- 1Department of Immunology and Microbiology, Indiana University School of Medicine, Indianapolis, IN, USA
- 2Prestage Department of Poultry Science, North Carolina State University, Raleigh, NC, USA
In the ancient anaerobic environment, ferrous iron (Fe2+) was one of the first metal cofactors. Oxygenation of the ancient world challenged bacteria to acquire the insoluble ferric iron (Fe3+) and later to defend against reactive oxygen species (ROS) generated by the Fenton chemistry. To acquire Fe3+, bacteria produce low-molecular weight compounds, known as siderophores, which have extremely high affinity for Fe3+. However, during infection the host restricts iron from pathogens by producing iron- and siderophore-chelating proteins, by exporting iron from intracellular pathogen-containing compartments, and by limiting absorption of dietary iron. Ferric Uptake Regulator (Fur) is a transcription factor which utilizes Fe2+ as a corepressor and represses siderophore synthesis in pathogens. Fur, directly or indirectly, controls expression of enzymes that protect against ROS damage. Thus, the challenges of iron homeostasis and defense against ROS are addressed via Fur. Although the role of Fur as a repressor is well-documented, emerging evidence demonstrates that Fur can function as an activator. Fur activation can occur through three distinct mechanisms (1) indirectly via small RNAs, (2) binding at cis regulatory elements that enhance recruitment of the RNA polymerase holoenzyme (RNAP), and (3) functioning as an antirepressor by removing or blocking DNA binding of a repressor of transcription. In addition, Fur homologs control defense against peroxide stress (PerR) and control uptake of other metals such as zinc (Zur) and manganese (Mur) in pathogenic bacteria. Fur family members are important for virulence within bacterial pathogens since mutants of fur, perR, or zur exhibit reduced virulence within numerous animal and plant models of infection. This review focuses on the breadth of Fur regulation in pathogenic bacteria.
Introduction
Transition metals are essential elements in biological systems. Metabolic pathways, DNA synthesis, RNA synthesis, and protein synthesis are dependent on the availability of the appropriate metal cofactor. In support of this, all cells have designated gene products that transport metals to maintain cellular function; however, certain essential metals cause the formation of toxic reactive oxygen species (ROS). In the earliest description of what is now known as the Fenton reaction, iron (Fe) was shown to act catalytically in the oxidation of tartaric acid (Fenton, 1894). The Fenton reaction produces the hydroxyl radical (HO.), a ROS capable of oxidizing macromolecules and lipids (Imlay et al., 1988; Lloyd et al., 1997). Therefore, cells must tightly regulate the concentration of Fe to avoid ROS-mediated cell damage.
Bacteria sense their environment and alter expression of genes that promote survival. This is accomplished by transcription factors that regulate expression of beneficial or detrimental genes. In order to acquire Fe in Fe-limiting environments, bacteria and fungi synthesize and secrete low molecular weight compounds, called siderophores, which have high affinity for binding Fe3+. Most siderophores are produced by the non-ribosomal peptide synthesis (NRPS) pathway and an example is the siderophore enterochelin. The final steps of the pathway are executed by the action of the Ent proteins (encoded by the entD, entF, and entCEBA genes) (Gehring et al., 1998; Salvail et al., 2010). Aerobactin, another siderophore, is sequentially produced by the proteins IucD, IucB, IucA, and IucC (Figure 1A) that are expressed in an operon (iucABCD). Aerobactin is an example of a siderophore not produced by the NRPS pathway. Transcriptional control of both siderophores is regulated by the concentration of intracellular Fe2+ (Bagg and Neilands, 1987b); when intracellular Fe2+ is low, the model bacterial organism, Escherichia coli induces siderophore production (Brot and Goodwin, 1968; Bryce and Brot, 1971). The Fe-bound siderophores are subsequently transported into the cell to satisfy an Fe2+ requirement. Because Fe2+ transcriptionally controls expression of gene products that promote iron acquisition, Fe2+ was predicted to be a corepressor for a DNA-binding protein. Isolation of a mutant of Salmonella enterica subsp. enterica serovar Typhimurium (S. Typhimurium) that constitutively expresses iron uptake proteins supported this hypothesis (Ernst et al., 1978). A mutation in Ferric Uptake Regulator (Fur) encoded by the fur gene was identified in E. coli mutants that exhibited constitutive expression of iron uptake genes (Hantke, 1981, 1984; Bagg and Neilands, 1985). Fur is a DNA-binding protein that recognizes specific DNA sequences, utilizes Fe2+ or Mn2+ as a corepressor, and blocks transcription of target genes (Bagg and Neilands, 1987a; De Lorenzo et al., 1987). Not surprisingly, the transcriptional control of entD, entF, entCEBA, and iucABCD is negatively regulated by Fur (De Lorenzo et al., 1987; Brickman et al., 1990; Stojiljkovic et al., 1994; Tsolis et al., 1995; Bjarnason et al., 2003; McHugh et al., 2003; Troxell et al., 2011a).
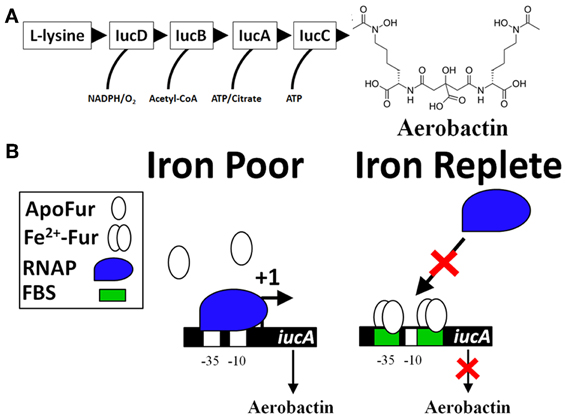
Figure 1. The classic model of Fur repression of iron acquisition (iucA as an example). (A) Biosynthesis of the siderophore aerobactin requires several genes located in an operon (iucABCD, iutA). Expression of the initial gene, iucA, is Fur-repressed (De Lorenzo et al., 1987) and production of aerobactin is known to be produced by virulent strains of bacteria, especially strains causing disease in avian hosts (i.e., Avian pathogenic E. coli or APEC) (Lafont et al., 1987; Xiong et al., 2012; Ling et al., 2013). The sequential enzymatic activity of IucD, IucB, IucC, and IucA convert L-lysine into aerobactin, a potent Fe-scavenging siderophore. (B) There are two Fur-binding sites (FBS) for Fe-dependent regulation of iucA. Both FBS are located within the P1 promoter (overlapping the −35 and also the −10 sites). Under conditions of Fe-deprivation (left panel), there is increased transcription (signified by a +1) of the iucABCD genes whose protein products form a biosynthetic pathway that produces aerobactin. Under Fe-replete conditions (right panel), Fur binds to DNA at the FBS (green box) and blocks access of the −35 and −10 sites by RNA polymerase (RNAP, blue shape).
The collective work supports a simple model for the molecular mechanism of Fur repression that consists of Fur binding to cis regulatory elements of a gene and preventing the binding of the RNA polymerase holoenzyme (RNAP) (Figure 1B) (De Lorenzo et al., 1987; Escolar et al., 1999, 2000; Hantke, 2001; Lee and Helmann, 2007; Carpenter et al., 2009). As a transcriptional repressor, Fur-Fe2+ homodimer binds to the operator site of a target promoter (Ernst et al., 1978; Bagg and Neilands, 1985, 1987a; Neilands, 1993; Escolar et al., 1997, 1998). However, Fur can form a multimeric complex with DNA sequences extending beyond the operator site (Escolar et al., 2000; Baichoo and Helmann, 2002; Lavrrar et al., 2002). Initial studies defined the Fur-binding site (the Fur box) as an ≈19 bp DNA sequence with dyad symmetry, GATAATGATAATCATTATC (Calderwood and Mekalanos, 1987, 1988; De Lorenzo et al., 1987; Stojiljkovic et al., 1994). Insertion of this sequence into an operator site in the promoter of a non-Fe2+ regulated gene results in derepression under Fe2+-limiting conditions (Calderwood and Mekalanos, 1988). In an elegant approach to define Fur regulated genes within bacteria, a high copy number plasmid containing randomly cloned DNA sequences from Gram positive and negative bacteria were transformed into an E. coli strain that harbored a single copy of a fhuF::lacZ reporter fusion (Hantke, 1987). Fur represses transcription of the fhuF gene, which encodes a protein involved in the acquisition of Fe3+ (Hantke, 1983, 1987). If the cloned DNA fragment on the high copy number plasmid contains a Fur-binding site, then Fur proteins will be titrated away from the promoter of fhuF resulting in derepression of the fhuF::lacZ fusion, which can be qualitatively detected during growth on MacConkey agar plates or quantified by a β-galactosidase assay. This assay is called the Fur titration assay (FURTA) and has been used to study Fur regulation for nearly 20 years (Stojiljkovic et al., 1994; Tsolis et al., 1995; Baumler et al., 1996; Fassbinder et al., 2000; Osorio et al., 2004; Haraszthy et al., 2006; Jackson et al., 2010; Tanabe et al., 2010). In toto, these works solidified the role of Fur as a Fe2+-dependent transcriptional repressor. However, global gene expression studies have identified numerous genes that require Fur for expression (Foster and Hall, 1992; D'Autreaux et al., 2002; Bjarnason et al., 2003; McHugh et al., 2003; Troxell et al., 2011a).
Multifactorial Roles of Fe2+-Fur Regulation in Bacteria
Fur is required for the expression of several proteins within the tricarboxylic acid cycle (TCA) and the Fe2+-dependent superoxide dismutase (SodB) (Hantke, 1987; Gruer and Guest, 1994; Dubrac and Touati, 2000, 2002). The disruption of the TCA cycle within fur mutants may have a relevant role for the regulation of virulence since mutations within the TCA cycle alter virulence expression in Staphylococcus epidermidis and Vibrio cholera (Sadykov et al., 2008; Minato et al., 2013). In addition, disruption of the TCA cycle reduces S. Typhimurium virulence in mice (Tchawa Yimga et al., 2006; Bowden et al., 2010). The role of Fur in TCA cycle regulation is an example of how Fur regulation is multifactorial; fur mutants exhibit many phenotypes not just enhanced expression of siderophores. The molecular mechanism for the Fur's positive activation in the TCA cycle and SodB went unexplained until a landmark publication determined the importance of a highly conserved small untranslated RNA (sRNA) named ryhB in activation by Fur (Masse and Gottesman, 2002). ryhB is directly repressed by Fur (Vassinova and Kozyrev, 2000; Masse and Gottesman, 2002) and base pairs with target mRNAs, such as sodB and the succinate dehydrogenase operon sdhCDAB, which results in degradation of the mRNAs thereby reducing expression of the gene products (Figure 2A). Deletion of ryhB in a Δfur results in restoration of expression of TCA proteins, SodB, and growth on succinate or fumarate minimal medium (Masse and Gottesman, 2002). Because regulation by ryhB requires the RNA chaperone protein, Hfq, deletion of hfq in Δfur also restores expression of many Fur activated genes (Masse and Gottesman, 2002; Ellermeier and Slauch, 2008; Troxell et al., 2011a). ryhB homologs have a role in virulence, are Fur-repressed, and are encoded in the genomes of several Gram negative pathogens (i.e., Klebsiella pneumoniae, Shigella, Vibrio cholera, Yersinia pestis, Salmonella, Pseudomonas aeruginosa, Neisseria meningitidis, and Neisseria gonorrhoeae) (Wilderman et al., 2004; Davis et al., 2005; Mey et al., 2005a; Oglesby et al., 2005; Mellin et al., 2007; Murphy and Payne, 2007; Ellermeier and Slauch, 2008; Ducey et al., 2009; Metruccio et al., 2009; Troxell et al., 2011a; Deng et al., 2012; Huang et al., 2012; Kim and Kwon, 2013; Leclerc et al., 2013). Indirect positive regulation by Fur through negative regulation of the negative regulator, ryhB, is the most studied molecular mechanism for Fe2+-dependent activation of gene expression; however, additional evidence demonstrates that Fur may regulate virulence through more complicated mechanisms.
