group of Clp protease subunits, which include the ClpP proteolytic subunit, four ATPase regulatory subunits; ClpX, ClpC, ClpL, ClpE, and three identical ClpA ATPase paralogs. Characterization of the ClpP serine protease in GBS reported its critical involvement in growth regulation under stress conditions (Nair et al., 2003). Moreover, elevated transcriptions of clpE and clpL in GBS during the more acidic stationary phase have been observed (Sitkiewicz and Musser, 2009).
Metal Ion Homeostasis
Metal ion homeostasis in streptococci plays a central role for colonization and invasion. It is involved in enabling adhesins to interact with host surfaces, supporting streptococcal growth in nutrient limiting niches, tolerating oxidative stress, evading host innate immune defenses and producing biofilms (Nobbs et al., 2009). Of special interest in this regard are manganese and iron transporters. Both ions are crucial for bacterial pathogenesis and are known to be highly restricted inside the host. Several investigations propose that the uptake and response to metal ions, such as Mn2+ and Fe2+, is vital for virulence and physiology of pathogenic streptococci (Kloosterman et al., 2008). Cytoplasmic Mn2+ can help in protecting bacteria against oxidative stress, and the induction of Mn2+ uptake by H2O2 has been observed in many bacteria (Horsburgh et al., 2002). Iron represents an essential cofactor for the function of several enzymes. It acts as a catalyst in electron transport processes (Somerville and Proctor, 2009; Diaz-Ochoa et al., 2014) and could indirectly play a role in the anti-oxidative response (Echave et al., 2003).
Connections between acid stress and metal ion homeostasis have been addressed in several studies. Transcriptional analyses of acid tolerance responses in streptococci have reported a remarkable up-regulation of different metal-ion transporters. In S. pneumoniae (Martin-Galiano et al., 2005) elevated expression levels of genes belonging to the psaABC and putative fatDCBE operons were observed which are involved in manganese (Dintilhac et al., 1997; Johnston et al., 2006) and iron transport (Hoskins et al., 2001). Similarly, the expression of sloR could be linked to the acid tolerance response in S. mutans (Gong et al., 2009) (Idone et al., 2003; Dunning et al., 2008). The sloR gene encodes a metal-dependent transcriptional regulator of the sloABCR operon, which is responsible for Mn and Fe uptake (Paik et al., 2003). Genes encoding metal ion transporters of nickel, Fe2+ and Mn2+, as well as genes encoding ABC transporter systems for the import of zinc and molybdenum, were found to be up-regulated in response to acid stress. The authors attributed this to an increased need for essential metals during acid stress. Likewise, for GBS, Santi et al. (2009) reported higher expression levels of several Mn2+ and Fe2+ transporters upon shifting from pH 7 to pH 5.5. These included members of the fhuCDBG operon coding for a siderophore-dependent iron transporter (Clancy et al., 2006), the putative iron-compound ABC transporter operon (Glaser et al., 2002; Tettelin et al., 2005) and members of the mtsABC operon (Glaser et al., 2002; Tettelin et al., 2005) which represents of of the Mn2+ and Fe2+ transporters (Bray et al., 2009) in GBS.
Another metal ion transporter reported by Santi et al. (2009) to be up-regulated in response to acid stress is the mntH gene coding for a metal ion NRAMP transporter of GBS. This transporter is predicted to participate in Mn2+ and Fe2+ homeostasis (Glaser et al., 2002; Tettelin et al., 2005). Interestingly, a recent investigation indicated a direct connection between acid stress and the expression of the NRAMP transporter mntH in GBS (Shabayek et al., 2016). MntH was characterized as crucial for GBS growth and survival under low pH conditions and essential for prolonged intracellular survival inside acidic host compartments. In line with these observations, global transcription studies in GBS (Di Palo et al., 2013) reported mntH to be regulated by the CovRS system which has been shown to be directly implicated in the GBS response to acid stimuli. Furthermore, previously bacterial NRAMP homologs have been characterized as pH-dependent transporters of divalent metal ions with preference for Mn2+ and Fe2+ in the following bacterial species E. coli, S. typhimurium, Mycobacterium tuberculosis and B. subtilis (Agranoff et al., 1999; Kehres et al., 2000; Makui et al., 2000; Que and Helmann, 2000).
Oxidative and Acidic Stress
Aerobic conditions are potentially harmful for bacteria due to the formation of highly reactive oxygen species (ROS), such as superoxide anions, hydrogen peroxide and hydroxyl radicals. ROS are toxic leading to irreversible DNA and protein damage (Fridovich, 1978; Imlay and Linn, 1988) and subsequent microbial killing. The primary microbial defense mechanisms to counteract these lethal effects is the release of superoxide dismutases (SODs) (Rolfe et al., 1978). The Mn-dependent SOD, encoded by sodA gene, is a major mechanism for ROS detoxification especially in streptococci, which lack a catalase (Poyart et al., 1998). Interestingly, several reports have shown sodA to play a significant role in acid resistance and acid-adaptive responses, underlining important overlaps between oxidative and acidic stress. Wilkins et al. (2002) demonstrated SOD of S. mutans among the up-regulated proteins in response to low pH. Similarly, SOD induction was also found in S. oralis when cells are cultured at pH 5.2 vs. pH 7 (Wilkins et al., 2003). The correlation of SOD expression with acidic conditions does not seem to be restricted to streptococci. A sodA mutant of S. aureus was shown to be more sensitive to acid stress in comparison to the parental strain (Clements et al., 1999). Moreover, SOD was reported to be crucial for acid tolerance in Vibrio vulnificus where sodA mutants suffered higher killing rates at pH 5 than at pH 7.5 in comparison to the wild-type strain (Kim et al., 2005).
In GBS with a lack of catalase activity SOD is fundamental in neutralizing oxidative stress (Poyart et al., 1998). Despite a previous investigation reporting SOD to be down-regulated upon exposure to low pH in GBS (Santi et al., 2009), there is evidence for the indirect involvement of the manganese dependent SOD in acid resistance. MntH, belonging to the NRAMP family of Mn transporters has been identified as essential for GBS growth under low pH conditions and in response to ROS (Shabayek et al., 2016). Both sodA and mntH were found to be induced at low pH in this recent investigation. These results support an interpretation were GBS SOD activity is dependent on MntH activity to provide optimum intracellular levels of manganese, its metal ion cofactor. A similar correlation between Mn homeostasis and SOD activity was reported earlier for Brucella abortus (Anderson et al., 2009) and S. pyogenes (Janulczyk et al., 2003).
