- 1Department of Biological Sciences, Ohio University, Athens, OH, USA
- 2Department of Biomedical Sciences, Heritage College of Osteopathic Medicine, Ohio University, Athens, OH, USA
Survival of Shigella within the host is strictly dependent on the ability of the pathogen to acquire essential nutrients, such as iron. As an innate immune defense against invading pathogens, the level of bio-available iron within the human host is maintained at exceeding low levels, by sequestration of the element within heme and other host iron-binding compounds. In response to sequestration mediated iron limitation, Shigella produce multiple iron-uptake systems that each function to facilitate the utilization of a specific host-associated source of nutrient iron. As a mechanism to balance the essential need for iron and the toxicity of the element when in excess, the production of bacterial iron acquisition systems is tightly regulated by a variety of molecular mechanisms. This review summarizes the current state of knowledge on the iron-uptake systems produced by Shigella species, their distribution within the genus, and the molecular mechanisms that regulate their production.
Introduction
Shigella is a genus of Gram negative, facultative anaerobic, pathogenic enterobacteria, composed of four species (S. boydii, S. sonnei, S. flexneri, and S. dysenteriae), each a causative agent of dysentery in humans. Like all invading pathogens, Shigella experiences a wide variety of environmental conditions during transmission and throughout the course of a natural infection. The survival of Shigella, and thus its ability to cause disease, is strictly dependent on the ability of the organism to acquire iron from each encountered environment. The strict requirement for iron stems from the fact that this element is an essential co-factor of several enzymes involved in basic biological processes such as DNA replication and respiration. While iron is essential, too much of the element is toxic to a bacterium (Imlay et al., 1988). To balance the necessity and potential toxicity of the element, iron homeostasis must be precisely maintained, a requirement that is facilitated, at least in part, by regulation of bacterial iron-acquisition systems.
As an innate immune defense against invading pathogens, iron within the human body is sequestrated within iron binding compounds and proteins, generating a concentration of bioavailable iron of approximately 10−24 M, a concentration that is far below the 10−7 M required for the survival of most bacteria (Andrews et al., 2003; Raymond et al., 2003). In response to this iron limitation, pathogenic bacteria have evolved several systems to utilize the various sources of iron present within the infected host. Regulating the production of specific iron acquisition systems in response to environmental conditions allows the bacterium to both efficiently utilize the iron present within a given environment and to avoid iron-mediated toxicity. This review briefly summarizes the current state of knowledge regarding the function and regulation of iron-uptake systems in Shigella species.
Iron-uptake Systems in Shigella
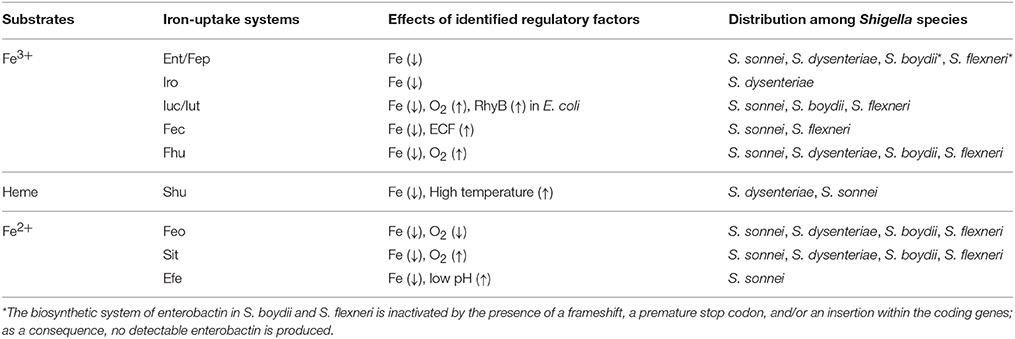
The currently identified Shigella iron-uptake systems can be grouped into three broad categories based on the form of iron that they facilitate the utilization of: (1) systems for the utilization of ferric iron (Fe3+), (2) systems for the utilization of heme-bound iron, and (3) systems for the utilization of ferrous iron (Fe2+). Each of the four Shigella species contains multiple iron uptake systems, however, the combination of iron acquisition systems present vary by species. Each identified Shigella iron-uptake system and their distribution among Shigella species is presented below and is summarized in Table 1.