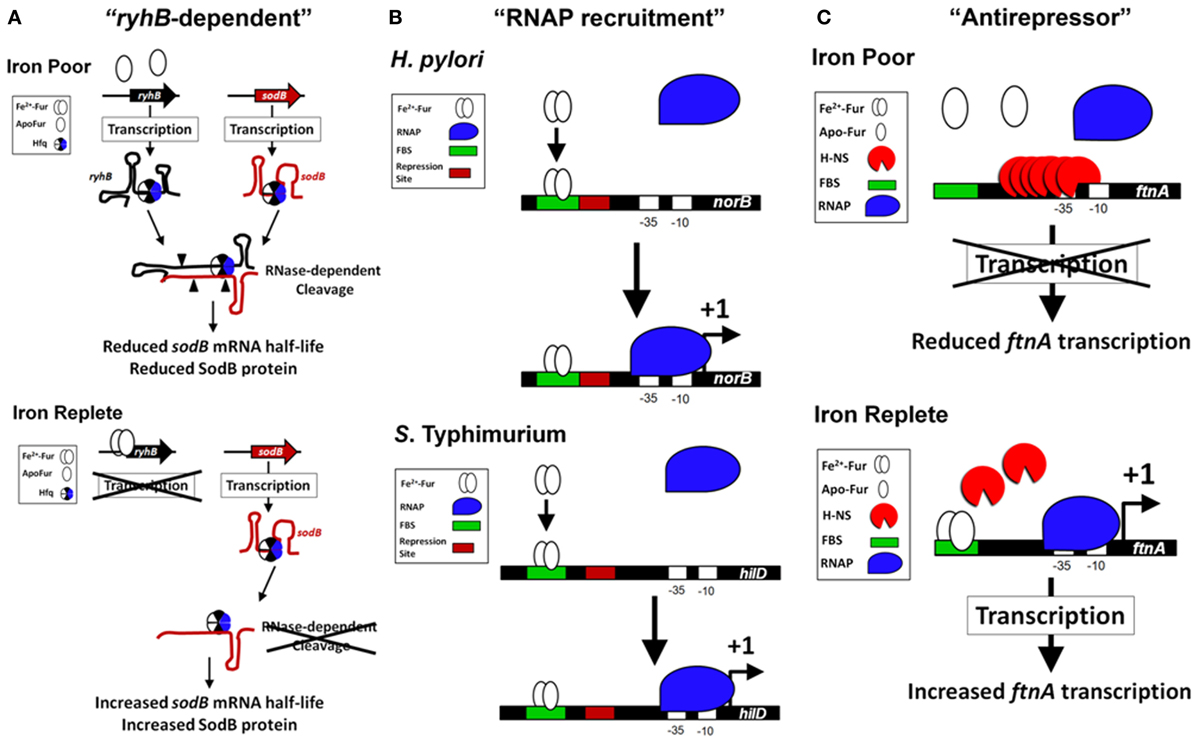
Figure 2. Models of the Fur-dependent activation of gene expression in bacteria. (A) Fur activation through “ryhB-dependent” mechanism (SodB as an example). Fur is indirectly required for the expression of the FeSOD (SodB) in bacteria through the sRNA ryhB (Masse and Gottesman, 2002; Ellermeier and Slauch, 2008). Under conditions of Fe2+ depletion (top panel), Fur is unable to directly repress transcription of the sRNA ryhB (or its paralog). This results in an increase in the level of ryhB within the cell. The RNA chaperone Hfq binds to ryhB and to the target mRNA of sodB (Afonyushkin et al., 2005; Urban and Vogel, 2007), which through the RNase-dependent cleavage (cleavage sites are signified by filled triangles) reduces the half-life of sodB mRNA and reduces SodB protein within the cell. The Fur activation of sodB is diminished in the absence of Hfq or ryhB (Masse and Gottesman, 2002; Ellermeier and Slauch, 2008; Troxell et al., 2011a). When Fur is activated during Fe2+ replete conditions (bottom panel), transcription of ryhB is blocked, which increases the half-life of sodB mRNA allowing for enhanced production of SodB protein and FeSOD activity. (B) Fur activation through “RNAP recruitment” mechanism (Examples from S. Typhimurium and H. Pylori). In vitro transcription assays with H. pylori norB regulatory sequences (Delany et al., 2004) and S. Typhimurium hilD regulatory sequences (Teixido et al., 2011) demonstrate an active Fur-Fe2+ binding to a FBS (signified with a green box) that promotes increased binding of the RNAP (signified with a blue shape) to the promoter and transcription of the target gene (signified with a +1). In both examples, the regulatory sequences of norB and hilD contain a repression site (signified with a red box) that may overlap the FBS (an ArsR-binding site with norB) or be located immediately downstream of the FBS (an H-NS binding site with hilD). If Fur-Fe2+ physically contacts the RNAP is unknown. (C) Fur activation through “antirepressor” mechanism (FtnA as an example). In E. coli, expression of the ftnA gene is Fur activated, but independent of the “ryhB-dependent” activation. Under Fe2+ poor conditions, H-NS binds upstream of the ftnA gene and represses transcription (top panel). When Fur is activated, Fur-Fe2+ binds to several FBS located upstream of ftnA, which prevents H-NS nucleation at the ftnA promoter and repressing transcription (bottom panel). In this example, Fur is required to block H-NS binding and can physically remove H-NS from the upstream regulatory site, which allows for ftnA expression.
For example, in S. Typhimurium, transcription of the virulence factor hilA requires Fe2+ through Fur-dependent regulation (Thompson et al., 2006; Ellermeier and Slauch, 2008; Troxell et al., 2011b). Recently, we demonstrated enhanced transcription of hns in Δfur and in a modified chromatin immunoprecipitation (ChIP) assay we determined that Fur bound the upstream regulatory region of hns in a metal-dependent manner (Troxell et al., 2011b). H-NS is known to repress transcription of hilA (Olekhnovich and Kadner, 2006). H-NS is a protein associated with the bacterial nucleoid and is also known as OsmZ, BglY, and PilG (Defez and De Felice, 1981; Spears et al., 1986; May et al., 1990). Deletion of fur and hns resulted in Fur-independent activation of hilA, which supports the indication that Fur regulation of hilA was indirect through H-NS (Troxell et al., 2011b). Furthermore, Fur is not required for expression of Fur-activated genes when the repressor H-NS is absent (Nandal et al., 2010; Troxell et al., 2011b) and Fur and H-NS appeared to recognize similar DNA sequences throughout the bacterial chromosome (Prajapat and Saini, 2012). In another example of the multifactorial role of Fur in bacteria, a recent study shows that Fur represses transcription of the vvhA gene, which encodes the major haemolysin of Vibrio vulnificus, yet haemolytic activity and VvhA protein level were reduced in Δfur (Lee et al., 2013). Two metal-dependent proteases are responsible for degradation of VvhA, VvpE, and VvpM and transcription of vvpE is under negative regulation by Fur. Through genetic and biochemical approaches, it was shown that VvpE and VvpM exhibited enhanced activity in Δfur resulting in reduction of the VvhA protein (Lee et al., 2013). Clearly, it can be appreciated from these two examples that the influence of Fur within the cell is global and typically involves multiple layers of regulation. Nevertheless, recent evidence indicates Fur may have a more direct role for activation of gene expression in bacteria (Figure 2).
Mechanisms of Activation of Gene Expression Via DNA Binding by Fur: Location, Location, Location
Global gene expression studies have identified genes that require Fur for expression (Foster and Hall, 1992; D'Autreaux et al., 2002; Bjarnason et al., 2003; McHugh et al., 2003; Troxell et al., 2011a). Earlier work demonstrated a unique mechanism for Fur activation in N. meningitidis that involves Fur directly binding to cis regulatory elements upstream of a Fur-activated gene (Delany et al., 2004). Unlike Fur-repressed genes that possess a characteristic Fur-binding site overlapping the RNAP-binding site, Fur-activated genes [norB, pan1 (aniA), and nuoA] contain Fur boxes located ≈100 bp upstream of the transcriptional start site, while the Fur-repressed tbp contains a Fur box that overlaps with the RNAP-binding site. The Fur box and activation of norB, which encodes a protein responsible for protection against NO (Anjum et al., 2002), is conserved in N. gonorrhoeae (Isabella et al., 2008). Moreover, in Helicobacter pylori, Fur activates expression of oorB, which encodes a 2-oxoglutarate:acceptor oxidoreductase (Hughes et al., 1998), by directly binding to a cis regulatory elements located 130 bp upstream of the transcriptional start site (Gilbreath et al., 2012). The importance of OorB in virulence is demonstrated by the significant reduction in colonization of the chicken gut by a ΔoorB mutant strain of Campylobacter jejuni (Weerakoon et al., 2009). In V. cholera, Fur activates expression of the outer membrane porin, ompT, through binding a Fur box located 90 bp upstream of the transcriptional start site (Craig et al., 2011). In S. Typhimurium, transcription of the virulence factor hilD is activated by Fur through a Fur box located nearly 200 bp upstream of the transcriptional start site (Teixido et al., 2011). HilD is an AraC/XylS-type DNA-binding protein that regulates transcription of important virulence factors within S. Typhimurium and is required for infection (Ellermeier et al., 2005). Importantly, the sequence of the Fur box site for activated genes is virtually identical to the Fur box of repressed genes. Collectively, the molecular evidence suggests the location of the Fur box in proximity to the RNAP-binding site determines the ability of Fur to activate gene expression.
How does Fur activate gene expression? In vitro transcription experiments demonstrate that Fur can activate transcription of a target gene even though the Fur boxes are located ≈100 and 200 bp upstream of the transcriptional start site, respectively (Delany et al., 2004; Teixido et al., 2011). This example of Fur activation is rare, but may involve enhanced recruitment of RNAP to the promoter of target genes (“RNAP recruitment” activation model, Figure 2B). Surprisingly, addition of the Fur protein to the in vitro transcription assay stimulated the production of hilD mRNA, which suggests improved recruitment of RNAP to the promoter of hilD even though the Fur box is nearly 200 bp upstream of the transcriptional start site (Teixido et al., 2011). While deletion of fur reduces transcription of hilD (Teixido et al., 2011) overexpression of Fur results in little increased activation of the hilD promoter contrary to overexpression of a direct activator HilC, which increases hilD's promoter activity by ≈5-fold (Ellermeier and Slauch, 2008). These results indicate the role of Fur in direct transcriptional activation of a target gene is complex.
Transcriptional activators that bind upstream of the RNAP-binding site have been shown to interact with the C-terminal domain of the α subunit (α-CTD) of RNAP, which promotes transcription of the target gene (Ishihama, 1992; Busby and Ebright, 1994; Ebright and Busby, 1995; Murakami et al., 1997; Hochschild and Dove, 1998). Contact between activators and α-CTD is inhibited when the upstream activator binding site is ≥100 bp upstream of the transcriptional start site (Murakami et al., 1997). Thus, transcription factor binding sites located further than 100 bp upstream of the transcriptional start site are unlikely to interact physically with the α-CTD of RNAP. However, oligomerization of the Fur protein at Fur boxes is known to occur (De Lorenzo et al., 1987; Tardat and Touati, 1993; Escolar et al., 2000; Nandal et al., 2010; Teixido et al., 2011), which suggests Fur proteins may extend to interact with other proteins nearby. Whether Fur contacts the RNAP is not known, but emerging in vivo evidence indicates there is another plausible molecular mechanism for Fur-dependent activation through binding DNA at a distal regulatory site.
