
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 25 July 2017
Sec. Bacteria and Host
Volume 7 - 2017 | https://doi.org/10.3389/fcimb.2017.00342
This article is part of the Research Topic Role of Iron in Bacterial Pathogenesis View all 13 articles
 Yingjie Li1,2
Yingjie Li1,2 Qingjun Ma1,2*
Qingjun Ma1,2*The hemorrhagic septicemic disease vibriosis caused by Vibrio anguillarum shows noticeable similarities to invasive septicemia in humans, and in this case, the V. anguillarum–host system has the potential to serve as a model for understanding native eukaryotic host–pathogen interactions. Iron acquisition, as a fierce battle occurring between pathogenic V. anguillarum and the fish host, is a pivotal step for virulence. In this article, advances in defining the roles of iron uptake pathways in growth and virulence of V. anguillarum have been summarized, divided into five aspects, including siderophore biosynthesis and secretion, iron uptake, iron release, and regulation of iron uptake. Understanding the molecular mechanisms of iron acquisition will have important implications for the pathogenicity of this organism.
The Gram-negative bacterium Vibrio anguillarum is a pathogen that causes vibriosis with lethal hemorrhagic septicemia in aquatic animals worldwide (Toranzo et al., 2005). Although up to 23 O serotypes of V. anguillarum are identified in the European serotyping system, with most serotypes encompassing free-living environmental strains (Pedersen et al., 1999), only serotypes O1, O2, and partial O3 are found to be implicated in vibriosis outbreaks (Toranzo et al., 2005). Many studies have been performed in an attempt to understand the virulence mechanism in V. anguillarum. Several main virulence factors have been recognized by using genetic approaches, including iron acquisition components (Naka and Crosa, 2011), hemolysins (Hirono et al., 1996; Rodkhum et al., 2005; Rock and Nelson, 2006; Li et al., 2008; Xu et al., 2011; Mou et al., 2013), metalloproteases (Milton et al., 1992; Yang et al., 2007; Varina et al., 2008; Mo et al., 2010), chemotaxis and motility (O'Toole et al., 1999; Ormonde et al., 2000), exopolysaccharides (Croxatto et al., 2007), and lipopolysaccharides (Welch and Crosa, 2005). Among them, iron uptake systems are a critical component for infection of the host fish leading to disease (Wolf and Crosa, 1986).
V. anguillarum, like most other organisms, has an absolute requirement for iron to synthesize a large number of crucial enzymes, which are involved in many fundamental cellular processes, such as cytochromes for cell respiration, ribonucleotide reductase for the biosynthesis of DNA precursors, and enzymes for the tricarboxylic acid (TCA) cycle (Crosa et al., 2004). However, due to the low solubility of iron (~10−18 M) at physiological pH in aerobic environments, ferric iron mainly forms insoluble hydroxides, whereas a cytoplasmic iron concentration of ~10−6 M is required for bacterial growth (Hantke, 1981). Therefore, iron is suggested to be the growth-limiting factor in ocean environments (Martin et al., 1991). To respond to this selective pressure, bacteria have evolved numerous mechanisms for iron acquisition, including transport of iron from the mammalian iron carriers, transferrin and heme, and synthesis of small ferric iron-binding molecules, known as siderophores. Some of these iron transport systems are conserved among all Vibrio species, reflecting their common ancestry, while other acquisition systems appear to have been developed by horizontal transfer, such as the anguibactin transport system that is mainly specific to V. anguillarum. It has been shown that V. anguillarum harbors a number of genes encoding for iron uptake and regulation, which are essential for its virulence beyond simple iron chelation (Lemos and Osorio, 2007; Naka et al., 2013b). In this article, we describe the developments in understanding the molecular mechanisms of iron acquisition systems in V. anguillarum, divided into the following aspects: siderophore biosynthesis and secretion, iron uptake, iron release, and regulation of iron uptake.
Two different siderophore-dependent systems have been identified in V. anguillarum strains. One is mediated by a 65 kb pJM1 plasmid, which contains most of the genes encoding for biosynthesis and transport proteins of the siderophore anguibactin (Naka et al., 2013b). This anguibactin system is only found in pathogenic plasmid-bearing strains of serotype O1. The other system, existing in all serotype O2 strains tested thus far, and some plasmidless serotype O1 strains, synthesizes a catecholated-type siderophore, vanchrobactin (Alice et al., 2005; Balado et al., 2006).
The structure of anguibactin is unique in containing both catechol and hydroxamate metal-chelating functional groups (Actis et al., 1986), derived from 2,3-dihydroxybenzoic acid (DHBA) and N-hydro-histamine, respectively. Utilizing the chorismate, plasmid-carrying V. anguillarum strain 775 generates a repertoire of molecules through a ribosome-independent process and finally synthesizes anguibactin. This biosynthesis is controlled by a number of genetic determinants. To date, more than 10 different genes have been described and yield anguibactin-related phenotypes in plasmid-carrying V. anguillarum strains when mutated by genetic approaches (Table S1). Most are located on the plasmid pJM1 or pJM1-like plasmids while some are on the chromosomes (Figure 1). First, chorismate is catalyzed stepwise by a series of proteins, AngC/VabC (isochrorismate synthase), AngB/VabB (isochrorismatase; Du et al., 2017), and VabA (2,3-dihydro-2,3-dehydroxybenzoate dehydrogenase), to synthesize DHBA (Figure 2; Alice et al., 2005; Balado et al., 2008). In later steps, phosphopantetheine transferase AngD is required to transfer a phosphopantetheinyl moiety to a serine residue of AngB and AngM (Balado et al., 2008). AngE/VabE (2,3-dehydroxybenzoate-AMP ligase) activates DHBA to form acyl adenylate and further transfers it to the free thiol of the phosphopantetheine AngB (Liu et al., 2004; Alice et al., 2005). AngC, AngB, and AngE are pJM1-encoded proteins while VabC, VabB, VabA, and VabE are chromosomally encoded with VabC, VabB, and VabE showing functional redundancy with AngC, AngB, and AngE, respectively. AngN catalyzes this DHBA thioester to combine with cysteine, which is activated by AngR and tethered by AngM, thereby producing a dihydroxyphenyl-thiazoline-thioester. AngH (histidine decarboxylase; Tolmasky et al., 1995; Barancin et al., 1998) and possibly AngU (Naka et al., 2013b) modify the histidine to form N-hydro-histamine, which is then transferred to dihydroxyphenyl-thiazoline-thioester to yield anguibactin. It is interesting to note that homologs of these plasmid-located genes are all present on the chromosome of Vibrio harveyi, which is also able to produce anguibactin, suggesting that the plasmid-mediated anguibactin system might originate from V. harveyi or vice versa (Naka et al., 2013a,b).
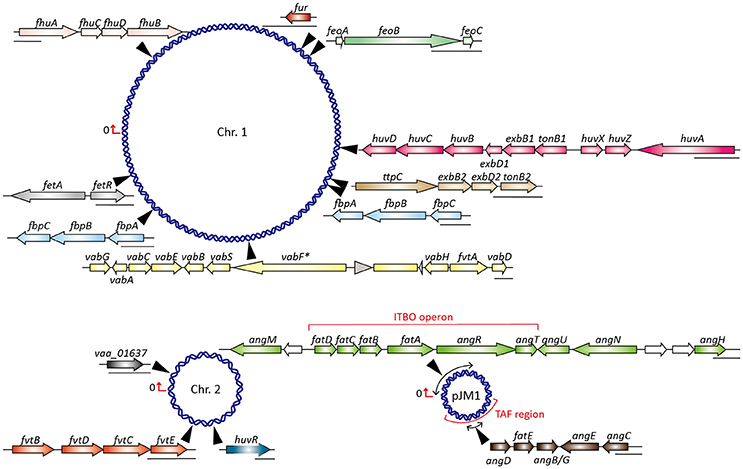
Figure 1. Map of V. anguillarum 775 chromosomes and plasmid pJM1 showing locations and molecular organization of known and putative iron transport genes. Chr., chromosome; ITBO, iron transport biosynthesis operon; TAF, trans-acting factor; vabF* indicates vabF is inactivated by the insertion sequence RS1.
