- 1Department of Preventive Veterinary, College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, China
- 2Key Lab of Food Quality and Safety of Jiangsu Province-State Key Laboratory Breeding Base, Institute of Food Safety, Jiangsu Academy of Agricultural Sciences, Nanjing, China
Free-living protozoa affect the survival and virulence evolution of pathogens in the environment. In this study, we explored the fate of Aeromonas hydrophila when co-cultured with the bacteriovorous ciliate Tetrahymena thermophila and investigated bacterial gene expression associated with the co-culture. Virulent A. hydrophila strains were found to have ability to evade digestion in the vacuoles of this protozoan. In A. hydrophila, a total of 116 genes were identified as up-regulated following co-culture with T. thermophila by selective capture of transcribed sequences (SCOTS) and comparative dot-blot analysis. A large proportion of these genes (42/116) play a role in metabolism, and some of the genes have previously been characterized as required for bacterial survival and replication within macrophages. Then, we inactivated the genes encoding methionine sulfoxide reductases, msrA, and msrB, in A. hydrophila. Compared to the wild-type, the mutants ΔmsrA and ΔmsrAB displayed significantly reduced resistance to predation by T. thermophila, and 50% lethal dose (LD50) determinations in zebrafish demonstrated that both mutants were highly attenuated. This study forms a solid foundation for the study of mechanisms and implications of bacterial defenses.
Introduction
Aeromonas hydrophila, a Gram-negative ubiquitous bacterium with diverse host specificity, is distributed widely in aquatic environments (Daskalov, 2006; Janda and Abbott, 2010). Aeromonas infection has been linked to major die-offs and fish kills and has thus resulted in significant economic losses around the world for decades (Pang et al., 2015). In addition, this bacterium has been proposed to cause a variety of serious illnesses in other cold-blooded species and humans (Janda and Abbott, 2010). The pathogenesis of A. hydrophila is multifactorial and is likely mediated by virulence factors such as adhesins, exotoxins, extracellular enzymes, secretion systems, iron acquisition systems, and quorum-sensing systems (Tomas, 2012). Notably, environmental factors, such as predation by heterotrophic protists, have a dramatic effect on the virulence evolution of pathogens (Erken et al., 2013). However, the mechanism underlying this has not been investigated in A. hydrophila.
A. hydrophila can be isolated from numerous aquatic environments, such as drinking water, groundwater, wastewater, rivers, lakes, ponds, and sewage in various stages of treatment (Janda and Abbott, 2010). The free-living ciliate Tetrahymena is commonly found in the same aquatic environments (Valster et al., 2009). Evidence increasingly supports interactions between Tetrahymena and microbial pathogens. King et al. (1988) reported that many bacterial pathogens can resist the grazing protozoan Tetrahymena pyriformis. After predation by Tetrahymena species, Legionella pneumophila (Berk et al., 2008; Hojo et al., 2012) and Salmonella enterica (Brandl et al., 2005) are released in a viable form in vesicles or pellets from the protozoa. Due to the presence of a membrane around the vesicle, the bacterial cells within the vesicles are more resistant to disinfectants than those remaining free in suspension (Brandl et al., 2005). Ciliates thus may act as a reservoir for potentially pathogenic bacteria (Brandl et al., 2005). Grazing by phagotrophic protists is an important course of microbial mortality in aquatic environments (Pernthaler, 2005). To resist this predation, virulence factors in many bacterial species may have evolved for anti-predator defense (Ahmed et al., 2010; Erken et al., 2013).
Rahman et al. (2008) indicated that amoebae present in aquatic environments play an important role as reservoirs for Aeromonas species. We have previously demonstrated that the hypervirulence phenotype of A. hydrophila can survive efficiently within T. thermophila (Li et al., 2011; Pang et al., 2012). All of this evidence indicates an important link between Aeromonas and grazing protozoa. The question then arose as to which bacterial genes were involved in the anti-predator defense. In this study, we investigated the fate of A. hydrophila strains after co-culture with T. thermophila and used selective capture of transcribed sequences (SCOTS) to identify the genes that were preferentially expressed by A. hydrophila upon interaction with this protozoan. Additionally, we evaluated the role of the msr genes of A. hydrophila, which encode methionine sulfoxide reductases, in the response to predation by T. thermophila.
Materials and Methods
Strains and Culture Conditions
Seven virulent A. hydrophila strains (NJ-35, XY-16, NJ-34, CS-43, NJ-1, XX-14, and NJ-37), and five avirulent A. hydrophila strains (NJ-28, JH-19, NJ-3, CS-34, and JH-17; Pang et al., 2012), were used in this study (Table 1). The nucleotide sequence of the complete genome of NJ-35 has been deposited in GenBank (accession number CP006870). The bacterial strains were routinely cultured in Luria broth (LB) containing 1% NaCl, 1% peptone, and 0.5% yeast extract at 28°C. T. thermophila SB210 (Eisen et al., 2006) was obtained from Dr. Miao Wei, Institute of Hydrobiology, China Academy of Sciences. The genome sequence of T. thermophila SB210 has been deposited in GenBank under accession number GCA_000261185.1. T. thermophila SB210 was grown axenically in SPP medium (2% protease peptone, 0.1% yeast extract, 0.2% glucose, 0.003% EDTA-Fe) at 28°C and maintained in 5 mL of ultrapure water containing soybean. A. hydrophila and T. thermophila were co-cultured in TBSS (2 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2,and 1 mM Tris [pH 6.8–7.2]). All reagents used in this study were supplied by Sigma (St. Louis, MO, USA) unless otherwise indicated.
Survival of A. hydrophila in T. thermophila Vacuoles
To track the survival of A. hydrophila in T. thermophila, seven virulent strains and five avirulent strains were intrinsically labeled with green fluorescent protein (GFP) by electroporation of the plasmid pWSK129-gfp (Li et al., 2011). Then, 5000:1 co-cultures of A. hydrophila and T. thermophila were used to investigate their interaction (Pang et al., 2012). Before co-culture, T. thermophila SB210 with an initial inoculum of 103 cells/mL was grown in 50 mL of SPP medium at 28°C for 36 h, when the cultures entered stationary phase. The cells were washed twice with TBSS, counted using a hemacytometer, and then diluted in TBSS to a concentration of 2 × 105 cells/mL. A. hydrophila was incubated in 5 mL of LB medium at 28°C for 12 h until stationary-phase growth using an initial inoculum of 107 cells/mL, washed twice with TBSS, and then adjusted to 1 × 109 CFU/mL using TBSS. Five hundred microliters of A. hydrophila suspension was mixed with an equal volume of T. thermophila cells and incubated at 28°C for 12 h without shaking. The bacterial cells in T. thermophila were observed by laser scanning confocal microscopy (LSCM, Zeiss LSM710). In addition, co-cultures were prepared for transmission electron microscopy (TEM, Hitachi H-7650) by pelleting the cells and immediately fixing them with 2.5% glutaraldehyde (Solarbio, Beijing, China) for 2 h at 4°C. TEM observation was performed as described by Serratrice et al. (2014).
The LIVE/DEAD BacLight Bacterial Viability Kit (Invitrogen, New York, USA) was used to measure the proportion of viable bacterial cells contained in the vacuoles of T. thermophila. Propidium iodide (PI, Molecular Probes) was added at a final concentration of 7.5 μM to each of three replicate tubes containing the cultures, and the tubes were incubated for 15 min at 28°C in the dark. Then, the cultures were washed twice in TBSS and fixed in 2.5% glutaraldehyde. Ten microliters of each replicate suspension was placed on slides and observed by LSCM using the GFP and PI channels. For each sample, 100 vacuoles were examined, and the numbers of green (viable) and red (dead) fluorescent bacterial cells per vacuole were counted. Vacuoles were examined from three replicate tubes.
