
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 27 April 2017
Sec. Microbiome in Health and Disease
Volume 7 - 2017 | https://doi.org/10.3389/fcimb.2017.00144
This article is part of the Research TopicParasite Infections: From Experimental Models To Natural SystemsView all 21 articles
Parasitic nematode infections are widespread in nature, affecting humans as well as wild, companion, and livestock animals. Most parasitic nematodes inhabit the intestines of their hosts living in close contact with the intestinal microbiota. Many species also have tissue migratory life stages in the absence of severe systemic inflammation of the host. Despite the close coexistence of helminths with numerous microbes, little is known concerning these interactions. While the environmental niche is considerably different, the free-living nematode Caenorhabditis elegans (C. elegans) is also found amongst a diverse microbiota, albeit on decaying organic matter. As a very well characterized model organism that has been intensively studied for several decades, C. elegans interactions with bacteria are much more deeply understood than those of their parasitic counterparts. The enormous breadth of understanding achieved by the C. elegans research community continues to inform many aspects of nematode parasitology. Here, we summarize what is known regarding parasitic nematode-bacterial interactions while comparing and contrasting this with information from work in C. elegans. This review highlights findings concerning responses to bacterial stimuli, antimicrobial peptides, and the reciprocal influences between nematodes and their environmental bacteria. Furthermore, the microbiota of nematodes as well as alterations in the intestinal microbiota of mammalian hosts by helminth infections are discussed.
Parasitic nematodes are responsible for widespread morbidity in humans and animals. It is estimated that approximately 1.5 billion people are infected with one or more of these organisms (Hotez et al., 2008; World Health Organization, 2017) which also pose a considerable burden for animal production (Eijck and Borgsteede, 2005; Nganga et al., 2008). The extraordinary success of these parasites speaks to their ability to withstand a multitude of stresses such as host immune pressures and infectious challenges from microbes. Most parasitic nematodes inhabit the intestines of their hosts, co-existing with numerous microbial species. In studying these dynamics, researchers have focused extensively on the host-parasite relationship; more recently, the role of the microbiota as a major third party in the relationship is better appreciated due to diverse and far-reaching influences in health and disease (Donaldson et al., 2016). Much attention in these studies is given to host immune mechanisms while the interactions between nematodes and their microbial environments are largely overlooked. Due to many technical and biological challenges associated with studying parasites, these questions can be very difficult to address. The free-living nematode Caenorhabditis elegans (C. elegans) is frequently found in nature amongst a diverse microbiota on decaying organic matter (Frézal and Félix, 2015). C. elegans has been very well characterized and its interactions with bacteria studied in considerable detail. As such, these findings might be conveyed to parasitic nematodes and greatly inform our understanding of how parasites interact with the host-microbiota, as many immune-related pathways and responses may be conserved (Tarr, 2012; Rosso et al., 2013). This review will summarize current knowledge regarding how parasitic nematodes and their microbial environments may influence each other, supplemented with insights from work in C. elegans.
Studies on intestinal nematodes report a range of alterations to the composition of the microbiota, ranging from increased or decreased diversity, dysbiosis, or in some cases no identifiably significant changes (summarized in Tables 1, 2). Responses by C. elegans to numerous microbes have also been studied. Accordingly, parasitic helminths illuminate our understanding of how microbial communities can be altered by the presence of nematodes whereas studies of C. elegans can explain how microbial populations impact nematode physiology, reproduction and growth; considered together, one can better understand how these multifactorial biological systems operate. Herein, we focus most of our discussion on selected members of the genera Trichuris and Ascaris, as well as the rodent parasite Heligmosomoides polygyrus bakeri and the free-living C. elegans. These organisms have been selected due to their importance to human and animal health along with the abundance of available data in the literature, though additional studies from other helminths are considered where appropriate.
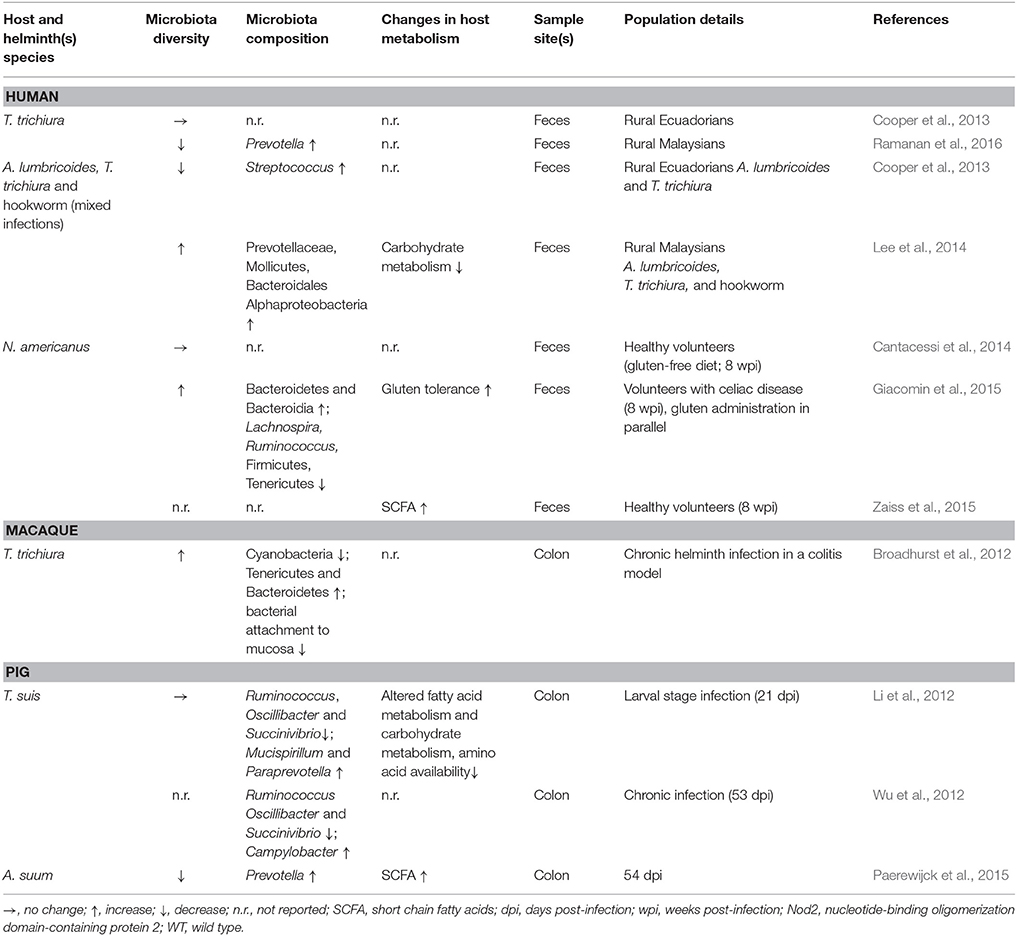
Table 1. Effects of helminth infection on host microbiota and metabolism in humans, macaques, and pigs.
The whipworms Trichuris trichiura, T. suis, and T. muris infect humans, pigs, and mice respectively. The life cycles of these species are comparable and infection begins with ingestion of developed eggs (Bethony et al., 2006; Klementowicz et al., 2012). Hatching occurs within hours of ingestion, liberating L1 larvae which invade the intestinal wall and undergo successive molts to the L4 stage by 3 weeks post-infection (pi) and finally develop into mature adults by 12 weeks pi (Bethony et al., 2006). Trichurids inhabit the most dense and diverse microbial environments of their hosts: the cecum and colon (Klementowicz et al., 2012). They can survive here for 1–2 years with individual females laying up to 5,000 eggs per day (Bethony et al., 2006).
The roundworms A. lumbricoides and the closely related A. suum infect humans and pigs respectively. As with Trichuriasis, Ascariasis also spreads via the fecal-oral route in humans as well as in pigs (Dold and Holland, 2011). Within 3 h, ingested eggs containing L3 larvae hatch and by 18 h pi the larvae begin their tissue migratory phase, passing through the liver after invading the cecum and proximal colon (Murrell et al., 1997). The larvae reach the lungs and pharynx by days six to eight pi (Roepstorff et al., 1997), are swallowed and can then be found in the small intestine as L4 stage larvae before further developing into mature adults. Adult Ascarids reside in the small intestine for around 1 year with individual females producing hundreds of thousands of eggs per day (Dold and Holland, 2011).
The trichostrongyloid H. p. bakeri is amongst the most common helminth parasites of rodents and a well characterized model for chronic intestinal nematode infection (Wu et al., 2012). Infection is initiated by ingestion of L3 larvae which migrate into the mucosa of the small intestine. By day three pi the larvae develop into L4 within the mucosa before returning to the lumen of the duodenum by day nine pi where they develop into, and remain as, egg-laying adults for approximately 12 weeks in the wood mouse (Apodemus sylvaticus) and more than twice as long in BALB/c laboratory mice (Robinson et al., 1989; Gregory et al., 1990; Behnke et al., 2009).
C. elegans is a free-living, bacterivorous nematode found predominantly in humid temperate areas (Félix and Duveau, 2012; Frézal and Félix, 2015). These nematodes can be identified in feeding and reproductive stages in rotting fruit and herbaceous stems. The life cycle is characterized by freshly hatched L1 stage larvae undergoing multiple molts before reaching adulthood in as little as 3 days. Under stressful conditions including crowding, limited food, and heat stress, L1 individuals will pursue an alternate life cycle through a pre-dauer L2 (L2d) stage, followed by a non-feeding, stress-resistant alternate L3 stage called “dauer.” Dauer individuals can survive for several months without food before reentering a relatively normal development cycle when conditions are more favorable. In the laboratory, the reference N2 strain is typically cultured on a diet of the rather innocuous Escherichia coli OP50. In contrast to controlled laboratory conditions, C. elegans shares its natural habitat with a variety of organisms, including bacteria, phages, fungi, isopods, arthropods, and other nematodes (Félix and Duveau, 2012). Additionally, worms can be found in states of starvation or constipation in their natural environments (Barrière and Félix, 2005).
The mammalian intestine is home to approximately 3.8 × 1013 microbes from all three domains of life, archaea, bacteria, and eukaryotes, collectively referred to as the microbiota (Sender et al., 2016). The different regions of the intestinal tract form divergent habitats, varying in bacterial type and density. In humans, an estimated 103–104 microbial cells/mL intestinal content reside in the small intestine (Sender et al., 2016). Considerably more bacterial diversity and density is found in the large intestine with as many as 1011 cells/mL contents representing thousands of different species (Zoetendal et al., 2008; Sender et al., 2016). Interestingly, microbial density and diversity of intestinal regions are inversely correlated with concentrations of host-derived antimicrobials; specialized secretory Paneth cells are a key source of α-defensins, lysozymes, and C-type lectins and are particularly prominent in the proximal intestine, the intestinal site of lowest microbial richness (Bevins and Salzman, 2011; Donaldson et al., 2016). Additionally, persistence of common gut commensals is partially mediated by resistance to host defense molecules (Cullen et al., 2015). Whether these factors influence helminth niche selection remains to be determined. Various conditions such as age, diet, health status and genetic background can impact the host microbiota composition. Maturation of the gut microbiota is characterized by increasing diversification with age while age-related changes are also due to exposure to a more varied diet (David et al., 2014; Odamaki et al., 2016). Analysis of human fecal samples can detect rapid, diet-mediated changes in microbial composition. For example, consumption of animal products increases the abundance of bile-tolerant species and decreases abundance of species that metabolize plant-derived polysaccharides (David et al., 2014). Dysbiosis, a potentially pathogenic imbalance of microbial communities, can be precipitated by exposure to pharmaceutical substances such as antimicrobials as well as the presence of infectious agents (Donaldson et al., 2016). Acute antibiotic treatment can drastically perturb the gut microbiota, decreasing species diversity (Dethlefsen et al., 2008). Alarmingly, while some changes appear reversible, other alterations can be detected in fecal samples even years after a short course of antibiotic treatment (Jernberg et al., 2007). A major clinical implication of a disturbed gut microbiota is elevated risk of enteric infection, such as with Clostridium difficile, which itself is associated with decreased microbial diversity (Milani et al., 2016). Many studies have now shown that intestinal nematodes influence their microbial niches as they establish themselves as part of their wider environment within the host; therefore, the role of helminths in dysbiosis is an area of active investigation (Figure 1).
The gut microbiota of humans, pigs, and mice is dominated by two of the 29 known bacterial phyla: Bacteroidetes and Firmicutes, with lower abundance phyla differing between hosts and including Actinobacteria, Deferribacteres, Proteobacteria, Spirochaetes, Tenericutes, and Verrucomicrobia (Leser et al., 2002; Eckburg et al., 2005; Consortium, 2012; Nguyen et al., 2015; Weldon et al., 2015). The diversity of the microbiota is thought to reflect the health of the intestine, with far reaching implications for the overall health of the host; greater species diversity contributes to healthy metabolic and immune functioning. Dysbiosis is associated with a reduction in intestinal biodiversity, predisposing to the outgrowth of particularly harmful bacterial species (Carding et al., 2015). The literature is mixed with respect to whether or not helminths cause dysbiosis, and some reports have indicated beneficial effects in therapeutic settings. The few studies assessing the influence of helminths on intestinal microbial communities are a mix of clinical observations and animal experiments, employing different analytical tools and acquiring microbiota samples from different sources. Hence, it is difficult to draw meaningful and generalizable conclusions from the available evidence. Still, through a careful reading of the literature, some common threads emerge.
Analysis of fecal samples from children in rural Ecuador revealed widespread helminth infection (Cooper et al., 2013). Children co-infected with both T. trichiura and A. lumbricoides appeared to have a decreased microbial diversity compared to uninfected children and children with T. trichiura single infections who did not differ from uninfected individuals. Interestingly, in a subset of children with mixed infections the authors also reported a higher abundance of Streptococcus spp., not usually dominant in healthy individuals. Taken together, these data suggest dysbiosis in the presence of A. lumbricoides, while T. trichiura single infections did not result in drastic alterations to the fecal microbiota. Another indication of dysbiosis associated with Ascaris infection comes from a study of A. suum-infected pigs which showed worm burden-dependent decreased bacterial diversity compared to control animals (Paerewijck et al., 2015). One study in humans found no difference in community structure of fecal samples in healthy volunteers infected with Necator americanus (Cantacessi et al., 2014). In contrast, a study of fecal samples from helminth-infected (T. trichiura, A. lumbricoides, hookworm) individuals in rural Malaysia found a positive association between helminth-colonization and microbiota diversity (Lee et al., 2014). Burrowed in the epithelia, T. trichiura likely interacts with the mucosal microbiota, the composition of which is known to differ considerably from fecal communities (Eckburg et al., 2005). Combined with the obstacles to sampling the small intestine, the site of A. lumbricoides and N. americanus infections, it is difficult to draw firm conclusions from these human studies regarding beneficial or harmful effects of helminth infections with respect to intestinal microbial composition. Though in the case of Ascaris, the local effects seen in the porcine small intestine (Paerewijck et al., 2015) correspond with the distal effects seen in human feces (Cooper et al., 2013).