Ferric Iron Utilization Systems
To utilize Fe3+, Shigella have evolved several systems to synthesize, secrete, and uptake siderophores, a group of compounds with high affinity for iron that functionally compete for iron bound by iron-sequestrating factors within the infected host (Carrano and Raymond, 1979; Hider and Kong, 2010). In Shigella, the combination of siderophores that are synthesized and utilized varies by species, but the uptake process is generally conserved (Figure 1). Specifically, transport of each Fe3+-siderophore into the bacterium is initiated by binding of the complex to a specific outer-membrane receptor. Once bound by its receptor, the complex is transported across the outer-membrane, passed to a periplasmic binding protein (PBP), and finally transported across the inner-membrane by the activity of an ABC permease complex. Compared to that of the outer-membrane receptors, PBPs have lower substrate specificity, thus one PBP can facilitate the transportation of multiple siderophores with similar chemical structures (Miethke and Marahiel, 2007). Transport of any cargo across a membrane requires energy. Anchored within the inner-membrane, the TonB/ExbB/ExbD complex transduces energy generated by the proton gradient to a given iron-binding outer-membrane receptor to provide the energy required to transport the associated cargo across the outer-membrane (Larsen et al., 1997). ATP hydrolysis generates the energy needed for the subsequent transport of the cargo across the inner-membrane. Once inside the bacterial cell, iron is either released from the siderophore for utilization, or bound within storage proteins for future use. Each identified Shigella siderophore and its transport system is discussed below.
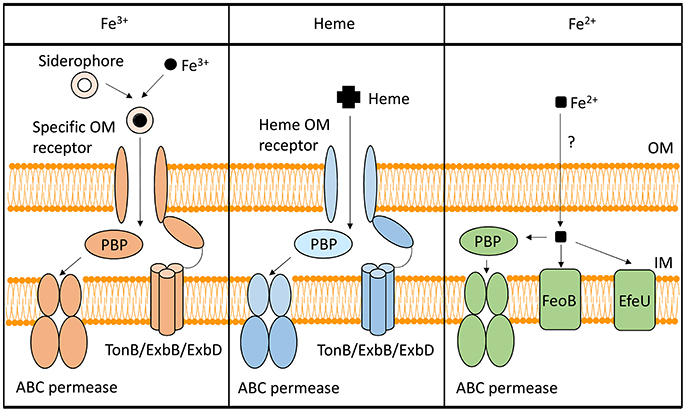
Figure 1. Iron-uptake systems in Shigella. This figure is a schematic of the Shigella iron-uptake systems categorized into three broad groups based on the form of iron or iron-containing compounds being utilized. These three groups are (1) systems for the utilization of ferric iron (Fe3+), (2) systems for the utilization of heme (Heme), and (3) systems for the uptake of ferrous iron (Fe2+). Within the ferrous iron uptake systems, the PBP represents SitA, while the ABC permease represents SitBCD. “PBP” stands for periplasmic binding protein.
Enterobactin
Discovered in 1970, enterobactin is the siderophore with the highest known affinity for Fe3+ (~1049) (O'Brien et al., 1970; Pollack and Neilands, 1970; Loomis and Raymond, 1991). Genes involved in the synthesis, secretion, and uptake of enterobactin are encoded in a single locus, with ent genes encoding factors involved in the synthesis/secretion of the siderophore and fep genes encoding factors composing the uptake system (Laird et al., 1980). Specifically, fepA encodes a TonB-dependent outer-membrane receptor, fepB encodes a PBP, and fepCDG encodes proteins constituting the ABC permease complex (Ozenberger et al., 1987). All four Shigella species contain the ent/fep locus, however some of these genes are inactivated in S. boydii and in some strains of S. flexneri, due to the presence of a frameshift, a premature stop codon, and/or an insertion mutation (Payne, 1980; Payne et al., 1983).
Due to its high iron-binding affinity, enterobactin can chelate the element from various host iron-binding factors (Carrano and Raymond, 1979). For example, a recent study demonstrates that enterobactin can overcome the sequestration of iron by ATP within the intracellular environment, and as such would promote survival of the pathogen within a macrophage (Tatano et al., 2015). When present in the extracellular environment however, bacteria often induce host cells to produce and secrete lipocalin-2, a protein that specifically binds enterobactin, thus preventing the bacterium from utilizing the iron bound within it (Flo et al., 2004).