Other strategies used to prevent ROS formation in streptococci include NADH oxidases. There are two genes, nox-1 encoding a H2O2-forming NADH oxidase which promotes the reduction of oxygen to H2O2 whereas nox-2 encodes a H2O-forming NADH oxidase which catalyzes the reduction of oxygen to water without forming harmful ROS intermediates (Higuchi et al., 1993, 1999). Both genes were described in S. mutans (Higuchi et al., 1993, 1999), however, only nox-2 gene was investigated in most other streptococci including GBS (Gibson et al., 2000; Yu et al., 2001; Yamamoto et al., 2006; Ge et al., 2016). Yamamoto et al. (2006), characterized Nox-2 as the main NADH oxidase that is required for aerobic growth and oxidative stress in conventional culture media in GBS. Moreover, addition of antioxidants did not alleviate aerobic growth defect in the GBS nox-2 mutant, however, it could be restored by the addition of exogenous unsaturated fatty acids. Previous reports in streptococci have related membrane fatty acid composition with acid tolerance (Kajfasz and Quivey, 2011; Quivey et al., 2016). Even more, concurrent acid and oxidative stresses have been shown to be associated with elevated unsaturated fatty acid abundance (Derr et al., 2012). Taken together, these data suggest that the aerobic growth defect of the GBS nox-2 mutant is due to the impaired ability to cope with the simultaneous acid stress induced by lactic acid, a growth by-product, as confirmed by the altered membrane fatty acid composition. Aerobic growth in GBS is fermentive and results mainly in acid production in the absence of heme and quinone (Yamamoto et al., 2005). In agreement, the nox-2 mutant of S. sanguinis was shown to be more sensitive to oxidative stress and acid stress (Ge et al., 2016). Furthermore, cultures of S. mutans UA159 exposed to concurrent acid and oxidative stresses demonstrated elevated levels of nox-2 transcription than under either stress alone (Baker et al., 2014, 2015b). Unfortunately, studies characterizing the direct involvement of Nox-2 in GBS acid stress are not available.
The transcription of nox have been shown to be modulated by rex which encodes the redox-sensing regulator Rex. This is a NAD+/NADH sensing transcription factor that is active when bound to NAD+ and inactive when bound to NADH. It senses the NAD+/NADH ratio in the cell and controls its regulon accordingly (Bitoun and Wen, 2016). The Rex regulator is ubiquitously conserved across Gram-positive bacteria (Ravcheev et al., 2012; Bitoun and Wen, 2016). It has been characterized in S. mutans (Bitoun et al., 2011), E. faecalis (Vesic and Kristich, 2013), and recently in S. pneumoniae (Luong et al., 2015). Although genome sequencing deduced Rex to be present in GBS (Bitoun and Wen, 2016), it has not yet been characterized.
Cytochrome bd quinol oxidase (CydABCD) is another system that is involved in protection against oxidative stress. This system has been shown to be essential for the respiratory metabolism and establishing a full respiration chain in GBS when the surrounding environment supplies heme and quinone. Aeration alone had a slight effect on aerobic growth in comparison to static conditions in GBS as indicated by Yamamoto et al. (2005). Interestingly, the authors observed that aerobic growth in GBS was mainly fermentive resulting in lower pH culture values in the absence of heme and quinone. They also observed that cydA expression was induced late in growth. In concordance, Santi et al. (2009) reported all the four subunits of the Cytochrome bd quinol oxidase system to be upregulated in response to acid stress.
Oxidative stress tolerance in streptococci is also driven by alkayl hydroperoxidase (AhpCF) and thiol peroxidase (Tpx) (Lemos et al., 2011; Papadimitriou et al., 2016). In S. mutans, peroxidase activity is mainly achieved through the AhpC-AhpF complex where AhpF (or Nox-1) acts as a dehydrogenase that reduces NADH and delivers electrons to AhpC. In addition, AhpC converts peroxidase into water or alchohol (Lemos et al., 2011). Transcriptional profile analysis displayed the upregulation of ahpCF upon transition from steady-state growth at pH 7 to steady-state growth at pH 5 in S. mutans (Baker et al., 2015a). Surprisingly, AhpC was characterized as a heme-binding protein that is required for complete respiration activity in GBS. It was demonstrated to be involved in managing intracellular heme (Lechardeur et al., 2010). A direct contribution of AhpC in GBS acid tolerance has not been reported. Thiol peroxidase (Tpx) is a member of the peroxiredoxin family of antioxidant enzymes, which collect electrons from a reducing system containing thioredoxin and thioredoxine reductase (La Carbona et al., 2007). Genome-wide transcription analysis in GBS demonstrated tpx gene to be induced in response to acid stress (Santi et al., 2009). This is consistent with the observation of significantly diminished intracellular survival of a E. faecalis tpx mutant inside mouse peritoneal macrophages in comparison to mutants for the NADH peroxidase npr and the alkayl hydroperoxidase ahpCF genes (La Carbona et al., 2007). However, Tpx has not been characterized in GBS and remains to be investigated.
Conclusion
In GBS a wealth of data has accumulated from genome sequencing projects, genome-wide transcription analysis and proteome mapping resulting in the large-scale identification of genes and proteins that are induced in response to low-pH environments. However, care must be taken when interpreting these results, to differentiate between genes and proteins with a direct contribution in the pH stress response and more unspecific effects. To validate these results, more conventional techniques need to be employed, such as the construction of mutants and addressing the consequences of these mutations on an individual basis. Unfortunately, functional studies to investigate GBS acid responses are still rare. Nonetheless, this review shows that genomic and proteomics-based approaches have identified many acid-inducible genes, such as the ADI system genes, F1F0-ATPase genes, genes of the glycine betaine system, genes encoding Clp chaperones, metal-ion transporters especially for Mn2+ and Fe2+, the Mn-dependent SOD, cytochrome bd quinol oxidase and thiol peroxidase. These genes can be targeted in further mutagenesis studies to assess and confirm their physiological significance for GBS in coping with acid stress. As discussed above, a successful scenario was recently shown when proofing the direct contribution of the mntH NRAMP transporter in GBS acid stress that was first mentioned in an earlier genome-wide transcription study.
Author Contributions
BS and SS both designed and wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the Egyptian Ministry of Higher Education and Scientific Research for the financial support of a PhD scholarship to SS.
References
Acikgoz, Z. C., Gamberzade, S., Gocer, S., and Ceylan, P. (2005). Inhibitor effect of vaginal lactobacilli on group B streptococci. Mikrobiyol. Bul. 39, 17–23.