Animal studies of helminthiases can offer more depth compared to human studies, accounting for the limitations associated with sampling only the fecal microbiota. During the larval stage, T. suis appears not to disrupt the porcine colonic microbiota (Li et al., 2012); however, chronic infections in pigs demonstrate worm burden-dependent disruption (Wu et al., 2012). T. suis also appears to promote Campylobacter infection in pigs, intensifying colitis disease severity (Mansfield and Urban, 1996; Mansfield et al., 2003; Wu et al., 2012). Chronic T. muris infections in mice considerably decrease overall microbial diversity, an effect that appears reversible upon worm clearance (Holm et al., 2015; Houlden et al., 2015). In these studies, the murine microbiota also showed a shift away from Bacteroidetes in favor of Firmicutes (Holm et al., 2015; Houlden et al., 2015). A study of wild mice has also observed an increased Firmicutes/Bacteroidetes ratio in helminth-infected individuals (Kreisinger et al., 2015) whereas mice experimentally infected with the small intestinal nematode Nippostrongylus brasiliensis showed a decrease of Firmicutes while increasing Bacteroidetes (Fricke et al., 2015). Though N. brasiliensis decreases the Firmicutes/Bacteroidetes ratio, it also promotes the reduction of segmented filamentous bacteria (SFB) which are thought to prevent colonization by bacterial pathogens, as it was shown in SFB-colonized mice with enhanced resistance to the pathogenic Citrobacter rodentium (Ivanov et al., 2009; Fricke et al., 2015). Mice with chronic T. muris infections (Holm et al., 2015; Houlden et al., 2015) as well as those with acute H. p. bakeri infections (Rausch et al., 2013) have higher abundance of Enterobacteriaceae, a family shown to overgrow during intestinal inflammation (Lupp et al., 2007) and strongly correlated with Crohn's disease (Gevers et al., 2014) and C. difficile infection (Milani et al., 2016) in humans. These data indicate a propensity for helminths to associate with a simplified microbiota; however, whether these observations are generalizable across helminth infections in diverse mammalian hosts remains to be determined; nonetheless, these animal experiments are suggestive of helminths promoting dysbiosis in their hosts.
Despite the evidence for dysbiosis, a fairly consistent finding across various helminth infections is an increased abundance of Lactobacillaceae (Walk et al., 2010; Rausch et al., 2013; Reynolds et al., 2014; Fricke et al., 2015; Holm et al., 2015; Houlden et al., 2015; Ramanan et al., 2016), a family composed primarily of Lactobacillus spp., currently under intense investigation for use as probiotics (Walter, 2008; Salvetti et al., 2012). Additionally, studies employing intestinal nematodes as therapeutic interventions to treat intestinal inflammatory conditions have shown beneficial outcomes (Broadhurst et al., 2012; Giacomin et al., 2015; Ramanan et al., 2016). Macaques with idiopathic chronic diarrhea show signs characteristic of dysbiosis such as decreased diversity, increased bacterial attachment to the intestinal mucosa, and increased abundance of Enterobacteriaceae (Broadhurst et al., 2012). Bacterial attachment and abundance of Enterobacteriaceae were effectively decreased by T. trichiura infection, while bacterial diversity was restored, indicating a protective effect of helminth infection in this setting. Similarly, mice deficient in Nod2 are susceptible to Crohn's disease and can be colonized by inflammatory Bacteroides spp., while T. muris infection protects against pathogenic colonization (Ramanan et al., 2016). In patients with celiac disease, low-level infection with N. americanus increases gluten tolerance (Croese et al., 2015), while also increasing microbial richness when combined with gluten consumption (Giacomin et al., 2015). Taken together, these findings indicate that intestinal nematodes possess great therapeutic potential and may promote microbial restoration in conditions of pre-existing dysbiosis.
From taxonomic data referring to the microbiota composition alone it is challenging to conclude whether intestinal nematodes contribute to or ameliorate dysbiosis. As such, the influence of helminths on gut metabolic profiles may offer more clues. Methodological and technological advances allow for metabolic profiling of gut microbiota, along with identification and quantification of metabolites of interest (Vernocchi et al., 2016). Such experiments can be quite informative, though they are currently far less abundant than those reporting taxonomic changes. Metabolomic analysis of the colonic microbiota of pigs infected with T. suis demonstrated reduced capacity to metabolize and utilize carbohydrates relative to control animals, an effect seen in acute and chronic infections (Li et al., 2012; Wu et al., 2012). Interestingly, T. suis-infected pigs also showed signs of altered fatty acid metabolism, including higher levels of oleic acid relative to control pigs (Li et al., 2012). Notably, oleic acid possesses antibacterial activity (Dilika et al., 2000) and may therefore influence microbial composition during Trichuris infection. Mice chronically infected with T. muris also showed signs of reduced carbohydrate and amino acid metabolism and nutrient uptake compared to naïve animals, features which likely contribute to decreased weight gain during helminth infection (Houlden et al., 2015). Observational analysis of helminth-infected humans has attributed decreased carbohydrate metabolism to Ascaris infection (Lee et al., 2014). In contrast, wild mice infected with helminths showed mixed effects with respect to metabolomic shifts (Kreisinger et al., 2015). Notably, wild mice with H. p. bakeri appeared to have an increased capacity to metabolize carbohydrates; however, metabolites were not measured and mouse weights were not reported. Together, these observations suggest helminth-mediated microbiota changes tend to reduce metabolic capability in the gut, ultimately manifesting as nutritional deficiencies.
While dietary carbohydrate utilization may be impaired by intestinal nematodes, other experiments demonstrate potential benefits of helminth infection such as increased abundance of intestinal short-chain fatty acids (SCFAs), typically produced from microbial processing of indigestible oligosaccharides (Paerewijck et al., 2015; Zaiss et al., 2015; Vernocchi et al., 2016). SCFAs can serve as energy sources and anti-inflammatory compounds and are therefore under investigation for their therapeutic potential (Vernocchi et al., 2016). Increased SCFAs have been detected during infection with H. p. bakeri in mice, A. suum in pigs, and N. americanus in humans (Paerewijck et al., 2015; Zaiss et al., 2015). Furthermore, H. p. bakeri and A. suum can produce SCFAs directly (Tielens et al., 2010; Zaiss et al., 2015). Reduced weight gain associated with helminth infection may also prove to be therapeutic in different circumstances, as illustrated by Yang et al. in a study of obese mice and mice fed obesity-inducing diets (Yang et al., 2013). In agreement with other observations, N. brasiliensis infection attenuated weight gain. Importantly, helminth infection decreased adiposity and hepatic lipid storage while improving glycemic control, implying a reversal of metabolic disease. These studies indicate that microbiota alterations and metabolomic changes associated with intestinal nematodes may offer certain benefits.
The mixed observations described herein highlight the difficulties of generalizing findings from these studies, simultaneously hinting at the massive potential to apply these insights in diverse therapeutic settings. Animal studies designed to induce strong immune responses are typically a result of a high infection dose and thus high worm burdens and may not reflect average worm burdens seen in nature. Indeed, some of the aforementioned animal experiments demonstrate microbiota changes suggestive of dysbiosis (Li et al., 2012; Wu et al., 2012; Holm et al., 2015; Houlden et al., 2015). In contrast, observations from natural settings or studies inducing low-level helminth infections suggest less dramatic changes (Cantacessi et al., 2014; Kreisinger et al., 2015). Additionally, when intestinal nematodes are introduced into individuals with intestinal inflammation or metabolic abnormalities, they appear beneficial (Broadhurst et al., 2012; Yang et al., 2013; Giacomin et al., 2015). Taken together, these data emphasize co-evolution of helminths, gut microbiota, and mammalian hosts, such that helminths can be considered less strictly as intruders but rather members of the gut macrobiota (Gause and Maizels, 2016). In order to establish themselves in the mammalian intestine, rather than being solely beneficial or causing outright harm, nematodes must shape the niche such that a new ecological balance is found. This task is aided by some microbes and challenged by others, and the insights gained from understanding these dynamics can provide opportunities to better understand and perhaps manipulate incredibly complex biological systems. It should be noted that many potential mechanisms have been identified which act in concert to shape the intestinal microbiota. Immune mechanisms such as modifying the balance between Th1/Th17 vs. Th2 responses as well as the induction of regulatory T cells have been characterized and discussed elsewhere (Reynolds et al., 2015; Gause and Maizels, 2016; Zaiss and Harris, 2016). Furthermore, the bulk of work done thus far has focused primarily on bacteria and far less is known about the archaeal and fungal components of intestinal microbial communities. In addition to metabolomic factors touched upon here, important considerations which are relatively under-studied and not frequently discussed are the direct signals between helminths and microbes. These reciprocal interactions are considered below.
The study of intestinal parasites is particularly amenable to understanding alterations to microbial communities of the gut, though the microbial influence on helminths is relatively difficult to assess in these systems. As a model organism with many available genetic tools, C. elegans can bridge some of the knowledge gaps encountered when studying parasitic nematodes. When recovered from compost or isopods, which worms may use as vectors to shuttle between food sources, C. elegans is frequently found in the dauer stage (Félix and Duveau, 2012). In contrast, proliferating populations of nematodes can be found in microbe-dense environments, particularly rotting fruits or vegetation. A study by Samuel et al. characterized the natural bacterial microenvironment of C. elegans (Samuel et al., 2016). The most prevalent bacterial phyla in decaying apples were Proteobacteria, Bacteroidetes, Firmicutes, and Actinobacteria. The Enterobacteriaceae family within Proteobacteria encompassed the most abundant genera detected, while Lactococcus, Lactobacillus, Acetobacter, Gluconobacter, and Gluconoacetobacter were also quite prevalent. In some samples, Escherichia spp. were also identified, though they were not particularly abundant. Considering the immensity of bacterial diversity coming from 29 extant phyla (Yarza et al., 2014), there is considerable overlap in the microbial environments of parasitic and free-living nematodes. Experiments assessing the effect of C. elegans on an experimental microbiota could be designed, whereby the nematodes are introduced into a defined microbial environment and followed over time to assess changes to bacterial constituents. Such studies would be analogous, albeit profoundly simplified, to studies assessing gut microbial changes in helminth infections and could shed light on the underlying mechanisms contributing to compositional alterations. Akin to some reports of parasitic nematodes, observations from rotting fruit suggest that growth of C. elegans is enhanced amongst a simpler microbiota (Samuel et al., 2016).
Many studies have demonstrated the susceptibility of the C. elegans intestine to a multitude of pathogens (Couillault and Ewbank, 2002), including Salmonella enterica serovar Typhimurium (S. typhimurium) (Lee et al., 2015), Enterococcus faecalis (Sim and Hibberd, 2016), Burkholderia spp. (Lee et al., 2013), Staphylococcus aureus (JebaMercy and Balamurugan, 2012), Proteus mirabilis (JebaMercy and Balamurugan, 2012), and Pseudomonas aeruginosa (Dai et al., 2015). Beyond posing a threat to the nematode intestine, bacteria may also damage and infect the nematode surface. In experimental settings, Yersinia spp. can form biofilms that block the oral cavity of the worms, resulting in starvation and death (Tan and Darby, 2004). Wild nematodes are subject to cuticular infection by Microbacterium nematophilum (O'Rourke et al., 2006) and the fungal pathogen Drechmeria coniospora (Engelmann et al., 2011). Additionally, Elizabethkingia are capable of digesting the nematode cuticle components keratin and collagen (Riffel et al., 2003; Félix and Duveau, 2012). C. elegans is also infected by intracellular pathogens such as the microsporidian parasite Nematocida parisii (Troemel et al., 2008) and the Orsay virus (Félix et al., 2011). Though studies have demonstrated the ability of bacteria to colonize parasitic nematodes, no information is available concerning which microbes might be beneficial or harmful for them. In contrast, numerous studies in C. elegans have shown beneficial and harmful effects of specific bacterial strains. Alphaproteobacteria and Lactococcus spp. tend to be beneficial and promote C. elegans proliferation while Bacteroidetes and Gammaproteobacteria seem to impair nematode physiology and induce stress/immune responses (Samuel et al., 2016). It stands to reason that similar interactions exist between bacteria and parasitic nematodes and that the composition of the host microbiota would then influence the health of parasitic nematodes in the gut. Interestingly, nematode fecundity is subject to various stresses; the host immune response can decrease helminth fecundity, exemplified by reduced egg production by H. p. bakeri during a type 2 helper T cell-mediated immune response (Strandmark et al., 2016). As bacteria influence C. elegans fecundity (Szewczyk et al., 2006), it is possible that communities within the microbiota may also influence the reproductive capacity of parasites. Larger populations of proliferating C. elegans are found in more highly degraded apples containing larger microbial populations, thereby providing more nutritional sources for the nematodes. Also, apples with higher populations of worms had microbiota which were more similar to each other than apples with lower/no worms. Apples containing proliferating populations of C. elegans also had significantly fewer bacterial species and diversity than apples with non-proliferating populations. Most importantly, proliferation was associated with Alphaproteobacteria and the absence of potential pathogens from Gammaproteobacteria and Bacteroidetes. Perhaps there is a general preference by nematodes for decreased abundance of several Bacteroidetes organisms, given the observations in parasite-host microbiota studies discussed previously. As Lactobacillaceae are more predictive of successful H. p. bakeri establishment in mice (Reynolds et al., 2014), Enterobacteriaceae and Acetobacteraceae are predictive for larger proliferating C. elegans populations in rotting apples, while Gammaproteobacteria show an inverse relationship (Samuel et al., 2016). It is not fully understood how intestinal nematodes establish in specific habitats such as the cecum and colon in the case of Trichurids, or the small intestine in the case of Ascarids and H. p. bakeri; however, some speculate the choice is based on the abundance of nutrients or the relative lack of harmful stimuli (Davey, 1964; Bansemir and Sukhdeo, 1994, 2001). While determining the nature of habitat selection in these settings may prove experimentally elusive, the available data suggest that parasitic nematodes, as with their free-living counterparts such as C. elegans, have advantageous associations with some microbial species and detrimental interactions with others.
