
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 01 February 2017
Sec. Molecular Bacterial Pathogenesis
Volume 7 - 2017 | https://doi.org/10.3389/fcimb.2017.00024
 Yinjuan Guo1†
Yinjuan Guo1† Shanshan Wang1†
Shanshan Wang1† Lingling Zhan1
Lingling Zhan1 Ye Jin1
Ye Jin1 Jingjing Duan1
Jingjing Duan1 Zhihao Hao1
Zhihao Hao1 Jingnan Lv1
Jingnan Lv1 Xiuqin Qi1
Xiuqin Qi1 Liang Chen2
Liang Chen2 Barry N. Kreiswirth2
Barry N. Kreiswirth2 Liangxing Wang3*
Liangxing Wang3* Fangyou Yu1*
Fangyou Yu1*A distinctive syndrome caused by hypermucoviscous Klebsiella pneumoniae (HMKP) including pyogenic liver abscess (PLA) is now becoming a globally emerging disease. In the present study, 22.8% (84/369) of K. pneumoniae clinical isolates associated with various types of invasive infections were identified as HMKP, with 45.2% associated with PLA. Multivariate regression analysis showed that male patients with 41–50 years, PLA, diabetes mellitus, and hypertension were independent risk factors for HMKP infections. K2 (42.9%, 36/84) was the most common capsular serotype among HMKP isolates, followed by K1 (23.8%, 20/84). Seventy-five percentage of K1 HMKP isolates were associated with PLA, while K2 HMKP isolates accounted for more types of invasive infections. The positive rates of iutA, mrkD, aerobactin, iroN, and rmpA among HMKP isolates were significantly higher than those among non-HMKP isolates (p < 0.05). There was a correlation between magA, ybtS, alls, and wcaG and K1 isolates. Interestingly, mrkD was exclusively detected among HMKP (32.1%, 27/84) and K2 isolates (65.9%, 27/41). All K1 and K2 HMKP and non-HMKP isolates were positive for rmpA. Aerobactin was found among 95.0 and 97.5% of K1 and K2 isolates. ST23 was found to be the most prevalent ST among 69 HMKP isolates with K1, K2, K5, K20, and K57 (27.5%, 19/69) and was only found among K1 isolates. ST65 was the second most prevalent ST (26.1%, 18/69) and was also only found among K2 isolates. ST23-K1 HMKP isolates (84.2%, 16/19) were associated with PLA, while ST65-K2 isolates were correlated with more types of infections relative to ST23-K1 isolates. PFGE results showed that the homology of 84 HMKP isolates was diverse. Only five PFGE clusters with more than 75% similarity accounted for more than three isolates. These five PFGE clusters only accounted for 35 (41.7%, 35/84) isolates. In conclusion, our study first found that hypertension and male patients with 41–50 years old were independent risk factors. The composition of ST types and PFGE clusters among K. pneumoniae K2 isolates was more diverse than K1 isolates. K1 and K2 HMKP isolates had respective specific profiles of virulence-associated genes.
Klebsiella pneumoniae is a frequently-isolated and well-established bacterial pathogen responsible for numerous infections in hospitals and long-term care facilities worldwide (Shon et al., 2013). Infections caused by K. pneumoniae can involve lung, urinary tract, surgical sites, abdominal cavity, intra-vascular devices, soft tissues, and subsequent bacteremia (Shon et al., 2013). Between the mid-1980s and 1990s, reports from Taiwan described a distinctive syndrome of community-acquired invasive K. pneumoniae infections, mainly in the form of pyogenic liver abscesses (PLA; Liu et al., 1986; Cheng et al., 1991; Wang et al., 1998). These infections are often complicated by devastating metastatic infections including endophthalmitis and meningitis in younger and healthy individuals (Struve et al., 2015). The K. pneumoniae strains associated with these serious infections are defined as hyper-virulent (hypermucoviscous) K. pneumonia (hvKP), with a distinct hypermucoviscosity phenotype when grown on agar plates (Struve et al., 2015). Although these infections were reported initially within Southeast Asia including Taiwan, South Korea, and Japan, an increasing number of cases were reported from North America, Europe, South America, the Middle East, Australia, Africa and China, indicates that this unique syndrome is becoming a globally emerging disease (Fang et al., 2005; Siu et al., 2012; Shon et al., 2013; Bialek-Davenet et al., 2014; Yang et al., 2014; Struve et al., 2015; Prokesch et al., 2016). A combination of clinical and bacterial phenotypic features is used for distinguishing hvKP from classic K. pneumoniae (cKP) strains. The hvKP isolates characteristically express a distinct hypermucoviscosity phenotype determined by string test when grown on agar plates and most of these isolates belong to capsular serotype K1 and K2 (Fang et al., 2007; Shon and Russo, 2012; Cheng et al., 2015). A number of putative virulence factors, mainly including mucoviscosity-associated gene A (magA) and regulator of mucoid phenotype A (rmpA), have been found to be associated with hvKP (Fang et al., 2004; Yu et al., 2006). Recently, aerobactin accounting for increased siderophore production was found to be a major virulence determinant and new defining trait for hvKP (Russo et al., 2014). A multiplex PCR assay targeting seven virulence factors and the wzi gene specific for the K1 and K2 K. pneumoniae was developed for the surveillance of emerging highly virulent strains (Compain et al., 2014; Russo et al., 2014). Alarmingly, increasing studies reported that multi-drug-resistant, even carbapenem-resistant hvKP isolates have been emerged (Yang et al., 2014; Yao et al., 2015; Zhang et al., 2015), which is becoming an important threat to public health. Recently, many studies from China described the prevalence, clinical presentations and epidemiology of hvKP isolates (Liu et al., 2014; Yang et al., 2014; Qu et al., 2015; Yao et al., 2015; Sun et al., 2016; Zhao et al., 2016). However, the limitations of these studies included limited number of K. pnuemoniae involved, short span of investigation and focusing a specific infection. In the present study, we investigated the prevalence, clinical characteristics, and molecular epidemiology of hypermucoviscous Klebsiella pneumoniae (HMKP) isolates with hypermucoviscosity phenotype determined by the string test among 369 K. pneumoniae isolates associated with various invasive infections from January 2013 to October 2015.