Experimental Infection, RNA Extraction, cDNA Synthesis, and Amplification
For RNA extraction, 1 × 105 T. thermophila cells were co-incubated with 5 × 108 CFU of A. hydrophila NJ-35 without shaking for 12 h at 28°C. This time point was selected on the basis of our previous study, which demonstrated that virulent A. hydrophila strains survived better than avirulent A. hydrophila strains when co-cultured with T. thermophila, particularly after co-culture for 12 h (Pang et al., 2012). In considering the MOI, our preliminary dose-response studies showed that, the number of A. hydrophila which was taken up by an average protis nearly reached saturation at the MOI of 5000 (data not shown). After incubation, the co-cultures were centrifuged at 100 g for 1 min. Then, the obtained pellet was washed twice with TBSS by centrifugation at 200 g for 1 min and 400 g for 1 min, respectively. The final pellet was resuspended in 1 mL of TBSS and further treated with gentamycin (100 μg/mL) for 1 h to kill the remaining extracellular or adherent bacteria. Samples were then centrifuged at 400 g for 1 min and the harvested Tetrahymena cells were washed once in TBSS. The Tetrahymena pellet was then resuspended in 1 mL TBSS containing 1% Triton X-100 for 10 min at 37°C to release ingested bacteria. The suspension containing lysed Tetrahymena cells was centrifuged at 4000 g for 5 min at 4°C to provide the ingested bacteria. Control bacteria without Tetrahymena were incubated in TBSS for the same time, and then collected by centrifugation to provide protozoa-unexposed bacterial cells. Total RNA was extracted using TRIzol reagent (Invitrogen) from two samples containing equal numbers of A. hydrophila differing only in the presence (protozoa-exposed RNA) or absence (protozoa-unexposed RNA) of T. thermophila. The RNA was subsequently treated with DNase I (Fermentas) for 1 h at 37°C. The integrity, purity and concentration of the RNA were determined by agarose gel electrophoresis, PCR and A260/A280 spectrophotometer readings, respectively. The total RNA isolated from protozoa-exposed or protozoa-unexposed bacteria was converted to first-strand cDNA by random priming with Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's specifications. The primers had a defined 5′ terminal sequence and a 3′ random hexamer, and different terminal sequences were used for protozoa-exposed (SCOTS-N6-01) and protozoa-unexposed RNA (SCOTS-N6-02; Froussard, 1992). The second strand of cDNA was synthesized using Klenow fragment (Fermentas). Then, the cDNA libraries were amplified by PCR with 25 cycles of amplification (95°C for 30 s, 66°C for 60 s, and 72°C for 60 s).
Selective Capture of Transcribed Sequences (SCOTS)
Bacterial transcripts were then separated from host cDNA by SCOTS as described previously (Guo et al., 2014). Briefly, denatured, biotinylated, and sonicated A. hydrophila genomic DNA (gDNA) fragments (0.6 μg) were mixed with 5 μg of sonicated ribosomal DNA (from plasmid pMD18-T16S, pMD18-T23S1, and pMD18-T23S2) to pre-block rRNA encoding regions on the gDNA. For each round of SCOTS, a sample of the mixture (8 μL) was denatured by incubation at 98°C for 3 min. The mixture was incubated at 64°C for 30 min, and 2 μL of 1 M NaCl was then added. At the same time, 2 μL of 1 M NaCl was added to the total amplified cDNA of bacteria exposed or unexposed to T. thermophila in 8 μL of 10 mM EPPS-1 mM EDTA. The denatured cDNA mixture was added to the biotinylated gDNA–rDNA pre-hybridized mixture, and hybridization was continued at 64°C for 24 h. Bacterial cDNA that was hybridized to biotinylated gDNA was then captured by binding hybrids to streptavidin-coated magnetic beads (Dynal M280). The captured cDNA was eluted, precipitated, and amplified by PCR using the defined primers SCOTS01 (protozoa-exposed) or SCOTS02 (protozoa-unexposed). For each condition, three rounds of capture were performed, and the normalized cDNA was obtained. The primers used in this study are showed in Supplementary Table 1.
Competitive Enrichment
To preferentially enrich for protozoa-exposed expressing transcripts, enrichment of cDNA was conducted to capture hybridizations. A total of 0.6 μg of A. hydrophila NJ-35 chromosome was pre-blocked with both 5 μg of rDNA and 5 μg of denatured triple-SCOTS normalized protozoa-unexposed cDNA. Then, 5 μg of triple-SCOTS normalized protozoa-exposed cDNA was denatured and re-annealed for 30 min at 64°C to remove abundant transcripts. The cDNA and blocked gDNA samples were combined and hybridized for 20 h at 64°C. Hybrids were collected using Dynal streptavidin-coated magnetic beads. The captured cDNA was eluted, precipitated, and amplified using the protozoa-exposed library-specific defined primer SCOTS01. After three rounds of this enrichment procedure, the cDNAs were ligated into the pMD18-T vector (TaKaRa, Dalian, China).
Southern Hybridization for Primary Verification and Sequence Analysis
To eliminate false-positive sequences that escaped the subtraction process, southern hybridization was used for primary verification. Cloned inserts obtained from protozoa-exposed-specific cDNA libraries were amplified by PCR with SCOTS01 primers. PCR amplicons of positive SCOTS clones were transferred to a positively charged membrane (Roche, Mannheim, Germany). Samples of gDNA and cDNA mixtures generated from protozoa-exposed strain NJ-35 and protozoa-unexposed strain NJ-35 were used as probes, followed by labeling with DIG-dUTP (Roche). Dot blot hybridization analysis using DIG Easy Hyb (Roche) was performed according to the manufacturer's instructions. The clones that hybridized positively with the protozoa-exposed probes but negatively with the protozoa-unexposed probes were termed SCOTS clones. Then, the inserts of positive cDNA clones were sequenced by GENEWIZ, Inc., and the nucleotide sequences were queried using BLASTn implemented in BLAST+ (version 2.2.29; ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/) against the genome of A. hydrophila NJ-35. To classify the functions of the preferentially expressed genes, BLASTp implemented in BLAST+ (version 2.2.29) was used to align the amino acid sequences against the COGs database (updated 2014), and some genes related to bacterial virulence were classified according to a previous study (Pang et al., 2015).
Secondary Verification Using Quantitative Reverse Transcription-PCR (qRT-PCR)
To further validate the SCOTS results, we randomly selected 14 genes to measure the level of expression by qRT-PCR. RNA extraction from protozoa-exposed strain NJ-35 and protozoa-unexposed strain NJ-35 was performed as described above. The altered expression levels of 14 genes in protozoa-exposed strain NJ-35 and protozoa-unexposed strain NJ-35 were examined individually. The cDNA was synthesized in triplicate using Superscript II with random hexamers (Invitrogen) according to the manufacturer's instructions. The QuantiTect SybrGreen PCR kit (Qiagen, Valencia, USA) was used for qRT-PCR in an ABI PRISM 7300 Fast Real-time PCR machine. For each sample, a no-reverse transcription reaction was performed as a no template control (NTC). The primers used are described in Supplementary Table 1. For each qRT-PCR run, the calculated cycle threshold (CT) was normalized to the CT of the internal control 16S rDNA amplified from the corresponding sample, and the fold-change was calculated using the 2−ΔΔCT method as previously described (Livak and Schmittgen, 2001).