All animals, including nematodes, live in association with a multitude of other species. The host-microbe relationship has gained a lot of attention in recent studies, with various reciprocal interactions ranging from parasitism, to commensalism and mutualism, seen as potent forces guiding the co-evolution of species. In addition to influencing the rich and diverse microbial environments that they inhabit, nematodes inevitably also harbor their own microbiota that is likely essential for normal development and physiology, as is the case for other organisms. Though the data are limited, studies have documented the ability of bacteria to colonize Ascaris, H. p. bakeri, as well as C. elegans.
Earlier studies demonstrated nematode-associated bacterial communities in A. suum (Hsu et al., 1986; Shahkolahi and Donahue, 1993). One study aimed at identifying the source of serotonin present in nematodes, which generally lack the enzymes necessary for de novo synthesis of serotonin from tryptophan, revealed 4 × 109 bacteria per gram of nematode intestine (Hsu et al., 1986). In addition to E. coli, Enterobacter, Klebsiella, Acinetobacter, Citrobacter, Pseudomonas, Aeromonas, and Shigella were identified among gram-negatives while Staphylococcus, Streptococcus, Corynebacterium, and Bacillus were identified amongst gram-positive organisms. The species identified suggest that the nematode's intestinal microbiota may have been derived from that of the host. Another study demonstrated that antibiotic treatment of ex vivo cultured A. suum reduced, but did not eliminate, the bacterial load carried by the worms and determined that the posterior portion of the intestine harbored the highest number of culturable bacteria (Shahkolahi and Donahue, 1993). Studies that employ more modern methods while sampling site-specific host- and parasite-associated microbial communities could greatly enhance our understanding of the selectivity and development of an Ascaris-specific microbiota.
Another study showed that A. lumbricoides nematodes isolated from cholera patients were colonized by Vibrio cholerae (Nalin and McLaughlin, 1976). Worms were retrieved from patients after being spontaneously passed or after de-worming and a majority of worms were colonized by V. cholerae of the same serotypes found in stool samples of cholera patients. Serial sectioning of the worms revealed colonization along the entire length of the intestine, from oral cavity to anus. The bacteria were recovered and viable even after worms were in culture in saline for 6 days. The authors reported retrieval of one hookworm which also revealed two colonies of bacteria which were not V. cholerae. The authors reasoned that pathogenic bacteria may survive inside parasites when parasites are passed in the stool, as happens commonly during a cholera infection, thereby contributing to environmental spread of microbial pathogens. Further, the authors discussed the possibility of pathogenic microbes reaching the reproductive tract of female nematodes during copulation and adhering to egg shells which may also serve as a vehicle for transmission. There was no indication in this study that V. cholerae are pathogenic to Ascarids and further study in this area could reveal interesting associations between different enteric infections. Taken together, these findings highlight the clinical relevance of understanding interactions between intestinal nematodes and environmental microbes.
Walk et al. provided evidence for a H. p. bakeri-associated microbiota when they sampled L3 and adult nematodes (Walk et al., 2010), finding only 28 16S sequences obtained from larvae, concluding that they are associated with few bacteria; however, the L3-associated microbiota was completely unique and consisted of 6 bacterial families, unlike the adult-associated microbiota which was very similar to the ileum of the murine host. As H. p. bakeri hatch outside of the host, they likely enter their hosts with a distinct microbiota which changes over time in the new environment. This L3-specific microbiota may also serve as a source of non-indigenous microbes for the mammalian host.
A special microbe-helminth relationship is also well documented for filarial nematodes in which the gram-negative intracellular bacterium Wolbachia is obligatory for normal larval growth and development, embryogenesis, and survival of adult worms (Taylor et al., 2005). Such endosymbiotic relationships probably emerged from ancestral infections of the host nematode by free-living bacteria, concomitant with gene losses and genome rearrangements on both sides during coevolution (Masson et al., 2016). So far, such an endosymbiotic relationship has not been detected in other parasitic nematode species, but parallels to other microbe-nematode partnerships may be drawn. For example, Ascaris requires and absorbs Vitamin B12 from microbial sources (Zam and Martin, 1969). Wolbachia displays cell tropism and is restricted to somatic tissues in adult male worms, whereas females also harbor bacteria in the germline (Kozek, 1977; Taylor et al., 1999). The level of infection varies substantially during filarial development, where shortly after transmission to the mammalian host a dramatic increase in the bacterial population occurs (McGarry et al., 2004). As bacterial loads within individual worms differ, Taylor et al. hypothesized that higher levels of Wolbachia infection within a worm may potentially confer selective advantages in terms of filarial development or fecundity (Taylor et al., 2013). Transcriptomic analysis of the Wolbachia genome of Onchocerca ochengi indicated that Wolbachia may have a mitochondrion-like function in the soma, generating ATP for the nematode host (Darby et al., 2012). Hence, recent trials aim at utilizing antibiotics such as tetracycline (Hoerauf et al., 1999), and other chemotherapeutics targeting Wolbachia as a novel tool for the treatment of filarial infection and disease, reviewed in Taylor et al. (2014).
C. elegans nematodes isolated from the wild are observed to harbor live bacteria in their intestine which may exist as commensals or which may proliferate and cause obstruction and pathology of the worm gut (Félix and Duveau, 2012). Worms isolated from the wild also carry a distinct and diverse microbiota when compared to other nematode species in the same environment (Dirksen et al., 2016). Dirksen et al. reported a microbiota rich in unclassified Proteobacteria from the family Enterobacteriaceae, as well as members of the genera Pseudomonas, Stenotrophomonas, Ochrobactrum, and Sphingomonas. When isolated worms were enriched on plates of E. coli OP50, their microbiota maintained considerable similarity to freshly isolated samples despite 3 weeks in culture on E. coli plates, suggestive of a C. elegans-specific microbiota and closely developed microbial-host community. This study did not mention any changes in overall microbial population sizes over time. In the same study, a subset of 14 bacterial isolates were chosen in order to cultivate nematodes on an experimental microbiota with bacterial frequencies mirroring the nematode-associated microbiota of worms isolated from the wild. Three genotypes of C. elegans, including two natural isolates (MY316 and MY379) and the laboratory N2 strain, were cultured from hatched sterilized eggs through to adulthood. The investigators found significant influences of genotype and life stage on the microbial composition of the nematodes. Interestingly, certain bacteria including Ochrobactrum MYb71 and Stenotrophomonas MYb57 were enriched in the nematode samples relative to the agar plates. The effect was quite pronounced in the case of Ochrobactrum which was present in only trace amounts in the agar plates but represented as much as 20% of the nematode-associated bacterial community. Ochrobactrum was also able to persist in the intestine even under starvation conditions without being used as a food source or eliminated during the ensuing stress response, which can include upregulation of antimicrobial effectors such as lysozymes (Uno et al., 2013). This bacterial strain may be a prominent symbiont for C. elegans as it seems to use the nematode as an environmental niche in the absence of apparent fitness costs to the worm. The experimental microbiota was also shown to enhance growth and nematode population size relative to worms cultured on E. coli. While detailed analysis of this sort is absent for parasitic nematodes, the ability of microbes to colonize these worms supports the idea of worm-associated microbial communities, providing benefits similar to those seen in C. elegans and other organisms, such as supplying nutrients and providing protection from pathogens (Cabreiro and Gems, 2013; Watson et al., 2014; Lee et al., 2015; Dirksen et al., 2016).
Living in intimate association with microbes, nematodes are subject to diverse microbial influences. Beneficial effects of microbes on nematodes may include nutrition, promotion of longevity, protection from infection by other microbes, and contributions to a hospitable environment (Cabreiro and Gems, 2013; MacNeil and Walhout, 2013). From the perspective of mammalian hosts, the question arises whether a specific microbial environment might significantly influence susceptibility to helminth infection. Further, microbes might also be a source of competition, stress, and disease for nematodes.
The microbial environment can profoundly impact establishment and propagation of parasitic nematode infections by influencing egg hatching and reproductive success. T. muris eggs may require the presence of selected bacterial species to induce hatching, as demonstrated using murine cecal explants as well as E. coli, S. typhimurium, S. aureus, and P. aeruginosa (Hayes et al., 2010). Live bacteria appear to be necessary for these effects as heat inactivation prevented hatching, while bacteriostatic gentamycin treatment did not. It was proposed that physical contact between the bacteria and eggs is required as Type 1 fimbriae facilitate E. coli-induced hatching, though additional mechanisms likely exist. The authors also reported that mice pre-treated with antibiotics had significantly lower worm burdens at day 21 pi compared to untreated animals, indicating a significant role for bacterial communities in parasitic nematode establishment. Interestingly, bacteria-induced T. muris egg hatching seems to occur efficiently with members of Proteobacteria (E. coli) and Firmicutes (Enterococcus caccae, Streptococcus hyointestinalis, Lactobacillus reuteri, and Lactobacillus amylovorus). It is possible that many different bacterial species contribute via different mechanisms for optimal T. muris egg hatching (Vejzagić et al., 2015).
Lactobacillaceae abundance in the duodenum positively correlates with susceptibility to H. p. bakeri infection (Reynolds et al., 2014). Low-level vancomycin treatment prior to nematode infection did not significantly reduce total bacteria but elevated abundance of Lactobacillaceae and Enterobacteriaceae while reducing Eubacterium/Clostridium species in the fecal microbiota. This was associated with H. p. bakeri persistence in the host. H. p. bakeri infection also elevated duodenal Lactobacillaceae and Enterobacteriaceae. Administration of Lactobacillus taiwanensis enhanced susceptibility to H. p. bakeri infection and worm fecundity, thought to be due to the induction of immunosuppressive regulatory T cells. This study demonstrated a reciprocal interaction whereby Lactobacillus spp. promote nematode establishment which then promote growth of Lactobacilli. Similar to T. muris infections discussed above, studies in germfree mice revealed higher H. p. bakeri worm burdens in conventionally raised mice, implicating the host microbiota as a key part of the parasite's environmental niche (Wescott, 1968). Bacterial populations also influence the host's immune status which can have a profound influence on the intestinal environment for parasite establishment, though specific immune variables and bacterial species are still being identified (Cattadori et al., 2016). These data suggest not only that the microbiota is essential for parasite development, but also that particular bacteria facilitate helminth establishment.
While C. elegans and other free-living nematodes are known to subsist on microbes, the food sources of intestinal nematodes are less well established. The digested remains of yeasts can be found in the intestines of C. elegans isolated from the wild, demonstrating the capability of these nematodes to use eukaryotic cells as a food source in addition to bacteria (Félix and Duveau, 2012). There is corresponding evidence suggestive of consumption of intestinal epithelia by A. suum in pigs as well as H. p. bakeri in mice (Davey, 1964; Bansemir and Sukhdeo, 1994). Freshly isolated Ascaris nematodes were found to accumulate eukaryotic cellular material in their buccal cavities, thought to be of host origin (Davey, 1964). Labeling studies sought to determine whether H. p. bakeri accumulates ingesta, host blood, or host tissue material and found an accumulation of host tissue components rather than blood or ingesta (Bansemir and Sukhdeo, 1994). These studies did not assess the intestinal contents of the worms and these data do not preclude the ability of intestinal nematodes to digest bacterial cells, especially considering bacteria are counted amongst the intestinal contents of A. suum and A. lumbricoides as discussed previously. Further insights using modern methods could be provided by studies designed to specifically assess uptake and digestion of microbes by intestinal nematodes.
Bacteria can promote or hinder nematode proliferation by various means. From serving as direct food sources or by producing essential nutrients, microbes are required for nematode growth. Furthermore, specific species are well documented to promote helminth establishment while themselves being reinforced by nematodes, as discussed. One could imagine that intentional manipulation of microbial variables could influence parasite establishment and might possibly impact how parasitic diseases are treated in the future. In the case of C. elegans, characterizing its interactions with microbes beyond E. coli OP50 serves to improve this powerful model and enhances its utility to investigators with diverse objectives. Together, these diverse systems complement each other in elucidating the varied interactions between nematodes and microbes, thereby setting a stable foundation upon which future research can build.
Nematodes are confronted with a tremendous and highly diverse microbial environment, presenting many infectious challenges. Alterations in the intestinal microbiota during helminth infection, coupled with studies demonstrating distinctly beneficial or harmful outcomes to C. elegans physiology in response to various bacteria, illustrate the ability of nematodes to sense their microbial environments and possibly discriminate between microbial species. Parasitic nematodes must have evolved particular strategies to overcome colonization by microbes, likely employing selected antibacterial defense mechanisms to coexist with the host microbiota. C. elegans has become an important model organism for the study of innate immune defense against pathogenic bacteria (Schulenburg et al., 2004; Irazoqui et al., 2010). The C. elegans immune system is seemingly of ancient origin, and there exist homologs of nearly all of its components in other organisms, including humans (Schulenburg et al., 2008). Consequently, sensing of the microbial environment might follow conserved or comparable mechanisms in parasitic nematodes as well.
C. elegans uses different protective mechanisms when confronted with potential pathogens, such as avoidance behavior (Meisel and Kim, 2014) and activation of stress and immune responses (Samuel et al., 2016) leading to the production and release of defense molecules with antimicrobial activities (Mallo et al., 2002). Identification of microbe-associated molecular patterns is followed by signal transduction via several signaling cascades, culminating in transcriptional alterations, reviewed in Rosso et al. (2013) and Kim and Ewbank (2015). While the pathogen-recognition receptors responsible for initiating immune responses are not entirely known, candidates include F-box proteins, lectins, follicle stimulating hormone receptor homolog-1, scavenger receptors, and the Toll-like receptor TOL-1 (Kim and Ewbank, 2015). Stress and immune responses illustrate an overlap between microbial and abiotic stresses; the main responses involved include: autophagy, insulin-like receptor (DAF-2), mitogen-activated protein kinases (MAPK), transforming growth factor-β-like (DBL-1), programmed cell death pathways, and unfolded protein responses (UPR) (Kim and Ewbank, 2015). Perhaps surprisingly, C. elegans lacks a homolog of NF-κB, an essential transcription factor in the innate immune response of many invertebrate and vertebrate species (Vallabhapurapu and Karin, 2009). Other proteins regulating transcription of infection and stress-modulated genes have been identified, including cyclic AMP-dependent transcription factor-7 (ATF-7), forkhead box O (FOXO) ortholog DAF-16, GATA transcription factors (ELT-2, ELT-3), helix loop helix-30 (HLH-30), heat-shock factor-1 (HSF-1), NF-E2-related factor SKN-1, signal transducer and activator of transcription STA-2, X-box binding protein-1 (XBP-1), and basic leucine zipper domain transcription factor ZIP-2 (Kim and Ewbank, 2015). As indicated, the intricacies of these responses have been reviewed elsewhere; as such, only examples of particular interest are highlighted here.
