
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol. , 24 December 2015
Sec. Microbiome in Health and Disease
Volume 5 - 2015 | https://doi.org/10.3389/fcimb.2015.00099
This article is part of the Research Topic Use of saliva in diagnosis of Periodontitis: cumulative use of bacterial and host-derived biomarkers View all 10 articles
In the pathogenesis of periodontitis, an infection-induced inflammatory disease of the tooth-supporting tissues, there is a complex interaction between the subgingival microbiota and host tissues. A periodontal diagnostic tool for detecting the initiation and progression of the disease, monitoring the response to therapy, or measuring the degree of susceptibility to future disease progression has been of interest for a long time. The value of various enzymes, proteins, and immunoglobulins, which are abundant constituents of saliva, as potential biomarkers has been recognized and extensively investigated for periodontal diseases. Gingival defensins and cathelicidins are small cationic antimicrobial peptides that play an important role in innate immune response. However, their applicability as salivary biomarkers is still under debate. The present review focuses on proteomic biomarkers and antimicrobial peptides, in particular, to be used at early phases of periodontitis.
Periodontitis, an infection-induced inflammatory disease of the tooth-supporting tissues, is initiated by the formation of pathogenic biofilms at and under the gingival margin. Out of more than 700 resident bacterial species of the oral cavity, about a half can be found in subgingival biofilms in both healthy and diseased sites (Teles et al., 2006). Periodontitis-associated pathogens and their toxins in bacterial biofilms perturb gingival epithelial cells triggering a sequence of inflammatory and immune responses. The initial inflammatory response aims to limit bacterial invasion by promoting the infiltration of neutrophils and macrophages to the site of bacterial challenge. This infiltration is achieved by the secretion of proinflammatory cytokines and chemokines from gingival epithelial cells and fibroblasts (Preshaw and Taylor, 2011). After their migration to inflamed tissues, leukocytes suppress bacterial invasion by their oxygen dependent or independent mechanisms. Subsequently, T- and B-cells emerge to the site of infection and secrete immunoglobulins as an antigen-specific response. If the host defense fails to suppress the level of infection by eliminating pathogens, continuing inflammation finally ends up in alveolar bone destruction (Bartold and Narayanan, 2006; Preshaw and Taylor, 2011). In the course of the disease, a number of matrix metalloproteases (MMPs), MMP-8, MMP-9, and MMP-13, in particular, are produced and activated by host cells in a cascade leading to degradation of gingival tissues and alveolar bone (Sorsa et al., 2004). Osteoclastogenesis is connected to an increased expression of Receptor Activator for Nuclear Factor κB Ligand (RANKL) and a decreased expression of osteoprotegerin in osteoblast cells. A consensus report of the 7th European Workshop on Periodontology recently highlighted interleukin (IL)-1β, IL-6, tumor necrosis factor (TNF)-α, and RANKL as important players in the periodontitis network (Kinane et al., 2011). Therefore, it does not seem an exaggeration to argue that rather than the bacterial infection, the magnitude of the inflammatory response raised against pathogens seems to be the determinant for developing a destructive periodontal disease (Page and Kornman, 1997; Van Dyke, 2007; Silva et al., 2008). Without an adequate therapy, chronic inflammation may result in the destruction in the attachment between the tooth and the gingival tissue, formation of periodontal pockets, alveolar bone loss and, eventually, tooth loss (Darveau, 2010). Yet, this complex interplay between the subgingival microbiota and host tissues is not the same for everyone; several factors, such as smoking, age, systemic disease, and genetic susceptibility, modify the formation and progression of periodontal diseases (Marsh et al., 2011).
In general, dental clinicians are looking for a diagnostic tool, preferably a non-invasive one, to determine the current status of periodontal disease, to monitor the response to therapy, and to measure the degree of susceptibility to future disease progression (Giannobile et al., 2009). Conventional diagnostic tools, such as probing pocket depths, bleeding on probing, and clinical attachment level, are inadequate to identify patients who are at risk for disease progression (Goodson, 1992). Saliva has a major importance in the maintenance of oral health, and, during the past two decades, it has been considered a potential specimen to detect oral and systemic diseases (Ji and Choi, 2015). Saliva as a diagnostic fluid has been evaluated for detecting caries (Bratthall and Hänsel Petersson, 2005), periodontitis (Christodoulides et al., 2007; Gursoy et al., 2011), oral and breast cancer (Li et al., 2004; Streckfus and Bigler, 2005), and hepatitis (Ohnishi and Daikuhara, 2003).
With the development of different—omics technologies, analysis of saliva has become particularly fascinating, not only in dentistry but also in general medicine (Zhang et al., 2009; Cuevas-Córdoba and Santiago-García, 2014). By analyzing an array of constituents present in saliva, it is possible to estimate the risk of disease onset, to monitor disease progression, and to evaluate therapeutic efficacy of oral infections as well as oropharyngeal lesions (Zhang et al., 2009).
