- Project Group P2, Robert Koch-Institute, Wernigerode, Germany
Before the advent of molecular biology methods, studies of pathogens were dominated by analyses of their metabolism. Development of molecular biology techniques then enabled the identification and functional characterisation of the fascinating toolbox of virulence factors. Increasing, genomic and proteomic approaches form the basis for a more systemic view on pathogens' functions in the context of infection. Re-emerging interest in the metabolism of pathogens and hosts further expands our view of infections. There is increasing evidence that virulence functions and metabolism of pathogens are extremely intertwined. Type three secretion systems (T3SSs) are major virulence determinants of many Gram-negative pathogens and it is the objective of this review to illustrate the intertwined relationship between T3SSs and the metabolism of the pathogens deploying them.
Introduction
In the introduction to a review on “Metabolism of Microorganisms as related to their pathogenicity” published more than 50 years ago, Panos and Ajl stated, “The problem of how certain aspects of the metabolism of a bacterial cell are related to its pathogenicity is an exceedingly complex and difficult one” (Panos and Ajl, 1963). As will be outlined here, our present view on this subject could hardly be formulated more to the point. Elsewhere in their review, Panos and Ajl state that “This review is not intended to be an exhaustive coverage of the voluminous literature available on this subject,” indicating that many forgotten treasures could be raised from that past literature, including important details that newcomers to the field might not know (Panos and Ajl, 1963). Indeed, as will be also illustrated here, many of these early studies can now be linked to more recent ones, mounting increasingly complex regulatory networks in which functions of metabolism and virulence are closely intertwined. Other early studies might serve as stimuli for reinvestigation using state-of-the-art methodology.
It is obvious that a pathogen requires metabolic functions in order to establish an infection and that these metabolic functions adapt to nutritional situation(s) during infection. Consequently, the metabolism of pathogens during infection can differ substantially from that of the in vitro situation (Munoz-Elias and McKinney, 2006). It is also known that sensing specific nutrients together with other environmental conditions that are indicative of a host environment stimulates production of virulence factors in many pathogens (Poncet et al., 2009). Further, global regulatory networks such as the stringent response system, which responds to various nutritional and metabolic stresses, or the carbon catabolite repression and carbon storage regulator systems, respectively, play an important role in the control of virulence traits (Romeo, 1998; Gorke and Stulke, 2008; La et al., 2008; Poncet et al., 2009; Dalebroux et al., 2010; Heroven et al., 2012). Though important, these aspects are not the focus of this review, are treated elsewhere and will only be mentioned briefly. The question we address is whether there is anything beyond this global and well-established relationship in the interrelatedness of virulence functions and metabolism in bacterial pathogens.
It will become apparent in this review that the link between virulence and metabolism is so important in many pathogens that we are probably only seeing the tip of the iceberg. Using the example of the type three secretion system (T3SS), a prototypic virulence determinant of many Gram-negatives, we will show in particular that not only metabolism controls and underpins production and functioning of virulence factors but also that, conversely, virulence factors can modulate metabolic functions of pathogens in a coordinate way, as for example, in Yersinia (Moncla et al., 1983; Du et al., 2009; Schmid et al., 2009) or pave the way for novel metabolic options as in Salmonella (Winter et al., 2010) and Pseudomonas (Dacheux et al., 2001b). The following criteria served as indicators to identify similar phenomena in other pathogens: (i) Can it be assumed or is it known that deletion of a T3SS component results in changes of metabolism? (ii) Is there evidence that such metabolic changes are due to a specific interference between T3SS and metabolic functions rather than due to global metabolic effects (e.g., relief from a global metabolic burden due to the loss of a virulence plasmid). (iii) Assuming global effects in general, studies of specific metabolic functions required for full virulence will not been considered unless the metabolic functions can be linked to specific T3SS components (e.g., deletion of a metabolic function specifically interferes with T3SS gene expression). Examples of interrelatedness between T3SS functions and metabolism will be followed by discussion of common principles and their impact on our view of bacterial pathogenesis.
Yersinia: Cross-Talk Between a Type Three Secretion System and Metabolism
Studies linking metabolism and virulence in Yersinia pestis (originally called Pasteurella pestis), the causative agent of plague, go back more than 50 years. After Devignat and Schoetter (1942) reported in 1942 that virulent strains of Y. pestis become avirulent upon continued aerobic culturing at 37°C, Fukui et al. (1957) showed that loss of virulence was accompanied by a loss of the so-called V-antigen (Bacon and Burrows, 1956). Only decades later did it become clear that V-antigen (LcrV) was a key component of the plasmid-encoded Yersinia type three secretion system (T3SS), a major virulence determinant shared by Y. pestis and the enteropathogenic species Y. pseudotuberculosis and Y. enterocolitica. It was this virulence plasmid, termed pCD in Y. pestis and pYV in Y. pseudotuberculosis and Y. enterocolitica, that was rapidly lost at 37°C in vitro (Cornelis et al., 1998). Higuchi and Carlin (Higuchi and Carlin, 1958) found that virulent strains grew faster than avirulent types at 37°C but not at 27°C. A search for media promoting growth of virulent strains of Y. pestis revealed a critical role of calcium ions preventing the emergence of avirulent variants (Kupferberg and Higuchi, 1958). Shortly after, Delwiche et al. (1959) noted that bicarbonate, aspartate and glutamate also prevented loss of virulence. How can these early observations be interpreted in the light of the current knowledge and do they point to a specific interrelationship between virulence and metabolism in Yersinia? First, production and assembly of the Yersinia T3SS machinery occur only at 37°C (Yother et al., 1986), and a low concentration of calcium ions triggers the massive secretion of T3SS substrate proteins called Yops into the culture supernatant (Heesemann et al., 1984). It seemed reasonable to assume that the poor growth of pathogenic Yersinia at 37°C under low calcium conditions could be explained by the heavy metabolic burden imposed by the massive production and secretion of Yops (Ramamurthi and Schneewind, 2002). This interpretation was supported by the fact that loss of the virulence plasmid encoding the T3SS relieves from this burden but the effect of bicarbonate, aspartate and glutamate could not be explained.
The fact that bicarbonate prevented the loss of virulence in Y. pestis (Delwiche et al., 1959) stimulated Baugh et al. (1964) to compare carbon dioxide fixation reactions in extracts from virulent and avirulent strains. Although Baugh et al. did not observe any significant difference (Baugh et al., 1964), it seems now likely that their hypothesis was correct, since there is a direct linkage between carbon dioxide fixation and T3SS in Yersinia (Schmid et al., 2009). Specifically, two regulatory components of the Y. enterocolitica T3SS, YscM1 and YscM2, bind directly to the phosphoenolpyruvate carboxylase (PEPC) of Yersinia. PEPC catalyses the carboxylation of phosphoenolpyruvate (PEP) to form oxaloacetate, a replenishing or so-called anaplerotic reaction. The PEPC reaction replenishes the tricarboxylic acid (TCA) cycle that, besides generating reduction equivalents, provides building blocks for amino acid synthesis. More precisely, oxaloacetate is used to form aspartate and other amino acids of the aspartate family, while α-ketoglutarate is used to form glutamate and related amino acids. It is important to remember that aspartate and glutamate both suppress growth restriction in Y. pestis (Delwiche et al., 1959), and that activation of the Yersinia T3SS requires glutamate, glutamine, aspartate, or asparagine (Lee et al., 2001). Furthermore, alkaline pH, which favors formation of bicarbonate from CO2, as well as bicarbonate, mitigates growth restriction of pathogenic Yersinia (Ogg et al., 1958; Delwiche et al., 1959). So, is the interaction between the T3SS regulators YscM1/YscM2 and the CO2-fixing PEPC the key to understanding Yersinia growth cessation under conditions leading to T3SS activation?
