
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 25 August 2017
Sec. Molecular Bacterial Pathogenesis
Volume 7 - 2017 | https://doi.org/10.3389/fcimb.2017.00382
This article is part of the Research Topic Role of Iron in Bacterial Pathogenesis View all 13 articles
 Yunqing Guo1,2
Yunqing Guo1,2 Di Hu1,2
Di Hu1,2 Jie Guo1,2
Jie Guo1,2 Xiaowen Li1,2
Xiaowen Li1,2 Jinyue Guo1,2
Jinyue Guo1,2 Xiliang Wang1,2
Xiliang Wang1,2 Yuncai Xiao1,2
Yuncai Xiao1,2 Hui Jin1,3
Hui Jin1,3 Mei Liu1,2
Mei Liu1,2 Zili Li1,2
Zili Li1,2 Dingren Bi1,2,3*
Dingren Bi1,2,3* Zutao Zhou1,2*
Zutao Zhou1,2*Riemerella anatipestifer, an avian pathogen, has resulted in enormous economic losses to the duck industry globally. Notwithstanding, little is known regarding the physiological, pathogenic and virulence mechanisms of Riemerella anatipestifer (RA) infection. However, the role of Ferric uptake regulator (Fur) in the virulence of R. anatipestifer has not, to date, been demonstrated. Using a genetic approach, unmarked gene deletion system, we evaluated the function of fur gene in the virulence of R. anatipestifer. For this purpose, we constructed a suicide vector containing pheS as a counter selectable marker for unmarked deletion of fur gene to investigate its role in the virulence. After successful transformation of the newly constructed vector, a mutant strain was characterized for genes regulated by iron and Fur using RNA-sequencing and a comparison was made between wild type and mutant strains in both iron restricted and enriched conditions. RNA-seq analysis of the mutant strain in a restricted iron environment showed the downregulation and upregulation of genes which were involved in either important metabolic pathways, transport processes, growth or cell membrane synthesis. Electrophoretic mobility shift assay was performed to identify the putative sequences recognized by Fur. The putative Fur-box sequence was 5′-GATAATGATAATCATTATC-3′. Lastly, the median lethal dose and histopathological investigations of animal tissues also illustrated mild pathological lesions produced by the mutant strain as compared to the wild type RA strain, hence showing declined virulence. Conclusively, an unmarked gene deletion system was successfully developed for RA and the role of the fur gene in virulence was explored comprehensively.
Riemerella anatipestifer (R. anatipestifer, RA) is non-spore forming, non-motile, Gram-negative, rod-shaped bacterium belonging to the family Flavobacteriaceae. Other than ducks, RA can also affect the majority of poultry including turkeys, geese, which has resulted in significant economic losses to the poultry industry worldwide. Infection leads to polyserositis and septicaemia often with neurological symptoms. At least 21 serotypes have been identified in different countries, with serotypes 1, 2, and 10 most prevalent in China (Loh et al., 1992; Cheng et al., 2003). Due to extensive genomic divergences, even within a given serotype, there is often limited cross protection and variation of virulence (Higgins et al., 2000). Presently, little is known regarding RA pathogenesis, although a number of attempts have been made to explore the molecular mechanisms underlying virulence. In previous studies, the role of outer membrane protein A (OmpA), TonB dependent receptor 1 (TbdR1), TonB family protein (TbfA), siderophore interacting protein (Sip) and CAMP cohemolysin have all been proposed as virulence associated factors (Crasta et al., 2002; Hu et al., 2011; Lu et al., 2013; Tu et al., 2014; Liu et al., 2016). All of these studies were based on gene knockout, which result in modified expression of downstream genes, known as the polar effect. As a genetic analysis tool, unmarked gene deletion system is advantageous over gene knockout strategy, as well as able to provide a more accurate estimation of gene expression and has a limited polar effect. Indeed, in many species of bacteria, such as Enterococcus faecalis, Burkholderia family, Streptococcus mutans, Bacillus amyloliquefaciens, unmarked gene deletion system has been established to elucidate molecular mechanisms of pathogenesis and virulence (Kristich et al., 2007; Barrett et al., 2008; Xie et al., 2011; Zhou et al., 2016). To the best of our knowledge, no data is available on unmarked gene deletion system in RA. The development and application of such strategies will accelerate our understanding of the mechanism of pathogenesis, virulence and antibiotic resistance in RA.
Earlier studies have established the role of Ferric uptake regulator (Fur) proteins in virulence in a variety of bacterial species (Ernst et al., 2005a; Haraszthy et al., 2006; Yuhara et al., 2008; Porcheron and Dozois, 2015; Pi et al., 2016). Fur is a regulator of transcription in bacteria, involved in iron homeostasis, acid resistance, oxidative stress and virulence (Bijlsma et al., 2002; Ernst et al., 2005b; Mathieu et al., 2016). Iron is an essential element in various metabolic pathways of bacteria and eukaryotic host (Holmes et al., 2005). To date, the function of fur gene in virulence of RA has not been demonstrated in any previous study. In this novel study, the role of fur gene in virulence of RA has been examined by adopting unmarked gene deletion system. Having observed the limitations of other counter-selectable markers, pheS is an appropriate non-antibiotic resistance counter-selectable marker. Previously, the applications of the mutant E. coli pheS gene (A294G), the mutant E. faecalis pheS gene (A312G), the mutant Burkholderia pheS gene (A294G), and the mutant S. mutans pheS gene (A314G) were successful for allelic replacement in those organisms (Kast and Hennecke, 1991; Ibba et al., 1994; Kristich et al., 2007; Barrett et al., 2008; Xie et al., 2011). Therefore, we postulated the role of pheS gene for this purpose.
In summary, in this study, we engineered a suicide vector pRE-lacZ-mpheS-spc, using mutated pheS as a counter-selectable marker and lacZ to select a fur gene deletion mutant RA-YM Δfur. This is the first successful attempt to construct mutant RA using an unmarked gene deletion system. The RA-YM Δfur complemented strain was constructed to confirm virulence of the wild type strain, compared with the mutant. Lastly, using whole genome transcriptional sequencing, genes regulated by the fur gene were screened out in mutant and wild types. Moreover, the predictive sequence of Fur-box of RA was analyzed.
The bacterial strains and plasmids used in this study, and their relevant characteristics are described in Table 1. R. anatipestifer strains were grown at 37°C in tryptic soy broth (TSB) (Difco, Detroit, USA) in an atmosphere of 5% CO2, E. coli strains were cultured at 37°C in Luria Bertani broth (Sigma-Aldrich, St. Louis, USA). Both R. anatipestifer strains and E. coli strains included in this study were obtained from laboratory stocks of the Department of Veterinary Microbiology and Immunology of Huazhong Agricultural University, China. Where necessary, the following antibiotics were added in to the selection media: ampicillin (Amp), 100 mg/mL; spectinomycin (Spc), 100 mg/mL; kanamycin (Kan), 100 mg/mL; and medium was supplemented with 2, 6-diaminopimelic acid (DAP), 100 mg/mL; 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal), 20 mg/mL; Isopropylβ-D-1-thiogalactopyranoside (IPTG), 20 mg/mL; 4-chloro-DL-phenylalanine (cPhe), 0.2% (w/v).
