- Department of Microbial Genetics, Faculty of Science, Interfaculty Institute of Microbiology and Infection Medicine Tübingen (IMIT), University of Tübingen, Tübingen, Germany
Toxin-antitoxin (TA) systems are small genetic elements ubiquitous in prokaryotic genomes that encode toxic proteins targeting various vital cellular functions. Typically, toxin activity is controlled by adjacently encoded protein or RNA antitoxins and unleashed as a consequence of genetic fluctuations or stressful conditions. Whereas some TA systems interfere with replication or cell wall synthesis, most of them influence transcriptional and post-transcriptional gene regulation. Antitoxin proteins often act as DNA binding transcriptional regulators and many TA toxins exhibit endoribonuclease activity to selectively degrade different RNA species and thus alter gene expression patterns. Some TA RNases cleave tRNA, tmRNAs or rRNAs, whereas most commonly mRNAs either in association with the ribosome or as free transcripts, are targeted. Examples are provided on how TA toxins differentially shape gene expression in bacterial pathogens by creating specialized ribosomes or by altering the transcriptome and how this may be tied in the control of pathogenicity factors.
About Toxin-Antitoxin Systems
Toxin-antitoxin (TA) systems are small and frequently bicistronic elements that are widely distributed throughout bacterial and archaeal genomes. They generally encode a low molecular weight protein which interferes with vital cellular functions and another protein or RNA molecule that keeps the toxin activity in check. The first TA loci have been characterized as plasmid addiction systems on E. coli episomes. There, cells which suffered stochastic plasmid loss in the course of cell division were found to be eradicated by a stable TA toxin whose less stable cognate antitoxin was no longer produced. For example, the hok-sok system ensures faithful partitioning of the plasmid R1 via the membrane pore forming Hok toxin (Gerdes et al., 1986), whereas the F-plasmid is stabilized in a population through the gyrase inhibitor activity of the ccdAB locus (Ogura and Hiraga, 1983; Jaffé et al., 1985). Dozens of further TA systems have been identified since (Pandey and Gerdes, 2005; Makarova et al., 2009; Fozo et al., 2010; Blower et al., 2012; Aakre et al., 2013) and according to the mode of action and chemical nature of the antitoxin, they to date fall into at least five types, as reviewed recently (Schuster and Bertram, 2013). Toxin activity of type I TA systems is post-transcriptionally controlled by a small non-coding RNA antitoxin, whereas in Type II TA systems, the toxin protein is inactivated by direct interaction with the antitoxin protein. More recently, type III, IV, and V TA systems have been described, albeit represented by only few validated examples (Fineran et al., 2009; Masuda et al., 2012; Wang et al., 2012). Whereas type IV and V TA systems are genetically similar to type II systems, type III TA toxin proteins are bound by non-coding RNA antitoxins in the inactive state. Only very recently a new type of TA system was identified which features an unstable toxin protein that is degraded by Clp in association with the cognate antitoxin (Aakre et al., 2013). According to this novel mode of activity control, it might be justified to define it as a novel type VI TA system (Markovski and Wickner, 2013).
Generally, TA toxin activity is unleashed by an imbalance in favor of free toxin molecules, generated either by stochastic fluctuations in the TA systems' gene expression or cellular stress (Christensen et al., 2003; Vogel et al., 2004; Dörr et al., 2010; Maisonneuve et al., 2013). Antitoxin proteins of type II TA systems are frequently proteolyzed by Clp or Lon factors (Brzozowska and Zielenkiewicz, 2013) and hence toxin-antitoxin stoichiometries are shifted. The abundance of TA systems in prokaryotic chromosomes has sparked vivid discussions about their physiological roles beyond plasmid maintenance, including functions in programmed cell death, bacterial growth impediment, defense against foreign genetic material, or (fine) tuning of the physiological activity to shift cells into dormant, drug-tolerant states (Magnuson, 2007; Lewis, 2010; Gerdes and Maisonneuve, 2012). TA toxins target a variety of vital cellular structures and functions within bacteria such as membrane integrity, replication, cell wall synthesis, ribosome assembly, and translation-factors, with RNA cleavage as the most prevalent mode of action (Schuster and Bertram, 2013). Type II TA systems have been grouped into fuzzy numbers of families and superfamilies (Gerdes et al., 2005; Leplae et al., 2011) and TA RNases can further be distinguished according to their ribosome-dependency (Yamaguchi and Inouye, 2011). Recent reviews highlight the mechanisms of RNA maturation and degradation and the involvement of TA encoded RNA interferases (Yamaguchi and Inouye, 2009; Rochat et al., 2013). We here provide examples on how TA systems impact the flow of genetic information in pathogenic bacteria, with a focus on post-transcriptional regulation via TA toxin RNase activity (summarized in Figure 1).