The Yersinia virulence plasmid-encoded T3SS comprises about 45 components (Cornelis et al., 1998). The regulatory gene lcrQ in Y. pestis and Y. pseudotuberculosis was presumably duplicated in Y. enterocolitica to give yscM1, encoding a protein 99% identical to LcrQ, and, yscM2, whose product is almost 60% identical to YscM1 and LcrQ. YscM1 and YscM2 are functionally redundant in the control of yop gene expression (Stainier et al., 1997). However, while both were shown to interact with PEPC, only YscM1 influenced PEPC activity in vitro (Schmid et al., 2009). In fact, YscM1 is a potent inhibitor of PEPC. What is more, YscM1 (LcrQ) and YscM2 are also T3SS secretion substrates and are injected into host cells (Pettersson et al., 1996; Cambronne et al., 2000). Their status as secretion substrates is crucial since induction of type III secretion (accompanied by growth cessation) decreases intracellular YscM/LcrQ levels (Pettersson et al., 1996). In other words, how can inhibition of PEPC activity by YscM1/LcrQ explain growth restriction given that the YscM1/LcrQ level in Yersinia is low under these conditions? Perhaps this has to do with the complex protein/protein interaction network in which these regulators are intertwined with at least seven possible interaction partners besides PEPC (Cambronne et al., 2000; Swietnicki et al., 2004; Dittmann et al., 2007; Wilharm et al., 2007; Li et al., 2014a,b). Moreover, there is also evidence that PEPC is not the only junction between the YscM proteins and metabolism in Yersinia.
Stable isotope labeling experiments with universally 13C-labeled glucose followed by mass spectrometry analyses of the labeling patterns of amino acids were performed to assess the influence of PEPC, YscM1 and YscM2 on carbon fluxes during Yop secretion (Schmid et al., 2009). The data showed that PEPC replenishes the oxaloacetate pool of the TCA cycle under Yop secretion conditions. Deletion of either yscM1 or yscM2 caused slight changes of carbon fluxes in glycolysis and/or the Entner-Doudoroff pathway, the pentose phosphate pathway, the TCA cycle and amino acid biosynthesis. Collectively, these results suggest that not only the PEPC reaction but also one or several other central metabolic reactions are modulated via yscM1/yscM2. Strikingly, while purified YscM1 and YscM2 behaved differently with respect to PEPC inhibition in vitro, as mentioned above, the yscM1 and yscM2 deletion strains were indistinguishable with respect to 13C labeling patterns.
Although it is evident that much has to be done to get a clearer picture of the interrelatedness of virulence and metabolism in Yersinia, it is already reasonable to speculate on possible functions of such a cross-talk. Yersinia type III secretion is demanding with respect to both synthesis of Yops and provision of energy for their secretion (Wilharm et al., 2007; Schmid et al., 2009). Thus, it seems plausible that functioning of the T3SS under in vivo conditions requires the coordinated adaptation of metabolic pathways. Due to their bifunctionality, YscM/LcrQ proteins are ideally suited to coordinate yop gene expression and metabolic requirements. Since YscM/LcrQ interact with multiple T3SS components, and in particular chaperones of the Yop effectors, they might integrate information on the status of the T3SS, e.g., by sensing if the chaperones are charged with Yops, and transduce this information into control of both Yop production and the anaplerotic PEPC reaction, thereby balancing amino acid biosynthesis and energy provision.
A crucial role of the Yersinia T3SS is to prevent phagocytosis. The T3SS is therefore fully assembled at 37°C and a pre-synthesized pool of Yops is “ready-to-go” for microinjection. As the extracellular milieu of the host is rich in calcium, growth restriction will not occur and Yersinia is able to replicate. Upon phagocytic attack, a very rapid and concerted reaction is pivotal for Yersinia. It seems plausible that maintenance of energy charge for translocation of Yop effectors and replenishment of Yops have to be prioritized over continued replication under these conditions. Sensing an attacking cell might thus trigger a growth cessation programe, accompanied by reprogramming of metabolic pathways in which the PEPC-YscM/LcrQ interaction plays a role.
In support, Meng et al. (2010) demonstrated that phosphoenolpyruvate synthase and glutaminase change phosphorylation/modification status upon induction of the T3SS in Y. enterocolitica.
Aspartase: A Key to Understanding Yersinia Virulence
Fully virulent epidemic isolates of Y. pestis exhibit a lower aspartase activity than attenuated Y. pestis isolates and the enteropathogenic Y. pseudotuberculosis and Y. enterocolitica (Dreyfus and Brubaker, 1978; Bearden et al., 2009; Bearden and Brubaker, 2010). Aspartase (aspartate ammonia lyase, AspA) catalyses the deamination of aspartate to yield fumarate, an intermediate of the tricarboxylic acid (TCA) cycle. This reaction can thus be regarded as anaplerotic, and its absence from Y. pestis results in a net loss of carbon excreted in the form of aspartate (Dreyfus and Brubaker, 1978; Bearden and Brubaker, 2010). Fowler and Brubaker (1994) pointed out a specific role of carbon dioxide fixation reactions in relation to the aspartase deficiency identified in Y. pestis (Dreyfus and Brubaker, 1978). Carbon dioxide fixing reactions such as formation of the TCA-cycle intermediate oxaloacetate from phosphoenolpyruvate by the phosphoenolpyruvate carboxylase (PEPC, see previous section) might be especially important to replenish the TCA cycle and to compensate for the aspartase deficiency in Y. pestis. The aspartase deficiency might explain why growth restriction associated with low calcium response (LCR) is more prominent in Y. pestis compared to the enteropathogenic Yersinia (Bearden and Brubaker, 2010). According to Brubaker and colleagues, a key to explaining the exceptional virulence of epidemic Y. pestis compared to attenuated enzootic strains and to the enteropathogenic Yersinia is to understand how aspartase deficiency increases the virulence of Y. pestis (Viola et al., 2008; Bearden et al., 2009; Bearden and Brubaker, 2010). In support of a direct linkage between virulence and aspartase in Y. pestis, aspA is down-regulated in a mutant strain lacking the T3SS regulator gene lcrG (Du et al., 2009).
Uptake and Degradation of Fatty Acids Depends on the Presence of Virulence Plasmids in Y. pestis
The cross-talk between virulence functions and metabolism in Y. pestis is presumably even more complicated. Moncla et al. (1983) demonstrated constitutive uptake and degradation of fatty acids in Y. pestis via β–oxidation and the glyoxylate shunt. Intriguingly, fatty acid uptake was found to depend on the T3SS-encoding plasmid pCD1 as well as on the small virulence plasmid encoding the plasminogen activator (Pla) and pesticin (Pst). Unfortunately, these interesting findings were not followed-up. However, transcriptional profiling of wild-type Y. pestis and a strongly attenuated mutant lacking lcrG indicated up-regulation of aceA and aceB, encoding the key enzymes of the glyoxylate shunt, isocitrate lyase and malate synthase, in the lcrG mutant, which supports the interrelationship between T3SS and fatty acid metabolism (Du et al., 2009). Furthermore, differential expression of aceA and aceB upon a shift from 26 to 37°C was reported in two studies (Han et al., 2004; Motin et al., 2004), supporting a specific role of fatty acid metabolism during infection.
Another illustration of the complexity of the network is the fact that acetyl-CoA, fatty acids and fatty acyl-CoA are allosteric activators of PEPC, and β–oxidation of fatty acids, which leads to acetyl-CoA, requires oxaloacetate as an acceptor (Morikawa et al., 1980; Sauer and Eikmanns, 2005). Furthermore, L-malate, a product of the glyoxylate shunt reactions, is an allosteric inhibitor of PEPC.
