
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 30 August 2016
Sec. Bacteria and Host
Volume 6 - 2016 | https://doi.org/10.3389/fcimb.2016.00091
This article is part of the Research Topic Evasion of innate immunity by bacterial pathogens View all 10 articles
The intracellular pathogen Salmonella enterica servovar Typhimurium (S.typhimurium) modulates the host cell's phosphoinositide (PI) metabolism to establish its intracellular replicative niche, the Salmonella-containing vacuole (SCV). Upon invasion, phosphoinositide 3-phosphate (PI(3)P) and other early endosomal markers are rapidly recruited to and remain associated with the SCV throughout its early maturation. While the phosphoinositide 3-phosphatase myotubularin 4 (MTMR4) has an established role in regulating autophagy and cellular PI(3)P-content, two processes associated with the intracellular survival of S. typhimurium, a direct role for MTMR4 in Salmonella biology has not been examined. Here we demonstrate that GFP-tagged MTMR4 is recruited to the SCV and infection of cells depleted of endogenous MTMR4 results in a decrease in viable intracellular Salmonella. This reflects a significant increase in the proportion of SCVs with compromised integrity, which targets the compartment for autophagy and consequent bacterial cell death. These findings highlight the importance of PI(3)P regulation to the integrity of the SCV and reveal a novel role for the myotubularins in bacterial pathogenesis.
Salmonella is a Gram-negative facultative intracellular pathogen and a major cause of disease in humans (Haraga et al., 2008). The two predominant human pathogenic strains are Salmonella enterica serovar typhi, responsible for typhoid fever, and S. enterica servoar typhimurium, a causative agent of human gastroenteritis (Haraga et al., 2008). Once ingested, Salmonellae traverse the digestive tract and invade non-phagocytic epithelial cells lining the intestinal walls. Bacterial effector proteins are translocated directly into the host cells by one of two Type 3 Secretion Systems (T3SSs), to manipulate host membrane trafficking and cytoskeletal elements, initiating macropinocytosis and uptake of the pathogen into the cell (Kubori et al., 1998; Zhou and Galan, 2001). Whilst at least 40 effector proteins are translocated by the Salmonella Pathogenicity Island 1 (SPI1)-T3SS, prominent amongst these is SopB, a phosphatidylinositol phosphatase with sequence similarity to both mammalian phosphatidylinositol 4-phosphatase and phosphatidylinositol 5-phosphatase (Norris et al., 1998).
Phosphotidylinositols are an important class of lipid signaling molecules that can be singly or multiply phosphorylated on their inositol group to yield 7 spatio-temporally regulated phoshoinositides that are integral to a variety of cellular processes (Balla, 2013). Of these, phosphatidylinositol 3-phosphate (PI(3)P) and phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2) are primarily responsible for the regulation of traffic within the endosomal pathways (Di Paolo and De Camilli, 2006; Kerr et al., 2010). Following invasion, Salmonella alters the encompassing macropinosome to generate a replicative niche known as the Salmonella-containing vacuole (SCV). Concomittantly, the Salmonella Pathogenicity Island 2 T3SS (SPI2-T3SS) is activated, facilitating pathogen survival and replication (Figueira and Holden, 2012). Early in development, the SCV associates with organelles of the endosomal system, acquiring markers such as EEA1, SNX1 (Bujny et al., 2008), PI(3)P, and Rab5 (Dai et al., 2007; Bakowski et al., 2010). PI(3)P in particular is crucial to the stability and integrity of the SCV as intracellular Salmonella treated with PI(3)-kinase inhibitor, wortmannin, escape from the SCV and replicate within the cytoplasm unchallenged (Brumell et al., 2002; Scott et al., 2002). In the later stages of infection, SCV maturation is characterized by the formation of tubular protrusions called Salmonella Induced Filaments (SIFs), as well as the loss of PI(3)P and the acquisition of late endosomal markers such as LAMP1 and Rab7 (Knodler and Steele-Mortimer, 2005). Perturbing this maturation through inhibition of the phosphoinositide 5-kinase, PIKfyve, blocks SIF formation and has significant impact on the intracellular growth of the pathogen (Kerr et al., 2010). These observations highlight the tight spatiotemporal coordination of PI(3)P and PI(3,5)P2 on the SCV and demonstrate that alteration of either will ultimately influence the intracellular fate of the pathogen.
The 3′-dephosphorylation of PI(3)P and PI(3,5)P2 to phosphatidylinositol (PI) and phosphatidylinositol 5-phosphate (PI(5)P) is governed by the myotubularin (MTMR) family (Robinson and Dixon, 2006). The 14 myotubularins are characterized by the presence of a signature phosphatase domain, for which 6 are inactive due to mutations within the catalytic site. Of the 8 with an active phosphatase domain, MTMR3 and MTMR4 are further distinguished by a C-terminal FYVE domain (Lorenzo et al., 2006) but only MTMR4 is localized to PI(3)P-containing early endosomes, with MTMR3 localized to the cytosol (Lorenzo et al., 2006; Naughtin et al., 2010). MTMR4 can dephosphorylate PI(3)P in vitro and is recruited to the both early and recycling endosomes where it has been shown to influence the PI(3)P levels on these organelles (Zhao et al., 2001; Lorenzo et al., 2006; Naughtin et al., 2010). The capacity for MTMR4 to dephosphorylate PI(3,5)P2 remains unclear with the only evidence that MTMR4 immunoprecipitates appear to dephosphorylate PI(3,5)P2 (Naughtin et al., 2010).
Here we demonstrate that RNAi-mediated depletion of MTMR4 perturbs the intracellular growth of S. typhimurium, further highlighting the pathogen's dependence upon the 3-PIs. Specifically, we demonstrate that MTMR4 is recruited to PI(3)P-rich SCVs and report dysregulation of PI(3)P dependent stages of SCV maturation in MTMR4-depleted cells. This destabilizes the SCV leading to release of the pathogen into the cytoplasm. Concommitantly, we observe an elevation in the autophagy pathway and ultimately the destruction of the cytoplasmic Salmonella by the host cell innate immune system.
HA-MTMR4, GFP-MTMR4, GFP-MTMR3, GFP-LC3, myc-2*ML1N were as described previously in Walker et al. (2001), Birmingham et al. (2006), Lorenzo et al. (2006), Naughtin et al. (2010) and Li et al. (2013). mCherry-2*ML1N was obtained by performing restriction digest using restriction enzymes BglII and EcoRI on myc-2*ML1N to obtain the open reading frame of 2*ML1N and subcloned into mCherry-C1 following standard protocols. Monoclonal antibodies against EEA1 (610457, 1:100) and SNX1 (611483,1:100) were supplied by BD Bioscience. Monoclonal antibodies against the haemagglutinin epitope (MMS-101P, 1:1000) (YPYDVPDYA), myc epiptope (9B11, 1:2000), galectin-8 (ab109519,1:100) and p62 (R&D, MAB8028, 1:100) were supplied by Cell Signaling Technology, Abcam and R&D systems respectively. Monocloncal antibodies against β-tubulin (T3526, 1:2000) was purchased from Sigma Aldrich. Rabbit polyclonal antibodies against LC3 (NB100-2220) and GFP (A-6455, 1:500) were purchased from Novus biological and Molecular Probes (Invitrogen). Secondary antibodies and dextran-fluorescent conjugates were purchased from Life Technologies. S. Typhimurium strains used include the wild-type strain SL1344 (Hoiseth and Stocker, 1981) and the isogenic derivatives RFP-SL1344 (Birmingham et al., 2006) and SopB-deficient SL1344 (Mallo et al., 2008). Rapamycin, Wortmannin and 3-MA were purchased from Sigma Aldrich.