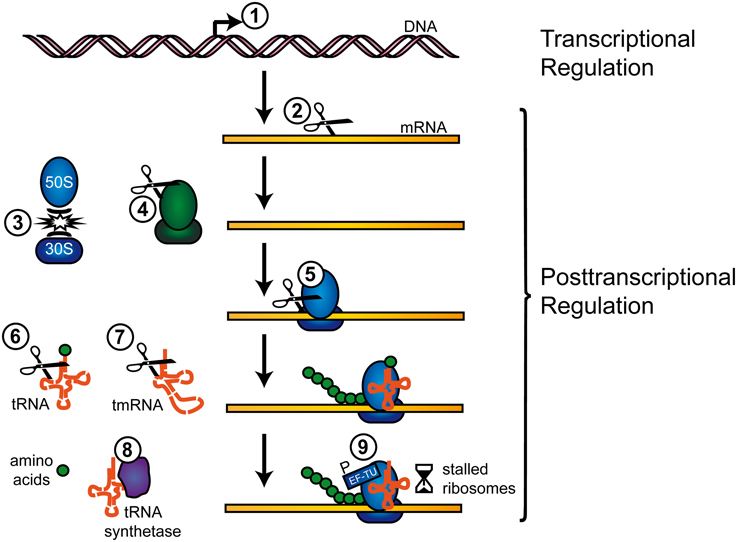
Figure 1. Regulation of transcription and translation by TA systems. TA systems can regulate gene expression on a transcriptional and post-transcriptional level, as follows (examples in parentheses): (1) Canonical transcriptional gene regulation by DNA binding (MqsR). (2) Cleavage of free mRNA transcripts (MazF, ChpBK, PemK, HicA). (3) Inhibition of ribosome association (RatA, MazF). (4) Cleavage of rRNAs (MazF, VapC). (5) Cleavage of ribosome associated mRNAs (RelE, YoeB). (6) Cleavage of tRNAs (VapC). (7) Cleavage of tmRNAs (HicA, RelE). (8) Inhibition of tRNA synthetase (HipA). (9) Phosphorylation of EF-Tu (Doc). Further information on the mechanisms is available in the respective sections.
VapC Homologs Target a Multitude of Substrates in Different Bacteria
Type II TA systems of the VapBC type are highly abundant in prokaryotes and frequently, multiple paralogs of them are found in bacterial genomes (Pandey and Gerdes, 2005; Jørgensen et al., 2009). In Mycobacteria, the number of identified VapBC type TA systems ranges between one in M. smegmatis to more than 40 in the pathogen M. tuberculosis (Pandey and Gerdes, 2005). A hallmark of the VapC toxins is the PIN domain, which is typically associated with Mg2+ dependent ribonuclease activity (Arcus et al., 2004). Sharp et al. (2012) determined the ACGC or AC[AU]GC motifs of mRNAs as targets of the VapC-mt4 protein of M. tuberculosis and reported inhibition of translation mostly by binding to ssRNA instead of cleaving it. Experiments with the VapC20 (Rv2549c) homolog of M. tuberculosis in vitro and in E. coli have shown cleavage of the 23S ribosomal RNA between G and A in a prominent loop region (Winther et al., 2013). By contrast, purified VapC of M. smegmatis exhibits RNase activity preferentially at AUAU and AUAA sites (McKenzie et al., 2012). Microarray studies demonstrated differential expression of almost 3% of the M. smegmatis genome upon overexpression of the toxin with a striking downregulation of carbohydrate metabolism genes. The identification of VapC as a post-transcriptional regulator of glycerol consumption in mycobacteria highlights the influence of TA systems in bacterial metabolism. In addition, Winther and Gerdes (2009) were able to link induction of vapC expression in Salmonella enterica serovar Typhimurium to the stringent response. In contrast to analyzed homologs from Mycobacteria, this VapC protein and the one encoded on the Shigella flexneri 2a virulence plasmid pMYSH6000 exclusively target tRNAfMet by cleaving within the anticodon stem-loop. Together with the RelE-type toxin YoeB (see below), the concomitant stimulation of translation initiation at elongator codons in lieu of canonical start codons may globally affect the cellular translation program (Winther and Gerdes, 2011).
The HicAB TA Homologs
Features like induction by the stringent response and cleavage of tmRNA, required to rescue stalled ribosomes (Keiler et al., 1996), are shared by the E. coli hicAB TA system, which can also be activated during carbon starvation by a Lon protease dependent process. The HicA toxin furthermore cleaves selected mRNAs such as transcripts of dksA, and rpoD encoding regulators or subunits of the RNA polymerase or ompA, an outer membrane protein (Jørgensen et al., 2009). An orthologous system from the opportunistic pathogen Acinetobacter baumannii also targets tmRNA and a set of mRNAs when expressed in E. coli (Jurėnaitė et al., 2013). To date, it is unclear if and how the hicAB locus regulates differential gene expression in the host-cell, but it is likely that some genes are post-transcriptionally influenced by HicA-dependent cleavage.