Specific biomarkers have been identified from saliva, reflecting the three key features of pathogenic processes in periodontal disease, i.e., infection-induced inflammation, collagen degradation, and bone turnover (Zhang et al., 2009). Host- and bacteria-derived enzymes, proteins, and other inflammatory mediators appear to hold great promise as salivary biomarkers for the diagnosis of periodontal disease (Giannobile et al., 2009). In infected periodontal tissues, numerous cytokines are secreted as part of the innate response by resident cells (epithelial cells, fibroblasts) and neutrophils. There is strong evidence to suggest that salivary IL-1β is a relevant biomarker of periodontitis, while no significant association has been found between salivary TNF-α or IL-6 levels and the presence of periodontitis (Gursoy et al., 2009; Ebersole et al., 2013; Taylor, 2014). Host-derived MMPs are considered initiators of the extracellular matrix degradation associated with periodontitis (Sorsa et al., 1990; Uitto et al., 1990). Especially MMP-8, a neutrophil collagenase, has the potential to be used as a biomarker of periodontal destruction. In a number of studies, it has been shown that MMP-8 activity is elevated in saliva of periodontitis patients as compared to their periodontally healthy controls, regardless of study subjects' smoking status (Sorsa et al., 2004; Miller et al., 2006; Gursoy et al., 2010).
The oral cavity with its various surfaces offers an open environment, which allows a constant exposure of microorganisms to be colonized and, in favorable circumstances, this can lead to disease (Diamond et al., 2008). Bacterial infection in the mouth is resisted by the stratified squamous epithelium, which acts as a mechanical barrier, and saliva provides a mechanical rinsing action (Gorr, 2012). The oral epithelium and saliva are the most central defense systems in the mouth. These two defense systems do not act as passive protection mechanisms, but both of them contain several types of antimicrobial peptides, including histatins, defensins, and hCAP18/LL-37 (De Smet and Contreras, 2005). Antimicrobial peptides are small cationic peptides with a broad spectrum of antimicrobial activity. A complex mixture of over 45 antimicrobial proteins and peptides are found in oral fluids (Denny et al., 2008); of these, 13 are up-regulated in periodontal disease, while 11 are down-regulated. Defensins and hCAP18/LL-37 belong to key components of the mucosal antimicrobial defense (Hosokawa et al., 2006; Gorr, 2012).
Defensins are divided into subfamilies of α- and β-defensins (Table 1). These defensins differ in their cysteine motifs, but share a similar secondary structure, and both of them are rich in cationic residues (Beckloff and Diamond, 2008). Four types of α-defensins [Human Neutrophilic Peptide (HNP) 1-4] are found predominantly in neutrophils (Ganz et al., 1985), whereas human β-defensins (hBDs) are mainly produced by epithelial cells (Gursoy and Könönen, 2012). The number of hBDs has been suggested to be over 20 based on genomic targeting (Lehrer, 2011). Of the four hBDs known so far, hBD 1-3 are expressed and secreted in the human oral cavity (Dale and Krisanaprakornkit, 2001; Vardar-Sengul et al., 2007). The sole human cathelicidin, hCAP18/LL-37, was initially identified in infiltrating neutrophils in the oral cavity (Dale and Krisanaprakornkit, 2001). However, it has been observed in the salivary glands and gingival epithelium as well (Woo et al., 2003; Gursoy et al., 2012).
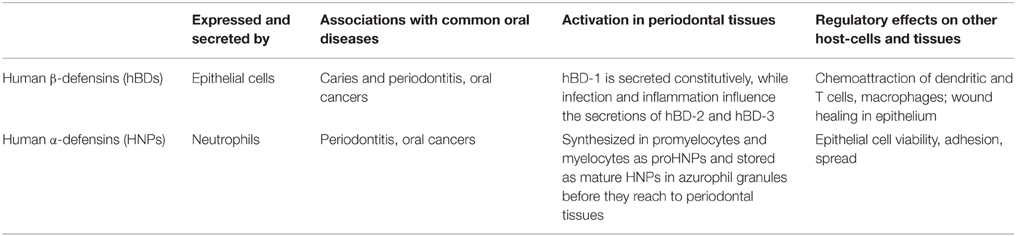
Table 1. Comparisons between human β-defensins (hBDs) and human α-defensins (human neutrophil peptides, HNPs).
