- Bacterial Infections and Antimicrobial Therapies Group, Institute for Bioengineering of Catalonia, Barcelona, Spain
Ribonucleotide reductase (RNR) is a key enzyme that mediates the synthesis of deoxyribonucleotides, the DNA precursors, for DNA synthesis in every living cell. This enzyme converts ribonucleotides to deoxyribonucleotides, the building blocks for DNA replication, and repair. Clearly, RNR enzymes have contributed to the appearance of genetic material that exists today, being essential for the evolution of all organisms on Earth. The strict control of RNR activity and dNTP pool sizes is important, as pool imbalances increase mutation rates, replication anomalies, and genome instability. Thus, RNR activity should be finely regulated allosterically and at the transcriptional level. In this review we examine the distribution, the evolution, and the genetic regulation of bacterial RNRs. Moreover, this enzyme can be considered an ideal target for anti-proliferative compounds designed to inhibit cell replication in eukaryotic cells (cancer cells), parasites, viruses, and bacteria.
Introduction
All organisms require active DNA synthesis prior to cell division. A first prerequisite for DNA synthesis is the balanced supply of the different deoxyribonucleotide triphosphates (dNTPs). The only biochemical pathway for de novo dNTP synthesis is the reaction catalyzed through the enzyme ribonucleotide reductase (RNR), which converts the four ribonucleotides triphosphates (NTPs) into their corresponding dNTPs through the reduction of the C2′-OH bond (see Figure 1).
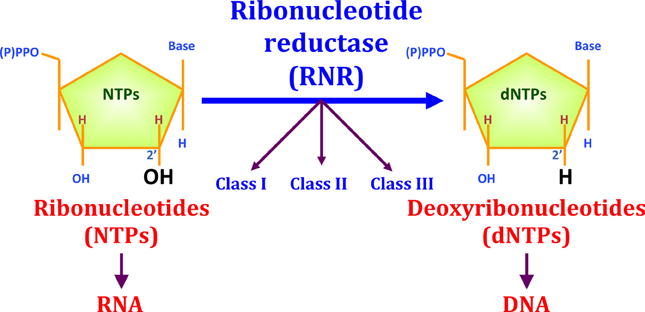
Figure 1. The reduction of ribonucleotides to deoxyribonucleotides by RNR. Three different RNR classes (I, II, and III) have been described for this enzyme family. RNR is important for evolution, as this enzyme played an important role during the transition from an RNA to a DNA world. RNR enzymes catalyze the reduction of the ribose C2′-OH to C2′-H.
Ribonucleotide Reductase (RNR): Structure and Mechanisms
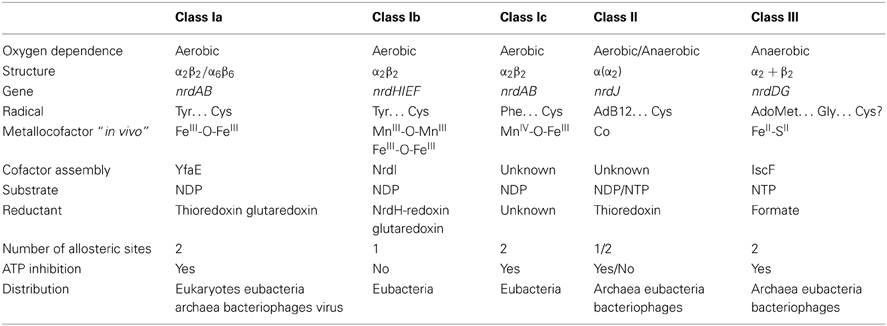
RNR uses radical chemistry to catalyze the reduction of each NTP. How the enzyme generates this radical, the type of cofactor and metal required, the three-dimensional structure of this enzyme complex and the dependence of oxygen are all characteristics that are considered when classifying RNRs. Currently, three different RNR classes have been described (I, II, and III), and class I is further subdivided into Ia, Ib, and Ic (see Table 1). All three RNR classes share a common three-dimensional protein structure at the catalytic subunit and a highly conserved α/β barrel structure in the active site of the enzyme. In addition, the two potential allosteric centers (specificity and activity) are highly conserved among the different RNR classes, although in class Ib, and some class II RNRs activity allosteric site is absent (reviewed in Nordlund and Reichard, 2006; Hofer et al., 2012).
Reduction of the four different nucleotides (ATP/CTP/GTP/TTP) occurs at a single active site in each polypeptide chain, therefore the tight regulation of dNTP levels is important for each dividing cell. Unbalanced dNTP levels could lead to increased mutation rates (Mathews, 2006). Thus, one of the most important aspects of the dNTP supply required for DNA synthesis and repair is the tight regulation of RNRs at different levels, including the allosteric regulation of enzyme activity, transcriptional regulation, and cell cycle-specific proteolysis in mammalian cells.
RNR activity is controlled at two different levels: substrate specificity, in which the binding of different nucleotides results in the reduction of each specific NTP at the active site, and enzymatic activity, in which the binding of ATP, or dATP respectively activates or inhibits enzymatic activity (Figure 2). Extensive reviews concerning allosteric regulation at the biochemical level have previously been published (Reichard, 1997; Nordlund and Reichard, 2006; Hofer et al., 2012; Ahmad and Dealwis, 2013), thus this review will focus on the bacterial RNR rather than the eukaryotic RNR.
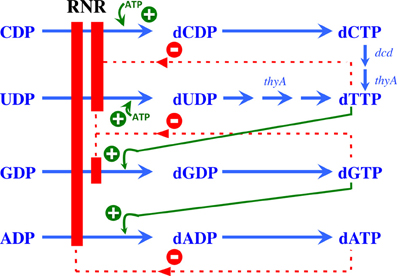
Figure 2. Allosteric regulation of RNR. Model showing the allosteric regulation of class Ia RNRs. The binding of ATP at the substrate specificity site activates the enzyme, promoting the reduction of CDP and UDP to dCDP and dUDP, respectively. Once formed, dTTP promotes the reduction of GDP to dGDP, which in turn induces the reduction of ADP to dADP. A high dATP concentration inhibits the overall activity of this enzyme through binding to the allosteric activity site. Green plus symbols indicate stimulation of RNR reduction, and red minus symbols indicate inhibition.