Catabolite Repression Controls type III Effector Production in Yersinia
Zhan et al. (2008, 2009) demonstrated that cyclic AMP receptor protein (CRP) controls expression of the sycO-ypkA-yopJ operon in Y. pestis. This operon encodes two effectors of the T3SS, YopJ (YopP in Y. enterocolitica) and YpkA (YopO in Y. enterocolitica), together with SycO, the YpkA chaperone. The cAMP/CRP complex, but not CRP alone, binds to the promoter region of sycO-ypkA-yopJ, suggesting that high cAMP levels repress the sycO-ypkA-yopJ operon (Zhan et al., 2009). Zhan et al. (2008) defined a minimal CRP regulon by identifying all genes affected by the crp deletion. Besides ypkA the 37 genes or operons of the minimal CRP regulon include those encoding pesticin (pst) and plasminogen activator (pla), which are on the small virulence plasmid. Strikingly, expression of aspartase gene aspA was also strongly influenced by crp (Zhan et al., 2008). While Δcrp substantially increased ypkA expression, it dramatically lowered expression of pst, pla and aspA, suggesting these inversely regulated factors are required under completely different physiological conditions. These results indicate the effector YpkA (and possibly also YopJ) is only engaged under certain physiological conditions, since other yop genes are not part of the CRP regulon (Zhan et al., 2008). It is also worth mentioning that SycO interacts with YscM1 and regulates Yop production (Dittmann et al., 2007). SycO may thus also interfere with the PEPC-YscM1 regulatory network.
Taken together, a densely interwoven regulatory network connects the T3SS along with other virulence factors to the central carbon metabolism in Yersinia. The relationships depicted above are graphically summarized in Figure 1. It is important to note that very recently a study on the evolution of Yersinia identified that different from the previous view (Carniel, 2002; Wren, 2003) acquisition of the pYV virulence plasmid occurred independently in the Y. enterocolitica and Y. pseudotuberculosis/Y. pestis lineages after their separation (Reuter et al., 2014). Consequently, the regulatory networks in which the T3SSs are embedded can differ for the two lineages as can the specific interrelations between metabolism and the respective T3SS.
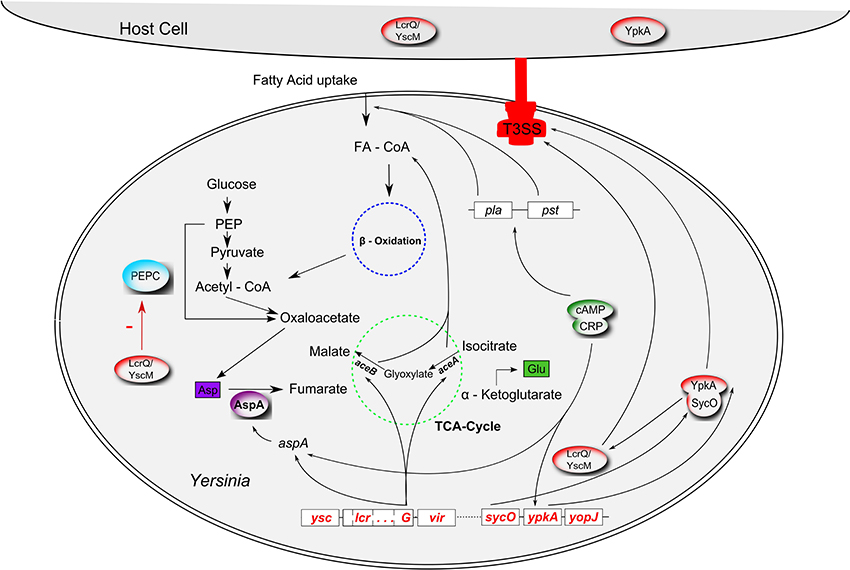
Figure 1. Interrelations between virulence factors and metabolism in Yersinia. The type 3 secretion system (T3SS; red), encoded by the pCD virulence plasmid (called pYV in Y. enterocolitica) interferes with the anaplerotic enzyme phosphoenolpyruvate carboxylase (PEPC, blue) via the T3SS regulator LcrQ (YscM1 in Y. enterocolitica; inhibitor of PEPC); lcrG, another component of the low calcium response regulon, controls transcription of aceAB and aspA. The glyoxylate cycle enzymes isocitrate lyase and malate synthase, encoded by aceAB, fulfill an anaplerotic function required for fatty acid degradation (FA, fatty acid; FA-CoA, fatty acyl-CoA). AspA, an aspartase generating fumarate from aspartate can also be regarded as an anaplerotic enzyme. AspA seems to be catalytically inactive in Y. pestis. In an unknown way, plasmid pCD seems to be involved in fatty acid uptake together with another virulence plasmid, pPCP. Production of the pPCP-encoded virulence factors Pla (plasminogen activator) and Pst (pesticin) is controlled by cAMP (cyclic adenosine monophosphate) via the cAMP receptor protein CRP (“catabolite repression”). The cAMP/CRP complex represses the T3SS effector gene ypkA (yopO in Y. enterocolitica). SycO, the chaperone of YpkA, directly interacts with LcrQ/YscM1. Cross-talk between fatty acid metabolism, glyoxylate shunt and the PEPC reaction also occurs via allosteric effectors (FA, FA-CoA, malate; interactions not illustrated for reasons of clarity).
Pseudomonas aeruginosa T3SS: A Hunting Weapon?
The opportunistic pathogen Pseudomonas aeruginosa deploys a T3SS with many constituents highly homologous to that of the Yersinia T3SS. Their regulatory networks differ considerably, however, because Pseudomonas lacks a homolog of the Yersinia LcrQ/YscM regulators. Another important difference concerns the use of the respective T3SS. While immune evasion and avoidance of collateral damage seems to be a major issue for Yersinia, the situation seems less clear for P. aeruginosa. The pore-forming activity of the P. aeruginosa T3SS results in macrophage oncosis and release of a chemoattractant from these oncotic cells (Dacheux et al., 2001b). This leads to rapid accumulation of pseudomonads around the cells, a phenomenon called pack swarming. It seems plausible that leakage of the oncotic cells is exploited by the bacteria for their nutrition and, thus, that the Pseudomonas T3SS is deployed both for immune evasion and as a weapon to hunt for nutrients.
Several studies have shown that production of the T3SS of Pseudomonas aeruginosa is affected by the metabolic state of the cells. The pyruvate dehydrogenase operon aceAB (please note the confusing nomenclature, these genes are unrelated to genes with identical names encoding the isocitrate lyase and malate synthase enzymes of the glyoxylate shunt) is required for maximal production of the T3SS (Dacheux et al., 2002). While the growth defect of aceAB mutants could be restored by acetate supplementation, the associated defect in T3SS production could be not, suggesting an interaction between aceAB gene products and the T3SS signaling cascade, rather than a global metabolic defect restricting expression of the T3SS genes.
Under T3SS-inducing conditions, adenylate cyclases CyaA and CyaB control expression of the T3SS genes via the cAMP binding protein Vfr (Wolfgang et al., 2003; Smith et al., 2004; Rietsch and Mekalanos, 2006), but global catabolite repression control in Pseudomonas is not critically dependent on Vfr (Suh et al., 2002), a close homolog of the cAMP receptor protein (CRP) of E. coli. A metabolic signal derived from acetyl-CoA and controlling the P. aeruginosa T3SS might also control activity of CyaA and/or CyaB (Rietsch and Mekalanos, 2006).
Also of interest here is the fact that histidine utilization in P. aeruginosa interferes with production and translocation of the T3SS cytotoxic effector ExoS (Rietsch et al., 2004), indicated by the observation that a transposon insertion leading to loss of ExoS-dependent cytotoxicity causes over-expression of several histidine utilization (hut) genes. Rietsch and colleagues showed that the loss of cytotoxicity could be suppressed by excluding histidine from the medium and by deletion of several hut genes. Another suppressing mutation was localized in cbrA, encoding the sensor of a two-component system involved in sensing of- and responding to carbon-nitrogen imbalance. Such an imbalance might result from excessive histidine catabolism. Tryptophan catabolism also interferes with type III secretion in P. aeruginosa (Shen et al., 2008). This linkage might be explained by the role of tryptophan as a precursor of quorum sensing-like signaling molecules, given that expression of the P. aeruginosa T3SS genes is cell density-dependent.
