- 1Liggins Institute, The University of Auckland, Auckland, New Zealand
- 2Department of Infectious Diseases, Counties Manukau Health, Auckland, New Zealand
- 3Gravida: National Centre for Growth and Development, Auckland, New Zealand
Key Points
• The microbiome has been implicated in the development of obesity.
• Conventional therapeutic methods have limited effectiveness for the treatment of obesity and prevention of related complications.
• Gut microbiome transplantation may represent an alternative and effective therapy for the treatment of obesity.
Obesity has reached epidemic proportions. Despite a better understanding of the underlying pathophysiology and growing treatment options, a significant proportion of obese patients do not respond to treatment. Recently, microbes residing in the human gastrointestinal tract have been found to act as an “endocrine” organ, whose composition and functionality may contribute to the development of obesity. Therefore, fecal/gut microbiome transplantation (GMT), which involves the transfer of feces from a healthy donor to a recipient, is increasingly drawing attention as a potential treatment for obesity. Currently the evidence for GMT effectiveness in the treatment of obesity is preliminary. Here, we summarize benefits, procedures, and issues associated with GMT, with a special focus on obesity.
Introduction
Obesity has recently been identified as a disease by the American Medical Association with >33% of the world's adult population (20 years and older) overweight or obese (World Health Organization, 2014). Sadly, this is projected to increase to the point where up to 57.8% of the world's population aged 20 and over is either overweight or obese (World Health Organization, 2014). There are various causative factors that contribute to the development of obesity including genetics (Wang et al., 2011a), low levels of physical activity and exercise, poor diet and other unhealthy behaviors. Obesity is a major risk factor for diabetes, hypertension, and metabolic syndrome. Despite the promotion of numerous strategies for the prevention and treatment of obesity, most patients are refractory to treatment. Thus, new approaches are currently being sought to reduce the financial, social, and health consequences of the obesity epidemic.
The human gut contains an extensive population of microbes (the gut microbiome) that effectively constitute a microbial “endocrine organ” (Cani and Delzenne, 2007; Clarke et al., 2014). Recent research has implicated these microbes as having a significant role in the development of obesity (Bäckhed et al., 2004; Ley et al., 2005, 2006b; Turnbaugh et al., 2006, 2009; Backhed et al., 2007; Zhang et al., 2009), diabetes (Larsen et al., 2010; Qin et al., 2012), and cardiovascular disease (Ordovas and Mooser, 2006; Wang et al., 2011b; Howitt and Garrett, 2012; Tang and Hazen, 2014). Therefore, environmental effects on these microbes and our ability to manipulate them in a controlled manner are under increasing scrutiny.
Fecal/gut microbiome transplantation (GMT; Box 1) has been suggested as a new method of altering the gut microbiota that may lead to beneficial metabolic changes (Smits et al., 2013). Modifications of the host's microflora by GMT were first performed in the 1950s to treat pseudomembranous colitis now known to be due to Clostridium difficile infection (CDI) (Eiseman et al., 1958). Since then, GMT has been successfully used for CDI treatment and is increasingly considered the treatment of choice for chronic pseudomembranous colitis (Gough et al., 2011). Despite the fact that GMT has been shown to improve insulin sensitivity in adults with features of metabolic syndrome (Vrieze et al., 2012), its application as a therapy for other conditions, including obesity, is still experimental. As such, it is still unclear how, when, or under which circumstances GMT should be performed. Here, we will address the procedures, benefits, and issues associated with GMT, with a special focus on obesity.
Box 1. FMT vs. GMT.
Through-out this manuscript we refer to gut microbiome transplantation (GMT) and not fecal microbiome transplantation (FMT). Predominant amongst our reasons for this minor change in terminology is the public attitude and perception of products and treatments derived from feces as being “dirty” or “unhygienic” (Brandt, 2012; Leslie et al., 2014). These prejudices are ingrained and continually reinforced by the testing and notifications of fecal contamination of public drinking and bathing sources that form part of a public system to identify and prevent disease outbreaks. Moreover, the eating of feces (i.e., coprophagia) is recognized as a symptom of mental health disorders (Zeitlin and Polivy, 1995). Collectively, these conscious and sub-conscious prejudices combine to reduce the potential acceptability of fecal transfers. Therefore, in order for microbiome transfer to be implemented as a widespread treatment for chronic and non-acute disorders, it must be promoted in a way that minimizes the fecal stigma. We propose that the first step in this journey is the use of the term GMT.
The Human Host
A human being is more than the sum of their “own cells.” Rather, the ~10 trillion human cells that we each contain constitute < 10% of the cells within our bodies with the remaining ~100 trillion cells, that reside in and on the human body, being of microbial origin (Ley et al., 2006a). As a consequence of this, our ~20,000 human genes (Yang et al., 2009) are vastly outnumbered by the human microbiome's 2 to 20 million microbial genes (at least 100 times the number of human genes; Knight, 1993). These microbial genes (99%) are mostly encoded by the bacteria within the human gut (Qin et al., 2010). It is now becoming increasingly clear that these microbial communities interact with the human host at many levels, which include the local and systemic gut and immune function (Macpherson and Harris, 2004).
The microbes comprising the human microbiome generally have a symbiotic relationship with the host. The human intestine provides them with a supply of nutrition and a relatively stable living environment. In return, microbes play a vital role in our body by synthesizing metabolites (e.g., vitamin K, thiamine, biotin, folic acid, vitamin B12; Gorbach, 1996), digesting non-starch polysaccharides into additional nutrients for the human host (Vercellotti et al., 1977), providing a physical barrier in the form of a biofilm to boost the immune system, and protecting from pathogens (Mazmanian et al., 2005). Moreover, intestinal microbes may be also an important factor for brain development (Diaz Heijtz et al., 2011), metabolic function, and hormones and neurochemicals production (Lyte, 2013).
The Development of Human Gut Microbiome
The human gut is generally considered to be sterile in utero (Ley et al., 2006a; Maynard et al., 2012), with microbial colonization beginning during delivery when newborns come into contact with maternal womb, vaginal, fecal, and skin microbes (Lee and Polin, 2003). However, meconium of healthy neonates, collected within 2 h of delivery from healthy mothers, has been shown to contain microbes (e.g., E. fecalis, S. epidermidis, and E. coli; Jiménez et al., 2008). This has led to the promotion of hypotheses that bacteria from the maternal gut are transferred to amniotic fluid, possibly via the circulation (Kornman and Loesche, 1980), and through swallowing of amniotic fluid into the fetal gut (Goldenberg et al., 2008; Neu and Rushing, 2011). Given that a fetus swallows 400–500 ml of amniotic fluid per day late in gestation (Goldenberg et al., 2008; Neu and Rushing, 2011), only low numbers of microbes would be required within the amniotic fluid to facilitate microbial colonization of the fetal gut. This mechanism of fetal colonization is supported by the detection of microbes and microbial products within amniotic fluid isolated from healthy mothers (Li et al., 2014). Finally, microbes have been isolated from the umbilical cord (Jiménez et al., 2005) and placenta (Aagaard et al., 2014) of healthy infants (without infection or inflammation). Collectively these observations are consistent with the hypothesis that fetus is colonized by microbes before birth.
Mode of delivery (e.g., vaginal delivery or cesarean section) has been observed to have a significant impact on the microbiota within the newborn gut (Dominguez-Bello et al., 2010; Neu and Rushing, 2011). Interestingly, children born by cesarean section have a greater risk of obesity in later childhood, suggesting a causal link between early gut bacterial colonization and later obesity (Blustein and Liu, 2015). Cesarean section has been associated with a greater likelihood of C. difficile and lower number of Bacteroides spp. colonization (Penders et al., 2005, 2006). Gestational age of newborns (e.g., were they born prematurely, at term or post-term) also correlates with gut microflora composition. The gut of preterm infants contains higher levels of C. difficile compared to full term infants (Penders et al., 2006). Moreover, data obtained from short-term stool culture have shown that colonization by Bifidobacterium and Lactobacillus is delayed in preterm infants, whereas colonization by potentially pathogenic bacteria (especially E. coli) is increased (Westerbeek et al., 2006; Butel et al., 2007).
