- Department of Microbiology and Immunology, University of Louisville, Louisville, KY, USA
Preservation of health from infectious diseases depends upon both mucosal and systemic immunity via the collaborative effort of innate and adaptive immune responses. The proficiency of host immunity stems from robust defense mechanisms—physical barriers and specialized immune cells—and a failure of these mechanisms leads to pathology. Intriguingly, immunocompetence to pathogens can be shaped by the gut microbiome as recent publications highlight a dynamic interplay between the gut microbiome and host susceptibility to infection. Modulation of host immunity to enteric pathogens has long been studied where gut bacteria shape multiple facts of both innate and adaptive immunity. Conversely, the impact of gut commensals on host immunity to extra-gastrointestinal (GI) tract infections has only recently been recognized. In this context, the gut microbiome can augment host immunity to extra-GI tract bacterial, viral, and parasitic pathogens. This review explores the research that affords insight into the role of the gut microbiome in various infectious diseases, with a particular emphasis on extra-GI tract infections. A better understanding of the link between the gut microbiome and infectious disease will be critical for improving global health in the years ahead.
Introduction
Infectious diseases plague millions of people every year and are the leading cause of death in low-income and developing nations (World Health Organization, 2014; see Box 1). Prevention and treatment of infectious diseases requires a comprehensive understanding of the factors contributing to transmission and risk of infection. Understanding host biological and immunological factors, namely the development, function, and distribution of immune cells, associated with pathogens is important for the development of novel medical interventions and treatments. Yet, we now know the gut microbiome—the collective group of bacteria, viruses, fungi and their associated genes abiding within the intestinal tract—must also be considered as playing a significant role in modulating the host immune response. This microbial community in the gastrointestinal tract is mainly bacterial, containing four main phyla: Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria (Eckburg et al., 2005). The composition is relatively stable at the phylum level, but lower taxonomic levels can vary quite widely between two individuals, with differences being shaped by factors such as delivery (i.e., vaginal vs. cesarean), diet and antibiotic use among many others (reviewed by Honda and Littman, 2012; Chevalier et al., 2015; Mikkelsen et al., 2015). It has long been speculated that the number of organisms in the gut is between 1014 and 1015, resulting in approximately 10-times more microbial cells than human cells; however, a recent calculation determined there are about 1013 bacterial cells in humans resulting in the microbe to human ratio closer to 1:1 (Sender et al., 2016). From birth, the gut microbiome contributes greatly to the development of the neonatal immune system as germ-free (GF) mice have underdeveloped immune systems compared to conventionally raised mice (Guarner and Malagelada, 2003). Even more, the gut microbiome influences host health throughout the lifespan as it is linked to various inflammatory and autoimmune conditions, such as irritable bowel syndrome (Hong and Rhee, 2014), inflammatory bowel disease (De Cruz et al., 2012), ulcerative colitis (Flanagan et al., 2011), obesity (Turnbaugh et al., 2006), type 2 diabetes mellitus (van Olden et al., 2015), metabolic syndrome (Vijay-Kumar et al., 2010), and multiple sclerosis (Miyake et al., 2015). The gut microbiome also modulates the immune response to infectious agents both within and outside of the GI tract. In this review we first establish the relationship between the gut flora and the host immune response by discussing several, of the many, intra-GI tract infections affected by the gut microbiota. From there we move into a discussion of the role of the gut microbiome in extra-GI tract infections.
Box 1. Worldwide burden of infectious disease—does the gut microbiome contribute?
Among the most prevalent infectious diseases are HIV/AIDS, malaria, and tuberculosis (TB) as these contribute to one-third of deaths in developing nations and territories (Murray et al., 2013). HIV, human immunodeficiency virus, causes significant mortality and morbidity as evidenced by data collected in 2014, which estimated that 1.2 million individuals died due to an HIV infection, or associated illness, and a total of 36.9 million individuals lived with the disease (10 Facts, World Health Organization, 2015). Additionally, nearly 2 million individuals were infected with HIV in 2014, which illustrates the need for improved methods of prevention in communities where HIV transmission is highest, such as sub-Saharan Africa (HIV/AIDS, World Health Organization, 2015). In addition to HIV, malaria is a leading cause of mortality and morbidity when compared to other infectious agents. This parasitic disease is caused by Plasmodium spp. and has a global disease burden of 198 million cases and 584,000 deaths across all continents, respectively (World Malaria Report, World Health Organization, 2014). Most of these reported deaths occurred in sub-Saharan Africa in children under the age of 5 years old (World Malaria Report, World Health Organization, 2014). Another infectious disease that currently afflicts two billion individuals across the globe is tuberculosis (TB), caused by the bacterium Mycobacterium tuberculosis, with nearly 9 million cases of infection and 1.5 million TB-associated deaths reported in 2013 (Glaziou et al., 2015).
Interactions Between the Gut Microbiome and Gastrointestinal Tract Infections
Clostridium difficile
Clostridium difficile infection (CDI)1 is a prominent health care-associated illness that primarily impacts the health of individuals on long-term antibiotic regimens and individuals receiving inpatient care at hospitals or nursing homes (Agha, 2012). Though this gram-positive bacterium is found in the intestinal tract of humans as part of the normal gut microbiota, it can cause disease when the gut microbiota community becomes dysbiotic (Antharam et al., 2013; Koenigsknecht et al., 2015). During dysbiosis C. difficile is able to interact with the epithelial cell lining of the gut and promote a pro-inflammatory response that causes numerous clinical symptoms associated with a C. difficile infection, such as diarrhea, abdominal pain and sensitivity, low-grade fever, anorexia, and bloody stools (Leber et al., 2015).
The composition of the gut bacterial community is predictive of the degree of C. difficile colonization. More specifically, Porphyromonadaceae, Lachnospiraceae, Lactobacillus, Alistipes, and Turicibacter are associated with colonization resistance, while Escherichia and Streptococcus are associated with increased susceptibility to C. difficile growth (Schubert et al., 2015). Consistent with these observations, colonization of GF mice with Lachnospiraceae resulted in a significant decline in C. difficile colonization; however GF mice colonized with E. coli did not provide resistance to C. difficile. These studies demonstrate the ability of certain bacterial species within the gut to inhibit or promote CDI, however there remains much to be learned about the specific gut microbes that promote resistance and susceptibility to CDI. Beyond the bacterial species that inhibit or promote CDI it is also important to determine the physiological mechanisms that explain how differences in the gut community impact susceptibility to CDI as the metabolic profile of the gut microbiota certainly influences susceptibility to CDI (Theriot et al., 2014; Buffie et al., 2015). Antibiotic treatment induces significant changes in gut bacterial populations, which in turn influences metabolite levels (Theriot et al., 2014). Following antibiotic treatment there are changes in the abundance of primary and secondary bile acids, glucose, free fatty acids, dipeptides, and sugar alcohols (Theriot et al., 2014). C. difficile exploits these changes and is capable of using metabolites that increase in abundance (e.g., mannitol, fucturse, sorbitol, etc.) as carbon sources for growth. Conversely, certain metabolites confer resistance to CDI. Bacteria such as Clostridium scindens that encode the 7α-hydroxysteroid dehydrogenase enzyme, which is needed for secondary bile acid biosynthesis (e.g., deoxycholate and lithocholate) inhibit the growth of C. difficile (Buffie et al., 2015).