Antimicrobial peptides play a major role in the innate host defense. HNPs, hCAP18/LL-37, and hBDs exhibit a broad-spectrum antimicrobial activity against Gram-positive and -negative bacteria, fungi, and enveloped viruses (Gomes Pde and Fernandes, 2010). Like HNPs, hBDs are considered to exert their antibacterial effect by permeabilizing the bacterial cellular membrane. In addition to their direct antimicrobial activity, both HNPs and hBDs exhibit numerous other biological activities (Dommisch and Jepsen, 2015). HNP 1-3 and hBD 1-3 have a selective chemotactic activity for a variety of host defense cells like immature dendritic cells and mast cells (Soruri et al., 2007). They function as both proinflammatory and anti-inflammatory agents in the periodontal disease pathogenesis (Bowdish et al., 2006). Besides their antimicrobial and immune regulatory functions, hBDs contribute to the healing process of wounds (Niyonsaba et al., 2007) and HNP-1 regulates epithelial cell adhesion and spread (Gursoy et al., 2013). While initially isolated as an antimicrobial peptide, hCAP18/LL-37 has been proposed to play additional roles in inflammation. hCAP18/LL-37 demonstrates a chemotactic activity for neutrophils, monocytes, and some T-cells. Furthermore, hCAP18/LL-37 affects dendritic cell maturation (Kai-Larsen and Agerberth, 2008). Taken together, these multiple activities of antimicrobial peptides suggest that they play an important, multifunctional role in host defense (Gorr and Abdolhosseini, 2011).
It is considered that HNPs, hBDs, and hCAP18/LL-37 have the same function in health and disease, but in a coordinated manner. Periodontal infection and inflammation is thought to affect the expression of each antimicrobial peptide (Gursoy and Könönen, 2012). Up-regulated expression of hBDs has been demonstrated in infections, inflammatory stimulations, and keratinocyte differentiation. In non-inflamed gingival tissues, both hBD-1 and hBD-2 are expressed; their levels are highest at gingival margin close to dental plaque (Yilmaz et al., 2015). During the inflammatory state, these peptides are expressed also in the sulcular epithelium (Dale and Krisanaprakornkit, 2001). The expression of hBD-3 is primarily located in the basal layer of healthy gingival tissues, but it is extended toward superficial layers of the gingival epithelium in periodontitis (Lu et al., 2005; Yilmaz et al., 2015). Although a number of studies have described expression levels and localizations of hCAP18/LL-37, hBDs, and HNPs in healthy and inflamed gingival tissues (Krisanaprakornkit et al., 2000; Dale and Krisanaprakornkit, 2001; Dommisch et al., 2005; Hosokawa et al., 2006; Kuula et al., 2008; Brancatisano et al., 2011; Yilmaz et al., 2015), their relation to the initiation and progression of periodontal disease is still poorly understood.
The main sources of antimicrobial peptides in the oral cavity are the gingival epithelium and neutrophils, although salivary glands also secrete some amounts of defensins and hCAP18/LL-37 (Mathews et al., 1999; Mizukawa et al., 1999). By using high performance liquid chromatography, it has been demonstrated that healthy adults have a mean value of 0.5–0.9 mg/ml HNP-1 in whole saliva (Goebel et al., 2000). As a large number of neutrophils continuously enter the oral cavity through the junctional epithelium, it is possible that HNPs are mainly derived from these neutrophils. Immunohistochemical staining for HNP 1-3 showed the presence of peptides in parts of the ductal cells in submandibular glands and in minor salivary glands, while no HNP was detected in an individual major salivary gland (Mizukawa et al., 1999). This indicates that ductal cells can be a source of HNPs in saliva (Abiko and Saitoh, 2007). The sources of hBDs in saliva are assumed to be the oral epithelium and salivary glands. hBD-1, -2, and -3 mRNA have been detected in salivary glands, including the parotid, submandibular, and minor glands, as well as the oral epithelium (Bonass et al., 1999; Dunsche et al., 2001). The mean concentrations of hBD-1 and -2 in whole saliva of healthy subjects are around 150 ng/ml (Mathews et al., 1999), while that of hBD-3 is about 730 ng/ml (Abiko and Saitoh, 2007). In humans, hCAP18/LL-37 is mainly secreted by neutrophils and it is present in saliva at concentrations of 0.14–3 μg/ml (Gorr, 2009).
Saliva carries a significant amount of antimicrobial peptides as part of its defense mechanism but also impairs their antimicrobial functions. As an example, saliva can reduce the antibacterial activities of hBD 1-3 and hCAP18/LL-37 by 20–50% in in vitro conditions (Mineshiba et al., 2003). This in vitro effect is generally explained by the salt concentration of saliva. However, this is probably unlikely because of the low salt concentrations in saliva. Moreover, hBD activity in saliva may get affected by proteases and redox enzymes. On the one hand, proteases, at least in in vitro conditions, affect the activity and concentration of antimicrobial peptides (Kuula et al., 2008), thereby may reduce their value to be used as salivary biomarkers of periodontal disease. On the other hand, defensins are reduced by thioredoxin reductases to their active forms. For instance, glutaredoxin can reduce hBD-1 to its antibacterial form (Jaeger et al., 2013). The activation or inactivation by other proteins in saliva can have a significant effect on the use of antimicrobial peptides as biomarkers, since a selected method for analysis may detect only one form of the peptide, depending on the antibody chosen. Therefore, interactions of antimicrobial peptides with other proteins in saliva should be thoroughly analyzed (Wilson et al., 1999).