For the construction of suicide vector pRE-lacZ-mpheS-spc, a 3.9 kb fragment of pRE was amplified from suicide vector pRE112 using primer S1L (Table 1) (introducing BamHI, SalI, and NdeI site) and S2R (Table 1) (introducing XhoI and BamHI site); then digested with BamHI enzyme and ligated to generate circular pRE which contained the essential components of the conjugational transfer. As RA cannot catabolize sucrose, the selected marker SacB was removed. The mutated pheS gene (mpheS) and the sequence of the multiple cloning sites were engineered into pUC57 to generate pUC57-mpheS and pUC57-MCS by the GenScript Corporation (Nanjing, China). This pUC57-mpheS vector contained the mutated R. anatipestifer pheS gene with altered DNA sequences (Supplemental Figure 1), which was driven by an upstream PS12 promoter of the R. anatipestifer rpsL gene. The 1.1 kb spc cassette was amplified from plasmid pIC333 using primers spcL and spcR (Table 1), mpheS was amplified from plasmid pUC57-mpheS using primers pheS1and pheS2 (Table 1). The spc cassette was then fused with the mpheS fragment using overlap PCR (introducing KpnI and XbaI site). The fragment of mpheS-spc was inserted into pMD18T to generate pMD18T-mpheS-spc. The pMD18T-mpheS-spc was digested with KpnI and XbaI enzymes, and 2.2 kb PS12-mpheS-spc fragment was ligated into pUC57-MCS and digested with KpnI and XbaI to obtain pUC57-MCS-mpheS-spc. The pRE and pUC57-MCS-mpheS-spc were digested with XhoI and XbaI and ligated to generate pRE-mpheS-spc. The next step was to amplify a 3.3 kb fragment of lacZ from E. coli BL21 genome using primer lacZR and lacZL (Table 1) (introducing a BamHI site). A 135 bp fragment of PS12 promoter was amplified from RA-YM using primer rpsL-LacZ (introducing a XhoI site) and rpsR-LacZ (Table 1) and fused to lacZ fragment by overlap PCR using rpsL-LacZ and lacZR primers. pRE-mpheS-spc and lacZ fragments were digested with BamHI and XhoI and ligated to obtain 9 kb pRE-lacZ-mpheS-spc.
The shuttle plasmid pRES-JXrep-spc was constructed in several steps including the amplification of 2.5 kb fragment of replicon region and replicase gene (Genebank: KY806579) with primers rep1 and rep2 (Table 1) (introducing a XhoI site). The wild type plasmid of R. anatipestifer strain RA-JX was used as a template. A 1.1 kb spc cassette was amplified from plasmid pIC333 using primers spcL1 and spcR1 (Table 1), thereby introducing a BamHI site. The replicon region of RA-JX was joined with the spc cassette using overlap PCR. The JXrep-spc fragment was inserted into pMD18T to obtain pMD18T-JXrep-spc. In the next step, plasmid pRE and pMD18T-JXrep-spc were digested with XhoI and BamHI and ligated to generate the shuttle plasmid pRES-JXrep-spc.
To obtain the suicide vector pRE-lacZ-mpheS-spc-fur for the deletion of whole fur gene from R. anatipestifer RA-YM strain, upstream (738 bp) and downstream (802 bp) DNA fragments were amplified using primers Fur-L1 and Fur-L2 (Table 1) (introducing SphI site), Fur-R1 and Fur-R2 (Table 1) (introducing KpnI site), respectively. The two fragments were joined together by overlap PCR. The LR fragment and pRE-lacZ-mpheS-spc were digested with SphI and KpnI; 1.5 kb LR fragment was inserted into pRE-lacZ-mpheS-spc to generate the suicide vector pRE-lacZ-mpheS-spc-fur. E. coli strain x7213 was used as a donor in conjugation step to introduce the suicide vector pRE-lacZ-mpheS-spc-fur into RA-YM strain as described previously (Hu et al., 2011). For phenotypic detection of mutant strains, conjugation filters were plated on tryptic soya agar (TSA) containing 100 μg/mL Spc. Colonies were then grown on TSA containing cPhe (0.2%) and X-gal (40 μg/mL). Appearance of white colonies confirmed successful construction of deletion mutant strains. For identification of recombinants carrying the chromosomal fur gene deletion, colonies were analyzed using PCR primers FurL1 and FurR2 to determine presence of wild-type or mutant allele at the target locus. The wild-type and deleted alleles could be differentiated on the basis of size of amplicon by agarose gel electrophoresis.
Similarly, for generation of complemented mutant strain, shuttle vector pRES-JXrep-spc-fur was constructed by amplification of the promoter sequence (171 bp) and the coding sequence (486 bp) of fur gene. The promoter sequence and the coding sequence were amplified using primers Promoter-fur1 and Promoter-fur2 (introducing KpnI site, Table 1), primers Fur-inL and Fur-inR (introducing SphI site, Table 1). The two fragments were joined together by overlap PCR. The plasmid pRES-JXrep-spc and fur gene fragment were digested with SphI and KpnI, then the fragment of 657 bp was inserted into plasmid pRES-JXrep-spc to obtain shuttle vector pRES-JXrep-spc-fur. The E. coli strain x7213 was used as donor in conjugation transfer of shuttle vector into the RA-YM Δfur strain (Hu et al., 2011). The phenotypic identification of complemented mutant strain was conducted on TSA plates containing Spc 100 μg/mL. Furthermore, PCR reaction was performed using primers Fur inL and Fur inR to ensure that recombinant strains were harboring shuttle vectors.
The colonies of the wild-type and the RA-YM Δfur mutant were suspended into tryptic soya broth (TSB) and incubated overnight with shaking at 37°C to an OD600 of 0.2. FeCl3 (Sigma-Aldrich) or 2, 2-Dipyridyl (2, 2-DP, Sigma-Aldrich) was added to the bacterial suspension to produce a final concentration of 200 and 30 μM, as iron restricted and iron rich conditions, respectively and incubated at 37°C until the OD600 reached 0.8. Total RNA was extracted from bacteria solution using Bacterial RNA Kit (OMEGA, Norcross, USA) following the guidelines. Extracted RNA was purified with RNase-free DNase (Promega, Wisconsin, USA) at 37°C for 30 min to remove impurities of DNA, the DNA-free purified RNA was examined by 1% agarose gel electrophoresis. Purified RNA (23S rRNA and 16S rRNA) was sent to Huada Gene Center (Shenzhen, China) for RNA sequencing. All RNA samples were performed in two independent biological replicates (BioProject: SRP106941).
RT-qPCR was performed to quantify the expression of genes regulated by Fur. Primers were designed with Primer 5.0 software. RNA was extracted from wild-type and RA-YM Δfur strains grown in iron-restricted and iron-rich medium. RNA was reverse transcribed to cDNA using PrimerScript RT regent Kit with gDNA Eraser (Takara, Dalian, China). Real-time PCR reaction was performed using SYBR Premix (Takara, Dalian, China). Each reaction was performed in triplicate. Relative quantification of gene expression was calculated according to 2−ΔΔCt method, RA-YM 16S rRNA was used as reference gene for normalized expression for each RNA sample.
The fragment of fur gene was amplified using the primers Fur1 and Fur2 (introducing the BamHI and XhoI sites); the fragment and vector pET-28a were digested with BamHI and XhoI and restricted fragment was ligated into the expression vector pET-28a to generate the expression vector pET-28a-fur. The expression plasmid was then transformed into competent cells of E. coli BL21 (DE3). Then the Fur protein was purified with an ÄKTA Purifier (GE His Trap FF, USA).
EMSA was performed with the Lightshift Chemiluminescent EMSA Kit (Thermo fisher scientific, Waltham, USA). The reaction was incubated at 30°C for 1 h, then loaded into 6% non-denaturing polyacrylamide gel electrophoretic and exposured. The reaction mixture (20 μL) contained 1 μg biotin labeled DNA fragment, 2 μL binding buffer, 1 μL KCl, 1 μL MgCl2, 1 μL glycerol, 1 μL NP-40 and 1 μL Poly(dI-dC) and desired concentration of Fur protein, the final concentration of Fur protein were 0, 0.1, 1, and 10 μg in four lanes. 16S rDNA was used as a negative control. DNA fragments to be identified were amplified by biotin labeled primer (Sangon, Shanghai, China). The length of DNA fragments ranged from 350 to 420 bp.
One-day-old Cherry Valley ducklings obtained from the Wuhan Duck Farm (Wuhan, China) housed in cages under 12-h light/dark cycle, at controlled temperature (28–30°C) and free access to food and water during the whole course of this study. Care and maintenance of all animals were in line with the standards of Institutional Animal Care. This experiment was approved by the Institutional Animal Experimental Committee of the Veterinary Faculty of Huazhong Agricultural University.
To determine the role of fur in virulence, the median lethal dose (LD50) of the deletion mutant RA-YM Δfur strain, the complemented mutant RA-YM Δfur strain and the wild-type RA-YM was measured using the Reed–Muench method (Reed and Muench, 1938). For each wild type, mutant and complemented strains, 12-day-old ducklings were evenly divided into five groups (10 ducklings/group). All five groups were injected intramuscularly with 1.0 × 104, 1.0 × 105, 1.0 × 106, 1.0 × 107, and 1.0 × 108 colony forming units (CFU) of wild type strain, respectively. Similarly, mutant and complemented strains were injected to respective groups of ducklings for the evaluation of LD50. Moribund ducklings were killed humanely and counted as dead. Dead ducklings were identified for the presence of RA. Mortality of the ducklings was recorded daily for a period of 10 days.
