
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell. Infect. Microbiol. , 11 November 2014
Sec. Parasite and Host
Volume 4 - 2014 | https://doi.org/10.3389/fcimb.2014.00165
This article is part of the Research Topic New perspectives in evolution: The superorganisms, symbiopoiesis and integration of systems. View all 7 articles
Schistosomes, a class of parasitic trematode worms, cause schistosomiasis. Accumulating evidence suggests that microRNAs (miRNAs)—small, non-coding RNAs that are known to play critical regulatory roles in many organisms—may be involved in schistosome development and sexual maturation, as well as the pathogenesis of schistosomiasis. Schistosoma miRNAs, such as Bantam and miR-10, may be involved in the pathological processes of schistosomiasis, and recent studies suggest that schistosome-specific miRNAs (e.g., Bantam, miR-3479-3p) in the bloodstream of a final host could be used as biomarkers for schistosomiasis diagnosis. Furthermore, aberrant miRNAs, such as miR-223 and miR-454, can be produced by a host in response to schistosome infection, and these miRNAs may contribute to the pathogenesis of schistosomiasis-associated liver injury. Here, we summarize recent progress evaluating the relationship between schistosome miRNAs and schistosomiasis and discuss how these miRNAs can mediate the pathogenesis of schistosomiasis and be used as biomarkers for schistosomiasis diagnosis.
Schistosomiasis is a parasitic disease found worldwide and is caused primarily by worms of the genus Schistosoma. The main disease-causing species are S. haematobium, S. mansoni, and S. japonicum. Over 200 million people are infected with this parasite worldwide, leading to 300,000 deaths annually, mostly in the developing world (Colley et al., 2014). In China, schistosomiasis japonica is a serious public health problem (Zhou et al., 2005a,b). Unfortunately, there is no available vaccine for schistosomiasis. Praziquantel is the only drug that is widely used to treat schistosomiasis, but it is ineffective in preventing reinfection and there are concerns regarding the development of praziquantel resistance following large-scale and repeated use (Bergquist et al., 2008; Wang et al., 2012b). Consequently, a deeper understanding of the mechanisms of schistosome development and the pathogenesis of schistosomiasis may aid in the development of novel strategies to control this disease.
MicroRNAs (miRNAs), a class of small non-coding RNAs, can regulate a wide range of pivotal biological processes such as development, cell proliferation and differentiation, cell death, metabolism, and signal transduction in many organisms (Ambros, 2003; Carrington and Ambros, 2003; Miska, 2005). Schistosomes have a complex developmental cycle with diverse life stages, which suggests that gene expression in these organisms is regulated accurately and precisely (Gomes et al., 2009; Zhou et al., 2009; De Souza Gomes et al., 2011). Since miRNAs act as critical post-transcriptional regulators in many organisms, studies have been actively carried out to determine the role miRNAs play in schistosomes. These studies support our notion that miRNAs could be important regulators for schistosome development and potential therapeutic targets against schistosomiasis (Cheng and Jin, 2012). In addition, recent findings have provided new information regarding the roles of miRNAs in schistosome development and the pathogenesis of schistosomiasis. Here, we summarize recent research evaluating miRNAs in schistosomes and explore how schistosome miRNAs can serve as mediators of the pathogenesis of schistosomiasis. In addition, we also discuss the use of miRNAs as biomarkers for schistosomiasis diagnosis.
To date, 79 mature miRNAs in S. japonicum and 225 mature miRNAs in S. mansoni have been documented in miRBase (Version 21). Although schistosome miRNAs have been summarized previously (Cheng and Jin, 2012), recent research has further investigated miRNA profiles in schistosomes, the results of these studies are summarized in Table 1.
Briefly, miRNA profiles at several stages of S. japonicum development have been characterized; miRNAs present in lung-stage schistosomula, hepatic-stage schistosomula, adult males, adult females, and eggs of this species were documented, and 199 novel miRNAs with stage specificities were found (Cai et al., 2011). Additional research evaluating miRNA profiles in S. japonicum eggs have indicated that sja-miR-71b-5p, sja-miR-71, sja-miR-1, sja-miR-36-3p, and sja-miR-124-3p were the most abundant miRNAs at this developmental stage (Cai et al., 2013a). Concurrent with research in S. japonicum, 112 miRNAs (including 84 novel miRNA families) were reported in adult worms of S. mansoni (Marco et al., 2013). Among them, 26 miRNAs were shown to be conserved in other flatworms such as Schmidtea mediterranea and Dugesia japonica (Marco et al., 2013). In addition, a more recent study reported a total of 2258 miRNAs identified in S. japonicum collected from single- and double-sex infected mice (Sun et al., 2014); however, the authors did not provide information about the number of evolutionary conserved miRNAs. Moreover, in our recent work, five pathogen-specific miRNAs were identified in the plasma of rabbits infected with S. japonicum, including four known miRNAs (Bantam, miR-3479, miR-10, and miR-3096), and one novel miRNA (sja-miR-8185) (Cheng et al., 2013).