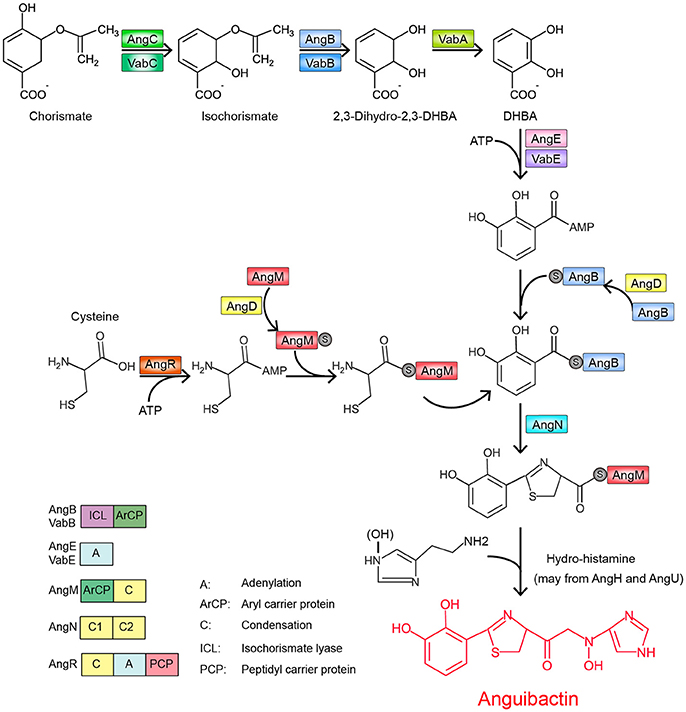
Figure 2. Proposed pathway of anguibactin biosynthesis in V. anguillarum plasmid-carrying strains (modified from Naka et al., 2013b).
Plasmidless O1 strains and those belonging to a number of other V. anguillarum serotypes synthesize a chromosome-mediated siderophore, vanchrobactin (Lemos et al., 1988; Soengas et al., 2008). In a similar way to anguibactin synthesis, DHBA of these V. anguillarum strains is also produced from chorismate by the sequential activities of VabC, VabB, and VabA. However, how vanchrobactin is synthesized from the DHBA precursor remains obscure, even though some genes have been found to be indispensable for this process, including vabB, vabD, vabE, and vabF (Balado et al., 2006, 2008). According to well-studied pathways for synthesis of anguibactin and vibriobactin, a siderophore produced by Vibrio cholerae, late steps for vanchrobactin formation in V. anguillarum have been proposed during which DHBA is assembled (Figure 3; Balado et al., 2006). Specifically, VabD contributes to transfer of a phosphopantetheinyl moiety to the aryl carrier domain of VabB and the peptidyl carrier domain of VabF, respectively. Like anguibactin formation, VabE activates DHBA and arginine to yield acyl adenylates, and then delivers them to VabB and VabF, respectively. The DHBA-VabB is combined with arginine by the condensation domain of VabF to form (2,3-dihydroxybenzoyl)argininate. Finally, the condensation domain of VabF in (2,3-dihydroxybenzoyl)argininate-VabF may be loaded with VabE-activated serine, which adheres to the peptidyl carrier domain of VabF to produce vanchrobactin. Although similar roles of AngB and VabB are proposed in these processes, the aryl carrier protein (ArCP) domain of VabB is not able to complement the function of the AngB ArCP domain (Di Lorenzo et al., 2011), suggesting different but unknown roles may occur between AngB and VabB during anguibactin and vanchrobactin formation.
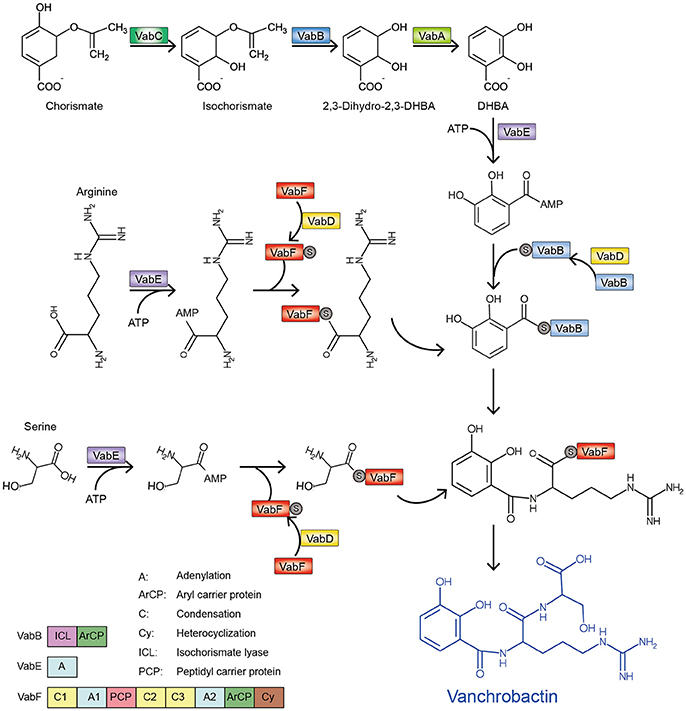
Figure 3. Proposed pathway of vanchrobactin biosynthesis in V. anguillarum plasmidless strains (modified from Balado et al., 2006).
Siderophore secretion is an essential step in iron uptake, yet the mechanisms of this process remain largely unknown. Two siderophore export systems have been found so far, including the ATP-dependent efflux pump and the major facilitator superfamily protein (MFS)-mediated efflux pump. PvdRT-OpmQ is the first ATP-dependent export system to be identified in Pseudomonas aeruginosa (reviewed by Schalk and Guillon, 2013). Schalk and colleagues found that this system exports not only the newly synthesized mature siderophore pyoverdine but also pyoverdine that has already delivered iron into the bacterium (Hannauer et al., 2010; Yeterian et al., 2010). In addition, PvdRT-OpmQ can secrete unwanted metal-pyoverdine complexes into the periplasm of P. aeruginosa (Hannauer et al., 2012). By using the respective protein sequences from P. aeruginosa as a query in BLASTP analysis, only genes encoding for PvdR and PvdT are identified (Table S2) while the ompQ gene is absent in the genomes of the sequenced V. anguillarum strains, suggesting that the MFS system rather than PvdRT-OpmQ may participate in siderophore secretion in V. anguillarum.
The secretion of enterobactin in Escherichia coli is the best-studied paradigm of siderophore export via the MFS system (Horiyama and Nishino, 2014). Based on this, a proposed siderophore-export pathway is depicted in Figure 4, and putative genes involved in this process are listed in Table S2. First, siderophores are exported to the periplasm from the cytoplasm via the MSF VabS, a homolog of E. coli EntS that has been shown to transport enterobactin across the cytoplasmic membrane (Furrer et al., 2002). Subsequently, the resistance-nodulation-cell division (RND) family proteins, which have been found to play a role in multidrug resistance in many microbes including V. cholerae strains (Rahman et al., 2007; Bina et al., 2008), capture the periplasmic siderophores and secrete them to the environment via the outer membrane channel TolC. In agreement with this model, a RND efflux system recently described in V. cholerae plays an essential role in maintenance of cellular homeostasis by secreting the siderophore vibriobactin (Kunkle et al., 2017), which again indicates that a MFS-mediated efflux pump system might be used for siderophore export in V. anguillarum.
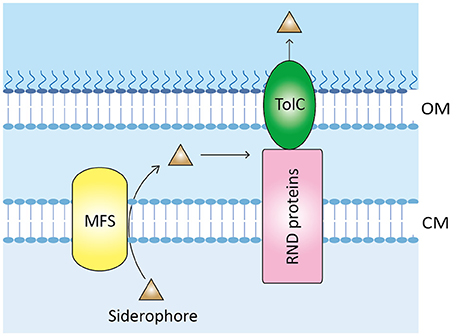
Figure 4. Proposed model of siderophore export in V. anguillarum. When produced in the cytoplasm, siderophores such as anguibactin or vanchrobactin are exported to the periplasmic space by MFS protein. Subsequently, RND proteins capture and further secrete the siderophores to the environment through the outer membrane protein TolC. OM, outer membrane; CM, cytoplasmic membrane; MFS, major facilitator superfamily protein; RND, resistance-nodulation-cell division.