Collectively, several studies suggest that virulence gene expression and the nutritional situation of P. aeruginosa are coordinated during infection. It is interesting to note that the broad metabolic adaptation manifested by P. aeruginosa associated with long-term colonization of cystic fibrosis (CF) patients is accompanied by a down-regulation of ExoS production (Hogardt et al., 2007; Hoboth et al., 2009). In the light of the interdependence of metabolism and virulence, down-regulation of ExoS in this case might be explained by metabolic adjustments that lead to altered regulation of T3SS gene expression. In line with this explanation, ExoS down-regulation in late CF isolates can be overcome by overproduction of the T3SS transcriptional activator ExsA (Dacheux et al., 2001a; Hoboth et al., 2009). Finally, since P. aeruginosa T3SS is apparently designed to provide nutrients to the bacteria, coordination between metabolic states and expression of virulence factor genes is intuitive.
Salmonella: T3SS-Mediated Recruitment of Nutrients for My Brother
Tetrathionate has long been used as a supplement to media for enrichment of Salmonella (Muller, 1923). However, only in recent years has the biological context of tetrathionate usage by Salmonella been elucidated in more detail, revealing an intricate relation to T3SS-based virulence. Salmonella species causing typhoid fever and gastroenteritis deploy two T3SSs encoded in Salmonella pathogenicity islands SPI-1 and SPI-2, here termed T3SS-1 and T3SS-2, respectively. With reference to the link between pathogenicity and metabolism, a sub-population of Salmonella Typhimurium seems to use the T3SS-1 altruistically but self-destructive to the nutritional benefit of the surviving subpopulation (Ackermann et al., 2008). During colonization of the mouse gut, only a subpopulation of S. Typhimurium expresses the T3SS-1 genes (15% of the salmonellae found in the lumen), which then invades the gut tissue and triggers an inflammatory response. While the invading bacteria seem to be killed, the subpopulation residing in the gut lumen is enabled to compete with the normal gut flora and presumably to acquire the released nutrients. Specifically, Winter et al. (2010) could show that the respiratory burst induced by the T3SS-1 results in formation of the alternative respiratory electron acceptor tetrathionate in the gut. As a consequence, tetrathionate then enables salmonellae to use ethanolamine as a substrate which is abundant and provided by the host yet does not promote growth effectively by fermentation (Thiennimitr et al., 2011) (Figure 2). It is interesting to note that the ttr genes required for tetrathionate respiration are encoded within SPI-2 in close proximity to T3SS-2 genes (Hensel et al., 1999) although it is hitherto unknown whether there is a linkage between ttr genes and T3SS-2 genes beyond that co-localization within SPI-2.
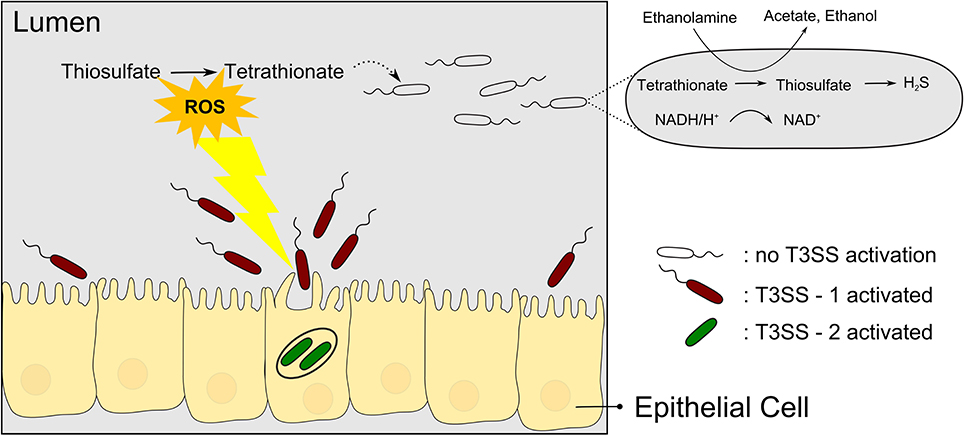
Figure 2. Salmonellae recruit new nutrients for their brothers by means of self-destructive deployment of a T3SS. T3SS-1 mediated invasion of the gut tissue is mostly a kamikaze mission but the inflammatory response induced comprises the release of reactive oxygen species (ROS) which results in the formation of tetrathionate, an alternative electron acceptor. Surviving salmonellae can then use tetrathionate to metabolize ethanolamine to proliferate and outcompete the normal gut flora.
It is worthwhile mentioning here that Y. enterocolitica also harbors the tetrathionate pathway suggesting that inflammatory responses are exploited in a similar way (Thomson et al., 2006; Winter et al., 2010).
Another layer of complexity is observable in Salmonella Typhimurium strains harboring the SopEΦ prophage. SopE, an effector protein translocated by T3SS-1, can induce increased levels of nitric oxide (NO) within the mouse gut, which as a net result of interaction with reactive oxygen species (ROS) can lead to increased nitrate levels, that is generation of another alternative electron acceptor that is preferred over tetrathionate and promoting growth in the gut lumen (Lopez et al., 2012).
Taken together, the Salmonella T3SS-1 is not only deployed to mediate entry into host cells by the manipulation of host cellular signaling but also to unlock novel nutritional resources, suggesting intimate cross-talk between the physiological state and the expression of virulence factor genes. In fact, the central regulator gene of the Salmonella T3SS-1, hilA, is activated by carbon source deprivation and (p)ppGpp (Pizarro-Cerda and Tedin, 2004; Song et al., 2004). Moreover, hilA and many other SPI-1 encoded genes are controlled by metabolic enzyme genes, adhE, encoding a bifunctional acetaldehyde-CoA/alcohol dehydrogenase and pflB, encoding pyruvate formate lyase I, of the pyruvate metabolism (Abernathy et al., 2013). Furthermore, expression of several virulence factor genes including those of T3SS–1, is controlled by short-chain fatty acids that might serve as nutrients in the intestine (El-Gedaily et al., 1997; Utley et al., 1998; Lawhon et al., 2002). The DNA-binding protein Fis might play a decisive role in coordinating metabolism and virulence in Salmonella (Kelly et al., 2004), since it controls expression of genes in several pathogenicity islands on the one hand and of genes encoding metabolic pathway components involved in fatty acid and acetate metabolism including the glyoxylate shunt on the other. The role of the glyoxylate shunt during infection in particular is unclear since it seems to depend on infection stages and differs between Salmonella serovars (Utley et al., 1998; Faucher et al., 2006; Tchawa Yimga et al., 2006; Eisenreich et al., 2010; Bowden et al., 2014). In Salmonella central metabolic enzymes including key enzymes of glycolysis, gluconeogenesis, TCA cycle and glyoxylate shunt are extensively acetylated to control metabolic fluxes, imposing yet another layer of complexity in understanding regulatory networks (Wang et al., 2010).
Very recently, another striking example of a direct interrelationship between a T3SS and metabolism was discovered in Salmonella (Maze et al., 2014). The phosphotransferase system (PTS) of bacteria catalyses the uptake and concomitant phosphorylation of sugars using phosphoenolpyruvate (PEP) as donor of the phosphoryl group (Deutscher et al., 2006). Now, the PTS component EIIAGlc was shown to associate with the SPI-2 encoded T3SS-2 (Maze et al., 2014). Moreover, effector secretion by the T3SS-2 required EIIAGlc and systemic Salmonella virulence was found to critically depend on EIIAGlc. Interestingly, however, the phosphorylation-dependent sugar transport function of EIIAGlc and its regulatory impact on adenylate cyclase activity did not account for the phenotype. Rather, a direct interaction between EIIAGlc and T3SS-2 was suggested (Maze et al., 2014). It remains to be determined whether this moonlighting function of EIIAGlc is of purely structural nature or whether it contributes to a coordination of metabolic and virulence functions.
All in all, the literature on Salmonella contains numerous arguments of a close interrelation between metabolism and virulence, notably referring to the T3SSs.