Although, the levels of single markers in saliva can be statistically distinguished between subjects with and without periodontitis, the large variation in their values between individuals make a prospective assignment difficult (Miller et al., 2010). Antimicrobial peptides are typically expressed in response to oral bacteria or bacterial toxins, which makes them suitable biomarkers for the diagnosis of periodontal disease (Gorr, 2009; Gorr and Abdolhosseini, 2011). Information on the association between salivary antimicrobial peptide concentrations and periodontal disease status is limited. Pereira et al. (2013) studied salivary levels of hBD-2 in 31 chronic periodontitis and 27 gingivitis patients, compared to 31 periodontally healthy controls, and detected elevated hBD-2 levels in chronic periodontitis patients. No relationship between the frequency of examined periodontopathogens and hBD-2 protein concentrations was found. Salazar et al. (2013) examined 20 periodontally healthy and 20 diseased subjects to identify periodontitis-associated changes in the proteome of the whole saliva. Twenty proteins, including HNP-1, were elevated in periodontitis patients in comparison to their controls (Salazar et al., 2013). It is important to note that peptide concentrations can be significantly diluted in saliva and, therefore, much lower than those in periodontal pockets and gingival tissues (Gorr, 2012). Salivary LL-37 concentrations have been demonstrated to correlate to periodontal tissue destruction in subjects with chronic periodontitis (Takeuchi et al., 2012).
Advances in genomic technologies offer hitherto unprecedented observations on complex human diseases. To date, however, there is only one study by Jaradat et al. (2013) where associations between the genomic copy number of hBD-2 and periodontitis are evaluated. According to their results, there is an association between decreased hBD-2 genomic copy numbers and severity periodontitis. With increasing information, it may be possible to avoid some of the limitations that currently exist in the use of gingival defensins as biomarkers of periodontitis. Moreover, the outcomes of genomic research would help in understanding clinically distinct diseases, for example Crohn's disease, and periodontitis, with a view on their shared molecular targets, such as hBD-2 (Keskin et al., 2015).
In this review, we evaluated the evidence on salivary antimicrobial peptides as biomarkers of periodontitis. These small peptides form the initial tissue response against infection and thus could function as an early diagnostic marker of periodontitis. However, in the use of antimicrobial peptides as biomarkers of periodontitis there are significant limitations to consider, and the majority of these limitations are not fully characterized (Figure 1). Firstly, antimicrobial peptides can aggregate in a concentration dependent manner (Brogden, 2005), and this may weaken the sensitivity of test methods, such as an enzyme-linked immunoassay (ELISA). It is also possible that host-derived and bacterial enzymes degrade antimicrobial peptides, decreasing the sensitivity of the methods depending on the antibody of choice. Further, binding to bacterial lipopolysaccharides and DNA may force salivary antimicrobials to accumulate in the pellet at the initial centrifugation of the sample. Finally, some antimicrobial peptides do not get out of the tissue, but accumulate in the cell cytoplasm and cell nucleus as recently described by Yilmaz et al. (2015). They may still be protective against invading bacteria, however, their accumulation tendency affects their concentrations in saliva that will not reflect the degree of inflammation.
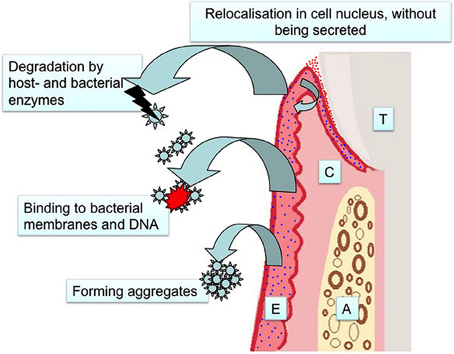
Figure 1. Limitations in the use of antimicrobial defensins as salivary biomarkers of periodontitis (T, Tooth; C, Connective Tissue; A, Alveolar Bone; E, Epithelium; red dots, α-defensins; blue dots, β-defensins).
Despite the limitations, it can be concluded that salivary antimicrobial peptides have potential to be considered as early markers of periodontitis. The entire human salivary proteome was reported by a consortium of three research groups, revealing that 1166 proteins are present in human saliva (Denny et al., 2008). In the pathogenesis of periodontitis, antimicrobial peptides cooperate with other inflammatory proteins and regulate distinct inflammatory pathways. Thus, a combinational approach, in which antimicrobial peptides are measured together with their activators or target proteins, will increase their value as diagnostic biomarkers. This aim may be achieved by newly developed omics technologies.