V. anguillarum strains contain several iron transport systems to sequester the different sources of iron, including anguibactin or vanchrobactin, heme, free Fe2+, free Fe3+, and ferrichrome, which are summarized in Figure 5.
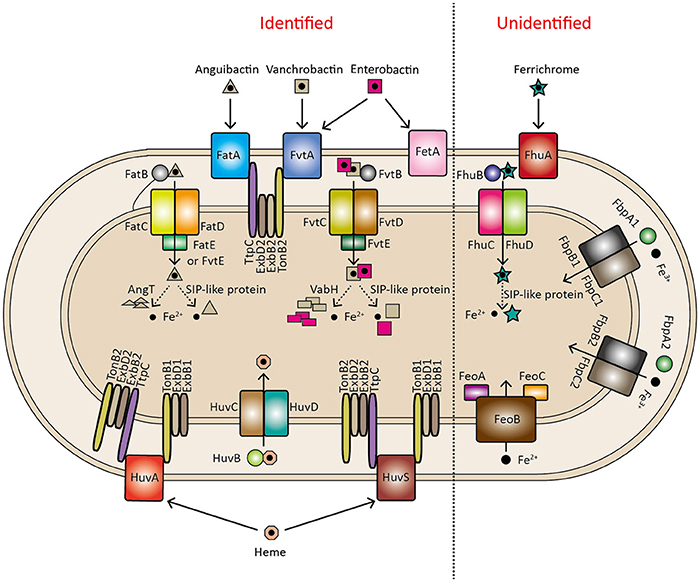
Figure 5. Identified and unidentified iron transport systems in V. anguillarum. A ferrisiderophore release model is proposed, indicated by a dashed line, including two mechanisms: degradation of the siderophore (achieved by AngT for anguibactin hydrolysis and VabH for vanchrobactin hydrolysis) and reduction of the ferrisiderophore [achieved by a siderophore-interacting protein (SIP)-like protein].
Once siderophores are produced and exported to the environment, they capture iron to form a ferric-siderophore complex, which is recognized by a specific transporter on the surface of the outer membrane. In V. anguillarum, ferric-anguibactin is translocated across the outer membrane via its specific transporter FatA, which is essential for ferric-anguibactin uptake (Walter et al., 1983; Actis et al., 1988; Lopez and Crosa, 2007; Lopez et al., 2007). This process requires the TonB2 system for energy transmission, which originates from the proton-motive force of the inner membrane (Stork et al., 2004).
The TonB2 complex is located across the inner membrane and comprises TonB2, ExbB2, ExbD2, and TtpC (TonB2 complex-associated transport protein C), all of which are indispensable for ferric-anguibactin import to the periplasmic space. Deletion of either of these genes completely abolishes ferric-anguibactin uptake, and they are thereby considered essential virulence factors for V. anguillarum since ferrisiderophore transport during iron uptake is a critical step for virulence (Stork et al., 2004, 2007). Notably, TtpC which shows homology to the TolR protein of E. coli, has been identified to be part of the TonB2 system in several Vibrio species, including V. anguillarum (Stork et al., 2007), V. cholerae (Stork et al., 2007), Vibrio alginolyticus (Wang et al., 2008), Vibrio parahaemolyticus (Kuehl and Crosa, 2010), and Vibrio vulnificus (Kuehl and Crosa, 2009). Despite a high similarity among all pathogenic vibrios studied thus far, the TtpC proteins likely play an important role in specific iron transport mediated by the TonB2 system, a conclusion drawn based on the observation that V. anguillarum ΔttpC cannot be complemented by the TtpC protein from V. cholerae (Stork et al., 2007; Kuehl and Crosa, 2009).
In addition to anguibactin transport, the TonB2 system is also involved in the uptake of vanchrobactin and the xenosiderophore (siderophore produced by other organisms) enterobactin. The transport of vanchrobactin and enterobactin is mediated by another outer membrane transporter, FvtA, which displays vanchrobactin-dependent expression (Balado et al., 2008, 2009; Naka et al., 2008). In the serotype O2 V. anguillarum strain RV22, loss of fvtA leads to impaired growth under iron-limiting conditions due to incapability of vanchrobactin transport in cells (Balado et al., 2009). Besides FvtA, during transport of the xenosiderophore enterobactin, it seems another transporter, FetA, is present that shows specific binding to enterobactin, and the expression of FetA is regulated by FetR (Naka and Crosa, 2012). This fetA-fetR cluster in V. anguillarum is speculated to derive from V. cholerae based on similar molecular organization and high similarity (Naka and Crosa, 2012).
Subsequently, when combined with the periplasmic lipoprotein FatB, ferric-anguibactin passes through the cytoplasmic membrane by using an ATP-binding cassette (ABC) transporter, which includes inner membrane permeases consisting of a heterodimer of FatC and FatD, and an ATPase FatE (Köster et al., 1991; Actis et al., 1995; Naka et al., 2010, 2013c). FatBCD are required for ferric-anguibactin transport while a fatE mutant is still capable of ferric-anguibactin transport due to the presence of its homolog FvtE (Naka et al., 2013c). Double deletion of fatE and fvtE significantly impairs ferric-anguibactin uptake (Naka et al., 2013c). Similarly, the passage of vanchrobactin through the inner membrane is also achieved in a stepwise manner by a periplasmic protein, FvtB, and an ABC transporter, FvtCDE, which together are essential for ferric-vanchrobactin or ferric-enterobactin import; deletion of either protein causes a defect in ferric-vanchrobactin and ferric-enterobactin transport (Naka et al., 2013c). Different from the specific involvement of FatE in the uptake of anguibactin, FvtE can transport both anguibactin and vanchrobactin/enterobactin (Naka et al., 2013c).
It is noteworthy that although fatDCBA encoding for ferric-anguibactin transport are located on the pJM1-type plasmid in serotype O1 strains, they are also present on the chromosomes of plasmidless vanchrobactin-synthesizing strains, albeit fatA is inactivated by an insertion sequence (Bay et al., 2007; Balado et al., 2009). In fact, gene clusters coding for vanchrobactin synthesis and uptake are ubiquitous in both anguibactin- and vanchrobactin-producing V. anguillarum strains (Balado et al., 2009), while plasmid-harboring strains do not produce vanchrobactin due to an inactivated chromosomal vabF gene (Figure 1; Naka et al., 2008). This is caused by the presence of the insertion sequence RS1, observed in all serotype O1 strains carrying a pJM1-type plasmid, and when RS1 is removed, pJM1-containing strains can simultaneously produce the siderophore vanchrobactin (Naka et al., 2008). However, because anguibactin appears to have a higher affinity for iron than does vanchrobactin, it is suggested to be the primary siderophore for iron import even though vanchrobactin is produced in the cell (Naka et al., 2008). Nevertheless, the minimal inhibitory concentration (MIC) values for iron chelators in plasmid-bearing strains seem to be lower than those of plasmidless V. anguillarum strains, which might be caused by higher stoichiometry of vanchrobactin in the plasmidless strains (Conchas et al., 1991).