Though much is still unknown about the relationship between CDI and the gut microbiome, the use of fecal-derived microbiota transplantation (FMT) to treat recurrent CDI has shown great promise in the clinic. For example, FMT has been shown to be well tolerated with an overall clinical cure rate of 89% (Hirsch et al., 2015). 68% of these patients responded to one dose of the orally administered capsules and nearly 21% of patients were cured of CDI following a second dose. In another study, 79% of patients with severe and refractory CDI were cured after intestinal microbiota transplantation (Zainah et al., 2015). These studies highlight the success and future potential of FMT for patients with persistent CDI.
Citrobacter rodentium
Citrobacter rodentium is a gram-negative murine pathogen that colonizes the intestinal mucosa and is used to model enteropathogenic and enterohemorrhagic E. coli as well as IBD. Citrobacter rodentium, along with pathogenic E. coli, depends on attachment and effacement of intestinal epithelial cells for colonization. These pathogens bind to intestinal epithelial cells and disrupt the brush border of the villi while forming an actin pedestal around the attached bacteria, leading to epithelial damage and effacement (Wong et al., 2012). Citrobacter rodentium attachment and colonization cause transmissible murine crypt hyperplasia, a form of colitis that induces thickening of the colonic mucosa and inflammation. Ultimately, this inflammation facilitates a loss of diversity and dysbiosis of the gut microbiota (Lupp et al., 2007).
As a gastrointestinal pathogen, C. rodentium affects the gut microbiome composition and is in turn affected by the presence or absence of the microbiota, promoting colonization resistance through strategies such as nutrient and niche competition (Lupp et al., 2007). To determine whether specific members of the gut microbiome, or the entire community, are required to prevent C. rodentium infection, GF mice were infected with C. rodentium followed by colonization with E. coli, Bacteroides vulgatus, or B. thetaiotaomicron. Colonization with E. coli, but neither of the Bacteroides species, was effective at outcompeting C. rodentium (Kamada et al., 2012). This points to specific commensals as important for resistance to C. rodentium, and not just the presence or absence of the gut microbiota as a whole. Additionally, certain gut bacteria like Lactobacillus plantarum can exert protective effects on gut integrity by stimulating TLR2 and thus upregulating tight-junction structures, enhancing epithelial barrier functions and reducing overall susceptibility to C. rodentium (Karczewski et al., 2010). Resistance to C. rodentium through the gut microbiome, as well as resistance to EHEC and EPEC, is also mediated through the presence of segmented filamentous bacteria (SFB), which promote Th17 immune responses in the intestine (Ivanov et al., 2009). During the immune response to C. rodentium, interleukin-22 (IL-22) is produced by T helper cells and leads directly to expression of antimicrobial peptides by Paneth cells that help control C. rodentium; these peptides include RegIIIβ and RegIIIγ (Zheng et al., 2008), as well as beta-defensins 1, 3, and 4 (Kamada et al., 2012). Toll-like receptors (TLRs) and Nod-like receptors (NLRs) contribute to protection against C. rodentium by sensing various microbe-associated molecular patterns (MAMPs) such as LPS and muramyl dipeptide and inducing inflammatory conditions in the gut (Manigold et al., 2000; Khan et al., 2006; Kanneganti et al., 2007). If innate immune sensing is impaired, such as in MyD88-deficient mice, control of C. rodentium by neutrophils is lost and bacteremia, colitis, and death ensue (Lebeis et al., 2007). Overall, there is a well-organized response to C. rodentium by the host, which is mediated by different members of the gut microbiota.
Toxoplasma gondii
Toxoplasmosis, an infectious disease caused by the parasite Toxoplasma gondii, prevails around the world and an estimated 60 million individuals within the United States are infected with this parasite (2013)2. Transmission of T. gondii occurs through various routes—congenital transmission, carnivorism, and fecal-oral transmission—with fecal-oral transmission serving as the primary source of widespread infection in the United States and across the globe (2013). Initial control of T. gondii infection depends on the efficacy of the mucosal immune response, which includes the collaborative efforts of innate immune defenses, chemokine and antimicrobial secretions, and antigen presentation for activation of the adaptive immune response (Benson et al., 2009). Among these factors, TLR11 is important for detecting the parasite and the production of protective IL-12 and IFN-γ response (Plattner et al., 2008; Yarovinsky et al., 2008).
In some species, including humans, TLR11 is a pseudogene (Roach et al., 2005), yet these hosts still produce a protective IL-12 response by activated dendritic cells (Gazzinelli et al., 1993; Tosh et al., 2016). Recent work has focused on unveiling the TLR11-independent mechanism by which human hosts control T. gondii infection (Benson et al., 2009). Protective immunity is, at least in part, attributed to gut bacteria that elicit a TLR11-independent, MyD88-dependent protective immune response following T. gondii infection. T. gondii infection causes intestinal damage, which results in commensal bacteria indirectly eliciting protective immunity. Following intestinal damage commensal bacteria activate dendritic cells through MyD88-depentent TLRs, primarily TLR2, TLR4, and TLR9, resulting in the production of IL-12 that confers resistance to T. gondii (Benson et al., 2009).