Further Examples
The examples presented and discussed above are systems in which the link between metabolism and T3SS has been most extensively studied. Interested readers will find further examples and hints in studies on the Aeromonas T3SS. The regulatory interplay between the pyruvate dehydrogenase complex and T3SS regulator genes aexT and aopN in A. hydrophila reported by Vilches et al. (2009) is strongly reminiscent of the interrelation between aceAB and T3SS in P. aeruginosa discovered by Dacheux et al. (2002) and discussed above.
In another example, enteropathogenic E. coli (EPEC) secrete glyceraldehyde-3-phosphate dehydrogenase (GAPDH) via a T3SS (Kenny and Finlay, 1995). GAPDH might be injected into the host cell cytosol and interfere with host cellular metabolism e.g., by inhibiting host cellular GAPDH complexes through formation of unproductive heterooligomers or by increasing fluxes within the glycolytic pathway. Alternatively, GAPDH has a moonlighting function independent of its enzymatic activity, fulfilling in bacteria and/or the host. For example, GAPDH is also secreted by different Streptococcus species, and exhibits ADP-ribosylating activity and immunomodulatory properties (Pancholi and Fischetti, 1992, 1993; Madureira et al., 2007). It is interesting to note in this context, that a T3SS effector, NleB, found in EPEC and EHEC (enterohaemorrhagic E. coli), was recently found to act as glycosyltransferase toward host cellular GAPDH to inhibit NF-κB activation, illustrating yet another moonlighting facet of GAPDH (Gao et al., 2013). It remains to be determined whether it is just by chance that EPEC secrete and possibly translocate GAPDH into host cells in which they also target host cellular GAPDH and whether these phenomena are first indications of an interlock of metabolism and T3SS on both the bacterial and the host cellular side.
Finally, AdhE, a bifunctional acetaldehyde-CoA dehydrogenase/alcohol dehydrogenase was shown to control expression of the EHEC T3SS (Beckham et al., 2014), a finding reminiscent of the relation between adhE and SPI-1 found in Salmonella (Baumler et al., 1994; Abernathy et al., 2013).
Conclusions and Perspectives
A Novel Interrelationship Between Virulence Functions and Metabolism
In very general terms, the environment within the host determines the physiological state of the pathogen, which in turn governs production and functioning of virulence factors. Our knowledge of the complexity of these regulatory networks is further increasing (as illustrated above; see Table 1 for summary). Our way of looking at bacterial pathogenesis is still dominated by the question of which virulence factors are produced and how metabolism controls virulence gene expression. However, virulence factors conversely control metabolic functions in a coordinated manner. Furthermore, there is evidence that virulence functions are deployed to manipulate host metabolism, potentially for the benefit of the metabolism in the pathogen. Clearly, we face a novel interrelationship between virulence and metabolic functions. However, do such phenomena reflect the exception or the rule?
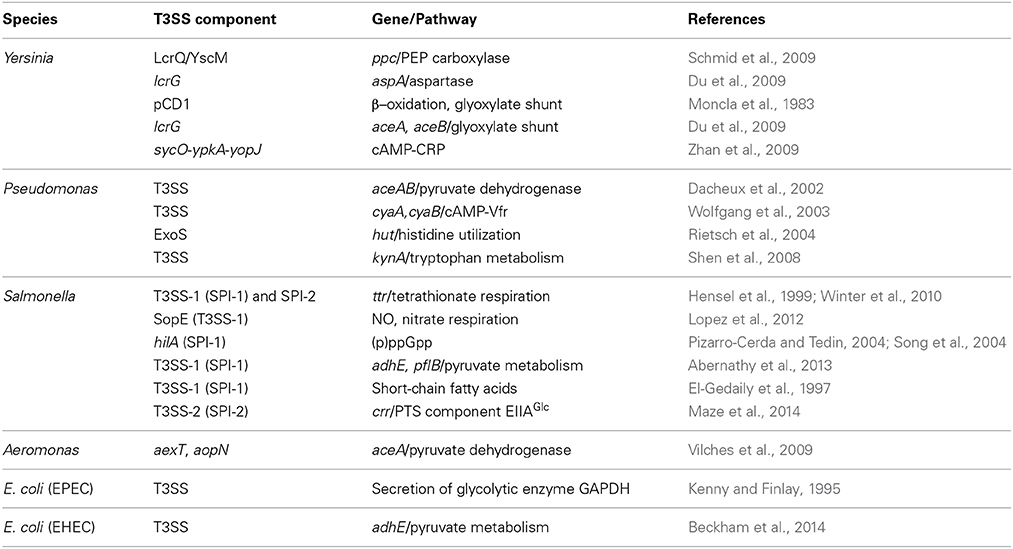
Table 1. Summary of the metabolic pathways impacted by T3SSs and/or vice versa in the order of appearance in this manuscript.
The multiple convergent evolution of dense regulatory networks to coordinate virulence and metabolic functions suggests strong selection pressures acting and indicates that such cross-talk could be the rule rather than the exception. From a cybernetic point of view, many regulator circuits seem plausible.
Cross-Talk Via Anaplerotic Functions: A Common Theme
From the point of view of the invader, infection usually means unbalanced diet and shortage. From in vitro studies it is known that growing on a limited number of carbon sources requires considerable reprogramming of metabolic fluxes via anaplerotic enzymes. From the above examples, it seems as if such anaplerotic enzymes are preferred nodal points for communication with virulence functions, lending support to the view that “unbalanced diet” is a condition frequently faced by pathogens.
Agrobacterium tumefaciens induces crown gall tumors on its plant hosts by means of a type 4 secretion system (T4SS). The virulence (vir) genes are localized on the Ti (tumor-inducing) virulence plasmid and are controlled by the two-component system VirAG. Deletion of pckA encoding the anaplerotic enzyme phosphoenolpyruvate carboxykinase (PckA) attenuates virulence through reduced vir gene expression (Liu et al., 2005) illustrating that such regulatory circuits are not restricted to pathogens running T3SSs.
This view is further underscored by the fact that anaplerotic reactions play a key role during infection in many pathogens with less well-defined armament. A crucial function of the glyoxylate shunt during infection, for example, has been demonstrated for Mycobacterium tuberculosis (McKinney et al., 2000) and for the fungal pathogen Candida albicans (Lorenz and Fink, 2001).
Metabolism of the Host
For reasons of clarity and scope, this review was confined mainly to the central carbon metabolism of bacterial pathogens and its interplay with virulence functions related to T3SSs. Availability of all kind of nutrients (not only of carbon), however, depends on host metabolism, which differs depending on localization within tissues and cells and, moreover, is not static but changes over the course of infection. As an example, sepsis is accompanied by a massive redistribution of carbon and nitrogen fluxes within the infected individual, accompanied by decreased glutamine uptake in the intestine and a switch from glutamine uptake state to release in the kidney. In contrast, proliferating lymphocytes increase glutamine utilization upon sepsis and the liver becomes the major glutamine consumer under these conditions (Karinch et al., 2001). Consequently, many pathogens will have to adapt their metabolism to rearrangements of the host metabolism.
Changes of host metabolism in the context of infection can be viewed as a host response or as directed manipulation of host metabolism by the pathogen. As discussed above, there are hints that virulence factors are deployed to manipulate host metabolism and to serve the release of nutrients. The function of virulence factors is often discussed in a biased way, neglecting that the primary goal of the pathogen is not to fool the host but to replicate in it. We expect that manipulating host metabolism, on the single host cell level as well as on the tissue, organ and systemic level will turn out to be a major issue of pathogens.