Heme usually exists abundantly but not freely as a chemical compound in host cells. Therefore, bacteria secrete exotoxins such as cytolysins, proteases, or hemolysins to release it for further uptake. In V. anguillarum, several hemolysin genes have been identified and some of them are involved in V. anguillarum virulence (Hirono et al., 1996; Rodkhum et al., 2005; Rock and Nelson, 2006; Li et al., 2008, 2013; Xu et al., 2011). After its release, free heme is captured by a putative outer membrane transporter, HuvA, which has been shown to be essential for heme uptake because the huvA mutant does not grow when heme is used as the sole iron source (Mazoy et al., 2003). However, compared with the wild type, the huvA mutant still maintains heme-binding activity under the conditions tested (Mazoy et al., 2003), indicating that HuvA is not the only protein capable of binding heme. In line with this, two heme-binding proteins with molecular masses of 39 and 37 kDa were isolated from V. anguillarum serotype O1 and O2 strains, respectively, which are completely different from the 79 kDa HuvA protein (Mazoy and Lemos, 1996b; Mazoy et al., 1996). Therefore, Mazoy et al. speculate that, besides HuvA, additional proteins are probably present in V. anguillarum and function in heme binding, but not in its transport (Mazoy et al., 2003; Lemos and Osorio, 2007). Like siderophore import, this process is also energy-dependent, where HuvA is energized by TonB systems. The difference is that not only is the TonB2 complex involved in heme utilization, but a TonB1 system composed of TonB1-ExbB1-ExbD1 is also available for energy supply (Stork et al., 2004). Deletion of all genes for these two TonB systems leads to a complete defect in heme uptake and thus avirulence to the host, suggesting a key role of TonB systems in V. anguillarum virulence (Stork et al., 2004). The recent structural studies of the Ton complex from E. coli provide a mechanistic insight into this complex (Celia et al., 2016). It is proposed that the functional unit of the Ton complex contains an ExbB pentamer, an ExbD dimer, and at least one TonB. Electrophysiology experiments suggest that the ExbB-ExbD forms pH-sensitive channels, by which the Ton complex likely harnesses the proton-motive force for energy production and transduction.
Bioinformatic studies indicate that genes of the two TonB systems exist ubiquitously among all Vibrio species, and moreover, a third TonB system is even observed in some vibrios and other marine organisms (Kuehl and Crosa, 2010; Kustusch et al., 2011). However, it remains unclear why so many TonB systems are present in the Vibrio species. The V. anguillarum strains lack HuvA but contain an alternative heme transporter, HuvS, which is able to restore heme transport in a huvA mutant (Mouriño et al., 2005). The observation that huvS and huvA possess similar flanking DNA sequences implies that horizontal transmission and recombination might have occurred and thus be responsible for this genetic diversity (Mouriño et al., 2005).
Furthermore, the huvA gene is located in a gene cluster coding for nine heme uptake-related proteins, including HuvA, HuvZ, HuvX, TonB1, ExbB1, ExbD1, HuvB, HuvC, and HuvD (Mouriño et al., 2004). The periplasmic heme-binding protein HuvB delivers periplasmic heme to an inner membrane complex consisting of a permease, HuvC, and an ATPase, HuvD, which subsequently transport heme into the cytosol (Mouriño et al., 2004). Therefore, HuvBCD are required for heme transport, and deletion of either gene results in heme transport deficiencies (Mouriño et al., 2004). HuvZ also plays an import role in heme uptake, and loss of huvZ severely affects the growth of cells when heme serves as the sole iron source (Mouriño et al., 2004). Little is known about the function of HuvZ in heme utilization, and in V. cholerae it is suggested to have a role in heme storage (Wyckoff et al., 2004). HuvX, a predicted intracellular heme delivery protein in V. cholerae (Sekine et al., 2016), is not required for heme uptake in the V. anguillarum 775 plasmidless avirulent strain because deletion of huvX does not cause obvious differences in growth and heme utilization compared with the wild type (Mouriño et al., 2004). However, it is still under debate whether heme utilization is indeed involved in V. anguillarum virulence in nature.
Besides the anguibactin/vanchrobactin and heme uptake systems, four operons encoding putative iron transport systems, including transport of unchelated ferrous (feoABC) and ferric iron (fbpABC1 and fbpABC2), and siderophore ferrichrome transport (fhuABCD), have been identified in the genomes of V. anguillarum strains (Figures 1, 5). The presence of different iron transport systems in V. anguillarum probably results from differences in growth conditions because siderophores can only promote growth under limited iron conditions that must be insufficient for iron uptake under all environmental conditions. In line with this, all the vibrios examined have been shown to have additional iron acquisition systems (Table S3; Payne et al., 2016).
The ferrous iron transporter FeoABC is speculated to be the most ancient iron transport system and widely found among bacterial species including Vibrio species. In V. cholerae, all feoABC genes are required for ferrous iron uptake although their functions have not been fully characterized (Wyckoff et al., 2006; Weaver et al., 2013). However, it is still unknown how ferrous iron passes through the outer membrane for transport by the Feo system in the periplasm. FbpABC, a ferric iron transporter, is also found in vibrios and has been shown to promote better growth at alkaline pH in V. cholerae (Peng et al., 2016). FhuABCD, responsible for siderophore ferrichrome utilization, have been demonstrated to be required for ferrichrome utilization in V. parahaemolyticus and V. cholerae (Rogers et al., 2000; Funahashi et al., 2009).
Notably, although it has been demonstrated that V. anguillarum is capable of using ferric citrate as the only iron source by a siderophore-independent mechanism (Mazoy et al., 1997), we do not find any genes involved in ferric citrate transport in the genome of V. anguillarum or other vibrios. In P. aeruginosa, iron delivered by citrate is suggested to enter the cell as Fe2+, and FeoB is further required for citrate-mediated Fe2+ uptake (Marshall et al., 2009). Therefore, it is plausible that FeoB of V. anguillarum may also play a role in Fe2+ uptake from ferric citrate to maintain iron homeostasis.
When iron–siderophore complexes are transported into the cell cytoplasm, bacteria can use different strategies to release iron:
– Via the reduction of ferric iron.
– Via the degradation or modification of siderophores.
– Via both iron reduction and siderophore degradation.
Genes predicted for iron reduction and siderophore degradation occur in the genome of V. anguillarum, and proposed pathways are shown in Figure 5. VabH, a putative cytoplasmic esterase, exhibits homology to E. coli Fes, which can hydrolyze enterobactin during iron release (Brickman and McIntosh, 1992). Therefore, VabH may serve as a vanchrobactin degradation enzyme during ferric-vanchrobactin compound dissociation. Similarly, a putative thioesterase gene, angT, may be involved in iron release from ferric-anguibactin. However, deletion of vabH or angT does not completely block siderophore uptake (Wertheimer et al., 1999; Balado et al., 2006), indicating that additional pathways for iron release may occur. In accordance with this, a nicotinamide adenine dinucleotide phosphate (NADPH)-dependent ferric reductase has been identified in the genomes of V. anguillarum strains. It shows high homology to the reported siderophore-interacting proteins (SIP) YqjH of E. coli and FscN of Thermobifida fusca, which are suggested to participate in iron reduction during ferrisiderophore dissociation (Miethke et al., 2011; Li et al., 2015). Furthermore, Mazoy and Lemos observed that ferric reductase activities of cell fractions are significantly increased in the presence of NADPH compared to its absence (Mazoy and Lemos, 1996a), indicating a functional NADPH-dependent ferric reductase may occur in V. anguillarum. In addition, they also found it is only in the cytoplasmic, but not in periplasmic or membrane faction, where ferric reductase activity is stimulated under iron-limiting conditions (Mazoy and Lemos, 1996a). This implies iron release from ferrisiderophore might happen in the cytoplasm. Further investigation of the role of SIP in the ferrisiderophore dissociation pathway will provide more detail to elucidate the strategies deployed by V. anguillarum to release iron for utilization.
As excess iron is lethal and may lead to oxidative damage to DNA when free Fe2+ reacts with hydrogen peroxide via the Fenton reaction (Imlay, 2002), tight regulation of iron transport is a prerequisite to meet, but not exceed, the requirement for iron. To date, several regulators have been identified in V. anguillarum to control the uptake of the iron, including the negative regulators Fur and an antisense RNA (RNAα), and the positive regulators AngR, TAFr, and anguibactin.