Class I
Class I RNRs are the best-known and most-studied enzymes. These enzymes comprise two homodimeric subunits, namely R1 (or α) and R2 (or β). The α subunit contains the catalytic subunit, containing the active site where nucleotide reduction occurs, and two allosteric sites, involved in the allosteric regulation of substrate specificity and general enzyme activity. The β subunit harbors the metallocofactor essential for the initiation of nucleotide reduction.
The active form of RNR in eukaryotes and prokaryotes comprises two proteins (R1 + R2 or α + β) associated in a dimeric or other oligomeric forms, such as αnβm (where n can be 2, 4 or 6 and m 1, 2, 3 or more). Several reviews and studies have previously described the structural basis for the allosteric regulation and cluster assembly of class I RNRs (Ando et al., 2011; Hofer et al., 2012; Ahmad and Dealwis, 2013; Tomter et al., 2013).
Class I RNRs can be further subdivided into class Ia, Ib, and Ic enzymes (see Table 1) based on the type of metal center required to generate the protein radical.
The nrdAB genes encode class Ia enzymes, which require a di-iron center (FeIII-O-FeIII) in the NrdB (β) subunit to generate the tyrosyl radical. The nrdHIEF genes encode the class Ib operon, with NrdE and NrdF encoding the α and β subunits, respectively, NrdI encoding a flavodoxin (Cotruvo and Stubbe, 2008; Roca et al., 2008b) and NrdH encoding a glutaredoxin-like protein (Jordan et al., 1997; Crona et al., 2011). NrdI encodes a specific protein involved in the biosynthesis and maintenance of the active metal center, and NrdH is the specific electron donor for the NrdEF enzyme system.
Class Ib RNRs harbor a di-manganese center (MnIII-O-MnIII) to generate the tyrosyl radical in vivo, although this radical can also be generated with a di-ferric center (FeIII-O-FeIII) (Cotruvo and Stubbe, 2010, 2011, 2012). The catalytic subunit NrdE also differs from that of other class I RNRs because this enzyme lacks the activity site at the N-terminal region of the protein.
Moreover, the nrdAB genes encode class Ic RNRs, which is distinguished from class Ia RNRs, as the protein radical is generated through a MnIV-O-FeIII center (Jiang et al., 2007a,b; Dassama et al., 2012).
During catalysis, the radical is formed in the β subunit in all three class I RNRs and subsequently transferred to the large subunit (α) via a long-range radical transfer pathway, generating a thiol radical at the active site of the enzyme, where two cysteines are ultimately responsible for NTP reduction (Ekberg et al., 1996; Nordlund and Reichard, 2006; Cotruvo and Stubbe, 2011). Furthermore, all class I RNRs requires the presence of oxygen for the generation of a radical under aerobic conditions.
Class II
Class II RNRs comprise a single α-chain polypeptide encoded by a single nrdJ gene. NrdJ harbors the active center and allosteric sites of the enzyme. This RNR class uses S-adenosylcobalamine (AdoCob) to generate the cysteinyl radical that substitutes the class I small protein (ß subunit) (Tamao and Blakley, 1973; Licht et al., 1996). This enzymatic reaction does not require oxygen, as this RNR class is completely oxygen independent. Class II RNRs harbor an allosteric specificity site, but lack the allosteric activity site, similar to class Ib RNRs (Eliasson et al., 1999; Larsson et al., 2010). An exceptional class II RNR has been identified in P. aeruginosa, and this enzyme differs from all hitherto known class II RNRs, as this enzyme is split and encoded by two consecutive open reading frames, namely nrdJa and nrdJb, separated by 16 bp (Torrents et al., 2005).
Class III
Class III RNRs comprise two homodimeric proteins encoded by nrdD and nrdG genes. NrdD is the large enzymatic catalytic subunit, harboring the active site and the two allosteric regulation sites, respectively determining substrate specificity and activity. Furthermore, the NrdG protein (known as activase) is responsible for generating the radical (Sun et al., 1993, 1995). Class III RNRs require the binding of S-adenosylmethionine (SAM) to a 4Fe-4S metal center located in the NrdG protein for radical formation (Eliasson et al., 1990). This interaction generates an extremely oxygen sensitive glycyl radical at the C-terminal domain of the NrdD protein (King and Reichard, 1995). Owing to the high sensitivity of the glycyl radical and the potential metal center oxidation in the presence of oxygen, this RNR class is only active under anaerobic conditions.
Experiments on the Lactococcus lactis class III RNR have shown that NrdD alone catalyzes the reduction of NTPs (Torrents et al., 2001). After activation of NrdD, NrdG was no longer required and dissociated from the complex. Activated NrdD protein can catalyze several rounds of NTP reduction in the presence of formate as an electron donor, with ATP as the allosteric effector and requiring Mg2+ and K+ to stimulate the reaction (Torrents et al., 2001). This enzymatic reaction is similar to that of the E. coli pyruvate formate lyase (PFL) enzyme system (Kessler and Knappe, 1996), in which PFL is the catalytic subunit component and PFL-activase initiates the system.
Distribution
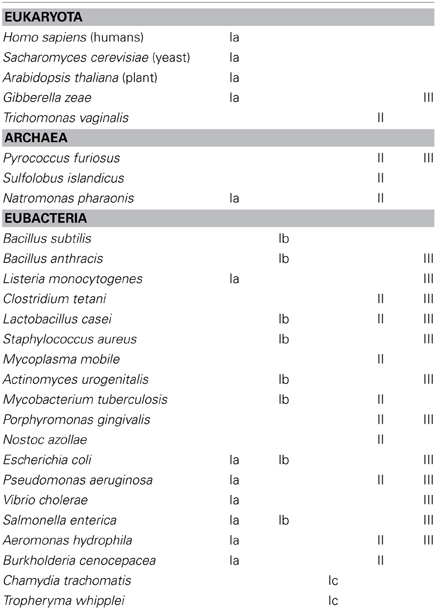
The distribution of the different RNR classes among the different organisms on Earth is remarkable. Several organisms have been sequenced and some encode different functionally redundant RNR classes. Moreover, a combination of different RNR classes can be encoded in the same organism. This occurrence is complex, and the RNR class is not always associated with the life style and phylogeny of the organism (Torrents et al., 2000, 2002; Lundin et al., 2009, 2010). Table 2 provides a short list of the RNR occurrence among eukaryotes, archaea, and eubacteria. A complete list of the distribution of all RNR classes is provided in the RNR database (RNRdb) (Lundin et al., 2009).