On comparing these known miRNAs across the Platyhelminthes, most were found to be conserved (Jin et al., 2013). In addition, some S. japonicum miRNAs (e.g., miR-1b, miR-124, miR-190, et al.) also displayed evolutionary conservation among Homo sapiens, Mus musculus, Caenorhabditis elegans, and Drosophila melanogaster (Cheng and Jin, 2012).
Different stages of schistosome development are associated with different categories of miRNAs, suggesting that miRNAs may be involved in the regulation of schistosome development (Krautz-Peterson and Skelly, 2008; Xue et al., 2008; Chen et al., 2010; Hao et al., 2010; Luo et al., 2010; Cai et al., 2011; Simoes et al., 2011). Several lines of evidence also indirectly imply that miRNAs may regulate the pathogenesis of schistosomiasis (Cheng et al., 2013; Hoy et al., 2014).
First, the key components of miRNA biogenesis, including the Ago proteins (Chen et al., 2010) and Dicer (Krautz-Peterson and Skelly, 2008; Luo et al., 2010), have been shown to be differentially expressed during different stages of schistosome development. Second, some of miRNAs such as miR-36, miR-71, bantam, miR-7, and others, have been shown to specifically express in a specific stage of S. japonicum (Xue et al., 2008; Hao et al., 2010; Cai et al., 2011). In addition, the similar phenomenon was also observed in S. mansoni (miR-4, miR-6, miR-9, miR-32, miR-125, miR-3, and miR-5 were expressed in adult worms only, and miR-20, miR-18, miR-22, miR-26, and bantam were expressed in schistosomula only) (Simoes et al., 2011). Third, bioinformatic analyses have indicated that several evolutionarily conserved miRNAs (miR-8, miR-1, miR-124, miR-71, and miR-195) in schistosomes may regulate phylogenetically conserved mRNA targets (Cheng and Jin, 2012); however, experimental confirmation of these results is needed.
Besides role of miRNAs in schistosome development, recent work within our laboratory implies that schistosome-derived miRNAs may regulate the pathogenesis of schistosomiasis within a final host (Cheng et al., 2013). In this study, we identified Schistosoma-specific Bantam miRNA within plasma derived from a host infected with S. japonicum. In Drosophila, it has been shown that Bantam can target a tumor-suppressor pathway, and that overexpression of Bantam led to cellular growth and the suppression of cellular death (Nolo et al., 2006). In addition, it was reported that human schistosomiasis is associated with increased risks for bladder (S. haematobium) and liver (S. mansoni, S. japonicum) cancers (Takemura et al., 1998; Gryseels et al., 2006). Given this research, we hypothesized that schistosome-specific miRNAs, such as Bantam, may be involved in the etiology of cancers in a host infected with schistosomes (Figure 1); however, further studies will need to be carried out to test this hypothesis.
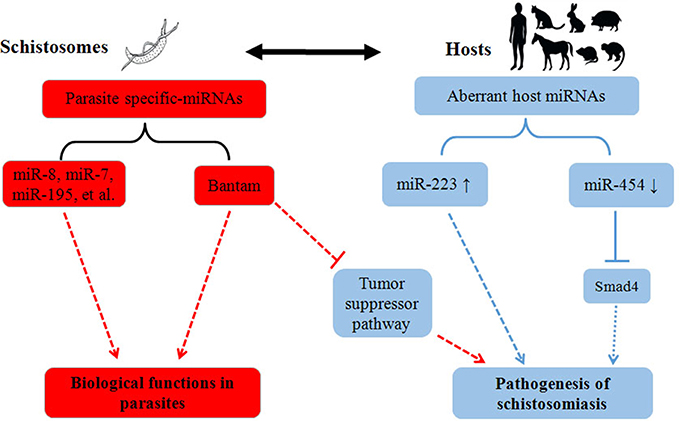
Figure 1. Summary of the roles of miRNAs in the pathogenesis of schistosomiasis. Parasite activities are depicted in red, and host activities are depicted in blue. The solid lines indicate the proven biological effects of miRNAs, and the dotted lines indicate their putative effects.