Inactivation and Complementation of msrA and msrB in A. hydrophila
The msrA mutant (ΔmsrA) was constructed via homologous recombination using the suicide plasmid pYAK1. Briefly, the primers msrA-up-F/msrA-up-R and msrA-down-F/msrA-down-R were designed to amplify two flanking sequences of the msrA gene by PCR. Then, the two segments were ligated by fusion PCR and inserted into pYAK1 to construct the recombinant plasmid pYAK-msrA using Escherichia coli SM10 as the host strain. Subsequently, parental mating was used to transfer the recombinant plasmid pYAK-msrA into strain NJ-35 (Ampr). The transconjugants with the first allelic exchange were selected on LB agar plates with ampicillin and chloramphenicol. Positive clones were transferred to LB for growth for 12 h and then transferred to LB agar plates containing 10% sucrose. The suspected ΔmsrA strain was verified by PCR. Using the same approach, the msrB deletion mutant and a double gene (msrA, msrB) deletion mutant were also constructed.
To complement the function of the deleted genes in the mutants, the complete ORFs of msrA and msrB were amplified from A. hydrophila genomic DNA to construct the pMMB-msrA and pMMB-msrB plasmids for genetic complementation. Then, the plasmids were introduced into ΔmsrA and ΔmsrB by conjugation using E. coli SM10 as the donor strain, and the complemented mutants CΔmsrA and CΔmsrB were selected on LB agar containing 100 μg/mL ampicillin and 34 μg/mL chloromycetin. The primers used for mutant construction are showed in Supplementary Table 1.
Assessment of Bacterial Resistance to Predation by T. thermophila
Bacterial resistance to predation was assessed by measuring the relative survival of bacteria after co-culture with T. thermophila (Pang et al., 2012). Briefly, T. thermophila SB210 was cultured at 28°C for 36 h in SPP medium until the stationary phase of growth using an initial inoculum of 103 cells/mL. Cells were diluted in TBSS to a concentration of 2 × 105 cells/mL. A. hydrophila was incubated in LB medium at 28°C for 12 h, washed twice with TBSS, and then adjusted to 1 × 109 CFU/mL using TBSS. Five hundred microliters of A. hydrophila suspension was mixed with the same volume of T. thermophila cells, and 200 μl of these mixed cell suspensions was transferred into each well of a 96-well plate. A. hydrophila suspensions and T. thermophila suspensions mixed with an equal volume of TBSS separately served as controls. TBSS served as the blank control. Plates were incubated for 12 h at 28°C without shaking, and the bacterial population was detected by measuring the absorbance at 450 nm (OD450) every 2 h. The absorbance of T. thermophila cells was negligible (Pang et al., 2012). The relative survival of bacteria was expressed as the OD450 value of bacteria remaining in co-culture with T. thermophila divided by that of bacteria grown alone at 12 h. Three independent measurements were performed in quadruplicate.
Determination of 50% Lethal Dose (LD50) in Zebrafish
Zebrafish weighing ~3 g were supplied by Pearl River Fishery Research Institute, Chinese Academic of Fishery Science. The animal-challenge experiment with A. hydrophila was performed as described previously (Pang et al., 2012). For each A. hydrophila strain, eight groups of 15 zebrafish were intraperitoneally injected with 0.02 mL of 10-fold serially diluted suspensions of bacteria (101–107 CFU) in PBS. Another 15 zebrafish (the control group) were injected with 0.02 mL of sterile PBS. The survival rates of the zebrafish were recorded daily for a period of 7 days post infection, and the LD50 values were calculated. Animal experiments were conducted according to animal welfare standards and approved by the Ethical Committee for Animal Experiments of Nanjing Agricultural University, China.
Statistical Analysis
Data were collected and analyzed using MS Excel 2010 and SPSS Statics v20.0 software. Relative survival of bacteria was analyzed by analysis of variance (ANOVA) followed by Turkey's multiple comparison test; The gene expression levels in protozoa-exposed A. hydrophila and protozoa-unexposed A. hydrophila were analyzed using a Student's t-test; P < 0.05 was considered a significant difference, whereas P < 0.01 was considered highly significant.
Results
Survival of A. hydrophila in T. thermophila
To investigate the fate of A. hydrophila in response to phagocytosis by T. thermophila, 12 A. hydrophila strains of different virulence were intrinsically labeled with GFP by transformation with the plasmid pWSK129-gfp. Then, LSCM was used to examine the predation of A. hydrophila by T. thermophila SB210. After the addition of bacteria to the T. thermophila suspensions, green food vacuoles could be observed in nearly all T. thermophila cells within 30 min, and T. thermophila fed readily on all A. hydrophila strains. Here, virulent strain NJ-35 and avirulent strain CS-34 were described as examples. As shown in Figure 1, after co-culture for 12 h, a high proportion of the cells of strain NJ-35 maintained their integrity and exhibited bright green fluorescent (Figure 1A), while strain CS-34 presented dispersed green fluorescent (Figure 1D).
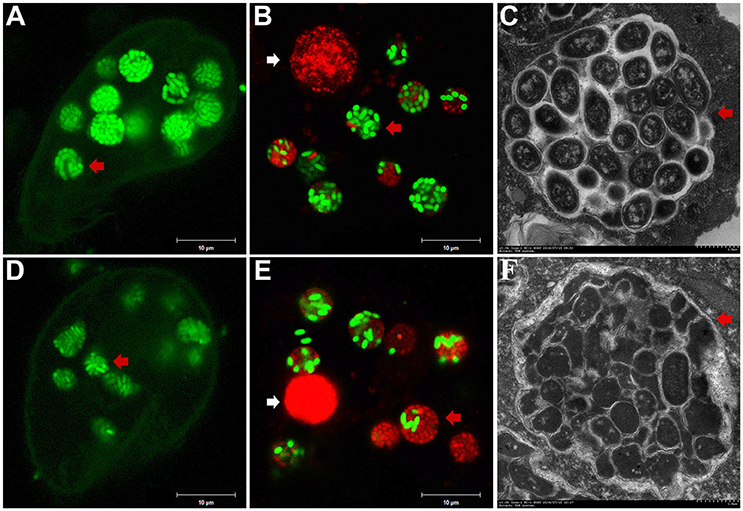
Figure 1. Survival of A. hydrophila in T. thermophila vacuoles after co-culture for 12 h. (A–C) show micrographs of virulent strain NJ-35 in T. Thermophila, and (D–F) show the micrographs of avirulent strain CS-34. (A,D) were acquired by LSCM (Zeiss LSM710) using a GFP channel and displayed by Gamma 0.45; (B,E) were acquired by LSCM (Zeiss LSM710) using GFP and PI channels; (C,F) were acquired by TEM (Hitachi H-7650). Viable and dead cells exhibit green and red fluorescence, respectively. The red arrow indicates bacterial cells in T. thermophila vacuoles. The white arrow indicates the nuclei of T. thermophila displaying red fluorescence when labeled by propidium iodide.