All authors contributed to the preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abiko, Y., and Saitoh, M. (2007). Salivary defensins and their importance in oral health and disease. Curr. Pharm. Des. 13, 3065–3072. doi: 10.2174/138161207782110417
Bartold, P. M., and Narayanan, A. S. (2006). Molecular and cell biology of healthy and diseased periodontal tissues. Periodontol 2000 40, 29–49. doi: 10.1111/j.1600-0757.2005.00140.x
Beckloff, N., and Diamond, G. (2008). Computational analysis suggests beta-defensins are processed to mature peptides by signal peptidase. Protein Pept. Lett. 15, 536–540. doi: 10.2174/092986608784567618
Bonass, W. A., High, A. S., Owen, P. J., and Devine, D. A. (1999). Expression of beta-defensin genes by human salivary glands. Oral Microbiol. Immunol. 14, 371–374. doi: 10.1034/j.1399-302X.1999.140607.x
Bowdish, D. M., Davidson, D. J., and Hancock, R. E. (2006). Immunomodulatory properties of defensins and cathelicidins. Curr. Top. Microbiol. Immunol. 306, 27–66. doi: 10.1007/3-540-29916-5_2
Brancatisano, F. L., Maisetta, G., Barsotti, F., Esin, S., Miceli, M., Gabriele, M., et al. (2011). Reduced human beta defensin 3 in individuals with periodontal disease. J. Dent. Res. 90, 241–245. doi: 10.1177/0022034510385686
Bratthall, D., and Hänsel Petersson, G. (2005). Cariogram–a multifactorial risk assessment model for a multifactorial disease. Community Dent. Oral Epidemiol. 33, 256–264. doi: 10.1111/j.1600-0528.2005.00233.x
Brogden, K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250. doi: 10.1038/nrmicro1098
Christodoulides, N., Floriano, P. N., Miller, C. S., Ebersole, J. L., Mohanty, S., Dharshan, P., et al. (2007). Lab-on-a-chip methods for point-of-care measurements of salivary biomarkers of periodontitis. Ann. N. Y. Acad. Sci. 1098, 411–428. doi: 10.1196/annals.1384.035
Cuevas-Córdoba, B., and Santiago-García, J. (2014). Saliva: a fluid of study for OMICS. OMICS 18, 87–97. doi: 10.1089/omi.2013.0064
Dale, B. A., and Krisanaprakornkit, S. (2001). Defensin antimicrobial peptides in the oral cavity. J. Oral Pathol. Med. 30, 321–327. doi: 10.1034/j.1600-0714.2001.300601.x
Darveau, R. P. (2010). Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8, 481–490. doi: 10.1038/nrmicro2337
Denny, P., Hagen, F. K., Hardt, M., Liao, L., Yan, W., Arellanno, M., et al. (2008). The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J. Proteome Res. 7, 1994–2006. doi: 10.1021/pr700764j
De Smet, K., and Contreras, R. (2005). Human antimicrobial peptides: defensins, cathelicidins and histatins. Biotechnol. Lett. 27, 1337–1347. doi: 10.1007/s10529-005-0936-5
Diamond, G., Beckloff, N., and Ryan, L. K. (2008). Host defense peptides in the oral cavity and the lung: similarities and differences. J. Dent. Res. 87, 915–927. doi: 10.1177/154405910808701011
Dommisch, H., Açil, Y., Dunsche, A., Winter, J., and Jepsen, S. (2005). Differential gene expression of human beta-defensins (hBD-1, -2, -3) in inflammatory gingival diseases. Oral Microbiol. Immunol. 20, 186–190. doi: 10.1111/j.1399-302X.2005.00211.x
Dommisch, H., and Jepsen, S. (2015). Diverse functions of defensins and other antimicrobial peptides in periodontal tissues. Periodontol. 2000 69, 96–110. doi: 10.1111/prd.12093
Dunsche, A., Açil, Y., Siebert, R., Harder, J., Schroder, J. M., and Jepsen, S. (2001). Expression profile of human defensins and antimicrobial proteins in oral tissues. J. Oral Pathol. Med. 30, 154–158. doi: 10.1034/j.1600-0714.2001.300305.x
Ebersole, J. L., Schuster, J. L., Stevens, J., Dawson, D. III, Kryscio, R. J., Lin, Y., et al. (2013). Patterns of salivary analytes provide diagnostic capacity for distinguishing chronic adult periodontitis from health. J. Clin. Immunol. 33, 271–279. doi: 10.1007/s10875-012-9771-3
Ganz, T., Selsted, M. E., Szklarek, D., Harwig, S. S., Daher, K., Bainton, D. F., et al. (1985). Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Invest. 76, 1427–1435. doi: 10.1172/JCI112120