Salmochelin
Due to their different chemical structures, some enterobactin derivatives are not recognized by lipocalin-2 and, therefore not surprisingly, have been found to contribute to virulence. One such enterobactin derivative is salmochelin, a siderophore produced by S. dysenteriae (Fischbach et al., 2006; Wyckoff et al., 2009). The iro locus contains genes encoding enzymes that transform enterobactin into salmochelin, the machinery required to secrete salmochelin, and a salmochelin specific TonB-dependent outer-membrane receptor (Hantke et al., 2003). Additional factors involved in salmochelin utilization, including the PBP and the ABC transporter, are the same as those used for the utilization of enterobactin (Müller et al., 2009).
Aerobactin
Some strains of S. flexneri, S. boydii, and S. sonnei produce aerobactin, a siderophore that has a different chemical structure from that of enterobactin and as a result, can also escape the sequestration by host protein lipocalin-2 (Lawlor and Payne, 1984; Flo et al., 2004). Aerobactin has been shown to promote the virulence of uropathogenic Escherichia coli, and to facilitate extracellular growth of Shigella (de Lorenzo and Martinez, 1988; Torres et al., 2001). iucABCD encodes the enzymes required for the synthesis of aerobactin and is found within a single locus along with iutA, a gene encoding the aerobactin-specific TonB-dependent outer-membrane receptor (Carbonetti and Williams, 1984). The remaining factors required for the utilization of aerobactin are encoded within the fhu locus, and are also utilized for the transportation of ferrichrome (see below; Köster and Braun, 1990).
Xenosiderophores
Like many other bacterial species, Shigella can utilize xenosiderophores, siderophores produced by other microorganisms (Payne, 1980). For example, ferrichrome, a fungal siderophore with a chemical structure similar to that of aerobactin is utilized by Shigella species. The utilization of ferrichrome is mediated by factors composing the Fhu system including FhuA, a ferrichrome-specific TonB-dependent outer-membrane receptor (Köster and Braun, 1990; Miethke and Marahiel, 2007).
Ferric-Dicitrate
Ferric iron can bind with citrate to form ferric-dicitrate. S. sonnei and at least one strain of S. flexneri can utilize ferric-dicitrate as a source of nutrient iron (Luck et al., 2001; Wyckoff et al., 2009). The utilization of ferric-dicitrate bound iron is mediated by factors encoded within the fec locus, including an outer-membrane receptor (FecA), a PBP (FecB), and an ABC transport complex (FecCDE) (Braun and Mahren, 2005).
Heme Utilization System
The Shigella heme-uptake (Shu) system was first identified in S. dysenteriae and is predicted to be present in some strains of S. sonnei (Wyckoff et al., 1998). Inactivation of shuA, a gene encoding the outer-membrane heme receptor, eliminates the ability of S. dysenteriae to utilize heme as a sole source of nutrient iron, suggesting that the Shu system is the only functional heme-utilization system in this species (Mills and Payne, 1997). In uropathogenic E. coli, inactivation of the orthologous gene (chuA) results in attenuation of virulence, a finding that directly identifies the heme receptor as a virulence factor in this closely related species (Torres et al., 2001). Additional components of the Shu system include a PBP (ShuT) and an inner-membrane ABC permease complex (ShuUV) (Wyckoff et al., 1998; Eakanunkul et al., 2005; Burkhard and Wilks, 2008). The process of heme-uptake is similar to that detailed above for the uptake of siderophores (Figure 1). Interestingly, the fact that Shigella species which lack the shu genes are able to utilize heme-bound iron suggests the existence of a yet unidentified heme-utilization system(s) (Payne et al., 2006).
Ferrous Iron Utilization Systems
Under anaerobic and/or acidic conditions, Fe2+ is the dominant form of the element. Three Fe2+-uptake systems have been identified in Shigella: the Feo system, the Sit system, and the Efe system (Kammler et al., 1993; Zhou et al., 1999; Jin et al., 2002; Große et al., 2006; Figure 1).
Feo System
Despite being the first Fe2+ utilization system identified in Shigella, details of the molecular mechanism(s) underlying the activity of the Feo system remain largely unknown (Kammler et al., 1993). Only three components of the Feo system have been identified to date: FeoA, FeoB, and FeoC. FeoB is an inner-membrane transporter with GTPase activity and a structure similar to that of a eukaryotic G protein (Marlovits et al., 2002). FeoC contains an oxygen-responsive [4Fe-4S] cluster and functions to promote proteolysis of FeoB in the presence of oxygen (Hsueh et al., 2013; Kim et al., 2015). While known to be required for the transport of Fe2+ by FeoB, the exact function of FeoA remains to be determined (Kim et al., 2012; Lau et al., 2013).