In Gram-negative bacteria, Fur is the major global regulator of iron metabolism, which serves as a sensor of intracellular iron concentration. It can bind to a Fe2+-bound dimer at a specific site, termed the Fur box, in the promoter region and thereby negatively regulate the transcription of corresponding genes (Troxell and Hassan, 2013). Therefore, it is not surprising that the Fur protein of V. anguillarum blocks or depresses the expression of most genes involved in iron acquisition systems, such as those coding for anguibactin and vanchrobactin synthesis (Salinas and Crosa, 1995; Chen et al., 1996; Di Lorenzo et al., 2004; Alice et al., 2005; Balado et al., 2008), TonB systems (Mouriño et al., 2006), and iron transport systems (Waldbeser et al., 1993; Tolmasky et al., 1994; Chen and Crosa, 1996; Chai et al., 1998; Mouriño et al., 2006; Balado et al., 2008; Naka and Crosa, 2012). In addition to Fur, an antisense RNA, RNAα, is capable of negatively modulating the expression of the fatA and fatB genes involved in iron transport by specifically binding to fatA and fatB mRNAs and thus repressing their transcription under iron-rich conditions (Waldbeser et al., 1995; Chen and Crosa, 1996). Moreover, the Fur protein is also crucial for RNAα synthesis and regulates its transcription initiation, which is independent of the iron status of the cell (Chen and Crosa, 1996).
AngR is a bifunctional protein, involved not only in the formation of anguibactin but also in the positive regulation of transport and biosynthesis genes (Salinas et al., 1989; Singer et al., 1991). Although AngR consists of two helix-turn-helix (HTH) regulatory motifs, only the first HTH is essential for gene regulation, as demonstrated by the finding that modulation of an angR deletion mutant is restored by a construct containing a frame shift and leaving only the first HTH motif (Wertheimer et al., 1999). These data again suggest that AngR plays an important role in both anguibactin synthesis and regulation of gene expression. The transcription of genes including the iron transport biosynthesis operon (ITBO; Figure 1) and angN is tightly controlled by AngR, and they display the highest expression level when iron is limited (Actis et al., 1995; Chen and Crosa, 1996; Di Lorenzo et al., 2008). The TAF (trans-acting factor) region shown in Figure 1 is essential for anguibactin biosynthesis and for maximal expression of the ITBO genes, which are achieved by two separate entities: one involved in anguibactin biosynthesis (TAFb) and the other in regulation (TAFr) (Tolmasky et al., 1988; Welch et al., 2000). Studies have demonstrated that TAFr and AngR work in a synergistic manner to modulate the level of anguibactin synthesis under iron-limiting conditions (Salinas et al., 1989; Salinas and Crosa, 1995). In addition, anguibactin by itself is also capable of increasing the transcriptional level of the ITBO, which reaches the highest level when AngR, TAFr, and anguibactin are all acting synergistically (Chen and Crosa, 1996).
Besides iron, many other environmental factors have been identified in vibrios, including V. anguillarum, that affect the expression of genes associated with iron transport, such as oxygen, temperature, carbon sources, and quorum sensing molecules (Mou et al., 2013; Payne et al., 2016). Therefore, an expanded search for sensors involved in other environmental signals may lead to a more complete picture of the strategies of iron regulation in V. anguillarum.
Despite the challenges of iron acquisition in various environments, such as competition with other organisms on host surfaces, or sequestration by the high-affinity iron-binding host proteins lactoferrin and transferrin, growth of V. anguillarum is proficient in both host and marine habitats, achieved by multiple iron transport systems. Moreover, there is increasing evidence for the position of iron uptake, especially siderophores, at the crux of the microbial infection process, thereby reducing the appeal of siderophores as antimicrobial targets. In this context, it is important to investigate the whole of the iron acquisition systems of V. anguillarum, which will not only provide new insights to explain the evolutionary origin of versatile iron transport systems but also supply more evidence to understand the pathogenicity of this organism.
However, despite the exciting developments in understanding siderophore synthesis and ferrisiderophore transport in some vibrios, such as V. anguillarum and V. cholerae, there are many unsolved aspects of the iron uptake systems. For example, the mechanisms that govern the synthesis of anguibactin or vanchrobactin are still not fully elucidated, and how proteins are regulated for anguibactin or vanchrobactin synthesis is not yet well understood. How is mature siderophore secreted to the environment, and how is the iron released to the periplasm or cytoplasm from the ferric-siderophore complex? To answer these questions, an effective way is required to monitor anguibactin or vanchrobactin in real time. Finally, the different iron uptake systems that are responsible for the utilization of diverse iron sources will need to be established at the genetic level in order to uncover the relationship between iron uptake and virulence.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
YL was supported by the China Postdoctoral Science Foundation (no. 2016M602200). QM was supported by grants from the “1000 Talents Program” of China, “100 Talents Program” of the Chinese Academy of Sciences, and “AoShan Talents Program” of Qingdao National Laboratory for Marine Science and Technology (no. 2015ASTP).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Wenteng Xu, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, is greatly acknowledged for language proofing.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00342/full#supplementary-material
Actis, L. A., Fish, W., Crosa, J. H., Kellerman, K., Ellenberger, S. R., Hauser, F. M., et al. (1986). Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(pJM1). J. Bacteriol. 167, 57–65. doi: 10.1128/jb.167.1.57-65.1986
Actis, L. A., Tolmasky, M. E., Crosa, L. M., and Crosa, J. H. (1995). Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol. Microbiol. 17, 197–204. doi: 10.1111/j.1365-2958.1995.mmi_17010197.x
Actis, L. A., Tolmasky, M. E., Farrell, D. H., and Crosa, J. H. (1988). Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid mediated iron transport system. J. Biol. Chem. 263, 2853–2860.
Alice, A. F., Lopez, C. S., and Crosa, J. H. (2005). Plasmid- and chromosome-encoded redundant and specific functions are involved in biosynthesis of the siderophore anguibactin in Vibrio anguillarum 775: a case of chance and necessity? J. Bacteriol. 187, 2209–2214. doi: 10.1128/JB.187.6.2209-2214.2005
Balado, M., Osorio, C. R., and Lemos, M. L. (2006). A gene cluster involved in the biosynthesis of vanchrobactin, a chromosome-encoded siderophore produced by Vibrio anguillarum. Microbiology 152, 3517–3528. doi: 10.1099/mic.0.29298-0
Balado, M., Osorio, C. R., and Lemos, M. L. (2008). Biosynthetic and regulatory elements involved in the production of the siderophore vanchrobactin in Vibrio anguillarum. Microbiology 154, 1400–1413. doi: 10.1099/mic.0.2008/016618-0
Balado, M., Osorio, C. R., and Lemos, M. L. (2009). FvtA is the receptor for the siderophore vanchrobactin in Vibrio anguillarum: utility as a route of entry for vanchrobactin analogues. Appl. Environ. Microbiol. 75, 2775–2783. doi: 10.1128/AEM.02897-08
Barancin, C. E., Smoot, J. C., Findlay, R. H., and Actis, L. A. (1998). Plasmid-mediated histamine biosynthesis in the bacterial fish pathogen Vibrio anguillarum. Plasmid 39, 235–244. doi: 10.1006/plas.1998.1345
Bay, L., Larsen, J. L., and Leisner, J. J. (2007). Distribution of three genes involved in the pJM1 iron-sequestering system in various Vibrio anguillarum serogroups. Syst. Appl. Microbiol. 30, 85–92. doi: 10.1016/j.syapm.2006.03.006
Bina, X. R., Provenzano, D., Nguyen, N., and Bina, J. E. (2008). Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 76, 3595–3605. doi: 10.1128/IAI.01620-07
Brickman, T. J., and McIntosh, M. A. (1992). Overexpression and purification of ferric enterobactin esterase from Escherichia coli. demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J. Biol. Chem. 267, 12350–12355.
Celia, H., Noinaj, N., Zakharov, S. D., Bordignon, E., Botos, I., Santamaria, M., et al. (2016). Structural insight into the role of the Ton complex in energy transduction. Nature 538, 60–65. doi: 10.1038/nature19757
Chai, S. H., Welch, T. J., and Crosa, J. H. (1998). Characterization of the interaction between Fur and the iron transport promoter of the virulence plasmid in Vibrio anguillarum. J. Biol. Chem. 273, 33841–33847. doi: 10.1074/jbc.273.50.33841
Chen, Q., and Crosa, J. H. (1996). Antisense RNA, Fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J. Biol. Chem. 271, 18885–18891. doi: 10.1074/jbc.271.31.18885
Chen, Q., Wertheimer, A. M., Tolmasky, M. E., and Crosa, J. H. (1996). The AngR protein and the siderophore anguibactin positively regulate the expression of iron-transport genes in Vibrio anguillarum. Mol. Microbiol. 22, 127–134. doi: 10.1111/j.1365-2958.1996.tb02662.x
Conchas, R. F., Lemos, M. L., Barja, J. L., and Toranzo, A. E. (1991). Distribution of plasmid- and chromosome-mediated iron uptake systems in Vibrio anguillarum strains of different origins. Appl. Environ. Microbiol. 57, 2956–2962.