From January 2013 to October 2015, a total of 369 K. pneumoniae isolates were consecutively isolated from the specimens of patients with various invasive infections at the first Affiliated Hospital of Wenzhou Medical University located in Wenzhou, east China. The K. pneumoniae isolates from the specimens of respiratory tract, urinary tract, and intestinal tract were excluded in this investigation, because it is difficult to discriminate invasive isolates from colonizing isolates. K. pneumoniae isolates were identified by Gram-staining and a VITEK-2 automated platform (bioMérieux, Marcy l'Etoile, France). Staphylococcus aureus ATCC25923, Escherichia coli ATCC25922, and Pseudomonas aeruginosa ATCC87253 were used as control strains for the identification of bacterial clinical isolates. Acquiring the information in the medical records of the patients and the K. pneumoniae isolates for research purposes were approved by the ethical committee of the first Affiliated Hospital of Wenzhou Medical University. Informed consents were obtained from the patients involved.
The susceptibility of the K. pneumoniae clinical isolates included to clinically often used antimicrobial agents was also determined using VITEK-2 automated platform (bioMérieux, Marcy l'Etoile, France) in accordance with the manufactory' s instructions. Escherichia coli ATCC25922 was used as control strain for determining the antimicrobial susceptibility for K. pneumoniae clinical isolates.
The K. pneumoniae invasive isolates with hypermucoviscosity phenotype determined by string test described previously were identified as HMKP (Shon et al., 2013). Briefly, when an inoculation loop or needle was able to generate a viscous string >5 mm in length by stretching K. pneumoniae colonies on Columbia blood agar plate (BIO-KONT, Wenzhou, China), the isolate was considered to be positive for the string test and was defined as HMKP.
K1, K2, K5, K20, K54, and K57 capsular serotypes were identified by PCR as described previously (Fang et al., 2007; Turton et al., 2010). Seventeen virulence-associated genes, including ybtS, ureA, uge, wabG, ycf, entB, iutA, vatD, magA, fimH, mrkD, aerobactin, iroN, kfuB, rmpA, wcaG, and alls, were determined by PCR using primers described previously (Yu et al., 2006, 2008; Turton et al., 2010; Candan and Aksöz, 2015) for all HMKP and 95 selected randomly non-HMKP isolates. The isolates with virulence-associated genes determined by PCR and DNA sequencing were selected as positive control strains for the subsequent PCR experiments.
MLST was performed on HMKP isolates by amplifying internal fragments of the seven standard housekeeping loci (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) as described previously (Diancourt et al., 2005). Sequence types (STs) were identified using the online database on the Pasteur Institute MLST website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
PFGE was performed on 84 HMKP isolates using XbaI digestion for 12 h at 37°C and electrophoresis for 20 h at 14°C, 120 angle, with switch times of 6 and 36 s at 6 V/cm, Bio-Rad CHEF III system. Comparison of the PFGE patterns was performed with Bionumerics software (Applied Maths, Sint-Martens-Latem, Belgium) using the Dice Similarity coefficient. More than 75 percentage of similarity was used for the threshold for clustering in PFGE pattern analysis.
Statistical analysis was performed using SPSS statistical software (version 19, IBM SPSS Statistics). The χ2 or Fisher's exact tests were used for categorical variables. Logistic regression with univariate model and multivariate model was used to identify variables associated with HMKP infections. P < 0.05 was considered statistically significant. Diabetes, hypertension, liver abscess, sex, age, and leukemia were used for the logistic regression model.
Three hundred and sixty-nine K. pneumoniae invasive isolates were obtained from clinical specimens including 129 blood (35.0%), 133 pus (35.4%), 51 drainage (13.6%), 2 pleural effusion (0.5%), 20 bile (5.3%), 8 ascites (2.1%), 9 tissue (2.4%), 13 catheter tip (3.5%), and 3 cerebrospinal fluid (0.8%). ≤60 and >60-year old patients accounted for 162 (43.9%) and 207 patients (56.1%). The male and female patients accounted for 62.1% (229) and 37.9% (140). Among 369 K. pneumoniae invasive isolates, 84 (22.8%) were positive for the string test and were identified as HMKP. The proportions of HMKP isolates among K. pneumoniae invasive isolates in 2013, 2014, and 2015 were 23.2% (23/99), 24.3% (33/136), and 20.9% (28/134), respectively, with a stable HMKP prevalence.