Sit System
The sit locus contains four genes: sitA encoding the PBP and sitBCD encoding components of the ABC permease complex (Zhou et al., 1999; Fisher et al., 2009). Interestingly, no outer-membrane receptor has been identified to date, leading to the hypothesis that Fe2+ is transported through the outer-membrane via non-specific porins and/or ion channels (Andrews et al., 2003). Interestingly, studies in Salmonella indicate that SitA has a higher affinity for manganese than for Fe2+, suggesting that the primary function of the Sit system might be to transport manganese (Kehres et al., 2002). It has been demonstrated however, that S. flexneri is able to use the Sit system as the sole iron-uptake system to survive and form plaques in a monolayer of eukaryotic cells, suggesting not only that the system can function to uptake iron but also that the Sit system has a direct role in pathogenesis (Runyen-Janecky et al., 2003). It remains a formal possibility that the Shigella Sit system functions to transport both manganese and iron. Interestingly, the E. coli orthologous of the Shigella manganese transporter MntH and zinc transporter YgiE (MntH and ZupT, respectively) have been shown to transport Fe2+ in addition to their specific substrates, findings that support the hypothesis that additional Fe2+ transport systems may exist in Shigella species (Kehres et al., 2000; Grass et al., 2005).
Efe System
Originally characterized in E. coli, genetic analysis demonstrates that the EfeUOB system, formally called YcdNOB, is encoded by genes present on a tricistronic transcript in at least one strain of S. sonnei (Große et al., 2006; Cao et al., 2007; Payne and Alexandra, 2010). efeU encodes the inner membrane permease and is homologous to the yeast Fe2+ transporter Ftr1p (Große et al., 2006). EfeO and EfeB are both periplasmic proteins necessary for Fe2+ transport, however their specific functions remain unknown.
Regulation of Iron-uptakes Systems
To ensure that a given iron acquisition system is optimally produced under the specific environmental condition(s) in which its function will be most advantageous to the pathogen, production of Shigella iron-uptake systems is tightly controlled by multiple regulatory factors via distinct molecular mechanisms (Table 1). The major regulatory mechanisms governing the production of Shigella iron acquisition systems are detailed below.
Regulation by Extracytoplasmic Function (ECF) Sigma Factors
The Shigella Fec system, utilized for the uptake of ferric-dicitrate, is regulated by the ECF sigma factor FecI (Lonetto et al., 1994; Braun and Mahren, 2005). Upon binding of ferric-dicitrate to FecA, the TonB-dependent outer-membrane receptor undergoes a conformational change resulting in the interaction of its N-terminal domain with the C-terminal domain of the trans-membrane anti-sigma factor FecR. The interaction of FecA with FecR results in the release of the alternative sigma factor FecI that, in turn, directs RNA polymerases to the alternative promoter region of the fecABCDE operon (Braun and Mahren, 2005). Such regulation results directly in increased production of the Fec system when Shigella is within an environment containing ferric citrate.
Regulation by Fur
Availability of iron influences the production of several Shigella iron-acquisition systems via Fur, an iron-responsive transcriptional regulator (Fleming et al., 1983; de Lorenzo et al., 1987; Wyckoff et al., 1998; Payne et al., 2006). The interaction of Fur with intracellular iron induces a conformational change in the protein that enables it to dimerize and bind DNA in a sequence specific manner (Troxell and Hassan, 2013). Iron-dependent binding of Fur at or near the promoter region of a target gene most often inhibits transcription by physically blocking binding of RNA polymerase (Troxell and Hassan, 2013).
Regulation by RhyB
RyhB, a Fur-repressed regulatory small RNA first identified in E. coli, plays an important role in achieving iron homeostasis in several bacterial species, including Shigella. RyhB functions to modulate the stability of specific target transcripts in response to iron availability within the environment (Massé and Gottesman, 2002). Specifically, under iron-poor conditions, Fur-mediated repression of RhyB is relieved, and once produced, the small RNA functions to repress the production of several factors including iron-containing enzymes, such as SodB and iron-storage proteins, such as ferritin, in both E. coli and Shigella (Massé and Gottesman, 2002; Murphy and Payne, 2007). In E. coli, RhyB also regulates the production of aerobactin, however the details of this regulatory mechanism, as well as the role that such a mechanism may play in controlling Shigella gene expression remains to be revealed (Porcheron et al., 2014).