Crosa, J. H., Payne, S. M., and Mey, A. R. (2004). Iron Transport in Bacteria. Washington, DC: ASM Press.
Croxatto, A., Lauritz, J., Chen, C., and Milton, D. L. (2007). Vibrio anguillarum colonization of rainbow trout integument requires a DNA locus involved in exopolysaccharide transport and biosynthesis. Environ. Microbiol. 9, 370–382. doi: 10.1111/j.1462-2920.2006.01147.x
Di Lorenzo, M., Poppelaars, S., Stork, M., Nagasawa, M., Tolmasky, M. E., and Crosa, J. H. (2004). A nonribosomal peptide synthetase with a novel domain organization is essential for siderophore biosynthesis in Vibio anguillarum. J. Bacteriol. 186, 7327–7336. doi: 10.1128/JB.186.21.7327-7336.2004
Di Lorenzo, M., Stork, M., and Crosa, J. H. (2011). Genetic and biochemical analyses of chromosome and plasmid gene homologues encoding ICL and ArCP domains in Vibrio anguillarum strain 775. Biometals 24, 629–643. doi: 10.1007/s10534-011-9416-7
Di Lorenzo, M., Stork, M., Naka, H., Tolmasky, M. E., and Crosa, J. H. (2008). Tandem heterocyclization domains in a nonribosomal peptide synthetase essential for siderophore biosynthesis in Vibrio anguillarum. Biometals 21, 635–648. doi: 10.1007/s10534-008-9149-4
Du, J. S., Deng, T., and Ma, Q. J. (2017). Crystal structures of the isochorismatase domains from Vibrio anguillarum. Biochem. Biophys. Res. Commun. 490, 827–833. doi: 10.1016/j.bbrc.2017.06.125
Funahashi, T., Tanabe, T., Shiuchi, K., Nakao, H., and Yamamoto, S. (2009). Identification and characterization of genes required for utilization of desferri-ferrichrome and aerobactin in Vibrio parahaemolyticus. Biol. Pharm. Bull. 32, 359–365. doi: 10.1248/bpb.32.359
Furrer, J. L., Sanders, D. N., Hook-Barnard, I. G., and McIntosh, M. A. (2002). Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44, 1225–1234. doi: 10.1046/j.1365-2958.2002.02885.x
Hannauer, M., Braud, A., Hoegy, F., Ronot, P., Boos, A., and Schalk, I. J. (2012). The PvdRT-OpmQ efflux pump controls the metal selectivity of the iron uptake pathway mediated by the siderophore pyoverdine in Pseudomonas aeruginosa. Environ. Microbiol. 14, 1696–1708. doi: 10.1111/j.1462-2920.2011.02674.x
Hannauer, M., Yeterian, E., Martin, L. W., Lamont, I. L., and Schalk, I. J. (2010). An efflux pump is involved in secretion of newly synthesized siderophore by Pseudomonas aeruginosa. FEBS Lett. 584, 4751–4755. doi: 10.1016/j.febslet.2010.10.051
Hantke, K. (1981). Regulation of ferric iron transport in Escherichia coli K12 isolation of a constitutive mutant. Mol. Gen. Genet. 182, 288–292. doi: 10.1007/BF00269672
Hirono, I., Masuda, T., and Aoki, T. (1996). Cloning and detection of the hemolysin gene of Vibrio anguillarum. Microb. Pathog. 21, 173–182. doi: 10.1006/mpat.1996.0052
Horiyama, T., and Nishino, K. (2014). AcrB, AcrD, and MdtABC multidrug efflux systems are involved in enterobactin export in Escherichia coli. PLoS ONE 9:e108642. doi: 10.1371/journal.pone.0108642
Imlay, J. A. (2002). How oxygen damages microbes: oxygen tolerance and obligate anaerobiosis. Adv. Microb. Physiol. 46, 111–153. doi: 10.1016/S0065-2911(02)46003-1
Köster, W. L., Actis, L. A., Waldbeser, L. S., Tolmasky, M. E., and Crosa, J. H. (1991). Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J. Biol. Chem. 266, 23829–23833.
Kuehl, C. J., and Crosa, J. H. (2009). Molecular and genetic characterization of the TonB2-cluster TtpC protein in pathogenic vibrios. Biometals 22, 109–115. doi: 10.1007/s10534-008-9194-z
Kuehl, C. J., and Crosa, J. H. (2010). The TonB energy transduction systems in Vibrio species. Future Microbiol. 5, 1403–1412. doi: 10.2217/fmb.10.90
Kunkle, D. E., Bina, X. R., and Bina, J. E. (2017). The Vibrio cholerae VexGH RND efflux system maintains cellular homeostasis by efluxing vibriobactin. mBio 8:e00126-17. doi: 10.1128/mBio.00126-17
Kustusch, R. J., Kuehl, C. J., and Crosa, J. H. (2011). Power plays: iron transport and energy transduction in pathogenic vibrios. Biometals 24, 559–566. doi: 10.1007/s10534-011-9437-2
Lemos, M. L., and Osorio, C. R. (2007). Heme, an iron supply for vibrios pathogenic for fish. Biometals 20, 615–626. doi: 10.1007/s10534-006-9053-8
Lemos, M. L., Salinas, P., Toranzo, A. E., Barja, J. L., and Crosa, J. H. (1988). Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J. Bacteriol. 170, 1920–1925. doi: 10.1128/jb.170.4.1920-1925.1988
Li, K. H., Chen, W. H., and Bruner, S. D. (2015). Structure and mechanism of the siderophore-interacting protein from the fuscachelin gene cluster of Thermobifida fusca. Biochemistry 54, 3989–4000. doi: 10.1021/acs.biochem.5b00354
Li, L., Mou, X. Y., and Nelson, D. R. (2013). Characterization of Plp, a phosphatidylcholine-specific phospholipase and hemolysin of Vibrio anguillarum. BMC Microbiol. 13:271. doi: 10.1186/1471-2180-13-271
Li, L., Rock, J. L., and Nelson, D. R. (2008). Identification and characterization of a repeat-in-toxin gene cluster in Vibrio anguillarum. Infect. Immun. 76, 2620–2632. doi: 10.1128/IAI.01308-07
Liu, Q., Ma, Y., Wu, H. Z., Shao, M. F., Liu, H. F., and Zhang, Y. X. (2004). Cloning, identification and expression of an entE homologue angE from Vibrio anguillarum serotype O1. Arch. Microbiol. 181, 287–293. doi: 10.1007/s00203-004-0652-x
Lopez, C. S., Alice, A. F., Chakraborty, R., and Crosa, J. H. (2007). Identification of amino acid residues required for ferric-anguibactin transport in the outer-membrane receptor FatA of Vibrio anguillarum. Microbiology 153, 570–584. doi: 10.1099/mic.0.2006/001735-0