A431 cells and Hela cells stably expressing GFP-LAMP1 (Kehl and Hensel, 2015) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal bovine serium with 2 mM L-glutamine (Invitrogen) in humidified air/atmosphere (5% CO2) at 37°C. Cells were transfected using Lipofectamine 2000 as per manufacturer's instructions (Life Technologies)
pGIPZ small hairpin RNA (shRNA) plasmids (Thermo Scientific) used included non-silencing control shRNA: RHS4346; human MTMR4 shRNAs (shRNA #1: V2LHS_375815), were supplied by the Institute for Molecular Bioscience Life Science Automation (LISA) Facility. A431 cells were transduced with shRNA particles in the presence of 8 μg/ml polybrene (Milipore) for 4 h in serum free media before incubating them overnight in normal growth media. Transduced cells were selected for with DMEM containing 1 μg/ml puromycin (Sigma Aldrich) for 1 week.
RNA from cells was extracted according to the manufacturer's directions (Sigma Aldrich). One microgram total RNA was used to produce cDNA using oligdT primers and Superscript III (Invitrogen). Quantitative RT-PCR was conducted on samples following shRNA treatment and analyzed in triplicate. The housekeeping gene GAPDH was used as an internal control to calculate the ΔCT for each sample. Primers for MTMR4 were targeted. MTMR4 expression was quantified using TaqMan gene expression assays as per manufacturer instructions (Applied Biosystems) in 96 well plate format.
Endosomes were labeled with fluorescent dextran by culturing live cells in the presence of 100 μg/ml dextran conjugated to tetramethylrhodamine for 15 min before being washed thoroughly with excess media and fixed for further analysis.
Salmonella infections were performed as previously described (Yang et al., 2015). Briefly, S. typhimurium (SL1344) was cultured in Luria Broth (LB) overnight at 37°C with shaking (225 rpm) followed by dilution into 3 ml of fresh LB (1:31) and continued culture for a further 3.5 h. 1 ml of the resultant culture was then centrifuged at 13,000 rpm for 1 min and resuspended in 10 ml of phosphate-buffered saline (PBS) and the optical density (OD) measured. The appropriate bacterial dilution was prepared in normal growth media and HeLa cells infected with a multiplicity of infection (MOI) of 1.0 unless otherwise stated. After 30 min of infection, the cells were washed three times and incubated in normal media containing 100 μg/ml gentamicin. At 1.5 h p.i., the gentamicin concentration was reduced to 10 μg/ml for the remaining duration of the experiment.
Colony forming unit assay was performed as previously described (Kerr et al., 2010). Briefly, infected cells were washed 3 times with PBS and lysed in 1X PBS containing 0.25% SDS. The resultant lysate were serially diluted and plated onto LB agar plates containing 100 μg/ml Ampicillin. The plates were incubated at 37°C for 24 h and resultant colonies counted to determine the number of Colony Forming Units (CFU).
For live imaging, cells were seeded on 35 mm glass-bottom MatTek dishes and subsequently imaged on a Zeiss LSM 710 confocal microscope using a 63 X or 100 X oil immersion objective and maintained at 37°C and imaged over the time-course in a 5% CO2 humidified chamber.
Cells fixed with 4% paraformaldehyde were permeabilized with 0.1% triton X-100 for 10 min, and blocked in 2% BSA (in 1xPBS). Primary antibodies were incubated for 1 h at room temperature. After incubation, cells were washed three times with blocking reagent and incubated with Alexa-Fluor-488-conjucated, Alexa-Fluor-547-conjugated and Alex-Fluor-647-conjugated secondary antibodies (1:400) Cells were also labeled with DAPI, washed three times with 1xPBS and mounted onto glass slides using mounting Media. Single-plane or stacked images were obtained using a Zeiss LSM 710 upright Microscope and a 40 X oil or 63 X oil immersion objective.
Cell lysate samples were subjected to bicinchoninic acid (BCA) assay (Thermo Scientific) to determine total protein concentration. Twenty microgram of protein per sample were resolved by SDS-PAGE transferred onto Immobilon-P membrane (Millipore) according to the manufacturer's instructions. Western immunoblotting using ECL was performed as per manufacturer's instructions.
Intracellular Salmonella growth was analyzed and quantified by FIJI. Briefly, the “subtract background” functionality was applied and images thresholded to an intensity of 155 (8-bit image) and quantified through the “Analyze Particles” feature. The resultant particle count is represented in arbitrary units. All results were tabulated and presented using GraphPad software version 6.
Maximum projections images were captured using a Zeiss LSM710 confocal laser scanning microscope under 63 × magnification and the number of endosomal markers positive SCVs were quantified using FIJI. Endosomal markers overlapping SCVs were scored as positive. All results were tabulated in GraphPad Prism software version 6 and represented as a percentage out of 100.
Data were analyzed using Student's T-test, performing pairwise analysis to control where appropriate. A p-value of less than 0.05 was considered statistically significant. All analyses were performed using GraphPad Prism software version 6.
The transition of early to late endosomal markers as the SCV matures implies the conversion of PI(3)P to PI(3,5)P2. We demonstrated that inhibition of PIKfyve disrupts the formation of SIFs and the intracellular replication of S. typhimurium (Kerr et al., 2010), however, due to the absence of a PI(3,5)P2-specific probe at the time, we were unable to directly monitor the presence of PI(3,5)P2 accumulation on the SCV. Recently a PI(3,5)P2-specific probe was reported (Li et al., 2013) and we therefore sought to monitor the association of PI(3,5)P2 with the SCV. Hela cells stably expressing GFP-LAMP1 were transfected with mCherry-2*ML1N and infected with late log-phase S. typhimurium (WT-SL1344). Time-lapse videomicroscopy revealed a concomitant accumulation of mCherry-2*ML1N and LAMP1 on the SCV from 60 min post-infection (p.i.) (Figure 1A). The final stages of SCV maturation include the formation of extensive LAMP1-positive tubular structures known as SIFs, with which mCherry-2*ML1N remains associated at 6 h p.i (Figure 1B), validating the model that the SCV transitions from a PI(3)P-positive compartment to become PI(3,5)P2-positive as it matures. Given that we demonstrated that the SCV is only transiently enriched with PI(3)P (Kerr et al., 2010), we speculated that concurrent with its conversion to PI(3,5)P2, SCV-PI(3)P may also be dephosphorylated to PI by the endosome-associated myotubularin phosphatase MTMR4 to ensure complete maturation of the SCV.