Giannobile, W. V., Beikler, T., Kinney, J. S., Ramseier, C. A., Morelli, T., and Wong, D. T. (2009). Saliva as a diagnostic tool for periodontal disease: current state and future directions. Periodontol. 2000 50, 52–64. doi: 10.1111/j.1600-0757.2008.00288.x
Goebel, C., Mackay, L. G., Vickers, E. R., and Mather, L. E. (2000). Determination of defensin HNP-1, HNP-2, and HNP-3 in human saliva by using LC/MS. Peptides 21, 757–765. doi: 10.1016/S0196-9781(00)00205-9
Gomes Pde, S., and Fernandes, M. H. (2010). Defensins in the oral cavity: distribution and biological role. J. Oral Pathol. Med. 39, 1–9. doi: 10.1111/j.1600-0714.2009.00832.x
Goodson, J. M. (1992). Diagnosis of periodontitis by physical measurement: interpretation from episodic disease hypothesis. J. Periodontol. 63, 373–382. doi: 10.1902/jop.1992.63.4s.373
Gorr, S. U. (2009). Antimicrobial peptides of the oral cavity. Periodontol. 2000 51, 152–180. doi: 10.1111/j.1600-0757.2009.00310.x
Gorr, S. U. (2012). Antimicrobial peptides in periodontal innate defense. Front. Oral Biol. 15, 84–98. doi: 10.1159/000329673
Gorr, S. U., and Abdolhosseini, M. (2011). Antimicrobial peptides and periodontal disease. J. Clin. Periodontol. 38(Suppl. 11), 126–141. doi: 10.1111/j.1600-051X.2010.01664.x
Gursoy, U. K., and Könönen, E. (2012). Understanding the roles of gingival beta-defensins. J. Oral. Microbiol. 4:1527. doi: 10.3402/jom.v4i0.15127
Gursoy, U. K., Könönen, E., Luukkonen, N., and Uitto, V. J. (2013). Human neutrophil defensins and their effect on epithelial cells. J. Periodontol. 84, 126–133. doi: 10.1902/jop.2012.120017
Gursoy, U. K., Könönen, E., Pradhan-Palikhe, P., Tervahartiala, T., Pussinen, P. J., Suominen-Taipale, L., et al. (2010). Salivary MMP-8, TIMP-1, and ICTP as markers of advanced periodontitis. J. Clin. Periodontol. 37, 487–493. doi: 10.1111/j.1600-051X.2010.01563.x
Gursoy, U. K., Könönen, E., Pussinen, P. J., Tervahartiala, T., Hyvärinen, K., Suominen, A. L., et al. (2011). Use of host- and bacteria-derived salivary markers in detection of periodontitis: a cumulative approach. Dis. Markers 30, 299–305. doi: 10.1155/2011/621484
Gursoy, U. K., Könönen, E., Uitto, V. J., Pussinen, P. J., Hyvärinen, K., Suominen-Taipale, L., et al. (2009). Salivary interleukin-1beta concentration and the presence of multiple pathogens in periodontitis. J. Clin. Periodontol. 36, 922–927. doi: 10.1111/j.1600-051X.2009.01480.x
Gursoy, U. K., Pöllänen, M., Könönen, E., and Uitto, V. J. (2012). A novel organotypic dento-epithelial culture model: effect of Fusobacterium nucleatum biofilm on B-defensin-2, -3, and LL-37 expression. J. Periodontol. 83, 242–247. doi: 10.1902/jop.2011.110177
Hosokawa, I., Hosokawa, Y., Komatsuzawa, H., Goncalves, R. B., Karimbux, N., Napimoga, M. H., et al. (2006). Innate immune peptide LL-37 displays distinct expression pattern from beta-defensins in inflamed gingival tissue. Clin. Exp. Immunol. 146, 218–225. doi: 10.1111/j.1365-2249.2006.03200.x
Jaeger, S. U., Schroeder, B. O., Meyer-Hoffert, U., Courth, L., Fehr, S. N., Gersemann, M., et al. (2013). Cell-mediated reduction of human β-defensin 1: a major role for mucosal thioredoxin. Mucosal Immunol. 6, 1179–1190. doi: 10.1038/mi.2013.17
Jaradat, S. W., Hoder-Przyrembel, C., Cubillos, S., Krieg, N., Lehmann, K., Piehler, S., et al. (2013). Beta-defensin-2 genomic copy number variation and chronic periodontitis. J. Dent. Res. 92, 1035–1040. doi: 10.1177/0022034513504217
Ji, S., and Choi, Y. (2015). Point-of-care diagnosis of periodontitis using saliva: technically feasible but still a challenge. Front. Cell. Infect. Microbiol. 5:65. doi: 10.3389/fcimb.2015.00065
Kai-Larsen, Y., and Agerberth, B. (2008). The role of the multifunctional peptide LL-37 in host defense. Front. Biosci. 13, 3760–3767. doi: 10.2741/2964
Keskin, M., Zeidán-Chuliá, F., Gursoy, M., Könönen, E., Rautava, J., and Gursoy, U. K. (2015). Two cheers for Crohn's disease and periodontitis: beta-defensin-2 as an Actionable target to intervene on two clinically distinct diseases. OMICS 19, 443–450. doi: 10.1089/omi.2015.0077