To further analyze bacterial survival in vacuoles, GFP fluorescence in combination with PI viability staining was used in this study. GFP and PI exhibited good segregation of fluorescent labels in a mixed population of viable (green) and dead (red) cells. However, because the bacterial cells used in this study were labeled with GFP, some of the cells were yellow because of simultaneous red and green fluorescence, consistent with a previous study (Brandl et al., 2005). Such cells were relatively few (no more than 3%) and were not included in the counts. Compared to strain CS-34 (Figure 1E), more viable bacterial cells of strain NJ-35 (Figure 1B) were observed when co-cultured with T. thermophila. In addition, TEM observations also revealed that the intracellular NJ-35 remained morphologically intact (Figure 1C), whereas most of the intracellular CS-34 exhibited an irregular shape (Figure 1F).
To support the speculation that virulent A. hydrophila strains may be able to evade digestion in the vacuoles of T. thermophila, the survival rates of seven virulent strains (NJ-35, XY-16, NJ-34, CS-43, NJ-1, XX-14, and NJ-37) and five avirulent strains (NJ-28, JH-19, NJ-3, CS-34, and JH-17) in vacuoles were calculated. As shown in Figure 2, after 12 h of co-culture, the survival rates of bacterial cells per vacuole in the virulent A. hydrophila groups, except strain NJ-37, were all higher than those of avirulent A. hydrophila groups. These findings indicated that virulent A. hydrophila strains may have a better ability to evade digestion in T. thermophila vacuoles.
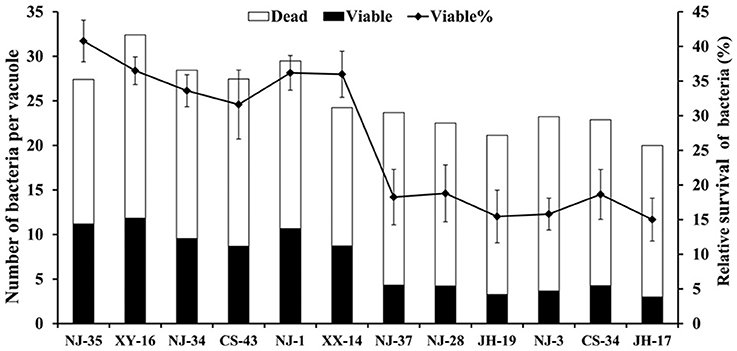
Figure 2. Survival rate of A. hydrophila strains with different virulence in T. thermophila vacuoles. Viable and dead bacterial cells in 100 T. thermophila vacuoles were counted using Zeiss LSM710, and the survival rate of the bacteria was expressed as the number of viable bacteria divided by the number of total bacteria per vacuole.
Selective Capture of A. hydrophila Transcripts
For identification of the genes that are differentially expressed by A. hydrophila NJ-35 when grown in protozoa-exposed and protozoa-unexposed environments, SCOTS (Figures 3A,B) was used in this study. After primary verification by southern hybridization (Figures 3C,D), a total of 288 positive SCOTS clones in the protozoa-exposed group were obtained and subjected to further sequence analysis. Subsequently, 256 available sequences were obtained. Among the 256 sequences, 26 sequences were unidentifiable “junk” DNA, and the remaining 230 sequences were identified as 116 genes since some of the sequences were the same. As shown in Table 2, these 116 genes were characterized into five functional categories: (1) Forty-two genes were involved in metabolism, such as amino acid transport, inorganic ion transport, energy production, carbohydrate transport, and metabolism. Genes such as panF, gltD, oppA, purF, napA, thyA, mgtA, cysE, and norV may endow the bacteria with ability to uptake multiple forms of nutrients or similar metabolites; (2) Twenty-two genes, including rstB, msrA, msrB, clpP, and clpA, encoded proteins responsible for cellular processes and signaling, including cell membrane biogenesis, post-translational modification, and signal transduction mechanisms; (3) Twenty genes, including dnaA and rpoC, were involved in information storage and processing, including transcription, replication, recombination, and repair; (4) Eighteen genes encoded proteins that can be characterized as virulence-associated factors, such as the type 6 secretion system (T6SS) effector proteins hemolysin co-regulated protein (Hcp) and valine glycine repeat G (VgrG), and proteins involved in motility and adhesion; (5) The remaining 14 genes were poorly characterized, and eight encoded hypothetical proteins.
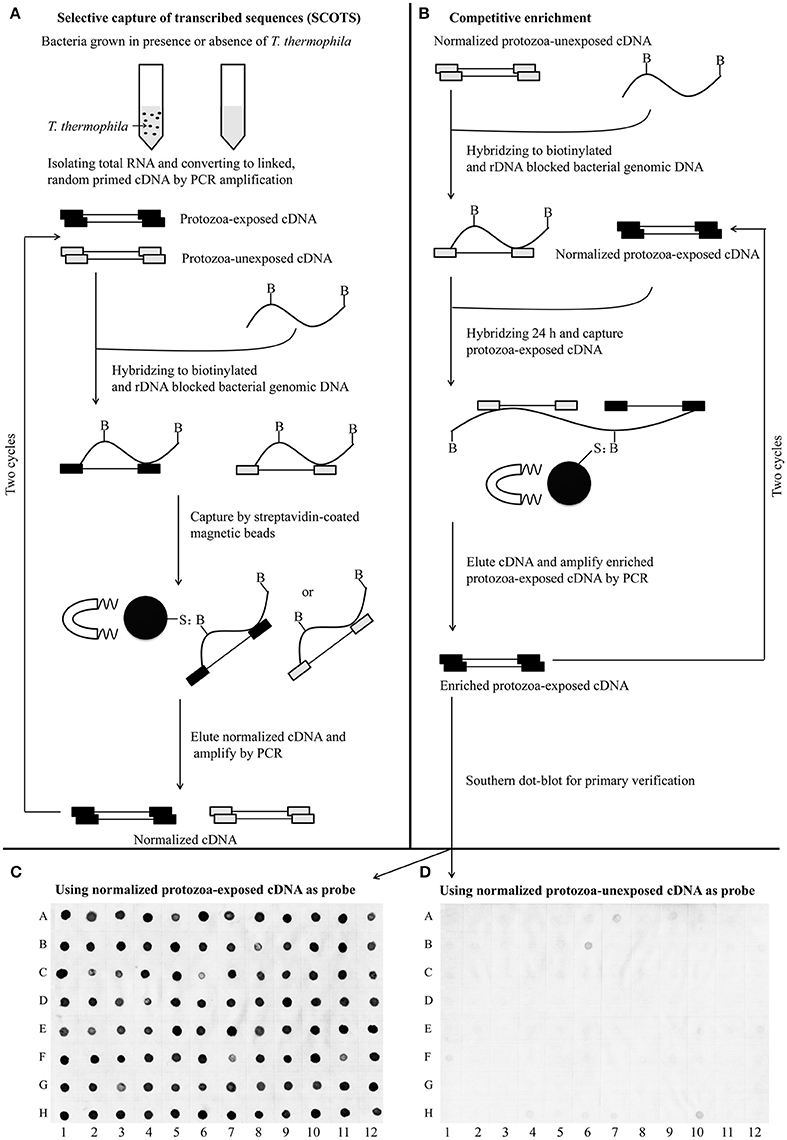
Figure 3. Schematic presentation of the SCOTS approach followed by Southern dot-blot analysis. (A) Normalization of protozoa-exposed cDNA and protozoa-unexposed cDNA; (B) competitive enrichment of protozoa-exposed expressing transcripts; (C,D) Southern dot-blot analysis of SCOTS clones using probes generated from normalized protozoa-exposed cDNA and protozoa-unexposed cDNA, respectively. The schematic presentations (A,B) were designed as described by An and Grewal (2012), with some modifications.
