
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 17 February 2015
Sec. Molecular Bacterial Pathogenesis
Volume 5 - 2015 | https://doi.org/10.3389/fcimb.2015.00014
This article is part of the Research Topic Biofilm formation by staphylococci and streptococci: Structural, functional and regulatory aspects and implications for pathogenesis View all 12 articles
Staphylococcus epidermidis is a usually harmless commensal bacterium highly abundant on the human skin. Under defined predisposing conditions, most importantly implantation of a medical device, S. epidermidis, however, can switch from a colonizing to an invasive life style. The emergence of S. epidermidis as an opportunistic pathogen is closely linked to the biofilm forming capability of the species. During the past decades, tremendous advance regarding our understanding of molecular mechanisms contributing to surface colonization has been made, and detailed information is available for several factors active during the primary attachment, accumulative or dispersal phase of biofilm formation. A picture evolved in which distinct factors, though appearing to be redundantly organized, take over specific and exclusive functions during biofilm development. In this review, these mechanisms are described in molecular detail, with a highlight on recent insights into multi-functional S. epidermidis cell surface proteins contributing to surface adherence and intercellular adhesion. The integration of distinct biofilm-promoting factors into regulatory networks is summarized, with an emphasis on mechanism that could allow S. epidermidis to flexibly adapt to changing environmental conditions present during colonizing or invasive life-styles.
Staphylococcus epidermidis, member of the group of coagulase-negative staphylococci, belongs to the commensal skin flora of every human individual (Kloos, 1980, 1997; Kloos and Schleifer, 1986). In its natural niche, the species is of significant importance for maintenance of local homoeostasis—a role that is so far understood only superficially (Grice and Segre, 2011). Only recently light was shed onto the potential importance of S. epidermidis to interfere with colonization with Staphylococcus aureus via expression of a serine-type protease termed Esp, thereby interfering with colonization mechanisms of S. aureus, e.g., biofilm formation (Iwase et al., 2010; Sugimoto et al., 2013). Given the tremendous abundance of S. epidermidis on the skin, it is not surprising that S. epidermidis is the most common cause of contamination in clinical specimens, and it is a challenge for medical microbiologist to reliably identify true invasive isolates (Mack et al., 2006). Improved abilities to discriminate between clinically relevant and contaminating S. epidermidis isolates is of utmost importance, as the species is today one of the most important bacteria related to hospital acquired infections. If the invasive behavior of S. epidermidis follows a clear pathogenic strategy that extends its colonizing abilities (Mack et al., 2009), or merely can be regarded as an accident during which mechanisms usually contributing to commensalism turn into virulence features (Otto, 2009), is still a matter of debate. Nevertheless, without doubt S. epidermidis is a true opportunistic pathogen that only causes disease in patients with predisposing factors. These include individual patient characteristics (i.e., premature birth, congenital immune defects) or concomitant medical conditions—like HIV infection, immunosuppression after bone marrow or solid organ transplantation and chemotherapy related neutropenia (Goldmann and Pier, 1993; Rupp and Archer, 1994). Most significantly, S. epidermidis is the leading organism isolated from foreign material related infections (FMRI) (Darouiche, 2004; Geipel and Herrmann, 2005) such as infected prosthetic joints, central venous catheters (CVC), cerebrospinal fluid shunts, intracardiac devices, artificial heart valves, and vascular grafts (Mack et al., 2006; Rogers et al., 2009). Use of implanted medical devices increases in number, and certainly, this will further propel the importance of S. epidermidis as an important human pathogen.
S. epidermidis is responsible for the vast majority of nosocomial catheter related blood stream infections (CRBSI) in the United States (Wisplinghoff et al., 2003; Hidron et al., 2008) and also in German intensive care units (ICUs) (Geffers and Gastmeier, 2011). Evaluation of a multicenter international data collection calculated a risk of 6.8 CRBSI per 1000 central line-days in ICUs (Rosenthal et al., 2014). Results from the Surveillance and Control of Pathogens of Epidemiological Importance (SCOPE) indicate that coagulase-negative staphylococci are the most frequent cause of nosocomial blood stream infections (Wisplinghoff et al., 2003). Species discrimination identified eighty percent of CoNS from these infections as S. epidermidis (Jukes et al., 2010).
CoNS rank as third most common infective agent in native (NVIE) and first in prosthetic valve infective endocarditis (PVIE) clearly demonstrating the importance of CoNS in these clinical entities (Murdoch et al., 2009). Among CoNS isolates, about eighty percent were identified as S. epidermidis (Chu et al., 2004, 2009). Evaluation of the results of the International Collaboration on Endocarditis database showed a significantly higher rate of complicated clinical courses of PVIE due to CoNS with respect to heart failure compared to S. aureus or viridans streptococci (Lalani et al., 2006).
S. epidermidis is a significant cause of infections of prosthetic joint implants. The lack of non-invasive curative treatment options for joint implant infections often necessitates surgical intervention including replacement surgery. In the UK, CoNS and S. epidermidis were isolated in 36% of total hip and 49% of total knee arthroplasty infections (Phillips et al., 2006; Nickinson et al., 2010). In another study of infected total hip and knee arthroplasties about 77% of the isolated CoNS were confirmed S. epidermidis (Rohde et al., 2007).
First evidence suggesting a pathogenetic link between foreign-material implantation and S. epidermidis infection came from early electron microscopic analysis of explanted central venous catheters. Here, bacteria were found to colonize artificial material in large agglomerations, embedded into an amorphous material (Peters et al., 1982). While this specific mode of growth was first referred to as “slime,” today it is termed biofilm formation (Götz, 2002). In fact, there is significant evidence connecting the biofilm mode of growth to the general persistent course of S. epidermidis foreign-material infections (Scherr et al., 2014a), and to the regular failure to eradicate infections by antimicrobial therapies (Lewis, 2005; Mack et al., 2009; Otto, 2009; Rohde et al., 2010). The latter aspect has been subject to extensive review recently (Lewis, 2010). Although S. epidermidis infections are regarded as prototypic biofilm infections (Costerton et al., 1999; Otto, 2009), it must be noted that it is by far not clear that biofilms observed under in vitro conditions indeed correlate with the biofilm growth evident in vivo. While this is most probably the case in central venous catheter (Peters et al., 1981) or cerebrospinal fluid shunt (Kockro et al., 2000) infections, there is some doubt that this model can be easily transferred to infections occurring at the interface of an implant and the surrounding tissues, e.g., prosthetic joint infections (Broekhuizen et al., 2008; Zaat et al., 2010). Clearly, much needs to be learned with regard to the exact spatial organization of S. epidermidis in implant infections, e.g., by making use of ex vivo or in vivo imaging approaches.
Traditionally, the process of biofilm formation is divided into at least three steps. During the phase of primary attachment, bacteria adhere to the surface to be colonized, while during the accumulative phase, bacteria initiate the establishment of a three dimensional, multi-cellular and multi-layered architecture in which, intriguingly, most bacteria do not have direct contact to the surface (Mack et al., 2009; Otto, 2009; Rohde et al., 2010). S. epidermidis, then, is able to disassemble the biofilm structure again, and liberated cells are believed to allow S. epidermidis to colonize additional body sites. The process of biofilm formation needs a wide range of functional activities, ranging from molecules mediating binding to native or conditioned (i.e., host extracellular matrix covered) surfaces, over glue-like factors fostering cell-cell aggregation, to activities that break down matrix components (Otto, 2009). The tremendous diversity of specific functional requirements during biofilm formation is on the bacterial side significantly mirrored by the expression of a plethora of different, highly specialized factors characterized by very distinct profiles of biological functions. Thus, related to their specialized functions during S. epidermidis biofilm morphogenesis, specific factors are assigned to groups representing mechanisms being active either during primary attachment or during biofilm accumulation, respectively.
Tight binding of bacteria to foreign-materials is a pivotal step toward establishment of a device-associated infection. Not unexpectedly, factors specifically involved in mediating bacterial–surface interactions were identified and further characterized. Some genetic evidence suggests that bacterial binding to unmodified polystyrene is fostered by the S. epidermidis autolysin AtlE (Heilmann et al., 1997). AtlE is a 115 kDa protein, which belongs to a group of bacterial peptidoglycan (PGN)-hydrolases playing a pivotal role in the degradation of the bacterial cell wall (Biswas et al., 2006). The protein consists of an N-terminal signal peptide, a propeptide, a catalytic domain with N-acetylmuramyl-L-alanine amidase activity, three repeats (R1–3), and a C-terminal catalytic domain with N-acetylglucosaminidase activity (Schlag et al., 2010; Zoll et al., 2010). In addition to its general role in cell wall turnover, AtlE also is of importance for binding to unmodified polystyrene, as demonstrated by the defect of an atlE::Tn917 transposon mutant of S. epidermidis O-47 that lost its ability to adhere to plastic surfaces (Heilmann et al., 1997). Intriguingly, the importance of distinct domains for recruitment of AtlE to the bacterial cell wall (i.e., internal repeats) and enzymatic activities have been identified (Zoll et al., 2010). High resolution structural information is available, however so far it remains unclear, which exact AtlE domains are relevant for the primary attachment process and stable bacterial surface binding. In fact, at present it appears possible that expression and functional activation of AtlE induces significant changes in cell surface hydrophobicity, and thus, the AtlE effect on primary attachment might be secondary (Otto, 2014). In addition, a significant role of AtlE in eDNA mediated S. epidermidis biofilm formation is apparent (Qin et al., 2007; Christner et al., 2012).
While interactions between S. epidermidis and unmodified artificial surfaces most likely does not involve specific receptor-ligand interactions, it is well-known that S. epidermidis, similar to S. aureus, expresses cell surface proteins that mediate specific interactions with host extracellular matrix (ECM) components (Patti et al., 1994). Proteins with ECM-binding activity are believed to be of significant importance for the initiation of a device infection, since foreign materials become, as soon as they are inserted into the body, covered by ECM material (e.g., FN, fibronectin; Fg, fibrinogen; Vn, vitronectin; Cn, collagen) (Arrecubieta et al., 2006; Mack et al., 2009). In fact, S. epidermidis can use AtlE to adhere to surface organized Vn, while the lipase GehD is involved into interactions with collagen (Bowden et al., 2002). In addition to these proteins, for which their enzymatic activities might be of primary importance for S. epidermidis physiology, S. epidermidis also expresses proteins with a primary and dedicated function in bacterial-ECM interactions. These proteins belong to the group of serine-aspartate repeat (Sdr) proteins (McCrea et al., 2000), a widely investigated protein family of microbial surface components recognizing adhesive matrix molecules (MSCRAMM) (Josefsson et al., 1998; Foster et al., 2014). In S. epidermidis, three Sdr proteins referred to as SdrF, SdrG, and SdrH have been identified (Josefsson et al., 1998). SdrG, a LPXTG-motif containing protein covalently attached to the bacterial cell surface, is crucial for S. epidermidis adherence to fibrinogen-coated surfaces. It is therefore also being referred to as fibrinogen binding protein of S. epidermidis (Fbe) (Nilsson et al., 1998; Pei et al., 1999; Hartford et al., 2001). The gene encoding Fbe/SdrG is common in clinical S. epidermidis isolates (Nilsson et al., 1998; Rohde et al., 2004, 2007). Fbe/SdrG protein contains five distinct regions: an N-terminal export motif sequence, an A region that contains the Fg binding activity, a B region of so far unknown function, and the R region containing serine-aspartate repeat sequences. Fbe/SdrG specifically binds to a peptide sequence of 14 amino acids found in the N-terminus of the β-chain of Fg (Ponnuraj et al., 2003), and structural analysis of the interaction revealed a unique “dock, lock, and latch” mechanism ensuring a particularly strong interaction (Bowden et al., 2008; Herman et al., 2014). SdrF, sharing overall organizational similarity with SdrG, has been shown to mediate S. epidermidis binding to collagen I (Arrecubieta et al., 2007). In contrast to Fg-binding properties of Fbe/SdrG, the collagen binding epitopes of SdrF are located within the B repeat region (Arrecubieta et al., 2007). So far no specific functionality has been attributed to the N-terminal A domain of SdrF. A single B domain repeat of SdrF was sufficient to interact with collagen I, and apparently, this binding occurs via interactions with the α1 and α2 chains of type I collagen (Arrecubieta et al., 2007). Using a Lactococcus lactis heterologous expression system and a murine infection model evidence was generated that SdrF may contribute to cardiac assist device driveline infections (Arrecubieta et al., 2009). SdrF also mediates binding to unmodified Dacron surfaces covering drivelines. In contrast, L. lactis expressing GehD bound only weakly to driveline surfaces (Arrecubieta et al., 2009). Anti-SdrF inhibited S. epidermidis 9491 binding in the in vivo model only by roughly 50%, indicating that additional S. epidermidis collagen binding factors may be involved (Arrecubieta et al., 2009).