Infectious risks could be mitigated and resources preserved if pathogens were simply not encountered. Avoidance behavior is facilitated by the sole C. elegans toll-like receptor, TOL-1 (Pujol et al., 2001), and is characterized by the detection of specific microbial products, such as serrawettin W2 produced by Serratia marcescens, resulting in worms migrating away from the offending agents (Pradel et al., 2007). Unlike other organisms, C. elegans may not rely heavily on toll-like signaling for immune defense, though reports on this are conflicting (Pujol et al., 2001; Couillault et al., 2004; Tenor and Aballay, 2008). Other mechanisms such as aerotaxis also contribute to pathogen avoidance, as demonstrated by C. elegans avoidance of P. aeruginosa (Reddy et al., 2009). Lectins, carbohydrate-binding proteins implicated in multiple facets of immunity in a diverse range of species, may sense or neutralize microbes (Zelensky and Gready, 2005). Finally, there is evidence for an important role for scavenger receptors such as cell death abnormal-1 (CED-1) and scavenger (SCAV) proteins in pathogen recognition and resistance (Nicholas and Hodgkin, 2004; Means et al., 2009). Comparable studies of differential activation of recognition receptors in parasites by microbial communities from different intestinal regions might provide insights into how parasitic nematodes recognize threats and whether or not activation of such pathways impacts their niche selection.
In addition to sensing microbes and their toxins, organisms induce immunity-related genes due to disruptions of core physiologic processes by pathogens. C. elegans employs this surveillance immunity in response to inhibited translation, cellular damage, and mitochondrial stress, all of which can result from infection and exposure to microbial toxins (Pukkila-Worley, 2016). Transcriptional responses to these stresses include activation of immune signaling and xenobiotic metabolic pathways (Pukkila-Worley, 2016).
Toxins produced by several microbes, including diphtheria toxin from Corynebacterium diphtheria (Collier, 2001), cholix toxin from Vibrio cholerae (Jørgensen et al., 2008), and exotoxin A (ToxA) from P. aeruginosa (Dunbar et al., 2012), interfere with protein translation. When non-pathogenic E. coli engineered to express ToxA are fed to C. elegans, immune pathways required for resistance to the toxin, especially MAPK pathways, are upregulated by the worms (McEwan et al., 2012). Also, the aminoglycoside antibiotic hygromycin B blocks elongation of the amino acid chain during protein translation like ToxA (McEwan et al., 2012). Taken together, C. elegans likely responds to translation inhibition by ToxA rather than to the protein itself. An RNAi screen showed that disrupting core processes, including translation, activatesbasic leucine zipper domain transcription factor ZIP-2-dependent immune signaling, similar to responses seen during P. aeruginosa infection of C. elegans and following exposure to ToxA (Dunbar et al., 2012). More recently, another transcription factor called CCAAT-enhancer-binding-protein-2 (CEBP-2) was identified to act in concert with ZIP-2 to promote defense by C. elegans against P. aeruginosa, ToxA, and in response to inhibition of translation and other core processes (Reddy et al., 2016).
In addition to surveillance of disrupted physiologic processes, C. elegans also induces immune responses after injury of its epidermis by microbial infection and sterile wounding (Pujol et al., 2008a). During infection with D. coniospora, a fungal pathogen which damages the nematode cuticle, 4-hydoxyphenyllactic acid (HPLA) acts as a damage-associated molecular pattern recognized by the G protein-coupled receptor DCAR-1 which is required for AMP expression during fungal infection (Zugasti et al., 2014). Another study showed that epidermal damage liberates the STAT-like transcription factor STA-2, triggering AMP production (Zhang et al., 2015). These findings exemplify the ability of epithelial barriers to surveil physical damage and respond by activating innate immune pathways.
Microbial infection can often lead to mitochondrial stress; a survey of C. elegans responses to bacteria isolated from the nematode's natural environment found that 101 strains of the >550 isolates tested induced the mitochondrial stress reporter promoter hsp-6::GFP (Samuel et al., 2016). Interestingly, mitochondrial disruption also induces drug-detoxification enzymes belonging to the cytochrome P450 superfamily (CYPs) and enzymes involved in glucuronidation (Liu et al., 2014) as well as infection response gene-1 (irg-1) which is also induced during P. aerugionsa infection in a ZIP-2-dependent manner (Estes et al., 2010; Dunbar et al., 2012). Another toxin of P. aeruginosa, pyoverdin, disrupts the mitochondria of C. elegans. The nematodes respond by a specialized form of autophagy called mitophagy, whereby damaged mitochondria are degraded to resist P. aeruginosa-mediated killing (Kirienko et al., 2015).
By sensing various forms of cellular stress and damage, C. elegans is able to launch an integrated response which promotes survival and longevity (Pukkila-Worley, 2016). These responses can be induced by microbes and xenobiotics, activating immune and detoxification pathways (Melo and Ruvkun, 2012; Pukkila-Worley et al., 2014; Pukkila-Worley, 2016). As helminths may employ similar strategies to deal with microbial and xenobiotic stresses, the overlap between immune and drug detoxification responses is of particular interest. Various contributing factors have been identified for the development of anthelmintic resistance, including resistance alleles in detoxification genes, parasites not exposed to treatments, and underdosing (Vercruysse et al., 2011). The concept of surveillance immunity raises the notion that elements within the host microbiota prime helminths to resist anthelmintic treatments. Whether certain microbial species of the host intestine can promote drug resistance by activating xenobiotic detoxification within intestinal nematodes has not yet been studied.
As illustrated by many studies, MAPK pathways have a fundamental function in C. elegans stress and immune responses, including the p38 PMK-1 and extracellular signal-regulated kinase (ERK) pathways. Mutations at multiple levels of the NSY-1-SEK-1-PMK-1 pathway increase susceptibility to P. aeruginosa without impacting growth on E. coli OP50 (Kim et al., 2002). PMK-1 is activated by the toll-interleukin-1 receptor domain adaptor protein TIR-1 which is required for resisting killing by numerous microbial pathogens (Couillault et al., 2004). Phosphorylation of the ATF-7 transcription factor by PMK-1 activates transcription of lectins and candidate antimicrobial peptides (AMPs), conferring intestinal resistance to various species, including P. aeruginosa, S. marcescens, and E. faecalis (Shivers et al., 2010). Small molecule-mediated stimulation of the PMK-1 pathway and subsequent activation of ATF-7 can also increase lifespan in the presence of E. faecalis (Pukkila-Worley et al., 2012). Growth of C. elegans on soil-derived non-pathogenic bacteria, Bacillus megaterium and Pseudomonas mendocina also enhanced resistance to P. aeruginosa in a PMK-1 dependent fashion (Montalvo-Katz et al., 2013). Additionally, the PMK-1 pathway is active in epidermal infections as shown by induced expression of AMPs in response to D. coniospora as well in response to epidermal injury (Pujol et al., 2008a,b). Another MAPK pathway involving ERK signaling has been shown to be protective against rectal infection by M. nematophilum, (Gravato-Nobre et al., 2005). Based on these observations, one could speculate that MAPK pathways are also essential for helminth survival amongst numerous microbes.
Insulin-like signaling via DAF-2 highlights the intersection between immunity and metabolism. DAF-2 signaling inhibits DAF-16 activation by preventing its localization to the nucleus. Loss of function mutations of daf-2 confer pathogen resistance and longevity by allowing activation of the DAF-16 transcription factor (Kenyon et al., 1993; Garsin et al., 2003). DAF-16 activation also increases resistance to non-microbial insults, such as heat stress and oxidative stress (Barsyte et al., 2001). Intriguingly, C. elegans can be primed to withstand pathogens and heat stress when exposed to pathogens during development, increasing the worm's lifespan (Leroy et al., 2012). DAF-16 activation appears to increase antimicrobial gene transcription as well as stress and detoxification genes, placing this transcription factor at the nexus of stress and immune signaling (McElwee et al., 2003). Autophagy, a lysosomal degradation pathway, is also implicated in longevity and its inhibition increases intracellular replication of S. typhimurium in C. elegans intestinal epithelia (Jia et al., 2009).
As intestinal nematodes are typically much larger than C. elegans, with female Ascaris worms growing up to 35 cm in length (Dold and Holland, 2011), the surface area available for attachment and infection by microbes is extensive. Furthermore, these nematodes are much longer-lived than C. elegans. Stress responses, especially autophagy, may be of particular importance in these organisms, though this area remains virtually unexplored. An intriguing study by Voronin et al. identified autophagy as a bactericidal mode of action in filarial worms targeting the bacterium Wolbachia (Voronin et al., 2012). The activation of autophagy coincided with the onset of rapid bacterial growth and expansion, showing that in spite of their mutualistic association, the nematode may recognize Wolbachia as a stressor and respond to regulate bacterial abundance (Taylor et al., 2013). Further studies assessing antibacterial responses by intestinal nematodes are largely lacking; however, two reports have shed some light on the issue. Adult female A. suum nematodes challenged with heat-inactivated E. coli respond by increasing transcription of two AMP families, the Ascaris suum antibacterial factors (ASABF) and the Cecropins (Pillai et al., 2003, 2005). Worms were challenged by injection of heat-killed bacteria into the pseudocoelom and appeared to demonstrate tissue-specific responses. While these data suggest an inducible defense system in A. suum, it would be relevant to study tissue-specific AMP expression using live bacterial cultures introduced externally, without wounding the worms, coupled with modern transcriptomic methods. Such an experiment would preserve pathogen-associated molecular patterns without invoking pathogen-independent, epithelial-damage responses, thereby allowing identification of transcriptional changes induced specifically by infection.
After sensing microbes, typical responses by nematodes include the production and release of factors possessing antimicrobial activity (Table 3). Among the different effectors of innate immunity, AMPs constitute the most ancient gene-encoded antimicrobial tools of eukaryotes (Zasloff, 2002). Nematodes produce different families of AMPs, many of which have been well characterized in C. elegans, while some have also been identified in parasitic nematodes. Antimicrobial activity has been experimentally demonstrated for many of these factors. AMPs can be classified based on their amino acid constituents and structural characteristics: α-helices, β–sheets, extended, and looped (Melo et al., 2009). Many of the best-characterized AMPs tend to be small, cationic, amphipathic molecules thought to act by membrane disruption. While many candidate immune effectors have been proposed in the literature for C. elegans, reviewed in Kim and Ewbank (2015), here we focus on effectors with corresponding data from parasitic nematodes. AMPs such as antibacterial factors and cecropins are discussed along with larger proteins involved in the immune response including lectins, nemapores, and lysozymes. We will then conclude this section by discussing antimicrobial activities of helminth-derived products.
First identified in A. suum, ABFs are present in at least 25 nematode species, including seven ASABFs produced by Ascaris and six ABFs produced by C. elegans (Tarr, 2012). Sequence analysis reported by Tarr shows that other parasitic nematode species expressing ABFs include the human hookworm N. americanus, the ruminant nematode Haemonchus contortus, and the rodent model parasite N. brasiliensis (Tarr, 2012). Body fluid isolated from the pseudocoelem of A. suum demonstrated bactericidal activity, particularly against gram positive organisms (Wardlaw et al., 1994; Kato, 1995). Kato et al. isolated ASABF-α (Kato and Komatsu, 1996) and subsequently demonstrated broad-spectrum antibacterial and weak antifungal activity of a recombinant form of the peptide (Zhang et al., 2000). The observed activity was rapid, killing S. aureus in under 1 min, and could be inhibited by increasing salt concentrations (Zhang et al., 2000). Andersson et al. isolated ASABF-β and γ from A. lumbricoides which also showed antimicrobial activity, especially against gram positives (Andersson et al., 2003). ABFs were the first AMPs described in C. elegans, based on sequence similarity to their Ascarid counterparts and also demonstrated antimicrobial activity (Kato et al., 2002). ABFs belong to the cysteine-stabilized-αβ (CS-αβ) group of AMPs, are cationic, and contain a cysteine-stabilized α-helix and two β-sheets (Kato and Komatsu, 1996; Tarr, 2012). Transcripts for ASABFs have been detected in all tissues of A. suum, with some members upregulated in the body wall, intestine, and ovaries after bacterial challenge (Pillai et al., 2003). Expression patterns and anatomical localization of ABFs in C. elegans are indicative of roles in defense and digestion (Kato et al., 2002). For example, ABF-1 and 2 are detected in the pharynx, and ABF-1 and 3 in the intestine of C. elegans. The expression of some ABFs is inducible by different pathogens: ABF-2 by S. typhimurium (Alegado and Tan, 2008), ABF-1 and 2 by Cryptococcus neoformans, and ABF-3 by S. aureus (Alper et al., 2007).
In contrast to ABFs which seem to be widely distributed amongst nematodes, the cecropin family of AMPs has only been found in three Ascarids: A. suum, A. lumbricoides, and Toxocara canis (Pillai et al., 2005; Tarr, 2012). The first described nematode cecropin, Cecropin P1, was originally thought to be of porcine origin due to its isolation from pig intestine (Lee et al., 1989). Subsequently, its production was correctly attributed to A. suum (Andersson et al., 2003). Andersson et al. also isolated Cecropin P1 from A. lumbricoides and demonstrated antibacterial activity against gram negative organisms (Andersson et al., 2003). Pillai et al. demonstrated broad-spectrum antimicrobial activity of all four Ascaris cecropins as well as bacterial inducibility as detected for ASABFs (Pillai et al., 2005). Cecropins are typically 31–39 amino acids in length, strongly basic, cationic, α-helical peptides which are thought to disrupt microbial membranes by first laying on the membrane surface before reorientation and insertion into the membrane leading to disrupted lipid packing and subsequent membrane disintegration (Sipos et al., 1992; Gazit et al., 1995).