Agranoff, D., Monahan, I. M., Mangan, J. A., Butcher, P. D., and Krishna, S. (1999). Mycobacterium tuberculosis expresses a novel pH-dependent divalent cation transporter belonging to the Nramp family. J. Exp. Med. 190, 717–724. doi: 10.1084/jem.190.5.717
Ahn, S.-J., Wen, Z. T., and Burne, R. A. (2006). Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 74, 1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006
Aldunate, M., Tyssen, D., Johnson, A., Zakir, T., Sonza, S., Moench, T., et al. (2013). Vaginal concentrations of lactic acid potently inactivate HIV. J. Antimicrob. Chemother. 68, 2015–2025. doi: 10.1093/jac/dkt156
Anderson, E. S., Paulley, J. T., Gaines, J. M., Valderas, M. W., Martin, D. W., Menscher, E., et al. (2009). The manganese transporter MntH is a critical virulence determinant for Brucella abortus 2308 in experimentally infected mice. Infect. Immun. 77, 3466–3474. doi: 10.1128/IAI.00444-09
Baker, J. L., Abranches, J., Faustoferri, R. C., Hubbard, C. J., Lemos, J. A., Courtney, M. A., et al. (2015a). Transcriptional profile of glucose-shocked and acid-adapted strains of Streptococcus mutans. Mol. Oral Microbiol. 30, 496–517. doi: 10.1111/omi.12110
Baker, J. L., Derr, A. M., Faustoferri, R. C., and Quivey, R. G. Jr. (2015b). Loss of NADH Oxidase Activity in Streptococcus mutans leads to rex-mediated overcompensation in NAD+ regeneration by lactate dehydrogenase. J. Bacteriol. 197, 3645–3657. doi: 10.1128/JB.00383-15
Baker, J. L., Derr, A. M., Karuppaiah, K., MacGilvray, M. E., Kajfasz, J. K., Faustoferri, R. C., et al. (2014). Streptococcus mutans NADH oxidase lies at the intersection of overlapping regulons controlled by oxygen and NAD+ levels. J. Bacteriol. 196, 2166–2177. doi: 10.1128/JB.01542-14
Bitoun, J. P., Nguyen, A. H., Fan, Y., Burne, R. A., and Wen, Z. T. (2011). Transcriptional repressor Rex is involved in regulation of oxidative stress response and biofilm formation by Streptococcus mutans. FEMS Microbiol. Lett. 320, 110–117. doi: 10.1111/j.1574-6968.2011.02293.x
Bitoun, J. P., and Wen, Z. T. (2016). Transcription factor Rex in regulation of pathophysiology in oral pathogens. Mol. Oral Microbiol. 31, 115–124. doi: 10.1111/omi.12114
Bore, E., Langsrud, S., Langsrud, O., Rode, T. M., and Holck, A. (2007). Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology 153, 2289–2303. doi: 10.1099/mic.0.2007/005942-0
Boskey, E. R., Cone, R. A., Whaley, K. J., and Moench, T. R. (2001). Origins of vaginal acidity: high D/L lactate ratio is consistent with bacteria being the primary source. Hum. Reprod. 16, 1809–1813. doi: 10.1093/humrep/16.9.1809
Boskey, E. R., Telsch, K. M., Whaley, K. J., Moench, T. R., and Cone, R. A. (1999). Acid production by vaginal flora in vitro is consistent with the rate and extent of vaginal acidification. Infect. Immun. 67, 5170–5175.
Brabin, L., Roberts, S. A., Fairbrother, E., Mandal, D., Higgins, S. P., Chandiok, S., et al. (2005). Factors affecting vaginal pH levels among female adolescents attending genitourinary medicine clinics. Sex. Transm. Infect. 81, 483–487. doi: 10.1136/sti.2005.014621
Bray, B. A., Sutcliffe, I. C., and Harrington, D. J. (2009). Expression of the MtsA lipoprotein of Streptococcus agalactiae A909 is regulated by manganese and iron. Antonie Van Leeuwenhoek 95, 101–109. doi: 10.1007/s10482-008-9291-6
Burne, R. A., and Marquis, R. E. (2000). Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol. Lett. 193, 1–6. doi: 10.1111/j.1574-6968.2000.tb09393.x
Clancy, A., Loar, J. W., Speziali, C. D., Oberg, M., Heinrichs, D. E., and Rubens, C. E. (2006). Evidence for siderophore-dependent iron acquisition in group B streptococcus. Mol. Microbiol. 59, 707–721. doi: 10.1111/j.1365-2958.2005.04974.x
Clements, M. O., Watson, S. P., and Foster, S. J. (1999). Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J. Bacteriol. 181, 3898–3903.
Cornacchione, P., Scaringi, L., Fettucciari, K., Rosati, E., Sabatini, R., Orefici, G., et al. (1998). Group B streptococci persist inside macrophages. Immunology 93, 86–95. doi: 10.1046/j.1365-2567.1998.00402.x
Cotter, P. D., and Hill, C. (2003). Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67, 429–453. doi: 10.1128/MMBR.67.3.429-453.2003
Cumley, N. J., Smith, L. M., Anthony, M., and May, R. C. (2012). The CovS/CovR acid response regulator is required for intracellular survival of group B Streptococcus in macrophages. Infect. Immun. 80, 1650–1661. doi: 10.1128/IAI.05443-11
Derr, A. M., Faustoferri, R. C., Betzenhauser, M. J., Gonzalez, K., Marquis, R. E., and Quivey, R. G. Jr. (2012). Mutation of the NADH oxidase gene (nox) reveals an overlap of the oxygen- and acid-mediated stress responses in Streptococcus mutans. Appl. Environ. Microbiol. 78, 1215–1227. doi: 10.1128/AEM.06890-11
Diaz-Ochoa, V. E., Jellbauer, S., Klaus, S., and Raffatellu, M. (2014). Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis. Front. Cell. Infect. Microbiol. 4:2. doi: 10.3389/fcimb.2014.00002
Dintilhac, A., Alloing, G., Granadel, C., and Claverys, J. P. (1997). Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Mol. Microbiol. 25, 727–739. doi: 10.1046/j.1365-2958.1997.5111879.x
Di Palo, B., Rippa, V., Santi, I., Brettoni, C., Muzzi, A., Metruccio, M. M., et al. (2013). Adaptive response of group B streptococcus to high glucose conditions: new insights on the CovRS regulation network. PLoS ONE 8:e61294. doi: 10.1371/journal.pone.0061294
Dong, Y., Chen, Y. Y., Snyder, J. A., and Burne, R. A. (2002). Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl. Environ. Microbiol. 68, 5549–5553. doi: 10.1128/AEM.68.11.5549-5553.2002
Dunning, D. W., McCall, L. W., Powell, W. F. Jr., Arscott, W. T., McConocha, E. M., McClurg, C. J., et al. (2008). SloR modulation of the Streptococcus mutans acid tolerance response involves the GcrR response regulator as an essential intermediary. Microbiology 154, 1132–1143. doi: 10.1099/mic.0.2007/012492-0
D'Urzo, N., Martinelli, M., Pezzicoli, A., De Cesare, V., Pinto, V., Margarit, I., et al. (2014). Acidic pH strongly enhances in vitro biofilm formation by a subset of hypervirulent ST-17 Streptococcus agalactiae strains. Appl. Environ. Microbiol. 80, 2176–2185. doi: 10.1128/AEM.03627-13
Echave, P., Tamarit, J., Cabiscol, E., and Ros, J. (2003). Novel antioxidant role of alcohol dehydrogenase E from Escherichia coli. J. Biol. Chem. 278, 30193–30198. doi: 10.1074/jbc.M304351200