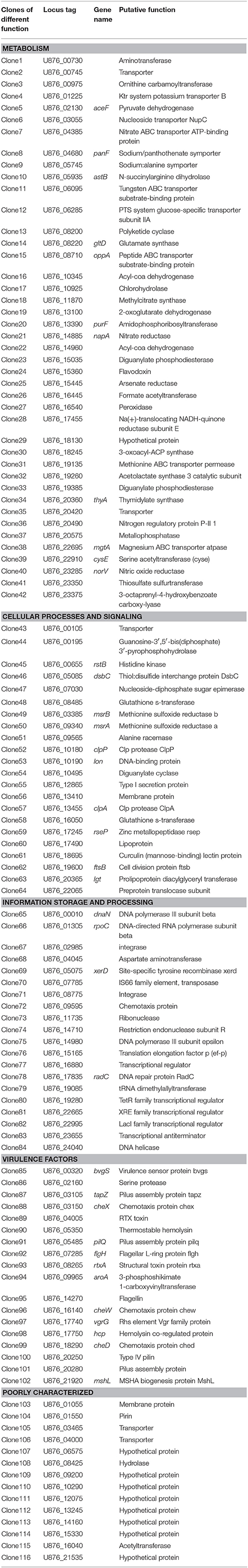
Table 2. Genes identified by SCOTS that were differentially expressed in T. thermophila-exposed A. hydrophila.
Validation of SCOTS Results by qRT-PCR
The results of the SCOTS experiments were confirmed by qRT-PCR. Fourteen genes (clpP, dsbC, flgH, hcp, lgt, lon, msrA, msrB, norV, purF, rstB, rtxA, U876_13245, and vgrG) belonging to different functional categories were chosen and validated. As shown in Figure 4, compared with the protozoa-unexposed group, the expression levels of all 14 genes were up-regulated significantly in protozoa-exposed A. hydrophila except for lgt (1.22-fold change, P = 0.83). Among the remaining 13 genes, the expression levels of three genes dsbC, hcp, and msrA ranged from 1.64- to 1.89-fold (P < 0.05), whereas the expression levels of other 10 genes all changed more than two-fold (P < 0.05). The high coincidence rate (92.9%) of qRT-PCR with SCOTS indicates the reliability of the SCOTS results.
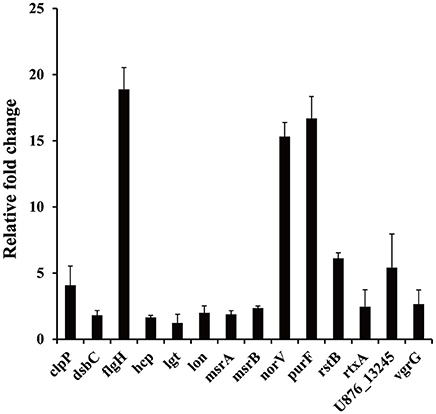
Figure 4. Relative expression of genes in protozoa-exposed A. hydrophila compared to protozoa-unexposed A. hydrophila. Data are presented as relative fold changes with protozoa-unexposed A. hydrophila as the control and all fold changes are normalized to 16S rDNA. Relative fold changes were calculated using the 2−ΔΔCt method, where ΔΔCt = (Ctgene of interest − Ctcontrol gene)protozoa-exposed group − (Ctgene of interest − Ctcontrol gene)protozoa-unexposed group. Error bars represent standard deviations from three independent experiments.
Effect of msr Inactivation on Resistance of A. hydrophila to Predation by T. thermophila
To further validate the SCOTS results and also determine whether msrA and msrB play important roles during co-culture of A. hydrophila strains with T. thermophila, the mutants ΔmsrA, ΔmsrB, and ΔmsrAB were constructed by homologous replacement in strain NJ-35. The relative survivals of the wild-type and mutant strains after co-culture with T. thermophila are shown in Figure 5. Compared to the wild-type strain, the relative survivals of strains ΔmsrA and ΔmsrB were 17.77% lower (P < 0.01) and 8.46% lower (P < 0.05), respectively. ΔmsrAB exhibited obviously lower relative survival (30.35%) than the wild-type strain (P < 0.01). However, the relative survivals of the complemented strains CΔmsrA and CΔmsrB were restored to the level of the wild-type strain. These results suggest that msrA and msrB play important roles in the resistance of A. hydrophila to protozoan predation.
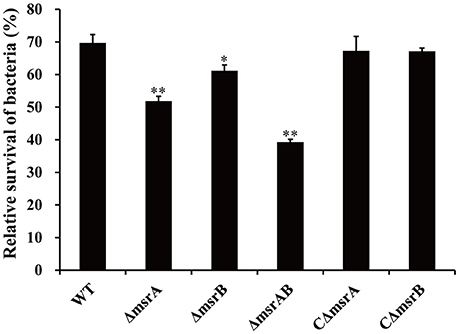
Figure 5. Relative survival of a wild-type A. hydrophila and its msr gene mutant derivatives after co-culture with T. thermophila. WT represents the wild-type strain NJ-35. The relative survival of bacteria was expressed as the OD450 value of A. hydrophila co-cultured with T. thermophila divided by that of A. hydrophila grown alone at 12 h. The error bars represent standard deviations from four independent experiments performed in quadruplicate. *P < 0.05 or **P < 0.01 indicates significantly different relative survival compared with the WT group.
Effect of msr Inactivation on the Virulence of A. hydrophila in Zebrafish
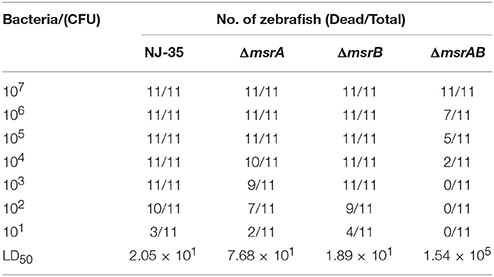
To investigate the roles of msrA and msrB in the virulence of A. hydrophila, zebrafish were injected intraperitoneally with the wild-type or mutant strains. The mortality of zebrafish was recorded daily over a period of 7 days following infection. As shown in Table 3, the LD50 value of the msrA mutant (7.68 × 101 CFU) was nearly four-fold higher than that of the wild-type strain (2.05 × 101 CFU), indicating a significant reduction in the virulence of the mutant. However, the LD50 value of the msrB mutant (1.89 × 101 CFU) was similar to that of the wild-type strain, suggesting that msrB is not essential for the virulence of A. hydrophila in zebrafish. The simultaneous inactivation of msrA and msrB caused a more significant reduction in A. hydrophila virulence, and the LD50 value of the msrAB mutant (1.54 × 105 CFU) was more than 2000-fold higher than that of the wild-type strain. These results suggest that MsrA plays an important role in the virulence of A. hydrophila in zebrafish and that a synergistic relationship may exist between MsrA and MsrB.