Effect of the Gut Microbiome on Extra-Gastrointestinal Tract Infections and Vice Versa
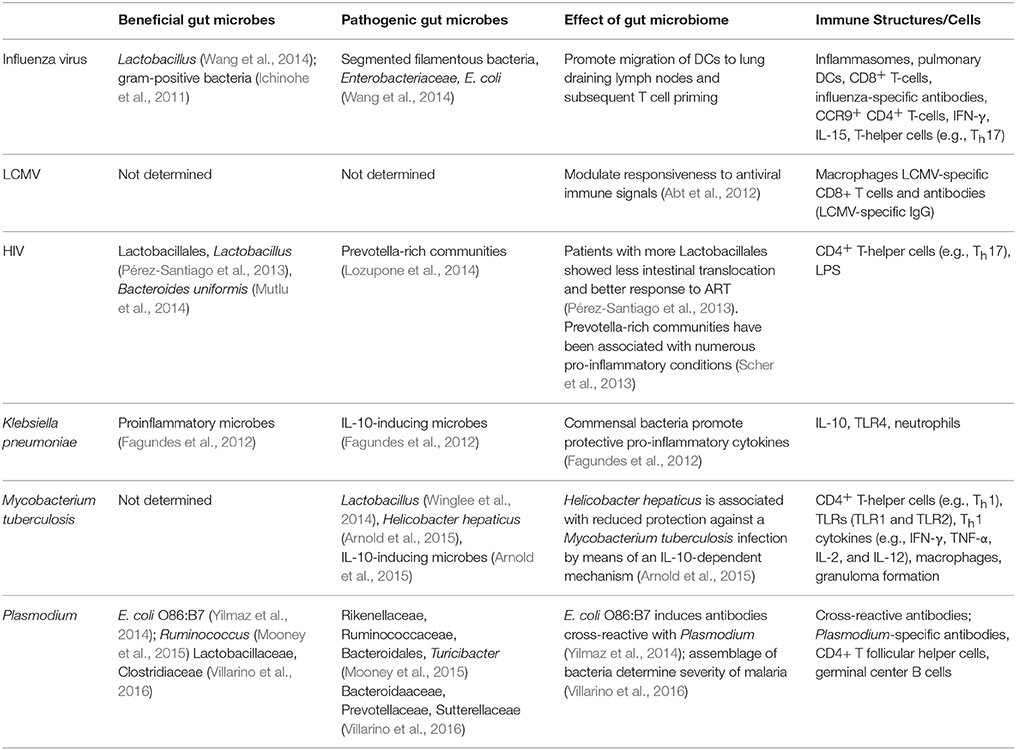
In addition to the gut microbiota modulating host immunity to intra-GI tract pathogens, increasing evidence demonstrates its impact on infectious diseases beyond the GI tract (Table 1). Conversely, extra-GI tract infections also appear to impact the composition of the gut microbiota. In the following section we will discuss current literature specifically focused on understanding the role of the gut microbiome in modulating the host immune response to extra-GI tract infections.
Influenza Virus
Flu is an infectious disease in the respiratory tract that is caused by influenza virus (Monto et al., 2000). Infected individuals may develop mild symptoms or severe disease that in some cases leads to death (Monto, 1987). The common symptoms associated with influenza virus include fever, cough, pharyngitis, myalgia, and fatigue (Barik, 2012). Additionally, many patients experience abnormalities in gastrointestinal function, such as vomiting, diarrhea, and nausea, though these symptoms are more common in children than adults (Baden et al., 2009). The seasonal influenza vaccine moderates the clinical burden associated with this virus, but the efficacy of the vaccine depends on the strain of virus dominating the flu season and the number of people vaccinated. A more effective approach for preventing and treating infection warrants an exhaustive understanding of the host-pathogen interaction.
Protection against an influenza infection is associated with the diversity and health of the gut microbiome. Recent findings suggest an important role of gram-positive bacterial populations within the gut and possibly within the nasal tract in fostering an adequate immune response to a respiratory influenza infection (Ichinohe et al., 2011). The gut bacteria provided protection in part by initiating an appropriate inflammatory response through activation of inflammasomes. In the absence of inflammasome activation, lung dendritic cells exhibited reduced migration to the lung draining lymph node resulting in reduced activation of CD8+ T cells. These results correlated with impaired expansion of influenza-specific T cells, influenza-specific antibodies, and elevated viral titers (Ichinohe et al., 2011).
Mice treated with broad-spectrum antibiotics to diminish gut bacteria populations followed by infection with influenza have exacerbated weight loss, a greater drop in blood oxygen saturation, elevated viral titers in the lung, higher epithelial cell necrosis, and more cell death in both the bronchiolar lumen and the bronchiole alveolar lavage (BAL) fluid (Abt et al., 2012). Even more, GF mice experienced significantly more weight loss, a weaker ability to clear influenza, diminished virus-specific antibody titers, and more severe bronchiole epithelial degeneration when compared to outcomes for conventional mice (Abt et al., 2012). Consistent with the work by Ichinohe et al. mice treated with broad-spectrum antibiotics also had fewer influenza-specific antibodies and influenza-specific T cells. Together, these findings suggest a modulatory role of the gut microbiome in eliciting a strong host immune response to influenza.
Gastrointestinal damage that transpires as a result of a respiratory influenza infection is a perplexing phenomenon that is poorly understood. A recent report by Wang et al. demonstrated pronounced changes in the gut bacterial assemblage (2014). Populations that are significantly changed as a result of an influenza infection included a decrease in Lactobacillus and increases in both segmented filamentous bacteria (SFB) and Enterobacteriaceae (Wang et al., 2014). Influenza-induced gastroenteritis was caused by lung-derived CD4+ T cell-induced dysbiosis that resulted in the expansion of Th17 cells and subsequent intestinal injury. Specifically, during influenza CCR9+ CD4+ T cells are recruited from the lung to intestinal tissues. CD4+ T cell-produced IFN-γ alters the gut microbiota composition, including an increase in E. coli that resulted in the production of IL-15. Increased production of IL-15 stimulated in situ differentiation of CD4+ T cells into Th17 cells that caused intestinal damage.
LCMV
Lymphocytic choriomeningitis virus (LCMV) is an Arenavirus that is a naturally occurring rodent pathogen and is often used to study host-pathogen interactions. There are several strains of LCMV that differ in their ability to cause acute vs. chronic infections. Using a strain of LCMV that takes about 1-month to clear post-infection Abt et al. determined how the gut microbiome impacted the host immune response following systemic infection (Abt et al., 2012). Mice treated with antibiotics to deplete the gut microbiome prior to LCMV infection elicited an impaired adaptive immune response to LCMV infection. Specifically, antibiotic treated mice had decreased expansion of LCMV-specific CD8+ T cells, with an elevated expression of inhibitory receptors, and decreased titers of LCMV-specific IgG compared to mice that did not receive antibiotics (Abt et al., 2012). Consistent with these observations, the impaired adaptive immune response in antibiotic treated mice correlated with delayed clearance of LCMV. In addition to changes in the adaptive immune system, this report demonstrated the gut microbiota primes macrophages to respond to interferon stimulation (Abt et al., 2012). Whereas macrophages from antibiotic treated mice have an impaired ability to respond to LCMV infection, the viral titers in antibiotic treated and control mice were similar on day 7 post-infection. Therefore, it is not clear to what effect gut microbiome-dependent priming of macrophages contributes to the delayed clearance of LCMV observed at later time points. One possibility is that the impaired activity of macrophages contributes to the diminished adaptive immune response; alternatively the gut microbiota may modulate adaptive immune responses through another mechanism. Taken together with the previous observations on influenza, the gut microbiome can affect responses to extra-gastrointestinal tract viral infections that have no direct interactions with mucosal surfaces.