Falagas, M., Betsi, G. I., and Athanasiou, S. (2007). Probiotics for the treatment of women with bacterial vaginosis. Clin. Microbiol. Infect. 13, 657–664. doi: 10.1111/j.1469-0691.2007.01688.x
Faralla, C., Metruccio, M. M., De Chiara, M., Mu, R., Patras, K. A., Muzzi, A., et al. (2014). Analysis of two-component systems in group B Streptococcus shows that RgfAC and the novel FspSR modulate virulence and bacterial fitness. mBio 5:e00870-14. doi: 10.1128/mBio.00870-14
Farley, M. M. (2001). Group B streptococcal disease in nonpregnant adults. Clin. Infect. Dis. 33, 556–561. doi: 10.1086/322696
Firon, A., Tazi, A., Da Cunha, V., Brinster, S., Sauvage, E., Dramsi, S., et al. (2013). The Abi-domain protein Abx1 interacts with the CovS histidine kinase to control virulence gene expression in group B Streptococcus. PLoS Pathog. 9:e1003179. doi: 10.1371/journal.ppat.1003179
Flannagan, R. S., Cosio, G., and Grinstein, S. (2009). Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7, 355–366. doi: 10.1038/nrmicro2128
Foster, J. W., and Hall, H. K. (1990). Adaptive acidification tolerance response of Salmonella typhimurium. J. Bacteriol. 172, 771–778. doi: 10.1128/jb.172.2.771-778.1990
Fridovich, I. (1978). The biology of oxygen radicals. Science 201, 875–880. doi: 10.1126/science.210504
Ge, X., Yu, Y., Zhang, M., Chen, L., Chen, W., Elrami, F., et al. (2016). Involvement of NADH oxidase in competition and endocarditis virulence in Streptococcus sanguinis. Infect. Immun. 84, 1470–1477. doi: 10.1128/IAI.01203-15
Gibson, C. M., Mallett, T. C., Claiborne, A., and Caparon, M. G. (2000). Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182, 448–455. doi: 10.1128/JB.182.2.448-455.2000
Glaser, P., Rusniok, C., Buchrieser, C., Chevalier, F., Frangeul, L., Msadek, T., et al. (2002). Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45, 1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x
Gong, Y., Tian, X. L., Sutherland, T., Sisson, G., Mai, J., Ling, J., et al. (2009). Global transcriptional analysis of acid-inducible genes in Streptococcus mutans: multiple two-component systems involved in acid adaptation. Microbiology 155, 3322–3332. doi: 10.1099/mic.0.031591-0
Goodson, M., and Rowbury, R. J. (1989). Habituation to normally lethal acidity by prior growth of Escherichia coli at a sub-lethal acid pH value. Lett. Appl. Microbiol. 8, 77–79. doi: 10.1111/j.1472-765X.1989.tb00227.x
Griswold, A., Chen, Y. Y., Snyder, J. A., and Burne, R. A. (2004). Characterization of the arginine deiminase operon of Streptococcus rattus FA-1. Appl. Environ. Microbiol. 70, 1321–1327. doi: 10.1128/AEM.70.3.1321-1327.2004
Gruening, P., Fulde, M., Valentin-Weigand, P., and Goethe, R. (2006). Structure, regulation, and putative function of the arginine deiminase system of Streptococcus suis. J. Bacteriol. 188, 361–369. doi: 10.1128/JB.188.2.361-369.2006
Hamilton, I. R., and Buckley, N. D. (1991). Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol. Immunol. 6, 65–71. doi: 10.1111/j.1399-302X.1991.tb00453.x
Henderson, B., Allan, E., and Coates, A. R. M. (2006). Stress wars: the direct role of host and bacterial molecular chaperones in bacterial infection. Infect. Immun. 74, 3693–3706. doi: 10.1128/IAI.01882-05
Hering, S., Sieg, A., Kreikemeyer, B., and Fiedler, T. (2013). Kinetic characterization of arginine deiminase and carbamate kinase from Streptococcus pyogenes M49. Protein Expr. Purif. 91, 61–68. doi: 10.1016/j.pep.2013.07.002
Higuchi, M., Shimada, M., Yamamoto, Y., Hayashi, T., Koga, T., and Kamio, Y. (1993). Identification of two distinct NADH oxidases corresponding to H2O2-forming oxidase and H2O-forming oxidase induced in Streptococcus mutans. J. Gen. Microbiol. 139, 2343–2351. doi: 10.1099/00221287-139-10-2343
Higuchi, M., Yamamoto, Y., Poole, L. B., Shimada, M., Sato, Y., Takahashi, N., et al. (1999). Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J. Bacteriol. 181, 5940–5947.
Ho, Y. R., Li, C. M., Yu, C. H., Lin, Y. J., Wu, C. M., Harn, I. C., et al. (2013). The enhancement of biofilm formation in group B streptococcal isolates at vaginal pH. Med. Microbiol. Immunol. 202, 105–115. doi: 10.1007/s00430-012-0255-0
Horsburgh, M. J., Wharton, S. J., Karavolos, M., and Foster, S. J. (2002). Manganese: elemental defence for a life with oxygen. Trends Microbiol. 10, 496–501. doi: 10.1016/S0966-842X(02)02462-9
Hoskins, J., Alborn, W. E. Jr., Arnold, J., Blaszczak, L. C., Burgett, S., DeHoff, B. S., et al. (2001). Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183, 5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001
Idone, V., Brendtro, S., Gillespie, R., Kocaj, S., Peterson, E., Rendi, M., et al. (2003). Effect of an orphan response regulator on Streptococcus mutans sucrose-dependent adherence and cariogenesis. Infect. Immun. 71, 4351–4360. doi: 10.1128/IAI.71.8.4351-4360.2003
Imlay, J., and Linn, S. (1988). DNA damage and oxygen radical toxicity. Science 240, 1302–1309. doi: 10.1126/science.3287616
Janulczyk, R., Ricci, S., and Bjorck, L. (2003). MtsABC is important for manganese and iron transport, oxidative stress resistance, and virulence of Streptococcus pyogenes. Infect. Immun. 71, 2656–2664. doi: 10.1128/IAI.71.5.2656-2664.2003
Jayaraman, G. C., Penders, J. E., and Burne, R. A. (1997). Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol. Microbiol. 25, 329–341. doi: 10.1046/j.1365-2958.1997.4671835.x
Johnston, J. W., Briles, D. E., Myers, L. E., and Hollingshead, S. K. (2006). Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect. Immun. 74, 1171–1180. doi: 10.1128/IAI.74.2.1171-1180.2006
Jordan, I. K., Rogozin, I. B., Wolf, Y. I., and Koonin, E. V. (2002). Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res. 12, 962–968. doi: 10.1101/gr.87702
Juarez Tomas, M. S., Ocana, V. S., Wiese, B., and Nader-Macias, M. E. (2003). Growth and lactic acid production by vaginal Lactobacillus acidophilus CRL 1259, and inhibition of uropathogenic Escherichia coli. J. Med. Microbiol. 52, 1117–1124. doi: 10.1099/jmm.0.05155-0
Kajfasz, J. K., and Quivey, R. G. Jr. (2011). “Stress responses of lactic acid bacteria to acid stress,” in Stress Responses of Lactic Acid Bacteria, ed M. P. Doyle (New York, NY: Springer), 23–54.