A comparative analysis of bacterial load in the blood of ducklings infected with mutants and wild type was made. Blood and target organs (brain, liver, heart and spleen) were collected at 24 and 48 h post-inoculation (five ducklings per group at each time-point). The target organs were homogenized with PBS to obtain supernatant. Blood and supernatant were plated on TSB agar plates for bacterial count with a 10-fold dilution method. In addition, the degree of lesions developing on the liver, spleen, heart and brain by the wild-type and the Δfur mutant strains were also recorded. For pathological investigations, all tissues were immersed in 10% formalin solution, embedded in paraffin section and stained with hematoxylin and eosin (H E). The pathological findings of the wild-type and the RA-YM Δfur mutant were compared.
As homologous recombination follows a two-step procedure, the selection of the R. anatipestifer Δfur mutant was carried out in two steps (Stibitz, 1994). The selection of mutant with Spc resistance was initially carried out, followed by the expression of mpheS gene. The function of lacZ gene could directly confirm whether the plasmid had been excised. The pheS gene was engineered by substituting alternative bases at numerous positions. The sequence similarity rate between wild-type pheS and mpheS was 71%. However, no difference was observed in amino acid sequence with the exception of the A301G mutation. In this study, the mpheS gene was driven by the promoter of RA rpsL gene. The first process was obtained by growing RA-YM strains on TSA medium containing Spc resistance. The first process obtained the merodiploid strains, which harbored the suicide vector. Then, the merodiploid strains were screened on TSA medium containing 0.2% cPhe and X-gal to obtain the deletion mutant. The merodiploid strains grew on the plate containing Spc but had no growth on the agar plate with 0.2% cPhe. The wild strains could be grown on the plate contains cPhe (Figures 1A2,A3). In addition, the merodiploid strains appeared as blue colonies while the wild type strain was of a white color on the plate containing X-gal (Figures 1A4,A5). This finding demonstrated the effectiveness of the counter-selectable markers pheS in RA. A suicide vector pRE-lacZ-mpheS-spc-fur, containing mutated pheS as a counter-selectable marker (Figure 1A1) was constructed and successfully transformed into RA-YM strain to generate RA-YM Δfur strain.
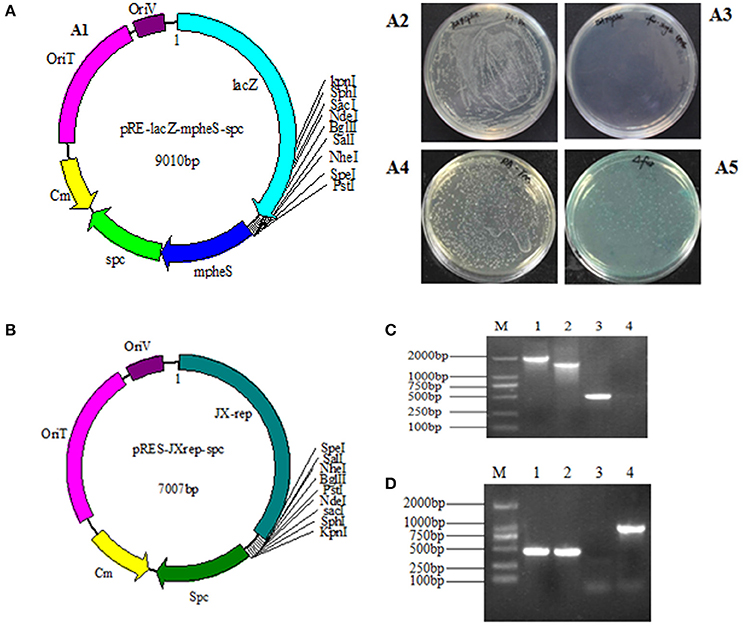
Figure 1. (A1) The map of plasmid pRE-lacZ-mpheS-spc containing resistance gene spc, counter-selectable marker mpheS and lacZ; (A2,A3) show the growth of the wild type strain and the merodiploid strains on the TSA plate with 0.2% cPhe respectively. (A4) The color of the RA-YM on X-gal plate was white. (A5) The color of the merodiploid strains on X-gal plate was blue. (B) The map of the complemented shuttle plasmid pRES-JXrep-spc. (C) The PCR amplification of the RA-YM Δfur deletion mutant strain and wild type RA-YM strain. Lane M: DL2000 DNA Marker; Lane 1: LR fragment amplification from RA-YM; Lane 2: LR fragment amplification from RA-YM Δfur deletion mutant strain; Lan 3: Amplification of fur gene from RA-YM; Lane 4: Amplification of fur gene from RA-YM Δfur deletion mutant strain; (D) The PCR amplification of RA-YM Δfur mutant complemented strain. Lane M: DL2000 DNA Marker; Lan 1: Amplification of fur gene wild type RA-YM; Lane 2: Amplification of fur gene RA-YM Δfur complemented mutant strain; Lane 3: Amplification of spc gene from wild type RA-YM; Lane 4: spc gene amplification from the RA-YM Δfur complementary mutant strain.
To determine whether the plasmid replicated and deleted fur gene after homologous recombination, no PCR amplification of fur gene and smaller LR fragment size (1,540 bp) from RA-YM Δfur strain as compared to larger LR fragment size (2,008 bp) from RA-YM strain (Figure 1C) confirmed the plasmid activity and recombination. Similarly, development of a recombinant RA-YM Δfur complemented mutant strain was confirmed by PCR amplification of fur and spc genes as shown in Figure 1D.
A comparison of gene expression regulated by iron and/or Fur in wild type (WT) and mutant (Δfur) strains grown in iron rich (+Fe) and iron restricted condition(−Fe) was exclusively established and comparison of RA-YM Δfur deletion mutant strain with wild type RA-YM was also performed. In our experiments, the significance of differentially expressed genes was estimated by the false discovery rate (FDR) and was considered significant if FDR < 0.001 and the |log2Ratio| >1(Ernst et al., 2005a; Ledala et al., 2010). In total, 25 genes were downregulated and 45 genes were upregulated by iron when grown in iron-restricted conditions, in both parent and mutant. Seventeen genes were directly regulated by Fur. The expression of eight genes randomly selected regulated by iron and Fur was confirmed by real-time PCR in RA-YM and the Δfur mutant (Figure 7). The real-time PCR result was in accordance to the transcriptional data.
Exclusively, 25 genes were downregulated when grown without iron in medium in both wild type RA-YM and RA-YM Δfur deletion mutant strains. Furthermore, ratio (WT-Fe/WT+Fe ratio and Δfur-Fe/Δfur+Fe ratio) of gene expression were calculated in both iron-restricted (−Fe) and iron-rich (+Fe) conditions, which were ≥2. Of the 25 downregulated genes, five genes encoded proteins which acted as transporters; six genes encoded enzymes which participated in tricarboxylic acid cycle; six were involved in oxidation-reduction; six genes encoded hypothetical proteins and two genes actively participated in amino acid biosynthesis (Table 2).
Similarly, 45 genes were upregulated by iron both in the parent and the Δfur mutant strain when iron was restricted. Gene ratio (WT-Fe/WT+Fe ratio and Δfur-Fe/Δfur+Fe ratio) was calculated ≥2. Among the 45 upregulated genes, nine genes participated in amino acids and cofactor biosynthesis; six were involved in cell envelope and surface structure formation; eight genes were involved in protein synthesis; transport and binding protein were encoded by six genes; three genes encoded regulators; cellular processes were regulated by three genes, and ten encoded hypothetical proteins (Table 3).
In almost all bacteria, Fur acted as a negative regulator. Genes directly regulated by Fur were observed to be upregulated when iron was restricted and when fur was mutated. The ratio (WT-Fe/WT+Fe ratio, Δfur-Fe/Δfur+Fe ratio and Δfur+Fe/WT+Fe ratio) of 17 genes regulated by Fur was ≥2 (Table 4). Of the 17 genes directly regulated by Fur, five genes contributed in iron acquisition; two were involved in oxidation-reduction; one gene participated in activation of type IX secretion system (T9SS); the functions of six genes remained unknown, and three encoded hypothetical proteins (Table 4).