Roles of Fur and H-NS in the regulation of FtnA
Fe2+ activates expression of the Fe-storage gene ftnA in a Fur-dependent manner (Masse and Gottesman, 2002; Velayudhan et al., 2007). Overexpression of ryhB results in the down regulation of many Fe-cofactored proteins (i.e., SodB) and increases the intracellular Fe2+ concentration resulting in enhanced Fur activation (Masse et al., 2005; Jacques et al., 2006). This is known as the “iron-sparing” response (Gaballa et al., 2008). Masse et al. theorized that Fur may negatively regulate a negative regulator of ftnA, which would manifest as a Fur activation. Evidence to support this theory was demonstrated by work from Simon C. Andrews' lab, which showed that Fur binds to a distal regulator site upstream of the RNAP-binding site in the promoter of ftnA to physically remove the histone-like protein, H-NS, which mediates repression of ftnA (Nandal et al., 2010). Unlike the activation of norB and hilD, Fur was not required for transcription of ftnA using in vitro transcription assays (Nandal et al., 2010). H-NS repressed transcription of ftnA and Fur was only required to relieve this repression. The role of Fur as an antirepressor in the activation of ftnA is supported with in vivo evidence: (1) fur is not required for ftnA expression in the absence of hns; and (2) ftnA expression is not reduced by Fe2+-chelation in Δhns (Nandal et al., 2010). Fur activation of gene expression by this mode represents a 3rd type of activation, the “antirepressor” activation model (Figure 2C). In vivo evidence supports the antirepressor model as a major mechanism for Fur-dependent activation of gene expression. Evidence for the antirepressor model is evident in N. gonorrhoeae because the Fur-binding site upstream of norB is not required for activation of expression when the norB repressor, ArsR, is deleted (Isabella et al., 2008). Thus, Fur antirepressor activity is an emerging model of Fur activation through DNA binding.
Fur Controls Defenses Against ROS
During bacterial infection the host responds to non-self molecules and initiates a potent antimicrobial response. However, bacterial pathogens are well-adapted to defending against the host antimicrobial response. In many bacterial pathogens the defense against ROS requires the Fur protein. Enzymatic defense against ROS occurs by the rapid enzymatic dismutation of superoxide (O2−) by superoxide dismutases (SODs) and detoxification of H2O2 by hydroperoxidases [i.e., the heme containing peroxidase/catalase (HPI), and the heme containing catalase (HPII)]. Unlike most pathogenic bacteria, S. Typhimurium contains 6 genes whose gene products are devoted toward degradation of H2O2. HPI (encoded by katG), HPII (encoded by katE), a Mn-dependent catalase (encoded by katN), an NADH-dependent alkyl peroxidase system (encoded by ahpCF), and two thiol specific peroxidases (encoded by tsaA and tpx). HPII and KatN are under positive regulation by the alternative σ factor RpoS, whereas HPI is induced by the redox sensing regulator OxyR during hydrogen peroxide stress (Tartaglia et al., 1989; Ivanova et al., 1994; Robbe-Saule et al., 2001; Vazquez-Torres, 2012). In addition, OxyR activates expression of ahpC (Storz et al., 1989; Tartaglia et al., 1989) and also fur (Zheng et al., 1999; Varghese et al., 2007). Regulation of tsaA appears Fur-independent (Delany et al., 2001) and there is a lack of evidence for whether Fe2+ and perhaps Fur regulate tpx. Deletion of any single gene or in combinations does not influence virulence; only the combined deletion of 5 out of the 6 genes results in reduced virulence signifying the importance of redundant H2O2 scavengers to virulence (Hebrard et al., 2009; Horst et al., 2010). As evident from studies in other bacterial pathogens, there are profound redundancies that contribute to resistance to H2O2 and virulence in vivo (Cosgrove et al., 2007; Lindgren et al., 2007; Soler-Garcia and Jerse, 2007). Because SODs and H2O2-degrading enzymes require certain metals as cofactors for enzymatic function and because Fur is a redox sensing protein (Fleischhacker and Kiley, 2011), it is not surprising that Fur is involved in the regulation of defenses against ROS.
SODs and HPI/HPII require the appropriate cofactors; Fe2+ is required for FeSOD (SodB) and Mn2+ for MnSOD (SodA) whereas heme is required for HPI and HPII function (Keele et al., 1970; Yost and Fridovich, 1973; Hassan and Fridovich, 1978; Claiborne and Fridovich, 1979; Claiborne et al., 1979). Fur directly represses transcription of the gene encoding the MnSOD (sodA) and indirectly activates expression of the gene encoding the FeSOD (sodB; Niederhoffer et al., 1990; Tardat and Touati, 1991; Beaumont and Hassan, 1993). This indirect control of sodB requires the RNA chaperone Hfq or ryhB (Masse and Gottesman, 2002; Ellermeier and Slauch, 2008; Troxell et al., 2011a). In addition, Fur controls HPI/HPII activity in a complex manner that may depend on the ability of Fur to regulate biosynthesis of the heme cofactor (Hamza et al., 2000; Benov and Sequeira, 2003; Hoerter et al., 2005; Mey et al., 2005a; Gaballa et al., 2008) (R. Saah and H. M. Hassan, unpublished data). Surprisingly, despite the enhanced transcription of sodA in Δfur, a corresponding increase in MnSOD activity was not observed due to the increased [Fe2+] in the mutant. Indeed, increase in MnSOD activity in Δfur was only discernible upon supplementation of the growth medium with excess [Mn2+] in order to outcompete the available Fe2+ for the active site of MnSOD (Hassan and Schrum, 1994; Schrum and Hassan, 1994; Troxell et al., 2011a). Thus, with respect to O−2 defense Δfur behaves phenotypically like ΔsodAΔsodB under Fe2+ replete conditions. The Fur regulation of Mn2+ transport is well-documented (Patzer and Hantke, 2001; Kehres et al., 2002; Guedon et al., 2003; Ikeda et al., 2005; Runyen-Janecky et al., 2006; Perry et al., 2012). Furthermore, because katN encodes a Mn-containing catalase and is activated by RpoS (Robbe-Saule et al., 2001) and repressed by H-NS (Beraud et al., 2010), it is likely that Fur is involved in katN expression in S. Typhimurium. Thus, the modulation of the intracellular Mn2+ concentration will undoubtedly influence protection against ROS and likely virulence. In support of this, numerous studies have demonstrated the importance of Mn2+ in the regulation of virulence and infectivity (Boyer et al., 2002; Corbin et al., 2008; Anderson et al., 2009; Ouyang et al., 2009; Ogunniyi et al., 2010; Wu et al., 2010; Champion et al., 2011; Kehl-Fie et al., 2011; Damo et al., 2013; Troxell et al., 2013). Likewise, additional members of the Fur family of metal-dependent transcription factors either bind Mn2+ directly and/or regulate Mn2+ transport.
Fe2+ Sequestration by the Host
Because Fur requires Fe2+ as a corepressor the availability of this metal controls Fur activity. Moreover, the Fe2+-Fur complex is inactivated by ROS and reactive nitrogen species (RNS) (D'Autreaux et al., 2002; Varghese et al., 2007), both of which are generated by the host during infection. Humans and other higher eukaryotes produce numerous proteins that sequester free Fe2+ and heme to deprive the pathogens of iron and meanwhile prevent the toxic formation of ROS. A potent antimicrobial response, including ROS production, produced by innate cells of the host's immune system is activated in response to detection of pathogen-associated molecular patterns (PAMPs) during bacterial infection. Innate cell activation by PAMPs initiates the synthesis of large amounts of Fe2+ sequestering proteins to limit the available Fe pool for the pathogen, known as “nutritional immunity” (Kehl-Fie and Skaar, 2010; Hood and Skaar, 2012) and activates signaling pathways that causes the host to reduce dietary absorption of Fe that is known as “the anemia of inflammation.” In addition, the host responds to infection by increasing the body temperature (the febrile response) as a means to inhibit bacterial growth. The antimicrobial host factors produced during activation of nutritional immunity can be inhibited by the addition of Fe (Weinberg, 1974). Furthermore, the febrile response to bacterial pathogens is antimicrobial, in part, due to the reduced ability of bacteria to acquire Fe2+ at febrile temperatures (Kluger and Rothenburg, 1979).
Anemia of inflammation by the host in response to infection has been known for more than 60 years (Cartwright et al., 1946; Greenberg et al., 1947; Wintrobe et al., 1947) and the host protein, hepcidin, controls this response (Nicolas et al., 2002; Nemeth et al., 2004a,b). In addition, hepcidin is a host factor that strongly reduces the absorption of dietary Fe (Shayeghi et al., 2005; Drakesmith and Prentice, 2012; Prentice et al., 2012). Because Fe2+ is required for cellular function within nearly all cells, limiting the availability of Fe2+ starves pathogens for Fe2+ and weakens the pathogens' ability to combat antimicrobial responses by the host. Not surprisingly, there is fierce competition for accessibility of Fe2+ during infection. Phagocytosis of the intracellular pathogen S. Typhimurium by macrophages enhances expression of the Fe2+ export protein ferroportin, which limits the available Fe2+ during intracellular residence of S. Typhimurium (Nairz et al., 2007). Expression of ferroportin correlates directly with reduced bacterial burden of several intracellular pathogens (Paradkar et al., 2008). Thus, the host responds to infection by sequestering Fe2+ from the local environment of pathogens, limits the absorption of dietary Fe resulting in a very Fe2+ limiting host environment, and restricts available Fe2+ within the phagosome.
An important host factor that controls bacterial infection is the natural resistance-associated macrophage protein 1 (NRAMP1, also known as SLC11A1) and several research groups determined the contribution of the SLC11A1 locus to severity of infection within animal models (Plant and Glynn, 1976; Bradley, 1977; Skamene et al., 1982; Brown et al., 2013). S. Typhimurium lacking fur are avirulent within mice with a functional NRAMP1, whereas the isogenic parent is fully virulent. Mice lacking a functional NRAMP1 are partially resistant to infection with Δfur demonstrating that Fur function is important for virulence, in part, independent of the host NRAMP1 function (Troxell et al., 2011b). Evidence indicates that Fur is functional within an unstimulated macrophage cell-line expressing either a functional or mutated NRAMP1 (Taylor et al., 2009). The NRAMP1 protein is a highly conserved transporter of divalent cations and is expressed within phagocytic cells (Cellier et al., 1995; Canonne-Hergaux et al., 2002; Cellier, 2012); NRAMP1 functions as a transporter of manganese (Mn2+), Fe2+, or cobalt (Co) and is important for acidification of the phagosome (Hackam et al., 1998; Jabado et al., 2000; Forbes and Gros, 2003). Furthermore, NRAMP1 promotes additional host factors of the antimicrobial response including production of nitric oxide (NO) (Fritsche et al., 2003, 2008; Nairz et al., 2009) and production of lipocalin-2 (also called siderocalin), which binds to bacterial siderophores thereby sequestering bacterial Fe2+ acquisition proteins (Fritsche et al., 2012). However, bacteria have evolved a counter defense mechanism by producing salmochelins, which are structurally distinct from enterochelin and therefore not susceptible to binding by lipocalin-2 (Smith, 2007). RNS and NO perturb Fur-Fe2+ function within pathogens (Mukhopadhyay et al., 2004; Richardson et al., 2006; Bourret et al., 2008). NO is a crucial factor in the antimicrobial response and its production is regulated by Fe2+ (Weiss et al., 1994; Melillo et al., 1997; Dlaska and Weiss, 1999). Consequently, the inability to generate NO increases the Fe2+ content within macrophages, splenic cells, and hepatocytes thereby increasing disease severity in animal models of infection (Nairz et al., 2013). This signifies the importance of NRAMP1 in the ability to sequester Fe2+ from pathogens and in general antimicrobial response.
Control of Virulence by the Fur Family of Transcriptional Regulators
The Fur protein contributes to virulence in animal models for numerous bacterial pathogens (Table 1). Although the precise mechanism for the observed attenuation of fur mutants is not clear, evidence indicates that a reduction in the activity of enzymes required for protection against ROS may be involved. Furthermore, virulence factors within the fur mutants exhibit altered expression or activity, which may additionally contribute to a decrease in virulence. Because Fur also controls expression or activity of enzymes within the TCA cycle, fur mutants are defective in the utilization of several carbon sources (i.e., succinate, etc.), which may contribute to the inability of fur mutants to cause disease within animal hosts.