HIV
One of the most prevalent infections impacting global health is human immunodeficiency virus (HIV), which is caused by a retrovirus that invades and infects immune cells of its host, specifically helper T cells, macrophages, and dendritic cells (Kumar et al., 2015). The primary targets of HIV are helper T cells, CD4+ T cells, which leads to a progressive loss of these cells over the course of an HIV infection. Eventually, host CD4+ T cells drop to a level that cell-mediated immunity of the host ceases, and the likelihood of opportunistic infections substantially increases (Pollock et al., 2015).
Recent observations found that individuals with HIV also have an increased risk for intestinal dysbiosis, culminating in conditions such as diarrhea, colitis, or even metabolic syndrome (Lozupone et al., 2013). In the gut, HIV affects the mucosal immune system by infecting and depleting the Th17 cells in the gut-associated lymphoid tissue (GALT), beginning during the acute phase of infection and continuing throughout the course of the disease (Lozupone et al., 2013). Recent work explored differences among patients with early HIV infection, and the findings ultimately suggested that patients with higher proportions of bacteria from the order Lactobacillales, which contains Lactobacillus species, showed lower markers of bacterial translocation, such as LPS in the blood (Pérez-Santiago et al., 2013). Additionally, higher abundance of Lactobacillales in patients was positively associated with CD4+ T cell count and was negatively associated with viral load, indicating that bacteria from Lactobacillales could in some way modulate the infectivity or pathology of HIV infection (Pérez-Santiago et al., 2013). As HIV progresses, translocation of bacteria and bacterial products can occur; for example, LPS can be more readily detected in circulation during HIV infection (Brenchley et al., 2006; Cassol et al., 2010). Translocation during HIV is also associated with higher levels of immune activation and inflammation, along with a poorer response to anti-retroviral therapy (ART) (Brenchley et al., 2006; Cassol et al., 2010). Overall, there is evidence that HIV can induce inflammation and gut permeability during the course of infection, but this pathology can also be partially ameliorated by certain gut bacteria.
Klebsiella pneumoniae
Klebsiella pneumoniae is a gram-negative human commensal and is commonly found as part of the oral and gut microbiota. Whereas K. pneumoniae is a normal oral commensal, nosocomial infections in the lower respiratory tract can cause significant diseases, especially in patients that are immunocompromised and those with inadequate respiratory immunity and function (Tsay et al., 2002; Jung et al., 2012). It has been observed that GF mice are more susceptible to pulmonary K. pneumoniae infection than conventionally raised mice (Fagundes et al., 2012). Following intratracheal infection with K. pneumoniae GF mice produced elevated levels of anti-inflammatory IL-10, reduced neutrophil influx into the lung, and elevated lung bacterial burden and bacteremia compared to control mice. Treatment with TLR ligands, specifically TLR4 stimulation, was sufficient to rescue control of K. pneumoniae infection in GF mice. These observations clearly identify a role for commensal flora in shaping the host immune response to pulmonary K. pneumoniae infection, however these data do not differentiate the contribution of the lung microbiome vs. the gut microbiome. Indeed, the lung microbiome is involved in other disease states, such as disease progression in cystic fibrosis and asthma (Daniels et al., 2013; Huang et al., 2014).
Mycobacterium tuberculosis
Tuberculosis (TB) is an infectious disease caused by Mycobacterium tuberculosis, an aerobic bacterium that invades the respiratory tract of human hosts. The bacterium primarily establishes infection within the lungs of the host, but the infection can remain latent for the entirety of the host’s lifetime as nearly one-third of the world’s population has latent TB (Glaziou et al., 2015). Although, extensive research has been conducted on the environmental and immunological factors that contribute to TB, the contribution of the gut microbiome to TB and the effect of TB on the gut microbiome are relatively unexplored. Winglee and colleagues conducted a longitudinal study in a mouse model for TB in order to track changes in the gut microbiome prior to, during, and subsequent to an aerosol infection of M. tuberculosis (2014). Overall, significant changes in the resident microbiome occurred between infection and death; in particular, there was a significant loss in gut bacteria diversity initially that was followed by a recovery in diversity (Winglee et al., 2014). Of note, these changes in the gut bacteria community occurred very rapidly, within 6 days after M. tuberculosis infection. Given the ability of an influenza infection to modulate gut bacteria populations and intestinal inflammation it would be interesting to know if similar mechanisms are at play following pulmonary TB infection.
Plasmodium
Malaria is a parasitic infectious disease caused by infection with Plasmodium species. In 2013, there were at least 198 million infections and 584,000 deaths attributed to Plasmodium infections, with the majority occurring in sub-Saharan Africa (World Malaria Report, World Health Organization, 2014). The life cycle of Plasmodium is complex, involving developmental stages in the human host as well as in the insect vector, the Anopheles mosquito. Transmission to the human host is initiated when infected mosquitoes deposit sporozoites while taking blood meals; sporozoites enter the bloodstream and migrate to the liver where they infect hepatocytes (Mota et al., 2001; Gueirard et al., 2010). Inside the infected hepatocytes sporozoites differentiate into the asexual merozoites, which are released and establish cyclical infection of red blood cells (RBCs) causing the symptomatic blood stage of infection.
Recent publications have identified links between the gut microbiome and the different developmental stages of Plasmodium infections in vertebrate hosts. Beginning with the initial stage of infection, antibodies elicited against glycans present on gut bacteria are able to cross-react with sporozoites. Many microbes, including Plasmodium, are able to glycosylate proteins with the Galα1-3Galβ1-4GlcNAc-R glycan (α-gal) (Yilmaz et al., 2014). Importantly, humans do not use that glycosylation pattern and are capable of generating antibody responses directed against α-gal (Yilmaz et al., 2014). The gut pathobiont Escherichia coli O86:B7 expresses α-gal that elicits α-gal-specific antibodies that cross-react with α-gal expressed on Plasmodium sporozoites (Yilmaz et al., 2014). These antibodies prevent the ability of sporozoites to emigrate from the skin and infect hepatocytes. Consistent with this observation, there was a positive correlation between protection from Plasmodium falciparum infection and the presence of α-gal-specific IgM, but not α-gal-specific IgG, in humans (Yilmaz et al., 2014). Of note, the α-gal-specific antibodies did not have any impact on Plasmodium infections when sporozoites were injected directly intravenously or when the pre-erythrocytic stage was bypassed and mice were directly infected with parasitized RBCs. These data suggest that certain gastrointestinal bacteria could provide protection against the early stage of the parasite life cycle in humans. Similarly, the composition of the stool bacteria community in Malian children correlated with susceptibility to P. falciparum infection, however it did not correlate with progression to febrile malaria (Yooseph et al., 2015). The consistency of this report with that of Yilmaz et al. (susceptibility to sporozoite infection but not parasitized RBC infection) raises the possibility that perhaps Malian children resistant to P. falciparum infection harbored higher levels of α-gal-specific IgM titers.