The putative Fur binding sequence and the distance from the start condon of the genes regulated by Fur is shown in Table 5. Promoter sequences were analyzed using software RegPredict and ClustalW for identification of putative binding sequence of Fur protein which was 19 bp long and sequence was predicted as 5′-ATTTAGAATTATTCTAAAT-3′ (Figure 8A). Therefore, the Fur binding sequence might be located within 100 bp of the translation initiation codon of the regulated genes (Table 5). To verify the putative role of the Fur-box sequence, the promoters of hmuR, sprT, RAYM_01847, RAYM_09824 were selected for electrophoretic mobility shift assay (EMSA). Our findings illustrated that purified Fur protein could bind to the DNA fragment containing the putative Fur-box (Figure 8B).
The LD50 values of RA-YM, RA-YM Δfur deletion mutant and RA-YM Δfur complemented strain were recorded as 2.0 × 106 CFU, 1.6 × 108 CFU, 1.2 × 107CFU, respectively. LD50 counted for wild type as compared to RA-YM Δfur deletion mutant was approximately 80 times higher, whereas no significant difference was observed as in the case of wild type in comparison to RA-YM Δfur complemented mutant that was six times higher. Due to slight difference between wild type and RA-YM Δfur complemented mutant, a further comparison was established only between wild type and RA-YM Δfur deletion mutant strains. As the fur gene was disrupted in RA-YM Δfur deletion mutant strains resulted in attenuation of virulence of RA. However, virulence to ducklings was partially restored when the mutant was complemented with the plasmid pRES-JXrep-spc.
Microbiological analysis of heart, brain, liver and spleen showed that bacterial load was higher in wild type RA-YM strain compared to RA-YM Δfur deletion mutant strain at 24 and 48 h post-infection collection, a detailed comparison is shown in Figures 2A,B. Correspondingly, the pathological investigations illustrated that lesions were more significant in wild type as compared to mutant strains. The epicardial tissue of ducklings infected with wild type bacteria consisted of a higher degree of fibrinous exudate and inflammatory cell infiltration as compared to infection with mutant pathogens after 24 and 48 h (Figure 3). The lesions in brain tissue after both 24 and 48 h, the subarachnoid space was examined where mild inflammatory cell infiltration was noted in case of RA-YM Δfur deletion mutant strain as compared to wild type RA-YM strains (Figure 4). Similarly, a large number of hepatocytes expressed fatty degeneration and slight fibrotic effusion when infected with wild type RA-YM bacteria, whereas, such lesions were hardly observed in ducklings infected with RA-YM Δfur mutant bacteria after 24 h inoculation. A higher degree of fibrotic effusions in liver tissue was observed after 48 h of infection with wild type pathogens, in comparison to mutant pathogens. Collectively, severe hepatic congestion was noticed in ducklings infected with wild type RA-YM strains (Figure 5). Likely, both splenomegaly and congestion of spleen was observed in ducklings after 24 and 48 h in case of infection with wild type pathogens, whereas only splenomegaly was noticed in case of infection with mutant pathogens (Figure 6). Conclusively, all groups of ducklings, inoculated with wild type RA-YM strains, were severely infected in comparison to those inoculated with RA-YM Δfur deletion mutant strains. The control group of ducklings showed no significant pathological lesions.
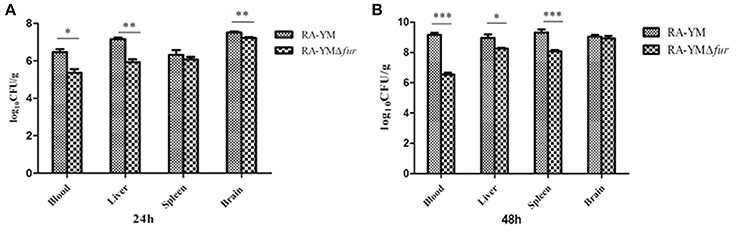
Figure 2. Graphical presentation of bacterial load in blood, liver, spleen, and brain of ducklings infected with wild type RA-YM and RA-YM Δfur deletion mutant strain. The error bars represent mean ± standard deviation from five ducks. (A) The tissue burden of the group infected with wild type and RA-YM Δfur deletion mutant after 24 h. (B) The tissue burden of the group infected with wild type and RA-YM Δfur deletion mutant after 48 h.
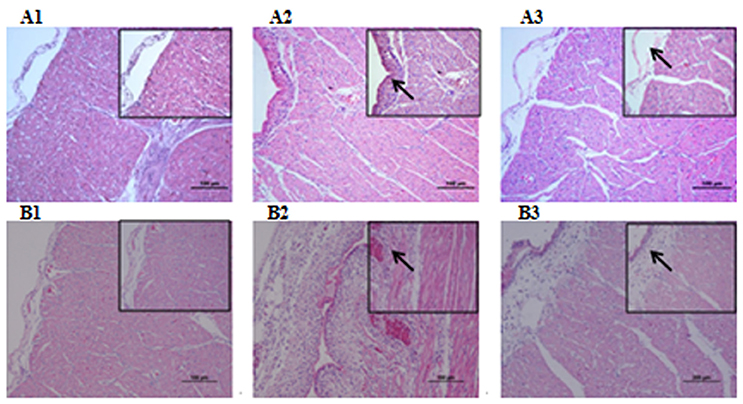
Figure 3. Histopathological diagram of heart. (A1) The blank control group after 24 h, (A2) The group with wild type RA-YM strain after 24 h, (A3) The group with RA-YM Δfur deletion mutant strain after 24 h. (B1) The blank control group after 48 h, (B2) The group with wild type RA-YM strain after 48 h, (B3) The group with RA-YM Δfur deletion mutant strain after 48 h.
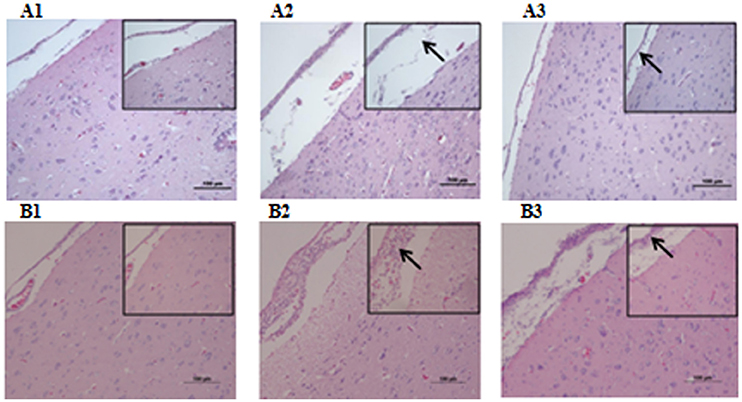
Figure 4. Histopathological diagram of brain (A1) The blank control group after 24 h, (A2) The group with wild type RA-YM strain after 24 h, (A3) The group with RA-YM Δfur deletion mutant strain after 24 h. (B1) The blank control group after 48 h, (B2) The group with wild type RA-YM strain after 48 h, (B3) The group with RA-YM Δfur deletion mutant strain after 48 h.
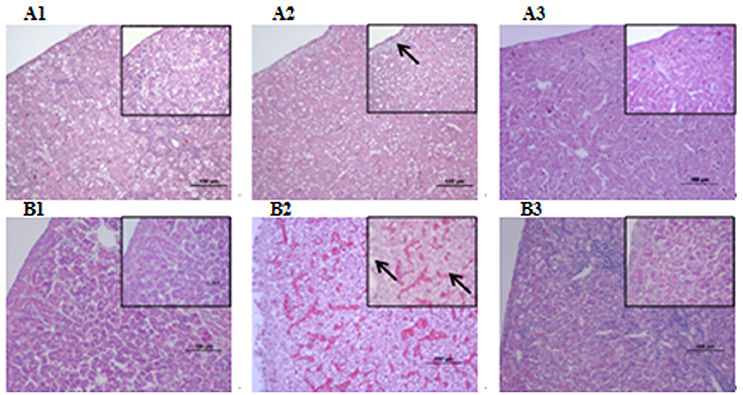
Figure 5. Histopathological diagram of liver (A1) The blank control group after 24 h, (A2) The group with wild type RA-YM strain after 24 h, (A3) The group with RA-YM Δfur deletion mutant strain after 24 h. (B1) The blank control group after 48 h, (B2) The group with wild type RA-YM strain after 48 h, (B3) The group with RA-YM Δfur deletion mutant strain after 48 h.