Extensive work has addressed the role of extracellular DNA (eDNA) in S. epidermidis and S. aureus biofilm formation. Data confirm that eDNA is a structural component of the biofilm matrix in both species, although evidence anticipates that eDNA has, at least partially, different functions in both species. Several independent studies have demonstrated that eDNA is released through increased cell lysis (Allesen-Holm et al., 2006; Rice et al., 2007; Christner et al., 2012). In S. epidermidis autolysis is determined to a large extent by the activity of the major autolysin AtlE (Biswas et al., 2006). A role for eDNA in S. epidermidis 1457 during primary attachment was deduced from observations showing that addition of DNase I abrogated bacterial attachment to glass surfaces. These findings were confirmed in additional, genetically independent S. epidermidis backgrounds (i.e., RP62A) (Qin et al., 2007). In extent to its effect on primary attachment, eDNA functions as an intercellular adhesin contributing to the stabilization of biofilms (Whitchurch et al., 2002). Based on the finding that DNase I has biofilm disintegrating activity when added within the first 6 h of biofilm accumulation currently it is believed that eDNA mediated intercellular adhesion is critical especially during the early accumulative phase (Qin et al., 2007). A role of eDNA in earlier stages of staphylococcal biofilm formation has recently been underpinned by observations showing that during S. aureus surface colonization under flow conditions eDNA, while not having impact on primary attachment, is critical during the transition from attachment to accumulation (Moormeier et al., 2014). It should again be stressed that functional differences of eDNA during S. epidermidis and S. aureus biofilm formation are apparent (especially with respect to the function during accumulation) (Izano et al., 2008; Christner et al., 2012), and observations in one species cannot easily be extrapolated to the other. This is especially true for the role of eDNA as a target during biofilm detachment events. By saeRS regulated expression of nuclease Nuc S. aureus can remodel the biofilm ultrastructure and control the release of bacteria from established biofilms (Mann et al., 2009; Olson et al., 2013). The lack of nuclease activity questions if this biofilm-escape mechanism is, in addition to agr-mediated biofilm dispersal (Vuong et al., 2003; Wang et al., 2011a), of relevance in S. epidermidis.
The hallmark of the accumulative phase is expression of intercellular adhesive properties, ultimately leading to cell aggregation and subsequent development of a multicellular, multilayered biofilm architecture (Costerton et al., 1995). Parallel to the discovery of factors with dedicated functions during primary attachment, the nature of intercellular adhesins, functioning as the “biofilm glue,” was partially unraveled. Based on the early electron microscopic studies, showing S. epidermidis cells embedded in an amorphous extracellular matrix (Peters et al., 1981), focus was set onto the biochemical analysis of biofilm matrix components. These efforts ultimately resulted in the discovery of the polysaccharide intercellular adhesin (PIA), which is at present the most extensively studied intercellular adhesin (Mack et al., 1994b).
Structural analysis of PIA and comparison of PIA isolated from S. epidermidis and S. aureus has been recently reviewed (Mack et al., 2013). The structure of PIA was first described for biofilm-forming S. epidermidis 1457 and RP62A. PIA was extracted from the cells by sonication after the strains had been cultured in trypticase soy broth, which revealed the existence of both a major polysaccharide I (>80%), and a minor polysaccharide II (<20%), which are structurally closely related and could be separated due to differing ionic properties (Mack et al., 1996). Chemical analyses and NMR spectroscopy have demonstrated that polysaccharide I is a linear homoglycan of β-1,6-linked 2-amino-2-deoxy-D-glucopyranosyl residues. Approximately 80–85% of them are N-acetylated; the rest are non-N-acetylated and carry a positive charge. Polysaccharide II of PIA has a lower proportion of de-N-acetylated 2-amino-2-deoxy-D-glucopyranosyl residues and is modified by ester-linked succinate residues rendering it anionic (Mack et al., 1996). Despite a high apparent molecular weight indicated by elution in the void volume of Sephadex G200 (Mack et al., 1994a, 1996) or Sephacryl S300 columns (C. Fischer and D. Mack, unpublished results), the ratio of reducing terminal sugar residues to total sugar residues was shown by methylation analysis to be 1:130, implying an average Mr of 30,000 for PIA polysaccharide chains (Mack et al., 1996). This implies aggregation of PIA polysaccharide chains in solution. PIA was shown to function also as the hemagglutinin of S. epidermidis (Rupp and Archer, 1992; Fey et al., 1999; Mack et al., 1999). Production of a functionally active PIA molecule requires expression of all four icaADBC genes (Gerke et al., 1998). The process has been the subject of detailed study in recombinant strains of S. carnosus which expressed different combinations of the icaADBC genes and with UDP-GlcNAc as a sugar donor (Gerke et al., 1998). IcaA belongs to the glycosyltransferase 2 family. It is an integral membrane protein with 412 aa and four predicted transmembrane domains (Heilmann et al., 1996; Gerke et al., 1998; Gill et al., 2005), and directs the synthesis of β-1,6-linked GlcNAc oligosaccharides of up to 20 GlcNAc units. IcaD is required for full activity of IcaA in vitro. It is a 101 aa integral membrane protein with two potential membrane spanning domains: it may be a chaperone directing folding and membrane insertion of IcaA and may act as a link between IcaA and IcaC (Gerke et al., 1998). Also essential for the synthesis of fully functioning PIA is IcaC, a 355 aa integral membrane protein with 10 predicted transmembrane domains, which may be involved in externalization and elongation of the growing polysaccharide (Gerke et al., 1998). IcaB is a member of the polysaccharide deacetylase family, including, for example, chitin deacetylases or the chitooligosaccharide deacetylase NodB of Rhizobium melioti. In its mature form it is a 259 aa secreted protein with a predicted signal sequence, responsible for the de-N-acetylation of PIA, and crucial for PIA activity in biofilm formation and for virulence in S. epidermidis (Vuong et al., 2004a). In ΔicaB-mutants, where the icaB gene has been deleted, PIA is poorly retained on the cell surface as it does not contain non-N-acetylated GlcNAc (Vuong et al., 2004a).
Early observations made by biochemical analysis of biofilm matrix extracts not only showed the presence of(poly-)saccharides but pointed toward the additional presence of proteins and nucleic acids (Hussain et al., 1991). Specific proteins have been identified and characterized. Apart from biofilm associated protein Bap (Tormo et al., 2005a), that is only rarely found in invasive S. epidermidis from human infections (Tormo et al., 2005a; Rohde et al., 2007; Piessens et al., 2012), SesC has been proposed to play a role in biofilm formation (Shahrooei et al., 2009). SesC is an LPXTG motif-containing 68 kDa surface protein of S. epidermidis distantly related to clumping factor A of S. aureus and is expressed more strongly in biofilm-associated as compared to planktonic S. epidermidis 1457 and 10b cells in vitro and in vivo (Shahrooei et al., 2009; Lam et al., 2014). Rabbit anti-SesC inhibited biofilm formation of a number of S. epidermidis isolates in vitro, which may be related to changes in primary attachment to fibrinogen-coated surfaces in the presence of anti-SesC. All of 105 S. epidermidis isolates recovered from nose swabs or infections were in possession of the sesC (Shahrooei et al., 2009). Passive and active immunization using SesC as a target protein was shown to decrease S. epidermidis biofilm formation in an in vivo model of central venous catheter infections (Shahrooei et al., 2012). A specific role of SesC as an intercellular adhesin in biofilm accumulation remains to be demonstrated.
Although having partially additional enzymatic functions (e.g., AltE, GehD), it is a common feature of many factors contributing to S. epidermidis surface colonization that they carry out functions either during the primary attachment or accumulative phase of biofilm formation (Rohde et al., 2006; Mack et al., 2009; Otto, 2009). With the increasing interest in protein factors contributing to staphylococcal biofilm accumulation, it became apparent that, at least in S. aureus, many factors (e.g., FnBPA, ClfA) must be regarded as multifunctional proteins not having an exclusive role in either primary attachment or accumulation (Foster et al., 2014). This concept of multifunctional proteins with important roles during several phases of biofilm formation and surface colonization is now also evolving in S. epidermidis, with the accumulation associated protein (Aap) and the extracellular matrix binding protein (Embp) being the most prominent factors.
Embp and its ortholog in S. aureus designated Ebh were almost simultaneously identified during studies aiming at identifying S. epidermidis or S. aureus protein factors with Fn binding activities (Clarke et al., 2002; Williams et al., 2002). By using a phage display approach a phage was isolated, which exhibited fibronectin binding activity and contained a DNA fragment from a 30,500 bp open reading frame (ORF) coding for a 10,203 aa protein that was referred to as extracellular matrix binding protein Embp (Williams et al., 2002) (Figure 1). Using bioinformatics the architecture of Embp was predicted to consist of an N-terminal YSIRK-motif containing export signal (aa 58–84), followed by an unordered region of approximately 2500 amino acids (aa 85–2586). The overall architecture of Embp mainly is characterized by 21 repetitive “Found In Various Architectures” (FIVAR) repeats (aa 2587–4500) and 38 alternating “G-related Albumine-binding” (GA) motifs and FIVAR repeats—termed the FIVAR-GA repeats—(aa 4501–9443) that span roughly 7000 amino acids in the central proportion of the Embp protein. Finally the C-terminus consists of four domains of unknown function (DUF1542) (aa 9444–9841), followed by a potential transmembrane motive (aa 10,070–10,088) (Christner et al., 2010) (Figure 1). Although the S. aureus homolog Ebh displays at least functional homologies with respect to fibronectin binding activity (Clarke et al., 2002) some functional predictions found in Ebh, e.g., an N-terminal hyperosmolarity resistence domain (Kuroda et al., 2008; Tanaka et al., 2008) have not been identified in the Embp. The overall structural organization of the Ebh protein seems to be more or less identical to Embp, but in detail gradual differences are apparent, e.g., only seven FIVAR motifs but 12 additional FIVAR-GA repeats and four additional DUF1542 repeats compared to Embp were predicted (Tanaka et al., 2008). In fact, it appears that among different S. aureus strains the number of repetitive modules (FIVAR and FIVAR-GA) within Ebh is variable, while available sequence data shows no variability of these features in Embp from S. epidermidis RP62A, ATCC12228 or 1585.
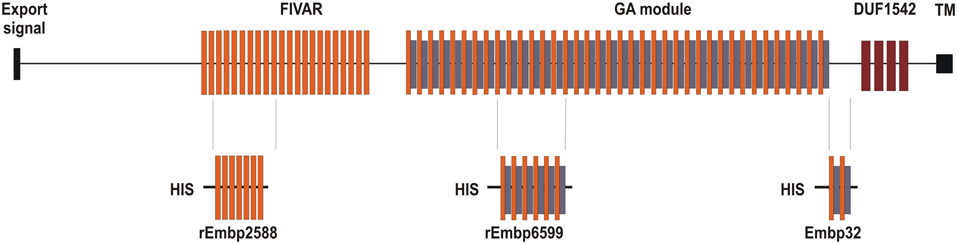
Figure 1. Schematic representation of Embp and relative position of recombinant proteins for which functional data are available. Embp possesses an N-terminal YSIRK motif containing export signal, which is followed by a region of unordered conformation. The central part is comprised of the FIVAR or FIVAR-GA repeats, while the C-terminal end comprises up by four Domains of Unknown Function (DUF) and a putative transmembrane (TM) region (Christner et al., 2010). Embp32 is a recombinant Embp fragment (aa 9180–9421) from S. epidermidis NCTC11047 spanning four alternating FIVAR and FIVAR-GA repeats and that exhibits fibronectin binding capacity (Williams et al., 2002). rEmbp2588 (aa 2588–3187) contains seven FIVAR-only repeat units, while rEmbp6599 (aa 6599–7340) consists of six G-related albumin binding (GA) modules intercalated by FIVAR repeats. The rEmbp2588 and rEmbp6599 both bind to surface immobilized fibronectin (Christner et al., 2010).