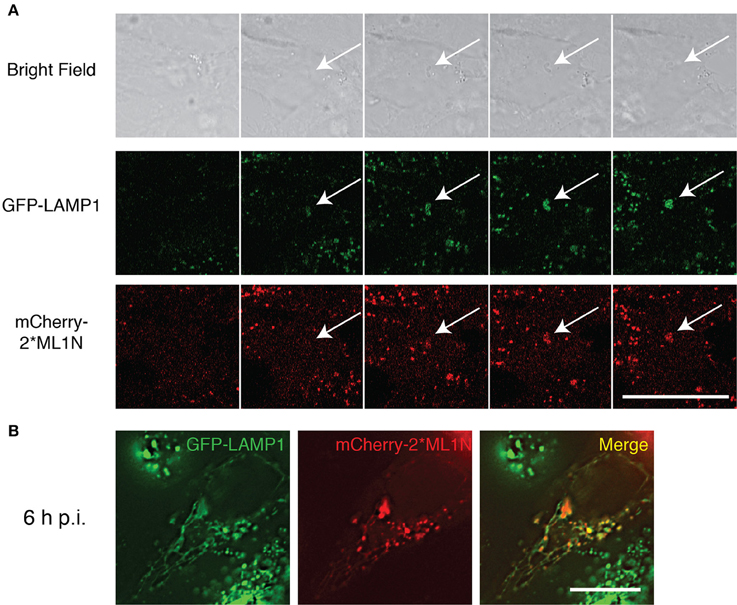
Figure 1. Recruitment of PI(3,5)P2 to the SCV and Sifs. (A) Time-lapse video microscopy of WT-SL1344 infected HeLa cells stably expressing GFP-LAMP1 and transiently expressing mCherry 2*ML1N. Montage representative of a 90 min movie captured using Zeiss LSM710 FCS scanning confocal microscope. (B) HeLa cells stably expressing GFP-LAMP1 cells and transiently expressing mCherry-2*ML1N were infected with WT-SL1344 and imaged 6 h p.i live on Zeiss LSM710 FCS scanning confocal microscope. Scale bar: 20 μm. White arrows indicate SCV.
To examine how MTMR4 regulates intracellular PI(3)P and/or PI(3,5)P2 concentrations, shRNA-MTMR4 depleted A431 cells were generated and validated using RT-PCR (Figure 2A). MTMR4 knockdown cells, as well as those overexpressing HA-MTMR4, were fixed and immunolabelled for endogenous PI(3)P-binding EEA1 (Simonsen et al., 1998) or the PI(3,5)P2-binding probe Myc-2*ML1N (Li et al., 2013). Cells depleted for MTMR4 presented a 25% increase in the total number of EEA1-positive puncta relative to non-silencing control cells (Figures 2B,C). In contrast, HA-MTMR4 overexpression decreased the total number of EEA1-positive puncta by 30% relatively to the mock-transfected cells (Figures 2D,E). To determine if the observed increase in EEA1 labeling in MTMR4-depleted cells reflected perturbation in overall number of endosomes, a 10,000 MW dextran-tetramethylrhodamine (TR) uptake assay was performed. A 30 min pulse-chase revealed no significant difference in the number of dextran-TR positive endosomes in the MTMR4-depleted cells when compared to the non-silencing control cells, indicating no significant impact on the number of endosomes formed within these cells (Figures 2F,G). These observations are consistant with the documented activity of MTMR4 (Lorenzo et al., 2006; Naughtin et al., 2010). Several members of the myotubularins family have been implicated to regulate PI(3,5)P2 (Robinson and Dixon, 2006) althought to date no direct evidence exists that PI(3,5)P2 is a substrate for MTMR4. To determine if MTMR4 modulates intracellular PI(3,5)P2, MTMR4 depleted or control cells were singly or co-transfected with either myc-2*ML1N and/or HA-MTMR4. No differences in fluorescent intensities and subcellular distributions of myc-2*ML1N in either cells depleted for MTMR4 or ectopically expressing MTMR4 were observed (Figures 2H–K). Taken together, these results indicate that MTMR4 negatively regulates PI(3)P content on early endosomes, but has no direct role in regulating PI(3,5)P2 distribution in the cell.
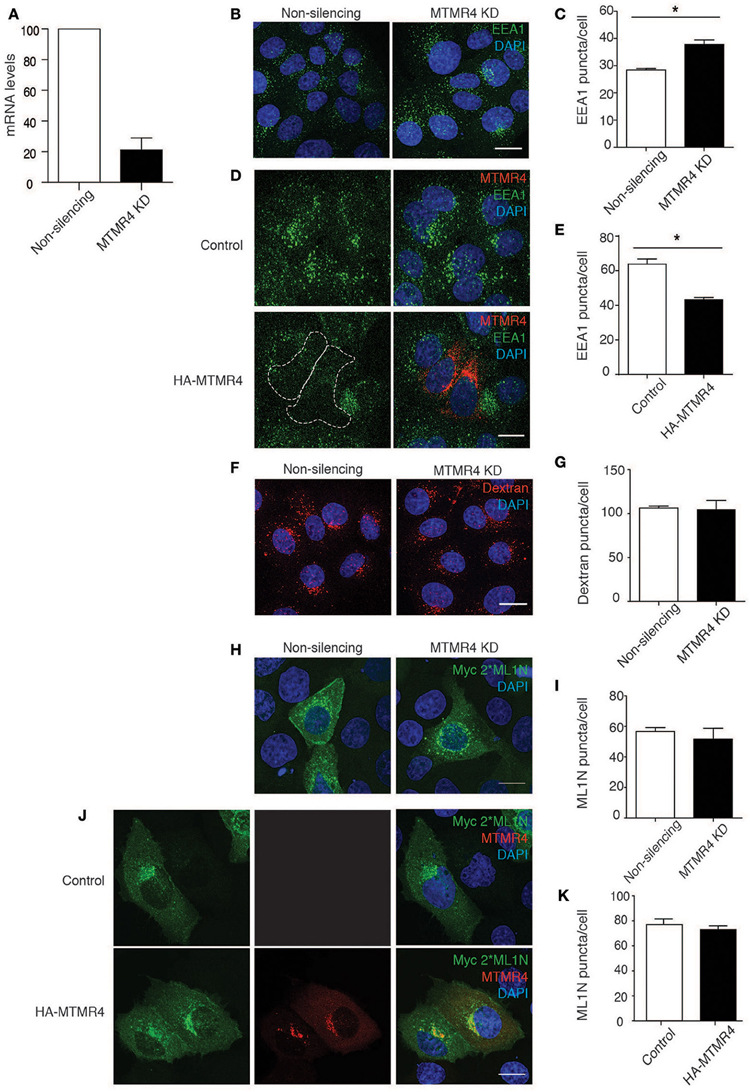
Figure 2. MTMR4 regulates PI(3)P endosomal levels but not PI(3,5)P2. (A) A431 cells were transduced with shRNA targeting MTMR4 and the knockdown efficiently confirmed using RT-PCR. (B) MTMR4-depleted cells were fixed and immunolabelled with EEA1, counterstained with DAPI. (C) The number of EEA1 puncta was analyzed. (D) A431 cells were seeded and transfected with HA-MTMR4 or HA-empty vector, fixed, and immunofluorescence performed using antibodies against EEA1. (E) Quantification of EEA1 puncta following overexpression of HA-MTMR4. (F) MTMR4-depleted cells were pulsed with dextran conjugated to tetramethylrhodamine for 30 min before thorough washing with excess media and fixed, counterstained with DAPI. (G) Quantification of dextran puncta. (H) MTMR4 depleted cells were transfected with Myc-2*ML1N construct, immunolabelled and quantified in (I). (J) A431 cells were co-transfected with MTMR4 or empty vector and Myc-2*ML1N, fixed, immunolabeled and puncta quantified in (K). Data representative of three independent experiments (N = 3; at least 25 cells counted per condition; Error bar denote ± S.E.M., *P < 0.05, Scale bar: 20 μm).