Whereas α-gal-specific antibodies had no effect on the blood stage infection, it has been shown that differences in the composition of the gut microbiome in mice modulates susceptibility to the blood stage infection (Villarino et al., 2016). In this study, C57BL/6 mice from different commercial vendors (Jackson Labs (Jax), Taconic Biosciences (Tac), National Cancer Institute/Charles River Labs (NCI), and Harlan/Envigo (Har)) were infected with P. yoelii 17XNL, a rodent-specific Plasmodium species. Following infection the severity of malaria was substantially more pronounced in NCI and Har mice compared to Jax and Tac mice. 16S rRNA sequencing from GI samples showed that P. yoelii-resistant Jax and Tac mice were enriched for Lactobacillaceae and Clostridiaceae, while P. yoelii-susceptible NCI and Har mice were enriched for Bacteroidaceae, Prevotellaceae, and Sutterellaceae. Moreover, colonization of GF mice with cecal content from severe malaria-resistant Jax or-susceptible NCI mice transferred the susceptibility phenotypes, respectively. The report also demonstrated that the gut microbiome could be modulated to decrease susceptibility to severe malaria (Villarino et al., 2016). Examination of the immune response following P. yoelii infection showed an increase in the number of germinal center B cells and accelerated antibody class switching in Jax mice compared to NCI mice, which correlated with decreased parasite burden in the Jax mice (Villarino et al., 2016). Taken together, these publications show that the gut microbiome affects Plasmodium infections at different points in its life cycle through distinct mechanisms.
Recent reports also demonstrated that the dialog between the gut microbiota and Plasmodium is bi-directional. Following infection of mice with P. yoelii there is an initial decrease in the alpha diversity and overall abundance of fecal bacteria that returns to pre-infection levels following resolution of the infection (Mooney et al., 2015). The mechanism responsible for this change in gut bacterial composition following P. yoelii infection is not known, however it does correlate with sequestration of infected red blood cells in the intestinal vasculature and infiltration of inflammatory immune cells. Similarly, in C57BL/6 mice infected with Plasmodium berghei ANKA, which causes experimental cerebral malaria, it has been observed that dysbiosis of the gut microbiota, sequestration of red blood cells in the intestinal vasculature, and intestinal lesions occur during infection, however these differences were not observed in BALB/c mice that are resistant to experimental cerebral malaria (Taniguchi et al., 2015). It remains unclear whether dysbiosis of the gut microbiota observed following Plasmodium infection is an effect of the infection, or if the dysbiosis contributes to the severity of malaria. Of note, changes in bacterial diversity have important implications for the role of Plasmodium in the pathogenicity of Salmonella enterica serotype Typhimurium. Plasmodium infections, in both mice and humans, increase susceptibility to invasive bacterial infections, in particular non-typhoid Salmonella (Scott et al., 2011; Cunnington et al., 2012; Chau et al., 2013; Mooney et al., 2014). Mooney et al. colonized GF mice with cecal contents from either P. yoelii-infected or uninfected control mice followed by Salmonella infection; mice that received cecal content from P. yoelii infected mice had a higher burden of Salmonella in feces than control mice (Mooney et al., 2015). Therefore, not only does Plasmodium infection induce dysbiosis of the gut microbiota, but this dysbiosis also leaves the host susceptible to enteric pathogen infections. However, a mechanism for how this occurs has not been elucidated.
Conclusion
While this review has highlighted recent insight into gut microbiome-dependent control of extra-GI tract infections, there is still much that is unknown. One of the biggest obstacles facing the field is the lack of specific mechanisms that have been elucidated. With the identification of each mechanism, the potential for future therapeutics increases drastically. For example, it has been observed that in mice, short chain fatty acids (SCFAs) such as propionate can circulate to the bone marrow and induce generation of antigen-presenting cells (APCs) that induce diminished Th2 response and thus ablates allergic airway disease (Trompette et al., 2014). An excellent source of SCFAs is fermentation of dietary fiber by the gut microbiome, particularly members of Bacteroidetes and Bifidobacteria (Trompette et al., 2014). Thus, uncovering mechanistic interactions between the gut microbiome and the host in the context of extra-GI tract infections should be a top priority.
Although, decades of research have linked the gut flora with susceptibility to enteric pathogens, research in recent years has now demonstrated that the gut microbiota also impact the host immune response to extra-GI tract infections. This effect may not be universal, yet we should not be surprised to learn of other systemic infections that are modulated by gut commensals. It is also becoming increasingly clear that systemic infections can also alter the composition of the gut microbiota. Collectively, these observations point toward novel therapeutic interventions for pathogens, such as Plasmodium, that have evaded the development of effective vaccines. Therefore, it is essential we gain a greater understanding of how the gut microbiota communicates with the immune system to affect the outcome of systemic infections.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Yousef Abu-Kwaik for critical review of this article. This work was supported by grants from the NIH (1R21AI113386) and the American Cancer Society (Research Scholar Grant, RSG-14-057-01-MPC) to NS.