Kinane, D. F., Preshaw, P. M., Loos, B. G., and Working Group 2 of Seventh European Workshop On Periodontology (2011). Host-response: understanding the cellular and molecular mechanisms of host-microbial interactions–consensus of the Seventh European Workshop on Periodontology. J. Clin. Periodontol. 38(Suppl. 11), 44–48. doi: 10.1111/j.1600-051X.2010.01682.x
Krisanaprakornkit, S., Kimball, J. R., Weinberg, A., Darveau, R. P., Bainbridge, B. W., and Dale, B. A. (2000). Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68, 2907–2915. doi: 10.1128/IAI.68.5.2907-2915.2000
Kuula, H., Salo, T., Pirilä, E., Hagström, J., Luomanen, M., Gutierrez-Fernandez, A., et al. (2008). Human beta-defensin-1 and -2 and matrix metalloproteinase-25 and -26 expression in chronic and aggressive periodontitis and in peri-implantitis. Arch. Oral Biol. 53, 175–186. doi: 10.1016/j.archoralbio.2007.09.010
Lehrer, R. I. (2011). Immunology: peptide gets in shape for self-defence. Nature 469, 309–310. doi: 10.1038/469309a
Li, Y., St John, M. A., Zhou, X., Kim, Y., Sinha, U., Jordan, R. C., et al. (2004). Salivary transcriptome diagnostics for oral cancer detection. Clin. Cancer Res. 10, 8442–8450. doi: 10.1158/1078-0432.CCR-04-1167
Lu, Q., Samaranayake, L. P., Darveau, R. P., and Jin, L. (2005). Expression of human beta-defensin-3 in gingival epithelia. J. Periodont. Res. 40, 474–481. doi: 10.1111/j.1600-0765.2005.00827.x
Marsh, P. D., Moter, A., and Devine, D. A. (2011). Dental plaque biofilms: communities, conflict and control. Periodontol. 2000 55, 16–35. doi: 10.1111/j.1600-0757.2009.00339.x
Mathews, M., Jia, H. P., Guthmiller, J. M., Losh, G., Graham, S., Johnson, G. K., et al. (1999). Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 67, 2740–2745.
Miller, C. S., Foley, J. D., Bailey, A. L., Campell, C. L., Humphries, R. L., Christodoulides, N., et al. (2010). Current developments in salivary diagnostics. Biomark. Med. 4, 171–189. doi: 10.2217/bmm.09.68
Miller, C. S., King, C. P. Jr., Langub, M. C., Kryscio, R. J., and Thomas, M. V. (2006). Salivary biomarkers of existing periodontal disease: a cross-sectional study. J. Am. Dent. Assoc. 137, 322–329. doi: 10.14219/jada.archive.2006.0181
Mineshiba, F., Takashiba, S., Mineshiba, J., Matsuura, K., Kokeguchi, S., and Murayama, Y. (2003). Antibacterial activity of synthetic human B defensin-2 against periodontal bacteria. J. Int. Acad. Periodontol. 5, 35–40.
Mizukawa, N., Sugiyama, K., Ueno, T., Mishima, K., Takagi, S., and Sugahara, T. (1999). Defensin-1, an antimicrobial peptide present in the saliva of patients with oral diseases. Oral Dis. 5, 139–142. doi: 10.1111/j.1601-0825.1999.tb00078.x
Niyonsaba, F., Ushio, H., Nakano, N., Ng, W., Sayama, K., Hashimoto, K., et al. (2007). Antimicrobial peptides human beta-defensins stimulate epidermal keratinocyte migration, proliferation and production of proinflammatory cytokines and chemokines. J. Invest. Dermatol. 127, 594–604. doi: 10.1038/sj.jid.5700599
Ohnishi, T., and Daikuhara, Y. (2003). Hepatocyte growth factor/scatter factor in development, inflammation and carcinogenesis: its expression and role in oral tissues. Arch. Oral Biol. 48, 797–804. doi: 10.1016/S0003-9969(03)00180-8
Page, R. C., and Kornman, K. S. (1997). The pathogenesis of human periodontitis: an introduction. Periodontol. 2000 14, 9–11. doi: 10.1111/j.1600-0757.1997.tb00189.x
Pereira, A. L., Franco, G. C., Cortelli, S. C., Aquino, D. R., Costa, F. O., Raslan, S. A., et al. (2013). Influence of periodontal status and periodontopathogens on levels of oral human beta-defensin-2 in saliva. J. Periodontol. 84, 1445–1453. doi: 10.1902/jop.2012.120321
Preshaw, P. M., and Taylor, J. J. (2011). How has research into cytokine interactions and their role in driving immune responses impacted our understanding of periodontitis? J. Clin. Periodontol. 38(Suppl. 11), 60–84. doi: 10.1111/j.1600-051X.2010.01671.x