There are additional transcription factors within the Fur family that require alternative metals to control gene regulation and virulence. First discovered by work in B. subtilis within the lab of John Helmann (Bsat et al., 1998; Mongkolsuk and Helmann, 2002), PerR is widespread in other bacteria and contributes to virulence within pathogens (Van Vliet et al., 1999; Horsburgh et al., 2001a; Rea et al., 2004, 2005; Gryllos et al., 2008). The DNA-binding activity of PerR is sensitive to relevant concentrations of H2O2 and upon metal-dependent oxidation results in derepression of target genes (Lee and Helmann, 2006). PerR homodimers are detected as two forms, one which contains two ions of Zn2+/Fe2+ per monomer and one which contains two ions of Zn2+/Mn2+ per monomer. Only the Zn/Fe form is sensitive to H2O2-induced derepression and, as expected, PerR regulates genes whose protein products detoxify H2O2 (Herbig and Helmann, 2001; Lee and Helmann, 2006). Thus, the H2O2-sensing of PerR is directly influenced by the Mn2+:Fe2+ ratio within the cell. Maintenance of the Mn2+:Fe2+ ratio is an important aspect within bacterial pathogens (Veyrier et al., 2011).
Zinc (Zn2+) uptake regulator (Zur) is a Fur family regulator that responds to Zn2+ and was discovered by two groups working with E. coli and Bacillus subtilis (B. subtilis) (Gaballa and Helmann, 1998; Patzer and Hantke, 1998). As expected for a Fur homolog, Zur represses transcription of Zn2+ uptake when bound to the corepressor Zn2+ (Patzer and Hantke, 2000; Gaballa and Helmann, 2002). Because ribosomal proteins utilize Zn2+ for activity Zur also represses transcription of genes involved in mobilization of Zn2+ by ribosomal protein paralogs, which may allow for protein synthesis under conditions of Zn2+ limitation known as the “failsafe” model (Maciag et al., 2007; Natori et al., 2007; Gabriel and Helmann, 2009). The Zur protein or Zn2+ uptake systems have an important role for bacterial pathogens, which demonstrate the importance of Zn2+ acquisition during infection (Campoy et al., 2002; Ammendola et al., 2007; Sabri et al., 2009; Smith et al., 2009; Desrosiers et al., 2010; Pesciaroli et al., 2011; Corbett et al., 2012; Dowd et al., 2012; Gielda and Dirita, 2012). The ability to acquire Zn2+ by bacterial pathogens is likely a broad requirement among bacterial pathogens during infection. More recently, a Fur-homolog was characterized as a Mn2+-dependent DNA-binding protein (Mur). This regulator, originally isolated from Rhizobium leguminosarum (Diaz-Mireles et al., 2004, 2005; Bellini and Hemmings, 2006), utilizes Mn2+ as a corepressor. In contrast to Fur and Zur, the role of Mur in bacterial pathogenesis is less understood. However, genes regulated by Mur are important for virulence in the pathogen Brucella abortus (Anderson et al., 2009; Menscher et al., 2012), which indicates Mur function may be important to virulence.
Conclusions
The Fur family of transcriptional regulators control virulence, defense against ROS, and transport of Fe2+, Zn2+, and Mn2+. Because of the anemia of inflammation and nutritional immunity exerted by the host during infection, metals are in low abundance in response to infection. In this metal-poor environment, the demetaleted Fur would allow for efficient acquisition of iron and enhances the fitness of the pathogen; however, deletion of fur most often results in partial or complete attenuation within animal models of infection. Notably, deletion of fur results in reduced expression of active enzymes responsible for defense against ROS, reduced expression of key metabolic pathways, and reduced expression of important virulence factors. This signifies that Fur's critical contribution to virulence may not be due to its classical role as a transcriptional repressor of metal acquisition (Figure 1), but to its complex role as a transcriptional activator of virulence (Figure 2).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Bryan Troxell was supported by NIH T32 AI060519. This work was supported in part by grants from: USDA-NIFA 2012-68003-19621, NC Biotech. Center, and NCSU Chancellor's Innovation Funds to Hosni M. Hassan. We thank Mathew D. Koci for critically reading the manuscript.
References
Afonyushkin, T., Vecerek, B., Moll, I., Blasi, U., and Kaberdin, V. R. (2005). Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 33, 1678–1689. doi: 10.1093/nar/gki313
Ammendola, S., Pasquali, P., Pistoia, C., Petrucci, P., Petrarca, P., Rotilio, G., et al. (2007). High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect. Immun. 75, 5867–5876. doi: 10.1128/IAI.00559-07
Anderson, E. S., Paulley, J. T., Gaines, J. M., Valderas, M. W., Martin, D. W., Menscher, E., et al. (2009). The manganese transporter MntH is a critical virulence determinant for Brucella abortus 2308 in experimentally infected mice. Infect. Immun. 77, 3466–3474. doi: 10.1128/IAI.00444-09
Anjum, M. F., Stevanin, T. M., Read, R. C., and Moir, J. W. (2002). Nitric oxide metabolism in Neisseria meningitidis. J. Bacteriol. 184, 2987–2993. doi: 10.1128/JB.184.11.2987-2993.2002
Bagg, A., and Neilands, J. B. (1985). Mapping of a mutation affecting regulation of iron uptake systems in Escherichia coli K-12. J. Bacteriol. 161, 450–453.
Bagg, A., and Neilands, J. B. (1987a). Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26, 5471–5477. doi: 10.1021/bi00391a039
Bagg, A., and Neilands, J. B. (1987b). Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol. Rev. 51, 509–518.
Baichoo, N., and Helmann, J. D. (2002). Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184, 5826–5832. doi: 10.1128/JB.184.21.5826-5832.2002
Baumler, A. J., Tsolis, R. M., Van Der Velden, A. W., Stojiljkovic, I., Anic, S., and Heffron, F. (1996). Identification of a new iron regulated locus of Salmonella typhi. Gene 183, 207–213. doi: 10.1016/S0378-1119(96)00560-4
Beaumont, M. D., and Hassan, H. M. (1993). Characterization of regulatory mutations causing anaerobic derepression of the sodA gene in Escherichia coli K12: cooperation between cis- and trans-acting regulatory loci. J. Gen. Microbiol. 139, 2677–2684. doi: 10.1099/00221287-139-11-2677
Bellini, P., and Hemmings, A. M. (2006). In vitro characterization of a bacterial manganese uptake regulator of the fur superfamily. Biochemistry 45, 2686–2698. doi: 10.1021/bi052081n
Benov, L., and Sequeira, F. (2003). Escherichia coli deltafur mutant displays low HPII catalase activity in stationary phase. Redox Rep. 8, 379–383. doi: 10.1179/135100003225003357
Beraud, M., Kolb, A., Monteil, V., D'Alayer, J., and Norel, F. (2010). A proteomic analysis reveals differential regulation of the sigma(S)-dependent yciGFE(katN) locus by YncC and H-NS in Salmonella and Escherichia coli K-12. Mol. Cell. Proteomics 9, 2601–2616. doi: 10.1074/mcp.M110.002493
Bjarnason, J., Southward, C. M., and Surette, M. G. (2003). Genomic profiling of iron-responsive genes in Salmonella enterica serovar typhimurium by high-throughput screening of a random promoter library. J. Bacteriol. 185, 4973–4982. doi: 10.1128/JB.185.16.4973-4982.2003
Bourret, T. J., Porwollik, S., McClelland, M., Zhao, R., Greco, T., Ischiropoulos, H., et al. (2008). Nitric oxide antagonizes the acid tolerance response that protects Salmonella against innate gastric defenses. PLoS ONE 3:e1833. doi: 10.1371/journal.pone.0001833
Bowden, S. D., Ramachandran, V. K., Knudsen, G. M., Hinton, J. C., and Thompson, A. (2010). An incomplete TCA cycle increases survival of Salmonella Typhimurium during infection of resting and activated murine macrophages. PLoS ONE 5:e13871. doi: 10.1371/journal.pone.0013871
Boyer, E., Bergevin, I., Malo, D., Gros, P., and Cellier, M. F. (2002). Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70, 6032–6042. doi: 10.1128/IAI.70.11.6032-6042.2002
Bradley, D. J. (1977). Regulation of Leishmania populations within the host. II. genetic control of acute susceptibility of mice to Leishmania donovani infection. Clin. Exp. Immunol. 30, 130–140.
Brickman, T. J., Ozenberger, B. A., and McIntosh, M. A. (1990). Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J. Mol. Biol. 212, 669–682. doi: 10.1016/0022-2836(90)90229-F
Brot, N., and Goodwin, J. (1968). Regulation of 2,3-dihydroxybenzoylserine synthetase by iron. J. Biol. Chem. 243, 510–513.
Brown, D. E., Libby, S. J., Moreland, S. M., McCoy, M. W., Brabb, T., Stepanek, A., et al. (2013). Salmonella enterica Causes more severe inflammatory disease in C57/BL6 Nramp1G169 Mice Than Sv129S6 Mice. Vet. Pathol. 50, 867–876. doi: 10.1177/0300985813478213
Bryce, G. F., and Brot, N. (1971). Iron transport in Escherichia coli and its relation to the repression of 2,3-dihydroxy-N-benzoyl-L-serine synthetase. Arch. Biochem. Biophys. 142, 399–406. doi: 10.1016/0003-9861(71)90503-0
Bsat, N., Herbig, A., Casillas-Martinez, L., Setlow, P., and Helmann, J. D. (1998). Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29, 189–198. doi: 10.1046/j.1365-2958.1998.00921.x
Bury-Mone, S., Thiberge, J. M., Contreras, M., Maitournam, A., Labigne, A., and De Reuse, H. (2004). Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53, 623–638. doi: 10.1111/j.1365-2958.2004.04137.x
Busby, S., and Ebright, R. H. (1994). Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79, 743–746. doi: 10.1016/0092-8674(94)90063-9
Calderwood, S. B., and Mekalanos, J. J. (1987). Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J. Bacteriol. 169, 4759–4764.
Calderwood, S. B., and Mekalanos, J. J. (1988). Confirmation of the Fur operator site by insertion of a synthetic oligonucleotide into an operon fusion plasmid. J. Bacteriol. 170, 1015–1017.
Campoy, S., Jara, M., Busquets, N., Perez De Rozas, A. M., Badiola, I., and Barbe, J. (2002). Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar typhimurium virulence. Infect. Immun. 70, 4721–4725. doi: 10.1128/IAI.70.8.4721-4725.2002
Canonne-Hergaux, F., Calafat, J., Richer, E., Cellier, M., Grinstein, S., Borregaard, N., et al. (2002). Expression and subcellular localization of NRAMP1 in human neutrophil granules. Blood 100, 268–275. doi: 10.1182/blood.V100.1.268
Carpenter, B. M., Whitmire, J. M., and Merrell, D. S. (2009). This is not your mother's repressor: the complex role of fur in pathogenesis. Infect. Immun. 77, 2590–2601. doi: 10.1128/IAI.00116-09
Cartwright, G. E., Lauritsen, M. A., Jones, P. J., Merrill, I. M., and Wintrobe, M. M. (1946). The anemia of infection. i. hypoferremia, hypercupremia, and alterations in porphyrin metabolism in patients. J. Clin. Invest. 25, 65–80. doi: 10.1172/JCI101690
Cellier, M., Prive, G., Belouchi, A., Kwan, T., Rodrigues, V., Chia, W., et al. (1995). Nramp defines a family of membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 92, 10089–10093. doi: 10.1073/pnas.92.22.10089
Cellier, M. F. (2012). Nramp: from sequence to structure and mechanism of divalent metal import. Curr. Top. Membr. 69, 249–293. doi: 10.1016/B978-0-12-394390-3.00010-0
Champion, O. L., Karlyshev, A., Cooper, I. A., Ford, D. C., Wren, B. W., Duffield, M., et al. (2011). Yersinia pseudotuberculosis mntH functions in intracellular manganese accumulation, which is essential for virulence and survival in cells expressing functional Nramp1. Microbiology 157, 1115–1122. doi: 10.1099/mic.0.045807-0
Claiborne, A., and Fridovich, I. (1979). Purification of the o-dianisidine peroxidase from Escherichia coli B. Physicochemical characterization and analysis of its dual catalatic and peroxidatic activities. J. Biol. Chem. 254, 4245–4252.