MazEF, a Jack-of-All-Trades System
mazEF belongs to the best characterized TA systems and encodes a MazF toxin and a MazE antitoxin protein. MazF homologs have been identified in a large number of bacteria (Mittenhuber, 1999) and archaea, with the E. coli system representing the far most extensively studied one (Aizenman et al., 1996). E. coli MazF is an ACA sequence specific endoribonuclease, which upon induction cleaves the bulk of all cellular mRNAs (Zhang et al., 2003). Accordingly, as much as 90% of the encoded proteins are no longer produced, leading to growth impediment (Yamaguchi and Inouye, 2009). While it had previously been assumed that ribosomal RNAs and tRNAs are protected from MazF cleavage by ribosomal proteins and secondary structures (Zhang et al., 2005a). Vesper et al. (2011) demonstrated that 16S rRNAs are processed by E. coli MazF, presumably in response to stressful conditions. Thereby the anti-Shine-Dalgarno (anti-SD) sequence of the 16S rRNA is removed, yielding so called “stress ribosomes”. These are capable of translating a set of leaderless mRNAs with clipped SD sequences as another result of sequence specific MazF activity. This orthogonal translation system produces about 50 proteins which are associated with population heterogeneity in E. coli cultures that leaves only few survivors behind (Amitai et al., 2009). The controversial role of the mazEF system is reflected by two opposite camps. One suggests an involvement in programmed cell death (Aizenman et al., 1996; Amitai et al., 2004; Engelberg-Kulka et al., 2006), whereas the other provides evidence for the modulation of physiological activities for a defined time-frame only (Pedersen et al., 2002; Gerdes and Maisonneuve, 2012). The proficiency of E. coli MazF and a homologous TA toxin ChpBK, which exhibits less stringent ACA specificity (Zhang et al., 2005b), appear to be enhanced by a pentapeptide called extracellular death factor (EDF) (Kolodkin-Gal et al., 2007; Belitsky et al., 2011). Functional analogs thereof are also produced by P. aeruginosa and B. subtilis (Kumar et al., 2013). Mycobacterium tuberculosis bears at least seven mazEF operons, four of which were validated to encode functional mRNA interferases, targeting different three- or five-base consensus sequences (Zhu et al., 2006, 2008). These MazF proteins were proposed to alter gene expression through selective RNA degradation by each paralog addressing different targets. These also include RNA species beyond mRNA, as in case of MazF-mt6. It cleaves 23S rRNA of M. tuberculosis in a bulge region that is part of an association interface between the 30S and the 50S subunits. This leads to an mRNA independent global shut-down of translation activity (Schifano et al., 2013). Interestingly, MazF-mt7 (Rv1495) can physically interact with the M. tuberculosis DNA topoisomerase I (MtbTopA), which inhibits nucleic acid cleavage activity of both enzymes. This interaction exemplifies an additional regulatory function of a TA interferase beyond ribonuclease activity (Huang and He, 2010). Heterologous expression of MazF-mt7 in M. smegmatis causes growth cessation, which, together with findings on the MazF-mt6 system, provides further indications for the involvement of TA systems in mycobacterial long-term dormancy. Among the Gram positive pathogens, mazEF systems were also identified in Clostridium difficile, Streptococcus mutans, and Staphylococcus aureus (Fu et al., 2009; Zhu et al., 2009; Syed et al., 2011; Rothenbacher et al., 2012). Relative abundances of target sites may serve as gene specific indicators for cleavage sensitivity, direct evidence is mostly lacking and secondary structures and RNA associated factors can influence accessibility. This has striking implications for post-transcriptional regulation of factors associated with host-pathogen interactions. Recently, a pemIK system generally related to mazEF (Zhang et al., 2004), was identified on two staphylococcal plasmids and in the chromosomes of other Staphylococcus species (Bukowski et al., 2013). Whereas PemIKSa fulfils the classical TA systems' role of ensuring plasmid propagation, its toxin component PemKSa, an UAUU specific endoribonuclease, has also been speculated to post-transcriptionally control chromosomally located S. aureus genes. Transcripts biased for a low number of UAUU stretches include ORFs of various virulence factors, whereas numerous transporter genes harbor more target sequences than statistically predicted. The finding that the transcript encoding the PemISa antitoxin is resistant to PemKSa cleavage led the authors to propose a mechanism for reinstating TA homeostasis after stressful conditions (Bukowski et al., 2013).
Ribosome Dependent RNases of The RelE Family
The RelE toxins of the relBE TA family interact with the ribosome and cleave mRNAs at the ribosomal A-site (Pedersen et al., 2003). E. coli RelE preferentially targets the trimeric RNA motifs UAG, UCG, and CAG. Interestingly, tmRNA can also be a substrate of RelE, which suggests a role in stress regulation (Christensen and Gerdes, 2003). YafQ toxins, which are structurally similar to RelE family RNases, associate with the 50S ribosomal subunit and specifically restrict selected mRNAs at AAA[AG] consensus sequences. Since the AAA lysin codon is particularly overrepresented at codon +2 in secretory proteins (Zalucki et al., 2007), RelE activity may arrest their translation (Prysak et al., 2009). More recently, also in vivo cleavage of the transcripts lpp (lipoprotein), acpP (acyl carrier protein) and hns (DNA condensing and supercoiling protein) at AAG, GAA, and ACA was demonstrated (Armalytė et al., 2012). Likewise, the E. coli YoeB toxin binds to the 50S ribosomal subunit and cleaves the lpp and ompA (porin) mRNA in vivo, three nucleotides downstream of the start codon (Zhang and Inouye, 2009). The YefM-YoeB system promotes colonization of the bladder by uropathogenic E. coli (Norton and Mulvey, 2012) and is also found in the chromosome of Gram positive pathogens such as Staphylococcus aureus, Streptococcus pneumoniae, and Mycobacterium tuberculosis (Cherny and Gazit, 2004; Nieto et al., 2007; Yoshizumi et al., 2009). A homologous TA system, termed axe-txe, has been identified on Enterococcus plasmids before (Grady and Hayes, 2003; Halvorsen et al., 2011) and Txe was shown to cleave lpp mRNA at the first base after an AUG start codon when expressed in E. coli. Apart from a number of in vivo and in vitro assays which demonstrate the ribosome dependency for these TA toxins, a holistic picture of which genes are affected under specific conditions is yet to be painted for most of these systems. An exception is the RelE family toxin HigB from the higBA TA system, which also cleaves RNA in association with the ribosome (Hurley and Woychik, 2009). Schuessler et al. (2013) analyzed an M. tuberculosis strain overexpressing higB by comparative RNAseq in order to define the entire set of RNAs targeted by the toxin. The relative abundances of tmRNA and of 32 different mRNAs was decreased, of which many are controlled by regulators associated with iron acquisition and stress response (Keiler et al., 1996). It should be noted that although these approaches reveal the complete set of directly and indirectly regulated genes, they do not disclose the targeted nucleotide sequence of the respective loci.