Footnotes
1. ^Clostridium difficile infection [Online]. Centers for Disease Control and Prevention. Available online at: http://www.cdc.gov/HAI/organisms/cdiff/Cdiff_infect.html
2. ^Parasites – Toxoplasmosis (Toxoplasma infection) [Online]. Centers for Disease Control and Prevention. Available online at: http://www.cdc.gov/parasites/toxoplasmosis/gen_info/faqs.html
References
(2013). Toxoplasmosis Frequently Asked Questions (FAQs) [Online]. Centers for Disease Control and Prevention. Available online at: http://www.cdc.gov/parasites/toxoplasmosis/gen_info/faqs.html
10 Facts, World Health Organization. (2015). 10 Facts on HIV/AIDS [Online]. World Health Organization. Available online at: http://www.who.int/features/factfiles/hiv/en/
Abt, M. C., Osborne, L. C., Monticelli, L. A., Doering, T. A., Alenghat, T., Sonnenberg, G. F., et al. (2012). Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170. doi: 10.1016/j.immuni.2012.04.011
Agha, M. (2012). Epidemiology and pathogenesis of C. difficile and MRSA in the light of current NHS control policies: a policy review. Ann. Med. Surg. (Lond.) 1, 39–43. doi: 10.1016/s2049-0801(12)70012-2
Antharam, V. C., Li, E. C., Ishmael, A., Sharma, A., Mai, V., Rand, K. H., et al. (2013). Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 51, 2884–2892. doi: 10.1128/jcm.00845-13
Arnold, I. C., Hutchings, C., Kondova, I., Hey, A., Powrie, F., Beverley, P., et al. (2015). Helicobacter hepaticus infection in BALB/c mice abolishes subunit-vaccine-induced protection against M. tuberculosis. Vaccine 33, 1808–1814. doi: 10.1016/j.vaccine.2015.02.041
Baden, L. R., Drazen, J. M., Kritek, P. A., Curfman, G. D., Morissey, S., and Campion, E. W. (2009). H1N1 Influenza a disease—information for health professionals. New Engl. J. Med. 360, 2666–2667. doi: 10.1056/NEJMe0903992
Benson, A., Pifer, R., Behrendt, C. L., Hooper, L. V., and Yarovinsky, F. (2009). Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe. 6, 187–196. doi: 10.1016/j.chom.2009.06.005
Brenchley, J. M., Price, D. A., Schacker, T. W., Asher, T. E., Silvestri, G., Rao, S., et al. (2006). Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12, 1365–1371. doi: 10.1038/nm1511
Buffie, C. G., Bucci, V., Stein, R. R., McKenney, P. T., Ling, L., Gobourne, A., et al. (2015). Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208. doi: 10.1038/nature13828
Cassol, E., Malfeld, S., Mahasha, P., van der Merwe, S., Cassol, S., Seebregts, C., et al. (2010). Persistent microbial translocation and immune activation in HIV-1-Infected South Africans receiving combination antiretroviral therapy. J. Infect. Dis. 202, 723–733. doi: 10.1086/655229
Chau, J. Y., Tiffany, C. M., Nimishakavi, S., Lawrence, J. A., Pakpour, N., Mooney, J. P., et al. (2013). Malaria-associated L-arginine deficiency induces mast cell-associated disruption to intestinal barrier defenses against nontyphoidal Salmonella bacteremia. Infect. Immun. 81, 3515–3526. doi: 10.1128/iai.00380-13
Chevalier, C., Stojanović, O., Colin, D. J., Suarez-Zamorano, N., Tarallo, V., Veyrat-Durebex, C., et al. (2015). Gut microbiota orchestrates energy homeostasis during cold. Cell 163, 1360–1374. doi: 10.1016/j.cell.2015.11.004
Cunnington, A. J., de Souza, J. B., Walther, M., and Riley, E. M. (2012). Malaria impairs resistance to Salmonella through heme- and heme oxygenase-dependent dysfunctional granulocyte mobilization. Nat. Med. 18, 120–127. doi: 10.1038/nm.2601
Daniels, T. W., Rogers, G. B., Stressmann, F. A., van der Gast, C. J., Bruce, K. D., Jones, G. R., et al. (2013). Impact of antibiotic treatment for pulmonary exacerbations on bacterial diversity in cystic fibrosis. J. Cyst. Fibros. 12, 22–28. doi: 10.1016/j.jcf.2012.05.008
De Cruz, P., Prideaux, L., Wagner, J., Ng, S. C., McSweeney, C., Kirkwood, C., et al. (2012). Characterization of the gastrointestinal microbiota in health and inflammatory bowel disease. Inflamm. Bowel. Dis. 18, 372–390. doi: 10.1002/ibd.21751
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Fagundes, C. T., Amaral, F. A., Vieira, A. T., Soares, A. C., Pinho, V., Nicoli, J. R., et al. (2012). Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J. Immunol. 188, 1411–1420. doi: 10.4049/jimmunol.1101682
Flanagan, P., Campbell, B. J., and Rhodes, J. M. (2011). Bacteria in the pathogenesis of inflammatory bowel disease. Biochem. Soc. Trans. 39, 1067–1072. doi: 10.1042/bst0391067
Gazzinelli, R. T., Hieny, S., Wynn, T. A., Wolf, S., and Sher, A. (1993). Interleukin 12 is required for the T-lymphocyte-independent induction of interferon gamma by an intracellular parasite and induces resistance in T-cell-deficient hosts. Proc. Natl. Acad. Sci. U.S.A. 90, 6115–6119.
Glaziou, P., Sismanidis, C., Floyd, K., and Raviglione, M. (2015). Global epidemiology of tuberculosis. Cold Spring Harb. Perspect. Med. 5:a017798. doi: 10.1101/cshperspect.a017798
Guarner, F., and Malagelada, J. R. (2003). Gut flora in health and disease. Lancet 361, 512–519. doi: 10.1016/s0140-6736(03)12489-0
Gueirard, P., Tavares, J., Thiberge, S., Bernex, F., Ishino, T., Milon, G., et al. (2010). Development of the malaria parasite in the skin of the mammalian host. Proc. Natl. Acad. Sci. U.S.A. 107, 18640–18645. doi: 10.1073/pnas.1009346107
Hirsch, B. E., Saraiya, N., Poeth, K., Schwartz, R. M., Epstein, M. E., and Honig, G. (2015). Effectiveness of fecal-derived microbiota transfer using orally administered capsules for recurrent Clostridium difficile infection. BMC Infect. Dis. 15:191. doi: 10.1186/s12879-015-0930-z
HIV/AIDS, World Health Organization. (2015). HIV/AIDS Key Facts Sheet [Online]. World Health Organization. Available online at: http://www.who.int/mediacentre/factsheets/fs360/en/
Honda, K., and Littman, D. R. (2012). The Microbiome in Infectious Disease and Inflammation. Annu. Rev. Immunol. 30, 759–795. doi: 10.1146/annurev-immunol-020711-074937
Hong, S. N., and Rhee, P. L. (2014). Unraveling the ties between irritable bowel syndrome and intestinal microbiota. World J. Gastroenterol. 20, 2470–2481. doi: 10.3748/wjg.v20.i10.2470
Huang, Y. J., Sethi, S., Murphy, T., Nariya, S., Boushey, H. A., and Lynch, S. V. (2014). Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J. Clin. Microbiol. 52, 2813–2823. doi: 10.1128/jcm.00035-14