Crystallization of two 126 amino acid FIVAR-GA repeats from EbhA of S. aureus MU50 (termed EbhA-R7-R8) revealed a triple α-helical structure interconnected by a continuous alpha helical string displaying an elongated shape (Sakamoto et al., 2008; Tanaka et al., 2008). In fact, the corresponding FIVAR-GA repeat of Embp exhibits a very similar, if not identical structure (Büttner, Perbandt, Rohde, unpublished results). Additional preliminary structural analysis of repetitive FIVAR regions (Büttner, Perband, Rohde, unpublished results) or DUF1542 repeats (Linke et al., 2012) suggest that overall Embp constitutes an elongated rod-like conformation.
In collections of clinical significant S. epidermidis isolates embp was detected in more than 90% of strains (Rohde et al., 2004, 2007). In addition, evidence for the expression of Embp in vivo resulted from investigations showing the presence of anti-Embp antibodies in patients with confirmed S. epidermidis prosthetic joint infections (Mack, Büttner, Rohde, unpublished). Strikingly, using a flow cell model of biofilm formation, anti-Embp antibodies were shown to inhibit S. epidermidis 1457 biofilm formation (Lam et al., 2014), making Embp a potential candidate for preventive strategies (Götz, 2004).
Experimental evidence primarily suggested a role of Embp in primary attachment. Importantly, over-expression of Embp did not alter binding to unmodified polystyrene, but only boosted bacterial adherence to Fn-coated surfaces (Christner et al., 2010), and Embp–Fn interactions were necessary for biofilm accumulation on plastic surfaces that otherwise did not promote bacterial binding (Christner et al., 2010). Results from phage display suggested that FIVAR-GA repeats were relevant to the Fn-binding activity of Embp (Clarke et al., 2002; Williams et al., 2002). This assumption was later validated by biochemical analysis showing direct evidence for interactions between a recombinant protein containing FIVAR-GA repeats (Christner et al., 2010). In addition, these studies also found evidence that FIVAR-modules alone are capable of binding to immobilized Fn (Christner et al., 2010) (Figure 1). The vast majority of bacterial Fn-binding proteins bind to Fn via interactions with the N-terminal Fn type I domains (Bingham et al., 2008; Chagnot et al., 2012). This is especially true for the S. aureus Fn binding protein FnBPA (Meenan et al., 2007). Although the exact mechanism of Embp–Fn interaction awaits definitive molecular analysis, it is already clear that Embp uses an mechanism for Fn interactions independent of type I Fn modules, but involving Fn type III modules located at the C-terminus of Fn (Bustanji et al., 2003), most likely FnIII12−14 (Christner et al., 2010). This type of interaction has only rarely been described in bacterial pathogens (Kingsley et al., 2004; Dabo et al., 2006).
In addition to its function in primary attachment, Embp is also functional as an intercellular adhesin. The intercellular adhesive properties and biofilm inducing activity of Embp was first detected in a laboratory derived strain S. epidermidis 1585v that, by a spontaneous chromosomal rearrangement, overexpressed a truncated isoform of Embp referred to as Embp1 (Christner et al., 2010). A transposon insertion within Embp1 resulted in abolished biofilm formation. By placing an inducible promoter in front of the wild-type embp, a biofilm inducing effect of full length Embp became apparent (Christner et al., 2010), proving the intercellular adhesive properties of Embp. Of notice, up-regulation of embp is also associated with resistance against up-take by professional phagocytes (Schommer et al., 2011). Studies on the overall impact of Embp on S. epidermidis cell wall assembly and its relation to immune-escape will shed light on the question if in this species, the giant protein carries similar functions as compared to Ebh in S. aureus (Cheng et al., 2014).
While at the time of its identification Embp appeared primarily as a factor mediating primary attachment, Aap was initially, and as already suggested by its designation, thought to confer intercellular adhesion and thereby to contribute to biofilm accumulation. Aap is a covalently linked, cell wall associated protein consisting of an A- and a B-domain (Rohde et al., 2005; Gruszka et al., 2012; Conrady et al., 2013; Schaeffer et al., 2015). The 584 aa A domain harbors a N-terminal export signal, several imperfect, 16 amino acid repeats, and a globular 212 amino acid region with predicted α-helical and β-sheet content. The 212 amino acid region is highly conserved between Aap and its S. aureus ortholog SasG, and bioinformatical analysis predicts that this domain possesses lectin-like activity (Schaeffer et al., 2015). The B domain consists of a varying number of repetitive 128 amino acid repeats (Rohde et al., 2007). Variations of B repeats not only exist between independent S. epidermidis strains (e.g., reference strain RP62A possess 13 repeats, while S. epidermidis 1457 only harbors seven repeats) (Monk and Archer, 2007; Schaeffer et al., 2015), but are also encountered in clonally identical clinical isolates subsequently recovered during the course of device infections from individual patients (Rohde et al., 2007). This observation lead to the hypothesis that Aap B repeat variations could represent a mechanism contributing to S. epidermidis immune escape through modification of major cell surface epitopes (Rohde et al., 2007).
Aap can be detected on the bacterial cell wall, where it is most likely retained by covalent linkage to the peptidoglycan via its C-terminal gram-positive anchor region (Hussain et al., 1997; Rohde et al., 2005; Schommer et al., 2011; Conlon et al., 2014). A more detailed analysis using confocal microscopy demonstrated that within living, three dimensional S. epidermidis biofilms, Aap strictly localizes to the bacterial cell surface, while only minimal amounts are released into the biofilm matrix itself (Schommer et al., 2011). These results are underpinned and extended by electron-microscopic studies showing that Aap forms elongated fibers that project 120 nm away from the cell wall in localized tufts (Banner et al., 2007). Recently, using a structural biology approach, the molecular basis of this intriguing spatial organization was determined. An X-ray crystallography derived high resolution model of different recombinant proteins from the B region of S. epidermidis Aap (Conrady et al., 2013) or S. aureus SasG (Gruszka et al., 2012) showed that each B repeat consists of two regions, an approximately 80 aa G5 domain and an approximately 50 aa linker region referred to as E-region that interconnects repetitive G5 domains (Gruszka et al., 2012). The G5 domains each comprise two successive three-stranded β-sheets connected by triple-helix-like regions, while the E region is composed of two β-sheets (Gruszka et al., 2012; Conrady et al., 2013). E sequences fold cooperatively and form interlocking interfaces with G5 domains in a head-to-tail fashion, resulting in a contiguous, elongated, monomeric structure. Although E and G5 domains lack a compact hydrophobic core, G5 domain and multidomain constructs thereof have thermodynamic stabilities only slightly lower than globular proteins of similar size, explaining why Aap could form protruding fibers even under harsh environmental conditions (Gruszka et al., 2012; Conrady et al., 2013).
The functional importance of Aap for S. epidermidis biofilm formation was first recognized during studies in which chemically derived, biofilm-negative mutant M7 of S. epidermidis RP62A was analyzed (Hussain et al., 1997). Mutant M7 failed to express Aap on the cell surface, and antibodies raised against Aap were able to inhibit biofilm formation in biofilm-positive parent strain RP62A (Hussain et al., 1997). Later, Aap was independently picked up in experiments in which cell surface proteins of a clinically significant but biofilm-negative S. epidermidis wild-type strain 5179 were compared with those isolated from a laboratory derived, biofilm-positive revertant of that strain, referred to as 5179-R1. In protein preparations from 5179-R1 reduced amounts of full length Aap were detected, while in parallel, a shorter, roughly 140 kDa Aap isoform became apparent (Rohde et al., 2005). By using mass spectrometry and N-terminal sequencing evidence was created that the 140 kDa isoform mainly consists of repetitive B domain, mapping to aa 596 of the mature Aap protein. Rabbit antiserum raised against recombinantly expressed B domain inhibited biofilm formation by strain 5179-R1, not only directly supporting the idea of a functional involvement of Aap, but moreover indicating the crucial importance of B domain during this process. Indeed, genetic studies corroborated this hypothesis, showing that in trans expression of B domain in biofilm-negative S. epidermidis 1585 and surrogate host S. carnosus TM300 was sufficient to induce a biofilm-positive phenotype (Rohde et al., 2005). Importantly, expression of B domain did not alter the primary adherence properties, but resulted in cell aggregation, showing that in fact, Aap can be regarded as an intercellular adhesin (Rohde et al., 2005; Conrady et al., 2008; Geoghegan et al., 2010). Indeed, the importance of B domain for intercellular adhesion was also described for SasG in an S. aureus background (Geoghegan et al., 2010). The intercellular adhesive properties can be partially explained by Zinc-dependent homodimerization of B domains (Conrady et al., 2008, 2013), however, some evidence suggests the existence of additional, heterotypic interactions involved in Aap B domain mediated biofilm accumulation (Decker et al., unpublished results).
Importantly, expression of full length mature Aap is not sufficient to mediate intercellular adhesion during the second, accumulative phase of biofilm formation, but to become functionally active as an intercellular adhesin, Aap requires proteolytic processing, resulting in the removal of the A domain (Rohde et al., 2005; Geoghegan et al., 2010). Since Aap processing does not regularly occur under in vitro growth conditions (Rohde et al., 2005; Schaeffer et al., 2015), this phenomenon is a reasonable explanation for the finding that Aap-expressing S. epidermidis not necessarily form a biofilm (Rohde et al., 2007).
Although the intercellular adhesive Aap properties were recognized first, there is now mounting evidence supporting a significant role of Aap also in primary attachment to natural epithelial cells or artificial surfaces (Macintosh et al., 2009; Conlon et al., 2014; Schaeffer et al., 2015). Intriguingly, early work already pointed toward a role of domain A in this process, in which Aap B domain most likely is of only minor functional importance (Macintosh et al., 2009). Binding of Aap expressing S. epidermidis NCTC 11047 to squamous epithelial cells was partially inhibited by the addition of recombinant Aap A domain, as was binding of several additional, Aap-positive clinical S. epidermidis strains (Macintosh et al., 2009). Moreover, in trans expression of A domain in surrogate host L. lactis enabled the bacteria to more efficiently adhere to corneocytes as compared to a L. lactis strain that expressed B domain alone (Macintosh et al., 2009), thus providing genetic evidence for a potential role of A domain in colonization of natural skin surfaces. In extension to the work of Macintosh and co-workers, the role of Aap A domain in primary attachment was further refined in two studies addressing the question as to which extent the A domain could also contribute to colonization of abiotic surfaces (Conlon et al., 2014; Schaeffer et al., 2015). In a clinical S. epidermidis isolate CSF41498, expressing an unprocessed mature, i.e., A domain containing Aap on the surface, attachment to polystyrene was almost completely abolished after deletion of aap. In addition, attachment was significantly inhibited by an antiserum raised against Aap A domain, whereas anti-Aap domain B antiserum had no significant impact on adherence capacities of that strain (Conlon et al., 2014). Intriguingly, deletion of aap in S. epidermidis 1457, expressing a processed Aap isoform devoid of the A domain, did not render the adherence capacities of that strain (Schaeffer et al., 2015). Moreover, anti-Aap A domain antiserum had no effect on binding to the surface of S. epidermidis 1457, supporting the idea that indeed, the presence of A domain, i.e., expression of a mature unprocessed Aap, is essential for Aap-mediated surface adherence. Indeed, in trans expression of full length Aap in 1457Δaap resulted in improved bacterial binding (Schaeffer et al., 2015). Direct genetic evidence for an involvement of Aap A domain in staphylococcal plastic adherence is demonstrated by in trans expression of A domain fused to the C-terminal cell wall anchor in S. carnosus TM300, which significantly improved bacterial binding as compared to the wild-type strain or a strain expressing the Aap B domain (Schaeffer et al., 2015). Thus, a new picture of Aap evolved in which the protein plays a bifunctional role in both, the very early primary attachment phase as well as the later accumulative phases of biofilm formation, and that Aap's inherent functionalities are represented by separated and structurally distinct domains within the protein (Rohde et al., 2005; Macintosh et al., 2009; Conlon et al., 2014; Schaeffer et al., 2015).