In addition to schistosome-derived miRNAs, recent studies also suggest that host miRNAs may be involved in regulating the pathogenesis of schistosomiasis (Figure 1). Integrative analysis of miRNA and mRNA profiles in murine liver suggested that miRNAs, and their corresponding mRNA targets, may function coordinately to regulate the hepatic pathology of a host during schistosome infection (Cai et al., 2013b). Furthermore, miRNA profiles in different tissues (liver, spleen, and lung) of S. japonicum-infected mice implied that miRNAs may be involved in the regulation of several signaling pathways—such as the MAPK, insulin, Toll-like receptor, and TGF-β pathways—during schistosome infection (Han et al., 2013). In a recent study, miR-223 was primarily found in Kupffer cells in the liver, and the expression of this miRNA was dramatically elevated in liver cells of schistosome-infected mice (He et al., 2013); it is possible that the expression levels of miR-223 could reflect the extent of pathological changes in the liver of an infected host. Moreover, miR-454, a target miRNA of Smad4, was found to be down-regulated in S. japonicum-induced murine liver fibrosis models, while α-SMA and Smad4 were up-regulated (Zhu et al., 2014); these results suggested that the down-regulation of miR-454 may be involved in the pathogenesis of liver fibrosis in S. japonicum-infected mice. Despite these findings, the functions of miR-223 and miR-454 need to be further investigated at different biological levels, in order to fully understand the roles that these miRNAs play in the pathogenesis of schistosomiasis.
Since certain miRNAs have been shown to potentially play an important role in the pathogenesis of schistosomiasis, the usage of the antisense technology blocking specific miRNAs and/or an augment method for improving the expressions of some miRNAs may have a potential to provide alternative strategies for schistosomiasis control as optimism have been demonstrated from miRNA-based anti-tumor therapy (Krutzfeldt et al., 2005; Ma et al., 2010; Wang et al., 2012a).
Early diagnosis of schistosomiasis is critical to control this disease. However, no reliable biomarker exists for early diagnosis of schistosomiasis. Circulating miRNAs, which are present in a stable form in the plasma or serum of an infected host, have been considered ideal biomarkers for the diagnosis of some cancers. It is possible that such circulating miRNAs could also serve as biomarkers for schistosomiasis diagnosis.
As mentioned above, five pathogen-specific miRNAs in the circulation of a S. japonicum-infected host were identified in our study and the subsequent validation study indicated that there is a correlation between the abundance of schistosome-specific miRNAs in the plasma of schistosome-infected mice and the number of S. japonicum cercariae in the inoculum (Cheng et al., 2013). More recently, eleven schistosome-specific miRNAs were identified in mice infected with S. mansoni (Hoy et al., 2014); three of these parasite-derived miRNAs (Bantam, miR-3479-3p, and miR-277) may be potential biomarkers for schistosomiasis diagnosis. Similarly, we detected three miRNAs (Bantam, miR-3479, and miR-10) in the plasma of mice infected with S. japonicum (Cheng et al., 2013). Collectively, these results suggest that parasite-specific miRNAs in the circulation of a host may be potential biomarkers for schistosomiasis diagnosis. However, it remains to be determined whether this kind of pathogen specific circulating miRNAs is associated with specific species in schistosomes.
Apart from Schistosoma-specific miRNAs, aberrant host miRNAs associated with schistosome infection may also be potential indicators of schistosomiasis. A recent study indicated that miR-223 was significantly up-regulated in the serum of mice infected with S. japonicum and returned to near normal levels on praziquantel treatment (He et al., 2013), implying that up-regulated murine miR-223 may be a biomarker for Schistosoma infection. However, the possibility that altered serum levels of miR-223 are caused by other diseases or the pathogenesis of other organs needs to be assessed.