Systematic Investigation of Cross-Talk Between Virulence and Metabolism: Toward a Systemic View of Infection
Much of the above discussion is based on incidental observations. So, how should we examine cross-talk between metabolism and virulence functions more systematically? As a first step, literature from the pioneers studying pathogens may still store many valuable hints to follow-up. Further, it is important to screen the transcriptomics, proteomics and metabolomics datasets to identify novel interrelationships. Clearly, a comprehensive view will require other “omics” such as “interactomics” and “fluxosomics,” including data on the host. Integration of these data to form a penetrable view of infection will be a major intellectual challenge. Feeding systems biologists with “omics” data is a must to drive modeling toward a systemic view of infection. It is however clear that applicability of modern systems biology approaches is still limited from an economic and data handling perspective but also with regard to sensitivity as well as temporal and spatial resolution. For example, bacterial pathogens practice division of labor as do cells of their hosts so that the different events on which we sum up in most experiments cannot be resolved and the results misguide us. Moreover, type three secretion is not only a very complex but also a highly dynamic process which poses a specific challenge to the temporal resolution of our methodology. Single cell analysis is now rapidly developing on the “omics” level and at least DNA and RNA sequencing will become broadly available (Pan, 2014) to resolve some of the heterogeneity that we are faced with.
In conclusion, cohesive use of physiology and molecular biology is needed to enlighten the complex events of infection: we cannot understand infection without understanding of metabolism of host and the pathogen.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work in the authors' lab is supported by the DFG priority programme 1316 “Host-adapted metabolism of bacterial pathogens.”
References
Abernathy, J., Corkill, C., Hinojosa, C., Li, X., and Zhou, H. (2013). Deletions in the pyruvate pathway of Salmonella Typhimurium alter SPI1-mediated gene expression and infectivity. J. Anim. Sci. Biotechnol. 4, 5. doi: 10.1186/2049-1891-4-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ackermann, M., Stecher, B., Freed, N. E., Songhet, P., Hardt, W. D., and Doebeli, M. (2008). Self-destructive cooperation mediated by phenotypic noise. Nature 454, 987–990. doi: 10.1038/nature07067
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bacon, G. A., and Burrows, T. W. (1956). The basis of virulence in Pasteurella pestis: an antigen determining virulence. Br. J. Exp. Pathol. 37, 481–493.
Baugh, C. L., Lanham, J. W., and Surgalla, M. J. (1964). Effects of bicarbonate on growth of pasteurella pestis. ii. carbon dioxide fixation into oxalacetate by cell-free extracts. J. Bacteriol. 88, 553–558.
Baumler, A. J., Kusters, J. G., Stojiljkovic, I., and Heffron, F. (1994). Salmonella typhimurium loci involved in survival within macrophages. Infect. Immun. 62, 1623–1630.
Bearden, S. W., and Brubaker, R. R. (2010). Recent findings regarding maintenance of enzootic variants of Yersinia pestis in sylvatic reservoirs and their significance in the evolution of epidemic plague. Vector Borne Zoonotic Dis. 10, 85–92. doi: 10.1089/vbz.2009.0043
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bearden, S. W., Sexton, C., Pare, J., Fowler, J. M., Arvidson, C. G., Yerman, L., et al. (2009). Attenuated enzootic (pestoides) isolates of Yersinia pestis express active aspartase. Microbiology 155, 198–209. doi: 10.1099/mic.0.021170-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beckham, K. S., Connolly, J. P., Ritchie, J. M., Wang, D., Gawthorne, J. A., Tahoun, A., et al. (2014). The metabolic enzyme AdhE controls the virulence of Escherichia coli O157:H7. Mol. Microbiol. 93, 199–211. doi: 10.1111/mmi.12651
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Bowden, S. D., Hopper-Chidlaw, A. C., Rice, C. J., Ramachandran, V. K., Kelly, D. J., and Thompson, A. (2014). Nutritional and metabolic requirements for the infection of HeLa cells by Salmonella enterica serovar Typhimurium. PLoS ONE 9:e96266. doi: 10.1371/journal.pone.0096266
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cambronne, E. D., Cheng, L. W., and Schneewind, O. (2000). LcrQ/YscM1, regulators of the Yersinia yop virulon, are injected into host cells by a chaperone-dependent mechanism. Mol. Microbiol. 37, 263–273. doi: 10.1046/j.1365-2958.2000.01974.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Carniel, E. (2002). Plasmids and pathogenicity islands of Yersinia. Curr. Top. Microbiol. Immunol. 264, 89–108. doi: 10.1007/978-3-642-56031-6_6
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cornelis, G. R., Boland, A., Boyd, A. P., Geuijen, C., Iriarte, M., Neyt, C., et al. (1998). The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62, 1315–1352.
Dacheux, D., Attree, I., and Toussaint, B. (2001a). Expression of ExsA in trans confers type III secretion system-dependent cytotoxicity on noncytotoxic Pseudomonas aeruginosa cystic fibrosis isolates. Infect. Immun. 69, 538–542. doi: 10.1128/IAI.69.1.538-542.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dacheux, D., Epaulard, O., De Groot, A., Guery, B., Leberre, R., Attree, I., et al. (2002). Activation of the Pseudomonas aeruginosa type III secretion system requires an intact pyruvate dehydrogenase aceAB operon. Infect. Immun. 70, 3973–3977. doi: 10.1128/IAI.70.7.3973-3977.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dacheux, D., Goure, J., Chabert, J., Usson, Y., and Attree, I. (2001b). Pore-forming activity of type III system-secreted proteins leads to oncosis of Pseudomonas aeruginosa-infected macrophages. Mol. Microbiol. 40, 76–85. doi: 10.1046/j.1365-2958.2001.02368.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dalebroux, Z. D., Svensson, S. L., Gaynor, E. C., and Swanson, M. S. (2010). ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 74, 171–199. doi: 10.1128/MMBR.00046-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Delwiche, E. A., Fukui, G. M., Andrews, A. W., and Surgalla, M. J. (1959). Environmental conditions affecting the population dynamics and the retention of virulence of Pasteurella pestis: the role of carbon dioxide. J. Bacteriol. 77, 355–360.
Deutscher, J., Francke, C., and Postma, P. W. (2006). How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031. doi: 10.1128/MMBR.00024-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Devignat, R., and Schoetter, M. (1942). Le bacille de yersin en milieu aere. Rec. Trav. Sci. Med. Congo Belge 1, 161–181.
Dittmann, S., Schmid, A., Richter, S., Trulzsch, K., Heesemann, J., and Wilharm, G. (2007). The Yersinia enterocolitica type three secretion chaperone SycO is integrated into the Yop regulatory network and binds to the Yop secretion protein YscM1. BMC Microbiol. 7:67. doi: 10.1186/1471-2180-7-67
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dreyfus, L. A., and Brubaker, R. R. (1978). Consequences of aspartase deficiency in Yersinia pestis. J. Bacteriol. 136, 757–764.
Du, Z., Tan, Y., Yang, H., Qiu, J., Qin, L., Wang, T., et al. (2009). Gene expression profiling of Yersinia pestis with deletion of lcrG, a known negative regulator for Yop secretion of type III secretion system. Int. J. Med. Microbiol. 299, 355–366. doi: 10.1016/j.ijmm.2008.10.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Eisenreich, W., Dandekar, T., Heesemann, J., and Goebel, W. (2010). Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8, 401–412. doi: 10.1038/nrmicro2351
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
El-Gedaily, A., Paesold, G., Chen, C. Y., Guiney, D. G., and Krause, M. (1997). Plasmid virulence gene expression induced by short-chain fatty acids in Salmonella dublin: identification of rpoS-dependent and rpo-S-independent mechanisms. J. Bacteriol. 179, 1409–1412.
Faucher, S. P., Porwollik, S., Dozois, C. M., McClelland, M., and Daigle, F. (2006). Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. U.S.A. 103, 1906–1911. doi: 10.1073/pnas.0509183103
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fowler, J. M., and Brubaker, R. R. (1994). Physiological basis of the low calcium response in Yersinia pestis. Infect. Immun. 62, 5234–5241.
Fukui, G. M., Ogg, J. E., Wessman, G. E., and Surgalla, M. J. (1957). Studies on the relation of cultural conditions and virulence of Pasteurella pestis. J. Bacteriol. 74, 714–717.