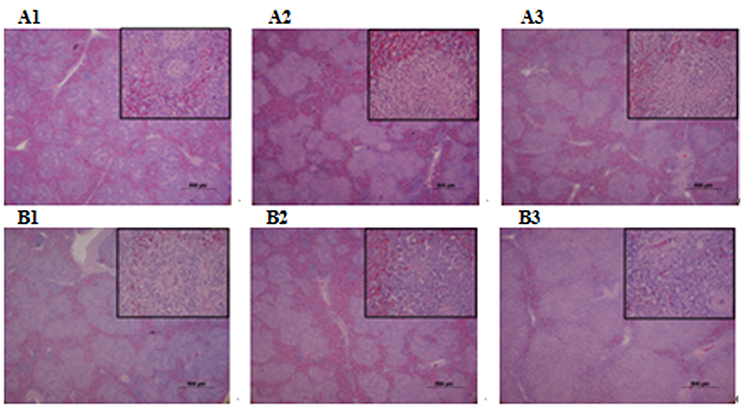
Figure 6. Histopathological diagram of spleen. (A1) The blank control group after 24 h, (A2) The group with wild type RA-YM strain after 24 h, (A3) The group with RA-YM Δfur deletion mutant strain after 24 h. (B1) The blank control group after 48 h, (B2) The group with wild type RA-YM strain after 48 h, (B3) The group with RA-YM Δfur deletion mutant strain after 48 h.
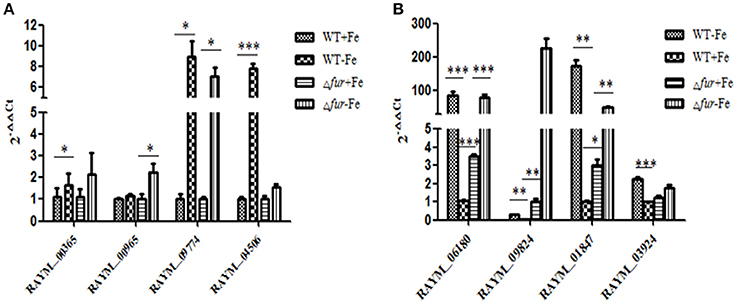
Figure 7. Real-Time PCR analysis of the Fur-and Iron- regulated genes. (A) The genes (RAYM_00365, RAYM_00965, RAYM_09774, RAYM_04506) regulated by iron. (B) The genes (RAYM_06180, RAYM_09824, RAYM_01847, RAYM_03924) regulated by Fur.
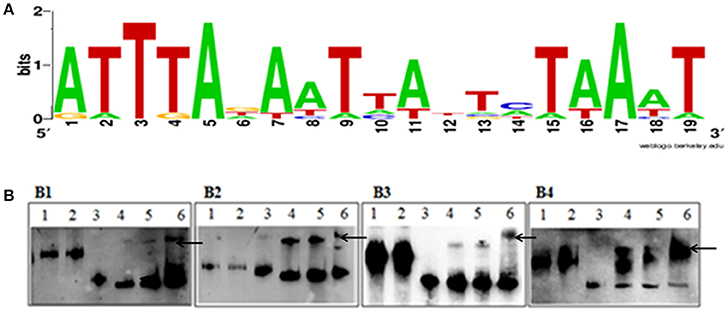
Figure 8. (A) Sequence logo of the Fur box of RA-YM. The binding sequence was listed by using WEBLOGO program. (B) EMSA of the Fur protein and its putative target promoters. (B1) Lane 1, DNA fragment of 16S rRNA; Lane 2, DNA fragment of 16S rRNA and Fur protein; Lane 3, DNA fragment of RAYM_01847; Lane 4, DNA fragment of RAYM_01847 and Fur protein (0.1 μg); Lane 5, DNA fragment of RAYM_01847 and Fur protein (1 μg); Lane 6, DNA fragment of RAYM_01847 and Fur protein (10 μg); (B2) Lane 1, DNA fragment of 16S rRNA; Lane 2, 1 DNA fragment of 16s rRNA and Fur protein; Lane 3, DNA fragment of RAYM_03924; Lane 4, DNA fragment of RAYM_03924 and Fur protein (0.1 μg); Lane 5, DNA fragment of RAYM_03924 and Fur protein (1 μg); Lane 6, DNA fragment of RAYM_03924 and Fur protein (10 μg); (B3) Lane 1, DNA fragment of 16S rRNA; Lane 2, DNA fragment of 16S rRNA and Fur protein; Lane 3, DNA fragment of RAYM_06180; Lane 4, DNA fragment of RAYM_06180 and Fur protein (0.1 μg); Lane 5, DNA fragment of RAYM_06180 and Fur protein (1 μg); Lane 6, DNA fragment of RAYM_06180 and Fur protein (10 μg); (B4) Lane 1, DNA fragment of 16S rRNA; Lane 2, DNA fragment of 16S rRNA and Fur protein; Lane 3, DNA fragment of RAYM_09824; Lane 4, DNA fragment of RAYM_09824 and Fur protein (0.1 μg); Lane 5, DNA fragment of RAYM_09824 and Fur protein (1 μg); Lane 6, DNA fragment of RAYM_09824 and Fur protein (10 μg); each lane the concentration of DNA fragment was 1 μg. The band which marked by arrows shows the Fur protein and DNA fragment complex.
In the present study, we employed the suicide vector pRE-lacZ-mpheS-spc-fur to construct an unmarked mutant RA-YM Δfur successfully. Using this technique, the traditional method of mutant development by inducing antibiotic resistance can be circumvented, and influence on the expression of downstream genes can be minimized in some cases. However, the expression of the fur downstream gene RAYM_04841 remained unchanged. The counter-selectable marker pheS in combination with lacZ have been successfully developed for unmarked gene deletion in RA. Previously, pheS had been also successfully used in Burkholderia (Barrett et al., 2008) and E. faecalis (Kristich et al., 2007), and S. mutans (Xie et al., 2011). The technique was developed to determine the role of fur in the pathogenicity of RA. Indeed, the virulence of RA-YM Δfur mutant strain was attenuated in comparison to wild type and virulence was partially restored when RA-YM Δfur mutant strain was complemented with the plasmid pRES-JXrep-spc. Moreover, bacterial load in different tissues was significantly decreased in RA-YM Δfur mutant infection as compared to wild type strain. Similarly significantly mild lesions were observed in case of RA-YM Δfur mutant infection in comparison to wild type infection. Conclusively, it was observed that Fur regulated virulence factors of RA infection. Indeed, the role of Fur with respect to virulence has been previously examined in Vibrio cholerae (Mey et al., 2005) and Staphylococcus aureus (Johnson et al., 2011). Consequently, findings of this study depicted the role of fur in virulence of RA.