Claiborne, A., Malinowski, D. P., and Fridovich, I. (1979). Purification and characterization of hydroperoxidase II of Escherichia coli B. J. Biol. Chem. 254, 11664–11668.
Corbett, D., Wang, J., Schuler, S., Lopez-Castejon, G., Glenn, S., Brough, D., et al. (2012). Two zinc uptake systems contribute to the full virulence of Listeria monocytogenes during growth in vitro and in vivo. Infect. Immun. 80, 14–21. doi: 10.1128/IAI.05904-11
Corbin, B. D., Seeley, E. H., Raab, A., Feldmann, J., Miller, M. R., Torres, V. J., et al. (2008). Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319, 962–965. doi: 10.1126/science.1152449
Cosgrove, K., Coutts, G., Jonsson, I. M., Tarkowski, A., Kokai-Kun, J. F., Mond, J. J., et al. (2007). Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J. Bacteriol. 189, 1025–1035. doi: 10.1128/JB.01524-06
Craig, S. A., Carpenter, C. D., Mey, A. R., Wyckoff, E. E., and Payne, S. M. (2011). Positive regulation of the Vibrio cholerae porin OmpT by iron and fur. J. Bacteriol. 193, 6505–6511. doi: 10.1128/JB.05681-11
Curtiss, R. 3rd. Wanda, S. Y., Gunn, B. M., Zhang, X., Tinge, S. A., Ananthnarayan, V., et al. (2009). Salmonella enterica serovar typhimurium strains with regulated delayed attenuation in vivo. Infect. Immun. 77, 1071–1082. doi: 10.1128/IAI.00693-08
Damo, S. M., Kehl-Fie, T. E., Sugitani, N., Holt, M. E., Rathi, S., Murphy, W. J., et al. (2013). Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 110, 3841–3846. doi: 10.1073/pnas.1220341110
D'Autreaux, B., Touati, D., Bersch, B., Latour, J. M., and Michaud-Soret, I. (2002). Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc. Natl. Acad. Sci. U.S.A. 99, 16619–16624. doi: 10.1073/pnas.252591299
Davis, B. M., Quinones, M., Pratt, J., Ding, Y., and Waldor, M. K. (2005). Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 187, 4005–4014. doi: 10.1128/JB.187.12.4005-4014.2005
De Lorenzo, V., Wee, S., Herrero, M., and Neilands, J. B. (1987). Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169, 2624–2630.
Defez, R., and De Felice, M. (1981). Cryptic operon for beta-glucoside metabolism in Escherichia coli K12: genetic evidence for a regulatory protein. Genetics 97, 11–25.
Delany, I., Pacheco, A. B., Spohn, G., Rappuoli, R., and Scarlato, V. (2001). Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur repressor protein. J. Bacteriol. 183, 4932–4937. doi: 10.1128/JB.183.16.4932-4937.2001
Delany, I., Rappuoli, R., and Scarlato, V. (2004). Fur functions as an activator and as a repressor of putative virulence genes in Neisseria meningitidis. Mol. Microbiol. 52, 1081–1090. doi: 10.1111/j.1365-2958.2004.04030.x
Deng, Z., Meng, X., Su, S., Liu, Z., Ji, X., Zhang, Y., et al. (2012). Two sRNA RyhB homologs from Yersinia pestis biovar microtus expressed in vivo have differential Hfq-dependent stability. Res. Microbiol. 163, 413–418. doi: 10.1016/j.resmic.2012.05.006
Desrosiers, D. C., Bearden, S. W., Mier, I. Jr. Abney, J., Paulley, J. T., Fetherston, J. D., et al. (2010). Znu is the predominant zinc importer in Yersinia pestis during in vitro growth but is not essential for virulence. Infect. Immun. 78, 5163–5177. doi: 10.1128/IAI.00732-10
Diaz-Mireles, E., Wexler, M., Sawers, G., Bellini, D., Todd, J. D., and Johnston, A. W. (2004). The Fur-like protein Mur of Rhizobium leguminosarum is a Mn(2+)-responsive transcriptional regulator. Microbiology 150, 1447–1456. doi: 10.1099/mic.0.26961-0
Diaz-Mireles, E., Wexler, M., Todd, J. D., Bellini, D., Johnston, A. W., and Sawers, R. G. (2005). The manganese-responsive repressor Mur of Rhizobium leguminosarum is a member of the Fur-superfamily that recognizes an unusual operator sequence. Microbiology 151, 4071–4078. doi: 10.1099/mic.0.28342-0
Dlaska, M., and Weiss, G. (1999). Central role of transcription factor NF-IL6 for cytokine and iron-mediated regulation of murine inducible nitric oxide synthase expression. J. Immunol. 162, 6171–6177.
Dowd, G. C., Casey, P. G., Begley, M., Hill, C., and Gahan, C. G. (2012). Investigation of the role of ZurR in the physiology and pathogenesis of Listeria monocytogenes. FEMS Microbiol. Lett. 327, 118–125. doi: 10.1111/j.1574-6968.2011.02472.x
Drakesmith, H., and Prentice, A. M. (2012). Hepcidin and the iron-infection axis. Science 338, 768–772. doi: 10.1126/science.1224577
Dubrac, S., and Touati, D. (2000). Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182, 3802–3808. doi: 10.1128/JB.182.13.3802-3808.2000
Dubrac, S., and Touati, D. (2002). Fur-mediated transcriptional and post-transcriptional regulation of FeSOD expression in Escherichia coli. Microbiology 148, 147–156.
Ducey, T. F., Jackson, L., Orvis, J., and Dyer, D. W. (2009). Transcript analysis of nrrF, a Fur repressed sRNA of Neisseria gonorrhoeae. Microb. Pathog. 46, 166–170. doi: 10.1016/j.micpath.2008.12.003
Ebanks, R. O., Goguen, M., Knickle, L., Dacanay, A., Leslie, A., Ross, N. W., et al. (2013). Analysis of a ferric uptake regulator (Fur) knockout mutant in Aeromonas salmonicida subsp. salmonicida. Vet. Microbiol. 162, 831–841. doi: 10.1016/j.vetmic.2012.10.038
Ebright, R. H., and Busby, S. (1995). The Escherichia coli RNA polymerase alpha subunit: structure and function. Curr. Opin. Genet. Dev. 5, 197–203. doi: 10.1016/0959-437X(95)80008-5
Ellermeier, C. D., Ellermeier, J. R., and Slauch, J. M. (2005). HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57, 691–705. doi: 10.1111/j.1365-2958.2005.04737.x
Ellermeier, J. R., and Slauch, J. M. (2008). Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J. Bacteriol. 190, 476–486. doi: 10.1128/JB.00926-07
Ernst, J. F., Bennett, R. L., and Rothfield, L. I. (1978). Constitutive expression of the iron-enterochelin and ferrichrome uptake systems in a mutant strain of Salmonella typhimurium. J. Bacteriol. 135, 928–934.
Escolar, L., De Lorenzo, V., and Perez-Martin, J. (1997). Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol. Microbiol. 26, 799–808. doi: 10.1046/j.1365-2958.1997.6211987.x
Escolar, L., Perez-Martin, J., and De Lorenzo, V. (1998). Coordinated repression in vitro of the divergent fepA-fes promoters of Escherichia coli by the iron uptake regulation (Fur) protein. J. Bacteriol. 180, 2579–2582.
Escolar, L., Perez-Martin, J., and De Lorenzo, V. (1999). Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181, 6223–6229.
Escolar, L., Perez-Martin, J., and De Lorenzo, V. (2000). Evidence of an unusually long operator for the fur repressor in the aerobactin promoter of Escherichia coli. J. Biol. Chem. 275, 24709–24714. doi: 10.1074/jbc.M002839200
Fassbinder, F., Van Vliet, A. H., Gimmel, V., Kusters, J. G., Kist, M., and Bereswill, S. (2000). Identification of iron-regulated genes of Helicobacter pylori by a modified fur titration assay (FURTA-Hp). FEMS Microbiol. Lett. 184, 225–229. doi: 10.1111/j.1574-6968.2000.tb09018.x
Fenton, H. (1894). Oxidation of tartaric acid in presence of iron. J. Chem. Soc. Trans. 65, 899–911. doi: 10.1039/ct8946500899
Fleischhacker, A. S., and Kiley, P. J. (2011). Iron-containing transcription factors and their roles as sensors. Curr. Opin. Chem. Biol. 15, 335–341. doi: 10.1016/j.cbpa.2011.01.006
Forbes, J. R., and Gros, P. (2003). Iron, manganese, and cobalt transport by Nramp1 (Slc11a1) and Nramp2 (Slc11a2) expressed at the plasma membrane. Blood 102, 1884–1892. doi: 10.1182/blood-2003-02-0425
Foster, J. W., and Hall, H. K. (1992). Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174, 4317–4323.
Fritsche, G., Dlaska, M., Barton, H., Theurl, I., Garimorth, K., and Weiss, G. (2003). Nramp1 functionality increases inducible nitric oxide synthase transcription via stimulation of IFN regulatory factor 1 expression. J. Immunol. 171, 1994–1998.
Fritsche, G., Nairz, M., Libby, S. J., Fang, F. C., and Weiss, G. (2012). Slc11a1 (Nramp1) impairs growth of Salmonella enterica serovar typhimurium in macrophages via stimulation of lipocalin-2 expression. J. Leukoc. Biol. 92, 353–359. doi: 10.1189/jlb.1111554
Fritsche, G., Nairz, M., Werner, E. R., Barton, H. C., and Weiss, G. (2008). Nramp1-functionality increases iNOS expression via repression of IL-10 formation. Eur. J. Immunol. 38, 3060–3067. doi: 10.1002/eji.200838449
Gaballa, A., and Helmann, J. D. (1998). Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport operons in Bacillus subtilis. J. Bacteriol. 180, 5815–5821.