The proportions of HMKP among K. pneumoniae isolates from different specimens were as follows: blood, 17.1% (22/129); pus, 32.3% (43/133); drainage, 23.5% (12/51); pleural effusion, 100% (2/2); bile, 5% (1/20); ascites, 25% (2/8); tissue, 11.1% (1/9); and catheter tip, 7.7% (1/13), respectively. These HMKP isolates were associated with various types of invasive infections including PLA (37, 44.0%), bacteremia (21 including seven associated with PLA, 25.0%), abdominal infections (13, 14.8%), skin and soft tissue infections (SSTIs; nine including two causing necrotic fascilitis, 10.7%), and lung abscess (4, 4.8%).
The clinical characteristics of patients with HMKP and non-HMKP infections were showed in Table 1. In the present study, there were no differences between the proportions of HMKP among male and female patients regardless of age, as well as among patients with >60 years old and ≤60 years old regardless of sex. Further investigation found that HMKP prevalence in male patients with 41–50 years old was 40.6% (13/32), while no HMKP was found in female patients with the same age. Univariate analysis revealed that male patients with 41–50 years (OR = 2.554) was found to be statistically significant risk factors for HMKP infections. Multivariate regression analysis further demonstrated that male patients with 41–50 years [aOR = 2.482, 95%CI(1.069–5.760)] was an independent risk factor for HMKP infections (Table 2). Diabetes mellitus was found among 29.8% (25/84) of patients with HMKP infections and 16.8% (48/285) of patients with HMKP infections. Multivariate analysis showed diabetes mellitus [aOR = 3.417, 95%CI(1.632–7.155)] was an independent risk factor for HMKP infections (Table 2).
Of 84 HMKP isolates, capsular serotype K1 (23.8%, 20/84), K2 (42.9%, 36/84), K5 (2.4%, 2/84), K20 (4.8%, 4/84), and K57 (9.6%, 8/84) were identified, while K54 was not found in any of the isolates tested. Fourteen HMKP isolates (16.7%) were not typed successfully and were defined as K-non-typable isolates. K1 prevalence among HMKP isolates (23.8%, 20/84) was similar to that among non-HMKP isolates (23.2%, 22/95). However, more HMKP isolates belonged to K2 relative to non-HMKP isolates (6.3%, 6/95). Among 38 HMKP isolates associated with PLA, 15 (37.5%), 14 (35.0%), 4 (10.0%), and 1 (2.5%) belonged to K1, K2, K57, and K20, respectively. The capsular serotypes of 22 HMKP isolates associated with bacteremia were K1 (18.2%, 4/22), K2 (22.7%, 5/22), K57 (18.2%, 4/22), K20 (9.1%, 2/22), and K2 (4.5%, 1/22) and K-non-typable capsular serotype (27.3%, 6/22). 6, 2, and 1 of 9 HMKP causing SSTIs belonged to K2, K1, and K5. Among 13 isolates associated with abdominal infections, 3, 2, 2, and 1 belonged to K2, K20, K57, and K1, respectively. Three K2 and one K57 were associated with lung abscess.
HMKP isolates were more susceptible to clinically often used antimicrobial agents relative to non-HMKP isolates (Table 3). All HMKP and non-HMKP isolates were resistant to ampicillin. The resistance rates of 84 HMKP isolates to cefotetan, cefepime, ertapenem, imipenem, piperacillin/tazobactam, and trimethoprim-sulfamethoxazole were 4.8% (4/84). 7.1% (6/84) HMKP isolates were resistant to aztreonam, ceftazidime, and ciprofloxacin. 5.9% (5/84) HMKP isolates were resistant to ciprofloxacin, levofloxacin, and gentamicin. 8 (9.5%), 7(8.3%), 3(3.6%), and 3(3.6%) HMKP isolates were resistant to ampicillin/sulbatam, ceftriaxone, tobramycin, and amikacin, respectively. In the present study, three HMKP isolates were resistant to all antimicrobial agents except trimethoprim-sulfamethoxazole and four carbapenem-resistant HMKP isolates were also found. The resistance rates of HMKP isolates to amicillin/sulbatam, aztreonam, ceftriaxone, ceftazidime, ciprofloxacin, levofloxacin, tobramycin, gentamicin, and trimethoprim-sulfamethoxazole were significantly lower than non-HMKP isolates (p < 0.05; Table 3).
The positive rates of 17 virulence-associated genes tested among HMKP, non-HMKP, K1 and K2 isolates were showed in Table 4. The virulence-associated genes with more than 50% of positive rates among HMKP and non-HMKP isolates tested included entB (91.7 and 94.7%), fimH (77.4 and 89.5%), uge (90.5 and 91.6%), ureA (97.6 and 94.7%), wabG (96.3 and 94.7%), and ycf (90.5 and 93.7%). The positive rates of iutA, mrkD, aerobactin, iroN, and rmpA among HMKP isolates were significantly higher than those among non-HMKP isolates (p < 0.05). Ninety-file percentage of K1 HMKP isolates harbored magA, while this important virulence-associated gene was not found in any of K2 isolates, indicating that there was a strongly correlation between magA gene and K1 HMKP isolates (p < 0.01). Interestingly, mrkD was exclusively detected among HMKP (32.1%, 27/84) and K2 isolates (65.9%, 27/41). The positive rates of ybtS, alls, and wcaG among K1 isolates were significantly higher than those among K2 isolates (p < 0.01). All K5, K20, and K57 isolates were negative for alls. All K5 and K57 isolates were negative for wcaG. The positive rates of the remaining genes tested were similar between K1 and K2 isolates.