Complex eukaryotic organisms only encode class Ia RNRs, thus limiting their survival in aerobic environments. Nevertheless, some unicellular algae (e.g., Euglena gracilis) also encode an active class II RNR (Torrents et al., 2006a), but the number of eukaryotic organisms with this atypical RNR occurrence is low. Furthermore, some fungi genomes show class III RNR sequences (e.g., Gibberella zeae), but surprisingly, these enzymes are atypical compared with other class III RNRs, and further studies are needed to elucidate whether this RNR class is functional (Torrents et al., 2008).
Any potential RNR combination can be identified in eubacteria and archaea. Some bacteria encode only one RNR class, such as Bacillus subtillis (class Ib), while other bacteria encode two RNR classes, such as Staphylococcus aureus (class Ib and III) (Kirdis et al., 2007), or a higher level of complexity, such as in Pseudomonas aeruginosa, which encodes three different RNR classes (Ia, II, and III) (Jordan et al., 1999; Torrents et al., 2005; Sjöberg and Torrents, 2011).
Ribonucleotide reductases function under a range of different environmental conditions; indeed, the type of RNR class will impact the adaptability of microorganisms to different environmental conditions. Bacteria encoding more than one RNR class can survive in a wide range of ecological niches (Figure 3). For example, class II or III RNRs facilitate survival under anaerobic conditions. Furthermore, the presence of class II RNRs facilitates the survival of bacteria in the transition from aerobic to anaerobic environments, or vice versa, and also at the interface of both oxygenic conditions. Indeed, the proper functionality of class II RNRs depends on S-adenosylcobalamine (vitamin B12) availability, as previously observed in P. aeruginosa; thus, vitamin B12 is a limiting cofactor for the activity of this RNR class (Torrents et al., 2005; Sjöberg and Torrents, 2011). Class I RNRs, and no other RNR classes, require an aerobic environment, as these enzymes are oxygen dependent. In addition, bacteria encoding both class Ia and Ib RNRs facilitates survival under environmental conditions in which iron is limiting, as class Ib RNRs substitute iron from manganese in the metal center (Martin and Imlay, 2011). Moreover, it has been shown that class Ib RNRs are highly expressed and essential for E. coli biofilm formation, under nutrient-limited conditions (Cendra et al., 2012) or high oxidative stress (Monje-Casas et al., 2001), indicating the use of class Ib RNRs under special growth conditions.
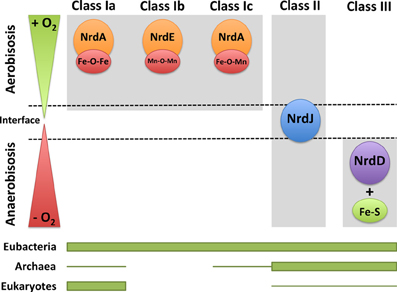
Figure 3. RNRs as survival markers under aerobic or anaerobic environments. RNR classes can be activated under different environmental conditions in which one organism survives depending on the oxygen availability (aerobic, anaerobic, and in the interface). The distribution of each RNR class in the three domains of life (eubacteria, archaea, and eukaryotes) is shown. The thicker line represents higher RNR class occurrence, and the thinner line corresponds to the occurrence in only a few organisms.
RNR Evolution
The maintenance of life on Earth depends on the ability to reproduce. Reproduction requires an accurate and stable storage system for the genetic information in all organisms, including viruses. It has been recently suggested that the RNA molecule, with auto-replicative capacity, is the primary primitive molecule for the genetic information storage. Despite the wide acceptance of this idea, there are arguments against the concept of an RNA world that cannot be underestimated.
The transition from an RNA to a DNA-protein world, which currently exists, implies the existence of ribosome-based translation, a genetic code and the replacement of RNA with DNA molecule as the genetic material. The results from recent studies concerning the characterization of RNRs and the current knowledge of the evolution of organisms on Earth could help determine which type of RNR enzyme is most similar to the ancestral RNR (ur-RNR), and this information will lead to a better understanding of the transition from a RNA to a DNA world (Reichard, 1993; Stubbe, 2000; Stubbe et al., 2001).
Today, three different RNR classes have been described, with little apparent similarity between them in terms of primary protein sequence (approximately 10–20% similarity). Thus, it could be assumed that each RNR class appeared independently from each other over time. But, surprisingly, there is a great similarity the reaction mechanism, allosteric regulation and three-dimensional structure (tertiary structure) of these enzymes, suggesting a potential common origin (Logan et al., 1999; Sintchak et al., 2002; Torrents et al., 2002).
Based on the geological history of the Earth and trace element content of the sediments, early stages of life originated in an anaerobic environment (Large et al., 2014). Under this restrictive condition, class III RNRs were, most likely, the primitive form from which all other RNR enzymes were derived, and this enzyme class is might be the ancestral RNR (ur-reductase). Moreover, the enzymatic activation of class III RNR requires S-adenosylmethionine (SAM), one of the most ancestral molecules, with few steps required for its biosynthesis (Frey et al., 2008). NrdG protein harbors an Fe-S cluster for glycyl radical generation, and this cluster is considered among the most ancient, ubiquitous, and functionally diverse among biological prosthetic groups (Beinert et al., 1997). Furthermore, this RNR class uses formate as the external reductant, which is a simple molecule compared with the electron donors used for other RNR classes (thioredoxin, glutaredoxin, or NrdH). Class II RNRs also function under anaerobic conditions, but other factors determine whether these enzymes act as ur-reductases. Class II RNRs use S-adenosylcobalamine (AdoCob) for radical generation, which is more structurally complex than SAM. However, other studies have suggested that class II RNRs represent an ancient ur-RNR class (Poole et al., 2002). Certainly, the appearance of oxygen on Earth was key to this evolutionary process, culminating in the existence of the different RNR classes observed today.