Discussion
Hahn and Höfle (2001) reported that predation by protozoa can influence bacterial populations. Once preyed by protozoa, most microbes are digested as food, but some microbes appear to be resistant to protozoa digestion and can even replicate within protozoa. Several bacterial pathogens, including E. coli (King et al., 1988), L. pneumophila (Berk et al., 2008; Hojo et al., 2012), S. enterica (Brandl et al., 2005; Rehfuss et al., 2011), and Listeria monocytogenes (Pushkareva and Ermolaeva, 2010), have been shown to be resistant to destruction in digestive vacuoles of Tetrahymena. In this study, we observed that T. thermophila fed readily on A. hydrophila strains, however, LSCM and TEM observations and the survival rate of A. hydrophila in vacuoles indicated that the virulent strains were able to survive in T. thermophila vacuoles. Thus, Tetrahymena may represent an unappreciated reservoir for the hypervirulence phenotype of A. hydrophila. In this regard, previous reports have demonstrated that exposure to rumen protozoa leads to the selection of Salmonella strains with enhanced virulence traits (Rasmussen et al., 2005; Brewer et al., 2011). Therefore, protozoa may not only serve as a protective reservoir but also select for virulence traits.
We hypothesize that the survival of pathogenic A. hydrophila within Tetrahymena necessitates the expression of bacterial genes that are unlikely to be expressed in a protozoa-unexposed environment. In this study, 116 preferentially expressed genes were identified in A. hydrophila in response to phagocytosis by Tetrahymena using SCOTS. Genes involved in metabolism accounted for 36.2% (42/116) of differentially up-regulated genes in protozoa-exposed bacteria, including enzymes associated with amino acid transport, inorganic ion transport, energy production, carbohydrate transport, and metabolism. It is not surprising that A. hydrophila may alter its metabolism to obtain available nutrient and energy sources to adapt to the intracellular niche in Tetrahymena. Interestingly, some of these genes, including panF, gltD, oppA, purF, napA, thyA, mgtA, cysE, and norV, have been known to be associated with bacterial virulence or resistance in other bacteria. For instance, in Moraxella catarrhalis, an oppA mutant exhibited marked impairment in its capacity to persist in the respiratory tract compared to wild-type in a mouse pulmonary clearance model (Yang et al., 2011). Similarly, the mutation of the transport domain of the oppA gene in Mycobacterium avium resulted in bacterial attenuation in both macrophages and in mice (Danelishvili et al., 2014). The gene purF, which encodes amidophosphoribosyltransferase, was identified as a novel virulence factor in Francisella tularensis by screening a library of corresponding transposon mutants for replication in RAW264.7 macrophages (Llewellyn et al., 2011). In Staphylococcus aureus, inactivation of thyA, which is involved in thymidylate synthesis, strongly attenuated bacterial virulence in Caenorhabditis elegans and mouse models (Kriegeskorte et al., 2014). Another gene, norV, which encodes nitric oxide reductase, was observed to contribute to the survival of enterohemorrhagic E. coli (EHEC) O157 within macrophages (Shimizu et al., 2012). This obvious alteration of expression in these metabolism-related genes may be required for nutrient acquisition and virulence of A. hydrophila when exposed to T. thermophila.
In this study, 18 virulence-related genes were up-regulated in protozoa-exposed A. hydrophila. The structural toxin protein (RtxA) can disrupt the actin cytoskeleton of HeLa cells, resulting in a rounding phenotype and hence contributing to host cell apoptosis (Suarez et al., 2012). The gene aroA encodes 3-phosphoshikimate 1-carboxyvinyltransferase, and its inactivation has been reported to attenuate A. hydrophila virulence (Hernanz Moral et al., 1998; Vivas et al., 2004). In addition, Hcp and VgrG, two known T6SS effectors of A. hydrophila, were also identified in protozoa-exposed A. hydrophila. T6SS has been identified in 25% of sequenced Gram-negative genomes and is involved in virulence and host associations in these bacterial species (Pukatzki et al., 2007). Efficient colonization is critical for bacterial virulence, and both pili and flagella contribute to colonization in A. hydrophila (Tomas, 2012). In this study, genes responsible for the formation of type IV pili (tapZ, pilQ, and mshL) and flagella (flgH) were identified. Moreover, cheX, cheW, and cheD, which encode chemotaxis protein, were obtained using SCOTS. Antunez-Lamas et al. (2009) reported that the genes involved in the chemotactic signal transduction system and in the structure of the flagellar motor play important roles in the pathogenicity of Dickeya dadantii. In A. hydrophila, chemotaxis is not necessary for pathogenicity but may be a necessary parameter for this bacterium to become an obligate pathogen (Seshadri et al., 2006). The overall up-regulation of virulence genes in protozoa-exposed environments may explain why the virulent A. hydrophila strains had a greater ability to evade digestion by T. thermophila. Additionally, from an evolutionary perspective, the identification of the common virulence factors in protozoan and vertebrate hosts indicates the universality of virulence implicated in the infectious process in the evolutionarily divergent hosts.
Additionally, 22 genes, including rstB, msrA, msrB, clpP, and clpA, which are involved in cellular processes and signaling, were also identified. RstB encodes the sensor kinase and acts on the PhoQ sensor to control the expression of PhoP-regulated genes in Salmonella (Nam et al., 2010). The response regulator PhoP and its partner sensor PhoQ constitute the PhoP/PhoQ two-component system, which governs virulence, mediates the adaptation to Mg2+-limiting environments, and regulates other physiological processes of Salmonella (Groisman, 2001). Thus, RstB indirectly controls the virulence of Salmonella. The ATP-dependent caseinolytic proteases (Clp) are important in the resistance of pathogenic bacteria against environmental stresses and host immune defenses. ClpP is the proteolytic subunit, and ClpA acts as both a chaperone and an ATPase driving the degradation of damaged or improperly folded proteins. The clpA and clpP mutants of Helicobacter pylori exhibit increased sensitivity to oxidative stress, in addition to reduced survival in human macrophages (Loughlin et al., 2009). In addition, the ClpP protein is required for the stress tolerance of Actinobacillus pleuropneumoniae (Xie et al., 2013).
Methionine sulfoxide reductases (Msrs) are key enzymes in repairing ROS-mediated damage to proteins and include mainly MsrA and MsrB (Sansom et al., 2013). As the best characterized Msr, MsrA plays a role in resistance to oxidative stress and virulence in a number of bacteria, including Mycobacterium species (St. John et al., 2001; Douglas et al., 2004), S. aureus (Singh and Moskovitz, 2003), Salmonella typhimurium (Denkel et al., 2011), and E. coli (St. John et al., 2001). In this study, both msrA and msrB were up-regulated in A. hydrophila during co-culture with T. thermophila. To determine the role of the two genes in response to phagocytosis by Tetrahymena, we constructed the mutants ΔmsrA, ΔmsrB, and ΔmsrAB. Single and double inactivation of msrA and msrB significantly reduced the resistance of A. hydrophila to predation by T. thermophila. These findings indicate that msrA and msrB were required for A. hydrophila to resist predatory protozoans. Moreover, the msr genes have previously been characterized as required for bacterial survival and replication within macrophages (Douglas et al., 2004; Sansom et al., 2013). These findings suggest that the mechanisms responsible for survival within the phagosomes of protozoa and macrophages may be similar. In addition, we observed that the deletion of msrA resulted in significantly reduced virulence in zebrafish, whereas the virulence of the msrB mutant was essentially unaffected. Notably, the double deletion of the msr genes (ΔmsrAB) resulted in an extreme reduction of virulence (2000-fold higher LD50 value than ΔmsrA strain), suggesting a synergistic effect of these two genes on bacterial virulence.