Currently, circulating miRNA detection mostly relies on qRT-PCR (Cheng, 2014). Additionally, several recent studies also documented that the sensitivity of miRNA detection could be significantly improved by using novel target miRNA amplification methods such as RAKE assay (Nelson et al., 2004), rolling-circle amplification (Cheng et al., 2009), and DNA concatamers-based amplification (Hong et al., 2013) and also the combination of new signal detection techniques including a total internal reflection fluorescence microscope (Ho et al., 2014) or an electrochemical device (Hong et al., 2013; Ren et al., 2013). In future, it may be necessary to evaluate these methods for schistosomiasis diagnosis by detecting circulating miRNAs.
Recent studies have indicated that miRNAs play important roles in schistosome development and their parasitism. Although analyses of miRNA profiles have provided valuable information regarding the mechanisms underlying the pathogenesis of schistosomiasis, and suggested potential novel applications in diagnostics, our current understanding of these molecular mechanisms remains limited. Consequently, it is necessary to characterize the functions of schistosome-specific miRNAs and clarify their potential as diagnostic markers for schistosomiasis. Since miRNAs exert their effect by targeting protein-encoding mRNAs, verifying miRNA targets in both hosts and parasites is essential for understanding the mechanisms of host-parasite interactions, and developing novel therapeutic agents for schistosomiasis.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This study was supported, in part or in whole, by the National Natural Science Foundation of China (Grant No. 30901068), the Science and Technology Commission of Shanghai Municipality of China (Grant Nos. 10410703400 and 10PJ1412300), and the Shanghai Talent Developing Foundation of China (Grant No. 2009032).
Ambros, V. (2003). MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113, 673–676. doi: 10.1016/S0092-8674(03)00428-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bergquist, R., Utzinger, J., and McManus, D. P. (2008). Trick or treat: the role of vaccines in integrated schistosomiasis control. PLoS Negl. Trop. Dis. 2:e244. doi: 10.1371/journal.pntd.0000244
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cai, P. F., Hou, N., Piao, X. Y., Liu, S., Liu, H., Yang, F., et al. (2011). Profiles of small non-coding RNAs in Schistosoma japonicum during development. PLoS Negl. Trop. Dis. 5:e1256. doi: 10.1371/journal.pntd.0001256
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cai, P. F., Piao, X. Y., Hao, L. L., Liu, S., Hou, N., Wang, H., et al. (2013a). A deep analysis of the small non-coding RNA population in Schistosoma japonicum eggs. PLoS ONE 8:e64003. doi: 10.1371/journal.pone.0064003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cai, P. F., Piao, X. Y., Liu, S., Hou, N., Wang, H., and Chen, Q. J. (2013b). MicroRNA-gene expression network in murine liver during Schistosoma japonicum infection. PLoS ONE 8:e67037. doi: 10.1371/journal.pone.0067037
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carrington, J. C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301, 336–338. doi: 10.1126/science.1085242
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chen, J., Yang, Y. P., Guo, S. X., Peng, J. B., Liu, Z. P., Li, J. K., et al. (2010). Molecular cloning and expression profiles of Argonaute proteins in Schistosoma japonicum. Parasitol. Res. 107, 889–899. doi: 10.1007/s00436-010-1946-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, G. (2014). Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv. Drug Deliv. Rev. doi: 10.1016/j.addr.2014.09.001. [Epub ahead of print].