Oxygen-Dependent Regulation
Within a given environment, the amount of oxygen influences the relative abundance of Fe3+ and Fe2+; thus it is reasonable that bacterial iron-uptake systems are also regulated in response to environmental oxygen levels. Two regulatory systems with over-lapping, yet non-identical regulons control the production of Shigella bacterial iron acquisition systems in response to oxygen status: the two component system ArcAB, and the DNA-binding regulator Fnr (Carpenter and Payne, 2014).
ArcAB System
Within the ArcAB two-component system, ArcB is a membrane-anchored kinase and ArcA is a DNA-binding response regulator (Carpenter and Payne, 2014). Following auto-phosphorylation of ArcB, a process that occurs only under anaerobic conditions, the activated kinase phosphorylates ArcA. Once phosphorylated, ArcA functions to directly alter the expression of specific target genes (Carpenter and Payne, 2014). Under aerobic conditions, the auto-phosphorylation of ArcB is inhibited, ultimately resulting in the lack of ArcA activation and thus altered expression of ArcA-regulated genes. As a global regulator, ArcA not only modulates the production of factors involved in iron-uptake, but also that of the iron-responsive regulator Fur (Boulette and Payne, 2007).
Fnr
As a DNA-binding protein, the regulatory mechanism underlying the activity of Fnr is similar to that underlying the activity of Fur. Unlike Fur however, the conformational change controlling the regulatory activity of Fnr is determined directly by the oxidative status of its [4Fe-4S] cluster, and thus indirectly by the oxygen status of the environment (Carpenter and Payne, 2014). With increased levels of intracellular oxygen, the [4Fe-4S] cluster within Fnr is oxidized into [2Fe-2S], a transition that results into the loss of DNA-binding ability, and thus regulatory activity, of the protein.
Temperature-Dependent Regulation
A change in environmental temperature to that encountered within the human body (37°C) is an important signal that can indicate a transition from the non-host environment to the human host. As such, temperature influences the expression of many Shigella virulence-associated genes, including that of shuA (Tobe et al., 1991; Kouse et al., 2013). Production of ShuA, an outer-membrane heme receptor, is subject to temperature-dependent post-transcriptional regulation via the activity of an RNA thermometer located within the 5′ untranslated region of shuA (Kouse et al., 2013). RNA thermometers regulate translation of the gene in which they are housed by the formation of an inhibitory structure that physically occludes the ribosomal binding site at relatively low temperatures. At relatively high temperatures, such as that encountered within the human host, the inhibitory structure within an RNA thermometer is destabilized, the ribosomal binding site is exposed and translation proceeds. While ShuA is currently the only Shigella iron acquisition factor known to be regulated by the activity of an RNA thermometer, the full impact of this temperature-dependent regulatory mechanism on iron utilization by these pathogens remains to be determined.
Future Prospective
Given the importance of iron homeostasis to survival, the mechanisms by which bacteria acquire iron from the environment and the regulatory mechanisms controlling the production of bacterial iron acquisition systems have long been the subject of active investigation. As highlighted above, the importance of several iron utilization systems to virulence has been proven in other pathogenic bacteria, however, exactly how these factors, and their associated regulation, contribute to Shigella virulence remains to be fully elucidated. Moreover, whether the presence of a specific combination of iron uptake systems in a given Shigella species contributes to differences in survival within host or non-host environments, differences in geographic distribution and/or differences in pathogenesis would be a potentially revealing analysis, but one that can only be completed after all the iron acquisition systems in these pathogenic organisms are recognized. Finally, future investigations to identify additional Shigella iron acquisition systems and to understand the role of these systems in Shigella virulence could lead to the development of novel therapeutics designed to disrupt iron acquisition, and by doing so eliminate or reduce the ability of these bacterial pathogens to cause human disease.
Author Contributions
Wrote the article: YW and EM. Edited article: YW and EM. Generated figure: YW. Generated table: YW.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Ohio University and the American Heart Association for funding of ongoing research (AHA-13GBIA16310001).