The present study is the first to characterize gene expression in A. hydrophila under phagocytosis by Tetrahymena. In this study, 116 genes were identified as up-regulated, including genes associated with metabolism, cellular process and signaling, information storage and processing, virulence factors, as well as some genes whose functions are currently unknown. Because protozoa share many features with mammalian phagocytes, particularly macrophages (Jacobs et al., 2006; Cosson and Soldati, 2008), a better understanding of protozoa-bacteria interactions will provide fascinating glimpses into host-pathogen relationships. This study will be a starting point for investigating the co-evolution of bacteria and protozoa. Future functional characterization of the genes identified in this study will deepen our understanding of the epidemiology of an infectious disease and the development of procedures for its control.
Author Contributions
YL, MP, and XL conceived the study and drafted the paper; MP, XL, and JL performed the experiments; CG, SG, and HD helped with the experiments; CG and CL provided valuable suggestions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Nature Science Foundation of China (31372454, 31072151), Aquatic Three New Projects in Jiangsu Province (D2015-13), Graduate Student Research and Innovation Program of Jiangsu Province (CXZZ13_0310), and Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2016.00199/full#supplementary-material
References
Abolghait, S. K. (2013). Suicide plasmid-dependent IS1-element untargeted integration into Aeromonas veronii bv. sobria generates brown pigment-producing and spontaneous pelleting mutant. Curr. Microbiol. 67, 91–99. doi: 10.1007/s00284-013-0335-4
Ahmed, N., Adiba, S., Nizak, C., van Baalen, M., Denamur, E., and Depaulis, F. (2010). From grazing resistance to pathogenesis: the coincidental evolution of virulence factors. PLoS ONE 5:e11882. doi: 10.1371/journal.pone.0011882
An, R., and Grewal, P. S. (2012). Selective capture of transcribed sequences: a promising approach for investigating bacterium-insect interactions. Insects 3, 295–306. doi: 10.3390/insects3010295
Antunez-Lamas, M., Cabrera-Ordonez, E., Lopez-Solanilla, E., Raposo, R., Trelles-Salazar, O., Rodriguez-Moreno, A., et al. (2009). Role of motility and chemotaxis in the pathogenesis of Dickeya dadantii 3937 (ex Erwinia chrysanthemi 3937). Microbiology 155, 434–442. doi: 10.1099/mic.0.022244-0
Berk, S. G., Faulkner, G., Garduno, E., Joy, M. C., Ortiz-Jimenez, M. A., and Garduno, R. A. (2008). Packaging of live Legionella pneumophila into pellets expelled by Tetrahymena spp. does not require bacterial replication and depends on a Dot/Icm-mediated survival mechanism. Appl. Environ. Microbiol. 74, 2187–2199. doi: 10.1128/AEM.01214-07
Brandl, M. T., Rosenthal, B. M., Haxo, A. F., and Berk, S. G. (2005). Enhanced survival of Salmonella enterica in vesicles released by a soilborne Tetrahymena species. Appl. Environ. Microbiol. 71, 1562–1569. doi: 10.1128/AEM.71.3.1562-1569.2005
Brewer, M. T., Xiong, N., Dier, J. D., Anderson, K. L., Rasmussen, M. A., Franklin, S. K., et al. (2011). Comparisons of Salmonella conjugation and virulence gene hyperexpression mediated by rumen protozoa from domestic and exotic ruminants. Vet. Microbiol. 151, 301–306. doi: 10.1016/j.vetmic.2011.03.011
Cosson, P., and Soldati, T. (2008). Eat, kill or die: when amoeba meets bacteria. Curr. Opin. Microbiol. 11, 271–276. doi: 10.1016/j.mib.2008.05.005
Danelishvili, L., Stang, B., and Bermudez, L. E. (2014). Identification of Mycobacterium avium genes expressed during in vivo infection and the role of the oligopeptide transporter OppA in virulence. Microb. Pathog. 76, 67–76. doi: 10.1016/j.micpath.2014.09.010
Daskalov, H. (2006). The importance of Aeromonas hydrophila in food safety. Food Control 17, 474–483. doi: 10.1016/j.foodcont.2005.02.009
Denkel, L. A., Horst, S. A., Rouf, S. F., Kitowski, V., Bohm, O. M., Rhen, M., et al. (2011). Methionine sulfoxide reductases are essential for virulence of Salmonella typhimurium. PLoS ONE 6:e26974. doi: 10.1371/journal.pone.0026974
Douglas, T., Daniel, D. S., Parida, B. K., Jagannath, C., and Dhandayuthapani, S. (2004). Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J. Bacteriol. 186, 3590–3598. doi: 10.1128/JB.186.11.3590-3598.2004
Eisen, J. A., Coyne, R. S., Wu, M., Wu, D., Thiagarajan, M., Wortman, J. R., et al. (2006). Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 4:e286. doi: 10.1371/journal.pbio.0040286
Erken, M., Lutz, C., and McDougald, D. (2013). The rise of pathogens: predation as a factor driving the evolution of human pathogens in the environment. Microb. Ecol. 65, 860–868. doi: 10.1007/s00248-013-0189-0
Froussard, P. (1992). A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 20:2900. doi: 10.1093/nar/20.11.2900
Groisman, E. A. (2001). The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183, 1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001
Guo, C. M., Chen, R. R., Kalhoro, D. H., Wang, Z. F., Liu, G. J., Lu, C. P., et al. (2014). Identification of genes preferentially expressed by highly virulent piscine Streptococcus agalactiae upon interaction with macrophages. PLoS ONE 9:e87980. doi: 10.1371/journal.pone.0087980
Hahn, M. W., and Höfle, M. G. (2001). Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35, 113–121 doi: 10.1111/j.1574-6941.2001.tb00794.x
Hernanz Moral, C., Flano del Castillo, E., Lopez Fierro, P., Villena Cortes, A., Anguita Castillo, J., Cascon Soriano, A., et al. (1998). Molecular characterization of the Aeromonas hydrophila aroA gene and potential use of an auxotrophic aroA mutant as a live attenuated vaccine. Infect. Immun. 66, 1813–1821.
Hojo, F., Sato, D., Matsuo, J., Miyake, M., Nakamura, S., Kunichika, M., et al. (2012). Ciliates expel environmental Legionella-Laden pellets ro stockpile food. Appl. Environ. Microbiol. 78, 5247–5257. doi: 10.1128/AEM.00421-12
Jacobs, M. E., DeSouza, L. V., Samaranayake, H., Pearlman, R. E., Siu, K. W., and Klobutcher, L. A. (2006). The Tetrahymena thermophila phagosome proteome. Eukaryot. Cell 5, 1990–2000. doi: 10.1128/EC.00195-06
Janda, J. M., and Abbott, S. L. (2010). The genus aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
King, C. H., Shotts, E. B. Jr., Wooley, R. E., and Porter, K. G. (1988). Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54, 3023–3033.