In the current study, we also recorded the expression of the genes downregulated by iron under iron-restricted conditions. Among those genes, six downregulated genes were involved in regulation of tricarboxylic acid (TCA) cycle, which play an important role in metabolism, energy generation and synthesis of precursors (Vuoristo et al., 2016). Under iron-restricted conditions, certain key enzymes of the TCA cycle of RA-YM Δfur strain were downregulated, which included succinate dehydrogenase (SDH) subunit (RAYM_01977, RAYM_01982, RAYM_01987), fumarate hydratase (RAYM_00925) and aconitase. SDH is involved in the respiratory chain and Krebs cycle of bacteria (Yankovskaya et al., 2003). Similarly, glyoxylate bypass pathways are also repressed in Yersinia pestis iron-restricted conditions (Pieper et al., 2010). In previous reports, downregulation of SDH, fumurase and aconitase were reported in E. coli (Massé et al., 2005) and Bacillus subtilis (Gaballa et al., 2008) in iron-sparing conditions. This phenomenon may be accounted for as the bacteria utilize an alternative iron-independent pathway of the TCA cycle and repressed numerous of iron-containing proteins under iron–restricted conditions. Moreover, cysteine synthase A (cysK, a significant enzyme of cysteine biosynthesis) and serine O-acetyltransferase (cysE, catalyzes the acetylation of L-serine to O-acetyl-L-serine) involved in amino biosynthesis were also downregulated due to iron deficiency. This inhibition regulates the conversion of available serine to siderophore, enterobactin, which thereby increases iron acquisition (Salvail et al., 2010). The cytochrome c oxidase (Cco) family related to oxidation-reduction was also inhibited in response to iron limitation, which is comprised of four subunits, CcoN, CcoO, CcoP, and CcoQ, which act as the terminal enzyme of respiratory chain (Ahn et al., 2015; Steininger et al., 2016). The Cco family, a member of heme-copper oxidase superfamily, may play a role in iron- restricted conditions. In Pseudomonas stutzeri, the Cco family has also been reported as an essential element for nitrogen-fixing (Nyquist et al., 2001; Xie et al., 2014). Our data showed that nitrogen-fixing associated genes (RAYM_07584, RAYM_07589) were repressed in iron-restricted conditions. In conclusion, iron-sparing responses, which means the repression of iron-dependent genes when iron is deficient, was the vital reason of the genes downregulated by iron in iron restricted conditions.
Among genes induced by iron, the genes (RAYM_00510, RAYM_00515) associated with iron acquisition were upregulated, which included Ferrous iron transport protein A (FeoA) and Ferrous iron transport protein B (FeoB). The feoB gene encodes an inner membrane Fe (II) transporter in multiple bacteria, such as E. coli and V. cholerae whereas feoA gene was demonstrated crucial for FeoB uptake of Fe(II) (Marlovits et al., 2002; Kim et al., 2012). Other genes involved in the iron-acquisition system were also upregulated, including Ferritin (RAYM_01160), ABC transporter related protein (RAYM_06607), TonB-dependent receptor (RAYM_04481). This is an apparent response to iron starvation in bacteria, which has also been demonstrated in Klebsiella pneumonia and Listeria monocytogenes (Ledala et al., 2010; Lin et al., 2011). Similarly, a group of genes contributed to the biosynthetic process of iron-sulfur (Fe-S) were also regulated by iron. In bacteria, Fe-S machinery is comprised of a nitrogen-fixing NifU domain protein (RAYM_01100), SUF system protein (RAYM_01495, RAYM_06457, RAYM_06467, RAYM_06507). Similar changes of NIF and SUF systems have been confirmed in E. coli in previous studies (Outten et al., 2004). Moreover, the genes Phosphoserine aminotransferase (RAYM_04219), D-3-phosphoglycerate-dehydrogenase (RAYM_04224) controlling the shikimate pathway were upregulated. D-3-phosphoglycerate-dehydrogenase (pabB) converts chorismate to 4-amino-4-deoxychorismate (ADC) and phosphoserine aminotransferase (pabC) converts ADC to p-aminobenzoate (PABA) and pyruvate (Green et al., 1992). Shikimate pathway catalyzes serine to form siderophore, enterobactin (Prévost et al., 2007). The upregulation of the pabB and pabC in iron-limited conditions results in high levels of both aromatic amino acids and phenolate siderophore (Lemaître et al., 2014). Moreover, in our data, sigma factor protein (RAYM _00365) was upregulated by iron. Iron-starvation sigmas, a extracytoplamic function (ECFs) subfamily, have been demonstrated previously to play a role in iron acquisition in P. aeruginosa (Visca et al., 2002). In conclusion, the genes induced by iron are involved in iron-acquisition, some metabolic pathways and several transcriptional regulation factors. The series of regulatory responses under low-iron conditions resulted in increasing iron acquisition.
Among the genes regulated by Fur under iron-restricted conditions, Hmu system, comprised of hmuY (RAYM_06175) and hmuR (RAYM_06180), was observed to be regulated by Fur. The homologous Fur-box sequence has been identified upstream of the hmuY start codon in Porphyromonas gingivalis (Simpson et al., 2000). HmuR, the TonB-dependent receptor for ferric, has previously been illustrated to be regulated by Fur in Y. pestis (Branger et al., 2010). Furthermore, genes (RAYM_01847, RAYM_09824, and RAYM_09779) encoding TonB-dependent outer membrane proteins are also regulated by Fur under iron-restricted conditions. The accessory proteins of TonB system can transduce energy in Gram-negative bacteria (Postle and Kadner, 2003; Lim et al., 2012). Our data confirmed that TonB-ExbB-ExbD system was a significant component for ferric enterobactin acquisition, which was previously reported in Campylobacter (Zeng et al., 2013). In addition, RAYM_00450 (oxidoreductase), RAYM_03864 (3-hydroxyacyl-CoA dehydrogenase) were also regulated by Fur which was demonstrated in previous study as Fur regulated the response to oxidative stress in C. jejuni (Holmes et al., 2005). Interestingly, sprT gene (RAYM_03924), a component of the type IX secretion system (T9SS) was also regulated by Fur. Parallel findings were recorded in Flavobacterium johnsoniae (Kharade and McBride, 2015). T9SS was also characterized as a novel protein secretion system mediated outer membrane translocation to the cell surface in P. gingivalis (de Diego et al., 2016). In conclusion, Fur regulated processes included iron acquisition, oxidation and reduction, and regulation of some components of T9SS.
Fur has been reported to act as a negative regulator. It was demonstrated that Fur protein, together with iron, can bind to a consensus sequence, resulting in transcriptional inhibition (Baichoo and Helmann, 2002) which had been described in E. coli, P. aeruginosa, Helicobacter pylori, and H. hepaticus (Escolar et al., 1998; Vasil and Ochsner, 1999; Belzer et al., 2007; Pich et al., 2012). The Fur-box consensus sequence 5′-GATAATGATAATCATTATC-3, was slightly different among bacteria. It was identified as an adjacent hexamer unit of the sequence 5′-GATTAT-3′ or three repeat of the NATWAT (Lavrrar and McIntosh, 2003). Typically, the Fur binding sequence was located within 150 bp of the translation initiation codon of the regulated genes (Grifantini et al., 2003). The putative Fur-box sequence of RA-YM in our study was predicted as 5′-ATTTAGAATTATTCTAAAT- 3′, and the sequences might be located within 100 bp of the translation initiation codon of regulated genes which could be reasoned due to the unique promoter of the strain. RA-YM belongs to the Flavobacterium, where promoter have −7 and −33 consensus elements, whereas the promotor of E. coli has −10 and −35 consensus elements (Chen et al., 2007).
In summary, our work showed that pheS acted as effective counter-selectable marker for conjugal transfer. We successfully constructed an unmarked deletion mutant of RA with the suicide vector pRE-lacZ-mpheS-spc. In addition, we elucidated the role of the fur gene in virulence of RA. Furthermore, we screened out the genes regulated by iron and Fur. The putative Fur-box sequence of RA was also predicted. Conclusively, this was a comprehensive study on the metabolism of R. anatipestifer which may help facilitate the control of this pathogen.