Salazar, M. G., Jehmlich, N., Murr, A., Dhople, V. M., Holtfreter, B., Hammer, E., et al. (2013). Identification of periodontitis associated changes in the proteome of whole human saliva by mass spectrometric analysis. J. Clin. Periodontol. 40, 825–832. doi: 10.1111/jcpe.12130
Silva, N., Dutzan, N., Hernandez, M., Dezerega, A., Rivera, O., Aguillon, J. C., et al. (2008). Characterization of progressive periodontal lesions in chronic periodontitis patients: levels of chemokines, cytokines, matrix metalloproteinase-13, periodontal pathogens and inflammatory cells. J. Clin. Periodontol. 35, 206–214. doi: 10.1111/j.1600-051X.2007.01190.x
Sorsa, T., Suomalainen, K., and Uitto, V. J. (1990). The role of gingival crevicular fluid and salivary interstitial collagenases in human periodontal diseases. Arch. Oral. Biol. 35(Suppl), 193S–196S. doi: 10.1016/0003-9969(90)90156-5
Sorsa, T., Tjäderhane, L., and Salo, T. (2004). Matrix metalloproteinases (MMPs) in oral diseases. Oral Dis. 10, 311–318. doi: 10.1111/j.1601-0825.2004.01038.x
Soruri, A., Grigat, J., Forssmann, U., Riggert, J., and Zwirner, J. (2007). beta-Defensins chemoattract macrophages and mast cells but not lymphocytes and dendritic cells: CCR6 is not involved. Eur. J. Immunol. 37, 2474–2486. doi: 10.1002/eji.200737292
Streckfus, C., and Bigler, L. (2005). The use of soluble, salivary c-erbB-2 for the detection and post-operative follow-up of breast cancer in women: the results of a five-year translational research study. Adv. Dent. Res. 18, 17–24. doi: 10.1177/154407370501800105
Takeuchi, Y., Nagasawa, T., Katagiri, S., Kitagawara, S., Kobayashi, H., Koyanagi, T., et al. (2012). Salivary levels of antibacterial peptide (LL-37/hCAP-18) and cotinine in patients with chronic periodontitis. J. Periodontol. 83, 766–762. doi: 10.1902/jop.2011.100767
Taylor, J. J. (2014). Protein biomarkers of periodontitis in saliva. ISRN Inflamm. 2014:593151. doi: 10.1155/2014/593151
Teles, R. P., Haffajee, A. D., and Socransky, S. S. (2006). Microbiological goals of periodontal therapy. Periodontol. 2000 42, 180–218. doi: 10.1111/j.1600-0757.2006.00192.x
Uitto, V. J., Suomalainen, K., and Sorsa, T. (1990). Salivary collagenase. Origin, characteristics and relationship to periodontal health. J. Periodontal. Res. 25, 135–142. doi: 10.1111/j.1600-0765.1990.tb01035.x
Van Dyke, T. E. (2007). Control of inflammation and periodontitis. Periodontol. 2000 45, 158–166. doi: 10.1111/j.1600-0757.2007.00229.x
Vardar-Sengul, S., Demirci, T., Sen, B. H., Erkizan, V., Kurulgan, E., and Baylas, H. (2007). Human beta defensin-1 and -2 expression in the gingiva of patients with specific periodontal diseases. J. Periodont. Res. 42, 429–437. doi: 10.1111/j.1600-0765.2006.00964.x
Wilson, C. L., Ouellette, A. J., Satchell, D. P., Ayabe, T., López-Boado, Y. S., Stratman, J. L., et al. (1999). Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286, 113–117. doi: 10.1126/science.286.5437.113
Woo, J. S., Jeong, J. Y., Hwang, Y. J., Chae, S. W., Hwang, S. J., and Lee, H. M. (2003). Expression of cathelicidin in human salivary glands. Arch. Otolaryngol. Head Neck Surg. 129, 211–214. doi: 10.1001/archotol.129.2.211
Yilmaz, D., Güncü, G. N., Könönen, E., Baris, E., Çaglayan, F., and Gursoy, U. K. (2015). Overexpressions of hBD-2, hBD-3, and hCAP18/LL-37 in gingiva of diabetics with periodontitis. Immunobiology 220, 1219–1226 doi: 10.1016/j.imbio.2015.06.013
Keywords: saliva, biomarker, periodontitis, antimicrobial peptides
Citation: Güncü GN, Yilmaz D, Könönen E and Gürsoy UK (2015) Salivary Antimicrobial Peptides in Early Detection of Periodontitis. Front. Cell. Infect. Microbiol. 5:99. doi: 10.3389/fcimb.2015.00099
Received: 16 September 2015; Accepted: 11 December 2015;
Published: 24 December 2015.
Edited by:
Richard Lamont, University of Louisville, USACopyright © 2015 Güncü, Yilmaz, Könönen and Gürsoy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Güliz N. Güncü, Z3VsaXpAaGFjZXR0ZXBlLmVkdS50cg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.