MqsRA Modulates E. coli Stress Response
Whereas many TA systems have been solely characterized in vitro or by heterologous expression in vivo, a distinct physiological role of the type II TA system MqsRA (Yamaguchi et al., 2009) and the modulation of the E. coli stress response has been established (Wang et al., 2011). Apart from MsqA functioning as a transcriptional stress regulator, the MqsR toxin acts as a sequence-specific RNA-interferase. In vivo primer extensions revealed cleavage of three different native E. coli transcripts at the GCU triplet upon artificial induction of MqsR. All of the GCU sequences in ompF mRNA were cleaved without exception although mRNAs form secondary and tertiary structures and can associate with proteins. It was speculated that the MqsRA system could also have important implications for uropathogenic strains switching from a motile non-adhesive state in the urine to a recalcitrant biofilm or other types of non-motile bacterial communities (Hadjifrangiskou et al., 2011).
As shown recently, MqsRA also controls another TA system, termed ghoST (Wang et al., 2013). The small GhoT toxin protein inserts into the membrane resulting in lysed (“ghosts”) or drug tolerant persister cells (Cheng et al., 2013). Notably, ghoT mRNA lacks the primary MqsR target site GCU (Yamaguchi et al., 2009), but instead is specifically cleaved by the GhoS antitoxin. GhoS in turn targets another 20 mRNAs involved in nucleotide precursor anabolism (Wang et al., 2012). Cleavage of toxin mRNA has so far exclusively been described for GhoST, which represents a type V TA system member with extraordinary activities of self-control.
Additional Modes of Post-Transcriptional Regulation
Apart from gene regulation based upon RNA degradation, a number of TA systems exert post-transcriptional regulation by targeting downstream processes.
Like mycobacterial MazF-mt6 (see above), the RatA toxin, encoded by the ratAB locus from E. coli, inhibits the association of the ribosomal subunits. Although already assembled ribosomes are unaffected, synthesis of new proteins is impeded (Zhang and Inouye, 2011).
The PhD/Doc system blocks general translation by binding to the 30S ribosome subunit and thus inhibiting translation elongation (Liu et al., 2008). As found recently, the toxicity of the Doc-type protein Fic is mediated by phosphorylation of the elongation factor EF-Tu, which in turn prevents binding to aminoacetylated tRNAs (Castro-Roa et al., 2013).
The prominent E. coli hipAB TA system, which is involved in a high persister phenotype, was thought to phosphorylate EF-Tu to nonspecifically inhibit overall translation (Schumacher et al., 2009). Newest results, however, indicate that translation inhibition is caused by impeding the glutamyl-tRNA synthetase, thus leading to an accumulation of uncharged tRNAs and therefore translation arrest (Germain et al., 2013). Although these systems are presumably non-selective regulators due to the unspecific shutdown of translation in general, they may regulate translation of total transcriptomes in stress related situations.
Closing Remarks
Gone are the days when TA systems were considered to solely act as plasmid stabilizers. Instead, a multitude of roles for these genetic elements in the modulation of bacterial physiology has emerged. It became evident that TA systems target numerous intracellular structures and processes in prokaryotes among which the modulation of translational cessation seems to be of prime importance. First approaches have been taken to decipher the entirety of transcripts (including small regulatory RNAs) affected by TA RNases throughout different bacteria (Kim et al., 2010; Schuessler et al., 2013). In this regard, secondary structures of RNA molecules, factors interacting with RNAs such as Hfq or other RNA chaperones (Vogel and Luisi, 2011), as well as activity modulators of TA system RNases, including peptides (Kolodkin-Gal et al., 2007; Kumar et al., 2013) or hierarchically organization of TA systems may need to be considered in greater detail (Winther and Gerdes, 2009; Wang et al., 2012). Most importantly, to elucidate regulatory networks, TA systems must be studied in their cellular and environmental context, as heterologous expression in E. coli and activities observed in vitro most likely reflect only part of the extent of regulation. Numerous clinical isolates of S. aureus and P. aeruginosa have been demonstrated to harbor and express TA systems (Williams et al., 2011), which permits refined in vivo analyses without the need for spurious noise due to artificial induction. Another recently discovered antitoxin-like protein in an enterohaemorrhagic E. coli phage can directly undermine the native TA systems and thus nicely underlines the importance of homologous in vivo experiments (Otsuka and Yonesaki, 2012).
Insights into extended roles of type II antitoxins are emerging. Besides controlling toxin activity by protein-protein interaction and transcriptional autoregulation, some antitoxins have been proven to additionally act as transcription factors controlling other regulons (Kim et al., 2010; Lin et al., 2013). Recent findings underscore the hypothesis that pathogenic bacteria contain higher numbers of TA systems than non-pathogenic relatives (Georgiades and Raoult, 2011). A recent publication furthermore showed a direct effect of a newly discovered Salmonella TA system (sehAB) and virulence in a mouse model (De la Cruz et al., 2013). Taken together, TA systems provide interesting targets for antibacterial strategies (Williams and Hergenrother, 2012), which clearly merits further research in this dynamic field.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Volkmar Braun for critically reading the manuscript. Work in the authors' lab was supported by grant BE4038/5 within the priority programme SPP1617 “Phenotypic heterogeneity and sociobiology of bacterial populations” of the Deutsche Forschungsgemeinschaft (DFG). Funding for the open access charge was provided by the Open Access Publishing Fund of the University of Tübingen.