During infancy, diet is one of the many contributors to the development of gut microbiome (Koenig et al., 2011). The importance of diet is reinforced by observations that breast-fed infants have more Bifidobacteria than formula-fed infants (Koenig et al., 2011). By contrast, formula-fed infants have a lower microbial density, yet higher diversity of other microbial species compared to breast-fed infants (Harmsen et al., 2000; Koenig et al., 2011). After the introduction of solid food into the diet, at weaning, an adult-like microbial ecology begins to develop within the gut (Fanaro et al., 2007).
By 3–4 years of age, the gut microbiome composition is dominated by two phyla (>90% of bacteria): Firmicutes, which are pro-inflammatory and obesogenic, and Bacteroidetes, which protect from these effects (Cani and Delzenne, 2007; Clarke et al., 2014). Once established, the gut microbiota remains relatively stable throughout the life of healthy adults albeit subject to temporary modifications (Palmer et al., 2007). There are two broad groups of influences on the gut microbiome: dynamic factors (diet and drugs) and less dynamic factors (genetic, early events/exposures, and lifestyle factors). Diet contributes to dynamic changes in gut microbiome and influences approximately half of the microbial population activity (Zhang et al., 2010). Conversely, other factors tend to maintain the activity of the microbial population. However, microbial composition undergoes changes in the elderly (Tiihonen et al., 2010), which include increases in the levels of Lactobacilli, Coliforms, Clostridium, and Enterococci and a decrease in the number of Bifidobacterium (Mitsuoka, 1990). The presence of imbalance in the composition of the gut microbiota at all ages, which is also known as “dysbiosis,” is associated with obesity development (Bäckhed et al., 2004; Ley et al., 2005; Turnbaugh et al., 2009).
In otherwise healthy individuals, diet quality is the major modulator of the gut microbiota, accounting for 57% of host gut bacterial variation (Zhang et al., 2010). Diet-induced changes to gut microbial content are relatively rapid, occurring over 3–4 days and are readily reversible (Walker et al., 2011). Modification of gut microbiome can also be achieved by use of prebiotics and probiotics, and antibiotics (Walker et al., 2011; Binns, 2013; Modi et al., 2014). Prebiotics and probiotics appear to support a more favorable gut environment (Binns, 2013). However, these supplements need to be consumed regularly to maintain changes in gut microbiota (Binns, 2013), as it is unclear how long these changes last in the gut. Short- and long-term modifications of gut microbiome can also result from antibiotics, which reduce diversity by promoting the elimination of some bacterial species and antibiotic resistance by horizontal transfer within the remaining flora (Modi et al., 2014). Alcohol consumption also affects composition of gut microflora (Mutlu et al., 2012), with chronic alcohol consumption causing microbial dysbiosis, a reduction in the number of Bacteroidetes and an increase in the numbers of Proteobacteria present in the gut (Mutlu et al., 2012). Alterations in gut microbiome in alcoholic subjects correlate with increased levels of serum pro-inflammatory toxins (Mutlu et al., 2012). However, a recent study on microbiome development showed that microbial metabolites and their metabolic pathways are constant from birth, although microbial diversity increases with age and becomes more consistent from the age of 3 years (Kostic et al., 2015).
Composition of Human Microbiome
Each individual has their own unique microbial population whose composition is affected by host genetic make-up, history of exposure to microbes, age, diet, environment, and geographical location (The Human Microbiome Project Consortium, 2012; Ursell et al., 2012; Yatsunenko et al., 2012). Moreover, even within an individual there are a myriad of distinct environments each of which is colonized by different microorganisms (e.g., skin, oral cavity, gastrointestinal, respiratory, and urogenital tracts; Gerritsen et al., 2011). It is universally accepted that the high surface area and availability of nutrients make the gut an ideal site for microbial growth (Gebbers and Laissue, 1989; Sekirov et al., 2010). However, the gut microbiota composition changes at different sites within the gut (Zoetendal et al., 2002) and even within the different layers of the gut epithelium (Swidsinski et al., 2005). Despite this complexity, the ease of collection and the high microbial content (Hütter et al., 2012) mean that fecal matter is generally used to study “the gut microbiome.” Therefore, despite the fact that the numbers of bacteria are several orders of magnitude larger in the distal colon, which seems to have a relatively uniform composition of microbes (Whitman et al., 1998; Eckburg et al., 2005; Ley et al., 2006a; Gerritsen et al., 2011), this does not reflect the situation throughout the entire gut. As such, it must be borne in mind that fecal bacteria do not necessarily inform on the composition of the microbiome within the distinct environments that are present throughout the gut and are characterized by differing levels of pH, oxygen levels, and food transit rates.
How do We Characterize the Microbiota?
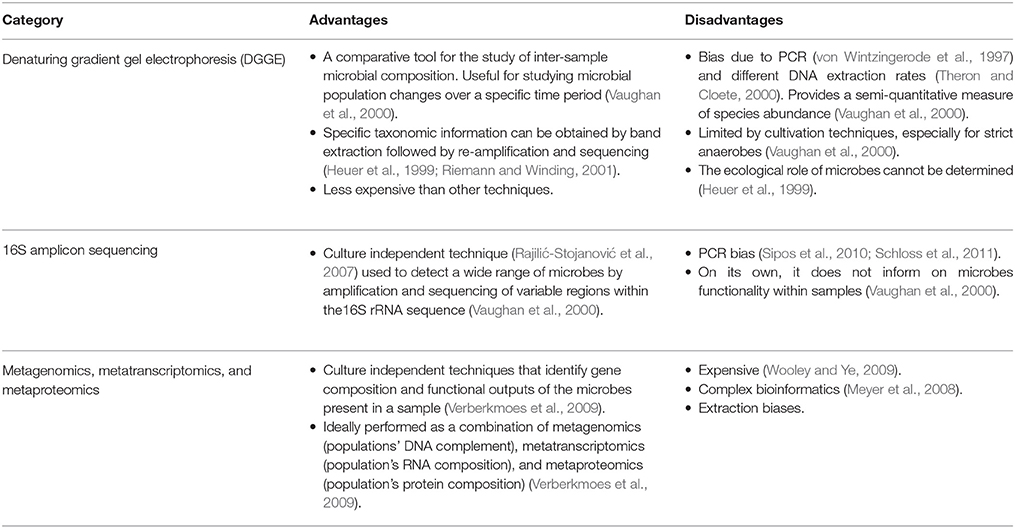
The application of metagenomic techniques (Kim et al., 2013) to the study of the composition, functional capacity, ecology, and integration of human microbiota with human cellular metabolism (Tremaroli and Bäckhed, 2012) is increasing our knowledge of how this “microbial organ” integrates into the human system. Metagenomic techniques overcome limitations of conventional bacterial cell culture and other molecular techniques that have been applied to the study of the gut microbiome (Table 1; Aslam et al., 2010). The Human Microbiome Consortium, the European Consortium of the Meta-HIT and the International Human Microbiome Consortium are currently developing and applying these techniques to understand microbial effects on human health and diseases (Kim et al., 2013).
Conventional techniques for the identification and characterization of microbial communities are mostly culture dependent and are unable to easily identify all of the microorganisms present and functional contributions that specific microorganisms make to the complex biological environments in which they exist (Verberkmoes et al., 2009). Despite their expense, metagenomic studies overcome many of these limitations.
Connecting the Gut Microbiome to Obesity and Cardio-Metabolic Disorders
Four bacterial phyla (i.e., Firmicutes, Bacteroidetes, Proteobacteria, and Actionobacteria) account for the majority of the bacteria present in the human gut (Khanna and Tosh, 2014). Typically ~60% of the bacteria present in the human gut belong to the gram positive Bacteroidetes or gram negative Firmicutes phyla (Bäckhed et al., 2005). The most commonly found gut bacteria genera in adults are Bifidobacterium, Lactobacillus, Bacteroides, Clostridium, Escherichia, Streptococcus, and Ruminococcus (Conlon and Bird, 2015). Individually and collectively, these bacteria produce a vast range of microbial products that include enzymes for carbohydrate metabolism (Xu et al., 2003), short chain fatty acids (SCFA) (Bergman, 1990), lipopolysaccharide (LPS) (Munford, 2008), and secondary bile acids (Nicholson et al., 2012). These microbial products can enter into the human circulation where they contribute to energy flux in the human, or cause inflammation and other complications (Tehrani et al., 2012; Trompette et al., 2014).