The detailed analysis of functional molecules contributing to S. epidermidis biofilm formation has clearly revealed that primary attachment as well as biofilm accumulation is mediated by redundantly organized factors with remarkable exclusive properties, but certainly broadly overlapping functional characteristics. The latter aspect is especially true for molecules active during the accumulative phase: production of PIA, Aap or Embp all efficiently induce cell aggregation, ultimately leading to assembly of a biofilm consortium. However, given the common observation that in clinical S. epidermidis isolates genes encoding for the PIA-synthesis machinery (icaADBC), aap, embp, and additional adherence-associated factors are all widely distributed, with a large percentage of strains carrying all three genes (Frebourg et al., 2000; Galdbart et al., 2000; Klug et al., 2003; Arciola et al., 2004; Rohde et al., 2004, 2007; Petrelli et al., 2006), the question is put forward if and how these mechanisms interact, and which consequences co-expression of distinct intercellular adhesins might have for S. epidermidis biofilm accumulation.
Indeed, the idea that distinct S. epidermidis intercellular adhesins cooperate during biofilm assembly is supported by epidemiological work identifying that strains being positive for icaADBC and aap appear to form stronger biofilms compared to strains being positive only for icaADBC or aap (Stevens et al., 2008). Bioinformatic analysis of aap and embp indeed suggests that direct interaction with PIA might be possible. The G5 domains of the Aap B domain are postulated to have N-acetylglucosamine binding activity (Bateman et al., 2005), and similarly, FIVAR regions of Embp are likewise predicted to have sugar binding potential (Christner et al., 2010), making the direct interaction between proteinacous intercellular adhesins and N-acetylglucosamine containing PIA possible. However, so far no experimental data are available that would support this important and interesting hypothesis. At present, it rather appears that parallel expression of specific intercellular adhesins introduces functional redundancy into biofilm accumulation, i.e., the intercellular adhesive properties of distinct adhesins functionally substitute for each other. Experiments, in which the susceptibility of biofilms of Embp-producing S. epidermidis strain 1585v against treatment with proteases was tested, revealed that, while being naturally sensitive against protease activity, additional in trans expression of icaADBC and PIA production protected the S. epidermidis 1585v biofilm from proteolytic breakdown (Christner et al., 2010). On the other hand, while PIA-dependent biofilms are readily disrupted by PIA-degrading enzyme DspB (Kaplan et al., 2004; Rohde et al., 2007), parallel expression of a proteinacous intercellular adhesin Embp and PIA rescued a biofilm-positive phenotype even in the presence of DspB (Christner et al., 2010). Functional substitution is also evident for Aap and PIA: the inactivation of aap in a PIA-producing genetic background had no apparent effect on biofilm formation in S. epidermidis 1457 as tested by conventional crystal violet biofilm assays or confocal laser scanning microscopy, probably because biofilm formation is already maximal in the various assay systems when PIA is expressed alone (Schaeffer et al., 2015). On the other hand, in trans expression of Aap B domain, being sufficient for induction of cell aggregation, induces biofilm formation in a PIA-negative, icaADBC mutant 1457-M10 (Henke and Rohde, unpublished results).
The interpretation of functional redundancy in intercellular adhesive molecules as a simple means allowing S. epidermidis to form as robust biofilms as possible, however, might be an inappropriate oversimplification of their actual functional importance during different S. epidermidis live styles, ranging between colonization and (foreign-material associated) host invasion. In that respect, it is important to acknowledge that a more detailed morphological analysis of PIA-, Aap- or Embp dependent biofilms revealed that these biofilm types differ significantly in their morphological properties. While in PIA-dependent biofilms, S. epidermidis cells are embedded into meshwork of PIA-containing extracellular matrix fibers, Aap production induces formation of densely packed cell layers that evenly cover the surface. In Embp-dependent biofilms, bacteria produce small amounts of Embp-containing extracellular matrix structures, however, these biofilms differ from PIA-dependent cell consortia by the lack of towers and clusters (Schommer et al., 2011). Overall, PIA-dependent biofilms are significantly more stable against washing procedures as compared to protein-dependent biofilms, indicating their inherent, pronounced mechanical robustness. Thus, the specific biological properties of a given intercellular adhesin could constitute a way how S. epidermidis can cope with varying challenges during colonization and infection (Schommer et al., 2011). Analysis of invasive S. epidermidis strains from various types of infections supports the idea that in fact, the ability to differentially make use of specific intercellular adhesins equips S. epidermidis to specifically adapt to changing environments with potential fundamentally different requirements, e.g., presence of mechanical or osmotic stress, or exposure to effectors of the host immune system (Otto, 2014). Specifically, S. epidermidis strains from central venous catheter infections, i.e., a situation with significant exposure to mechanical stress and cellular and soluble factors of innate immunity (e.g., complement factors) are more likely to carry icaADBC compared to strains from prosthetic joint infections, i.e., an infection setting characterized by static conditions at the implant–tissue interface (Ziebuhr et al., 1997; Arciola et al., 2004; Rohde et al., 2004, 2007; Stevens et al., 2008; Mack et al., 2013).
Additional evidence suggesting that usage of specific intercellular adhesins indeed follows an adaptive program results from the observation that PIA-dependent biofilm formation on one hand, and Aap- and Embp-dependent biofilm formation on the other, are, at least under in vitro growth conditions, mutually exclusive (Rohde et al., 2005, 2007; Christner et al., 2010). In fact, under standard growth conditions in trypticase soy broth (TSB), neither Aap nor Embp-dependent biofilm formation would have been identified, since here Aap is not properly processed, while embp is not expressed at all (Rohde et al., 2005; Christner et al., 2010). The characterization of these intercellular adhesins only was possible by analysis of laboratory derived, spontaneous mutants or by introduction of artificial promoters allowing for inducible gene expression, respectively (Christner et al., 2010; Rohde et al., 2005). Thus, PIA- and Aap- or Embp-dependent modes of biofilm formation are obviously under the control of opposed regulatory circuits. For PIA-dependent biofilm formation, a very detailed picture of how icaADBC expression is integrated into a complex superimposed regulatory network has evolved. Others have recently reviewed this field in detail (Mack et al., 2004; Cue et al., 2012). In brief, several regulators of icaADBC expression and PIA synthesis were identified (Xu et al., 2006; Sadykov et al., 2011; Wang et al., 2011b), with sigma factor B and staphylococcal accessory regulator SarA being the most prominent (Fluckiger et al., 1998; Knobloch et al., 2001, 2004; Tormo et al., 2005b; Handke et al., 2007). Especially, negative regulator of icaADBC expression, IcaR, has been elucidated in great detail (Conlon et al., 2002). However, the regulation of PIA-independent mechanism of S. epidermidis biofilm formation remained obscure.
More recently, in an attempt to shed light onto potential negative regulators interfering with biofilm formation in clinical S. epidermidis isolates under in vitro conditions, a transposon mutant library established in biofilm-negative S. epidermidis 1585 was screened for biofilm-positive mutants (Christner et al., 2012). Independent biofilm-positive mutants were identified, and further analysis showed that all carried Tn917 insertions in sarA. Intriguingly, inactivation of sarA induced a strong up-regulation of embp expression, and additional experimental work proved that Embp was necessary for 1585ΔsarA biofilm formation. Moreover, higher eDNA amounts were present in biofilms of 1585ΔsarA, and this finding was related to increased autolysis, itself being a result of over-production of metalloprotease SepA (Lai et al., 2007) and subsequent processing and functional activation of AtlE (Christner et al., 2012). Of notice, protease-mediated autolysin processing, augmented autolysis and subsequent eDNA release and biofilm formation has also been described in Enterococcus faecalis as part of a fratricidal mechanism (Thomas et al., 2008, 2009).
The results of the study by Christner and co-workers appear to contradict previous findings, showing that SarA is a positive regulator of S. epidermidis biofilm formation by augmenting icaADBC expression (Tormo et al., 2005b; Handke et al., 2007). However, a more detailed analysis of SarA function in PIA-positive background of S. epidermidis 1457 showed that in this strain, inactivation of sarA does not completely abolish biofilm formation (Handke et al., 2007). Even inactivation of icaADBC in 1457ΔsarA did not render the strain biofilm-negative, and in fact, the biofilm of 1457-M10ΔsarA is Embp- and eDNA-dependent (Christner et al., 2012). In conclusion, the study by Christner and co-workers establishes a key role for SarA in controlling the mode of biofilm formation in S. epidermidis: up-regulation of sarA shifts S. epidermidis toward production of a PIA-dependent type of biofilm, whereas down-regulation of sarA supports formation of PIA-independent types of biofilm formation (Figure 2). Clearly, the regulation involves transcriptional effects with direct consequences for intercellular aggregation (i.e., embp and icaADBC up- or down-regulation), but also post-translational mechanisms of regulation exerted via up- or down-regulation of metalloprotease SepA with subsequent proteolytic processing and functional modification of cell surface proteins (e.g., AtlE). Importantly, through the latter mode of action SarA could also influence Aap-dependent biofilm formation by boosting proteolytic removal and functional activation of Aap B-domain (Figure 2). In the future it will be of major interest as to which extent the SarA regulatory circuit and interrelated additional regulators, e.g., agr (Vuong et al., 2004b), ssrAB (Wu et al., 2015), saeRS (Lou et al., 2011), or codY (Batzilla et al., 2006) as well as the levels of proteolytic activity influence the balanced formation of PIA-dependent or -independent types of biofilm formation.
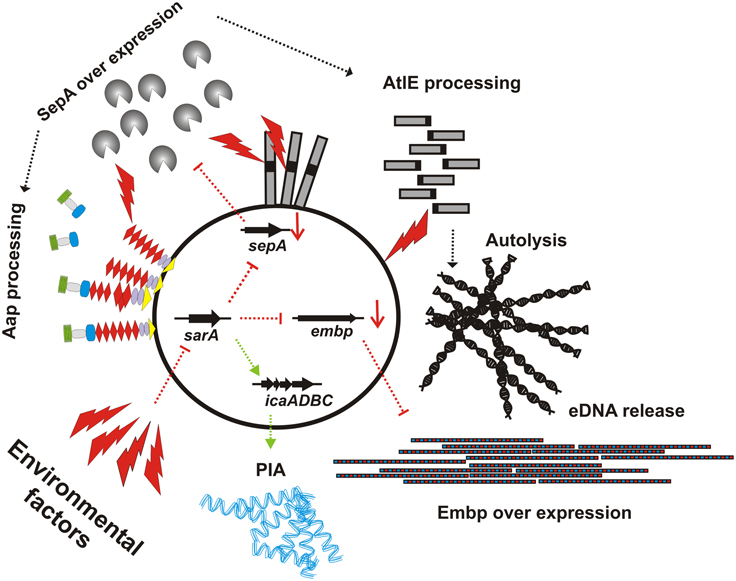
Figure 2. Schematic representation of SarA effects on expression of independent intercellular adhesins. While in trypticase soy broth, sarA is expressed, leading to icaADBC expression and PIA dependent biofilm formation, down-regulation of sarA leads to an de-repression of embp expression, allowing for maintenance of a biofilm-positive phenotype despite icaADBC down-regulation and loss of PIA production. Parallel to embp, following inactivation of sarA, metalloprotease sepA is up-regulated, leading to increased AtlE processing, autolysis and subsequent eDNA release. Potentially, the over-production of SepA also contributes to Aap-dependent biofilm accumulation by proteolytically processing of the mature protein, resulting in the removal of it's A domain. Environmental (potentially also host) factors that repress sarA expression are unknown, as are the pathways via which sarA itself is regulated.