During Salmonella infection, PI(3)P has been observed to first accumulate on membrane ruffles and subsequently remains associated with the SCV throughout its early biogenesis (Scott et al., 2002). To determine if MTMR4 is associated with the nascent SCV, A431 cells were transfected with GFP-MTMR4 or GFP-MTMR3 for 18 h before being infected with a wildtype S. typhimurium strain (SL1344) expressing RFP (RFP-SL1344) at a MOI of 1. Time-lapse video microscopy demonstrated that GFP-MTMR4 but not GFP-MTMR3 associated with the nascent SCV as early as 15 min p.i. (Figure 3A). To dissect the mechanism of MTMR4 recruitment further, we infected GFP-MTMR4 expressing cells with a SopB-deletion mutant strain reported to be deficient in accumulation of PI(3)P on its SCV (Mallo et al., 2008). Indeed, MTMR4 was not recruited to the ΔSopB-SL1344 SCVs even at 1 h p.i (Figure 3B).
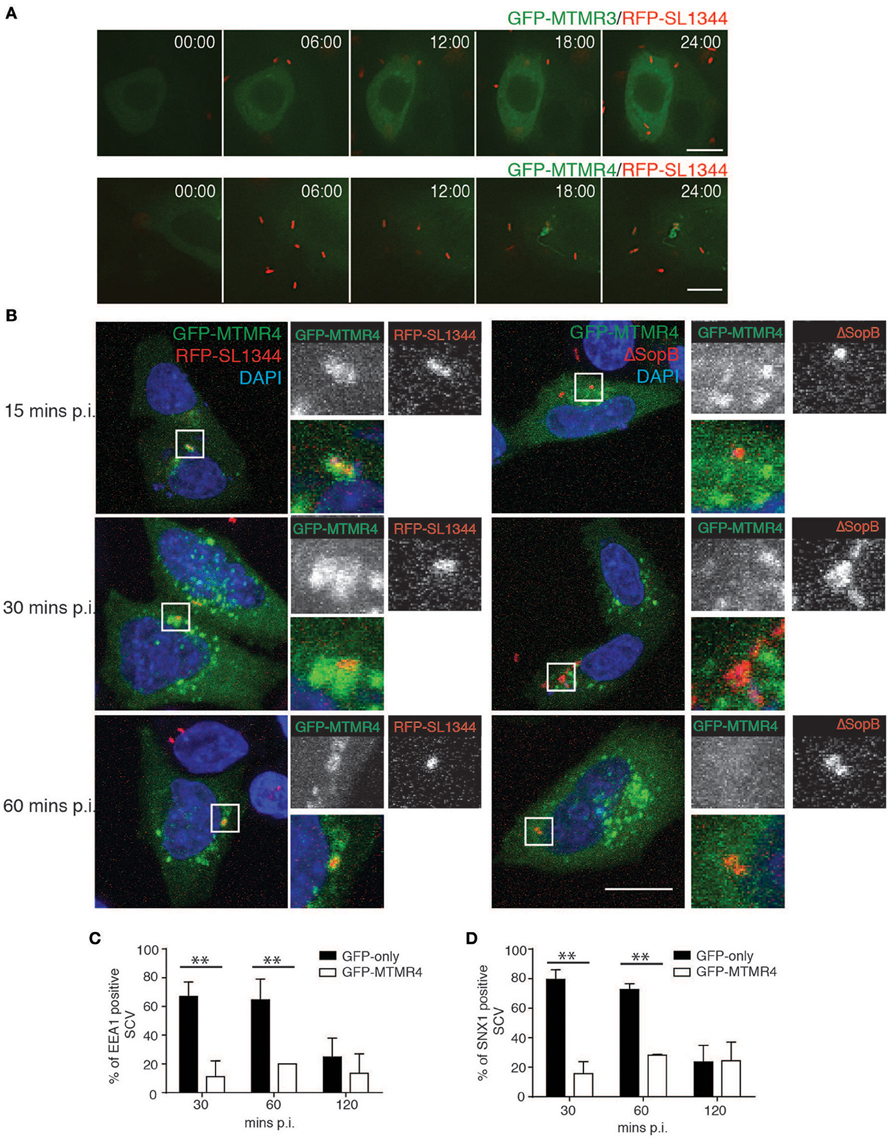
Figure 3. MTMR4 is associated with the SCV. (A) Time-lapse video microscopy of A431 cells transfected with GFP-MTMR4 or GFP-MTMR3 and infected with RFP-SL1344. Montage is representative of a 25 min movie captured using Nikon Deconvolution microscope. (B) A431 cells were transfected with GFP-MTMR4 and infected with either RFP-SL1344 or ΔSopB-SL1344. MTMR4-positive Salmonella were then probed for the presence of EEA1 or SNX1 and quantified over a time course assay (C,D) respectively. (N = 3; Error bar denote mean ± S.D., Scale bar: 20 μm, **p < 0.005).