Kajfasz, J. K., Martinez, A. R., Rivera-Ramos, I., Abranches, J., Koo, H., Quivey, R. G. Jr., et al. (2009). Role of Clp proteins in expression of virulence properties of Streptococcus mutans. J. Bacteriol. 191, 2060–2068. doi: 10.1128/JB.01609-08
Kakinuma, Y. (1998). Inorganic cation transport and energy transduction in Enterococcus hirae and other streptococci. Microbiol. Mol. Biol. Rev. 62, 1021–1045.
Kappes, R. M., Kempf, B., and Bremer, E. (1996). Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178, 5071–5079. doi: 10.1128/jb.178.17.5071-5079.1996
Kappes, R. M., Kempf, B., Kneip, S., Boch, J., Gade, J., Meier-Wagner, J., et al. (1999). Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32, 203–216. doi: 10.1046/j.1365-2958.1999.01354.x
Kawada-Matsuo, M., Shibata, Y., and Yamashita, Y. (2009). Role of two component signaling response regulators in acid tolerance of Streptococcus mutans. Oral Microbiol. Immunol. 24, 173–176. doi: 10.1111/j.1399-302X.2008.00485.x
Kehres, D. G., Zaharik, M. L., Finlay, B. B., and Maguire, M. E. (2000). The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol. Microbiol. 36, 1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x
Kempf, B., and Bremer, E. (1995). OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270, 16701–16713. doi: 10.1074/jbc.270.28.16701
Kempf, B., and Bremer, E. (1998). Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 170, 319–330. doi: 10.1007/s002030050649
Kim, J. S., Sung, M. H., Kho, D. H., and Lee, J. K. (2005). Induction of manganese-containing superoxide dismutase is required for acid tolerance in Vibrio vulnificus. J. Bacteriol. 187, 5984–5995. doi: 10.1128/JB.187.17.5984-5995.2005
Klinzing, D. C., Ishmael, N., Dunning Hotopp, J. C., Tettelin, H., Shields, K. R., Madoff, L. C., et al. (2013). The two-component response regulator LiaR regulates cell wall stress responses, pili expression and virulence in group B Streptococcus. Microbiology 159, 1521–1534. doi: 10.1099/mic.0.064444-0
Kloosterman, T. G., Witwicki, R. M., van der Kooi-Pol, M. M., Bijlsma, J. J., and Kuipers, O. P. (2008). Opposite effects of Mn2+ and Zn2+ on PsaR-mediated expression of the virulence genes pcpA, prtA, and psaBCA of Streptococcus pneumoniae. J. Bacteriol. 190, 5382–5393. doi: 10.1128/JB.00307-08
Kobayashi, H., Suzuki, T., and Unemoto, T. (1986). Streptococcal cytoplasmic pH is regulated by changes in amount and activity of a proton-translocating ATPase. J. Biol. Chem. 261, 627–630.
Konto-Ghiorghi, Y., Mairey, E., Mallet, A., Dumenil, G., Caliot, E., Trieu-Cuot, P., et al. (2009). Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422. doi: 10.1371/journal.ppat.1000422
Kuhnert, W. L., Zheng, G., Faustoferri, R. C., and Quivey, R. G. Jr. (2004). The F-ATPase operon promoter of Streptococcus mutans is transcriptionally regulated in response to external pH. J. Bacteriol. 186, 8524–8528. doi: 10.1128/JB.186.24.8524-8528.2004
La Carbona, S., Sauvageot, N., Giard, J. C., Benachour, A., Posteraro, B., Auffray, Y., et al. (2007). Comparative study of the physiological roles of three peroxidases (NADH peroxidase, Alkyl hydroperoxide reductase and Thiol peroxidase) in oxidative stress response, survival inside macrophages and virulence of Enterococcus faecalis. Mol. Microbiol. 66, 1148–1163. doi: 10.1111/j.1365-2958.2007.05987.x
Lamy, M. C., Zouine, M., Fert, J., Vergassola, M., Couve, E., Pellegrini, E., et al. (2004). CovS/CovR of group B streptococcus: a two-component global regulatory system involved in virulence. Mol. Microbiol. 54, 1250–1268. doi: 10.1111/j.1365-2958.2004.04365.x
Landwehr-Kenzel, S., and Henneke, P. (2014). Interaction of Streptococcus agalactiae and cellular innate immunity in colonization and disease. Front. Immunol. 5:519. doi: 10.3389/fimmu.2014.00519
Lechardeur, D., Fernandez, A., Robert, B., Gaudu, P., Trieu-Cuot, P., Lamberet, G., et al. (2010). The 2-Cys peroxiredoxin alkyl hydroperoxide reductase c binds heme and participates in its intracellular availability in Streptococcus agalactiae. J. Biol. Chem. 285, 16032–16041. doi: 10.1074/jbc.M109.024505
Le Doare, K., and Heath, P. T. (2013). An overview of global GBS epidemiology. Vaccine 31(Suppl. 4), D7–D12. doi: 10.1016/j.vaccine.2013.01.009
Lemos, J. A., and Burne, R. A. (2002). Regulation and physiological significance of ClpC and ClpP in Streptococcus mutans. J. Bacteriol. 184, 6357–6366. doi: 10.1128/JB.184.22.6357-6366.2002
Lemos, J. A. C., Chen, Y.-Y. M., and Burne, R. A. (2001). Genetic and physiologic analysis of the groE operon and role of the HrcA repressor in stress gene regulation and acid tolerance in Streptococcus mutans. J. Bacteriol. 183, 6074–6084. doi: 10.1128/JB.183.20.6074-6084.2001
Lemos, J. A., Tsakalidou, E., and Papadimitriou, K. (2011). “Stress responses of Streptococci,” in Stress Responses of Lactic Acid Bacteria, ed M. P. Doyle (New York, NY: Springer), 251–303.