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, G., and Jin, Y. (2012). MicroRNAs: potentially important regulators for schistosome development and therapeutic targets against schistosomiasis. Parasitology 139, 669–679. doi: 10.1017/S0031182011001855
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, G. F., Luo, R., Hu, C., Cao, J., and Jin, Y. X. (2013). Deep sequencing-based identification of pathogen-specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum. Parasitology 140, 1751–1761. doi: 10.1017/S0031182013000917
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cheng, Y. Q., Zhang, X., Li, Z. P., Jiao, X. X., Wang, Y. C., and Zhang, Y. L. (2009). Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification. Angew. Chem. Int. Edit. 48, 3268–3272. doi: 10.1002/anie.200805665
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Colley, D. G., Bustinduy, A. L., Secor, W. E., and King, C. H. (2014). Human schistosomiasis. Lancet 383, 2253–2264. doi: 10.1016/S0140-6736(13)61949-2
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
De Souza Gomes, M. D., Muniyappa, M. K., Carvalho, S. G., Guerra-Sa, R., and Spillane, C. (2011). Genome-wide identification of novel microRNAs and their target genes in the human parasite Schistosoma mansoni. Genomics 98, 96–111. doi: 10.1016/j.ygeno.2011.05.007
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gomes, M. S., Cabral, F. J., Jannotti-Passos, L. K., Carvalho, O., Rodrigues, V., Baba, E. H., et al. (2009). Preliminary analysis of miRNA pathway in Schistosoma mansoni. Parasitol. Int. 58, 61–68. doi: 10.1016/j.parint.2008.10.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gryseels, B., Polman, K., Clerinx, J., and Kestens, L. (2006). Human schistosomiasis. Lancet 368, 1106–1118. doi: 10.1016/S0140-6736(06)69440-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Han, H. X., Peng, J. B., Hong, Y., Zhang, M., Han, Y. H., Liu, D. D., et al. (2013). MicroRNA expression profile in different tissues of BALB/c mice in the early phase of Schistosoma japonicum infection. Mol. Biochemi.Parasitol. 188, 1–9. doi: 10.1016/j.molbiopara.2013.02.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hao, L. L., Cai, P. F., Jiang, N., Wang, H., and Chen, Q. J. (2010). Identification and characterization of microRNAs and endogenous siRNAs in Schistosoma japonicum. BMC Genomics 11:55. doi: 10.1186/1471-2164-11-55
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
He, X., Sai, X., Chen, C., Zhang, Y. B., Xu, X. D., Zhang, D. M., et al. (2013). Host serum miR-223 is a potential new biomarker for Schistosoma japonicum infection and the response to chemotherapy. Parasit. Vectors 6, 272. doi: 10.1186/1756-3305-6-272
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ho, S. L., Chan, H. M., Wong, R. N., and Li, H. W. (2014). Self-assembling protein platform for direct quantification of circulating microRNAs in serum with total internal reflection fluorescence microscopy. Anal. Chim. Acta 823, 61–68. doi: 10.1016/j.aca.2014.03.020
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hong, C. Y., Chen, X., Liu, T., Li, J., Yang, H. H., Chen, J. H., et al. (2013). Ultrasensitive electrochemical detection of cancer-associated circulating microRNA in serum samples based on DNA concatamers. Biosens. Bioelectron. 50, 132–136. doi: 10.1016/j.bios.2013.06.040
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hoy, A. M., Lundie, R. J., Ivens, A., Quintana, J. F., Nausch, N., Forster, T., et al. (2014). Parasite-derived MicroRNAs in host serum as novel biomarkers of helminth infection. PLoS Negl. Trop. Dis. 8:e2701. doi: 10.1371/journal.pntd.0002701
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jin, X. L., Lu, L. X., Su, H. L., Lou, Z. Z., Wang, F., Zheng, Y. D., et al. (2013). Comparative analysis of known miRNAs across platyhelminths. FEBS J. 280, 3944–3951. doi: 10.1111/febs.12395
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Krautz-Peterson, G., and Skelly, P. J. (2008). Schistosoma mansoni: the dicer gene and its expression. Exp. Parasitol. 118, 122–128. doi: 10.1016/j.exppara.2007.06.013
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Krutzfeldt, J., Rajewsky, N., Braich, R., Rajeev, K. G., Tuschl, T., Manoharan, M., et al. (2005). Silencing of microRNAs in vivo with antagomirs. Nature 438, 685–689. doi: 10.1038/nature04303
Luo, R., Xue, X. Y., Wang, Z. X., Sun, J., Zou, Y., and Pan, W. Q. (2010). Analysis and characterization of the genes encoding the Dicer and Argonaute proteins of Schistosoma japonicum. Parasit. Vectors 3:90. doi: 10.1186/1756-3305-3-90
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ma, L., Reinhardt, F., Pan, E., Soutschek, J., Bhat, B., Marcusson, E. G., et al. (2010). Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat. Biotechnol. 28, U341–U367. doi: 10.1038/nbt.1618