To examine the potential for MTMR4 in Salmonella invasion, an invasion assay was performed by infecting cells ectopically expressing GFP-MTMR4 with RFP-SL1344 at a MOI~5 for 10 min before fixing the cells. Anti-Salmonella LPS antibodies were used to probe for extracellular Salmonella in unpermeabalised cells, which revealed no difference the invasion rates between MTMR4 overexpressing (34.4% ± 1.99% internalized bacteria) or control cells (34.61 ± 1.66% internalized bacteria), confirming that MTMR4 does not play a role in bacterial invasion. Although, the early stages of an SCV appears to share properties in common with constitutive macropinosomes, including recruitment of early endosomal markers, SNXs, and PI(3)P, it is significantly modified by secreted Salmonella effectors so that the PI(3)P accumulates and remains elevated for up to 90 min p.i on individual SCVs (Hernandez et al., 2004). We next assessed the impact MTMR4 has on the early maturation of the SCV. GFP-MTMR4 expressing cells were infected with RFP-SL1344 for 30 min, washed and incubated with 100 μg/ml of gentamycin for 1 h to kill extracellular Salmonella before incubating the cells further in 10 μg/ml gentamycin. The samples were fixed, immunolabelled with antibodies specific for EEA1 or SNX1 followed by a fluorescently-conjugated secondary antibody and examined using confocal microscopy to quantify recruitment of these proteins to individual SCV (Figures 3C,D). At 30 and 60 min p.i, the majority of SCVs containing RFP-SL1344 in GFP-expressing control cells were associated with early maturation markers EEA1 and SNX1 which then decreased by 120 min as the SCV undergoes maturation. In contrast, ectopic expression of GFP-MTMR4 resulted in the MTMR4-positive SCVs remaining void of any early maturation markers throughout the 120 min time period examined (Figures 3C,D). Here we demonstrate that the elevated expression of MTMR4 decreases the normal recruitment of PI(3)P-dependent endosomal proteins during the early SCV maturation stages presumably due to the excess phosphatase efficiently converting PI(3)P to PI.
Because MTMR4 was recruited to the SCV and modulation of its level of expression impacted on SCV early maturation, we next determined if specific depletion of MTMR4 had an impact on intracellular Salmonella growth. Cells depleted for MTMR4 were infected with RFP-SL1344 and prepared for immunofluorescent investigation or harvested for colony forming unit (CFU) assays at specific times post-infection (Figure 4). No significant difference was observed 1 h p.i, indicating that depletion of MTMR4 does not impair Salmonella invasion. A significant reduction in viable bacteria was observed in the MTMR4 knockdown cells as early as 4 h p.i through to 12 h p.i. (Figure 4C). Immunofluorescent analysis confirmed a decrease in total number of intracellular Salmonella in MTMR4 depleted cells 12 h p.i (Figures 4A,B). The apparent incongruence between the CFU and immunofluorescence assays earlier in the infection suggests that a proportion of the Salmonella in MTMR4-depleted cells are no longer viable.
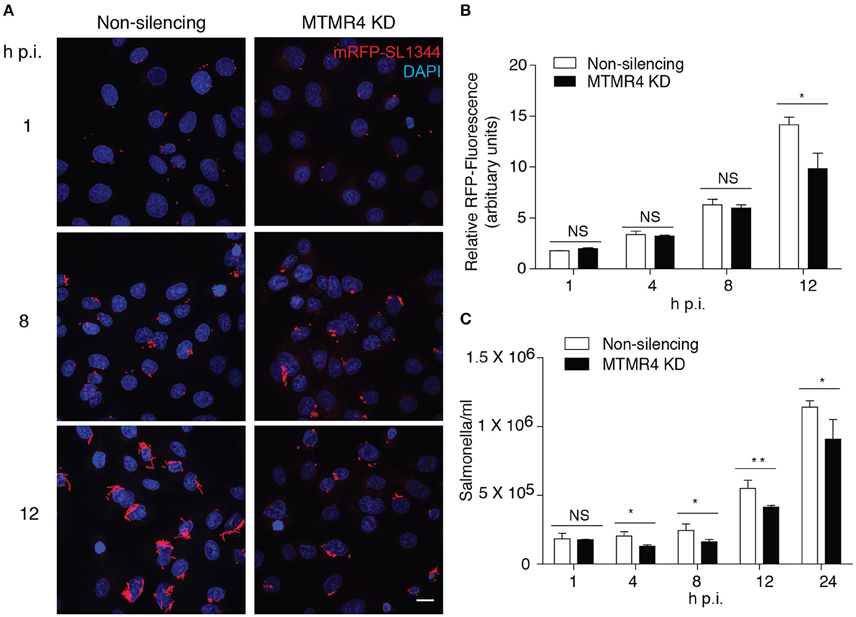
Figure 4. MTMR4 depletion renders cells less permissible to SCV growth. (A) MTMR4-depleted A431 and control cells were infected with RFP-SL1344. Intracellular replication was monitored by measuring the relative area occupied by RFP-fluorescence in (B) infected cells and colony forming unit (C) assay. (N = 3, Error bar denote mean ± S.D., Scale bar 20 μm, *p < 0.05, **p < 0.005).
Ordinarily, Salmonella maintains the SCV such that it matures, acquires LAMP1 from 1 h p.i and and forms SIFs that expand the compartment to accommodate the replicating bacteria within. Birmingham et al. (2006) reported that a small subpopulation of Salmonella is unable to establish such a stable replicative niche, and are exposed to the cytoplasm after damaging the SCV with their SPI1 components. The exposed bacteria are consequently recognized and destroyed by the cell's autophagic system (Birmingham et al., 2006). Several active MTMR family members have recently been implicated as negative regulators of autophagy (Walker et al., 2001; Vergne et al., 2009; Zou et al., 2012). As such, we questioned if the bactericidal effect observed in MTMR4-depleted cells could be attributed to elevated autophagy. This was confirmed by both western immunoblotting of endogenous LC3 and fluorescence microscopy of GFP-LC3. Because the shRNA constructs used to generate non-silencing and MTMR4 knockdown cells also encode turbo-GFP, antibodies against the GFP-fused to LC3 and appropriate secondary antibodies were used to monitor the subcellular distribution of GFP-LC3 rather than its native fluorescence. In both cases, MTMR4 knockdown cells presented elevated autophagy when compared to the control cells (Figure 5A) as well as the increased endogenous LC3 band intensity by western immunoblotting (Figure 5B). Within HeLa cells, the accepted non-phagocytic model used in Salmonella research, LC3 is observed as a single band with the soluble form of LC3-II not observed (Klionsky et al., 2016). Therefore, we are not able to directly monitor variations in the ratio of soluble LC3 with lipidated LC3-II to monitor autophagic flux in these cells. To verify the identity of the GFP-LC3-positive puncta, cells were treated with inhibitors of autophagy (wortmannin or 3-methyladenine (3-MA); Lindmo and Stenmark, 2006) and inducers of autophagy (rapamycin; Jung et al., 2010). Consistent with expectation, the number of GFP-LC3 positive puncta observed in the MTMR4 knockdown cells was decreased when autophagy was inhibited and the number of GFP-LC3 puncta within control cells rose to meet that present in DMSO-treated MTMR4 knockdown cells when autophagy was induced with rapamycin. The loss of GFP-LC3 punctate structures following treatment with wortmannin is consistent with a global increase in autophagy under resting conditions in MTMR4 knockdown cells. We speculate that this elevates the defensive potential of these cells making the environment less hospitable to the pathogen.