References
Aakre, C. D., Phung, T. N., Huang, D., and Laub, M. T. (2013). A bacterial toxin inhibits DNA replication elongation through a direct interaction with the β sliding clamp. Mol. Cell 52, 617–628. doi: 10.1016/j.molcel.2013.10.014
Aizenman, E., Engelberg-Kulka, H., and Glaser, G. (1996). An Escherichia coli chromosomal “addiction module” regulated by guanosine-3′, 5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. U.S.A. 93, 6059–6063. doi: 10.1073/pnas.93.12.6059
Amitai, S., Kolodkin-Gal, I., Hananya-Meltabashi, M., Sacher, A., and Engelberg-Kulka, H. (2009). Escherichia coli MazF leads to the simultaneous selective synthesis of both “death proteins” and “survival proteins.” PLoS Genet. 5:e1000390. doi: 10.1371/journal.pgen.1000390
Amitai, S., Yassin, Y., and Engelberg-Kulka, H. (2004). MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 186, 8295–8300. doi: 10.1128/JB.186.24.8295-8300.2004
Arcus, V. L., Backbro, K., Roos, A., Daniel, E. L., and Baker, E. N. (2004). Distant structural homology leads to the functional characterization of an archaeal PIN domain as an exonuclease. J. Biol. Chem. 279, 16471–16478. doi: 10.1074/jbc.M313833200
Armalytė, J., Jurėnaitė, M., Beinoravičiūtė, G., Teišerskas, J., and Sužiedėlienė, E. (2012). Characterization of Escherichia coli dinJ-yafQ toxin-antitoxin system using insights from mutagenesis data. J. Bacteriol. 194, 1523–1532. doi: 10.1128/JB.06104-11
Belitsky, M., Avshalom, H., Erental, A., Yelin, I., Kumar, S., London, N., et al. (2011). The Escherichia coli extracellular death factor EDF induces the endoribonucleolytic activities of the toxins MazF and ChpBK. Mol. Cell 41, 625–635. doi: 10.1016/j.molcel.2011.02.023
Blower, T. R., Short, F. L., Rao, F., Mizuguchi, K., Pei, X. Y., Fineran, P. C., et al. (2012). Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. 40, 6158–6173. doi: 10.1093/nar/gks231
Brzozowska, I., and Zielenkiewicz, U. (2013). Regulation of toxin-antitoxin systems by proteolysis. Plasmid 70, 33–41. doi: 10.1016/j.plasmid.2013.01.007
Bukowski, M., Lyzen, R., Helbin, W. M., Bonar, E., Szalewska-Palasz, A., Wegrzyn, G., et al. (2013). A regulatory role for Staphylococcus aureus toxin-antitoxin system PemIKSa. Nat. Commun. 4, 2012. doi: 10.1038/ncomms3012
Castro-Roa, D., Garcia-Pino, A., De Gieter, S., van Nuland, N. A., Loris, R., and Zenkin, N. (2013). The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 9, 811–817. doi: 10.1038/nchembio.1364
Cheng, H. Y., Soo, V. W., Islam, S., McAnulty, M. J., Benedik, M. J., and Wood, T. K. (2013). Toxin GhoT of the GhoT/GhoS TA system damages the cell membrane to reduce ATP and to reduce growth under stress. Environ. Microbiol. doi: 10.1111/1462-2920.12373. [Epub ahead of print].
Cherny, I., and Gazit, E. (2004). The YefM antitoxin defines a family of natively unfolded proteins: implications as a novel antibacterial target. J. Biol. Chem. 279, 8252–8261. doi: 10.1074/jbc.M308263200
Christensen, S. K., and Gerdes, K. (2003). RelE toxins from bacteria and archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48, 1389–1400. doi: 10.1046/j.1365-2958.2003.03512.x
Christensen, S. K., Pedersen, K., Hansen, F. G., and Gerdes, K. (2003). Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332, 809–819. doi: 10.1016/S0022-2836(03)00922-7
De la Cruz, M. A., Zhao, W., Farenc, C., Gimenez, G., Raoult, D., Cambillau, C., et al. (2013). A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog 9:e1003827. doi: 10.1371/journal.ppat.1003827
Dörr, T., Vulić, M., and Lewis, K. (2010). Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8:e1000317. doi: 10.1371/journal.pbio.1000317
Engelberg-Kulka, H., Amitai, S., Kolodkin-Gal, I., and Hazan, R. (2006). Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2:e135. doi: 10.1371/journal.pgen.0020135
Fineran, P. C., Blower, T. R., Foulds, I. J., Humphreys, D. P., Lilley, K. S., and Salmond, G. P. (2009). The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U.S.A. 106, 894–899. doi: 10.1073/pnas.0808832106
Fozo, E. M., Makarova, K. S., Shabalina, S. A., Yutin, N., Koonin, E. V., and Storz, G. (2010). Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 38, 3743–3759. doi: 10.1093/nar/gkq054
Fu, Z., Tamber, S., Memmi, G., Donegan, N. P., and Cheung, A. L. (2009). Overexpression of MazFsa in Staphylococcus aureus induces bacteriostasis by selectively targeting mRNAs for cleavage. J. Bacteriol. 191, 2051–2059. doi: 10.1128/JB.00907-08
Georgiades, K., and Raoult, D. (2011). Genomes of the most dangerous epidemic bacteria have a virulence repertoire characterized by fewer genes but more toxin-antitoxin modules. PloS ONE 6:e17962. doi: 10.1371/journal.pone.0017962
Gerdes, K., and Maisonneuve, E. (2012). Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 66, 103–123. doi: 10.1146/annurev-micro-092611-150159
Gerdes, K., Bech, F. W., Jørgensen, S. T., Løbner-Olesen, A., Rasmussen, P. B., Atlung, T., et al. (1986). Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 5, 2023–2029.