References
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Boulette, M. L., and Payne, S. M. (2007). Anaerobic regulation of Shigella flexneri virulence: ArcA regulates fur and iron acquisition genes. J. Bacteriol. 189, 6957–6967. doi: 10.1128/JB.00621-07
Braun, V., and Mahren, S. (2005). Transmembrane transcriptional control (surface signalling) of the Escherichia coli Fec type. FEMS Microbiol. Rev. 29, 673–684. doi: 10.1016/j.femsre.2004.10.001
Burkhard, K. A., and Wilks, A. (2008). Functional characterization of the Shigella dysenteriae heme ABC transporter. Biochemistry 47, 7977–7979. doi: 10.1021/bi801005u
Cao, J., Woodhall, M. R., Alvarez, J., Cartron, M. L., and Andrews, S. C. (2007). EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 65, 857–875. doi: 10.1111/j.1365-2958.2007.05802.x
Carbonetti, N. H., and Williams, P. H. (1984). A cluster of five genes specifying the aerobactin iron uptake system of plasmid ColV-K30. Infect. Immun. 46, 7–12.
Carpenter, C., and Payne, S. M. (2014). Regulation of iron transport systems in Enterobacteriaceae in response to oxygen and iron availability. J. Inorg. Biochem. 133, 110–117. doi: 10.1016/j.jinorgbio.2014.01.007
Carrano, C. J., and Raymond, K. N. (1979). Ferric ion sequestering agents. 2. Kinetics and mechanism of iron removal from transferrin by enterobactin and synthetic tricatechols. J. Am. Chem. Soc. 101, 5401–5404. doi: 10.1021/ja00512a047
de Lorenzo, V., and Martinez, J. L. (1988). Aerobactin production as a virulence factor: a reevaluation. Eur. J. Clin. Microbiol. Infect. Dis. 7, 621–629.
de Lorenzo, V., Wee, S., Herrero, M., and Neilands, J. B. (1987). Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169, 2624–2630.
Eakanunkul, S., Lukat-Rodgers, G. S., Sumithran, S., Ghosh, A., Rodgers, K. R., Dawson, J. H., et al. (2005). Characterization of the periplasmic heme-binding protein ShuT from the heme uptake system of Shigella dysenteriae. Biochemistry 44, 13179–13191. doi: 10.1021/bi050422r
Fischbach, M. A., Lin, H., Zhou, L., Yu, Y., Abergel, R. J., Liu, D. R., et al. (2006). The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proc. Natl. Acad. Sci. U.S.A. 103, 16502–16507. doi: 10.1073/pnas.0604636103
Fisher, C. R., Davies, N. M. L. L., Wyckoff, E. E., Feng, Z., Oaks, E. V., and Payne, S. M. (2009). Genetics and virulence association of the Shigella flexneri Sit iron transport system. Infect. Immun. 77, 1992–1999. doi: 10.1128/IAI.00064-09
Fleming, T. P., Nahlik, M. S., and McIntosh, M. A. (1983). Regulation of enterobactin iron transport in Escherichia coli: characterization of ent::Mu d(Apr lac) operon fusions. J. Bacteriol. 156, 1171–1177.
Flo, T. H., Smith, K. D., Sato, S., Rodriguez, D. J., Holmes, M. A., Strong, R. K., et al. (2004). Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature 432, 917–921. doi: 10.1038/nature03104
Grass, G., Franke, S., Taudte, N., Nies, D. H., Kucharski, L. M., Maguire, M. E., et al. (2005). The metal permease ZupT from Escherichia coli is a transporter with a broad substrate spectrum. J. Bacteriol. 187, 1604–1611. doi: 10.1128/JB.187.5.1604-1611.2005
Große, C., Scherer, J., Koch, D., Otto, M., Taudte, N., and Grass, G. (2006). A new ferrous iron-uptake transporter, EfeU (YcdN), from Escherichia coli. Mol. Microbiol. 62, 120–131. doi: 10.1111/j.1365-2958.2006.05326.x
Hantke, K., Nicholson, G., Rabsch, W., and Winkelmann, G. (2003). Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. U.S.A. 100, 3677–3682. doi: 10.1073/pnas.0737682100
Hider, R. C., and Kong, X. (2010). Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637. doi: 10.1039/b906679a
Hsueh, K.-L., Yu, L.-K., Chen, Y.-H., Cheng, Y.-H., Hsieh, Y.-C., Ke, S.-C., et al. (2013). FeoC from Klebsiella pneumoniae contains a [4Fe-4S] cluster. J. Bacteriol. 3, 4726–4734. doi: 10.1128/JB.00687-13
Imlay, J. A., Chin, S. M., and Linn, S. (1988). Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240, 640–642. doi: 10.1126/science.2834821
Jin, Q., Yuan, Z., Xu, J., Wang, Y., Shen, Y., Lu, W., et al. (2002). Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30, 4432–4441. doi: 10.1093/nar/gkf566
Kammler, M., Schön, C., and Hantke, K. (1993). Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175, 6212–6219.