Gao, X., Wang, X., Pham, T. H., Feuerbacher, L. A., Lubos, M. L., Huang, M., et al. (2013). NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-kappaB activation. Cell Host Microbe 13, 87–99. doi: 10.1016/j.chom.2012.11.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gorke, B., and Stulke, J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6, 613–624. doi: 10.1038/nrmicro1932
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Han, Y., Zhou, D., Pang, X., Song, Y., Zhang, L., Bao, J., et al. (2004). Microarray analysis of temperature-induced transcriptome of Yersinia pestis. Microbiol. Immunol. 48, 791–805. doi: 10.1111/j.1348-0421.2004.tb03605.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heesemann, J., Algermissen, B., and Laufs, R. (1984). Genetically manipulated virulence of Yersinia enterocolitica. Infect. Immun. 46, 105–110.
Hensel, M., Hinsley, A. P., Nikolaus, T., Sawers, G., and Berks, B. C. (1999). The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32, 275–287. doi: 10.1046/j.1365-2958.1999.01345.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Heroven, A. K., Bohme, K., and Dersch, P. (2012). The Csr/Rsm system of Yersinia and related pathogens: a post-transcriptional strategy for managing virulence. RNA Biol. 9, 379–391. doi: 10.4161/rna.19333
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Higuchi, K., and Carlin, C. E. (1958). Studies on the nutrition and physiology of Pasteurella pestis. II. A defined medium for the growth of Pasteurella pestis. J. Bacteriol. 75, 409–413.
Hoboth, C., Hoffmann, R., Eichner, A., Henke, C., Schmoldt, S., Imhof, A., et al. (2009). Dynamics of adaptive microevolution of hypermutable Pseudomonas aeruginosa during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 200, 118–130. doi: 10.1086/599360
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hogardt, M., Hoboth, C., Schmoldt, S., Henke, C., Bader, L., and Heesemann, J. (2007). Stage-specific adaptation of hypermutable Pseudomonas aeruginosa isolates during chronic pulmonary infection in patients with cystic fibrosis. J. Infect. Dis. 195, 70–80. doi: 10.1086/509821
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Karinch, A. M., Pan, M., Lin, C. M., Strange, R., and Souba, W. W. (2001). Glutamine metabolism in sepsis and infection. J. Nutr. 131, 2535S–2538S; discussion 2550S–2531S.
Kelly, A., Goldberg, M. D., Carroll, R. K., Danino, V., Hinton, J. C., and Dorman, C. J. (2004). A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150, 2037–2053. doi: 10.1099/mic.0.27209-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kenny, B., and Finlay, B. B. (1995). Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 92, 7991–7995. doi: 10.1073/pnas.92.17.7991
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kupferberg, L. L., and Higuchi, K. (1958). Role of calcium ions in the stimulation of growth of virulent strains of Pasteurella pestis. J. Bacteriol. 76, 120–121.
La, M. V., Raoult, D., and Renesto, P. (2008). Regulation of whole bacterial pathogen transcription within infected hosts. FEMS Microbiol. Rev. 32, 440–460. doi: 10.1111/j.1574-6976.2008.00103.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lawhon, S. D., Maurer, R., Suyemoto, M., and Altier, C. (2002). Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol. Microbiol. 46, 1451–1464. doi: 10.1046/j.1365-2958.2002.03268.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lee, V. T., Mazmanian, S. K., and Schneewind, O. (2001). A program of Yersinia enterocolitica type III secretion reactions is activated by specific signals. J. Bacteriol. 183, 4970–4978. doi: 10.1128/JB.183.17.4970-4978.2001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, L., Yan, H., Feng, L., Li, Y., Lu, P., Hu, Y., et al. (2014a). LcrQ blocks the role of LcrF in regulating the Ysc-Yop type III secretion genes in Yersinia pseudotuberculosis. PLoS ONE 9:e92243. doi: 10.1371/journal.pone.0092243
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Li, Y., Li, L., Huang, L., Francis, M. S., Hu, Y., and Chen, S. (2014b). Yersinia Ysc-Yop type III secretion feedback inhibition is relieved through YscV-dependent recognition and secretion of LcrQ. Mol. Microbiol. 91, 494–507. doi: 10.1111/ mmi.12474
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Liu, P., Wood, D., and Nester, E. W. (2005). Phosphoenolpyruvate carboxykinase is an acid-induced, chromosomally encoded virulence factor in Agrobacterium tumefaciens. J. Bacteriol. 187, 6039–6045. doi: 10.1128/JB.187.17.6039-6045.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lopez, C. A., Winter, S. E., Rivera-Chavez, F., Xavier, M. N., Poon, V., Nuccio, S. P., et al. (2012). Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. MBio 3:e00143-12. doi: 10.1128/mBio.00143-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Lorenz, M. C., and Fink, G. R. (2001). The glyoxylate cycle is required for fungal virulence. Nature 412, 83–86. doi: 10.1038/35083594
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Madureira, P., Baptista, M., Vieira, M., Magalhaes, V., Camelo, A., Oliveira, L., et al. (2007). Streptococcus agalactiae GAPDH is a virulence-associated immunomodulatory protein. J. Immunol. 178, 1379–1387. doi: 10.4049/jimmunol.178.3.1379
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Maze, A., Glatter, T., and Bumann, D. (2014). The central metabolism regulator EIIA(Glc) switches Salmonella from growth arrest to acute virulence through activation of virulence factor secretion. Cell Rep. 7, 1426–1433. doi: 10.1016/j.celrep.2014.04.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McKinney, J. D., Honer Zu Bentrup, K., Munoz-Elias, E. J., Miczak, A., Chen, B., Chan, W. T., et al. (2000). Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738. doi: 10.1038/35021074
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Meng, L., Yang, S. H., Kim, T.-J., and Suh, J.-W. (2010). PpsA, TufA, and glutaminase were modulated under Yop secretion conditions in Yersinia enterocolitica. J. Korean Soc. Appl. Biol. Chem. 53, 540–544. doi: 10.3839/jksabc.2010.083
Moncla, B. J., Hillier, S. L., and Charnetzky, W. T. (1983). Constitutive uptake and degradation of fatty acids by Yersinia pestis. J. Bacteriol. 153, 340–344.
Morikawa, M., Izui, K., Taguchi, M., and Katsuki, H. (1980). Regulation of Escherichia coli phosphoenolpyruvate carboxylase by multiple effectors in vivo. Estimation of the activities in the cells grown on various compounds. J. Biochem. 87, 441–449.
Motin, V. L., Georgescu, A. M., Fitch, J. P., Gu, P. P., Nelson, D. O., Mabery, S. L., et al. (2004). Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 186, 6298–6305. doi: 10.1128/JB.186.18.6298-6305.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Muller, L. (1923). Un nouveau milieu d'enrichissement pour la recherche du bacille typhique at paratyphique. C. R. Seances Soc. Biol. Fil. 89, 434–437.
Munoz-Elias, E. J., and McKinney, J. D. (2006). Carbon metabolism of intracellular bacteria. Cell. Microbiol. 8, 10–22. doi: 10.1111/j.1462-5822.2005.00648.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ogg, J. E., Friedman, S. B., Andrews, A. W., and Surgalla, M. J. (1958). Factors influencing the loss of virulence in Pasteurella pestis. J. Bacteriol. 76, 185–191.