Certainly, the highly artificial character of studies related to the function of SarA makes it difficult to draw general conclusions on the relevance of this mechanism during in vivo biofilm accumulation. In that respect, it becomes clearer that that more appropriate models of biofilm formation are generally needed in order to reflect the in vivo situation during infection and colonization. These models must take into account the potential inappropriateness of TSB as a growth medium that obviously introduces a significant bias toward PIA-dependent biofilm formation into the experimental system. The finding that embp, but also additional factors involved in S. epidermidis biofilm formation (e.g., Fbe/SdrG) are only expressed in the presence of serum (Sellman et al., 2005, 2008; Christner et al., 2010), clearly argues for experimental set-ups that mimic environmental conditions in ecological niches usually encountered by S. epidermidis, e.g., within the vestibulum nasi (Krismer et al., 2014) or on the skin (Ohnemus et al., 2007; Olson et al., 2014).
In general, studies aiming at elucidating the clinical relevance of biofilm formation and the relative contribution of specific factors to foreign-material colonization and establishment of a chronic persistent infection are either studied using cell culture models (Vuong et al., 2004a,c; Schommer et al., 2011), Caenorhabditis elegans (Begun et al., 2007) or animal models of device infections, e.g., central venous catheter or prosthetic device infection models (Wang et al., 2011a; Odekerken et al., 2013; Scherr et al., 2014b; Schaeffer et al., 2015). In early studies on the importance of biofilm formation in vivo using animal models and genetically unrelated, biofilm-positive or biofilm-negative S. epidermidis isolates failed to demonstrate conclusive evidence that biofilm forming isolates are more virulent compared to biofilm-negative strains. However, subsequent studies using genetically defined, isogenic pairs of biofilm-positive wild-type and biofilm-negative mutants were more conclusive. In a subcutaneous catheter infection model in mice and a central venous catheter infection model in rats biofilm-positive, PIA-producing S. epidermidis 1457 was more virulent than its isogenic biofilm-negative transductant 1457-M10 (Rupp et al., 1999a,b). An AtlE-deletion mutant of this strain was also attenuated (Rupp et al., 2001). In a rat central venous catheter model, expression of icaRADBC in icaADBC-negative S. epidermidis strains also led to increased virulence (Li et al., 2005). A Caenorhabditis elegans infection model was used to study biofilm-positive S. epidermidis 9142, showing attenuation of an icaA insertion mutant compared to its isogenic, PIA producing parent strain 9142 (Begun et al., 2007). Virulence was restored to wild-type in the biofilm-negative mutant by complementation with cloned icaADBC (Begun et al., 2007). However, in a collection of S. epidermidis infective endocarditis isolates, PIA expression and pathogenicity for C. elegans was not closely associated (Monk et al., 2008). More recently, using a catheter infection model in which realistic colonization modalities were chosen (i.e., infection after catheter insertion and not usage of pre-colonized materials) the inactivation of icaADBC had no apparent effect on colonization, while aap inactivation almost completely abolished the ability of S. epidermidis to establish an infection (Schaeffer et al., 2015).
A significant reason for the impaired pathogenicity of icaADBC-negative mutants in animals models is the improved ability of the innate immune system to clear biofilm-negative S. epidermidis (Schommer et al., 2011). There is significant experimental evidence from cell culture assays that indeed biofilm positive strain 1457 was less susceptible to killing by antimicrobial peptides and also displayed decreased phagocytosis and killing by polymorphonuclear granulocytes (PMNs) compared to its isogenic icaA mutant 1457-M10 (Vuong et al., 2004c). When S. epidermidis 1457 was either grown in a static biofilm or planktonic culture, the organism grown in a biofilm was less susceptible to phagocytic killing after opsonisation with normal human serum, as was an isogenic biofilm-negative icaA-insertion mutant (Kristian et al., 2008). PIA-dependent biofilm formation also interferes with host complement activation. Biofilm-positive wild-type bacteria pre-opsonised with normal human serum were more resistant to complement-dependent killing than the corresponding isogenic biofilm-negative bacteria (Kristian et al., 2008). There is, moreover, evidence that S. epidermidis biofilm formation interferes with phagocytic up-take and with pro-inflammatory activation of macrophages. This effect was irrespective of the intercellular adhesin used (Schommer et al., 2011). These phenotypes clearly could additionally contribute to the chronic persistent, low-grade inflammatory course of a S. epidermidis infection.
It is important to stress that, since S. epidermidis is an opportunistic pathogen, mechanisms of pathogenicity which are important in some types of device-related infection might be less crucial in others. For example, in the guinea pig tissue cage model (Zimmerli et al., 1982) there was no difference in virulence between a biofilm-positive wild-type S. epidermidis 1457 and its isogenic icaA-insertion mutant, and no difference between icaADBC-positive and -negative clinical isolates (Francois et al., 2003; Chokr et al., 2007). Nonetheless PIA was expressed in vivo in the tissue cages, and when animals were infected with both strains at the same time, the wild-type out-competed the mutant (Fluckiger et al., 2005). This may be because phagocytes are severely impaired in tissue cages (Zimmerli et al., 1984), masking the expected advantage of the wild-type.
S. epidermidis produces a number of pro-inflammatory peptides called phenol-soluble modulins (PSMs), which are produced in a strictly agr-controlled manner (Mehlin et al., 1999; Yao et al., 2005). PSM-δ rapidly lyses neutrophils, supporting the idea that the peptide is of relevance to the pathogenesis of S. epidermidis. However, PSM-δ is expressed only at low levels by S. epidermidis 1457, in line with a low overall cytolytic activity of S. epidermidis (Cheung et al., 2010). PSM-δ is expressed only at very low levels in S. epidermidis 1457 biofilms as compared to planktonic cells (Wang et al., 2011a). PSM-β peptides promote S. epidermidis biofilm structuring and detachment in vitro and dissemination of infection during catheter colonization in vivo, thereby providing the first mechanism of biofilm detachment in S. epidermidis (Wang et al., 2011a).
Over the past two decades, significant progress has been made in our understanding of the specific pathogenic nature of S. epidermidis in foreign material-associated infections. A molecular picture evolved showing that S. epidermidis virulence is linked to biofilm formation, a phenotype that depends on a wide variety of different factors which carry distinct and tightly regulated functions during surface colonization and interactions with host immune responses. Thus, S. epidermidis biofilm research has reached a turning point, at which on one hand additional in vitro evidence for the involvement of dedicated mechanisms in surface colonization can easily be accumulated, but on the other hand the question of the in vivo relevance of a given factor or process certainly arises. Thus, a major future challenge will be to translate findings from highly artificial, simple in vitro biofilm analysis systems into complex (organ-) models that more appropriately reflect the in vivo infection settings. Moreover, it is of urgent importance to validate findings from in vitro models in relevant animal models of device infections. These approaches should involve not only state of the art molecular biology, biochemical and immunological methods but also time-resolved in vivo and ex vivo imaging technologies, allowing to create a more distinct picture of the invasive S. epidermidis life style in different and extremely variable environmental conditions. Using the new armament of technologies, including three-dimensional cell culture techniques and tissue engineering, efforts are necessary to study the role of S. epidermidis as a beneficial skin commensal more intensively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Allesen-Holm, M., Barken, K. B., Yang, L., Klausen, M., Webb, J. S., Kjelleberg, S., et al. (2006). A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 59, 1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arciola, C. R., Campoccia, D., Gamberini, S., Rizzi, S., Donati, M. E., Baldassarri, L., et al. (2004). Search for the insertion element IS256 within the ica locus of Staphylococcus epidermidis clinical isolates collected from biomaterial-associated infections. Biomaterials 25, 4117–4125. doi: 10.1016/j.biomaterials.2003.11.027
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arrecubieta, C., Asai, T., Bayern, M., Loughman, A., Fitzgerald, J. R., Shelton, C. E., et al. (2006). The role of Staphylococcus aureus adhesins in the pathogenesis of ventricular assist device-related infections. J. Infect. Dis. 193, 1109–1119. doi: 10.1086/501366
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arrecubieta, C., Lee, M. H., Macey, A., Foster, T. J., and Lowy, F. D. (2007). SdrF, a Staphylococcus epidermidis surface protein, binds type I collagen. J. Biol. Chem. 282, 18767–18776. doi: 10.1074/jbc.M610940200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Arrecubieta, C., Toba, F. A., von Bayern, M., Akashi, H., Deng, M. C., Naka, Y., et al. (2009). SdrF, a Staphylococcus epidermidis surface protein, contributes to the initiation of ventricular assist device driveline-related infections. PLoS. Pathog. 5:e1000411. doi: 10.1371/journal.ppat.1000411
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Banner, M. A., Cunniffe, J. G., Macintosh, R. L., Foster, T. J., Rohde, H., Mack, D., et al. (2007). Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation-associated protein. J. Bacteriol. 189, 2793–2804. doi: 10.1128/JB.00952-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bateman, A., Holden, M. T., and Yeats, C. (2005). The G5 domain: a potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics 21, 1301–1303. doi: 10.1093/bioinformatics/bti206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Batzilla, C. F., Rachid, S., Engelmann, S., Hecker, M., Hacker, J., and Ziebuhr, W. (2006). Impact of the accessory gene regulatory system (Agr) on extracellular proteins, codY expression and amino acid metabolism in Staphylococcus epidermidis. Proteomics 6, 3602–3613. doi: 10.1002/pmic.200500732
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Begun, J., Gaiani, J. M., Rohde, H., Mack, D., Calderwood, S. B., Ausubel, F. M., et al. (2007). Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS. Pathog. 3:e57. doi: 10.1371/journal.ppat.0030057
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bingham, R. J., Rudino-Pinera, E., Meenan, N. A., Schwarz-Linek, U., Turkenburg, J. P., Hook, M., et al. (2008). Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc. Natl. Acad. Sci. U.S.A. 105, 12254–12258. doi: 10.1073/pnas.0803556105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Biswas, R., Voggu, L., Simon, U. K., Hentschel, P., Thumm, G., and Gotz, F. (2006). Activity of the major staphylococcal autolysin Atl. FEMS Microbiol. Lett. 259, 260–268. doi: 10.1111/j.1574-6968.2006.00281.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bowden, M. G., Heuck, A. P., Ponnuraj, K., Kolosova, E., Choe, D., Gurusiddappa, S., et al. (2008). Evidence for the “dock, lock, and latch” ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG. J. Biol. Chem. 283, 638–647. doi: 10.1074/jbc.M706252200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bowden, M. G., Visai, L., Longshaw, C. M., Holland, K. T., Speziale, P., and Hook, M. (2002). Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J. Biol. Chem. 277, 43017–43023. doi: 10.1074/jbc.M207921200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Broekhuizen, C. A., Schultz, M. J., van der Wal, A. C., Boszhard, L., de Boer, L., Vandenbroucke-Grauls, C. M., et al. (2008). Tissue around catheters is a niche for bacteria associated with medical device infection. Crit. Care Med. 36, 2395–2402. doi: 10.1097/CCM.0b013e3181818268
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bustanji, Y., Arciola, C. R., Conti, M., Mandello, E., Montanaro, L., and Samori, B. (2003). Dynamics of the interaction between a fibronectin molecule and a living bacterium under mechanical force. Proc. Natl. Acad. Sci. U.S.A. 100, 13292–13297. doi: 10.1073/pnas.1735343100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chagnot, C., Listrat, A., Astruc, T., and Desvaux, M. (2012). Bacterial adhesion to animal tissues: protein determinants for recognition of extracellular matrix components. Cell. Microbiol. 14, 1687–1696. doi: 10.1111/cmi.12002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, A. G., Missiakas, D., and Schneewind, O. (2014). The giant protein Ebh is a determinant of Staphylococcus aureus cell size and complement resistance. J. Bacteriol. 196, 971–981. doi: 10.1128/JB.01366-13