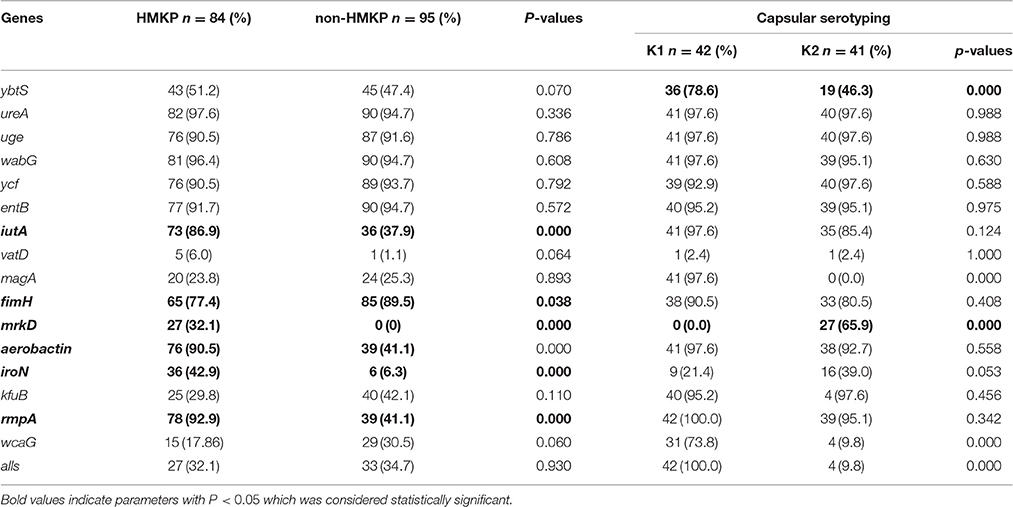
Table 4. The proportions of virulence-associated genes among 84 HMKP, 95 non-HMKP, K1 and K2 isolates.
MLST was performed on the HMKP isolates with K1, K2, K5, K20, and K57 (70 isolates) and not on the K-non-typable isolates (14 isolates). Among 70 isolates tested, 21 STs were identified and one K2 isolate with seven loci not match STs in MLST database was defined as non-typable. ST23 was the most prevalent ST (27.1%, 19/70), followed by ST65 (25.7%, 18/70). The STs accounting for four isolates (5.7%, 4/70) were ST375, ST86, and ST268. ST412 was found in three isolates (4.3%, 3/70). ST25 and ST592 were identified among two isolates (2.9%, 2/70). The remaining STs were found only one isolate (Figure 1). ST23 was only found among K1 isolates, with ST23 accounting for all K1 isolates except one isolate belonging to single locus variant of ST23, ST 1265 (Figure 1). Similarly, ST65 was found exclusively among K2 isolates tested (Figure 1).

Figure 1. Clonal relatedness of 84 HMKP isolates. ps, pus; bl, blood; su, drainage; pf, pleural effusion; bi, bile; ti, tissue; ab, ascites; ca, catheter tip.
Our study also found that ST23-K1 HMKP isolates (78.9%, 15/19) were strongly associated with PLA (p < 0.05), while ST65-K2 isolates were correlated with more types of infections relative to ST23-K1 isolates, with 44.4% (8/18) associated with PLA. ST86, ST375, ST25, and ST380 were also exclusively identified among K2 isolates. Not only was ST268 exclusively found among K20 isolates, but also all four K20 isolates belonged to ST268. Three ST412 and two ST592 isolates belonged to K57. Among 20 K1 isolates, only ST23 (19 isolates) and its single locus variant, ST 1265 (one isolate) were identified. On the contrast, 11 STs and an unknown ST were identified among 36 K2 HMKP isolates, including ST65 (50.0%, 18/36), ST86 (11.1%, 4/36), ST375 (11.1%, 4/36), and ST25 (5.6%, 2/36; Figure 1). There were five STs including ST412, ST592, ST1336, ST161, and ST660 among eight K57 isolates. Two STs, ST54, and ST686, were found among two K5 isolates. Nine isolates causing SSTIs belonged to six STs (ST54, ST23, ST375, ST65, ST86, and ST1106). Six STs (ST268, ST412, ST161, ST23, ST875, and ST65) were found among eight MLST-typed isolates which were associated with abdominal infections. Among 16 isolates typed by MLST which were associated with bacteremia, 10 STs were identified, with no prevalent ST. Although 12 STs were found among 34 MLST-typed isolates causing PLA, 44.1% (15/34) and 23.5% (8/34) belonged to ST23-K1 and ST65-K2.