Genetic Regulation
An unbalanced pool of dNTPs leads to an increase in the mutation rate and the loss of DNA replication fidelity (Wheeler et al., 2005; Mathews, 2006). Thus, RNR activity must be highly regulated at either the enzymatic activity level through allosteric regulation, as previously described, or at the transcriptional level.
Most RNRs, in both microorganisms and eukaryotic cells, are transcriptionally regulated depending on the cell cycle, as DNA replication primarily occurs during cell division where a high concentration of different dNTPs is needed. Here, the transcriptional regulation of RNRs in microorganisms, particularly eubacteria, is discussed. Several other studies have previously addressed eukaryotic RNR transcriptional regulation (Hakansson et al., 2006; Nordlund and Reichard, 2006; Thelander, 2007).
Transcriptional regulation is complex in organisms encoding more than one RNR class, as this process is essential to achieve adequate coordination of the expression of the different nrd genes to ensure proper enzyme concentration and provide balanced dNTPs levels. For example, in Escherichia coli, class Ia RNRs are active during growth under laboratory conditions, while class III enzymes are active under anaerobic conditions (Boston and Atlung, 2003; Roca et al., 2008a) and class Ib RNRs are important during growth in iron-deficient medium (Martin and Imlay, 2011) and biofilm formation (Cendra et al., 2012). However, Pseudomonas aeruginosa also express class Ia RNRs under laboratory growth conditions (Jordan et al., 1999; Torrents et al., 2006b) and class II and III RNRs are expressed during infection (Sjöberg and Torrents, 2011).
Several transcription factors (activators and inhibitors) have been implicated for regulation the expression of each RNR class during bacterial growth under certain environmental conditions. For example IciA, FIS, and DnaA are transcriptional activators for E. coli class Ia RNRs, while H-NS and NrdR act as a transcriptional repressors (Torrents et al., 2007; Cendra et al., 2012, 2013). Class Ib RNRs are transcriptionally activated through FUR (Vassinova and Kozyrev, 2000; McHugh et al., 2003). Moreover, class III RNRs are transcriptionally regulated through the FNR protein (Boston and Atlung, 2003; Roca et al., 2008a).
In 2007 and 2008, comprehensive reviews were published (Herrick and Sclavi, 2007; Torrents et al., 2008) concerning the transcriptional regulation of certain RNR classes in E. coli and other bacteria; however, in this chapter, the focus will be on the recently described transcriptional regulator NrdR, implicated in the regulation of all three RNR classes.
NrdR—a global ribonucleotide reductase regulator
Recent publications have described the transcriptional regulation of different RNR classes; however, little is known about how the expression of different RNRs might be coordinated in microorganisms encoding several RNR classes. A novel transcriptional factor, NrdR, has recently been implicated in the regulation of all three RNR classes through binding to conserved NrdR boxes in the promoter regions of almost all genes encoding RNRs (Grinberg et al., 2006; Torrents et al., 2007; Mowa et al., 2009; Panosa et al., 2010; Case et al., 2011). All currently known eubacteria encode and nrdR gene except that Rickettsia, Helicobacter, and Desulfovibrio.
The results obtained from a recent study conducted at Tel Aviv University (Israel) indicated a role for NrdR in the transcriptional regulation of different RNR genes encoded in a single organism. This group was the first to identify an open reading frame in Streptomyces coelicolor, encoding NrdR, a protein that binds to the promoter regions of class Ia and II RNRs (Borovok et al., 2004). Subsequently, a computational study comparing all RNR promoter regions at the genomic level revealed a highly conserved palindromic sequence, named the NrdR-box, with the consensus sequence acaCwAtATaTwGtg (Rodionov and Gelfand, 2005). This NrdR-box has since been identified in eubacterial genomes and is absent in archaea and eukaryotes. Typically two copies of the NrdR-box are consistently detected in the promoter region of different nrd genes, with specific spacing corresponding to an integral number of turns in the double DNA helix.
Currently, NrdR has been classified as a member of a highly conserved family of proteins confined exclusively to prokaryotes, eubacteria, and some archaea (Lundin et al., 2009). NrdR is a 150–200-amino acid protein harboring two protein domains: an N-terminal zinc finger like DNA-binding domain and a C-terminal ATP-cone domain that binds nucleotides (Figure 4A). The N-terminus contains a zinc ribbon motif for binding to the upstream regulatory regions of the nrd genes (NrdR-box). The ATP domain is similar to the allosteric activity domain observed in some RNRs. The Streptomyces coelicolor NrdR protein is the best characterized (Grinberg et al., 2006, 2009). Studies have demonstrated that the ATP-cone domain alone is essential for nucleotide binding and important for the correct three-dimensional NrdR structure and DNA binding to the NrdR-box. One hypothesis is that NrdR regulates RNR transcription by acting as ATP/dATP-dependent regulator that controls the oligomeric state of NrdR. Thus, a native NrdR oligomer, likely comprising eight subunits, binds ATP or dATP, and the zing finger domain is free to bind to the DNA target site to repress RNR gene expression. When DNA replicates, the cellular levels of dNTPs are decreased, NrdR is depleted of dATP, and the ATP-cone domain undergoes a change from an oligomeric state to a dimeric protein, abolishing its DNA binding activity and increasing nrd transcription (Figure 4B).
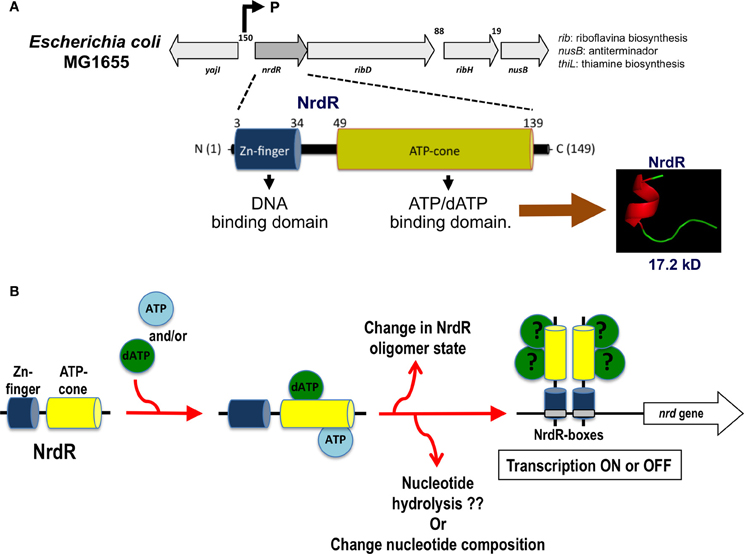
Figure 4. nrdR operon organization and hypothetical mechanism of transcription regulation by NrdR. (A) Structure of the nrdR operon and functional protein domains for the NrdR transcriptional regulator. (B) Hypothetical mechanism for the transcriptional regulation of the nrd gene through NrdR. Depending on which nucleotide is bound and the oligomerization state of the NrdR, this transcription factor can modulate the expression of the nrd genes through binding to the specific NrdR-box.