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheung, G. Y., Rigby, K., Wang, R., Queck, S. Y., Braughton, K. R., Whitney, A. R., et al. (2010). Staphylococcus epidermidis strategies to avoid killing by human neutrophils. PLoS. Pathog. 6:e1001133. doi: 10.1371/journal.ppat.1001133
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chokr, A., Leterme, D., Watier, D., and Jabbouri, S. (2007). Neither the presence of ica locus, nor in vitro-biofilm formation ability is a crucial parameter for some Staphylococcus epidermidis strains to maintain an infection in a guinea pig tissue cage model. Microb. Pathog. 42, 94–97. doi: 10.1016/j.micpath.2006.09.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Christner, M., Franke, G. C., Schommer, N. N., Wendt, U., Wegert, K., Pehle, P., et al. (2010). The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol. Microbiol. 75, 187–207. doi: 10.1111/j.1365-2958.2009.06981.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Christner, M., Heinze, C., Busch, M., Franke, G., Hentschke, M., Bayard, D. S., et al. (2012). sarA negatively regulates Staphylococcus epidermidis biofilm formation by modulating expression of 1 MDa extracellular matrix binding protein and autolysis-dependent release of eDNA. Mol. Microbiol. 86, 394–410. doi: 10.1111/j.1365-2958.2012.08203.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chu, V. H., Cabell, C. H., Abrutyn, E., Corey, G. R., Hoen, B., Miro, J. M., et al. (2004). Native valve endocarditis due to coagulase-negative staphylococci: report of 99 episodes from the International Collaboration on Endocarditis Merged Database. Clin. Infect. Dis. 39, 1527–1530. doi: 10.1086/424878
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chu, V. H., Miro, J. M., Hoen, B., Cabell, C. H., Pappas, P. A., Jones, P., et al. (2009). Coagulase-negative staphylococcal prosthetic valve endocarditis–a contemporary update based on the International Collaboration on Endocarditis: prospective cohort study. Heart 95, 570–576. doi: 10.1136/hrt.2008.152975
Clarke, S. R., Harris, L. G., Richards, R. G., and Foster, S. J. (2002). Analysis of Ebh, a 1.1-megadalton cell wall-associated fibronectin-binding protein of Staphylococcus aureus. Infect. Immun. 70, 6680–6687. doi: 10.1128/IAI.70.12.6680-6687.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conlon, B. P., Geoghegan, J. A., Waters, E. M., McCarthy, H., Rowe, S. E., Davies, J. R., et al. (2014). Role for the A domain of unprocessed accumulation-associated protein (Aap) in the attachment phase of the Staphylococcus epidermidis biofilm phenotype. J. Bacteriol. 196, 4268–4275. doi: 10.1128/JB.01946-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conlon, K. M., Humphreys, H., and O'Gara, J. P. (2002). Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol. Lett. 216, 171–177. doi: 10.1111/j.1574-6968.2002.tb11432.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conrady, D. G., Brescia, C. C., Horii, K., Weiss, A. A., Hassett, D. J., and Herr, A. B. (2008). A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U.S.A. 105, 19456–19461. doi: 10.1073/pnas.0807717105
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Conrady, D. G., Wilson, J. J., and Herr, A. B. (2013). Structural basis for Zn2+-dependent intercellular adhesion in staphylococcal biofilms. Proc. Natl. Acad. Sci. U.S.A. 110, E202–E211. doi: 10.1073/pnas.1208134110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Costerton, J. W., Lewandowski, Z., Caldwell, D. E., Korber, D. R., and Lappin-Scott, H. M. (1995). Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745. doi: 10.1146/annurev.mi.49.100195.003431
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Costerton, J. W., Stewart, P. S., and Greenberg, E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322. doi: 10.1126/science.284.5418.1318
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cue, D., Lei, M. G., and Lee, C. Y. (2012). Genetic regulation of the intercellular adhesion locus in staphylococci. Front Cell Infect. Microbiol. 2:38. doi: 10.3389/fcimb.2012.00038
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dabo, S. M., Confer, A. W., Anderson, B. E., and Gupta, S. (2006). Bartonella henselae Pap31, an extracellular matrix adhesin, binds the fibronectin repeat III13 module. Infect. Immun. 74, 2513–2521. doi: 10.1128/IAI.74.5.2513-2521.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Darouiche, R. O. (2004). Treatment of infections associated with surgical implants. N. Engl. J. Med. 350, 1422–1429. doi: 10.1056/NEJMra035415
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fey, P. D., Ulphani, J. S., Götz, F., Heilmann, C., Mack, D., and Rupp, M. E. (1999). Characterization of the relationship between polysaccharide intercellular adhesin and hemagglutination in Staphylococcus epidermidis. J. Infect. Dis. 179, 1561–1564. doi: 10.1086/314762
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fluckiger, U., Ulrich, M., Steinhuber, A., Doring, G., Mack, D., Landmann, R., et al. (2005). Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 73, 1811–1819. doi: 10.1128/IAI.73.3.1811-1819.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fluckiger, U., Wolz, C., and Cheung, A. L. (1998). Characterization of a sar homolog of Staphylococcus epidermidis. Infect. Immun. 66, 2871–2878.
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., and Hook, M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62. doi: 10.1038/nrmicro3161
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Francois, P., Tu Quoc, P. H., Bisognano, C., Kelley, W. L., Lew, D. P., Schrenzel, J., et al. (2003). Lack of biofilm contribution to bacterial colonisation in an experimental model of foreign body infection by Staphylococcus aureus and Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 35, 135–140. doi: 10.1016/S0928-8244(02)00463-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Frebourg, N. B., Lefebvre, S., Baert, S., and Lemeland, J. F. (2000). PCR-Based assay for discrimination between invasive and contaminating Staphylococcus epidermidis strains. J. Clin. Microbiol. 38, 877–880.
Galdbart, J. O., Allignet, J., Tung, H. S., Ryden, C., and El, S. N. (2000). Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J. Infect. Dis. 182, 351–355. doi: 10.1086/315660
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geffers, C., and Gastmeier, P. (2011). Nosocomial infections and multidrug-resistant organisms in Germany: epidemiological data from KISS (the Hospital Infection Surveillance System). Dtsch. Arztebl. Int. 108, 87–93. doi: 10.3238/arztebl.2011.0087
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Geipel, U., and Herrmann, M. (2005). The infected implant: bacteriology. Unfallchirurg 108, 961–975.
Geoghegan, J. A., Corrigan, R. M., Gruszka, D. T., Speziale, P., O'Gara, J. P., Potts, J. R., et al. (2010). Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J. Bacteriol. 192, 5663–5673. doi: 10.1128/JB.00628-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gerke, C., Kraft, A., Süssmuth, R., Schweitzer, O., and Götz, F. (1998). Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273, 18586–18593. doi: 10.1074/jbc.273.29.18586
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gill, S. R., Fouts, D. E., Archer, G. L., Mongodin, E. F., Deboy, R. T., Ravel, J., et al. (2005). Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187, 2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Goldmann, D. A., and Pier, G. B. (1993). Pathogenesis of infections related to intravascular catheterization. Clin. Microbiol. Rev. 6, 176–192.
Götz, F. (2002). Staphylococcus and biofilms. Mol. Microbiol. 43, 1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Götz, F. (2004). Staphylococci in colonization and disease: prospective targets for drugs and vaccines. Curr. Opin. Microbiol. 7, 477–487. doi: 10.1016/j.mib.2004.08.014
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Grice, E. A., and Segre, J. A. (2011). The skin microbiome. Nat. Rev. Microbiol. 9, 244–253. doi: 10.1038/nrmicro2537
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gruszka, D. T., Wojdyla, J. A., Bingham, R. J., Turkenburg, J. P., Manfield, I. W., Steward, A., et al. (2012). Staphylococcal biofilm-forming protein has a contiguous rod-like structure. Proc. Natl. Acad. Sci. U.S.A. 109, E1011–E1018. doi: 10.1073/pnas.1119456109
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Handke, L. D., Slater, S. R., Conlon, K. M., O'Donnell, S. T., Olson, M. E., Bryant, K. A., et al. (2007). SigmaB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can. J. Microbiol. 53, 82–91. doi: 10.1139/w06-108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hartford, O., O'Brien, L., Schofield, K., Wells, J., and Foster, T. J. (2001). The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology 147, 2545–2552.
Heilmann, C., Hussain, M., Peters, G., and Götz, F. (1997). Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24, 1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heilmann, C., Schweitzer, O., Gerke, C., Vanittanakom, N., Mack, D., and Götz, F. (1996). Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Herman, P., El-Kirat-Chatel, S., Beaussart, A., Geoghegan, J. A., Foster, T. J., and Dufrene, Y. F. (2014). The binding force of the staphylococcal adhesin SdrG is remarkably strong. Mol. Microbiol. 93, 356–368. doi: 10.1111/mmi.12663
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hidron, A. I., Edwards, J. R., Patel, J., Horan, T. C., Sievert, D. M., Pollock, D. A., et al. (2008). NHSN annual update: antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29, 996–1011. doi: 10.1086/591861
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hussain, M., Hastings, J. G., and White, P. J. (1991). Isolation and composition of the extracellular slime made by coagulase-negative staphylococci in a chemically defined medium. J. Infect. Dis. 163, 534–541. doi: 10.1093/infdis/163.3.534
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hussain, M., Herrmann, M., von Eiff, C., Perdreau-Remington, F., and Peters, G. (1997). A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65, 519–524.
Iwase, T., Uehara, Y., Shinji, H., Tajima, A., Seo, H., Takada, K., et al. (2010). Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349. doi: 10.1038/nature09074
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Izano, E. A., Amarante, M. A., Kher, W. B., and Kaplan, J. B. (2008). Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl. Environ. Microbiol. 74, 470–476. doi: 10.1128/AEM.02073-07
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Josefsson, E., McCrea, K. W., Ni, E. D., O'Connell, D., Cox, J., Hook, M., et al. (1998). Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144(Pt 12), 3387–3395. doi: 10.1099/00221287-144-12-3387
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jukes, L., Mikhail, J., Bome-Mannathoko, N., Hadfield, S. J., Harris, L. G., El-Bouri, K., et al. (2010). Rapid differentiation of Staphylococcus aureus, Staphylococcus epidermidis and other coagulase-negative staphylococci and meticillin susceptibility testing directly from growth-positive blood cultures by multiplex real-time PCR. J. Med. Microbiol. 59, 1456–1461. doi: 10.1099/jmm.0.023168-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kaplan, J. B., Ragunath, C., Velliyagounder, K., Fine, D. H., and Ramasubbu, N. (2004). Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 48, 2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kingsley, R. A., Keestra, A. M., de Zoete, M. R., and Baumler, A. J. (2004). The ShdA adhesin binds to the cationic cradle of the fibronectin 13FnIII repeat module: evidence for molecular mimicry of heparin binding. Mol. Microbiol. 52, 345–355. doi: 10.1111/j.1365-2958.2004.03995.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kloos, W. E. (1980). Natural populations of the genus Staphylococcus. Annu. Rev. Microbiol. 34, 559–592. doi: 10.1146/annurev.mi.34.100180.003015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kloos, W. E. (1997). “Taxonomy and systematics of staphylococci indigenous to humans,” in The Staphylococci in Human Disease, eds K. B. Crossley, and G. L. Archer. (New York, NY: Churchill Livingston), 113–137.
Kloos, W. E., and Schleifer, K. H. (1986). “Genus IV. Staphylococcus,” in Bergey's Manual of Systematic Bacteriology, Vol. 2, ed H. A. Sneath (Baltimore, MD: Williams and Wilkins), 1013–1035.
Klug, D., Wallet, F., Kacet, S., and Courcol, R. J. (2003). Involvement of adherence and adhesion Staphylococcus epidermidis genes in pacemaker lead-associated infections. J. Clin. Microbiol. 41, 3348–3350. doi: 10.1128/JCM.41.7.3348-3350.2003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Knobloch, J. K., Jager, S., Horstkotte, M. A., Rohde, H., and Mack, D. (2004). RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor sigmaB by repression of the negative regulator gene icaR. Infect. Immun. 72, 3838–3848. doi: 10.1128/IAI.72.7.3838-3848.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Knobloch, J. K. M., Bartscht, K., Sabottke, A., Rohde, H., Feucht, H. H., and Mack, D. (2001). Biofilm formation of Staphylococcus epidermidis depends on RsbU, a functional activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183, 2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kockro, R. A., Hampl, J. A., Jansen, B., Peters, G., Scheihing, M., Giacomelli, R., et al. (2000). Use of scanning electron microscopy to investigate the prophylactic efficacy of rifampin-impregnated CSF shunt catheters. J. Med. Microbiol. 49, 441–450.