Gaballa, A., and Helmann, J. D. (2002). A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol. Microbiol. 45, 997–1005. doi: 10.1046/j.1365-2958.2002.03068.x
Gaballa, A., Antelmann, H., Aguilar, C., Khakh, S. K., Song, K. B., Smaldone, G. T., et al. (2008). The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 11927–11932. doi: 10.1073/pnas.0711752105
Gabriel, S. E., and Helmann, J. D. (2009). Contributions of Zur-controlled ribosomal proteins to growth under zinc starvation conditions. J. Bacteriol. 191, 6116–6122. doi: 10.1128/JB.00802-09
Gancz, H., Censini, S., and Merrell, D. S. (2006). Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74, 602–614. doi: 10.1128/IAI.74.1.602-614.2006
Gehring, A. M., Mori, I., and Walsh, C. T. (1998). Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37, 2648–2659. doi: 10.1021/bi9726584
Gielda, L. M., and Dirita, V. J. (2012). Zinc competition among the intestinal microbiota. MBio 3, e00171-12. doi: 10.1128/mBio.00171-12
Gilbreath, J. J., West, A. L., Pich, O. Q., Carpenter, B. M., Michel, S., and Merrell, D. S. (2012). Fur activates expression of the 2-oxoglutarate oxidoreductase genes (oorDABC) in Helicobacter pylori. J. Bacteriol. 194, 6490–6497. doi: 10.1128/JB.01226-12
Greenberg, G. R., Ashenbrucker, H., Lauritsen, M., Worth, W., Humphreys, S. R., and Wintrobe, M. M. (1947). The anemia of infection. V. fate of injected radioactive iron in the presence of inflammation. J. Clin. Invest. 26, 121–125. doi: 10.1172/JCI101784
Gruer, M. J., and Guest, J. R. (1994). Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology 140(Pt 10), 2531–2541. doi: 10.1099/00221287-140-10-2531
Gryllos, I., Grifantini, R., Colaprico, A., Cary, M. E., Hakansson, A., Carey, D. W., et al. (2008). PerR confers phagocytic killing resistance and allows pharyngeal colonization by group A Streptococcus. PLoS Pathog. 4:e1000145. doi: 10.1371/journal.ppat.1000145
Guedon, E., Moore, C. M., Que, Q., Wang, T., Ye, R. W., and Helmann, J. D. (2003). The global transcriptional response of Bacillus subtilis to manganese involves the MntR, Fur, TnrA and sigmaB regulons. Mol. Microbiol. 49, 1477–1491. doi: 10.1046/j.1365-2958.2003.03648.x
Hackam, D. J., Rotstein, O. D., Zhang, W., Gruenheid, S., Gros, P., and Grinstein, S. (1998). Host resistance to intracellular infection: mutation of natural resistance-associated macrophage protein 1 (Nramp1) impairs phagosomal acidification. J. Exp. Med. 188, 351–364. doi: 10.1084/jem.188.2.351
Hamza, I., Qi, Z., King, N. D., and O'Brian, M. R. (2000). Fur-independent regulation of iron metabolism by Irr in Bradyrhizobium japonicum. Microbiology 146(Pt 3), 669–676.
Hantke, K. (1981). Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182, 288–292. doi: 10.1007/BF00269672
Hantke, K. (1983). Identification of an iron uptake system specific for coprogen and rhodotorulic acid in Escherichia coli K12. Mol. Gen. Genet. 191, 301–306. doi: 10.1007/BF00334830
Hantke, K. (1984). Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol. Gen. Genet. 197, 337–341. doi: 10.1007/BF00330982
Hantke, K. (1987). Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K 12: fur not only affects iron metabolism. Mol. Gen. Genet. 210, 135–139. doi: 10.1007/BF00337769
Hantke, K. (2001). Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4, 172–177. doi: 10.1016/S1369-5274(00)00184-3
Haraszthy, V. I., Jordan, S. F., and Zambon, J. J. (2006). Identification of Fur-regulated genes in Actinobacillus actinomycetemcomitans. Microbiology 152, 787–796. doi: 10.1099/mic.0.28366-0
Harrison, A., Santana, E. A., Szelestey, B. R., Newsom, D. E., White, P., and Mason, K. M. (2013). Ferric uptake regulator and its role in the pathogenesis of nontypeable Haemophilus influenzae. Infect. Immun. 81, 1221–1233. doi: 10.1128/IAI.01227-12
Hassan, H. M., and Fridovich, I. (1978). Regulation of the synthesis of catalase and peroxidase in Escherichia coli. J. Biol. Chem. 253, 6445–6420.
Hassan, H. M., and Schrum, L. W. (1994). Roles of manganese and iron in the regulation of the biosynthesis of manganese-superoxide dismutase in Escherichia coli. FEMS Microbiol. Rev. 14, 315–323. doi: 10.1111/j.1574-6976.1994.tb00105.x
Hebrard, M., Viala, J. P., Meresse, S., Barras, F., and Aussel, L. (2009). Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J. Bacteriol. 191, 4605–4614. doi: 10.1128/JB.00144-09
Herbig, A. F., and Helmann, J. D. (2001). Roles of metal ions and hydrogen peroxide in modulating the interaction of the Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41, 849–859. doi: 10.1046/j.1365-2958.2001.02543.x
Hochschild, A., and Dove, S. L. (1998). Protein-protein contacts that activate and repress prokaryotic transcription. Cell 92, 597–600. doi: 10.1016/S0092-8674(00)81126-5
Hoerter, J. D., Arnold, A. A., Ward, C. S., Sauer, M., Johnson, S., Fleming, T., et al. (2005). Reduced hydroperoxidase (HPI and HPII) activity in the Deltafur mutant contributes to increased sensitivity to UVA radiation in Escherichia coli. J. Photochem. Photobiol. B. 79, 151–157. doi: 10.1016/j.jphotobiol.2005.01.003
Hood, M. I., and Skaar, E. P. (2012). Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10, 525–537. doi: 10.1038/nrmicro2836
Horsburgh, M. J., Clements, M. O., Crossley, H., Ingham, E., and Foster, S. J. (2001a). PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69, 3744–3754. doi: 10.1128/IAI.69.6.3744-3754.2001
Horsburgh, M. J., Ingham, E., and Foster, S. J. (2001b). In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and Is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183, 468–475. doi: 10.1128/JB.183.2.468-475.2001
Horst, S. A., Jaeger, T., Denkel, L. A., Rouf, S. F., Rhen, M., and Bange, F. C. (2010). Thiol peroxidase protects Salmonella enterica from hydrogen peroxide stress in vitro and facilitates intracellular growth. J. Bacteriol. 192, 2929–2932. doi: 10.1128/JB.01652-09
Huang, S. H., Wang, C. K., Peng, H. L., Wu, C. C., Chen, Y. T., Hong, Y. M., et al. (2012). Role of the small RNA RyhB in the Fur regulon in mediating the capsular polysaccharide biosynthesis and iron acquisition systems in Klebsiella pneumoniae. BMC Microbiol. 12:148. doi: 10.1186/1471-2180-12-148
Hughes, N. J., Clayton, C. L., Chalk, P. A., and Kelly, D. J. (1998). Helicobacter pylori porCDAB and oorDABC genes encode distinct pyruvate:flavodoxin and 2-oxoglutarate:acceptor oxidoreductases which mediate electron transport to NADP. J. Bacteriol. 180, 1119–1128.
Ikeda, J. S., Janakiraman, A., Kehres, D. G., Maguire, M. E., and Slauch, J. M. (2005). Transcriptional regulation of sitABCD of Salmonella enterica serovar Typhimurium by MntR and Fur. J. Bacteriol. 187, 912–922. doi: 10.1128/JB.187.3.912-922.2005
Imlay, J. A., Chin, S. M., and Linn, S. (1988). Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240, 640–642. doi: 10.1126/science.2834821
Isabella, V., Wright, L. F., Barth, K., Spence, J. M., Grogan, S., Genco, C. A., et al. (2008). cis- and trans-acting elements involved in regulation of norB (norZ), the gene encoding nitric oxide reductase in Neisseria gonorrhoeae. Microbiology 154, 226–239. doi: 10.1099/mic.0.2007/010470-0
Ishihama, A. (1992). Role of the RNA polymerase alpha subunit in transcription activation. Mol. Microbiol. 6, 3283–3288. doi: 10.1111/j.1365-2958.1992.tb02196.x
Ivanova, A., Miller, C., Glinsky, G., and Eisenstark, A. (1994). Role of rpoS (katF) in oxyR-independent regulation of hydroperoxidase I in Escherichia coli. Mol. Microbiol. 12, 571–578. doi: 10.1111/j.1365-2958.1994.tb01043.x
Jabado, N., Jankowski, A., Dougaparsad, S., Picard, V., Grinstein, S., and Gros, P. (2000). Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J. Exp. Med. 192, 1237–1248. doi: 10.1084/jem.192.9.1237
Jackson, L. A., Ducey, T. F., Day, M. W., Zaitshik, J. B., Orvis, J., and Dyer, D. W. (2010). Transcriptional and functional analysis of the Neisseria gonorrhoeae Fur regulon. J. Bacteriol. 192, 77–85. doi: 10.1128/JB.00741-09
Jacobsen, I., Gerstenberger, J., Gruber, A. D., Bosse, J. T., Langford, P. R., Hennig-Pauka, I., et al. (2005). Deletion of the ferric uptake regulator Fur impairs the in vitro growth and virulence of Actinobacillus pleuropneumoniae. Infect. Immun. 73, 3740–3744. doi: 10.1128/IAI.73.6.3740-3744.2005
Jacques, J. F., Jang, S., Prevost, K., Desnoyers, G., Desmarais, M., Imlay, J., et al. (2006). RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol. Microbiol. 62, 1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x
Keele, B. B. Jr. McCord, J. M., and Fridovich, I. (1970). Superoxide dismutase from Escherichia coli B. A new manganese-containing enzyme. J. Biol. Chem. 245, 6176–6181.
Kehl-Fie, T. E., Chitayat, S., Hood, M. I., Damo, S., Restrepo, N., Garcia, C., et al. (2011). Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 10, 158–164. doi: 10.1016/j.chom.2011.07.004
Kehl-Fie, T. E., and Skaar, E. P. (2010). Nutritional immunity beyond iron: a role for manganese and zinc. Curr. Opin. Chem. Biol. 14, 218–224. doi: 10.1016/j.cbpa.2009.11.008
Kehres, D. G., Janakiraman, A., Slauch, J. M., and Maguire, M. E. (2002). Regulation of Salmonella enterica serovar Typhimurium mntH transcription by H(2)O(2), Fe(2+), and Mn(2+). J. Bacteriol. 184, 3151–3158. doi: 10.1128/JB.184.12.3151-3158.2002
Kim, J. N., and Kwon, Y. M. (2013). Genetic and phenotypic characterization of the RyhB regulon in Salmonella Typhimurium. Microbiol. Res. 168, 41–49. doi: 10.1016/j.micres.2012.06.007
Kluger, M. J., and Rothenburg, B. A. (1979). Fever and reduced iron: their interaction as a host defense response to bacterial infection. Science 203, 374–376. doi: 10.1126/science.760197
Lafont, J. P., Dho, M., D'Hauteville, H. M., Bree, A., and Sansonetti, P. J. (1987). Presence and expression of aerobactin genes in virulent avian strains of Escherichia coli. Infect. Immun. 55, 193–197.