A recent study reported a more complex mechanism of NrdRs nucleotide binding through controlled complex allosteric mechanisms. A model has been proposed, in which NrdR selectively binds nucleotide triphosphates, which are subsequently hydrolyzed to monophosphates to regulate the NrdR oligomeric state and DNA binding (McKethan and Spiro, 2013).
Hence, NrdR might be a transcriptional regulator with complex cooperative and allosterically regulated nucleotide binding mechanism that finely tunes the expression of the ribonucleotide reductase gene in response to cellular nucleotides.
Ribonucleotide Reductase as a Biomedical Target
RNRs are essential enzymes for all organisms, as these proteins provide the dNTPs needed for chromosome replication and DNA damage repair. The contribution of RNR to these key reactions makes this enzyme a perfect target for the design of compounds that inhibit cell growth in either cells with altered cell cycles (cancer cells) or during viral, bacterial or protozoan infections (Torrents et al., 2008).
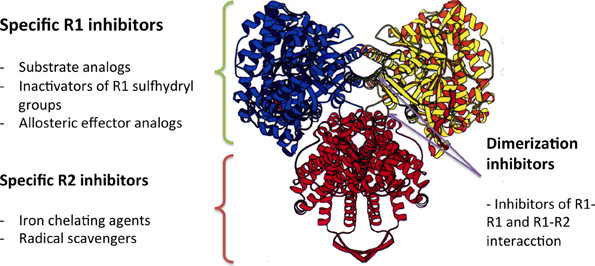
RNRs are complex enzymes, and several inhibitors of these enzymes have been described and classified according to their mode of action (Figure 5). The first group of inhibitors identified interacted directly with the catalytic subunit (R1), alternating enzymatic activity through the irreversible binding to the active site of the enzyme (substrate analogs or inactivators of sulfhydryl groups) or to allosteric binding sites. The inhibitors in the second group interact with the activator subunit (ß) of class I RNRs, and have been identified as either chelating agents of the dinuclear iron center or radical scavengers. The third group of inhibitors comprises antisense inhibitors, which are antisense RNA molecules that bind to mRNA encoding the components of the enzyme. These antisense inhibitors have shown promising results in some cancers. Moreover, there are inhibitors that prevent the dimerization of the enzyme components, which are peptides that mimic the interaction between the catalytic and activator subunit (R1-R2) or the interactions between the catalytic subunits themselves (R1-R1) (Torrents et al., 2008; Wijerathna et al., 2011; Basu and Sinha, 2012; Bhave et al., 2013; Liu et al., 2013; Zhou et al., 2013).
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part through grants from the Ministerio de Economia y Competitividad (BFU2011-24066 and CSD2008-00013), ERA-NET PathoGenoMics (BIO2008-04362-E), Generalitat de Catalunya (2009SGR66), and Associació Catalana de Fibrosis Quísica. Eduard Torrents was supported through funding from the Ramón y Cajal and I3 programs from the Ministerio de Ciencia e Innovación. The author would like to thank Pep Astola, Aida Baelo, Anna Crespo, and Lucas Pedraz for critically reading this manuscript.
References
Ahmad, M. F., and Dealwis, C. G. (2013). The structural basis for the allosteric regulation of ribonucleotide reductase. Prog. Mol. Biol. Trans. Sci. 117, 389–410. doi: 10.1016/B978-0-12-386931-9.00014-3
Ando, N., Brignole, E. J., Zimanyi, C. M., Funk, M. A., Yokoyama, K., Asturias, F. J., et al. (2011). Structural interconversions modulate activity of Escherichia coli ribonucleotide reductase. Proc. Natl. Acad. Sci. U.S.A. 108, 21046–21051. doi: 10.1073/pnas.1112715108
Basu, A., and Sinha, B. N. (2012). Radical scavengers as ribonucleotide reductase inhibitors. Curr. Top. Med. Chem. 12, 2827–2842. doi: 10.2174/1568026611212240009
Beinert, H., Holm, R. H., and Munck, E. (1997). Iron-sulfur clusters: nature's modular, multipurpose structures. Science 277, 653–659. doi: 10.1126/science.277.5326.653
Bhave, S., Elford, H., and Mcvoy, M. A. (2013). Ribonucleotide reductase inhibitors hydroxyurea, didox, and trimidox inhibit human cytomegalovirus replication in vitro and synergize with ganciclovir. Antiviral Res. 100, 151–158. doi: 10.1016/j.antiviral.2013.07.016
Borovok, I., Gorovitz, B., Yanku, M., Schreiber, R., Gust, B., Chater, K., et al. (2004). Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol. Microbiol. 54, 1022–1035. doi: 10.1111/j.1365-2958.2004.04325.x
Boston, T., and Atlung, T. (2003). FNR-mediated oxygen-responsive regulation of the nrdDG operon of Escherichia coli. J. Bacteriol. 185, 5310–5313. doi: 10.1128/JB.185.17.5310-5313.2003
Case, E. D., Akers, J. C., and Tan, M. (2011). CT406 encodes a chlamydial ortholog of NrdR, a repressor of ribonucleotide reductase. J. Bacteriol. 193, 4396–4404. doi: 10.1128/JB.00294-11
Cendra, M., Juarez, A., Madrid, C., and Torrents, E. (2013). H-NS is a novel transcriptional modulator of the ribonucleotide reductase genes in Escherichia coli. J. Bacteriol. 195, 4255–4263. doi: 10.1128/JB.00490-13