The gut microbial composition is distinctive in obese individuals, and tends to show reduced complexity (Turnbaugh et al., 2009). For example, obese mice have reduced numbers of Bacteroidetes and increased numbers of Firmicutes when compared to lean mice (Ley et al., 2005). These changes in gut microbial populations have significant implications for energy homeostasis, as a 20% increase in Firmicutes and a corresponding 20% decrease in Bacteroidetes is estimated to provide an additional 150 kcal of energy per day to an adult human (Jumpertz et al., 2011). Lactobacillus numbers have also been observed to increase in obese people, while anorexic patients show higher numbers of Methanobrevibacter smithii (Armougom et al., 2009).
Early research into the relationship between the gut microbiome and obesity has used 16S ribosomal RNA (rRNA) gene sequences to examine microbial diversity in obese and lean individuals. Numerous studies have found phylum-wide differences in lean or obese individuals (Eckburg et al., 2005; Ley et al., 2006b; Frank et al., 2007). However, findings on the relative proportions of the main phyla in obese and lean individuals are contradictory (Duncan et al., 2008; Turnbaugh et al., 2009; Schwiertz et al., 2010; Bervoets et al., 2013; Colson et al., 2013; Ferrer et al., 2013). Meta-analysis has shown that the microbial changes associated with obesity are not simply phylum based but are the result of a collection of numerous small differences within the overall population structure (Walters et al., 2014). Therefore, it is important to look at the overall composition of the gut microbial population structure as an indicator of obesity rather than simply the proportion of Bacteroidetes to Firmicutes.
Type 2 diabetes has also been linked with gut microbiota that differ from that found in a healthy individual (Larsen et al., 2010; Qin et al., 2012). Patients with type 2 diabetes have reduced level of butyrate-producing bacteria and more pathogenic bacteria (Qin et al., 2012). These patients also show more Betaproteobacteria and reduced Firmicutes and Clostridia levels compared to healthy subjects (Larsen et al., 2010). Furthermore, a correlation has been observed between Bacteroidetes to Firmicutes ratio and plasma glucose concentration in type 2 diabetic and obese patients (Larsen et al., 2010; Schwiertz et al., 2010). With these observations, it is clear that manipulating the microbiome composition may represent a novel approach for preventing and treating obesity and related alterations.
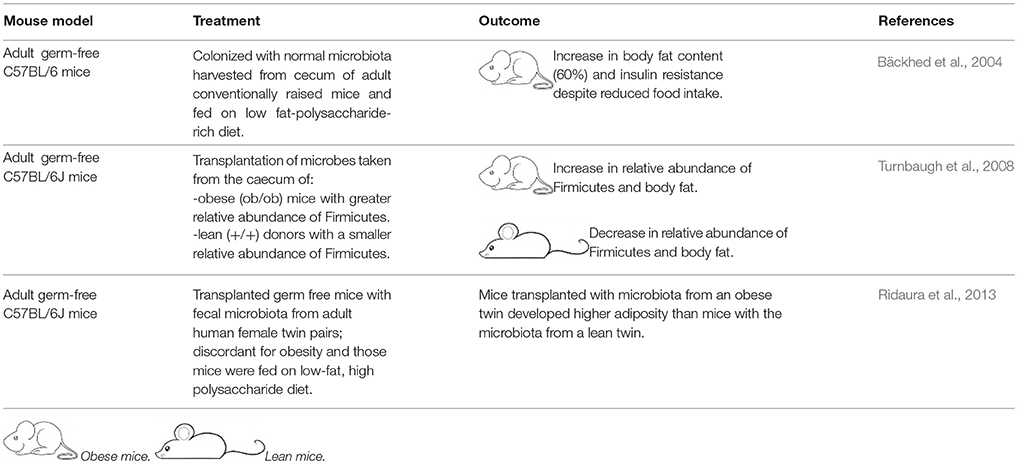
Several recent in-depth reviews provide detailed information about potential mechanisms through which the microbiome is linked to the development of obesity (Hartstra et al., 2014; Gérard, 2015). The association between characterization of an altered gut microbiome in obese or diabetic subjects does not demonstrate cause and effect. However, there are indications that the gut microbiome actively contributes to the development of obesity. Specifically, Backhed et al. compared the fat mass of germ-free and conventionally raised mice, and showed that intestinal microbes are able to control fat storage (Bäckhed et al., 2004). Similarly, Turnbaugh et al. introduced an “obesogenic microbiota” to germ-free mice and found that mice with obesogenic microbes developed more body fat than those with “lean microbiota” (Turnbaugh et al., 2006).
Various mechanisms have been proffered to explain the association of an “obese microbiota” with higher fat content in mice (Bäckhed et al., 2004; Ley et al., 2005; Turnbaugh et al., 2006; Hartstra et al., 2014). Most simply, microbial mediated degradation of dietary fiber to SCFA contributes additional calories to the host (Bäckhed et al., 2004; Hartstra et al., 2014). In addition, SCFAs, notably butyrate, facilitates enhanced insulin sensitivity and fatty acid oxidation in muscle and reduced hepatic lipogenesis as well as increased satiety (Hartstra et al., 2014). The way in which butyrate leads to these changes is unclear, however it is likely to involve the activation of the G protein coupled receptors GPR41 and GPR43, which are involved in glucose metabolism (Hartstra et al., 2014). Moreover, SCFA and bacterial lipopolysaccharides activate Toll-Like receptor 4 (TLR4) and signal intracellular inflammatory pathways related to the induction of insulin resistance and increased adiposity (Tsukumo et al., 2007; Tehrani et al., 2012).
Turnbaugh et al. observed a higher content of SCFAs (e.g., butyrate and acetate) in the large intestine of obese mice (Turnbaugh et al., 2006) consistent with a mechanism that involves increased absorption of SCFA (Bäckhed et al., 2004). In addition, comparisons of normal mice on a high-fat diet with germ free mice on the same diet have demonstrated that the gut microbes can reduce the expression of host fasting-induced adipose factor/angiopoietin-like protein-4, a lipoprotein lipase inhibitor (Ley et al., 2005). Reduced expression of fasting-induced adipose factor increases lipoprotein lipase activity and triglycerides storage in hepatic cells (Bäckhed et al., 2004), again contributing to alterations to patterns and levels of fat deposits. Despite these potential mechanisms, the exact contribution(s) that changes in the proportions of Firmicutes to Bacteroides species make to the development of obesity remains unknown (Ley et al., 2006b). More work is required to more accurately understand the contributions of the many proposed mechanisms linking the gut microbiome with obesity, particularly in humans.
Can we Manipulate the Microbiome to Prevent and Treat Obesity and its Related Complications?
Lifestyle modifications are an important part of obesity management. However, lifestyle interventions (such as diet and exercise) have not consistently led to appreciable weight loss (Golan et al., 1998). Furthermore, pharmacotherapy may have negative impacts on the physiology and psychology of obese patients (Collins, 1988; Hill et al., 1994; Hill, 2007). Surgical interventions (e.g., Bariatric surgery) can be effective for short term-to-medium-term weight management in severely obese patients (Gloy et al., 2013). However, there are significant risks associated with surgical interventions [e.g., dumping syndrome (rapid gastric empting), micronutrient malabsorption, cholelithiasis, and hypoglycaemia] (Puzziferri et al., 2014; Tack and Deloose, 2014) and the treatment is expensive (Encinosa et al., 2005). Therefore, new approaches for the prevention and treatment of obesity are required. GMT represents an excellent and economic (Encinosa et al., 2005) option for individuals who are unable to lose weight by lifestyle measures, or those who cannot undergo surgical treatment.
As gut microbes have been implicated in the development of obesity (Turnbaugh et al., 2006), replacement of a microbial population (“bad” microbes) that promotes obesity with a population that promotes a healthy state (“good” microbes) may represent a possible treatment. The question remains: how do you change the entire flora of an individual at once? GMT with fecal bacteria transferred from unaffected individuals to affected recipients has been suggested as a promising method of altering and improving gastrointestinal microbiota and human health (Aroniadis and Brandt, 2013; Smits et al., 2013).