Ichinohe, T., Pang, I. K., Kumamoto, Y., Peaper, D. R., Ho, J. H., Murray, T. S., et al. (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 108, 5354–5359. doi: 10.1073/pnas.1019378108
Ivanov, I. I., Atarashi, K., Manel, N., Brodie, E. L., Shima, T., Karaoz, U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. doi: 10.1016/j.cell.2009.09.033
Jung, Y., Lee, M. J., Sin, H. Y., Kim, N. H., Hwang, J. H., Park, J., et al. (2012). Differences in characteristics between healthcare-associated and community-acquired infection in community-onset Klebsiella pneumoniae bloodstream infection in Korea. BMC Infect. Dis. 12:239. doi: 10.1186/1471-2334-12-239
Kamada, N., Kim, Y. G., Sham, H. P., Vallance, B. A., Puente, J. L., Martens, E. C., et al. (2012). Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329. doi: 10.1126/science.1222195
Kanneganti, T. D., Lamkanfi, M., and Núñez, G. (2007). Intracellular NOD-like receptors in host defense and disease. Immunity 27, 549–559. doi: 10.1016/j.immuni.2007.10.002
Karczewski, J., Troost, F. J., Konings, I., Dekker, J., Kleerebezem, M., Brummer, R. J., et al. (2010). Regulation of human epithelial tight junction proteins by Lactobacillus plantarum in vivo and protective effects on the epithelial barrier. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G851–G859. doi: 10.1152/ajpgi.00327.2009
Khan, M. A., Ma, C., Knodler, L. A., Valdez, Y., Rosenberger, C. M., Deng, W., et al. (2006). Toll-like receptor 4 contributes to colitis development but not to host defense during Citrobacter rodentium infection in mice. Infect. Immun. 74, 2522–2536. doi: 10.1128/iai.74.5.2522-2536.2006
Koenigsknecht, M. J., Theriot, C. M., Bergin, I. L., Schumacher, C. A., Schloss, P. D., and Young, V. B. (2015). Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect. Immun. 83, 934–941. doi: 10.1128/iai.02768-14
Kumar, N. A., Cheong, K., Powell, D. R., da Fonseca Pereira, C., Anderson, J., Evans, V. A., et al. (2015). The role of antigen presenting cells in the induction of HIV-1 latency in resting CD4(+) T-cells. Retrovirology 12, 76. doi: 10.1186/s12977-015-0204-2
Lebeis, S. L., Bommarius, B., Parkos, C. A., Sherman, M. A., and Kalman, D. (2007). TLR signaling mediated by MyD88 is required for a protective innate immune response by neutrophils to Citrobacter rodentium. J. Immunol. 179, 566–577. doi: 10.4049/jimmunol.179.1.566
Leber, A., Viladomiu, M., Hontecillas, R., Abedi, V., Philipson, C., Hoops, S., et al. (2015). Systems modeling of interactions between mucosal immunity and the gut microbiome during clostridium difficile infection. PLoS ONE 10:e0134849. doi: 10.1371/journal.pone.0134849
Lozupone, C. A., Li, M., Campbell, T. B., Flores, S. C., Linderman, D., Gebert, M. J., et al. (2013). Alterations in the Gut Microbiota associated with HIV-1 Infection. Cell Host Microbe 14, 329–339. doi: 10.1016/j.chom.2013.08.006
Lozupone, C. A., Rhodes, M. E., Neff, C. P., Fontenot, A. P., Campbell, T. B., and Palmer, B. E. (2014). HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut. Microbes 5, 562–570. doi: 10.4161/gmic.32132
Lupp, C., Robertson, M. L., Wickham, M. E., Sekirov, I., Champion, O. L., Gaynor, E. C., et al. (2007). Host-Mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of enterobacteriaceae. Cell Host Microbe 2, 119–129. doi: 10.1016/j.chom.2007.06.010
Manigold, T., Bocker, U., Traber, P., Dong-Si, T., Kurimoto, M., Hanck, C., et al. (2000). Lipopolysaccharide/endotoxin induces IL-18 via CD14 in human peripheral blood mononuclear cells in vitro. Cytokine 12, 1788–1792. doi: 10.1006/cyto.2000.0783
Mikkelsen, K. H., Frost, M., Bahl, M. I., Licht, T. R., Jensen, U. S., Rosenberg, J., et al. (2015). Effect of antibiotics on Gut Microbiota, Gut hormones and glucose metabolism. PLoS ONE 10:e0142352. doi: 10.1371/journal.pone.0142352
Miyake, S., Kim, S., Suda, W., Oshima, K., Nakamura, M., Matsuoka, T., et al. (2015). Dysbiosis in the Gut Microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV Clusters. PLoS ONE 10:e0137429. doi: 10.1371/journal.pone.0137429
Monto, A. S. (1987). Influenza: quantifying morbidity and mortality. Am. J. Med. 82(Suppl. 1), 20–25.
Monto, A. S., Gravenstein, S., Elliott, M., Colopy, M., and Schweinle, J. (2000). Clinical signs and symptoms predicting influenza infection. Arch Intern Med. 160, 3243–3247. doi: 10.1001/archinte.160.21.3243
Mooney, J. P., Butler, B. P., Lokken, K. L., Xavier, M. N., Chau, J. Y., Schaltenberg, N., et al. (2014). The mucosal inflammatory response to non-typhoidal Salmonella in the intestine is blunted by IL-10 during concurrent malaria parasite infection. Mucosal. Immunol. 7, 1302–1311. doi: 10.1038/mi.2014.18
Mooney, J. P., Lokken, K. L., Byndloss, M. X., George, M. D., Velazquez, E. M., Faber, F., et al. (2015). Inflammation-associated alterations to the intestinal microbiota reduce colonization resistance against non-typhoidal Salmonella during concurrent malaria parasite infection. Sci. Rep. 5:14603. doi: 10.1038/srep14603
Mota, M. M., Pradel, G., Vanderberg, J. P., Hafalla, J. C., Frevert, U., Nussenzweig, R. S., et al. (2001). Migration of Plasmodium sporozoites through cells before infection. Science 291, 141–144. doi: 10.1126/science.291.5501.141
Murray, C. J. L., Ortblad, K. F., Guinovart, C., Lim, S. S., Wolock, T. M., Roberts, D. A., et al. (2013). Global, regional, and national incidence and mortality for HIV, tuberculosis, and malaria during 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 384, 1005–1070. doi: 10.1016/S0140-6736(14)60844-8
Mutlu, E. A., Keshavarzian, A., Losurdo, J., Swanson, G., Siewe, B., Forsyth, C., et al. (2014). A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog 10:e1003829. doi: 10.1371/journal.ppat.1003829
Pérez-Santiago, J., Gianella, S., Massanella, M., Spina, C. A., Karris, M. Y., Var, S. R., et al. (2013). Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. Aids 27, 1921–1931. doi: 10.1097/QAD.0b013e3283611816
Plattner, F., Yarovinsky, F., Romero, S., Didry, D., Carlier, M. F., Sher, A., et al. (2008). Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe. 3, 77–87. doi: 10.1016/j.chom.2008.01.001