Krismer, B., Liebeke, M., Janek, D., Nega, M., Rautenberg, M., Hornig, G., et al. (2014). Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS. Pathog. 10:e1003862. doi: 10.1371/journal.ppat.1003862
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kristian, S. A., Birkenstock, T. A., Sauder, U., Mack, D., Götz, F., and Landmann, R. (2008). Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J. Infect. Dis. 197, 1028–1035. doi: 10.1086/528992
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kuroda, M., Tanaka, Y., Aoki, R., Shu, D., Tsumoto, K., and Ohta, T. (2008). Staphylococcus aureus giant protein Ebh is involved in tolerance to transient hyperosmotic pressure. Biochem. Biophys. Res. Commun. 374, 237–241. doi: 10.1016/j.bbrc.2008.07.037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lai, Y., Villaruz, A. E., Li, M., Cha, D. J., Sturdevant, D. E., and Otto, M. (2007). The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol. Microbiol. 63, 497–506. doi: 10.1111/j.1365-2958.2006.05540.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lalani, T., Kanafani, Z. A., Chu, V. H., Moore, L., Corey, G. R., Pappas, P., et al. (2006). Prosthetic valve endocarditis due to coagulase-negative staphylococci: findings from the International Collaboration on Endocarditis Merged Database. Eur. J. Clin. Microbiol. Infect. Dis. 25, 365–368. doi: 10.1007/s10096-006-0141-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lam, H., Kesselly, A., Stegalkina, S., Kleanthous, H., and Yethon, J. A. (2014). Antibodies to PhnD inhibit staphylococcal biofilms. Infect. Immun. 82, 3764–3774. doi: 10.1128/IAI.02168-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lewis, K. (2005). Persister cells and the riddle of biofilm survival. Biochemistry (Mosc.) 70, 267–274. doi: 10.1007/s10541-005-0111-6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lewis, K. (2010). Persister cells. Annu. Rev. Microbiol. 64, 357–372. doi: 10.1146/annurev.micro.112408.134306
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, H., Xu, L., Wang, J., Wen, Y., Vuong, C., Otto, M., et al. (2005). Conversion of Staphylococcus epidermidis strains from commensal to invasive by expression of the ica locus encoding production of biofilm exopolysaccharide. Infect. Immun. 73, 3188–3191. doi: 10.1128/IAI.73.5.3188-3191.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Linke, C., Siemens, N., Oehmcke, S., Radjainia, M., Law, R. H., Whisstock, J. C., et al. (2012). The extracellular protein factor Epf from Streptococcus pyogenes is a cell surface adhesin that binds to cells through an N-terminal domain containing a carbohydrate-binding module. J. Biol. Chem. 287, 38178–38189. doi: 10.1074/jbc.M112.376434
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lou, Q., Zhu, T., Hu, J., Ben, H., Yang, J., Yu, F., et al. (2011). Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation. BMC Microbiol. 11:146. doi: 10.1186/1471-2180-11-146
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Macintosh, R. L., Brittan, J. L., Bhattacharya, R., Jenkinson, H. F., Derrick, J., Upton, M., et al. (2009). The terminal A domain of the fibrillar accumulation-associated protein (Aap) of Staphylococcus epidermidis mediates adhesion to human corneocytes. J. Bacteriol. 191, 7007–7016. doi: 10.1128/JB.00764-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mack, D., Becker, P., Chatterjee, I., Knobloch, J. K. M., Peters, G., Rohde, H., et al. (2004). Mechanisms of biofilm formation in Staphylococcus epidermidis and Staphylococcus aureus: functional molecules, regulatory circuits, and adaptive responses. Int. J. Med. Microbiol. 294, 203–212. doi: 10.1016/j.ijmm.2004.06.015
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mack, D., Davies, A. P., Harris, L. G., Jeeves, R., Pascoe, B., Knobloch, J. K. M., et al. (2013). “Staphylococcus epidermidis in biomaterial associated infections,” in Biomaterials Associated Infections: Immunological Aspects and Antimicrobial Strategies, eds T. F. Moriatry, S. A. Zaat, and H. J. Busscher (New York, NY: Springer), 25–56.
Mack, D., Davies, A. P., Harris, L. G., Knobloch, J. K., and Rohde, H. (2009). Staphylococcus epidermidis biofilms: functional molecules, relation to virulence, and vaccine potential. Top. Curr. Chem. 288, 157–182. doi: 10.1007/128_2008_19
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mack, D., Fischer, W., Krokotsch, A., Leopold, K., Hartmann, R., Egge, H., et al. (1996). The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178, 175–183.
Mack, D., Horstkotte, M. A., Rohde, H., and Knobloch, J. K. M. (2006). “Coagulase-negative staphylococci,” in Biofilms, Infection, and Antimicrobial Therapy, eds J. L. Pace, M. E. Rupp, and R. G. Finch (Boca Raton, FL: CRC Press), 109–153.
Mack, D., Nedelmann, M., Krokotsch, A., Schwarzkopf, A., Heesemann, J., and Laufs, R. (1994a). Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62, 3244–3253.
Mack, D., Riedewald, J., Rohde, H., Magnus, T., Feucht, H. H., Elsner, H. A., et al. (1999). Essential functional role of the polysaccharide intercellular adhesin of Staphylococcus epidermidis in hemagglutination. Infect. Immun. 67, 1004–1008.
Mack, D., Siemssen, N., and Laufs, R. (1994b). Identification of a cell cluster associated antigen specific for plastic-adherent Staphylococcus epidermidis which is functional related to intercellular adhesion. Zentralbl. Bakteriol. Suppl. 26, 411–413.
Mann, E. E., Rice, K. C., Boles, B. R., Endres, J. L., Ranjit, D., Chandramohan, L., et al. (2009). Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PloS ONE 4:e5822. doi: 10.1371/journal.pone.0005822
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCrea, K. W., Hartford, O., Davis, S., Eidhin, D. N., Lina, G., Speziale, P., et al. (2000). The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology 146(Pt 7), 1535–1546.
Meenan, N. A., Visai, L., Valtulina, V., Schwarz-Linek, U., Norris, N. C., Gurusiddappa, S., et al. (2007). The tandem beta-zipper model defines high affinity fibronectin-binding repeats within Staphylococcus aureus FnBPA. J. Biol. Chem. 282, 25893–25902. doi: 10.1074/jbc.M703063200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mehlin, C., Headley, C. M., and Klebanoff, S. J. (1999). An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J. Exp. Med. 189, 907–918. doi: 10.1084/jem.189.6.907
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Monk, A. B., and Archer, G. L. (2007). Use of outer surface protein repeat regions for improved genotyping of Staphylococcus epidermidis. J. Clin. Microbiol. 45, 730–735. doi: 10.1128/JCM.02317-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Monk, A. B., Boundy, S., Chu, V. H., Bettinger, J. C., Robles, J. R., Fowler, V. G. Jr., et al. (2008). Analysis of the genotype and virulence of Staphylococcus epidermidis isolates from patients with infective endocarditis. Infect. Immun. 76, 5127–5132. doi: 10.1128/IAI.00606-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Moormeier, D. E., Bose, J. L., Horswill, A. R., and Bayles, K. W. (2014). Temporal and stochastic control of Staphylococcus aureus biofilm development. MBio 5:e01341-14. doi: 10.1128/mBio.01341-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Murdoch, D. R., Corey, G. R., Hoen, B., Miro, J. M., Fowler, V. G. Jr. Bayer, A. S., et al. (2009). Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: the International Collaboration on Endocarditis-Prospective Cohort Study. Arch. Intern. Med. 169, 463–473. doi: 10.1001/archinternmed.2008.603
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nickinson, R. S., Board, T. N., Gambhir, A. K., Porter, M. L., and Kay, P. R. (2010). The microbiology of the infected knee arthroplasty. Int. Orthop. 34, 505–510. doi: 10.1007/s00264-009-0797-y
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nilsson, M., Frykberg, L., Flock, J. I., Pei, L., Lindberg, M., and Guss, B. (1998). A fibrinogen-binding protein of Staphylococcus epidermidis. Infect. Immun. 66, 2666–2673.
Odekerken, J. C., Arts, J. J., Surtel, D. A., Walenkamp, G. H., and Welting, T. J. (2013). A rabbit osteomyelitis model for the longitudinal assessment of early post-operative implant infections. J. Orthop. Surg. Res. 8, 38. doi: 10.1186/1749-799X-8-38
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ohnemus, U., Kohrmeyer, K., Houdek, P., Rohde, H., Wladykowski, E., Vidal, S., et al. (2007). Regulation of epidermal tight-junctions (TJ) during infection with exfoliative toxin-negative Staphylococcus strains. J. Invest. Dermatol.
Olson, M. E., Nygaard, T. K., Ackermann, L., Watkins, R. L., Zurek, O. W., Pallister, K. B., et al. (2013). Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect. Immun. 81, 1316–1324. doi: 10.1128/IAI.01242-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Olson, M. E., Todd, D. A., Schaeffer, C. R., Paharik, A. E., Van Dyke, M. J., Buttner, H., et al. (2014). The Staphylococcus epidermidis agr quorum-sensing system: signal identification, cross talk, and importance in colonization. J. Bacteriol. 196, 3482–3493. doi: 10.1128/JB.01882-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Otto, M. (2009). Staphylococcus epidermidis–the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7, 555–567. doi: 10.1038/nrmicro2182
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Otto, M. (2014). Physical stress and bacterial colonization. FEMS Microbiol. Rev. 38, 1250–1270. doi: 10.1111/1574-6976.12088
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Patti, J. M., Allen, B. L., McGavin, M. J., and Höök, M. (1994). MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48, 585–617. doi: 10.1146/annurev.mi.48.100194.003101
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pei, L., Palma, M., Nilsson, M., Guss, B., and Flock, J. I. (1999). Functional studies of a fibrinogen binding protein from Staphylococcus epidermidis. Infect. Immun. 67, 4525–4530.
Peters, G., Locci, R., and Pulverer, G. (1981). Microbial colonization of prosthetic devices. II. Scanning electron microscopy of naturally infected intravenous catheters. Zentralbl. Bakteriol. Mikrobiol. Hyg. B 173, 293–299.
Peters, G., Locci, R., and Pulverer, G. (1982). Adherence and growth of coagulase-negative staphylococci on surfaces of intravenous catheters. J. Infect. Dis. 146, 479–482. doi: 10.1093/infdis/146.4.479
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Petrelli, D., Zampaloni, C., D'Ercole, S., Prenna, M., Ballarini, P., Ripa, S., et al. (2006). Analysis of different genetic traits and their association with biofilm formation in Staphylococcus epidermidis isolates from central venous catheter infections. Eur. J. Clin. Microbiol. Infect. Dis. 25, 773–781. doi: 10.1007/s10096-006-0226-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Phillips, J. E., Crane, T. P., Noy, M., Elliott, T. S., and Grimer, R. J. (2006). The incidence of deep prosthetic infections in a specialist orthopaedic hospital: a 15-year prospective survey. J. Bone Joint Surg. Br. 88, 943–948. doi: 10.1302/0301-620X.88B7.17150
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Piessens, V., De Vliegher, S., Verbist, B., Braem, G., Van Nuffel, A., De Vuyst, L., et al. (2012). Characterization of coagulase-negative staphylococcus species from cows' milk and environment based on bap, icaA, and mecA genes and phenotypic susceptibility to antimicrobials and teat dips. J. Dairy Sci. 95, 7027–7038. doi: 10.3168/jds.2012-5400
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ponnuraj, K., Bowden, M. G., Davis, S., Gurusiddappa, S., Moore, D., Choe, D., et al. (2003). A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 115, 217–228. doi: 10.1016/S0092-8674(03)00809-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Qin, Z., Ou, Y., Yang, L., Zhu, Y., Tolker-Nielsen, T., Molin, S., et al. (2007). Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 153, 2083–2092. doi: 10.1099/mic.0.2007/006031-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rice, K. C., Mann, E. E., Endres, J. L., Weiss, E. C., Cassat, J. E., Smeltzer, M. S., et al. (2007). The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 104, 8113–8118. doi: 10.1073/pnas.0610226104
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rogers, K. L., Fey, P. D., and Rupp, M. E. (2009). Coagulase-negative staphylococcal infections. Infect. Dis. Clin. North Am. 23, 73–98. doi: 10.1016/j.idc.2008.10.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rohde, H., Burandt, E. C., Siemssen, N., Frommelt, L., Burdelski, C., Wurster, S., et al. (2007). Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28, 1711–1720. doi: 10.1016/j.biomaterials.2006.11.046
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rohde, H., Burdelski, C., Bartscht, K., Hussain, M., Buck, F., Horstkotte, M. A., et al. (2005). Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55, 1883–1895. doi: 10.1111/j.1365-2958.2005.04515.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rohde, H., Frankenberger, S., Zähringer, U., and Mack, D. (2010). Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur. J. Cell Biol. 89, 103–111. doi: 10.1016/j.ejcb.2009.10.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rohde, H., Kalitzky, M., Kroger, N., Scherpe, S., Horstkotte, M. A., Knobloch, J. K., et al. (2004). Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J. Clin. Microbiol. 42, 5614–5619. doi: 10.1128/JCM.42.12.5614-5619.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rohde, H., Mack, D., Christner, M., Burdelski, C., Franke, G. C., and Knobloch, J. K. (2006). Pathogenesis of staphylococcal device-related infections: from basic science to new diagnostic, therapeutic and prophylactic approaches. Rev. Med. Microbiol. 17, 45–54. doi: 10.1097/01.revmedmi.0000244134.43170.83
Rosenthal, V. D., Maki, D. G., Mehta, Y., Leblebicioglu, H., Memish, Z. A., Al-Mousa, H. H., et al. (2014). International Nosocomial Infection Control Consortium (INICC) report, data summary of 43 countries for 2007–2012. Device-associated module. Am. J. Infect. Control. 42, 942–956. doi: 10.1016/j.ajic.2014.05.029
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rupp, M. E., and Archer, G. L. (1992). Hemagglutination and adherence to plastic by Staphylococcus epidermidis. Infect. Immun. 60, 4322–4327.