GMT uses live microorganisms as a potential intervention that “confers a beneficial health effect on the host.” Thus, the fecal samples can be considered a probiotic (Park and Bae, 2015). However, unlike typical probiotics, GMT doesn't modify the recipient's gut flora using microorganisms associated with fermentation. Instead, GMT modifies the recipient's gut flora using a community of organisms that was isolated from a healthy gut—that is the same biological niche. This approach is essential for the modification of the gut flora in obesity because of the multiplicity of small, yet predictive, differences between the flora of obese and lean individuals (Walters et al., 2014).
GMT is not new. In the fourth century A.D., Chinese patients suffering from severe diarrhea were given oral fecal suspensions (Zhang et al., 2012). Likewise, in the sixteenth century stool was used to treat diarrhea, fever, vomiting and constipation (Zhang et al., 2012). In modern times (1958), fecal enemas have been used as a cure of human pseudo-membranous colitis (Eiseman et al., 1958). The use of GMT as a treatment for any disease, except recurrent C. difficile (CD) infection, requires an approved investigational new drug (IND) permit according to the US Food and Drug Administration (FDA) (Moore et al., 2014). As such, most studies of the effects of GMT on obesity have been limited to mice (Table 2).
We contend that, when considering the potential efficacy of the GMT approach for obesity, it is more appropriate to reflect on the meta-analyses of the effectiveness of fecal transfers in the treatment of C. difficile and inflammatory bowel disease (Kassam et al., 2013; Colman and Rubin, 2014). Until recently, there was no consistently effective treatment for recurrent C. difficile infection, which leads to considerable morbidity, including chronic diarrhea, colitis, and toxic megacolon, as well as a reported mortality of up to 38% (Hota et al., 2012). However, GMT is being increasingly viewed as the treatment of choice for recurrent C. difficile infection. Moreover, meta-analyses of clinical trials have consistently demonstrated that gut microbiome transfer is efficacious and safe [IBD, pooled cure rate 36.2% (95% CI 17.4–60.4%); C. difficile, pooled cure rate 89.1% (95% CI 84–93%)] (Kassam et al., 2013; Colman and Rubin, 2014). Finally, a recent study in patients with C. difficile colitis has shown that gut microbiome transfer causes a significant shift in composition from the diseased state to one equivalent to that seen for healthy individuals by the human microbiome project (Weingarden et al., 2015). As such, gut microbiome transfer holds significant promise as a treatment for the rapid and concerted modification of an unhealthy flora.
GMT is now being considered for a wider range of disorders, including severe obesity and type 2 diabetes mellitus. To date, investigation of the therapeutic benefit of GMT in adult obesity or type 2 diabetes has been limited to a single pilot study. Vrieze et al. performed a short-term GMT study in nine treated and nine control middle-aged men with metabolic syndrome (Vrieze et al., 2012), with transfer via a naso-duodenal tube. Six weeks after GMT, treated subjects had an impressive 75% increase in insulin sensitivity. Furthermore, GMT was associated with favorable changes to gut microbiota that included greater bacterial diversity and a 2.5-fold increase in butyrate-producing bacteria (Vrieze et al., 2012). However, the study was not continued long enough to evaluate the full potential of therapy, notably on body weight, and composition.
Whilst gut microbiome transfer in humans offers so much promise, it is not clear yet whether it actually leads to significant weight loss. Moreover, the duration of the effect, treatment composition, and mode of delivery required to achieve optimum weight loss must be established. There are currently 17 clinical trials registered (USA, Europe, and Australia) to test the efficacy of GMT as a clinical treatment, mostly for C. difficile infection. Only two of these trials are looking at GMT as a means of treating obesity. However, the reverse effect (lean to obese) has been demonstrated as the result of use of an overweight donor for the treatment of recurrent C. difficile infection (Alang and Kelly, 2015). It remains clear that there are considerable practical and safety issues that need to be considered and overcome before GMT can be used as a routine clinical or non-clinical intervention (Box 2).
Box 2. Practical and safety issues of GMT.
• Choice of donor (Andrews et al., 1995; Jakobsson et al., 2010; Bakken et al., 2011; Pérez-Cobas et al., 2013; Viaud et al., 2013; Kostic et al., 2014; Panda et al., 2014)
∘ Related, unrelated or universal? There is debate over the relative merits of using related or unrelated donors (Bakken et al., 2011).
∘ Once chosen, donors must be screened for: conditions associated with microbial dysbiosis (e.g., metabolic syndrome, morbid obesity, chronic fatigue syndrome, inflammatory bowel syndrome, irritable bowel syndrome, chronic diarrhea or constipation, GI malignancy, CD toxins); intestinal pathogens (e.g., Giardia, Cryptosporidium, Isopora and Rotavirus, Hepatitis A, B and C, HIV, Syphilis, and Helicobater pylori); antibiotic use within the previous 3 months; immunosuppressive treatments and anti-cancer agents; high risk-sexual behaviors; illegal drug use; recent travel to areas with endemic diarrhea, or recent body piercings/tattoos.
• Donor feces preparation (Berg et al., 1988; Lund-Tønnesen et al., 1998; Persky and Brandt, 2000; Mueller et al., 2006; Kostic et al., 2014)
∘ The use of fresh or frozen feces.
∘ It is unclear if the solvent (saline, non-bacteriostatic milk, yoghurt, or water), method of homogenization (hand stirring, shaking, or blender), or filtration (coffee filter, gauze pad, or steel strainer) make a difference to transfer efficiency (Persky and Brandt, 2000; Borody et al., 2015).
∘ There is currently no recommended standardized amount of feces suggested for use in GMT.
• Route of administration and site of inoculation (Lund-Tønnesen et al., 1998; Mueller et al., 2006; Yang et al., 2009; Silverman et al., 2010; Borody and Khoruts, 2012; Kostic et al., 2014)
∘ Retention enemas/naso-gastric tube/naso-jejunal tube/upper tract endoscopy (esophagogastroduodenoscopy)/colonoscopy/self-administered enemas.
What are the Challenges Associated with GMT
GMT is a promising treatment for antibiotic resistant C. difficile infection. However, the use of GMT as a treatment for metabolic diseases such as obesity or type 2 diabetes is only experimental (Bäckhed et al., 2004; Turnbaugh et al., 2008; Vrieze et al., 2012; Ridaura et al., 2013). There is still much to be learnt about the GMT method that includes: characteristics of the ideal donor, delivery formulation (e.g., in solution, encapsulation), mode of administration (e.g., oral, nasojejunal, or rectal), duration of benefit and, thus, frequency of treatment (Figure 1).
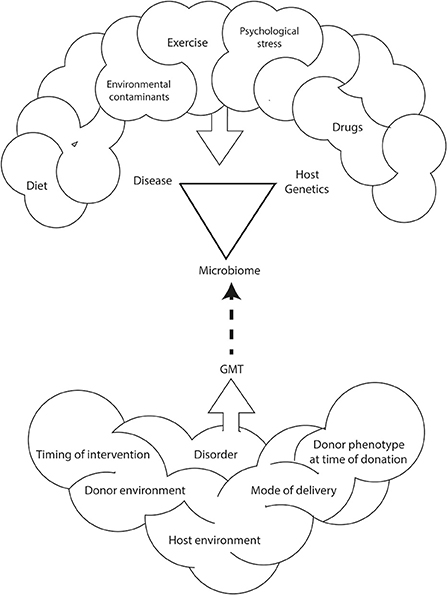
Figure 1. Environmental and genetic interactions between the host and the host's microbiome impact the development and incidence of obesity and related disorders. This relationship is also affected by diet, exercise, psychological stress, and environmental contaminants. As such, methods for human microbiome manipulation, including GMT, may represent a revolutionary approach for the treatment of non-communicable diseases including obesity.
Limited data suggests that GMT is a safe treatment (Borody and Khoruts, 2012; Vrieze et al., 2012; Van Nood et al., 2013) that has not currently been found to be associated with the development of new infections or diseases (Brandt et al., 2012). Therefore, further studies are required to monitor the long-term side-effects of GMT on both donors and recipients. These studies should also test the theoretical and practical benefits and side-effects of using fecal transplants as a treatment for obesity. These include: (1) the cost, ease of intervention, and relative safety of the non-invasive GMT as opposed to gastric by-pass surgery and pharmaceutical interventions; (2) the chances that GMT causes non-specific short- and long-term side-effects similar to those caused by pharmaceutical interventions; and (3) the psychological stress associated with the procedure (e.g., effects of performance anxiety on the donor, Brandt, 2013).