Proteins containing lectin domains are conserved across Metazoans, including nematodes (Zelensky and Gready, 2005). Harcus et al. isolated one C-type lectin (CTL) from H. p. bakeri and two from N. brasiliensis (Harcus et al., 2009). Moreover, lectin-like carbohydrate-binding activity has previously been reported in the intestine of A. suum (Cuperlović et al., 1987) and lectins have been identified in excreted and secreted products (ESPs) of A. suum larvae (Wang et al., 2013). In general, CTLs are capable of binding carbohydrates and function in pathogen recognition and neutralization (Zelensky and Gready, 2005). CTLs from H. p. bakeri and N. brasiliensis are predominantly expressed by gut-dwelling adults rather than larvae; while Harcus et al. made a compelling case for host-immunomodulation by their reported CTLs, a dual role in pathogen neutralization or recognition is worth testing for parasite-derived lectins (Harcus et al., 2009). Parasite-derived CTL molecules localize in the nematode cuticle but could also be found in ESPs (Page et al., 1992). The CTLs detected in parasitic nematodes were most similar to the C. elegans-CTLs clec-48, -49, and -50. Clec-50 is expressed in the C. elegans intestine and upregulated in response to the bacterium S. marcescens (Mallo et al., 2002). Though Harcus et al. did not report antimicrobial activity for the parasite-derived molecules, studies in C. elegans suggest they may have a role in defense. In addition to clec-50, clec-49 is also upregulated in response to S. marcescens (Engelmann et al., 2011) and mutants deficient in this protein and another CTL, clec-39, have increased susceptibility to, and reduced survival and fecundity during, S. marcescens infection compared to wild-type worms (Miltsch et al., 2014). Furthermore, recombinant clec-39 and clec-49 bind S. marcescens in the absence of bactericidal activity (Miltsch et al., 2014). Additionally, in response to M. nematophilum, CTLs were found to be the most up-regulated protein class in C. elegans (O'Rourke et al., 2006). While nematode-derived lectins have not yet been shown to possess bactericidal activity, the mammalian lectin RegIIIγ is antibacterial and promotes commensalism by maintaining segregation between the microbiota and the host epithelium in mice (Vaishnava et al., 2011). Though many questions remain, the limited evidence available is suggestive of a role for lectins during infectious challenges and it is possible that parasite-derived lectins may confer resistance to different bacteria as has been demonstrated for C. elegans.
Lysozymes are polysaccharide hydrolases, targeting bacterial peptidoglycan leading to cell lysis (Ellison and Giehl, 1991; Monzingo et al., 1996), and have been widely found in different nematode species. Lysozymes are encoded in the genomes of Ascarids, N. americanus, H. contortus, N. brasiliensis, Brugia malayi, and Wuchereria bancrofti, though missing from the Trichurids included in the sequence analysis by Tarr (2012). Lysozymes were also detected in the ESPs of A. suum (Wang et al., 2013) and H. p. bakeri (Hewitson et al., 2011), though further study is required to determine antibacterial activities of these helminth-lysozymes. 15 lysozymes were detected in C. elegans, divided into 10 protist-type (lys-1–lys-10) and five invertebrate-type (ilys-1–ilys-5), so named for sequence similarities to other organisms (Schulenburg and Boehnisch, 2008). Some lysozymes are markedly upregulated by C. elegans in response to bacterial infection (Mallo et al., 2002; O'Rourke et al., 2006; Gravato-Nobre et al., 2016). Lysozymes expressed in the C. elegans intestine (Schulenburg and Boehnisch, 2008) have been proposed to act in concert with caenopores (discussed in the next section) in digestion and immunity (Bányai and Patthy, 1998). For example, the C. elegans invertebrate lysozyme ILYS-3 is needed for physiological pharyngeal grinder function and for defense against bacterial pathogens (Gravato-Nobre et al., 2016). It was also shown that ILYS-3 is induced by danger signals generated both by bacterial pathogens and starvation (Gravato-Nobre et al., 2016). In healthy C. elegans, intestinal ilys-3 expression undergoes a post-developmental regulatory oscillation: levels increase after L1 hatching, decline after the L2 transition, and increase again after L4 transition becoming abundantly expressed in the intestine of adult worms (Gravato-Nobre et al., 2016). Notably, coelomocytes of C. elegans also express ilys-3 (Gravato-Nobre et al., 2016) and are scavenger cells that endocytose fluid from the pseudocoelom (Fares and Greenwald, 2001). The role of coelomocytes in parasitic nematodes is not well understood and they have not been studied in immunity. Given the demonstrated importance of lysozymes for C. elegans and the wide distribution of this protein family across species, it is expected that they support helminth survival amongst the host-microbiota.
Known as caenopores in C. elegans, nemapores contain a saposin domain and share similarity with protozoan amoebapores and mammalian NK-lysin and granulysin (Leippe, 1995; Roeder et al., 2010). As with ABFs, nemapores are also cysteine-rich, but contain multiple α-helices (Mysliwy et al., 2010). Nemapores are widely distributed and in addition to C. elegans, have been found in 46 nematode species including Ascarids, Trichurids, and the human filarial worms B. malayi and W. bancrofti (Tarr, 2012). Only caenopores have been studied experimentally, with caenopore-1 and 5 demonstrating antimicrobial activity (Bányai and Patthy, 1998; Roeder et al., 2010). Caenopore-5 is constitutively expressed in the intestine, while caenopore-3 is induced by starvation as well as the bacterial strains B. megaterium and Micrococcus luteus, suggesting functions in nutrition and immune defense (Roeder et al., 2010). Interestingly, the free-living soil bacteria B. megaterium and M. luteus are both frequently found with Rhabditid nematodes (Coolon et al., 2009) and may be present in the environment of C. elegans. As with lysozymes, experimental evidence of nemapore function in parasitic nematodes is currently lacking.
In addition to studies demonstrating antimicrobial activities of specific helminth-derived molecules described here, activity can also be detected from the ESPs of parasites. In one study, ESPs from the filarial worm O. ochengi were analyzed revealing 36 candidate AMPs (Eberle et al., 2015). Of the 36 candidates, the investigators were able to unambiguously attribute bactericidal activity against E. coli to three peptides while the other 33 peptides had been assessed as part of peptide mixtures. Antibacterial activity has also been shown for ESPs from adult T. suis worms (Abner et al., 2001). Different bacterial strains were tested, including S. aureus, E. coli, and C. jejuni. The observed activity was shown to be due to small (<10 kDa), boiling-resistant molecules, indicative of AMPs. Interestingly, pore-forming proteins have also been isolated from T. trichiura and T. muris, indicating that antimicrobial activity of nematode ESPs is likely due to multiple components of varying sizes and mechanisms (Drake et al., 1994). As nematodes produce a variety of antimicrobial factors, innate immune responses to microbes by helminths likely offer protection using several different strategies.
Antibiotic resistance is a rapidly escalating problem with devastating consequences for human and animal health. Drug resistance mechanisms described for microbes and helminths foreshadow the emergence of immense clinical and economic challenges associated with previously treatable infectious diseases. Meanwhile, inflammatory and metabolic diseases also inflict great suffering and are a source of considerable strain on healthcare systems; therefore, novel solutions are sought after to combat these difficulties. As nematodes have evolved over millions of years in a diverse microbial environment, a better understanding of how nematodes interact with microbial populations may offer innovative strategies for treating human and animal diseases. Given the importance of the microbiota and the ability of helminths to influence microbial communities in the gut (Figure 2), helminth infections and helminth products are being studied for their role in dysbiosis. Preliminary data suggest potential benefits of helminths in inflammatory diseases and in individuals with metabolic abnormalities. An additional application of the impact of nematodes on microbes is the development of new antimicrobials. AMPs are under investigation for their therapeutic potential and an understanding of how antimicrobial effectors are used by organisms living amongst diverse microbial populations can guide development efforts. In elucidating the impacts of different bacteria on nematodes, one can anticipate the discovery of novel anthelmintic targets. Moreover, these studies also provide insights into conserved innate immune mechanisms. The divergent environments and experimental systems of parasitic and free-living nematodes offer unique tools to investigators, the combination of which enhance our comprehension of nematode biology and evolutionary ecology.
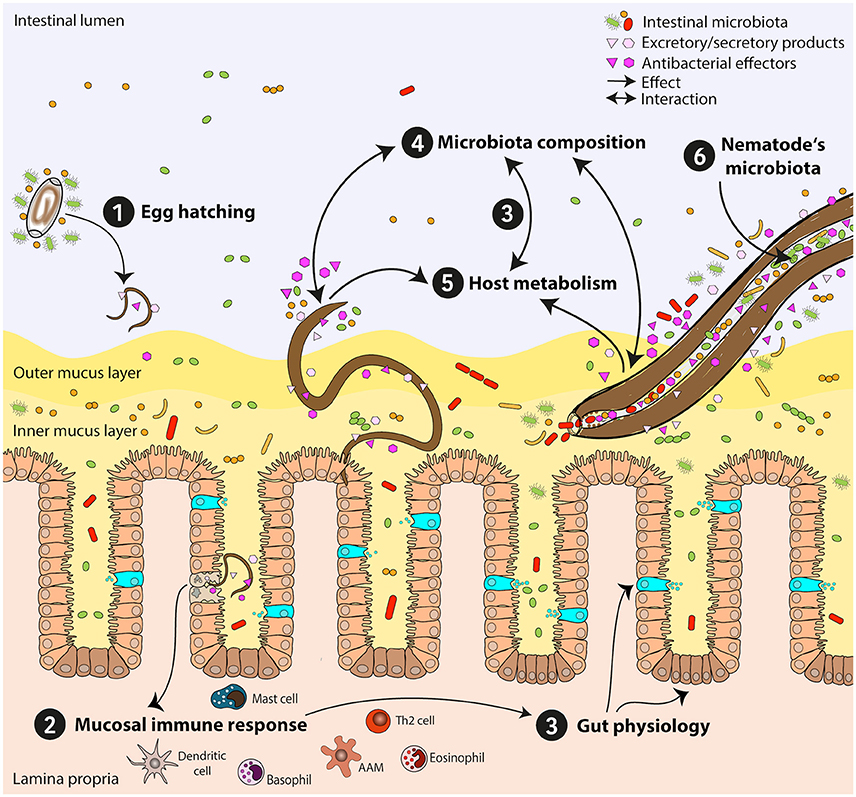
Figure 2. Mutual influences of intestinal nematodes and host-gut bacteria. Establishment and persistence of intestinal nematodes in the host's gut are affected by bacterial communities and lead to substantial changes of the gut microbiota. Here, nematode-microbiota interactions and their impact on the host immune response and physiology are exemplified. (1) Egg hatching: Interaction of eggs with the intestinal microbiota is needed for some species, enabling larvae to hatch. (2) Mucosal immune response: Attachment to the epithelium and tissue migratory phase might lead to bacterial translocation and manipulation of immune responses. The anti-helminth immune response is predominantly a T helper type 2 response. (3) Gut physiology: Immune responses induce changes in gut physiology via induction of goblet cell hyperplasia, mucus production, and epithelial turnover, leading to changes in the host microbiota and its metabolome. Specific subsets of bacteria directly influence host physiology through their metabolic activities (e.g., short chain fatty acids-producing bacteria). (4) Microbiota composition: Intestinal nematodes modify intestinal microbial communities via different mechanisms: a) directly via secretion of antibacterial molecules and/or excretory/secretory products, b) indirectly by metabolic and physiologic shifts influencing the gut milieu. Chronic infections often lead to reduction in bacterial diversity and outgrowth of specific bacterial species beneficial for parasite survival. (5) Host metabolism: Intestinal nematodes modify host metabolism and nutrient uptake, e.g., alter amino acid, fatty acid, and carbohydrate metabolism, with subsequent influence on gut physiology, immune reactivity, and intestinal microbiota composition. (6) Nematode's microbiota: Nematode-associated bacterial communities might reflect the host microbiota and may also serve as transmission vehicles for pathogenic bacteria. Th2 cell, T helper type 2 cell; AAM, alternatively activated macrophage.
All authors listed have made substantial contributions to the planning and writing of this work. All authors have approved this work for publication.