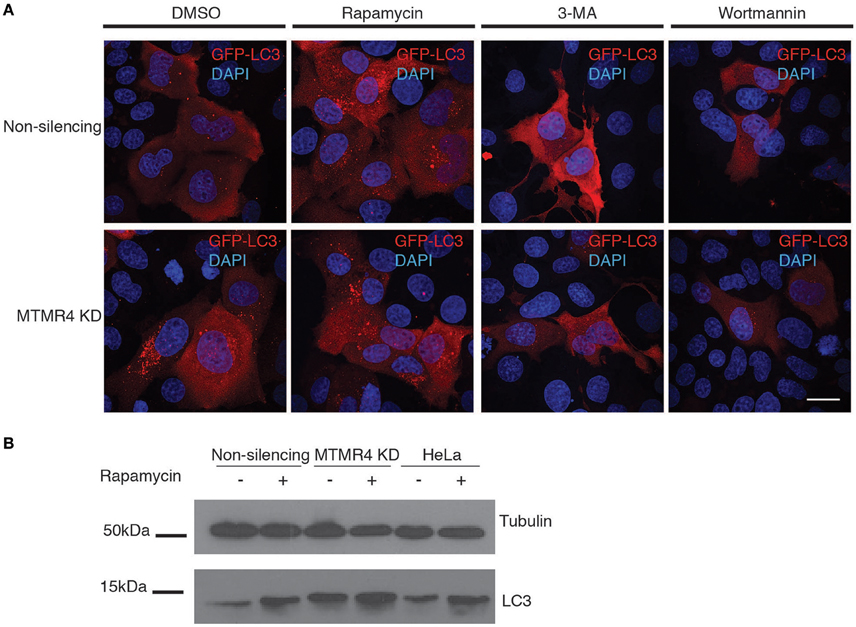
Figure 5. Depletion of MTMR4 enhances autophagy. (A) MTMR4-depleted HeLa and control cells were transfected with GFP-LC3, and treated with DMSO, 100 nM rapamycin, 100 nM wortmannin, or 20 mM 3-MA. Coverslips were then fixed and immunolabelled using antibodies against GFP. (B) Representative blot of MTMR4 knockdown and control whole cell lysates were harvested and 20 μg of proteins subjected to western immunoblotting against LC3 and tubulin. Scale bar 20 μm.
We subsequently extended these observations into the context of Salmonella infection. First, we determined the association rate of galectin-8, a β-galectoside-binding lectin that has been shown to accumulate on damaged bacteria-containing vacuoles (Thurston et al., 2012). A time course assay on S. typhimurium-infected MTMR4-depleted cells was performed and probed for endogenous levels of galectin-8 associated with the SCV using specific antibodies. Although, there were no differences between MTMR4 depleted and control cells at 1 h p.i, at 3 h p.i, galectin-8 positive SCVs in knockdown cells were observed at higher frequency when compared to the proportion of galectin-8 positive SCVs in control cells which rapidly dropped to background levels (~5%; Figure 6A).
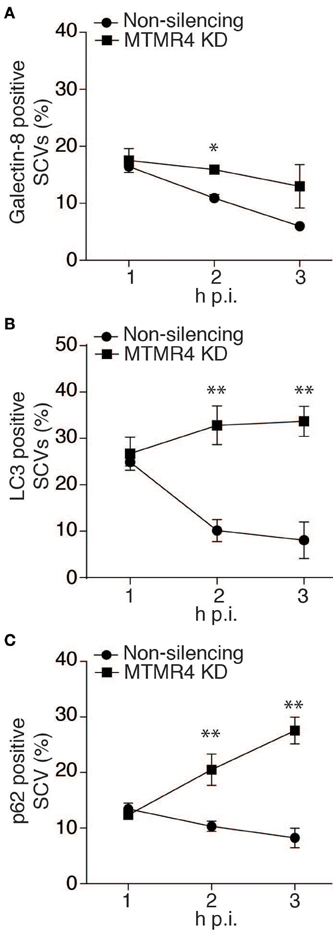
Figure 6. The proportion of damaged SCVs increases in MTMR4 depleted cells. MTMR4-depleted A431 cells and non-silencing control cells were infected with RFP-SL1344 or WT-SL1344. Cells were fixed at stipulated time points and probed for association autophagic markers. Quantification of the percentage of (A) galectin-8+, (B) recruitment of GFP-LC3+ or (C) p62+ bacteria. (N = 3, Error bar denote mean ± S.D., *p < 0.05, **p < 0.005).
Having shown an increased proportion of damaged SCVs in MTMR4 depleted cells, we next wanted to quantify the number of SCVs being recognized by the autophagy pathway. To achieve this, MTMR4-depleted cells infected with RFP-SL1344 or WT-SL1344 were examined to determine the proportion of SCVs positive for GFP-LC3 or endogenous p62. Similar to what was observed with galectin-8, when compared with control cells, there was an increase proportion of SCVs with both LC3 and p62 in MTMR4 depleted cells (Figures 6B,C). LC3 association with the SCV remained elevated (~30%) in MTMR4-depleted cells relative to control cells from 2 to 3 h p.i. The association of p62 with SCVs rapidly increased from 1 h p.i. (~13%) to 3 h p.i (~27%). Taken together, these findings demonstrate that the depletion of MTMR4 leads to an increased number of SCVs being damaged and the bacteria being targeted by the host cell's autophagic pathway.
The SCV undergoes a maturation process similar to that of an endosome, but stops short of fusing with lysosomes (Eswarappa et al., 2010). To achieve this, Salmonella modulates host cell phosphoinositide metabolism to alter the cell's membrane trafficking pathways(Kerr et al., 2012). Previously, we demonstrated that the SCV is transiently enriched for PI(3)P and that perturbation of phosphatidylinositide 5-kinase activity interferes with the later stages of SCV maturation and the intracellular replication of the pathogen (Kerr et al., 2010). Here we go onto demonstrate the accumulation of PI(3,5)P2 on the maturing SCV and set out to investigate the potential role for PI(3)-phosphatases in SCV maturation.
MTMR4 is one of a large family of 3-phosphatases that have been reported to hydrolyse PI(3)P and PI(3,5)P2 (Lorenzo et al., 2006; Naughtin et al., 2010). We find that when MTMR4 is ectopically expressed, the proportion of PI(3)P-positive endosomes, as monitored by recruitement of EEA1, decreases while RNAi mediated depletion of MTMR4 leads to a corresponding increase in the proportion of EEA1-positive endosomes. No change in PI(3,5)P2-positive endosomes, as monitored using a PI(3,5)P2-specific probe (Li et al., 2013) was observed. This is potentially due to PI(3)P being the preferential substrate of MTMR4 or alternatively MTMR4 membrane recruitement via its C-terminal FYVE domain (Lorenzo et al., 2006) precludes it from been associated with PI(3,5)P2 positive membranes.
Interestingly, despite MTMR3 and MTMR4 each encoding a FYVE domain (Lorenzo et al., 2005, 2006), only MTMR4 was observed to be recruited to the nascent SCV. This recruitment was dependent upon PI(3)P as ΔSopB Salmonella mutants which are unable to generate PI(3)P on their SCVs through the recruitment of the class III PI 3-kinase, Vps34 (Hernandez et al., 2004; Mallo et al., 2008), did not recruit MTMR4. Additionally, no association between MTMR4 and the SCV was observed in the later stages of the infection when PI(3)P levels are much reduced (Kerr et al., 2010).