Gerdes, K., Christensen, S. K., and Lobner-Olesen, A. (2005). Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382. doi: 10.1038/nrmicro1147
Germain, E., Castro-Roa, D., Zenkin, N., and Gerdes, K. (2013). Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52, 248–254. doi: 10.1016/j.molcel.2013.08.045
Grady, R., and Hayes, F. (2003). Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol. Microbiol. 47, 1419–1432. doi: 10.1046/j.1365-2958.2003.03387.x
Hadjifrangiskou, M., Kostakioti, M., and Hultgren, S. J. (2011). Antitoxins: therapy for stressed bacteria. Nat. Chem. Biol. 7, 345–347. doi: 10.1038/nchembio.584
Halvorsen, E. M., Williams, J. J., Bhimani, A. J., Billings, E. A., and Hergenrother, P. J. (2011). Txe, an endoribonuclease of the enterococcal Axe-Txe toxin-antitoxin system, cleaves mRNA and inhibits protein synthesis. Microbiology 157, 387–397. doi: 10.1099/mic.0.045492-0
Huang, F., and He, Z. G. (2010). Characterization of an interplay between a Mycobacterium tuberculosis MazF homolog, Rv1495 and its sole DNA topoisomerase I. Nucleic Acids Res. 38, 8219–8230. doi: 10.1093/nar/gkq737
Hurley, J. M., and Woychik, N. A. (2009). Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J. Biol. Chem. 284, 18605–18613. doi: 10.1074/jbc.M109.008763
Jaffé, A., Ogura, T., and Hiraga, S. (1985). Effects of the ccd function of the F plasmid on bacterial growth. J. Bacteriol. 163, 841–849.
Jørgensen, M. G., Pandey, D. P., Jaskolska, M., and Gerdes, K. (2009). HicA of Escherichia coli defines a novel family of translation-independent mRNA interferases in bacteria and archaea. J. Bacteriol. 191, 1191–1199. doi: 10.1128/JB.01013-08
Jurėnaitė, M., Markuckas, A., and Sužiedėlienė, E. (2013). Identification and characterization of type II toxin-antitoxin systems in the opportunistic pathogen Acinetobacter baumannii. J. Bacteriol. 195, 3165–3172. doi: 10.1128/JB.00237-13
Keiler, K. C., Waller, P. R., and Sauer, R. T. (1996). Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993. doi: 10.1126/science.271.5251.990
Kim, Y., Wang, X., Zhang, X. S., Grigoriu, S., Page, R., Peti, W., et al. (2010). Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 12, 1105–1121. doi: 10.1111/j.1462-2920.2009.02147.x
Kolodkin-Gal, I., Hazan, R., Gaathon, A., Carmeli, S., and Engelberg-Kulka, H. (2007). A linear pentapeptide is a quorum-sensing factor required for mazEF-mediated cell death in Escherichia coli. Science 318, 652–655. doi: 10.1126/science.1147248
Kumar, S., Kolodkin-Gal, I., and Engelberg-Kulka, H. (2013). Novel quorum-sensing peptides mediating interspecies bacterial cell death. mBio 4, e00314–00313. doi: 10.1128/mBio.00314-13
Leplae, R., Geeraerts, D., Hallez, R., Guglielmini, J., Drèze, P., and Van Melderen, L. (2011). Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 39, 5513–5525. doi: 10.1093/nar/gkr131
Lewis, K. (2010). Persister cells. Annu. Rev. Microbiol. 64, 357–372. doi: 10.1146/annurev.micro.112408.134306
Lin, C. Y., Awano, N., Masuda, H., Park, J. H., and Inouye, M. (2013). Transcriptional Repressor HipB Regulates the multiple promoters in Escherichia coli. J. Mol. Microbiol. Biotechnol. 23, 440–447. doi: 10.1159/000354311
Liu, M., Zhang, Y., Inouye, M., and Woychik, N. A. (2008). Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. Proc. Natl. Acad. Sci. U.S.A. 105, 5885–5890. doi: 10.1073/pnas.0711949105
Magnuson, R. D. (2007). Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189, 6089–6092. doi: 10.1128/JB.00958-07
Maisonneuve, E., Castro-Camargo, M., and Gerdes, K. (2013). (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150. doi: 10.1016/j.cell.2013.07.048
Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2009). Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct. 4:19. doi: 10.1186/1745-6150-4-19
Markovski, M., and Wickner, S. (2013). Preventing bacterial suicide: a novel toxin-antitoxin strategy. Mol. Cell 52, 611–612. doi: 10.1016/j.molcel.2013.11.018
Masuda, H., Tan, Q., Awano, N., Wu, K. P., and Inouye, M. (2012). YeeU enhances the bundling of cytoskeletal polymers of MreB and FtsZ, antagonizing the CbtA (YeeV) toxicity in Escherichia coli. Mol. Microbiol. 84, 979–989. doi: 10.1111/j.1365-2958.2012.08068.x
McKenzie, J. L., Robson, J., Berney, M., Smith, T. C., Ruthe, A., Gardner, P. P., et al. (2012). A VapBC toxin-antitoxin module is a posttranscriptional regulator of metabolic flux in mycobacteria. J. Bacteriol. 194, 2189–2204. doi: 10.1128/JB.06790-11
Mittenhuber, G. (1999). Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J. Mol. Microbiol. Biotechnol. 1, 295–302.