YG designed, performed the experimentation, data analysis and wrote the manuscript. DH, JG, XL, and JYG performed experimentations. XW, YX, HJ, ML, and ZL designed and contributed to experimental work. ZZ and DB designed, analyzed the data and revised the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Natural Science Foundation of China (31201933) and the Fundamental Research Founds for the Central Universities (52902-0900206127).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00382/full#supplementary-material
Ahn, Y. O., Lee, H. J., Kaluka, D., Yeh, S. R., Rousseau, D. L., Ädelroth, P., et al. (2015). The two transmembrane helices of CcoP are sufficient for assembly of the cbb3-type heme-copper oxygen reductase from Vibrio cholerae. Biochim. Biophys. Acta 1847, 1231–1239. doi: 10.1016/j.bbabio.2015.06.013
Baichoo, N., and Helmann, J. D. (2002). Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184, 5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002
Barrett, A. R., Kang, Y., Inamasu, K. S., Son, M. S., Vukovich, J. M., and Hoang, T. T. (2008). Genetic tools for allelic replacement in Burkholderia species. Appl. Environ. Microbiol. 74, 4498–4508. doi: 10.1128/AEM.00531-08
Belzer, C., van schendel, B. A. M., Kuipers, E. J., Kusters, J. G., and van Vliet, A. H. M. (2007). Iron-responsive repression of urease expression in Helicobacter hepaticus is mediated by the transcriptional regulator Fur. Infect. Immun. 75, 745–752. doi: 10.1128/IAI.01163-06
Bijlsma, J. J. E., Waidner, B., Vliet, A. H., Hughes, N. J., Häg, S., Bereswill, S., et al. (2002). The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect. Immun. 70, 606–611. doi: 10.1128/IAI.70.2.606-611.2002
Branger, C. G., Sun, W., Torres-Escobar, A., Perry, R., Roland, K. L., Fetherston, J., et al. (2010). Evaluation of Psn, HmuR and a modified LcrV protein delivered to mice by live attenuated Salmonella as a vaccine against bubonic and pneumonic Yersinia pestis challenge. Vaccine 29, 274–282. doi: 10.1016/j.vaccine.2010.10.033
Chen, S., Bagdasarian, M. M., Kaufman, M. G., Bates, A. K., and Walker, E. D. (2007). Mutational Analysis of the ompA Promoter from Flavobacterium johnsoniae. J. Bacteriol.189, 5108–5118. doi: 10.1128/JB.00401-07
Cheng, A., Wang, M., Chen, X., Zhu, D., Huang, C., Fei, L., et al. (2003). Epidemiology and new serotypes of Riemerella anatipestifer isolated from ducks in China and studies on their pathogenic characteristics. Chinese J. Vet. Sci. 78, 1469–1473.
Crasta, K. C., Chua, K. L., Subramaniam, S., Frey, J., Loh, H., and Tan, H. M. (2002). Identification and characterization of CAMP cohemolysin as a potential virulence factor of Riemerella anatipestifer. J. Bacteriol. 184, 1932–1939. doi: 10.1128/JB.184.7.1932-1939.2002
de Diego, I., Ksiazek, M., Mizgalska, D., Koneru, L., Golik, P., Szmigielski, B., et al. (2016). The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal β-sandwich domain. Sci Rep. 6:23123. doi: 10.1038/srep23123
Ernst, F. D., Bereswill, S., Waidner, B., Stoof, J., Mäder, U., Kusters, J. G., et al. (2005a). Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151, 533–546. doi: 10.1099/mic.0.27404-0
Ernst, F. D., Homuth, G., Stoof, J., Mäder, U., Waidner, B., Kuipers, E. J., et al. (2005b). Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase sodb is mediated by fur. J. Bacteriol. 187, 3687–3692. doi: 10.1128/JB.187.11.3687-3692.2005
Escolar, L., Pérez-Martín, J., and de Lorenzo, V. (1998). Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283, 537–547. doi: 10.1006/jmbi.1998.2119
Gaballa, A., Antelmann, H., Aguilar, C., Khakh, S. K., Song, K. B., Smaldone, G. T., et al. (2008). The Bacillus subtilis Iron-Sparing Response Is Mediated by a Fur-Regulated Small RNA and Three Small, Basic Proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 11927–11932. doi: 10.1073/pnas.0711752105
Green, J. M., Merkel, W. K., and Nichols, B. P. (1992). Characterization and sequence of Escherichia coli pabC, the gene encoding aminodeoxychorismate lyase, a pyridoxal phosphate-containing enzyme. J. Bacteriol. 174, 5317–5323. doi: 10.1128/jb.174.16.5317-5323.1992
Grifantini, R., Sebastian, S., Frigimelica, E., Draghi, M., Bartolini, E., Muzzi, A., et al. (2003). Identification of iron-activated and-repressed Fur-dependent genes by transcriptome analysis of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. U.S.A. 100, 9542–9547. doi: 10.1073/pnas.1033001100
Haraszthy, V. I., Jordan, S. F., and Zambon, J. J. (2006). Identification of Fur-regulated genes in Actinobacillus actinomycetemcomitans. Microbiology 152, 787–796. doi: 10.1099/mic.0.28366-0
Higgins, D. A., Henry, R. R., and Kounev, Z. V. (2000). Duck immune responses to Riemerella anatipestifer vaccines. Dev. Comp. Immunol. 24, 153–167. doi: 10.1016/S0145-305X(99)00070-1
Holmes, K., Mulholland, F., Pearson, B. M., Pin, C., McNicholl-Kennedy, J., Ketley, J. M., et al. (2005). Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151, 243–257. doi: 10.1099/mic.0.27412-0
Hu, Q., Han, X., Zhou, X., Ding, C., Zhu, Y., and Yu, S. (2011). OmpA is a virulence factor of Riemerella anatipestifer. Vet. Microbiol. 150, 278–283. doi: 10.1016/j.vetmic.2011.01.022
Ibba, M., Kast, P., and Hennecke, H. (1994). Substrate specificity is determined by amino acid binding pocket size in Escherichia coli phenylalanyl-tRNA synthetase. Biochem. 33, 7107–7112.
Johnson, M., Sengupta, M., Purves, J., Tarrant, E., Williams, P. H., Cockayne, A., et al. (2011). Fur is required for the activation of virulence gene expression through the induction of the sae regulatory system in Staphylococcus aureus. Int. J. Med. Microbiol. 301, 44–52. doi: 10.1016/j.ijmm.2010.05.003
Kast, P., and Hennecke, H. (1991). Amino acid substrate specificity of Escherichia coli phenylalanyl-tRNA synthetase altered by distinct mutations. J. Mol. Biol. 222, 99–124. doi: 10.1016/0022-2836(91)90740-w
Kharade, S. S., and McBride, M. J. (2015). Flavobacterium johnsoniae PorV is required for secretion of a subset of proteins targeted to the type IX secretion system. J. Bacteriol. 197, 147–158. doi: 10.1128/JB.02085-14
Kim, H., Lee, H., and Shin, D. (2012). The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem. Biophys. Res. Commun. 423, 733–738. doi: 10.1016/j.bbrc.2012.06.027
Kristich, C. J., Chandler, J. G., and Dunny, G. M. (2007). Development of a host-genotype-independent counterselectable marker and a high-frequency conjugative delivery system and their use in genetic analysis of Enterococcus faecalis. Plasmid 57, 131–144. doi: 10.1016/j.plasmid.2006.08.003
Lavrrar, J. L., and McIntosh, M. A. (2003). Architecture of a Fur binding site: a comparative analysis. J. Bacteriol. 185, 2194–2202. doi: 10.1128/JB.185.7.2194-2202.2003
Ledala, N., Sengupta, M., Muthaiyan, A., Wilkinson, B. J., and Jayaswal, R. K. (2010). Transcriptomic response of Listeria monocytogenes to iron limitation and fur mutation. Appl. Environ. Microbiol. 76, 406–416.