Strikingly, we find that depletion of MTMR4 destabilized the SCV and led to a concomitant depletion in intracellular bacterial viability. Multiple studies have offered explanations as to why Salmonella chooses to reside within a vacuole and modulate trafficking instead of proliferating freely in the nutrient rich cytosol. Perhaps most compelling amongst these is that the SCV provides the means to avoid the myriad of host defenses that serve as a protective net for the host cell in the event of an infection (Nakagawa et al., 2004; Birmingham et al., 2006; Thurston et al., 2012). Autophagy in particular has been demonstrated to target cytosolic Salmonella for degradation within autophagolysosomes (Birmingham et al., 2006). A recent siRNA-based screen determined that depletion of MTMR6, MTMR7, and MTMR14 lead to increased levels of autophagic vacuoles as well as an upregulation of long-lived protein proteolysis (Vergne et al., 2009). MTMR3 has also been implicated in regulating constitutive autophagy initiation and autophagosome size in epithelial cells (Taguchi-Atarashi et al., 2010). It is therefore perhaps not so surprising that depletion of MTMR4, which induces increased cellular concentrations of PI(3)P-positive organelles, also leads to enhanced autophagy. This enhanced state of autophagy likely contributes to the increased capacity of the MTMR4-depleted cells to eliminate intracellular Salmonella. Recent studies have made conflicting conclusions about the role of autophagy with regards to the intracellular viability of Salmonella (Yu et al., 2014; Kreibich et al., 2015). Using CFU assays to monitored viable bacteria 6 h post-infection, Yu et al. (2014) observed decreased viability in HeLa cells when autophagy was inhibited using RNAi-depletion of LC3 while Kreibich et al. (2015) observed elevated viable bacteria in autophagy impaired Atg5−/− MEFs. The differences between these studies may reflect multiple functions associated with the individual autophagy components manipulated, cell types used or the differences in MOI used.It should also be noted that these targeted factors demonstrated to directly regulate autophagy whilst, as demonstrated in our findings, MTMR4 regulates both cellular PI(3)P and autophagy, both of which are recognized to be intrinsic to intracellular survival of Salmonella.
Taken together we propose a model where by PI(3)P turnover on the SCV is a tightly regulated process coordinated through the action of both bacterial effectors like SopB as well as the host factors, PIKfyve and MTMR4. Disregulation of this turnover through MTMR4-depletion both destabilizes the SCV and elevates cellular autophagy leading to destruction of the pathogen. MTMR4 is therefore a novel host factor that plays a vital role in Salmonella pathogenesis.
All authors contributed to the conception, design, interpretation of the findings, drafting, and preparation of the manuscript. WT and MK executed the experiments.
This work was supported by funding from the National Health and Medical Research Council (NHMRC) of Australia (606788). MK is supported by an Australian Research Council Discovering Early Career Researcher Award (DE120102321). RT is supported by NHMRC Senior Research Fellowship (APP1041929).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Microscopy was carried out at the Australian Cancer Research Foundation (ACRF)/Institute for Molecular Bioscience (IMB) Dynamic Imaging Facility for Cancer Biology. We would like to thank Christina Mitchell (Monash University, Australia) for supplying HA-MTMR4 and Michael Clague (University of London, United Kingdom) for supplying GFP-MTMR3 and GFP-MTMR4. We would like to thank Nat F. Brown (University of British Colombia, Canada) for supplying SopB-deficient Salmonella strains.
Bakowski, M. A., Braun, V., Lam, G. Y., Yeung, T., Do Heo, W., Meyer, T., et al. (2010). The phosphoinositide phosphatase SopB manipulates membrane surface charge and trafficking of the Salmonella Containing Vacuole. Cell Host Microbe 7, 453–462. doi: 10.1016/j.chom.2010.05.011
Balla, T. (2013). Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137. doi: 10.1152/physrev.00028.2012
Birmingham, C. L., Smith, A. C., Bakowski, M. A., Yoshimori, T., and Brumell, J. H. (2006). Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 281, 11374–11383. doi: 10.1074/jbc.M509157200
Brumell, J. H., Tang, P., Zaharik, M. L., and Finlay, B. B. (2002). Disruption of the Salmonella-containing vacuole leads to increased replication of Salmonella enterica serovar typhimurium in the cytosol of epithelial cells. Infect. Immun. 70, 3264–3270. doi: 10.1128/IAI.70.6.3264-3270.2002
Bujny, M. V., Ewels, P. A., Humphrey, S., Attar, N., Jepson, M. A., and Cullen, P. J. (2008). Sorting nexin-1 defines an early phase of Salmonella-containing vacuole-remodeling during Salmonella infection. J. Cell Sci. 121, 2027–2036. doi: 10.1242/jcs.018432
Dai, S., Zhang, Y., Weimbs, T., Yaffe, M. B., and Zhou, D. (2007). Bacteria-generated PtdIns(3)P recruits VAMP8 to facilitate phagocytosis. Traffic 8, 1365–1374. doi: 10.1111/j.1600-0854.2007.00613.x
Di Paolo, G., and De Camilli, P. (2006). Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657. doi: 10.1038/nature05185
Eswarappa, S. M., Negi, V. D., Chakraborty, S., Sagar, B. K. C., and Chakravortty, D. (2010). Division of the Salmonella-containing vacuole and depletion of acidic lysosomes in Salmonella-infected host cells are novel strategies of Salmonella enterica to avoid lysosomes. Infect. Immun. 78, 68–79. doi: 10.1128/IAI.00668-09
Figueira, R., and Holden, D. W. (2012). Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology 158, 1147–1161. doi: 10.1099/mic.0.058115-0
Haraga, A., Ohlson, M. B., and Miller, S. I. (2008). Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6, 53–66. doi: 10.1038/nrmicro1788
Hernandez, L. D., Hueffer, K., Wenk, M. R., and Galán, J. E. (2004). Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science 304, 1805–1807. doi: 10.1126/science.1098188
Hoiseth, S. K., and Stocker, B. A. D. (1981). Aromatic-dependent salmonella-typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239. doi: 10.1038/291238a0
Jung, C. H., Ro, S. H., Cao, J., Otto, N. M., and Kim, D. H. (2010). mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295. doi: 10.1016/j.febslet.2010.01.017
Kehl, A., and Hensel, M. (2015). Live cell imaging of intracellular Salmonella enterica. Methods Mol. Biol. 1225, 199–225. doi: 10.1007/978-1-4939-1625-2_13
Kerr, M. C. C., Castro, N. A., Karunaratne, S., and Teasdale, R. D. (2012). “The phosphoinositides: key regulators of Salmonella containing vacuole (SCV) trafficking and identity,” in Salmonella - Distribution, Adaptation, Control Measures and Molecular Technologies, eds B. A. Annous and J. B. Gurtler (Rijeka: InTech), 251–264.