Lavrrar, J. L., Christoffersen, C. A., and McIntosh, M. A. (2002). Fur-DNA interactions at the bidirectional fepDGC-entS promoter region in Escherichia coli. J. Mol. Biol. 322, 983–995. doi: 10.1016/S0022-2836(02)00849-5
Leclerc, J. M., Dozois, C. M., and Daigle, F. (2013). Role of the Salmonella enterica serovar Typhi Fur regulator and small RNAs RfrA and RfrB in iron homeostasis and interaction with host cells. Microbiology 159, 591–602. doi: 10.1099/mic.0.064329-0
Lee, H. J., Kim, J. A., Lee, M. A., Park, S. J., and Lee, K. H. (2013). Regulation of haemolysin (VvhA) production by ferric uptake regulator (Fur) in Vibrio vulnificus: repression of vvhA transcription by Fur and proteolysis of VvhA by Fur-repressive exoproteases. Mol. Microbiol. 88, 813–826. doi: 10.1111/mmi.12224
Lee, J. W., and Helmann, J. D. (2006). The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440, 363–367. doi: 10.1038/nature04537
Lee, J. W., and Helmann, J. D. (2007). Functional specialization within the Fur family of metalloregulators. Biometals 20, 485–499. doi: 10.1007/s10534-006-9070-7
Lindgren, H., Shen, H., Zingmark, C., Golovliov, I., Conlan, W., and Sjostedt, A. (2007). Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect. Immun. 75, 1303–1309. doi: 10.1128/IAI.01717-06
Ling, J., Pan, H., Gao, Q., Xiong, L., Zhou, Y., Zhang, D., et al. (2013). Aerobactin synthesis genes iucA and iucC contribute to the pathogenicity of avian pathogenic Escherichia coli O2 strain E058. PLoS ONE 8:e57794. doi: 10.1371/journal.pone.0057794
Lloyd, R. V., Hanna, P. M., and Mason, R. P. (1997). The origin of the hydroxyl radical oxygen in the Fenton reaction. Free Radic. Biol. Med. 22, 885–888. doi: 10.1016/S0891-5849(96)00432-7
Maciag, A., Dainese, E., Rodriguez, G. M., Milano, A., Provvedi, R., Pasca, M. R., et al. (2007). Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189, 730–740. doi: 10.1128/JB.01190-06
Masse, E., and Gottesman, S. (2002). A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 4620–4625. doi: 10.1073/pnas.032066599
Masse, E., Vanderpool, C. K., and Gottesman, S. (2005). Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187, 6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005
May, G., Dersch, P., Haardt, M., Middendorf, A., and Bremer, E. (1990). The osmZ (bglY) gene encodes the DNA-binding protein H-NS (H1a), a component of the Escherichia coli K12 nucleoid. Mol. Gen. Genet. 224, 81–90. doi: 10.1007/BF00259454
McHugh, J. P., Rodriguez-Quinones, F., Abdul-Tehrani, H., Svistunenko, D. A., Poole, R. K., Cooper, C. E., et al. (2003). Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278, 29478–29486. doi: 10.1074/jbc.M303381200
Melillo, G., Taylor, L. S., Brooks, A., Musso, T., Cox, G. W., and Varesio, L. (1997). Functional requirement of the hypoxia-responsive element in the activation of the inducible nitric oxide synthase promoter by the iron chelator desferrioxamine. J. Biol. Chem. 272, 12236–12243. doi: 10.1074/jbc.272.18.12236
Mellin, J. R., Goswami, S., Grogan, S., Tjaden, B., and Genco, C. A. (2007). A novel fur- and iron-regulated small RNA, NrrF, is required for indirect fur-mediated regulation of the sdhA and sdhC genes in Neisseria meningitidis. J. Bacteriol. 189, 3686–3694. doi: 10.1128/JB.01890-06
Menscher, E. A., Caswell, C. C., Anderson, E. S., and Roop, R. M. 2nd. (2012). Mur regulates the gene encoding the manganese transporter MntH in Brucella abortus 2308. J. Bacteriol. 194, 561–566. doi: 10.1128/JB.05296-11
Metruccio, M. M., Fantappie, L., Serruto, D., Muzzi, A., Roncarati, D., Donati, C., et al. (2009). The Hfq-dependent small noncoding RNA NrrF directly mediates Fur-dependent positive regulation of succinate dehydrogenase in Neisseria meningitidis. J. Bacteriol. 191, 1330–1342. doi: 10.1128/JB.00849-08
Mey, A. R., Craig, S. A., and Payne, S. M. (2005a). Characterization of Vibrio cholerae RyhB: the RyhB regulon and role of ryhB in biofilm formation. Infect. Immun. 73, 5706–5719. doi: 10.1128/IAI.73.9.5706-5719.2005
Mey, A. R., Wyckoff, E. E., Kanukurthy, V., Fisher, C. R., and Payne, S. M. (2005b). Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect. Immun. 73, 8167–8178. doi: 10.1128/IAI.73.12.8167-8178.2005
Minato, Y., Fassio, S. R., Wolfe, A. J., and Hase, C. C. (2013). Central metabolism controls transcription of a virulence gene regulator in Vibrio cholerae. Microbiology 159, 792–802. doi: 10.1099/mic.0.064865-0
Mongkolsuk, S., and Helmann, J. D. (2002). Regulation of inducible peroxide stress responses. Mol. Microbiol. 45, 9–15. doi: 10.1046/j.1365-2958.2002.03015.x
Mukhopadhyay, P., Zheng, M., Bedzyk, L. A., Larossa, R. A., and Storz, G. (2004). Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. U.S.A. 101, 745–750. doi: 10.1073/pnas.0307741100
Murakami, K., Owens, J. T., Belyaeva, T. A., Meares, C. F., Busby, S. J., and Ishihama, A. (1997). Positioning of two alpha subunit carboxy-terminal domains of RNA polymerase at promoters by two transcription factors. Proc. Natl. Acad. Sci. U.S.A. 94, 11274–11278. doi: 10.1073/pnas.94.21.11274
Murphy, E. R., and Payne, S. M. (2007). RyhB, an iron-responsive small RNA molecule, regulates Shigella dysenteriae virulence. Infect. Immun. 75, 3470–3477. doi: 10.1128/IAI.00112-07
Nairz, M., Fritsche, G., Crouch, M. L., Barton, H. C., Fang, F. C., and Weiss, G. (2009). Slc11a1 limits intracellular growth of Salmonella enterica sv. Typhimurium by promoting macrophage immune effector functions and impairing bacterial iron acquisition. Cell. Microbiol. 11, 1365–1381. doi: 10.1111/j.1462-5822.2009.01337.x
Nairz, M., Schleicher, U., Schroll, A., Sonnweber, T., Theurl, I., Ludwiczek, S., et al. (2013). Nitric oxide-mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in Salmonella infection. J. Exp. Med. 210, 855–873. doi: 10.1084/jem.20121946
Nairz, M., Theurl, I., Ludwiczek, S., Theurl, M., Mair, S. M., Fritsche, G., et al. (2007). The co-ordinated regulation of iron homeostasis in murine macrophages limits the availability of iron for intracellular Salmonella typhimurium. Cell. Microbiol. 9, 2126–2140. doi: 10.1111/j.1462-5822.2007.00942.x
Nandal, A., Huggins, C. C., Woodhall, M. R., McHugh, J., Rodriguez-Quinones, F., Quail, M. A., et al. (2010). Induction of the ferritin gene (ftnA) of Escherichia coli by Fe(2+)-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol. Microbiol. 75, 637–657. doi: 10.1111/j.1365-2958.2009.06977.x
Natori, Y., Nanamiya, H., Akanuma, G., Kosono, S., Kudo, T., Ochi, K., et al. (2007). A fail-safe system for the ribosome under zinc-limiting conditions in Bacillus subtilis. Mol. Microbiol. 63, 294–307. doi: 10.1111/j.1365-2958.2006.05513.x
Nemeth, E., Rivera, S., Gabayan, V., Keller, C., Taudorf, S., Pedersen, B. K., et al. (2004a). IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J. Clin. Invest. 113, 1271–1276.
Nemeth, E., Tuttle, M. S., Powelson, J., Vaughn, M. B., Donovan, A., Ward, D. M., et al. (2004b). Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306, 2090–2093. doi: 10.1126/science.1104742
Nicolas, G., Chauvet, C., Viatte, L., Danan, J. L., Bigard, X., Devaux, I., et al. (2002). The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J. Clin. Invest. 110, 1037–1044.
Niederhoffer, E. C., Naranjo, C. M., Bradley, K. L., and Fee, J. A. (1990). Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J. Bacteriol. 172, 1930–1938.
Oglesby, A. G., Murphy, E. R., Iyer, V. R., and Payne, S. M. (2005). Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Mol. Microbiol. 58, 1354–1367. doi: 10.1111/j.1365-2958.2005.04920.x
Ogunniyi, A. D., Mahdi, L. K., Jennings, M. P., McEwan, A. G., McDevitt, C. A., Van Der Hoek, M. B., et al. (2010). Central role of manganese in regulation of stress responses, physiology, and metabolism in Streptococcus pneumoniae. J. Bacteriol. 192, 4489–4497. doi: 10.1128/JB.00064-10
Olekhnovich, I. N., and Kadner, R. J. (2006). Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357, 373–386. doi: 10.1016/j.jmb.2006.01.007
Osorio, C. R., Lemos, M. L., and Braun, V. (2004). Identification of Fur regulated genes in the bacterial fish pathogen Photobacterium damselae ssp. piscicida using the Fur titration assay. Biometals 17, 725–733. doi: 10.1007/s10534-004-1652-7
Ouyang, Z., He, M., Oman, T., Yang, X. F., and Norgard, M. V. (2009). A manganese transporter, BB0219 (BmtA), is required for virulence by the Lyme disease spirochete, Borrelia burgdorferi. Proc. Natl. Acad. Sci. U.S.A. 106, 3449–3454. doi: 10.1073/pnas.0812999106
Palyada, K., Threadgill, D., and Stintzi, A. (2004). Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186, 4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004
Paradkar, P. N., De Domenico, I., Durchfort, N., Zohn, I., Kaplan, J., and Ward, D. M. (2008). Iron depletion limits intracellular bacterial growth in macrophages. Blood 112, 866–874. doi: 10.1182/blood-2007-12-126854
Patzer, S. I., and Hantke, K. (1998). The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28, 1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x
Patzer, S. I., and Hantke, K. (2000). The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J. Biol. Chem. 275, 24321–24332. doi: 10.1074/jbc.M001775200
Patzer, S. I., and Hantke, K. (2001). Dual repression by Fe(2+)-Fur and Mn(2+)-MntR of the mntH gene, encoding an NRAMP-like Mn(2+) transporter in Escherichia coli. J. Bacteriol. 183, 4806–4813. doi: 10.1128/JB.183.16.4806-4813.2001
Perry, R. D., Craig, S. K., Abney, J., Bobrov, A. G., Kirillina, O., Mier, I. Jr., et al. (2012). Manganese transporters Yfe and MntH are Fur-regulated and important for the virulence of Yersinia pestis. Microbiology 158, 804–815. doi: 10.1099/mic.0.053710-0
Pesciaroli, M., Aloisio, F., Ammendola, S., Pistoia, C., Petrucci, P., Tarantino, M., et al. (2011). An attenuated Salmonella enterica serovar Typhimurium strain lacking the ZnuABC transporter induces protection in a mouse intestinal model of Salmonella infection. Vaccine 29, 1783–1790. doi: 10.1016/j.vaccine.2010.12.111
Plant, J., and Glynn, A. A. (1976). Genetics of resistance to infection with Salmonella typhimurium in mice. J. Infect. Dis. 133, 72–78. doi: 10.1093/infdis/133.1.72
Prajapat, M. K., and Saini, S. (2012). Interplay between Fur and HNS in controlling virulence gene expression in Salmonella typhimurium. Comput. Biol. Med. 42, 1133–1140. doi: 10.1016/j.compbiomed.2012.09.005
Prentice, A. M., Doherty, C. P., Abrams, S. A., Cox, S. E., Atkinson, S. H., Verhoef, H., et al. (2012). Hepcidin is the major predictor of erythrocyte iron incorporation in anemic African children. Blood 119, 1922–1928. doi: 10.1182/blood-2011-11-391219
Rea, R., Hill, C., and Gahan, C. G. (2005). Listeria monocytogenes PerR mutants display a small-colony phenotype, increased sensitivity to hydrogen peroxide, and significantly reduced murine virulence. Appl. Environ. Microbiol. 71, 8314–8322. doi: 10.1128/AEM.71.12.8314-8322.2005
Rea, R. B., Gahan, C. G., and Hill, C. (2004). Disruption of putative regulatory loci in Listeria monocytogenes demonstrates a significant role for Fur and PerR in virulence. Infect. Immun. 72, 717–727. doi: 10.1128/IAI.72.2.717-727.2004
Richardson, A. R., Dunman, P. M., and Fang, F. C. (2006). The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol. Microbiol. 61, 927–939. doi: 10.1111/j.1365-2958.2006.05290.x
Robbe-Saule, V., Coynault, C., Ibanez-Ruiz, M., Hermant, D., and Norel, F. (2001). Identification of a non-haem catalase in Salmonella and its regulation by RpoS (sigmaS). Mol. Microbiol. 39, 1533–1545. doi: 10.1046/j.1365-2958.2001.02340.x
Runyen-Janecky, L., Dazenski, E., Hawkins, S., and Warner, L. (2006). Role and regulation of the Shigella flexneri sit and MntH systems. Infect. Immun. 74, 4666–4672. doi: 10.1128/IAI.00562-06
Sabri, M., Houle, S., and Dozois, C. M. (2009). Roles of the extraintestinal pathogenic Escherichia coli ZnuACB and ZupT zinc transporters during urinary tract infection. Infect. Immun. 77, 1155–1164. doi: 10.1128/IAI.01082-08
Sadykov, M. R., Olson, M. E., Halouska, S., Zhu, Y., Fey, P. D., Powers, R., et al. (2008). Tricarboxylic acid cycle-dependent regulation of Staphylococcus epidermidis polysaccharide intercellular adhesin synthesis. J. Bacteriol. 190, 7621–7632. doi: 10.1128/JB.00806-08
Salvail, H., Lanthier-Bourbonnais, P., Sobota, J. M., Caza, M., Benjamin, J. A., Mendieta, M. E., et al. (2010). A small RNA promotes siderophore production through transcriptional and metabolic remodeling. Proc. Natl. Acad. Sci. U.S.A. 107, 15223–15228. doi: 10.1073/pnas.1007805107
Santander, J., Golden, G., Wanda, S. Y., and Curtiss, R. 3rd. (2012). Fur-regulated iron uptake system of Edwardsiella ictaluri and its influence on pathogenesis and immunogenicity in the catfish host. Infect. Immun. 80, 2689–2703. doi: 10.1128/IAI.00013-12
Schrum, L. W., and Hassan, H. M. (1994). The effects of fur on the transcriptional and post-transcriptional regulation of MnSOD gene (sodA) in Escherichia coli. Arch. Biochem. Biophys. 309, 288–292. doi: 10.1006/abbi.1994.1115
Shayeghi, M., Latunde-Dada, G. O., Oakhill, J. S., Laftah, A. H., Takeuchi, K., Halliday, N., et al. (2005). Identification of an intestinal heme transporter. Cell 122, 789–801. doi: 10.1016/j.cell.2005.06.025
Sheehan, B. J., Bosse, J. T., Beddek, A. J., Rycroft, A. N., Kroll, J. S., and Langford, P. R. (2003). Identification of Actinobacillus pleuropneumoniae genes important for survival during infection in its natural host. Infect. Immun. 71, 3960–3970. doi: 10.1128/IAI.71.7.3960-3970.2003
Skamene, E., Gros, P., Forget, A., Kongshavn, P. A., St Charles, C., and Taylor, B. A. (1982). Genetic regulation of resistance to intracellular pathogens. Nature 297, 506–509. doi: 10.1038/297506a0
Smith, K. D. (2007). Iron metabolism at the host pathogen interface: lipocalin 2 and the pathogen-associated iroA gene cluster. Int. J. Biochem. Cell Biol. 39, 1776–1780. doi: 10.1016/j.biocel.2007.07.003
Smith, K. F., Bibb, L. A., Schmitt, M. P., and Oram, D. M. (2009). Regulation and activity of a zinc uptake regulator, Zur, in Corynebacterium diphtheriae. J. Bacteriol. 191, 1595–1603. doi: 10.1128/JB.01392-08
Soler-Garcia, A. A., and Jerse, A. E. (2007). Neisseria gonorrhoeae catalase is not required for experimental genital tract infection despite the induction of a localized neutrophil response. Infect. Immun. 75, 2225–2233. doi: 10.1128/IAI.01513-06
Spears, P. A., Schauer, D., and Orndorff, P. E. (1986). Metastable regulation of type 1 piliation in Escherichia coli and isolation and characterization of a phenotypically stable mutant. J. Bacteriol. 168, 179–185.