Cendra, M., Juarez, A., and Torrents, E. (2012). Biofilm modifies expression of ribonucleotide reductase genes in Escherichia coli. PLoS ONE 7:e46350. doi: 10.1371/journal.pone.0046350
Cotruvo, J. A. Jr., and Stubbe, J. (2008). NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Proc. Natl. Acad. Sci. U.S.A. 105, 14383–14388. doi: 10.1073/pnas.0807348105
Cotruvo, J. A. Jr., and Stubbe, J. (2010). An active dimanganese(III)-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Biochemistry 49, 1297–1309. doi: 10.1021/bi902106n
Cotruvo, J. A., and Stubbe, J. (2011). Class I ribonucleotide reductases: metallocofactor assembly and repair in vitro and in vivo. Annu. Rev. Biochem. 80, 733–767. doi: 10.1146/annurev-biochem-061408-095817
Cotruvo, J. A. Jr., and Stubbe, J. (2012). Metallation and mismetallation of iron and manganese proteins in vitro and in vivo: the class I ribonucleotide reductases as a case study. Metallomics 4, 1020–1036. doi: 10.1039/c2mt20142a
Crona, M., Torrents, E., Rohr, A. K., Hofer, A., Furrer, E., Tomter, A. B., et al. (2011). NrdH-redoxin protein mediates high enzyme activity in manganese-reconstituted ribonucleotide reductase from Bacillus anthracis. J. Biol. Chem. 286, 33053–33060. doi: 10.1074/jbc.M111.278119
Dassama, L. M., Boal, A. K., Krebs, C., Rosenzweig, A. C., and Bollinger, J. M. Jr. (2012). Evidence that the beta subunit of Chlamydia trachomatis ribonucleotide reductase is active with the manganese ion of its manganese(IV)/iron(III) cofactor in site 1. J. Am. Chem. Soc. 134, 2520–2523. doi: 10.1021/ja211314p
Ekberg, M., Sahlin, M., Eriksson, M., and Sjoberg, B. M. (1996). Two conserved tyrosine residues in protein R1 participate in an intermolecular electron transfer in ribonucleotide reductase. J. Biol. Chem. 271, 20655–20659. doi: 10.1074/jbc.271.34.20655
Eliasson, R., Fontecave, M., Jornvall, H., Krook, M., Pontis, E., and Reichard, P. (1990). The anaerobic ribonucleoside triphosphate reductase from Escherichia coli requires S-adenosylmethionine as a cofactor. Proc. Natl. Acad. Sci. U.S.A. 87, 3314–3318. doi: 10.1073/pnas.87.9.3314
Eliasson, R., Pontis, E., Jordan, A., and Reichard, P. (1999). Allosteric control of three B12-dependent (class II) ribonucleotide reductases. Implications for the evolution of ribonucleotide reduction. J. Biol. Chem. 274, 7182–7189. doi: 10.1074/jbc.274.11.7182
Frey, P. A., Hegeman, A. D., and Ruzicka, F. J. (2008). The radical SAM superfamily. Crit. Rev. Biochem. Mol. Biol. 43, 63–88. doi: 10.1080/10409230701829169
Grinberg, I., Shteinberg, T., Gorovitz, B., Aharonowitz, Y., Cohen, G., and Borovok, I. (2006). The Streptomyces NrdR transcriptional regulator is a Zn ribbon/ATP cone protein that binds to the promoter regions of class Ia and class II ribonucleotide reductase operons. J. Bacteriol. 188, 7635–7644. doi: 10.1128/JB.00903-06
Grinberg, I., Shteinberg, T., Hassan, A. Q., Aharonowitz, Y., Borovok, I., and Cohen, G. (2009). Functional analysis of the Streptomyces coelicolor NrdR ATP-cone domain: role in nucleotide binding, oligomerization, and DNA interactions. J. Bacteriol. 191, 1169–1179. doi: 10.1128/JB.01145-08
Hakansson, P., Hofer, A., and Thelander, L. (2006). Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J. Biol. Chem. 281, 7834–7841. doi: 10.1074/jbc.M512894200
Herrick, J., and Sclavi, B. (2007). Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol. Microbiol. 63, 22–34. doi: 10.1111/j.1365-2958.2006.05493.x
Hofer, A., Crona, M., Logan, D. T., and Sjöberg, B. M. (2012). DNA building blocks: keeping control of manufacture. Crit. Rev. Biochem. Mol. Biol. 47, 50–63. doi: 10.3109/10409238.2011.630372
Jiang, W., Hoffart, L. M., Krebs, C., and Bollinger, J. M. Jr. (2007a). A manganese(IV)/iron(IV) intermediate in assembly of the manganese(IV)/iron(III) cofactor of Chlamydia trachomatis ribonucleotide reductase. Biochemistry 46, 8709–8716. doi: 10.1021/bi700906g
Jiang, W., Yun, D., Saleh, L., Barr, E. W., Xing, G., Hoffart, L. M., et al. (2007b). A manganese(IV)/iron(III) cofactor in Chlamydia trachomatis ribonucleotide reductase. Science 316, 1188–1191. doi: 10.1126/science.1141179
Jordan, A., Aslund, F., Pontis, E., Reichard, P., and Holmgren, A. (1997). Characterization of Escherichia coli NrdH. A glutaredoxin-like protein with a thioredoxin-like activity profile. J. Biol. Chem. 272, 18044–18050. doi: 10.1074/jbc.272.29.18044
Jordan, A., Torrents, E., Sala, I., Hellman, U., Gibert, I., and Reichard, P. (1999). Ribonucleotide reduction in Pseudomonas species: simultaneous presence of active enzymes from different classes. J. Bacteriol. 181, 3974–3980.
Kessler, D., and Knappe, J. (1996). “Anaerobic dissimilation of pyruvate,” in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds F. C. Neidhardt et al. (Washington, DC: American Society for MIcrobiology Press), 199–205.