Len, A. C., Harty, D. W., and Jacques, N. A. (2004). Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150, 1339–1351. doi: 10.1099/mic.0.27008-0
Lévesque, C. M., Mair, R. W., Perry, J. A., Lau, P. C. Y., Li, Y. H., and Cvitkovitch, D. G. (2007). Systemic inactivation and phenotypic characterization of two-component systems in expression of Streptococcus mutans virulence properties. Lett. Appl. Microbiol. 45, 398–404. doi: 10.1111/j.1472-765X.2007.02203.x
Li, Y.-H., Lau, P. C. Y., Tang, N., Svensäter, G., Ellen, R. P., and Cvitkovitch, D. G. (2002). Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184, 6333–6342. doi: 10.1128/JB.184.22.6333-6342.2002
Lund, P., Tramonti, A., and De Biase, D. (2014). Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol. Rev. 38, 1091–1125. doi: 10.1111/1574-6976.12076
Luong, T. T., Kim, E. H., Bak, J. P., Nguyen, C. T., Choi, S., Briles, D. E., et al. (2015). Ethanol-induced alcohol dehydrogenase E (AdhE) potentiates pneumolysin in Streptococcus pneumoniae. Infect. Immun. 83, 108–119. doi: 10.1128/IAI.02434-14
Maisey, H. C., Doran, K. S., and Nizet, V. (2008a). Recent advances in understanding the molecular basis of group B streptococcus virulence. Expert Rev. Mol. Med. 10:e27. doi: 10.1017/S1462399408000811
Maisey, H. C., Quach, D., Hensler, M. E., Liu, G. Y., Gallo, R. L., Nizet, V., et al. (2008b). A group B streptococcal pilus protein promotes phagocyte resistance and systemic virulence. FASEB J. 22, 1715–1724. doi: 10.1096/fj.07-093963
Makui, H., Roig, E., Cole, S. T., Helmann, J. D., Gros, P., and Cellier, M. F. (2000). Identification of the Escherichia coli K-12 nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol. Microbiol. 35, 1065–1078. doi: 10.1046/j.1365-2958.2000.01774.x
Martinez, A. R., Abranches, J., Kajfasz, J. K., and Lemos, J. A. (2010). Characterization of the Streptococcus sobrinus acid-stress response by interspecies microarrays and proteomics. Mol. Oral Microbiol. 25, 331–342. doi: 10.1111/j.2041-1014.2010.00580.x
Martín-Galiano, A. J., Ferrándiz, M. J., and De La Campa, A. G. (2001). The promoter of the operon encoding the F0F1-ATPase of Streptococcus pneumoniae is inducible by pH. Mol. Microbiol. 41, 1327–1338. doi: 10.1046/j.1365-2958.2001.02597.x
Martin-Galiano, A. J., Overweg, K., Ferrandiz, M. J., Reuter, M., Wells, J. M., and de la Campa, A. G. (2005). Transcriptional analysis of the acid tolerance response in Streptococcus pneumoniae. Microbiology 151, 3935–3946. doi: 10.1099/mic.0.28238-0
Matsui, R., and Cvitkovitch, D. (2010). Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol. 5, 403–417. doi: 10.2217/fmb.09.129
Melin, P. (2011). Neonatal group B streptococcal disease: from pathogenesis to preventive strategies. Clin. Microbiol. Infect. 17, 1294–1303. doi: 10.1111/j.1469-0691.2011.03576.x
Mereghetti, L., Sitkiewicz, I., Green, N. M., and Musser, J. M. (2008). Extensive adaptive changes occur in the transcriptome of Streptococcus agalactiae (group B streptococcus) in response to incubation with human blood. PLoS ONE 3:e3143. doi: 10.1371/journal.pone.0003143
Nair, S., Poyart, C., Beretti, J. L., Veiga-Fernandes, H., Berche, P., and Trieu-Cuot, P. (2003). Role of the Streptococcus agalactiae ClpP serine protease in heat-induced stress defence and growth arrest. Microbiology 149, 407–417. doi: 10.1099/mic.0.25783-0
Nascimento, M. M., Lemos, J. A. C., Abranches, J., Gonçalves, R. B., and Burne, R. A. (2004). Adaptive acid tolerance response of Streptococcus sobrinus. J. Bacteriol. 186, 6383–6390. doi: 10.1128/JB.186.19.6383-6390.2004
Nobbs, A. H., Lamont, R. J., and Jenkinson, H. F. (2009). Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450. doi: 10.1128/MMBR.00014-09
Obis, D., Guillot, A., Gripon, J. C., Renault, P., Bolotin, A., and Mistou, M. Y. (1999). Genetic and biochemical characterization of a high-affinity betaine uptake system (BusA) in Lactococcus lactis reveals a new functional organization within bacterial ABC transporters. J. Bacteriol. 181, 6238–6246.
Ohkuma, S., and Poole, B. (1978). Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. U.S.A. 75, 3327–3331. doi: 10.1073/pnas.75.7.3327
Paik, S., Brown, A., Munro, C. L., Cornelissen, C. N., and Kitten, T. (2003). The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J. Bacteriol. 185, 5967–5975. doi: 10.1128/JB.185.20.5967-5975.2003
Papadimitriou, K., Alegria, A., Bron, P. A., de Angelis, M., Gobbetti, M., Kleerebezem, M., et al. (2016). Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 80, 837–890. doi: 10.1128/MMBR.00076-15
Papadimitriou, K., Pratsinis, H., Nebe-von-Caron, G., Kletsas, D., and Tsakalidou, E. (2007). Acid tolerance of Streptococcus macedonicus as assessed by flow cytometry and single-cell sorting. Appl. Environ. Microbiol. 73, 465–476. doi: 10.1128/AEM.01244-06
Park, S. E., Jiang, S., and Wessels, M. R. (2012). CsrRS and environmental pH regulate group B streptococcus adherence to human epithelial cells and extracellular matrix. Infect. Immun. 80, 3975–3984. doi: 10.1128/IAI.00699-12
Patras, K. A., Wang, N. Y., Fletcher, E. M., Cavaco, C. K., Jimenez, A., Garg, M., et al. (2013). Group B Streptococcus CovR regulation modulates host immune signalling pathways to promote vaginal colonization. Cell. Microbiol. 15, 1154–1167. doi: 10.1111/cmi.12105
Perichon, B., Szili, N., du Merle, L., Rosinski-Chupin, I., Gominet, M., Bellais, S., et al. (2017). Regulation of PI-2b pilus expression in hypervirulent Streptococcus agalactiae ST-17 BM110. PLoS ONE 12:e0169840. doi: 10.1371/journal.pone.0169840
Poolman, B., Blount, P., Folgering, J. H., Friesen, R. H., Moe, P. C., and van der Heide, T. (2002). How do membrane proteins sense water stress? Mol. Microbiol. 44, 889–902. doi: 10.1046/j.1365-2958.2002.02894.x
Poyart, C., Quesne, G., Coulon, S., Berche, P., and Trieu-Cuot, P. (1998). Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36, 41–47.