Lopez, C. S., and Crosa, J. H. (2007). Characterization of ferric-anguibactin transport in Vibrio anguillarum. Biometals 20, 393–403. doi: 10.1007/s10534-007-9084-9
Marshall, B., Stintzi, A., Gilmour, C., Meyer, J. M., and Poole, K. (2009). Citrate-mediated iron uptake in Pseudomonas aeruginosa: involvement of the citrate-inducible FecA receptor and the FeoB ferrous iron transporter. Microbiology 155, 305–315. doi: 10.1099/mic.0.023531-0
Martin, J. H., Gordon, R. M., and Fitzwater, S. E. (1991). Iron limitation? The case for iron. Limnol. Oceanogr. 36, 1793–1802. doi: 10.4319/lo.1991.36.8.1793
Mazoy, R., Botana, L. M., and Lemos, M. L. (1997). Iron uptake from ferric citrate by Vibrio anguillarum. FEMS Microbiol. Lett. 154, 145–150. doi: 10.1111/j.1574-6968.1997.tb12636.x
Mazoy, R., and Lemos, M. L. (1996a). Ferric-reductase activities in whole cells and cell fractions of Vibrio (Listonella) anguillarum. Microbiol-UK 142, 3187–3193. doi: 10.1099/13500872-142-11-3187
Mazoy, R., and Lemos, M. L. (1996b). Identification of heme-binding proteins in the cell membranes of Vibrio anguillarum. FEMS Microbiol. Lett. 135, 265–270. doi: 10.1111/j.1574-6968.1996.tb07999.x
Mazoy, R., Osorio, C. R., Toranzo, A. E., and Lemos, M. L. (2003). Isolation of mutants of Vibrio anguillarum defective in haeme utilisation and cloning of huvA, a gene coding for an outer membrane protein involved in the use of haeme as iron source. Arch. Microbiol. 179, 329–338. doi: 10.1007/s00203-003-0529-4
Mazoy, R., Vazquez, F., and Lemos, M. L. (1996). Isolation of heme-binding proteins from Vibrio anguillarum using affinity chromatography. FEMS Microbiol. Lett. 141, 19–23. doi: 10.1111/j.1574-6968.1996.tb08357.x
Miethke, M., Hou, J., and Marahiel, M. A. (2011). The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry 50, 10951–10964. doi: 10.1021/bi201517h
Milton, D. L., Norqvist, A., and Wolfwatz, H. (1992). Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio anguillarum. J. Bacteriol. 174, 7235–7244. doi: 10.1128/jb.174.22.7235-7244.1992
Mo, Z. L., Guo, D. S., Mao, Y. X., Ye, X. H., Zou, Y. X., Xiao, P., et al. (2010). Identification and characterization of the Vibrio anguillarum prtV gene encoding a new metalloprotease. Chin. J. Oceanol. Limnol. 28, 55–61. doi: 10.1007/s00343-010-9246-4
Mou, X. Y., Spinard, E. J., Driscoll, M. V., Zhao, W. J., and Nelson, D. R. (2013). H-NS is a negative regulator of the two hemolysin/cytotoxin gene clusters in Vibrio anguillarum. Infect. Immun. 81, 3566–3576. doi: 10.1128/IAI.00506-13
Mouriño, S., Osorio, C. R., and Lemos, M. L. (2004). Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J. Bacteriol. 186, 6159–6167. doi: 10.1128/JB.186.18.6159-6167.2004
Mouriño, S., Osorio, C. R., Lemos, M. L., and Crosa, J. H. (2006). Transcriptional organization and regulation of the Vibrio anguillarum heme uptake gene cluster. Gene 374, 68–76. doi: 10.1016/j.gene.2006.01.014
Mouriño, S., Rodriguez-Ares, I., Osorio, C. R., and Lemos, M. L. (2005). Genetic variability of the heme uptake system among different strains of the fish pathogen Vibrio anguillarum: identification of a new heme receptor. Appl. Environ. Microbiol. 71, 8434–8441. doi: 10.1128/aem.71.12.8434-8441.2005
Naka, H., Actis, L. A., and Crosa, J. H. (2013a). The anguibactin biosynthesis and transport genes are encoded in the chromosome of Vibrio harveyi: a possible evolutionary origin for the pJM1 plasmid-encoded system of Vibrio anguillarum? Microbiology 2, 182–194. doi: 10.1002/mbo3.65
Naka, H., and Crosa, J. H. (2011). Genetic determinants of virulence in the marine fish pathogen Vibrio anguillarum. Fish Pathol. 46, 1–10. doi: 10.3147/jsfp.46.1
Naka, H., and Crosa, J. H. (2012). Identification and characterization of a novel outer membrane protein receptor FetA for ferric enterobactin transport in Vibrio anguillarum 775 (pJM1). Biometals 25, 125–133. doi: 10.1007/s10534-011-9488-4
Naka, H., Liu, M. Q., Actis, L. A., and Crosa, J. H. (2013b). Plasmid- and chromosome-encoded siderophore anguibactin systems found in marine vibrios: biosynthesis, transport and evolution. Biometals 26, 537–547. doi: 10.1007/s10534-013-9629-z
Naka, H., Liu, M. Q., and Crosa, J. H. (2013c). Two ABC transporter systems participate in siderophore transport in the marine pathogen Vibrio anguillarum 775 (pJM1). FEMS Microbiol. Lett. 341, 79–86. doi: 10.1111/1574-6968.12092
Naka, H., Lopez, C. S., and Crosa, J. H. (2008). Reactivation of the vanchrobactin siderophore system of Vibrio anguillarum by removal of a chromosomal insertion sequence originated in plasmid pJM1 encoding the anguibactin siderophore system. Environ. Microbiol. 10, 265–277. doi: 10.1111/j.1462-2920.2007.01450.x
Naka, H., Lopez, C. S., and Crosa, J. H. (2010). Role of the pJM1 plasmid-encoded transport proteins FatB, C and D in ferric anguibactin uptake in the fish pathogen Vibrio anguillarum. Environ. Microbiol. Re. 2, 104–111. doi: 10.1111/j.1758-2229.2009.00110.x
Ormonde, P., Horstedt, P., O'toole, R., and Milton, D. L. (2000). Role of motility in adherence to and invasion of a fish cell line by Vibrio anguillarum. J. Bacteriol. 182, 2326–2328. doi: 10.1128/JB.182.8.2326-2328.2000
O'Toole, R., Lundberg, S., Fredriksson, S. A., Jansson, A., Nilsson, B., and Wolf-Watz, H. (1999). The chemotactic response of Vibrio anguillarum to fish intestinal mucus is mediated by a combination of multiple mucus components. J. Bacteriol. 181, 4308–4317.