Nieto, C., Cherny, I., Khoo, S. K., de Lacoba, M. G., Chan, W. T., Yeo, C. C., et al. (2007). The yefM-yoeB toxin-antitoxin systems of Escherichia coli and Streptococcus pneumoniae: functional and structural correlation. J. Bacteriol. 189, 1266–1278. doi: 10.1128/JB.01130-06
Norton, J. P., and Mulvey, M. A. (2012). Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog. 8:e1002954. doi: 10.1371/journal.ppat.1002954
Ogura, T., and Hiraga, S. (1983). Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. U.S.A. 80, 4784–4788. doi: 10.1073/pnas.80.15.4784
Otsuka, Y., and Yonesaki, T. (2012). Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol. Microbiol. 83, 669–681. doi: 10.1111/j.1365-2958.2012.07975.x
Pandey, D. P., and Gerdes, K. (2005). Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976. doi: 10.1093/nar/gki201
Pedersen, K., Christensen, S. K., and Gerdes, K. (2002). Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45, 501–510. doi: 10.1046/j.1365-2958.2002.03027.x
Pedersen, K., Zavialov, A. V., Pavlov, M. Y., Elf, J., Gerdes, K., and Ehrenberg, M. (2003). The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140. doi: 10.1016/S0092-8674(02)01248-5
Prysak, M. H., Mozdzierz, C. J., Cook, A. M., Zhu, L., Zhang, Y., Inouye, M., et al. (2009). Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 71, 1071–1087. doi: 10.1111/j.1365-2958.2008.06572.x
Rochat, T., Bouloc, P., and Repoila, F. (2013). Gene expression control by selective RNA processing and stabilization in bacteria. FEMS Microbiol. Lett. 344, 104–113. doi: 10.1111/1574-6968.12162
Rothenbacher, F. P., Suzuki, M., Hurley, J. M., Montville, T. J., Kirn, T. J., Ouyang, M., et al. (2012). Clostridium difficile MazF toxin exhibits selective, not global, mRNA cleavage. J. Bacteriol. 194, 3464–3474. doi: 10.1128/JB.00217-12
Schifano, J. M., Edifor, R., Sharp, J. D., Ouyang, M., Konkimalla, A., Husson, R. N., et al. (2013). Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. U.S.A. 110, 8501–8506. doi: 10.1073/pnas.1222031110
Schuessler, D. L., Cortes, T., Fivian-Hughes, A. S., Lougheed, K. E., Harvey, E., Buxton, R. S., et al. (2013). Induced ectopic expression of HigB toxin in Mycobacterium tuberculosis results in growth inhibition, reduced abundance of a subset of mRNAs and cleavage of tmRNA. Mol. Microbiol. 90, 195–207. doi: 10.1111/mmi.12358
Schumacher, M. A., Piro, K. M., Xu, W., Hansen, S., Lewis, K., and Brennan, R. G. (2009). Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323, 396–401. doi: 10.1126/science.1163806
Schuster, C. F., and Bertram, R. (2013). Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 340, 73–85. doi: 10.1111/1574-6968.12074
Sharp, J. D., Cruz, J. W., Raman, S., Inouye, M., Husson, R. N., and Woychik, N. A. (2012). Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. J. Biol. Chem. 287, 12835–12847. doi: 10.1074/jbc.M112.340109
Syed, M. A., Koyanagi, S., Sharma, E., Jobin, M. C., Yakunin, A. F., and Lévesque, C. M. (2011). The chromosomal mazEF locus of Streptococcus mutans encodes a functional type II toxin-antitoxin addiction system. J. Bacteriol. 193, 1122–1130. doi: 10.1128/JB.01114-10
Vesper, O., Amitai, S., Belitsky, M., Byrgazov, K., Kaberdina, A. C., Engelberg-Kulka, H., et al. (2011). Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147, 147–157. doi: 10.1016/j.cell.2011.07.047
Vogel, J., and Luisi, B. F. (2011). Hfq and its constellation of RNA. Nat. Rev. Microbiol. 9, 578–589. doi: 10.1038/nrmicro2615
Vogel, J., Argaman, L., Wagner, E. G., and Altuvia, S. (2004). The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 14, 2271–2276. doi: 10.1016/j.cub.2004.12.003
Wang, X., Kim, Y., Hong, S. H., Ma, Q., Brown, B. L., Pu, M., et al. (2011). Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7, 359–366. doi: 10.1038/nchembio.560
Wang, X., Lord, D. M., Cheng, H. Y., Osbourne, D. O., Hong, S. H., Sanchez-Torres, V., et al. (2012). A new type V toxin-antitoxin system where mRNA for toxin GhoT is cleaved by antitoxin GhoS. Nat. Chem. Biol. 8, 855–861. doi: 10.1038/nchembio.1062
Wang, X., Lord, D. M., Hong, S. H., Peti, W., Benedik, M. J., Page, R., et al. (2013). Type II toxin/antitoxin MqsR/MqsA controls type V toxin/antitoxin GhoT/GhoS. Environ. Microbiol. 15, 1734–1744. doi: 10.1111/1462-2920.12063
Williams, J. J., and Hergenrother, P. J. (2012). Artificial activation of toxin-antitoxin systems as an antibacterial strategy. Trends Microbiol. 20, 291–298. doi: 10.1016/j.tim.2012.02.005