Quach, D., van Sorge, N. M., Kristian, S. A., Bryan, J. D., Shelver, D. W., and Doran, K. S. (2009). The CiaR response regulator in group B Streptococcus promotes intracellular survival and resistance to innate immune defenses. J. Bacteriol. 191, 2023–2032. doi: 10.1128/JB.01216-08
Que, Q., and Helmann, J. D. (2000). Manganese homeostasis in Bacillus subtilis is regulated by MntR, a bifunctional regulator related to the diphtheria toxin repressor family of proteins. Mol. Microbiol. 35, 1454–1468. doi: 10.1046/j.1365-2958.2000.01811.x
Quivey, R. G., Faustoferri, R. C., Santiago, B., Baker, J., Cross, B., and Xiao, J. (2016). “Acid-adaptive responses of Streptococcus mutans, and mechanisms of integration with oxidative stress,” in Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria, ed F. J. d. Bruijn (Hoboken, NJ: John Wiley and Sons, Inc.), 897–910.
Quivey, R. G., Kuhnert, W. L., and Hahn, K. (2001). Genetics of acid adaptation in oral streptococci. Crit. Rev. Oral Biol. Med. 12, 301–314. doi: 10.1177/10454411010120040201
Ravcheev, D. A., Li, X., Latif, H., Zengler, K., Leyn, S. A., Korostelev, Y. D., et al. (2012). Transcriptional regulation of central carbon and energy metabolism in bacteria by redox-responsive repressor Rex. J. Bacteriol. 194, 1145–1157. doi: 10.1128/JB.06412-11
Rinaudo, C. D., Rosini, R., Galeotti, C. L., Berti, F., Necchi, F., Reguzzi, V., et al. (2010). Specific involvement of pilus type 2a in biofilm formation in group B streptococcus. PLoS ONE 5:e9216. doi: 10.1371/journal.pone.0009216
Rolfe, R. D., Hentges, D. J., Campbell, B. J., and Barrett, J. T. (1978). Factors related to the oxygen tolerance of anaerobic bacteria. Appl. Environ. Microbiol. 36, 306–313.
Rosini, R., and Margarit, I. (2015). Biofilm formation by Streptococcus agalactiae: influence of environmental conditions and implicated virulence factors. Front. Cell. Infect. Microbiol. 5:6. doi: 10.3389/fcimb.2015.00006
Ruiz, F. O., Gerbaldo, G., Asurmendi, P., Pascual, L. M., Giordano, W., and Barberis, I. L. (2009). Antimicrobial activity, inhibition of urogenital pathogens, and synergistic interactions between lactobacillus strains. Curr. Microbiol. 59, 497–501. doi: 10.1007/s00284-009-9465-0
Ruiz, F. O., Gerbaldo, G., Garcia, M. J., Giordano, W., Pascual, L., and Barberis, I. L. (2012). Synergistic effect between two bacteriocin-like inhibitory substances produced by lactobacilli strains with inhibitory activity for Streptococcus agalactiae. Curr. Microbiol. 64, 349–356. doi: 10.1007/s00284-011-0077-0
Russell, D. G., Vanderven, B. C., Glennie, S., Mwandumba, H., and Heyderman, R. S. (2009). The macrophage marches on its phagosome: dynamic assays of phagosome function. Nat. Rev. Immunol. 9, 594–600. doi: 10.1038/nri2591
Samen, U., Gottschalk, B., Eikmanns, B. J., and Reinscheid, D. J. (2004). Relevance of peptide uptake systems to the physiology and virulence of Streptococcus agalactiae. J. Bacteriol. 186, 1398–1408. doi: 10.1128/JB.186.5.1398-1408.2004
Santi, I., Grifantini, R., Jiang, S. M., Brettoni, C., Grandi, G., Wessels, M. R., et al. (2009). CsrRS regulates group B Streptococcus virulence gene expression in response to environmental pH: a new perspective on vaccine development. J. Bacteriol. 191, 5387–5397. doi: 10.1128/JB.00370-09
Schulz, C., Gierok, P., Petruschka, L., Lalk, M., Mader, U., and Hammerschmidt, S. (2014). Regulation of the arginine deiminase system by ArgR2 interferes with arginine metabolism and fitness of Streptococcus pneumoniae. mBio 5:e01858-14. doi: 10.1128/mBio.01858-14
Shabayek, S., Bauer, R., Mauerer, S., Mizaikoff, B., and Spellerberg, B. (2016). A streptococcal NRAMP homologue is crucial for the survival of Streptococcus agalactiae under low pH conditions. Mol. Microbiol. 100, 589–606. doi: 10.1111/mmi.13335
Sitkiewicz, I., Green, N. M., Guo, N., Bongiovanni, A. M., Witkin, S. S., and Musser, J. M. (2009). Transcriptome adaptation of group B Streptococcus to growth in human amniotic fluid. PLoS ONE 4:e6114. doi: 10.1371/journal.pone.0006114
Sitkiewicz, I., and Musser, J. M. (2009). Analysis of growth-phase regulated genes in Streptococcus agalactiae by global transcript profiling. BMC Microbiol. 9:32. doi: 10.1186/1471-2180-9-32
Smith, A. J., Quivey, R. G. Jr., and Faustoferri, R. C. (1996). Cloning and nucleotide sequence analysis of the Streptococcus mutans membrane-bound, proton-translocating ATPase operon. Gene 183, 87–96. doi: 10.1016/S0378-1119(96)00502-1
Somerville, G. A., and Proctor, R. A. (2009). At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol. Mol. Biol. Rev. 73, 233–248. doi: 10.1128/MMBR.00005-09
Svensäter, G., Larsson, U. B., Greif, E. C. G., Cvitkovitch, D. G., and Hamilton, I. R. (1997). Acid tolerance response and survival by oral bacteria. Oral Microbiol. Immunol. 12, 266–273. doi: 10.1111/j.1399-302X.1997.tb00390.x
Svensater, G., Sjogreen, B., and Hamilton, I. R. (2000). Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology 146(Pt 1), 107–117. doi: 10.1099/00221287-146-1-107
Tamura, G. S., Kuypers, J. M., Smith, S., Raff, H., and Rubens, C. E. (1994). Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect. Immun. 62, 2450–2458.