Lemaître, C., Bidet, P., Benoist, J. F., Schlemmer, D., Sobral, E., d'Humières, C., et al. (2014). The ssbL gene harbored by the ColV plasmid of an Escherichia coli neonatal meningitis strain is an auxiliary virulence factor boosting the production of siderophores through the shikimate pathway. J. Bacteriol. 196, 1343–1349. doi: 10.1128/JB.01153-13
Lim, C. K., Hassan, K. A., Tetu, S. G., Loper, J. E., and Paulsen, I. T. (2012). The effect of iron limitation on the transcriptome and proteome of Pseudomonas fluorescens Pf-5. PLoS ONE 7:e39139. doi: 10.1371/journal.pone.0039139
Lin, C., Wu, C., Chen, Y., Lai, Y., Chi, C., Lin, J., et al. (2011). Fur regulation of the capsular polysaccharide biosynthesis and iron-acquisition systems in Klebsiella pneumoniae CG43. Microbiology 157, 419–429. doi: 10.1099/mic.0.044065-0
Liu, M., Wang, M., Zhu, D., Wang, M., Jia, R., Chen, S., et al. (2016). Investigation of TbfA in Riemerella anatipestifer using plasmid-based methods for gene over-expression and knockdown. Sci Rep. 6:37159. doi: 10.1038/srep37159
Loh, H., Teo, T. P., and Tan, H. C. (1992). Serotypes of 'Pasteurella' anatipestifer isolates from ducks in Singapore: a proposal of new serotypes. Avian Pathol. 21, 453–459. doi: 10.1080/03079459208418863
Lu, F., Miao, S., Tu, J., Ni, X., Xing, L., Yu, H., et al. (2013). The role of TonB-dependent receptor TbdR1 in Riemerella anatipestifer in iron acquisition and virulence. Vet. Microbiol. 167, 713–718. doi: 10.1016/j.vetmic.2013.08.020
Marlovits, T. C., Haase, W., Herrmann, C., Aller, S. G., and Unger, V. M. (2002). The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. U.S.A. 99, 16243–16248. doi: 10.1073/pnas.242338299
Massé, E., Vanderpool, C. K., and Gottesman, S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005
Mathieu, S., Cissé, C., Vitale, S., Ahmadova, A., Degardin, M., Pérard, J., et al. (2016). From peptide aptamers to inhibitors of FUR, bacterial transcriptional regulator of iron homeostasis and virulence. ACS Chem. Biol. 11, 2519–2528. doi: 10.1021/acschembio.6b00360
Mey, A. R., Wyckoff, E. E., Kanukurthy, V., Fisher, C. R., and Payne, S. M. (2005). Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect. Immun. 73, 8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005
Nyquist, R. M., Heitbrink, D., Bolwien, C., Wells, T. A., Gennis, R. B., and Heberle, J. (2001). Perfusion-induced redox differences in cytochrome c oxidase: ATR/FT-IR spectroscopy. FEBS Lett. 505, 63–67. doi: 10.1016/S0014-5793(01)02769-7
Outten, F. W., Djaman, O., and Storz, G. (2004). A suf operon requirement for Fe–S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52, 861–872. doi: 10.1111/j.1365-2958.2004.04025.x
Pi, H., Patel, S. J., Argüello, J. M., and Helmann, J. D. (2016). The Listeria monocytogenes Fur-regulated virulence protein FrvA is an Fe(II) efflux P1B4 -type ATPase. Mol. Microbiol. 100, 1066–1079. doi: 10.1111/mmi.13368
Pich, O. Q., Carpenter, B. M., Gilbreath, J. J., and Merrell, D. S. (2012). Detailed analysis of Helicobacter pylori Fur-regulated promoters reveals a Fur box core sequence and novel Fur-regulated genes. Mol. Microbiol. 84, 921–941. doi: 10.1111/j.1365-2958.2012.08066.x
Pieper, R., Huang, S. T., Parmar, P. P., Clark, D. J., Alami, H., Fleischmann, R. D., et al. (2010). Proteomic analysis of iron acquisition, metabolic and regulatory responses of Yersinia pestis to iron starvation. BMC Microbiol. 10:30. doi: 10.1186/1471-2180-10-30
Porcheron, G., and Dozois, C. M. (2015). Interplay between iron homeostasis and virulence: Fur and RyhB as major regulators of bacterial pathogenicity. Vet. Microbiol. 179, 2–14. doi: 10.1016/j.vetmic.2015.03.024
Postle, K., and Kadner, R. J. (2003). Touch and go: tying TonB to transport. Mol. Microbiol. 49, 869–882. doi: 10.1046/j.1365-2958.2003.03629.x
Prévost, K., Salvail, H., Desnoyers, G., Jacques, J. F., Phaneuf, E., and Massé, E. (2007). The small RNA RyhB activates the translation of shiA mRNA encoding a permease of shikimate, a compound involved in siderophore synthesis. Mol. Microbiol. 64, 1260–1273. doi: 10.1111/j.1365-2958.2007.05733.x
Reed, L. J., and Muench, H. (1938). A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27, 493–497. doi: 10.1093/oxfordjournals.aje.a118408
Salvail, H., Lanthier-Bourbonnais, P., Sobota, J. M., Caza, M., Benjamin, J. A., Mendieta, M. E., et al. (2010). A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc. Natl. Acad. Sci. U.S.A. 107, 15223–15228. doi: 10.1073/pnas.1007805107
Simpson, W., Olczak, T., and Genco, C. A. (2000). Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol. 182, 5737–5748. doi: 10.1128/JB.182.20.5737-5748.2000
Steininger, C., Reiner-Rozman, C., Schwaighofer, A., Knoll, W., and Naumann, R. L. C. (2016). Kinetics of cytochrome c oxidase from R. sphaeroides initiated by direct electron transfer followed by tr-SEIRAS. Bioelectrochemistry. 112, 1–8. doi: 10.1016/j.bioelechem.2016.06.005
Stibitz, S. (1994). Use of conditionally counterselectable suicide vectors for allelic exchange. Meth. Enzymol. 235, 458–465. doi: 10.1016/0076-6879(94)35161-9
Tu, J., Lu, F., Miao, S., Ni, X., Jiang, P., Yu, H., et al. (2014). The siderophore-interacting protein is involved in iron acquisition and virulence of Riemerella anatipestifer strain CH3. Vet. Microbiol. 168, 395–402. doi: 10.1016/j.vetmic.2013.11.027
Vasil, M. L., and Ochsner, U. A. (1999). The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34, 399–413. doi: 10.1046/j.1365-2958.1999.01586.x
Visca, P., Leoni, L., Wilson, M. J., and Lamont, I. L. (2002). Iron transport and regulation, cell signalling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45, 1177–1190. doi: 10.1046/j.1365-2958.2002.03088.x
Vuoristo, K. S., Mars, A. E., Sanders, J. P. M., Eggink, G., and Weusthuis, R. A. (2016). Metabolic engineering of TCA cycle for production of chemicals. Trends Biotechnol. 34, 191–197. doi: 10.1016/j.tibtech.2015.11.002
Xie, H., Buschmann, S., Langer, J. D., Ludwig, B., and Michel, H. (2014). Biochemical and biophysical characterization of the two isoforms of cbb3-type cytochrome c oxidase from Pseudomonas stutzeri. J. Bacteriol. 196, 472–482. doi: 10.1128/JB.01072-13
Xie, Z., Okinaga, T., Qi, F., Zhang, Z., and Merritt, J. (2011). Cloning-independent and counterselectable markerless mutagenesis system in Streptococcus mutans. Appl. Environ. Microbiol. 77, 8025–8033. doi: 10.1128/AEM.06362-11
Yankovskaya, V., Horsefield, R., Törnroth, S., Luna-Chavez, C., Miyoshi, H., Léger, C., et al. (2003). Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 299, 700–704. doi: 10.1126/science.1079605
Yuhara, S., Komatsu, H., Goto, H., Ohtsubo, Y., Nagata, Y., and Tsuda, M. (2008). Pleiotropic roles of iron-responsive transcriptional regulator Fur in Burkholderia multivorans. Microbiology 154, 1763–1774. doi: 10.1099/mic.0.2007/015537-0
Zeng, X., Xu, F., and Lin, J. (2013). Specific TonB-ExbB-ExbD energy transduction systems required for ferric enterobactin acquisition in Campylobacter. FEMS Microbiol. Lett. 347, 83–91. doi: 10.1111/1574-6968.12221
Keywords: Riemerella anatipestifer, fur, pheS, unmarked gene deletion system, virulence, Fur-box, RNA-seq
Citation: Guo Y, Hu D, Guo J, Li X, Guo J, Wang X, Xiao Y, Jin H, Liu M, Li Z, Bi D and Zhou Z (2017) The Role of the Regulator Fur in Gene Regulation and Virulence of Riemerella anatipestifer Assessed Using an Unmarked Gene Deletion System. Front. Cell. Infect. Microbiol. 7:382. doi: 10.3389/fcimb.2017.00382
Received: 15 May 2017; Accepted: 09 August 2017;
Published: 25 August 2017.
Edited by:
Susu M. Zughaier, Emory University, United StatesReviewed by:
Teresa Olczak, University of Wrocław, PolandCopyright © 2017 Guo, Hu, Guo, Li, Guo, Wang, Xiao, Jin, Liu, Li, Bi and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dingren Bi, YmlkaW5ncmVuQG1haWwuaHphdS5lZHUuY24=
Zutao Zhou, enR6aG91QG1haWwuaHphdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.