PFGE results showed that the homology of 84 HMKP isolates was diverse (Figure 1). Only five PFGE clusters with more than 75% similarity accounted for more than three isolates. These five PFGE clusters only accounted for 35 (41.7%, 35/84) isolates. PFGE cluster A accounted for six isolates including four ST23-K1, one ST65-K2, and one ST86-K2 isolate. PFGE cluster B accounted for three isolates including two ST23-K1 and one K-non-typable isolate. PFGE cluster C accounted for 11 isolates including nine ST23-K1, one ST1265-K1, and one K-non-typable isolate. PFGE cluster D accounted for 12 isolates including 11 ST65-K2 and one K-non-typable isolate. PFGE cluster E accounted for three ST375-K2 isolates.
A recent multicenter study from China showed that the prevalence of HMKP among K. pneumoniae isolates causing various types infections including PLA, bloodstream infections, hospital-acquired pneumonia, and intra-abdominal infections in10 cities of China during February to July 2013 were 37.8% (Zhang et al., 2016). Another Chinese study conducted in a single center in Beijing, China, showed that the prevalence of HMKP was 33% (Yang et al., 2014). Yan et al. reported that 14 (28.6%) of 49 K. pneumoniae isolates associated with ventilator-associated pneumonia from a university hospital in China from January 2014 to December 2014 were HMKP (Yan et al., 2016). The HMKP prevalence of our study was lower than those Chinese reports mentioned above, but significantly higher than the reports from a teaching hospital in Spain between 2007 and 2013 (5.4%, 53/878; Cubero et al., 2016) and a surveillance study in Alberta, Canada (8.2%; Peirano et al., 2013). Sun et al. reported 81.6% (31/38) of K. pneumoniae causing PLA were HMKP determined by the string test (Sun et al., 2016), which was significantly higher than that in our investigation. A report from Taiwan also showed that 90% of K. pneumoinae associated PLA were HMKP (Yu et al., 2008). However, in another Chinese study, only 28.9% (13/45) of K. pneumoniae isolates causing PLA were HMKP exhibiting hypermucoviscosity phenotype (Qu et al., 2015), which was lower than that in the present study. These data indicated that the proportion of PLA caused by HMKP showed a varied geographic distribution. Li et al. found that neither age nor sex was associated with hypermucoviscosity phenotype determined by string test (Yang et al., 2014). Zhang et al. also found age and sex were not correlated with HMKP (Zhang et al., 2016). However, HMKP prevalence in male ≤60-year old patients was significantly higher than that in female ≤60-year old patients in the present study. Our further investigation indicated that male patients with 41–50 years predisposed to HMKP infections. Diabetes mellitus has been considered as a significant risk factor for HMKP infection (Cheng et al., 1991; Wang et al., 1998; Shon et al., 2013; Zhang et al., 2016). However, other studies did not found this correlation (Yu et al., 2007; Liu et al., 2014; Yang et al., 2014; Yan et al., 2016). Zhang et al. reported that the proportion (33.8%) of patients with cancer among patients with HMKP infections was significantly higher than that (18.8%) among patients with cKP infections and showed that cancer (OR = 2.285) appeared to be independent variable associated with HMKP by multivariate analysis. However, in the present study, only two patients (2.4%) with HMKP infections were found to have solid tissue cancer while 9.2% of patients with non-HMKP infections had solid tissue cancer. Interestingly, we also found that there was an association between hypertension and HMKP infections (11.9 vs. 0.70%) and hypertension (OR = 7.333) was an independent risk factor for HMKP infections determined by multivariate regression analysis. Leukemia (OR = 0.190) was negatively correlated with HMKP infections (p < 0.05) determined by univariate analysis, but not by multivariate analysis.
Although HMKP isolates are usually more susceptible to clinically often used antimicrobial agents relative to non-HMKP isolates, more and more HMKP isolates were found to be multi-resistant to antimicrobial agents, even to carbapenems (Yang et al., 2014; Yao et al., 2015; Wei et al., 2016). In the present study, four HMKP isolates were found to be resistant to carbapenem. Acquisition of hyper virulence and carbapenem resistance poses major problems in the management of K. pneumoniae infection.