Payne, S. M., Mey, A. R., and Wyckoff, E. E. (2016). Vibrio iron transport: evolutionary adaptation to life in multiple environments. Microbiol. Mol. Biol. Rev. 80, 69–90. doi: 10.1128/MMBR.00046-15
Pedersen, K., Grisez, L., Van Houdt, R., Tiainen, T., Ollevier, F., and Larsen, J. L. (1999). Extended serotyping scheme for Vibrio anguillarum with the definition and characterization of seven provisional O-serogroups. Curr. Microbiol. 38, 183–189. doi: 10.1007/PL00006784
Peng, E. D., Wyckoff, E. E., Mey, A. R., Fisher, C. R., and Payne, S. M. (2016). Nonredundant roles of iron acquisition systems in Vibrio cholerae. Infect. Immun. 84, 511–523. doi: 10.1128/IAI.01301-15
Rahman, M. M., Matsuo, T., Ogawa, W., Koterasawa, M., Kuroda, T., and Tsuchiya, T. (2007). Molecular cloning and characterization of all RND-Type efflux transporters in Vibrio cholerae non-O1. Microbiol. Immunol. 51, 1061–1070. doi: 10.1111/j.1348-0421.2007.tb04001.x
Rock, J. L., and Nelson, D. R. (2006). Identification and characterization of a hemolysin gene cluster in Vibrio anguillarum. Infect. Immun. 74, 2777–2786. doi: 10.1128/IAI.74.5.2777-2786.2006
Rodkhum, C., Hirono, I., Crosa, J. H., and Aoki, T. (2005). Four novel hemolysin genes of Vibrio anguillarum and their virulence to rainbow trout. Microb. Pathog. 39, 109–119. doi: 10.1016/j.micpath.2005.06.004
Rogers, M. B., Sexton, J. A., Decastro, G. J., and Calderwood, S. B. (2000). Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J. Bacteriol. 182, 2350–2353. doi: 10.1128/JB.182.8.2350-2353.2000
Salinas, P. C., and Crosa, J. H. (1995). Regulation of angR, a gene with regulatory and biosynthetic functions in the pJM1 plasmid-mediated iron uptake system of Vibrio anguillarum. Gene 160, 17–23. doi: 10.1016/0378-1119(95)00213-P
Salinas, P. C., Tolmasky, M. E., and Crosa, J. H. (1989). Regulation of the iron uptake system in Vibrio anguillarum: evidence for a cooperative effect between two transcriptional activators. Proc. Natl. Acad. Sci. U.S.A. 86, 3529–3533. doi: 10.1073/pnas.86.10.3529
Schalk, I. J., and Guillon, L. (2013). Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: implications for metal homeostasis. Environ. Microbiol. 15, 1661–1673. doi: 10.1111/1462-2920.12013
Sekine, Y., Tanzawa, T., Tanaka, Y., Ishimori, K., and Uchida, T. (2016). Cytoplasmic heme-binding protein (HutX) from Vibrio cholerae is an intracellular heme transport protein for the heme-degrading enzyme, HutZ. Biochemistry 55, 884–893. doi: 10.1021/acs.biochem.5b01273
Singer, J. T., Schmidt, K. A., and Reno, P. W. (1991). Polypeptides p40, pOM2, and pAngR are required for iron uptake and for virulence of the marine fish pathogen Vibrio anguillarum 775. J. Bacteriol. 173, 1347–1352. doi: 10.1128/jb.173.3.1347-1352.1991
Soengas, R. G., Larrosa, M., Balado, M., Rodriguez, J., Lemos, M. L., and Jimenez, C. (2008). Synthesis and biological activity of analogues of vanchrobactin, a siderophore from Vibrio anguillarum serotype O2. Org. Biomol. Chem. 6, 1278–1287. doi: 10.1039/b719713f
Stork, M., Di Lorenzo, M., Mourino, S., Osorio, C. R., Lemos, M. L., and Crosa, J. H. (2004). Two tonB systems function in iron transport in Vibrio anguillarum, but only one is essential for virulence. Infect. Immun. 72, 7326–7329. doi: 10.1128/IAI.72.12.7326-7329.2004
Stork, M., Otto, B. R., and Crosa, J. H. (2007). A novel protein, TtpC, is a required component of the TonB2 complex for specific iron transport in the pathogens Vibrio anguillarum and Vibrio cholerae. J. Bacteriol. 189, 1803–1815. doi: 10.1128/JB.00451-06
Tolmasky, M. E., Actis, L. A., and Crosa, J. H. (1988). Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J. Bacteriol. 170, 1913–1919. doi: 10.1128/jb.170.4.1913-1919.1988
Tolmasky, M. E., Actis, L. A., and Crosa, J. H. (1995). A histidine decarboxylase gene encoded by the Vibrio anguillarum plasmid pJM1 is essential for virulence: histamine is a precursor in the biosynthesis of anguibactin. Mol. Microbiol. 15, 87–95. doi: 10.1111/j.1365-2958.1995.tb02223.x
Tolmasky, M. E., Wertheimer, A. M., Actis, L. A., and Crosa, J. H. (1994). Characterization of the Vibrio anguillarum fur gene: role in regulation of expression of the FatA outer membrane protein and catechols. J. Bacteriol. 176, 213–220. doi: 10.1128/jb.176.1.213-220.1994
Toranzo, A. E., Magarinos, B., and Romalde, J. L. (2005). A review of the main bacterial fish diseases in mariculture systems. Aquaculture 246, 37–61. doi: 10.1016/j.aquaculture.2005.01.002
Troxell, B., and Hassan, H. M. (2013). Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Varina, M., Denkin, S. M., Staroscik, A. M., and Nelson, D. R. (2008). Identification and characterization of epp, the secreted processing protease for the Vibrio anguillarum EmpA metalloprotease. J. Bacteriol. 190, 6589–6597. doi: 10.1128/JB.00535-08
Waldbeser, L. S., Chen, Q. A., and Crosa, J. H. (1995). Antisense RNA regulation of the fatB iron transport protein gene in Vibrio anguillarum. Mol. Microbiol. 17, 747–756. doi: 10.1111/j.1365-2958.1995.mmi_17040747.x
Waldbeser, L. S., Tolmasky, M. E., Actis, L. A., and Crosa, J. H. (1993). Mechanisms for negative regulation by iron of the fatA outer membrane protein gene expression in Vibrio anguillarum 775. J. Biol. Chem. 268, 10433–10439.
Walter, M. A., Potter, S. A., and Crosa, J. H. (1983). Iron uptake system mediated by Vibrio anguillarum plasmid pJM1. J. Bacteriol. 156, 880–887.
Wang, Q. Y., Liu, Q., Cao, X. D., Yang, M. J., and Zhang, Y. X. (2008). Characterization of two TonB systems in marine fish pathogen Vibrio alginolyticus: their roles in iron utilization and virulence. Arch. Microbiol. 190, 595–603. doi: 10.1007/s00203-008-0407-1
Weaver, E. A., Wyckoff, E. E., Mey, A. R., Morrison, R., and Payne, S. M. (2013). FeoA and FeoC are essential components of the Vibrio cholerae ferrous iron uptake system, and FeoC interacts with FeoB. J. Bacteriol. 195, 4826–4835. doi: 10.1128/JB.00738-13
Welch, T. J., Chai, S. H., and Crosa, J. H. (2000). The overlapping angB and angG genes are encoded within the trans-acting factor region of the virulence plasmid in Vibrio anguillarum: essential role in siderophore biosynthesis. J. Bacteriol. 182, 6762–6773. doi: 10.1128/JB.182.23.6762-6773.2000
Welch, T. J., and Crosa, J. H. (2005). Novel role of the lipopolysaccharide O1 side chain in ferric siderophore transport and virulence of Vibrio anguillarum. Infect. Immun. 73, 5864–5872. doi: 10.1128/IAI.73.9.5864-5872.2005
Wertheimer, A. M., Verweij, W., Chen, Q., Crosa, L. M., Nagasawa, M., Tolmasky, M. E., et al. (1999). Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect. Immun. 67, 6496–6509.
Wolf, M. K., and Crosa, J. H. (1986). Evidence for the role of a siderophore in promoting Vibrio anguillarum infections. J. Gen. Microbiol. 132, 2949–2952. doi: 10.1099/00221287-132-10-2949
Wyckoff, E. E., Mey, A. R., Leimbach, A., Fisher, C. F., and Payne, S. M. (2006). Characterization of ferric and ferrous iron transport systems in Vibrio cholerae. J. Bacteriol. 188, 6515–6523. doi: 10.1128/JB.00626-06
Wyckoff, E. E., Schmitt, M., Wilks, A., and Payne, S. M. (2004). HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 186, 4142–4151. doi: 10.1128/JB.186.13.4142-4151.2004
Xu, Z., Wang, Y., Han, Y., Chen, J., and Zhang, X. H. (2011). Mutation of a novel virulence-related gene mltD in Vibrio anguillarum enhances lethality in zebra fish. Res. Microbiol. 162, 144–150. doi: 10.1016/j.resmic.2010.08.003
Yang, H., Chen, J. X., Yang, G. P., Zhang, X. H., Li, Y., and Wang, M. (2007). Characterization and pathogenicity of the zinc metalloprotease EmpA of Vibrio anguillarum expressed in Escherichia coli. Curr. Microbiol. 54, 244–248. doi: 10.1007/s00284-006-0495-6
Keywords: Vibrio anguillarum, siderophore biosynthesis, siderophore secretion, iron uptake, iron release, iron acquisition mechanism
Citation: Li Y and Ma Q (2017) Iron Acquisition Strategies of Vibrio anguillarum. Front. Cell. Infect. Microbiol. 7:342. doi: 10.3389/fcimb.2017.00342
Received: 03 May 2017; Accepted: 11 July 2017;
Published: 25 July 2017.
Edited by:
Pierre Cornelis, Vrije Universiteit Brussel, BelgiumReviewed by:
Manuel L. Lemos, Universidade de Santiago de Compostela, SpainCopyright © 2017 Li and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingjun Ma, cW1hQHFkaW8uYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.