Williams, J. J., Halvorsen, E. M., Dwyer, E. M., DiFazio, R. M., and Hergenrother, P. J. (2011). Toxin-antitoxin (TA) systems are prevalent and transcribed in clinical isolates of Pseudomonas aeruginosa and methicillin-resistant Staphylococcus aureus. FEMS Microbiol. Lett. 322, 41–50. doi: 10.1111/j.1574-6968.2011.02330.x
Winther, K. S., and Gerdes, K. (2009). Ectopic production of VapCs from Enterobacteria inhibits translation and trans-activates YoeB mRNA interferase. Mol. Microbiol. 72, 918–930. doi: 10.1111/j.1365-2958.2009.06694.x
Winther, K. S., and Gerdes, K. (2011). Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 108, 7403–7407. doi: 10.1073/pnas.1019587108
Winther, K. S., Brodersen, D. E., Brown, A. K., and Gerdes, K. (2013). VapC20 of Mycobacterium tuberculosis cleaves the Sarcin-Ricin loop of 23S rRNA. Nat. Commun. 4, 2796. doi: 10.1038/ncomms3796
Yamaguchi, Y., and Inouye, M. (2009). mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog. Mol. Biol. Transl. Sci. 85, 467–500. doi: 10.1016/S0079-6603(08)00812-X
Yamaguchi, Y., and Inouye, M. (2011). Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9, 779–790. doi: 10.1038/nrmicro2651
Yamaguchi, Y., Park, J. H., and Inouye, M. (2009). MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU-specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284, 28746–28753. doi: 10.1074/jbc.M109.032904
Yoshizumi, S., Zhang, Y., Yamaguchi, Y., Chen, L., Kreiswirth, B. N., and Inouye, M. (2009). Staphylococcus aureus YoeB homologues inhibit translation initiation. J. Bacteriol. 191, 5868–5872. doi: 10.1128/JB.00623-09
Zalucki, Y. M., Power, P. M., and Jennings, M. P. (2007). Selection for efficient translation initiation biases codon usage at second amino acid position in secretory proteins. Nucleic Acids Res. 35, 5748–5754. doi: 10.1093/nar/gkm577
Zhang, J., Zhang, Y., Zhu, L., Suzuki, M., and Inouye, M. (2004). Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 279, 20678–20684. doi: 10.1074/jbc.M314284200
Zhang, Y., and Inouye, M. (2009). The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J. Biol. Chem. 284, 6627–6638. doi: 10.1074/jbc.M808779200
Zhang, Y., and Inouye, M. (2011). RatA (YfjG), an Escherichia coli toxin, inhibits 70S ribosome association to block translation initiation. Mol. Microbiol. 79, 1418–1429. doi: 10.1111/j.1365-2958.2010.07506.x
Zhang, Y., Zhang, J., Hara, H., Kato, I., and Inouye, M. (2005a). Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 280, 3143–3150. doi: 10.1074/jbc.M411811200
Zhang, Y., Zhu, L., Zhang, J., and Inouye, M. (2005b). Characterization of ChpBK, an mRNA interferase from Escherichia coli. J. Biol. Chem. 280, 26080–26088. doi: 10.1074/jbc.M502050200
Zhang, Y., Zhang, J., Hoeflich, K. P., Ikura, M., Qing, G., and Inouye, M. (2003). MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12, 913–923. doi: 10.1016/S1097-2765(03)00402-7
Zhu, L., Inoue, K., Yoshizumi, S., Kobayashi, H., Zhang, Y., Ouyang, M., et al. (2009). Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 191, 3248–3255. doi: 10.1128/JB.01815-08
Zhu, L., Phadtare, S., Nariya, H., Ouyang, M., Husson, R. N., and Inouye, M. (2008). The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 69, 559–569. doi: 10.1111/j.1365-2958.2008.06284.x
Keywords: toxin-antitoxin system, TA system, RNase, gene regulation, translation inhibition, pathogenicity, review
Citation: Bertram R and Schuster CF (2014) Post-transcriptional regulation of gene expression in bacterial pathogens by toxin-antitoxin systems. Front. Cell. Infect. Microbiol. 4:6. doi: 10.3389/fcimb.2014.00006
Received: 10 December 2013; Accepted: 13 January 2014;
Published online: 29 January 2014.
Edited by:
Wyndham W. Lathem, Northwestern University Feinberg School of Medicine, USAReviewed by:
Elizabeth Fozo, University of Tennessee, USABenedykt Wladyka, Jagiellonian University, Poland
Copyright © 2014 Bertram and Schuster. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ralph Bertram, Department of Microbial Genetics, Faculty of Science, Interfaculty Institute of Microbiology and Infection Medicine Tübingen (IMIT), University of Tübingen, Waldhäuser Str. 70/8, 72076 Tübingen, Germany e-mail:cmFscGguYmVydHJhbUB1bmktdHVlYmluZ2VuLmRl
 Ralph Bertram
Ralph Bertram Christopher F. Schuster
Christopher F. Schuster