Pan, X. (2014). Single cell analysis: from technology to biology and medicine. Single Cell Biol. 3:106. doi: 10.4172/2168-9431.1000106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pancholi, V., and Fischetti, V. A. (1992). A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176, 415–426. doi: 10.1084/jem.176.2.415
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pancholi, V., and Fischetti, V. A. (1993). Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc. Natl. Acad. Sci. U.S.A. 90, 8154–8158. doi: 10.1073/pnas.90.17.8154
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Panos, C., and Ajl, S. J. (1963). Metabolism of microorganisms as related to their pathogenicity. Annu. Rev. Microbiol. 17, 297–328. doi: 10.1146/annurev.mi.17.100163.001501
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pettersson, J., Nordfelth, R., Dubinina, E., Bergman, T., Gustafsson, M., Magnusson, K. E., et al. (1996). Modulation of virulence factor expression by pathogen target cell contact. Science 273, 1231–1233. doi: 10.1126/science.273.5279.1231
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pizarro-Cerda, J., and Tedin, K. (2004). The bacterial signal molecule, ppGpp, regulates Salmonella virulence gene expression. Mol. Microbiol. 52, 1827–1844. doi: 10.1111/j.1365-2958.2004.04122.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Poncet, S., Milohanic, E., Maze, A., Nait Abdallah, J., Ake, F., Larribe, M., et al. (2009). Correlations between carbon metabolism and virulence in bacteria. Contrib. Microbiol. 16, 88–102. doi: 10.1159/000219374
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ramamurthi, K. S., and Schneewind, O. (2002). Type iii protein secretion in yersinia species. Annu. Rev. Cell Dev. Biol. 18, 107–133. doi: 10.1146/annurev.cellbio.18.012502.105912
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reuter, S., Connor, T. R., Barquist, L., Walker, D., Feltwell, T., Harris, S. R., et al. (2014). Parallel independent evolution of pathogenicity within the genus Yersinia. Proc. Natl. Acad. Sci. U.S.A. 111, 6768–6773. doi: 10.1073/pnas.1317161111
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rietsch, A., and Mekalanos, J. J. (2006). Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol. Microbiol. 59, 807–820. doi: 10.1111/j.1365-2958.2005.04990.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Rietsch, A., Wolfgang, M. C., and Mekalanos, J. J. (2004). Effect of metabolic imbalance on expression of type III secretion genes in Pseudomonas aeruginosa. Infect. Immun. 72, 1383–1390. doi: 10.1128/IAI.72.3.1383-1390.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Romeo, T. (1998). Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29, 1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sauer, U., and Eikmanns, B. J. (2005). The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol. Rev. 29, 765–794. doi: 10.1016/j.femsre.2004.11.002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schmid, A., Neumayer, W., Trulzsch, K., Israel, L., Imhof, A., Roessle, M., et al. (2009). Cross-talk between type three secretion system and metabolism in Yersinia. J. Biol. Chem. 284, 12165–12177. doi: 10.1074/jbc.M900773200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Shen, D. K., Filopon, D., Chaker, H., Boullanger, S., Derouazi, M., Polack, B., et al. (2008). High-cell-density regulation of the Pseudomonas aeruginosa type III secretion system: implications for tryptophan catabolites. Microbiology 154, 2195–2208. doi: 10.1099/mic.0.2007/013680-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Smith, R. S., Wolfgang, M. C., and Lory, S. (2004). An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect. Immun. 72, 1677–1684. doi: 10.1128/IAI.72.3.1677-1684.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Song, M., Kim, H. J., Kim, E. Y., Shin, M., Lee, H. C., Hong, Y., et al. (2004). ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J. Biol. Chem. 279, 34183–34190. doi: 10.1074/jbc.M313491200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Stainier, I., Iriarte, M., and Cornelis, G. R. (1997). YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol. Microbiol. 26, 833–843. doi: 10.1046/j.1365-2958.1997.6281995.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Suh, S. J., Runyen-Janecky, L. J., Maleniak, T. C., Hager, P., Macgregor, C. H., Zielinski-Mozny, N. A., et al. (2002). Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology 148, 1561–1569.
Swietnicki, W., O'brien, S., Holman, K., Cherry, S., Brueggemann, E., Tropea, J. E., et al. (2004). Novel protein-protein interactions of the Yersinia pestis type III secretion system elucidated with a matrix analysis by surface plasmon resonance and mass spectrometry. J. Biol. Chem. 279, 38693–38700. doi: 10.1074/jbc.M405217200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tchawa Yimga, M., Leatham, M. P., Allen, J. H., Laux, D. C., Conway, T., and Cohen, P. S. (2006). Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 74, 1130–1140. doi: 10.1128/IAI.74.2.1130-1140.2006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thiennimitr, P., Winter, S. E., Winter, M. G., Xavier, M. N., Tolstikov, V., Huseby, D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 108, 17480–17485. doi: 10.1073/pnas.1107857108
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Thomson, N. R., Howard, S., Wren, B. W., Holden, M. T., Crossman, L., Challis, G. L., et al. (2006). The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206. doi: 10.1371/journal.pgen.0020206
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Utley, M., Franklin, D. P., Krogfelt, K. A., Laux, D. C., and Cohen, P. S. (1998). A Salmonella typhimurium mutant unable to utilize fatty acids and citrate is avirulent and immunogenic in mice. FEMS Microbiol. Lett. 163, 129–134. doi: 10.1111/j.1574-6968.1998.tb13036.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Vilches, S., Jimenez, N., Tomas, J. M., and Merino, S. (2009). Aeromonas hydrophila AH-3 type III secretion system expression and regulatory network. Appl. Environ. Microbiol. 75, 6382–6392. doi: 10.1128/AEM.00222-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Viola, R. E., Yerman, L., Fowler, J. M., Arvidson, C. G., and Brubaker, R. R. (2008). A missense mutation causes aspartase deficiency in Yersinia pestis. Microbiology 154, 1271–1280. doi: 10.1099/mic.0.2007/015529-0
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, Q., Zhang, Y., Yang, C., Xiong, H., Lin, Y., Yao, J., et al. (2010). Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007. doi: 10.1126/science.1179687
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wilharm, G., Dittmann, S., Schmid, A., and Heesemann, J. (2007). On the role of specific chaperones, the specific ATPase, and the proton motive force in type III secretion. Int. J. Med. Microbiol. 297, 27–36. doi: 10.1016/j.ijmm.2006.10.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Winter, S. E., Thiennimitr, P., Winter, M. G., Butler, B. P., Huseby, D. L., Crawford, R. W., et al. (2010). Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429. doi: 10.1038/nature09415
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wolfgang, M. C., Lee, V. T., Gilmore, M. E., and Lory, S. (2003). Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4, 253–263. doi: 10.1016/S1534-5807(03)00019-4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wren, B. W. (2003). The yersiniae–a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 1, 55–64. doi: 10.1038/nrmicro730
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yother, J., Chamness, T. W., and Goguen, J. D. (1986). Temperature-controlled plasmid regulon associated with low calcium response in Yersinia pestis. J. Bacteriol. 165, 443–447.
Zhan, L., Han, Y., Yang, L., Geng, J., Li, Y., Gao, H., et al. (2008). The cyclic AMP receptor protein, CRP, is required for both virulence and expression of the minimal CRP regulon in Yersinia pestis biovar microtus. Infect. Immun. 76, 5028–5037. doi: 10.1128/IAI.00370-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zhan, L., Yang, L., Zhou, L., Li, Y., Gao, H., Guo, Z., et al. (2009). Direct and negative regulation of the sycO-ypkA-ypoJ operon by cyclic AMP receptor protein (CRP) in Yersinia pestis. BMC Microbiol. 9:178. doi: 10.1186/1471-2180-9-178
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: virulence, metabolism, type three secretion system, T3SS, cross-talk, Yersinia, Salmonella, Pseudomonas
Citation: Wilharm G and Heider C (2014) Interrelationship between type three secretion system and metabolism in pathogenic bacteria. Front. Cell. Infect. Microbiol. 4:150. doi: 10.3389/fcimb.2014.00150
Received: 21 July 2014; Paper pending published: 31 August 2014;
Accepted: 07 October 2014; Published online: 27 October 2014.
Edited by:
Thomas Dandekar, University of Wuerzburg, GermanyReviewed by:
Knut Ohlsen, University of Wuerzburg, GermanySamuel Wagner, Tübingen University, Germany
Copyright © 2014 Wilharm and Heider. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gottfried Wilharm, Project Group P2, Robert Koch-Institute, Bereich Wernigerode, Burgstr. 37, D-38855 Wernigerode, Germany e-mail:d2lsaGFybWdAcmtpLmRl
 Gottfried Wilharm
Gottfried Wilharm Christine Heider
Christine Heider