The psychological stresses and social stigma associated with feces mean that some patients find GMT to be an unappealing treatment (Zipursky et al., 2012). However, a survey of CDI patients found that regardless of GMT's unappealing nature, patients are willing to try it (Zipursky et al., 2012). Whether this willingness to try GMT as a treatment would translate to obese patients is yet to be determined. However, if GMT is shown to be an effective treatment for obesity then there will inevitably be greater refinement of the transplanted microbiota into a more palatable and optimally efficacious formulation.
Conclusion
Changes in the ratio of different gut microbial species have been associated with onset and development of several disorders, including obesity (Ley et al., 2005). It can be assumed that gut microbiota impacts on host metabolism through the promotion of increased uptake of monosaccharides, storage of triglyceride, digestion of dietary fiber (Bäckhed et al., 2004), and synthesis of hormonal precursors (Hartstra et al., 2014). Use of GMT to treat several disorders (e.g., chronic C. difficile infection) has already been established. However, it remains to be determined if GMT may be successful also for other diseases, such as obesity and its related complications. Based on the available evidence, GMT may represent a novel and successful intervention that could potentially transform the management of severe obesity in children and adults. Randomized controlled trials are required to confirm outcomes, efficacy and long-term safety of GMT in the treatment of obesity. The role of specific bacteria/species and combinations of intestinal microbiota should be clearly addressed beyond simply the change in body fat, ideally through longitudinal analysis of the meta-genomic, -proteomic, and -transcriptomic composition of donor and recipient's gut microbial content, before and after GMT. This characterization of GMT effects must include determining whether the process simply changes the composition of the existing microbial population or if it results in the complete transplantation of a non-obese microbial population. In conclusion, GMT represents a very real and potentially revolutionary treatment for obesity.
Author Contributions
TJ wrote the manuscript. VC contributed to the writing of the manuscript. DH commented on the manuscript. WC and JO conceived, directed, and contributed to the writing of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work in WC and JO laboratories is funded by Gravida: National Centre for Growth and Development. TJ is supported by a University of Auckland Scholarship. VC is the recipient of a Pfizer Australasian Paediatric Endocrine Care (APEC) Research Grant 2015.
References
Aagaard, K., Ma, J., Antony, K. M., Ganu, R., Petrosino, J., and Versalovic, J. (2014). The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra65. doi: 10.1126/scitranslmed.3008599
Alang, N., and Kelly, C. R. (2015). Weight gain after fecal microbiota transplantation. Open Forum Infect. Dis. 2:ofv004. doi: 10.1093/ofid/ofv004
Andrews, P., Borody, T., Shortis, N., and Thompson, S. (1995). Bacteriotherapy for chronic constipation - a long term follow-up. Gastroenterology 108, A563.
Armougom, F., Henry, M., Vialettes, B., Raccah, D., and Raoult, D. (2009). Monitoring bacterial community of human gut microbiota reveals an increase in lactobacillus in obese patients and methanogens in anorexic patients. PLoS ONE 4:e7125. doi: 10.1371/journal.pone.0007125
Aroniadis, O. C., and Brandt, L. J. (2013). Fecal microbiota transplantation: past, present and future. Curr. Opin. Gastroenterol. 29, 79–84. doi: 10.1097/MOG.0b013e32835a4b3e
Aslam, Z., Yasir, M., Khaliq, A., Matsui, K., and Chung, Y. R. (2010). Too much bacteria still unculturable. Crop Environ. 1, 59–60.
Bäckhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A. 101, 15718–15723. doi: 10.1073/pnas.0407076101
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Backhed, F., Manchester, J. K., Semenkovich, C. F., and Gordon, J. I. (2007). Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. U.S.A. 104, 979–984. doi: 10.1073/pnas.0605374104
Bakken, J. S., Borody, T., Brandt, L. J., Brill, J. V., Demarco, D. C., Franzos, M. A., et al. (2011). Treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 9, 1044–1049. doi: 10.1016/j.cgh.2011.08.014
Berg, R. D., Wommack, E., and Deitch, E. A. (1988). Immunosuppression and intestinal bacterial overgrowth synergistically promote bacterial translocation. Arch Surg. 123, 1359–1364. doi: 10.1001/archsurg.1988.01400350073011
Bergman, E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590.
Bervoets, L., Van Hoorenbeeck, K., Kortleven, I., Van Noten, C., Hens, N., Vael, C., et al. (2013). Differences in gut microbiota composition between obese and lean children: a cross-sectional study. Gut Pathog. 5:10. doi: 10.1186/1757-4749-5-10
Binns, N. (2013). Probiotics, Prebiotics and the GUT Microbiota. Available online at: http://www.ilsi.org/Europe/Publications/Prebiotics-Probiotics.pdf
Blustein, J., and Liu, J. (2015). Time to consider the risks of caesarean delivery for long term child health. BMJ 350:h2410. doi: 10.1136/bmj.h2410
Borody, T. J., and Khoruts, A. (2012). Fecal microbiota transplantation and emerging applications. Nat. Rev. Gastroenterol. Hepatol. 9, 88–96. doi: 10.1038/nrgastro.2011.244
Borody, T., Peattie, D., and Mitchell, S. (2015). Fecal microbiota transplantation: expanding horizons for Clostridium difficile Infections and Beyond. Antibiotics 4, 254–266. doi: 10.3390/antibiotics4030254
Brandt, L. J. (2012). Editorial commentary: fecal microbiota transplantation: patient and physician attitudes. Clin. Infect. Dis. 55, 1659–1660. doi: 10.1093/cid/cis812
Brandt, L. J. (2013). American journal of gastroenterology lecture: intestinal microbiota and the role of fecal microbiota transplant (FMT) in treatment of C. difficile infection. Am. J. Gastroenterol. 108, 177–185. doi: 10.1038/ajg.2012.450
Brandt, L. J., Aroniadis, O. C., Mellow, M., Kanatzar, A., Kelly, C., Park, T., et al. (2012). Long-term follow-up of colonoscopic fecal microbiota transplant for recurrent Clostridium difficile infection. Am. J. Gastroenterol. 107, 1079–1087. doi: 10.1038/ajg.2012.60
Butel, M.-J., Suau, A., Campeotto, F., Magne, F., Aires, J., Ferraris, L., et al. (2007). Conditions of bifidobacterial colonization in preterm infants: a prospective analysis. J. Pediatr. Gastroenterol. Nutr. 44, 577–582. doi: 10.1097/MPG.0b013e3180406b20
Cani, P. D., and Delzenne, N. M. (2007). Gut microflora as a target for energy and metabolic homeostasis. Curr. Opin. Clin. Nutr. Metab. Care 10, 729–734. doi: 10.1097/MCO.0b013e3282efdebb
Clarke, G., Stilling, R. M., Kennedy, P. J., Stanton, C., Cryan, J. F., and Dinan, T. G. (2014). Minireview: Gut microbiota: the neglected endocrine organ. Mol. Endocrinol. 28, 1221–1238. doi: 10.1210/me.2014-1108
Collins, M. E. (1988). Education for healthy body weight: helping adolescents balance the cultural pressure for thinness. J. Sch. Health 58, 227–231. doi: 10.1111/j.1746-1561.1988.tb05870.x
Colman, R. J., and Rubin, D. T. (2014). Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J. Crohns. Colitis 8, 1569–1581. doi: 10.1016/j.crohns.2014.08.006
Colson, P., Fancello, L., Gimenez, G., Armougom, F., Desnues, C., Fournous, G., et al. (2013). Evidence of the megavirome in humans. J. Clin. Virol. 57, 191–200. doi: 10.1016/j.jcv.2013.03.018
Conlon, M. A., and Bird, A. R. (2015). The impact of diet and lifestyle on gut microbiota and human health. Nutrients 7, 17–44. doi: 10.3390/nu7010017
Diaz Heijtz, R., Wang, S., Anuar, F., Qian, Y., Björkholm, B., Samuelsson, A., et al. (2011). Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci. U.S.A. 108, 3047–3052. doi: 10.1073/pnas.1010529108
Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N., et al. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. U.S.A. 107, 11971–11975. doi: 10.1073/pnas.1002601107
Duncan, S. H., Lobley, G. E., Holtrop, G., Ince, J., Johnstone, A. M., Louis, P., et al. (2008). Human colonic microbiota associated with diet, obesity and weight loss. Int. J. Obes. (Lond.) 32, 1720–1724. doi: 10.1038/ijo.2008.155
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Eiseman, B., Silen, W., Bascom, G. S., and Kauvar, A. J. (1958). Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery 44, 854–859.