In contrast to previous reports with K1 being the most prevalent capsular serotype among HMKP isolates (Liu et al., 2014; Yang et al., 2014; Qu et al., 2015; Yan et al., 2016; Zhang et al., 2016), our study showed that K2 was the most common capsular serotype and K2 HMKP isolates were associated with more types of invasive infections than K1 isolates. In the present study, 95.2% of K. pneumoniae isolates with hypermucoviscosity phenotype were positive for rmpA, especially all K1, K2, K5, K20, and K57 isolates positive for rmpA, which was similar to a multicenter investigation from China (97.7%; Zhang et al., 2016), but was significantly higher than another report from Beijing, China (55%; Zhang et al., 2016). Surprisingly, all 14 isolates with K-non-tyable capsular serotypes were negative for rmpA. Previous studies showed that there was a correlation between rmpA gene and virulence in terms of abscess formation for HMKP isolates (Yu et al., 2006; Yang et al., 2014; Yan et al., 2016; Zhang et al., 2016). However, a recent study from China showed that all K. pneumoniae isolates causing PLA were positive for rmpA, regardless of hypermucoviscos phenotype(Qu et al., 2015). In our investigation, 41.1% of non-HMKP isolates were found to be positive for rmpA. The hypermucoviscosity phenotype in K. pneumoniae isolates is associated with the carriage of chromosomally encoded magA, which is characteristic of the K1 capsular operon (Fang et al., 2004, 2005; Chuang et al., 2006). magA has been described as the causative gene for K. pneumoniae isolates associated with PLA and septic metastatic complications(Fang et al., 2005; Chuang et al., 2006). Yan et al. reported that all K. pneumoniae isolates including HMKP and non-HMKP causing ventilator-associated pneumonia were positive for mrkD (Yan et al., 2016). Another study showed that 96.3% of K. pneumoniae isolates were positive for mrkD (El Fertas-Aissani et al., 2013). However, our study found that mrkD was only found among HMKP isolates and only among K2 isolates. To the best of our knowledge, this is the first report of the association between mrkD and K2 HMKP isolates. Our study also showed alls, ybtS, and wcaG were correlated with K1 isolates. Although Yu et al. reported that there was a strong association between kfuB and alls and K1 isolates, with all K1 isolates positive for kfuB and alls and all K2 isolates negative for these two genes (Yu et al., 2008), kfuB and alls were found among 8.5 and 11.1% of K2 HMKP isolates in the present study. aerobactin is important virulence determinant for hvKP even used as the marker for the identification of hvKP, instead of hypermucoviscosity phenotype determined by the string test (Zhang et al., 2016). In the present study, 95.1 and 41.1% of HMKP and non-HMKP isolates were found to harbor aerobactin, indicating that hypermucoviscosity phenotype as the marker for the identification of hvKP is controversial.
ST23 has been found to be the most commonly described ST among HMKP isolates and is strongly correlated with capsular serotype K1 and liver abscess (Turton et al., 2007; Chung et al., 2008; Shon et al., 2013). Similar to previous reports, our data showed that ST23 was the most prevalent among K1 isolates. ST57 and ST82 are also found to be associated with the K1 serotype and PLA (Brisse et al., 2009; Merlet et al., 2012). However, these two STs were not found in the present study. Previous studies showed that ST268 was only found among K20 isolates (Liu et al., 2014; Lin et al., 2015; Yan et al., 2015, 2016; Zhang et al., 2016). In the present study, we also found that there was an association between ST268 and K20 isolates. Eight different STs including ST65, ST66, ST86, ST373, ST374, ST375, ST380, and ST434 was found among capsular serotype K2 isolates from Singapore, Hong Kong, and Taiwan (Lin et al., 2014). Eleven STs were identified among K2 isolates associated with different types of infections from Taiwan, among which ST65 was the most common (n = 10), followed by ST86, ST373, and ST375 (Liao et al., 2014). In the present study, 16 (80%) of 20 K1 isolates belonged to PFGE cluster B or C and no K1 isolates belonged to PFGE cluster A, D, E, F, or G. However, K2 isolates belonged to more PFGE clusters including cluster A, B, D, E, and F. Our data support further the evidence that the composition of ST types and PFGE clusters among K. pneumoniae K2 isolates was more diverse than K1 isolates. Our data also indicated that the distributions of STs among HMKP isolates causing bacteremia, abdominal infections and SSTIs were more diverse than that associated with PLA.
In conclusion, our study first found that hypertension and male patients with 41–50 years old were independent risk factors. The composition of ST types and PFGE clusters among K. pneumoniae K2 isolates was more diverse than K1 isolates. K1 and K2 HMKP isolates had respective specific profiles of virulence-associated genes.