Kehres, D. G., Janakiraman, A., Slauch, J. M., and Maguire, M. E. (2002). SitABCD is the alkaline Mn2+ transporter of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184, 3159–3166. doi: 10.1128/JB.184.12.3159
Kehres, D. G., Zaharik, M. L., Finlay, B. B., and Maguire, M. E. (2000). The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol. Microbiol. 36, 1085–1100. doi: 10.1046/j.1365-2958.2000.01922.x
Kim, H., Lee, H., and Shin, D. (2012). The FeoA protein is necessary for the FeoB transporter to import ferrous iron. Biochem. Biophys. Res. Commun. 423, 733–738. doi: 10.1016/j.bbrc.2012.06.027
Kim, H., Lee, H., and Shin, D. (2015). Lon-mediated proteolysis of the FeoC protein prevents Salmonella enterica from accumulating the Fe(II) transporter FeoB under high-oxygen conditions. J. Bacteriol. 197, 92–98. doi: 10.1128/JB.01826-14
Köster, W., and Braun, V. (1990). Iron (III) hydroxamate transport into Escherichia coli. Substrate binding to the periplasmic FhuD protein. J. Biol. Chem. 265, 21407–21410.
Kouse, A. B., Righetti, F., Kortmann, J., Narberhaus, F., and Murphy, E. R. (2013). RNA-mediated thermoregulation of iron-acquisition genes in Shigella dysenteriae and pathogenic Escherichia coli. PLoS ONE 8:e63781. doi: 10.1371/journal.pone.0063781
Laird, A. J., Ribbons, D. W., Woodrow, G. C., Young, I. G., and Young, G. (1980). Bacteriophage Mu-mediated gene transposition and in vitro cloning of the enterochelin gene cluster of Escherichia coli. Gene 11, 347–357.
Larsen, R. A., Foster-Hartnett, D., McIntosh, M. A., and Postle, K. (1997). Regions of Escherichia coli TonB and FepA proteins essential for in vivo physical interactions. J. Bacteriol. 179, 3213–3221.
Lau, C. K. Y., Ishida, H., Liu, Z., and Vogel, H. J. (2013). Solution structure of Escherichia coli FeoA and its potential role in bacterial ferrous iron transport. J. Bacteriol. 195, 46–55. doi: 10.1128/JB.01121-12
Lawlor, K. M., and Payne, S. M. (1984). Aerobactin genes in Shigella spp. J. Bacteriol. 160, 266–272.
Lonetto, M. A., Brown, K. L., Rudd, K. E., and Buttner, M. J. (1994). Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase σ factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. U.S.A. 91, 7573–7577. doi: 10.1073/pnas.91.16.7573
Loomis, L. D., and Raymond, K. N. (1991). Solution equilibria of enterobactin and metal-enterobactin complexes. Inorg. Chem. 30, 906–911. doi: 10.1021/ic00005a008
Luck, S. N., Turner, S. A., Rajakumar, K., Sakellaris, H., and Adler, B. (2001). Ferric Dicitrate Transport System (Fec) of Shigella flexneri 2a YSH6000 is encoded on a novel pathogenicity island carrying multiple antibiotic resistance genes. Infect. Immun. 69, 6012–6021. doi: 10.1128/IAI.69.10.6012
Marlovits, T. C., Haase, W., Herrmann, C., Aller, S. G., and Unger, V. M. (2002). The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. U.S.A. 99, 16243–16248. doi: 10.1073/pnas.242338299
Massé, E., and Gottesman, S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625. doi: 10.1073/pnas.032066599
Miethke, M., and Marahiel, M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451. doi: 10.1128/MMBR.00012-07
Mills, M., and Payne, S. M. (1997). Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect. Immun. 65, 5358–5363.