Encinosa, W. E., Bernard, D. M., Steiner, C. A., and Chen, C.-C. (2005). Use and costs of bariatric surgery and prescription weight-loss medications. Health Aff. (Millwood). 24, 1039–1046. doi: 10.1377/hlthaff.24.4.1039
Fanaro, S., Chierici, R., Guerrini, P., and Vigi, V. (2007). Intestinal microflora in early infancy: composition and development. Acta Paediatr. 92, 48–55. doi: 10.1111/j.1651-2227.2003.tb00646.x
Ferrer, M., Ruiz, A., Lanza, F., Haange, S. B., Oberbach, A., Till, H., et al. (2013). Microbiota from the distal guts of lean and obese adolescents exhibit partial functional redundancy besides clear differences in community structure. Environ. Microbiol. 15, 211–226. doi: 10.1111/j.1462-2920.2012.02845.x
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., and Pace, N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785. doi: 10.1073/pnas.0706625104
Gebbers, J. O., and Laissue, J. A. (1989). Immunologic structures and functions of the gut. Schweiz. Arch. Tierheilkd. 131, 221–238.
Gérard, P. (2015). Gut microbiota and obesity. Cell. Mol. Life Sci. 73, 147–162. doi: 10.1007/s00018-015-2061-5
Gerritsen, J., Smidt, H., Rijkers, G. T., and de Vos, W. M. (2011). Intestinal microbiota in human health and disease: the impact of probiotics. Genes Nutr. 6, 209–240. doi: 10.1007/s12263-011-0229-7
Gloy, V. L., Briel, M., Bhatt, D. L., Kashyap, S. R., Schauer, P. R., Mingrone, G., et al. (2013). Bariatric surgery versus non-surgical treatment for obesity: a systematic review and meta-analysis of randomised controlled trials. BMJ 347:f5934. doi: 10.1136/bmj.f5934
Golan, M., Weizman, A., Apter, A., and Fainaru, M. (1998). Parents as the exclusive agents of change in the treatment of childhood obesity. Am. J. Clin. Nutr. 67, 1130–1135.
Goldenberg, R. L., Culhane, J. F., Iams, J. D., and Romero, R. (2008). Epidemiology and causes of preterm birth. Lancet 371, 75–84. doi: 10.1016/S0140-6736(08)60074-4
Gorbach, S. L. (1996). “Microbiology of the gastrointestinal tract,” in Medical Microbiology, 4th Edn., ed T. Samuel Baron (Galveston, TX: University of Texas Medical Branch). Available online at: http://www.ncbi.nlm.nih.gov/books/NBK7670/
Gough, E., Shaikh, H., and Manges, A. R. (2011). Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 53, 994–1002. doi: 10.1093/cid/cir632
Harmsen, H. J., Wildeboer-Veloo, A. C., Raangs, G. C., Wagendorp, A. A., Klijn, N., Bindels, J. G., et al. (2000). Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J. Pediatr. Gastroenterol. Nutr. 30, 61–7. doi: 10.1097/00005176-200001000-00019
Hartstra, A. V., Bouter, K. E. C., Bäckhed, F., and Nieuwdorp, M. (2014). Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care 38, 159–165. doi: 10.2337/dc14-0769
Heuer, H., Hartung, K., Wieland, G., Kramer, I., and Braunschweig, D. (1999). Polynucleotide probes that target a hypervariable region of 16S rRNA genes to identify bacterial isolates corresponding to bands of community fingerprints. 65, 1045–1049.
Hill, A. J. (2007). Causes and consequences of dieting and anorexia. Proc. Nutr. Soc. 52, 211–218. doi: 10.1079/PNS19930053
Hill, A. J., Draper, E., and Stack, J. (1994). A weight on children's minds: body shape dissatisfactions at 9-years old. Int. J. Obes. Relat. Metab. Disord. 18, 383–389.
Hota, S. S., Achonu, C., Crowcroft, N. S., Harvey, B. J., Lauwers, A., and Gardam, M. A. (2012). Determining mortality rates attributable to Clostridium difficile infection. Emerg. Infect. Dis. 18, 305–307. doi: 10.3201/eid1802.101611
Howitt, M. R., and Garrett, W. S. (2012). A complex microworld in the gut: gut microbiota and cardiovascular disease connectivity. Nat. Med. 18, 1188–1189. doi: 10.1038/nm.2895
Hütter, J. F., Schweickhardt, C., Hunneman, D. H., Piper, H. M., and Spieckermann, P. G. (2012). A new method for studying the incorporation of nonesterified fatty acids into cardiac lipids by using deuterium-labelled palmitate. Basic Res. Cardiol. 83, 87–93. doi: 10.1007/BF01907108
Jakobsson, H. E., Jernberg, C., Andersson, A. F., Sjölund-Karlsson, M., Jansson, J. K., and Engstrand, L. (2010). Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 5:e9836. doi: 10.1371/journal.pone.0009836
Jiménez, E., Fernández, L., Marín, M. L., Martín, R., Odriozola, J. M., Nueno-Palop, C., et al. (2005). Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr. Microbiol. 51, 270–274. doi: 10.1007/s00284-005-0020-3
Jiménez, E., Marín, M. L., Martín, R., Odriozola, J. M., Olivares, M., Xaus, J., et al. (2008). Is meconium from healthy newborns actually sterile? Res. Microbiol. 159, 187–193. doi: 10.1016/j.resmic.2007.12.007
Jumpertz, R., Le, D. S., Turnbaugh, P. J., Trinidad, C., Bogardus, C., Gordon, J. I., et al. (2011). Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 94, 58–65. doi: 10.3945/ajcn.110.010132
Kassam, Z., Lee, C. H., Yuan, Y., and Hunt, R. H. (2013). Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am. J. Gastroenterol. 108, 500–508. doi: 10.1038/ajg.2013.59
Khanna, S., and Tosh, P. K. (2014). A clinician's primer on the role of the microbiome in human health and disease. Mayo Clin. Proc. 89, 107–114. doi: 10.1016/j.mayocp.2013.10.011
Kim, B.-S., Jeon, Y.-S., and Chun, J. (2013). Current status and future promise of the human microbiome. Pediatr. Gastroenterol. Hepatol. Nutr. 16, 71–79. doi: 10.5223/pghn.2013.16.2.71
Knight, R. (1993). Bacterial Communities as a Theme Linking. Human, Animal and Environmental Health. Available online at: http://iom.nationalacademies.org/~/media/87E9D588D5C44B9999E6839D46D298B6.ashx
Koenig, J. E., Spor, A., Scalfone, N., Fricker, A. D., Stombaugh, J., Knight, R., et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl.), 4578–4585. doi: 10.1073/pnas.1000081107
Kornman, K. S., and Loesche, W. J. (1980). The subgingival microbial flora during pregnancy. J. Periodontal Res. 15, 111–122. doi: 10.1111/j.1600-0765.1980.tb00265.x
Kostic, A. D., Gevers, D., Siljander, H., Vatanen, T., Hyötyläinen, T., Hämäläinen, A.-M., et al. (2015). The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273. doi: 10.1016/j.chom.2015.01.001
Kostic, A. D., Xavier, R. J., and Gevers, D. (2014). The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499. doi: 10.1053/j.gastro.2014.02.009
Larsen, N., Vogensen, F. K., van den Berg, F. W. J., Nielsen, D. S., Andreasen, A. S., Pedersen, B. K., et al. (2010). Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 5:e9085. doi: 10.1371/journal.pone.0009085
Lee, J. S., and Polin, R. A. (2003). Treatment and prevention of necrotizing enterocolitis. Semin. Neonatol. 8, 449–459. doi: 10.1016/S1084-2756(03)00123-4
Leslie, P., Tzimas, D., Price, J., Mone, A., Hirsh, J., Poles, M., et al. (2014). “Perceptions of fecal microbiota transplantation: factors that predict acceptance,” in James W. Freston Conference, 35–37. Available online at: http://www.gastro.org/pressroom/James_W._Freston_Abstract_Factors_that_Predict_Acceptance.pdf
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075. doi: 10.1073/pnas.0504978102
Ley, R. E., Peterson, D. A., and Gordon, J. I. (2006a). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848. doi: 10.1016/j.cell.2006.02.017
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006b). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Li, M., Wang, M., and Donovan, S. M. (2014). Early development of the gut microbiome and immune-mediated childhood disorders. Semin. Reprod. Med. 32, 74–86. doi: 10.1055/s-0033-1361825
Lund-Tønnesen, S., Berstad, A., Schreiner, A., and Midtvedt, T. (1998). [Clostridium difficile-associated diarrhea treated with homologous feces]. Tidsskr. den Nor. lægeforening Tidsskr. Prakt. Med. ny række 118, 1027–1030.