YG, SW, LZ, YJ, JD, ZH, JL, XQ isolated bacteria and performed the laboratory measurements. FY and LW made substantial contributions to conception and design. LC and BK revised the manuscript critically for important intellectual content. LC and JL participated in experimental design and data analysis. FY drafted the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported in part by National Institutes of Health (NIH) Grant R01AI090155 (to BK) and R21AI117338 (to LC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Bialek-Davenet, S., Criscuolo, A., Ailloud, F., Passet, V., Jones, L., Delannoy-Vieillard, A. S., et al. (2014). Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg. Infect. Dis. 20, 1812–1820. doi: 10.3201/eid2011.140206
Brisse, S., Fevre, C., Passet, V., Issenhuth-Jeanjean, S., Tournebize, R., Diancourt, L., et al. (2009). Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS ONE 4:e4982. doi: 10.1371/journal.pone.0004982
Candan, E. D., and Aksöz, N. (2015). Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim. Pol. 62, 867–874. doi: 10.18388/abp.2015_1148
Cheng, D. L., Liu, Y. C., Yen, M. Y., Liu, C. Y., and Wang, R. S. (1991). Septic metastatic lesions of pyogenic liver abscess. Their association with Klebsiella pneumoniae bacteremia in diabetic patients. Arch. Intern. Med. 151, 1557–1559. doi: 10.1001/archinte.1991.00400080059010
Cheng, K. C., Lee, M. F., Chuang, Y. C., and Yu, W. L. (2015). First description of lung abscess caused by ST23 clone capsule genotype K1 Klebsiella pneumoniae. J. Formos. Med. Assoc. 114, 379–380. doi: 10.1016/j.jfma.2013.08.008
Chuang, Y. P., Fang, C. T., Lai, S. Y., Chang, S. C., and Wang, J. T. (2006). Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 193, 645–654. doi: 10.1086/499968
Chung, D. R., Lee, H. R., Lee, S. S., Kim, S. W., Chang, H. H., Jung, S. I., et al. (2008). Evidence for clonal dissemination of the serotype K1 Klebsiella pneumoniae strain causing invasive liver abscesses in Korea. J. Clin. Microbiol. 46, 4061–4063. doi: 10.1128/JCM.01577-08
Compain, F., Babosan, A., Brisse, S., Genel, N., Audo, J., Ailloud, F., et al. (2014). Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J. Clin. Microbiol. 52, 4377–4380. doi: 10.1128/JCM.02316-14
Cubero, M., Grau, I., Tubau, F., Pallarés, R., Dominguez, M. A., Liñares, J., et al. (2016). Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin. Microbiol. Infect. 22, 154–160. doi: 10.1016/j.cmi.2015.09.025
Diancourt, L., Passet, V., Verhoef, J., Grimont, P. A., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
El Fertas-Aissani, R., Messai, Y., Alouache, S., and Bakour, R. (2013). Virulence profiles and antibiotic susceptibility patterns of Klebsiella pneumoniae strains isolated from different clinical specimens. Pathol. Biol. 61, 209–216. doi: 10.1016/j.patbio.2012.10.004
Fang, C. T., Chuang, Y. P., Shun, C. T., Chang, S. C., and Wang, J. T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Fang, C. T., Lai, S. Y., Yi, W. C., Hsueh, P. R., Liu, K. L., and Chang, S. C. (2007). Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin. Infect. Dis. 45, 284–293. doi: 10.1086/519262
Fang, F. C., Sandler, N., and Libby, S. J. (2005). Liver abscess caused by magA+ Klebsiella pneumoniae in North America. J. Clin. Microbiol. 43, 991–992. doi: 10.1128/JCM.43.2.991-992.2005
Liao, C. H., Huang, Y. T., Chang, C. Y., Hsu, H. S., and Hsueh, P. R. (2014). Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur. J. Clin. Microbiol. Infect. Dis. 33, 365–369. doi: 10.1007/s10096-013-1964-z
Lin, J. C., Koh, T. H., Lee, N., Fung, C. P., Chang, F. Y., Tsai, Y. K., et al. (2014). Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut. Pathog. 6:21. doi: 10.1186/1757-4749-6-21
Lin, Y. T., Wang, Y. P., Wang, F. D., and Fung, C. P. (2015). Community-onset Klebsiella pneumoniae pneumonia in Taiwan: clinical features of the disease and associated microbiological characteristics of isolates from pneumonia and nasopharynx. Front. Microbiol. 9:122. doi: 10.3389/fmicb.2015.00122
Liu, Y. C., Cheng, D. L., and Lin, C. L. (1986). Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch. Intern. Med. 146, 1913–1916. doi: 10.1001/archinte.1986.00360220057011
Liu, Y. M., Li, B. B., Zhang, Y. Y., Zhang, W., Shen, H., Li, H., et al. (2014). Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob. Agents Chemother. 58, 5379–5385. doi: 10.1128/AAC.02523-14
Merlet, A., Cazanave, C., Dutronc, H., de Barbeyrac, B., Brisse, S., and Dupon, M. (2012). Primary liver abscess due to CC23-K1 virulent clone of Klebsiella pneumoniae in France. Clin. Microbiol. Infect. 18, E338–E339. doi: 10.1111/j.1469-0691.2012.03953.x
Peirano, G., Pitout, J. D., Laupland, K. B., Meatherall, B., and Gregson, D. B. (2013). Population-based surveillance for hypermucoviscosity Klebsiella pneumoniae causing community-acquired bacteremia in Calgary, Alberta. Can. J. Infect. Dis. Med. Microbiol. 24, e61–e64.