Stojiljkovic, I., Baumler, A. J., and Hantke, K. (1994). Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236, 531–545. doi: 10.1006/jmbi.1994.1163
Storz, G., Jacobson, F. S., Tartaglia, L. A., Morgan, R. W., Silveira, L. A., and Ames, B. N. (1989). An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J. Bacteriol. 171, 2049–2055.
Tanabe, T., Funahashi, T., Moon, Y. H., Tamai, E., and Yamamoto, S. (2010). Identification and characterization of a Vibrio mimicus gene encoding the heme/hemoglobin receptor. Microbiol. Immunol. 54, 606–617.
Tardat, B., and Touati, D. (1991). Two global regulators repress the anaerobic expression of MnSOD in Escherichia coli::fur (ferric uptake regulation) and Arc (aerobic respiration control). Mol. Microbiol. 5, 455–465. doi: 10.1111/j.1365-2958.1991.tb02129.x
Tardat, B., and Touati, D. (1993). Iron and oxygen regulation of Escherichia coli MnSOD expression: competition between the global regulators Fur and ArcA for binding to DNA. Mol. Microbiol. 9, 53–63. doi: 10.1111/j.1365-2958.1993.tb01668.x
Tartaglia, L. A., Storz, G., and Ames, B. N. (1989). Identification and molecular analysis of oxyR-regulated promoters important for the bacterial adaptation to oxidative stress. J. Mol. Biol. 210, 709–719. doi: 10.1016/0022-2836(89)90104-6
Taylor, C. M., Osman, D., and Cavet, J. S. (2009). Differential expression from two iron-responsive promoters in Salmonella enterica serovar Typhimurium reveals the presence of iron in macrophage-phagosomes. Microb. Pathog. 46, 114–118. doi: 10.1016/j.micpath.2008.11.001
Tchawa Yimga, M., Leatham, M. P., Allen, J. H., Laux, D. C., Conway, T., and Cohen, P. S. (2006). Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 74, 1130–1140. doi: 10.1128/IAI.74.2.1130-1140.2006
Teixido, L., Carrasco, B., Alonso, J. C., Barbe, J., and Campoy, S. (2011). Fur activates the expression of Salmonella enterica pathogenicity island 1 by directly interacting with the hilD operator in vivo and in vitro. PLoS ONE 6:e19711. doi: 10.1371/journal.pone.0019711
Thompson, A., Rolfe, M. D., Lucchini, S., Schwerk, P., Hinton, J. C., and Tedin, K. (2006). The bacterial signal molecule, ppGpp, mediates the environmental regulation of both the invasion and intracellular virulence gene programs of Salmonella. J. Biol. Chem. 281, 30112–30121. doi: 10.1074/jbc.M605616200
Torres, V. J., Attia, A. S., Mason, W. J., Hood, M. I., Corbin, B. D., Beasley, F. C., et al. (2010). Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect. Immun. 78, 1618–1628. doi: 10.1128/IAI.01423-09
Troxell, B., Fink, R. C., Porwollik, S., McClelland, M., and Hassan, H. M. (2011a). The Fur regulon in anaerobically grown Salmonella enterica sv. Typhimurium: identification of new Fur targets. BMC Microbiol. 11:236. doi: 10.1186/1471-2180-11-236
Troxell, B., Sikes, M. L., Fink, R. C., Vazquez-Torres, A., Jones-Carson, J., and Hassan, H. M. (2011b). Fur negatively regulates hns and is required for the expression of HilA and virulence in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193, 497–505. doi: 10.1128/JB.00942-10
Troxell, B., Ye, M., Yang, Y., Carrasco, S. E., Lou, Y., and Yang, X. F. (2013). Manganese and zinc regulate virulence determinants in Borrelia burgdorferi. Infect. Immun. 81, 2743–2752. doi: 10.1128/IAI.00507-13
Tsolis, R. M., Baumler, A. J., Stojiljkovic, I., and Heffron, F. (1995). Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J. Bacteriol. 177, 4628–4637.
Urban, J. H., and Vogel, J. (2007). Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 35, 1018–1037. doi: 10.1093/nar/gkl1040
Van Vliet, A. H., Baillon, M. L., Penn, C. W., and Ketley, J. M. (1999). Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181, 6371–6376.
Varghese, S., Wu, A., Park, S., Imlay, K. R., and Imlay, J. A. (2007). Submicromolar hydrogen peroxide disrupts the ability of Fur protein to control free-iron levels in Escherichia coli. Mol. Microbiol. 64, 822–830. doi: 10.1111/j.1365-2958.2007.05701.x
Vassinova, N., and Kozyrev, D. (2000). A method for direct cloning of fur-regulated genes: identification of seven new fur-regulated loci in Escherichia coli. Microbiology 146(Pt 12), 3171–3182.
Vazquez-Torres, A. (2012). Redox active thiol sensors of oxidative and nitrosative stress. Antioxid. Redox Signal. 17, 1201–1214. doi: 10.1089/ars.2012.4522
Velayudhan, J., Castor, M., Richardson, A., Main-Hester, K. L., and Fang, F. C. (2007). The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol. Microbiol. 63, 1495–1507. doi: 10.1111/j.1365-2958.2007.05600.x
Veyrier, F. J., Boneca, I. G., Cellier, M. F., and Taha, M. K. (2011). A novel metal transporter mediating manganese export (MntX) regulates the Mn to Fe intracellular ratio and Neisseria meningitidis virulence. PLoS Pathog. 7:e1002261. doi: 10.1371/journal.ppat.1002261
Wang, H. R., Hu, Y. H., Zhang, W. W., and Sun, L. (2009). Construction of an attenuated Pseudomonas fluorescens strain and evaluation of its potential as a cross-protective vaccine. Vaccine 27, 4047–4055. doi: 10.1016/j.vaccine.2009.04.023
Weerakoon, D. R., Borden, N. J., Goodson, C. M., Grimes, J., and Olson, J. W. (2009). The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb. Pathog. 47, 8–15. doi: 10.1016/j.micpath.2009.04.009
Weinberg, E. D. (1974). Iron and susceptibility to infectious disease. Science 184, 952–956. doi: 10.1126/science.184.4140.952
Weiss, G., Werner-Felmayer, G., Werner, E. R., Grunewald, K., Wachter, H., and Hentze, M. W. (1994). Iron regulates nitric oxide synthase activity by controlling nuclear transcription. J. Exp. Med. 180, 969–976. doi: 10.1084/jem.180.3.969
Wilderman, P. J., Sowa, N. A., Fitzgerald, D. J., Fitzgerald, P. C., Gottesman, S., Ochsner, U. A., et al. (2004). Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U.S.A. 101, 9792–9797. doi: 10.1073/pnas.0403423101
Wintrobe, M. M., Greenberg, G. R., Humphreys, S. R., Ashen-Brucker, H., Worth, W., and Kramer, R. (1947). The Anemia of Infection. Iii. The uptake of radio-active iron in iron-deficient and in pyridoxine-deficient pigs before and after acute inflammation. J. Clin. Invest. 26, 103–113. doi: 10.1172/JCI101781
Wu, H. J., Seib, K. L., Srikhanta, Y. N., Edwards, J., Kidd, S. P., Maguire, T. L., et al. (2010). Manganese regulation of virulence factors and oxidative stress resistance in Neisseria gonorrhoeae. J. Proteomics 73, 899–916. doi: 10.1016/j.jprot.2009.12.001
Xiong, L., Ling, J., Gao, Q., Zhou, Y., Li, T., Gao, S., et al. (2012). Construction of iucB and iucBiutA mutants of avian pathogenic Escherichia coli and evaluation of their pathogenicity. Vet. Microbiol. 159, 420–431. doi: 10.1016/j.vetmic.2012.04.024
Yost, F. J. Jr., and Fridovich, I. (1973). An iron-containing superoxide dismutase from Escherichia coli. J. Biol. Chem. 248, 4905–4908.
Keywords: Ferric Uptake Regulator, iron, oxidative stress, gene regulation, pathogenic bacteria
Citation: Troxell B and Hassan HM (2013) Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. doi: 10.3389/fcimb.2013.00059
Received: 24 July 2013; Accepted: 18 September 2013;
Published online: 02 October 2013.
Edited by:
Mathieu F. Cellier, Institut National de la Recherche Scientifique, CanadaReviewed by:
John Helmann, Cornell University, USAKlaus Hantke, Universität Tübingen, Germany
Caroline Genco, Boston University School of Medicine, USA
Copyright © 2013 Troxell and Hassan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hosni M. Hassan, Prestage Department of Poultry Science, North Carolina State University, 334C Scott Hall, Campus Box 7608, Raleigh, NC 27695-7608, USA e-mail:aG1oYXNzYW5AbmNzdS5lZHU=
†Present address: Bryan Troxell, Prestage Department of Poultry Science, North Carolina State University, Raleigh, USA