Lyte, M. (2013). Microbial endocrinology in the microbiome-gut-brain axis: how bacterial production and utilization of neurochemicals influence behavior. PLoS Pathog. 9:e1003726. doi: 10.1371/journal.ppat.1003726
Macpherson, A. J., and Harris, N. L. (2004). Interactions between commensal intestinal bacteria and the immune system. Nat. Rev. Immunol. 4, 478–485. doi: 10.1038/nri1373
Maynard, C. L., Elson, C. O., Hatton, R. D., and Weaver, C. T. (2012). Reciprocal interactions of the intestinal microbiota and immune system. Nature 489, 231–241. doi: 10.1038/nature11551
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118. doi: 10.1016/j.cell.2005.05.007
Meyer, F., Paarmann, D., D'souza, M., Olson, R., Glass, E. M., Kubal, M., et al. (2008). The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386
Mitsuoka, T. (1990). Bifidobacteria and their role in human health. J. Ind. Microbiol. 6, 263–267. doi: 10.1007/BF01575871
Modi, S. R., Collins, J. J., and Relman, D. A. (2014). Antibiotics and the gut microbiota. J. Clin. Invest. 124, 4212–4218. doi: 10.1172/JCI72333
Moore, T., Rodriguez, A., and Bakken, J. S. (2014). Fecal microbiota transplantation: a practical update for the infectious disease specialist. Clin. Infect. Dis. 58, 541–545. doi: 10.1093/cid/cit950
Mueller, S., Saunier, K., Hanisch, C., Norin, E., Alm, L., Midtvedt, T., et al. (2006). Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study. Appl. Environ. Microbiol. 72, 1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006
Munford, R. S. (2008). Sensing gram-negative bacterial lipopolysaccharides: a human disease determinant? Infect. Immun. 76, 454–465. doi: 10.1128/iai.00939-07
Mutlu, E. A., Gillevet, P. M., Rangwala, H., Sikaroodi, M., Naqvi, A., Engen, P. A., et al. (2012). Colonic microbiome is altered in alcoholism. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G966–G978. doi: 10.1152/ajpgi.00380.2011
Neu, J., and Rushing, J. (2011). Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clin. Perinatol. 38, 321–331. doi: 10.1016/j.clp.2011.03.008
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. doi: 10.1126/science.1223813
Ordovas, J. M., and Mooser, V. (2006). Metagenomics: the role of the microbiome in cardiovascular diseases. Curr. Opin. Lipidol. 17, 157–161. doi: 10.1097/01.mol.0000217897.75068.ba
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A., and Brown, P. O. (2007). Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. doi: 10.1371/journal.pbio.0050177
Panda, S., El khader, I., Casellas, F., López Vivancos, J., García Cors, M., Santiago, A., et al. (2014). Short-term effect of antibiotics on human gut microbiota. PLoS ONE 9:e95476. doi: 10.1371/journal.pone.0095476
Park, S., and Bae, J.-H. (2015). Probiotics for weight loss: a systematic review and meta-analysis. Nutr. Res. 35, 566–575. doi: 10.1016/j.nutres.2015.05.008
Penders, J., Thijs, C., Vink, C., Stelma, F. F., Snijders, B., Kummeling, I., et al. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521. doi: 10.1542/peds.2005-2824
Penders, J., Vink, C., Driessen, C., London, N., Thijs, C., and Stobberingh, E. E. (2005). Quantification of Bifidobacterium spp., Escherichia coli and Clostridium difficile in faecal samples of breast-fed and formula-fed infants by real-time PCR. FEMS Microbiol. Lett. 243, 141–147. doi: 10.1016/j.femsle.2004.11.052
Pérez-Cobas, A. E., Gosalbes, M. J., Friedrichs, A., Knecht, H., Artacho, A., Eismann, K., et al. (2013). Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 62, 1591–1601. doi: 10.1136/gutjnl-2012-303184
Persky, S. E., and Brandt, L. J. (2000). Treatment of recurrent Clostridium difficile-associated diarrhea by administration of donated stool directly through a colonoscope. Am. J. Gastroenterol. 95, 3283–3285. doi: 10.1111/j.1572-0241.2000.03302.x
Puzziferri, N., Roshek, T. B. III., Mayo, H. G., Gallagher, R., Belle, S. H., and Livingston, E. H. (2014). Long-term follow-up after bariatric surgery: a systematic review. JAMA 312, 934–942. doi: 10.1001/jama.2014.10706
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450
Rajilić-Stojanović, M., Smidt, H., and de Vos, W. M. (2007). Diversity of the human gastrointestinal tract microbiota revisited. Environ. Microbiol. 9, 2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x
Ridaura, V. K., Faith, J. J., Rey, F. E., Cheng, J., Duncan, A. E., Kau, A. L., et al. (2013). Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341:1241214. doi: 10.1126/science.1241214
Riemann, L., and Winding, A. (2001). Community dynamics of free-living and particle-associated bacterial assemblages during a freshwater phytoplankton bloom. Microb. Ecol. 42, 274–285. doi: 10.1007/s00248-001-0018-8
Schloss, P. D., Gevers, D., and Westcott, S. L. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 6:e27310. doi: 10.1371/journal.pone.0027310
Schwiertz, A., Taras, D., Schäfer, K., Beijer, S., Bos, N. A., Donus, C., et al. (2010). Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 18, 190–195. doi: 10.1038/oby.2009.167
Sekirov, I., Russell, S. L., Antunes, L. C. M., and Finlay, B. B. (2010). Gut microbiota in health and disease. Physiol. Rev. 90, 859–904. doi: 10.1152/physrev.00045.2009
Silverman, M. S., Davis, I., and Pillai, D. R. (2010). Success of self-administered home fecal transplantation for chronic Clostridium difficile infection. Clin. Gastroenterol. Hepatol. 8, 471–473. doi: 10.1016/j.cgh.2010.01.007
Sipos, R., Székely, A., Révész, S., and Márialigeti, K. (2010). Addressing PCR biases in environmental microbiology studies. Methods Mol. Biol. 599, 37–58. doi: 10.1007/978-1-60761-439-5_3
Smits, L. P., Bouter, K. E. C., de Vos, W. M., Borody, T. J., and Nieuwdorp, M. (2013). Therapeutic potential of fecal microbiota transplantation. Gastroenterology 145, 946–953. doi: 10.1053/j.gastro.2013.08.058
Swidsinski, A., Loening-Baucke, V., Lochs, H., and Hale, L. P. (2005). Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J. Gastroenterol. 11, 1131–1140. doi: 10.3748/wjg.v11.i8.1131
Tang, W. H., and Hazen, S. L. (2014). The contributory role of gut microbiota in cardiovascular disease. J. Clin. Investig. 124, 4204. doi: 10.1172/JCI72331
The Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214. doi: 10.1038/nature11234
Tack, J., and Deloose, E. (2014). Complications of bariatric surgery: dumping syndrome, reflux and vitamin deficiencies. Best Pract. Res. Clin. Gastroenterol. 28, 741–749. doi: 10.1016/j.bpg.2014.07.010
Tehrani, A. B., Nezami, B. G., Gewirtz, A., and Srinivasan, S. (2012). Obesity and its associated disease: a role for microbiota? Neurogastroenterol. Motil. 24, 305–311. doi: 10.1111/j.1365-2982.2012.01895.x
Theron, J., and Cloete, T. E. (2000). Molecular techniques for determining microbial diversity and community structure in natural environments. Crit. Rev. Microbiol. 26, 37–57. doi: 10.1080/10408410091154174
Tiihonen, K., Ouwehand, A. C., and Rautonen, N. (2010). Human intestinal microbiota and healthy ageing. Ageing Res. Rev. 9, 107–116. doi: 10.1016/j.arr.2009.10.004
Tremaroli, V., and Bäckhed, F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249. doi: 10.1038/nature11552
Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. doi: 10.1038/nm.3444
Tsukumo, D. M. L., Carvalho-Filho, M. A., Carvalheira, J. B. C., Prada, P. O., Hirabara, S. M., Schenka, A. A., et al. (2007). Loss-of-function mutation in Toll-like receptor 4 prevents diet-induced obesity and insulin resistance. Diabetes 56, 1986–1998. doi: 10.2337/db06-1595
Turnbaugh, P. J., Backhed, F., Fulton, L., and Gordon, J. I. (2008). Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223. doi: 10.1016/j.chom.2008.02.015
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. doi: 10.1038/nature07540
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Ursell, L. K., Clemente, J. C., Rideout, J. R., Gevers, D., Caporaso, J. G., and Knight, R. (2012). The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J. Allergy Clin. Immunol. 129, 1204–1208. doi: 10.1016/j.jaci.2012.03.010
Van Nood, E., Vrieze, A., Nieuwdorp, M., Fuentes, S., Zoetendal, E. G., de Vos, W. M., et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368, 407–415. doi: 10.1056/NEJMoa1205037
Vaughan, E. E., Schut, F., Heilig, H. G., Zoetendal, E. G., de Vos, W. M., and Akkermans, A. D. (2000). A molecular view of the intestinal ecosystem. Curr. Issues Intest. Microbiol. 1, 1–12.