King, D. S., and Reichard, P. (1995). Mass spectrometric determination of the radical scission site in the anaerobic ribonucleotide reductase of Escherichia coli. Biochem. Biophys. Res. Commun. 206, 731–735. doi: 10.1006/bbrc.1995.1103
Kirdis, E., Jonsson, I. M., Kubica, M., Potempa, J., Josefsson, E., Masalha, M., et al. (2007). Ribonucleotide reductase class III, an essential enzyme for the anaerobic growth of Staphylococcus aureus, is a virulence determinant in septic arthritis. Microb. Pathog. 43, 179–188. doi: 10.1016/j.micpath.2007.05.008
Large, R. R., Halpin, J. A., Danyushevsky, L. V., Maslennikov, V. V., Bull, S. W., Long, J. A., et al. (2014). Trace element content of sedimentary pyrite as a new proxy for deep-time ocean-atmosphere evolution. Earth Planet. Sci. Lett. 389, 209–220. doi: 10.1016/j.epsl.2013.12.020
Larsson, K. M., Logan, D. T., and Nordlund, P. (2010). Structural basis for adenosylcobalamin activation in AdoCbl-dependent ribonucleotide reductases. ACS Chem. Biol. 5, 933–942. doi: 10.1021/cb1000845
Licht, S., Gerfen, G. J., and Stubbe, J. (1996). Thiyl radicals in ribonucleotide reductases. Science 271, 477–481. doi: 10.1126/science.271.5248.477
Liu, R., Zhang, H., Yuan, M., Zhou, J., Tu, Q., Liu, J. J., et al. (2013). Synthesis and biological evaluation of apigenin derivatives as antibacterial and antiproliferative agents. Molecules 18, 11496–11511. doi: 10.3390/molecules180911496
Logan, D. T., Andersson, J., Sjöberg, B. M., and Nordlund, P. (1999). A glycyl radical site in the crystal structure of a class III ribonucleotide reductase. Science 283, 1499–1504. doi: 10.1126/science.283.5407.1499
Lundin, D., Gribaldo, S., Torrents, E., Sjöberg, B. M., and Poole, A. M. (2010). Ribonucleotide reduction—horizontal transfer of a required function spans all three domains. BMC Evol. Biol. 10:383. doi: 10.1186/1471-2148-10-383
Lundin, D., Torrents, E., Poole, A. M., and Sjöberg, B. M. (2009). RNRdb, a curated database of the universal enzyme family ribonucleotide reductase, reveals a high level of misannotation in sequences deposited to Genbank. BMC Genomics 10:589. doi: 10.1186/1471-2164-10-589
Martin, J. E., and Imlay, J. A. (2011). The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Mol. Microbiol. 80, 319–334. doi: 10.1111/j.1365-2958.2011.07593.x
Mathews, C. K. (2006). DNA precursor metabolism and genomic stability. FASEB J. 20, 1300–1314. doi: 10.1096/fj.06-5730rev
McHugh, J. P., Rodriguez-Quinones, F., Abdul-Tehrani, H., Svistunenko, D. A., Poole, R. K., Cooper, C. E., et al. (2003). Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278, 29478–29486. doi: 10.1074/jbc.M303381200
McKethan, B. L., and Spiro, S. (2013). Cooperative and allosterically controlled nucleotide binding regulates the DNA binding activity of NrdR. Mol. Microbiol. 90, 278–289. doi: 10.1111/mmi.12364
Monje-Casas, F., Jurado, J., Prieto-Alamo, M. J., Holmgren, A., and Pueyo, C. (2001). Expression analysis of the nrdHIEF operon from Escherichia coli. Conditions that trigger the transcript level in vivo. J. Biol. Chem. 276, 18031–18037. doi: 10.1074/jbc.M011728200
Mowa, M. B., Warner, D. F., Kaplan, G., Kana, B. D., and Mizrahi, V. (2009). Function and regulation of class I ribonucleotide reductase-encoding genes in mycobacteria. J. Bacteriol. 191, 985–995. doi: 10.1128/JB.01409-08
Nordlund, P., and Reichard, P. (2006). Ribonucleotide reductases. Annu. Rev. Biochem. 75, 681–706. doi: 10.1146/annurev.biochem.75.103004.142443
Panosa, A., Roca, I., and Gibert, I. (2010). Ribonucleotide reductases of Salmonella typhimurium: transcriptional regulation and differential role in pathogenesis. PLoS ONE 5:e11328. doi: 10.1371/journal.pone.0011328
Poole, A. M., Logan, D. T., and Sjoberg, B. M. (2002). The evolution of the ribonucleotide reductases: much ado about oxygen. J. Mol. Evol. 55, 180–196. doi: 10.1007/s00239-002-2315-3
Reichard, P. (1993). From RNA to DNA, why so many ribonucleotide reductases? Science 260, 1773–1777. doi: 10.1126/science.8511586
Reichard, P. (1997). The evolution of ribonucleotide reduction. Trends Biochem. Sci. 22, 81–85. doi: 10.1016/S0968-0004(97)01003-7
Roca, I., Ballana, E., Panosa, A., Torrents, E., and Gibert, I. (2008a). Fumarate and nitrate reduction (FNR) dependent activation of the Escherichia coli anaerobic ribonucleotide reductase nrdDG promoter. Int. Microbiol. 11, 49–56.