Müller, S. I., Valdebenito, M., and Hantke, K. (2009). Salmochelin, the long-overlooked catecholate siderophore of Salmonella. BioMetals 22, 691–695. doi: 10.1007/s10534-009-9217-4
Murphy, E. R., and Payne, S. M. (2007). RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect. Immun. 75, 3470–3477. doi: 10.1128/IAI.00112-07
O'Brien, I. G., Cox, G. B., and Gibson, F. (1970). Biologically active compounds containing 2,3-dihydroxybenzoic acid and serine formed by Escherichia coli. Biochim. Biophys. Acta 201, 453–460. doi: 10.1016/0304-4165(70)90165-0
Ozenberger, B. A., Schrodt Nahlik, M., and McIntosh, M. A. (1987). Genetic organization of multiple fep genes encoding ferric enterobactin transport functions in Escherichia coli. J. Bacteriol. 169, 3638–3646.
Payne, S. M. (1980). Synthesis and utilization of siderophores by Shigella flexneri. J. Bacteriol. 143, 1420–1424.
Payne, S. M., and Alexandra, M. R. (2010). “Iron uptake in Shigella and Escherichia coli,” in Iron Uptake and Homeostasis in Microorganisms, eds P. Cornelis and S. C. Andrews (Norfolk: Caister Academic Press), 87–100.
Payne, S. M., Niesel, D. W., Peixotto, S. S., and Lawlor, K. M. (1983). Expression of hydroxamate and phenolate siderophores by Shigella flexneri. J. Bacteriol. 155, 949–955.
Payne, S. M., Wyckoff, E. E., Murphy, E. R., Oglesby, A. G., Boulette, M. L., and Davies, N. M. L. (2006). Iron and pathogenesis of Shigella: iron acquisition in the intracellular environment. Biometals 19, 173–180. doi: 10.1007/s10534-005-4577-x
Pollack, J. R., and Neilands, J. B. (1970). Enterobactin, an iron transport compound from Salmonella typhimurium. Biochem. Biophys. Res. Commun. 38, 989–992. doi: 10.1016/0006-291X(70)90819-3
Porcheron, G., Habib, R., Houle, S., Caza, M., Lépine, F., Daigle, F., et al. (2014). The small RNA RyhB contributes to siderophore production and virulence of uropathogenic Escherichia coli. Infect. Immun. 82, 5056–5068. doi: 10.1128/IAI.02287-14
Raymond, K. N., Dertz, E. A., and Kim, S. S. (2003). Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U.S.A. 100, 3584–3588. doi: 10.1073/pnas.0630018100
Runyen-Janecky, L. J., Reeves, S. A., Gonzales, E. G., and Payne, S. M. (2003). Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect. Immun. 71, 1919–1928. doi: 10.1128/IAI.71.4.1919-1928.2003
Tatano, Y., Kanehiro, Y., Sano, C., Shimizu, T., and Tomioka, H. (2015). ATP exhibits antimicrobial action by inhibiting bacterial utilization of ferric ions. Sci. Rep. 5:8610. doi: 10.1038/srep08610
Tobe, T., Nagai, S., Okada, N., Adler, B., Yoshikawa, M., and Sasakawa, C. (1991). Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol. Microbiol. 5, 887–893.
Torres, A. G., Redford, P., Welch, R. A., and Payne, S. M. (2001). TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69, 6179–6185. doi: 10.1128/IAI.69.10.6179-6185.2001
Troxell, B., and Hassan, H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Wyckoff, E. E., Boulette, M. L., and Payne, S. M. (2009). Genetics and environmental regulation of Shigella iron transport systems. Biometals 22, 43–51. doi: 10.1007/s10534-008-9188-x
Wyckoff, E. E., Duncan, D., Torres, A. G., Mills, M., Maase, K., and Payne, S. M. (1998). Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28, 1139–1152.
Keywords: iron acquisition, Shigella, regulation, pathogenicity, virulence
Citation: Wei Y and Murphy ER (2016) Shigella Iron Acquisition Systems and their Regulation. Front. Cell. Infect. Microbiol. 6:18. doi: 10.3389/fcimb.2016.00018
Received: 20 November 2015; Accepted: 25 January 2016;
Published: 09 February 2016.
Edited by:
William D. Picking, University of Kansas, USAReviewed by:
Charles Martin Dozois, Institut National de la Recherche Scientifique, CanadaElizabeth Fozo, University of Tennessee, USA
Copyright © 2016 Wei and Murphy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erin R. Murphy, bXVycGh5ZUBvaGlvLmVkdQ==
 Yahan Wei
Yahan Wei Erin R. Murphy
Erin R. Murphy