Verberkmoes, N. C., Russell, A. L., Shah, M., Godzik, A., Rosenquist, M., Halfvarson, J., et al. (2009). Shotgun metaproteomics of the human distal gut microbiota. ISME J. 3, 179–189. doi: 10.1038/ismej.2008.108
Vercellotti, J. R., Salyers, A. A., Bullard, W. S., and Wilkins, D. (1977). Breakdown of mucin and plant polysaccharides in the human colon. Can. J. Biochem. 55, 1190–1196. doi: 10.1139/o77-178
Viaud, S., Saccheri, F., Mignot, G., Yamazaki, T., Daillère, R., Hannani, D., et al. (2013). The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976. doi: 10.1126/science.1240537
Vrieze, A., Van Nood, E., Holleman, F., Salojärvi, J., Kootte, R. S., Bartelsman, J. F. W. M., et al. (2012). Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913–916.e7. doi: 10.1053/j.gastro.2012.06.031
Walker, A. W., Ince, J., Duncan, S. H., Webster, L. M., Holtrop, G., Ze, X., et al. (2011). Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 5, 220–230. doi: 10.1038/ismej.2010.118
Walters, W. A., Xu, Z., and Knight, R. (2014). Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 588, 4223–4233. doi: 10.1016/j.febslet.2014.09.039
Wang, K., Li, W.-D., Zhang, C. K., Wang, Z., Glessner, J. T., Grant, S. F. A., et al. (2011a). A genome-wide association study on obesity and obesity-related traits. PLoS ONE 6:e18939. doi: 10.1371/journal.pone.0018939
Wang, Z., Klipfell, E., Bennett, B. J., Koeth, R., Levison, B. S., Dugar, B., et al. (2011b). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63. doi: 10.1038/nature09922
Weingarden, A., González, A., Vázquez-Baeza, Y., Weiss, S., Humphry, G., Berg-Lyons, D., et al. (2015). Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 3, 10. doi: 10.1186/s40168-015-0070-0
Westerbeek, E. A. M., van den Berg, A., Lafeber, H. N., Knol, J., Fetter, W. P. F., and van Elburg, R. M. (2006). The intestinal bacterial colonisation in preterm infants: a review of the literature. Clin. Nutr. 25, 361–368. doi: 10.1016/j.clnu.2006.03.002
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583. doi: 10.1073/pnas.95.12.6578
von Wintzingerode, F., Göbel, U. B., and Stackebrandt, E. (1997). Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21, 213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x
Wooley, J. C., and Ye, Y. (2009). Metagenomics: facts and artifacts, and computational challenges*. J. Comput. Sci. Technol. 25, 71–81. doi: 10.1007/s11390-010-9306-4
World Health Organization (2014). Global Status Report on Noncommunicable Diseases. World Health Organization. Available online at: http://apps.who.int/iris/bitstream/10665/148114/1/9789241564854_eng.pdf?ua=1
Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., et al. (2003). A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076. doi: 10.1126/science.1080029
Yang, X., Xie, L., Li, Y., and Wei, C. (2009). More than 9,000,000 unique genes in human gut bacterial community: estimating gene numbers inside a human body. PLoS ONE 4:e6074. doi: 10.1371/journal.pone.0006074
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Zeitlin, S. B., and Polivy, J. (1995). Coprophagia as a manifestation of obsessive-compulsive disorder: a case report. J. Behav. Ther. Exp. Psychiatry 26, 57–63.
Zhang, C., Zhang, M., Wang, S., Han, R., Cao, Y., Hua, W., et al. (2010). Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 4, 232–241. doi: 10.1038/ismej.2009.112
Zhang, F., Luo, W., Shi, Y., Fan, Z., and Ji, G. (2012). Should we standardize the 1,700-year-old fecal microbiota transplantation? Am. J. Gastroenterol. 107, 1755; author reply p.1755–6. doi: 10.1038/ajg.2012.251
Zhang, H., DiBaise, J. K., Zuccolo, A., Kudrna, D., Braidotti, M., Yu, Y., et al. (2009). Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. U.S.A. 106, 2365–2370. doi: 10.1073/pnas.0812600106
Zipursky, J. S., Sidorsky, T. I., Freedman, C. A., Sidorsky, M. N., and Kirkland, K. B. (2012). Patient attitudes toward the use of fecal microbiota transplantation in the treatment of recurrent Clostridium difficile infection. Clin. Infect. Dis. 55, 1652–1658. doi: 10.1093/cid/cis809
Zoetendal, E. G., von Wright, A., Vilpponen-Salmela, T., Ben-Amor, K., Akkermans, A. D. L., and de Vos, W. M. (2002). Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl. Environ. Microbiol. 68, 3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002
Keywords: gut microbiome transplantation, microbiome, microbiota, obesity, treatment
Citation: Jayasinghe TN, Chiavaroli V, Holland DJ, Cutfield WS and O'Sullivan JM (2016) The New Era of Treatment for Obesity and Metabolic Disorders: Evidence and Expectations for Gut Microbiome Transplantation. Front. Cell. Infect. Microbiol. 6:15. doi: 10.3389/fcimb.2016.00015
Received: 22 October 2015; Accepted: 25 January 2016;
Published: 19 February 2016.
Edited by:
Daniel Hassett, University of Cincinnati, College of Medicine, USAReviewed by:
Marina Santic', University of Rijeka, CroatiaV. K. Viswanathan, The University of Arizona, USA
Lily Q. Dong, University of Texas Health Science Center at San Antonio, USA
Copyright © 2016 Jayasinghe, Chiavaroli, Holland, Cutfield and O'Sullivan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wayne S. Cutfield, dy5jdXRmaWVsZEBhdWNrbGFuZC5hYy5ueg==;
Justin M. O'Sullivan, anVzdGluLm9zdWxsaXZhbkBhdWNrbGFuZC5hYy5ueg==
 Thilini N. Jayasinghe
Thilini N. Jayasinghe Valentina Chiavaroli
Valentina Chiavaroli David J. Holland2
David J. Holland2 Wayne S. Cutfield
Wayne S. Cutfield Justin M. O'Sullivan
Justin M. O'Sullivan