Roca, I., Torrents, E., Sahlin, M., Gibert, I., and Sjöberg, B. M. (2008b). NrdI essentiality for class Ib ribonucleotide reduction in Streptococcus pyogenes. J. Bacteriol. 190, 4849–4858. doi: 10.1128/JB.00185-08
Rodionov, D. A., and Gelfand, M. S. (2005). Identification of a bacterial regulatory system for ribonucleotide reductases by phylogenetic profiling. Trends Genet. 21, 385–389. doi: 10.1016/j.tig.2005.05.011
Sintchak, M. D., Arjara, G., Kellogg, B. A., Stubbe, J., and Drennan, C. L. (2002). The crystal structure of class II ribonucleotide reductase reveals how an allosterically regulated monomer mimics a dimer. Nat. Struct. Biol. 9, 293–300. doi: 10.1038/nsb774
Sjöberg, B. M., and Torrents, E. (2011). Shift in ribonucleotide reductase gene expression in Pseudomonas aeruginosa during infection. Infect. Immun. 79, 2663–2669. doi: 10.1128/IAI.01212-10
Stubbe, J. (2000). Ribonucleotide reductases: the link between an RNA and a DNA world? Curr. Opin. Struct. Biol. 10, 731–736. doi: 10.1016/S0959-440X(00)00153-6
Stubbe, J., Ge, J., and Yee, C. S. (2001). The evolution of ribonucleotide reduction revisited. Trends Biochem. Sci. 26, 93–99. doi: 10.1016/S0968-0004(00)01764-3
Sun, X., Eliasson, R., Pontis, E., Andersson, J., Buist, G., Sjöberg, B. M., et al. (1995). Generation of the glycyl radical of the anaerobic Escherichia coli ribonucleotide reductase requires a specific activating enzyme. J. Biol. Chem. 270, 2443–2446. doi: 10.1074/jbc.270.6.2443
Sun, X., Harder, J., Krook, M., Jornvall, H., Sjöberg, B. M., and Reichard, P. (1993). A possible glycine radical in anaerobic ribonucleotide reductase from Escherichia coli: nucleotide sequence of the cloned nrdD gene. Proc. Natl. Acad. Sci. U.S.A. 90, 577–581. doi: 10.1073/pnas.90.2.577
Tamao, Y., and Blakley, R. L. (1973). Direct spectrophotometric observation of an intermediate formed from deoxyadenosylcobalamin in ribonucleotide reduction. Biochemistry 12, 24–34. doi: 10.1021/bi00725a005
Thelander, L. (2007). Ribonucleotide reductase and mitochondrial DNA synthesis. Nat. Genet. 39, 703–704. doi: 10.1038/ng0607-703
Tomter, A. B., Zoppellaro, G., Andersen, N. H., Hersleth, H.-P., Hammerstad, M., Røhr, Å K., et al. (2013). Ribonucleotide reductase class I with different radical generating clusters. Coord. Chem. Rev. 257, 3–26. doi: 10.1016/j.ccr.2012.05.021
Torrents, E., Aloy, P., Gibert, I., and Rodriguez-Trelles, F. (2002). Ribonucleotide reductases: divergent evolution of an ancient enzyme. J. Mol. Evol. 55, 138–152. doi: 10.1007/s00239-002-2311-7
Torrents, E., Eliasson, R., Wolpher, H., Graslund, A., and Reichard, P. (2001). The anaerobic ribonucleotide reductase from Lactococcus lactis. Interactions between the two proteins NrdD and NrdG. J. Biol. Chem. 276, 33488–33494. doi: 10.1074/jbc.M103743200
Torrents, E., Grinberg, I., Gorovitz-Harris, B., Lundstrom, H., Borovok, I., Aharonowitz, Y., et al. (2007). NrdR controls differential expression of the Escherichia coli ribonucleotide reductase genes. J. Bacteriol. 189, 5012–5021. doi: 10.1128/JB.00440-07
Torrents, E., Jordan, A., Karlsson, M., and Gibert, I. (2000). Occurrence of multiple ribonucleotide reductase classes in gamma-proteobacteria species. Curr. Microbiol. 41, 346–351. doi: 10.1007/s00284000
Torrents, E., Poplawski, A., and Sjöberg, B. M. (2005). Two proteins mediate class II ribonucleotide reductase activity in Pseudomonas aeruginosa: expression and transcriptional analysis of the aerobic enzymes. J. Biol. Chem. 280, 16571–16578. doi: 10.1074/jbc.M501322200
Torrents, E., Sahlin, M., and Sjöberg, B.-M. (2008). “The ribonucleotide reductase family- genetics and genomics,” in Ribonucleotide Reductases, ed K. K. Andersson (Hauppauge, NY: Nova Science Publishers), 17–77.
Torrents, E., Trevisiol, C., Rotte, C., Hellman, U., Martin, W., and Reichard, P. (2006a). Euglena gracilis ribonucleotide reductase: the eukaryote class II enzyme and the possible antiquity of eukaryote B12 dependence. J. Biol. Chem. 281, 5604–5611. doi: 10.1074/jbc.M512962200
Torrents, E., Westman, M., Sahlin, M., and Sjöberg, B. M. (2006b). Ribonucleotide reductase modularity: atypical duplication of the ATP-cone domain in Pseudomonas aeruginosa. J. Biol. Chem. 281, 25287–25296. doi: 10.1074/jbc.M601794200
Vassinova, N., and Kozyrev, D. (2000). A method for direct cloning of fur-regulated genes: identification of seven new fur-regulated loci in Escherichia coli. Microbiology 146(Pt. 12), 3171–3182.
Wheeler, L. J., Rajagopal, I., and Mathews, C. K. (2005). Stimulation of mutagenesis by proportional deoxyribonucleoside triphosphate accumulation in Escherichia coli. DNA Repair (Amst) 4, 1450–1456. doi: 10.1016/j.dnarep.2005.09.003
Wijerathna, S. R., Ahmad, M. F., Xu, H., Fairman, J. W., Zhang, A., Kaushal, P. S., et al. (2011). Targeting the large subunit of human ribonucleotide reductase for cancer chemotherapy. Pharmaceuticals (Basel) 4, 1328–1354. doi: 10.3390/ph4101328
Keywords: ribonucleotide reductase, evolution, gene regulation, DNA synthesis, NrdR, transcriptional regulation, anaerobiosis
Citation: Torrents E (2014) Ribonucleotide reductases: essential enzymes for bacterial life. Front. Cell. Infect. Microbiol. 4:52. doi: 10.3389/fcimb.2014.00052
Received: 11 March 2014; Paper pending published: 03 April 2014;
Accepted: 08 April 2014; Published online: 28 April 2014.
Edited by:
Martin John McGavin, University of Western Ontario, CanadaReviewed by:
Michael L. Vasil, University of Colorado School of Medicine, USAGuangchun Bai, Albany Medical College, USA
Copyright © 2014 Torrents. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Eduard Torrents, Bacterial Infections and Antimicrobial Therapies Group, Institute for Bioengineering of Catalonia, Baldiri Reixac 15-21, 08028, Barcelona, Spain e-mail:ZXRvcnJlbnRzQGliZWNiYXJjZWxvbmEuZXU=
 Eduard Torrents
Eduard Torrents