Prokesch, B. C., Tekippe, M., Kim, J., Raj, P., Tekippe, E. M., and Greenberg, D. E. (2016). Primary osteomyelitis caused by hypervirulent Klebsiella pneumoniae. Lancet Infect. Dis. 16, e190–e195. doi: 10.1016/S1473-3099(16)30021-4
Qu, T. T., Zhou, J. C., Jiang, Y., Shi, K. R., Li, B., Shen, P., et al. (2015). Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect. Dis. 15, 161. doi: 10.1186/s12879-015-0899-7
Russo, T. A., Olson, R., Macdonald, U., Metzger, D., Maltese, L. M., Drake, E. J., et al. (2014). Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect. Immun. 82, 2356–2367. doi: 10.1128/IAI.01667-13
Shon, A. S., Bajwa, R. P., and Russo, T. A. (2013). Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4, 107–118. doi: 10.4161/viru.22718
Shon, A. S., and Russo, T. A. (2012). Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol 7, 669–671. doi: 10.2217/fmb.12.43
Siu, L. K., Yeh, K. M., Lin, J. C., Fung, C. P., and Chang, F. Y. (2012). Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect. Dis. 12, 881–887. doi: 10.1016/S1473-3099(12)70205-0
Struve, C., Roe, C. C., Stegger, M., Stahlhut, S. G., Hansen, D. S., Engelthaler, D. M., et al. (2015). Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio 6, e00630. doi: 10.1128/mBio.00630-15
Sun, Y., Wu, H., and Shen, D. (2016). Clinical and Molecular analysis of Klebsiella pneumoniae causing liver abscess in China. J. Mol. Microbiol. Biotechnol. 26, 245–251. doi: 10.1159/000444367
Turton, J. F., Englender, H., Gabriel, S. N., Turton, S. E., Kaufmann, M. E., and Pitt, T. L. (2007). Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J. Med. Microbiol. 56, 593–597. doi: 10.1099/jmm.0.46964-0
Turton, J. F., Perry, C., Elgohari, S., and Hampton, C. V. (2010). PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J. Med. Microbiol. 59, 541–547. doi: 10.1099/jmm.0.015198-0
Wang, J. H., Liu, Y. C., Lee, S. S., Yen, M. Y., Chen, Y. S., Wang, J. H., et al. (1998). Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 26, 1434–1438. doi: 10.1086/516369
Wei, D.-D., Wan, L.-G., Deng, Q., and Liu, Y. (2016). Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in Mainland China. Diagn. Microbiol. Infect. Dis. 85, 192–194. doi: 10.1016/j.diagmicrobio.2015.03.012
Yan, J. J., Zheng, P. X., Wang, M. C., Tsai, S. H., Wang, L. R., and Wu, J. J. (2015). Allocation of Klebsiella pneumoniae bloodstream isolates into four distinct groups by ompk36 typing in a taiwanese university hospital. J. Clin. Microbiol. 53, 3256–3263. doi: 10.1128/JCM.01152-15
Yan, Q., Zhou, M., Zou, M., and Liu, W. E. (2016). Hypervirulent Klebsiella pneumoniae induced ventilator-associated pneumonia in mechanically ventilated patients in China. Eur. J. Clin. Microbiol. Infect. Dis. 35, 387–396. doi: 10.1007/s10096-015-2551-2
Yang, Z., Liu, W., Cui, Q., Niu, W., Li, H., Zhao, X., et al. (2014). Prevalence and detection of Stenotrophomonas maltophilia carrying metallo-beta-lactamase blaL1 in Beijing, China. Front. Microbiol. 5:692. doi: 10.3389/fmicb.2014.00692
Yao, B., Xiao, X., Wang, F., Zhou, L., Zhang, X., and Zhang, J. (2015). Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int. J. Infect. Dis. 37, 107–112. doi: 10.1016/j.ijid.2015.06.023
Yu, V. L., Hansen, D. S., Ko, W. C., Sagnimeni, A., Klugman, K. P., von Gottberg, A., et al. (2007). Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg. Infect. Dis. 13, 986–993. doi: 10.3201/eid1307.070187
Yu, W. L., Ko, W. C., Cheng, K. C., Lee, C. C., Lai, C. C., and Chuang, Y. C. (2008). Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn. Microbiol. Infect. Dis. 62, 1–6. doi: 10.1016/j.diagmicrobio.2008.04.007
Yu, W. L., Ko, W. C., Cheng, K. C., Lee, H. C., Ke, D. S., Lee, C. C., et al. (2006). Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin. Infect. Dis. 42, 1351–1358. doi: 10.1086/503420
Zhang, Y., Zeng, J., Liu, W., Zhao, F., Hu, Z., Zhao, C., et al. (2015). Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J. Infect. 71, 553–560. doi: 10.1016/j.jinf.2015.07.010
Zhang, Y., Zhao, C., Wang, Q., Wang, X., Chen, H., Li, H., et al. (2016). High prevalence of hypervirulent Klebsiella pneumoniae infection in China: geographic distribution, clinical characteristics and antimicrobial resistance. Antimicrob. Agents Chemother. 60, 6115–6120. doi: 10.1128/AAC.01127-16
Zhao, J., Chen, J., Zhao, M., Qiu, X., Chen, X., Zhang, W., et al. (2016). Multilocus sequence types and virulence determinants of hypermucoviscosity-positive Klebsiella pneumoniae isolated from community-acquired infection cases in Harbin, North China. Jpn. J. Infect. Dis. doi: 10.7883/yoken.JJID.2015.321
Keywords: Klesiella pneumonia, hypermucoviscosity, characteristics, epidemiology
Citation: Guo Y, Wang S, Zhan L, Jin Y, Duan J, Hao Z, Lv J, Qi X, Chen L, Kreiswirth BN, Wang L and Yu F (2017) Microbiological and Clinical Characteristics of Hypermucoviscous Klebsiella pneumoniae Isolates Associated with Invasive Infections in China. Front. Cell. Infect. Microbiol. 7:24. doi: 10.3389/fcimb.2017.00024
Received: 14 September 2016; Accepted: 18 January 2017;
Published: 01 February 2017.
Edited by:
David Dockrell, University of Sheffield, UKReviewed by:
Chris Whitehouse, United States Food and Drug Administration, USACopyright © 2017 Guo, Wang, Zhan, Jin, Duan, Hao, Lv, Qi, Chen, Kreiswirth, Wang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangxing Wang, Mzg4MDVAMTYzLmNvbQ==
Fangyou Yu, d3pqeHlmeUAxNjMuY29t
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.