Kerr, M. C., Wang, J. T. H., Castro, N. A., Hamilton, N. A., Town, L., Brown, D. L., et al. (2010). Inhibition of the PtdIns(5) kinase PIKfyve disrupts intracellular replication of Salmonella. EMBO J. 29, 1331–1347. doi: 10.1038/emboj.2010.28
Klionsky, D. J., Abdelmohsen, K., Abe, A., Abedin, M. J., Abeliovich, H., Acevedo Arozena, A., et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. doi: 10.1080/15548627.2015.1100356
Knodler, L. A., and Steele-Mortimer, O. (2005). The Salmonella effector PipB2 affects late endosome/lysosome distribution to mediate sif extension. Mol. Biol. Cell 16, 4108–4123. doi: 10.1091/mbc.E05-04-0367
Kreibich, S., Emmenlauer, M., Fredlund, J., Rämö, P., Münz, C., Dehio, C., et al. (2015). Autophagy proteins promote repair of endosomal membranes damaged by the Salmonella type three secretion system 1. Cell Host Microbe 18, 527–537. doi: 10.1016/j.chom.2015.10.015
Kubori, T., Matsushima, Y., Nakamura, D., Uralil, J., Lara-Tejero, M., Sukhan, A., et al. (1998). Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605. doi: 10.1126/science.280.5363.602
Li, X., Wang, X., Zhang, X., Zhao, M., Tsang, W. L., Zhang, Y., et al. (2013). Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc. Natl. Acad. Sci. 110, 21165–21170. doi: 10.1073/pnas.1311864110
Lindmo, K., and Stenmark, H. (2006). Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119, 605–614. doi: 10.1242/jcs.02855
Lorenzo, O., Urbe, S., and Clague, M. J. (2005). Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J. Cell Sci. 118, 2005–2012. doi: 10.1242/jcs.02325
Lorenzo, O., Urbe, S., and Clague, M. J. (2006). Systematic analysis of myotubularins: heteromeric interactions, subcellular localisation and endosome-related functions. J. Cell Sci. 119, 2953–2959. doi: 10.1242/jcs.03040
Mallo, G. V., Espina, M., Smith, A. C., Terebiznik, M. R., Aleman, A., Finlay, B. B., et al. (2008). SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34. J. Cell Biol. 182, 741–752. doi: 10.1083/jcb.200804131
Nakagawa, I., Amano, A., Mizushima, N., Yamamoto, A., Yamaguchi, H., Kamimoto, T., et al. (2004). Autophagy defends cells against invading Group A Streptococcus. Science 306, 1037–1040. doi: 10.1126/science.1103966
Naughtin, M. J., Sheffield, D. A., Rahman, P., Hughes, W. E., Gurung, R., Stow, J. L., et al. (2010). The myotubularin phosphatase MTMR4 regulates sorting from early endosomes. J. Cell Sci. 123, 3071–3083. doi: 10.1242/jcs.060103
Norris, F. A., Wilson, M. P., Wallis, T. S., Galyov, E. E., and Majerus, P. W. (1998). SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc. Natl. Acad. Sci. U.S.A. 95, 14057–14059. doi: 10.1073/pnas.95.24.14057
Robinson, F. L., and Dixon, J. E. (2006). Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 16, 403–412. doi: 10.1016/j.tcb.2006.06.001
Scott, C. C., Cuellar-Mata, P., Matsuo, T., Davidson, H. W., and Grinstein, S. (2002). Role of 3-phosphoinositides in the maturation of Salmonella-containing vacuoles within host cells. J. Biol. Chem. 277, 12770–12776. doi: 10.1074/jbc.M110399200
Simonsen, A., Lippé, R., Christoforidis, S., Gaullier, J. M., Brech, A., Callaghan, J., et al. (1998). EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394, 494–498. doi: 10.1038/28879
Taguchi-Atarashi, N., Hamasaki, M., Matsunaga, K., Omori, H., Ktistakis, N. T., Yoshimori, T., et al. (2010). Modulation of Local PtdIns3P Levels by the PI Phosphatase MTMR3 regulates constitutive autophagy. Traffic 11, 468–478. doi: 10.1111/j.1600-0854.2010.01034.x
Thurston, T. L. M., Wandel, M. P., von Muhlinen, N., Foeglein, A., and Randow, F. (2012). Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418. doi: 10.1038/nature10744
Vergne, I., Roberts, E., Elmaoued, R. A., Tosch, V., Delgado, M. A., Proikas-Cezanne, T., et al. (2009). Control of autophagy initiation by phosphoinositide 3-phosphatase jumpy. EMBO J. 28, 2244–2258. doi: 10.1038/emboj.2009.159
Walker, D. M., Urbe, S., Dove, S. K., Tenza, D., Raposo, G., and Clague, M. J. (2001). Characterization of MTMR3: an inositol lipid 3-phosphatase with novel substrate specificity. Curr. Biol. 11, 1600–1605. doi: 10.1016/S0960-9822(01)00501-2
Yang, Z., Soderholm, A., Lung, T. W. F., Giogha, C., Hill, M. M., Brown, N. F., et al. (2015). SseK3 Is a Salmonella Effector That Binds TRIM32 and Modulates the Host's NF-kappa B Signalling Activity. PLoS ONE 10:e138529. doi: 10.1371/journal.pone.0138529
Yu, H. B., Croxen, M. A., Marchiando, A. M., Ferreira, R. B., Cadwell, K., Foster, L. J., et al. (2014). Autophagy facilitates Salmonella replication in HeLa cells. MBio 5, e00865–e00814. doi: 10.1128/mBio.00865-14
Zhao, R. X., Qi, Y., Chen, J., and Zhao, Z. H. J. (2001). FYVE-DSP2, a FYVE domain-containing dual specificity protein phosphatase that dephosphorylates phosphotidylinositol 3-phosphate. Exp. Cell Res. 265, 329–338. doi: 10.1006/excr.2001.5185
Zhou, D., and Galan, J. (2001). Salmonella entry into host cells: the work in concert of type III secreted effector proteins. Microbes Infect. 3, 1293–1298. doi: 10.1016/S1286-4579(01)01489-7
Zou, J., Zhang, C. F., Marjanovic, J., Kisseleva, M. V., Majerus, P. W., and Wilson, M. P. (2012). Myotubularin-related protein (MTMR) 9 determines the enzymatic activity, substrate specificity, and role in autophagy of MTMR8. Proc. Natl. Acad. Sci. U.S.A. 109, 9539–9544. doi: 10.1073/pnas.1207021109
Keywords: Salmonella, Phosphotidylinositols, myotubularin, autophagy, Salmonella-containing vacuole
Citation: Teo WX, Kerr MC and Teasdale RD (2016) MTMR4 Is Required for the Stability of the Salmonella-Containing Vacuole. Front. Cell. Infect. Microbiol. 6:91. doi: 10.3389/fcimb.2016.00091
Received: 12 June 2016; Accepted: 15 August 2016;
Published: 30 August 2016.
Edited by:
Souhaila Al Khodor, Sidra Medical and Research Center, QatarReviewed by:
Jiri Stulik, University of Defence, Czech RepublicCopyright © 2016 Teo, Kerr and Teasdale. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rohan D. Teasdale, ci50ZWFzZGFsZUB1cS5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.