The work was supported by the German Research Foundation: GRK 2046 (AM and SH) and HA2542/3-2 (SH).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abner, S. R., Parthasarathy, G., Hill, D. E., and Mansfield, L. S. (2001). Trichuris suis: detection of antibacterial activity in excretory-secretory products from adults. Exp. Parasitol. 99, 26–36. doi: 10.1006/expr.2001.4643
Alegado, R. A., and Tan, M.-W. (2008). Resistance to antimicrobial peptides contributes to persistence of salmonella typhimurium in the C. Elegans INTESTINE. Cell. Microbiol. 10, 1259–1273. doi: 10.1111/j.1462-5822.2008.01124.x
Alper, S., McBride, S. J., Lackford, B., Freedman, J. H., and Schwartz, D. A. (2007). Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol. Cell. Biol. 27, 5544–5553. doi: 10.1128/MCB.02070-06
Andersson, M., Boman, A., and Boman, H. H. (2003). Ascaris nematodes from pig and human make three antibacterial peptides: isolation of cecropin P1 and two ASABF peptides. Cell. Mol. Life Sci. 60, 599–606. doi: 10.1007/s000180300051
Bansemir, A. D., and Sukhdeo, M. V. (1994). The food resource of adult Heligmosomoides Polygyrus in the small intestine. J. Parasitol. 80, 24–28. doi: 10.2307/3283340
Bansemir, A. D., and Sukhdeo, M. V. (2001). Intestinal distribution of worms and host ingesta in Nippostrongylus brasiliensis. J. Parasitol. 87, 1470–1472. doi: 10.1645/0022-3395(2001)087[1470:IDOWAH]2.0.CO;2
Bányai, L., and Patthy, L. (1998). Amoebapore homologs of Caenorhabditis elegans. Biochim. Biophys. Acta 1429, 259–264. doi: 10.1016/S0167-4838(98)00237-4
Barrière, A., and Félix, M. (2005). High local genetic diversity and low outcrossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 15, 1176–1184. doi: 10.1016/j.cub.2005.06.022
Barsyte, D., Lovejoy, D. A., and Lithgow, G. J. (2001). Longevity and heavy metal resistance in Daf-2 and age-1 long-lived mutants of Caenorhabditis elegans. FASEB J. 15, 627–634. doi: 10.1096/fj.99-0966com
Behnke, J. M., Menge, D. M., and Noyes, H. (2009). Heligmosomoides bakeri: a model for exploring the biology and genetics of resistance to chronic gastrointestinal nematode infections. Parasitology 136, 1565–1580. doi: 10.1017/S0031182009006003
Bethony, J., Brooker, S., Albonico, M., Geiger, S. M., Loukas, A., Diemert, D., et al. (2006). Soil-transmitted helminth infections: ascariasis, trichuriasis, and hookworm. Lancet 367, 1521–1532. doi: 10.1016/S0140-6736(06)68653-4
Bevins, C. L., and Salzman, N. H. (2011). Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat. Rev. Microbiol. 9, 356–368. doi: 10.1038/nrmicro2546
Broadhurst, M. J., Ardeshir, A., Kanwar, B., Mirpuri, J., Gundra, U. M., Leung, J. M., et al. (2012). Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathog. 8:e1003000. doi: 10.1371/journal.ppat.1003000
Cabreiro, F., and Gems, D. (2013). Worms need microbes too: microbiota, health and aging in Caenorhabditis elegans. EMBO Mol. Med. 5, 1300–1310. doi: 10.1002/emmm.201100972
Cantacessi, C., Giacomin, P., Croese, J., Zakrzewski, M., Sotillo, J., McCann, L., et al. (2014). Impact of experimental hookworm infection on the human gut microbiota. J. Infect. Dis. 210, 1431–1434. doi: 10.1093/infdis/jiu256
Carding, S., Verbeke, K., Vipond, D. T., Corfe, B. M., and Owen, L. J. (2015). Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 26:26191. doi: 10.3402/mehd.v26.26191
Cattadori, I. M., Sebastian, A., Hao, H., Katani, R., Albert, I., Eilertson, K. E., et al. (2016). Impact of helminth infections and nutritional constraints on the small intestine microbiota. PLoS ONE 11:e0159770. doi: 10.1371/journal.pone.0159770
Collier, R. J. (2001). Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon 39, 1793–1803. doi: 10.1016/S0041-0101(01)00165-9
Consortium, T. H. M. P. (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Coolon, J. D., Jones, K. L., Todd, T. C., Carr, B. C., and Herman, M. A. (2009). Caenorhabditis elegans genomic response to soil bacteria predicts environment-specific genetic effects on life history traits. PLoS Genet. 5:e1000503. doi: 10.1371/journal.pgen.1000503
Cooper, P., Walker, A. W., Reyes, J., Chico, M., Salter, S. J., Vaca, M., et al. (2013). Patent human infections with the whipworm, Trichuris trichiura, are not associated with alterations in the faecal microbiota. PLoS ONE 8:e76573. doi: 10.1371/journal.pone.0076573
Couillault, C., and Ewbank, J. J. (2002). Diverse bacteria are pathogens of Caenorhabditis elegans infect. Immun. 70, 4705–4707. doi: 10.1128/IAI.70.8.4705-4707.2002
Couillault, C., Pujol, N., Reboul, J., Sabatier, L., Guichou, J.-F., Kohara, Y., et al. (2004). TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5, 488–494. doi: 10.1038/ni1060
Croese, J., Giacomin, P., Navarro, S., Clouston, A., McCann, L., Dougall, A., et al. (2015). Experimental hookworm infection and gluten microchallenge promote tolerance in celiac disease. J. Allergy Clin. Immunol. 135, 508–516. doi: 10.1016/j.jaci.2014.07.022
Cullen, T. W., Schofield, W. B., Barry, N. A., Putnam, E. E., Rundell, E. A., Trent, M. S., et al. (2015). Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347, 170–175. doi: 10.1126/science.1260580
Cuperlović, M., Movsesijan, M., and Hajduković, L. (1987). Ascaris lumbricoides: lectin activity in the gastrointestinal system. Exp. Parasitol. 63, 237–239. doi: 10.1016/0014-4894(87)90167-6
Dai, L.-L., Gao, J.-X., Zou, C.-G., Ma, Y.-C., and Zhang, K.-Q. (2015). mir-233 modulates the unfolded protein response in C. elegans during Pseudomonas aeruginosa infection. PLoS Pathog. 11:e1004606. doi: 10.1371/journal.ppat.1004606
Darby, A. C., Armstrong, S. D., Bah, G. S., Kaur, G., Hughes, M. A., Kay, S. M., et al. (2012). Analysis of gene expression from the Wolbachia genome of a filarial nematode supports both metabolic and defensive roles within the symbiosis. Genome Res. 22, 2467–2477. doi: 10.1101/gr.138420.112
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
Dethlefsen, L., Huse, S., Sogin, M. L., and Relman, D. A. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280. doi: 10.1371/journal.pbio.0060280
Dilika, F., Bremner, P. D., and Meyer, J. J. (2000). Antibacterial activity of linoleic and oleic acids isolated from Helichrysum pedunculatum: a plant used during circumcision rites. Fitoterapia 71, 450–452. doi: 10.1016/S0367-326X(00)00150-7
Dirksen, P., Marsh, S. A., Braker, I., Heitland, N., Wagner, S., Nakad, R., et al. (2016). The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 14:38. doi: 10.1186/s12915-016-0258-1
Dold, C., and Holland, C. V. (2011). Ascaris and ascariasis. Microbes Infect. 13, 632–637. doi: 10.1016/j.micinf.2010.09.012
Donaldson, G. P., Lee, S. M., and Mazmanian, S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32. doi: 10.1038/nrmicro3552
Drake, L., Korchev, Y., Bashford, L., Djamgoz, M., Wakelin, D., Ashall, F., et al. (1994). The major secreted product of the whipworm, Trichuris, is a pore-forming protein. Proc. Biol. Sci. 257, 255–261. doi: 10.1098/rspb.1994.0123
Dunbar, T. L., Yan, Z., Balla, K. M., Smelkinson, M. G., and Troemel, E. R. (2012). C. elegans detects pathogen-induced translational inhibition to activate immune signaling. Cell Host Microbe 11, 375–386. doi: 10.1016/j.chom.2012.02.008
Eberle, R., Brattig, N. W., Trusch, M., Schlüter, H., Achukwi, M. D., Eisenbarth, A., et al. (2015). Isolation, identification and functional profile of excretory-secretory peptides from Onchocerca ochengi. Acta Trop. 142, 156–166. doi: 10.1016/j.actatropica.2014.11.015
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Eijck, I. A., and Borgsteede, F. H. (2005). A survey of gastrointestinal pig parasites on free-range, organic and conventional pig farms in The Netherlands. Vet. Res. Commun. 29, 407–414. doi: 10.1007/s11259-005-1201-z
Ellison, R. T., and Giehl, T. J. (1991). Killing of gram-negative bacteria by lactoferrin and lysozyme. J. Clin. Invest. 88, 1080–1091. doi: 10.1172/JCI115407
Engelmann, I., Griffon, A., Tichit, L., Montañana-Sanchis, F., Wang, G., Reinke, V., et al. (2011). A comprehensive analysis of gene expression changes provoked by bacterial and fungal infection in C. elegans. PLoS ONE 6:e19055. doi: 10.1371/journal.pone.0019055
Estes, K. A., Dunbar, T. L., Powell, J. R., Ausubel, F. M., and Troemel, E. R. (2010). bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 107, 2153–2158. doi: 10.1073/pnas.0914643107
Fares, H., and Greenwald, I. (2001). Genetic analysis of endocytosis in Caenorhabditis elegans: coelomocyte uptake defective mutants. Genetics 159, 133–145.
Félix, M.-A., Ashe, A., Piffaretti, J., Wu, G., Nuez, I., Bélicard, T., et al. (2011). Natural and experimental infection of Caenorhabditis nematodes by novel viruses related to nodaviruses. PLoS Biol. 9:e1000586. doi: 10.1371/journal.pbio.1000586
Félix, M.-A., and Duveau, F. (2012). Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biol. 10:59. doi: 10.1186/1741-7007-10-59
Frézal, L., and Félix, M.-A. (2015). C. elegans outside the Petri dish. Elife 4:e05849. doi: 10.7554/eLife.05849
Fricke, W. F., Song, Y., Wang, A.-J., Smith, A., Grinchuk, V., Mongodin, E., et al. (2015). Type 2 immunity-dependent reduction of segmented filamentous bacteria in mice infected with the helminthic parasite Nippostrongylus brasiliensis. Microbiome 3:40. doi: 10.1186/s40168-015-0103-8
Garsin, D. A., Villanueva, J. M., Begun, J., Kim, D. H., Sifri, C. D., Calderwood, S. B., et al. (2003). Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science 300:1921. doi: 10.1126/science.1080147
Gause, W. C., and Maizels, R. M. (2016). Macrobiota - helminths as active participants and partners of the microbiota in host intestinal homeostasis. Curr. Opin. Microbiol. 32, 14–18. doi: 10.1016/j.mib.2016.04.004
Gazit, E., Boman, A., Boman, H. G., and Shai, Y. (1995). Interaction of the mammalian antibacterial peptide cecropin P1 with phospholipid vesicles. Biochemistry (Mosc.) 34, 11479–11488. doi: 10.1021/bi00036a021
Gevers, D., Kugathasan, S., Denson, L. A., Vázquez-Baeza, Y., Van Treuren, W., Ren, B., et al. (2014). The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15, 382–392. doi: 10.1016/j.chom.2014.02.005
Giacomin, P., Zakrzewski, M., Croese, J., Su, X., Sotillo, J., McCann, L., et al. (2015). Experimental hookworm infection and escalating gluten challenges are associated with increased microbial richness in celiac subjects. Sci. Rep. 5:13797. doi: 10.1038/srep13797
Gravato-Nobre, M. J., Nicholas, H. R., Nijland, R., O'Rourke, D., Whittington, D. E., Yook, K. J., et al. (2005). Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 171, 1033–1045. doi: 10.1534/genetics.105.045716
Gravato-Nobre, M. J., Vaz, F., Filipe, S., Chalmers, R., and Hodgkin, J. (2016). The invertebrate lysozyme effector ILYS-3 is systemically activated in response to danger signals and confers antimicrobial protection in C. elegans. PLoS Pathog. 12:e1005826. doi: 10.1371/journal.ppat.1005826
Gregory, R. D., Keymer, A. E., and Clarke, J. R. (1990). Genetics, sex and exposure: the ecology of Heligmosomoides polygyrus (Nematoda) in the wood mouse. J. Anim. Ecol. 59, 363–378. doi: 10.2307/5178
Harcus, Y., Nicoll, G., Murray, J., Filbey, K., Gomez-Escobar, N., and Maizels, R. M. (2009). C-type lectins from the nematode parasites Heligmosomoides polygyrus and Nippostrongylus brasiliensis. Parasitol. Int. 58, 461–470. doi: 10.1016/j.parint.2009.08.011
Hayes, K. S., Bancroft, A. J., Goldrick, M., Portsmouth, C., Roberts, I. S., and Grencis, R. K. (2010). Exploitation of the intestinal microflora by the parasitic nematode Trichuris muris. Science 328, 1391–1394. doi: 10.1126/science.1187703
Hewitson, J. P., Harcus, Y., Murray, J., van Agtmaal, M., Filbey, K. J., Grainger, J. R., et al. (2011). Proteomic analysis of secretory products from the model gastrointestinal nematode Heligmosomoides polygyrus reveals dominance of Venom Allergen-Like (VAL) proteins. J. Proteomics 74, 1573–1594. doi: 10.1016/j.jprot.2011.06.002
Hoerauf, A., Nissen-Pähle, K., Schmetz, C., Henkle-Dührsen, K., Blaxter, M. L., Büttner, D. W., et al. (1999). Tetracycline therapy targets intracellular bacteria in the filarial nematode Litomosoides sigmodontis and results in filarial infertility. J. Clin. Invest. 103, 11–18. doi: 10.1172/JCI4768
Holm, J. B., Sorobetea, D., Kiilerich, P., Ramayo-Caldas, Y., Estellé, J., Ma, T., et al. (2015). Chronic Trichuris muris infection decreases diversity of the intestinal microbiota and concomitantly increases the abundance of Lactobacilli. PLoS ONE 10:e0125495. doi: 10.1371/journal.pone.0125495
Hotez, P. J., Brindley, P. J., Bethony, J. M., King, C. H., Pearce, E. J., and Jacobson, J. (2008). Helminth infections: the great neglected tropical diseases. J. Clin. Invest. 118, 1311–1321. doi: 10.1172/JCI34261
Houlden, A., Hayes, K. S., Bancroft, A. J., Worthington, J. J., Wang, P., Grencis, R. K., et al. (2015). Chronic Trichuris muris Infection in C57BL/6 mice causes significant changes in host microbiota and metabolome: effects reversed by pathogen clearance. PLoS ONE 10:e0125945. doi: 10.1371/journal.pone.0125945
Hsu, S. C., Johansson, K. R., and Donahue, M. J. (1986). The bacterial flora of the intestine of Ascaris suum and 5-hydroxytryptamine production. J. Parasitol. 72, 545–549. doi: 10.2307/3281505
Irazoqui, J. E., Urbach, J. M., and Ausubel, F. M. (2010). Evolution of host innate defence: insights from C. elegans and primitive invertebrates. Nat. Rev. Immunol. 10, 47–58. doi: 10.1038/nri2689
Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. doi: 10.1016/j.cell.2009.09.033
JebaMercy, G., and Balamurugan, K. (2012). Effects of sequential infections of Caenorhabditis elegans with Staphylococcus aureus and Proteus mirabilis. Microbiol. Immunol. 56, 825–835. doi: 10.1111/j.1348-0421.2012.00509.x
Jernberg, C., Löfmark, S., Edlund, C., and Jansson, J. K. (2007). Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66. doi: 10.1038/ismej.2007.3
Jia, K., Thomas, C., Akbar, M., Sun, Q., Adams-Huet, B., Gilpin, C., et al. (2009). Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc. Natl. Acad. Sci. U.S.A. 106, 14564–14569. doi: 10.1073/pnas.0813319106
Jørgensen, R., Purdy, A. E., Fieldhouse, R. J., Kimber, M. S., Bartlett, D. H., and Merrill, A. R. (2008). Cholix Toxin, a novel ADP-ribosylating factor from Vibrio cholerae. J. Biol. Chem. 283, 10671–10678. doi: 10.1074/jbc.M710008200
Kato, Y. (1995). Humoral defense of the nematode Ascaris suum: antibacterial, bacteriolytic and agglutinating activities in the body fluid. Zoolog. Sci. 12, 225–230. doi: 10.2108/zsj.12.225
Kato, Y., Aizawa, T., Hoshino, H., Kawano, K., Nitta, K., and Zhang, H. (2002). abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem. J. 361, 221–230. doi: 10.1042/bj3610221
Kato, Y., and Komatsu, S. (1996). ASABF, a novel cysteine-rich antibacterial peptide isolated from the nematode Ascaris suum. Purification, primary structure, and molecular cloning of cDNA. J. Biol. Chem. 271, 30493–30498. doi: 10.1074/jbc.271.48.30493
Kenyon, C., Chang, J., Gensch, E., Rudner, A., and Tabtiang, R. (1993). A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464. doi: 10.1038/366461a0
Kim, D. H., and Ewbank, J. J. (2015). Signaling in the innate immune response. WormBook doi: 10.1895/wormbook.1.83.2. [Epub ahead of print].