Rupp, M. E., and Archer, G. L. (1994). Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19, 231–243. doi: 10.1093/clinids/19.2.231
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rupp, M. E., Fey, P. D., Heilmann, C., and Götz, F. (2001). Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J. Infect. Dis. 183, 1038–1042. doi: 10.1086/319279
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rupp, M. E., Ulphani, J. S., Fey, P. D., Bartscht, K., and Mack, D. (1999a). Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67, 2627–2632.
Rupp, M. E., Ulphani, J. S., Fey, P. D., and Mack, D. (1999b). Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect. Immun. 67, 2656–2659.
Sadykov, M. R., Hartmann, T., Mattes, T. A., Hiatt, M., Jann, N. J., Zhu, Y., et al. (2011). CcpA coordinates central metabolism and biofilm formation in Staphylococcus epidermidis. Microbiology 157, 3458–3468. doi: 10.1099/mic.0.051243-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sakamoto, S., Tanaka, Y., Tanaka, I., Takei, T., Yu, J., Kuroda, M., et al. (2008). Electron microscopy and computational studies of Ebh, a giant cell-wall-associated protein from Staphylococcus aureus. Biochem. Biophys. Res. Commun. 376, 261–266. doi: 10.1016/j.bbrc.2008.08.117
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schaeffer, C. R., Woods, K. M., Longo, G. M., Kiedrowski, M. R., Paharik, A. E., Buttner, H., et al. (2015). Accumulation-associated protein enhances Staphylococcus epidermidis biofilm formation under dynamic conditions and is required for infection in a rat catheter model. Infect. Immun. 83, 214–226. doi: 10.1128/IAI.02177-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scherr, T. D., Heim, C. E., Morrison, J. M., and Kielian, T. (2014a). Hiding in plain sight: interplay between Staphylococcal biofilms and host immunity. Front Immunol. 5:37. doi: 10.3389/fimmu.2014.00037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scherr, T. D., Lindgren, K. E., Schaeffer, C. R., Hanke, M. L., Hartman, C. W., and Kielian, T. (2014b). Mouse model of post-arthroplasty Staphylococcus epidermidis joint infection. Methods Mol. Biol. 1106, 173–181. doi: 10.1007/978-1-62703-736-5_16
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schlag, M., Biswas, R., Krismer, B., Kohler, T., Zoll, S., Yu, W., et al. (2010). Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol. Microbiol. 75, 864–873. doi: 10.1111/j.1365-2958.2009.07007.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schommer, N. N., Christner, M., Hentschke, M., Ruckdeschel, K., Aepfelbacher, M., and Rohde, H. (2011). Staphylococcus epidermidis uses distinct mechanisms of biofilm formation to interfere with phagocytosis and activation of mouse macrophage-like cells 774A.1. Infect. Immun. 79, 2267–2276. doi: 10.1128/IAI.01142-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sellman, B. R., Howell, A. P., Kelly-Boyd, C., and Baker, S. M. (2005). Identification of immunogenic and serum binding proteins of Staphylococcus epidermidis. Infect. Immun. 73, 6591–6600. doi: 10.1128/IAI.73.10.6591-6600.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sellman, B. R., Timofeyeva, Y., Nanra, J., Scott, A., Fulginiti, J. P., Matsuka, Y. V., et al. (2008). Expression of Staphylococcus epidermidis SdrG increases following exposure to an in vivo environment. Infect. Immun. 76, 2950–2957. doi: 10.1128/IAI.00055-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shahrooei, M., Hira, V., Khodaparast, L., Khodaparast, L., Stijlemans, B., Kucharikova, S., et al. (2012). Vaccination with SesC decreases Staphylococcus epidermidis biofilm formation. Infect. Immun. 80, 3660–3668. doi: 10.1128/IAI.00104-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shahrooei, M., Hira, V., Stijlemans, B., Merckx, R., Hermans, P. W., and Van Eldere, J. (2009). Inhibition of Staphylococcus epidermidis biofilm formation by rabbit polyclonal antibodies against the SesC protein. Infect. Immun. 77, 3670–3678. doi: 10.1128/IAI.01464-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stevens, N. T., Tharmabala, M., Dillane, T., Greene, C. M., O'Gara, J. P., and Humphreys, H. (2008). Biofilm and the role of the ica operon and aap in Staphylococcus epidermidis isolates causing neurosurgical meningitis. Clin. Microbiol. Infect. 14, 719–722. doi: 10.1111/j.1469-0691.2008.02012.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sugimoto, S., Iwamoto, T., Takada, K., Okuda, K., Tajima, A., Iwase, T., et al. (2013). Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J. Bacteriol. 195, 1645–1655. doi: 10.1128/JB.01672-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tanaka, Y., Sakamoto, S., Kuroda, M., Goda, S., Gao, Y. G., Tsumoto, K., et al. (2008). A helical string of alternately connected three-helix bundles for the cell wall-associated adhesion protein Ebh from Staphylococcus aureus. Structure 16, 488–496. doi: 10.1016/j.str.2007.12.018
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thomas, V. C., Hiromasa, Y., Harms, N., Thurlow, L., Tomich, J., and Hancock, L. E. (2009). A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 72, 1022–1036. doi: 10.1111/j.1365-2958.2009.06703.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thomas, V. C., Thurlow, L. R., Boyle, D., and Hancock, L. E. (2008). Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 190, 5690–5698. doi: 10.1128/JB.00314-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tormo, M. A., Knecht, E., Götz, F., Lasa, I., and Penades, J. R. (2005a). Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology 151, 2465–2475. doi: 10.1099/mic.0.27865-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tormo, M. A., Marti, M., Valle, J., Manna, A. C., Cheung, A. L., Lasa, I., et al. (2005b). SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 187, 2348–2356. doi: 10.1128/JB.187.7.2348-2356.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vuong, C., Gerke, C., Somerville, G. A., Fischer, E. R., and Otto, M. (2003). Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188, 706–718. doi: 10.1086/377239
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vuong, C., Kocianova, S., Voyich, J. M., Yao, Y., Fischer, E. R., DeLeo, F. R., et al. (2004a). A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 279, 54881–54886. doi: 10.1074/jbc.M411374200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vuong, C., Kocianova, S., Yao, Y., Carmody, A. B., and Otto, M. (2004b). Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190, 1498–1505. doi: 10.1086/424487
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vuong, C., Voyich, J. M., Fischer, E. R., Braughton, K. R., Whitney, A. R., DeLeo, F. R., et al. (2004c). Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 6, 269–275. doi: 10.1046/j.1462-5822.2004.00367.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, R., Khan, B. A., Cheung, G. Y., Bach, T. H., Jameson-Lee, M., Kong, K. F., et al. (2011a). Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Invest. 121, 238–248. doi: 10.1172/JCI42520
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, X., Niu, C., Sun, G., Dong, D., Villaruz, A. E., Li, M., et al. (2011b). ygs is a novel gene that influences biofilm formation and the general stress response of Staphylococcus epidermidis. Infect. Immun. 79, 1007–1015. doi: 10.1128/IAI.00916-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C., and Mattick, J. S. (2002). Extracellular DNA required for bacterial biofilm formation. Science 295, 1487. doi: 10.1126/science.295.5559.1487
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Williams, R. J., Henderson, B., Sharp, L. J., and Nair, S. P. (2002). Identification of a Fibronectin-Binding Protein from Staphylococcus epidermidis. Infect. Immun. 70, 6805–6810. doi: 10.1128/IAI.70.12.6805-6810.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wisplinghoff, H., Seifert, H., Tallent, S. M., Bischoff, T., Wenzel, R. P., and Edmond, M. B. (2003). Nosocomial bloodstream infections in pediatric patients in United States hospitals: epidemiology, clinical features and susceptibilities. Pediatr. Infect. Dis. J. 22, 686–691. doi: 10.1097/01.inf.0000078159.53132.40
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wu, Y., Wu, Y., Zhu, T., Han, H., Liu, H., Xu, T., et al. (2015). Staphylococcus epidermidis SrrAB regulates bacterial growth and biofilm formation differently under oxic and microaerobic conditions. J. Bacteriol. 197, 459–476. doi: 10.1128/JB.02231-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, L., Li, H., Vuong, C., Vadyvaloo, V., Wang, J., Yao, Y., et al. (2006). Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 74, 488–496. doi: 10.1128/IAI.74.1.488-496.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yao, Y., Sturdevant, D. E., Villaruz, A., Xu, L., Gao, Q., and Otto, M. (2005). Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect. Immun. 73, 1856–1860. doi: 10.1128/IAI.73.3.1856-1860.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zaat, S., Broekhuizen, C., and Riool, M. (2010). Host tissue as a niche for biomaterial-associated infection. Future Microbiol. 5, 1149–1151. doi: 10.2217/fmb.10.89
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ziebuhr, W., Heilmann, C., Götz, F., Meyer, P., Wilms, K., Straube, E., et al. (1997). Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65, 890–896.
Zimmerli, W., Lew, P. D., and Waldvogel, F. A. (1984). Pathogenesis of foreign body infection. Evidence for a local granulocyte defect. J. Clin. Invest. 73, 1191–1200. doi: 10.1172/JCI111305
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zimmerli, W., Waldvogel, F. A., Vaudaux, P., and Nydegger, U. E. (1982). Pathogenesis of foreign body infection: description and characteristics of an animal model. J. Infect. Dis. 146, 487–497. doi: 10.1093/infdis/146.4.487
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zoll, S., Patzold, B., Schlag, M., Gotz, F., Kalbacher, H., and Stehle, T. (2010). Structural basis of cell wall cleavage by a staphylococcal autolysin. PLoS. Pathog. 6:e1000807. doi: 10.1371/journal.ppat.1000807
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Staphylococcus epidermidis biofilm formation, regulation, PIA, Aap, Embp, AtlE, primary attachment, biofilm accumulation
Citation: Büttner H, Mack D and Rohde H (2015) Structural basis of Staphylococcus epidermidis biofilm formation: mechanisms and molecular interactions. Front. Cell. Infect. Microbiol. 5:14. doi: 10.3389/fcimb.2015.00014
Received: 15 December 2014; Accepted: 26 January 2015;
Published online: 17 February 2015.
Edited by:
Pietro Speziale, University degli Studi di Pavia, ItalyReviewed by:
Chia Lee, University of Arkansas for Medical Sciences, USACopyright © 2015 Büttner, Mack and Rohde. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Holger Rohde, Institut für Medizinische Mikrobiologie, Virologie und Hygiene, Universitätsklinikum Hamburg-Eppendorf, Martinistrasse 52, D-20246 Hamburg, Germany e-mail:cm9oZGVAdWtlLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.