Pollock, K. M., Montamat-Sicotte, D. J., Cooke, G. S., Kapembwa, M. S., Kon, O. M., Grass, L., et al. (2015). Differences in antigen-specific CD4+ responses to opportunistic infections in HIV infection. Immun. Inflamm. Dis. 3, 141–153. doi: 10.1002/iid3.50
Roach, J. C., Glusman, G., Rowen, L., Kaur, A., Purcell, M. K., Smith, K. D., et al. (2005). The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. U.S.A. 102, 9577–9582. doi: 10.1073/pnas.0502272102
Scher, J. U., Sczesnak, A., Longman, R. S., Segata, N., Ubeda, C., Bielski, C., et al. (2013). Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2:e01202. doi: 10.7554/eLife.01202
Schubert, A. M., Sinani, H., and Schloss, P. D. (2015). Antibiotic-induced alterations of the murine Gut Microbiota and subsequent effects on Colonization Resistance against Clostridium difficile. MBio 6, e00974. doi: 10.1128/mBio.00974-15
Scott, J. A., Berkley, J. A., Mwangi, I., Ochola, L., Uyoga, S., Macharia, A., et al. (2011). Relation between falciparum malaria and bacteraemia in Kenyan children: a population-based, case-control study and a longitudinal study. Lancet 378, 1316–1323. doi: 10.1016/s0140-6736(11)60888-x
Sender, R., Fuchs, S., and Milo, R. (2016). Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340. doi: 10.1016/j.cell.2016.01.013
Taniguchi, T., Miyauchi, E., Nakamura, S., Hirai, M., Suzue, K., Imai, T., et al. (2015). Plasmodium berghei ANKA causes intestinal malaria associated with dysbiosis. Sci. Rep. 5:15699. doi: 10.1038/srep15699
Theriot, C. M., Koenigsknecht, M. J., Carlson, P. E. Jr. Hatton, G. E., Nelson, A. M., Li, B., et al. (2014). Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5, 3114. doi: 10.1038/ncomms4114
Tosh, K. W., Mittereder, L., Bonne-Annee, S., Hieny, S., Nutman, T. B., Singer, S. M., et al. (2016). The IL-12 response of primary human dendritic cells and monocytes to Toxoplasma gondii is stimulated by phagocytosis of live parasites rather than host cell invasion. J. Immunol. 196, 345–356. doi: 10.4049/jimmunol.1501558
Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. doi: 10.1038/nm.3444
Tsay, R. W., Siu, L. K., Fung, C. P., and Chang, F. Y. (2002). Characteristics of bacteremia between community-acquired and nosocomial Klebsiella pneumoniae infection: risk factor for mortality and the impact of capsular serotypes as a herald for community-acquired infection. Arch. Intern. Med. 162, 1021–1027. doi: 10.1001/archinte.162.9.1021
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
van Olden, C., Groen, A. K., and Nieuwdorp, M. (2015). Role of intestinal microbiome in lipid and glucose metabolism in diabetes mellitus. Clin. Ther. 37, 1172–1177. doi: 10.1016/j.clinthera.2015.03.008
Vijay-Kumar, M., Aitken, J. D., Carvalho, F. A., Cullender, T. C., Mwangi, S., Srinivasan, S., et al. (2010). Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231. doi: 10.1126/science.1179721
Villarino, N. F., LeCleir, G. R., Denny, J. E., Dearth, S. P., Harding, C. L., Sloan, S. S., et al. (2016). Composition of the gut microbiota modulates the severity of malaria. Proc. Natl. Acad. Sci. U.S.A. 113, 2235–2240. doi: 10.1073/pnas.1504887113
Wang, J., Li, F., Wei, H., Lian, Z. X., Sun, R., and Tian, Z. (2014). Respiratory influenza virus infection induces intestinal immune injury via microbiota-mediated Th17 cell-dependent inflammation. J. Exp. Med. 211, 2397–2410. doi: 10.1084/jem.20140625
Winglee, K., Eloe-Fadrosh, E., Gupta, S., Guo, H., Fraser, C., and Bishai, W. (2014). Aerosol mycobacterium tuberculosis infection causes rapid loss of diversity in gut microbiota. PLoS ONE 9:e97048. doi: 10.1371/journal.pone.0097048
Wong, A. R., Raymond, B., Collins, J. W., Crepin, V. F., and Frankel, G. (2012). The enteropathogenic E. coli effector EspH promotes actin pedestal formation and elongation via WASP-interacting protein (WIP). Cell Microbiol. 14, 1051–1070. doi: 10.1111/j.1462-5822.2012.01778.x
World Health Organization (2014). Influenza Virus Infection in Humans (February 2014) [Online]. World Health Organization. Available online at: http://www.who.int/influenza/human_animal_interface/virology_laboratories_and_vaccines/influenza_virus_infections_humans_feb14.pdf?ua=1
World Malaria Report, World Health Organization. (2014). World Malaria Report 2014 Summary [Online]. World Health Organization. Available online at: http://www.who.int/malaria/publications/world_malaria_report_2014/wmr-2014-summary-eng.pdf
Yarovinsky, F., Hieny, S., and Sher, A. (2008). Recognition of Toxoplasma gondii by TLR11 prevents parasite-induced immunopathology. J. Immunol. 181, 8478–8484. doi: 10.4049/jimmunol.181.12.8478
Yilmaz, B., Portugal, S., Tran, T. M., Gozzelino, R., Ramos, S., Gomes, J., et al. (2014). Gut microbiota elicits a protective immune response against malaria transmission. Cell 159, 1277–1289. doi: 10.1016/j.cell.2014.10.053
Yooseph, S., Kirkness, E. F., Tran, T. M., Harkins, D. M., Jones, M. B., Torralba, M. G., et al. (2015). Stool microbiota composition is associated with the prospective risk of Plasmodium falciparum infection. BMC Genomics 16:631. doi: 10.1186/s12864-015-1819-3
Zainah, H., Hassan, M., Shiekh-Sroujieh, L., Hassan, S., Alangaden, G., and Ramesh, M. (2015). Intestinal microbiota transplantation, a simple effective treatment for severe refractory Clostridium difficile infection. Dig. Dis. Sci. 60, 181–185. doi: 10.1007/s10620-014-3296-y
Keywords: gut microbiome, Infectious Disease Medicine, Cross-talk, influenza, human, Plasmodium
Citation: Denny JE, Powell WL and Schmidt NW (2016) Local and Long-Distance Calling: Conversations between the Gut Microbiota and Intraand Extra-Gastrointestinal Tract Infections. Front. Cell. Infect. Microbiol. 6:41. doi: 10.3389/fcimb.2016.00041
Received: 12 January 2016; Accepted: 22 March 2016;
Published: 11 April 2016.
Edited by:
Brian J. Akerley, University of Mississippi Medical Center, USAReviewed by:
Geanncarlo Lugo-Villarino, Centre National de la Recherche Scientifique, FranceEmily Mallick, Brown University, USA
Copyright © 2016 Denny, Powell and Schmidt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nathan W. Schmidt, bmF0aGFuLnNjaG1pZHRAbG91aXN2aWxsZS5lZHU=
†These authors have contributed equally to this work.
 Joshua E. Denny
Joshua E. Denny Whitney L. Powell
Whitney L. Powell Nathan W. Schmidt
Nathan W. Schmidt