Kim, D. H., Feinbaum, R., Alloing, G., Emerson, F. E., Garsin, D. A., Inoue, H., et al. (2002). A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626. doi: 10.1126/science.1073759
Kirienko, N. V., Ausubel, F. M., and Ruvkun, G. (2015). Mitophagy confers resistance to siderophore-mediated killing by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 112, 1821–1826. doi: 10.1073/pnas.1424954112
Klementowicz, J. E., Travis, M. A., and Grencis, R. K. (2012). Trichuris muris: a model of gastrointestinal parasite infection. Semin. Immunopathol. 34, 815–828. doi: 10.1007/s00281-012-0348-2
Kozek, W. J. (1977). Transovarially-transmitted intracellular microorganisms in adult and larval stages of Brugia malayi. J. Parasitol. 63, 992–1000. doi: 10.2307/3279832
Kreisinger, J., Bastien, G., Hauffe, H. C., Marchesi, J., and Perkins, S. E. (2015). Interactions between multiple helminths and the gut microbiota in wild rodents. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 370:20140295. doi: 10.1098/rstb.2014.0295
Lee, J., Choe, J., Kim, J., Oh, S., Park, S., Kim, S., et al. (2015). Heat-killed Lactobacillus spp. cells enhance survivals of Caenorhabditis elegans against Salmonella and Yersinia infections. Lett. Appl. Microbiol. 61, 523–530. doi: 10.1111/lam.12478
Lee, J. Y., Boman, A., Sun, C. X., Andersson, M., Jörnvall, H., Mutt, V., et al. (1989). Antibacterial peptides from pig intestine: isolation of a mammalian cecropin. Proc. Natl. Acad. Sci. U.S.A. 86, 9159–9162. doi: 10.1073/pnas.86.23.9159
Lee, S. C., Tang, M. S., Lim, Y. A. L., Choy, S. H., Kurtz, Z. D., Cox, L. M., et al. (2014). Helminth colonization is associated with increased diversity of the gut microbiota. PLoS Negl. Trop. Dis. 8:e2880. doi: 10.1371/journal.pntd.0002880
Lee, S.-H., Wong, R.-R., Chin, C.-Y., Lim, T.-Y., Eng, S.-A., Kong, C., et al. (2013). Burkholderia pseudomallei suppresses Caenorhabditis elegans immunity by specific degradation of a GATA transcription factor. Proc. Natl. Acad. Sci. U.S.A. 110, 15067–15072. doi: 10.1073/pnas.1311725110
Leippe, M. (1995). Ancient weapons: NK-lysin, is a mammalian homolog to pore-forming peptides of a protozoan parasite. Cell 83, 17–18. doi: 10.1016/0092-8674(95)90229-5
Leroy, M., Mosser, T., Manière, X., Alvarez, D. F., and Matic, I. (2012). Pathogen-induced Caenorhabditis elegans developmental plasticity has a hormetic effect on the resistance to biotic and abiotic stresses. BMC Evol. Biol. 12:187. doi: 10.1186/1471-2148-12-187
Leser, T. D., Amenuvor, J. Z., Jensen, T. K., Lindecrona, R. H., Boye, M., and Møller, K. (2002). Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68, 673–690. doi: 10.1128/AEM.68.2.673-690.2002
Li, R. W., Wu, S., Li, W., Navarro, K., Couch, R. D., Hill, D., et al. (2012). Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis. Infect. Immun. 80, 2150–2157. doi: 10.1128/IAI.00141-12
Liu, Y., Samuel, B. S., Breen, P. C., and Ruvkun, G. (2014). Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature 508, 406–410. doi: 10.1038/nature13204
Lupp, C., Robertson, M. L., Wickham, M. E., Sekirov, I., Champion, O. L., Gaynor, E. C., et al. (2007). Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2, 119–129. doi: 10.1016/j.chom.2007.06.010
MacNeil, L. T., and Walhout, A. J. (2013). Food, pathogen, signal. Worm 2:e26454. doi: 10.4161/worm.26454
Mallo, G. V., Kurz, C. L., Couillault, C., Pujol, N., Granjeaud, S., Kohara, Y., et al. (2002). Inducible antibacterial defense system in C. elegans. Curr. Biol. CB 12, 1209–1214. doi: 10.1016/S0960-9822(02)00928-4
Mansfield, L., Gauthier, D. T., Abner, S. R., Jones, K. M., Wilder, S. R., and Urban, J. F. (2003). Enhancement of disease and pathology by synergy of Trichuris suis and Campylobacter jejuni in the colon of immunologically naive swine. Am. J. Trop. Med. Hyg. 68, 70–80. doi: 10.4269/ajtmh.2003.68.70
Mansfield, L. S., and Urban, J. F. (1996). The pathogenesis of necrotic proliferative colitis in swine is linked to whipworm induced suppression of mucosal immunity to resident bacteria. Vet. Immunol. Immunopathol. 50, 1–17. doi: 10.1016/0165-2427(95)05482-0
Masson, F., Zaidman-Rémy, A., and Heddi, A. (2016). Antimicrobial peptides and cell processes tracking endosymbiont dynamics. Philos. Trans. R. Soc. B 371:20150298. doi: 10.1098/rstb.2015.0298
McElwee, J., Bubb, K., and Thomas, J. H. (2003). Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell 2, 111–121. doi: 10.1046/j.1474-9728.2003.00043.x
McEwan, D. L., Kirienko, N. V., and Ausubel, F. M. (2012). Host translational inhibition by Pseudomonas aeruginosa exotoxin a triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 11, 364–374. doi: 10.1016/j.chom.2012.02.007
McGarry, H. F., Egerton, G. L., and Taylor, M. J. (2004). Population dynamics of Wolbachia bacterial endosymbionts in Brugia malayi. Mol. Biochem. Parasitol. 135, 57–67. doi: 10.1016/j.molbiopara.2004.01.006
Means, T. K., Mylonakis, E., Tampakakis, E., Colvin, R. A., Seung, E., Puckett, L., et al. (2009). Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors SCARF1 and CD36. J. Exp. Med. 206, 637–653. doi: 10.1084/jem.20082109
Meisel, J. D., and Kim, D. H. (2014). Behavioral avoidance of pathogenic bacteria by Caenorhabditis elegans. Trends Immunol. 35, 465–470. doi: 10.1016/j.it.2014.08.008
Melo, J. A., and Ruvkun, G. (2012). Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149, 452–466. doi: 10.1016/j.cell.2012.02.050
Melo, M. N., Ferre, R., and Castanho, M. A. R. B. (2009). Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 7, 245–250. doi: 10.1038/nrmicro2095
Milani, C., Ticinesi, A., Gerritsen, J., Nouvenne, A., Lugli, G. A., Mancabelli, L., et al. (2016). Gut microbiota composition and Clostridium difficile infection in hospitalized elderly individuals: a metagenomic study. Sci. Rep. 6:25945. doi: 10.1038/srep25945
Miltsch, S. M., Seeberger, P. H., and Lepenies, B. (2014). The C-type lectin-like domain containing proteins Clec-39 and Clec-49 are crucial for Caenorhabditis elegans immunity against Serratia marcescens infection. Dev. Comp. Immunol. 45, 67–73. doi: 10.1016/j.dci.2014.02.002
Montalvo-Katz, S., Huang, H., Appel, M. D., Berg, M., and Shapira, M. (2013). Association with soil bacteria enhances p38-dependent infection resistance in Caenorhabditis elegans. Infect. Immun. 81, 514–520. doi: 10.1128/IAI.00653-12
Monzingo, A. F., Marcotte, E. M., Hart, P. J., and Robertas, J. D. (1996). Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat. Struct. Mol. Biol. 3, 133–140. doi: 10.1038/nsb0296-133
Murrell, K. D., Eriksen, L., Nansen, P., Slotved, H. C., and Rasmussen, T. (1997). Ascaris suum: a revision of its early migratory path and implications for human ascariasis. J. Parasitol. 83, 255–260. doi: 10.2307/3284450
Mysliwy, J., Dingley, A. J., Stanisak, M., Jung, S., Lorenzen, I., Roeder, T., et al. (2010). Caenopore-5: the three-dimensional structure of an antimicrobial protein from Caenorhabditis elegans. Dev. Comp. Immunol. 34, 323–330. doi: 10.1016/j.dci.2009.11.003
Nalin, D. R., and McLaughlin, J. (1976). Colonization of ascaris lumbricoides by V. cholerae. J. Parasitol. 62, 839–841. doi: 10.2307/3278979
Nganga, C. J., Karanja, D. N., and Mutune, M. N. (2008). The prevalence of gastrointestinal helminth infections in pigs in Kenya. Trop. Anim. Health Prod. 40, 331–334. doi: 10.1007/s11250-007-9112-3
Nguyen, T. L. A., Vieira-Silva, S., Liston, A., and Raes, J. (2015). How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 8, 1–16. doi: 10.1242/dmm.017400
Nicholas, H. R., and Hodgkin, J. (2004). Responses to infection and possible recognition strategies in the innate immune system of Caenorhabditis elegans. Mol. Immunol. 41, 479–493. doi: 10.1016/j.molimm.2004.03.037
O'Rourke, D., Baban, D., Demidova, M., Mott, R., and Hodgkin, J. (2006). Genomic clusters, putative pathogen recognition molecules, and antimicrobial genes are induced by infection of C. elegans with M. nematophilum. Genome Res. 16, 1005–1016. doi: 10.1101/gr.50823006
Odamaki, T., Kato, K., Sugahara, H., Hashikura, N., Takahashi, S., Xiao, J.-Z., et al. (2016). Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 16:90. doi: 10.1186/s12866-016-0708-5
Paerewijck, O., Vlaminck, J., Li, R. W., Urban, J., Zaiss, M. M., Harris, N. L., et al. (2015). The Influence of an Infection with Ascaris Suum on the Intestinal Microbiota of the Host. IPVS Belgian Branch Studiedag, Abstracts. International Pig Veterinary Society (IPVS). Belgian Branch. Abstract retrieved from Ghent University Academic Bibliography. Available online at: http://hdl.handle.net/1854/LU-7277973
Page, A. P., Hamilton, A. J., and Maizels, R. M. (1992). Toxocara canis: monoclonal antibodies to carbohydrate epitopes of secreted (TES) antigens localize to different secretion-related structures in infective larvae. Exp. Parasitol. 75, 56–71. doi: 10.1016/0014-4894(92)90122-Q
Pillai, A., Ueno, S., Zhang, H., and Kato, Y. (2003). Induction of ASABF (Ascaris suum antibacterial factor)-type antimicrobial peptides by bacterial injection: novel members of ASABF in the nematode Ascaris suum. Biochem. J. 371, 663–668. doi: 10.1042/bj20021948
Pillai, A., Ueno, S., Zhang, H., Lee, J. M., and Kato, Y. (2005). Cecropin P1 and novel nematode cecropins: a bacteria-inducible antimicrobial peptide family in the nematode Ascaris suum. Biochem. J. 390, 207–214. doi: 10.1042/BJ20050218
Pradel, E., Zhang, Y., Pujol, N., Matsuyama, T., Bargmann, C. I., and Ewbank, J. J. (2007). Detection and avoidance of a natural product from the pathogenic bacterium Serratia marcescens by Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 104, 2295–2300. doi: 10.1073/pnas.0610281104
Pujol, N., Cypowyj, S., Ziegler, K., Millet, A., Astrain, A., Goncharov, A., et al. (2008a). Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr. Biol. CB 18, 481–489. doi: 10.1016/j.cub.2008.02.079
Pujol, N., Link, E. M., Liu, L. X., Kurz, C. L., Alloing, G., Tan, M. W., et al. (2001). A reverse genetic analysis of components of the Toll signaling pathway in Caenorhabditis elegans. Curr. Biol. CB 11, 809–821. doi: 10.1016/S0960-9822(01)00241-X
Pujol, N., Zugasti, O., Wong, D., Couillault, C., Kurz, C. L., Schulenburg, H., et al. (2008b). Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog. 4:e1000105. doi: 10.1371/journal.ppat.1000105
Pukkila-Worley, R. (2016). Surveillance immunity: an emerging paradigm of innate defense activation in Caenorhabditis elegans. PLoS Pathog. 12:e1005795. doi: 10.1371/journal.ppat.1005795
Pukkila-Worley, R., Feinbaum, R., Kirienko, N. V., Larkins-Ford, J., Conery, A. L., and Ausubel, F. M. (2012). Stimulation of host immune defenses by a small molecule protects C. elegans from bacterial infection. PLoS Genet. 8:e1002733. doi: 10.1371/journal.pgen.1002733
Pukkila-Worley, R., Feinbaum, R. L., McEwan, D. L., Conery, A. L., and Ausubel, F. M. (2014). The evolutionarily conserved mediator subunit MDT-15/MED15 links protective innate immune responses and xenobiotic detoxification. PLoS Pathog. 10:e1004143. doi: 10.1371/journal.ppat.1004143
Ramanan, D., Bowcutt, R., Lee, S. C., Tang, M. S., Kurtz, Z. D., Ding, Y., et al. (2016). Helminth infection promotes colonization resistance via type 2 immunity. Science 352, 608–612. doi: 10.1126/science.aaf3229
Rausch, S., Held, J., Fischer, A., Heimesaat, M. M., Kühl, A. A., Bereswill, S., et al. (2013). Small intestinal nematode infection of mice is associated with increased enterobacterial loads alongside the intestinal tract. PLoS ONE 8:e74026. doi: 10.1371/journal.pone.0074026
Reddy, K. C., Andersen, E. C., Kruglyak, L., and Kim, D. H. (2009). A Polymorphism in npr-1 Is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323, 382–384. doi: 10.1126/science.1166527
Reddy, K. C., Dunbar, T. L., Nargund, A. M., Haynes, C. M., and Troemel, E. R. (2016). The C. elegans CCAAT-enhancer-binding protein gamma is required for surveillance immunity. Cell Rep. 14, 1581–1589. doi: 10.1016/j.celrep.2016.01.055
Reynolds, L. A., Finlay, B. B., and Maizels, R. M. (2015). Cohabitation in the intestine: interactions among helminth parasites, bacterial microbiota, and host immunity. J. Immunol. 195, 4059–4066. doi: 10.4049/jimmunol.1501432
Reynolds, L. A., Smith, K. A., Filbey, K. J., Harcus, Y., Hewitson, J. P., Redpath, S. A., et al. (2014). Commensal-pathogen interactions in the intestinal tract: Lactobacilli promote infection with, and are promoted by, helminth parasites. Gut Microbes 5, 522–532. doi: 10.4161/gmic.32155
Riffel, A., Lucas, F., Heeb, P., and Brandelli, A. (2003). Characterization of a new keratinolytic bacterium that completely degrades native feather keratin. Arch. Microbiol. 179, 258–265. doi: 10.1007/s00203-003-0525-8
Robinson, M., Wahid, F., Behnke, J. M., and Gilbert, F. S. (1989). Immunological relationships during primary infection with Heligmosomoides polygyrus, Nematospiroides dubius): dose-dependent expulsion of adult worms. Parasitology 98, 115–124. doi: 10.1017/S0031182000059758
Roeder, T., Stanisak, M., Gelhaus, C., Bruchhaus, I., Grötzinger, J., and Leippe, M. (2010). Caenopores are antimicrobial peptides in the nematode Caenorhabditis elegans instrumental in nutrition and immunity. Dev. Comp. Immunol. 34, 203–209. doi: 10.1016/j.dci.2009.09.010
Roepstorff, A., Eriksen, L., Slotved, H. C., and Nansen, P. (1997). Experimental Ascaris suum infection in the pig: worm population kinetics following single inoculations with three doses of infective eggs. Parasitology 115, 443–452. doi: 10.1017/S0031182097001480
Rosso, M. N., Pujol, N., and Ewbank, J. J. (2013). “Innate immunity in Caenorhabditis elegans and other nematodes,” in Parasitic Nematodes: Molecular Biology, Biochemistry and Immunology, eds M. W. Kennedy and W. Harnett (Wallingford: CABI), 42–66.