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marco, A., Kozomara, A., Hui, J. H. L., Emery, A. M., Rollinson, D., Griffiths-Jones, S., et al. (2013). Sex-biased expression of MicroRNAs in Schistosoma mansoni. PLoS Negl. Trop. Dis. 7:e2402. doi: 10.1371/journal.pntd.0002402
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Miska, E. A. (2005). How microRNAs control cell division, differentiation and death. Curr. Opin. Genet. Dev. 15, 563–568. doi: 10.1016/j.gde.2005.08.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nelson, P. T., Baldwin, D. A., Scearce, L. M., Oberholtzer, J. C., Tobias, J. W., and Mourelatos, Z. (2004). Microarray-based, high-throughput gene expression profiling of microRNAs. Nat. Methods 1, 155–161. doi: 10.1038/nmeth717
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nolo, R., Morrison, C. M., Tao, C. Y., Zhang, X. W., and Halder, G. (2006). The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16, 1895–1904. doi: 10.1016/j.cub.2006.08.057
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ren, Y. Q., Deng, H. M., Shen, W., and Gao, Z. Q. (2013). A highly sensitive and selective electrochemical biosensor for direct detection of MicroRNAs in serum. Anal. Chem. 85, 4784–4789. doi: 10.1021/ac400583e
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Simoes, M. C., Lee, J., Djikeng, A., Cerqueira, G. C., Zerlotini, A., da Silva-Pereira, R. A., et al. (2011). Identification of Schistosoma mansoni microRNAs. BMC Genomics 12:47. doi: 10.1186/1471-2164-12-47
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sun, J., Wang, S., Li, C., Ren, Y., and Wang, J. (2014). Novel expression profiles of microRNAs suggest that specific miRNAs regulate gene expression for the sexual maturation of female Schistosoma japonicum after pairing. Parasit. Vectors 7:177. doi: 10.1186/1756-3305-7-177
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Takemura, Y., Kikuchi, S., and Inaba, Y. (1998). Epidemiologic study of the relationship between schistosomiasis due to Schistosoma japonicum and liver cancer/cirrhosis. Am. J. Trop. Med. Hyg. 59, 551–556
Wang, Q. Z., Lv, Y. H., Gong, Y. H., Li, Z. F., Xu, W., Diao, Y., et al. (2012a). Double-stranded Let-7 mimics, potential candidates for cancer gene therapy. J. Physiol. Biochem. 68, 107–119. doi: 10.1007/s13105-011-0124-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, W., Wang, L., and Liang, Y. S. (2012b). Susceptibility or resistance of praziquantel in human schistosomiasis: a review. Parasitol. Res. 111, 1871–1877. doi: 10.1007/s00436-012-3151-z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xue, X. Y., Sun, J., Zhang, Q. F., Wang, Z. X., Huang, Y. F., and Pan, W. Q. (2008). Identification and characterization of novel MicroRNAs from Schistosoma japonicum. PLoS ONE 3:e4034. doi: 10.1371/journal.pone.0004034
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, X. N., Wang, L. Y., Chen, M. G., Wu, X. H., Jiang, Q. W., Chen, X. Y., et al. (2005a). The public health significance and control of schistosomiasis in China—then and now. Acta Trop. 96, 97–105. doi: 10.1016/j.actatropica.2005.07.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, X. N., Wang, L. Y., Chen, M. G., Wang, T. P., Guo, J. G., Wu, X. H., et al. (2005b). An economic evaluation of the national schistosomiasis control programme in China from 1992 to 2000. Acta Trop. 96, 255–265. doi: 10.1016/j.actatropica.2005.07.026
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhou, Y., Zheng, H., Chen, Y., Zhang, L., Wang, K., Guo, J., et al. (2009). The Schistosoma japonicum genome reveals features of host–parasite interplay. Nature 460, 345–351. doi: 10.1038/nature08140
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhu, D., He, X., Duan, Y., Chen, J., Wang, J., Sun, X., et al. (2014). Expression of microRNA-454 in TGF-β1-stimulated hepatic stellate cells and in mouse livers infected with Schistosoma japonicum. Parasit. Vectors 7:148. doi: 10.1186/1756-3305-7-148
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: schistosome, microRNA, development, pathogenesis, biomarkers, schistosomiasis
Citation: Zhu L, Liu J and Cheng G (2014) Role of microRNAs in schistosomes and schistosomiasis. Front. Cell. Infect. Microbiol. 4:165. doi: 10.3389/fcimb.2014.00165
Received: 23 July 2014; Accepted: 23 October 2014;
Published online: 11 November 2014.
Edited by:
Emiliano J. Salvucci, Consejo Nacional de Investigaciones Científicas y Técnicas, ArgentinaReviewed by:
Charles Kelly, King's College London, UKCopyright © 2014 Zhu, Liu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guofeng Cheng, Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Key Laboratory of Animal Parasitology, Ministry of Agriculture, 518 Ziyue Road, Shanghai 200241, China e-mail:Y2hlbmdnZmVuZ0BzaHZyaS5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.