Tamura, G. S., Nittayajarn, A., and Schoentag, D. L. (2002). A glutamine transport gene, glnQ, is required for fibronectin adherence and virulence of group B streptococci. Infect. Immun. 70, 2877–2885. doi: 10.1128/IAI.70.6.2877-2885.2002
Tazi, A., Disson, O., Bellais, S., Bouaboud, A., Dmytruk, N., Dramsi, S., et al. (2010). The surface protein HvgA mediates group B streptococcus hypervirulence and meningeal tropism in neonates. J. Exp. Med. 207, 2313–2322. doi: 10.1084/jem.20092594
Teatero, S., Ramoutar, E., McGeer, A., Li, A., Melano, R. G., Wasserscheid, J., et al. (2016). Clonal Complex 17 group B Streptococcus strains causing invasive disease in neonates and adults originate from the same genetic pool. Sci. Rep. 6:20047. doi: 10.1038/srep20047
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., et al. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U.S.A. 102, 13950–13955. doi: 10.1073/pnas.0506758102
Tettelin, H., Masignani, V., Cieslewicz, M. J., Eisen, J. A., Peterson, S., Wessels, M. R., et al. (2002). Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U.S.A. 99, 12391–12396. doi: 10.1073/pnas.182380799
Tomoyasu, T., Tabata, A., Imaki, H., Tsuruno, K., Miyazaki, A., Sonomoto, K., et al. (2012). Role of Streptococcus intermedius DnaK chaperone system in stress tolerance and pathogenicity. Cell Stress Chaperones 17, 41–55. doi: 10.1007/s12192-011-0284-4
Valenti-Weigand, P., Benkel, P., Rohde, M., and Chhatwal, G. S. (1996). Entry and intracellular survival of group B streptococci in J774 macrophages. Infect. Immun. 64, 2467–2473.
van de Guchte, M., Serror, P., Chervaux, C., Smokvina, T., Ehrlich, S., and Maguin, E. (2002). Stress responses in lactic acid bacteria. Antonie Van Leeuwenhoek 82, 187–216. doi: 10.1023/A.1020631532202
Verani, J. R., McGee, L., and Schrag, S. J. (2010). Prevention of perinatal group B streptococcal disease–revised guidelines from CDC, (2010). MMWR Recomm. Rep. 59, 1–36.
Vesic, D., and Kristich, C. J. (2013). A Rex family transcriptional repressor influences H2O2 accumulation by Enterococcus faecalis. J. Bacteriol. 195, 1815–1824. doi: 10.1128/JB.02135-12
Walker, J. E., Saraste, M., and Gay, N. J. (1984). The unc operon. nucleotide sequence, regulation and structure of ATP-synthase. Biochim. Biophys. Acta 768, 164–200. doi: 10.1016/0304-4173(84)90003-X
Wei, Y., Zeng, X., Yuan, Y., Jiang, H., Zheng, Y., Tan, Y., et al. (2011). DNA microarray analysis of acid-responsive genes of Streptococcus suis serotype 2. Ann. Microbiol. 61, 505–510. doi: 10.1007/s13213-010-0165-6
Wilkins, J. C., Beighton, D., and Homer, K. A. (2003). Effect of acidic pH on expression of surface-associated proteins of Streptococcus oralis. Appl. Environ. Microbiol. 69, 5290–5296. doi: 10.1128/AEM.69.9.5290-5296.2003
Wilkins, J. C., Homer, K. A., and Beighton, D. (2002). Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions. Appl. Environ. Microbiol. 68, 2382–2390. doi: 10.1128/AEM.68.5.2382-2390
Wilson, C. B., and Weaver, W. M. (1985). Comparative susceptibility of group B streptococci and Staphylococcus aureus to killing by oxygen metabolites. J. Infect. Dis. 152, 323–329. doi: 10.1093/infdis/152.2.323
Wong, P., and Houry, W. A. (2004). Chaperone networks in bacteria: analysis of protein homeostasis in minimal cells. J. Struct. Biol. 146, 79–89. doi: 10.1016/j.jsb.2003.11.006
Yamamoto, Y., Pargade, V., Lamberet, G., Gaudu, P., Thomas, F., Texereau, J., et al. (2006). The group B Streptococcus NADH oxidase Nox-2 is involved in fatty acid biosynthesis during aerobic growth and contributes to virulence. Mol. Microbiol. 62, 772–785. doi: 10.1111/j.1365-2958.2006.05406.x
Yamamoto, Y., Poyart, C., Trieu-Cuot, P., Lamberet, G., Gruss, A., and Gaudu, P. (2005). Respiration metabolism of group B Streptococcus is activated by environmental haem and quinone and contributes to virulence. Mol. Microbiol. 56, 525–534. doi: 10.1111/j.1365-2958.2005.04555.x
Yang, Q., Porter, A. J., Zhang, M., Harrington, D. J., Black, G. W., and Sutcliffe, I. C. (2012). The impact of pH and nutrient stress on the growth and survival of Streptococcus agalactiae. Antonie Van Leeuwenhoek 102, 277–287. doi: 10.1007/s10482-012-9736-9
Yang, Q., Zhang, M., Harrington, D. J., Black, G. W., and Sutcliffe, I. C. (2010). A proteomic investigation of Streptococcus agalactiae grown under conditions associated with neonatal exposure reveals the upregulation of the putative virulence factor C protein beta antigen. Int. J. Med. Microbiol. 300, 331–337. doi: 10.1016/j.ijmm.2010.01.001
Yang, Q., Zhang, M., Harrington, D. J., Black, G. W., and Sutcliffe, I. C. (2011). A proteomic investigation of Streptococcus agalactiae reveals that human serum induces the C protein beta antigen and arginine deiminase. Microbes Infect. 13, 757–760. doi: 10.1016/j.micinf.2011.03.001
Yates, R. M., Hermetter, A., Taylor, G. A., and Russell, D. G. (2007). Macrophage activation downregulates the degradative capacity of the phagosome. Traffic 8, 241–250. doi: 10.1111/j.1600-0854.2006.00528.x
Yu, J., Bryant, A. P., Marra, A., Lonetto, M. A., Ingraham, K. A., Chalker, A. F., et al. (2001). Characterization of the Streptococcus pneumoniae NADH oxidase that is required for infection. Microbiology 147, 431–438. doi: 10.1099/00221287-147-2-431
Keywords: Streptococcus agalactiae, acid resistance, low pH, molecular mechanisms, stress response
Citation: Shabayek S and Spellerberg B (2017) Acid Stress Response Mechanisms of Group B Streptococci. Front. Cell. Infect. Microbiol. 7:395. doi: 10.3389/fcimb.2017.00395
Received: 26 March 2017; Accepted: 23 August 2017;
Published: 07 September 2017.
Edited by:
Wolfgang Eisenreich, Technische Universität München, GermanyReviewed by:
Peter Valentin-Weigand, University of Veterinary Medicine Hannover, GermanySven Hammerschmidt, University of Greifswald, Germany
Copyright © 2017 Shabayek and Spellerberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara Spellerberg, YmFyYmFyYS5zcGVsbGVyYmVyZ0B1bmlrbGluaWstdWxtLmRl
 Sarah Shabayek
Sarah Shabayek Barbara Spellerberg
Barbara Spellerberg