Salvetti, E., Torriani, S., and Felis, G. E. (2012). The genus lactobacillus. probiotics antimicrob. Proteins 4, 217–226. doi: 10.1007/s12602-012-9117-8
Samuel, B. S., Rowedder, H., Braendle, C., Félix, M.-A., and Ruvkun, G. (2016). Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl. Acad. Sci. U.S.A. 113, E3941–E3949. doi: 10.1073/pnas.1607183113
Schulenburg, H., and Boehnisch, C. (2008). Diversification and adaptive sequence evolution of Caenorhabditis lysozymes (Nematoda: Rhabditidae). BMC Evol. Biol. 8:114. doi: 10.1186/1471-2148-8-114
Schulenburg, H., Hoeppner, M. P., Weiner, J. III., and Bornberg-Bauer, E. (2008). Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology 213, 237–250. doi: 10.1016/j.imbio.2007.12.004
Schulenburg, H., Kurz, C. L., and Ewbank, J. J. (2004). Evolution of the innate immune system: the worm perspective. Immunol. Rev. 198, 36–58. doi: 10.1111/j.0105-2896.2004.0125.x
Sender, R., Fuchs, S., and Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533
Shahkolahi, A. M., and Donahue, M. J. (1993). Bacterial flora, a possible source of serotonin in the intestine of adult female Ascaris suum. J. Parasitol. 79, 17–22. doi: 10.2307/3283271
Shivers, R. P., Pagano, D. J., Kooistra, T., Richardson, C. E., Reddy, K. C., Whitney, J. K., et al. (2010). Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet. 6:e1000892. doi: 10.1371/journal.pgen.1000892
Sim, S., and Hibberd, M. L. (2016). Caenorhabditis elegans susceptibility to gut Enterococcus faecalis infection is associated with fat metabolism and epithelial junction integrity. BMC Microbiol. 16:6. doi: 10.1186/s12866-016-0624-8
Sipos, D., Andersson, M., and Ehrenberg, A. (1992). The structure of the mammalian antibacterial peptide cecropin P1 in solution, determined by proton-NMR. Eur. J. Biochem. 209, 163–169. doi: 10.1111/j.1432-1033.1992.tb17273.x
Strandmark, J., Steinfelder, S., Berek, C., Kühl, A. A., Rausch, S., and Hartmann, S. (2016). Eosinophils are required to suppress Th2 responses in Peyer's patches during intestinal infection by nematodes. Mucosal Immunol. doi: 10.1038/mi.2016.93. [Epub ahead of print].
Szewczyk, N. J., Udranszky, I. A., Kozak, E., Sunga, J., Kim, S. K., Jacobson, L. A., et al. (2006). Delayed development and lifespan extension as features of metabolic lifestyle alteration in C. elegans under dietary restriction. J. Exp. Biol. 209, 4129–4139. doi: 10.1242/jeb.02492
Tan, L., and Darby, C. (2004). A movable surface: formation of yersinia sp. biofilms on motile Caenorhabditis elegans. J. Bacteriol. 186, 5087–5092. doi: 10.1128/JB.186.15.5087-5092.2004
Tarr, D. E. K. (2012). Distribution and characteristics of ABFs, cecropins, nemapores, and lysozymes in nematodes. Dev. Comp. Immunol. 36, 502–520. doi: 10.1016/j.dci.2011.09.007
Taylor, M. J., Bandi, C., and Hoerauf, A. (2005). Wolbachia bacterial endosymbionts of filarial nematodes. Adv. Parasitol. 60, 245–284. doi: 10.1016/S0065-308X(05)60004-8
Taylor, M. J., Bilo, K., Cross, H. F., Archer, J. P., and Underwood, A. P. (1999). 16S rDNA phylogeny and ultrastructural characterization of Wolbachia intracellular bacteria of the filarial nematodes Brugia malayi, B. pahangi, and Wuchereria bancrofti. Exp. Parasitol. 91, 356–361. doi: 10.1006/expr.1998.4383
Taylor, M. J., Hoerauf, A., Townson, S., Slatko, B. E., and Ward, S. A. (2014). Anti-Wolbachia drug discovery and development: safe macrofilaricides for onchocerciasis and lymphatic filariasis. Parasitology 141, 119–127. doi: 10.1017/S0031182013001108
Taylor, M. J., Voronin, D., Johnston, K. L., and Ford, L. (2013). Wolbachia filarial interactions. Cell. Microbiol. 15, 520–526. doi: 10.1111/cmi.12084
Tenor, J. L., and Aballay, A. (2008). A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep. 9, 103–109. doi: 10.1038/sj.embor.7401104
Tielens, A. G. M., van Grinsven, K. W. A., Henze, K., van Hellemond, J. J., and Martin, W. (2010). Acetate formation in the energy metabolism of parasitic helminths and protists. Int. J. Parasitol. 40, 387–397. doi: 10.1016/j.ijpara.2009.12.006
Troemel, E. R., Félix, M.-A., Whiteman, N. K., Barrière, A., and Ausubel, F. M. (2008). Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 6:e309. doi: 10.1371/journal.pbio.0060309
Uno, M., Honjoh, S., Matsuda, M., Hoshikawa, H., Kishimoto, S., Yamamoto, T., et al. (2013). A fasting-responsive signaling pathway that extends life span in C. elegans. Cell Rep. 3, 79–91. doi: 10.1016/j.celrep.2012.12.018
Vaishnava, S., Yamamoto, M., Severson, K. M., Ruhn, K. A., Yu, X., Koren, O., et al. (2011). The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258. doi: 10.1126/science.1209791
Vallabhapurapu, S., and Karin, M. (2009). Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 27, 693–733. doi: 10.1146/annurev.immunol.021908.132641
Vejzagić, N., Adelfio, R., Keiser, J., Kringel, H., Thamsborg, S. M., and Kapel, C. M. O. (2015). Bacteria-induced egg hatching differs for Trichuris muris and Trichuris suis. Parasit. Vectors 8:371. doi: 10.1007/s00436-015-4476-1
Vercruysse, J., Albonico, M., Behnke, J. M., Kotze, A. C., Prichard, R. K., McCarthy, J. S., et al. (2011). Is anthelmintic resistance a concern for the control of human soil-transmitted helminths? Int. J. Parasitol. Drugs Drug Resist. 1, 14–27. doi: 10.1016/j.ijpddr.2011.09.002
Vernocchi, P., Del Chierico, F., and Putignani, L. (2016). Gut Microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Front. Microbiol. 7:1144. doi: 10.3389/fmicb.2016.01144
Voronin, D., Cook, D. A. N., Steven, A., and Taylor, M. J. (2012). Autophagy regulates Wolbachia populations across diverse symbiotic associations. Proc. Natl. Acad. Sci. U.S.A. 109, E1638–E1646. doi: 10.1073/pnas.1203519109
Walk, S. T., Blum, A. M., Ewing, S. A.-S., Weinstock, J. V., and Young, V. B. (2010). Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus inflamm. Bowel Dis. 16, 1841–1849. doi: 10.1002/ibd.21299
Walter, J. (2008). Ecological role of Lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl. Environ. Microbiol. 74, 4985–4996. doi: 10.1128/AEM.00753-08
Wang, T., Van Steendam, K., Dhaenens, M., Vlaminck, J., Deforce, D., Jex, A. R., et al. (2013). Proteomic analysis of the excretory-secretory products from larval stages of Ascaris suum reveals high abundance of glycosyl hydrolases. PLoS Negl. Trop. Dis. 7:e2467. doi: 10.1371/journal.pntd.0002467
Wardlaw, A. C., Forsyth, L. M., and Crompton, D. W. (1994). Bactericidal activity in the pig roundworm Ascaris suum. J. Appl. Bacteriol. 76, 36–41. doi: 10.1111/j.1365-2672.1994.tb04412.x
Watson, E., MacNeil, L. T., Ritter, A. D., Yilmaz, L. S., Rosebrock, A. P., Caudy, A. A., et al. (2014). Interspecies systems biology uncovers metabolites affecting C. elegans gene expression and life history traits. Cell 156, 759–770. doi: 10.1016/j.cell.2014.01.047
Weldon, L., Abolins, S., Lenzi, L., Bourne, C., Riley, E. M., and Viney, M. (2015). The gut microbiota of wild mice. PLoS ONE 10:e0134643. doi: 10.1371/journal.pone.0134643
Wescott, R. B. (1968). Experimental Nematospiroides dubius infection in germfree and conventional mice. Exp. Parasitol. 22, 245–249. doi: 10.1016/0014-4894(68)90099-4
World Health Organization (2017). Fact Sheet: Soil-Transmitted Helminth Infections [WWW Document]. WHO. Available online at: http://www.who.int/mediacentre/factsheets/fs366/en/ (accessed 3.27.17).
Wu, S., Li, R. W., Li, W., Beshah, E., Dawson, H. D., and Urban, J. F. (2012). Worm burden-dependent disruption of the porcine colon microbiota by Trichuris suis infection. PLoS ONE 7:e35470. doi: 10.1371/journal.pone.0035470
Yang, Z., Grinchuk, V., Smith, A., Qin, B., Bohl, J. A., Sun, R., et al. (2013). Parasitic nematode-induced modulation of body weight and associated metabolic dysfunction in mouse models of obesity. Infect. Immun. 81, 1905–1914. doi: 10.1128/IAI.00053-13
Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F. O., Ludwig, W., Schleifer, K.-H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645. doi: 10.1038/nrmicro3330
Zaiss, M. M., and Harris, N. L. (2016). Interactions between the intestinal microbiome and helminth parasites. Parasite Immunol. 38, 5–11. doi: 10.1111/pim.12274
Zaiss, M. M., Rapin, A., Lebon, L., Dubey, L. K., Mosconi, I., Sarter, K., et al. (2015). The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 43, 998–1010. doi: 10.1016/j.immuni.2015.09.012
Zam, S. G., and Martin, W. E. (1969). Binding of 60Co-vitamin B12 by Ascaris suum intestine. J. Parasitol. 55, 480–485. doi: 10.2307/3277283
Zasloff, M. (2002). Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. doi: 10.1038/415389a
Zelensky, A. N., and Gready, J. E. (2005). The C-type lectin-like domain superfamily. FEBS J. 272, 6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x
Zhang, H., Yoshida, S., Aizawa, T., Murakami, R., Suzuki, M., Koganezawa, N., et al. (2000). In vitro antimicrobial properties of recombinant ASABF, an antimicrobial peptide isolated from the nematode Ascaris suum. Antimicrob. Agents Chemother. 44, 2701–2705. doi: 10.1128/AAC.44.10.2701-2705.2000
Zhang, Y., Li, W., Li, L., Li, Y., Fu, R., Zhu, Y., et al. (2015). Structural damage in the C. elegans epidermis causes release of STA-2 and induction of an innate immune response. Immunity 42, 309–320. doi: 10.1016/j.immuni.2015.01.014
Zoetendal, E. G., Rajilic-Stojanovic, M., and de Vos, W. M. (2008). High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57, 1605–1615. doi: 10.1136/gut.2007.133603
Keywords: nematode, helminth, microbiota, antimicrobial peptides, antibiotic resistance
Citation: Midha A, Schlosser J and Hartmann S (2017) Reciprocal Interactions between Nematodes and Their Microbial Environments. Front. Cell. Infect. Microbiol. 7:144. doi: 10.3389/fcimb.2017.00144
Received: 22 February 2017; Accepted: 07 April 2017;
Published: 27 April 2017.
Edited by:
Lorenza Putignani, Bambino Gesù Ospedale Pediatrico (IRCCS), ItalyReviewed by:
Mario M. Zaiss, Universitätsklinikum Erlangen, GermanyCopyright © 2017 Midha, Schlosser and Hartmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Susanne Hartmann, c3VzYW5uZS5oYXJ0bWFubkBmdS1iZXJsaW4uZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.