Kriegeskorte, A., Block, D., Drescher, M., Windmuller, N., Mellmann, A., Baum, C., et al. (2014). Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. MBio 5, e01447–e01414. doi: 10.1128/mBio.01447-14
Li, J., Zhang, X. L., Liu, Y. J., and Lu, C. P. (2011). Development of an Aeromonas hydrophila infection model using the protozoan Tetrahymena thermophila. FEMS Microbiol. Lett. 316, 160–168. doi: 10.1111/j.1574-6968.2010.02208.x
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Llewellyn, A. C., Jones, C. L., Napier, B. A., Bina, J. E., and Weiss, D. S. (2011). Macrophage replication screen identifies a novel Francisella hydroperoxide resistance protein involved in virulence. PLoS ONE 6:e24201. doi: 10.1371/journal.pone.0024201
Loughlin, M. F., Arandhara, V., Okolie, C., Aldsworth, T. G., and Jenks, P. J. (2009). Helicobacter pylori mutants defective in the clpP ATP-dependant protease and the chaperone clpA display reduced macrophage and murine survival. Microb. Pathog. 46, 53–57. doi: 10.1016/j.micpath.2008.10.004
Morales, V. M., Backman, A., and Bagdasarian, M. (1991). A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97, 39–47. doi: 10.1016/0378-1119(91)90007-X
Nam, D., Choi, E., Kweon, D. H., and Shin, D. (2010). The RstB sensor acts on the PhoQ sensor to control expression of PhoP-regulated genes. Mol. Cells 30, 363–368. doi: 10.1007/s10059-010-0126-8
Pang, M., Jiang, J., Xie, X., Wu, Y., Dong, Y., Kwok, A. H., et al. (2015). Novel insights into the pathogenicity of epidemic Aeromonas hydrophila ST251 clones from comparative genomics. Sci. Rep. 5:9833. doi: 10.1038/srep09833
Pang, M. D., Lin, X. Q., Hu, M., Li, J., Lu, C. P., and Liu, Y. J. (2012). Tetrahymena: an alternative model host for evaluating virulence of Aeromonas strains. PLoS ONE 7:e48922. doi: 10.1371/journal.pone.0048922
Park, K. S., Ono, T., Rokuda, M., Jang, M. H., Okada, K., Iida, T., et al. (2004). Functional characterization of two type III secretion systems of Vibrio parahaemolyticus. Infect. Immun. 72, 6659–6665. doi: 10.1128/IAI.72.11.6659-6665.2004
Pernthaler, J. (2005). Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3, 537–546. doi: 10.1038/nrmicro1180
Pukatzki, S., Ma, A. T., Revel, A. T., Sturtevant, D., and Mekalanos, J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513. doi: 10.1073/pnas.0706532104
Pushkareva, V. I., and Ermolaeva, S. A. (2010). Listeria monocytogenes virulence factor Listeriolysin O favors bacterial growth in co-culture with the ciliate Tetrahymena pyriformis, causes protozoan encystment and promotes bacterial survival inside cysts. BMC Microbiol. 10:26. doi: 10.1186/1471-2180-10-26
Rahman, M., Abd, H., Romling, U., Sandstrom, G., and Möllby, R. (2008). Aeromonas–Acanthamoeba interaction and early shift to a viable but nonculturable state of Aeromonas by Acanthamoeba. J. Appl. Microbiol. 104, 1449–1457. doi: 10.1111/j.1365-2672.2007.03687.x
Rasmussen, M. A., Carlson, S. A., Franklin, S. K., McCuddin, Z. P., Wu, M. T., and Sharma, V. K. (2005). Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect. Immun. 73, 4668–4675. doi: 10.1128/IAI.73.8.4668-4675.2005
Rehfuss, M. Y., Parker, C. T., and Brandl, M. T. (2011). Salmonella transcriptional signature in Tetrahymena phagosomes and role of acid tolerance in passage through the protist. ISME J. 5, 262–273. doi: 10.1038/ismej.2010.128
Sansom, F. M., Tang, L., Ralton, J. E., Saunders, E. C., Naderer, T., and McConville, M. J. (2013). Leishmania major methionine sulfoxide reductase A is required for resistance to oxidative stress and efficient replication in macrophages. PLoS ONE 8:e56064. doi: 10.1371/journal.pone.0056064
Serratrice, N., Cubizolle, A., Ibanes, S., Mestre-Frances, N., Bayo-Puxan, N., Creyssels, S., et al. (2014). Corrective GUSB transfer to the canine mucopolysaccharidosis VII cornea using a helper-dependent canine adenovirus vector. J. Control Release 181, 22–31. doi: 10.1016/j.jconrel.2014.02.022
Seshadri, R., Joseph, S. W., Chopra, A. K., Sha, J., Shaw, J., Graf, J., et al. (2006). Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188, 8272–8282. doi: 10.1128/JB.00621-06
Shimizu, T., Tsutsuki, H., Matsumoto, A., Nakaya, H., and Noda, M. (2012). The nitric oxide reductase of enterohaemorrhagic Escherichia coli plays an important role for the survival within macrophages. Mol. Microbiol. 85, 492–512. doi: 10.1111/j.1365-2958.2012.08122.x
Singh, V. K., and Moskovitz, J. (2003). Multiple methionine sulfoxide reductase genes in Staphylococcus aureus: expression of activity and roles in tolerance of oxidative stress. Microbiology 149, 2739–2747. doi: 10.1099/mic.0.26442-0
St. John, G., Brot, N., Ruan, J., Erdjument-Bromage, H., Tempst, P., Weissbach, H., et al. (2001). Peptide methionine sulfoxide reductase from Escherichia coli and Mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl. Acad. Sci. U.S.A. 98, 9901–9906. doi: 10.1073/pnas.161295398
Suarez, G., Khajanchi, B. K., Sierra, J. C., Erova, T. E., Sha, J., and Chopra, A. K. (2012). Actin cross-linking domain of Aeromonas hydrophila repeat in toxin A (RtxA) induces host cell rounding and apoptosis. Gene 506, 369–376. doi: 10.1016/j.gene.2012.07.012
Tomas, J. M. (2012). The main Aeromonas pathogenic factors. ISRN Microbiol. 2012:256261. doi: 10.5402/2012/256261
Valster, R. M., Wullings, B. A., Bakker, G., Smidt, H., and van der Kooij, D. (2009). Free-living protozoa in two unchlorinated drinking water supplies, identified by phylogenic analysis of 18S rRNA gene sequences. Appl. Environ. Microbiol. 75, 4736–4746. doi: 10.1128/AEM.02629-08
Vivas, J., Carracedo, B., Riano, J., Razquin, B. E., Lopez-Fierro, P., Acosta, F., et al. (2004). Behavior of an Aeromonas hydrophila aroA live vaccine in water microcosms. Appl. Environ. Microbiol. 70, 2702–2708 doi: 10.1128/aem.70.5.2702-2708.2004
Xie, F., Zhang, Y., Li, G., Zhou, L., Liu, S., and Wang, C. (2013). The ClpP protease is required for the stress tolerance and biofilm formation in Actinobacillus pleuropneumoniae. PLoS ONE 8:e53600. doi: 10.1371/journal.pone.0053600
Keywords: Aeromonas hydrophila, Tetrahymena, phagocytosis, SCOTS, msr genes
Citation: Pang M, Lin X, Liu J, Guo C, Gao S, Du H, Lu C and Liu Y (2016) Identification of Aeromonas hydrophila Genes Preferentially Expressed after Phagocytosis by Tetrahymena and Involvement of Methionine Sulfoxide Reductases. Front. Cell. Infect. Microbiol. 6:199. doi: 10.3389/fcimb.2016.00199
Received: 25 June 2016; Accepted: 13 December 2016;
Published: 26 December 2016.
Edited by:
Martin G. Klotz, Queens College, City University of New York, USAReviewed by:
Christian Berens, Friedrich Loeffler Institute, GermanyWeihua Chu, China Pharmaceutical University, China
Copyright © 2016 Pang, Lin, Liu, Guo, Gao, Du, Lu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongjie Liu, bGl1eW9uZ2ppZUBuamF1LmVkdS5jbg==
 Maoda Pang1,2
Maoda Pang1,2 Hechao Du
Hechao Du Yongjie Liu
Yongjie Liu